Abstract
Because mouse models play a crucial role in biomedical research related to the human nervous system, understanding the similarities and differences between mouse and human brain is of fundamental importance. Studies comparing transcription in human and mouse have come to varied conclusions, in part because of their relatively small sample sizes or underpowered methodologies. To better characterize gene expression differences between mouse and human, we took a systems-biology approach by using weighted gene coexpression network analysis on more than 1,000 microarrays from brain. We find that global network properties of the brain transcriptome are highly preserved between species. Furthermore, all modules of highly coexpressed genes identified in mouse were identified in human, with those related to conserved cellular functions showing the strongest between-species preservation. Modules corresponding to glial and neuronal cells were sufficiently preserved between mouse and human to permit identification of cross species cell-class marker genes. We also identify several robust human-specific modules, including one strongly correlated with measures of Alzheimer disease progression across multiple data sets, whose hubs are poorly-characterized genes likely involved in Alzheimer disease. We present multiple lines of evidence suggesting links between neurodegenerative disease and glial cell types in human, including human-specific correlation of presenilin-1 with oligodendrocyte markers, and significant enrichment for known neurodegenerative disease genes in microglial modules. Together, this work identifies convergent and divergent pathways in mouse and human, and provides a systematic framework that will be useful for understanding the applicability of mouse models for human brain disorders.
Keywords: neurodegenerative disease, systems biology, evolution, metaanalysis, gene expression
Although certain disease mutations cause comparable phenotypes in mouse and human, the effects in mouse of any given disease-causing mutation can diverge wildly from human, especially in the case of neurological disorders. For example, whereas mouse knock-in models of Huntington disease (HD) display many of the same behavioral and pathological features seen in human HD (1), none of the three highly penetrant, dominantly transmitted causes of Alzheimer disease (AD), alone, produce AD-like pathology consisting of both plaques and tangles in mouse brain (2). Given the multifactorial, complex genetic nature of most human neurological and psychiatric diseases, such species differences are unlikely to be caused by one single pathway or gene. Therefore, understanding the molecular basis of phenotypic differences between mouse and human caused by even a single mutation is likely to require using a systems biology approach and a genome-wide view.
Previously, we demonstrated that the human brain transcriptome has a reproducible, higher-order organization, and that knowing this structure permits significant functional insights (3). So, to systematically appreciate potential species similarities and differences, we created and compared separate transcriptional networks from mouse and human brain tissue. Such a characterization in mouse brain could orient future brain-related studies, by providing a priori knowledge regarding the similarity of specific gene expression patterns between human and mouse. We created networks by merging data from many microarray studies, and then applying weighted gene coexpression network analysis (WGCNA) (4–7). WGCNA elucidates the higher-order relationships between genes based on their coexpression relationships, delineating modules of biologically related genes and permitting a robust view of transcriptome organization (3, 5, 8, 9). Within groups of highly coexpressed genes (“modules”) that comprise the core functional units of the transcriptional network, WGCNA also identifies the most highly connected, or most central genes within each module, referred to as “hubs.” We find that both gene expression and connectivity—the summation of coexpression relationships for each gene with all other genes—tend to be preserved between species. Furthermore, we find that many modules in human show preserved expression patterns in mouse. This includes, for example, all modules associated with core cellular processes, such as ribosomal and mitochondrial function, consistent with previous results (10, 11).
We also find many between-species differences that provide insight into human disease. First, we identify a human-specific module that was originally associated with AD progression in an earlier study of aging and AD (7), as well as another related human-specific module containing GSK3β and tau, both of which are implicated in AD and other dementias (12). Next, we find that significant changes in network position between the species may reflect a gene's relationship to human-specific disease phenotypes. For example, the AD-associated gene presenilin 1 (PSEN1) exhibits poor between-species network preservation, showing strong transcriptional coexpression with oligodendrocyte markers in human alone, suggesting that its role in adult human brain has significantly diverged from its role in mouse. We also find evidence for clustering of neurodegenerative disease related genes within microglial modules, highlighting the potential role of this glial cell type in human neurodegeneration. To the best of our knowledge this is the first metaanalysis to focus solely on brain-specific data, and can therefore provide unique insight into similarities and differences in transcriptional patterns between the human and mouse brain.
Results
Constructing the Mouse and Human Networks.
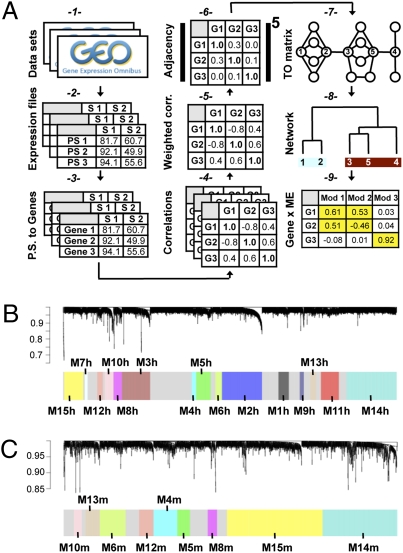
We reasoned that comparison of coexpression networks between mouse and human could provide valuable insight into human brain disorders. We sought to compile inclusive coexpression networks representing a general survey of brain transcription in both species (Fig. 1A shows a schematic of network creation, and the SI Text includes a glossary of network related terms). After careful data filtering and preprocessing to eliminate outliers (3) (Materials and Methods and SI Text), our analysis included 1,066 samples from 18 human and 20 mouse data sets, representing various diseases, brain regions, study designs, and Affymetrix platforms (Table S1). For each species, we created a network from these data, first by calculating weighted Pearson correlation matrices corresponding to gene expression, then by following the standard procedure of WGCNA to create the two networks. Briefly, weighted correlation matrices were transformed into matrices of connection strengths using a power function (5). These connection strengths were then used to calculate topological overlap (TO), a robust and biologically meaningful measurement that encapsulates the similarity of two genes’ coexpression relationships with all other genes in the network (5, 13). Hierarchical clustering based on TO was used to group genes with highly similar coexpression relationships into modules. In all, we found 15 modules in the human network (Fig. 1B) and nine modules in the mouse network (Fig. 1C), which were used to guide our final module characterizations (Materials and Methods).
Fig. 1.
Creation of mouse and human networks. (A) Flowchart for network creation. Data sets were collected from GEO (1) and preprocessed separately (2) creating similarly scaled expression files with the best probe set (P.S.) chosen for each gene and the outlier samples removed (3) (SI Text). After calculating Pearson correlation matrices separately for each data set (4), these matrices were combined to form a single weighted correlation (corr.) matrix for each species (5). Networks were created from the weighted correlation matrices using WGCNA, by first calculating adjacency matrices (6), then calculating TO (7) and using these values to hierarchically cluster genes into coexpression modules (8) (Materials and Methods). Final module assignments were made based on MM (9). (B and C) (Upper) Cluster dendrograms in the human (B) and mouse (C) metaanalyses group genes into distinct modules (step 8). The y-axes correspond to distance (1 − TO). (Lower) Dynamic tree cutting was used to determine modules, generally by dividing the dendrogram at significant branch points. Modules with significant overlap were assigned the same labels.
Global Similarities Between Mouse and Human Brain Transcription.
We first compared general network properties to ensure that our networks were reasonably matched. Both gene expression and connectivity between networks were significantly preserved between the species (R = 0.60, P < 10−400 for expression; R = 0.27, P < 10−70 for connectivity; Fig. S1). Expression levels were more preserved than connectivity, consistent with our previous results, indicating that connectivity is a more sensitive measure of evolutionary divergence than differential expression (4). We also found greater between-species expression correlations than previous microarray studies of mouse and human brain (14) as well as liver, testes, and muscle (15) (R of approximately 0.45 for all studies). These results suggest that our large data sets allow us to recognize interspecies transcriptional similarities as well as, or better than, previous methods. As a further validation of predicted network interactions on a global level, we compared gene–gene connectivity based on TO to known protein–protein interactions (PPIs), demonstrating a linear relationship between TO and PPIs in both human and mouse (Fig S2 and SI Text). Genes with high TO were much more likely to interact, consistent with previous results from multiple species (3, 16).
Many Mouse and Human Network Modules Are Highly Similar.
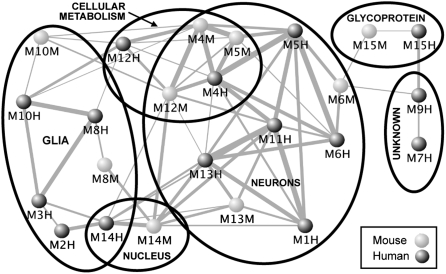
To assess gene coexpression preservation between the species on a module-by-module basis, we first calculated the module membership (MM)—a measure of how well each gene correlates with the first principal component of gene expression within a module, termed the module eigengene (ME; Materials and Methods) (3, 17). We then imposed a threshold based on MM values (R > 0.2, P < 10−13) to make final module assignments (Materials and Methods). Using this method, each module contained an exact number of assigned genes, and many genes were assigned to multiple modules, albeit with different strengths. We observed a high degree of between-species module preservation (Fig. 2 and Materials and Methods). In fact, all mouse modules showed significant overlap, in terms of gene members, with at least one human module, whereas there were multiple human-specific modules (Table 1 and Fig. 1 B and C). Gene-by-eigengene tables containing MM and initial module characterizations for all genes in both networks are available at our website (www.genetics.ucla.edu/labs/horvath/CoexpressionNetwork/MouseHumanBrain).
Fig. 2.
There is high module overlap both within and between-species. Furthermore, modules with significant overlap tend also to have similar functional characterizations (also see Table 1 and Table S2). Dots correspond to modules from the mouse (light) and human (dark) networks. Line widths are scaled based on the significance of overlap between corresponding modules. Position of the dots and length of the lines are arbitrary to aid visualization.
Table 1.
Characterization and preservation of mouse and human network modules
|
P value |
||||||||
| Human module | Mouse module | Overlap number* | Overlap P value | Top CTX module | Top CTX module characterization | Human | Mouse | PreservationZ-score |
| M1h | — | — | — | M17 | PvalB+ interneurons | 8.1E-148 | — | 2.86 |
| M2h | — | — | — | M9 | Oligodendrocytes | <1E-300 | — | 5.79 |
| M3h | — | — | — | M15 | Astrocytes | 8.1E-300 | — | 0.86 |
| M4h | M4m | 163 (34) | 2.6E-80 | M7 | Mitochondria | 1.5E-243 | 1.9E-41 | 17.21 |
| M5h | M5m | 137 (43) | 5.5E-41 | M7 | Mitochondria | 1.2E-255 | 2.5E-57 | 6.72 |
| M6h | M6m | 58 (16) | 4.8E-18 | M10 | Glutamatergic synapse | 2.9E-102 | 3.7E-62 | 4.19 |
| M7h | — | — | — | M11 | Unknown | 4.0E-91 | — | 0.32 |
| M8h | M8m | 26 (7) | 2.8E-08 | M5 | Microglia | 1.1E-169 | 1.6E-06‡ | 2.80 |
| M9h | — | — | — | M19 | Unknown | 1.6E-19 | — | 0.42 |
| M10h | M10m | 6 (1) | 9.9E-04 | M4 | Microglia | 8.6E-120 | N/S | 4.15 |
| M11h | — | — | — | M16 | Neuron | 3.5E-278 | — | 11.73 |
| M12h | M12m | 38 (7) | 5.7E-18 | M2 | Ribosome | 5.1E-68 | 4.7E-09 | 12.84 |
| M13h | M13m | 92 (29) | 1.6E-25 | M16 | Neuron | 1.1E-160 | 2.2E-33 | 2.10 |
| M14h | M14m | 200 (61) | 2.1E-65 | M18 | Nucleus† | 1.0E-233 | 1.0E-107 | 8.90 |
| M15h | M15m | 15 (6) | 4.2E-02 | M19 | Unknown | 6.1E-151 | 1.9E-11‡ | 1.23 |
Columns 5–7 represent the module from the CTX network in ref. 3 showing the highest overlap with each module in the human network, along with associated characterization. Significance of overlap with the corresponding mouse module is presented in column 8. Column 9 measures module preservation (bolded Z-scores indicate significant preservation). P values are corrected for multiple comparisons.
*The expected number of overlapping genes is presented in parentheses.
†Module characterization was inferred using other means.
‡Overlap with a network other than CTX in ref. 3 (M8m = CN network, M15m = CTX95 network).
Modules identified here in mouse and human were both validated and annotated by comparison with a previous analysis of the human brain transcriptome in cortex (3). This analysis showed not only that all modules in our human network overlapped significantly with those identified previously, but also that these modules consist of biologically meaningful gene groups (Table 1, SI Text, and Materials and Methods). Annotations using Gene Ontology (GO) and Ingenuity pathway analysis (IPA) also showed high concordance between matched human and mouse modules (Table S2). Furthermore, modules associated with basic cellular components showed the most significant between-species preservation, as measured both by the number of overlapping genes and by a summary measure of module preservation between networks (Table 1 and SI Text) (18). Finally, markers for at least one neuronal module (M11h) showed higher between-species preservation than markers for astrocytes or microglia, consistent with current knowledge of glial divergence between the species, in that astroglia in human are more numerous and of higher complexity than in other mammals (19).
Interspecies Convergence (Marker Genes) and Divergence for Cell Types.
Previous work from our group has found that coexpression is a viable method for determining cell type markers (3). To establish interspecies markers in this analysis, in essence providing additional confirmation of human–mouse preservation at the cellular level, we first chose one module for each major cell type based on annotation (Fig. 2, Table 1, and Table S2). We then identified the top ranked genes based on the significance of MM in both human and mouse networks (SI Text). The top 12 marker genes for oligodendrocyte (M2h), neuron (M13h), astrocyte (M3h), and microglia (M10h) were chosen as a starting point for comparison (Table S3; a more extensive table is presented as Table S4). These markers were compared with published mouse (20–22) and human (3, 23) marker genes for each cell type, and validated by showing highly significant overlap (P < 10−10 for each cell type). To provide further validation we also ran the converse assessment, measuring to what extent our networks correctly identified well-established marker genes, again finding generally positive results (SI Text and Table S5). Finally, we provide supplementary tables to allow readers to further screen for interspecies marker genes (Table S4) or identify genes with significant between-species divergence (Table S6).
Differences in Mouse and Human Modules Provide Insight into AD.
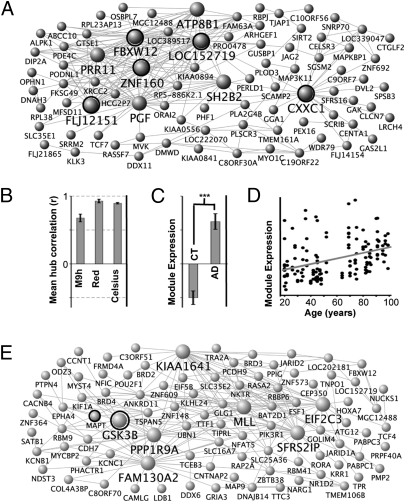
There is great precedent for the importance of studying the molecular evolutionary basis of phenotypic differences between humans and other mammalian species on the transcriptional level (24, 25). We reasoned that differences in network organization could provide a basis for better understanding diseases enriched in human populations, such as AD. With this in mind, we identified one highly human-specific module, M9h, showing significant overlap with a module recently identified to correlate highly with AD progression in another human data set (P < 10−13; compare Fig. 3A vs. figure 3D in ref. 7). Both of these modules had four matching hub genes (FBXW12, LOC152719/ZNF721, FLJ12151, and ZNF160). Data from the Celsius database, a unique resource encompassing many Affymetrix microarrays, confirms high coexpression of probe sets representing these four hubs (Fig. 3B) (26). Although three of these hubs are of unknown function, ZNF160 is a known transcriptional repressor of TLR4, which contributes to amyloid peptide–induced microglial toxicity (27), suggesting a possible known molecular link between M9h and AD.
Fig. 3.
Human-specific modules provide insight into AD mechanisms. (A) Network depiction of M9h shows that M9h shares four hubs in common with the red module from ref. 7, which contained genes whose expression increased with AD progression; and one hub in common with a corresponding module in a second study of AD (29) (Fig. S3A). Dots correspond to genes and lines to connections, with the top 250 connections in M9h shown. Larger genes correspond to hubs, which have at least 15 connections. (B) These hubs show extremely high coexpression in both studies and in the large Celsius database. Error bars represent SD of between-hub correlation values. (C) The corresponding module from ref. 29 shows significantly higher expression in AD than in control (CT). Bars represent mean ME values over all CT/AD data sets and error bars represent SE. ***P < 10−11. (D) M9h is also reproduced in a study of aging (28), in which the corresponding module (Fig. S3B) shows positive correlation with age. Points represent ME expression for individual samples plotted against age. The gray line indicates the line of best fit of the data. P < 10−5, R = 0.37. (E) Network depiction of M7h, which is poorly characterized but contains both MAPT and GSK3B (a prominent tau kinase). Labeling is as in A.
We performed two confirmatory analyses to ensure that the correlation of this module with AD progression was not an artifact of the specific samples or microarray platforms chosen: (i) an analysis of human aging in the hippocampus, entorhinal cortex, superior frontal gyrus, and postcentral gyrus using data from Affymetrix microarrays (28); and (ii) an analysis of human AD in the temporal cortex using Illumina arrays (29) (SI Text). We found that, not only were modules corresponding to M9h present in both analyses (Fig. S3), but that they also showed significant positive correlation with both age and AD progression (Fig. 3 C and D). Further, we found that CXXC1 is a common hub to both M9h and the corresponding module in Webster et al. (Fig S3A) (29). CXXC1 is a DNA binding protein that binds the polyglutamine protein TBP (30), which is known to accumulate in neurofibrillary tangles in AD (31). By using independent data sets run on different brain regions, on different platforms, and in different labs, these results confirm the likely role of this module and its reproduced hubs in human disease. A list of the top genes across M9h and its related modules is presented in Table S7.
Interestingly, two other genes related to AD and frontotemporal dementia (FTD) in humans, GSK3β and tau (12), were also relatively central genes in another human specific module, M7h (Fig. 3E). Not only does M7h correspond to a module from Oldham et al. (3) based on number of overlapping genes (Table 1), but GSK3B was also found to be a hub gene in both modules (see figure S4K in ref. 3), further implicating both this hub and this module in disease processes. Thus, although both M7h and M9h fail to show significant functional annotation (Table 1 and Table S2) and contain many genes whose functions are unknown in the nervous system, these two human-specific modules provide key targets for furthering our understanding of neurodegenerative dementias.
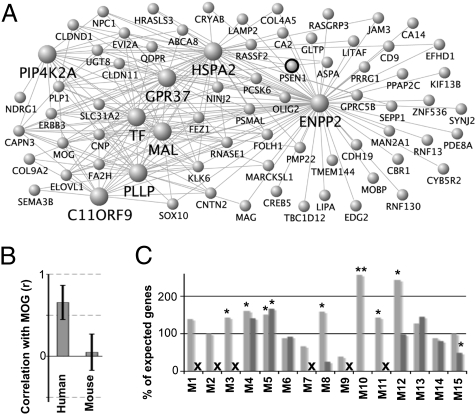
Next, we assessed how transcriptional differences between mouse and human brain networks can provide insight into disease at the level of individual genes. Orthologous genes showing discordant expression patterns may indicate divergent regulation or novel functions between species (32), and may be important regulators of brain function (14). We identified 67 validated, human-specific marker genes for cell type (SI Text and Table S6), of which PSEN1—one of three known genes whose mutation causes familial AD (2)—is centrally positioned in the human, but not the mouse, oligodendrocyte module (Fig. 4A). To quantify this observation, we measured the correlation between PSEN1 and myelin oligodendrocyte glycoprotein (MOG; the top interspecies marker for oligodendrocytes and a known myelin sheath surface protein) in each data set and compared the results between species (Fig. 4B). PSEN1 and MOG showed consistent positive correlation only in human, providing strong evidence for species-specific function and regulation of PSEN1. Expression patterns of other prominent AD-related genes are presented in SI Text.
Fig. 4.
Multiple glial cell types show disease-relevant, human-specific properties. (A) PSEN1 has high connectivity in M2h (the human oligodendrocyte module). Labeling is as in Fig 3A. (B) PSEN1 and MOG show strong positive correlation in all human data sets, but minimal correlation in any mouse data set. Error bars represent SD of correlation values in each network. (C) Most modules contain excess dementia-related genes, with microglial modules showing the greatest between-species differences. The x axis corresponds to mouse and human module labels (“X” indicates no mouse module). The y axis corresponds to the percent of observed dementia DGs relative to the number expected by chance (100%). *P < 0.05; **P < 0.001.
Systematic Evaluation of Neurodegenerative Disease and AD Genes.
To assess the distribution of disease genes (DGs) in both networks more systematically, we accessed a public curated list (Jackson Labs) (33) of approximately 5,000 known DGs (more precisely, genes known to cause disease phenotypes in humans and/or mouse when mutated). From this list, we found the subset of DGs related to neurodegeneration or dementia (dementia DGs; SI Text). We then compared the module assignments of these dementia DGs between species. In human, we found that most modules associated with cell types or basic cellular processes showed significant over-representation of dementia DGs (Fig. 4C). While most corresponding mouse modules showed similar enrichments, both microglial mouse modules (M8m, M10m) contained very few such genes. We confirmed this result for AD by measuring the overlap between each module and a published list of AD genes (34). Only the two human microglial modules showed significant enrichment for AD genes (M8h, P = 0.002; M10h, P = 0.02), providing evidence of another important species difference in glial cells. This result is particularly striking given increasing evidence of a causal role for neuroinflammation in AD pathogenesis, and considering that the microglia is the resident innate neural immune cell (35). These collective results reaffirm the idea that neuronal cell death is only one small part of the overall biological changes occurring with the progression of AD, and likely other dementias.
Observed Network Differences Are Not Due to Confounding Factors.
Because the initial human samples involved several brain regions and diseases not represented in the mouse samples, it was possible that the human-specific modules were inadvertently related to these disease samples. To control for this possibility, we constructed additional mouse and human networks using only the subset of “control” (nondisease) microarrays from cortex of both species, finding similarly preserved modules and the same between-species differences identified in the larger network analysis (SI Text). Another potential confounder was agonal state, which differs between mice and humans. To control for this factor, we assessed the modules identified here for enrichment in genes previously associated with agonal state in humans (SI Text) (36). Agonal state genes were not concentrated in the human-specific modules; rather, highly preserved modules between mouse and human (including the mitochondrial modules) showed the most significant enrichment for agonal state genes, indicating that agonal state is not a significant source of the observed between-species differences (SI Text).
Discussion
Implications for AD and Neurodegenerative Disease Research.
We have used a systems biology approach to find a number of between-species transcriptional differences relevant to neurodegenerative disease research, especially dementia (Results and SI Text). This includes a human specific module related to AD progression, which implicates many genes of previously unknown nervous system function in dementia. These genes include at least three hub genes with zinc finger motifs (ZNF160, CXXC1, and LOC152719/ZNF721) that are likely involved in transcriptional regulation—in some cases with disease-related genes—providing a new window of investigation into AD pathophysiology.
At the level of genes with known function, we highlight PSEN1 as an example, as (i) mutations in human PSEN1 cause a dominant, highly penetrant form of AD, but only limited pathology is seen in mouse mutants; and (ii) in the present study, PSEN1 shows high correlation with oligodendrocyte markers only in human (Fig. 4 A and B). Recent studies suggest myelin dysfunction contributes to a wide range of psychiatric disorders, such as schizophrenia and depression, and may be involved in normal cognitive function, learning, and IQ (reviewed in ref. 37). In AD, there is growing genetic evidence that myelin integrity may play an early role, both in humans and in animal models (reviewed in ref. 38)—evidence that is supported by recent imaging data (39). In particular, oligodendrocytes located in brain regions most vulnerable to AD pathology myelinate many axonal segments, and are thought to be susceptible to AD risk factors, such as head injury and high cholesterol (38). Dysfunction of these cells could then lead to a progressive disruption of cell communication followed by neurodegeneration in a predictable sequence. The current results are consistent with this theory, suggesting that one of the likely many distinctions between AD in mouse models and humans is related to evolutionary changes in expression patterns of PSEN1 in the context of neuron–oligodendrocyte interactions.
Combining WGCNA and Coexpression Metaanalysis in Brain.
Several studies comparing human and mouse transcription have been published (10, 15, 40–43), coming to different conclusions about divergence of the transcriptome. One reason for this may be that changes in gene expression levels are not as sensitive as network position (connectivity) to evolutionary divergence (3, 4). This comprehensive network-based metaanalysis thus has a number of advantages over traditional transcriptional analyses, leading to more reliable results than in previous studies. First, we limit our data only to arrays run on brain tissue. Many genes are expressed in more than one tissue (43, 44); therefore, such filtering emphasizes transcriptional correlations based on brain-specific gene functions. Second, we include data from multiple studies across array platforms, resulting in more functionally relevant coexpression relationships (26, 45). Our unbiased preprocessing steps lead to much higher comparability than previous studies, as measured by correlation of ranked expression between species (Fig. S1). Third, we compare data across species. Between-species coexpression preservation has been shown to prioritize DG selection under genetic disease loci (40) and to categorize the function of poorly-characterized genes better than coexpression in a single species (11). Finally, our data are combined at the level of correlation matrices (rather than gene expression levels), which minimizes issues arising from between-study comparisons (46) and highlights coexpression relationships (47). Following this approach, we constructed networks using WGCNA, a method proven to produce functionally relevant modules in a wide variety of situations (3–8). Together, these strategies result in highly reproducible networks, lending credence to the claim that our results are biologically relevant and may provide important insights into disease.
Limitations and Future Work.
Because this study represents a relatively new approach, it is important to discuss potential limitations. First, we were able to include only the 4,527 genes in common between networks, whereas nearly 80% of all gene transcripts are thought to be expressed in mouse and human brain (20). With improvements in sequencing technology, future data sets should allow for a more complete comparative analysis. Second, differences between human and mouse networks may be a result of a number of factors. The majority of network differences are likely to be genuinely biological, rather than caused by confounders such as agonal state or unbalanced sample selection. We have addressed these issues by creating smaller networks using only control microarrays from cortex, and have also shown that differences in agonal state do not account for the between-species differences (Results and SI Text).
Our results suggest that, alongside behavioral and physiological profiling, gene expression analysis could be a useful tool for evaluating mouse models of human neurodegenerative and neuropsychiatric disease. First, we identify several similarities between the mouse and human transcriptomes, providing useful resources for the study of mouse model systems. We also find multiple human-specific modules associated with dementia, including one that may play a role in AD progression and another containing the tau gene, which is mutated in the related condition, FTD (48). More generally, our methods can determine coexpression patterns and extensive between-species comparisons for any gene in which expression data are available. For example, Creutzfeld–Jakob disease is caused by mutations in the prion protein (PrP), which, when inoculated into mice, recapitulate human neurodegenerative phenotypes with more fidelity than single AD mutations, consistent with the strong interspecies module preservation observed here for PrP (PRNP—the gene for PrP—is in M14 in both networks). Conversely, several genes involved in autism (49) show significant between-species differences [i.e., CNTNAP2 is in the human-specific module M7h (Fig. 3E) and CYFIP1 is a human-specific hub in the microglia module (Table S6)], whereas others appear well preserved across species (i.e., FMR1 is an interspecies hub in M14; Table S4). Similar analyses for genes involved in other disorders could be conducted (Tables S4 and S6). Overall, our results suggest that information is present in transcriptional data that should be used to aid in the understanding of neurodegenerative and neuropsychiatric disorders, and the corresponding mouse models developed to study these diseases.
Materials and Methods
Data Set Acquisition and Network Formation.
Mouse and human microarray data sets were downloaded from the Gene Expression Omnibus (GEO) (50). As our goal was to compile an extensive set of comparable data, we collected as many relevant data sets as we could find, and then subjected these data to a stringent but unbiased filtering process (SI Text); we included only brain samples from experiments run on Affymetrix platforms in our analysis, then removed data sets with disproportionately low average within-species expression or connectivity correlation. These studies represent various diseases, brain regions, study designs, Affymetrix platforms, and sample sizes, and therefore represent a general survey of brain transcription (Table S1). From these expression data we followed the protocols of WGCNA (3, 5) to create within-species consensus networks for human and mouse (as described in Results and SI Text; modified from ref. 47). This left a total of 9,778 genes in the human analysis and 6,368 genes in the mouse analysis. Fig. 1A summarizes this entire methodology.
Module Formation and Characterization.
For the initial module characterization, all but the top 5,000 connected genes in the human network (3,000 in mouse) were excluded to decrease noise, leaving the most informative genes for network formation (SI Text). Genes were hierarchically clustered and modules were determined by using a dynamic tree-cutting algorithm (Fig. 1 B and C) (51). Module identifiers in the mouse network were then changed to match the most similar module in the human network based on gene overlap (3). For both networks, MM was calculated, then a threshold (R > 0.2; P < 10−13) was used to establish final module assignments as described in Results. These specific values were chosen such that on average each module contains approximately 5% of the genes present in our networks. This thresholding procedure allowed us to measure module overlap with any other lists using the hypergeometric distribution. Modules were graphically depicted using the program VisANT (52) as previously described (4, 7). These network depictions show the 250 strongest reciprocal within-module gene–gene interactions (i.e., connections) as measured by TO. A gene was considered a hub if it had at least 15 depicted connections. It is important to note that with the module definitions based on MM, some genes can be members of multiple modules.
Network Comparisons.
For all between-species network comparisons, human orthologs of the mouse gene were used as proxy, and only the 4,527 genes common to both networks were included. Nearly all of our comparative analyses involving cross-tabulations were done using a hypergeometric distribution, whereby we tested if the number of overlapping genes between one category and a given module was significantly large. For example, this strategy was used to assess module overlap (Fig. 2 and Table 1), to confirm interspecies markers for cell type (Table S3), and to compare modules against a list of known dementia DGs (Fig. 4C and SI Text) (33). In the case of our comparisons with modules from ref. 3, CTX modules were defined with a cutoff MM significance of P < 0.05.
Supplementary Material
Acknowledgments
We thank Peter Langfelder for providing valuable code and conversation regarding statistical issues related to this study, Michael Oldham for valuable discussions, and Brent Bill, Jeff Goodenbour, Jennifer Lowe, and Irina Voineagu for reading the final manuscript for clarity. This work was supported by National Research Service Award F31 AG031649 from the National Institute on Aging (NIA) (to J.A.M.); National Institutes of Health (NIH) Grants U19 AI063603-01 and P01 HL028481 (to S.H.); NIH/National Institute of Mental Health Merit Award R37 MH 60233-09S1 (to D.H.G); NIH/NIA Award R01 AG26938-05 (to D.H.G); and Consortium for Frontotemporal Dementia Research Award 108400 (to D.H.G).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.0914257107/-/DCSupplemental.
References
- 1.Lin CH, et al. Neurological abnormalities in a knock-in mouse model of Huntington's disease. Hum Mol Genet. 2001;10:137–144. doi: 10.1093/hmg/10.2.137. [DOI] [PubMed] [Google Scholar]
- 2.Oddo S, et al. Triple-transgenic model of Alzheimer's disease with plaques and tangles: Intracellular Abeta and synaptic dysfunction. Neuron. 2003;39:409–421. doi: 10.1016/s0896-6273(03)00434-3. [DOI] [PubMed] [Google Scholar]
- 3.Oldham MC, et al. Functional organization of the transcriptome in human brain. Nat Neurosci. 2008;11:1271–1282. doi: 10.1038/nn.2207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Oldham MC, Horvath S, Geschwind DH. Conservation and evolution of gene coexpression networks in human and chimpanzee brains. Proc Natl Acad Sci USA. 2006;103:17973–17978. doi: 10.1073/pnas.0605938103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zhang B, Horvath S. A general framework for weighted gene co-expression network analysis. Stat Appl Genet Mol Biol. 2005;4:e17. doi: 10.2202/1544-6115.1128. [DOI] [PubMed] [Google Scholar]
- 6.Horvath S, et al. Analysis of oncogenic signaling networks in glioblastoma identifies ASPM as a molecular target. Proc Natl Acad Sci USA. 2006;103:17402–17407. doi: 10.1073/pnas.0608396103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Miller JA, Oldham MC, Geschwind DH. A systems level analysis of transcriptional changes in Alzheimer's disease and normal aging. J Neurosci. 2008;28:1410–1420. doi: 10.1523/JNEUROSCI.4098-07.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Konopka G, et al. Human-specific transcriptional regulation of CNS development genes by FOXP2. Nature. 2009;462:213–217. doi: 10.1038/nature08549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Winden KD, et al. The organization of the transcriptional network in specific neuronal classes. Mol Syst Biol. 2009;5:291. doi: 10.1038/msb.2009.46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bergmann S, Ihmels J, Barkai N. Similarities and differences in genome-wide expression data of six organisms. PLoS Biol. 2004;2:E9. doi: 10.1371/journal.pbio.0020009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Stuart JM, Segal E, Koller D, Kim SK. A gene-coexpression network for global discovery of conserved genetic modules. Science. 2003;302:249–255. doi: 10.1126/science.1087447. [DOI] [PubMed] [Google Scholar]
- 12.Geschwind DH. Tau phosphorylation, tangles, and neurodegeneration: The chicken or the egg? Neuron. 2003;40:457–460. doi: 10.1016/s0896-6273(03)00681-0. [DOI] [PubMed] [Google Scholar]
- 13.Ravasz E, Somera AL, Mongru DA, Oltvai ZN, Barabási AL. Hierarchical organization of modularity in metabolic networks. Science. 2002;297:1551–1555. doi: 10.1126/science.1073374. [DOI] [PubMed] [Google Scholar]
- 14.Strand AD, et al. Conservation of regional gene expression in mouse and human brain. PLoS Genet. 2007;3:e59. doi: 10.1371/journal.pgen.0030059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Xing Y, Ouyang Z, Kapur K, Scott MP, Wong WH. Assessing the conservation of mammalian gene expression using high-density exon arrays. Mol Biol Evol. 2007;24:1283–1285. doi: 10.1093/molbev/msm061. [DOI] [PubMed] [Google Scholar]
- 16.Ge H, Liu Z, Church GM, Vidal M. Correlation between transcriptome and interactome mapping data from Saccharomyces cerevisiae. Nat Genet. 2001;29:482–486. doi: 10.1038/ng776. [DOI] [PubMed] [Google Scholar]
- 17.Horvath S, Dong J. Geometric interpretation of gene coexpression network analysis. PLOS Comput Biol. 2008;4:e1000117. doi: 10.1371/journal.pcbi.1000117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Langfelder P, Horvath S. WGCNA: An R package for weighted correlation network analysis. BMC Bioinformatics. 2008;9:559. doi: 10.1186/1471-2105-9-559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Oberheim NA, Wang X, Goldman S, Nedergaard M. Astrocytic complexity distinguishes the human brain. Trends Neurosci. 2006;29:547–553. doi: 10.1016/j.tins.2006.08.004. [DOI] [PubMed] [Google Scholar]
- 20.Lein ES, et al. Genome-wide atlas of gene expression in the adult mouse brain. Nature. 2007;445:168–176. doi: 10.1038/nature05453. [DOI] [PubMed] [Google Scholar]
- 21.Cahoy JD, et al. A transcriptome database for astrocytes, neurons, and oligodendrocytes: A new resource for understanding brain development and function. J Neurosci. 2008;28:264–278. doi: 10.1523/JNEUROSCI.4178-07.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gan L, et al. Identification of cathepsin B as a mediator of neuronal death induced by Abeta-activated microglial cells using a functional genomics approach. J Biol Chem. 2004;279:5565–5572. doi: 10.1074/jbc.M306183200. [DOI] [PubMed] [Google Scholar]
- 23.Albright AV, González-Scarano F. Microarray analysis of activated mixed glial (microglia) and monocyte-derived macrophage gene expression. J Neuroimmunol. 2004;157:27–38. doi: 10.1016/j.jneuroim.2004.09.007. [DOI] [PubMed] [Google Scholar]
- 24.Khaitovich P, Enard W, Lachmann M, Pääbo S. Evolution of primate gene expression. Nat Rev Genet. 2006;7:693–702. doi: 10.1038/nrg1940. [DOI] [PubMed] [Google Scholar]
- 25.King MC, Wilson AC. Evolution at two levels in humans and chimpanzees. Science. 1975;188:107–116. doi: 10.1126/science.1090005. [DOI] [PubMed] [Google Scholar]
- 26.Day A, Carlson MR, Dong J, O'Connor BD, Nelson SF. Celsius: A community resource for Affymetrix microarray data. Genome Biol. 2007;8:R112. doi: 10.1186/gb-2007-8-6-r112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Takahashi K, Sugi Y, Hosono A, Kaminogawa S. Epigenetic regulation of TLR4 gene expression in intestinal epithelial cells for the maintenance of intestinal homeostasis. J Immunol. 2009;183:6522–6529. doi: 10.4049/jimmunol.0901271. [DOI] [PubMed] [Google Scholar]
- 28.Berchtold NC, et al. Gene expression changes in the course of normal brain aging are sexually dimorphic. Proc Natl Acad Sci USA. 2008;105:15605–15610. doi: 10.1073/pnas.0806883105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Webster JA, et al. NACC-Neuropathology Group Genetic control of human brain transcript expression in Alzheimer disease. Am J Hum Genet. 2009;84:445–458. doi: 10.1016/j.ajhg.2009.03.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Keshava Prasad TS, et al. Human Protein Reference Database—2009 update. Nucleic Acids Res. 2009;37(database issue):D767–D772. doi: 10.1093/nar/gkn892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Reid SJ, et al. TBP, a polyglutamine tract containing protein, accumulates in Alzheimer's disease. Brain Res Mol Brain Res. 2004;125:120–128. doi: 10.1016/j.molbrainres.2004.03.018. [DOI] [PubMed] [Google Scholar]
- 32.Pao SY, Lin WL, Hwang MJ. In silico identification and comparative analysis of differentially expressed genes in human and mouse tissues. BMC Genomics. 2006;7:86. doi: 10.1186/1471-2164-7-86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Bult CJ, Eppig JT, Kadin JA, Richardson JE, Blake JA. Mouse Genome Database Group The Mouse Genome Database (MGD): Mouse biology and model systems. Nucleic Acids Res. 2008;36(database issue):D724–D728. doi: 10.1093/nar/gkm961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bertram L, McQueen MB, Mullin K, Blacker D, Tanzi RE. Systematic meta-analyses of Alzheimer disease genetic association studies: The AlzGene database. Nat Genet. 2007;39:17–23. doi: 10.1038/ng1934. [DOI] [PubMed] [Google Scholar]
- 35.Meda L, et al. Activation of microglial cells by beta-amyloid protein and interferon-gamma. Nature. 1995;374:647–650. doi: 10.1038/374647a0. [DOI] [PubMed] [Google Scholar]
- 36.Atz M, et al. Methodological considerations for gene expression profiling of human brain. J Neurosci Methods. 2007;163:295–309. doi: 10.1016/j.jneumeth.2007.03.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Fields RD. White matter in learning, cognition and psychiatric disorders. Trends Neurosci. 2008;31:361–370. doi: 10.1016/j.tins.2008.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Bartzokis G. Age-related myelin breakdown: A developmental model of cognitive decline and Alzheimer's disease. Neurobiol Aging. 2004;25:5–18. doi: 10.1016/j.neurobiolaging.2003.03.001. [DOI] [PubMed] [Google Scholar]
- 39.Ringman J, et al. Diffusion tensor imaging in preclinical and presymptomatic carriers of familial Alzheimer's disease mutations. Brain. 2007;130:1767–1776. doi: 10.1093/brain/awm102. [DOI] [PubMed] [Google Scholar]
- 40.Ala U, et al. Prediction of human disease genes by human-mouse conserved coexpression analysis. PLOS Comput Biol. 2008;4:e1000043. doi: 10.1371/journal.pcbi.1000043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Chen J, Xu H, Aronow BJ, Jegga AG. Improved human disease candidate gene prioritization using mouse phenotype. BMC Bioinformatics. 2007;8:392. doi: 10.1186/1471-2105-8-392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Tsaparas P, Mariño-Ramírez L, Bodenreider O, Koonin EV, Jordan IK. Global similarity and local divergence in human and mouse gene co-expression networks. BMC Evol Biol. 2006;6:70. doi: 10.1186/1471-2148-6-70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Yanai I, Graur D, Ophir R. Incongruent expression profiles between human and mouse orthologous genes suggest widespread neutral evolution of transcription control. OMICS. 2004;8:15–24. doi: 10.1089/153623104773547462. [DOI] [PubMed] [Google Scholar]
- 44.Su AI, et al. A gene atlas of the mouse and human protein-encoding transcriptomes. Proc Natl Acad Sci USA. 2004;101:6062–6067. doi: 10.1073/pnas.0400782101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Lee HK, Hsu AK, Sajdak J, Qin J, Pavlidis P. Coexpression analysis of human genes across many microarray data sets. Genome Res. 2004;14:1085–1094. doi: 10.1101/gr.1910904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Berg J, Lässig M. Cross-species analysis of biological networks by Bayesian alignment. Proc Natl Acad Sci USA. 2006;103:10967–10972. doi: 10.1073/pnas.0602294103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Langfelder P, Horvath S. Eigengene networks for studying the relationships between co-expression modules. BMC Syst Biol. 2007;1:54. doi: 10.1186/1752-0509-1-54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Lee VM, Goedert M, Trojanowski JQ. Neurodegenerative tauopathies. Annu Rev Neurosci. 2001;24:1121–1159. doi: 10.1146/annurev.neuro.24.1.1121. [DOI] [PubMed] [Google Scholar]
- 49.Bill BR, Geschwind DH. Genetic advances in autism: heterogeneity and convergence on shared pathways. Curr Opin Genet Dev. 2009;19:271–278. doi: 10.1016/j.gde.2009.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Edgar R, Domrachev M, Lash AE. Gene Expression Omnibus: NCBI gene expression and hybridization array data repository. Nucleic Acids Res. 2002;30:207–210. doi: 10.1093/nar/30.1.207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Langfelder P, Zhang B, Horvath S. Defining clusters from a hierarchical cluster tree: The Dynamic Tree Cut package for R. Bioinformatics. 2008;24:719–720. doi: 10.1093/bioinformatics/btm563. [DOI] [PubMed] [Google Scholar]
- 52.Hu Z, Mellor J, Wu J, DeLisi C. VisANT: An online visualization and analysis tool for biological interaction data. BMC Bioinformatics. 2004;5:17. doi: 10.1186/1471-2105-5-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.