Abstract
The development of a universal soybean (Glycine max [L.] Merr.) cytogenetic map that associates classical genetic linkage groups, molecular linkage groups, and a sequence-based physical map with the karyotype has been impeded due to the soybean chromosomes themselves, which are small and morphologically homogeneous. To overcome this obstacle, we screened soybean repetitive DNA to develop a cocktail of fluorescent in situ hybridization (FISH) probes that could differentially label mitotic chromosomes in root tip preparations. We used genetically anchored BAC clones both to identify individual chromosomes in metaphase spreads and to complete a FISH-based karyotyping cocktail that permitted simultaneous identification of all 20 chromosome pairs. We applied these karyotyping tools to wild soybean, G. soja Sieb. and Zucc., which represents a large gene pool of potentially agronomically valuable traits. These studies led to the identification and characterization of a reciprocal chromosome translocation between chromosomes 11 and 13 in two accessions of wild soybean. The data confirm that this translocation is widespread in G. soja accessions and likely accounts for the semi-sterility found in some G. soja by G. max crosses.
SOYBEAN is an important food worldwide for both humans and livestock. Soybean has the ability to fix atmospheric nitrogen through its symbiotic interaction with rhizobia and, therefore, has served as a model species to decipher the mechanism of plant–microbe interaction and nodule development. Soybean is a model crop for sustainable agriculture, due to its reduced need for added nitrogen fertilizer as well as its ability to supply fixed nitrogen in crop rotations. More recently, soybean has become a valuable resource for biodiesel, an increasingly important alternative to fossil fuels. Thus, the importance of soybean as a major crop justifies the investment of substantial resources toward sequencing of the genome and the development of functional genomic tools (Stacey et al. 2004; Jackson et al. 2006). A significant milestone was the recent completion of the complete genome sequence (Schmutz et al. 2010). Knowledge of the soybean genome promises to reveal significant new insights into important agronomic traits, such as plant–pathogen interactions, seed oil and protein biosynthesis, and environmental stress resistance.
Classical linkage groups (CLGs), consisting of genetic markers, were defined for soybean >10 years ago (Palmer and Shoemaker 1998). Subsequently, a single, integrated molecular genetic map was developed for soybean using molecular (e.g., SSR) markers, thereby defining 20 molecular linkage groups (MLGs) that then were associated with CLGs (Cregan et al. 1999; Mahama et al. 2002). More recently, a bacterial artificial chromosome (BAC)-based physical map was generated for soybean, which then was linked via a number of markers to the MLGs (Shoemaker et al. 2008). The dense soybean genetic map (Song et al. 2004; Choi et al. 2007), as well as the physical map, were significant aids during the assembly of the soybean genome sequence (Schmutz et al. 2010).
Karyotype maps have been developed for a number of major crop species, including wheat, rice, maize, barley, and tomato. These maps allow the association of CLGs, MLGs, and a sequence-based physical map with each of the physical chromosomes. The first cytological description of domesticated soybean, Glycine max [L.] Merr. (G. max) (2n = 40), was developed using pachytene chromosomes, which were numbered from 1 to 20 on the basis of total chromosome length, arm length ratios, and relative proportions of euchromatin and heterochromatin (Singh and Hymowitz 1988). Primary trisomic lines (2x + 1 = 41; Xu et al. 2000) were used to associate 11 of the 20 MLGs with cytologically numbered chromosomes (Cregan et al. 2001; Zou et al. 2003). However, this created a problem when the soybean genome sequencing was completed because 9 of the physical chromosomes were unassigned. This was resolved by assigning chromosome numbers on the basis of molecular and genetic recombination distances (centimorgans), which do not necessarily correspond to physical distances (micrometers) or cytological size. This approach may have resulted in the numbering of soybean chromosomes in direct contradiction to the original cytologically determined chromosome designations (Singh and Hymowitz 1988).
To resolve these potential problems and to provide a useful resource for correlating physical soybean chromosomes to the other available soybean mapping resources, we undertook efforts to generate a soybean karyotype map using fluorescent in situ hybridization (FISH) with chromosome-specific, pseudomolecule- and repeat-derived DNA probes. The cytological study of soybean metaphase chromosomes has been challenging for several reasons: first, the chromosomes are small, on the order of ∼1–2 μm in length (Sen and Vidyabhusan 1960; Clarindo et al. 2007); second, the 20 pairs of soybean chromosomes (Veatch 1934) make FISH-mapping challenging; and third, these 20 chromosome pairs show little morphological diversity. With the exception of a single acrocentric pair, soybean chromosomes are all metacentric or submetacentric (supporting information, Figure S1; Ahmad et al. 1984; Singh and Hymowitz 1988; Clarindo et al. 2007), making them difficult to distinguish in routine mitotic preparations. Moreover, the low mitotic index characteristic of soybean root meristems (Ahmad et al. 1983) renders chromosome preparation for karyotyping quite inefficient.
A powerful strategy that was developed for karyotyping maize (Kato et al. 2004) was to use a collection (“cocktail”) of fluorescent DNA probes, based on repetitive sequences whose targets are present in multiple chromosomes at various copy numbers and positions and whose collective hybridization pattern discriminates each chromosome. Because genomic repeat content and genome organization vary from organism to organism, development of a repeat-based FISH karyotyping cocktail requires identification and testing of candidate repeat probes in the species of interest. Based on DNA denaturation studies, ∼40–60% (Goldberg 1978; Gurley et al. 1979) of the soybean 1115 Mb/1C genome (Arumuganathan and Earle 1991) is composed of repetitive DNA. These repeats have been well cataloged in terms of diversity and abundance (Nunberg et al. 2006; Swaminathan et al. 2007), but not with regard to chromosome distribution. FISH studies (Lin et al. 2005) suggest that much of soybean's repetitive sequences are restricted to the large pericentromeric, heterochromatic blocks on each chromosome. In terms of composition, whole-genome shotgun and BAC-end sequencing (Nunberg et al. 2006), as well as 454 pyrosequencing-based studies (Swaminathan et al. 2007), show that the majority of soybean repetitive sequences are copia-like and gypsy-like long terminal repeat retrotransposons. However, due to their potential for cross-hybridization, probes made from these elements have limited utility in FISH karyotyping.
Retrotransposons represent the largest class of repeats in soybean because many different repeat families contribute to the total retrotransposon content. However, the most abundant sequences in soybean (Swaminathan et al. 2007) are members of the SB92 (Kolchinsky and Gresshoff 1995; Vahedian et al. 1995) repeat family. The STR120 (Morgante et al. 1997) repeat family also is highly abundant (Swaminathan et al. 2007). Each of these repeat classes exists as large tandem arrays, likely in the form of higher-order repeat units of slight variants of the main consensus repeat (Swaminathan et al. 2007). SB92 repeats were recently renamed CentGm repeats due to evidence (Gill et al. 2009) suggesting that they are soybean centromeric satellite repeats. At 91–92 bp, CentGm repeats are significantly smaller than many other centromeric repeats, such as the 180-bp pAL1 repeat of Arabidopsis (Martinez-Zapater et al. 1986), the 156-bp CentC of maize (Ananiev et al. 1998), the 155-bp CentO repeat of rice (Cheng et al. 2002), or the 171-bp alpha satellite repeat of humans (Wevrick and Willard 1989). The above repeats are the approximate size of a nucleosome unit, while the CentGm repeats are ∼1/2 of a nucleosome unit (Swaminathan et al. 2007). Yet like other centromeric repeats (Henikoff et al. 2001), CentGm elements are rapidly evolving. Southern blots detect the repeats in annual soybeans, G. max and G. soja Sieb. and Zucc. (G. soja), but not in related perennial Glycine species (Vahedian et al. 1995). Through FISH screening of high-copy repeats, such as CentGm, it should be possible to capitalize on variation in copy number, sequence, and localization in the formulation of a karyotyping cocktail for soybean.
Differential FISH labeling of soybean chromosomes would be the first step in karyotyping; specific chromosomes must then be cross-referenced with the genetic and physical maps. The whole-genome physical maps (Glyma1.01, at http://www.phytozome.net) generated by the Soybean Genome Sequencing Consortium (Schmutz et al. 2010) present an opportunity to identify such chromosome-specific probes. Because each BAC used in the physical mapping itself contains, or is locally assembled with other BACs containing mapped molecular markers, these clones represent a means to correlate pseudomolecules, and hence MLGs, with mitotic or meiotic chromosomes.
Our first objective was to screen soybean repetitive DNA sequences by FISH to identify repeats with sufficient diversity in copy number and localization to serve as the basis for a karyotyping cocktail for G. max cv. Williams 82 (G. max W82), the standard cultivar chosen for sequencing (Stacey et al. 2004; Jackson et al. 2006; Schmutz et al. 2010). Our second objective was to use pseudomolecule-derived BAC clones to identify individual chromosomes in metaphase spreads. Our third objective was to formulate karyotyping cocktails for G. max W82 and a G. soja accession. Our fourth objective was to characterize a chromosome exchange in two G. soja accessions to demonstrate the utility of the soybean karyotyping system.
MATERIALS AND METHODS
Informatic discovery and analysis of CentGm and SB86 genomic repeats:
The most abundant repeat units in the soybean genome were identified and their approximate copy number estimated, using a 454-sequence survey of randomly sheared genomic DNA (Swaminathan et al. 2007). In this previously published work, a catalog of 80,377 repeat consensus sequences (“contigs”) was generated and numbered in order of abundance in the soybean genome. Thus, contig 80,377 (a sequence that contains multiple slight variations of the CentGm1 repeat in tandem; i.e., a higher order repeat of CentGm1) is the single most abundant sequence in the soybean genome. Contig 80,377 and other repeats described here were identified using a noncognate assembly technique (Swaminathan et al. 2007), where randomly generated survey DNA sequences are assembled into putative repeat sequences. Sequences from different regions of the genome are assembled into a “noncognate” consensus of the repeat, where the number of genomic reads assembled into any one noncognate contig provides a means of estimating genomic copy number. Unlike whole-genome sequencing, where any sequences that assemble in a noncognate manner must be excluded, this method provides large contigs of higher-order units containing slight variations of small tandem repeats (such as CentGm1), as well as assemblies of larger repeat units such as transposons. As a result of their tendency to assemble in a noncognate manner, many of the sequences identified using this method are excluded from the soybean whole-genome assembly (Schmutz et al. 2010). To discover small tandem repeats within the larger assembled contigs produced by the assembly, mreps software (Kolpakov et al. 2003) was used to identify the contigs that consisted of tandemly repeated, slightly varied smaller sequence units. Three major tandem repeat families with estimated copy number above 1,000 per genome were identified: the SB92 class (CentGm-1 and CentGm-2), the STR101 class, and a repeat of 86 bp that has not been described previously, here termed the SB86 repeat. Using a Perl script (available from the authors) and a simple set of rules (each must be 50% GC, 25 bp long, and specifically identify one repeat subfamily), a set of fluorochrome-labeled oligonucleotide probes was designed for screening as karyotyping agents. Probes for the CentGm family of repeats were designed manually from a sequence alignment using the same rules. Probes that showed useful, discriminative patterns for karyotyping are described below.
Identification of low-repeat content BACs:
To identify potential BAC clones that would generate minimal background signal by cross-hybridization in FISH, clones with low-repeat content were identified computationally. BAC sequences were identified as regions of assembled sequence flanked by BAC-end pairs in genomic assembly scaffold sequences of G. max W82 (Schmutz et al. 2010). BAC-end matches were required to have the following characteristics: blastn (Altschul et al. 1990) with >99% identity and >90% query coverage, the end pairs have opposite orientations, and putative pairs are separated by >20 kb and <250 kb. BAC sequences were selected that had <4% repeat content, judged by the proportion of the BAC masked using RepeatMasker (http://repeatmasker.org) with a library of low-complexity sequences and 573 representative soybean transposable elements (Du et al. 2010). Additionally, preference was given to low-repeat BACs occurring near the ends of pseudomolecule assemblies. These BACs then were screened (data not shown) by FISH to identify one or two low-background BACs for each chromosome.
Oligonucleotide FISH probes:
We used the following fluorochrome-labeled oligonucleotide FISH probes (Invitrogen or Integrated DNA Technologies). CentGm-2-M: TTGCTCAGAGCTTCAACATTCAATT, labeled with cyanine 5 (Cy5), Texas red-615; CentGm-2-G: AAGCTCTGAGCAAATTCAAACGAC, labeled with fluorescein; CentGm-1-AF: CGAGAAATTCAAATGGTCATAACT, labeled with fluorescein or Texas red-615; CentGm-1-E: TTCACTCGGATGTCCGATTCGAGGA, labeled with fluorescein; CentGm-1-J2: TTCTCGAGAGCTTCCGTTGTTCAAT, labeled with Texas red; SB86-C: ATGTGATCTTTGTTATTTTCCCG, labeled with Cy5 or Texas red; and 18S-rDNA: AGAGCTCTTTCTTGATTCTATGGGTGGTGGT, labeled with Texas red-615.
BAC FISH probes:
To generate sufficient amounts of BAC DNA for FISH probe synthesis, BAC DNA was amplified by rolling circle amplification (RCA) (Berr and Schubert 2006). For each BAC, DNA was prepared from a 2-ml overnight Escherichia coli culture grown in LB medium supplemented with 20 μg/ml kanamycin using a Wizard Plus SV Miniprep DNA purification kit (Promega). Five to 20 ng of purified BAC DNA was used for RCA in a thermal cycler using thiophosphate-modified random hexamer primers (5′-NpNpNpNpSNpSN-3′, IDT). The RCA product was used to synthesize a fluorochrome-labeled FISH probe by nick translation as follows: each 20-μl (Texas red) probe synthesis reaction contained 2.5 μl (about 2 μg) of RCA product, 7.7 μl water, 2 μl 10X Nick Translation Buffer (500 mm Tris–HCl, pH 7.8, 50 mm magnesium chloride, 100 μg/ml bovine serum albumin (fraction V), and 100 mm beta-mercaptoethanol), 2 μl [-C]-dNTP mix (2 mm dATP, 2 mm dGTP, and 2 mm dTTP), 0.4 μl of 1 mm Texas red-5-dCTP (Perkin Elmer), 5 μl DNA polymerase I (10 units/μl; Invitrogen) and 0.4 μl DNAse I (0.1 units/μl; Roche). For fluorescein-labeled BAC probes, 1 mm Fluorescein-12-dUTP (Perkin Elmer) and [-T]-dNTP mix (2 mm dATP, 2 mm dCTP, and 2 mm dGTP,) were used instead of the corresponding solutions mentioned above. The reaction was incubated at 15° for 2 hr. Probes were purified using the Wizard SV gel and PCR clean-up kit (Promega). The purified probes, eluted in water, were dried in a Speedvac and resuspended in 10 μl of 2X SSC/1X TE solution.
Fluorescent in situ hybridizations and image processing:
FISH experiments involving only oligonucleotide probes were carried out using the pressurized nitrous oxide technique (Kato 1999) adapted from maize (Kato et al. 2004) to soybean, precisely as described in Gill et al. (2009). Experiments in which BAC probes were hybridized in combination with oligonucleotide probes used a procedure slightly modified as follows: For a single or multiple BAC probe FISH experiment, 0.5–2 μl of each BAC probe (above) was combined and brought to 8 μl using a 2X SSC/1X TE solution, incubated at 99° for 5 min and quenched for 5 min on ice. Two microliters of a 5X solution of oligonucleotide probe (or probe oligonucleotide cocktail) was added, and the complete probe mix was applied to a slide.
FISH images were collected as TIF format files and processed in Photoshop CS2 (Adobe Systems Incorporated), as described by Gill et al. (2009), with the following modifications: The 4′,6-Diamidino-2-phenylindole (DAPI) signal for each FISH experiment was collected as a separate image that was later reintroduced as a separate gray-scale layer into a second image containing only the fluorochrome-labeled probe signals. For images with both FISH probe(s) and DAPI signals, the gray-scale DAPI layer was adjusted in the “levels” menu to “lighten,” with an opacity of 25%. The signal for Cy5-labeled probes was pseudocolored blue. Raw image files were imported into Photoshop and the resolution was increased from 72 dpi to 200 dpi. Images were converted from 8-bit to 16-bit mode; using the levels menu, cytoplasmic background was subtracted using the “set black point” tool. At this point, for experiments using the four-component (CentGm-2-M, CentGm-2-G, CentGm-1-E, and CentGm-1-AF oligonucleotides) centromere-labeling cocktail, the centromere color of single chromosome pair (Gm20 or Gs20) was quite close to yellow. The color of this pair was manually adjusted to yellow by minor enhancement of the green channel in the levels menu. Because images with BAC probes were collected under conditions optimal for the (generally brighter) centromere signals, these images were slightly enhanced in the levels menu to visualize the (generally less bright) BAC signals. For panels in which chromosomes are arrayed as a chart (e.g., Figure 1C), areas corresponding to individual chromosomes were traced with the “magnetic lasso” tool, copied, rotated to perpendicular with the “rotate” tool, and finally aligned in a grid. Postmanipulation, all images were converted back to 8-bit mode and saved as .psd files.
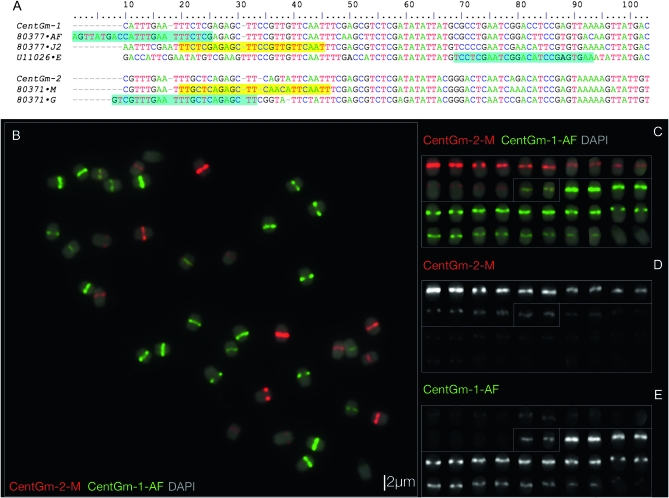
Figure 1.—
Sequence and localization analysis of CentGm repeats in G. max W82. (A) ClustalW alignment of consensus DNA sequences of the most abundant CentGm family repeats identified by Swaminathan et al. (2007), together with U11026, a CentGm variant identified by Vahedian et al. (1995), in addition to CentGm-1 and CentGm-2, monomers identified by Gill et al. (2009). Two major CentGm repeat subtypes are evident: a 92-bp class (upper) and a 91-bp subtype (lower). Oligonucleotide probes used to target individual repeats are indicated after the sequence names (e.g. “·AF,” with a corresponding probe name of CentGm-AF). Oligonucleotide probes identical to a given sequence are indicated by yellow background, whereas those that are reverse complements are indicated by aqua background. Probes ·AF and ·G would each hybridize across the junction of two monomers; thus, the sequences are duplicated for these regions. Oligonucleotides CentGm-1-AF and CentGm-1-J2 target 24- or 25-bp sequences, respectively, conserved among members of the 92-bp repeat subtype, and oligonucleotide CentGm-1-E targets a specific variant of the 92-bp subtype. Oligonucleotide CentGm-2-M targets a consensus 25-bp sequence conserved among a fraction of the 91-bp repeat subtypes, whereas oligonucleotide CentGm-2-G targets a consensus 25-bp sequence conserved among most of the abundant 91-bp variants. (B–E) FISH of G. max W82 chromosomes with fluorochrome-tagged oligonucleotide probes targeting 91- and 92-bp CentGm repeats. The following oligonucleotide probes (amounts) were used per slide: fluorescein-CentGm-1-AF (20 ng) Texas red-615-CentGm-2-M (20 ng). (B) Chromosome spread probed with a red-fluorochrome-tagged CentGm-2-M oligonucleotide and a green-fluorochrome-tagged CentGm-1-AF oligonucleotide. The DAPI signal is included as a 25% intensity gray-scale channel to delineate chromosomes. (C) Individual chromosomes from B arrayed generally by color and intensity of CentGm hybridization signal. (D) The red channel (CentGm-2-M), excluding DAPI, of the chromosome array in C. (E) The green channel (CentGm-1-AF), excluding DAPI, of the chromosome array in C. Three types of chromosomes are apparent: 7 pairs (upper chromosomes in C–E) that hybridize predominantly to the red CentGm-2-M probe, 12 pairs (lower chromosomes in C–E) that hybridize to the green CentGm-1-AF probe, and a single pair (boxed in center in C–E) that hybridizes equally to both probes. Bar in B is also valid for C–E.
RESULTS
A repetitive element foundation for the soybean karyotyping cocktail:
To identify probes for karyotyping metaphase soybean chromosomes, we focused on non-retroelement short tandem repeats identified by analysis of consensus repeat families. The repeat family consensus sequences originally were generated through 454 sequencing of soybean genomic DNA (Swaminathan et al. 2007). To screen the repeats from this analysis in the CentGm family, we generated a ClustalW sequence alignment of consensus sequences derived from the most abundant CentGm-related sequences in the soybean genome (Swaminathan et al. 2007), in addition to previously identified CentGm-related sequences (Vahedian et al. 1995; Gill et al. 2009). The alignment (Figure 1A) suggests that this repeat family does contain a 91-bp class and a 92-bp class, although other, rarer size variants are present, including 93 bp and 94 bp. This result is concordant with a recent, but independent study (Gill et al. 2009) that used whole-genome shotgun-derived soybean genomic sequences. To investigate the distribution of these repeats in chromosomes and to assess their utility in karyotyping, we designed fluorochrome-labeled oligonucleotide probes targeting conserved sequences within either the 91- or 92-bp classes and tested them using FISH on G. max W82 chromosomes (Figure 1, B–E). Probe CentGm-2-M, targeting a 25-bp sequence conserved among members of the CentGm-2 class, primarily hybridized to the centromeres of 8 pairs of chromosomes (Figure 1D); whereas probe CentGm-1-AF, targeting a 25-bp sequence conserved among members of the 92-bp class, primarily hybridized to the centromeres of 13 pairs of chromosomes (Figure 1E). A single chromosome pair hybridized to both probes under these hybridization conditions (Figure 1, C–E). A similar result (Figure S2) was obtained using a probe (CentGm-1-J2, Figure 1A) targeting a different conserved sequence among the 92-bp variants. The range of centromeres labeled (Figure 1C) by these probes makes the CentGm repeat probes a promising foundation for a karyotyping cocktail.
To further explore the potential diversity of CentGm monomer distribution in the genome, we designed and screened probes to additional CentGm repeat consensus or monomer sequences (Figure 1A). Two FISH examples are shown in Figure 2. When the CentGm-2-M probe (Cy5-labeled and pseudocolored blue) was hybridized in combination with the (fluorescein-labeled) CentGm-2-G probe that also targets members of the 91-bp class, there was substantial overlap in hybridization (Figure 2, A–C). However, the ratio of blue to green varied among the centromeres, presumably due to variable copy numbers of probe targets (variants) within different centromeres. In addition, the CentGm-2-G probe hybridized to more chromosomes than CentGm-2-M (Figure 2, B and C). A comparable, imperfectly overlapping pattern also was generated by hybridizing two oligonucleotide probes targeting variants of CentGm-1 (Figure 2, D–F). When the red fluorochrome-labeled CentGm-1-AF oligonucleotide, targeting a consensus sequence identified by genome survey (Swaminathan et al. 2007), was used in combination with a fluorescein-labeled CentGm-1-E oligonucleotide, which targeted a published single 92-bp CentGm-1 variant (Vahedian et al. 1995) not identified as a high-copy variant (Swaminathan et al. 2007), a range of centromere color/intensity again was produced (Figure 2D).
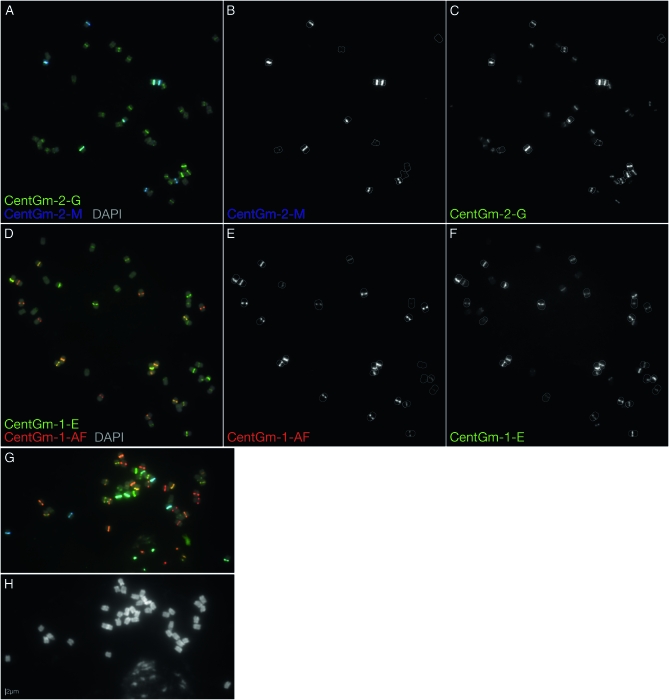
Figure 2.—
Localization of 91- and 92-bp CentGm repeat variants in G. max W82. (A–F) FISH of G. max W82 chromosomes with fluorochrome-tagged oligonucleotide probes targeting 91-bp (CentGm-2) and 92-bp (CentGm-1) CentGm repeats, either within the 91-bp class (A–C) or within the 92-bp class (D–F). (A) Cy5 (pseudocolored blue)-labeled CentGm-2-M oligonucleotide targets a subset of chromosomes that largely overlaps those targeted by the green-labeled CentGm-2-G oligonucleotide. (B) Gray-scale image of the CentGm-2-M fluorescent channel, excluding DAPI, of the chromosomes in A. All chromosomes (7 pairs are detectable with the Cy5-labeled CentGm-2-M probe at this exposure) with hybridization signal are outlined. (C) Gray-scale image of the CentGm-2-G fluorescent channel, excluding DAPI, of the chromosomes in A. The CentGm-2-M-hybridizing chromosomes from B are outlined. Several additional chromosome pairs are detected by the CentGm-2-G probe. (D) The red-labeled CentGm-1-AF probe targets a subset of chromosomes that largely overlaps those targeted by the green-labeled CentGm-1-E probe. (E) Gray-scale image of the CentGm-1-AF fluorescent channel, excluding DAPI, of the chromosomes in D. All chromosomes (13 pairs) with detectable hybridization signal are outlined. (F) Gray-scale image of the CentGm-1-E fluorescent channel, excluding DAPI, of the chromosomes in D. The CentGm-1-AF-hybridizing chromosomes from E are outlined. Several additional chromosome pairs are detected by the CentGm-1-E probe. (G and H) Four-component CentGm probe FISH cocktail. (G) FISH using a combination of four probes: two (CentGm-2-M and CentGm-2-G) for the 91-bp CentGm class and two (CentGm-1-AF and CentGm-1-E) for the 92-bp CentGm class. The following oligonucleotide probes (amounts) were used per slide: CY5-CentGm-2-M (7.5 ng), fluorescein-CentGm-2-G (20 ng), Texas red-615-CentGm-1-AF (0.01 ng), and fluorescein-CentGm-1-E (20 ng). (H) DAPI channel from the image in G. In addition to the condensed metaphase chromosomes, a portion of an intact nucleus is evident at the bottoms of G and H. In A, D, and G, DAPI signal is included as a 25% intensity gray-scale channel to delineate chromosomes. Bar in H is valid for all FISH panels.
Because each oligonucleotide pair (CentGm-2-M/CentGm-2-G, or CentGm-1-AF/CentGm-1-E) hybridized to a largely complementary subset of chromosomes (Figure 1), and because each of the two probes within a given CentGm class hybridized to differing degrees within a given subset (Figure 2), careful selection of fluorochrome color of the oligonucleotide probes permitted generation of diverse centromere labeling with a minimal number of probes. In Figure 2G, the two pairs of CentGm probes were combined in a single FISH experiment. The CentGm-2-M/CentGm-2-G pair labeled centromeres in the color range from blue to green, whereas the CentGm-1-AF/CentGm-1-E pair labeled centromeres in the complementary color range, from green to red.
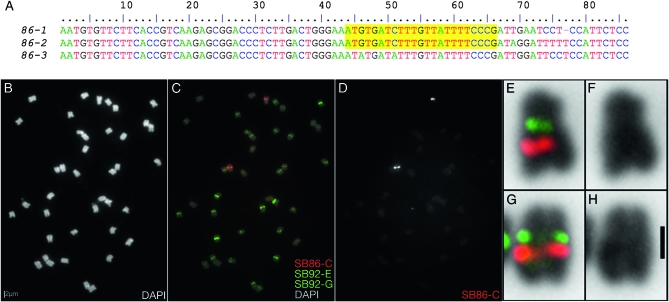
We next investigated a 86-bp repeat unit identified by analysis of the genome survey data. We term this repeat SB86; we estimate, on the basis of the number of survey reads, that it occurs in at least 5,000 copies per genome (Swaminathan et al. 2007) (Figure 3A). This repeat is present in substantially lower abundance genome-wide than the CentGm repeats, based on survey read depth. To assess the chromosomal localization of this repeat family, we designed an oligonucleotide probe, SB86-C, that targets a conserved sequence shared by most SB86 variants. The SB86-C probe primarily hybridized to a single chromosome pair (Figure 3, C and D) later identified (see below) as Gm01. The SB86-C probe hybridized to single spots at a proximal position on the long arm of the chromosome. Furthermore, the signal did not overlap CentGm probe hybridization, which localized to the primary constriction (Figure 3, E–H).
Figure 3.—
Characterization of a novel 86-bp repeat in G. max W82. (A) ClustalW alignment of consensus sequences of a novel 86-bp repeat, indicating the conserved sequence (SB86-C, highlighted yellow) targeted for FISH. (B) DAPI-stained chromosomes for FISH in C–H. (C) FISH using a Texas red-tagged oligonucleotide targeting a conserved sequence within the repeat, plus two fluorescein-tagged CentGm probes and 25% DAPI signal. (D) Gray-scale image of the SB86-C fluorescent channel, excluding DAPI, of the chromosomes in C. (E–H) Enlargements of the two chromosomes with strong SB86 probe hybridization from C, with the DAPI signal inverted to black. E and G include SB86 signal together with enhanced CentGm signals. The CentGm signals correspond to the primary constrictions of the two chromosomes, which are more evident in F and H, which show DAPI without the FISH channels. The following oligonucleotide probes (amounts) were used: Texas red-SB86-C (10 ng), fluorescein-CentGm-2-G (20 ng), and fluorescein-CentGm-1-E (20 ng). Bar in B is also valid for C and D. Bar in H, 1 μm and also valid for E–G.
The combination of four differentially labeled CentGm-related and SB86-related oligonucleotide probes generated sufficient diversity of centromere labeling to differentiate more than half of soybean's 20 chromosome pairs simply on the basis of centromere color/signal intensity combinations, leaving only a few small groups of similarly labeled chromosomes to distinguish through the development of chromosome-specific probes.
Chromosome identification and karyotyping cocktail development:
Our next objective was to use FISH probes made from BAC clones to identify chromosomes that were labeled by the five-component CentGm/SB86 oligonucleotide probe cocktail described above. To identify probes useful for karyotyping, BACs from each pseudomolecule were screened to identify those with low-repeat content. Euchromatin-derived BACs that contain even low numbers of repeats that are present at high copy number elsewhere in the genome generate significant background in FISH. Therefore, we screened BACs from terminal positions (generally within 5 Mb of either end) of each pseudomolecule against a recently developed database of 573 representative soybean repeats (Du et al. 2010; see materials and methods) and identified BACs with low total repeat content that also generated low background in FISH (Table 1 and data not shown). One or two BACs from each pseudomolecule were used in FISH in combination with the CentGm/SB86 oligonucleotide probe cocktail to correlate a given BAC/pseudomolecule with its cognate chromosome (Table 1, Figure 4 and data not shown). In many cases, a BAC hybridized to a chromosome that was uniquely labeled by the cocktail. Thus, many chromosomes could be identified solely on the basis of their centromere probe hybridization. Oligonucleotide cocktail labeling did result, however, in several groups of chromosomes with similar appearing centromeres. For these groups, centromere “coding” alone was insufficient to identify an individual chromosome. Therefore, to unambiguously resolve these sets of chromosomes, differential chromosome painting by BACs was used to distinguish them. For example, BACs from pseudomolecules corresponding to Gm07, Gm08, and Gm12 each hybridized to a different chromosome with a similarly labeled centromere (Figure 5 A–C). To distinguish these in a single chromosome spread, two Texas red-labeled BAC probes were used for Gm07, a single fluorescein-labeled BAC probe was used for Gm08, and no additional probe was used to identify Gm12. Thus, as a result of this analysis, at least one G. max W82 pseudomolecule-derived BAC clone was developed as a FISH probe for each chromosome.
TABLE 1.
BAC clones used for FISH-based chromosome assignment of G. max W82 chromosomes
| Chr | MLG | BAC | Short name | Marker | Map | cM | POS | ARM | First base | Last base | Insert | No. R/Rbp/R% | 2°fish | 2°BLAST |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Gm01 | D1a | GM_WBb0043P22 | BAC01A | Near BARC-024195-04791 | Map3 | 92.5 | HI | LA | 49,818,725 | 49,945,149 | 126,424 | 56/3,978/3.1 | Gm11 | Gm11 |
| Gm02 | D1b | GM_WBb0085H06 | BAC02A | Near BARC-021631-04160 | Map4 | 131.1 | HI | LA | 49,825,876 | 49,872,572 | 46,696 | 11/597/1.3 | Gm09 or Gm14 | Gm14 |
| Gm03 | N | GM_WBb0106L07 | BAC03A | Contains BARC-029409-06170 | Map4 | 92.7 | HI | LA | 46,297,776 | 46,409,382 | 111,607 | 41/3,862/3.5 | Gm19 | Gm19 |
| Gm04 | C1 | GM_WBb0139K20 | BAC04A | Near BARC-040387-07722 | Map4 | 75.2 | HI | LA | 44,101,333 | 44,121,856 | 20,523 | 5/257/1.3 | Gm06 | Gm06 |
| Gm05 | A1 | GM_WBb0173E04 | BAC05A | Near BARC-048987-10780 | Map4 | 0 | LO | SA | 281,063 | 389,159 | 108,096 | 44/3,440/3.2 | Gm17 | Gm17 |
| Gm05 | A1 | GM_WBb0040D01 | BAC05B | Contains BARC-013065-00437 | Map3 | 78.8 | HI | LA | 39,135,905 | 39,238,663 | 102,758 | 36/3.161/3.1 | (n.d) | Gm08 |
| Gm06 | C2 | GM_WBb0139N15 | BAC06A | Near BARC-064413-18929 | Map4 | 23.2 | LO | LA | 2,627,513 | 2,671,338 | 43,825 | 18/1,214/2.8 | Gm04 | Gm04 |
| Gm06 | C2 | GM_WBb0079P05 | BAC06B | Near Satt371 | Map3 | 127.9 | HI | SA | 48,947,618 | 48,997,768 | 50,150 | 25/1.986/4.0 | None | Gm12 |
| Gm07 | M | GM_WBb0076F20 | BAC07A | Near BARC-044075-08603 | Map3 | 8.6 | LO | SA | 1,506,830 | 1,627,146 | 120,316 | 33/2.288/1.9 | Gm08 or Gm12 | Gm08 |
| Gm07 | M | GM_WBb0114J08 | BAC07B | Near Satt336 | Map3 | 125.6 | HI | LA | 43,442,004 | 43,551,713 | 109,709 | 38/2,646/2.4 | Gm17 | Gm17 |
| Gm08 | A2 | GM_WBb0096N20 | BAC08A | Contains BARC-016861-02356 | Map3 | 45.3 | LO | LA | 8,690,272 | 8,788,538 | 98,266 | 22/1,587/1.6 | (n.d) | Gm05 |
| Gm09 | K | GM_WBb0124B19 | BAC09A | Near BARC-042081-08175 | Map3 | 86.7 | HI | (LA) | 44,014,970 | 44,121,216 | 106,246 | 34/2,566/2.4 | None | Gm18 |
| Gm10 | O | GM_WBb0170O19 | BAC10A | Near BARC-032467->08977 | Map4 | 120.4 | HI | LA | 50,083,977 | 50,183,107 | 99,130 | 31/1,899/1.9 | Gm20 | Gm20 |
| Gm11 | B1 | GM_WBb0021C24 | BAC11A | Near BARC-018583-02981 | Map4 | 2.5 | LO | LA | 1,839,546 | 1,870,687 | 31,141 | 14/944/3.0 | Gm01 | Gm01 |
| Gm12 | H | GM_WBb0139N13 | BAC12A | Contains BARC-050539-09728 | Map4 | 5.8 | LO | LA | 591,462 | 726,823 | 135,361 | 36/3,010/2.2 | Gm09 or Gm14 | Gm09 |
| Gm13 | F | GM_WBb0036C23 | BAC13A | Near BARC-016585-02149 | Map3 | 112.4 | HI | LA | 43,040,843 | 43,072,256 | 31,413 | 10/863/2.7 | Gm15 | Gm15 |
| Gm14 | B2 | GM_WBb0051I24 | BAC14A | Contains BARC-039561-07508 | Map3 | 96.5 | HI | LA | 48,863,899 | 48,908,609 | 44,710 | 17/1,523/3.4 | Gm17 | Gm17 |
| Gm15 | E | GM_WBb0045I07 | BAC15A | Contains BARC-020127-04472 | Map3 | 11.7 | LO | LA | 2,525,415 | 2,666,509 | 141,094 | 43/3,689/2.6 | Gm13 | Gm13 |
| Gm16 | J | GM_WBb0053C02 | BAC16A | Contains Sat_393 | Map3 | 90.5 | HI | LA | 37,055,360 | 37,172,436 | 117,076 | 43/2,772/2.4 | Gm09 or Gm14 | Gm09 |
| Gm17 | D2 | GM_WBb0092K07 | BAC17A | Near BARC-015033-01953 | Map3 | 14.6 | LO | LA | 3,276,988 | 3,302,405 | 25,417 | 10/700/2.8 | Gm13 | Gm13 |
| Gm18 | G | GM_WBb0149C22 | BAC18A | Contains BARC-014381-01340 | Map3 | 9.4 | LO | SA | 1,089,786 | 1,188,426 | 98,640 | 33/2,547/2.6 | Gm11 | Gm11 |
| Gm19 | L | GM_WBb0143B15 | BAC19A | Near BARC-014509-01567 | Map4 | 91.1 | HI | LA | 48,777,026 | 48,886,806 | 109,780 | 36/2,596/2.4 | Gm03 | Gm03 |
| Gm20 | I | GM_WBb0126K17 | BAC20A | Contains BARC-017125-02212 | Map3 | 110.6 | HI | LA | 46,462,795 | 46,629,738 | 166,943 | 59/4,199/2.5 | Gm10 | Gm10 |
Chr, pseudomolecule from which a BAC was derived; MLG, molecular linkage group to which a pseudomolecule was assigned; BAC, full BAC name; short name, BAC annotation used in subsequent figures; marker, molecular marker contained within or positioned near to the BAC; map, molecular linkage group map; Map3, data originates from Choi et al. (2007); Map4, data originates from Hyten et al. (2010a,b); cM, recombination distance at which the “marker” mapped in the MLG; POS, position of BAC clone within the sequence assembly of the pseudomolecule; LO, low position in bp; HI, high position in bp; ARM, in most of the pseudomolecules, positioning of the major centromere repeat array resulted in sequence assemblies in which chromosome arms had different lengths, thus, LA indicates a predicted BAC position on the long chromosome arm position; whereas SA indicates predicted short arm position. First base and last base, positions of the first and last bp, respectively, of the BAC within the pseudomolecule; insert, total length of the BAC clone in bp. For a given BAC clone, no. R/Rbp/R% indicates the number (no. R) of discrete repeat elements identified, the sum (in bp) of all repeat sequences (Rbp), and the total repeat content as a percentage (R%) of BAC insert length (i.e., the ratio of Rpb/insert). 2°fish, the chromosome on which a weaker FISH signal was detected during BAC mapping experiments (not shown). ND, not determined; none, no secondary signal was detected; 2°BLAST, the pseudomolecule of the second-best hit in a BLAST search of the genome using the full BAC sequence (the best match was always to the pseudomolecule in which the BAC sequence was assembled.
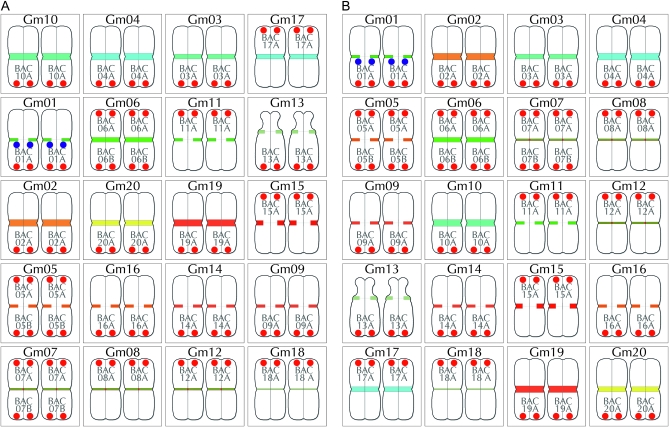
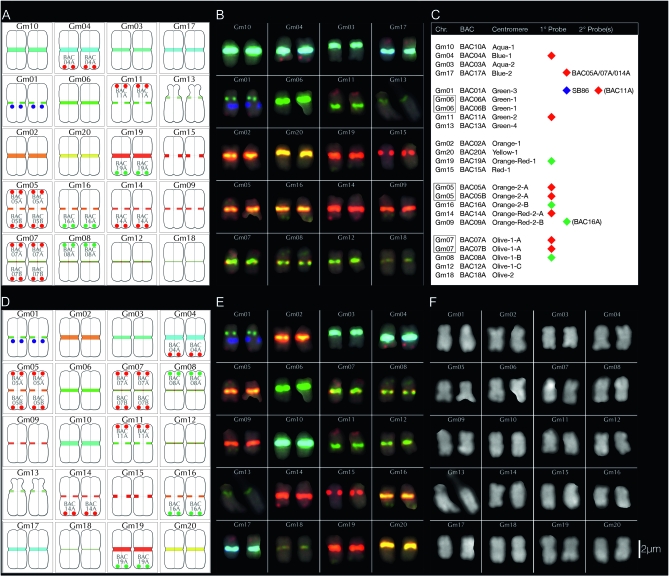
Figure 4.—
Summary of BACs used for chromosome assignments. Pseudomolecule-derived BACs were individually mapped by FISH onto G. max W82 chromosomes labeled with the CentGm/SB86 probe cocktail (Figure 2G). BAC signals (paired red dots) are labeled in shorthand (see Table 1) and positioned either at the top of a chromosome pair (for BACs at a low bp position in the pseudomolecule) or at the bottom (for BACs at a high position). The paired blue dots on Gm01 indicate SB86 repeat hybridization signal. With the exception of the satellite chromosome (Gm13), relative chromosome dimensions are not indicated. (A) Diagram in which chromosomes are arranged by color/intensity of FISH centromere signal. The top three rows contain chromosomes with unique CentGm/SB86 labeling, whereas the lower two rows contain groups of chromosomes with more similar centromere labeling. (B) Diagram in which chromosomes are arranged by chromosome number.
Figure 5.—
FISH-based karyotyping cocktail for G. max W82. The CentGm/SB86 oligonucleotide probe cocktail supplemented with 10 BAC probes permits discrimination of each of the 20 chromosome pairs. Chromosomes are arrayed by centromere color groups in A–C and by chromosome number in D–F. (A and D) are diagrams of the FISH images in B and E, respectively, in which centromere regions are depicted according to the color and size of CentGm probe hybridization signal under collection conditions optimized for centromere signals. In A and D, the blue dots on Gm01 indicate SB86-C probe hybridization. (C) Chart indicating how BAC probes were used to distinguish between members of chromosome groups with similar centromere signals. Chr, chromosome; BAC, BAC clone(s) used to identify an individual chromosome (see Figure 4); centromere, color (e.g., aqua) and relative intensity (e.g., 1 > 2A = 2B) of centromere signal. For example, Gm10, the brightest aqua-colored centromere, and Gm03, the second-brightest aqua-colored centromere are coded aqua-1 and aqua-2, respectively. 1°probe, the 10 BAC probes used in the cocktail and their respective fluorochrome colors; 2°probe, the presence of detectable secondary signal(s) on a given chromosome due to cross-hybridization by 1°probe probe(s). For example, BAC05A, BAC07A, and BAC014A, each used as a 1°probe to label their respective chromosomes, also cross-hybridize to Gm17; unlike these BACs, the secondary signals from BAC11A and BAC16A were not essential for the cocktail. SB86 indicates the SB86-C oligonucleotide probe. (F) DAPI channel of chromosomes in E; the short arm of Gm13 appears compact in the left chromosome and distended in the right chromosome. (G) Original image for chromosomes in B, E, and F; for DAPI channel and larger version of the image in G, see Figure S3. In B, E, and G, DAPI signal is included as a 25% intensity gray-scale channel to delineate chromosomes. The 15-probe karyotyping cocktail for G. max W82 used the following oligonucleotide probes (amount per slide): CY5-CentGm-2-M (7.5 ng), fluorescein-CentGm-2-G (20 ng), Texas red-615-CentGm-1-AF (0.01 ng), fluorescein-CentGm-1-E (20 ng), and CY5-SB86-C (20 ng) in combination with the following Texas red-labeled BAC probes (0.5 μl per slide) GM_WBb0021C24, GM_WBb0040D01, GM_WBb0051I24, GM_WBb0076F20, GM_WBb0114J08, GM_WBb0139K20, GM_WBb0173E04 and fluorescein-labeled BAC probes (1 μl per slide): GM_WBb0053C02, GM_WBb0096N20, and GM_WBb0143B15. Bar in F is also valid for B, E, and G.
Karyotyping cocktail for G. max W82:
On the basis of the above results, we were able to design a karyotyping cocktail for G. max W82 to simultaneously identify all 20 chromosome pairs. G. max is thought to be a diploidized tetraploid, following a genome duplication at ∼13 MYA (Blanc and Wolfe 2004; reviewed by Shoemaker et al. 2006; Schlueter et al. 2007).Therefore, most of the chromosome regions from which the BACs were derived are expected to have homeologous sequences elsewhere in the genome. Because these sequences often are detected as generally lower intensity signals when BACs are used in FISH (Pagel et al. 2004; Walling et al. 2006), we mapped secondary signals for most of the BAC probes by reexamining original or enhanced images from the mapping studies above (Table 1 and data not shown). This step was critical because a complete karyotyping cocktail would utilize combinations of BAC probes, whose individual signal intensity in FISH could vary. To verify the FISH mapping, we conducted BLAST searches at Soybase (http://soybase.org) for each BAC against the soybean genome. The second-best BLAST match was always a chromosome to which the BAC secondary signal was mapped (Table 1 and data not shown). Taking these secondary signals into account, we designed a cocktail consisting of 10 BAC probes (Figure 5C), 4 CentGm repeat-related probes, and the SB86 probe. This 15-component FISH cocktail permitted identification of each of the 20 pairs of G. max W82 chromosomes (Figure 5, B, E, and G; Figure S3).
Karyotyping in G. soja:
To determine whether the G. max W82-based CentGm/SB86 oligonucleotide probe cocktail would be useful to karyotype wild soybean accessions, we used the same strategy described above for G. max W82 to map W82 pseudomolecule-derived BACs to chromosomes of G. soja P.I. 464890B (Gong di No. 2019), a cultivar originating from Jilin province, in northeast China and maintained in the USDA Soybean Germplasm Collection (http://www.ars-grin.gov/). Although the cytogenetics of G. soja (2n = 40) were previously studied (Ahmad et al. 1984; Singh and Hymowitz 1988), most wild soybean accessions have not been mapped by molecular or classical genetic markers. Therefore, chromosome assignments based on positioned G. max W82 BACs should be viewed as tentative. With one exception (see below), each of the Gm chromosome-specific BAC probes tested hybridized to a single, specific chromosome pair in P.I. 464890B (data not shown). Overall, the range of centromere coloration by the oligonucleotide cocktail in P.I. 464890B bears a general resemblance to that of G. max W82 (Figure S4). There are comparable numbers of chromosomes in each color range, attesting to a conservation of CentGm repeat classes. However, the CentGm labeling generally was reduced in this accession, as evidenced by the increased exposure times required to achieve comparable image intensity. Despite the overall similarity in hybridization patterns, several chromosome pairs had distinctly different CentGm labeling. Compare (Figure 5 vs. Figure S4) for example, Gm01 and Gs01, or Gm02 and Gs02, which exhibited great differences in labeling intensities. Due to these differences, a different set of seven BACs was required to delineate the P.I. 464890B karyotype (Figure S4).
Characterizing a chromosome translocation in G. soja P.I. 464890B:
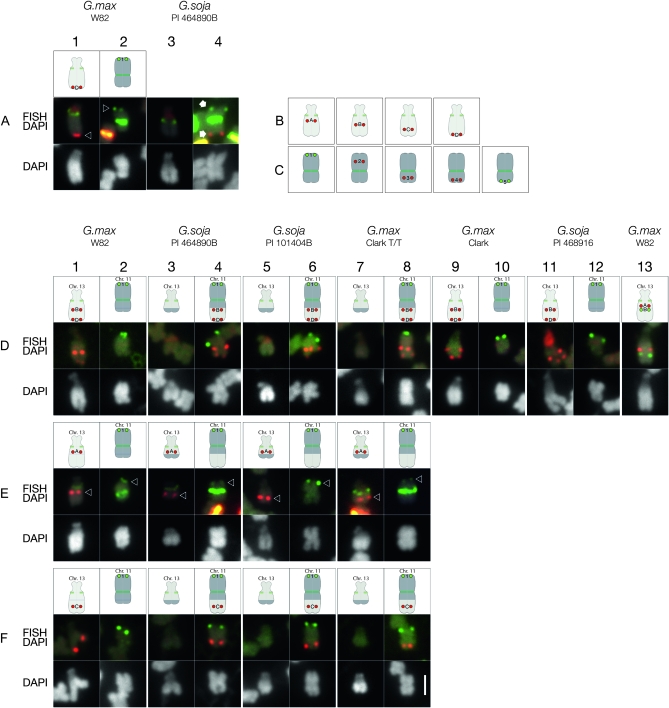
During the karyotyping of P.I. 464890B, BAC GM_WBb0036C23, which in G. max W82 hybridizes to the distal end of the long arm of Gm13 (Figure 6A, column 1), did not hybridize to the corresponding G. soja chromosome (Figure 6A, column 4), indicating that a chromosome exchange of some sort had occurred in this G. soja accession. In G. max, chromosome 13 has been very well characterized. It is the sole satellite chromosome pair (Palmer and Heer 1973; Singh and Hymowitz 1988), corresponding to molecular linkage group F (Cregan et al. 1999; Cregan et al. 2001) and classical genetic linkage groups 6 and 8 (Mahama et al. 2002). The short (satellite) arm of Gm13 contains the nucleolar organizer region (NOR) (Skorupska et al. 1989; Griffor et al. 1991) that is composed of arrays of genes encoding 18S−5.8S−28S ribosomal RNA (rDNA). Because the CentGm oligonucleotide cocktail labeling of centromeres in P.I. 464890B (Figure S4) was similar overall to that in G. max W82, the appearance of the chromosome to which the GM_WBb0036C23 probe hybridized suggested that this sequence was associated with the G. soja equivalent of Gm11. We therefore hypothesized that a translocation had occurred between chromosomes 11 and 13 in this accession relative to G. max W82. To test this hypothesis, we selected a series of BACs from G. max pseudomolecules representing chromosomes 11 and 13 (Figure 6, B and C; Table S1and Figure S5) to test in P.I. 464890B. Testing of Gm11 BAC probes in G. max W82 (Figure 7 and data not shown) verified the assembly of the pseudomolecule. In contrast, mapping of the Gm13 BACs suggested that the published sequence assembly for Gm13 (Schmutz et al. 2010) was disordered. Specifically, six BACs positioned within the first Mb of the assembly, hence on the short arm, hybridized instead to the proximal side of the long arm (Figure S5C). This is in a region of the assembly thought to be uncertain, containing only one marker in the first 0.6 Mb and with sparse BAC clone coverage (S. Cannon, personal communication). However, 12 additional BACs, spanning positions 26.4–43.1 Mb of the 44.4 Mb Gm13 pseudomolecule appeared to be colinear at the resolution of FISH (Figure S5D). For mapping in P.I. 464890B, we chose four Gm13 BACs (Figure 6B) and five Gm11 BACs (Figure 6C).
Figure 6.—
Mapping Gm13 pseudomolecule-derived BAC FISH probes in G. max and G. soja accessions. (A) In G. max W82, the karyotyping BAC probe for Gm13 (labeled “D” in the diagram, corresponding to the FISH panel in column 1) hybridizes to a distal position (arrowhead) on the long arm of Gm13, the satellite chromosome; whereas the BAC probe for Gm11 (labeled “1” in column 2), hybridizes to a distal position (arrowhead) on the long arm of Gm11. In P.I. 464890B (columns 3 and 4), both probes hybridize (arrows) to opposite ends of a single chromosome (column 4) that does not have a satellite. Additional FISH signals in these images are derived from centromere-hybridizing CentGm repeat probes and a ribosomal DNA probe (red signal on the satellite arms). (B) Gm13 pseudomolecule-derived BAC probes used for chromosome mapping: “A,” GM_WBc0172G08; “B,” GM_WBb0071K05; “C,” GM_WBb0102G11; and “D,” GM_WBb0036C23. (C) Gm11 pseudomolecule-derived BAC probes used for chromosome mapping: “1,” GM_WBb0021C24; “2,” GM_WBc0087G10; “3,” GM_WBc0230E21; “4,” GM_WBc0128E05; and “5,” GM_WBc0205I11. (D–F) Mapping Gm13 pseudomolecule-derived BAC FISH probes in G. max and G. soja accessions. In each experimental series (rows D, E, and F), Gm11 probe 1 (labeled in green), was hybridized in combination with one or two Gm13 probes (labeled in red). (D) Gm13 probes B and D hybridize (FISH/DAPI row) to a chromosome segment on the long arm of the satellite chromosome (Gm13) of G. max W82 (column 1), Clark (column 9), and P.I. 468916 (column 11). In P.I. 464890B (column 4), P.I. 101404B (column 6) and Clark T/T (column 8), these probes are associated with a chromosome targeted by Gm11 probe 1, rather than with the satellite chromosome. By DAPI staining, the long arm of the satellite chromosome appears shorter, and the arm opposite the Gm11-marked arm appears longer in P.I. 464890B (column 3), P.I. 101404B (column 5), and Clark T/T (column 7) than in the corresponding chromosomes of G. max W82 (column 1), Clark (column 9), and P.I. 468916 (column 11). (E) Gm13 probe A hybridizes (red signal, indicated by arrowhead) to a centromere-proximal position on the long arm of the satellite chromosome in G. max W82 (column 1), P.I. 464890B (column 3), P.I. 101404B (column 5), and Clark T/T (column 7). The signal from Gm11 probe 1 is indicated by arrowheads in columns 2, 4, 6, and 8. Additional FISH signals in the FISH/DAPI images are derived from centromere-hybridizing CentGm repeat probes. (F) Gm13 probe C, an additional Gm13 long arm marker positioned between BACs B and D in the Gm13 pseudomolecule, localizes to the long arm of Gm13 in G. max W82 (column 1), and to the Gm11 probe 1 target in P.I. 464890B (column 4), P.I. 101404B (column 6), and Clark T/T (column 8). The following oligonucleotide probes (amounts) were used: CY5-CentGm-2-M (7.5 ng), fluorescein-CentGm-2-G (20 ng), Texas red-615-CentGm-1-AF (0.01 ng), fluorescein-CentGm-1-E (20 ng), CY5-SB86-C (20 ng), and Texas red-615-rDNA (10 ng). Bar in row F, column 8, DAPI panel is 2 μm and valid for all FISH and DAPI panels.
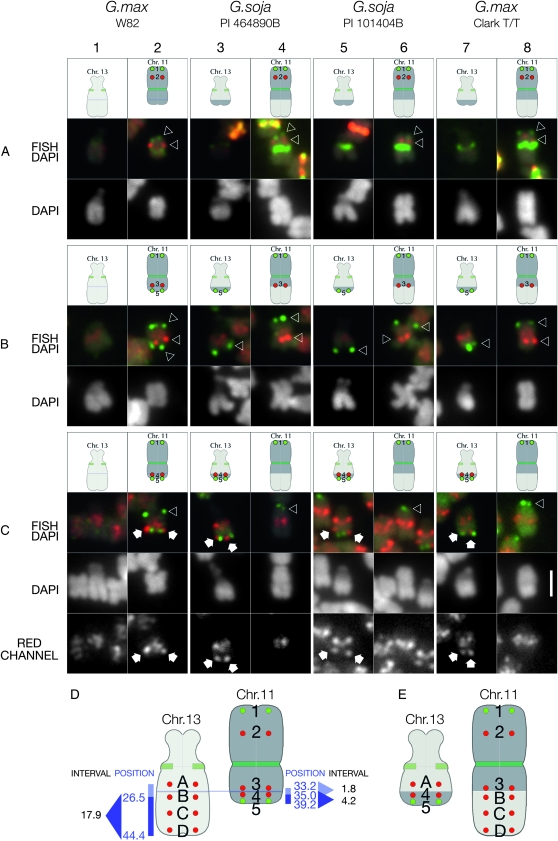
Figure 7.—
Mapping Gm11 pseudomolecule-derived BAC FISH probes in G. max and G. soja accessions. In each experimental series (rows A, B, and C), Gm11 BAC probe 1, (terminal upper probe, labeled in green), was hybridized in combination with one or two additional Gm11 probes to evaluate chromosome structure. (A) Gm11 probe 2 (red signals indicated by lower arrowhead) hybridizes approximately midway between Gm11 probe 1 (green signals indicated by upper arrowhead) and the centromere (targeted by CentGm repeat probes) in G. max W82 (column 2), P.I. 464890B (column 4), P.I. 101404B (column 6), and Clark T/T (column 8). No BAC-derived signal hybridizes to the satellite chromosome in any accession. (B) Gm11 probe 3 (red signals indicated by arrowheads) hybridizes at a centromere-proximal position on the arm opposite to the one with Gm11 probe 1 in G. max W82 (column 2), P.I. 464890B (column 4), P.I. 101404B (column 6), and Clark T/T (column 8). However, BAC probe 5, which hybridizes to a terminal position on the short arm of chromosome 11 in G. max W82 (green signals indicated by lower arrowhead in column 2), localizes to the long arm of chromosome 13 in P.I. 464890B (column 3), P.I. 101404B (column 5), and Clark T/T (column 7). The additional, low-level diffuse red signal in these images is the result of cross-hybridization of probe 3 with pericentromeric regions of all chromosomes. (C) Gm11 BAC probe 4, which is 1.8 Mb distal to probe 3 (above) in the Gm11 pseudomolecule, hybridizes (white arrows) close to probe 5 (lower green signals), near the terminus of the short arm of Gm11 in G. max W82 (column 2). However, in P.I. 464890B (column 3), P.I. 101404B (column 5), and Clark T/T (column 7), the BAC 4 probe hybridizes to a distal position on the long arm of chromosome 13. Due to the high repeat content of probe 4, there is significant cross-hybridization to pericentromeric regions of all chromosomes. Therefore, the red channel from each FISH/DAPI panel is included as a separate gray-scale image (RED CHANNEL) below. The only discrete, nonpericentromeric probe 4 hybridization is restricted to chromosome 11 in G. max W82 (column 2) and to chromosome 13 in P.I. 464890B (column 3), P.I. 101404B (column 5), and Clark T/T (column 7). (D) Summary of G. max W82 BAC probes used to map the reciprocal translocation between chromosomes 13 and 11 in other accessions. Position, the position in megabases of the central base pairs of a given BAC (Table S1for details) in the respective pseudomolecule for a given chromosome; interval, numbers represent the window in megabases between BAC probe position numbers. Gm13 BAC probe B is at 26.5 Mb, D is at 43.0 Mb (not indicated), and the 3′ end of the Gm13 pseudomolecule is at 44.4 Mb. The position of probe A is undetermined. Thus, a fragment of chromosome 13 at least 17.9 Mb long has exchanged with an ∼4.2-Mb fragment of chromosome 11 in P.I. 464890B, P.I. 101404B, and Clark T/T, resulting in the chromosomes in E. The following oligonucleotide probes (amounts) were used: CY5-CentGm-2-M (7.5 ng), fluorescein-CentGm-2-G (20 ng), Texas red-615-CentGm-1-AF (0.01 ng), fluorescein-CentGm-1-E (20 ng), CY5-SB86-C (20 ng), and Texas red-615-rDNA (10 ng). Bar in row C, column 8, DAPI panel is 2 μm and valid for all FISH and DAPI panels.
In P.I. 464890B, a Gm13-localized probe (A) hybridized to a proximal position on the chromosome arm opposite the satellite arm (Figure 6E, column 3), as it did in G. max W82 (Figure 6E, column 1). This suggested that the NOR chromosome of P.I. 464890B is chromosome 13, due to sequence contiguity from the NOR (verified with an 18S rDNA probe), through the centromere and into the long arm. However, a second Gm13 probe (B), tested in combination with a third probe (D), the most terminal Gm13 probe tested, did not hybridize to the NOR-containing chromosome in P.I. 464890B, as it did in G. max W82 (Figure 6D, column 1). Instead, they hybridized to the chromosome target of the Gm11 terminal long arm probe (Figure 6D, column 4). Furthermore, it is likely that the sequence between Gm13 BAC probes B and D in P.I. 464890B represents a continuous fragment because Gm13 BAC probe C, positioned intermediate between probes B and D in G. max W82 (Figure 6F, column 1), also hybridized to a position between the hybridization signals of BAC probes B and D in P.I. 464890B (Figure 6F, column 4). Together, these data suggest that a fragment distal to Gm13 probe A, spanning the sequences corresponding to BAC probes B through D, likely including the telomere, was translocated from chromosome 13 to another chromosome in P.I. 464890B. Furthermore, the chromosome exchange occurred between sequences corresponding to Gm13 BACs A and B (Figure 6D, column 13).
Since a chromosome 13 fragment was translocated in P.I. 464890B to a chromosome hybridizing to Gm11 BAC probe 1 (Figure 6A, column 4), we next determined whether the target chromosome was in fact the P.I. 464890B equivalent of chromosome 11 and also if the translocation involved a reciprocal exchange. Gm11 BAC probes 1, 2, and 3 hybridized to the distal long arm (Figure 7A, column 2, green probe), middle long arm (Figure 7A, column 2, red probe), and proximal short arm (Figure 7B, column 2, red probe), respectively, of chromosome 11 in G. max W82. In P.I. 464890B, Gm11 probes 1, 2, and 3 hybridized to the distal long arm (Figure 7A, column 4, green probe), middle long arm (Figure 7A, column 4, red probe), and proximal position on the arm to which the Gm13 probes B–D hybridized (Figure 6D, column 4, red probe). The apparent contiguity of hybridization of these Gm11 probes suggests that this is the P.I. 464890B chromosome 11.
To determine whether any part of chromosome 11 was translocated to chromosome 13 in P.I. 464890B, we screened several Gm11 BACs assembled on the 3′ end of the G. max W82 pseudomolecule (Figure 7, Table S1and data not shown). Gm11 BAC probe 5, which is at the 3′ end of the pseudomolecule, hybridized to the terminus of the short arm of Gm11 in G. max W82, as expected (Figure 7, B and C, column 2, green probe). In P.I. 464890B, however, this probe hybridized to the terminus of the long arm of chromosome 13 (Figure 7, B and C, column 3, green probe), indicating a reciprocal exchange between chromosomes 11 and 13. To determine how much of chromosome 11 DNA translocated, we “walked” up the short arm using FISH probes made from a series of BACs assembled between Gm11 BACs 3 and 5 (Table S1 and data not shown). We found that Gm11 BAC 4, which is located 1.8 Mb 3′ of BAC 3 and 4.2 Mb 5′ of BAC 5 in the Gm11 pseudomolecule, hybridized to the short arm of Gm11 as expected (Figure 7C, column 2, red probe). However, the probe hybridized to chromosome 13 in P.I. 464890B (Figure 7C, column 3, red probe). Thus, in P.I. 464890B, a fragment of chromosome 11, between sequences corresponding to Gm11 probes 4 and 5, translocated to chromosome 13. In summary (Figure 7, D and E), a segment of chromosome 13 at least 17.9 Mb long was reciprocally exchanged with an ∼4.2-Mb segment of chromosome 11 in P.I. 464890B.
Characterizing the chromosome translocation of Glycine soja P.I. 101404B:
One of the few known naturally occurring translocations in soybean (reviewed by Chung and Singh 2008) involves chromosome 13. G. soja P.I. 101404B is an accession from northeastern China that was originally identified (Williams 1948) as a translocation line because F1 hybrids of crosses of this accession with standard G. max cultivars exhibited pollen and ovule semi-sterility (Williams 1948; Palmer and Heer 1984). Semi-sterility is a hallmark of translocation heterozygotes in plants with 1:1 adjacent:alternate segregation in meiosis because 50% of the spores receive nonviable, duplication/deficiency chromosomes. This translocation subsequently was introgressed into the G. max cv. Clark to create Clark T/T, a near isogenic line homozygous for the translocation (Sadanaga and Newhouse 1982; Mahama et al. 1999). As expected for a reciprocal translocation, F1 progeny of a cross between CLARK T/T and a standard G. max cultivar exhibited 18 bivalents and a single quadrivalent in metaphase I (Mahama et al. 1999). The translocation breakpoint has been positioned genetically with respect to classical genetic markers, but only on chromosome 13 (Sadanaga and Grindeland 1984; Mahama and Palmer 2003); thus, a translocation partner for chromosome 13 has not been identified. Furthermore, the precise breakpoint has not been mapped either molecularly or cytologically for either chromosome involved in the translocation.
The translocation in P.I. 101404B appears to be quite common among Chinese G. soja accessions (Palmer et al. 1987). Thus, given the results obtained with P.I. 464890B, we hypothesized that the translocation previously reported in P.I. 101404B could be identical, or at least very similar to that in P.I. 464890B. We therefore extended our BAC-based mapping analysis to include the following lines: P.I. 101404B, Clark T/T, Clark, and P.I. 464916, a G. soja accession that does not have the translocation (Palmer et al. 1987). We repeated the BAC-based mapping using the same Gm11 and Gm13 probes (Figure 6, B and C). As expected, Gm13 BAC markers B and D, which bracket the chromosome fragment translocated to chromosome 11 in P.I. 464890B, hybridized to chromosome 13, not 11, in both Clark (Figure 6D, column 9) and P.I. 468916 (Figure 6D, column 11), verifying that these cultivars are both translocation negative. In contrast, in chromosome spreads of both Clark T/T, and P.I. 101404B, the localization patterns for the Gm11 and Gm13 BAC mapping markers were indistinguishable from those observed in P.I. 464890B (Figures 6 and 7, columns 3 and 4). For P.I. 101404B, see Figures 6 and 7, columns 5 and 6; for Clark T/T, see Figures 6 and 7, columns 7 and 8. We concluded that P.I. 101404B (and its introgressed line, Clark T/T) and P.I. 464890B share a cytologically similar translocation involving a reciprocal exchange between chromosomes 13 and 11.
DISCUSSION
The development of a FISH-based karyotype map for soybean provides a resource for direct correlation of genetic and sequence-based markers with the physical chromosomes. FISH cocktails composed solely of single-target probes (e.g., BACs and rDNAs) were used to karyotype other plant species, such as sorghum, (Kim et al. 2005) or potato (Dong et al. 2000). However, this approach did not provide sufficient labeling complexity for soybean because the large number of small chromosome arms severely constrain the space for differential probe hybridization. Instead, we were able to exploit sequence variation of a rapidly evolving satellite repeat, CentGm, to differentially label most soybean chromosomes. In the process, we also corroborated the finding of Gill et al. 2009, that the 91- and 92-bp CentGm subtypes are centromere-specific satellite sequences. Critical to this effort was the availability of a database of high-copy repeats for soybean (Swaminathan et al. 2007). Even low-coverage 454-pyrosequencing represents a rapid and cost-effective alternative to conventional cloning and sequencing to characterize the repetitive DNA content of a plant genomes (Macas et al. 2007; Swaminathan et al. 2007), whose size and repeat content vary widely (Hawkins et al. 2008). In particular, the results described here demonstrate that this is a powerful tool for detecting diversity in centromeric repeats within complex plant genomes, without the need for assembly, large insert clones, or physical mapping. When coupled with oligonucleotide-based FISH of metaphase chromosomes, global survey sequencing becomes a powerful technique to study the distribution of repetitive elements within an entire genome. The strategy also is particularly well suited to the study of high repeat content chromosomal loci, which can be recalcitrant to assembly into sequencing contigs. Since the survey was conducted (Swaminathan et al. 2007), advances in sequencing technology (Hudson 2008) have increased the potential power and depth of this technique while greatly decreasing its likely cost. Thus, this strategy now has potential as a method for large-scale analysis of centromere and repeat evolution across large numbers of related plant species.
The soybean physical map is anchored by high-density molecular markers, which relates these maps to the sequenced genome. Therefore, the use of single or combined low-repeat content pseudomolecule-derived BAC probes allowed unambiguous association of MLGs and pseudomolecules with chromosomes. As a result, we were able to develop a 15-component FISH karyotyping cocktail that permitted simultaneous identification of the 20 chromosome pairs in G. max W82, thereby finally integrating the karyotype with CLGs, MLGs, and a sequence-based physical map into a unified cytogenetic map for soybean.
A similar approach was used to generate a FISH karyotyping cocktail for use with wild G. soja accessions. G. soja is interfertile with G. max; it is thought to be the direct wild progenitor of cultivated soybean (G. max) and is widely distributed across East Asia (reviewed by Chung and Singh 2008). G. soja accessions represent a promising resource for agronomically valuable traits, such as disease and pest resistance, salt tolerance, and enhanced yield (e.g., Lee et al. 2009). Numerous molecular marker studies (e.g., Hyten et al. 2006; Nichols et al. 2007; Lee et al. 2008) demonstrated that North American soybean cultivars, such as W82, have much lower genetic diversity than G. soja. Hyten et al. (2006) suggested that this is likely due to two genetic bottlenecks that occurred during soybean domestication. The first occurred during the domestication of G. soja in Asia, which resulted in an ∼50% loss in genetic diversity in the resulting Asian landraces (Hyten et al. 2006). The second was an introduction bottleneck, well documented by pedigree analysis (Gizlice et al. 1994, 1996; Sneller 1994), in which only a small proportion of the available diversity of Asian G. max landraces was captured in the development of modern soybean cultivars (Hyten et al. 2006). During future exploitation of the genetic diversity of wild soybean accessions, FISH-based karyotyping should be able to complement molecular marker-assisted breeding in characterization of these largely cytologically unexplored wild accessions.
During the BAC mapping of G. soja P.I. 464890B, we arbitrarily defined G. max W82 chromosomes 11 and 13 as “normal” to simplify discussion. However, the study by Palmer et al. (1987) suggests that the translocation in P.I.s 464890B and 101404B probably represents the predominant form in wild soybean, at least in specific regions. In their survey study of G. soja, 21 of 26 accessions from China and 25 of 30 accessions from Russia each produced F1 progeny that exhibited semi-sterility when crossed to a standard North American G. max test cultivar. Furthermore, intercross testing between CLARK T/T (containing the P.I. 101404B translocation) and 12 of the above translocation lines produced fertile F1 plants, indicating that they shared similar or identical translocation chromosomes (Palmer et al. 1987). This study raises several interesting questions. The first is whether the forms of chromosomes 11 and 13 in G. soja accessions in fact represent the ancestral forms of these chromosomes. Addressing this question will require a more extensive survey because G. soja is diverse. Even within China, SSR marker analyses (e.g., Nichols et al. 2007) revealed distinct pools of genetic diversity according to geographical region. South Korean accessions appear to be comparably diverse (Lee et al. 2008). If the P.I. 464890B chromosome 11 and 13 configuration does appear to be more or less universal among G. soja (hence “wild type”), then G. max W82 should be considered a post-translocation type that may have arisen through direct selection. What is the frequency and geographical origin of this translocation in various, independent G. max breeding pools in Asia (Ude et al. 2003) and how might it have shaped soybean domestication? From a practical perspective, awareness of this chromosome configuration in domesticated and wild soybean stocks is important if only because any observed semi-sterility could be attributed to translocation heterozygosity, which would pose only a short-term breeding inconvenience.
The involvement of a chromosome containing a NOR in a translocation initially raised the possibility that the NOR itself had been mobilized from one chromosome to another, a phenomenon previously observed in Allium (Schubert and Wobus 1985; reviewed by Schubert 2007) and Oryza species (Chung et al. 2008). However, our mapping experiments with Gm13-pseudomolecule-derived BAC probes indicated that the NOR and surrounding chromosome structure was unaltered in the translocation. Repeats and transposable elements in particular, have been widely implicated in mediating both homologous and nonhomologous chromosome exchanges (Raskina et al. 2008). Regions of high repeat density may enhance recombination on chromosomes 11 and 13, creating potential hotspots for translocation. Defining the precise breakpoint requires additional FISH mapping and molecular analyses, including determination of potential synteny between the two chromosomes in this region, as well as a correction of the published Gm13 pseudomolecule assembly (Schmutz et al. 2010).
FISH-based karyotyping is a powerful approach for probing chromosome structure. The availability of a validated soybean karyotyping cocktail will lead to other important applications, such as characterization of trisomics (Singh and Hymowitz 1991; Xu et al. 2000), as well as a variety of available but largely uncharacterized translocation and inversion lines identified in soybean (reviewed by Chung and Singh 2008). Additional applications, such as characterization of deletion mutants and mapping of transgenes or transposable elements used for enhancer trapping, mutagenesis etc., should also be enhanced with this technology. Furthermore, chromosome-specific probes or karyotyping cocktails developed in this study can easily be applied to correlate mitotic chromosome numbers with pachytene chromosome numbers (Singh and Hymowitz 1988) for the nine chromosomes for which trisomics were unavailable for molecular mapping. Furthermore, due to the diversity in CentGm probe hybridization, it should be possible to track specific chromosomes in crosses of G. max with G. soja and to identify G. max chromosomes in crosses with other wild perennial relatives of the soybean, such as G. tomentella (Singh et al. 1998).
Acknowledgments
S.D.F. and G.S. gratefully acknowledge funding from the United States Department of Agriculture (USDA), grant 2008-34555-19305, and the National Center for Soybean Biotechnology; M.E.H. and K.V. received funding from the USDA, grant 2008-34488-19433; J.B. received funding from National Science Foundation grant DBI0423898, and S.C. received funding from the United Soybean Board. For soybean seeds, we thank Reid Palmer, Randall Nelson (USDA Soybean Germplasm Collection), and Perry Cregan. For BAC clones, we thank Xiaolei Wu and Scott Jackson. For use of plant culture facilities, we thank Zhanyuan Zhang. We thank Reid Palmer, Patrice Albert, Jeongmin Choi, Robert Gaeta, Christian Hans, Alexandre Berr, and members of the James Birchler and J. Chris Pires labs for helpful discussions; we also thank anonymous reviewers for their helpful comments.
Supporting information is available online at http://www.genetics.org/cgi/content/full/genetics.109.113753/DC1.
References
- Ahmad, Q. N., E. J. Britten and D. E. Byth, 1983. A quantitative method of karyotypic analysis applied to the soybean, Glycine max. Cytologia 48 879–892. [Google Scholar]
- Ahmad, Q. N., E. J. Britten and D. E. Byth, 1984. The karyotype of Glycine soja and its relationship to that of the soybean, Glycine max. Cytologia 49 645–658. [Google Scholar]
- Altschul, S. F., W. Gish, W. Miller, E. W. Myers and D. J. Lipman, 1990. Basic local alignment search tool. J. Mol. Biol. 215 403–410. [DOI] [PubMed] [Google Scholar]
- Ananiev, E. V., R. L. Phillips and H. W. Rines, 1998. Chromosome-specific molecular organization of maize (Zea mays L.) centromeric regions. Proc. Natl. Acad. Sci. USA 95 13073–13078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arumuganathan, K., and E. D. Earle, 1991. Nuclear DNA content of some important plant species. Plant Mol. Biol. Rep. 9 208–218. [Google Scholar]
- Assibi Mahama, A., K. S. Lewers and R. G. Palmer, 2002. Genetic linkage in soybean: Classical genetic linkage groups 6 and 8. Crop Sci. 42 1459–1464. [Google Scholar]
- Berr, A., and I. Schubert, 2006. Direct labelling of BAC-DNA by rolling-circle amplification. Plant J. 45 857–862. [DOI] [PubMed] [Google Scholar]
- Blanc, G., and K. H. Wolfe, 2004. Widespread paleopolyploidy in model plant species inferred from age distributions of duplicate genes. Plant Cell 16 1667–1678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng, Z., F. Dong, T. Langdon, S. Ouyang, C. R. Buell et al., 2002. Functional rice centromeres are marked by a aatellite aepeat and a centromere-specific retrotransposon. Plant Cell 14 1691–1704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi, I. Y., D. L. Hyten, L. K. Matukumalli, Q. Song, J. M. Chaky et al., 2007. A soybean transcript map: gene distribution, haplotype and single-nucleotide polymorphism analysis. Genetics 176 685–696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chung, G., and R. J. Singh, 2008. Broadening the genetic base of soybean: a multidisciplinary approach. Crit. Rev. Plant Sci. 27 295–341. [Google Scholar]
- Chung, M. C., Y. I. Lee, Y. Y. Cheng, Y. J. Chou and C. F. Lu, 2008. Chromosomal polymorphism of ribosomal genes in the genus Oryza. Theor. Appl. Genet. 116 745–753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clarindo, W. R., C. R. De Carvalho and B. M. G. Alves, 2007. Mitotic evidence for the tetraploid nature of Glycine max provided by high quality karyograms. Plant Syst. Evol. 265 101–107. [Google Scholar]
- Cregan, P. B., T. Jarvik, A. L. Bush, R. C. Shoemaker, K. G. Lark et al., 1999. An integrated genetic linkage map of the soybean genome. Crop Sci. 39 1464–1490. [Google Scholar]
- Cregan, P. B., K. P. Kollipara, S. J. Xu, R. J. Singh, S. E. Fogarty et al., 2001. Primary trisomics and SSR markers as tools to associate chromosomes with linkage groups in soybean. Crop Sci. 41 1262–1267. [Google Scholar]
- Dong, F., J. Song, S. K. Naess, J. P. Helgeson, C. Gebhardt et al., 2000. Development and applications of a set of chromosome-specific cytogenetic DNA markers in potato. Theor. Appl. Genet. 101 1001–1007. [Google Scholar]
- Du, J., D.T. Grant, Z. Tian, R. Nelson, L. Zhu et al., 2010. SoyTEdb: a comprehensive database of transposable elements in the soybean genome. BMC Genomics 11 113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gill, N., S. Findley, J. G. Walling, C. Hans, J. Ma et al., 2009. Molecular and chromosomal evidence for allopolyploidy in soybean. Plant Physiol. 151 1167–1174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gizlice, Z., T. E. Carter, Jr. and J. W. Burton, 1994. Genetic base for North American public soybean cultivars released between 1947 and 1988. Crop Sci. 34 1143–1151. [Google Scholar]
- Gizlice, Z., T. E. Carter, Jr., T. M. Gerig and J. W. Burton, 1996. Genetic diversity patterns in North American public soybean cultivars based on coefficient of parentage. Crop Sci. 36 753–765. [Google Scholar]
- Goldberg, R. B., 1978. DNA sequence organization in the soybean plant. Biochem. Genet. 16 45–68. [DOI] [PubMed] [Google Scholar]
- Griffor, M. C., L. O. Vodkin, R. J. Singh and T. Hymowitz, 1991. Fluorescent in situ hybridization to soybean metaphase chromosomes. Plant Mol. Biol. 17 101–109. [DOI] [PubMed] [Google Scholar]
- Gurley, W. B., A. G. Hepburn and J. L. Key, 1979. Sequence organization of the soybean genome. Biochim. Biophys. Acta 561 167–183. [DOI] [PubMed] [Google Scholar]
- Hawkins, J. S., C. E. Grover and J. F. Wendel, 2008. Repeated big bangs and the expanding universe: Directionality in plant genome size evolution. Plant Sci. 174 557–562. [Google Scholar]
- Henikoff, S., K. Ahmad and H. S. Malik, 2001. The centromere paradox: Stable inheritance with rapidly evolving DNA. Science 293 1098–1102. [DOI] [PubMed] [Google Scholar]
- Hudson, M. E., 2008. Sequencing breakthroughs for genomic ecology and evolutionary biology. Mol. Ecol. Res. 8 3–17. [DOI] [PubMed] [Google Scholar]
- Hyten, D. L., Q. Song, Y. Zhu, I. Y. Choi, R. L. Nelson et al., 2006. Impact of genetic bottlenecks on soybean genome diversity. Proc. Natl. Acad. Sci. USA 103 16666–16671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hyten, D. L., S. B. Cannon, Q. Song, N. T. Weeks, E. W. Fickus et al., 2010. a High-throughput SNP discovery through deep resequencing of a reduced representation library to anchor and orient scaffolds in the soybean whole genome sequence. BMC Genomics 11 38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hyten, D. L., I.-Y. Choi, Q. Song, J. E. Specht, T. E. Carter et al., 2010. b A high density integrated genetic linkage map of soybean and the development of a 1,536 Universal Soy Linkage Panel for QTL mapping. Crop Sci. 50 960–968. [Google Scholar]
- Jackson, S. A., D. Rokhsar, G. Stacey, R. C. Shoemaker, J. Schmutz et al., 2006. Toward a reference sequence of the soybean senome: a multiagency effort. Crop Sci. 46 S-55–S-61. [Google Scholar]
- Kato, A., 1999. Air drying method using nitrous oxide for chromosome counting in maize. Biotech. Histochem. 74 160–166. [DOI] [PubMed] [Google Scholar]
- Kato, A., J. C. Lamb and J. A. Birchler, 2004. Chromosome painting using repetitive DNA sequences as probes for somatic chromosome identification in maize. Proc. Natl. Acad. Sci. USA 101 13554–13559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim, J. S., P. E. Klein, R. R. Klein, H. J. Price, J. E. Mullet et al., 2005. Chromosome identification and nomenclature of Sorghum bicolor. Genetics 169 1169–1173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kolchinsky, A., and P. M. Gresshoff, 1995. A major satellite DNA of soybean is a 92-base pairs tandem repeat. Theor. Appl. Genet. 90 621–626. [DOI] [PubMed] [Google Scholar]
- Kolpakov, R., G. Bana and G. Kucherov, 2003. mreps: efficient and flexible detection of tandem repeats in DNA. Nucleic Acids Res. 31 3672–3678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee, J. D., J. K. Yu, Y. H. Hwang, S. Blake, Y. S. So et al., 2008. Genetic diversity of wild soybean (Glycine soja Sieb. and Zucc.) accessions from South Korea and other countries. Crop Sci. 48 606–616. [Google Scholar]
- Lee, J.-D., J. G. Shannon, T. D. Vuong and H. T. Nguyen, 2009. Inheritance of salt Tolerance in wild soybean (Glycine soja Sieb. and Zucc.) accession PI483463. J. Hered. 100 798–801. [DOI] [PubMed] [Google Scholar]
- Lin, J. Y., B. H. Jacobus, P. Sanmiguel, J. G. Walling, Y. Yuan et al., 2005. Pericentromeric regions of soybean (Glycine max L. Merr.) chromosomes consist of retroelements and tandemly repeated DNA and are structurally and evolutionarily labile. Genetics 170 1221–1230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Macas, J., P. Neumann and A. Navratilova, 2007. Repetitive DNA in the pea (Pisum sativum L.) genome: comprehensive characterization using 454 sequencing and comparison to soybean and Medicago truncatula. BMC Genomics 8 427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahama, A. A., and R. G. Palmer, 2003. Translocation breakpoints in soybean classical genetic linkage groups 6 and 8. Crop Sci. 43 1602–1609. [Google Scholar]
- Mahama, A. A., L. M. Deaderick, K. Sadanaga, K. E. Newhouse and R. G. Palmer, 1999. Cytogenetic analysis of translocations in soybean. J. Hered. 90 648–653. [Google Scholar]
- Martinez-Zapater, J. M., M. A. Estelle and C. R. Somerville, 1986. A highly repeated DNA sequence in Arabidopsis thaliana. Mol. Gen. Genet. 204 417–423. [Google Scholar]
- Morgante, M., I. Jurman, L. Shi, T. Zhu, P. Keim et al., 1997. The STR120 satellite DNA of soybean: organization, evolution and chromosomal specificity. Chromosome Res. 5 363–373. [DOI] [PubMed] [Google Scholar]
- Nichols, D. M., W. Lianzheng, Y. Pei, K. D. Glover and B. W. Diers, 2007. Variability among Chinese Glycine soja and Chinese and North American soybean genotypes. Crop Sci. 47 1289–1298. [Google Scholar]
- Nunberg, A., J. A. Bedell, M. A. Budiman, R. W. Citek, S. W. Clifton et al., 2006. Survey sequencing of soybean elucidates the genome structure, composition and identifies novel repeats. Funct. Plant Biol. 33 765–773. [DOI] [PubMed] [Google Scholar]
- Pagel, J., J. G. Walling, N. D. Young, R. C. Shoemaker and S. A. Jackson, 2004. Segmental duplications within the Glycine max genome revealed by fluorescence in situ hybridization of bacterial artificial chromosomes. Genome 47 764–768. [DOI] [PubMed] [Google Scholar]
- Palmer, R. G., and H. E. Heer, 1973. A root tip squash technique for soybean chromosomes. Crop Sci. 13 389–391. [Google Scholar]
- Palmer, R. G., and H. E. Heer, 1984. Agronomic characteristics and genetics of a chromosome interchange in soybean. Euphytica 33 651–663. [Google Scholar]
- Palmer, R. G., and R. C. Shoemaker, 1998. Soybean genetics, pp. 45–82 in Soybean Institute of Field and Vegetable Crops edited by M. Hrustic, M. Vidic and D. Jackovic. Soybean Institute of Field and Vegetable Crops, Novi Sad, Yugoslavia.
- Palmer, R. G., K. E. Newhouse, R. A. Graybosch and X. Delannay, 1987. Chromosome structure of the wild soybean: accessions from China and the Soviet Union of Glycine soja Sieb. & Zucc. J. Hered. 78 243–247. [Google Scholar]
- Raskina, O., J. C. Barber, E. Nevo and A. Belyayev, 2008. Repetitive DNA and chromosomal rearrangements: speciation-related events in plant genomes. Cytogenet. Genome Res. 120 351–357. [DOI] [PubMed] [Google Scholar]
- Sadanaga, K., and K. E. Newhouse, 1982. Identifying translocations in soybeans. Soybean Genet. Newsl. 9 129–130. [Google Scholar]
- Sadanaga, K., and R. L. Grindeland, 1984. Locating the w1 locus on the satellite chromosome in soybean. Crop Sci. 24 147–151. [Google Scholar]
- Schlueter, J. A., J. Y. Lin, S. D. Schlueter, I. F. Vasylenko-Sanders, S. Deshpande et al., 2007. Gene duplication and paleopolyploidy in soybean and the implications for whole genome sequencing. BMC Genomics 8 330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmutz, J., S. B. Cannon, J. A. Schlueter, J. Ma, T. Mitros et al., 2010. Genome sequence of the paleopolyploid soybean. Nature 463 178–183. [DOI] [PubMed] [Google Scholar]
- Schubert, I., 2007. Chromosome evolution. Curr. Opin. Plant Biol. 10 109–115. [DOI] [PubMed] [Google Scholar]
- Schubert, I., and U. Wobus, 1985. In situ hybridization confirms jumping nucleolus organizing regions in Allium. Chromosoma 92 143–148. [Google Scholar]
- Sen, N. K., and R. V. Vidyabhusan, 1960. Tetraploid soybeans. Euphytica 9 317–322. [Google Scholar]
- Shoemaker, R. C., J. Schlueter and J. J. Doyle, 2006. Paleopolyploidy and gene duplication in soybean and other legumes. Curr. Opin. Plant Biol. 9 104–109. [DOI] [PubMed] [Google Scholar]
- Shoemaker, R. C., D. Grant, T. Olson, W. C. Warren, R. Wing et al., 2008. Microsatellite discovery from BAC end sequences and genetic mapping to anchor the soybean physical and genetic maps. Genome 51 294–302. [DOI] [PubMed] [Google Scholar]
- Singh, R. J., and T. Hymowitz, 1988. The genomic relationship between Glycine max (L.) Merr. and G. soja Sieb. and Zucc. as revealed by pachytene chromosome analysis. Theor. Appl. Genet. 76 705–711. [DOI] [PubMed] [Google Scholar]
- Singh, R. J., and T. Hymowitz, 1991. Identification of four primary trisomics of soybean by pachytene chromosome analysis. J. Hered. 82 75–77. [Google Scholar]
- Singh, R. J., K. P. Kollipara and T. Hymowitz, 1998. Monosomic alien addition lines derived from Glycine max (L.) Merr. and G. tomentella Hayata: production, characterization, and breeding behavior. Crop Sci. 38 1483–1489. [Google Scholar]
- Skorupska, H., M. C. Albertsen, K. D. Langholz and R. G. Palmer, 1989. Detection of ribosomal RNA genes in soybean, Glycine max (L.) Merr., by in situ hybridization. Genome 32 1091–1095. [Google Scholar]
- Sneller, C. H., 1994. Pedigree analysis of elite soybean lines. Crop Sci. 34 1515–1522. [Google Scholar]
- Song, Q. J., L. F. Marek, R. C. Shoemaker, K. G. Lark, V. C. Concibido et al., 2004. A new integrated genetic linkage map of the soybean. Theor. Appl. Genet. 109 122–128. [DOI] [PubMed] [Google Scholar]
- Stacey, G., L. Vodkin, W. A. Parrott and R. C. Shoemaker, 2004. National Science Foundation-sponsored workshop report. Draft plan for soybean genomics. Plant Physiol. 135 59–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swaminathan, K., K. Varala and M. E. Hudson, 2007. Global repeat discovery and estimation of genomic copy number in a large, complex genome using a high-throughput 454 sequence survey. BMC Genomics 8 132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ude, G. N., W. J. Kenworthy, J. M. Costa, P. B. Cregan and J. Alvernaz, 2003. Genetic diversity of soybean cultivars from China, Japan, North America, and North American ancestral lines determined by amplified fragment length polymorphism. Crop Sci. 43 1858–1867. [Google Scholar]
- Vahedian, M., L. Shi, T. Zhu, R. Okimoto, K. Danna et al., 1995. Genomic organization and evolution of the soybean SB92 satellite sequence. Plant Mol. Biol. 29 857–862. [DOI] [PubMed] [Google Scholar]
- Veatch, C., 1934. Chromosomes of the soy bean. Bot. Gaz. 96 189. [Google Scholar]
- Walling, J. G., R. Shoemaker, N. Young, J. Mudge and S. Jackson, 2006. Chromosome-level homeology in paleopolyploid soybean (Glycine max) revealed through integration of genetic and chromosome maps. Genetics 172 1893–1900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wevrick, R., and H. F. Willard, 1989. Long-range organization of tandem arrays of alpha satellite DNA at the centromeres of human chromosomes: high-frequency array-length polymorphism and meiotic stability. Proc. Natl. Acad. Sci. USA 86 9394–9398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams, L. F., 1948. Inheritance in a species cross in soybean. Genetics 33 131–132. [PubMed] [Google Scholar]
- Xu, S. J., R. J. Singh, K. P. Kollipara and T. Hymowitz, 2000. Primary trisomics in soybean: origin, identification, breeding behavior and use in linkage mapping. Crop Sci. 40 1543–1551. [Google Scholar]
- Zou, J. J., R. J. Singh, J. Lee, S. J. Xu, P. B. Cregan et al., 2003. Assignment of molecular linkage groups to soybean chromosomes by primary trisomics. Theor. Appl. Genet. 107 745–750. [DOI] [PubMed] [Google Scholar]