Abstract
The innate antiviral response is mediated, at least in part, by Toll-like receptors (TLRs). TLR3 signaling is activated in response to viral infection, and the absence of TLR3 in mice significantly increases mortality after infection with enteroviruses that cause myocarditis and/or dilated cardiomyopathy. We screened TLR3 in patients diagnosed with enteroviral myocarditis/cardiomyopathy and identified a rare variant in one patient as well as a significantly increased occurrence of a common polymorphism compared with controls. Expression of either variant resulted in significantly reduced TLR3-mediated signaling after stimulation with synthetic double-stranded RNA. Furthermore, Coxsackievirus B3 infection of cell lines expressing mutated TLR3 abrogated activation of the type I interferon pathway, leading to increased viral replication. TLR3-mediated type I interferon signaling required cellular autophagy and was suppressed by 3-methyladenine and bafilomycin A1, by inhibitors of lysosomal proteolysis, and by reduced expression of Beclin 1, Atg5, or microtubule-associated protein 1 light chain 3β (MAP1LC3β). However, TLR3-mediated signaling was restored upon exogenous expression of Beclin 1 or a variant MAP1LC3β fusion protein refractory to RNA interference. These data suggest that individuals harboring these variants may have a blunted innate immune response to enteroviral infection, leading to reduced viral clearance and an increased risk of cardiac pathology.
Keywords: Diseases, Genetics/Human, Immunology/Innate Immunity, Immunology/Toll Receptors, Receptors/Toll-like, Viruses/Positive-strand RNA
Introduction
Virus-induced myocarditis is an important cause of morbidity and mortality (1, 2). The enteroviruses have been considered the most common etiologic agents; however, other viruses, such as the adenoviruses, have also been implicated (3). The advent of molecular hybridization and PCR techniques has directly demonstrated infection of the myocardium with these viruses and has provided evidence of a viral etiology for dilated cardiomyopathy (DCM)5 (1, 3). These findings further support the hypothesis that DCM is in some cases a long term sequela of acute or chronic myocarditis (4, 5), either due to the pathogenic effect of persistent viral replication or to an ongoing autoimmune process secondary to the viral infection.
The recognition of viruses or viral particles by the host triggers the activation of an innate immune response that is characterized by the production of mediators such as tumor necrosis factor-α, interleukins, interferons, and nitric oxide, all of which are toxic to replicating viruses (6). This initial antiviral response by the host is now known to be mediated at least in part by Toll-like receptors (TLRs) (5, 6). It has been shown that double-stranded (ds) RNA is the primary ligand for TLR3 (7). Although most TLR signaling pathways utilize the adaptor molecule MyD88 (myeloid differentiation primary response gene 88), TLR3 signals through the adaptor molecule called Toll-like receptor adaptor molecule 1 (Ticam1) or Toll/Interleukin 1 receptor domain-containing adapter-inducing interferon β (TRIF) (8, 9). Activation of this pathway triggers production of type I interferon, which orchestrates both the rapid response and long term resistance to viral infection.
We have previously shown that TLR3 plays an important role in the host innate response to infection with the cardiotropic virus, encephalomyocarditis virus (10). Mice deficient in TLR3 are more susceptible to encephalomyocarditis virus infection, resulting in significantly increased mortality compared with wild type mice. The TLR3-deficient mice had a higher viral load in the heart and liver, with a concomitant impaired expression of inflammatory cytokines and chemokines in the heart. Similar data have recently been reported in TLR3-deficient mice infected with Coxsackievirus B3 or B4 (11, 12). Taken together, data from these studies suggest that enteroviral infection leads to a TLR3-dependent innate stress response, which is involved in mediating protection against myocardial injury. Germane to our study, Zhang et al. (13) reported the identification of a TLR3 P554S variant in two patients with herpes simplex virus encephalitis. This variant significantly blunted TLR3 signaling and acted in a dominant-negative manner suggesting that its presence could contribute to the host susceptibility to infection and possibly determine clinical outcome.
Although it has been suggested that the genetic composition of the host likely plays a critical role in determining susceptibility to viral heart disease, nothing is known with respect to the effect that mutations or polymorphisms in TLR3 may have on susceptibility to viral myocarditis. In this study we screened patients with biopsy-proven enteroviral myocarditis or DCM for variations in the TLR3 gene.
EXPERIMENTAL PROCEDURES
Patient Enrollment
In this study patients with enterovirus-associated myocarditis or dilated cardiomyopathy were enrolled at the University Hospital Benjamin Franklin, Berlin, Germany. Right ventricular endomyocardial biopsies and blood samples were obtained from all patients after informed consent, as regulated by the Institutional Regulatory Boards of the University Hospital Benjamin Franklin and the Baylor College of Medicine. Biopsy samples for PCR analysis were immediately snap-frozen in liquid nitrogen and stored below −80 °C, whereas samples for histologic analysis were formalin-fixed and paraffin-embedded and then evaluated for myocardial inflammation, according to the “Dallas” classification (14). Inflammatory cardiomyopathy was diagnosed according to the criteria of Kühl et al. (15). The detection of enteroviral genomes in endomyocardial biopsy samples were performed by nested reverse transcription polymerase chain reaction, as described previously (3). Blood samples were collected and stored at −80 °C before shipment to the United States.
Mutation Analysis
DNA was isolated from blood samples using Qiagen DNA purification kits. PCR primers were designed to amplify across the entire TLR3 coding sequence in an exon-by-exon manner using primers complementary to intron sequences (primer sequences are available upon request; exons 4 and 5 were analyzed using 5 and 2 primer sets, respectively). Mutation analysis was performed for all PCR products (except exon 4d) by denaturing high performance liquid chromatography in a WAVETM DNA Fragment Analysis System (Transgenomic) according to the protocol for each PCR product developed using the WAVEMAKER software (Transgenomic) (16).
Exon 4d samples as well as those displaying abnormal WAVE patterns were analyzed by direct DNA sequencing using Big Dye terminator chemistry (Version 3.1: Applied Biosystems) according to the manufacturer's instructions followed by analysis on an ABI 3100 (Applied Biosystems). The presence and frequencies of variants in the control population was determined by direct DNA sequencing.
Expression of TLR3 Mutants and the Effect on dsRNA-induced NF-κB Signaling
A clone of human TLR3, with a C-terminal hemagglutinin tag (HA), was obtained from Invivogen (pUNO-hTLR3-HA). Mutants were prepared using the QuikChangeTM mutagenesis kit (Stratagene). All clones were re-sequenced to confirm the presence of the specific mutation and absence of cloning errors.
A reporter cell line was established by transfecting 293 cells with the plasmid pNIFTY-SEAP (Invivogen) using the Lipofectamine reagent (Invitrogen) according to the manufacturer's instructions followed by selection using the antibiotic Zeocin (100 μg/ml, Invivogen). This plasmid includes an engineered endothelial cell-leukocyte adhesion molecule (ELAM1, E-selectin) gene promoter that combines five NF-κB sites with the proximal ELAM1 promoter. The ELAM1 composite promoter drives the expression of the reporter gene SEAP (secreted embryonic alkaline phosphatase), which is induced in the presence of NF-κB.
The HEK293-SEAP reporter cell line was transfected with 8 μg of each of the TLR3 constructs or sham-transfected using Lipofectamine 2000TM (six T-25 flasks per construct) according to the manufacturer's instructions (Invitrogen). Twenty-four hours after transfection the cells of half of the flasks were stimulated with poly(I:C) (10 μg/ml; Invivogen) for 6 h. The cell culture supernatant was collected for SEAP assays, and the cell pellet was harvested to measure exogenous TLR3 expression. SEAP expression was measured in supernatant fractions of transfected cells using a SEAP Reporter Assay kit (final assay volume of 660 μl) as described by the manufacturer (Invivogen), except that the sample volume was increased to 60 μl, and the volume of water used was decreased to 30 μl.
The expression of TLR3 was confirmed by Western blot analysis of aliquots of the transfected cells using an anti-HA monoclonal antibody (Invivogen) 24 h post-transfection. Cell extracts were prepared in 0.3 ml Tris-HCl, pH 7.5, 1 mm dithiothreitol, 0.2 mm EDTA, and 0.5% Triton X-100 plus 2× Complete protease® inhibitors (Roche Applied Science) and 1 mm phenylmethylsulfonyl fluoride and quantitated using the Coomassie PlusTM Protein Assay Reagent as described by the manufacturer (Pierce). After dilution in Laemmli sample buffer and heating to 100 °C for 5 min, 10 or 30 μg of protein were loaded per lane. After separation on 10% high Tris (0.75 m Tris-HCl, pH 8.8) polyacrylamide gels at 100 V, the proteins were transferred to nitrocellulose membranes in buffer containing 20 mm Tris base, 193 mm glycine, and 1% methanol at 100 mA for 18 h at 4 °C. HA-tagged TLR3 was detected using a 1:1000 dilution of the HA antibody that was visualized using the Western LightningTM Chemiluminescence Reagent (PerkinElmer Life Sciences) and horseradish peroxidase-labeled goat anti-mouse IgG secondary antibodies (1:1000 dilution; ICN Biomedical). The amount of protein loaded per lane was normalized to the amount of α-tubulin present in the samples using a 1:1000 dilution of a monoclonal anti-α-tubulin IgM antibody (Santa Cruz Biotechnology) and detected with goat anti-mouse IgM (H+L) secondary antibodies (Calbiochem/EMD Biosciences) and chemiluminescence. Signals were quantitated by densitometry using the NIH Image 1.63 software package. SEAP activities were normalized to the expression of TLR3-HA and expressed relative to the activity detected in the unstimulated WT TLR3-expressing cells.
Activation of the Interferon-stimulated Response Element (ISRE) Promoter
Stable lines of HEK293 cells expressing wild type or mutant TLR3-HAs were established by transfecting cells with each of the TLR3 constructs as described above. Stable transfectants were selected with 6 μg/ml blasticidin S (Invivogen), and individual clones were isolated. Clones were expanded, and TLR3 expression was confirmed by Western blotting of total cell extracts using anti-HA antibodies as described above. The amount of TLR3 expressed in each cell line was quantitated by densitometry, and lines expressing similar levels of TLR3-HA protein were selected for further analyses.
Activation of ISRE promoter was determined by transfecting the TLR3-HA stable lines with the pISRE-LUC reporter or the control pCIS-CK plasmids (Stratagene) using Lipofectamine 2000TM. The pISRE-LUC reporter drives the synthesis of firefly luciferase under the control of five ISRE. Briefly, 4 × 104 WT- or mutant TLR3-HA-expressing cells were seeded per well of a 96-well plate and transfected 18 h later in triplicate with 0.2 μg of either reporter or control plasmid. The following day the cells in half the wells were stimulated with 10 μg/ml of poly(I:C) for 24 h, and the amount of luciferase produced was measured using 150 μl of the Bright-GloTM Luciferase Assay reagent (Promega) following the manufacturer's instructions. 150-μl samples from each well were collected and mixed with 50 μl of 4× Laemmli sample buffer. The amount of TLR3-HA in each sample was determined by running 20-μl samples on 10% SDS-PAGE gels followed by Western blotting with anti-HA antibodies and densitometry as described above. Luciferase activities were normalized to the expression of TLR3-HA and expressed relative to the activity detected in the unstimulated WT TLR3-expressing cells. A similar protocol was used to test the effect of 100 μm chloroquine, 2.5 mm NH4Cl, 100 nm bafilomycin A1, 10 mm 3-methyladenine, 100 μm pepstatin A (isovaleryl-Val-Val-(3S,4S)-4 amino-3-hydroxy-6-methylheptanoyl-Ala-(3S,4S)-4 amino-3-hydroxy-6-methylheptanoic acid) or 10 μm E64d ((2S,3S)-trans-epoxysuccinyl-l-leucylamido-3-methylbutane ethyl ester) (all from Sigma), except that WT and mutant TLR3-HA-expressing cells transfected with reporter plasmid were incubated with 25 μg/ml poly(I:C) for 24 h. Luciferase activity was then measured using 125 μl of Bright-GloTM Luciferase reagent and normalized to the amount of TLR3-HA in each well as described above.
Analysis of the effect of infection with Coxsackievirus B3 on ISRE activation was performed in an analogous manner. After transfection with the pISRE-LUC plasmid, 8 × 104 WT or mutant TLR3-HA-expressing cells were seeded per well of six 96-well plates (two plates for each time point). Eighteen hours later the media and cells from 8 wells total for each line were collected (0 h), and the remaining wells were infected with Coxsackievirus B3 (m.o.i. = 0.01 or 0.1) or sham infected (media) in a volume of 100 μl per well. Luciferase activity was measured 8-fold at 24, 48, and 72 h post-infection by adding 100 μl of the Bright-GloTM assay mixture to one of the 96-well plates in each time set. The reactions were stopped by mixing in 100 μl of Laemmli buffer and analyzed by SDS-PAGE, immunoblotting, and densitometry as described above. Cell viability was determined in the second 96-well plate of each time set using the CytoTox-GloTM Assay (Promega), which detects both the dead and live cell populations after cleavage of the luminogenic peptide substrate Ala-Ala-Phe-aminoluciferin. Luciferase activities were normalized to the expression of TLR3-HA and corrected for the percentage of live cells at each time point and then expressed relative to the activity measured in the uninfected lines.
Determination of Viral Titers
Stable COS7 cells expressing wild type, L412F, or P554S TLR3-HA were established by transfecting cells with each of the TLR3 constructs, as described above, followed by selection of stable transfectants in Dulbecco's modified Eagle's medium containing 10 μg/ml blasticidin S. The cells were infected with Coxsackievirus B3 (m.o.i. = 0.1) and grown for 24 and 48 h after which the culture media was collected and frozen at −80 °C until used. Non-transfected COS7 (or non-TLR3-expressing) cells were seeded in triplicate on 6-well plates at a density of 2.5 × 105 cells per well and grown overnight at 37 °C. Cells were incubated with serial dilutions of the viral supernatants for 2 h at 37 °C, the media was removed, and the cells were then overlaid with 2 ml of 1.25% agarose in Dulbecco's modified Eagle's medium. Cells were grown for 48 h and fixed with 20% trichloroacetic acid for 10 min. Upon removing the agarose plugs, the cell monolayers were stained with 0.01% crystal violet in 20% ethanol for 30 min at room temperature. The stained cells were rinsed with distilled water, and viral plaques were counted.
Localization of TLR3
To visualize protein localization, stable COS7 lines expressing wild type or mutant TLR3 were grown on coverslips and exposed to medium containing 1 mg/ml Alexa 568®-dextran conjugate (Invitrogen) for 60 min in the presence or absence of 25 μg/ml poly(I:C). Cells were washed briefly with phosphate-buffered saline (PBS) and permeabilized on ice for 5 min in 80 mm PIPES, pH 6.8, containing 5 mm EGTA, 1 mm MgCl2, and 0.05% saponin (Sigma). After a brief wash with ice-cold PBS, the cells were fixed on ice with 3% paraformaldehyde in PBS for 15 min. Free aldehyde groups were quenched with 50 mm NH4Cl in PBS for 15 min at room temperature before samples were blocked overnight at 4 °C with PBS containing 10% normal goat serum, 2% bovine serum albumin, 0.2% gelatin, 0.05% saponin, and 0.02% NaN3. Cells were then stained with monoclonal anti-HA diluted 1:20 in blocking buffer overnight at 4 °C. Cell-nuclei were counterstained with TO-PRO-3TM iodide (1:1000 dilution; Invitrogen), whereas anti-HA was visualized with Alexa 488® goat anti-mouse IgG (1:300 dilution; Invitrogen), incubated overnight at 4 °C, and observed by confocal microscopy. For autophagosome/amphisome and lysosome/autolysosome immunostaining, permeabilized and fixed cells were incubated with anti-HA and either a rabbit antibody to microtubule-associated protein 1 light chain β (MAP1LC3β) diluted 1:100 (Novus Biologicals LLC) or with goat anti-lysosomal-associated membrane protein 2 (LAMP-2) diluted 1:50 (Santa Cruz Biotechnology), respectively, visualized with Alexa 568® goat anti-rabbit IgG (1:200 dilution) or Alexa 488® donkey anti-goat IgG (1:300 dilution; Invitrogen) and observed by confocal microscopy.
Localization of TLR3 to amphisomes and autolysosomes was also determined after infection of the TLR3-expressing COS7 lines with Coxsackievirus B3 (m.o.i. = 0.1) for 3 h, after which the medium was removed; the cells were rinsed briefly with PBS and grown in fresh medium for 24 and 48 h. Cells were permeabilized, fixed, and stained with antibodies to the HA epitope, LAMP-2, or MAP1LC3β as described above.
RNA Interference of Human MAP1LC3β, Beclin 1, and Atg5
Knockdown of MAP1LC3β expression was performed with RNA duplexes from Santa Cruz Biotechnology and Integrated DNA Technologies directed to the coding (sc-43390A) or the 3′-untranslated region of the MAP1LC3β message (sc-43390B or sc-43390C) (see Table 1 for sequences). A siRNA duplex encoding two silent mutations within the cognate MAP1LC3β siRNA-A sequence was also used to rule out potential off-target silencing effects of the LC3β siRNA-A (Table 1). Briefly, confluent cultures of WT, Phe-412, and Ser-554 TLR3-HA-expressing 293 cells growing in 60-mm culture dishes were trypsinized, and ⅓ of the samples of each line were immediately transfected with 200 pmol of either wild type (LC3β siRNA-A, -B, or -C) or mutant MAP1LC3β siRNA duplex (Mut. LC3β-siRNA-A), or a scrambled siRNA that does not lead to the specific degradation of any known cellular mRNA (Santa Cruz Biotechnology) using 10 μl of Lipofectamine RNAiMAXTM (Invitrogen). Cells were grown for 72 h, after which the transfection with each siRNA duplex was repeated as described above. Forty-eight hours after the second siRNA transfection, the cells were harvested and transfected using 100 pmol of each siRNA duplex, 4 μg of pISRE-LUC reporter, and 10 μl of Lipofectamine 2000TM (Invitrogen) in 1 ml of reduced serum Opti-MEM® (Invitrogen) and 5.5 ml of antibiotic-free Dulbecco's modified Eagle's medium containing 10% fetal bovine serum. 100 μl of each cell suspension were then seeded in 8 wells of a 96-well plate, whereas the remaining cells were plated in a 60-mm culture dish. The following day, the cells in half the wells were stimulated with 25 μg/ml poly(I:C) for 24 h, and the amount of luciferase produced was measured and normalized to the amount of TLR3-HA as described above. Levels of MAP1LC3β protein were determined by Western blotting using a polyclonal antibody from Novus Biologicals.
TABLE 1.
Sequence of siRNA duplexes used in this study
Transfections were performed in 60-mm culture dishes using 100–200 pmol of siRNA and 10 μl of RNAiMAXTM Lipofectamine (MAP1LC3β siRNAs) or 10 μl of Lipofectamine 2000TM according to the manufacturer's instructions (see “Experimental Procedures” for details). Note that in all cases when a siRNA was co-transfected with the pISRE-LUC reporter, 20 μl of Lipofectamine 2000TM was used. Mutations introduced into specific siRNA sequences are underlined in bold face and follow general recommendations for design and experimental validation of siRNAs (65, 66).
| siRNA | Sequence |
|---|---|
| MAP1LC3β siRNA-A | Sense, 5′-CGGUGAUAAUAGAACGAUATT-3′ |
| Antisense, 5′-UAUCGUUCUAUUAUCACCGGG-3′ | |
| Mutant MAP1LC3β siRNA-A | Sense, 5′-CGGUAAUAAUAGAGCGAUATT-3′ |
| Antisense, 5′-UAUCGCUCUAUUAUUACCGGG-3′ | |
| MAP1LC3β siRNA-B | Sense, 5′-GGUUUGUUCUCUAGAUAGUTT-3′ |
| Antisense, 5′-ACUAUCUAGAGAACAAACC-3′ | |
| MAP1LC3β siRNA-C | Sense, 5′-CGUACGCUCUUUACAGAUATT-3′ |
| Antisense, 5′-UAUCUGUAAAGAGCGUACG-3′ | |
| Beclin 1 siRNA-A | Sense, 5′-CAGCUCAACGUCACUGAAATT-3′ |
| Antisense, 5′-UUUCAGUGACGUUGAGCUGAG-3′ | |
| Beclin 1 siRNA-B | Sense, 5′-GAGAUCUUAGAGCAAAUGATT-3′ |
| Antisense, 5′-UCAUUUGCUCUAAGAUCUCCA-3′ | |
| Beclin 1 siRNA-C | Sense, 5′-GGAUGACAGUGAACAGUUATT-3′ |
| Antisense, 5′-UAACUGUUCACUGUCAUCCTC-3 | |
| Mutant Beclin 1 siRNA-C | Sense, 5′-GGACGAUAGUGAACAAUUATT-3′ |
| Antisense, 5′-UAAUUGUUCACUAUCGUCCTC-3′ | |
| Atg5 siRNA-A | Sense, 5′-CAACUUGUUUCACGCUAUATT-3′ |
| Antisense, 5′-UAUAGCGUGAAACAAGUUGGA-3′ | |
| Mutant Atg5 siRNA-A | Sense, 5′-CAACUUGCUUUACUCUCUATT-3′ |
| Antisense, 5′-UAGAGAGUAAAGCAAGUUGGA-3′ | |
| Atg5 siRNA-B | Sense, 5′-GGAAUAUCCUGCAGAAGAATT-3′ |
| Antisense, 5′-UUCUUCUGCAGGAUAUUCCAT-3′ | |
| Mutant Atg5 siRNA-B | Sense, 5′-GGAGUACCCCGCCGAAGAATT-3′ |
| Antisense, 5′-UUCUUCGGCGGGGUACUCCAT-3′ |
Silencing of Atg5 and Beclin 1 expression was performed with commercial siRNA duplexes, sc-41445A and sc-41445B for Atg5 and sc-29797A-C for Beclin 1 (Santa Cruz Biotechnology and Integrated DNA Technologies) (see Table 1 for sequences). Duplexes encoding silent mutations within the cognate Atg5 siRNA-A and -B and the Beclin 1 siRNA-C sequences were also used to rule out potential off-target silencing effects of the Atg5 or Beclin 1 siRNAs (Table 1). Briefly, confluent cultures of WT TLR3-HA expressing HEK cells growing in 60-mm culture dishes were trypsinized and immediately transfected with 150 pmol of each siRNA duplex and 10 μl of Lipofectamine 2000TM in 1 ml of Opti-MEM® and 4 ml of antibiotic-free Dulbecco's modified Eagle's medium. Cells were grown for 48 h, after which they were transfected a second time as described above. Finally, the cells were transfected a third time, but in this case the transfection mixture included 150 pmol of siRNA duplex, 7.5 μg of pISRE-LUC reporter, and 20 μl of Lipofectamine 2000TM. Levels of the two proteins were determined by Western blotting using a monoclonal antibody to Beclin 1 and monoclonal anti-Atg5 antibodies (both from Santa Cruz Biotechnology).
Confocal Microscopy
Fluorescence confocal laser scanning microscopy was performed on an Olympus FV300 confocal microscope equipped with argon 488 and helium/neon 543/633 lasers using a 60× PlanApo 1.4 NA oil objective and Fluoview 5.0 software.
Statistical Analyses
Comparisons of genotype frequencies between patient and control groups were performed by χ2 analysis. Statistical comparisons of functional data were performed using t tests or one-way analysis of variance tests with the Bonferroni multiple comparison procedure. All analyses were performed using the statistical software SigmaStat v3.5. Statistical significance was set at p < 0.05.
RESULTS
Phenotypic Analysis
Fifty-seven individuals diagnosed with histologically proven myocarditis or with echocardiographically diagnosed DCM and with endomyocardial biopsy samples positive for enteroviral RNA were enrolled. Demographic data are presented in Table 2. The patient cohort included 38 males and 19 females, all Caucasian, and the age at diagnosis ranged from 27 to 77 years old.
TABLE 2.
Demographic and genotype data for the patient group
MC, myocarditis; DCMi, inflammatory DCM; PPCM, postpartum cardiomyopathy; M, male; F, female; L, homozygous leucine 412; L/F, heterozygous leucine 412 phenylalanine; F, homozygous phenylalanine 412; P554S, heterozygous proline 554 serine substitution.
| Patient number | Diagnosis | Age at DX | Sex | TLR3 L412F | TLR3 variants |
|---|---|---|---|---|---|
| Years | |||||
| 1 | MC | 36 | M | L | |
| 2 | MC | 29 | M | L | |
| 3 | DCM | 49 | F | L | |
| 4 | DCM | 46 | M | L | |
| 5 | MC | 31 | M | L/F | |
| 6 | MC | 37 | F | L/F | |
| 7 | MC | 33 | M | L | |
| 8 | DCM | 61 | M | L | |
| 9 | DCM | 75 | M | F | |
| 10 | MC | 54 | M | L | P554S |
| 11 | PPCM | 30 | F | L/F | |
| 12 | MC | 70 | F | L/F | |
| 13 | DCM | 55 | F | L | |
| 14 | DCM | 49 | M | L | |
| 15 | MC | 53 | F | L | |
| 16 | DCMi | 54 | M | F | |
| 17 | DCM | 61 | M | F | |
| 18 | DCM | 42 | M | L/F | |
| 19 | PPCM | 27 | F | L/F | |
| 20 | DCM | 67 | M | L/F | |
| 21 | DCM | 41 | M | L | |
| 22 | MC | 56 | M | L | |
| 23 | DCM | 52 | M | L | |
| 24 | MC | 39 | F | F | |
| 25 | DCMi | 57 | M | L | |
| 26 | MC | 55 | M | L | |
| 27 | MC | 57 | M | L | |
| 28 | MC | 49 | M | L/F | |
| 29 | DCMi | 54 | M | L/F | |
| 30 | DCM | 54 | M | L | |
| 31 | MC | 46 | M | L | |
| 32 | DCM | 40 | F | L | |
| 33 | MC | 41 | M | F | |
| 34 | DCMi | 32 | M | L | |
| 35 | DCM | 60 | M | L | |
| 36 | DCMi | 46 | F | L/F | |
| 37 | DCM | 43 | M | L | |
| 38 | DCM | 55 | M | L | |
| 39 | DCM | 58 | M | L | |
| 40 | DCMi | 52 | M | L | |
| 41 | DCM | 49 | M | L | |
| 42 | DCM | 34 | M | L | |
| 43 | DCM | 38 | F | L | |
| 44 | DCM | 50 | M | L | |
| 45 | DCM | 54 | F | L | |
| 46 | DCM | 66 | F | L | |
| 47 | DCM | 54 | M | L | |
| 48 | DCM | 70 | M | L/F | |
| 49 | MC | 29 | M | L/F | |
| 50 | MC | 56 | F | L/F | |
| 51 | DCM | 71 | F | L/F | |
| 52 | DCMi | 77 | F | L/F | |
| 53 | DCM | 51 | M | L/F | |
| 54 | MC | 51 | F | F | |
| 55 | MC | 51 | F | F | |
| 56 | DCM | 30 | M | F | |
| 57 | DCM | 42 | F | F |
Mutation Analysis
DNA sequences were compared with the TLR3 reference sequence, NM-003265. A common single nucleotide polymorphism (rs3775291:c.1235C→T: p.L412F) was detected in nearly half of the patient cohort (Table 2), including 9 (15.8%) individuals who were homozygous for phenylalanine. Analysis of DNA from 190 race (Caucasian)- and nationality (German)-matched anonymous blood donor controls identified 9 (4.7%) as being homozygous for Phe-412. Therefore, homozygous Phe-412 was detected in significantly more of the enterovirus-positive patients than in the controls (p = 0.012, Table 3). The frequencies of the heterozygous L412F and homozygous Leu412 genotypes were not significantly different between the patient and control groups. In addition, there was no significant difference in genotypes between male and female patients. To determine whether patients with significant cardiac inflammation were more likely to be homozygous for Phe-412, we compared frequencies between patients with myocarditis (21.1%) and those with other forms of cardiomyopathy (13.2%); this difference was not statistically significant.
TABLE 3.
Statistical analysis of genotype frequencies in Coxsackievirus B3-positive patients versus controls
Patients versus controls p = 0.012 (χ2 analysis).
| Patients | Controls | |
|---|---|---|
| Homozygous F412 | 9 | 9 |
| Homozygous Leu-412 or Heterozygous L412 F | 48 | 181 |
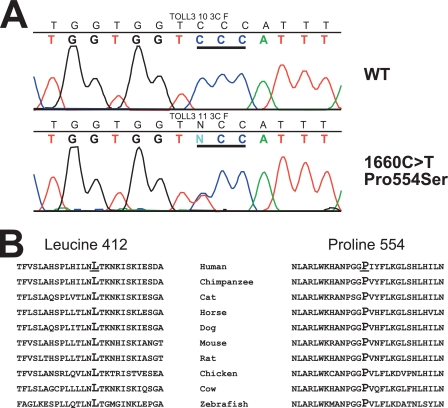
In addition, we have detected a rare variant (c.1660C→T: p.P554S) in one individual with Coxsackievirus B3 (CVB3) myocarditis (Fig. 1); this patient was homozygous for Leu-412. This substitution was not identified in the 190 German controls or in 174 US Caucasian controls. Both leucine 412 and proline 554 are highly conserved (Fig. 1B).
FIGURE 1.
Panel A, shown is mutation analysis of TLR3. The top panel shows the wild type exon 4 sequence from a control subject, whereas the bottom panel shows the detection of the heterozygous c.1660C>T (p.P554S) variant in patient 10. Codon 554 is underlined. Panel B, sequence conservation of leucine 412 and proline 554 is shown.
TLR3 Mutants Blunt Ligand-mediated TLR3 Signaling
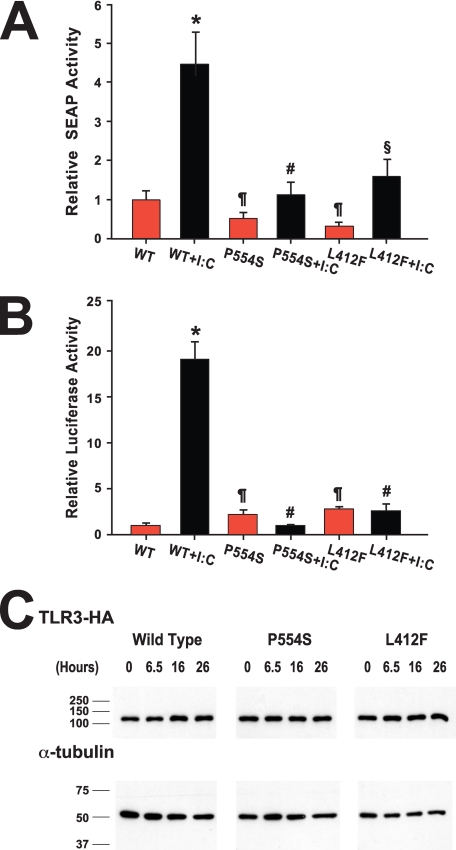
Because tissue samples from patients harboring the variants were not available for ex vivo studies of the effects of these variants on TLR3 function, we constructed TLR3 mutants encoding the P554S mutation and the L412F polymorphism, termed Ser-554 TLR3 and Phe-412 TLR3, respectively. The effect of these variants on TLR3 signaling was tested in a HEK293 cell line expressing an NF-κB-responsive product, SEAP, using the synthetic dsRNA mimic poly(I:C) as ligand. Both Phe-412 and Ser-554 TLR3 constructs had basal levels of activity statistically indistinguishable from the unstimulated WT TLR3 (Fig. 2A). Relative to unstimulated WT TLR3-expressing cells, SEAP activity in unstimulated cells was 0.5 ± 0.2 for Ser-554 TLR3 and 0.3 ± 0.1 for Phe-412 TLR3 (p > 0.05). After poly(I:C) stimulation, the WT TLR3 cells displayed significant increases in SEAP activity (p < 0.001). In contrast, SEAP activity in poly(I:C)-stimulated Ser-554 cells was statistically indistinguishable from unstimulated Ser-554-expressing cells or from unstimulated WT TLR3 cells (Fig. 2A). SEAP activity in Phe-412 TLR3 cells was significantly higher in stimulated cells compared with non-stimulated cells (p = 0.024). However, in all experiments the activity in Ser-554 or Phe-412 TLR3 cell lines was significantly lower after poly(I:C) stimulation than observed in WT TLR3 cells. In the experiment shown in Fig. 2A, SEAP activity was 4.2 ± 0.6 in poly(I:C)-stimulated WT TLR3, 1.1 ± 0.4 in Ser-554 TLR3 (p = 0.002 versus WT), and 1.4 ± 0.5 in Phe-412 TLR3 (p = 0.004 versus WT)-expressing cells.
FIGURE 2.
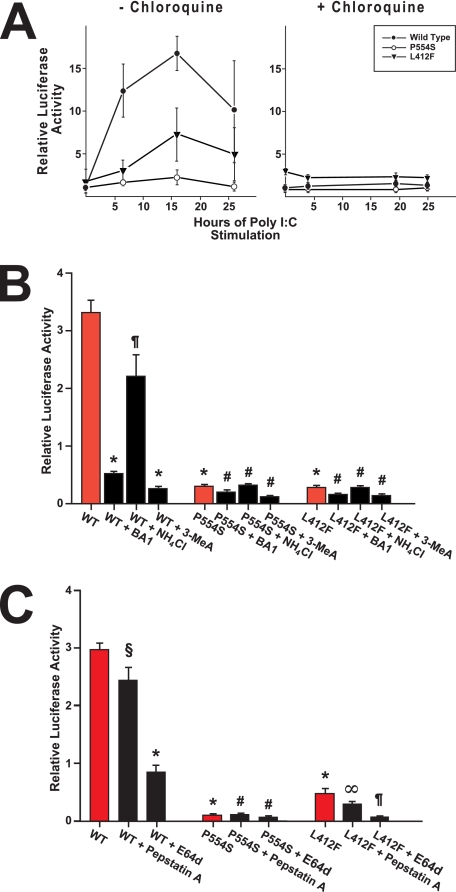
Analysis of the effect of TLR3 variants on NF-κB and type I interferon signaling in response to ligand. Panel A, cells expressing pNIFTY-SEAP were transfected with WT, Ser-554 (P554S), or Phe-412 (L412F) TLR3-HA and then stimulated with poly(I:C) (I:C: black bars) or vehicle (red bars). SEAP activities in the supernatant were measured, normalized to exogenous TLR3 expression, and expressed relative to unstimulated WT TLR3-expressing cells. *, p < 0.005 versus all other groups; #, p = NS versus all other groups except wild type + poly(I:C); ¶, p = NS versus unstimulated WT TLR3 cells; §, p = 0.024 versus unstimulated Phe-412 TLR3 cells. Panel B, stable cell lines expressing WT, Ser-554 (P554S), or Phe-412 (L412F) TLR3-HA were transfected with pISRE-LUC and then stimulated with poly(I:C) (I:C, black bars) or vehicle (red bars). Luciferase activities in the cell extracts were measured, normalized to exogenous TLR3 expression and expressed relative to unstimulated WT TLR3-expressing cells. *, p < 0.001 versus all other groups. #, p = NS versus all other groups except wild type + poly(I:C); ¶, p = NS versus unstimulated WT TLR3 cells. Panel C, stable TLR3-HA lines transfected with pISRE-LUC were stimulated with poly(I:C) for various times. Equal amounts of protein from each time point were separated by SDS-PAGE and analyzed by immunoblotting with monoclonal antibodies to the HA-epitope tag and α-tubulin.
To ensure that the aberrant signaling was not an artifact due to the transient expression of the TLR3 constructs, we generated stable cell lines expressing wild type or mutant TLR3. These cells were then transfected with a plasmid encoding luciferase under the control of a type I interferon responsive promoter. Similar results were obtained as those with NF-κB signaling. Both Phe-412 and Ser-554 TLR3 constructs had basal levels of activity statistically indistinguishable from the unstimulated WT TLR3 (Fig. 2B). In poly(I:C)-stimulated cells, relative luciferase activity was 19.1 ± 1.7 in WT TLR3, 0.9 ± 0.1 in Ser-554 TLR3 lines (p < 0.001 versus WT), and 2.6 ± 0.6 in Phe-412 TLR3 (p < 0.001 versus WT)-expressing cells. Both poly(I:C)-stimulated Phe-412- and Ser-554-expressing cells had luciferase levels that were statistically indistinguishable from unstimulated mutant cells (p > 0.05). The luciferase activity in each of the stimulated Ser-554 and Phe-412 lines also was indistinguishable from unstimulated WT cells. Analysis of more than one cell line for each mutant produced concordant results. Importantly, the differences in luciferase activity between WT and Ser-554 or Phe-412 TLR3-expressing cells was not due to differential expression of TLR3 variants as the steady-state level of TLR3 protein among the three cell lines was similar throughout a 24-h period of incubation in the presence of poly(I:C) (Fig. 2C).
TLR3 Mutants Reduce Coxsackievirus-mediated TLR3 Signaling
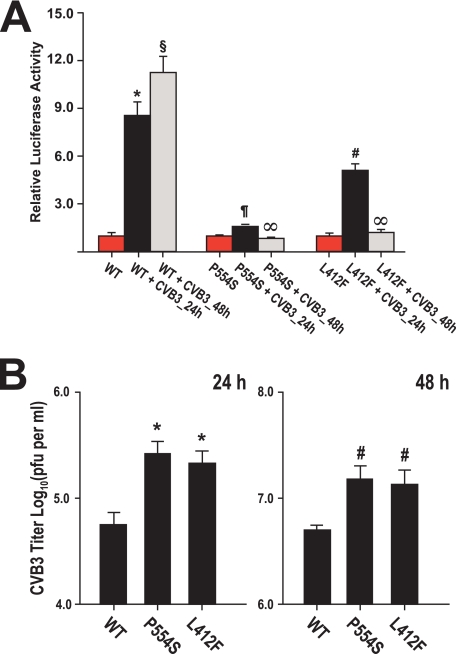
To demonstrate that the measured defects in TLR3 signaling were not related to the use of a synthetic ligand, we infected cells expressing WT or mutant TLR3 with Coxsackievirus B3 and measured the effect on activation of type I interferon signaling. Twenty-four hours after virus infection there was a 9- and 5-fold increase in signaling in WT and Phe-412 TLR3 cells, respectively, relative to uninfected cells (p < 0.001). The luciferase activity in WT cells was 8.6 ± 0.7, whereas in Phe-412, it was 5.1 ± 0.4 (Fig. 3A). By contrast, the activity in infected Ser-554 cells was statistically undistinguishable from uninfected cells. At 48 h post-infection the luciferase activity in WT cells was 11.3 ± 1.0 (p < 0.001 versus uninfected WT; p < 0.003 versus WT cells infected for 24 h), whereas the activity in Ser-554 and Phe-412 cells was undistinguishable from the activity in uninfected cells (p = NS versus uninfected Ser-554 or Phe-412 cells). When WT, Ser-554, and Phe 412 TLR3-expressing cells were infected with a lower amount of CVB3 (m.o.i. = 0.01), increased signaling was observed in the 3 cell lines at 48 h post-infection (supplemental Fig. 1). However, the luciferase activity in WT cells was more than 2-fold higher than in the mutant TLR3 lines (4.4 ± 0.4 versus 1.9 ± 0.2 in Ser-554 and 1.8 ± 0.3 in Phe-412; p < 0.002).
FIGURE 3.
Panel A, shown is analysis of the effect of TLR3 variants on type I interferon signaling after enterovirus infection (m.o.i. = 0.1). Stable cell lines expressing WT, Ser-554 (P554S) or Phe-412 (L412F) TLR3-HA were transfected with pISRE-LUC and then stimulated infected with Coxsackievirus B3. Luciferase activity was measured at 0 (red bars), 24 (black bars), and 48 h (gray bars) post-infection, normalized to exogenous TLR3 expression, and expressed relative to uninfected cells. The data shown are the means ± S.E. of eight independent measurements. *, p < 0.001 versus all other groups; §, p < 0.001 versus all other groups including WT cells infected for 24 h; ¶, p < 0.001 versus infected WT and Phe-412 cells infected for 24 h; p = NS versus uninfected Ser-554 and both Ser-554 and Phe-412 cells infected for 48 h; #, p < 0.001 versus uninfected Phe-412 and Phe-412 cells infected for 48 h; ∞, p = NS versus uninfected Ser-554 or Phe-412 cells. Panel B, a plaque assay measures the viral progeny titers in the culture media collected 24 (left) and 48 h (right) post-infection of COS7 stable lines expressing WT, Ser-554 (P554S), or Phe-412 (L412F) TLR3-HA with Coxsackievirus B3 (m.o.i. = 0.1). The data shown are the means ± S.E. of three independent experiments. *, p < 0.015 versus infected WT; ¶, p < 0.05; #, p < 0.015 versus infected WT cells.
The L412F and P554S TLR3 Mutations Increase Coxsackievirus B3 Replication
Activation of type I interferon signaling inversely correlated with Coxsackievirus B3 replication in infected WT, Phe-412, or Ser-554 TLR3-expressing cells. As shown in Fig. 3B, 24 h after infection with CVB3, the viral progeny titers in the supernatants collected from infected Ser-554 and Phe-412 TLR3-expressing cells were significantly higher than the CVB3 titer in the supernatants collected from infected WT cells: 5.3 ± 0.1 log10 pfu/ml from infected Ser-554 TLR3 and 5.3 ± 0.2 log10 pfu/ml from infected Phe-412 TLR3-expressing cells versus 4.6 ± 0.1 log10 pfu/ml from WT TLR3-expressing cells (p < 0.02 versus WT for both mutants). At 48 h after CVB3 infection, the viral titer in the supernatants from WT TLR3-expressing cells remained significantly lower than the titers in the supernatants collected from both mutant lines: 6.7 ± 0.04 log10 pfu/ml from infected WT TLR3-expressing cells versus 7.1 ± 0.1 log10 pfu/ml from infected Ser-554 TLR3 and 7.0 ± 0.2 log10 pfu/ml from infected Phe-412 TLR3 cells (p < 0.05 versus both mutants).
TLR3 Mutants Localize to Endosomes and Autophagic Compartments
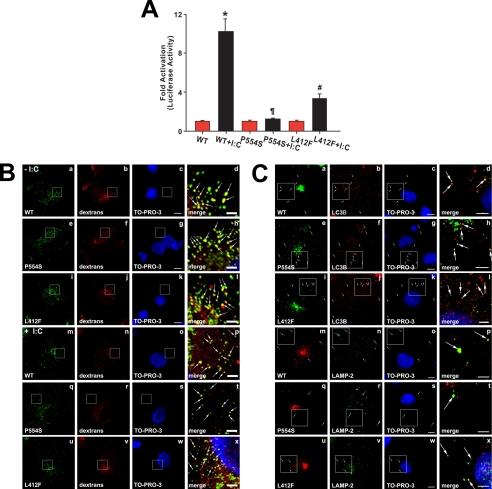
Signaling through TLR3 is initiated by binding of a dsRNA molecule ∼40 base pairs in length to two TLR3 ectodomains, thus bringing into apposition two Toll/interleukin-1 receptor domains on the cytoplasmic face of the endosome. Dimerization of the Toll/interleukin 1 receptor domain in turn enables the recruitment of the adaptor molecule TRIF (17, 18) and subsequent activation of the signaling cascade. To test whether TLR3 bearing the P554S or L412F mutations localize to endosomes, we generated stable COS7 cell lines expressing exogenous WT, Ser-554, or Phe-412 TLR3-HA. Transfection with the reporter pISRE-LUC revealed that the relative interferon signaling activity in these cells was similar to the response measured in 293 cells after poly(I:C) stimulation (compare Figs. 2B and 4A). The luciferase activity in WT TLR3-expressing COS7 cells increased 10-fold (p < 0.001 versus unstimulated WT), whereas the Ser-554 cells failed to be activated (p = NS versus unstimulated Ser-554), and the Phe-412 TLR3 COS7 cells displayed significantly reduced luciferase activity relative to WT TLR3 cells (3-fold activation: p < 0.001 versus unstimulated Phe-412 cells and p = 0.004 versus stimulated WT).
FIGURE 4.
Localization of wild type and mutant TLR3. Panel A, stable COS7 cells expressing WT, Ser-554 (P554S), or Phe-412 (L412F) TLR3-HA were transfected with pISRE-LUC and then stimulated with poly(I:C) (I:C, black bars) or vehicle (red bars). Luciferase activities in the cell extracts were measured, normalized to exogenous TLR3 expression, and expressed relative to unstimulated WT TLR3-expressing cells. The results are the mean ± S.E. of four independent measurements. *, p < 0.001 versus all other groups; ¶, p < 0.001 versus stimulated WT; #, p < 0.001 versus unstimulated Phe-412 cells. Panel B, stable COS7 cells expressing WT, Ser-554 (P554S), or Phe-412 (L412F) TLR3-HA were stimulated with poly(I:C) (+ I:C, panels m-x) or vehicle (− I:C, panels a–l) and exposed to medium containing Alexa 568® dextran conjugate for 30 min. Cells were permeabilized, fixed with 3% paraformaldehyde, and stained with an antibody against HA (to detect exogenous TLR3) and with TO-PRO-3TM iodide (to detect nuclei). The right panels show the merged and magnified images of the enclosed areas directly to the left. The white arrows indicate endocytic vesicles to which exogenous TLR3-HA variants localize. Bars, full-size images, 10 μm; enlarged images, 2.5 μm. Panel C, stable COS7 cells expressing WT, Ser-554 (P554S), or Phe-412 (L412F) TLR3-HA were stained with anti-HA, TO-PRO-3TM iodide, and either a polyclonal antibody to MAP1LC3β (to detect autophagosomes; LC3B, panels a–l) or to the lysosomal marker lysosome-associated membrane protein 2 (to detect lysosomes and autolysosomes: LAMP-2, panels m–x). Note that the shown images were obtained in the absence of poly(I:C). However, similar data were also obtained with cells incubated with poly(I:C)for up to 8 h. The enclosed areas are magnified and merged in the images to the right. The white arrows indicate autophagosomes or lysosomes in which exogenous TLR3-HA variants are present. Bars, full-size images, 10 μm; enlarged images, 5 μm.
Staining of the stable TLR3 COS7 lines with anti-HA revealed similar and predominantly punctate cytoplasmic staining (Fig. 4B). WT and mutant TLR3s were present in late endosomes/multivesicular bodies near the cell nucleus (supplemental Fig. 2) as well as on newly formed endocytic vesicles labeled with fluorescent dextran conjugate (Fig. 4B). Moreover, TLR3 staining in the three cell lines was similar in the presence (Fig. 4B, panels m–x) or absence (Fig. 4B, panels a–l) of poly(I:C), consistent with the localization of WT, Phe-412, or Ser-554 TLR3 to endosomes irrespective of their association with ligand.
Autophagy plays a role in both the innate and adaptive immune responses to promote the clearance of intracellular pathogens including viruses (19, 20). It has been reported that in mouse macrophages ligand-induced activation of either TLR1, TLR3, TLR4, TLR5, TLR6, or TLR7 induces autophagy (21–23). A hallmark of autophagosome formation is the incorporation of a lipidated form of MAP1LC3β into a double-membrane compartment that sequesters subcellular components and organelles (24). These so-called immature autophagosomes fuse with either early or late endosomes or with lysosomes to form amphisomes and autolysosomes, respectively (25). Staining of COS7 cells with anti-MAP1LC3β revealed that a fraction of WT, Ser-554, and Phe-412 TLR3 was localized to autophagosomes/amphisomes, and indeed lysosomes with or without the addition of poly(I:C)to the culture medium (Fig. 4C and data not shown), and moreover, the distribution of TLR3 proteins among endosomes and autophagic compartments was similar in cells expressing WT receptor or TLR3 bearing the L412F or P554S mutations. These data strongly support the conclusion that the changes in TLR3-mediated signaling observed with the mutant proteins cannot be explained simply by the mutant protein inability to localize to membrane compartments such as endosomes, amphisomes, or autolysosomes.
Inhibitors of Autophagy Blunt TLR3-mediated Type I Interferon Signaling
We stimulated WT and mutant TLR3-expressing cells with poly(I:C) in the presence of chloroquine, a pH-neutralizing amine that preferentially accumulates in acidified organelles such as endosomes and lysosomes and blocks membrane processes including endocytosis and late endosome/multivesicular body-to-lysosome fusion as well as the fusion of these compartments with autophagosomes and at various times then measured the amount of luciferase produced in the whole cell extracts. As shown in Fig. 5A, chloroquine ablated TLR3 signaling in both WT and mutant TLR3-expressing cells (Fig. 5A). It has been proposed that deprotonation of critical histidine residues upon elevation of endolysosomal pH interferes with dsRNA binding, thus explaining the inhibitory effect of chloroquine on TLR3 signaling (26). We tested this notion by incubating WT and mutant TLR3 cells in the presence of 3-methyladenine, an inhibitor of class III phosphatidylinositol 3-kinases required for autophagic sequestration that does not alter the endolysosomal pH (27–29). Stable 293 cells expressing WT or mutant TLR3-HA were transfected with the pISRE-LUC reporter and then stimulated with poly(I:C) in the presence of 3-methyladenine. After 24 h the amount of luciferase produced was measured in the total cell extracts and compared with the luciferase activity in unstimulated WT TLR3 cells (Fig. 5B). For comparison, the WT and mutant TLR3-expressing cells were also stimulated with poly(I:C) in the presence of bafilomycin A1 or ammonium chloride, both of which elevate lysosomal pH (30). In the presence of 3-methyladenine and bafilomycin A1, type I interferon signaling in the WT TLR3 cell line was inhibited 92 and 85%, respectively (p < 0.001 versus stimulated WT cells for both comparisons), whereas neither compound had a significant effect on signaling in the mutant TLR3 lines. By contrast, NH4Cl did not significantly inhibit interferon signaling in the WT TLR3-expressing cells (p = NS versus stimulated WT).
FIGURE 5.
Autophagy inhibitors blunt type I interferon signaling by TLR3. Panel A, chloroquine inhibits type I interferon signaling in response to ligand. Stable HEK cells expressing TLR3-HA variants were transfected with pISRE-LUC and then stimulated with poly(I:C) in the absence (left) or presence of chloroquine (right). Measured luciferase activities in the whole cell extracts were normalized to exogenous TLR3 expression and expressed relative to unstimulated WT TLR3-expressing cells. The results are the means ± S.E. of three independent measurements. Panel B, stable 293 cells expressing WT, Ser-554 (P554S), or Phe-412 (L412F) TLR3-HA were transfected with pISRE-LUC and then stimulated with poly(I:C) in the absence (red bars) or presence of either bafilomycin A1 (BA1) or NH4Cl or 3-methyladenine (3-MeA) (black bars). Luciferase activities in the cell extracts were measured, normalized to exogenous TLR3 expression, and expressed relative to unstimulated WT TLR3-expressing cells (1.00 ± 0.08, not shown). The results represent the mean ± S.E. of four measurements. *, p < 0.001 versus stimulated WT cells. ¶, p = NS versus stimulated WT; #, p = NS versus stimulated Ser-554 or Phe-412 cells. Panel C, inhibition of lysosomal proteolysis reduces TLR3 type I interferon signaling. Stable 293 cells expressing WT, Ser-554 (P554S), or Phe-412 (L412F) TLR3-HA were transfected with pISRE-LUC and then stimulated with poly(I:C) in the absence (red bars) or presence of pepstatin A or the thiol-protease inhibitor E64d (black bars). Normalized luciferase activities are expressed relative to unstimulated WT TLR3-expressing cells (1.00 ± 0.07, not shown). The results represent the means ± S.E. of four measurements. *, p < 0.001 versus stimulated WT; ¶, p < 0.001 versus stimulated Phe-412 cells; #, p = NS versus stimulated Ser-554 cells; §, p = NS versus stimulated WT; ∞, p = NS versus stimulated Phe-412 cells.
Inhibition of lysosomal proteolysis also decreased luciferase synthesis in response to poly(I:C) stimulation in both the WT and mutant TLR3 cell lines (Fig. 5C). In the presence of E64d, a broad cell-permeable inhibitor of thiol proteases that includes the majority of lysosomal cathepsins (31), the relative luciferase activity in WT TLR3-expressing cells decreased 71% (p < 0.001 versus stimulated WT cells) and 87% in Phe-412 TLR3-expressing cells (p < 0.001 versus stimulated Phe-412 cells). In Ser-554 TLR3-expressing cells the decrease (30%) did not reach statistical significance, but this may have reflected the fact that the initial signaling observed was barely above base line. Pepstatin A, an inhibitor of aspartic proteases, did not have a significant effect on interferon signaling by WT or mutant TLR3s (Fig. 5C). Overall, the results shown in Fig. 5 suggest that normal transit of TLR3 through the endolysosomal system may be required for signaling and that rather than being a consequence of TLR3 activation, autophagy may constitute a necessary step for signaling by TLR3.
Autophagy Components Are Required for Type I Interferon Signaling by TLR3
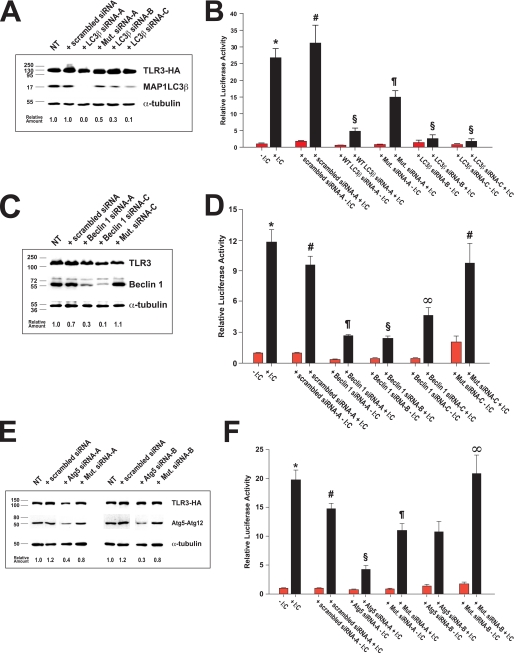
We further investigated the role of autophagy in WT TLR3 signaling by specifically decreasing the cellular levels of three proteins essential for autophagy using multiple small interfering RNAs (Table 1), thus limiting the flux of WT TLR3 through the autophagic pathway. RNA interference (RNAi) of MAP1LC3β, Beclin 1, or Atg5 resulted in significantly reduced amounts of these proteins being detected in the whole extracts of WT TLR3 cells (Fig. 6, A, C, and E; see also supplemental Fig. 5A) and was accompanied by a concomitant decrease (p < 0.001 versus stimulated WT cells) in type I interferon signaling after stimulation with poly(I:C) (Fig. 6, B, D, and F; see also Table 4). Consistent with these results, extracts from Ser-554 or Phe-412 TLR3 cells subjected to RNAi with a pool of three MAP1LC3β or Atg5 siRNAs exhibited a further decrease in signaling upon poly(I:C) stimulation (supplemental Fig. 3 and data not shown).
FIGURE 6.
Gene silencing of MAP1LC3β, Beclin 1, or Atg5 by RNA interference inhibits type I interferon signaling by TLR3. Panel A, levels of TLR3-HA, MAP1LC3β, and α-tubulin after MAP1LC3β knockdown are shown. Stable 293 cells expressing WT TLR3-HA were transfected with scrambled control siRNA-A (scrambled siRNA), one of three distinct siRNA duplexes targeting the mRNA of MAP1LC3β (LC3β siRNA-A, -B, or -C), or a version of MAP1LC3β duplex A encoding two silent mutations (Mut. siRNA-A). Equal amounts of protein (40 μg) from the whole cell extracts were then analyzed by SDS-PAGE and immunoblotting with monoclonal anti-HA, anti-α-tubulin, or a polyclonal antibody to MAP1LC3β. The amount of MAP1LC3β in each sample was determined by densitometry using NIH Image 1.63 and expressed relative to the amount of MAP1LC3β in non-transfected (NT) WT cells. Panel B, WT TLR3-HA-expressing cells were co-transfected with pISRE-LUC and either a scrambled control siRNA (control siRNA-A) or one of three MAP1LC3β siRNAs (LC3β siRNA-A, -B, or -C) or mutant LC3β siRNA-A duplex (Mut. siRNA-A) and then stimulated with poly(I:C) (+ I:C, black bars) or vehicle (red bars). The data represent the means ± S.E. of two independent experiments of quadruplicate measurements each. *, p < 0.001 versus all other groups except p = NS versus stimulated WT cells plus control siRNA-A; #, p < 0.001 versus all other groups except stimulated WT; §, p = NS versus other stimulated WT plus an LC3β siRNA except mutant siRNA-A; ¶, p < 0.001 versus all other groups. Panel C, levels of TLR3-HA, Beclin 1, and α-tubulin after Beclin 1 knockdown. WT TLR3-HA cells were transfected with scrambled siRNA-A or one of three siRNAs directed to Beclin 1 (siRNA-A, -B (see supplemental Fig. 5) or -C) or a version of siRNA-C encoding three silent mutations. Equal amounts of protein (30 μg) from the whole cell extracts were then analyzed by SDS-PAGE and immunoblotting with anti-HA, anti-α-tubulin, or a monoclonal antibody to Beclin 1. The amount of Beclin 1 in each sample was determined by densitometry and expressed relative to the amount of Beclin 1 in NT WT TLR3 cells. Panel D, WT TLR3-HA-expressing cells were transfected with pISRE-LUC and either control siRNA-A or one of three siRNA duplexes targeting Beclin 1 (siRNA-A, -B or -C) or mutant siRNA-C and then stimulated with poly(I:C) (+ I:C, black bars) or vehicle (red bars). The data represent the means ± S.E. of two or four independent experiments of eight replicate measurements each. *, p < 0.001 versus unstimulated WT cells or stimulated WT cells plus Beclin 1 siRNAs; p = NS versus stimulated WT plus control siRNA or WT treated with mutant siRNA-C (Mut. siRNA-C); #, p = NS versus stimulated WT; ¶, p < 0.001 versus all other groups except p = NS versus stimulated WT + siRNA-B; §, p < 0.001 versus all other groups except p = NS versus stimulated WT + siRNA-A; ∞, p < 0.001 versus all other groups. Panel E, levels of TLR3-HA, Atg5, and α-tubulin after Atg5 knockdown are shown. Cells expressing WT TLR3 were transfected with scrambled siRNA control or one of two duplexes targeting Atg5 (siRNA-A or -B) or modified versions of the cognate siRNAs encoding four silent mutations (Mut. siRNA-A or -B). Equal amounts of protein (30 μg) were analyzed by SDS-PAGE and immunoblotting with anti-HA, anti-α-tubulin, and monoclonal anti-Atg5 antibodies. The amount of Atg5 in the extracts was determined by densitometry and expressed relative to the amount of Atg5 in non-transfected cells. Note that the migration of Atg5 on SDS-PAGE gels is consistent with its presence in the extracts as the Atg5-Atg12 adduct (∼47 kDa). Panel F, relative luciferase activity in WT TLR3-HA-expressing cells co-transfected with pISRE-LUC reporter and cognate or mutant Atg5 siRNAs is shown. Twenty-four hours post-transfection the cells were stimulated with poly(I:C) (+ I:C, black bars) or vehicle (red bars). The data represent the means ± S.E. of two to four independent experiments of eight replicate measurements each. *, p < 0.001 versus all other groups except stimulated WT cells plus mutant siRNA-B; #, p < 0.001 versus stimulated WT + siRNA-A; p = 0.008 versus stimulated WT plus siRNA-B; p = NS versus stimulated WT + mutant siRNA-A; §, p < 0.001 versus all other groups except p = 0.002 versus stimulated WT cells treated with mutant siRNA-A; ¶, p = 0.002 versus stimulated WT plus siRNA-A; p = NS versus stimulated WT plus control siRNA-A; ∞, p < 0.001 versus stimulated WT + mutant siRNA-B. NT, “non-treated” with siRNA cells (transfected with pISRE-LUC reporter only).
TABLE 4.
TLR3-mediated interferon signaling after RNA interference of autophagy components
HEK cells expressing wild type TLR3-HA were transfected with the pISRE-LUC reporter and either a scrambled siRNA control duplex or a distinct siRNA duplex directed to MAPL1LC3β, Beclin 1, or Atg5 and then stimulated with 25 μg/ml poly(I:C) for 24 h. The amount of luciferase produced was measured in the whole cell extracts and expressed as the mean ± S.E. relative to the luciferase activity in unstimulated WT cells. Statistical comparisons of the measurements are listed in the legend to Figure 6.
| Gene target | siRNA duplex | Relative luciferase activity |
|
|---|---|---|---|
| −Poly I:C | +Poly I:C | ||
| MAP1LC3β | |||
| NTa | 1.0 ± 0.1 | 26.7 ± 2.6 | |
| Scrambled siRNA-A | 1.8 ± 0.2 | 31.2 ± 5.2 | |
| WT MAP1LC3β siRNA-A | 0.5 ± 0.1 | 4.8 ± 0.8 | |
| Mutant MAP1LC3β siRNA-A | 0.8 ± 0.1 | 15.0 ± 1.8 | |
| WT MAP1LC3β siRNA-B | 1.5 ± 0.5 | 2.7 ± 0.9 | |
| WT MAP1LC3β siRNA-C | 0.8 ± 0.2 | 1.9 ± 0.6 | |
| Beclin 1 | |||
| NT | 1.0 ± 0.1 | 11.9 ± 1.2 | |
| Scrambled siRNA-A | 1.0 ± 0.1 | 10.1 ± 0.9 | |
| Beclin 1 siRNA-A | 0.3 ± 0.1 | 2.8 ± 0.1 | |
| Beclin 1 siRNA-B | 0.5 ± 0.1 | 2.5 ± 0.2 | |
| Beclin 1 siRNA-C | 0.5 ± 0.1 | 4.7 ± 0.7 | |
| Mutant Beclin 1 siRNA-C | 2.1 ± 0.5 | 9.8 ± 1.9 | |
| Atg5 | |||
| NT | 1.0 ± 0.1 | 19.7 ± 1.6 | |
| Scrambled siRNA-A | 0.9 ± 0.1 | 11.4 ± 1.8 | |
| Atg5 siRNA-A | 0.8 ± 0.1 | 4.2 ± 0.6 | |
| Mutant Atg5 siRNA-A | 0.8 ± 0.1 | 11.1 ± 1.1 | |
| Atg5 siRNA-B | 1.4 ± 0.2 | 10.7 ± 1.8 | |
| Mutant Atg5 siRNA-B | 1.7 ± 0.3 | 20.8 ± 3.2 | |
a NT, non-treated with siRNA WT TLR3 cells were transfected with pISRE-LUC reporter only, whereas treated cells were subjected to two rounds of transfection with a specific siRNA duplex before co-transfection of the siRNA with the luciferase reporter.
To rule out the possibility that off-target silencing by the different siRNAs (32–34) might be the cause of the observed decrease in TLR3 signaling, we introduced silent mutations within the MAP1LC3β siRNA-A (Mut. siRNA-A in Fig. 6, A and B), Beclin 1 siRNA-C (Mut. siRNA-C in Fig. 6, C and D), and Atg5 siRNA-A or -B (Mut. siRNA-A or -B in Fig. 6, E and F) siRNA sequences and used the mutant siRNAs to transfect WT TLR3-expressing cells. The level of MAP1LC3β, Beclin 1, or Atg5 in extracts from WT cells treated with a mutant siRNA was significantly higher than the level of each protein in the extracts from WT TLR3 cells transfected with the corresponding wild type siRNA (Fig. 6). Furthermore, co-transfection of a mutant siRNA duplex and the pISRE-LUC reporter resulted in type I interferon signaling after poly(I:C) stimulation that was consistent with the level of MAP1LC3β, Beclin 1, or Atg5 in the cell extracts and that in most cases was statistically indistinguishable from the luciferase activity in stimulated WT cells or WT cells transfected with a scrambled control siRNA (Fig. 6, B, D, and F). Last, type I interferon signaling in WT TLR3 cells could be rescued after MAP1LC3β or Beclin 1 RNAi by exogenous expression of the recombinant proteins (supplemental Figs. 4 and 5). To this end we introduced the same silent mutations present in the Mut. LC3β siRNA-A duplex into a plasmid encoding human MAP1LC3β fused to the C terminus of enhanced green fluorescent protein (GFP; see Ref. 35 and supplementary Experimental Procedures). After treatment with wild type LC3β siRNA-A, WT TLR3-expressing cells were co-transfected with siRNA duplex and plasmid encoding either the wild type or the mutant GFP-MAP1LC3β. As shown in supplemental Fig. 4A, cells transfected with mutant GFP-MAP1LC3β plasmid expressed the fusion protein, whereas WT TLR3 cells transfected with the wild type GFP-MAP1LC3β-encoding plasmid did not. In both cases neither the WT nor the mutant plasmid had any effect on the knockdown of endogenous 17-kDa MAP1LC3β driven by the wild type siRNA.
We measured the type I interferon signaling activity of WT TLR3-expressing cells subjected to RNAi after transfection of either the wild type or the mutant GFP-MAP1LC3β-encoding plasmid. Cells were pretreated with LC3β siRNA-A for 72 h and then were transfected with the pISRE-LUC reporter, siRNA duplex and one of the two GFP-MAP1LC3β constructs. After transfection of the siRNA alone, the luciferase activity decreased more than 50% but was fully restored upon transfection of the mutant GFP-MAP1LC3β plasmid (supplemental Fig. 4B). By contrast, the activity in extracts from WT cells transfected with both duplex and wild type GFP-MAP1LC3β-encoding plasmid was statistically indistinguishable from the luciferase activity in WT cells transfected with siRNA alone. Moreover, when WT cells were transfected with Mut LC3β siRNA-A, luciferase reporter, and wild type GFP-MAP1LC3β, the cells exhibited a further increase in signaling relative to reporter-only control cells that correlated with the level of wild type fusion protein in the cell extract (data not shown). A similar approach was used to rescue the signaling activity of WT TLR3 cells after Beclin 1 silencing. The silent mutations present in mutant siRNA-C (Table 1) were introduced into a plasmid encoding WT Beclin 1 (36), and either the wild type or the mutant plasmid was transfected into cells treated with siRNA-C. Surprisingly, expression of mutant Beclin 1 rather than increased interferon signaling further reduced the luciferase activity, detected in the cell extracts, whereas expression of wild type protein restored activity (supplemental Fig. 5B). This result can be explained by the observation that elevated expression of Beclin 1 alone inhibited TLR3 signaling, and co-transfection of wild type siRNA-C- and WT Beclin 1-encoding plasmid resulted in expression levels for Beclin 1 similar to those found in non-transfected controls (supplemental Fig. 5A). Taken together, the decreased signaling observed in WT TLR3 cells treated with MAP1LC3β, Beclin 1, or Atg5 siRNAs as well as the restoration of luciferase synthesis in siRNA-treated WT cells expressing exogenous MAP1LC3β or Beclin 1 demonstrate that autophagy is essential for activation of TLR3-mediated type I interferon signaling in response to double-stranded RNA.
Activation of Autophagy after Coxsackievirus B3 Infection
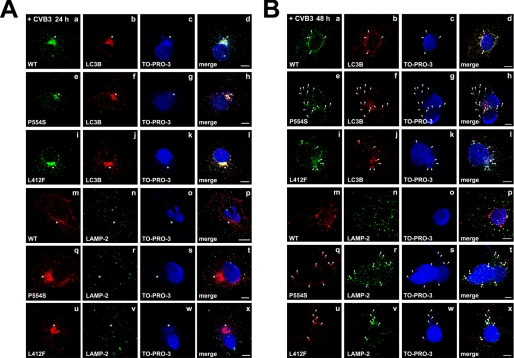
Many RNA viruses have evolved mechanisms to hijack the autophagy machinery and facilitate their own replication (35, 37). Recent studies by Wong et al. (38) have revealed that Coxsackievirus B3 induces autophagosome formation to enhance viral replication. Because our results implicate autophagy in TLR3 signaling, we observed by microscopy the effect of the P554S and L412F TLR3 mutations on autophagosome formation after CVB3 infection of COS7 TLR3 cell lines. Twenty-four hours after infection with Coxsackievirus B3, increased MAP1LC3β staining that overlapped with TLR3 in late endosomes/multivesicular bodies was observed in both the WT and mutant TLR3-expressing cells (compare Fig. 7, A, panels a–l with Fig. 4C). The observed staining was accompanied by DNA/RNA staining outside the nucleus consistent with CVB3 induction of autophagy and viral replication within autophagosomes (Fig. 7A, panels a–l). By contrast, lysosomal staining using LAMP-2 as a marker was present in individual vesicles dispersed throughout the cell (Fig. 7A, panels m–x). Forty-eight hours post-infection, TLR3 and MAP1LC3β staining no longer coalesced in a single juxtanuclear region but was present as multiple puncta distributed around the cell nucleus (Fig. 7B). The observed puncta also stained positive for LAMP-2 and nucleic acid, consistent with further fusion of autophagosomes and amphisomes with lysosomes (Fig. 7B, panels m–x). Although puncta that stained positive for TLR3, MAP1LC3β, and nucleic acid could be observed in the three cell lines, there were differences in the number of triple-labeled puncta per cell among WT and the Ser-554 or Phe-412 TLR3-expressing cells. In COS7 cells expressing WT TLR3, the number of triple-labeled puncta averaged 6.0 ± 1.0 per cell (n = 17 cells). By comparison, cells expressing Ser-554 and Phe-412 TLR3 had 16.0 ± 1.0 (n = 14 cells) and 11.0 ± 2.0 (n = 10 cells) triple-labeled puncta per cell, respectively (p < 0.01 versus WT for both). Furthermore, CVB3 continued to replicate more actively in the autolysosomes of Ser-554 and Phe-412 TLR3-expressing cells as shown by the higher number of TLR3- LAMP-2 nucleic acid triple-labeled puncta present in these cells relative to WT TLR3 COS7 cells (Fig. 7B, panels m–x). These data strongly suggest that the decreased signaling observed with the Ser-554 and Phe-412 TLR3 variants correlates with an increase in the number of autophagic compartments that support Coxsackievirus B3 replication.
FIGURE 7.
Panels A and B, activation of autophagy in stable cell lines expressing WT or mutant TLR3-HA in response to enterovirus infection is shown. Stable COS7 cells expressing WT, Ser-554 (P554S), or Phe-412 (L412F) TLR3-HA were infected with Coxsackievirus B3 (m.o.i. = 0.1) and grown for 24 (A) or 48 (B) h after infection. Cells were permeabilized, fixed with 3% paraformaldehyde, and stained with anti-HA (to detect exogenous TLR3), TO-PRO-3TM iodide (to detect nuclei), and either anti-MAP1LC3β (to detect autophagosomes; LC3B, panels a–l), or anti-LAMP-2 (to detect lysosomes and autolysosomes, panels m–x). The asterisks indicate sites of autophagosome-late endosome/multivesicular body fusion that support viral replication. The white arrowheads in Ser-554 and Phe-412 TLR3-expressing cells indicate autolysosomes visible at 48 h post-infection where CVB3 continues to replicate. Bars, 10 μm.
DISCUSSION
The results of this study show for the first time that genetic variations in TLR3 effect the host susceptibility to viral cardiomyopathies through a mechanism that involves inhibition of NF-κB and type I interferon signaling. Several lines of evidence support this statement. First, two genetic variants were identified, a rare non-synonymous substitution P554S in one patient with CVB3 myocarditis, and a common single nucleotide polymorphism, L412F, that was detected more frequently as homozygous for phenylalanine in the patient population by comparison with controls. Second, expression of both of these variants resulted in significant reductions in NF-κB and type I interferon signaling after stimulation with poly(I:C). Third, expression of both variants resulted in significant reductions in type I interferon signaling after infection with CVB3, the virus most commonly associated with viral cardiomyopathies. Finally, blunting of these signaling pathways resulted in increased viral replication. Although the scope of this study was not intended to delineate the full spectrum of mechanisms for the deleterious effects of these genetic variants, our results do suggest that the L412F and P554S TLR3 mutations allow for increase Coxsackievirus B3 replication, and their expression leads to an abnormal innate immune response.
Recently, Ranjith-Kumar et al. (39) also described the effect of the L412F polymorphism on TLR3 signaling. They reported a moderate reduction in the induction of NF-κB and type I interferon signaling after poly(I:C). However, signaling activity was not normalized to TLR3 expression. In our study, normalized activities show that this substitution results in a significant reduction in activity. These investigators also reported that the L412F variant was weakly dominant negative. We have confirmed this observation (data not shown), which is in agreement with the genetic data that only homozygous phenylalanine is associated with poor outcomes. Moreover, we have extended their observations by showing that Coxsackievirus B3 infection of cell lines expressing Phe-412 TLR3 results in a significant reduction in type I interferon activity when compared with infection of cells expressing the wild type receptor.
Zhang et al. (13) described the detection of the P554S variant in two patients with herpes simplex encephalitis. They showed that this variant, which was absent in 1581 healthy controls, results in impaired NF-κβ signaling and interferon regulatory factor-3 dimerization in response to poly(I:C). In the present study we also confirmed their observation that the Ser-554 TLR3 acts in a strongly dominant negative manner (data not shown). Although they showed that HSV-1 infection resulted in aberrant interferon production in fibroblasts isolated from the patients carrying the P554S variant, we have extended their observation to include defective signaling after CVB3 infection.
The magnitude of type I interferon induction in the WT TLR3 and Phe-412 cells after infection with CVB3 was similar to that observed with poly(I:C) stimulation (Fig. 3A). However, differences in signaling were not observed until 48 h after infection when a 10-fold lower concentration of CVB3 was used to infect the WT and mutant TLR3 lines (supplemental Fig. 1), suggesting that the CVB3 genomic RNA is not itself acting as the major ligand for TLR3. We speculate that TLR3 activation only occurs upon CVB3 replication, as has been shown for respiratory syncytial virus-induced TLR3 signaling (40). The requirement for enteroviral replication before TLR3 activation may explain the data of Zhang et al. (13), which showed that encephalomyocarditis virus infection of fibroblasts expressing either wild type or P554S TLR3 did not result in differences in interferon β and γ expression at 24 h after infection.
We have shown that the changes in TLR3 signaling observed with the Ser-554 and Phe-412 variants are unlikely to result from mislocalization of the mutated proteins as, in both cases, they appear to target to endosomes in a manner similar to wild type TLR3. Itoh et al. (41) have reported that the clathrin-dependent endocytic pathway mediates the cellular uptake of poly(I:C) and its delivery to endosomes. However, the mechanism whereby TLR3 is specifically targeted to and retained in endosomes is unknown. In this study we show that in stable COS7 transfectants endosomal targeting of WT or mutant TLR3 is not ligand-dependent.
By using a pharmacological inhibitor and siRNA knockdown approaches we have shown for the first time that autophagy plays an essential role in TLR3 mediated signaling. The autophagy pathway requires the concerted action of evolutionarily conserved proteins that coordinate the nucleation, expansion, and closure of the autophagosome and its subsequent fusion with lysosomes. Nucleation of autophagosomes is dependent on the class III phosphatidylinositol 3-kinase Vps34 complexed with Beclin 1 and other components that modulate their activity (42–45). Vesicle expansion involves proteolytic processing of MAP1LC3β by Atg4 to generate the cytosolic form LC3β-I (46, 47). Conjugation to phosphatidylethanolamine converts LC3β-I to the LC3β-II form required for association of MAP1LC3β with autophagosomal membranes (24, 48). The content of phosphatidylethanolamine-coupled MAP1LC3β governs the elongation of the isolation membrane and the size of autophagosomes (49); LC3β-II may also function to tether the forming isolation membrane to its cargo (50, 51). The function of LC3β-II depends on a second conjugation system that involves ligation of Atg12 to Atg5, an adduct that further interacts with Atg16 to form a multimeric complex that localizes to the outer isolation membrane during autophagosome expansion and presumably functions as an E3-like ligase for MAP1LC3β lipidation (52–54). Atg5 has also been implicated in immune responses independent of autophagosome formation (see Ref. 55 for a recent review). Autophagosomes can fuse with lysosomes directly, or they may fuse first with endosomes and multivesicular bodies to form amphisomes (56–59). In the present study we have shown that either elevating the luminal pH of endolysosomal compartments or preventing autophagic sequestration or inhibiting lysosomal proteolysis or reducing the cellular level of MAP1LC3β, Beclin 1, or Atg5 resulted in significant inhibition of TLR3 type I interferon signaling in response to poly(I:C) (Figs. 5 and 6). Therefore, our studies demonstrate that rather than being a consequence of TLR3 activation, autophagy is a requirement for TLR3 signaling.
The requirement for autophagy and its enhanced activity after CVB3 infection suggests that rather than beginning on the cytosolic face of endosomes, the TLR3-dependent signaling cascade likely originates on the cytosolic face of amphisomes and/or autolysosomes. Therefore, it could be expected that in the presence of poly(I:C) or after CVB3 infection the adaptor TRIF associates with TLR3-MAP1LC3β-positive compartments rather than with early or late endosomes. Further studies will be required to test this model, although the report by Shi and Kehrl that Beclin 1 binds TRIF (23) lends support to this hypothesis.
The crystal structure of TLR3 reveals that it forms a large horseshoe-shaped solenoid constituted by 23 leucine-rich repeats, which dimerizes upon binding a single dsRNA molecule. The two TLR3 ectodomains interact with the sugar-phosphate backbones on the dsRNA by means of both N-terminal and C-terminal binding sites on the glycan-free surface of each TLR3 ectodomain (26, 60–62). Critical residues for dsRNA binding have been identified at histidine 539 and asparagine 541 in the C-terminal binding site (60, 63), whereas histidine 39 and histidine 60 are essential for dsRNA binding by the N-terminal site (26, 62). Proline 554 is located adjacent to the C-terminal RNA binding site, and mutation to serine could introduce significant structural changes that preclude ligand-induced dimerization and/or prevent conformational changes that trigger downstream signaling. These two non-mutually exclusive hypotheses could explain why TLR3 bearing the P554S mutation failed to activate signaling in response to infection with Coxsackievirus B3 even though the mutant protein localized to autophagosomes (Fig. 7).
Leucine 412 is located in leucine-rich repeat 15 next to a glycosylated asparagine residue whose glycan moiety contacts the dsRNA (26). Mutation of asparagine 413 to alanine significantly reduces TLR3 signaling (64). Substitution of leucine 412 for phenylalanine could affect the glycosylation of asparagine 413 or hinder the interaction of the N-glycosyl moiety with dsRNA, explaining the reduced signaling activity of Phe-412 TLR3. Alternatively, based on the observation that the L412F mutation results in lower expression of Phe-412 TLR3 at the plasma membrane of transfected cells, Ranjith-Kumar et al. (39) have proposed that this variant is retained in the endoplasmic reticulum by the calnexin/calreticulin system of quality control. Our observation that lactacystin increases type I interferon signaling by Phe-412 TLR3 supports this hypothesis (data not shown). Thus, differences in the effects of the Phe-412 or Ser-554 variants on receptor trafficking, RNA binding, dimerization, and/or recruitment of downstream components could account for the differences in protein function with regard to whether variants act in a dominant negative fashion.
In conclusion, the findings of the present study indicate that variations in TLR3 alter the innate immune response and could influence host susceptibility to increased cardiac pathology as well as determine clinical outcomes after infection with cardiotropic viruses. Although our studies have focused on the effect of TLR3 variants in viral heart disease, Zhang et al. (13) have recently shown that the P554S variant also alters signaling in response to infection with HSV-1. Their findings indicate that TLR3 is essential for natural immunity to HSV-1 in the central nervous system. The development of animal models that express these mutant forms of TLR3 will help determine whether they indeed alter susceptibility to viral infection of the heart or central nervous system.
Supplementary Material
Acknowledgments
We are grateful to Dr. Mark Leppert (Dept. of Human Genetics, University of Utah), Dr. Robert Fujinami (Dept. of Pathology, University of Utah), and Dr. Martin Rechsteiner (Dept. of Biochemistry, University of Utah) for support and assistance.
This work was supported, in whole or in part, by National Institutes of Health Grant R21-HL091223 (NHLBI, to N. E. B. and J. G. V.). This work was supported by the American Heart Association (National Center and Texas Affiliate (to N. E. B.).

The on-line version of this article (available at http://www.jbc.org) contains supplemental “Experimental Procedures and Figs. 1–5.
- DCM
- dilated cardiomyopathy
- Atg5
- autophagy-related 5 homolog
- COS7
- African green monkey Cercopithecus aethiops kidney fibroblast-like cells
- CVB3
- Coxsackievirus B3
- E64d
- (2S,3S)-trans-epoxysuccinyl-l-leucylamido-3-methylbutane ethyl ester)
- ELAM1
- endothelial cell-leukocyte adhesion molecule
- HEK293 cells
- human embryonic kidney 293 cells
- HSV-1
- herpes simplex virus 1
- ISRE
- interferon-stimulated response element
- LAMP-2
- lysosomal membrane protein 2
- MAP1LC3β
- microtubule-associated protein l light chain 3β
- poly(I:C)
- polyinosine-polycytidylic acid
- SEAP
- secreted embryonic alkaline phosphatase
- siRNA
- small interfering double-stranded RNA
- TLR
- Toll-like receptor
- TRIF
- Toll/Interleukin 1 receptor domain-containing adapter-inducing interferon-β
- HA
- hemagglutinin tag
- PIPES
- piperazine-1,4-bis(2-ethanesulfonic acid
- NS
- not significant
- WT
- wild type
- PBS
- phosphate-buffered saline
- pfu
- plaque-forming unit
- GFP
- green fluorescent protein
- dsRNA
- double-stranded RNA.
REFERENCES
- 1.Bowles N. E., Ni J., Kearney D. L., Pauschinger M., Schultheiss H. P., McCarthy R., Hare J., Bricker J. T., Bowles K. R., Towbin J. A. (2003) J. Am. Coll. Cardiol. 42, 466–472 [DOI] [PubMed] [Google Scholar]
- 2.Bowles N. E., Vallejo J. (2003) Curr. Opin. Cardiol. 18, 182–188 [DOI] [PubMed] [Google Scholar]
- 3.Pauschinger M., Bowles N. E., Fuentes-Garcia F. J., Pham V., Kühl U., Schwimmbeck P. L., Schultheiss H. P., Towbin J. A. (1999) Circulation 99, 1348–1354 [DOI] [PubMed] [Google Scholar]
- 4.Goodwin J. F. (1983) in Myocarditis-cardiomyopathy (Just H., Schuster H. P. eds) pp. 7–11, Springer-Verlag, Berlin [Google Scholar]
- 5.Yajima T., Knowlton K. U. (2009) Circulation 119, 2615–2624 [DOI] [PubMed] [Google Scholar]
- 6.Muzio M., Polentarutti N., Bosisio D., Prahladan M. K., Mantovani A. (2000) J. Leukoc. Biol. 67, 450–456 [DOI] [PubMed] [Google Scholar]
- 7.Alexopoulou L., Holt A. C., Medzhitov R., Flavell R. A. (2001) Nature 413, 732–738 [DOI] [PubMed] [Google Scholar]
- 8.Yamamoto M., Sato S., Hemmi H., Hoshino K., Kaisho T., Sanjo H., Takeuchi O., Sugiyama M., Okabe M., Takeda K., Akira S. (2003) Science 301, 640–643 [DOI] [PubMed] [Google Scholar]
- 9.Oshiumi H., Matsumoto M., Funami K., Akazawa T., Seya T. (2003) Nat. Immunol. 4, 161–167 [DOI] [PubMed] [Google Scholar]
- 10.Hardarson H. S., Baker J. S., Yang Z., Purevjav E., Huang C. H., Alexopoulou L., Li N., Flavell R. A., Bowles N. E., Vallejo J. G. (2007) Am. J. Physiol. Heart Circ. Physiol. 292, H251–H258 [DOI] [PubMed] [Google Scholar]
- 11.Negishi H., Osawa T., Ogami K., Ouyang X., Sakaguchi S., Koshiba R., Yanai H., Seko Y., Shitara H., Bishop K., Yonekawa H., Tamura T., Kaisho T., Taya C., Taniguchi T., Honda K. (2008) Proc. Natl. Acad. Sci. U.S.A. 105, 20446–20451 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Richer M. J., Lavallée D. J., Shanina I., Horwitz M. S. (2009) PLoS ONE 4, e4127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zhang S. Y., Jouanguy E., Ugolini S., Smahi A., Elain G., Romero P., Segal D., Sancho-Shimizu V., Lorenzo L., Puel A., Picard C., Chapgier A., Plancoulaine S., Titeux M., Cognet C., von Bernuth H., Ku C. L., Casrouge A., Zhang X. X., Barreiro L., Leonard J., Hamilton C., Lebon P., Héron B., Vallée L., Quintana-Murci L., Hovnanian A., Rozenberg F., Vivier E., Geissmann F., Tardieu M., Abel L., Casanova J. L. (2007) Science 317, 1522–1527 [DOI] [PubMed] [Google Scholar]
- 14.Aretz H. T., Billingham M. E., Edwards W. D., Factor S. M., Fallon J. T., Fenoglio J. J., Jr., Olsen E. G., Schoen F. J. (1987) Am. J. Cardiovasc. Pathol. 1, 3–14 [PubMed] [Google Scholar]
- 15.Kühl U., Noutsias M., Seeberg B., Schultheiss H. P. (1996) Heart 75, 295–300 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bowles K. R., Abraham S. E., Brugada R., Zintz C., Comeaux J., Sorajja D., Tsubata S., Li H., Brandon L., Gibbs R. A., Scherer S. E., Bowles N. E., Towbin J. A. (2000) Genomics 67, 109–127 [DOI] [PubMed] [Google Scholar]
- 17.Matsumoto M., Funami K., Tanabe M., Oshiumi H., Shingai M., Seto Y., Yamamoto A., Seya T. (2003) J. Immunol. 171, 3154–3162 [DOI] [PubMed] [Google Scholar]
- 18.Funami K., Matsumoto M., Oshiumi H., Akazawa T., Yamamoto A., Seya T. (2004) Int. Immunol. 16, 1143–1154 [DOI] [PubMed] [Google Scholar]
- 19.Mizushima N., Levine B., Cuervo A. M., Klionsky D. J. (2008) Nature 451, 1069–1075 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Shoji-Kawata S., Levine B. (2009) Biochim. Biophys. Acta 1793, 1478–1484 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Delgado M. A., Elmaoued R. A., Davis A. S., Kyei G., Deretic V. (2008) EMBO J. 27, 1110–1121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Shi C. S., Kehrl J. H. (2008) J. Biol. Chem. 283, 33175–33182 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Xu Y., Jagannath C., Liu X. D., Sharafkhaneh A., Kolodziejska K. E., Eissa N. T. (2007) Immunity 27, 135–144 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kabeya Y., Mizushima N., Ueno T., Yamamoto A., Kirisako T., Noda T., Kominami E., Ohsumi Y., Yoshimori T. (2000) EMBO J. 19, 5720–5728 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Fader C. M., Colombo M. I. (2009) Cell Death Differ. 16, 70–78 [DOI] [PubMed] [Google Scholar]
- 26.Liu L., Botos I., Wang Y., Leonard J. N., Shiloach J., Segal D. M., Davies D. R. (2008) Science 320, 379–381 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Seglen P. O., Gordon P. B. (1982) Proc. Natl. Acad. Sci. U.S.A. 79, 1889–1892 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Blommaart E. F., Krause U., Schellens J. P., Vreeling-Sindelárová H., Meijer A. J. (1997) Eur. J. Biochem. 243, 240–246 [DOI] [PubMed] [Google Scholar]
- 29.Petiot A., Ogier-Denis E., Blommaart E. F., Meijer A. J., Codogno P. (2000) J. Biol. Chem. 275, 992–998 [DOI] [PubMed] [Google Scholar]
- 30.Dröse S., Altendorf K. (1997) J. Exp. Biol. 200, 1–8 [DOI] [PubMed] [Google Scholar]
- 31.Barrett A. J., Kembhavi A. A., Brown M. A., Kirschke H., Knight C. G., Tamai M., Hanada K. (1982) Biochem. J. 201, 189–198 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Saxena S., Jónsson Z. O., Dutta A. (2003) J. Biol. Chem. 278, 44312–44319 [DOI] [PubMed] [Google Scholar]
- 33.Jackson A. L., Bartz S. R., Schelter J., Kobayashi S. V., Burchard J., Mao M., Li B., Cavet G., Linsley P. S. (2003) Nat. Biotechnol. 21, 635–637 [DOI] [PubMed] [Google Scholar]
- 34.Scacheri P. C., Rozenblatt-Rosen O., Caplen N. J., Wolfsberg T. G., Umayam L., Lee J. C., Hughes C. M., Shanmugam K. S., Bhattacharjee A., Meyerson M., Collins F. S. (2004) Proc. Natl. Acad. Sci. U.S.A. 101, 1892–1897 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Jackson W. T., Giddings T. H., Jr., Taylor M. P., Mulinyawe S., Rabinovitch M., Kopito R. R., Kirkegaard K. (2005) PLoS Biol. 3, e156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Shibata M., Lu T., Furuya T., Degterev A., Mizushima N., Yoshimori T., MacDonald M., Yankner B., Yuan J. (2006) J. Biol. Chem. 281, 14474–14485 [DOI] [PubMed] [Google Scholar]
- 37.Wileman T. (2006) Science 312, 875–878 [DOI] [PubMed] [Google Scholar]
- 38.Wong J., Zhang J., Si X., Gao G., Mao I., McManus B. M., Luo H. (2008) J. Virol. 82, 9143–9153 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ranjith-Kumar C. T., Miller W., Sun J., Xiong J., Santos J., Yarbrough I., Lamb R. J., Mills J., Duffy K. E., Hoose S., Cunningham M., Holzenburg A., Mbow M. L., Sarisky R. T., Kao C. C. (2007) J. Biol. Chem. 282, 17696–17705 [DOI] [PubMed] [Google Scholar]
- 40.Rudd B. D., Burstein E., Duckett C. S., Li X., Lukacs N. W. (2005) J. Virol. 79, 3350–3357 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Itoh K., Watanabe A., Funami K., Seya T., Matsumoto M. (2008) J. Immunol. 181, 5522–5529 [DOI] [PubMed] [Google Scholar]
- 42.Sun Q., Fan W., Chen K., Ding X., Chen S., Zhong Q. (2008) Proc. Natl. Acad. Sci. U.S.A. 105, 19211–19216 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Itakura E., Kishi C., Inoue K., Mizushima N. (2008) Mol. Biol. Cell 19, 5360–5372 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zhong Y., Wang Q. J., Li X., Yan Y., Backer J. M., Chait B. T., Heintz N., Yue Z. (2009) Nat. Cell Biol. 11, 468–476 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Matsunaga K., Saitoh T., Tabata K., Omori H., Satoh T., Kurotori N., Maejima I., Shirahama-Noda K., Ichimura T., Isobe T., Akira S., Noda T., Yoshimori T. (2009) Nat. Cell Biol. 11, 385–396 [DOI] [PubMed] [Google Scholar]
- 46.Ichimura Y., Kirisako T., Takao T., Satomi Y., Shimonishi Y., Ishihara N., Mizushima N., Tanida I., Kominami E., Ohsumi M., Noda T., Ohsumi Y. (2000) Nature 408, 488–492 [DOI] [PubMed] [Google Scholar]
- 47.Mariño G., Uría J. A., Puente X. S., Quesada V., Bordallo J., López-Otín C. (2003) J. Biol. Chem. 278, 3671–3678 [DOI] [PubMed] [Google Scholar]
- 48.Kabeya Y., Mizushima N., Yamamoto A., Oshitani-Okamoto S., Ohsumi Y., Yoshimori T. (2004) J. Cell Sci. 117, 2805–2812 [DOI] [PubMed] [Google Scholar]
- 49.Xie Z., Nair U., Klionsky D. J. (2008) Mol. Biol. Cell 19, 3290–3298 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Pankiv S., Clausen T. H., Lamark T., Brech A., Bruun J. A., Outzen H., Øvervatn A., Bjørkøy G., Johansen T. (2007) J. Biol. Chem. 282, 24131–24145 [DOI] [PubMed] [Google Scholar]
- 51.Münz C. (2009) Annu. Rev. Immunol. 27, 423–449 [DOI] [PubMed] [Google Scholar]
- 52.Kuma A., Mizushima N., Ishihara N., Ohsumi Y. (2002) J. Biol. Chem. 277, 18619–18625 [DOI] [PubMed] [Google Scholar]
- 53.Hanada T., Noda N. N., Satomi Y., Ichimura Y., Fujioka Y., Takao T., Inagaki F., Ohsumi Y. (2007) J. Biol. Chem. 282, 37298–37302 [DOI] [PubMed] [Google Scholar]
- 54.Fujita N., Itoh T., Omori H., Fukuda M., Noda T., Yoshimori T. (2008) Mol. Biol. Cell 19, 2092–2100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Virgin H. W., Levine B. (2009) Nat. Immunol. 10, 461–470 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Gordon P. B., Seglen P. O. (1988) Biochem. Biophys. Res. Commun. 151, 40–47 [DOI] [PubMed] [Google Scholar]
- 57.Liou W., Geuze H. J., Geelen M. J., Slot J. W. (1997) J. Cell Biol. 136, 61–70 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Berg T. O., Fengsrud M., Strømhaug P. E., Berg T., Seglen P. O. (1998) J. Biol. Chem. 273, 21883–21892 [DOI] [PubMed] [Google Scholar]
- 59.Fader C. M., Sánchez D., Furlán M., Colombo M. I. (2008) Traffic 9, 230–250 [DOI] [PubMed] [Google Scholar]
- 60.Bell J. K., Askins J., Hall P. R., Davies D. R., Segal D. M. (2006) Proc. Natl. Acad. Sci. U.S.A. 103, 8792–8797 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Choe J., Kelker M. S., Wilson I. A. (2005) Science 309, 581–585 [DOI] [PubMed] [Google Scholar]
- 62.Pirher N., Ivicak K., Pohar J., Bencina M., Jerala R. (2008) Nat. Struct. Mol. Biol. 15, 761–763 [DOI] [PubMed] [Google Scholar]
- 63.Ranjith-Kumar C. T., Miller W., Xiong J., Russell W. K., Lamb R., Santos J., Duffy K. E., Cleveland L., Park M., Bhardwaj K., Wu Z., Russell D. H., Sarisky R. T., Mbow M. L., Kao C. C. (2007) J. Biol. Chem. 282, 7668–7678 [DOI] [PubMed] [Google Scholar]
- 64.Sun J., Duffy K. E., Ranjith-Kumar C. T., Xiong J., Lamb R. J., Santos J., Masarapu H., Cunningham M., Holzenburg A., Sarisky R. T., Mbow M. L., Kao C. (2006) J. Biol. Chem. 281, 11144–11151 [DOI] [PubMed] [Google Scholar]
- 65.Pei Y., Tuschl T. (2006) Nat. Methods 3, 670–676 [DOI] [PubMed] [Google Scholar]
- 66.Cullen B. R. (2006) Nat. Methods 3, 677–681 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.