Abstract
Mutations in the dynamin 2 gene have been identified in patients with autosomal dominant forms of centronuclear myopathy (CNM). Dynamin 2 is a ubiquitously expressed ∼100-kDa GTPase that assembles around the necks of vesiculating membranes and promotes their constriction and scission. It has also been implicated in regulation of the actin and microtubule cytoskeletons. At present, the cellular functions of dynamin 2 that are affected by CNM-linked mutations are not well defined, and the effects of these mutations on the physical and enzymatic properties of dynamin have been not examined. Here, we report the expression, purification, and characterization of four CNM-associated dynamin mutants. All four mutants display higher than wild-type GTPase activities, and more importantly, the mutants form high order oligomers that are significantly more resistant than wild-type dynamin 2 to disassembly by guanine nucleotides or high ionic strength. These observations suggest that the corresponding wild-type residues serve to prevent excessive or prolonged dynamin assembly on cellular membranes or inappropriate self-assembly in the cytoplasm. To our knowledge, this report contains the first identification of point mutations that enhance the stability of dynamin polymers without impairing their ability to bind and/or hydrolyze GTP. We envision that the formation of abnormally large and stable complexes of these dynamin mutants in vivo contributes to their role in CNM pathogenesis.
Keywords: Endocytosis, Mutant, Protein Self-assembly, Protein Stability, Skeletal Muscle, Centronuclear Myopathies, Dynamin 2, GTPase
Introduction
Autosomal dominant CNM2 is a congenital disorder that commonly results in muscle weakness and wasting, ptosis, and ophthalmoplegia (reviewed in Ref. 1). As the name implies, the most evident histopathological feature is the presence of a large number of muscle fibers of centrally (rather than peripherally) located nuclei. Other characteristics include a relative increase in the number of type I fibers, hypertrophy of these fibers, and the presence of sarcoplasmic strands distributed radially around the central nuclei. In 2005, Bitoun et al. (2) reported the identification of four mutations in the DNM2 (dynamin 2) gene in patients with autosomal dominant CNM, and an additional seven mutations have since been identified (3, 4). DNM2 is a ubiquitously expressed ∼100-kDa GTPase that assembles into helical polymers around the necks of vesiculating membranes, thereby providing force for their constriction and scission (reviewed in Refs. 5–8). In addition to its well characterized roles in endocytosis and Golgi budding, DNM2 is also implicated in regulation of the actin (9, 10) and microtubule (11, 12) cytoskeletons. The DNM2 molecule consists of five functional domains: an N-terminal catalytic domain; a so-called “middle domain” implicated in dynamin-dynamin interactions; a PH domain involved in phosphoinositide binding; a GTPase effector domain, which interacts with the catalytic domain and stimulates its GTPase activity; and a C-terminal proline/arginine-rich domain, which mediates interactions of dynamin with other proteins. Most of the currently known CNM-associated dynamin mutations are located in the middle and PH domains (4, 13), but others have recently been identified in the GTPase effector domain (3, 14).
Besides the observation that expression of some mutants inhibits endocytosis in cultured cells (14), there is no information regarding the consequences of these mutations on the physical and enzymatic properties of dynamin. To gain insight into the molecular basis of dynamin-dependent CNM pathogenesis, we have expressed, purified, and analyzed four CNM-linked DNM2 mutants, including three middle domain mutants that were identified in the original report (E368K, R369W, and R465W) (2), and a PH domain mutant (A618T) identified more recently (14). We found that all four mutants have a greater propensity to self-assemble than wild-type DNM2 and that the resulting structures are more resistant to depolymerization by guanine nucleotides or high ionic strength. The stability of these higher order oligomers in the presence of GTP is not due to defects in GTP binding and/or hydrolysis, as the mutants express higher than wild-type catalytic activities.
EXPERIMENTAL PROCEDURES
Materials
Phosphatidylcholine was from Calbiochem. PI(4,5)P2 was from Avanti Polar Lipids, Inc. (Alabaster, AL). [γ-32P]GTP was from PerkinElmer Life Sciences. QuikChange II XL site-directed mutagenesis reagent was from Stratagene (La Jolla, CA). Ni2+-nitrilotriacetic acid-agarose was from Qiagen (Valencia, CA). Reagents for protein assays and electrophoresis were from Bio-Rad. All other reagents, including GTP, buffers, charcoal-activated Norit A, and protease inhibitors, were from Sigma.
Generation of Dynamin Point Mutants
Wild-type dynamin 2 cDNA (rat isoform 2ba) with a C-terminal His6 tag was constructed and ligated to the pBacPAK8 plasmid (Clontech) as described previously (15). This construct was used as a template to introduce point mutations using the QuikChange site-directed mutagenesis kit (Stratagene) according to the manufacturer's protocol. The resulting E368K, R369W, R465W, A618T, and R361S plasmids, like that of wild-type DNM2, were cotransfected with BacPAK6 viral DNA digested with Bsu361 (Clontech) into Sf9 cells to produce recombinant baculoviruses. Recombinant viruses were plaque-purified and amplified by standard procedures.
Purification of Wild-type and Mutant Dynamins
Recombinant dynamin 2 and its mutants with a His6 tag were expressed in Sf9 cells and purified on Ni2+-nitrilotriacetic acid resin as described previously (15). Purified dynamins were dialyzed against 20 mm HEPES (pH 7.5), 0.3 m NaCl, 3 mm MgCl2, 1 mm EDTA, 0.5 mm dithiothreitol, and 0.2 mm phenylmethylsulfonyl fluoride. Aliquots of the protein were frozen in liquid nitrogen and stored at −70 °C. Immediately before use, the dynamin solutions were centrifuged at 213,000 × g for 15 min to remove any aggregated protein.
GTPase Assays
GTPase activities were measured by the release of 32Pi from [γ-32P]GTP after incubation at 37 °C in buffer containing 20 mm HEPES (pH 7.5), 2 mm MgCl2, 1 mm GTP, and various dynamin and NaCl concentrations. Reactions were initiated by addition of MgGTP and terminated by addition of 5% charcoal-activated Norit A in 50 mm NaH2PO4 at 4 °C according to Higashijima et al. (16). Charcoal was removed by centrifugation, and radioactivity in the supernatant was measured by scintillation counting.
Preparation of Phospholipid Vesicles
Phosphatidylcholine and PI(4,5)P2 in chloroform were mixed at a 9:1 molar ratio, dried under N2, and then dissolved in 20 mm HEPES (pH 7.5) and sonicated in a water bath sonicator (Model W185, Heat Systems Ultrasonics, Inc., Farmingdale, NY).
Turbidity Measurements
Samples of dynamin in 0.3 m NaCl were placed in quartz cuvettes and diluted with a prewarmed (37 °C) solution of 20 mm HEPES (pH 7.5) to obtain a final dynamin concentration of 1 μm and the NaCl concentrations indicated in the figure legends. Assembly was determined by measuring absorbance (350 nm) at 20-s intervals using a Beckman DU 650 spectrophotometer at 37 °C.
Sedimentation Equilibrium and Velocity Measurements
All analytical ultracentrifugation experiments were performed in a Beckman XL-I analytical ultracentrifuge using the An-60 Ti rotor and 1.2-cm path length centerpieces. Sedimentation equilibrium data (absorbance at 280 nm) were collected at 7000 rpm and 4 °C with a step size of 0.001 cm. Five readings were averaged for each scan in the final output. Loading protein concentrations were 0.8, 1.2, and 1.6 mg/ml, giving initial absorbances at 280 nm of ∼0.5, 0.75, and 1.0, respectively. All samples were in a solution containing 20 mm HEPES (pH 7.0), 1 mm EDTA, 5 mm MgCl2, 200 mm NaCl, 1 mm dithiothreitol, and 0.2 mm phenylmethylsulfonyl fluoride. Background absorbance was estimated by overspeeding at 40,000 rpm until a flat base line was obtained. Data were analyzed using the XL-I single program (Beckman) as described previously (17). Sedimentation velocity data were collected at 280 nm, 20 °C, and 35,000 rpm with a loading protein concentration of 1.8 mg/ml. Data were collected with a step size of 0.005 cm in the continuous mode. Five readings were averaged for each scan, and 40 scans were collected and analyzed using the second moment method in the XL-I data analysis software.
Other Methods
Protein concentration was determined as described by Bradford (18) using bovine serum albumin as a standard. SDS-PAGE was carried out according to the method of Laemmli (19) as modified by Matsudaira and Burgess (20).
RESULTS
Stabilization of DNM2 Polymers by the R369W Mutation
The first DNM2 mutations identified in CNM patients affected three residues, Glu368, Arg369, and Arg465, located in the middle domain (2). Because this domain mediates dynamin-dynamin interactions (21, 22), we expected these mutations to interfere with DNM2 self-association. To test this prediction, we chose to examine the R369W mutant based on its proximity to Arg361, which is required for self-assembly of the neuron-specific dynamin isoform, DNM1 (22). Self-assembly was initiated by dilution of dynamin into low salt buffer and then quantified using a high speed sedimentation assay. Fig. 1A shows that the preponderance of wild-type DNM2 sedimented upon dilution from 300 to 50 mm NaCl and that the sedimentable portion was significantly reduced in the presence of GTP. Contrary to our expectation, R369W self-assembled at least as well as wild-type DNM2 and, moreover, was far less sensitive to dissociation by GTP. For comparison, we also purified and analyzed the R361S mutant of DNM2. In the absence of GTP, R361S distributed almost equally between the pellet and the supernatant, and GTP induced further solubilization (Fig. 1A). The effect of the R361S mutation on DNM2 polymerization was less dramatic than reported for DNM1 (22),3 perhaps reflecting the higher tendency of DNM2 than DNM1 to self-associate (23). Arg369 is conserved in DNM1, which has a much lower propensity to self-assemble than DNM2 (23). Introduction of the R369W mutation in DNM1 did not increase its ability to self-associate (data not shown).
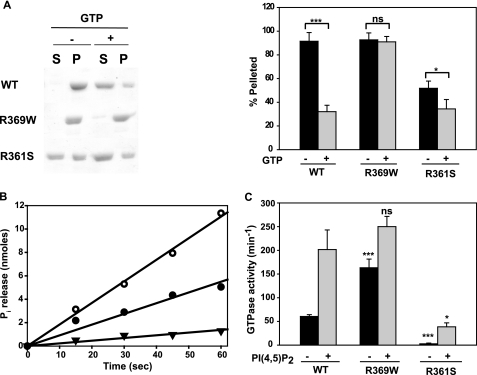
FIGURE 1.
Self-assembly and GTPase activities of wild-type DNM2, DNM2-R369W, and DNM2-R361S. A, sedimentation assay. Dynamin samples in 300 mm NaCl were diluted to obtain final concentrations of 50 mm NaCl and 1 μm dynamin. Following incubation at 37 °C for 15 min in the absence or presence of 1 mm GTP and 2 mm MgCl2, samples were centrifuged at 230,000 × g for 15 min at 30 °C. The distribution of protein into supernatants (S) and pellets (P) was analyzed by SDS-PAGE, followed by Coomassie Blue staining. Left panel, representative experiment; right panel, quantitative analysis of the data from at least three experiments, with each form of dynamin estimated by scanning the gels using a ScanJet 5300C, followed by analysis using NIH ImageJ. B, time course of GTP hydrolysis by 1 μm wild-type DNM2 (●), R369W (○), and R361S (▼) assayed at 50 mm NaCl. C, GTPase activities of 0.1 μm wild-type (WT) DNM2, R369W, and R361S in the absence (black bars) or presence (gray bars) of 2 μm PI(4,5)P2 assayed for 1 min at 50 mm NaCl. *, p < 0.05; ***, p < 0.001; ns, not significant.
The catalytically inactive K44A mutant of DNM1 forms polymers that are more resistant to disassembly by GTP than those of wild-type DNM1 (24). Therefore, we asked whether the relative stability of R369W polymers in the presence of GTP is likewise due to its impaired ability to bind and/or hydrolyze GTP. Fig. 1B shows that this is not the case, as R369W was actually twice as active as DNM2. In contrast, R361S expressed ∼5-fold lower activity than wild-type DNM2, corresponding to its lower tendency to self-associate.
Assembly of dynamins is facilitated by their binding to negatively charged scaffolds, including PI(4,5)P2-containing vesicles (15, 25–27). These scaffolds reduce the concentrations of dynamin required for polymerization and GTPase activation and stimulate GTPase activities to higher levels than can be achieved by dynamin alone. As shown in Fig. 1C, the GTPase activity of 0.1 μm wild-type DNM2 increased from ∼60 to ∼200 min−1 upon addition of PI(4,5)P2 vesicles. At the same dynamin concentration, the GTPase activity of R369W alone was 3-fold higher (∼150 min−1) than that of wild-type DNM2 alone, further demonstrating the enhanced ability of this mutant to self-assemble. However, PI(4,5)P2 vesicles stimulated the activity of R369W to approximately the same level as wild-type DNM2 (∼280 min−1). At 0.1 μm, R361S expressed negligible GTPase activity in the absence of PI(4,5)P2 and, similarly to DNM1-R361S (22), achieved levels of only ∼25–30 min−1 in its presence.
The enhanced stability of R369W polymers was confirmed using a turbidity assay. Wild-type DNM2 and R369W assembled rapidly and to approximately the same extent upon reduction of the NaCl concentration from 300 to 50 mm (Fig. 2A, left panel). Addition of GTP elicited a sharp (>70%) and rapid (<20 s) reduction in turbidity of solutions containing wild-type DNM2, whereas GTP had no effect on the turbidity of solutions containing R369W. The failure of GTP to disassemble R369W polymers was not due to its rapid hydrolysis, as GTPγS elicited similar results (data not shown). Although GTPγS stabilizes dynamin polymers in the presence of phospholipid scaffolds (28), this slowly hydrolyzable GTP analog is even more potent than GTP itself in disassembling DNM1 in the absence of a scaffold (24, 29).
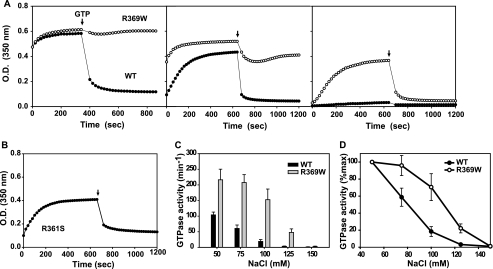
FIGURE 2.
Stability of self-assembled wild-type DNM2, DNM2-R369W, and DNM2-R361S. A, assembly and disassembly of dynamin polymers as measured by turbidity changes. Turbidity (350 nm) of solutions containing wild-type (WT) DNM2 (●) or R369W (○) at a final concentration of 1 μm was measured upon reduction of the NaCl concentration from 300 to 50 mm (left panel), 75 mm (middle panel), and 100 mm (right panel). After reaching plateau turbidities, GTP/MgCl2 was added to obtain final GTP and MgCl2 concentrations of 1 and 2 mm, respectively, as indicated by the arrows. B, turbidity changes of DNM2-R361S solution at 50 mm NaCl treated as described for A. C, GTPase activities of 1 μm wild-type DNM2 and R369W at different NaCl concentrations assayed for 1 min. D, data presented in C but expressed as the percent of activity measured at 50 mm NaCl. GTPase data represent the mean ± S.E. from analyses of triplicate measurements of three preparations of each construct.
The increased stability of R369W polymers was also observed at higher salt, as demonstrated by their higher turbidities compared with wild-type DNM2 at 75 and 100 mm NaCl (Fig. 2A, middle and right panels). Although R369W polymers were more susceptible to GTP-dependent disassembly at these higher salt concentrations, they were nevertheless more stable than wild-type dynamin polymers even under these conditions. Moreover, the R369W mutation increased the rate of dynamin polymerization, as evident at higher NaCl concentrations (Fig. 2A, middle and right panels). Consistent with this observation, R369W retained assembly-dependent GTPase activity at higher ionic strength compared with wild-type DNM2 (Fig. 2, C and D). These data suggest that the mutant may undergo self-assembly and carry out futile GTP hydrolysis in the cell cytoplasm. R361S also increased in turbidity upon dilution into low ionic strength buffers (Fig. 2B), although at a much lower rate than wild-type or R369W dynamins, and rapidly disassembled upon addition of GTP. The ability of R361S to self-assemble, albeit somewhat less extensively than wild-type DNM2, seems inconsistent with its low GTPase activity. We suggest that this mutant forms multimeric structures that are not sufficiently stable in the presence of guanine nucleotides to sustain multiple cycles of GTP hydrolysis.
Effect of Mutations on the Properties of Unassembled DNM2
In view of the dramatic and opposing effects of the R361S and R369W mutations on DNM2 polymerization and GTPase activity, we asked whether the mutations also affected the properties of unassembled dynamin. As shown in Table 1, the R369W mutation did not alter the oligomeric state or catalytic activity of unassembled dynamin. Data from sedimentation velocity (supplemental Fig. S1) and sedimentation equilibrium (supplemental Fig. S2) measurements indicated that both wild-type DNM2 and R369W are predominantly tetrameric at the concentrations examined (8–16-fold higher than those used for turbidity and GTPase experiments). Recovery of R369W following equilibrium ultracentrifugation was lower than that of wild-type DNM2, indicating that the mutant has a higher tendency to aggregate. Both analytical techniques revealed that R361S was present as a lower order oligomeric species than the other two dynamins, with an average molecular mass of 196 kDa. Thus, both the DNM1-R361S (22) and DNM2-R361S mutants are predominantly dimeric in the unassembled state. The GTPase activities of unassembled dynamins were very low and essentially identical.
TABLE 1.
Unassembled properties of DNM2 and its mutants
| DNM2 | R369W | R361S | |
|---|---|---|---|
| Sedimentation coefficient (s)a | 10.6 | 11.2 | 9.0 |
| Molecular mass (kDa)b | 327 | 336 | 196 |
| GTPase activity (min−1)c | 0.5 | 0.75 | 0.5 |
a Sedimentation velocity profiles are shown in supplemental Fig. S1. s = 10−13 s.
b Values are based on best single-species fit of sedimentation equilibrium data (supplemental Fig. S2).
c Activities were assayed in 200 mm NaCl.
Stabilization of DNM2 Polymers by Other CNM-linked Mutations
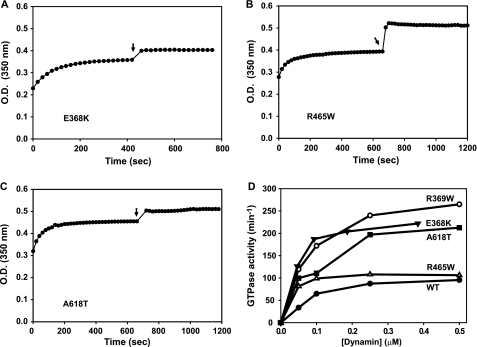
To determine whether stabilization of dynamin polymers is a unique property of the R369W mutation, we examined the effects of three other CNM-associated mutations: E368K and R465W in the middle domain and A618T in the C-terminal α-helical segment of the PH domain. As demonstrated by the turbidity assays shown in Fig. 3 (A–C), these mutants behaved similarly to R369W, achieving approximately wild-type plateau turbidity levels but failing to depolymerize upon addition of GTP. The slight increase in turbidity observed with these mutants upon addition of GTP was also evident in some measurements of R369W (data not shown). Like wild-type DNM2, all of the CNM-associated mutants exhibited concentration-dependent GTPase activation, but their activities increased more sharply as a function of protein concentration (Fig. 3D). Moreover, three of the mutants (E368K, R369W, and A618T) expressed maximal activities that were 2–2.5-fold higher compared with wild-type DNM2. Interestingly, these activities (∼250 min−1) were similar to those obtained in the presence of PI(4,5)P2 vesicles (Fig. 1C). Although the plateau activities of wild-type and R465W dynamins were similar, R465W was significantly more active than wild-type dynamin 2 at low concentrations (supplemental Fig. S3).
FIGURE 3.
Self-assembly and GTPase activities of CNM-associated dynamin 2 mutants. A–C, turbidity changes of solutions containing E368K, R465W, and A618T upon dilution to 50 mm NaCl, and addition of GTP/MgCl2 (arrows). Conditions and measurements were as described in the legend to Fig. 2. D, GTPase activities of wild-type (WT) dynamin 2 and CNM-associated dynamin 2 mutants assayed in 50 mm NaCl as a function of dynamin concentration. Data represent the means of triplicate measurements from at least two preparations of each form of dynamin.
DISCUSSION
The goal of this study was to determine how CNM mutations alter the in vitro properties of purified DNM2 to gain insights into the molecular basis for the disease. Interestingly, none of the four mutants that we examined showed impaired basal or stimulated GTPase activities, despite the reported ability of one of the mutants (R465W) to inhibit endocytosis upon overexpression in COS cells (14). However, they all shared one characteristic: stabilization of dynamin polymers in the presence of GTP, presumably reflecting abnormally strong dynamin-dynamin interactions. This observation suggests that the affected amino acid residues (Glu368, Arg369, Arg465, and Ala618) normally function to limit dynamin assembly, just as other residues (Arg361 and Arg399) were shown to be critical for promoting assembly (22). Thus, the dynamin molecule may have evolved to maintain an energetic balance that ensures timely and efficient assembly and disassembly during the process of membrane vesiculation. Disruption of this energetic balance may contribute to the inherited neuropathies and myopathies associated with DNM2 mutations.
Although three of the mutated residues (Glu368, Arg369, and Arg465) are located in the middle domain, which had previously been implicated in mediating dynamin assembly (21, 22), Ala618 is located in the PH domain, best characterized as the interaction site for phosphoinositides (30). Interestingly, the A618T mutation does not impair PI(4,5)P2-activated GTPase activity (data not shown). A potential role for the PH domain as a suppressor of dynamin assembly was suggested by reports demonstrating that deletion of this domain enhances the ability of dynamin 1 to polymerize (31–33). However, the specific regions within the PH domain that regulate dynamin self-association are unknown, as previous mutational analyses have been directed only at identifying residues that participate in lipid binding (25, 32, 34), membrane insertion (35), and binding to βγ-subunits of heterotrimeric G proteins (26, 36). Our finding that the A618T mutation stabilizes dynamin 2 polymers suggests that the C-terminal α-helix contributes to the inhibition of assembly by the PH domain.
Supplementary Material
Acknowledgment
We thank Derk Binns (Department of Pharmacology, University of Texas Southwestern Medical Center) for assistance with analytical ultracentrifugation.
This work was supported, in whole or in part, by National Institutes of Health Grant R01 GM076665.

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. S1–S3.
L. Wang, B. Barylko, C. Byers, J. A. Ross, D. M. Jameson, and J. P. Albanesi, unpublished data.
- CNM
- centronuclear myopathy
- PH
- pleckstrin homology
- PI(4,5)P2
- l-α-phosphatidylinositol 4,5-bisphosphate
- GTPγS
- guanosine 5′-O-(thiotriphosphate).
REFERENCES
- 1.Jungbluth H., Wallgren-Pettersson C., Laporte J. (2008) Orphanet J. Rare Dis. 3, 26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bitoun M., Maugenre S., Jeannet P. Y., Lacène E., Ferrer X., Laforêt P., Martin J. J., Laporte J., Lochmüller H., Beggs A. H., Fardeau M., Eymard B., Romero N. B., Guicheney P. (2005) Nat. Genet. 37, 1207–1209 [DOI] [PubMed] [Google Scholar]
- 3.Jungbluth H., Cullup T., Lillis S., Zhou H., Abbs S., Sewry C., Muntoni F. (2010) Neuromuscul. Disord. 20, 49–52 [DOI] [PubMed] [Google Scholar]
- 4.Durieux A. C., Prudhon B., Guicheney P., Bitoun M. (2010) J. Mol. Med. 88, 339–350 [DOI] [PubMed] [Google Scholar]
- 5.Hinshaw J. E. (2000) Annu. Rev. Cell Dev. Biol. 16, 483–519 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Allan V. J., Thompson H. M., McNiven M. A. (2002) Nat. Cell Biol. 4, E236–E242 [DOI] [PubMed] [Google Scholar]
- 7.Song B. D., Schmid S. L. (2003) Biochemistry 42, 1369–1376 [DOI] [PubMed] [Google Scholar]
- 8.Praefcke G. J., McMahon H. T. (2004) Nat. Rev. Mol. Cell Biol. l 5, 133–147 [DOI] [PubMed] [Google Scholar]
- 9.Schafer D. A. (2004) Traffic. 5, 463–469 [DOI] [PubMed] [Google Scholar]
- 10.Mooren O. L., Kotova T. I., Moore A. J., Schafer D. A. (2009) J. Biol. Chem. 284, 23995–24005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Thompson H. M., Cao H., Chen J., Euteneuer U., McNiven M. A. (2004) Nat. Cell Biol. 6, 335–342 [DOI] [PubMed] [Google Scholar]
- 12.Tanabe K., Takei K. (2009) J. Cell Biol. 185, 939–948 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bitoun M., Bevilacqua J. A., Prudhon B., Maugenre S., Taratuto A. L., Monges S., Lubieniecki F., Cances C., Uro-Coste E., Mayer M., Fardeau M., Romero N. B., Guicheney P. (2007) Ann. Neurol. 62, 666–670 [DOI] [PubMed] [Google Scholar]
- 14.Bitoun M., Durieux A. C., Prudhon B., Bevilacqua J. A., Herledan A., Sakanyan V., Urtizberea A., Cartier L., Romero N. B., Guicheney P. (2009) Hum. Mutat. 30, 1419–1427 [DOI] [PubMed] [Google Scholar]
- 15.Lin H. C., Barylko B., Achiriloaie M., Albanesi J. P. (1997) J. Biol. Chem. 272, 25999–26004 [DOI] [PubMed] [Google Scholar]
- 16.Higashijima T., Ferguson K. M., Smigel M. D., Gilman A. G. (1987) J. Biol. Chem. 262, 757–761 [PubMed] [Google Scholar]
- 17.Binns D. D., Barylko B., Grichine N., Atkinson M. A., Helms M. K., Jameson D. M., Eccleston J. F., Albanesi J. P. (1999) J. Protein Chem. 18, 277–290 [DOI] [PubMed] [Google Scholar]
- 18.Bradford M. M. (1976) Anal. Biochem. 72, 248–254 [DOI] [PubMed] [Google Scholar]
- 19.Laemmli U. K. (1970) Nature 227, 680–685 [DOI] [PubMed] [Google Scholar]
- 20.Matsudaira P. T., Burgess D. R. (1978) Anal. Biochem. 87, 386–396 [DOI] [PubMed] [Google Scholar]
- 21.Smirnova E., Shurland D. L., Newman-Smith E. D., Pishvaee B., van der Bliek A. M. (1999) J. Biol. Chem. 274, 14942–14947 [DOI] [PubMed] [Google Scholar]
- 22.Ramachandran R., Surka M., Chappie J. S., Fowler D. M., Foss T. R., Song B. D., Schmid S. L. (2007) EMBO J. 26, 559–566 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Warnock D. E., Baba T., Schmid S. L. (1997) Mol. Biol. Cell 8, 2553–2562 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Warnock D. E., Hinshaw J. E., Schmid S. L. (1996) J. Biol. Chem. 271, 22310–22314 [DOI] [PubMed] [Google Scholar]
- 25.Salim K., Bottomley M. J., Querfurth E., Zvelebil M. J., Gout I., Scaife R., Margolis R. L., Gigg R., Smith C. I., Driscoll P. C., Waterfield M. D., Panayotou G. (1996) EMBO J. 15, 6241–6250 [PMC free article] [PubMed] [Google Scholar]
- 26.Lin H. C., Gilman A. G. (1996) J. Biol. Chem. 271, 27979–27982 [DOI] [PubMed] [Google Scholar]
- 27.Barylko B., Binns D., Lin K. M., Atkinson M. A., Jameson D. M., Yin H. L., Albanesi J. P. (1998) J. Biol. Chem. 273, 3791–3797 [DOI] [PubMed] [Google Scholar]
- 28.Stowell M. H., Marks B., Wigge P., McMahon H. T. (1999) Nat. Cell Biol. 1, 27–32 [DOI] [PubMed] [Google Scholar]
- 29.Eccleston J. F., Binns D. D., Davis C. T., Albanesi J. P., Jameson D. M. (2002) Eur. Biophys. J. 31, 275–282 [DOI] [PubMed] [Google Scholar]
- 30.Lemmon M. A. (2007) Biochem. Soc. Symp. 74, 81–93 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Scaife R., Vénien-Bryan C., Margolis R. L. (1998) Biochemistry 37, 17673–17679 [DOI] [PubMed] [Google Scholar]
- 32.Vallis Y., Wigge P., Marks B., Evans P. R., McMahon H. T. (1999) Curr. Biol. 9, 257–260 [DOI] [PubMed] [Google Scholar]
- 33.Muhlberg A. B., Schmid S. L. (2000) Methods 20, 475–483 [DOI] [PubMed] [Google Scholar]
- 34.Achiriloaie M., Barylko B., Albanesi J. P. (1999) Mol. Cell. Biol. 19, 1410–1415 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ramachandran R., Pucadyil T. J., Liu Y. W., Acharya S., Leonard M., Lukiyanchuk V., Schmid S. L. (2009) Mol. Biol. Cell 20, 4630–4639 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Yang Z., Li H., Chai Z., Fullerton M. J., Cao Y., Toh B. H., Funder J. W., Liu J. P. (2001) J. Biol. Chem. 276, 4251–4260 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.