Abstract
Posttranscriptional gene regulation by microRNAs (miRNAs) has been implicated in the fine-tuning of TLR-mediated inflammatory response. The cytokine-inducible Src homology 2-containing protein (CIS), one member of the suppressors of cytokine signaling family of proteins, is an important negative regulator for inflammatory cytokine signaling. Using in vitro models using normal human biliary epithelial cells (cholangiocytes), we demonstrated that LPS stimulation or infection with the parasitic protozoan Cryptosporidium parvum induced expression of CIS protein without a change in CIS mRNA levels by activating the TLR signaling pathway. Of those miRNAs expressed in cholangiocytes, we found that targeting of the 3′-untranslated region of CIS by microRNA-98 (miR-98) or let-7 resulted in translational repression, but not CIS mRNA degradation. LPS stimulation or C. parvum infection decreased cholangiocyte expression of miR-98 and let-7. Down-regulation of miR-98 and let-7 relieved miRNA-mediated translational suppression of CIS and contributed to LPS- and C. parvum-stimulated CIS protein expression. Moreover, gain-of-function (by overexpression of CIS) and loss-of-function (by siRNA interference) studies revealed that CIS could enhance IκBα degradation and regulate NF-κB activation in cholangiocytes in response to LPS stimulation or C. parvum infection. Our data suggest that miR-98 and let-7 confer cholangiocyte expression of CIS in response to microbial challenge, a process that may be relevant to the regulation of TLR-mediated epithelial innate immune response.
Toll-like receptors recognize discrete pathogen-associated molecular patterns and activate a set of adaptor proteins (e.g., MyD88) leading to the nuclear translocation of transcription factors, such as NF-κB (1, 2). Activation of the TLR/NF-κB pathway initiates a series of host cell defense reactions against pathogens, including parasites (3). Nevertheless, sustained TLR/NF-κB signaling can have devastating effects on the host resulting in chronic inflammatory diseases and autoimmune disorders and aid in the pathogenesis of infectious human diseases (4, 5). In contrast, a delayed or insufficient response can lead to a failure to control infection. Thus, cells have developed multiple strategies for the feedback regulation of TLR/NF-κB signaling to fine-tune TLR-associated immune responses. Several endogenous feedback regulatory pathways have recently been identified to counter-regulate TLR signaling cascades and promote resolution of inflammation, such as activation of the armadillo-motif-containing protein, Toll-interacting protein, and A20 (6–8).
The cytokine-inducible Src homology 2-containing protein (CIS)4 and suppressors of cytokine signaling (SOCS) proteins are a family of intracellular proteins that have emerged as key physiological regulators of cytokine responses in various types of cells (9). Each CIS/SOCS protein has a Src homology (SH) 2 domain and a SOCS box (9). The SH2 domain binds to the phosphorylated tyrosine substrate residues and the E3 activity of CIS/SOCS proteins causes substrate ubiquitination leading to proteasome-mediated degradation. The best-characterized SOCS family members are CIS and SOCS1–3, which function in a classical negative-feedback loop inhibiting cytokine signaling by interacting with the JAK-STAT signaling cascades (9). Pathogen recognition via TLRs can also stimulate expression of CIS/SOCS proteins in host cells (10). CIS/SOCS proteins have been demonstrated to limit the extent of TLR signaling indirectly by inhibiting autocrine cytokine response in macrophages through IFN-γ and TNF-α (11). Recent studies also indicate that CIS/SOCS proteins may directly interact with the TLR signaling cascades and regulate TLR/NF-κB signaling via a mechanism distinct from an autocrine cytokine response. SOCS1 can interact with phosphorylated Mal, an adaptor molecule required for TLR2 and TLR4 signaling, resulting in Mal polyubiquitination and subsequent degradation (12). Interestingly, overexpression of SOCS1 in human respiratory epithelial cells enhances activation of NF-κB-dependent proinflammatory pathways (13). CIS-enhanced NF-κB activation has been reported in T cells (14, 15). Thus, CIS/SOCS proteins may play a role in the regulation of TLR/NF-κB signaling in cells in response to microbial challenge.
MicroRNAs (miRNAs) are endogenous RNAs of ~22 nucleotides that pair with the messages of protein-coding genes to direct posttranscriptional repression (16). These molecules identify target mRNAs based on complementarity with target 3′-untranslated regions (3′-UTRs) leading to translational repression and/or mRNA cleavage (17). Studies have revealed key roles for miRNAs in diverse regulatory pathways, including development timing control, cell differentiation, apoptosis, cell proliferation, organ development (16), and more recently, in immune regulation (18–20). We previously demonstrated that let-7 regulates TLR4 expression via translational suppression in human cholangiocytes, epithelial cells lining the biliary tree. We have also demonstrated let-7 involvement in the epithelial defense response to infection by Cryptosporidium parvum, a protozoan parasite that infects gastrointestinal epithelium and activation of TLR/NF-κB signaling in host cells (21, 22). MicroRNAs have been implicated in viral immune escape and antiviral defense (23). Induction of miR-155 during the macrophage inflammatory response suggests its potential involvement in regulation of inflammation (24).
In contrast to intestinal epithelial cells, human cholangiocytes express multiple TLRs and are susceptible to TLR activation, thus providing a good model for investigating regulation of TLR/NF-κB signaling in mucosal immunity (25–27). In this study, we found that microRNA-98 (miR-98) and let-7 regulate CIS protein expression via translational suppression in human cholangiocytes. Expression of CIS protein in cholangiocytes was up-regulated by LPS (an agonist of TLR4) or after exposure to C. parvum. This induced expression of CIS involves a relief of miRNA-mediated translation repression by miR-98 and let-7. Furthermore, expression of CIS correlated with an accelerated degradation of IκBα and enhanced NF-κB activation in cholangiocytes in response to LPS stimulation or C. parvum infection. The identification of this miRNA-mediated regulation of NF-κB signaling via CIS expression in cholangiocytes may represent a process relevant to the regulation of TLR/NF-κB signaling in general.
Materials and Methods
In vitro model of C. parvum infection and LPS stimulation
H69 cells are SV40-transformed normal human cholangiocytes originally derived from normal liver harvested for transplant (28, 29). Human intrahepatic biliary epithelial (HIBEpiC) cells are nonimmortalized isolated human cholangiocytes commercially available from ScienCell Research Laboratories (Carlsbad). HIBEpiC cells were grown on poly-L-lysine coated dishes and cultured using instructions and medium provided by the supplier. An in vitro model of human cholangiocyte infection by C. parvum oocysts was used as previously described (29). C. parvum oocysts of the Iowa strain were purchased from a commercial source (Bunch Grass Farm). Infection was done in DMEM/F-12 medium containing penicillin and streptomycin and hypochlorite treated C. parvum oocysts (29). Inactivated organisms (treated at 65°C for 30 min) were used for sham infection controls. Oocysts were added to achieve >90% infection of cell cultures with a cell/parasite ratio of 1:2 to 1:10 depending on the viability of oocysts confirmed by immunofluorescent staining as previously reported (28). Uninfected parasites were usually removed 4 h after incubation by washing with DMEM medium. For LPS stimulation, cells were exposed to culture medium containing LPS (Invivogen; final concentration 1 μg/ml). The proteasome inhibitor, MG132 (20 μM, Calbiochem), was used to inhibit proteasome-mediated degradation in cells (30).
Western blot analysis
Western blot analysis was performed (31) with Abs against CIS, IκBα, and β-actin from Santa Cruz Biotechnology and Sigma-Aldrich, respectively. CIS and IκBα levels were expressed as their ratio to β-actin (31).
Anti-miRs and miRNA precursors
To manipulate cellular levels of miRNAs, we used specific antisense oligonucleotides to miRNAs (anti-miRs) to inhibit miRNA function and specific miRNA precursors to increase miRNA expression as previously reported (21, 32). In experiments, H69 cells were transfected with 0–30 nM of miR-98 or let-7i precursors (Ambion), or anti-miR-98 or anti-let-7i (Ambion) using Lipofectamine 2000 (Invitrogen). Nonspecific oligonucleotides from Ambion were used as controls.
Plasmids
For plasmid constructs, the dominant negative (DN) functionally defective mutant of TLR4 was obtained from Dr. M. F. Smith (University of Virginia, Charlottesville, VA). MyD88-DN (a DN mutant of MyD88) was a gift from Prof. J. Tschopp (University of Lausanne, Lausanne, Switzerland). H69 cells stably transfected with TLR4-DN or MyD88-DN plasmid constructs were obtained by transfection followed by antibiotic selection as previously reported (21, 22). Primers used to amplify the open reading frame of human CIS (NM_145071) were: 5′-GGCTAGCACCATGGT CCTCTGCGTTCAGG-3′ (forward) and 5′-AGAATTCTCAGAGCTG GAAGGGGTACTGTC-3′ (reverse). PCR products were cloned into the NheI and EcoRI sites of the pcDNA3.1 (+) vector. H69 cells were transiently transfected with 0.25 μg of pcDNA-CIS or the control plasmid with the lipofectamine 2000 reagent and overexpression of CIS protein was confirmed by Western blot analysis. The HuSH 29mer shRNA-CIS and control constructs were purchased from Origene. IκBα was inserted into HindIII and EcoRI sites of the pcDNA.4/V5-His vector. PCR primers used to amplify IκBα cDNA were: 5′-TAAGCTTGCCATGGATGTTCCAG GCGGCCG-3′ (forward) and 5′-GGAATTCGTCTAACGTCAGACGC TGGC-3′ (reverse).
Real-time PCR
For quantitative analysis of CIS mRNA, total cellular RNA was isolated from cells using TRIzol (Invitrogen). Expression of CIS and GAPDH mRNA was analyzed using the TaqMan gene expression assay according to the manufacturer’s instructions (Applied Biosystems). The primer and probe sequences for the amplification of human CIS were: 5′-CCTACCTTCGGGAATCTGGCT-3′ (forward) and 5′-TGGCAT CTTCTGCAGGTGTT-3′ (reverse); 5′-FAM-TCCATTACGGCCAGCG AGGCC-TAMRA-3′ (probe) (Applied Biosystems). The primer and probe sequences for the amplification of GAPDH were as follows: 5′-GAAGGTGAAGGTCGGAGTC-3′ (forward); 5′-GAAGATGGTGAT GGGATTTC-3′ (reverse); 5′-VIC-CAAGCTTCCCGTTCTCAGCC-TAMRA-3′ (probe).
For real-time PCR analysis of miR-98, total RNA was isolated from cells with the mirVana miRNA Isolation kit (Ambion) and reverse transcribed (0.25 μg) using the Taqman MicroRNA Reverse Transcription Kit (Applied Biosystems). Comparative real-time PCR was performed by using Taqman Universal PCR Master Mix (Applied Biosystems) with the Applied Biosystems 7500 FAST real-time PCR System. Specific primers and probes for mature miR-98 and snRNA RNU6B (endogenous reference) were obtained from Applied Biosystems. All reactions were run in triplicate. Quantitation of miR-98 was performed normalizing with snRNA RNU6B and relative to a control (untreated cells) (33, 34).
Northern blot
Total RNAs harvested as above were run on a 15% Tris/Borate/EDTA (90 mM Tris/64.6 mM boric acid/2.5 mM EDTA (pH 8.3)) urea gel (Invitrogen) and transferred to a Nytran nylon transfer membrane (Ambion). A LNA DIG-probe of miRNA-98 (Exiqon) was hybridized using UltraHyb reagents (Ambion) according to the manufacturer’s instructions with blotted snRNA RNU6B as a control.
Luciferase reporter constructs and luciferase assay
Complementary 48-mer DNA oligonucleotides containing the putative miR-98 and let-7 target site within the 3′-UTR of human CIS were synthesized with flanking SpeI and HindIII restriction enzyme digestion sites (antisense, 5′-CTAGGTAATGACATTATACCTTTATTACCTCTTTATT TTATTACCTCT-3′; sense, 5′-AGCTAGAGGTAATAAAATAAAGAG GTAATAAAGGTATAATGTCATTAC-3′). The annealed oligonucleotides were ligated into the SpeI-HindIII sites of the pMIR-REPORT Luciferase vector (Ambion) for potential posttranscriptional luciferase regulation by miRNA interaction with the CIS 3′-UTR as we previously reported (21). As an additional control, a pMIR-REPORT Luciferase construct was generated containing CIS 3′-UTR with mutations (both ACCT to TGGA) in the putative seed regions for miR-98 and let-7 binding. Cells were transfected with each reporter construct, as well as miR-98 and let-7i antisense oligonucleotides or precursors, followed by assessment of luciferase activity 24 h after transfection. Luciferase activity was then measured and normalized to the expression of the control β-Gal construct as previously reported (21). In addition, the full sequence of IL-8 promoter was cloned into the pGL3-Basic Luciferase vector to transfect cells and monitor NF-κB activation (35).
Immunoprecipitation
Immunoprecipitation was used to detect the potential binding of CIS to IκBα as modified from previous studies by others (36, 37). In brief, cells transfected with the pcDNA-CIS and pcDNA.4/V5-His-IκBα vectors were grown to 95% confluence and exposed to C. parvum or LPS for 3 h in the presence of MG132. Cells were then lysed (lysis buffer: 20 mM Tris-HCl (pH 8.0), 150 mM NaCl, 1% Nonidet P-40, 20 μM MG132, 1 mM PMSF, 10 μg/ml leupeptin, and 2 μg/ml pepstatin) and 100 μg of lysate protein was incubated with anti-V5 Ab (Invitrogen) or the control IgG at 4°C overnight. Immune complexes were collected by direct binding to protein A-Sepharose. The immunoprecipitates were then blotted with specific Abs to CIS or IκBα (Santa Cruz Biotechnology).
Results
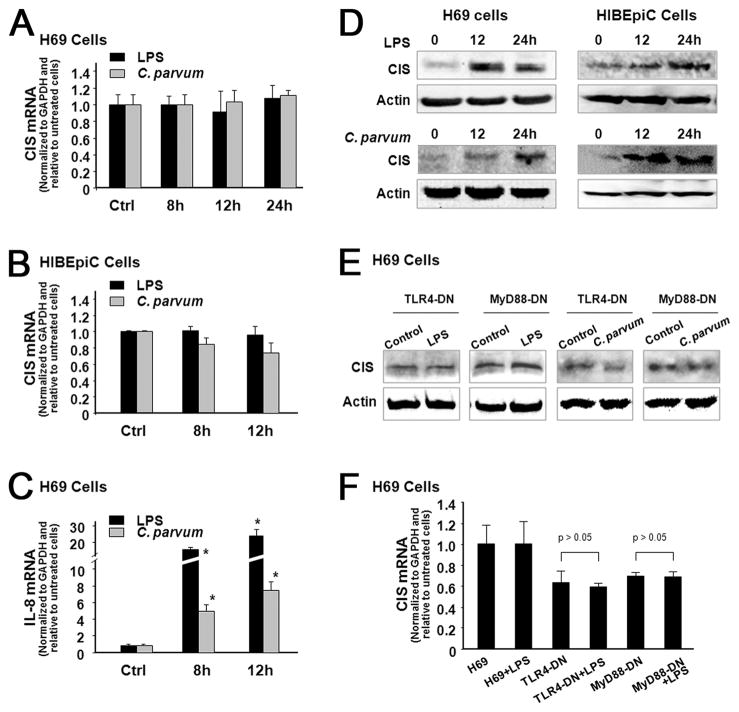
LPS Stimulation and C. parvum infection induce expression of CIS protein without a change in CIS mRNA levels in cholangiocytes by activation the TLR signaling pathway
We first assessed CIS expression in H69 and HIBEpiC cells in response to LPS or C. parvum infection. When H69 or HIBEpiC cells were exposed to LPS for up to 24 h (for H69) or 12 h (for HIBEpiC), no significant change of CIS mRNA levels was detected by real-time PCR analysis (Fig. 1, A and B). In contrast, as a positive control, a significant increase of IL-8 mRNA was found in cells following LPS stimulation or exposure to C. parvum (Fig. 1C), consistent with results from previous studies (31). Interestingly, a significant increase of CIS protein content was detectable in both H69 and HIBEpiC cells following LPS stimulation or C. parvum infection (Fig. 1D). To test whether TLR signals are involved in LPS- and C. parvum-induced CIS expression, we tested the expression of CIS in H69 cells stably transfected with the functionally defective DN mutants of TLR4 or MyD88 (21, 22). No increase of CIS protein was found in TLR4-DN or MyD88-DN cells following C. parvum infection or LPS stimulation compared with the control nontreated cells (Fig. 1E). Also, no change in CIS mRNA was detected in TLR4-DN or MyD88-DN cells following LPS stimulation or C. parvum infection (Fig. 1F).
FIGURE 1.
LPS stimulation and C. parvum infection induce expression of CIS protein without a change in CIS mRNA levels in cholangiocytes by activation the TLR signaling pathway. A and B, No significant change in CIS mRNA expression in human cholangiocytes in response to LPS stimulation and C. parvum infection. H69 (A) and HIBEpiC (B) cells were exposed to LPS or C. parvum oocysts followed by real-time PCR analysis for CIS mRNA. C, Expression of IL-8 mRNA in H69 cells in response to LPS stimulation and C. parvum infection. D, Up-regulation of CIS protein in human cholangiocytes following LPS stimulation and C. parvum infection. E, LPS- and C. parvum-induced CIS expression is TLR4/MyD88-dependent. H69 cells stably transfected with TLR4-DN or MyD88-DN were exposed to LPS or C. parvum followed by Western blot analysis for CIS. Western blots shown in D and E are representative from three independent experiments. β-actin was blotted as a loading control. F, Expression of CIS mRNA in TLR4-DN and MyD88-DN H69 cells in response to LPS stimulation as assessed by real-time PCR. *, p < 0.05 ANOVA vs nontreated controls.
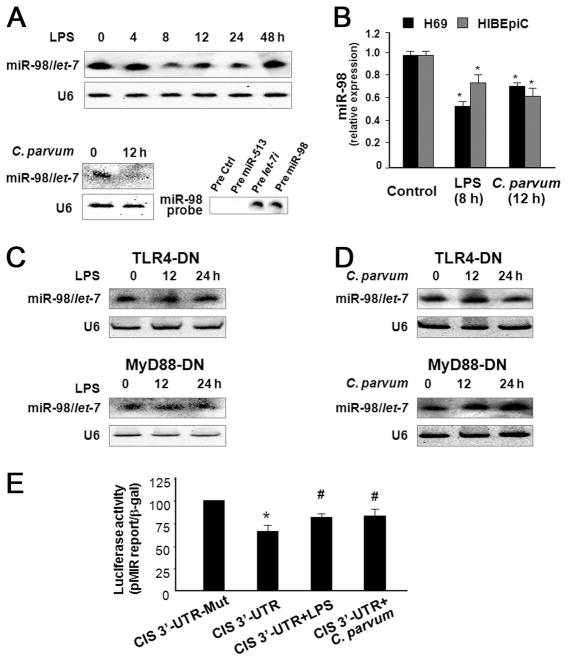
miR-98 and let-7 target CIS 3′-UTR resulting in translational suppression
The inconsistence of CIS expression between the message level and its protein level in both H69 and HIBEpiC cells following C. parvum infection or LPS stimulation suggests potential posttranscriptional regulation of CIS expression in cholangiocytes. To test whether miRNA-mediated posttranscriptional gene regulation is involved in this process, we used the algorithms Targetscan 4.2 program (38) to screen miRNAs expressed in H69 cells based on our previous microarray analysis (21). We found miR-98 and let-7 complementary to the CIS 3′-UTR with potential binding sites adjacent to each other in the CIS 3′-UTR, extending between nucleotide positions 1055 and 1093 (Fig. 2A).
FIGURE 2.
MicroRNA-98 and let-7 target CIS 3′-UTR causing translational suppression. A, The schematic of CIS mRNA shows two potential binding sites in the 3′-UTR for miR-98 and let-7. B, Binding of miR-98 and let-7 to the two potential binding sites in the CIS 3′-UTR results in translational suppression. CIS 3′-UTR sequence covering both potential binding sites was inserted into the pMIR-REPORT luciferase plasmid. A control plasmid with the mutant 3′-UTR sequence was also generated. H69 cells were transfected with the constructs and treated with the anti-miRs or miRNA precursors followed by luciferase analysis. C, miR-98 and let-7-mediated translational suppression of CIS requires both the potential binding sites for miR-98 and let-7 in the 3′-UTR. The two binding sites for miR-98 and/or let-7 in the CIS 3′-UTR were inserted separately to the pMIR-REPORT luciferase plasmid (3′-UTR Site A vs 3′-UTR Site B). Control plasmids with the mutant 3′-UTR sequence in each potential binding site were also generated. H69 cells were transfected with the reporter constructs for 24 h followed by luciferase analysis. Mut = mutant; *, p < 0.05 ANOVA vs the controls; #, p < 0.05 ANOVA vs CIS 3′-UTR transfected cells.
To test the potential targeting of CIS mRNA by miR-98 or let-7, we generated pMIR-REPORT luciferase constructs containing the CIS 3′-UTR with two putative let-7 and miR-98 binding sites (Fig. 2B). In addition, constructs with the ACCT to TGGA mutation at the putative binding sites were also generated and used as the controls (Fig. 2B). We then transfected H69 cells with these reporter constructs followed by assessment of luciferase activity 24 h after transfection. As shown in Fig. 2B, luciferase activity was significantly decreased in cells transfected with the CIS 3′-UTR construct containing both potential binding sites compared with the control vector. No change in luciferase activity was observed in cells transfected with the mutant CIS 3′-UTR construct, suggesting endogenous translational repression of the construct with the CIS 3′-UTR. In addition, anti-miR-98 or anti-let-7i markedly increased CIS 3′-UTR-associated luciferase reporter translation (Fig. 2B). In contrast, miR-98 and let-7i precursors did not significantly decrease luciferase reporter translation (Fig. 2B).
Because the binding sites for miR-98 and let-7 are adjacent to each other in the CIS 3′-UTR, we tested whether both are required for translational suppression. We generated pMIR-REPORT luciferase constructs containing the CIS 3′-UTR with the individual putative binding sites (Fig. 2C). When construct-driven luciferase activity was measured in transfected H69 cells no decrease in luciferase activity was found in cells containing only one of the putative binding sites vs those containing the control construct (Fig. 2C). Taken together, the above data suggest that miR-98 and let-7 target the CIS 3′-UTR resulting in posttranscriptional suppression of CIS in cholangiocytes which appears to require both adjacent binding sites.
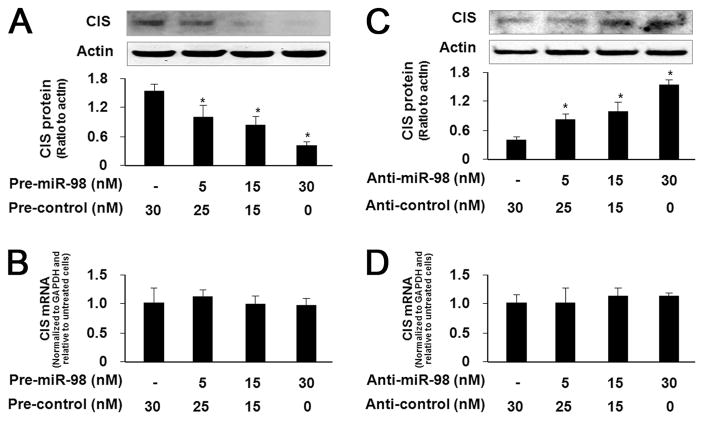
Manipulation of miR-98 function results in reciprocal alterations in CIS protein expression in H69 cells
To test whether miRNA-mediated translational repression of CIS is directly relevant to CIS protein expression, we treated H69 cells with anti-miR-98 or miR-98 precursor for 72 h and then measured CIS protein expression by Western blot. Transfection of H69 cells with the miR-98 precursor caused a dose-dependent decrease in CIS protein content (Fig. 3A). No significant change in CIS mRNA levels was found between the control cells and cells treated with miR-98 precursor (Fig. 3B), suggesting no affect on cellular CIS mRNA levels. Conversely, a dose-dependent increase in CIS protein content was identified in H69 cells treated with anti-miR-98 (Fig. 3C). However, no significant change in CIS mRNA levels was found between the control cells and cells treated with anti-miR-98 (Fig. 3D).
FIGURE 3.
Manipulation of miR-98 function results in reciprocal alterations in CIS protein expression in H69 cells. A and C, Transfection of miR-98 precursor or anti-miR-98 induces a dose-dependent decrease or increase, respectively, in CIS protein expression in H69 cells. H69 cells were treated with various dose of miR-98 precursor (A) or anti-miR-98 (C) followed by Western blot for CIS. A representative Western blot from three independent experiments is shown for A and C. Actin was also blotted to ensure equal loading. Densitometric levels of CIS signals were quantified and expressed as their ratio to actin. B and D, miR-98 precursor or anti-miR-98 transfection does not affect CIS mRNA levels. H69 cells were exposed to miR-98 precursor or anti-miR-98 followed by real-time PCR analysis for CIS mRNA. Pre = precursor. *, p < 0.05 ANOVA vs the nontreated control cells.
LPS stimulation and C. parvum infection decrease miR-98 and let-7 expression in a TLR4/MyD88-dependent manner resulting in relief of translational suppression of CIS
In our previous studies, we demonstrated that C. parvum infection and LPS stimulation decrease let-7 expression in H69 cells (21). Using a probe detecting both miR-98 and let-7 by Northern hybridization (Fig, 4A), we detected a significant decrease in miR-98/let-7 levels in H69 cells following LPS stimulation for up to 24 h or after exposed to C. parvum for 12 h (Fig. 4A). No decrease in miR-98/let-7 was found in cells 48 h after LPS stimulation (Fig. 4A). Furthermore, a significant decrease in miR-98 was confirmed in both H69 and HIBEpiC cells following LPS stimulation or C. parvum infection by real-time PCR analysis specific for miR-98 (Fig. 4B). In addition, no decrease in miR-98/let-7 was detected in TLR4-DN or MyD88-DN stably transfected H69 cells following LPS stimulation or C. parvum infection (Fig. 4, C and D), suggesting that LPS- and C. parvum-induced down-regulation of miR-98/let-7 requires activation of the TLR4/MyD88 signal pathway.
FIGURE 4.
LPS stimulation and C. parvum infection decrease expression of miR-98 and let-7 in a TLR4/MyD88-dependent manner resulting in relief of translational suppression of CIS in cholangiocytes. A, LPS stimulation and C. parvum infection decrease miR-98/let-7 expression in H69 cells by Northern blot analysis. RNU6B (U6) was used as a loading control. The miR-98 probe used for Northern blotting recognizes both precursors for miR-98 and let-7, but not a nonspecific precursor or the precursor for miR-513, confirming its specificity. B, LPS stimulation and C. parvum infection decrease miR-98 expression in both H69 and HIBEpiC cells as assessed by real-time PCR. *, p < 0.05 ANOVA vs the controls. C and D, LPS- and C. parvum-induced down-regulation of miR-98/let-7 expression is TLR4/MyD88-dependent. H69 cells stably transfected with TLR4-DN or MyD88-DN were exposed to LPS (C) or C. parvum (D) followed by Northern blotting. RNU6B (U6) was used as a loading control. E, Relief of miRNA-mediated translational suppression of CIS 3′-UTR in H69 cells following LPS stimulation or C. parvum infection. Cells were transfected with the pMIR-REPORT luciferase construct containing the CIS 3′-UTR with both the putative binding sites. Luciferase activity in cells after exposure to LPS or C. parvum was then measured and normalized to β-gal. Data represent three independent experiments. *, p < 0.05 ANOVA vs CIS 3′-UTR mutant control; #, p < 0.05 ANOVA vs CIS 3′-UTR transfected cells.
Because miR-98 and let-7 can target CIS 3′-UTR and induce translational suppression of CIS, C. parvum infection or LPS stimulation should induce a relief of miRNA-mediated CIS translation through down-regulation of miR-98 and let-7. To test this possibility, we transfected H69 cells with the pMIR-REPORT luciferase construct containing the CIS 3′-UTR with both the putative binding sites for let-7 and miR-98. Cells simultaneously exposed to LPS or C. parvum for 24 h showed a significant increase in CIS 3′-UTR-associated luciferase activity compared with the non-treated control (Fig. 4E). These data suggest that LPS stimulation or C. parvum infection can decrease miR-98 and let-7 expression to induce a relief of miRNA-mediated translational suppression of CIS in human cholangiocytes.
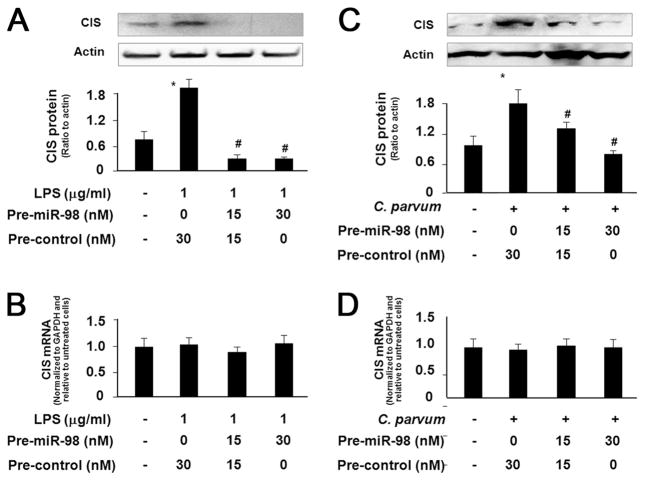
Transfection of miR-98 precursor abolishes C. parvum- and LPS-stimulated CIS protein expression
To confirm that relief of miRNA-mediated CIS translational repression is required for LPS/C. parvum-induced CIS protein expression, we transfected H69 cells with various doses of miR-98 precursors for 48 h and then exposed them to LPS or C. parvum for 24 h followed by Western blot analysis for CIS protein. The miR-98 precursor significantly inhibited up-regulation of CIS protein in H69 cells induced by LPS stimulation (Fig. 5A) or C. parvum infection (Fig. 5C) in a dose-dependent manner. Moreover, no significant change in CIS mRNA levels was found in the cells following LPS stimulation or C. parvum infection with or without the treatment by miR-98 precursor (Fig. 5, B and D). Thus, miR-98 precursor can abolish the up-regulation of CIS protein in cholangiocytes in response to LPS stimulation or C. parvum infection. Coupled with the down-regulation of miR-98 and let-7 in cells following LPS stimulation or C. parvum infection, the above data suggest that the relief of miR-98/let-7-mediated translational repression is required for LPS- and C. parvum-induced CIS protein expression.
FIGURE 5.
Transfection of miR-98 precursor abolishes LPS- and C. parvum-stimulated CIS protein expression. A and C, Transfection of miR-98 precursor inhibits up-regulation of CIS proteins in H69 cells following LPS stimulation (A) or C. parvum infection (C). Cells were transfected with the miR-98 precursor and then exposed to LPS or C. parvum followed by Western blotting for CIS. A representative Western blot from three independent experiments is shown in A and C. Densitometric levels of CIS signals were quantified and expressed as their ratio to actin. B and D, miR-98 precursor transfection does not affect CIS mRNA levels in cells following LPS stimulation or C. parvum infection. H69 cells were exposed to miR-98 precursor for 48 h and then exposed to LPS or C. parvum for 24 h followed by real-time PCR analysis for CIS mRNA. *, p < 0.05 ANOVA vs the nontreated cells; #, p < 0.05 ANOVA vs treated control cells.
CIS enhances NF-κB activation and binds to IκBα in cholangiocytes following LPS stimulation or C. parvum infection
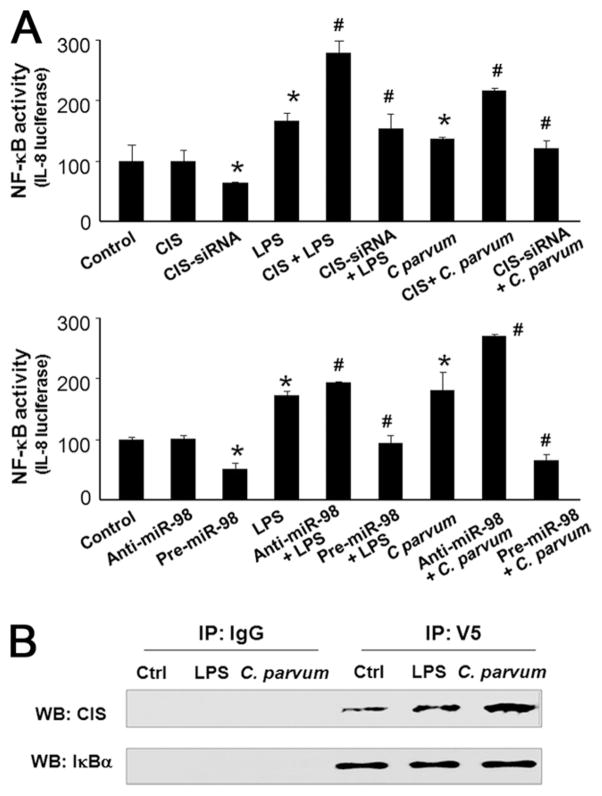
The CIS/SOCS proteins have emerged as key physiological negative regulators of cytokine responses (9). Therefore, we performed loss-of-function (by siRNA interference) and gain-of-function (by overexpression of CIS) studies in cholangiocytes. NF-κB activation in response to LPS stimulation or C. parvum infection was monitored by using a NF-κB driven IL-8 reporter construct as previously reported (34). Unexpectedly, we detected that knockdown of CIS through transfection of cells with a CIS siRNA significantly inhibited LPS- or C. parvum-induced IL-8 reporter activity (Fig. 6A). Overexpression of CIS increased LPS- or C. parvum-induced IL-8 reporter activity (Fig. 6A). No significant change in IL-8 reporter activity was found in cells overexpressing CIS but a slight decrease of reporter activity was detected in cells treated with the CIS siRNA (Fig. 6A). In addition, functional manipulation of miR-98 with miR-98 precursor or anti-miR-98 also caused reciprocal alterations in IL-8 reporter activity in response to LPS stimulation or C. parvum infection (Fig. 6A).
FIGURE 6.
CIS enhances NF-κB activation and binds to IκBα in cholangiocytes following LPS stimulation or C. parvum infection. A, Alteration of expression of CIS, as well as manipulation of miR-98 function, changes LPS- or C. parvum-stimulated NF-κB activation in H69 cells. Cells were transfected with pGL3-BasicIL-8 promoter and the plasmids (pcDNA-CIS or shRNA-CIS) and then exposed to LPS or C. parvum. Luciferase activity, reflecting NF-κB activation, was then monitored. For functional manipulation of miR-98, cells were transfected with pGL3-Basic-IL-8 promoter and miR-98 precursor or anti-miR-98 followed by exposure to LPS or C. parvum. *, p < 0.05 ANOVA vs the nontreated cells; #, p < 0.05 ANOVA vs treated control cells. B, Binding of CIS to IκBα in cells following LPS stimulation and C. parvum infection. Cells were transfected with pcDNA-CIS and pcDNA.4/V5-His-IκBα for 48 h, treated with 20 μM MG132 for 2 h, and then exposed to LPS or C. parvum for 3 h, followed by immunoprecipitation using the anti-V5 Ab or the control IgG and Western analysis for CIS and IκBα.
We further performed immunoprecipitation analysis to test whether CIS directly binds to IκBα. A proteasome inhibitor, MG132, was used to inhibit degradation of IκBα in cells cotransfected with CIS and V5-IκBα vectors as previously reported (36, 37). A V5 Ab was used to specifically precipitate IκBα followed by Western blotting for CIS and IκBα. As shown in Fig. 6B, a significant increase in CIS was detected in the immunoprecipitates from cells following LPS stimulation or C. parvum infection. No change in IκBα signal was observed in the immunoprecipitates. No IκBα or CIS was detected in the control IgG immunoprecipitates. Taken together, these results suggest a potential direct binding of CIS to IκBα in cells following LPS stimulation or C. parvum infection.
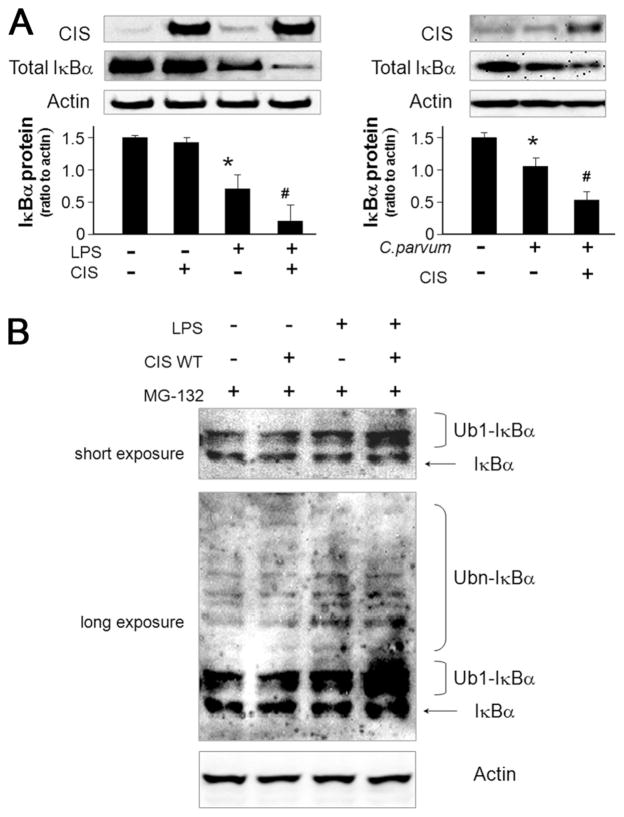
CIS enhances degradation of IκBα in cholangiocytes in response of LPS stimulation or C. parvum infection
To further test the potential mechanisms by which CIS enhances NF-κB activation, we overexpressed CIS in H69 cells and then measured cellular IκBα content following LPS stimulation or C. parvum infection. No significant change of IκBα was identified in cells overexpressing CIS compared with the control (Fig. 7A). However, a significant decrease of IκBα was detected in H69 cells after LPS stimulation for 2 h or exposure to C. parvum for 4 h. Cellular IκBα levels were further decreased in cells overexpressing CIS following LPS stimulation or C. parvum infection (Fig. 7A).
FIGURE 7.
CIS enhances degradation of IκBα induced by LPS stimulation or C. parvum infection in cholangiocytes. A, Overexpression of CIS increases LPS- or C. parvum-induced IκBα degradation in H69 cells. Cells were transfected with the pcDNA-CIS for 24 h and exposed to LPS (2 h) or C. parvum (4 h) followed by Western blot analysis for IκBα. B, Over-expression of CIS increases LPS-induced IκBα ubiquitination in H69 cells. Cells were transfected with pcDNA-CIS or the control vector for 48 h, treated with 20 μM MG132 for 2 h and then exposed to LPS for 2.5 h followed by Western analysis for IκBα. Experiments were performed in triplicate. Both a short time exposure and a longer time exposure of representative Western blots are shown. *, p < 0.05 ANOVA vs the non-treated cells; #, p < 0.05 ANOVA vs treated control cells.
To determine whether the decrease of IκBα occurred through altered ubiquitin-proteasome-mediated proteolytic degradation of IκBα, H69 cells were transfected with CIS or control vector for 48 h, incubated with MG132 for 2 h, stimulated by LPS for 2.5 h, and IκBα protein assessed by Western blot as previously reported by others (30). CIS overexpression did not alter ubiquitination of IκBα in nonstimulated cells. However, a significant increase in ubiquitination of IκBα was detected in cells following LPS stimulation (Fig. 7B), which was further increased with forced expression of CIS (Fig. 7B). Taken together, these data suggest that CIS may provide a feedback regulatory loop for NF-κB signaling via facilitation of IκBα degradation.
Discussion
In this study, we demonstrated that LPS stimulation and C. parvum infection induce CIS protein expression in human cholangiocytes through activation of the TLR/MyD88 pathway. Targeting of CIS 3′-UTR by miR-98 or let-7 results in posttranscriptional repression. Importantly, C. parvum- and LPS-induced CIS protein expression in cholangiocytes involves relief of miR-98/let-7-mediated posttranscriptional repression. Furthermore, forced expression of CIS is associated with an accelerated degradation of IκBα and enhanced NF-κB activation in cholangiocytes in response to LPS stimulation. These data suggest that miRNAs regulate CIS expression in cholangiocytes, a process that may be associated with the regulation of inflammatory responses in epithelial cells during microbial infection.
Despite recent significant progress, current understanding of the molecular basis underlying regulation of CIS/SOCS expression is still very limited. Most studies have focused on gene regulation at the transcriptional level (9). Emerging evidence suggests that miRNA-mediated posttranscriptional suppression is involved in the regulation of many genes. A recent report demonstrated that miR-155 targets SOCS1 to regulate T cell homeostasis (39). Our studies revealed that miR-98 and let-7 target the CIS 3′-UTR resulting in translational repression of CIS in cholangiocytes. Of those miRNAs expressed in human cholangiocytes (21, 40), we identified two adjacent potential binding sites for miR-98 or let-7 in the CIS 3′UTR. Using constructs containing the sequence with both potential binding sites, we demonstrated that miR-98 and let-7 mediate posttranscriptional suppression of CIS in nonstimulated cholangiocytes. Interestingly, it appears that the potential binding sites for both miR-98 and let-7 are required for the associated posttranscriptional suppression of CIS in cholangiocytes. The apparent requirement for both binding sites and whether they may function synergistically are currently under investigation.
Microbe-induced CIS/SOCS expression has been primarily demonstrated in immune cells. More recently, up-regulation of CIS/SOCS proteins has also been reported in epithelial cells in response to microbial infections (10). In this study, we found that LPS stimulation or C. parvum infection increased CIS expression in human cholangiocytes in a TLR4/MyD88-dependent manner. Interestingly, LPS stimulation and C. parvum infection appear not directly activate transcription of CIS gene. Instead, activation of TLR4/MyD88 signaling down-regulates miR-98 and let-7 expression and consequently, relieves miR-98/let-7-mediated translational suppression of CIS resulting in CIS protein expression. Our findings not only further support targeting of CIS by miRNAs but also raise the possibility that through miRNA-mediated posttranscriptional gene regulation, TLR signaling may regulate expression of those genes that are not activated at the transcriptional level.
CIS/SOCS proteins have classically been shown to be negative regulators of cytokine signaling. Each CIS/SOCS protein has a central SH2 domain and a carboxyterminal 40-amino acid module known as the SOCS box. The SOCS box interacts with elongin B, elongin C, cullin-5, and RING-box-2 to recruit ubiquitin transferase (41). Thus, CIS/SOCS proteins function as E3 ubiquitin ligases and mediate the degradation of the cytokine signaling complex resulting in negative feedback regulation (9). We demonstrated, in this study, that CIS may be involved in the feedback regulation of NF-κB signaling. We found that CIS promotes LPS-induced IκBα degradation and enhances NF-κB activity in cholangiocytes. Indeed, gain- or lose-of-function of CIS, as well as manipulation of miR-98 function, influences NF-κB activation in cells in response to LPS stimulation or C. parvum infection cells as monitored by the IL-8 luciferase reporter assay. Our results are consistent with previous studies on CIS-enhanced NF-κB activation in T cells (14, 15). Similarly, it was recently reported that forced expression of SOCS1 enhances NF-κB activity in cultured human respiratory epithelial cells (13). Although the underlying molecular mechanisms are currently unclear, CIS-mediated NF-κB activation may be associated with an increase of IκBα ubiquitination. Ubiquitination of IκBα is key to the regulation of NF-κB activity (42–44). Indeed, overexpression of CIS significantly decreased the IκBα level in LPS-treated cells while an increase in IκBα ubiquitination was detected in LPS-stimulated cells with forced expression of CIS. Thus, CIS may play a role in the feedback regulation of TLR/NF-κB signaling in epithelial cells in response to microbial challenge.
Although multiple strategies have been well documented for the fine-tuning of TLR/NF-κB signaling in epithelial cells, such regulation is currently limited to negative feedback loops (6–8). It remains to be determined how epithelial cells encounter these negative regulators and quickly restore their susceptibility for consistent microbial challenge (44). It is possible that positive feedback regulators are activated to encounter the negative regulators for a quick restoration of TLR/NF-κB pathway susceptibility. Therefore, fine-tuning of the TLR/NF-κB signaling dynamic may involve both negative and positive feedback regulators which function in concert to ensure finely controlled epithelial immunity against microbial infection (44). Results from this study suggest that CIS-associated IκBα degradation may be an important component of this positive feedback machinery in epithelial cells responding to microbial challenge.
Acknowledgments
We are grateful to Dr. M. F. Smith for providing the TLR4-DN construct and Prof. J. Tschopp for the MyD88-DN construct. We thank Dr. R. V. Goering for critical reading of the manuscript and Dr. X. Li for stimulating discussion.
Footnotes
This work was supported by National Institutes of Health Grants AI071321 and by the Nebraska Tobacco Settlement Biomedical Research Program LB692 (to X-M.C).
The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
Abbreviations used in this paper: CIS, cytokine-inducible Src homology 2-containing protein; SOCS, suppressors of cytokine signaling protein; SH, Src homology; miRNA, microRNA; UTR, untranslated region; HIBEpiC, human intrahepatic biliary epithelial; miR-98, microRNA-98; DN, dominant negative.
Disclosures
The authors have no financial conflict of interest.
References
- 1.Takeda K, Kaisho T, Akira S. Toll-like receptors. Annu Rev Immunol. 2003;21:335–376. doi: 10.1146/annurev.immunol.21.120601.141126. [DOI] [PubMed] [Google Scholar]
- 2.Iwasaki A, Medzhitov R. Toll-like receptor control of the adaptive immune responses. Nat Immunol. 2004;5:987–995. doi: 10.1038/ni1112. [DOI] [PubMed] [Google Scholar]
- 3.Hayden MS, Ghosh S. Shared principles in NF-κB signaling. Cell. 2008;132:344–362. doi: 10.1016/j.cell.2008.01.020. [DOI] [PubMed] [Google Scholar]
- 4.Karin M, Lawrence T, Nizet V. Innate immunity gone awry: linking microbial infections to chronic inflammation and cancer. Cell. 2006;124:823–835. doi: 10.1016/j.cell.2006.02.016. [DOI] [PubMed] [Google Scholar]
- 5.Abreu MT, Fukata M, Arditi M. TLR signaling in the gut in health and disease. J Immunol. 2005;174:4453–4460. doi: 10.4049/jimmunol.174.8.4453. [DOI] [PubMed] [Google Scholar]
- 6.Han J, Ulevitch RJ. Limiting inflammatory responses during activation of innate immunity. Nat Immunol. 2005;6:1198–1205. doi: 10.1038/ni1274. [DOI] [PubMed] [Google Scholar]
- 7.Lang T, Mansell A. The negative regulation of Toll-like receptor and associated pathways. Immunol Cell Biol. 2007;85:425–434. doi: 10.1038/sj.icb.7100094. [DOI] [PubMed] [Google Scholar]
- 8.Shibolet O, Podolsky DK. Negative regulation of Toll-like receptors and intestinal homeostasis: addition by subtraction. Am J Physiol. 2007;292:G1469–G1473. doi: 10.1152/ajpgi.00531.2006. [DOI] [PubMed] [Google Scholar]
- 9.Yoshimura A, Naka T, Kubo M. SOCS proteins, cytokine signalling and immune regulation. Nat Rev Immunol. 2007;7:454–465. doi: 10.1038/nri2093. [DOI] [PubMed] [Google Scholar]
- 10.Narayana Y, Balaji KN. NOTCH1 up-regulation and signaling involved in Mycobacterium bovis BCG-induced SOCS3 expression in macrophages. J Biol Chem. 2008;283:12501–12511. doi: 10.1074/jbc.M709960200. [DOI] [PubMed] [Google Scholar]
- 11.Baetz A, Frey M, Heeg K, Dalpke AH. Suppressor of cytokine signaling (SOCS) proteins indirectly regulate toll-like receptor signaling in innate immune cells. J Biol Chem. 2004;279:54708–54715. doi: 10.1074/jbc.M410992200. [DOI] [PubMed] [Google Scholar]
- 12.Mansell A, Smith R, Doyle SL, Gray P, Fenner JE, Crack PJ, Nicholson SE, Hilton DJ, O’Neill LA, Hertzog PJ. Suppressor of cytokine signaling 1 negatively regulates Toll-like receptor signaling by mediating Mal degradation. Nat Immunol. 2006;7:148–155. doi: 10.1038/ni1299. [DOI] [PubMed] [Google Scholar]
- 13.Pothlichet J, Chignard M, Si-Tahar M. Cutting edge: innate immune response triggered by influenza A virus is negatively regulated by SOCS1 and SOCS3 through a RIG-I/IFNAR1-dependent pathway. J Immunol. 2008;180:2034–2038. doi: 10.4049/jimmunol.180.4.2034. [DOI] [PubMed] [Google Scholar]
- 14.Li S, Chen S, Xu X, Sundstedt A, Paulsson KM, Anderson P, Karlsson S, Sjögren HO, Wang P. Cytokine-induced Src homology 2 protein (CIS) promotes T cell receptor-mediated proliferation and prolongs survival of activated T cells. J Exp Med. 2000;191:985–994. doi: 10.1084/jem.191.6.985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chen S, Anderson PO, Li L, Sjögren HO, Wang P, Li SL. Functional association of cytokine-induced SH2 protein and protein kinase C in activated T cells. Int Immunol. 2003;15:403–409. doi: 10.1093/intimm/dxg039. [DOI] [PubMed] [Google Scholar]
- 16.Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116:281–297. doi: 10.1016/s0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- 17.Ambros V. The functions of animal microRNAs. Nature. 2004;431:350–355. doi: 10.1038/nature02871. [DOI] [PubMed] [Google Scholar]
- 18.Sonkoly E, Ståhle M, Pivarcsi A. MicroRNAs and immunity: novel players in the regulation of normal immune function and inflammation. Semin Cancer Biol. 2008;18:131–140. doi: 10.1016/j.semcancer.2008.01.005. [DOI] [PubMed] [Google Scholar]
- 19.Asirvatham AJ, Gregorie CJ, Hu Z, Magner WJ, Tomasi TB. MicroRNA targets in immune genes and the Dicer/Argonaute and ARE machinery components. Mol Immunol. 2008;45:1995–2006. doi: 10.1016/j.molimm.2007.10.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Taganov KD, Boldin MP, Chang KJ, Baltimore D. NF-κB-dependent induction of microRNA miR-146, an inhibitor targeted to signaling proteins of innate immune responses. Proc Natl Acad Sci USA. 2006;103:12481–12486. doi: 10.1073/pnas.0605298103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chen XM, Splinter PL, O’Hara SP, LaRusso NF. A cellular miRNA, let-7i, regulates toll-like receptor 4 expression and contributes to cholangiocyte immune responses against Cryptosporidium parvum infection. J Biol Chem. 2007;282:28929–28938. doi: 10.1074/jbc.M702633200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Chen XM, Nelson JB, O’Hara SP, Splinter PL, Small AJ, Tietz PS, Limper AH, LaRusso NF. Multiple Toll-like receptors are expressed in human cholangiocytes and mediate host epithelial responses to Cryptoaporidium parvum via activation of NF-κB. J Immunol. 2005;175:7447–7456. doi: 10.4049/jimmunol.175.11.7447. [DOI] [PubMed] [Google Scholar]
- 23.Pedersen IM, Cheng G, Wieland S, Volinia S, Croce CM, Chisari FV, David M. Interferon modulation of cellular microRNAs as an antiviral mechanism. Nature. 2007;449:919–922. doi: 10.1038/nature06205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.O’Connell RM, Taganov KD, Boldin MP, Cheng G, Baltimore D. MicroRNA-155 is induced during the macrophage inflammatory response. Proc Natl Acad Sci USA. 2007;104:1604–1609. doi: 10.1073/pnas.0610731104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Harada K, Ohira S, Isse K, Ozaki S, Zen Y, Sato Y, Nakanuma Y. Lipopolysaccharide activates nuclear factor-κB through toll-like receptors and related molecules in cultured biliary epithelial cells. Lab Invest. 2003;83:1657–1667. doi: 10.1097/01.lab.0000097190.56734.fe. [DOI] [PubMed] [Google Scholar]
- 26.Chen XM, O’Hara SP, LaRusso NF. The immunobiology of cholangiocytes. Immunol Cell Biol. 2008;86:497–505. doi: 10.1038/icb.2008.37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Grubman SA, Perrone RD, Lee DW, Murray SL, Rogers LC, Wolkoff LI, Mulberg AE, Cherington V, Jefferson DM. Regulation of intracellular pH by immortalized human intrahepatic biliary epithelial cell lines. Am J Physiol. 1994;266:G1060–G1070. doi: 10.1152/ajpgi.1994.266.6.G1060. [DOI] [PubMed] [Google Scholar]
- 28.Chen XM, Levine SA, Tietz P, Krueger E, McNiven MA, Jefferson DM, Mahle M, LaRusso NF. Cryptosporidium parvum is cytopathic for cultured human biliary epithelia via an apoptotic mechanism. Hepatology. 1998;28:906–913. doi: 10.1002/hep.510280402. [DOI] [PubMed] [Google Scholar]
- 29.Verdon R, Keusch GT, Tzipor S, Grubman SA, Jefferson DM, Ward HD. An in vitro model of infection of human biliary epithelial cells by Cryptosporidium parvum. J Infect Dis. 1997;175:1268–1272. doi: 10.1086/593695. [DOI] [PubMed] [Google Scholar]
- 30.Ohtsubo M, Yasunaga S, Ohno Y, Tsumura M, Okada S, Ishikawa N, Shirao K, Kikuchi A, Nishitani H, Kobayashi M, Takihara Y. Poly-comb-group complex 1 acts as an E3 ubiquitin ligase for Geminin to sustain hematopoietic stem cell activity. Proc Natl Acad Sci USA. 2008;105:10396–10401. doi: 10.1073/pnas.0800672105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Horwich MD, Zamore PD. Design and delivery of antisense oligonucleotides to block microRNA function in cultured Drosophila and human cells. Nat Protoc. 2008;3:1537–1549. doi: 10.1038/nprot.2008.145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wu H, Neilson JR, Kumar P, Manocha M, Shankar P, Sharp PA, Manjunath N. miRNA profiling of naïve, effector and memory CD8 T cells. PLoS ONE. 2007;2:e1020. doi: 10.1371/journal.pone.0001020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Miller JE, Ghoshal K, Ramaswamy B, Roy S, Datta J, Shapiro CL, Jacob S, Majumder S. MicroRNA-221/222 confers tamoxifen resistance in breast cancer by targeting p27Kip1. J Biol Chem. 2008;283:29897–29903. doi: 10.1074/jbc.M804612200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Raingeaud J, Pierre J. Interleukin-4 downregulates TNFα-induced IL-8 production in keratinocytes. FEBS Lett. 2005;579:3953–3959. doi: 10.1016/j.febslet.2005.06.019. [DOI] [PubMed] [Google Scholar]
- 35.Lewis BP, I, Shih H, Jones-Rhoades MW, Bartel DP, Burgem CB. Prediction of mammalian microRNA targets. Cell. 2003;115:787–798. doi: 10.1016/s0092-8674(03)01018-3. [DOI] [PubMed] [Google Scholar]
- 36.Witherow DS, Garrison TR, William WE, Lefkowitz RJ. β-arrestin inhibits NF-κB activity by means of its interaction with the NF-κB inhibitor IκBα. Proc Natl Acad Sci USA. 2004;101:8603–8607. doi: 10.1073/pnas.0402851101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Aguilera C, Hoya-Arias R, Haegeman G, Espinosa L, Bigas A. Recruitment of IκBα to the hes1 promoter is associated with transcriptional repression. Proc Natl Acad Sci USA. 2004;101:16537–16542. doi: 10.1073/pnas.0404429101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Harris TA, Yamakuchi M, Ferlito M, Mendell JT, Lowenstein CJ. MicroRNA-126 regulates endothelial expression of vascular cell adhesion molecule 1. Proc Natl Acad Sci USA. 2008;105:1516–1521. doi: 10.1073/pnas.0707493105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lu LF, Thai TH, Calado DP, Chaudhry A, Kubo M, Tanaka K, Loeb GB, Lee H, Yoshimura A, Rajewsky K, Rudensky AY. Foxp3-dependent microRNA155 confers competitive fitness to regulatory T cells by targeting SOCS1 protein. Immunity. 2009;30:80–91. doi: 10.1016/j.immuni.2008.11.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Piessevaux J, De Ceuninck L, Catteeuw D, Peelman F, Tavernier J. Elongin B/C recruitment regulates substrate binding by CIS. J Biol Chem. 2008;283:21334–21346. doi: 10.1074/jbc.M803742200. [DOI] [PubMed] [Google Scholar]
- 41.Terzic J, Marinovic-Terzic I, Ikeda F, Dikic I. Ubiquitin signals in the NF-κB pathway. Biochem Soc Trans. 2007;35:942–945. doi: 10.1042/BST0350942. [DOI] [PubMed] [Google Scholar]
- 42.Tan P, Fuchs SY, Chen A, Wu K, Gomez C, Ronai Z, Pan ZQ. Recruitment of a ROC1-CUL1 ubiquitin ligase by Skp1 and HOS to catalyze the ubiquitination of IκBα. Mol Cell. 1999;3:527–533. doi: 10.1016/s1097-2765(00)80481-5. [DOI] [PubMed] [Google Scholar]
- 43.Roff M, Thompson J, Rodriguez MS, Jacque JM, Baleux F, Arenzana-Seisdedos F, Hay RT. Role of IκBα ubiquitination in signal-induced activation of NFκB in vivo. J Biol Chem. 1996;271:7844–7850. doi: 10.1074/jbc.271.13.7844. [DOI] [PubMed] [Google Scholar]
- 44.Mathes E, O’Dea EL, Hoffmann A, Ghosh G. NF-κB dictates the degradation pathway of IκBα. EMBO J. 2008;27:1357–1367. doi: 10.1038/emboj.2008.73. [DOI] [PMC free article] [PubMed] [Google Scholar]