Abstract
The Ca2+- and cAMP-responsive element-binding protein (CREB) and the related ATF-1 and CREM are stimulus-inducible transcription factors that link certain forms of cellular activity to changes in gene expression. They are attributed to complex integrative activation characteristics, but current biochemical technology does not allow dynamic imaging of CREB activation in single cells. Using fluorescence resonance energy transfer between mutants of green fluorescent protein we here develop a signal-optimized genetically encoded indicator that enables imaging activation of CREB due to phosphorylation of the critical serine 133. The indicator of CREB activation due to phosphorylation (ICAP) was used to investigate the role of the scaffold and anchoring protein AKAP79/150 in regulating signal pathways converging on CREB. We show that disruption of AKAP79/150-mediated protein kinase A anchoring or knock-down of AKAP150 dramatically reduces the ability of protein kinase A to activate CREB. In contrast, AKAP79/150 regulation of CREB via L-type channels may only have minor importance. ICAP allows dynamic and reversible imaging in living cells and may become useful in studying molecular components and cell-type specificity of activity-dependent gene expression.
Keywords: Calcium, Fluorescence, Fluorescence Resonance Energy Transfer (FRET), Gene Expression, Neurochemistry, CREB, GFP
Introduction
Changes in gene expression are a hallmark of long term adaptive processes in many cell types and are crucially involved in the conversion from short term to long term plasticity of neuronal circuits. Transcription factors of the cAMP-responsive element-binding protein (CREB)3 family have a prominent role in affecting such processes in various tissues and cell types (1–5). The family includes the homologous proteins CREB (6, 7), ATF-1 (8), and CREM (9). Upon stimulation a number of kinases phosphorylate the critical serine 133 residue within the kinase-inducible domain (KID) of CREB and the related transcription factors ATF-1 and CREM. Serine 133 phosphorylation leads to the recruitment of transcriptional coactivators, like CBP or its paralogue P300 that bind to the KID via the KID interaction domain (KIX) (10, 11). Interaction of CREB with CBP/P300 results in the assembly of the RNA-polymerase complex and in the expression of a large number of genes. Serine 133 phosphorylation is generally accepted as a key event in transcriptional regulation necessary although not in all cases sufficient, for CREB-mediated gene expression. Many studies focused on examining spatiotemporal profiles of CREB activation in cells and tissues by immunostainings using antibodies specific for serine 133 phosphorylation (for a selection, see Refs. 12–17). Few attempts were made to develop assays for real time imaging of CREB, a prerequisite for thorough analysis of the physiological events and the molecular dissection of signaling cascades converging on CREB (18, 19). Genetically encoded indicators based on mutants of the green fluorescent protein (GFP) have become valuable tools to monitor spatiotemporal patterns of biochemical signals in live cells (20–25). A number of indicators have been constructed that report activities of kinases such as PKA (26–29), calmodulin-dependent kinase II (CaMK II) (30, 31), MAP/extracellular signal-regulated kinase(ERK) kinase (32–35), gradients of cAMP (36, 37), or calcineurin activation (38). Here we generate a sensor that allows imaging activation of CREB in the nucleus of single live cells and use it to study the role of the anchor protein AKAP79/150 (AKAP79 is the human, AKAP150 the rodent variant) in CREB signaling. “A kinase anchor proteins” (AKAPs) have been identified as strategic scaffold proteins that regulate the architecture and spatiotemporal properties of signal transduction pathways (39). They do so by organizing the assembly of signaling complexes that recruit key enzymes such as kinases, phosphatases, and some of their substrates to distinct subcellular environments. AKAP79/150 is a multivalent anchor protein that simultaneously binds PKA, calcineurin, and protein kinase C (PKC) (40, 41), modulates L-type voltage-dependent calcium channels (42) and furthermore, interacts with AMPA and NMDA receptors at postsynaptic sites (43, 44). A role of AKAP79/150 in modulating gene expression through regulation of NFATc4 was recently demonstrated (41). The involvement of AKAP79/150-dependent signaling complexes in organizing pathways converging on CREB thus appeared as a suitable testing ground to demonstrate usefulness of ICAP.
EXPERIMENTAL PROCEDURES
Plasmid Construction
Amino acids 121–160 of the kinase-inducible domain (KID) and amino acids 586–662 of CREB-binding protein (CBP), including the linker GGSGGT, were generated by standard PCR from the full-length CREB cDNA. Both fragments were cloned into pRSET B (Invitrogen), the KID fragment with SphI/KpnI and the KIX fragment with KpnI/SacI, between CFP (BamHI/SphI) and Citrine (SacI/EcoRI). All cloning enzymes were purchased from New England Biolabs. Site-directed mutagenesis was done with the QuikChange Kit from Stratagene. For expression in mammalian cells the construct was subcloned into pcDNA3 (Invitrogen). Nucleus targeting was achieved by fusing the amino acid sequence PKKKRKVEDA to the N terminus of the CFP. The VIVIT-TagRFP construct was generated by standard PCR using the forward primer, 5′-GGA TCC ATG GCT GGC CCT CAT CCT GTG ATC GTG ATC ACC GGT CCT CAT GAA GAA ATG GTG TCT AAG GGC GAA GAGC and the reverse primer, 5′-GAATTC TTA ATT AAG TTT GTG CCC CAG TTT GC and cloned, as a BamHI/EcoRI fragment, to pcDNA3. The mCherry fluorescent protein was PCR amplified from pRSET-B-mCherry with BamHI/NotI. The pEGFP-N1-AKAP79-mCherry was constructed by replacing the yellow fluorescent protein gene in the pEGFP-N1-AKAP79 vector with mCherry.
Protein Expression, in Vitro Spectroscopy
Proteins were expressed in Escherichia coli BL21 and purified as described previously (45). Polyhistidine-tagged ICAP expressed using the plasmid vector pRSETB (Invitrogen) had a molecular mass of 71 kDa (versus 69.5 kDa for NLS-ICAP expressed in mammalian cells). ICAP in vitro fluorescence measurements were performed in a Cary Eclipse fluorometer (Varian, Darmstadt, Germany). The purified construct was treated with the catalytic subunit of PKA (New England Biolabs, 2.5–5 units/μl) and CaMK IV (Millipore, 2.5 units/μl) in the corresponding kinase reaction buffer at room temperature. ATP was purchased from Sigma. Samples were excited at 432 nm and emission was scanned from 440 to 600 nm. Traces shown are representative of at least three independent experiments. For Western blot analysis the purified proteins were separated by SDS-PAGE, transferred onto nitrocellulose membrane, and initially probed using an anti-GFP antibody (Research Diagnostics Inc., MA). The membrane was subsequently stripped of the probe via the use of a stripping buffer (62.5 mm Tris, 100 mm 2-mecaptoethanol, 2% SDS, pH 6.8) incubated at 50 °C for 30 min before labeling with an anti-phosphoserine 133 specific antibody (New England Biolabs).
Cell Culture and Imaging
HeLa cells were plated onto sterile glass bottom dishes and grown in Dulbecco's modified Eagle's medium (Invitrogen) supplemented with 10% fetal bovine serum at 37 °C in 6% CO2. Cells were transfected with Lipofectamine 2000 reagent (Invitrogen) and left for 24 h at 37 °C. Prior to Western blotting the cells were serum starved for 3 h in 0.5% fetal bovine serum followed by stimulation with 50 μm forskolin (Sigma) and 1 mm IBMX (Sigma) for 0, 2-, 5-, 10-, 30-, 60-, and 120-min time points. Additional untransfected HeLa cells were also stimulated with forskolin and IBMX as a further control. For Western blotting cells were lysed in a SDS lysis buffer, separated by SDS-PAGE, transferred onto nitrocellulose membrane, and initially probed with an anti-phosphoserine 133 specific antibody (New England Biolabs) before probing with an anti-glyceraldehyde-3-phosphate dehydrogenase (Sigma) loading control antibody. Western blots were quantified using ImageJ software and normalized to the 0-min control. Hippocampal neurons were prepared from 17–18-day-old rat embryos and transfected by calcium phosphate precipitation for 1–4 h. Neurons were imaged 1–2 days after transfection and before imaging were held at room temperature for 30 min. All biochemicals were purchased from Sigma and used in the indicated concentrations. Brain-derived neurotrophic factor (BDNF) was a gift of S. Lang, MPI for Neurobiology, Munich. Neurons were pretreated at least 30 min with the indicated inhibitor. U0126 and KN-62 were used at 10 μm, H-89 at 25 μm unless indicated differently. Nimodipine was used at 5 μm, St-Ht31 and St-Ht31pro at 10 μm. For knock-down of endogenous AKAP150 plasmids coding for short hairpin RNAs (shRNA) (AKAP 150 shRNA or control AKAP 150 shRNA, Santa Cruz Biotechnology, Santa Cruz, CA) were introduced into P1 hippocampal neurons using Lipofectin 2000-mediated transfection. Neurons were stained and analyzed 9–11 days after transfection. Fura-2-AM (Molecular Probes/Invitrogen) (5 μm) was dissolved in dimethyl sulfoxide/pluronic acid and cells were incubated 30 min with the solution, then washed and allowed to recover for 30 min. Cells were imaged on a Zeiss Axiovert 35M microsope with a charge-coupled device camera (CoolSnap, Roper Scientific, AZ). The imaging setup was controlled by Metafluor version 4.6 software (Universal Imaging, Puchheim, Germany). For ratio imaging, a 440/20 excitation filter, a 455-dichroic long-pass mirror, and two emission filters (485/35 for CFP, 535/25 for Citrine) operated in a filter wheel (Sutter instruments, CA) were used. For fura-2 imaging, 350 and 380 nm excitation filters and a 500/20 emission filter were used. Statistical analyses were performed using the two-tailed Student's t test. All error bars indicate mean ± S.E. Image processing was done using Image J.
Immunocytochemistry
DIV9–11 neurons were fixed in 4% paraformaldehyde, 4% sucrose in phosphate-buffered saline, permeabilized in 0.25% Triton X-100 in phosphate-buffered saline, and blocked in phosphate-buffered saline, 5% normal goat serum. Neurons were stained with rabbit polyclonal antibody to AKAP150 (1:300, Upstate, NY) and Alexa 568-conjugated goat anti-rabbit (1:500, Invitrogen) secondary antibody and coverslipped in Fluoromount (Sigma). All images were acquired as single layers using a laser-scanning confocal microscope (Leica SP2, Wetzlar, Germany) with a ×40 oil immersion objective.
RESULTS
Development of a Biosensor for CREB Activation
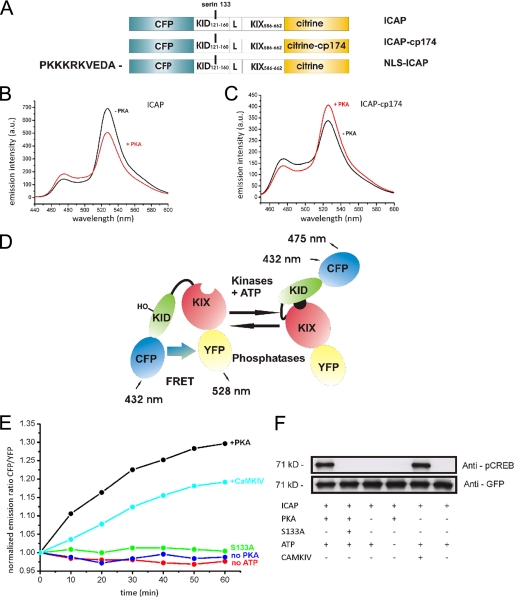
Our intention was to construct sensors that reliably report phosphorylation of the critical serine 133 by using FRET between mutants of GFP. The KIX domain (CREB interaction domain) of CBP was considered a specific module recognizing serine 133-phosphorylated KID and was incorporated into the sensor to initiate a specific conformational change upon phosphorylation of KID. Thus, to monitor CREB activation in living cells we fused amino acids 121–160 of the kinase-inducible domain (KID) of CREB, together with a flexible linker GGSGGT, to amino acids 586–662 of CBP (KIX), and sandwiched this construct between cyan fluorescent protein (CFP) and the yellow fluorescent protein variant Citrine (45) (Fig. 1A). This final variant, after a few tens of constructs to optimize signal strength, was called ICAP (indicator of CREB activation due to phosphorylation). We purified the protein and tested its properties in vitro (Fig. 1). ICAP started out with high FRET under basal conditions. Addition of recombinant PKA or CaMK IV reduced FRET, thus increasing the emission intensity of CFP and decreasing that of Citrine (Fig. 1, B and E). The maximal ratio change was 35–40%. Interestingly, substitution of Citrine with the circularly permuted variant Citrine cp174 (46–48) completely reverted the response behavior of ICAP. ICAP-cp174 showed lower FRET under basal conditions, which increased after phosphorylation with PKA, demonstrating that it is possible to tune the response behavior of genetically encoded sensors in more fundamental ways than previously expected (Fig. 1C). To confirm that the conformational change is specifically dependent on phosphorylation of serine 133 and on PKA and ATP, which are the components of the in vitro assay, we performed control experiments. In vitro kinase assays with one of the components excluded from the reaction or the crucial serine 133 mutated to alanine (S133A) did not result in a FRET change after 60 min in vitro (Fig. 1E). Western blot analysis using antibodies specific for phosphoserine 133-CREB verified that the sensor is phosphorylated in vitro under our assay conditions (Fig. 1F). Overall, these experiments show that the sensors are specifically activated in vitro by phosphorylation of serine 133 (data not shown). Further in vitro characterization addressing pH sensitivity and overall FRET efficiency within ICAP is provided in supplemenatal Fig. 1. Using a similar strategy, replacement of the KID domain of CREB within the sensor with the homologous KID domains of ATF-1 or CREM, resulted in functional sensors of ATF-1 or CREM activation. These sensors showed maximal ratio changes of 22–40 and 20–30%, respectively, when stimulated with PKA (supplemental Fig. 2).
FIGURE 1.
In vitro characterization of ICAP. A, schematic representation of constructs. A segment of the kinase-inducible domain (KID) of CREB was connected to the CREB-interaction domain (KIX) of CBP via a short linker (L). The KID-KIX fusion was then sandwiched between CFP and Citrine. Alternatively, a circularly permuted variant of Citrine was used as acceptor to generate ICAP-cp174. For transfection into mammalian cells a nuclear (NLS-ICAP) localization sequence was added to the N terminus. The KID domain of CREB could be replaced by those of ATF-1 and CREM, resulting in fluorescent sensors I-ATF and I-CREM. Maximal signals after phosphorylation with PKA are indicated as percent change in emission ratio. B and C, emission spectra of ICAP (B) and ICAP-cp174 (C) before (black line) and 60 min after phosphorylation (red line) of the sensor using protein kinase A. Note that ICAP-cp174 is reverted in its response behavior to phosphorylation. D, ICAP shows high FRET under resting conditions. Upon phosphorylation of the critical serine 133 (indicated as HO) within the KID domain, KID interacts with the KIX domain thereby reducing FRET from CFP to Citrine. E, in vitro time course of the ratio change in ICAP after phosphorylation with recombinant protein kinase A or CaMK IV. A ratio change was absent when either PKA or ATP was omitted from the assay or when the critical serine 133 was mutated to alanine. F, Western blot analysis of recombinant purified ICAP using antibodies specific for phosphorylated serine 133-KID (anti-pCREB) or GFP after various treatments.
Validation of ICAP in HeLa Cells and Hippocampal Neurons
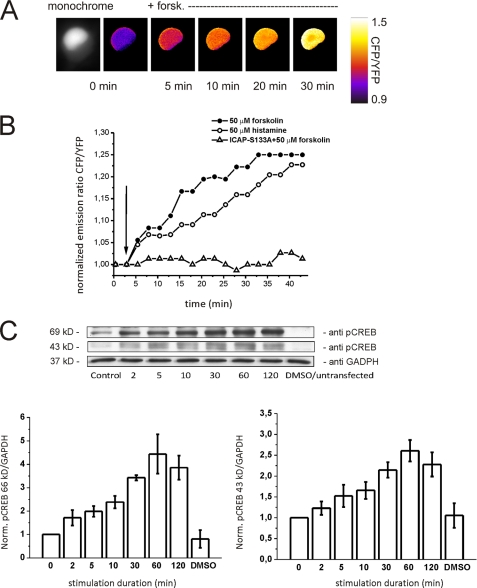
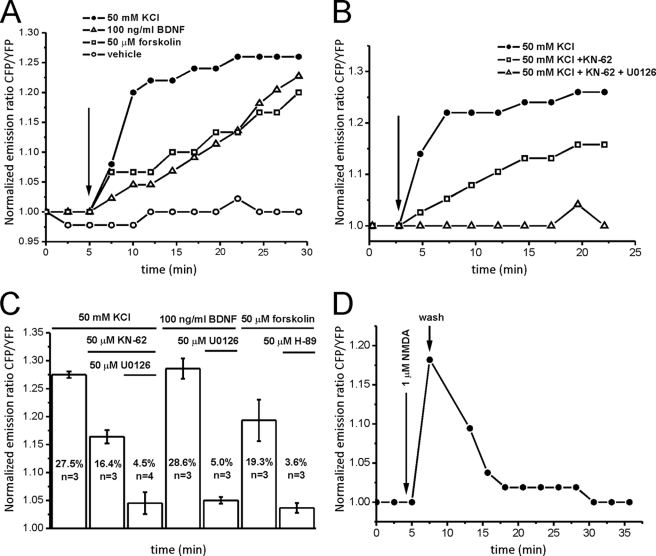
We fused a nuclear localization sequence to the sensor to specifically express ICAP in the nucleus of living cells (Figs. 1A and 2A). Transfection of the NLS-ICAP into hippocampal neurons (Fig. 2A) or HeLa cells reported activation of CREB by forskolin or histamine in a time-dependent manner (Fig. 2B), which was completely abolished by the S133A mutation within ICAP. To verify that the activation of ICAP corresponds well to the activation of endogenous CREB we performed Western blot analysis using lysates of forskolin-IBMX-stimulated HeLa cells transfected with ICAP. Endogenous CREB phosphorylation and ICAP phosphorylation could be well discriminated due to the difference in molecular mass of 43 and 69 kDa of endogenous CREB and the biosensor, respectively (Fig. 2C). The overall time course of phosphorylation of endogenous CREB and ICAP corresponded well. When transfected into primary hippocampal neurons NLS-ICAP reported three pathways with different activation kinetics converging on CREB, a fast high potassium-induced CREB activation and slower activations induced by forskolin or the neurotrophin BDNF (Fig. 3A). The high potassium-induced activation could be dissected in a fast component blocked by KN-62, an antagonist of calcium/calmodulin-dependent kinases, and a slower remaining activation blocked by UO126, an inhibitor of the MAP kinase pathway (Fig. 3, B and C). These results obtained with high potassium stimulation are in good agreement with previous studies that examined the time course of phosphorylation of endogenous CREB after depolarization and another demonstration that ICAP signals are an excellent measure for activation of endogenous CREB (16, 49, 50). Activation by forskolin was efficiently blocked by the PKA antagonist H-89 (Fig. 3C), whereas the BDNF-induced activation was inhibited by the MAP kinase blocker U0126 (Fig. 3C) (51–53). Time constants for the rise were 2.2 ± 0.42 min for the fast high potassium-mediated activation, and 47 ± 0.28 and 52.5 ± 0.43 min for the slow forskolin and BDNF-induced activations, respectively. Decay times were more heterogeneous in nature. In stimulations with high potassium, CREB activation persisted significantly after stimulations had ended. Stimulation with 1 μm NMDA, however, led to the rapid return of CREB to baseline levels within 5–10 min after wash-out of the drug (Fig. 3D), which has been attributed to induction of a CREB shut-off pathway (54) and was a good demonstration of the reversibility of ICAP signals in vivo. ICAP, furthermore, faithfully reported developmentally regulated CREB activation via GABAa and GABAb receptors and build-up and summation of CREB activation by succeeding repetitive calcium spikes (supplemental Figs. 3 and 4). To assess background signals due to bleaching or residual activation in the absence of stimulation we transfected NLS-ICAP into HeLa cells and imaged cells for 30 min in 30-s intervals. Under these conditions we observed a slight increase in CFP emission and decrease in Citrine emission presumably due to preferential bleaching of the acceptor protein (supplemental Fig. 5).
FIGURE 2.
Evaluation of ICAP in living cells. A, time lapse ratio image of a HeLa cell transfected with NLS-ICAP and stimulated with forskolin at the indicated time point (+forsk). Monochrome image displays the nucleus-targeted ICAP fluorescence (Citrine channel). A dim halo of background fluorescence is visible in the cytosol. Neurites were too dim to be visualized at these acquisition rates. B, CREB activation in HeLa cells stimulated with forskolin (50 μm) or histamine (50 μm). Cells transfected with NLS-ICAP S133A in which the critical serine 133 is mutated to alanine did not show any ratio changes after stimulation with forskolin, demonstrating the in vivo specificity of ICAP. C, comparison of ICAP phosphorylation (band at 69 kDa) and phosphorylation of endogenous CREB (band at 43 kDa) by Western blot in HeLa cells transfected with NLS-ICAP and stimulated with 50 μm forskolin and 1 mm IBMX. Anti-glyceraldehyde-3-phosphate dehydrogenase (GAPDH) was used as loading control. The band at 43 kDa is a double band as the antibody also recognizes phosphorylated ATF-1. Graphs provide quantifications (mean ± S.E.) of three independent determinations. DMSO, dimethyl sulfoxide; YFP, yellow fluorescent protein.
FIGURE 3.
Validation of ICAP in living hippocampal neurons. A, three pathways with different temporal features converge on CREB in primary hippocampal neurons. A fast high potassium-mediated CREB activation can be differentiated from slower activations by forskolin (50 μm) or the neurotrophin BDNF (100 ng/ml). B, the high potassium-mediated activation is dissected into a fast calcium-calmodulin kinase-mediated pathway that is blocked by KN-62 (50 μm) and a slower component mediated by the MAP kinase pathway that is blocked by U0126 (50 μm). C, summary of the pharmacology of CREB activation (mean ± S.E.) after stimulation with high potassium, forskolin, and BDNF. CREB activation was determined 30 min after stimulation in the presence or absence of the various compounds. D, bath application of NMDA (1 μm) leads to a fast activation that quickly returns to baseline levels after wash-out of the drug, demonstrating the in vivo reversibility of ICAP signals. YFP, yellow fluorescent protein.
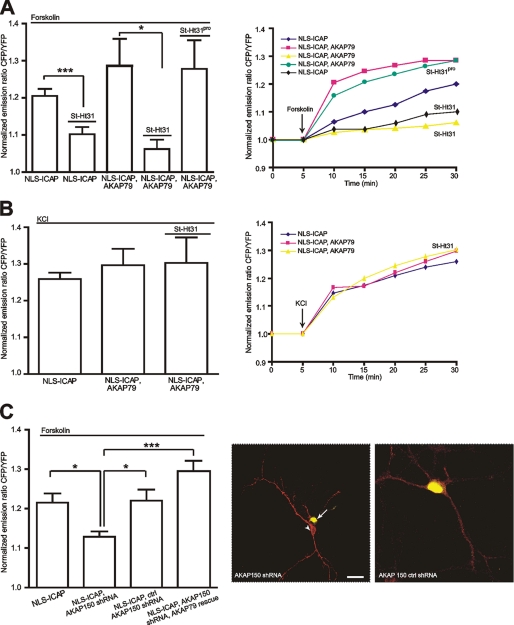
AKAP79/150 Anchoring of PKA and Calcineurin Controls CREB Activation in Hippocampal Neurons
AKAP79/150 has been identified as a major organizer of signaling events underlying excitatory neuronal plasticity. AKAP79/150 scaffold complexes have been localized to the membrane cytoskeleton and post-synaptic densities (55–57) of hippocampal neurons where they interact with and are implicated in the regulation of a plethora of partners such as NMDA and AMPA receptors or L-type calcium channels (Cav1.2) (4). We used ICAP to investigate to what extent AKAP79/150 influences CREB activation. Indeed, the scaffold protein had a significant effect on CREB (Fig. 4). Co-expression of AKAP79-mCherry with ICAP in hippocampal neurons initially appeared to enhance forskolin-induced CREB activation after 10 min of application. After 30 min, however, the effect was not statistically significant (Fig. 4A). To achieve the opposite effect of disrupting the anchoring of PKA to AKAP79/150 we applied the stearated peptide St-Ht31, which mimics the binding site of AKAP79/150 for PKA (58). It is membrane permeant and thus simply could be added to the extracellular solution. Disruption of PKA anchoring to AKAP79/150 dramatically reduced forskolin-induced CREB activation (n = 6) (Fig. 4A), thus demonstrating the need for strategic positioning of PKA to affect signaling to the nucleus. A biologically inactive analogue of St-Ht31 (St-Ht31pro) did not have an effect (n = 6, Fig. 4A). St-Ht31 application did not affect depolarization-induced CREB activation (n = 5) (Fig. 4B). As an alternative way to study involvement of AKAP79/150 in PKA-mediated CREB activation we knocked down expression of the rodent form of AKAP150 using shRNAs. When transfected into hippocampal neurons 1 day after plating this procedure efficiently inhibited expression of AKAP150, which typically is strongly induced in hippocampal neurons during the first week in vitro (59). Knockdown was verified by immunostaining at 9–11 days in vitro (Fig. 4C). In neurons lacking AKAP150 PKA-mediated CREB activation was significantly reduced, in good agreement with experiments using St-Ht31. PKA-mediated CREB activation could be restored by overexpressing the human homologue AKAP79, which is not susceptible to knock-down (Fig. 4C). A control shRNA that did not change AKAP150 expression levels detectably had no effect on PKA-mediated CREB activation (Fig. 4C).
FIGURE 4.
AKAP79/150 anchoring controls PKA-mediated activation of CREB in hippocampal neurons. A, left, mean ± S.E. CREB activation 30 min after application of forskolin (50 μm). AKAP79/150 overexpression (ICAP, AKAP79, n = 6) did not significantly increase mean forskolin-induced CREB activation compared with control (ICAP, n = 6). Disruption of PKA anchoring to AKAP79/150 using St-Ht31 (10 μm) dramatically reduced forskolin-induced CREB activation (n = 6). Right, averaged imaging traces under the indicated conditions. B, St-Ht31 did not affect depolarization-induced CREB activation. Left, mean ± S.E. CREB activation 30 min after depolarization with KCl (50 mm) (ICAP, n = 5). Overexpression of AKAP79/150 (ICAP, AKAP79; n = 5) did not enhance depolarization-induced CREB activation, nor did application of St-Ht31 (ICAP, AKAP 79, St-Ht31; n = 5) lead to a detectable block of depolarization-induced CREB activation. Right, averaged imaging traces after high potassium depolarization. *, p < 0.05. C, knockdown of endogenous AKAP150 using shRNA leads to reduced forskolin-induced CREB activation. CREB activation can be rescued by overexpression of the human homologue AKAP79. Left, mean ± S.E. CREB activation 30 min after forskolin (ICAP, n = 5, AKAP150 shRNA, ICAP, n = 10, AKAP150 ctrl shRNA, n = 5, AKAP150 shRNA, AKAP79 rescue, ICAP, n = 8), *, p < 0.05; ***, p < 0.001. Right, colocalization of anti-AKAP150 immunostaining (red) and NLS-ICAP fluorescence (yellow) to verify efficient knockdown of AKAP150 in neurons co-transfected with the corresponding shRNA. Note that neurons that have been co-transfected with ICAP and AKAP150 shRNA show no immunostaining (arrow), whereas the non-transfected neurons are stained (arrowheads). The right image demonstrates inefficiency of a control shRNA in knocking down AKAP150. Scale bar, 20 μm. YFP, yellow fluorescent protein.
Presumably AKAP79/150 serves to localize the regulatory subunit of PKA at neuronal membranes close to the neuronal sites of cAMP production. Neuronal adenylyl cyclases such as types 1 and 8 have been identified to localize to neuronal membranes including post-synaptic densities (60, 61) and thus presumably co-localize to a large degree with AKAP79/150 in these neurons. Interestingly, certain stimuli such as brief NMDA receptor stimulations can lead to a long lasting translocation of the AKAP79/150 complex and anchored PKA away from membranes and post-synaptic sites toward the cytosol (62). Controlling membrane and postsynaptic density localization of AKAP79/150 may thus be another mechanism to regulate PKA-induced signaling onto CREB.
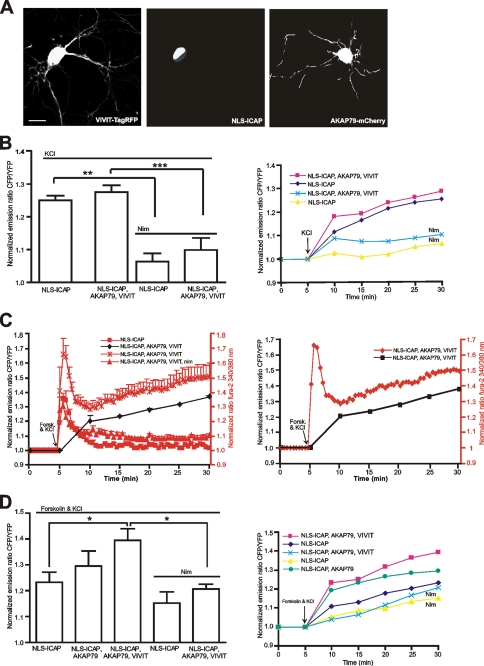
L-type calcium channels had been identified as the main calcium influx site leading to activation of CREB in the nucleus after depolarization (63). In agreement with this was the essential block of high potassium-induced ICAP response by the L-type calcium channel blocker nimodipine (5 μm, n = 10) (Fig. 5B). Overexpression of AKAP79/150 did not have any detectable effect on depolarization-induced CREB activation (n = 6) (Fig. 4B). For a further analysis of the AKAP79/150-controlled L-type calcium channel-induced CREB regulation we attempted to disrupt the association of calcineurin with AKAP79/150 and the L-type channel. For this purpose we employed a peptide termed VIVIT, which was identified to block calcineurin binding to AKAP79/150 via a PXIXIT motif without destroying phosphatase activity (41, 64, 65). We fused VIVIT to TagRFP (66) for visualization (Fig. 5A) and expressed it together with ICAP in hippocampal neurons. VIVIT had no detectable effect on the depolarization-induced ICAP response when coexpressed with ICAP alone (data not shown) and showed no statistically significant enhancement after triple transfection of VIVIT, AKAP79, and ICAP (n = 14) (Fig. 5B). Probably basal levels of PKA induced phosphorylation and current enhancement of Cav1.2 L-type channels were not high enough to be reflected in an increased CREB activation detected by ICAP. To reveal a possible modulatory role on CREB more clearly we depolarized neurons in the presence of forskolin (50 μm) to achieve a maximal possible activation of PKA while disrupting antagonistic action of calcineurin via expression of VIVIT. This treatment lead to enhanced calcium influx after depolarization as demonstrated by simultaneous imaging of calcium and CREB activation (Fig. 5C). Enhanced calcium influx was due to L-type channel-mediated action as it was efficiently blocked by nimodipine (5 μm, Fig. 5C). Under these conditions VIVIT significantly enhanced depolarization-induced CREB activation when AKAP79 was co-expressed with VIVIT and ICAP (Fig. 5, C and D), probably because more of the available pool of PKA had been recruited to assist in channel phosphorylation (n = 8). Anchored calcineurin therefore may have a subtle role in opposing CREB activation through its counteraction of PKA-mediated L-type calcium channel phosphorylation, a finding that is reminiscent of a mechanism identified to be important in signaling via NFATc4 (41). Compared with the drastic phenotype of disrupting AKAP79/150 anchoring of PKA on PKA-mediated CREB activation an antagonistic regulation of L-type calcium channels appears to be of minor importance for CREB using the depolarization-induced stimulus paradigm. Thus AKAP79/150 anchoring of PKA and calcineurin controls CREB activation mainly via direct effects on PKA, whereas regulation of L-type calcium channels through anchored PKA and calcineurin may present a more subtle form of modulation.
FIGURE 5.
Modulation of L-type calcium channel-mediated CREB activation through AKAP79/150 anchoring of PKA and Calcineurin. A, micrographs showing hippocampal neurons expressing VIVIT-TagRFP (left, excitation 545/30 nm, emission 620/60 nm), NLS-ICAP (middle, excitation 440/20 nm, emission 528/30 nm), and AKAP79-mCherry (right, excitation 545/30 nm, emission 620/60 nm). Scale bar, 10 μm. B, left, mean ± S.E. CREB activation 30 min after high potassium (50 mm) depolarization (ICAP, n = 5). Co-expression of AKAP79 and the VIVIT peptide (ICAP, AKAP79, and VIVIT, n = 14) did not significantly enhance depolarization-induced CREB activation. Depolarization-induced CREB activation and ICAP responses were effectively blocked by the L-type calcium channel antagonist nimodipine (5 μm) in the absence (ICAP, n = 4) and presence of AKAP79 and VIVIT (ICAP, AKAP79, and VIVIT, n = 10). Right, averaged imaging traces after high potassium stimulation under the indicated conditions. C, simultaneous calcium and ICAP imaging verifies modulation of L-type calcium channels. Left, averaged calcium imaging (red) and ICAP (black) traces for the indicated conditions (ICAP, n = 12; ICAP, AKAP79, and VIVIT, n = 6; ICAP, AKAP79, VIVIT, and nim, n = 4). Overexpression of AKAP79 and VIVIT lead to enhanced calcium influx after KCl/forskolin, which could be blocked with the specific L-type calcium channel blocker nimodipine (nim, 5 μm). Right, simultaneous imaging of calcium (red) and ICAP (black) in an individual hippocampal neuron expressing ICAP, VIVIT-TagRFP, and AKAP79. D, left, mean ± S.E. responses 30 min after high potassium depolarization (50 mm) in the presence of forskolin (50 μm). Co-expression of AKAP79 and VIVIT is required to achieve significantly enhanced CREB activation (ICAP, AKAP79, and VIVIT, n = 8). Nim indicates block by nimodipine. Right, averaged imaging traces under the indicated conditions. *, p < 0.05; **, p < 0.01; ***, p < 0.001.
DISCUSSION
Biochemical and histological methods have been widely used in determining activation of CREB transcription factors in many different cell types. Biochemical assays often require grinding up large amounts of tissue and averaging activation over many different cell types within a tissue, whereas immunostainings are more difficult to quantify and at best only provide snapshots at given time points. All of these techniques are performed on dead cells and tissues, which allows conclusions on possible physiological events leading to CREB activation only to be made posthumously. Two attempts were made previously to image CREB activation in live cells. One employed intermolecular FRET between GFPs (18) but due to small signals and the problem of co-transfecting two reporter constructs was not practical, whereas another approach used a highly sensitive β-lactamase-dependent reporter assay (19), which converts substrate in a non-reversible manner and cannot be targeted to subcellular organelles. Substrate loading into tissue to monitor CREB activation may be difficult. ICAP is a unimolecular FRET-based biosensor that is specific for phosphorylation of the critical serine 133 and was tuned to optimize the phosphorylation-dependent change in emission ratio (Fig. 1). In its overall architecture it is similar to other sensors of tyrosine kinase or PKA activity (26, 67) but in contrast to these sensors preserves the promiscuous phosphorylation of the KID domain by the various CREB kinases. It is targetable to the nucleus and mitochondria (data not shown) and can in principle be expressed in transgenic model organisms. By incorporating a circularly permuted acceptor protein we were able to construct the sensor in two configurations with opposing response properties, either increasing or decreasing FRET due to phosphorylation. This unexpected behavior demonstrates that it is possible to tune response properties of genetically encoded sensors in more ways than previously thought. ICAP was functional in primary hippocampal neurons and allowed for dynamic and reversible imaging of CREB activation. Three pathways with different activation time constants were readily imaged in these cells. Overall, signals imaged after high potassium depolarization, neurotrophin addition, or stimulation of adenylyl cyclases (Fig. 3) were in good agreement with previous studies focusing on phosphorylation of endogenous CREB and a confirmation that ICAP signals are an accurate measure of endogenous CREB activation (16).
We used ICAP to study the involvement of AKAP79/150 complexes in organizing signals converging on CREB within the nucleus of hippocampal neurons. AKAP79/150 was found to be important in regulating CREB activation, mainly through its binding of PKA. It is interesting to note that apart from AKAP79/150 there are numerous other AKAPs that are expressed in neurons (68–70) that may take on distinct or partially overlapping roles in compartmentalizing PKA activity. Recently, the non-AKAP microtubule-binding protein MAP2 was identified as a major anchoring protein for PKA (type II) in neurons (71). It was found to be crucial for regulating trafficking of the catalytic subunit between neuronal dendrites and spines. Moreover, elimination of MAP2 in transgenic mice lead to increased basal and decreased forskolin-induced CREB activation in tissues of knock-out mice (72). However, in these mice expression levels of PKA regulatory and catalytic subunits were also reported to be reduced, which leaves unclear whether the observed effects on CREB are due to lack of MAP2 anchoring or through regulation of PKA expression levels. The precise functional differences in regulating CREB signaling between AKAPs and MAP2 thus remain to be determined. Our new fluorescent reporter ICAP makes it possible to monitor CREB activation in live cells, thus hopefully opening up many avenues to explore the physiology underlying CREB activation in specific cell types and tissues.
Supplementary Material
Acknowledgments
We thank Beat Lutz, Roger Tsien, Marc Dell' Acqua, and Marc Montminy for providing DNA, and Anja Moritz and Birgit Kunkel for experimental help. Stephan Direnberger assisted with image processing.
This work was supported by the Max Planck Society.

The on-line version of this article (available at http://www..jbc.org) contains supplemental Figs. 1–5.
- CREB
- cAMP-responsive element-binding protein
- AKAP
- A kinase anchoring protein
- CBP
- CREB-binding protein
- CFP
- cyan fluorescent protein
- GFP
- green fluorescent protein
- FRET
- fluorescence (or Förster) resonance energy transfer
- ICAP
- indicator of CREB activation due to phosphorylation
- BDNF
- brain-derived neurotrophic factor
- PKA
- protein kinase A
- CaMK
- calmodulin kinase
- MAP
- mitogen-activated protein
- AMPA
- α-amino-3-hydroxyl-5-methyl-4-isoxazole-propionate
- NMDA
- N-methyl-d-aspartate
- IBMX
- isobutylmethylxanthine
- shRNA
- short hairpin RNA
- KID
- kinase-inducible domain
- KIX
- KID interaction domain.
REFERENCES
- 1.Shaywitz A. J., Greenberg M. E. (1999) Annu. Rev. Biochem. 68, 821–861 [DOI] [PubMed] [Google Scholar]
- 2.Mayr B., Montminy M. (2001) Nat. Rev. Mol. Cell. Biol. 2, 599–609 [DOI] [PubMed] [Google Scholar]
- 3.Lonze B. E., Ginty D. D. (2002) Neuron 35, 605–623 [DOI] [PubMed] [Google Scholar]
- 4.Conkright M. D., Montminy M. R. (2005) Trends Cell Biol. 15, 457–459 [DOI] [PubMed] [Google Scholar]
- 5.Carlezon W. A., Jr., Duman R. S., Nestler E. J. (2005) Trends Neurosci. 28, 436–445 [DOI] [PubMed] [Google Scholar]
- 6.Hoeffler J. P., Meyer T. E., Yun Y., Jameson J. L., Habener J. F. (1988) Science 242, 1430–1433 [DOI] [PubMed] [Google Scholar]
- 7.Gonzalez G. A., Yamamoto K. K., Fischer W. H., Karr D., Menzel P., Biggs W., 3rd, Vale W. W., Montminy M. R. (1989) Nature 337, 749–752 [DOI] [PubMed] [Google Scholar]
- 8.Hai T. W., Liu F., Coukos W. J., Green M. R. (1989) Genes Dev. 3, 2083–2090 [DOI] [PubMed] [Google Scholar]
- 9.Foulkes N. S., Borrelli E., Sassone-Corsi P. (1991) Cell 64, 739–749 [DOI] [PubMed] [Google Scholar]
- 10.Chrivia J. C., Kwok R. P., Lamb N., Hagiwara M., Montminy M. R., Goodman R. H. (1993) Nature 365, 855–859 [DOI] [PubMed] [Google Scholar]
- 11.Radhakrishnan I., Pérez-Alvarado G. C., Parker D., Dyson H. J., Montminy M. R., Wright P. E. (1997) Cell 91, 741–752 [DOI] [PubMed] [Google Scholar]
- 12.Liu F. C., Graybiel A. M. (1996) Neuron 17, 1133–1144 [DOI] [PubMed] [Google Scholar]
- 13.Liu F. C., Graybiel A. M. (1998) Proc. Natl. Acad. Sci. U.S.A. 95, 4708–4713 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Marie H., Morishita W., Yu X., Calakos N., Malenka R. C. (2005) Neuron 45, 741–752 [DOI] [PubMed] [Google Scholar]
- 15.Lee B., Butcher G. Q., Hoyt K. R., Impey S., Obrietan K. (2005) J. Neurosci. 25, 1137–1148 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wu G. Y., Deisseroth K., Tsien R. W. (2001) Proc. Natl. Acad. Sci. U.S.A. 98, 2808–2813 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wu G. Y., Deisseroth K., Tsien R. W. (2001) Nat. Neurosci. 4, 151–158 [DOI] [PubMed] [Google Scholar]
- 18.Mayr B. M., Canettieri G., Montminy M. R. (2001) Proc. Natl. Acad. Sci. U.S.A. 98, 10936–10941 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Spotts J. M., Dolmetsch R. E., Greenberg M. E. (2002) Proc. Natl. Acad. Sci. U.S.A. 99, 15142–15147 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Tsien R. Y. (1998) Annu. Rev. Biochem. 67, 509–544 [DOI] [PubMed] [Google Scholar]
- 21.Zhang J., Campbell R. E., Ting A. Y., Tsien R. Y. (2002) Nat. Rev. Mol. Cell Biol. 3, 906–918 [DOI] [PubMed] [Google Scholar]
- 22.Griesbeck O. (2004) Curr. Opin. Neurobiol. 14, 636–641 [DOI] [PubMed] [Google Scholar]
- 23.Miyawaki A. (2005) Neuron 48, 189–199 [DOI] [PubMed] [Google Scholar]
- 24.Yasuda R. (2006) Curr. Opin. Neurobiol. 16, 551–561 [DOI] [PubMed] [Google Scholar]
- 25.VanEngelenburg S. B., Palmer A. E. (2008) Curr. Opin. Chem. Biol. 12, 60–65 [DOI] [PubMed] [Google Scholar]
- 26.Zhang J., Ma Y., Taylor S. S., Tsien R. Y. (2001) Proc. Natl. Acad. Sci. U.S.A. 98, 14997–15002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Allen M. D., Zhang J. (2006) Biochem. Biophys. Res. Commun. 348, 716–721 [DOI] [PubMed] [Google Scholar]
- 28.Dunn T. A., Wang C. T., Colicos M. A., Zaccolo M., DiPilato L. M., Zhang J., Tsien R. Y., Feller M. B. (2006) J. Neurosci. 26, 12807–12815 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gervasi N., Hepp R., Tricoire L., Zhang J., Lambolez B., Paupardin-Tritsch D., Vincent P. (2007) J. Neurosci. 27, 2744–2750 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Takao K., Okamoto K., Nakagawa T., Neve R. L., Nagai T., Miyawaki A., Hashikawa T., Kobayashi S., Hayashi Y. (2005) J. Neurosci. 25, 3107–3112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lee S. J., Escobedo-Lozoya Y., Szatmari E. M., Yasuda R. (2009) Nature 458, 299–304 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Green H. M., Alberola-Ila J. (2005) BMC Chem. Biol. 5, 1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Fujioka A., Terai K., Itoh R. E., Aoki K., Nakamura T., Kuroda S., Nishida E., Matsuda M. (2006) J. Biol. Chem. 281, 8917–8926 [DOI] [PubMed] [Google Scholar]
- 34.Sato M., Kawai Y., Umezawa Y. (2007) Anal Chem. 79, 2570–2575 [DOI] [PubMed] [Google Scholar]
- 35.Harvey C. D., Ehrhardt A. G., Cellurale C., Zhong H., Yasuda R., Davis R. J., Svoboda K. (2008) Proc. Natl. Acad. Sci. U.S.A. 105, 19264–19269 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.DiPilato L. M., Cheng X., Zhang J. (2004) Proc. Natl. Acad. U.S.A. 101, 16513–16518 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Nikolaev V. O., Bünemann M., Hein L., Hannawacker A., Lohse M. J. (2004) J. Biol. Chem. 279, 37215–37218 [DOI] [PubMed] [Google Scholar]
- 38.Newman R. H., Zhang J. (2008) Mol. Biosyst. 4, 496–501 [DOI] [PubMed] [Google Scholar]
- 39.Wong W., Scott J. D. (2004) Nat. Rev. Mol. Cell Biol. 5, 959–970 [DOI] [PubMed] [Google Scholar]
- 40.Klauck T. M., Faux M. C., Labudda K., Langeberg L. K., Jaken S., Scott J. D. (1996) Science 271, 1589–1592 [DOI] [PubMed] [Google Scholar]
- 41.Oliveria S. F., Dell'Acqua M. L., Sather W. A. (2007) Neuron 55, 261–275 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Gao T., Yatani A., Dell'Acqua M. L., Sako H., Green S. A., Dascal N., Scott J. D., Hosey M. M. (1997) Neuron 19, 185–196 [DOI] [PubMed] [Google Scholar]
- 43.Colledge M., Dean R. A., Scott G. K., Langeberg L. K., Huganir R. L., Scott J. D. (2000) Neuron 27, 107–119 [DOI] [PubMed] [Google Scholar]
- 44.Bhattacharyya S., Biou V., Xu W., Schlüter O., Malenka R. C. (2009) Nat. Neurosci. 12, 172–181 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Griesbeck O., Baird G. S., Campbell R. E., Zacharias D. A., Tsien R. Y. (2001) J. Biol. Chem. 276, 29188–29194 [DOI] [PubMed] [Google Scholar]
- 46.Baird G. S., Zacharias D. A., Tsien R. Y. (1999) Proc. Natl. Acad. Sci. U.S.A. 96, 11241–11246 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Nagai T., Yamada S., Tominaga T., Ichikawa M., Miyawaki A. (2004) Proc. Natl. Acad. Sci. U.S.A. 101, 10554–10559 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Mank M., Reiff D. F., Heim N., Friedrich M. W., Borst A., Griesbeck O. (2006) Biophys. J. 90, 1790–1796 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Bito H., Deisseroth K., Tsien R. W. (1996) Cell 87, 1203–1214 [DOI] [PubMed] [Google Scholar]
- 50.Hardingham G. E., Arnold F. J., Bading H. (2001) Nat. Neurosci. 4, 261–267 [DOI] [PubMed] [Google Scholar]
- 51.Ginty D. D., Bonni A., Greenberg M. E. (1994) Cell 77, 713–725 [DOI] [PubMed] [Google Scholar]
- 52.Xing J., Ginty D. D., Greenberg M. E. (1996) Science 273, 959–963 [DOI] [PubMed] [Google Scholar]
- 53.Pizzorusso T., Ratto G. M., Putignano E., Maffei L. (2000) J. Neurosci. 20, 2809–2816 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Hardingham G. E., Fukunaga Y., Bading H. (2002) Nat. Neurosci. 5, 405–414 [DOI] [PubMed] [Google Scholar]
- 55.Dell'Acqua M. L., Faux M. C., Thorburn J., Thorburn A., Scott J. D. (1998) EMBO J. 17, 2246–2260 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Gomez L. L., Alam S., Smith K. E., Horne E., Dell'Acqua M. L. (2002) J. Neurosci. 22, 7027–7044 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Oliveria S. F., Gomez L. L., Dell'Acqua M. L. (2003) J. Cell Biol. 160, 101–112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Carr D. W., Hausken Z. E., Fraser I. D., Stofko-Hahn R. E., Scott J. D. (1992) J. Biol. Chem. 267, 13376–13382 [PubMed] [Google Scholar]
- 59.Robertson H. R., Gibson E. S., Benke T. A., Dell'Acqua M. L. (2009) J. Neurosci. 29, 7929–7943 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Wang H., Chan G. C., Athos J., Storm D. R. (2002) J. Neurochem. 83, 946–954 [DOI] [PubMed] [Google Scholar]
- 61.Conti A. C., Maas J. W., Jr., Muglia L. M., Dave B. A., Vogt S. K., Tran T. T., Rayhel E. J., Muglia L. J. (2007) Neuroscience 146, 713–729 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Smith K. E., Gibson E. S., Dell'Acqua M. L. (2006) J. Neurosci. 26, 2391–2402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Dolmetsch R. E., Pajvani U., Fife K., Spotts J. M., Greenberg M. E. (2001) Science 294, 333–339 [DOI] [PubMed] [Google Scholar]
- 64.Aramburu J., Yaffe M. B., López-Rodríguez C., Cantley L. C., Hogan P. G., Rao A. (1999) Science 285, 2129–2133 [DOI] [PubMed] [Google Scholar]
- 65.Li H., Rao A., Hogan P. G. (2004) J. Mol. Biol. 342, 1659–1674 [DOI] [PubMed] [Google Scholar]
- 66.Merzlyak E. M., Goedhart J., Shcherbo D., Bulina M. E., Shcheglov A. S., Fradkov A. F., Gaintzeva A., Lukyanov K. A., Lukyanov S., Gadella T. W., Chudakov D. M. (2007) Nat. Methods 4, 555–557 [DOI] [PubMed] [Google Scholar]
- 67.Ting A. Y., Kain K. H., Klemke R. L., Tsien R. Y. (2001) Proc. Natl. Acad. Sci. U.S.A. 98, 15003–15008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Bauman A. L., Goehring A. S., Scott J. D. (2004) Neuropharmacology 46, 299–310 [DOI] [PubMed] [Google Scholar]
- 69.Taskén K., Aandahl E. M. (2004) Physiol. Rev. 84, 137–167 [DOI] [PubMed] [Google Scholar]
- 70.Dell'Acqua M. L., Smith K. E., Gorski J. A., Horne E. A., Gibson E. S., Gomez L. L. (2006) Eur. J. Cell Biol. 85, 627–633 [DOI] [PubMed] [Google Scholar]
- 71.Zhong H., Sia G. M., Sato T. R., Gray N. W., Mao T., Khuchua Z., Huganir R. L., Svoboda K. (2009) Neuron 62, 363–374 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Harada A., Teng J., Takei Y., Oguchi K., Hirokawa N. (2002) J. Cell Biol. 158, 541–549 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.