1. Introduction
Nathans and Hogness [1] cloned the gene for the visual pigment rhodopsin in 1983, and found that the receptor contained seven hydrophobic membrane-spanning helices. At roughly the same time, the paradigm in the visual receptor field that rhodopsin functions as a light-activated Ca2+ channel was being overturned in favor of a competing model that rhodopsin is responsible for activating a heterotrimeric G-protein [2]. In 1986, a landmark paper by Strader, Lefkowitz and co-workers [3] reported the sequence of the β2-adrenergic receptor. They recognized that the β2-adrenergic receptor shares significant sequence homology with rhodopsin, including seven transmembrane (TM) segments. They proposed that the amine ligands of the adrenergic receptors intercalate into the TM helices and trigger receptor activation in the same manner as the retinal in rhodopsin. Because the receptors of the adenylate cyclase system were known to activate G-proteins [2,4,5], the similarity between the β2-adrenergic receptor and rhodopsin provided the first hint of the existence of a large G protein-coupled receptor (GPCR) family.
Early structural work in the emerging GPCR field was guided by two pioneering studies. In 1993, Schertler, Villa and Henderson [6] reported an electron density projection map based on cryo-EM images of rhodopsin that showed the seven-helix architecture is distinctly different from that of bacteriorhodopsin, which had been a favored template for modeling GPCRs. In 1996, Hubbell and Khorana [7] used site-directed spin labeling in combination with EPR spectroscopy to reveal that activation is mediated by rigid body rotation of the TM helices.
A breakthrough in the GPCR field came in 2000 with the determination of the rhodopsin crystal structure [8] (Fig. 1). The structure was solved of the dark, inactive conformation of the protein. The structure of the active state, however, has been more elusive. When illuminated, the crystals of rhodopsin crack and dissolve [9], presumably due to rigid body helix motions that accompany receptor activation. In 2007, the crystal structure of the β2-adrenergic receptor was solved with the inverse agonist carazolol bound in the active site [10]. The structure of this first ligand-activated GPCR confirmed that the overall seven-transmembrane helix architecture is similar to that of rhodopsin. Together the rhodopsin and β2-adrenergic receptor structures provide a high-resolution framework for understanding the structural features that are responsible for locking these receptors in the off conformation, and a starting point for investigating the mechanism of receptor activation.
Fig. 1.
Crystal structure of rhodopsin. The first crystal structure of a GPCR was obtained in 2000 by Palczewski and Okada [8]. Views of rhodopsin spanning the membrane bilayer (a) and from the extracellular (or intradiscal) surface of the membrane (b) show the seven transmembrane α-helix architecture and the position of the 11-cis retinal chromophore. Light-induced isomerization of the retinal to the all-trans configuration converts the chromophore to a full agonist. Motion of helices H5–H7 upon activation opens up the G-protein binding site on the intracellular side of the receptor. Helices H1–H4 form the tightly packed core of the receptor [228].
In this review, we describe the progress that has been made using NMR spectroscopy to characterize the structure and dynamics of GPCRs in membrane environments. NMR methods have been used extensively to establish the structural changes occurring upon the activation of rhodopsin, as well as several ligand-activated receptors. We describe below the importance of these receptors in terms of their cell biology and pharmacology, and then outline the role that NMR can play in answering questions of structure and function.
1.1 Cell biology
The 7-transmembrane helix GPCRs have evolved to recognize and transduce signals as diverse as light, Ca2+, small organic molecules and proteins. These receptors are found in both vertebrates and invertebrates, and are typically divided into six classes (Class A–F) based on sequence homology and functional similarity [11–13]. However, the classification is still open to debate. For example, on the basis of phylogeny, the human GPCRs have been divided into five families (Rhodopsin-like, Secretin, Adhesion, Glutamate, and Frizzled/Taste2) [14]. In this scheme, the Class A receptors correspond to the Rhodopsin-like family, but the Class B receptors are divided into the Secretin and Adhesion families. Nevertheless, in all classification schemes proposed to date, the lack of homology between classes or families suggests that nature has converged on the same seven transmembrane helix framework multiple times.
The Class A (Rhodopsin-like family) receptors respond to the presence of diverse stimuli ranging from light absorption to the binding of various ligands, which include small molecule amines and hormones. Class B (Secretin and Adhesion families) receptors are activated by peptides of the glucagon hormone family [15,16]. The Class C (Glutamate family) GPCRs are comprised of the metabotropic glutamate receptors. These receptors are characterized by a large N-terminal ligand binding domain [17], which appears to be structurally homologous to the amino terminal domain of the ligand-gated ionotropic glutamate receptors in postsynaptic neuronal membranes [18]. Pheromones (e.g. α-factor) secreted by Saccharomyces cerevisiae bind to Class D GPCRs (e.g. STE2) during the mating process. Similar mechanisms are involved in the mating of several fungi [19]. Class E receptors have been implicated in the chemotactic migration of slime mold and can potentially be exploited as antifungal targets [20,21]. Class F (Frizzled/smoothened/taste2 family) contains receptors in the Wnt signaling pathway [14], which perform indispensable roles in embryonic development [22].
The Class A receptors are by far the most populated class of GPCRs. In the GPCR database there are over 20,000 Class A sequences (http://www.gpcr.org/). In humans, 952 of 1061 GPCRs identified in the human genome are in Class A. Of the 952 human Class A receptors, most (509) are olfactory receptors. The remaining Class A GPCRs are subdivided into 18 subfamilies including the well studied visual and small molecule amine receptors, as well as hormone and peptide receptors. Despite the breadth of this group, there exists a degree of sequence conservation among these receptors. Furthermore, the Class A receptors share similar intracellular proteins (e.g. protein kinases, arrestins) that mediate receptor desensitization.
1.2. Pharmacology
Most drugs target four types of membrane proteins: Class A GPCRs (26.8%), nuclear receptors (13%), ligand-gated ion channels (7.9%) and voltage-gated ion channels (5.5%) [23]. There are at least three reasons for the predominance of GPCRs as drug targets. First, they are widely involved in most cellular processes (see Section 1.1 above). Second, GPCRs are located on the cell surface where they are accessible to drug binding. Third, clinical mutations in GPCRs are associated with various pathologies ranging from asthma and allergies to Parkinson’s disease [24,25]. These mutations can result in either an increase or a decrease in receptor activity. For example, in the visual system, mutations in rhodopsin can result in autosomal dominant retinitis pigmentosa, an inherited human disease that causes progressive retina degeneration due to the misfolding of the visual receptor, or congenital night blindness, which is due to constitutive receptor activation [26].
The amine subfamily of receptors (including the noradrenaline, dopamine, histamine, and 5-hydroxytryptamine receptors) is the largest drug target among GPCRs. Saunders [27] estimated that of the 35 top GPCR prescription drugs in 2003, there were 24 ligands targeting monoaminergic receptors. Inhibitors of the angiotensin-II receptor were a distant second in the number of drugs on the market. Over the past seven years, the drug targets have expanded well beyond this limited set. For example, CCR5 and CXCR4 and their cognate chemokine agonists have been implicated in various inflammatory and autoimmune conditions and in cancer. CXCR4 has also been shown to be crucial for embryonic development. Furthermore, CCR5 and CXCR4 are the major co-receptors used by HIV-1 for entry into host cells and specific entry inhibitors targeting these receptors have emerged as a new class of anti-HIV-1 drugs. Maraviroc (UK-427,857) is a potent antagonist of the CCR5 receptor that prevents HIV entry and is currently one of the only small molecule inhibitor available for HIV treatment [28].
The pharmacology of GPCRs has long been based on two-state receptor models [29,30]. In these models, ligands modulate the equilibrium between two distinct conformations (active and inactive) of the receptor. In this framework, constitutive activity can be explained by an agonist-independent conversion from an inactive (R) state to an active (R*) state. Recently, data suggesting the existence of multiple conformational states have emerged that calls for reevaluation of the two-state model. For example, biochemical data have shown that an activated receptor is often capable of coupling to a number of heterotrimeric G proteins [31], and that different signaling cascades can be activated from a single receptor depending on the nature of the bound agonist [32].
The challenge is to now unravel what appears to be a continuum of possible conformations, with varying degrees of activation selectively stabilized by different ligands [33]. A crucial step in this process is to correlate cell-based activity data with structural characterization of differentially activated receptors.
1.3. Structural biology
The X-ray crystal structures of rhodopsin [8,34,35] and the β2-adrenegic receptor [10,36,37] paved the way to the structures of other visual and ligand-activated GPCRs. Murakami and Kouyama [38] solved the structure of squid rhodopsin. Unlike the vertebrate visual receptors, the retinal chromophore within invertebrate squid rhodopsin does not bleach upon photoreaction, but remains bound and reisomerizes to regenerate the starting state of the receptor. For the ligand-activated receptors, crystal structures have been solved of the inactive state of the β1-adrenergic receptor [39] and subsequently, the adenosine A2a receptor [40] bound to high-affinity antagonists. Strategies to reduce conformational heterogeneity had to be implemented during the crystallization process for each of these receptors. Chimeric proteins were constructed to stabilize regions of predicted motion in the β2 adrenergic and adenosine A2A receptors. In the case of the β1 adrenergic receptor, mutations were introduced to increase its stability [39].
Fig. 1 presents the crystal structure of bovine rhodopsin solved in 2000 [8]. Higher resolution structures have subsequently been reported [34,35]. In addition, the structures of several of the photointermediates of rhodopsin have been determined, including bathorhodopsin [41,42], lumirhodopsin [43] and metarhodopsin I (Meta I) [44]. However, none of these structures differ appreciably from one another. The extracellular side of rhodopsin is folded into a well-organized structure consisting of the N-terminus and three extracellular loops (EL1, EL2 and EL3). The N-terminus is composed of 2 short β-strands (β1 and β2) and is glycosylated at Asn2 and Asn15. These first two strands adopt an antiparallel β-sheet fold oriented roughly parallel to the plane of the membrane. The most striking observation in the rhodopsin structure is that EL2 folds into a lid that encloses the retinal-binding site. The EL2 sequence consists of two short β-strands (β3 and β4) that are constrained by a conserved disulfide bond between Cys110 and Cys187 and by a salt bridge between Arg177 and Asp190.
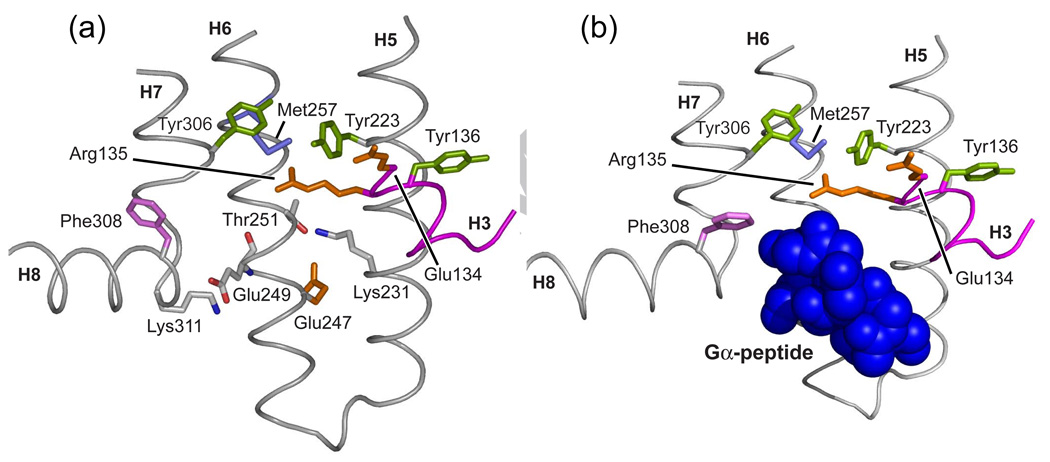
The seven TM helices (H1–H7) surround the retinal binding site in rhodopsin and the binding site for small molecule ligands in the ligand-activated receptors. Several of the seven TM helices feature prominent kinks due to the presence of conserved prolines. The most conserved residues in the Class A GPCRs are located in the TM helices suggesting that this region adopts a common structure. On the intracellular side of the TM helix bundle are two well-characterized microdomains associated with conserved structural motifs. The first is the (E/D)RY motif, corresponding to the Glu134-Arg135-Tyr136 sequence in rhodopsin. A salt bridge between Arg135 on H3 and Glu247 on H6, referred to as the “ionic lock”, must be broken upon receptor activation. The second motif is the conserved NPxxY sequence on helix H7. This motif is involved in disrupting the ionic lock upon activation [45,46].
The cytoplasmic side of rhodopsin is comprised of the C-terminus and three cytoplasmic loops (CL1, CL2 and CL3). The C-terminus emerges from H7 as a short amphipathic helix (H8) stretching from Lys311 to Leu321. The region involving the C-terminal ends of H1 and H7 are highly conserved suggesting a role in receptor structure and/or function. The C-terminal end of the amphipathic H8 is anchored to the membrane by palmitoylation of Cys322 and Cys323.
The X-ray crystal structures of GPCRs determined to date have provided a framework for understanding the wide range of biochemical and mutational data that has been amassed [47–51]. Thus far, there is scant crystallographic data on the active state of rhodopsin or any other GPCR. The recently published crystal structures of opsin [45,46] with and without the bound peptide from the C-terminus of the heterotrimeric G-protein, transducin, display a cytoplasmic conformation that retains features of the active state, metarhodopsin II (Meta II). However, the validity of opsin as a model for the active state has been questioned [52]. For example, a crystal structure of a photoactivated rhodopsin intermediate with an unprotonated Schiff base has been reported that differs considerably from the structure of opsin [53]. The structure of this intermediate does not show the helix movements characterized by EPR measurements, which may reflect an over-estimate of helix motion by EPR [53]. Alternatively, the structure may correspond to an inactive Meta II substate [46]. Unfortunately, the low resolution of the photoactivated intermediate (~4.2 Å) limits an analysis of the structure.
Outside of these crystallographic studies, there is a wide range of data from biophysical techniques aimed at the active state structure of rhodopsin and other GPCRs; these include electron paramagnetic resonance (EPR) [7,54–57], fluorescence [58–60], UV absorbance [61,62], Fourier transformed infrared (FTIR) [63–71], and resonance Raman spectroscopy [72–74]. These methods provide dynamic and time-resolved information on changes in the conformation of GPCRs upon activation.
In this review, we describe the progress made using NMR spectroscopy as a tool for probing GPCR structure and function. NMR spectroscopy has several advantages over existing methods for high-resolution structural studies. The major advantages over protein crystallography are that the measurements can be made in membrane environments and the composition of the membrane can be changed to investigate its role in activation. The current drawbacks of NMR spectroscopy for structural studies are the requirement for isotope labeling and limitations due to the molecular weight (see Section 2). Solution NMR studies have been used to characterize structures of isolated fragments of GPCRs that can be fully 13C, 15N-labeled (summarized in [75]). However, a more productive approach has been to use limited isotopic labeling to target specific regions of full length, functional receptors using the known crystal structures to guide the experimental design and analysis. These studies are the focus of this review.
NMR studies reported to date have been extremely complementary to the pioneering studies using EPR and FTIR spectroscopy. The EPR measurements have largely targeted the intracellular loops that can be readily spin labeled using site directed mutagenesis and thio-reactive spin labels. In contrast, NMR measurements have targeted the TM helix bundle and extracellular loops, particularly the second extracellular loop, which plays a key role in activation. FTIR spectroscopy, in combination with site directed mutagenesis, has been used extensively to identify changes in the protonation states and hydrogen-bonding strengths of amino acids in the activation pathway of rhodopsin. Recently, Ye et al. [76] have followed changes in characteristic vibrations of IR-active azido probes incorporated at different sites on the intracellular side of rhodopsin to probe changes in the electrostatic environment around Glu134 of the highly conserved ERY motif.
Despite the progress made over the past decade on GPCR structure and function, open questions remain. This review discusses many of the unresolved issues, and outlines the prospects for NMR spectroscopy in the future.
2. NMR approaches for probing GPCR structure
NMR spectroscopy provides a versatile approach to the study of GPCR structure and dynamics. In Section 2.1, we describe the methods for expressing and reconstituting GPCRs in a functional form for NMR measurements. In Sections 2.2 – 2.4, we describe the three key NMR interactions that yield structural information: chemical shifts, dipolar couplings and quadrupolar couplings. The chemical shift interaction leads to distinct NMR resonances for individual nuclei (e.g. 13Cα-resonance of Gly121) within a receptor, and can provide information on local environment, conformational changes and hydrogen-bonding. The dipolar interaction is used to measure internuclear distances and orientations. The quadrupolar interaction can span a very large frequency range and consequently is sensitive to molecular motion. For the non-specialist, the wide array and technical nature of different NMR methods can be daunting. However, a brief discussion of the chemical shift, dipolar and quadrupolar interactions provides a basis for understanding the most common NMR methods applied to GPCRs.
There are excellent reviews on both solution and solid-state NMR methods for studying membrane protein structure [77–82]. These reviews describe the details of the nuclear spin interactions that underlie the versatility of NMR spectroscopy.
2.1. Expression and reconstitution of functional GPCRs for NMR spectroscopy
The first structural studies on GPCRs by NMR involved the visual receptor rhodopsin, which can be isolated in milligram amounts from bovine rod cells. 13C NMR measurements were made on receptors regenerated with chemically synthesized retinal chromophores containing specific 13C labels [83–85]. 31P measurements were made on the phosphorylated C-terminus of rhodopsin [86], as well as on the headgroups of phospholipids to probe lipid-protein interactions [87,88].
The challenge for more detailed NMR structural studies of rhodopsin and other GPCRs has been the ability to express, isotope label and purify functional receptors. Heterologous over-expression of GPCRs, which is required for isotopic labeling and obtaining milligram quantities of receptor, still remains a challenging task [89,90]. GPCRs have been expressed in Escherichia coli, yeast, insect and mammalian cells [89,90]. In addition, cell free translation systems have been recently used for GPCR expression with some success [91,92].
Expression of GPCRs in E. coli has the advantage of low cost and short generation time. The lack of post-translational modifications can also provide an advantage because the receptors are homogenous. However, glycosylation and palmitoylation of GPCRs are often needed for proper folding and receptor function. For example, it has been demonstrated that glycosylation is important for ligand binding in several ligand-activated GPCRs [93–95]. In general, GPCRs are expressed in a protease-deficient expression strain, typically DH5R, BL21, CAG627, or KS474 [96]. Strains having auxiliary plasmids for encoding rare codon tRNAs or for promoting disulfide bond formation (e.g. Origami series), can also increase the expression level of eukaryotic proteins [97]. Furthermore, the BL21 E. coli cell line has been used to select for cells that tolerate the expression of membrane and toxic proteins [98]. The addition of glucose and/or the addition of the corresponding ligand have been shown to modestly improve overall expression levels [96].
In order to produce sustainable quantities of functional receptor, eukaryotic expression systems have been developed [89,99,100]. Mammalian cells contain the necessary co-receptors and membrane composition to provide the highest level of post-translational modifications for the correct folding of functional GPCRs, but at the cost of the most demanding culturing conditions. HEK293S cells have been used successfully for incorporating isotope labels into several different GPCRs [101–105]. The receptor is expressed in media containing specifically labeled amino acids. Selective incorporation has been attained for all of the amino acids except glutamine, glutamate, asparagine, aspartate, proline and alanine [106–108].
Insect cell cultures infected with a GPCR-containing recombinant DNA virus allow the production of foreign proteins [109,110]. This eukaryotic expression system offers the advantage that it can be readily adapted to high-density suspension culture for large-scale expression and allows many of the post-translational modifications present in mammalian systems. The Sf9 insect system has also been used for expressing isotope labeled rhodopsin [111–113].
Yeast expression systems combine the advantages of microbial growth (short generation times) and compartmentalized organelles allowing mammalian-like expression and trafficking (insertion into the ER membrane, transport through the Golgi apparatus, and subsequent fusion of the expressed GPCR with the plasma membrane). Yeast cells are capable of performing post-translational modifications, although the type and extent of the modifications can differ from mammalian systems. Recently, the yeast Pichia pastoris has been used to express the bradykinin receptor with significant post-translational modifications [114]. Functional analyses of expressed GPCRs can be performed in yeast [115,116] and mammalian cells [117] prior to purification.
An additional challenge associated with isotope enrichment with selectively labeled amino acids is one of amino acid catabolism. For example, scrambling of an amino acid label into other amino acids is possible, as was found in an HCNO experiment on rhodopsin produced using media enriched with α-15N-lysine and 1-13C-glycine. In this case, glycine and serine were found to interconvert via the enzymatic activity of serine hydroxymethyl transferase, and the 1-13C glycine label resulted in isotope enrichment of the 1-13C position of serine [118]. Similarly, selectively labeled alanine, asparagine, aspartic acid, proline, glutamatic acid and glutamine will result in isotope enrichment of other residues, and possibly reduced enrichment of the target residue.
For purification, receptors are typically solubilized in detergents and/or chaotropic agents. The efficiency of this step is influenced by several parameters, such as the type of detergent and its concentration, buffer composition (i.e. pH and salt concentration), temperature, presence of ligand and addition of osmoprotectants, such as glycerol [119]. Proteins expressed in E. coli typically form inclusion bodies and require refolding into a functional conformation [120–123]. The solubilized receptors are typically purified by size exclusion, ion exchange and/or affinity chromatography. Affinity chromatography using immobilized ligands provides a method of purification of functional receptor. Ligand binding and G-protein activation can also be used to estimate the amount of purified functional receptor. Several structural genomics networks involving membrane proteins track GPCR expression levels in different organisms along with information on specific ligand binding (see [124]). Comparison of the ligand binding levels of different purified GPCRs shows a wide range of values (10 pmol of ligand/mg of receptor to 287 pmol of ligand/mg of receptor [124]), and suggests that a measurement of specific ligand binding alone provides only a rough estimate of the amount of functional receptor.
An advantage of NMR methods over protein crystallography is the ability to study GPCRs reconstituted into membrane environments for structural measurements. The composition of the membrane for visual receptors has been shown to modulate the activity of the receptor [125,126]. The development of solubilized membrane patches, such as bicelles and nanodiscs, has opened up new approaches for NMR measurements in physiologically relevant membrane environments. These patches can be made by using short chain lipids (i.e. bicelles) [127] or by using apo-lipoproteins (i.e. nanodiscs) [128]. Nanodiscs have been used to solubilize a number of large membrane proteins, including the β2 adrenergic receptor [129]. Rienstra et al. have shown that membrane proteins reconstituted in this manner can be precipitated and are subsequently suitable for characterization via solid-state NMR spectroscopy [130].
One of the major questions in GPCR signaling is whether or not the organization of these receptors into dimers (or higher order oligomers) is necessary for signal transduction. In the case of rhodopsin, it has been shown that the monomer is the functional unit [131–133]. However, studies on the γ-aminobutyric acid (GABA)B-receptor [134,135], α1b-adrenoceptor [136], and olfactory receptors [137] have shown that hetero-and homo-oligomerization are important for receptor delivery to the cell surface and in some cases protein function. Nevertheless, the exact role of oligomerization in the function of these receptors is still unclear [138–140].
2.2. Chemical shifts
The measurement of chemical shifts and dipolar couplings is central to NMR studies in solution and in the solid-state. In solution, rapid isotropic motion of proteins averages the chemical shift interaction to its isotropic value and averages the dipolar interaction to zero. As a result, the solution NMR spectra of small proteins typically exhibit sharp, well-resolved resonances. GPCRs have molecular weights that are often >40 kDa and must be solubilized in detergents or membrane vesicles. The large size of the solubilized receptor leads to broadening of the NMR resonances due to loss of rapid, isotropic motion and reintroduction of the dipolar interaction. Nevertheless, progress has been made in using solution NMR methods to study GPCR structure.
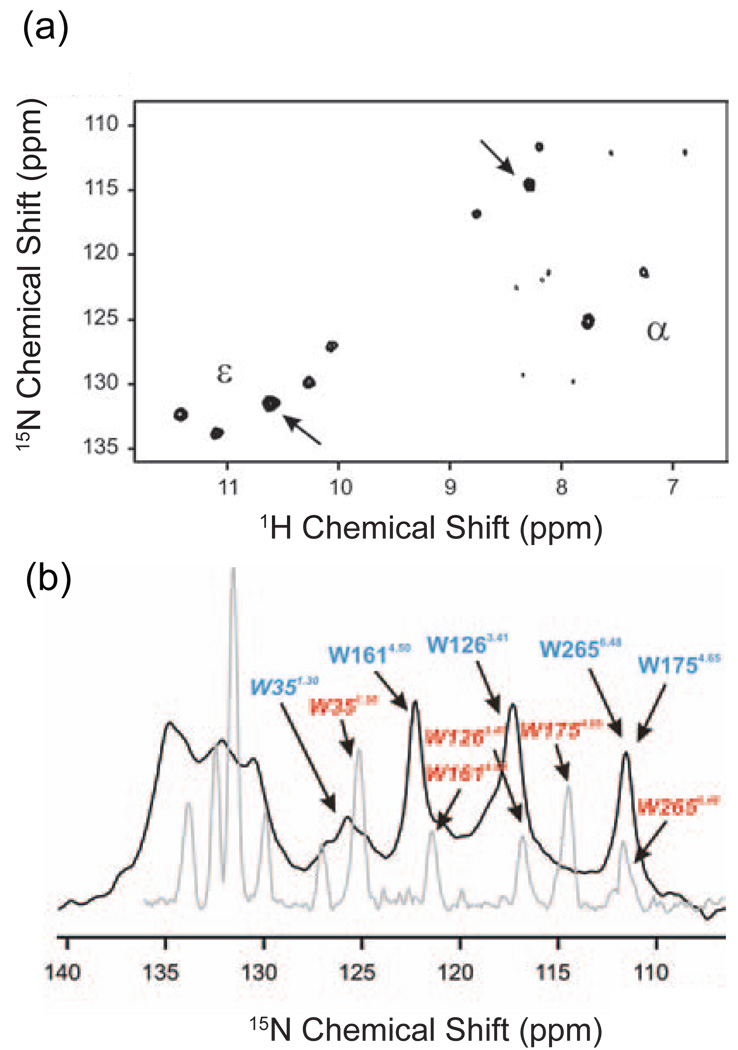
Fig. 2a presents the 1H,15N HSQC spectrum of selectively labeled, detergent-solubilized rhodopsin [141]. In this experiment, the backbone (α) and indole (ε) nitrogens of the five tryptophans in rhodopsin were labeled by incorporating α,ε-15N-tryptophan into defined media of HEK293S cells. The intensity of the cross peaks observed in Fig. 2a likely reflects mobility of the tryptophans [142]. The measurement of solvent accessibility through hydrogen-deuterium exchange and mobility through line widths and relaxation can be used to distinguish the location of surface and buried amino acids in membrane proteins and thus for the assignment of observed resonances to specific amino acids. The intense cross peaks marked with arrows in Fig. 2a disappeared when the sample was solubilized in D2O indicating rapid hydrogen-deuterium exchange. Of the five tryptophan residues in rhodopsin, Trp35 or Trp175 are likely to be solvent exposed because they are located at the helix-loop junctions on the periphery of the receptor. The other three tryptophans are located within the TM helix bundle.
Fig. 2.
Solution NMR spectroscopy of rhodopsin. (a) 1H, 15N HSQC solution NMR spectrum at 900 MHz of 15Nα,ε-tryptophan labeled rhodopsin [141]. The backbone 15Nα and indole 15Nε resonances of the five tryptophans are observed. In addition, weak 15Nα resonances are observed that may correspond to residues on the very flexible C-terminus of the receptor. (b) A single pulse 1D 15N solution NMR spectrum (grey) is overlaid with the corresponding 15N solid-state NMR spectrum obtained using cross polarization and MAS (black). The line widths in the solid-state NMR spectrum are only slightly broader than those obtained by solution NMR methods. The figure is adapted from Ref. [141].
1H spectra have also been reported for tryptophan indole N-H in the human thromboxane A2 receptor without specific labeling. In this case, the indole resonances were observed in 1H TOCSY spectra because of their unique position between 9.9 ppm and 10.2 ppm where there is no overlap with other 1H resonances. In these solution NMR studies, chemical shift changes in 3 of 9 tryptophans were observed between the ligand-free receptor and the receptor with either a bound antagonist or agonist [143]. Although the assignments of the tryptophan resonances have not been made, the NMR measurements suggest that the three tryptophans exhibiting chemical shift changes are involved in conformational transitions between the ligand-free state of the thromboxane A2 receptor and both the antagonist and agonist bound states.
Another approach for targeting specific amino acids through labeling strategies is to identify unique dipeptides within the receptor sequence. For instance, there is a unique Gly-Lys sequence in the C-terminal tail of rhodopsin (Gly324-Lys325). Klein-Seetharaman and colleagues labeled rhodopsin with 1-13C-glycine and α-15N-lysine [118]. Even though there are several glycines and lysines in rhodopsin, the labeling strategy yielded a single 13C-15N directly bonded pair, which resulted in a single 15N resonance being observed in a 2D HNCO experiment.
Solid-state NMR is characterized by the absence of isotropic motion and consequently is complementary to solution-state NMR. The chemical shift interaction in the solid-state is not averaged to its isotropic value as in solution and the dipolar interaction is non-zero. As a result, there is appreciable broadening of the NMR resonances. There are two very different strategies for narrowing the NMR line widths in the solid-state in order to observe individual resonances. The first strategy is the use of magic angle spinning (MAS) [144–147]. Both the chemical shift and the dipolar interaction have a 3cos2θ - 1 dependence on orientation with respect to the external magnetic field. Rapid mechanical rotation of the sample at the magic angle (θ = 54.7°) averages the chemical shift interaction to its isotropic value and the dipolar interaction to zero. The second strategy is to align the individual receptors relative to the magnetic field by layering lipid bilayers on glass slides or by incorporating receptors into membrane bicelles that have the ability to spontaneously orient in an external magnetic field [148]. Restricting the orientation of the receptor relative to the magnetic field narrows the frequency range of the chemical shift and dipolar interactions of a specific 13C or 15N nucleus, and consequently narrows the NMR resonance.
The ability of MAS to narrow the NMR resonances in a GPCR is shown in Fig. 2b, which compares the 1D 15N spectra of the backbone (α) and indole (ε) nitrogens of the five tryptophans in rhodopsin obtained in solution and in the solid-state using MAS. The chemical shifts and line widths in the two spectra are generally comparable. The solution-state NMR spectra were obtained in dodecyl-β-D-maltoside (DDM) micelles, while solid-state NMR spectra were obtained in dioleoylphosphatidylcholine (DOPC) lipid bilayers. The most appreciable difference between the solution and solid-state NMR spectra is in the position of the Trp175 resonance. The authors attribute this difference in chemical shift to the difference in the structure or environment of Trp175, which is exposed to solvent at the membrane-water interface, rather than being buried within the seven TM helix bundle. Importantly, these results show that the membrane environment can influence local GPCR structure, and provide a note of caution for structural studies of the ligand-activated GPCRs that are generally less stable than rhodopsin.
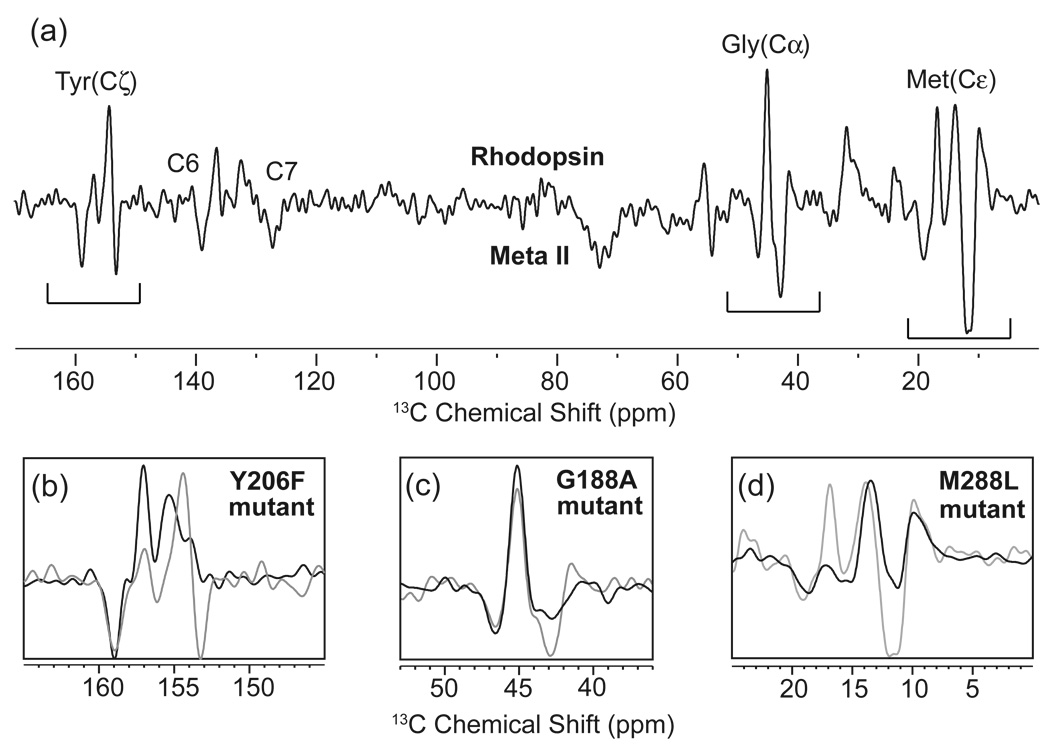
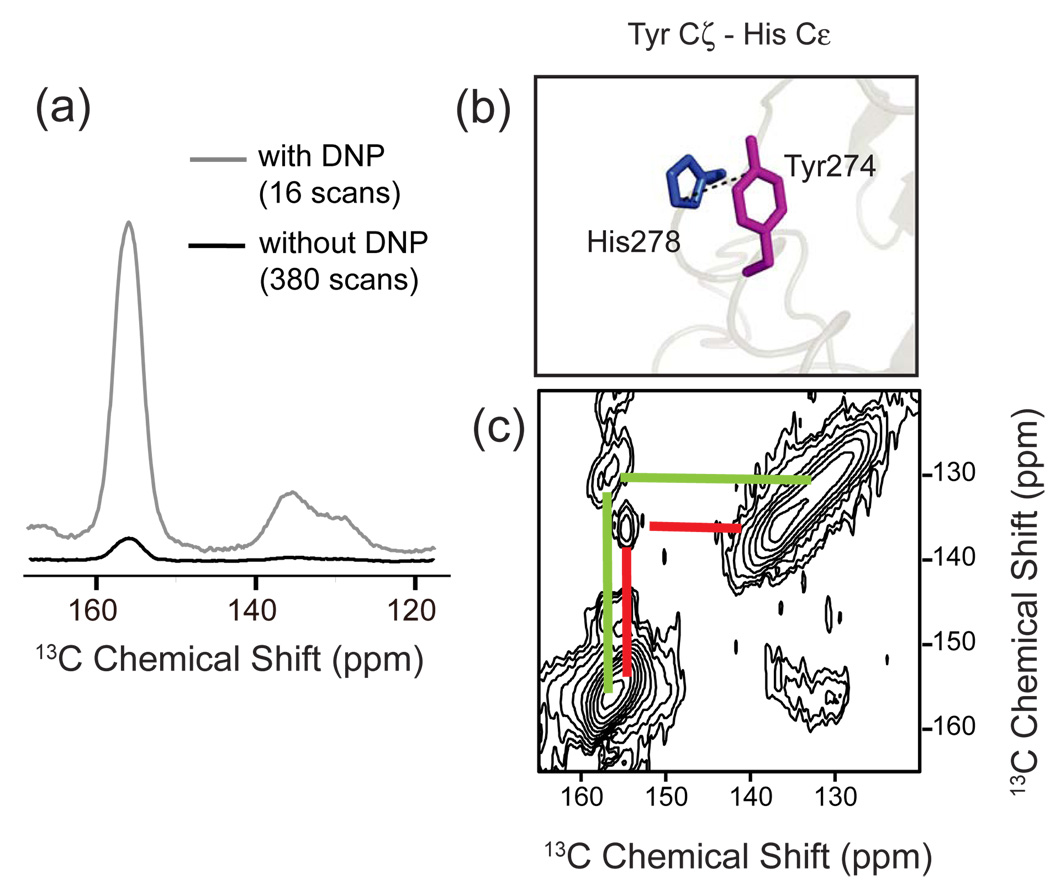
Difference spectroscopy provides a method to isolate the NMR resonances of amino acids whose chemical shift changes upon receptor activation. Fig. 3 presents the 13C NMR difference spectrum between the inactive and active states of rhodopsin using MAS. For this experiment, rhodopsin was isotopically labeled in HEK293S cells by introducing 13C-labeled tyrosine, glycine and methionine into the growth media. Changes in chemical shifts are observed for each type of amino acid upon activation. By comparing the wild type difference spectrum with difference spectra of site-specific mutants, the observed NMR resonances can be assigned to specific amino acids in the protein. For example, there are two distinct negative peaks in the region of the 13Cζ-resonances of tyrosine indicating that at least two tyrosines in the active Meta II intermediate have changed chemical environment. Herzfeld and colleagues [149] have shown that the 13Cζ-tyrosine chemical shift is sensitive to hydrogen bonding of the Cζ-OH hydroxyl group with a downfield shift reflecting an increase in hydrogen bonding. In Fig. 3b, the loss of the upfield tyrosine resonance in Meta II upon mutation of Tyr206 to phenylalanine allows one to assign this resonance to a specific amino acid. The upfield position of the chemical shift suggests that this tyrosine becomes more weakly hydrogen bonded upon activation [106].
Fig. 3.
Solid-state NMR measurements of chemical shifts in rhodopsin. (a) The 150 MHz 13C MAS difference spectrum is obtained by subtracting the spectrum of the active Meta II intermediate from the inactive dark state of rhodopsin. By correlating the difference spectrum of the wild-type receptor (grey) with difference spectra of receptors with site-specific mutations (black), residues undergoing changes in chemical shift can be assigned (b–d).
In summary, the chemical shift interaction provides resolution in both solution and solid-state NMR spectra. Measurements of chemical shifts can be used in several different ways. First, backbone 13C and 1H chemical shift measurements can be used to estimate protein secondary structure using TALOS, a database of chemical shifts compiled from proteins of known structure [150]. Second, backbone and side chain chemical shift (and relaxation) measurements provide information on protein mobility. In solution NMR spectra, the intensity of highly mobile resonances is enhanced, while in solid-state NMR obtained with cross polarization, the intensity of mobile resonances is reduced. Third, chemical shifts can be used to characterize differences in the binding of antagonists and agonists.
2.3. Dipolar couplings
Dipolar couplings represent the interaction between the dipole moments associated with individual nuclear spins. The dipolar coupling has the functional form, D = k (3cos2θ −1) γ1γ2/r3, where k is a constant, γ1 and γ2 are the gyromagnetic ratios of the two nuclei that are dipole coupled, and r is the distance between the coupled nuclei and θ is the angle between r and the magnetic field. The dependence of the hetero- and homonuclear dipole-dipole interactions on the internuclear distance between two spins and orientation of the interacting spins with respect to the external magnetic field can be exploited to derive useful structural constraints on GPCRs.
In solution, rapid isotropic motion averages the dipolar interactions to zero. The advantage of averaging the dipolar interaction is that the NMR line widths are narrowed and the spectral resolution is improved. The disadvantage is that information on the distance-dependent dipolar couplings is lost. Nevertheless, there are two ways to extract information on the dipolar interaction in solution NMR experiments. The first method is to measure the dipolar coupling indirectly through relaxation via the nuclear Overhauser effect (NOE). The second method is to partially orient the sample by restricting isotropic tumbling with filamentous Pf1 bacteriophages [151], dilute lipid bicelles [152], purple membrane fragments [153] or lamellar liquid-crystalline phases [154,155] that orient in a magnetic field. The residual dipolar couplings that result from partial orientation can be measured directly and used as constraints on the relative orientation of protein domains.
In solid-state NMR, when MAS is applied as the mechanism for obtaining high resolution spectra, the dipolar couplings with magnitudes on the order of or less than the MAS frequency are averaged to zero. Since residual motion can average the dipolar couplings even in spectra obtained using solid-state NMR methods, the NMR data are often collected at low temperature. Selective reintroduction of the dipolar interactions, while retaining the high resolution of MAS, allows one to measure internuclear distances using the 1/r3 distance dependence. In the past 20 years, many methods have been developed to reintroduce dipolar couplings under MAS conditions [156–162] in order to measure internuclear distances.
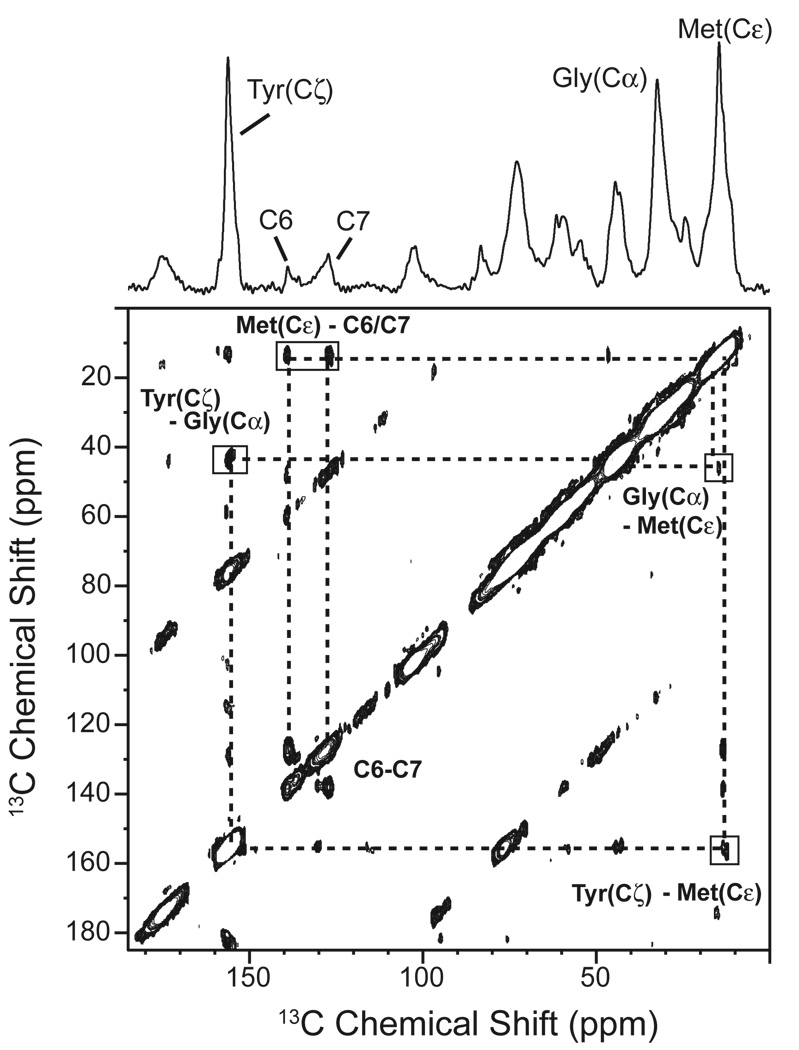
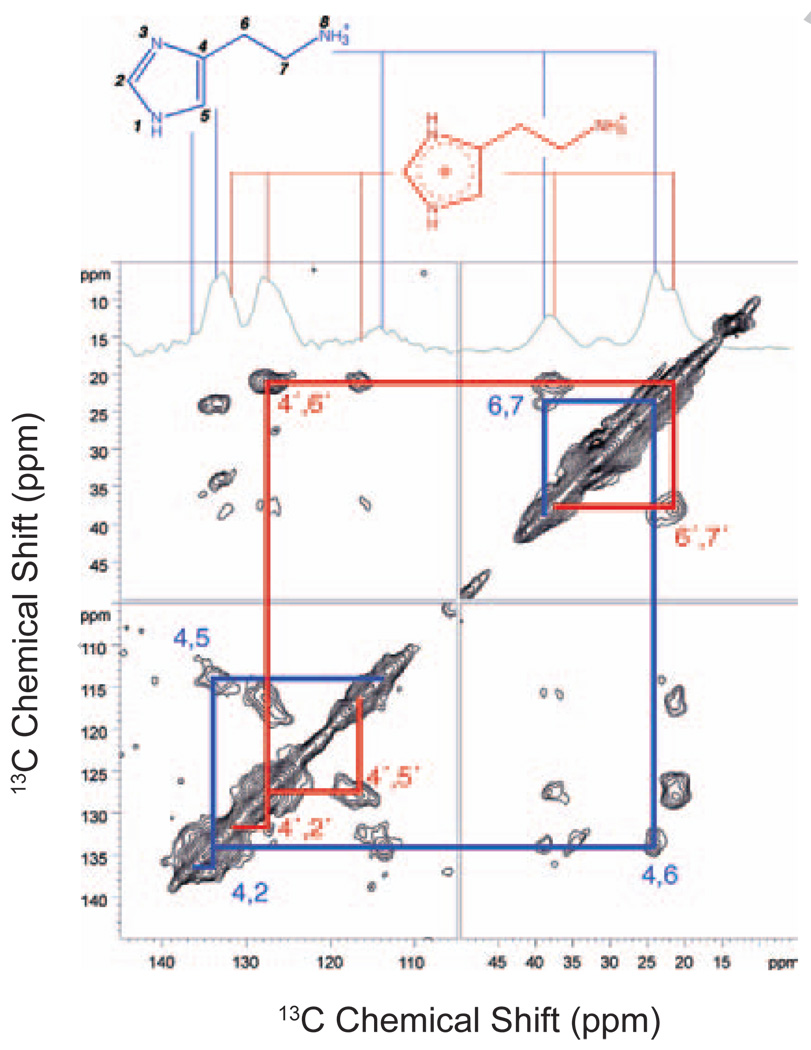
An example of a two-dimensional NMR experiment for measuring 13C…13C dipolar couplings is shown in Fig. 4. In this case, the method for reintroducing the dipolar couplings is referred to as dipolar-assisted rotational resonance (DARR) [162]. The spectrum shown is of rhodopsin containing the same 13C-labeled amino acids as in Fig. 3. The resonances along the diagonal correspond to those observed in the 1D spectrum. Off diagonal cross peaks are observed between the diagonal resonances when the corresponding 13C nuclei are separated by less than ~6 Å. The intensity of the cross peaks for unique sites can be related to the internuclear separation. In this example, the 13C6, 13C7 resonances are from unique sites on the retinal chromophore that are in close proximity to at least one methionine.
Fig. 4.
Solid-state NMR measurements of dipolar couplings in rhodopsin. The 150 MHz 13C 2D DARR NMR spectrum is shown of rhodopsin specifically labeled with 13Cα-glycine, 13Cζ-tyrosine and 13Cε-methionine and regenerated with 11-cis retinal 13C-labeled at the C6 and C7 positions on the retinal polyene chain. DARR is used to reintroduce dipolar couplings between specific 13C labeled sites while retaining the high-resolution afforded by MAS [162]. The 13C resonances introduced by specific labeling of the protein and retinal are observed along the diagonal. Off diagonal cross peaks correspond to 13C nuclei that are sufficiently close in space to transfer magnetization via the dipole-dipole interaction. By correlating the intensity of the observed cross peaks with internuclear distances obtained from the crystal structure of rhodopsin, the detection limit for the 2D DARR NMR experiment is ~6 – 6.5 Å.
The orientation dependence of the dipolar interaction is exploited in methods where the sample can be aligned relative to the external magnetic field. The alignment of membrane bilayers can be achieved by layering lipid bilayers on thin glass slides or by using bicelles that align spontaneously in an external magnetic field. Aligned samples exhibit the anisotropic interactions characteristic of the solid-state and provide useful information on the orientation, dynamics and overall topology of the membrane bound peptides and proteins. For example, helix tilt angles can be calculated with respect to the bilayer normal by correlating the backbone 15N chemical shift with the strength of the 15N−1H dipolar coupling using 2D NMR pulse sequences, such as PISEMA [163], HIMSELF [164] and SAMMY [165]. Recently, a new method involving porous anodic aluminum oxide filters has been used for aligning protein-containing membranes as tubular bilayers. Soubias et al. [166] successfully reconstituted rhodopsin in these nanopores in a functional form.
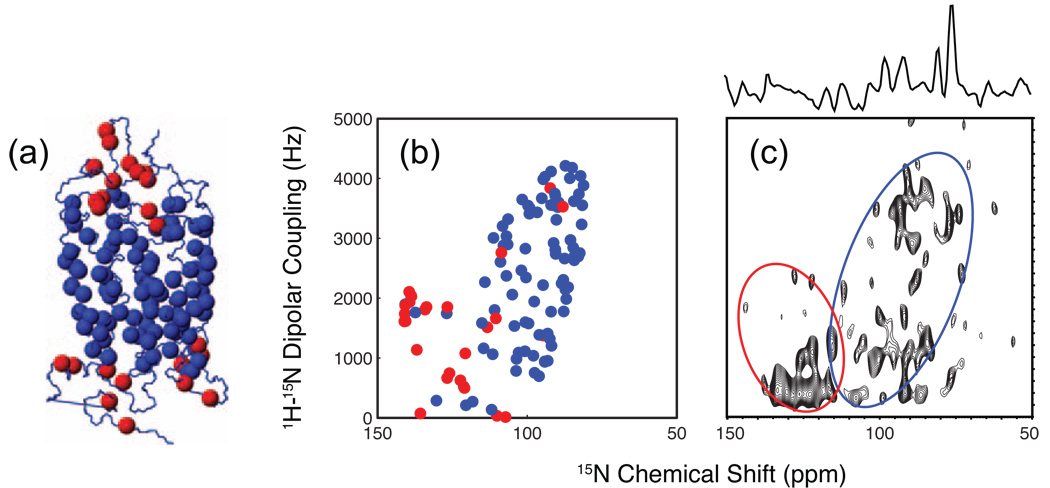
Fig. 5 presents the 2D NMR spectrum of rhodopsin 15N labeled with isoleucine, leucine, valine and phenylalanine, and reconstituted into 14-O-PC, 6-O-PC bicelles ((S. H. Park, M. Eilers, S.O. Smith and S. Opella, unpublished data). The bicelle samples spontaneously align in high magnetic fields. The labeling strategy resulted in 112 of 348 residues of rhodopsin being labeled with 15N and distributed into both the TM helices of the protein (blue spheres, Fig. 5a) and the loops (red spheres, Fig 5a). These two different regions can easily be distinguished in the 2D NMR spectrum. Similar experiments conducted on 15N isoleucine-enriched CXCR1 show that the resonances arising from residues in the helices and in the loops can be separated [167]. One advantage of this approach for structural studies on GPCRs is the ability to measure rigid body helix motion upon ligand binding by characterizing changes in helix orientation using “Pisa Wheels” [168] and “Dipolar Waves” [169] extracted from the 2D PISEMA-type spectra.
Fig. 5.
Solid-state NMR of NH orientations using aligned bicelles. (a) Crystal structure of rhodopsin (PDB code 1U19) showing the position of the 15N-labeled isoleucine, leucine, valine and phenylalanine in the loops (red) and transmembrane helices (blue). (b) Simulations of the separated local field SAMMY spectrum of rhodopsin plotting the amide 15N – 1H dipolar coupling as a function of the 15N chemical shift. Simulated resonances for the transmembrane (blue) and non-transmembrane (red) 15N-labeled amino acids are based on the crystal structure. The resulting cross peak pattern is indicative of elements of secondary structure. (c) Experimentally determined SAMMY spectrum of rhodopsin containing 15N labeled amino acids. The spectra were obtained at 45 °C in aligned (q = 3.2, 28% w/v lipid) bicelles using the SAMMY pulse sequence [165] and the following parameters: 1 ms mixing time, 5 ms acquisition time, 8s recycle delay, 52 kHz SPINAL 1H decoupling, 1024 scans, 750 MHz 1H frequency.
2.4. Quadrupolar couplings
While most NMR experiments of GPCRs make use of spin I = ½ nuclei (e.g. 1H, 13C, 15N and 31P), spin I > ½ nuclei exhibit quadrupolar couplings (e.g. 2H, 14N and 17O) that can also be exploited in studies of protein structure and dynamics. Coupling of the quadrupole moment with the electric field gradient at the nucleus gives rise to the quadrupolar interaction. For biological NMR studies, deuterium (spin I = 1) is the most widely used nucleus having a quadrupole moment. Deuterium is often easy to substitute for 1H nuclei in amino acids or small molecules, has a low natural abundance, and has a broad frequency range and orientation dependence that makes the deuterium line shape sensitive to molecular motion. 17O (I = 5/2) has not been as widely used for biological NMR, but has potential as a probe for water interactions within GPCRs.
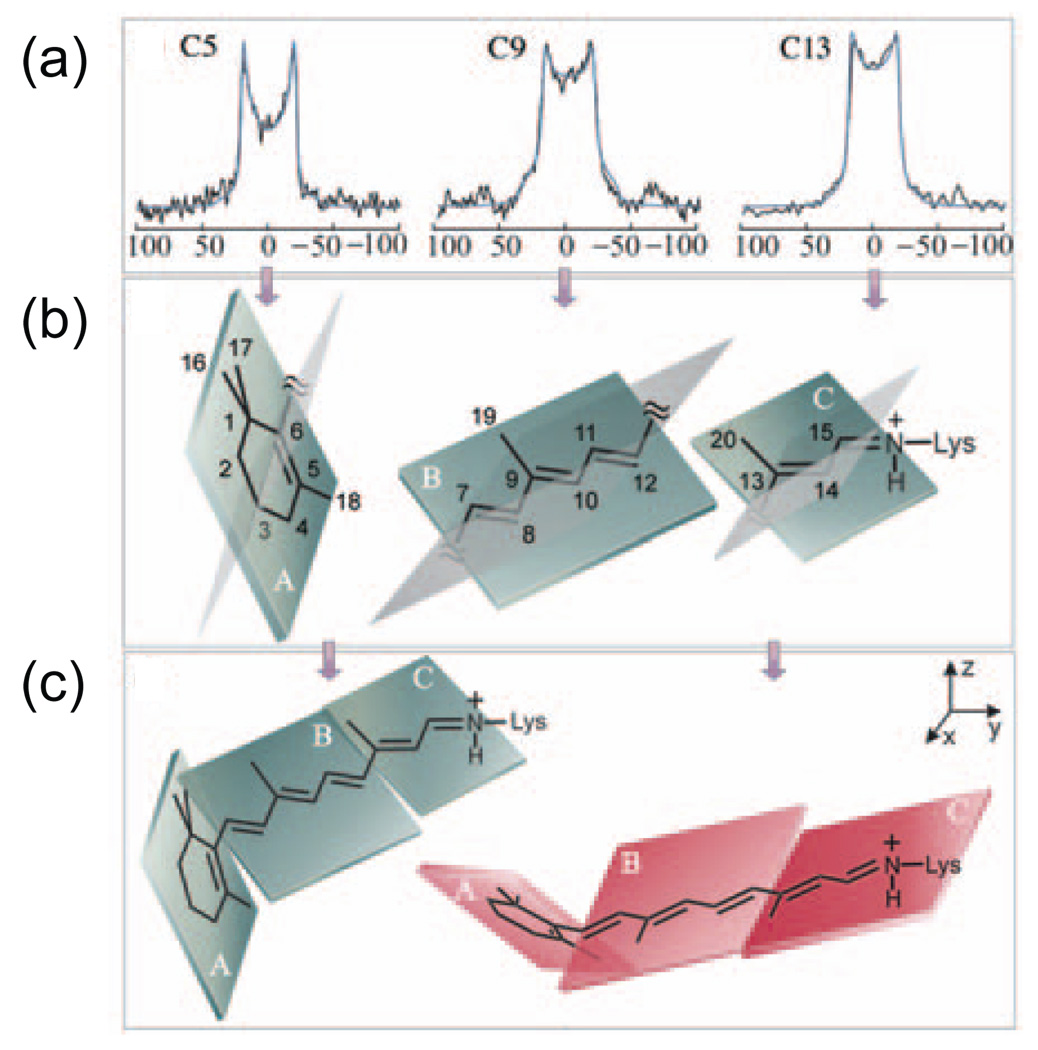
Deuterium NMR methods have been used extensively for studying the orientation and dynamics of the retinal chromophore in dark rhodopsin [170,171] and its photointermediates [172–174]. The strategy in these experiments is outlined in Fig. 6. The retinal chromophore can conceptually be broken down into three planes (Fig. 6b). One plane consists of the β-ionone ring, which is separated from the polyene chain by the C6–C7 single bond. The second plane stretches from the C6–C7 single bond to the C12-C13 single bond. The conjugated π-system keeps this region roughly planar. The third plane stretches from the C12-C13 single bond to the single bond connecting the C15 carbon of the retinal chromophore to the ε-nitrogen of Lys296. Site-specific 2H labels are incorporated into methyl groups attached to the C5, C9 and C13 carbons within these planes. The 2H line shapes shown in Fig. 6a reveal that the methyl groups hop rapidly about their three-fold axes. Simulation of the line shapes is used to establish the orientation of the C-CD3 bond relative to the membrane normal. These orientations, in turn, define the orientations of the planes in the inactive 11-cis chromophore (Fig. 6c). A number of conclusions can be drawn from these studies [171–174]. First, the retinal is twisted about both the C6–C7 and C12-C13 single bonds due to steric interactions between the retinal and the surrounding protein. Second, there must be an additional negative pre-twist about the C11=C12 double bond in order to accommodate the retinal within the rhodopsin binding pocket. Third, when the 2H line shapes in both the rhodopsin and Meta I states are considered, there are marked changes in the dynamics of all three methyl groups reflecting a change in the environment of the retinal.
Fig. 6.
Deuterium NMR spectroscopy of the retinal chromophore in rhodopsin. Retinals containing deuterated methyl groups attached to the C5, C9 and C13 carbons were regenerated into rhodopsin. The rhodopsin was reconstituted into POPC bilayers, which were oriented relative to the external magnetic field by ultracentrifugation onto ultrathin glass slides. (a) Experimental and simulated deuterium line shapes are shown for the C18, C19 and C20 methyl groups. The line shapes are sensitive to the orientations of the C5-CD3, C9-CD3 and C13-CD3 bonds relative to the external magnetic field. (b) The C5-CD3, C9-CD3 and C13-CD3 bond orientations can be used to orient specific regions of the retinal chromophore. In these studies, the retinal is divided into three planes containing the β-ionone ring, the polyene chain from C6–C12 and the polyene chain from C13 to the Nε-Lys single bond. Each plane contains one of the deuterated methyl groups. (c) The individual planes are assembled to form a twisted retinal chromophore by allowing rotation about the C5=C6–C7=C8 and C11=C12-C13=C14 torsional angles. The figure is adapted from Ref. [172] with permission from the American Chemical Society.
3. Ligand conformation and receptor interactions
The primary step of GPCR signaling begins with retinal isomerization in the visual receptors or ligand-binding in the ligand-activated receptors. As discussed in the introduction, the simple seven TM helix framework has evolved to recognize literally thousands of different ligands, which exhibit a wide range of biological activities. The two major questions involved with ligand-receptor interactions are 1) what determines ligand specificity and 2) how do ligand-receptor interactions modulate receptor activity? The question of specificity is highlighted by the sheer diversity of GPCRs and their activating ligands (see Section 1.1). The question of how ligands modulate receptor activity is central to understanding the mechanism of GPCR activation and to the development of drugs targeting GPCRs.
GPCRs without bound ligand typically exhibit a moderate level of basal activity. Ligand binding can modulate receptor activity from fully off to fully on, depending on the nature of the ligand. The level of activity is used to classify the ligand as an inverse, neutral, partial or full agonist. Inverse agonists (and antagonists) reduce the activity of a receptor to below its basal level. Neutral antagonists do not influence activity, but prevent other ligands from binding. Partial and full agonists activate the receptor to different degrees. Although they are unique GPCRs, the visual receptors provide several advantages for understanding the ligand-activated receptors. In Sections 3.1 and 3.2, we describe NMR studies on the retinal chromophore in visual receptors and on the ligands associated with ligand activated GPCRs, respectively. These studies outline how NMR measurements can 1) establish the conformation and protonation states of receptor-bound ligands, and 2) reveal specific receptor – ligand interactions.
3.1. Retinal conformation in the visual receptor rhodopsin
The retinal chromophore functions as a light-activated ligand and offers two significant advantages for structural studies as compared to other GPCRs. First, the retinal is covalently bound to the receptor. The covalent linkage through a protonated Schiff base (SB) to Lys296 insures 100% ligand occupancy. Second, the retinal functions as a pharmacological inverse agonist in the 11-cis configuration; binding of 11-cis retinal to the apo-protein opsin lowers the basal activity of the dark, inactive receptor to undetectable levels [175,176]. Light absorption rapidly converts the inverse agonist to a full agonist. Hence, for rhodopsin it is possible to isolate two well-defined, fully occupied states corresponding to the inactive and active receptor.
NMR measurements on the conformation of the 11-cis and all-trans chromophores in dark rhodopsin (inactive state) and the Meta II intermediate (active state), respectively, have provided structural insights into how the retinal functions as a light-activated ligand. First, light induces a rapid and selective isomerization of the C11=C12 double bond. As discussed in Section 2.4, deuterium measurements on the retinal chromophore have shown that the C20 methyl group is twisted out of the retinal plane and suggested that there is a small distortion or ‘pre-twist’ about the C11=C12 double bond [171,172]. These conformational distortions “prime” the retinal for isomerization in a specific direction.
Support for conformational distortions in the ground state structure of the retinal come from dipolar recoupling measurements. Measurements of the H-C10-C11-H torsion angle yield a value of 160° ± 10° in rhodopsin [177] and 180° ± 25° in the Meta I intermediate [178] indicating that this region has relaxed following isomerization. A twist about this bond in the ground state is consistent with data from rotational resonance experiments measuring the distance between the C20 methyl group and the polyene chain in rhodopsin and in the Meta I intermediate [179]. Importantly, these experiments illustrate the accuracy of torsion angle measurements within a GPCR ligand.
In investigating the conformation of the retinal chromophore in rhodopsin (or the conformation of ligands in the ligand-activated GPCRs), double quantum filtering (DQF) methods can greatly enhance the signal arising from enriched directly bonded 13C pairs relative to the natural abundance 13C signals originating from the lipid and surrounding protein. Because the natural abundance level of 13C is 1.1%, it is rare to have directly bonded 13C nuclei. The non-bonded 13C nuclei that are present at natural abundance give rise to very weak dipolar couplings due to the r−3 distance dependence of dipolar interaction. Against this background, the signal arising from enriched directly bonded 13C pairs dominates the DQF spectrum. Fig. 7 presents DQF spectra of retinal chromophores in rhodopsin that have been 13C-labeled at two directly bonded carbons: C9–C10, C11-C12, C12-C13, and C14-C15. The use of DQF was essential in these MAS NMR experiments for obtaining accurate chemical shifts of the bathorhodopsin intermediate since this state is in photo-equilibrium with rhodopsin and the 9-cis pigment isorhodopsin [180,181] and represents only ~35% of the sample. In this regard, it is worth noting that in the ligand-activated GPCRs, the use of DQF improves the ability to detect the receptor bound ligand (see Section 3.2). Fig. 7 shows that the 13C resonances from bathorhodopsin (marked with asterisks) can clearly be identified in these spectra devoid of natural abundance background signals. Measurement of the bathorhodopsin 13C chemical shifts reveals a large change (9.4 ppm) in the position of the 10-13C resonance. The bathorhodopsin chemical shifts [182] suggest that there is increased positive charge delocalization into the polyene chain and torsional strain in bathorhodopsin compared with rhodopsin. It is known that bathorhodopsin stores ~35 kcal/mol of the light energy absorbed by rhodopsin [183] and the NMR chemical shifts indicate that both electrostatic interactions and torsional strain are involved in the energy storage mechanism.
Fig. 7.
Double-quantum filtered 150 MHz 13C NMR spectra of the retinal chromophore in rhodopsin and bathorhodopsin [181]. Double quantum filtering can be used to eliminate the 13C background signal from rhodopsin and the membrane bilayer by inserting directly bonded 13C labels at the C9,C10 (a), C11,C12 (b), C12,C13 (c) and C14,C15 (d) positions of the retinal chromophore. The rhodopsin (top) and bathorhodopsin (bottom) spectra in each panel were both acquired at a temperature <120 K with 7 kHz MAS. The positions of the bathorhodopsin resonances are marked by asterisks. The figure is adapted from Ref. [181] with permission from the American Chemical Society.
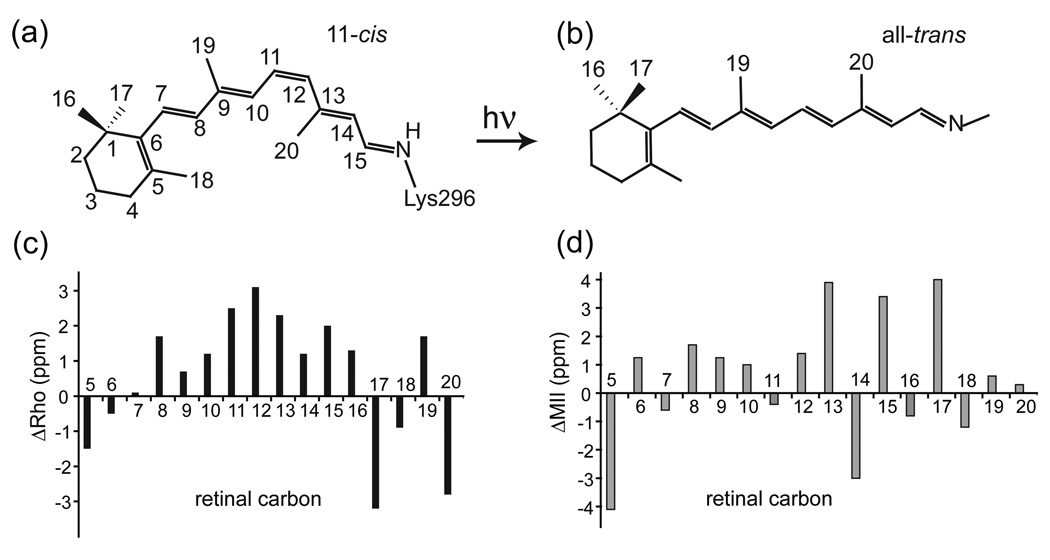
The ability of the retinal chemical shifts to reveal information on the environment of its receptor binding site is highlighted in Fig. 8c, which presents a comparison of the chemical shifts of the 11-cis retinal protonated SB in rhodopsin and 11-cis protonated SB model compound in solution. Significant differences in chemical shift are observed for the C8 to C13 positions [184,185]. The direction of the chemical shift differences reflects an increase in partial positive charge on these carbons. The most unusual difference is the chemical shift of C12 since charge delocalization along the conjugated polyene chain of the retinal typically leads to an alternation of charge where the odd numbered carbons (C5, C7, C9, C11, C13 and C15) stabilize partial positive charge and the even numbered carbons (C6, C8, C10, C12, C14) stabilize partial negative charge. When this chemical shift pattern was first observed in the early 1990s, it strongly suggested the presence of a negative charge near C12 [186,187]. The crystal structure of rhodopsin reported in 2000 revealed that the carboxyl group of Glu181 is positioned 4 Å from the retinal, with C12 being the closest point of contact [8]. In addition to the chemical shift differences along the conjugated chain, significant chemical shift differences are observed in the C16, C17, C18, C19 and C20 methyl groups due to steric contacts in the retinal binding site compared to solution [107,188].
Fig. 8.
Comparison of retinal protonated SB chemical shifts. (a,b) Structures of the 11-cis and all-trans retinal chromophores in the visual pigment rhodopsin. (c) Retinal 13C chemical shift differences (ΔRho) are plotted between rhodopsin and the 11-cis protonated SB model compound in solution (CDCl3). The differences in chemical shift provide a pharmacophore map of retinal-protein interactions. For example, the positive ΔRho values between C8 and C15 are caused by close interaction of the retinal with a negatively charged glutamate residue (Glu181) in the retinal binding pocket. (d) Retinal 13C chemical shift differences (ΔMII) are plotted between Meta II and the all-trans retinal SB model compound in solution (CD3OD). The largest differences in chemical shift are observed in the C13=C14–C15 region of the retinal. These highly polarized bonds likely facilitate Schiff base hydrolysis in the conversion of Meta II to opsin. The figure is adapted from Ref. [188] with permission from the American Chemical Society.
In Fig. 8d, the pattern of retinal chemical shifts in the all-trans chromophore of the active Meta II intermediate changes considerably from the pattern of chemical shifts observed in the 11-cis chromophore of rhodopsin. The largest changes are observed at the two ends of the retinal, namely the β-ionone ring (C5, C17) and in the C13-C14-C15 region of the retinal-lysine SB linkage. The large changes at C5 and C17 are due to structural changes in the β-ionone ring resulting from retinal isomerization within a tight receptor binding pocket, while the changes at C13-C15 are attributed to electrostatic interactions with the Glu113 carboxylic acid side chain, which becomes protonated upon receptor activation [107,188].
Direct 15N NMR measurements of the SB nitrogen, which links the retinal to Lys296 on TM helix H7, have provided additional structural insights into how the retinal functions as a light-activated ligand. The interaction of the protonated SB with its protein counterion, Glu113, is responsible for keeping the receptor off in the dark. Chemical shift measurements have been made on the protonated SB by incorporating 15N-labeled lysine into the protein. The observed 15N chemical shift of the protonated SB in rhodopsin is upfield of that in retinal model compounds and suggestive of a weaker counterion interaction [102]. This observation is consistent with an extensive hydrogen-bonding network that connects Glu113 to polar residues in EL2 and reduces its effective charge [189]. A “complex-counterion” structure was also observed for the retinal protonated SB and its counterion in bacteriorhodopsin [190] and may be responsible for the high Schiff base pKa needed to maintain the inactive state of the receptor [191]. 15N chemical shift measurements of the Meta II intermediate show a deprotonated SB with a chemical shift significantly upfield compared to model compounds and bacteriorhodopsin [188]. This unusual 15N chemical shift and a large downfield 13C chemical shift at the adjacent C15 position (Fig. 8d) indicate that the C15=N Schiff base bond is highly polarized with a significant partial positive charge localized on the C15 carbon [188]. This polarized structure is attributed to interaction with the protonated Glu113 side chain and may be responsible for the rapid hydrolysis of the retinal following activation and decay of the active Meta II intermediate [188].
3.2. Ligand binding and conformation in ligand-activated GPCRs
Ligand-activated receptors bind a diversity of ligands ranging from small organic compounds to large proteins. High-resolution structural studies on rhodopsin and the amine receptors show that small molecule ligands bind within the helical core of the receptor. Biochemical studies on GPCRs with peptide and protein ligands have suggested a wide range of binding modes. Progress in using NMR spectroscopy to probe the conformation and location of bound ligands has lagged behind the corresponding studies on the visual receptors, mainly because of the challenges associated with expression and isotope labeling discussed in Section 2.1. Nevertheless, NMR studies have been reported on GPCRs with both small molecule and peptide ligands.
In 2004, de Grip, de Groot and co-workers characterized the binding of histamine to the histamine H1 receptor using solid state NMR spectroscopy [192]. There are four Class A histamine GPCRs (H1-H4) that together regulate cellular processes ranging from digestion to inflammation. Binding of histamine to the H1 receptor is responsible for the constriction of bronchi in the lungs [193]. Fig. 9 presents 2D proton driven spin diffusion (PDSD) experiments with DQF to assign the 13C chemical shifts of histamine bound to the H1 receptor [194]. Using these methods, two ligand protonation states were identified and provided the basis of an activation mechanism involving proton transfer from the ligand to the receptor.
Fig. 9.
Solid-state NMR of 13C-labeled histamine bound to the H1-receptor. The 188 MHz 13C PDSD spectrum reveals cross peaks between 13C dipolar coupled resonances of the histamine ligand. A 1D double quantum filtered spectrum is shown at the top of the 2D plot. Double quantum filtering enhances the directly bonded 13C nuclei of the ligand and suppresses natural abundance 13C signal from the receptor. The figure is adapted from Ref. [194] with permission from the American Chemical Society.
Similar 2D NMR recoupling methods have been used to characterize the structure and interaction of peptide ligands. One example is the study of the tridecapeptide neurotensin [195]. This large (1.6 kDa) neurotransmitter has a regulatory role, modulating gastrointestinal hormones and dopaminergic signaling. Three different types of receptors have been identified that bind neurotensin. Two of these, the NTS1 and NTS2 receptors, belong to the Class A GPCR family; however they vary greatly in their affinity for neurotensin. Systems for the recombinant expression of NTS1 in E. coli have been described [196]. Resonance assignments of the ligand were obtained via DQF 13C – 13C dipolar recoupling experiments. In the presence of the NTS1 receptor, the conformation of 13C,15N-labeled neurotensin was found to be distinct from its conformation in solution [195].
The peptide subfamily of Class A GPCRs contains receptors that have both peptide and protein ligands. The two largest groups of receptors in this subfamily are the chemokine and melanocortin receptors. There are ~50 different human chemokine ligands and 20 chemokine receptors. The chemokine ligands and receptors are subdivided into 4 categories based on the pattern of cysteine residues in the ligand. These are designated CL#, CCL#, CXCL# and CX3CL# for the ligands and CR#, CCR#, CXCR# and CX3CR# for the receptors, where # corresponds to a number for a specific ligand or receptor [197]. Signaling via these 8–10 kDa protein ligands is associated with immune responses [198–201]. Solution NMR spectroscopy has been used extensively to characterize the structures of chemokine ligands (see [202]). These proteins generally have a disordered N-terminus, a long N-terminal loop (N-loop) terminated by a 310 helix, a three stranded anti-parallel β-sheet and C-terminal helix. The N-terminus and N-terminal loop often contain elements that interact with the receptor.
Cross saturation methods have been applied to CXCL12 (SDF-1α, old nomenclature) in complex with CXCR4 in detergent [203] in order to characterize ligand-receptor interactions. The authors demonstrated that several structural elements of CXCL12 are in close contact with CXCR4, including the N-terminus, the β1-strand, and one side of the central β-sheet. These data are consistent with a model in which a flexible segment of the ligand acts in concert with the N-terminus of the receptor in the first of a two-step model of chemokine binding [203].
4. Receptor structure and ligand induced conformational changes
Binding of a signaling ligand in a ligand-activated GPCR or isomerization of the retinal chromophore in the visual GPCRs triggers a number of conformational changes on the extracellular side of the receptor that induce structural changes on the intracellular side of the receptor. Although the ligands and extracellular ligand binding regions are diverse, amino acid conservation within the TM core of Class A GPCRs strongly argues that there is a common mechanism for relaying extracellular ligand binding to the formation of a G-protein binding site on the intracellular surface of GPCRs. In Section 4.1, we describe NMR studies on the molecular switches that appear to be present on the extracellular side of GPCRs. We focus on the visual receptor rhodopsin where the most progress has been made using NMR approaches. In Section 4.2, we describe NMR studies on the allosteric conformational changes that occur on the intracellular side of GPCRs upon activation. In Section 4.3, we describe how these structural changes facilitate the exposure of a heterotrimeric G-protein binding site.
4.1 Ligand-receptor interactions on the extracellular side of GPCRs
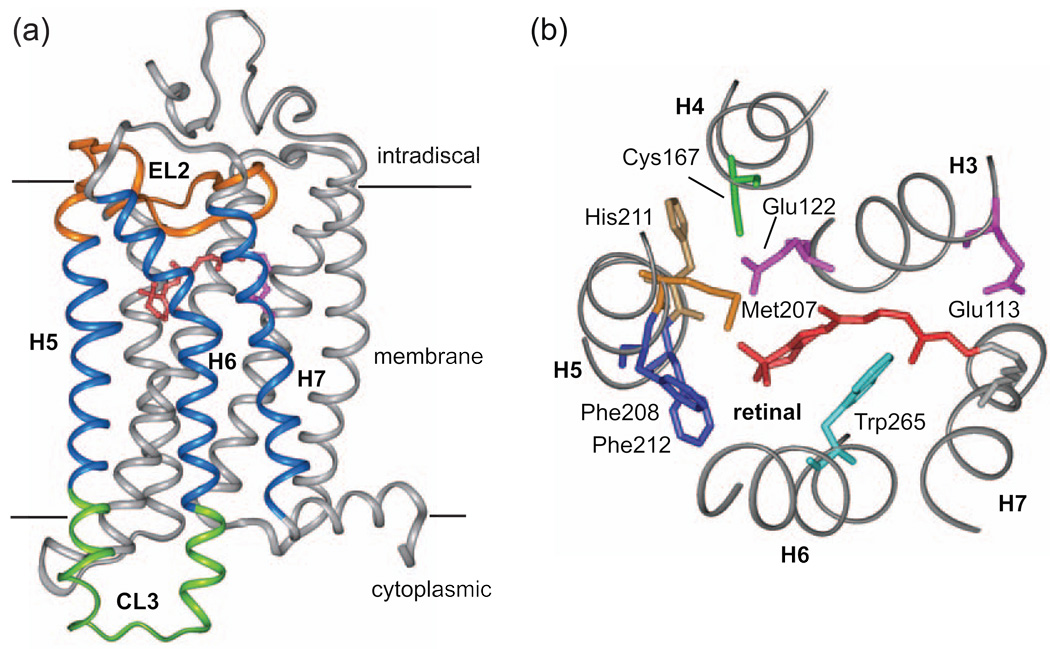
Fig. 10 presents a schematic of the steps that trigger activation of the visual receptor rhodopsin. On the extracellular side of the receptor there are at least four coupled events following retinal isomerization.
Rotation of Trp265 on H6 toward the extracellular surface of the receptor.
Proton transfer from the retinal protonated Schiff base to Glu113.
Displacement of extracellular loop 2 from the retinal binding site.
Rearrangement of the hydrogen-bonding network centered on Glu122 on H3.
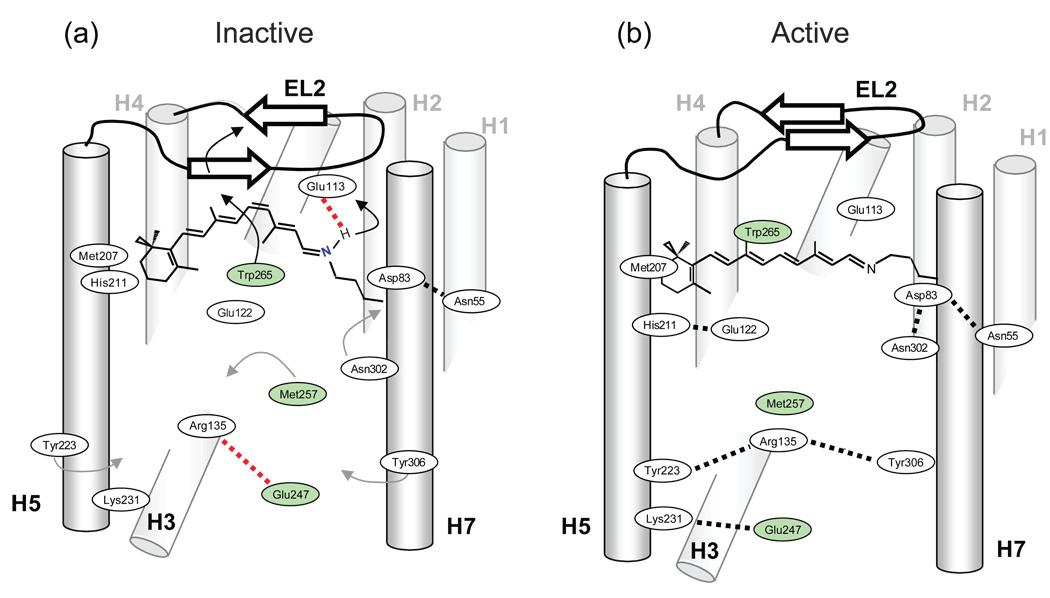
Fig. 10.
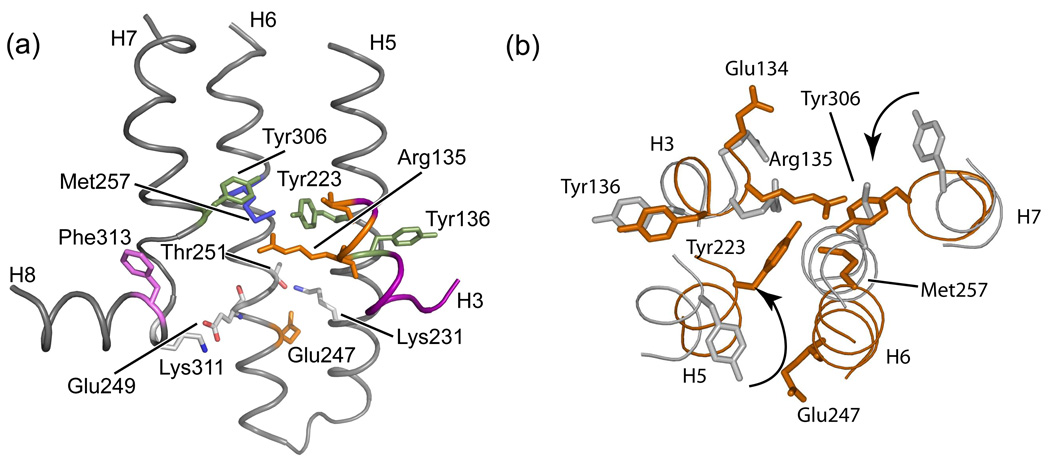
Molecular switches in the activation of rhodopsin. Retinal isomerization on the extracellular side of rhodopsin triggers several molecular switches that act in concert to disrupt the Arg135-Glu247 ionic lock on the intracellular side of the receptor. (a) Schematic of dark (inactive) rhodopsin showing the positions of key functional groups relative to the retinal chromophore. In the dark, rhodopsin is held in an inactive conformation by two salt bridges (dashed lines) that represent protonation switches. The first salt bridge is between the retinal protonated SB and the Glu113 counterion on the extracellular side of rhodopsin. The second salt bridge corresponds to the Arg135-Glu247 ionic lock on the intracellular side of rhodopsin. Residues on H6 (Glu247, Met257 and Trp265) are shaded. (b) Schematic of the active Meta II intermediate. On the extracellular side of the receptor, retinal isomerization leads to transfer of the retinal SB proton to Glu113 and motion of EL2. Motion of EL2 is coupled to the motion of H5 leading to the formation of an interhelical hydrogen bond between Glu122 and His211. In the dark, the retinal chromophore prevents motion of Trp265, the key residue in the rotamer toggle switch that drives the outward rotation of H6. Motion of Trp265 leads to a rearrangement in the hydrogen bonding interactions involving Asn302. On the intracellular side of rhodopsin, activation leads to the inward rotation of Tyr223 and Tyr306, which form inter-residue hydrogen bonds (grey dashed lines) with Arg135.
Solid-state NMR measurements of chemical shift and dipolar interactions have been used to characterize these molecular switches. These four areas are discussed below.
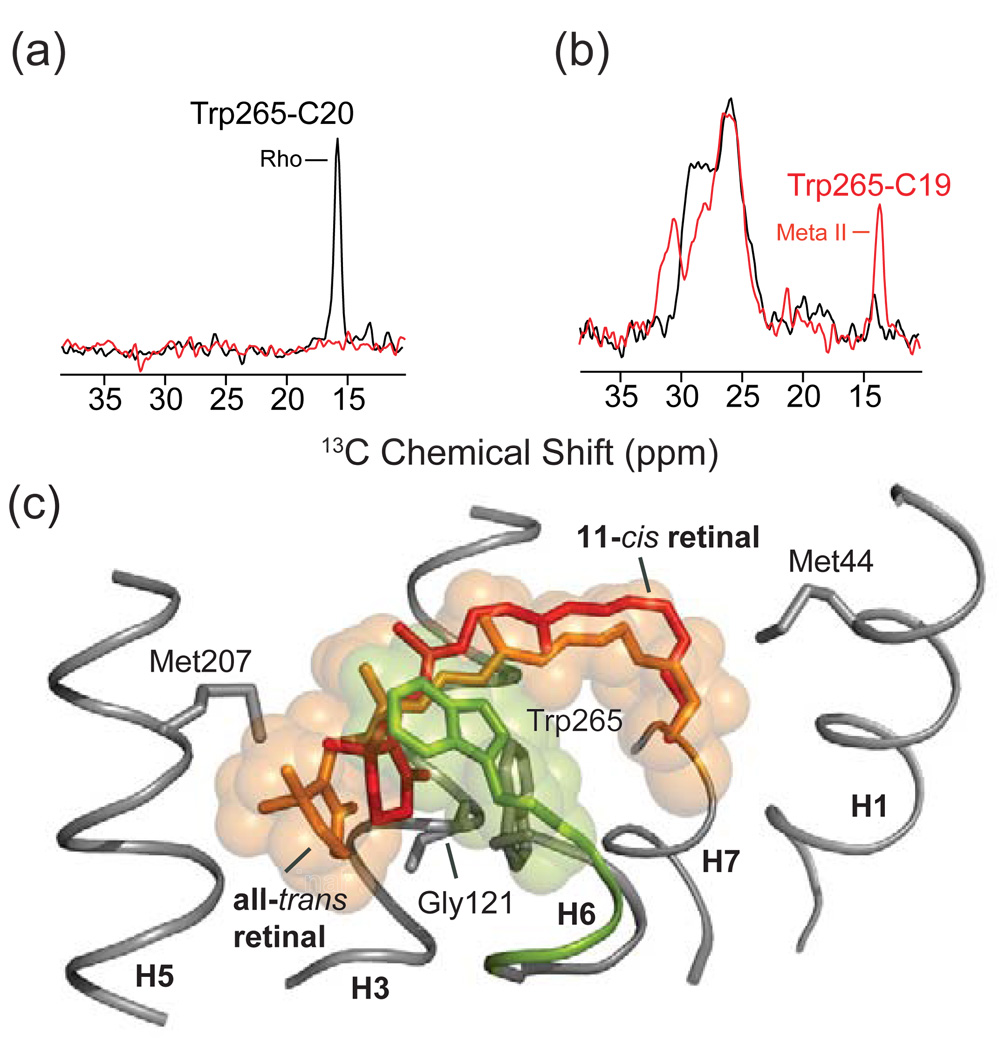
Rotation of Trp265 upon rhodopsin activation is part of the “rotamer-toggle switch” that was originally proposed by Shi et al. [204] for the dopamine receptor. Limiting the range of motion for the conserved Trp265 by the close packing of the β-ionone ring in rhodopsin is thought to be responsible for the extremely low level of basal activity observed in the dark [107,108]. This observation is consistent with a recent crystal structure of the inactive A2A adenosine receptor stabilized by a high affinity antagonist [40]. In this structure, the furan ring of the antagonist is in close contact with the conserved tryptophan on H6 and blocks rotation of the side chain.
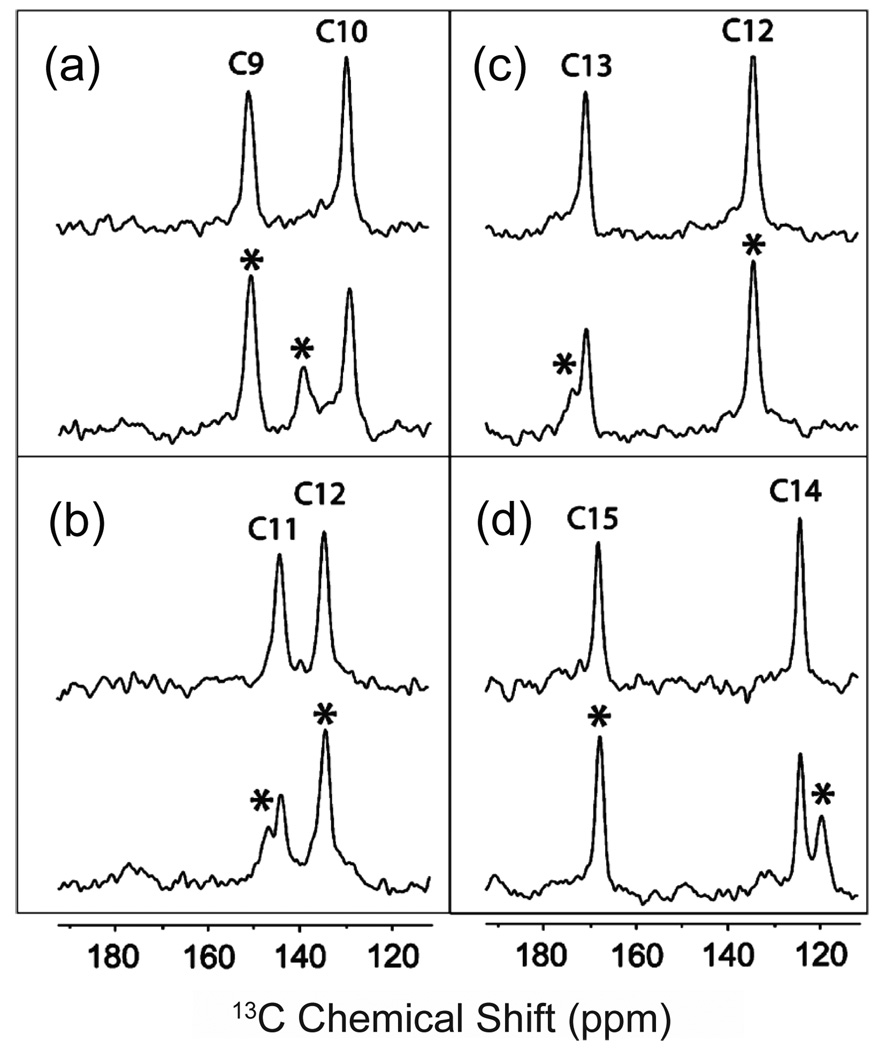
NMR measurements on the position of Trp265 have provided the first high-resolution structural data in support of the rotamer-toggle switch in rhodopsin [107,108]. Fig. 11 presents 2D DARR NMR data obtained on rhodopsin containing uniformly 13C labeled tryptophan and regenerated with retinal 13C labeled at either the 13C19 or the 13C20 methyl group [107,108]. In rhodopsin, the C20 methyl group is in van der Waals contact with Trp265 and a strong cross peak is observed in the 2D DARR NMR spectrum (Fig. 11a). Upon conversion to the active Meta II intermediate, this cross peak is lost. In contrast, a strong 2D DARR NMR cross peak is gained between Trp265 and the retinal C19 methyl group in Meta II (Fig. 11b). Fig. 11c shows an overlay of the 11-cis and all-trans-retinal chromophores in rhodopsin and Meta II, respectively, along with the position of the Trp265 side chain. The positions of the retinal and the Trp265 side chain in Meta II are established by solid-state NMR constraints [107,108], which provide input for guided MD simulations [205]. The NMR data reveal that there is a small translation of the retinal toward TM helix H5 in Meta II, i.e. in a direct away from Trp265. As the β-ionone ring of the retinal moves away from Trp265, the Trp265 side chain rotates toward the extracellular surface where it contacts the C19 retinal methyl group.
Fig. 11.
Molecular switches on the extracellular surface of rhodopsin: Trp265. Rows are shown from the 150 MHz 13C 2D DARR NMR spectra of rhodopsin labeled with U-13C-tryptophan and regenerated with either 13C20-labeled retinal (a) or 13C19-labeled retinal (b). The position of the Trp265 side chain changes between rhodopsin and Meta II as indicated by the Trp-retinal cross peaks. In (a), a cross peak is observed between the 13C20 methyl group and Trp side chain in rhodopsin, but not in Meta II, whereas in (b) a cross peak is observed between the 13C19 methyl group and tryptophan side chain in Meta II, but not in rhodopsin, as seen in the overlaid spectra. The β-ionone ring of 11-cis retinal is closely packed against the indole ring from the side chain of Trp265 as indicated in (c).
There are two protonation switches that control rhodopsin activation (marked by dashed lines in Fig. 10a). These two switches involve salt bridges that are buried within the TM region of the receptor and are neutralized upon activation. The first switch involves the transfer of the proton on the retinal protonated SB to its Glu113 counterion. NMR studies on this proton transfer were discussed in Section 3.1. The second switch involves the protonation of Glu134 on the intracellular side of the receptor. Glu134 is associated with the ionic lock (discussed in Section 1.3) between Arg135 on H3 and Glu247 on H6.
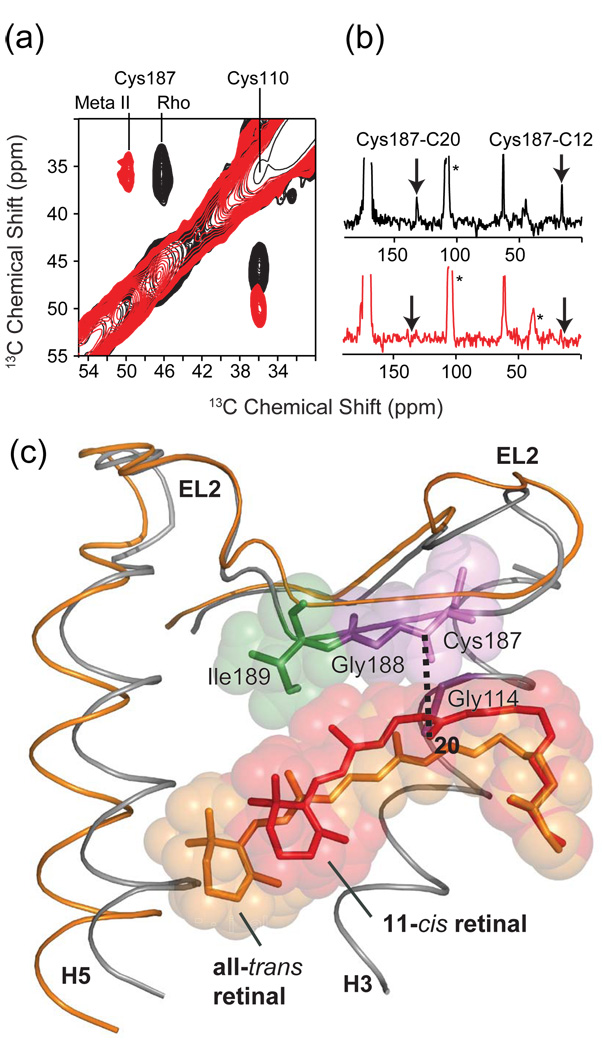
Deprotonation of the Schiff base, which is at the heart of the protonation switch on the extracellular side of rhodopsin, is driven by isomerization of the retinal. One of the major consequences of retinal isomerization and Schiff base deprotonation is the displacement of EL2 away from the retinal binding site (Fig. 12) [106]. The structure of EL2 in rhodopsin is stabilized by a number of polar residues that form a well-defined hydrogen bonded network. NMR chemical shift and distance measurements have been used to map out changes in the position and hydrogen bonding interactions involving EL2 (see Fig. 12a,b) [106]. For example, Fig. 12a shows the 2D DARR NMR spectrum of rhodopsin in the region of the 13C-labeled Cβ carbons of Cys110 and Cys187. The disulfide bond between these two cysteines connects H3 and EL2, and is conserved in the Class A GPCRs. The 13Cβ chemical shift of Cys187 changes considerably from 46.8 to 50.1 ppm upon rhodopsin activation, whereas the 13Cβ chemical shift of Cys110 remains the same. Dipolar coupling measurements between the retinal C12 and C20 carbons and the Cys187 Cβ carbon shows that the strong cross peaks observed in rhodopsin (arrows in Fig. 12b, top) are lost upon conversion to Meta II (Fig. 12b, bottom). In Fig. 12c, the retinal chromophores in rhodopsin and in Meta II are again overlaid to illustrate an important point, namely, that the C20 methyl group is oriented away from Cys187 in the rhodopsin crystal structure, yet a distinct C20-Cys187 cross peak is observed in Fig. 12b (top). Upon activation, the C20 methyl group rotates toward EL2 (and Cys187) in Meta II, yet the cross peak is lost. Parallel measurements between the retinal and Ser186, Gly188 and Ile189 on EL2 support the conclusion that EL2 moves away from the retinal binding site upon receptor activation [106]. Fig. 12c illustrates the displacement of EL2 on the basis of MD simulations guided by NMR distance constraints obtained on Meta II [205].
Fig. 12.
Solid-state NMR chemical shift and dipolar coupling measurements on EL2 in rhodopsin. (a) Chemical shift measurements from 150 MHz 13C 2D DARR NMR spectra of rhodopsin and Meta II showing the shift in the 13Cβ resonance of Cys187 on EL2. The Cys110 – Cys187 disulfide bond is conserved in the Class A GPCRs. The 13Cβ chemical shifts of disulfide linked cysteines are shifted by ~25 ppm from those of reduced cysteines. The chemical shift of Cys110 of ~36 ppm is characteristic of a cysteine in α-helical secondary structure, while the chemical shift of Cys187 between 46 ppm and 50 ppm is characteristic of extend β-structure. (b) Dipolar coupling measurements from 2D NMR spectra of rhodopsin (top) and Meta II (bottom) showing the loss of retinal-EL2 contacts upon the formation of Meta II. (c) Overlay of the crystal structure of rhodopsin and the structure of Meta II developed on the basis of NMR measurements and MD simulations [106]. EL2 is displaced from the retinal-binding pocket in the active Meta II state. The figure is adapted from Ref. [106].
The motion of EL2 leads to a rearrangement of the hydrogen-bonding network that stabilizes the position of EL2 in the dark [106]. One of the strengths of NMR is its ability to monitor hydrogen-bonding changes. For example, in Section 2.2 we presented the rhodopsin-Meta II difference spectrum and assigned one of the two distinct negative Meta II peaks in the region of the 13Cζ- tyrosine resonances to Tyr206. Similarly, the second tyrosine resonance can be assigned to Tyr191 [106]. Tyr191 is at a key position on EL2. The C19 methyl group packs against Tyr268 on H6, and Tyr191 on EL2 in rhodopsin. Counterclockwise rotation of the C19 methyl group results in a steric clash with Tyr191 and Tyr268, and may be responsible for a shift in hydrogen-bonding interactions involving EL2. In support of this model, removal of the C19 methyl group prevents receptor activation [206], while replacement of the retinal C19 methyl group with an ethyl or propyl group converts the 11-cis protonated SB chromophore from a potent inverse agonist into a partial agonist, with the amount of activity being proportional to the size of the substituent at the C19 position [207].
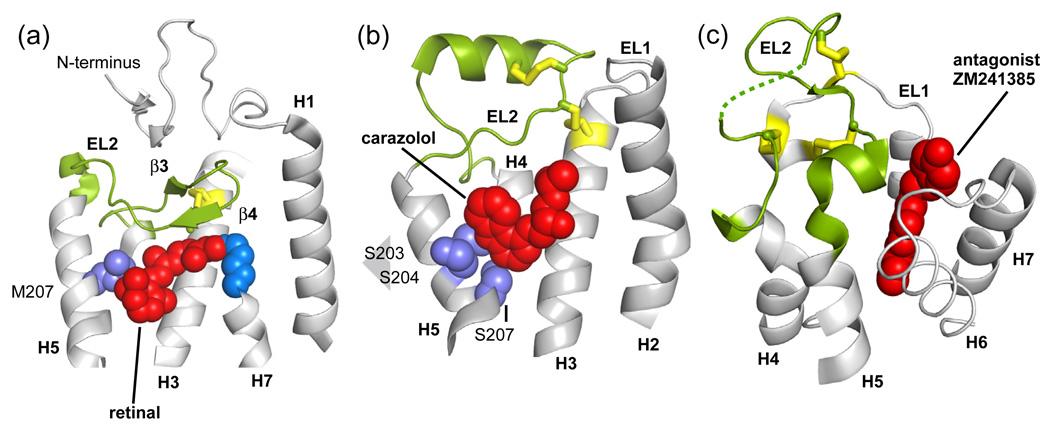
In contrast to rhodopsin, the recent crystal structures of the β1- and β2-adrenergic receptor [36,39] and the A2A adenosine receptor [40] reveal very different conformations of EL2 (see Fig. 13). The β2-adrenergic receptor crystal structure with a bound partial inverse agonist shows that EL2 does not cap the amine-binding site as in rhodopsin, but instead extends out into the extracellular space and contains an α-helix and an unusual pair of disulfide bonds [36]. The β2-adrenergic receptor structure, along with the observation that short loops may be correlated with constitutive active in GPCRs [106], raises the question of whether the role of EL2 as a stable cap is unique to rhodopsin due to the crucial requirement that visual pigments must have very low basal activity in the dark.
Fig. 13.
EL2 can adopt different conformations in Class A GPCRs. (a) In rhodopsin (PDB code 1U19), EL2 folds into two short β strands [34,35,38]. The 11-cis retinal (shown with van der Waals spheres) is closely packed against the β4 strand. (b) In the β2 adrenergic receptor (PDB code 2RH1) EL2 adopts an α-helix [10,39]. The binding pocket of carazolol, a partial inverse agonist, is not occluded by EL2 (c) In the adenosine A2A receptor (PDB code 3EML), EL2 lacks secondary structure [40] and does not contact the antagonist ZM241385.
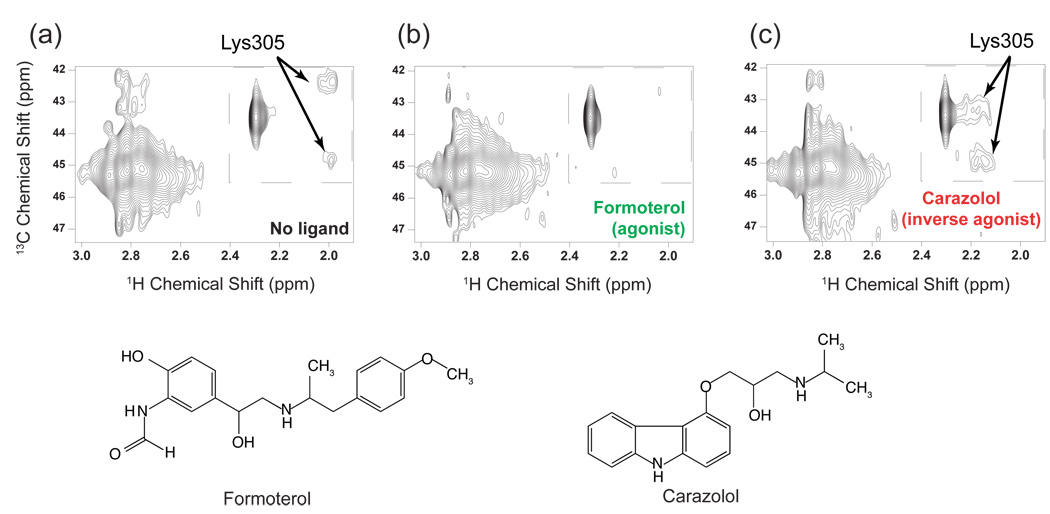
Recently, solution NMR has been used to characterize the role of EL2 in the β2 adrenergic receptor [208]. In a novel approach for specific 13C labeling of GCPRs, Kobilka and colleagues were able to monitor the local environment of a crucial salt bridge that tethers EL2 to EL3. In order to incorporate 13C-labels into the β2 adrenergic receptor, the lysine side chains were chemically di-methylated using a 13C-labeled methylating agent, dimethylamine. Di-methylation preserves the positive charge on lysine, and does not appear to alter the ligand-binding properties of the receptor. In the crystal structure of the β2 adrenergic receptor [10], Lys305 on EL3 interacts with Asp192 on EL2. This interaction is the only contact that prevents free access of ligand to the amine binding site within the TM helix bundle. Fig. 14 presents 2D heteronuclear 1H-13C spectra of the β2 adrenergic receptor with no ligand and in the presence of an agonist (formoterol) and inverse agonist (carazolol). Two peaks in the inset correspond to the methyl resonances of dimethyl-Lys305. The observation of resolved chemical shifts indicates that these methyl groups are in unique environments and the lysine side chain is not capable of chemical exchange. Upon binding of formoterol (Fig. 14b), an agonist with nanomolar affinity, the intensity of dimethyl Lys305 resonances is greatly reduced suggesting that the Lys305–Asp192 salt bridge is weakened in the active state. Replacement of formoterol with carazolol, an inverse agonist (Fig. 14c), re-establishes the Lys305 dimethyl resonances, which are now shifted upfield in the 1H dimension and have a larger separation in the 13C dimension.
Fig 14.
Solution NMR measurements of an EL2–EL3 salt bridge in the β2-adrenergic receptor. (a) The 800 MHz 1H HMQC spectrum of the unliganded β2 receptor obtained using saturation transfer differencing. (b) The same experiment as in (a) after adding a saturating concentration (320 mM) of the agonist (R,R)-formoterol. (c) The same experiment as in (b) after replacing formoterol with the inverse agonist carazolol by dialysis. The figure is adapted from Ref. [208].
The conformation of the EL2 in the A2A adenosine receptor crystal structure bound to an antagonist [40] is distinct from rhodopsin and the β1- and β2-adrenergic receptors. In the A2A adenosine receptor, EL2 extends into the extracellular space as a random coil and is constrained by three disulfide bonds to cysteines on EL1 (see Fig. 13). It is suggested that the rigid and open conformation of EL2 in the adrenergic and adenosine receptor constrained by the disulfide linkages provide solvent and small ligands with easy access to the ligand-binding pocket.
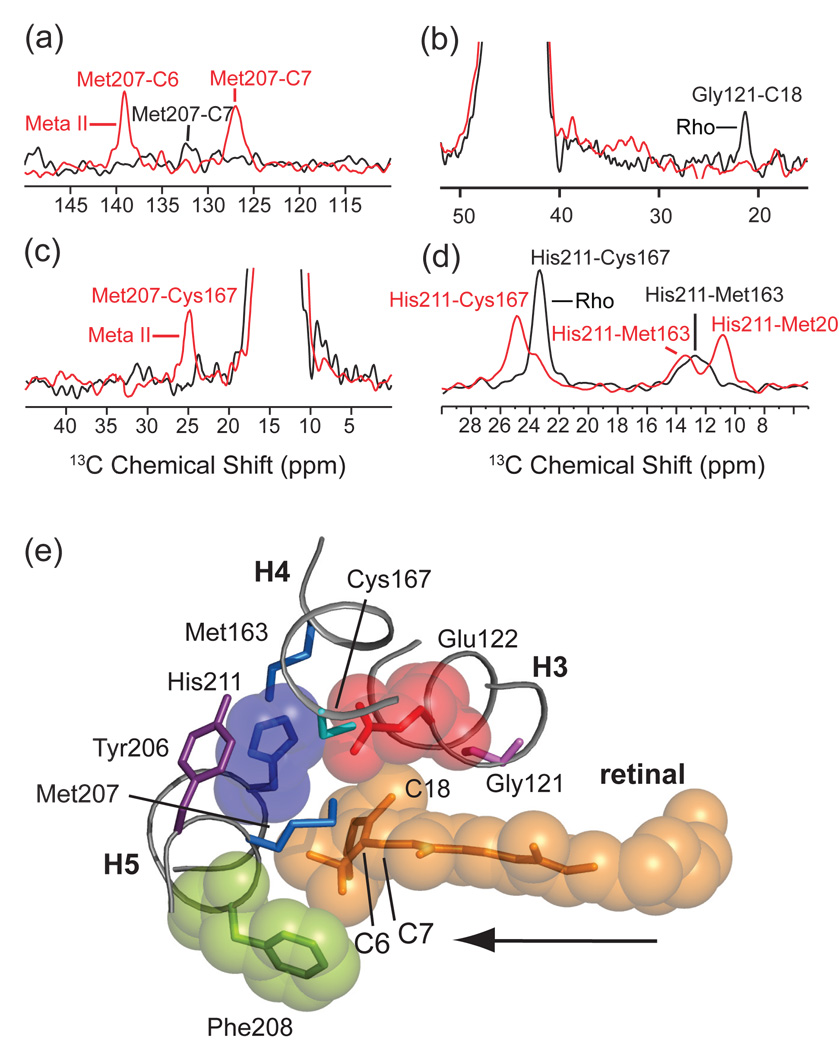
A fourth molecular switch on the extracellular side of rhodopsin is formed by a hydrogen bonding network centered on Glu122 and His211 (Fig. 15). The combination of retinal isomerization and displacement of EL2 appears to alter this network and lead to the motion of TM helix H5. NMR chemical shift measurements and distance measurements through 13C…13C dipolar couplings have been used to characterize the changes in this region of the receptor upon activation. First, Fig. 15a shows rows from the 13C 2D DARR spectra of rhodopsin and Meta II containing 6,7-13C retinal and 13Cε-methionine. There is a very weak cross peak between the terminal methyl group of Met207 and the C7 carbon of the retinal in rhodopsin. Activation leads to much stronger retinal – Met207 cross peaks in Meta II. The increase in cross peak intensity is interpreted in terms of a shift in the position of the retinal toward H5 (see Figs. 11c and 15e). Motion of the retinal within the binding site has been characterized by several retinal protein contacts. Fig. 15b shows the loss of a cross peak between the retinal C18 methyl group and Gly121 in the rhodopsin-to-Meta II transition. Second, Figs. 15c,d show changes in side chain interactions involving Met207 and His211 on H5. The loss of intensity in the His211-Cys167 cross peak reflects an increase in internuclear distance between these two amino acids, while the increase in the Met207-Cys167 cross peak reflects a decrease in internuclear distance.
Fig. 15.
Molecular switches on the extracellular surface of rhodopsin: Glu122-His211. Motion of the retinal chromophore toward H5 is revealed by an increase in NMR cross peak intensity between the retinal and Met207 (a) and by a decrease in cross peak intensity between the retinal and Gly121 (b). Panel (a) presents rows through the diagonal resonance of 3Cε-Met207 in rhodopsin and Meta II from 150 MHz 13C 2D DARR NMR spectra. Panel (b) presents rows through the diagonal resonance of 13Cα-Gly121 in rhodopsin and Meta II from 150 MHz 13C 2D DARR NMR spectra. Rearrangement of the hydrogen bonding network centered on His211 is revealed by changes in internuclear distance between His211 and surround in amino acids. In panel (c), rows are presented from 150 MHz 13C 2D DARR NMR spectra taken through the diagonal of 13Cε1-His211 in rhodopsin at 136.9 ppm and Meta II at 137.5 ppm. In rhodopsin, cross peaks are observed with 13Cβ-Cys167 at 23.7 ppm and with 13Cε-Met163 at 13.1 ppm. In Meta II, cross peaks with 13Cβ-Cys167 are observed at 25.3 ppm, with Met207 at 13.8 ppm and with Met163 at 11.2 ppm. On conversion to Meta II, the His211-Cys167 contact weakens, whereas the His211 side chain packs closer to the Met207 side chain on H5. In panel (d), a new cross peak appears between 13Cε-Met207 and 13Cβ-Cys167 in the formation of Meta II. (e) Guided molecular dynamics simulations show that the retinal chromophore in rhodopsin shifts toward H5 in Meta II. The retinal β-ionone ring contacts Glu122 and Met207 and leads to a change in a hydrogen bonding network involving H3, H4 and H5. The formation of a direct interaction between Glu122 and His211 stabilizes the active Meta II state. The figure is adapted from Ref. [106].
Fig. 15e shows the predicted position of the retinal chromphore in the active Meta II intermediate and is consistent with the distance measurements obtained by solid-state NMR. The net effect of the retinal – H5 interaction is a small rotation in the H5 helix, which leads to a change in the position of Tyr223 on the intracellular side of H5 (see Fig. 10 and discussion in the next section). An important contact that forms in Meta II is between the side chains of His211 and Glu122. The Glu122-His211 interaction is essential for stabilizing the Meta II state. To appreciate the importance of this contact, it is noteworthy that the Glu122-His211 pair is only conserved in the rod cell rhodopsin receptors responsible for dim light vision. The high sensitivity of these receptors relative to the cone cell receptors responsible for color vision is due to the stability of the active Meta II intermediate. Shichida and coworkers [209] found that substitution of Glu122 in rhodopsin with the corresponding amino acid in the green- or red-sensitive cone pigments converts the rate of Meta II decay into the that characteristic of the respective cone pigments. In contrast, when glutamate is substituted into the green-sensitive cone pigment, the rate Meta II decay is similar to that of rhodopsin. Using NMR, one can establish the protonation states and hydrogen bonding strengths of the residues forming the hydrogen bonding network shown in Fig. 15e. For example, the imidazole 15N resonances of His211 indicate that this histidine side chain is neutral in both rhodopsin and Meta II, but becomes more strongly hydrogen bonded in the Meta II once it interacts with the protonated Glu122 side chain [210].
Finally, the interaction between the retinal and H5 is analogous to the interaction between the ligands in the amine subfamily of Class A GPCRs and H5. In the amine receptors (such as the β2-adrenergic receptor), the amino acids corresponding to Met207, Phe208 and His211 in rhodopsin are conserved as serines. These serines hydrogen bond to hydroxyl groups on the catechol ring of the amine agonist ligand to stabilize an active receptor conformation [211–213], presumbably by leading to a conformational change in H5 in the same manner as described above for rhodopsin.
4.2. Allosteric structural changes in the transmembrane helices and intracellular loops
The molecular switches on the extracellular side of the receptor result in the following series of allosteric conformational changes on the intracellular side of the receptor (see Fig. 10).
Rearrangement of the hydrogen-bonding network involving Asn55, Asp83 and Asn302.
Rotation of Met257 toward Arg135.
Rotation of Tyr223 and Tyr306 toward Arg135.
Structural data on the intracellular switches comes mostly from the recent crystal structure of opsin [45,46], which retains many of the features characteristic of an active receptor. In this section, we discuss these switches and the role NMR may play in understanding key questions about how the extracellular conformational changes are relayed to the intracellular surface.
The first extracellular switch discussed above was the rotamer toggle switch that leads to a rotation of Trp265 on H6 toward the extracellular surface. Trp265 is hydrogen bonded in the dark state to Asn302 on H7 via structural waters that are observed in the high-resolution rhodopsin crystal structures [34]. 15N NMR and tryptophan fluorescence measurements both indicate that the indole NH becomes more weakly hydrogen bonded in Meta II [62,210]. This observation suggests that the hydrogen-bonding sphere of water surrounding Asn302 is disrupted. FTIR measurements have shown that a nearby conserved aspartic acid becomes more strongly hydrogen bonded upon activation and a working model that emerges from MD simulations is that the Asn302 side chain forms a hydrogen bonding contact with the Asp83 side chain on H2 [205]. NMR measurements may play a major role in characterizing the detailed changes in this conserved internal hydrogen bonding network upon activation and whether this is a conserved switch that triggers activation of the heterotrimeric G-protein.
The rotation of Trp265 (and correspondingly of H6) leads to an outward rotation of the intracellular end of H6. This rotation was first described by Hubbell and Khorana using results from EPR spectroscopy [7], and subsequently observed in the opsin crystal structure [45,46]. The outward rotation of H6 serves to move Met257 toward Arg135 and Glu247 away from Arg135 on H3. This rotation is shown in Fig. 16. The net result is a replacement of the charged Glu247 counterion with the hydrophobic Met257 side chain.
Fig. 16.
Intracellular ionic lock in rhodopsin. Views from the cytosolic surface of the rhodopsin (PDB code 1U19) (a) and opsin (PDB code 3CAP) (b) reveal the disruption of a salt bridge between Arg135 of the conserved E/DRY sequence and a glutamate side chain on H6 at position 247 upon activation. In concert with activation, the side chains of Tyr223 and Tyr306 on helices H5 and H7, respectively, rotate inward into close proximity to the guanidinium side chain of Arg135 as indicated in (b).
The displacement of H6 frees the guanidinium side chain of Arg135 from its salt bridge with Glu247. A striking feature of the opsin crystal structure is the observation that a highly conserved tyrosine residue on H5 (Tyr223) rotates into the helical bundle upon activation. A similar change in the position of the side chain of Tyr306 of the highly conserved NPxxY sequence places two phenol groups within hydrogen bonding distance of Arg135 (Fig. 10, dashed lines, Fig. 16b, arrows). This network of residues (Met257,Tyr223 and Tyr306) appears to be responsible for breaking the Arg135-Glu247 ionic lock and opening up the binding site for the heterotrimeric G-protein.
4.3. G protein-receptor interactions
The primary function of activated GPCRs is to catalyze the exchange of GTP for GDP in an intracellular heterotrimeric G protein. Receptor activation opens up a binding site on the intracellular surface of the receptor. However, there is only limited structural data on how the G-protein complex binds. The most detailed data comes from NMR and X-ray crystal structures of an 11-residue peptide corresponding to the C-terminus of the α subunit of transducin (GTα-CT). Transducin is the G protein that interacts with the visual receptors.
The structure of the GTα-CT peptide bound to Meta II was determined by transferred NOE measurements using solution NMR [214]. The peptide is disordered in solution, and binds weakly to rhodopsin upon light activation. The bound peptide has a helical turn followed by an open reverse turn centered at Gly348. By using a high magnetic field to align rhodopsin-containing unilamellar disks prepared from rod outer segments, Bax and coworkers were able to determine the orientation of two peptide NH groups in an 11-residue GTα peptide [215,216]. Peptides selectively labeled with 15N at Leu5 and Gly9 exhibited residual dipolar couplings that allowed the NH vectors of Leu5 and Gly9 to be oriented with respect to the disk normal [215,216].
Fig. 17a shows the structure of the intracellular region of opsin. Comparison with the same region in rhodopsin (Fig. 16a) shows several structural changes associated with an active conformation. These include rotation of Tyr223, Met257 and Tyr306 toward Arg135. This motion allows two additional salt-bridges to form on the intracellular surface of the receptor: Glu247 - Lys231 and Glu249 - Lys311. Together these changes open up a binding site for the C-terminus of transducin. Fig. 17b shows the position of the GTα-CT peptide bound in the opsin crystal structure [45].
Fig. 17.
G-protein binding site on the intracellular surface of rhodopsin. (a) View of the cytoplasmic region of opsin (PDB code 3CAP) in the region of the intracellular ionic lock. At pH 6.0, the opsin structure exhibits elements of the activated state of rhodopsin. (b) The same view of opsin co-crystallized with an 11-residue peptide corresponding to the C-terminus of transducin (PDB code 3DQB).
The results on the GTα-CT peptide have been extended to the full Gα subunit by Ridge and coworkers. They have demonstrated the ability to express milligram quantities of uniformly 15N-labeled G-protein α-subunits that can be reconstituted with Gβγ to form a functional heterotrimer [217]. They have further shown that the 15N-labeled Gα reconstituted heterotrimer forms functional complexes with light-activated rhodopsin solubilized in detergent and with a soluble mimic of activated rhodopsin [218].
5. Outlook
The diversity of GPCRs and their essential roles in cell biology indicate that these receptors are important targets for structural studies. This review highlights the progress that has been made to date using NMR to correlate structure with function, and outlines some of the unique advantages NMR has over other structural methods.
There are several areas where NMR can have an impact on understanding the structure and function of GPCRs: (i) ligand conformation and interactions; (ii) molecular mechanisms of activation switches; (iii) role of water in activation; and (iv) role of the membrane in activation.
Ligand conformation and interactions. One area where NMR promises to have a marked impact is in the determination of the conformation of bound ligands. As the crystal structures of additional GPCRs are determined, a key question will be to establish how different classes of ligands and newly developed pharmaceuticals bind to GPCRs and modulate their activity. In addition, allosteric GPCR modulators (compounds that bind in a different location from the activating ligand) are being developed that influence GPCR activity independent of the natural ligand. NMR studies on ligand conformation will require isotopic labeling only of the ligand.
Molecular mechanisms of activation switches. An emerging theme in GPCR activation is the presence of micro-switches that control activity [219,220]. The ability of NMR to characterize local structural features of GPCRs will allow one to characterize such switches.
Role of water in activation. An unresolved topic in the activation mechanism of GPCRs is the role of water. In rhodopsin, a large increase in the enthalpy of the Meta I – Meta II transition is compensated by a large increase in entropy. The nature of the entropy increase has not been established, but may be related to the release of water in the Meta I to Meta II transition [221]. Solid-state NMR methods have been developed to transfer magnetization from water to surrounding protein residues [222]. These methods may be useful for mapping out internal hydration of GPCRs in their inactive and active
Role of the membrane in activation. Unsaturation in the acyl chains and the type of lipid headgroup can influence the Meta I to Meta II transition. For example, NMR [223,224] and computational [225] studies provide evidence that ω-3 polyunsaturated chains, such as docosahexaenoic acid, preferentially solvate rhodopsin with the unsaturated chains oriented toward the TM helices. Brown and co-workers [125,226] suggested that the conformational change associated with Meta II increases the hydrophobic thickness of the receptor and that the phosphotidylethanolamine headgroup, which favors hexagonal phase lipids, will change the local curvature near the receptor to compensate for the hydrophobic mismatch between the receptor and the membrane. The recent crystal structures of opsin [45,46], which show a lengthening of TM helices H5 and H6, support this idea.
Finally, there will likely be advances in both expression and MR methodology that will move NMR structural studies forward. Dynamic Nuclear Polarization (DNP) is one such application, using microwave irradiation to polarize spin labels and transfer the resulting magnetization to NMR spins of interest. In 15N MAS NMR studies on intermediates in the bacteriorhodopsin photocycle, signal enhancements on the order of 40-fold were obtained with DNP [227]. When applied to a 13C-labeled rhodopsin sample, DNP results in a > 20-fold increase in sensitivity (Fig. 18). Fig. 18c shows the region of a 2D PDSD NMR spectrum of rhodopsin containing 13Cζ-tyrosine and 13Cε-histidine. The Tyr-His cross peaks were observed with good sensitivity with ~2 mgs of protein in less than 10 hours of data acquisition. As a result, the method opens up the possibility of structural studies on < 1 mg of expressed and functional GPCRs.
Fig. 18.
Dynamic nuclear polarization enhanced rhodopsin spectra. (a) 1D 100 MHz 13C MAS spectra of rhodopsin in the presence (grey) and absence (black) of microwave irradiation. DNP resulted in a >20 fold increase in the tyrosine Cζ signal. (b) Structure of rhodopsin showing the close proximity of Tyr274 and His278. (c) 100 MHz 13C 2D PDSD NMR spectrum of rhodopsin in the region of 13Cζ-Tyr and 13Cε-His cross peaks obtained using proton-driven spin diffusion. The DNP spectra were obtained in collaboration with Melanie Rosay (Bruker Instruments, Billerica, MA).
Acknowledgements
We thank Michael Brown, Malcolm Levitt, Stan Opella, and Harald Schwalbe for figures. We also thank Sang Ho Park for obtaining the SAMMY spectrum of 15N-labeled rhodopsin and Melanie Rosay for obtaining the PDSD spectra of 13C-labeled rhodopsin using DNP. The work on rhodopsin was supported by the National Institutes of Health through grants to Steven Smith (GM 41412) and Stan Opella (RO1EB005161 and P41EB002031).
Glossary of Abbreviations
- CL
cytoplasmic loop
- cryo-EM
cryogenic electron microscopy
- C-terminus
carboxy terminus
- DARR
dipolar-assisted rotational resonance
- DDM
dodecyl-β-D-maltoside
- DNA
deoxyribonucleic acid
- DNP
dynamic nuclear polarization
- DOPC
dioleoylphosphatidylcholine
- DQF
double quantum filtered
- EL
extracellular loop
- EPR
electron paramagnetic resonance
- ER
endoplasmic reticulum
- FTIR
Fourier transform infrared
- GPCR
G protein-coupled receptor
- H1–H7
transmembrane helices 1–7
- HEK
human embryonic kidney
- HIMSELF
heteronuclear isotropic mixing leading to spin exchange via the local field
- HIV
human immunodeficiency virus
- HSQC
heteronuclear single quantum coherence
- MAS
magic angle spinning
- Meta I
metarhodopsin I
- Meta II
metarhodopsin II
- NMR
nuclear magnetic resonance
- N-terminus
amino terminus
- PDSD
proton driven spin diffusion
- PISEMA
polarization inversion spin exchange at the magic angle
- SB
Schiff base
- TM
transmembrane
- TOCSY
total correlation spectroscopy
- UV
ultraviolet
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Nathans J, Hogness DS. Cell. 1983;34:807. doi: 10.1016/0092-8674(83)90537-8. [DOI] [PubMed] [Google Scholar]
- 2.Stryer L, Bourne HR. Annu. Rev. Cell Biol. 1986;2:391. doi: 10.1146/annurev.cb.02.110186.002135. [DOI] [PubMed] [Google Scholar]
- 3.Dixon RAF, Kobilka BK, Strader DJ, Benovic JL, Dohlman HG, Frielle T, Bolanowski MA, Bennett CD, Rands E, Diehl RE, Mumford RA, Slater EE, Sigal IS, Caron MG, Lefkowitz RJ, Strader CD. Nature. 1986;321:75. doi: 10.1038/321075a0. [DOI] [PubMed] [Google Scholar]
- 4.Ross EM, Gilman AG. Annu. Rev. Biochem. 1980;49:553. doi: 10.1146/annurev.bi.49.070180.002533. [DOI] [PubMed] [Google Scholar]
- 5.Huang C, Hepler JR, Chen LT, Gilman AG, Anderson RGW, Mumby SM. Mol. Biol. Cell. 1997;8:2365. doi: 10.1091/mbc.8.12.2365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Schertler GFX, Villa C, Henderson R. Nature. 1993;362:770. doi: 10.1038/362770a0. [DOI] [PubMed] [Google Scholar]
- 7.Farrens DL, Altenbach C, Yang K, Hubbell WL, Khorana HG. Science. 1996;274:768. doi: 10.1126/science.274.5288.768. [DOI] [PubMed] [Google Scholar]
- 8.Palczewski K, Kumasaka T, Hori T, Behnke CA, Motoshima H, Fox BA, Le Trong I, Teller DC, Okada T, Stenkamp RE, Yamamoto M, Miyano M. Science. 2000;289:739. doi: 10.1126/science.289.5480.739. [DOI] [PubMed] [Google Scholar]
- 9.Okada T, Le Trong I, Fox BA, Behnke CA, Stenkamp RE, Palczewski K. J. Struct.Biol. 2000;130:73. doi: 10.1006/jsbi.1999.4209. [DOI] [PubMed] [Google Scholar]
- 10.Rasmussen SGF, Choi HJ, Rosenbaum DM, Kobilka TS, Thian FS, Edwards PC, Burghammer M, Ratnala VRP, Sanishvili R, Fischetti RF, Schertler GFX, Weis WI, Kobilka BK. Nature. 2007;450:383. doi: 10.1038/nature06325. [DOI] [PubMed] [Google Scholar]
- 11.Kolakowski LF. Recept. Channels. 1994;2:1. [PubMed] [Google Scholar]
- 12.Bockaert J, Pin JP. EMBO J. 1999;18:1723. doi: 10.1093/emboj/18.7.1723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Foord SM, Bonner TI, Neubig RR, Rosser EM, Pin JP, Davenport AP, Spedding M, Harmar AJ. Pharmacol. Rev. 2005;57:279. doi: 10.1124/pr.57.2.5. [DOI] [PubMed] [Google Scholar]
- 14.Fredriksson R, Lagerström MC, Lundin LG, Schiöth HB. Mol. Pharmacol. 2003;63:1256. doi: 10.1124/mol.63.6.1256. [DOI] [PubMed] [Google Scholar]
- 15.Miller LJ, Dong M, Harikumar KG, Gao F. Biochem. Soc. Trans. 2007;35:709. doi: 10.1042/BST0350709. [DOI] [PubMed] [Google Scholar]
- 16.Markovic D, Grammatopoulos DK. Trends Biochem. Sci. 2009;34:443. doi: 10.1016/j.tibs.2009.06.002. [DOI] [PubMed] [Google Scholar]
- 17.Bräuner-Osborne H, Wellendorph P, Jensen AA. Curr. Drug Targets. 2007;8:169. doi: 10.2174/138945007779315614. [DOI] [PubMed] [Google Scholar]
- 18.Karakas E, Simorowski N, Furukawa H. EMBO J. 2009;28:3910. doi: 10.1038/emboj.2009.338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kothe E. Acta Microbiol. Immunol. Hung. 2008;55:125. doi: 10.1556/AMicr.55.2008.2.5. [DOI] [PubMed] [Google Scholar]
- 20.Klein PS, Sun TJ, Saxe CL, Kimmel AR, Johnson RL, Devreotes PN. Science. 1988;241:1467. doi: 10.1126/science.3047871. [DOI] [PubMed] [Google Scholar]
- 21.Johnson RL, Saxe CL, Gollop R, Kimmel AR, Devreotes PN. Genes Dev. 1993;7:273. doi: 10.1101/gad.7.2.273. [DOI] [PubMed] [Google Scholar]
- 22.Huang HC, Klein PS. Genome Biol. 2004;5:234. doi: 10.1186/gb-2004-5-7-234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Overington JP, Al-Lazikani B, Hopkins AL. Nat. Rev. Drug Discov. 2006;5:993. doi: 10.1038/nrd2199. [DOI] [PubMed] [Google Scholar]
- 24.Ma P, Zemmel R. Nat. Rev. Drug Discov. 2002;1:571. doi: 10.1038/nrd884. [DOI] [PubMed] [Google Scholar]
- 25.Lagerström MC, Schiöth HB. Nat. Rev. Drug Discov. 2008;7:339. doi: 10.1038/nrd2518. [DOI] [PubMed] [Google Scholar]
- 26.Rao VR, Oprian DD. Annu. Rev. Biophys. Biomol. Struct. 1996;25:287. doi: 10.1146/annurev.bb.25.060196.001443. [DOI] [PubMed] [Google Scholar]
- 27.Saunders J. Bioorg. Med. Chem. Lett. 2005;15:3653. doi: 10.1016/j.bmcl.2004.11.026. [DOI] [PubMed] [Google Scholar]
- 28.Dorr P, Westby M, Dobbs S, Griffin P, Irvine B, Macartney M, Mori J, Rickett G, Smith-Burchnell C, Napier C, Webster R, Armour D, Price D, Stammen B, Wood A, Perros M. Antimicrob. Agents Chemother. 2005;49:4721. doi: 10.1128/AAC.49.11.4721-4732.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.De Lean A, Stadel JM, Lefkowitz RJ. J. Biol. Chem. 1980;255:7108. [PubMed] [Google Scholar]
- 30.Samama P, Cotecchia S, Costa T, Lefkowitz RJ. J. Biol. Chem. 1993;268:4625. [PubMed] [Google Scholar]
- 31.Hermans E. Pharmacol. Ther. 2003;99:25. doi: 10.1016/s0163-7258(03)00051-2. [DOI] [PubMed] [Google Scholar]
- 32.Cordeaux Y, Nickolls SA, Flood LA, Graber SG, Strange PG. J. Biol. Chem. 2001;276:28667. doi: 10.1074/jbc.M008644200. [DOI] [PubMed] [Google Scholar]
- 33.Kobilka BK, Deupi X. Trends Pharmacol. Sci. 2007;28:397. doi: 10.1016/j.tips.2007.06.003. [DOI] [PubMed] [Google Scholar]
- 34.Li J, Edwards PC, Burghammer M, Villa C, Schertler GFX. J. Mol. Biol. 2004;343:1409. doi: 10.1016/j.jmb.2004.08.090. [DOI] [PubMed] [Google Scholar]
- 35.Okada T, Sugihara M, Bondar AN, Elstner M, Entel P, Buss V. J. Mol. Biol. 2004;342:571. doi: 10.1016/j.jmb.2004.07.044. [DOI] [PubMed] [Google Scholar]
- 36.Cherezov V, Rosenbaum DM, Hanson MA, Rasmussen SGF, Thian FS, Kobilka TS, Choi HJ, Kuhn P, Weis WI, Kobilka BK, Stevens RC. Science. 2007;318:1258. doi: 10.1126/science.1150577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hanson MA, Cherezov V, Griffith MT, Roth CB, Jaakola VP, Chien EYT, Velasquez J, Kuhn P, Stevens RC. Structure. 2008;16:897. doi: 10.1016/j.str.2008.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Murakami M, Kouyama T. Nature. 2008;453:363. doi: 10.1038/nature06925. [DOI] [PubMed] [Google Scholar]
- 39.Warne T, Serrano-Vega MJ, Baker JG, Moukhametzianov R, Edwards PC, Henderson R, Leslie AG, Tate CG, Schertler GFX. Nature. 2008;454:486. doi: 10.1038/nature07101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Jaakola VP, Griffith MT, Hanson MA, Cherezov V, Chien EYT, Lane JR, Ijzerman AP, Stevens RC. Science. 2008;322:1211. doi: 10.1126/science.1164772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Nakamichi H, Okada T. Angew. Chem. Int. Ed. Engl. 2006;45:4270. doi: 10.1002/anie.200600595. [DOI] [PubMed] [Google Scholar]
- 42.Schreiber M, Sugihara M, Okada T, Buss V. Angew. Chem. Int. Ed. Engl. 2006;45:4274. doi: 10.1002/anie.200600585. [DOI] [PubMed] [Google Scholar]
- 43.Nakamichi H, Okada T. Proc. Natl. Acad. Sci. U.S.A. 2006;103:12729. doi: 10.1073/pnas.0601765103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ruprecht JJ, Mielke T, Vogel R, Villa C, Schertler GFX. EMBO J. 2004;23:3609. doi: 10.1038/sj.emboj.7600374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Scheerer P, Park JH, Hildebrand PW, Kim YJ, Krauss N, Choe HW, Hofmann KP, Ernst OP. Nature. 2008;455:497. doi: 10.1038/nature07330. [DOI] [PubMed] [Google Scholar]
- 46.Park JH, Scheerer P, Hofmann KP, Choe HW, Ernst OP. Nature. 2008;454:183. doi: 10.1038/nature07063. [DOI] [PubMed] [Google Scholar]
- 47.Gether U. Endocr. Rev. 2000;21:90. doi: 10.1210/edrv.21.1.0390. [DOI] [PubMed] [Google Scholar]
- 48.Sakmar TP, Menon ST, Marin EP, Awad ES. Annu. Rev. Biophys. Biomol. Struct. 2002;31:443. doi: 10.1146/annurev.biophys.31.082901.134348. [DOI] [PubMed] [Google Scholar]
- 49.Palczewski K. Annu. Rev. Biochem. 2006;75:743. doi: 10.1146/annurev.biochem.75.103004.142743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Schwartz TW, Frimurer TM, Holst B, Rosenkilde MM, Elling CE. Annu. Rev. Pharmacol. Toxicol. 2006;46:481. doi: 10.1146/annurev.pharmtox.46.120604.141218. [DOI] [PubMed] [Google Scholar]
- 51.Rosenbaum DM, Rasmussen SGF, Kobilka BK. Nature. 2009;459:356. doi: 10.1038/nature08144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Angel TE, Chance MR, Palczewski K. Proc. Natl. Acad. Sci. U.S.A. 2009;106:8555. doi: 10.1073/pnas.0903545106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Salom D, Lodowski DT, Stenkamp RE, Le Trong I, Golczak M, Jastrzebska B, Harris T, Ballesteros JA, Palczewski K. Proc. Natl. Acad. Sci. U.S.A. 2006;103:16123. doi: 10.1073/pnas.0608022103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Altenbach C, Klein-Seetharaman J, Cai KW, Khorana HG, Hubbell WL. Biochemistry. 2001;40:15493. doi: 10.1021/bi011545o. [DOI] [PubMed] [Google Scholar]
- 55.Altenbach C, Cai KW, Klein-Seetharaman J, Khorana FG, Hubbell WL. Biochemistry. 2001;40:15483. doi: 10.1021/bi011546g. [DOI] [PubMed] [Google Scholar]
- 56.Knierim B, Hofmann KP, Ernst OP, Hubbell WL. Proc. Natl. Acad. Sci. U.S.A. 2007;104:20290. doi: 10.1073/pnas.0710393104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Altenbach C, Kusnetzow AK, Ernst OP, Hofmann KP, Hubbell WL. Proc. Natl. Acad. Sci. U.S.A. 2008;105:7439. doi: 10.1073/pnas.0802515105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Dunham TD, Farrens DL. J. Biol. Chem. 1999;274:1683. doi: 10.1074/jbc.274.3.1683. [DOI] [PubMed] [Google Scholar]
- 59.Yao XJ, Parnot C, Deupi X, Ratnala VRP, Swaminath G, Farrens D, Kobilka B. Nat. Chem. Biol. 2006;2:417. doi: 10.1038/nchembio801. [DOI] [PubMed] [Google Scholar]
- 60.Granier S, Kim S, Shafer AM, Ratnala VRP, Fung JJ, Zare RN, Kobilka B. J. Biol. Chem. 2007;282:13895. doi: 10.1074/jbc.M611904200. [DOI] [PubMed] [Google Scholar]
- 61.Chabre M, Breton J. Photochem. Photobiol. 1979;30:295. doi: 10.1111/j.1751-1097.1979.tb07150.x. [DOI] [PubMed] [Google Scholar]
- 62.Lin SW, Sakmar TP. Biochemistry. 1996;35:11149. doi: 10.1021/bi960858u. [DOI] [PubMed] [Google Scholar]
- 63.Fahmy K, Jäger F, Beck M, Zvyaga TA, Sakmar TP, Siebert F. Proc. Natl. Acad. Sci. U.S.A. 1993;90:10206. doi: 10.1073/pnas.90.21.10206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Rath P, DeCaluwe LLJ, Bovee-Geurts PHM, Degrip WJ, Rothschild KJ. Biochemistry. 1993;32:10277. doi: 10.1021/bi00090a001. [DOI] [PubMed] [Google Scholar]
- 65.Arnis S, Fahmy K, Hofmann KP, Sakmar TP. J. Biol. Chem. 1994;269:23879. [PubMed] [Google Scholar]
- 66.Fahmy K, Sakmar TP, Siebert F. Biochemistry. 2000;39:10607. doi: 10.1021/bi000912d. [DOI] [PubMed] [Google Scholar]
- 67.Vogel R, Siebert F, Mathias G, Tavan P, Fan GB, Sheves M. Biochemistry. 2003;42:9863. doi: 10.1021/bi034684+. [DOI] [PubMed] [Google Scholar]
- 68.Lehmann N, Alexiev U, Fahmy K. J. Mol. Biol. 2007;366:1129. doi: 10.1016/j.jmb.2006.11.098. [DOI] [PubMed] [Google Scholar]
- 69.Vogel R, Mahalingam M, Lüdeke S, Huber T, Siebert F, Sakmar TP. J. Mol. Biol. 2008;380:648. doi: 10.1016/j.jmb.2008.05.022. [DOI] [PubMed] [Google Scholar]
- 70.Mahalingam M, Martinez-Mayorga K, Brown MF, Vogel R. Proc. Natl. Acad. Sci. U.S.A. 2008;105:17795. doi: 10.1073/pnas.0804541105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Lüdeke S, Mahalingam M, Vogel R. Photochem. Photobiol. 2009;85:437. doi: 10.1111/j.1751-1097.2008.00490.x. [DOI] [PubMed] [Google Scholar]
- 72.Lin SW, Sakmar TP, Franke RR, Khorana HG, Mathies RA. Biochemistry. 1992;31:5105. doi: 10.1021/bi00137a003. [DOI] [PubMed] [Google Scholar]
- 73.Yan ECY, Kazmi MA, Ganim Z, Hou JM, Pan DH, Chang BSW, Sakmar TP, Mathies RA. Proc. Natl. Acad. Sci. U.S.A. 2003;100:9262. doi: 10.1073/pnas.1531970100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Kukura P, McCamant DW, Yoon S, Wandschneider DB, Mathies RA. Science. 2005;310:1006. doi: 10.1126/science.1118379. [DOI] [PubMed] [Google Scholar]
- 75.Costanzi S. J. Med. Chem. 2008;51:2907. doi: 10.1021/jm800044k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Ye SX, Huber T, Vogel R, Sakmar TP. Nat. Chem. Biol. 2009;5:397. doi: 10.1038/nchembio.167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Sanders CR, Sönnichsen F. Magn. Reson. Chem. 2006;44:S24. doi: 10.1002/mrc.1816. [DOI] [PubMed] [Google Scholar]
- 78.Kim HJ, Howell SC, Van Horn WD, Jeon YH, Sanders CR. Prog. Nucl. Magn. Reson. Spectrosc. 2009;55:335. doi: 10.1016/j.pnmrs.2009.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Poget SF, Girvin ME. Biochim. Biophys. Acta-Biomembr. 2007;1768:3098. doi: 10.1016/j.bbamem.2007.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.McDermott A, Polenova T. Curr. Opin. Struct. Biol. 2007;17:617. doi: 10.1016/j.sbi.2007.10.001. [DOI] [PubMed] [Google Scholar]
- 81.Nielsen NC, Malmendal A, Vosegaard T. Mol. Membr. Biol. 2004;21:129. doi: 10.1080/09687680410001693679. [DOI] [PubMed] [Google Scholar]
- 82.McDermott A. Ann. Rev. Biophys. 2009;38:385. doi: 10.1146/annurev.biophys.050708.133719. [DOI] [PubMed] [Google Scholar]
- 83.Smith SO, Palings I, Copie V, Raleigh DP, Courtin J, Pardoen JA, Lugtenburg J, Mathies RA, Griffin RG. Biochemistry. 1987;26:1606. doi: 10.1021/bi00380a018. [DOI] [PubMed] [Google Scholar]
- 84.Smith SO, Courtin J, de Groot H, Gebhard R, Lugtenburg J. Biochemistry. 1991;30:7409. doi: 10.1021/bi00244a007. [DOI] [PubMed] [Google Scholar]
- 85.Smith SO, de Groot H, Gebhard R, Lugtenburg J. Photochem. Photobiol. 1992;56:1035. doi: 10.1111/j.1751-1097.1992.tb09726.x. [DOI] [PubMed] [Google Scholar]
- 86.Getmanova E, Patel AB, Klein-Seetharaman J, Loewen MC, Reeves PJ, Friedman N, Sheves M, Smith SO, Khorana HG. Biochemistry. 2004;43:1126. doi: 10.1021/bi030120u. [DOI] [PubMed] [Google Scholar]
- 87.Brown MF, Deese AJ, Dratz EA. Meth. Enzymol. 1982;81:709. doi: 10.1016/s0076-6879(82)81098-7. [DOI] [PubMed] [Google Scholar]
- 88.Deese AJ, Dratz EA, Brown MF. FEBS Lett. 1981;124:93. doi: 10.1016/0014-5793(81)80061-0. [DOI] [PubMed] [Google Scholar]
- 89.Sarramegna V, Talmont R, Demange P, Milon A. Cell. Mol. Life. Sci. 2003;60:1529. doi: 10.1007/s00018-003-3168-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.McCusker EC, Bane SE, OMalley MA, Robinson AS. Biotechnol. Prog. 2007;23:540. doi: 10.1021/bp060349b. [DOI] [PubMed] [Google Scholar]
- 91.Kaiser L, Graveland-Bikker J, Steuerwald D, Vanberghem M, Herlihy K, Zhang SG. Proc. Natl. Acad. Sci. U.S.A. 2008;105:15726. doi: 10.1073/pnas.0804766105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Sansuk K, Balog CIA, van der Does AM, Booth R, de Grip WJ, Deelder AM, Bakker RA, Leurs R, Hensbergen PJ. J. Proteome Res. 2008;7:621. doi: 10.1021/pr7005654. [DOI] [PubMed] [Google Scholar]
- 93.Rensdomiano S, Reisine T. J. Biol. Chem. 1991;266:20094. [PubMed] [Google Scholar]
- 94.Russo D, Chazenbalk GD, Nagayama Y, Wadsworth HL, Rapoport B. Mol. Endocrinol. 1991;5:29. doi: 10.1210/mend-5-1-29. [DOI] [PubMed] [Google Scholar]
- 95.Kusui T, Benya RV, Battey JF, Jensen RT. Biochemistry. 1994;33:12968. doi: 10.1021/bi00248a005. [DOI] [PubMed] [Google Scholar]
- 96.Weiss HM, Grisshammer R. Eur. J. Biochem. 2002;269:82. doi: 10.1046/j.0014-2956.2002.02618.x. [DOI] [PubMed] [Google Scholar]
- 97.Prinz WA, Aslund F, Holmgren A, Beckwith J. J. Biol. Chem. 1997;272:15661. doi: 10.1074/jbc.272.25.15661. [DOI] [PubMed] [Google Scholar]
- 98.Miroux B, Walker JE. J. Mol. Biol. 1996;260:289. doi: 10.1006/jmbi.1996.0399. [DOI] [PubMed] [Google Scholar]
- 99.Mollaaghababa R, Davidson FF, Kaiser C, Khorana HG. Proc. Natl. Acad. Sci. U.S.A. 1996;93:11482. doi: 10.1073/pnas.93.21.11482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Magnin T, Fiez-Vandal C, Potier N, Coquard A, Leray I, Steffan T, Logez C, Alkhalfioui F, Pattus F, Wagner R. Protein Expr. Purif. 2009;64:1. doi: 10.1016/j.pep.2008.09.007. [DOI] [PubMed] [Google Scholar]
- 101.Reeves PJ, Thurmond RL, Khorana HG. Proc. Natl. Acad. Sci. U.S.A. 1996;93:11487. doi: 10.1073/pnas.93.21.11487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Eilers M, Reeves PJ, Ying WW, Khorana HG, Smith SO. Proc. Natl. Acad. Sci. U.S.A. 1999;96:487. doi: 10.1073/pnas.96.2.487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Eilers M, Ying WW, Reeves PJ, Khorana HG, Smith SO. Meth. Enzymol. 2002;343:212. doi: 10.1016/s0076-6879(02)43137-0. [DOI] [PubMed] [Google Scholar]
- 104.Reeves PJ, Kim JM, Khorana HG. Proc. Natl. Acad. Sci. U.S.A. 2002;99:13413. doi: 10.1073/pnas.212519199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Allen SJ, Ribeiro S, Horuk R, Handel TM. Protein Expr. Purif. 2009;66:73. doi: 10.1016/j.pep.2009.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Ahuja S, Hornak V, Yan ECY, Syrett N, Goncalves JA, Hirshfeld A, Ziliox M, Sakmar TP, Sheves M, Reeves PJ, Smith SO, Eilers M. Nat. Struct. Mol. Biol. 2009;16:168. doi: 10.1038/nsmb.1549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Ahuja S, Crocker E, Eilers M, Hornak V, Hirshfeld A, Ziliox M, Syrett N, Reeves PJ, Khorana HG, Sheves M, Smith SO. J. Biol. Chem. 2009;284:10190. doi: 10.1074/jbc.M805725200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Crocker E, Eilers M, Ahuja S, Hornak V, Hirshfeld A, Sheves M, Smith SO. J. Mol. Biol. 2006;357:163. doi: 10.1016/j.jmb.2005.12.046. [DOI] [PubMed] [Google Scholar]
- 109.Fraser MJ. Curr. Top. Microbiol. Immunol. 1992;158:131. doi: 10.1007/978-3-642-75608-5_6. [DOI] [PubMed] [Google Scholar]
- 110.Kost TA, Condreay JP, Jarvis DL. Nat. Biotechnol. 2005;23:567. doi: 10.1038/nbt1095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Creemers AFL, Klaassen CHW, Bovee-Geurts PHM, Kelle R, Kragl U, Raap J, De Grip WJ, Lugtenburg J, de Groot HJM. Biochemistry. 1999;38:7195. doi: 10.1021/bi9830157. [DOI] [PubMed] [Google Scholar]
- 112.Nisius L, Rogowski M, Vangelista L, Grzesiek S. Protein Expr. Purif. 2008;61:155. doi: 10.1016/j.pep.2008.06.001. [DOI] [PubMed] [Google Scholar]
- 113.Gether U, Ballesteros JA, Seifert R, Sanders-Bush E, Weinstein H, Kobilka BK. J. Biol. Chem. 1997;272:2587. doi: 10.1074/jbc.272.5.2587. [DOI] [PubMed] [Google Scholar]
- 114.Shukla AK, Haase W, Reinhart C, Michel H. Protein Expr. Purif. 2007;55:1. doi: 10.1016/j.pep.2007.02.021. [DOI] [PubMed] [Google Scholar]
- 115.Dowell SJ, Brown AJ. Recept. Channels. 2002;8:343. [PubMed] [Google Scholar]
- 116.Ladds G, Goddard A, Davey J. Trends Biotechnol. 2005;23:367. doi: 10.1016/j.tibtech.2005.05.007. [DOI] [PubMed] [Google Scholar]
- 117.Siehler S. Biotechnol. J. 2008;3:471. doi: 10.1002/biot.200800001. [DOI] [PubMed] [Google Scholar]
- 118.Klein-Seetharaman J, Reeves PJ, Loewen MC, Getmanova EV, Chung L, Schwalbe H, Wright PE, Khorana HG. Proc. Natl. Acad. Sci. U.S.A. 2002;99:3452. doi: 10.1073/pnas.052713999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Sarramegna V, Muller I, Milon A, Talmont F. Cell. Mol. Life. Sci. 2006;63:1149. doi: 10.1007/s00018-005-5557-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Kiefer H, Krieger J, Olszewski JD, Von Heijne G, Prestwich GD, Breer H. Biochemistry. 1996;35:16077. doi: 10.1021/bi9612069. [DOI] [PubMed] [Google Scholar]
- 121.Rogl H, Kosemund K, Kühlbrandt W, Collinson I. FEBS Lett. 1998;432:21. doi: 10.1016/s0014-5793(98)00825-4. [DOI] [PubMed] [Google Scholar]
- 122.Ma C, Marassi FM, Jones DH, Straus SK, Bour S, Strebel K, Schubert U, Oblatt-Montal M, Montal M, Opella SJ. Protein Sci. 2002;11:546. doi: 10.1110/ps.37302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Kiefer H. Biochim. Biophys. Acta-Biomembr. 2003;1610:57. doi: 10.1016/s0005-2736(02)00717-4. [DOI] [PubMed] [Google Scholar]
- 124.Lundstrom K. Bioorg. Med. Chem. Lett. 2005;15:3654. doi: 10.1016/j.bmcl.2005.05.041. [DOI] [PubMed] [Google Scholar]
- 125.Brown MF. Chem. Phys. Lipids. 1994;73:159. doi: 10.1016/0009-3084(94)90180-5. [DOI] [PubMed] [Google Scholar]
- 126.Ramon E, Marron J, del Valle L, Bosch L, Andres A, Manyosa J, Garriga P. Vision Res. 2003;43:3055. doi: 10.1016/j.visres.2003.08.009. [DOI] [PubMed] [Google Scholar]
- 127.Sanders CR, Landis GC. Biochemistry. 1995;34:4030. doi: 10.1021/bi00012a022. [DOI] [PubMed] [Google Scholar]
- 128.Li Y, Kijac AZ, Sligar SG, Rienstra CM. Biophys. J. 2006;91:3819. doi: 10.1529/biophysj.106.087072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Leitz AJ, Bayburt TH, Barnakov AN, Springer BA, Sligar SG. Biotechniques. 2006;40:601. doi: 10.2144/000112169. [DOI] [PubMed] [Google Scholar]
- 130.Kijac AZ, Li Y, Sligar SG, Rienstra CM. Biochemistry. 2007;46:13696. doi: 10.1021/bi701411g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Whorton MR, Jastrzebska B, Park PSH, Fotiadis D, Engel A, Palczewski K, Sunahara RK. J. Biol. Chem. 2008;283:4387. doi: 10.1074/jbc.M703346200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Banerjee S, Huber T, Sakmar TP. J. Mol. Biol. 2008;377:1067. doi: 10.1016/j.jmb.2008.01.066. [DOI] [PubMed] [Google Scholar]
- 133.Ernst OP, Gramse V, Kolbe M, Hofmann KP, Heck M. Proc. Natl. Acad. Sci. U.S.A. 2007;104:10859. doi: 10.1073/pnas.0701967104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Jones KA, Borowsky B, Tamm JA, Craig DA, Durkin MM, Dai M, Yao WJ, Johnson M, Gunwaldsen C, Huang LY, Tang C, Shen QR, Salon JA, Morse K, Laz T, Smith KE, Nagarathnam D, Noble SA, Branchek TA, Gerald C. Nature. 1998;396:674. doi: 10.1038/25348. [DOI] [PubMed] [Google Scholar]
- 135.White JH, Wise A, Main MJ, Green A, Fraser NJ, Disney GH, Barnes AA, Emson P, Foord SM, Marshall FH. Nature. 1998;396:679. doi: 10.1038/25354. [DOI] [PubMed] [Google Scholar]
- 136.Hague C, Uberti MA, Chen ZJ, Hall RA, Minneman KP. J. Biol. Chem. 2004;279:15541. doi: 10.1074/jbc.M314014200. [DOI] [PubMed] [Google Scholar]
- 137.Hague C, Uberti MA, Chen ZJ, Bush CF, Jones SV, Ressler KJ, Hall RA, Minneman KP. Proc. Natl. Acad. Sci. U.S.A. 2004;101:13672. doi: 10.1073/pnas.0403854101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.George SR, O'Dowd BF, Lee SR. Nat. Rev. Drug Discov. 2002;1:808. doi: 10.1038/nrd913. [DOI] [PubMed] [Google Scholar]
- 139.Angers S, Salahpour A, Bouvier M. Annu. Rev. Pharmacol. Toxicol. 2002;42:409. doi: 10.1146/annurev.pharmtox.42.091701.082314. [DOI] [PubMed] [Google Scholar]
- 140.Milligan G. Mol. Pharmacol. 2004;66:1. doi: 10.1124/mol.104.000497.. [DOI] [PubMed] [Google Scholar]
- 141.Werner K, Lehner IH, Dhiman K, Richter C, Glaubitz C, Schwalbe H, Klein-Seetharaman J, Khorana HG. J. Biomol. NMR. 2007;37:303. doi: 10.1007/s10858-007-9143-0. [DOI] [PubMed] [Google Scholar]
- 142.Klein-Seetharaman J, Yanamala NVK, Javeed F, Reeves PJ, Getmanova EV, Loewen MC, Schwalbe H, Khorana HG. Proc. Natl. Acad. Sci. U.S.A. 2004;101:3409. doi: 10.1073/pnas.0308713101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Ruan KH, Cervantes V, Wu JX. Biochemistry. 2009;48:3157. doi: 10.1021/bi801443g. [DOI] [PubMed] [Google Scholar]
- 144.Lowe IJ. Phys. Rev. Lett. 1959;2:285. [Google Scholar]
- 145.Andrew ER, Bradbury A, Eades RG. Nature. 1958;182:1659. [Google Scholar]
- 146.Schaefer J, Stejskal EO. J. Am. Chem. Soc. 1976;98:1031. [Google Scholar]
- 147.Maricq MM, Waugh JS. J. Chem. Phys. 1979;70:3300. [Google Scholar]
- 148.De Angelis AA, Jones DH, Grant CV, Park SH, Mesleh MF, Opella SJ. Meth. Enzymol. 2005;394:350. doi: 10.1016/S0076-6879(05)94014-7. [DOI] [PubMed] [Google Scholar]
- 149.Herzfeld J, Das Gupta SK, Farrar MR, Harbison GS, McDermott AE, Pelletier SL, Raleigh DP, Smith SO, Winkel C, Lugtenburg J, Griffin RG. Biochemistry. 1990;29:5567. doi: 10.1021/bi00475a022. [DOI] [PubMed] [Google Scholar]
- 150.Cornilescu G, Delaglio F, Bax A. J. Biomol. NMR. 1999;13:289. doi: 10.1023/a:1008392405740. [DOI] [PubMed] [Google Scholar]
- 151.Hansen MR, Mueller L, Pardi A. Nat. Struct. Biol. 1998;5:1065. doi: 10.1038/4176. [DOI] [PubMed] [Google Scholar]
- 152.Sanders CR, Schwonek JP. Biochemistry. 1992;31:8898. doi: 10.1021/bi00152a029. [DOI] [PubMed] [Google Scholar]
- 153.Koenig BW, Hu JS, Ottiger M, Bose S, Hendler RW, Bax A. J. Am. Chem. Soc. 1999;121:1385. [Google Scholar]
- 154.Prosser RS, Losonczi JA, Shiyanovskaya IV. J. Am. Chem. Soc. 1998;120:11010. [Google Scholar]
- 155.Rückert M, Otting G. J. Am. Chem. Soc. 2000;122:7793. [Google Scholar]
- 156.Raleigh DP, Levitt MH, Griffin RG. Chem. Phys. Lett. 1988;146:71. [Google Scholar]
- 157.Gullion T, Schaefer J. Detection of weak heteronuclear dipolar coupling by rotational-echo double-resonance NMR, in. In: Warren WS, editor. Advances in Magnetic Resonance; Conference on "High Resolution NMR in Solids"; January 19–21; San Diego, California, USA; London, England, UK: Academic Press, Inc; 1989. p. 57. [Google Scholar]
- 158.Bennett AE, Ok JH, Griffin RG, Vega S. J. Chem. Phys. 1992;96:8624. [Google Scholar]
- 159.Sodickson DK, Levitt MH, Vega S, Griffin RG. J. Chem. Phys. 1993;98:6742. [Google Scholar]
- 160.Lee YK, Kurur ND, Helmle M, Johannessen OG, Nielsen NC, Levitt MH. Chem. Phys. Lett. 1995;242:304. [Google Scholar]
- 161.Rienstra CM, Hatcher ME, Mueller LJ, Sun BQ, Fesik SW, Griffin RG. J. Am. Chem. Soc. 1998;120:10602. [Google Scholar]
- 162.Takegoshi K, Nakamura S, Terao T. Chem. Phys. Lett. 2001;344:631. [Google Scholar]
- 163.Ramamoorthy A, Opella SJ. Solid State Nucl. Magn. Reson. 1995;4:387. doi: 10.1016/0926-2040(95)00054-t. [DOI] [PubMed] [Google Scholar]
- 164.Dvinskikh SV, Yamamoto K, Ramamoorthy A. Chem. Phys. Lett. 2006;419:168. [Google Scholar]
- 165.Nevzorov AA, Opella SJ. J. Magn. Reson. 2003;164:182. doi: 10.1016/s1090-7807(03)00240-4. [DOI] [PubMed] [Google Scholar]
- 166.Soubias O, Polozov IV, Teague WE, Yeliseev AA, Gawrisch K. Biochemistry. 2006;45:15583. doi: 10.1021/bi061416d. [DOI] [PubMed] [Google Scholar]
- 167.Park SH, Prytulla S, De Angelis AA, Brown JM, Kiefer H, Opella SJ. J. Am. Chem. Soc. 2006;128:7402. doi: 10.1021/ja0606632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Marassi FM, Opella SJ. J. Magn. Reson. 2000;144:150. doi: 10.1006/jmre.2000.2035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Mesleh MF, Opella SJ. J. Magn. Reson. 2003;163:288. doi: 10.1016/s1090-7807(03)00119-8. [DOI] [PubMed] [Google Scholar]
- 170.Gröbner G, Burnett IJ, Glaubitz C, Choi G, Mason AJ, Watts A. Nature. 2000;405:810. doi: 10.1038/35015604. [DOI] [PubMed] [Google Scholar]
- 171.Salgado GFJ, Struts AV, Tanaka K, Fujioka N, Nakanishi K, Brown MF. Biochemistry. 2004;43:12819. doi: 10.1021/bi0491191. [DOI] [PubMed] [Google Scholar]
- 172.Salgado GFJ, Struts AV, Tanaka K, Krane S, Nakanishi K, Brown MF. J. Am. Chem. Soc. 2006;128:11067. doi: 10.1021/ja058738+. [DOI] [PubMed] [Google Scholar]
- 173.Struts AV, Salgad GFJ, Tanaka K, Krane S, Nakanishi K, Brown MF. J. Mol. Biol. 2007;372:50. doi: 10.1016/j.jmb.2007.03.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Brown MF, Martinez-Mayorga K, Nakanishi K, Salgado GFJ, Struts AV. Photochem. Photobiol. 2009;85:442. doi: 10.1111/j.1751-1097.2008.00510.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Cohen GB, Yang T, Robinson PR, Oprian DD. Biochemistry. 1993;32:6111. doi: 10.1021/bi00074a024. [DOI] [PubMed] [Google Scholar]
- 176.Vogel R, Siebert F. J. Biol. Chem. 2001;276:38487. doi: 10.1074/jbc.M105423200. [DOI] [PubMed] [Google Scholar]
- 177.Feng X, Verdegem PJE, Lee YK, Sandström D, Eden M, Bovee-Geurts P, Degrip WJ, Lugtenburg J, Degroot HJM, Levitt MH. J. Am. Chem. Soc. 1997;119:6853. [Google Scholar]
- 178.Feng X, Verdegem PJE, Eden M, Sandström D, Lee YK, Bovee-Geurts PHM, De Grip WJ, Lugtenburg J, de Groot HJM, Levitt MH. J. Biomol. NMR. 2000;16:1. doi: 10.1023/a:1008377231625. [DOI] [PubMed] [Google Scholar]
- 179.Verdegem PJE, Bovee-Geurts PHM, De Grip WJ, Lugtenburg J, de Groot HJM. Biochemistry. 1999;38:11316. doi: 10.1021/bi983014e. [DOI] [PubMed] [Google Scholar]
- 180.Concistrè M, Gansmüller A, McLean N, Johannessen OG, Montesinos IM, Bovee-Geurts PHM, Brown RCD, DeGrip WJ, Levitt MH. J. Am. Chem. Soc. 2009;131:6133. doi: 10.1021/ja809878c. [DOI] [PubMed] [Google Scholar]
- 181.Concistrè M, Gansmüller A, McLean N, Johannessen OG, Montesinos IM, Bovee-Geurts PHM, Verdegem P, Lugtenburg J, Brown RCD, DeGrip WJ, Levitt MH. J. Am. Chem. Soc. 2008;130:10490. doi: 10.1021/ja803801u. [DOI] [PubMed] [Google Scholar]
- 182.Gansmüller A, Concistrè M, McLean N, Johannessen OG, Marin-Montesinos I, Bovee-Geurts PHM, Verdegem P, Lugtenburg J, Brown RCD, DeGrip WJ, Levitt MH. Biochim. Biophys. Acta-Biomembr. 2009;1788:1350. doi: 10.1016/j.bbamem.2009.02.018. [DOI] [PubMed] [Google Scholar]
- 183.Cooper A. Nature. 1979;282:531. doi: 10.1038/282531a0. [DOI] [PubMed] [Google Scholar]
- 184.Smith SO, Palings I, Miley ME, Courtin J, de Groot H, Lugtenburg J, Mathies RA, Griffin RG. Biochemistry. 1990;29:8158. doi: 10.1021/bi00487a025. [DOI] [PubMed] [Google Scholar]
- 185.Verhoeven MA, Creemers AFL, Bovee-Geurts PHM, De Grip WJ, Lugtenburg J, de Groot HJM. Biochemistry. 2001;40:3282. doi: 10.1021/bi0023798. [DOI] [PubMed] [Google Scholar]
- 186.Han M, DeDecker BS, Smith SO. Biophys. J. 1993;65:899. doi: 10.1016/S0006-3495(93)81117-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 187.Han M, Smith SO. Biochemistry. 1995;34:1425. doi: 10.1021/bi00004a037. [DOI] [PubMed] [Google Scholar]
- 188.Ahuja S, Eilers M, Hirshfeld A, Yan ECY, Ziliox M, Sakmar TP, Sheves M, Smith SO. J. Am. Chem. Soc. 2009;131:15160. doi: 10.1021/ja9034768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 189.Okada T, Fujiyoshi Y, Silow M, Navarro J, Landau EM, Shichida Y. Proc. Natl. IAcad. Sci. U.S.A. 2002;99:5982. doi: 10.1073/pnas.082666399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 190.de Groot HJM, Smith SO, Courtin J, van den Berg E, Wnkel C, Lugtenburg J, Griffin RG, Herzfeld J. Biochemistry. 1990;29:6873. doi: 10.1021/bi00481a017. [DOI] [PubMed] [Google Scholar]
- 191.Steinberg G, Ottolenghi M, Sheves M. Biophys. J. 1993;64:1499. doi: 10.1016/S0006-3495(93)81518-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 192.Ratnala VRP, Swarts HGP, VanOostrum J, Leurs R, DeGroot HJM, Bakker RA, DeGrip WJ. Eur. J. Biochem. 2004;271:2636. doi: 10.1111/j.1432-1033.2004.04192.x. [DOI] [PubMed] [Google Scholar]
- 193.Miyamoto K, Iwase M, Nyui M, Arata S, Sakai Y, Gabazza EC, Kimura H, Homma I. Int. Arch. Allergy Immunol. 2006;140:215. doi: 10.1159/000093246. [DOI] [PubMed] [Google Scholar]
- 194.Ratnala VRP, Kiihne SR, Buda F, Leurs R, de Groot HJM, DeGrip WJ. J. Am. Chem. Soc. 2007;129:867. doi: 10.1021/ja0652262. [DOI] [PubMed] [Google Scholar]
- 195.Luca S, White JF, Sohal AK, Filippov DV, van Boom JH, Grisshammer R, Baldus M. Proc. Natl. Acad. Sci. U.S.A. 2003;100:10706. doi: 10.1073/pnas.1834523100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 196.White JF, Trinh LB, Shiloach J, Grisshammer R. FEBS Lett. 2004;564:289. doi: 10.1016/S0014-5793(04)00195-4. [DOI] [PubMed] [Google Scholar]
- 197.Murphy PM, Baggiolini M, Charo IF, Hebert CA, Horuk R, Matsushima K, Miller LH, Oppenheim JJ, Power CA. Pharmacol. Rev. 2000;52:145. [PubMed] [Google Scholar]
- 198.Rossi D, Zlotnik A. Annu. Rev. Immunol. 2000;18:217. doi: 10.1146/annurev.immunol.18.1.217. [DOI] [PubMed] [Google Scholar]
- 199.Olson TS, Ley K. Am. J. Physiol.-Regul. Integr. Comp. Physiol. 2002;283:R7. doi: 10.1152/ajpregu.00738.2001. [DOI] [PubMed] [Google Scholar]
- 200.Campbell JJ, Butcher EC. Curr. Opin. Immunol. 2000;12:336. doi: 10.1016/s0952-7915(00)00096-0. [DOI] [PubMed] [Google Scholar]
- 201.Gerard C, Rollins BJ. Nat. Immunol. 2001;2:108. doi: 10.1038/84209. [DOI] [PubMed] [Google Scholar]
- 202.Allen SJ, Crown SE, Handel TM. Annu. Rev. Immunol. 2007;25:787. doi: 10.1146/annurev.immunol.24.021605.090529. [DOI] [PubMed] [Google Scholar]
- 203.Kofuku Y, Yoshiura C, Ueda T, Terasawa H, Hirai T, Tominaga S, Hirose M, Maeda Y, Takahashi H, Terashima Y, Matsushima K, Shimada I. J. Biol. Chem. 2009;284:35240. doi: 10.1074/jbc.M109.024851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 204.Shi L, Liapakis G, Xu R, Guarnieri F, Ballesteros JA, Javitch JA. J. Biol. Chem. 2002;277:40989. doi: 10.1074/jbc.M206801200. [DOI] [PubMed] [Google Scholar]
- 205.Hornak V, Ahuja S, Eilers M, Goncalves JA, Sheves M, Reeves PJ, Smith SO. J. Mol. Biol. 2010;396:510. doi: 10.1016/j.jmb.2009.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 206.Corson DW, Cornwall MC, MacNichol EF, Tsang S, Derguini F, Crouch RK, Nakanishi K. Proc. Natl. Acad. Sci. U.S.A. 1994;91:6958. doi: 10.1073/pnas.91.15.6958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 207.Han M, Groesbeek M, Sakmar TP, Smith SO. Proc. Natl. Acad. Sci. U.S.A. 1997;94:13442. doi: 10.1073/pnas.94.25.13442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 208.Bokoch MP, Zou YZ, Rasmussen SGF, Liu CW, Nygaard R, Rosenbaum DM, Fung JJ, Choi HJ, Thian FS, Kobilka TS, Puglisi JD, Weis WI, Pardo L, Prosser RS, Mueller L, Kobilka BK. Nature. 2010;463:108. doi: 10.1038/nature08650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 209.Imai H, Kojima D, Oura T, Tachibanaki S, Terakita A, Shichida Y. Proc. Natl. Acad. Sci. U.S.A. 1997;94:2322. doi: 10.1073/pnas.94.6.2322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 210.Patel AB, Crocker E, Reeves PJ, Getmanova EV, Eilers M, Khorana HG, Smith SO. J. Mol. Biol. 2005;347:803. doi: 10.1016/j.jmb.2005.01.069. [DOI] [PubMed] [Google Scholar]
- 211.Liapakis G, Ballesteros JA, Papachristou S, Chan WC, Chen X, Javitch JA. J. Biol. Chem. 2000;275:37779. doi: 10.1074/jbc.M002092200. [DOI] [PubMed] [Google Scholar]
- 212.Strader CD, Candelore MR, Hill WS, Sigal IS, Dixon RAF. J. Biol. Chem. 1989;264:13572. [PubMed] [Google Scholar]
- 213.Marjamäki A, Pihlavisto M, Cockcroft V, Heinonen P, Savola JM, Scheinin M. Mol. Pharmacol. 1998;53:370. doi: 10.1124/mol.53.3.370. [DOI] [PubMed] [Google Scholar]
- 214.Kisselev OG, Kao J, Ponder JW, Fann YC, Gautam N, Marshall GR. Proc. Natl. Acad. Sci. U.S.A. 1998;95:4270. doi: 10.1073/pnas.95.8.4270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 215.Koenig BW, Mitchell DC, Konig S, Grzesiek S, Litman BJ, Bax A. J. Biomol. NMR. 2000;16:121. doi: 10.1023/a:1008378523816. [DOI] [PubMed] [Google Scholar]
- 216.Koenig BW, Kontaxis G, Mitchell DC, Louis JM, Litman BJ, Bax A. J. Mol. Biol. 2002;322:441. doi: 10.1016/s0022-2836(02)00745-3. [DOI] [PubMed] [Google Scholar]
- 217.Ridge KD, Marino JP, Ngo T, Ramon E, Brabazon DM, Abdulaev NG. Vision Res. 2006;46:4482. doi: 10.1016/j.visres.2006.07.024. [DOI] [PubMed] [Google Scholar]
- 218.Brabazon DM, Abdulaev NG, Marino JP, Ridge KD. Biochemistry. 2003;42:302. doi: 10.1021/bi0268899. [DOI] [PubMed] [Google Scholar]
- 219.Nygaard R, Frimurer TM, Holst B, Rosenkilde MM, Schwartz TW. Trends Pharmacol. Sci. 2009;30:249. doi: 10.1016/j.tips.2009.02.006. [DOI] [PubMed] [Google Scholar]
- 220.Ahuja S, Smith SO. Trends Pharmacol. Sci. 2009;30:494. doi: 10.1016/j.tips.2009.06.003. [DOI] [PubMed] [Google Scholar]
- 221.Mitchell DC, Litman BJ. J. Biol. Chem. 2000;275:5355. doi: 10.1074/jbc.275.8.5355. [DOI] [PubMed] [Google Scholar]
- 222.Etzkorn M, Martell S, Andronesi OC, Seidel K, Engelhard M, Baldus M. Angew. Chem. Int. Ed. Engl. 2007;46:459. doi: 10.1002/anie.200602139. [DOI] [PubMed] [Google Scholar]
- 223.Soubias O, Gawrisch K. J. Am. Chem. Soc. 2005;127:13110. doi: 10.1021/ja0538942. [DOI] [PubMed] [Google Scholar]
- 224.Soubias O, Teague WE, Gawrisch K. J. Biol. Chem. 2006;281:33233. doi: 10.1074/jbc.M603059200. [DOI] [PubMed] [Google Scholar]
- 225.Grossfield A, Feller SE, Pitman MC. Proc. Natl. Acad. Sci. U.S.A. 2006;103:4888. doi: 10.1073/pnas.0508352103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 226.Wiedmann TS, Pates RD, Beach JM, Salmon A, Brown MF. Biochemistry. 1988;27:6469. doi: 10.1021/bi00417a041. [DOI] [PubMed] [Google Scholar]
- 227.Mak-Jurkauskas ML, Bajaj VS, Hornstein MK, Belenky M, Griffin RG, Herzfeld J. Proc. Natl. Acad. Sci. U.S.A. 2008;105:883. doi: 10.1073/pnas.0706156105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 228.Liu W, Eilers M, Patel AB, Smith SO. J. Mol. Biol. 2004;337:713. doi: 10.1016/j.jmb.2004.02.001. [DOI] [PubMed] [Google Scholar]