Summary
Topoisomerase V is an archaeal type I topoisomerase that is unique among topoisomerases due to presence of both topoisomerase and DNA repair activities in the same protein. It is organized as an N-terminal topoisomerase domain followed by 24 tandem helix hairpin helix (HhH) motifs. Structural studies have shown that the active site is buried by the (HhH) motifs. Here we show that the N-terminal domain can relax DNA in the absence of any HhH motifs and that the HhH motifs are required for stable protein-DNA complex formation. Crystal structures of various topoisomerase V fragments show changes in the relative orientation of the domains mediated by a long bent linker helix, and these movements are essential for the DNA to enter the active site. Phosphate ions bound to the protein near the active site helped model DNA in the topoisomerase domain and shows how topoisomerase V may interact with DNA.
Introduction
DNA topoisomerases are enzymes found in all forms of life (bacteria, eukarya, and archaea) and they regulate the topological state of DNA inside the cell. They form a transient break in a single or double stranded DNA and allow the passage of another single or double DNA strand through the break, before resealing the break (Champoux, 2001) (Schoeffler and Berger, 2008). As a result of this, topoisomerases can relax supercoiled DNA, help in the segregation of DNA strands following replication, and lead to the formation and resolution of knots and catenates (Gellert, 1981). Topoisomerases participate in many aspects of DNA metabolism, such as replication, recombination, and transcription (Champoux, 2001). In addition, they are targets of various anti-cancerous drugs and anti-bacterial agents (Maxwell, 1999; Pommier, 1998; Rothenberg, 1997; Wang et al., 1997).
DNA topoisomerases are broadly classified into two types, type I and type II enzymes. Type I enzymes cleave a single strand of a DNA molecule and pass another single or double stranded DNA through the break before resealing the opening. Type II enzymes cleave both strands of a double stranded DNA in concert and pass another double stranded DNA through the break. Type I enzymes use the torsional energy stored in the supercoiled DNA to drive DNA relaxation and hence they do not require high energy cofactors, such as ATP, for their activity (Baker et al., 2009) Type II enzymes, on the other hand, require ATP and Mg2+ for their activity. Type I topoisomerases are further subdivided into three subtypes: IA, IB, and IC (Forterre et al., 2007). Type IA and IB enzymes have been studied extensively (Baker et al., 2009) and there is ample information available about their general mechanism of DNA relaxation and the mode of DNA binding. Type IC, on the other hand, is a relatively new subtype. Currently topoisomerase V is the only member of this family and it has been identified only in the Methanopyrus genus. Previously, topoisomerase V had been considered as a type IB enzyme based on its biochemical characteristics (Slesarev et al., 1993), but the crystal structure of an N-terminal 61 kDa of topoisomerase V (Topo-61) (Taneja et al., 2006) revealed a completely new fold without similarity to other topoisomerases or any other known protein. Furthermore, the orientation of the putative active site residues is also different from other type I topoisomerases, suggesting a different mechanism of cleavage and religation of DNA. These observations, together with the lack of sequence similarity, indicated that topoisomerase V defines a new subtype of type I enzymes (Forterre, 2006).
Topoisomerase V was identified in Methanopyrus kandleri, an extremophile isolated from a deep-water ‘black smoker’ chimney in the Gulf of California (Huber et al., 1989). The enzyme is active at very high temperatures (122°C) and high salt concentrations (0.65 M NaCl and 3.1 M potassium glutamate). The unusual characteristic of topoisomerase V is that it has both topoisomerase and DNA repair activities in the same polypeptide (Belova et al., 2001). Based on the sequence analysis of topoisomerase V, it has been predicted that the protein contains 24 helix-hairpin-helix (HhH) DNA binding motifs arranged as 12 (HhH)2 domains around the N-terminal topoisomerase domain (Belova et al., 2002) (Figure 1A). Some of these (HhH)2 domains are involved in the apurinic/apyrimidinic (AP) site-processing activity, but the exact location of the repair active site is not known yet. Topoisomerase V can relax both positively and negatively supercoiled DNA without the need for metal cations or high energy cofactors. Single molecule experiments have shown that topoisomerase V relaxes DNA by a constrained swiveling mechanism, relaxing around 12 turns of DNA per relaxation cycle (Taneja et al., 2007). Type IB enzymes, which also use a constrained swiveling mechanism for DNA relaxation, relax around 19 turns of DNA per relaxation cycle (Koster et al., 2005).
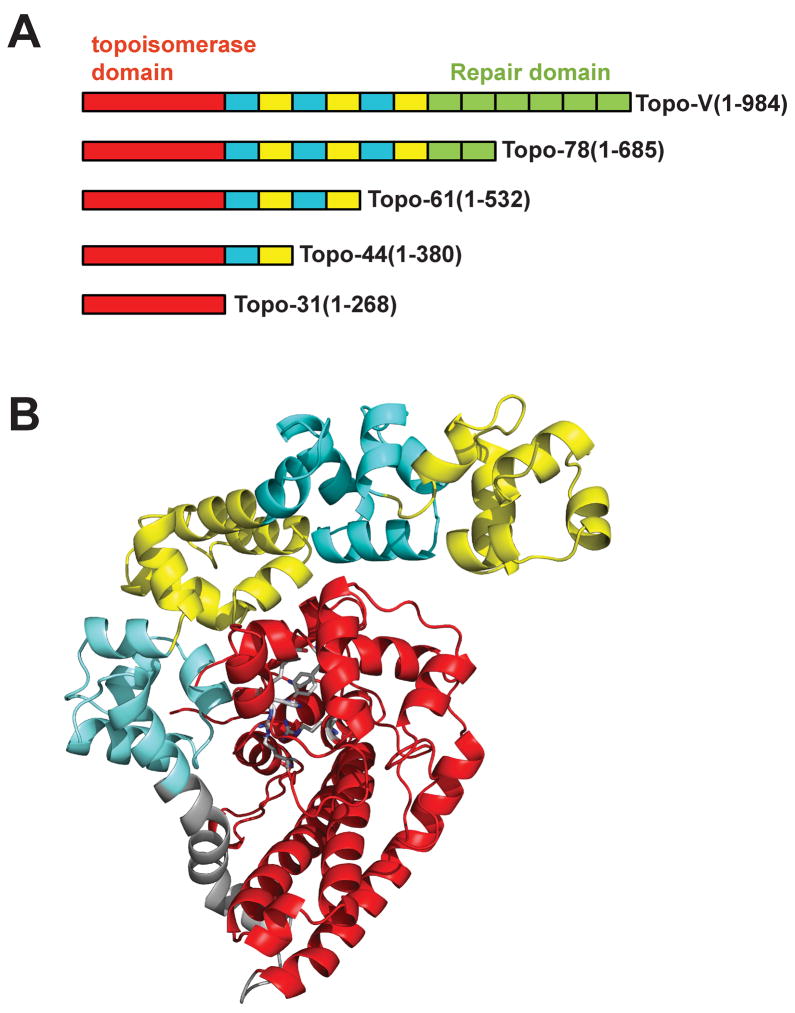
Figure 1. Organization of topoisomerase V.
Topoisomerase V is a multi-domain protein consisting of 24 helix-hairpin-helix (HhH) DNA binding motifs arranged as 12 (HhH)2 domains following the N-terminal topoisomerase domain. A) Schematic diagram of various topoisomerase V fragments. The topoisomerase domain is shown in red, the (HhH)2 domains are shown in alternating colors of cyan and yellow. The (HhH)2 domains with repair activity are shown in green. All fragments shown have topoisomerase activity, but only the full length protein and the Topo78 fragment have repair activity. B) Crystal structure of Topo-61 fragment (Taneja et al., 2006). The coloring scheme is the same as in Figure 1A, except that the linker helix is shown in grey.
The structure of Topo-61 showed that the topoisomerase domain is mainly alpha helical and that the first four (HhH)2 domains curl around the topoisomerase domain (Taneja et al., 2006) (Figure 1B). The topoisomerase and (HhH)2 domains are joined by a long bent helix, termed the “linker helix”. Three of the five putative active site residues are present in a helix-turn-helix (HTH) domain and the other two are present in an intervening loop and a helix. The active site residues are buried by the first (HhH)2 domain and it has been suggested that large conformational changes will be needed for the DNA to access the active site of topoisomerase V (Taneja et al., 2007). Here we present data that shows that the N-terminal 31 kDa fragment of topoisomerase V (Topo-31) has topoisomerase activity, consistent with previous predictions based on the structure. In addition, we show that the Topo-44 fragment (N-terminal 44 kDa fragment of topoisomerase V) can form a stable protein-DNA complex, emphasizing the need of the (HhH)2 domains for binding DNA. We determined a crystal structure of (Topo-31) fragment, which has only the topoisomerase domain, and three different crystal structures of the Topo-44 fragment, which includes the topoisomerase domain and three tandem HhH motifs. In all structures, the topoisomerase domain is very similar. In contrast, the structures of Topo-44 show conformational changes in the linker helix resulting in variable orientations of the (HhH)2 domains when compared to the Topo-61 structure. Phosphate ions are present in the vicinity of the topoisomerase active site in two of the Topo-44 structures. Some of the catalytic residues interact with the phosphate ions and may mimic contacts with DNA. These observations suggest that the movement of the (HhH)2 domains is mediated by the linker helix and helps expose the topoisomerase active site to facilitate DNA binding. In addition, the location of the phosphate ions suggests a possible path for the DNA and the way the active site residues interact with it.
Results
The topoisomerase domain can relax DNA
DNA relaxation assays using different topoisomerase V fragments showed that the topoisomerase domain alone is capable of relaxing DNA. Topoisomerase V fragments with different numbers of (HhH)2 domains, Topo-31, Topo-44, and Topo-78, were studied using relaxation assays. Topo-31 has no (HhH)2 domains, Topo-44 has one full and one partial (HhH)2 domain, while Topo-78 has eight full (HhH)2 domains, including a putative DNA repair domain. In addition to standard conditions, the effect of different pH conditions and presence of magnesium ions were also tested. The experiments show that Topo-31 is capable of relaxing DNA, despite the absence of the (HhH)2 domains (Figure 2B). A pH profile analysis for the DNA relaxation assays showed that Topo-78 relaxes DNA over a wider pH range (pH 5 to 9), while Topo-31 and Topo-44 relax DNA optimally at pH 5 (Figure 2A, 2B, 2C). In addition, magnesium is not required for the reaction, but stimulates it at all pH values (Figure 2B, 2C). Topo-78 can relax DNA to the same extent with lower amounts of protein (0.1 μg/reaction) compared to Topo-44 (~1.5 μg/reaction) and Topo-31 (~9 μg/reaction). This could be due to the enhanced DNA binding facilitated by the (HhH)2 domains. Together, these results suggest that, even though the (HhH)2 domains are dispensable for topoisomerase activity, they enhance DNA relaxation activity. In addition, the pH dependence of the DNA relaxation activity indicates that the reaction is likely to involve side chains with ionizable groups in the low pH range, such as glutamates. Finally, the magnesium independence of the reactions confirms that even the smallest fragments do not require metals for activity, although magnesium has a stimulatory effect. This may be due to favorable interactions of the cations with DNA.
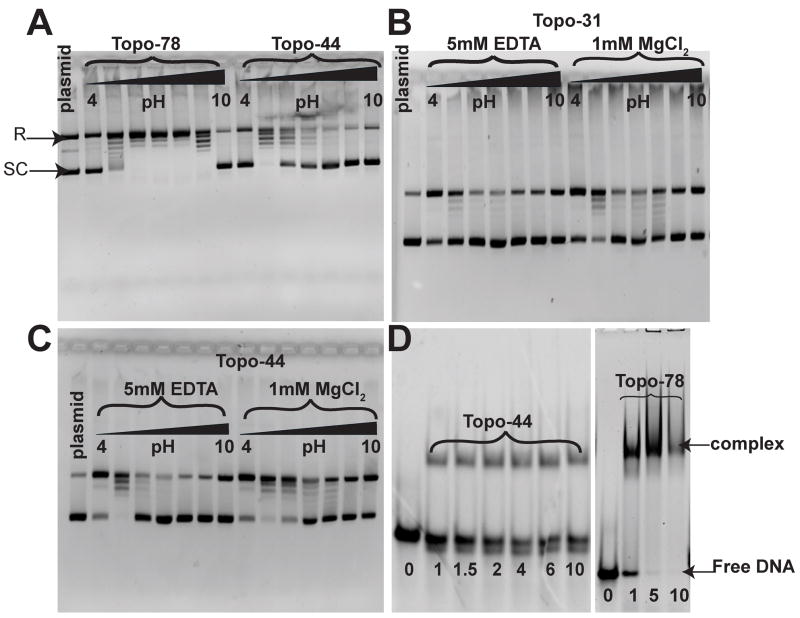
Figure 2. DNA relaxation activity and EMSA for Topo-31, Topo-44 and Topo-78 fragments of topoisomerase V.
A) pH profile of the DNA relaxation activity of Topo-78 and Topo-44 fragments. 0.2 μg of pUC19 DNA were incubated with 0.1 μg of Topo-78 or 1.5 μg of Topo-44 proteins at 65°C for 15 minutes for the relaxation reaction. The reaction buffer contained 50 mM of the appropriate buffer, 30 mM NaCl and 0.2 mM EDTA. Topo-78 relaxes DNA at a wider pH range (5 to 9) than Topo-44, which relaxes DNA efficiently only at pH 5. DNA relaxation activity of Topo-31 (B) and Topo-44 (C) fragments in the absence and presence of MgCl2. 0.2 μg of pUC19 DNA were incubated with 9 μg of Topo-31 or 1.5 μg of Topo-44 proteins at 65°C for 15 minutes for the relaxation reaction. The reaction buffer contained 50 mM of the appropriate buffer, 30 mM NaCl and 5 mM EDTA or 1 mM MgCl2. Both Topo-31 and Topo-44 fragments can relax DNA in the absence of MgCl2, but MgCl2 enhances the DNA relaxation activity of the topoisomerase V fragments. The black triangle in panels A, B and C represents increasing pH from 4 to 10 by one pH unit. D) EMSA of Topo-44 and Topo-78 fragments with a 39mer double stranded DNA. Both Topo-44 and Topo-78 form stable complexes with DNA, although Topo-78 seems to saturate DNA binding while Topo-44 does not. In addition, Topo-44 shows some cleavage of the DNA (bottom free DNA band), while the cleavage is not apparent in Topo-78. The numbers at the bottom represent the molar ratio of protein to DNA.
The (HhH)2 domains enhance DNA binding affinity
EMSA experiments with different fragments of topoisomerase V and DNA showed that (HhH)2 domains could help in the formation of a stable protein-DNA complex. Various topoisomerase V fragments (Topo-31, Topo-44, and Topo-78) and single and double stranded DNA were analyzed by EMSA experiments. Topo-44 and Topo-78 formed stable complexes with a 39mer double stranded DNA (Figure 2D), while no DNA binding was observed for the Topo-31 fragment (data not shown). These observations indicate that (HhH)2 domains are necessary for a stable protein-DNA complex and that as few as one and half (HhH)2 domains are enough for formation of a stable protein-DNA complex. EMSA with single stranded DNA showed that Topo-31 and Topo-44 cannot bind to single stranded DNA, while Topo-78 can bind to single stranded DNA (data not shown).
Overall Structures
The topoisomerase domain of topoisomerase V is a helical-rich compact domain that has no structural similarity to any other known protein. The only recognizable structural element is a HTH that contains some of the active site residues. Not surprisingly, the topoisomerase domain of the four structures (Topo-31, Topo-44 (Forms I, II, and III)) superimpose very well on each other and also to that from the Topo-61 structure. In the Topo-31 structure, two surface loops, residues 39-49 and 120-124, adopt a different conformation compared to the Topo-61 and Topo-44 structures. These two loops are not always visible in the Topo-44 structures, suggesting that they are mobile regions. The r.m.s.d. for the superposition of the topoisomerase core domain of all the new structures on to the Topo-61 structure range from 0.2 Å to 0.7 Å if the two mobile surface loops are not included (Figure 3A). In general, the topoisomerase domain remains unchanged and is identical in all structures. The (HhH)2 domains also remain largely unchanged, with r.m.s.d. for the superposition of only the (HhH)2 domains from the three Topo-44 crystal forms and equivalent domains in the Topo-61 structure ranging from 0.31 Å to 0.56 Å.
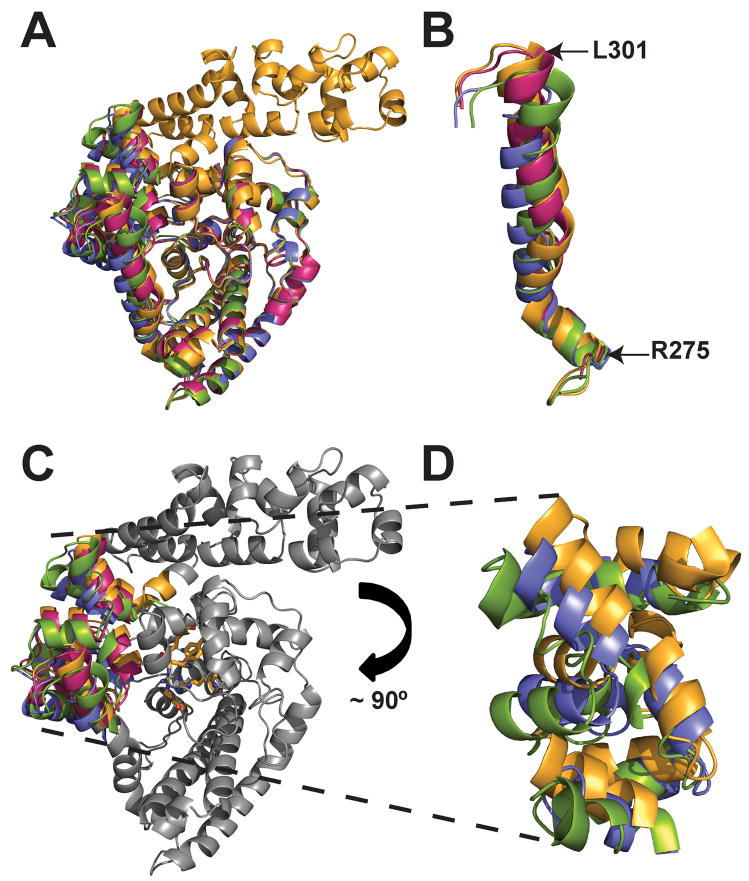
Figure 3. Structure of Topo-44 fragments.
A) Overlay of Form I (green), Form II (B subunit: blue), and Form III (B subunit: magenta) structures onto the Topo-61 structure (B subunit: orange). The topoisomerase domains superimpose very well for all the structures, while the linker helix and (HhH)2 domains show differences in orientation. B) Overlay of the linker helices of Form I, II, and III structures with that of Topo-61. The color scheme is same for all the figures unless mentioned otherwise. Note that the linker helices have the same orientation at the start and they change as they move further down the helix. C) Superposition of Form I, II, and III Topo-44 structures with that of Topo-61. Only the (HhH)2 domains are colored while the remaining parts are shown in gray for clarity. The active site residues are shown as orange sticks. Note that the (HhH)2 domains adopt different orientations in all the structures. D) Orientation of the (HhH)2 domains of Form I, II and Topo-61 structures. In Form I and II structures, the (HhH)2 domains are moved away from the topoisomerase domain. For clarity, the (HhH)2 domains of Form III are not shown. In panels C and D, the topoisomerase domains were superposed to emphasize the different orientation of the other domains.
The five crystallographically independent structures of Topo-44 (Form I, Form II A and B monomers, and Form III A and B monomers) were compared with each other and to the two crystallographically independent Topo-61 monomers to understand the conformational changes in the protein. The r.m.s.d. for the superposition of all the Topo-44 structures (residues 3-375) on to the Topo-61 fragment or on each other vary between 0.9 Å and 2.7 Å, with the majority above 1.5 Å, showing that in general the structures have slightly different conformations. As mentioned above, the different domains behave as rigid or almost rigid subunits and the only change in the structure is the relative orientation between the topoisomerase and the (HhH)2 domains. The change in orientation of the domains starts at the linker helix (residues 269-295), which acts as a hinge region, and follows into the (HhH)2 domains. At the start of the linker helix, the structures superimpose very well for all five Topo-44 and two Topo-61 structures. In the middle of the linker helix there is a kink after which the linker helix from all the structures shows different orientations (Figure 3B). The flexibility of the linker helix is also evident by the fact that the linker helix in the B subunit of Form III crystals appears in two alternate conformations. The change in the relative orientation of the (HhH)2 and topoisomerase domains (Figure 3C and 3D), suggests that these domains can adopt different orientations and these movements might be necessary for the DNA to access the active site.
The topoisomerase domain has a positively charged groove adjacent to the active site
The structure of the Topo-31 as well as the structures of the Topo-44 fragment reveals the presence of a positively charged groove in the protein that encompasses the active site region (shown later in Figure 6C). This charged groove had been observed before in the structure of the Topo-61 fragment, although several (HhH)2 motifs partially obstruct it (Taneja et al., 2006). The structure of the Topo-31 confirms the presence of the groove even in the absence of the (HhH)2 motifs. The groove is long and can be deep in some areas. It includes regions of the HTH motifs and extends all the way to the linker helix. All the residues forming the active site pentad point towards the groove. The active site tyrosine, Tyr226, is found near one of the ends of the groove, a region where it widens. The positively charged character of the groove and its presence by the active site strongly suggest that it may be involved in DNA binding.
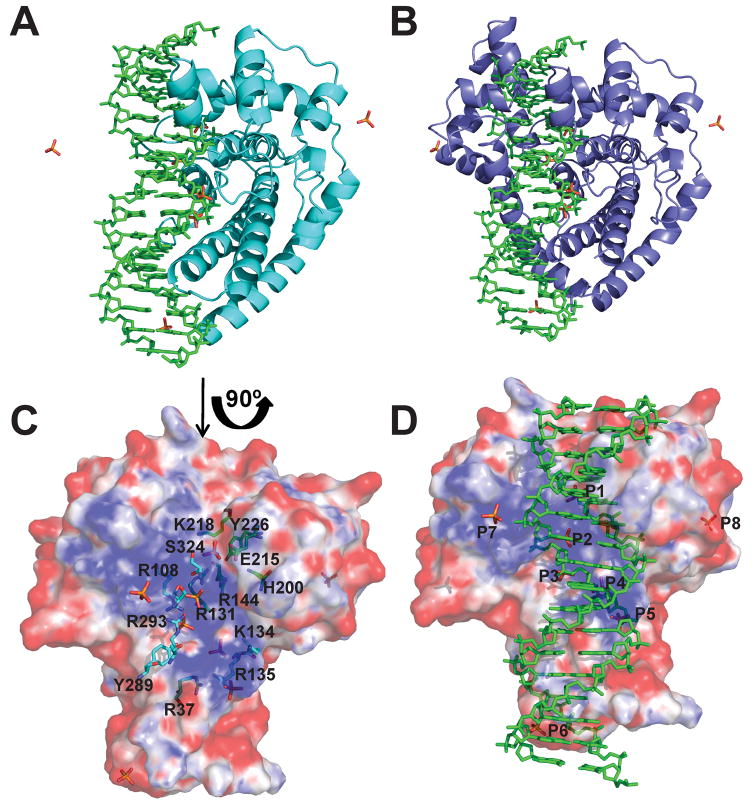
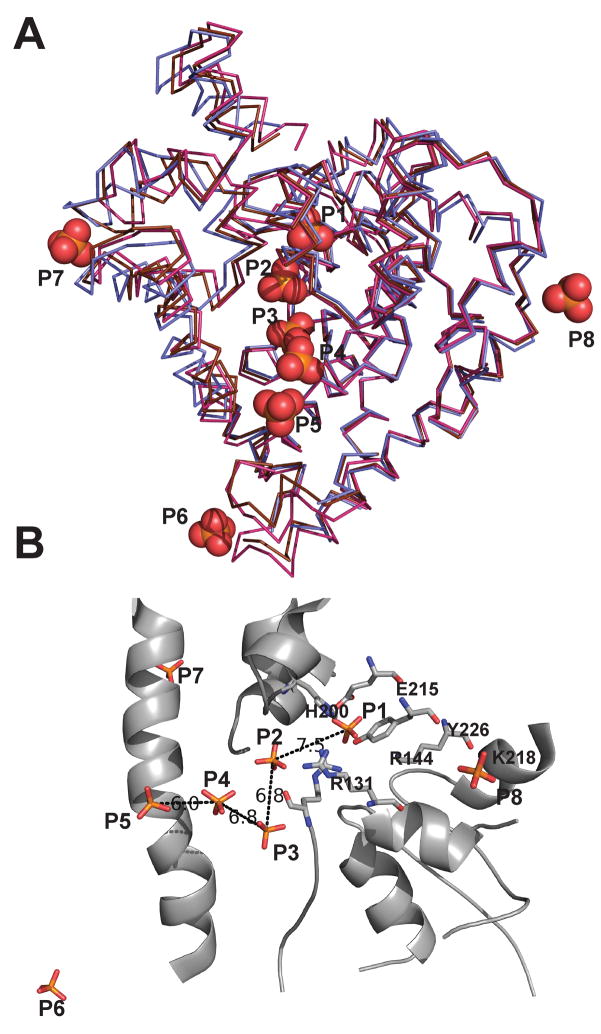
Figure 6. Model showing DNA bound to the topoisomerase domain.
A) Model of a 17-mer double stranded DNA bound to the Topo-31 structure (teal). The DNA is represented as green sticks, where as phosphate ions are represented as orange sticks. DNA binds along the DNA binding groove and five of the eight phosphate ions noted in the Topo-44 structures coincide with the DNA backbone. B) Model of Topo-44 (Form II, B subunit: blue) binding to 17-mer double stranded DNA. Note that the linker helix and the (HhH)2 domains interfere with DNA binding to the topoisomerase domain and are likely to move away to allow binding. C) Electrostatic surface representation of the Topo-31 structure. The positively charged DNA binding groove is clearly visible and the phosphate ions are bound in this groove. The orientation corresponds to a 90° rotation of the one shown in Figure 6A in the direction of the arrow. Note that the DNA binding groove goes from one end of the molecule to the other and it is narrower at one end (start of the linker helix) and wider at the other end. The putative active site residues (green sticks) are located at the wider end of the groove. Other residues lining the groove and interacting with the phosphate ions are shown as cyan sticks. D) Electrostatic surface representation of Topo-31 with phosphate ions (orange) and DNA (green). Three phosphate ions (P3, P4, and P5) coincide with the phosphates of one of the DNA strands, where as P1 coincides with a phosphate of the opposite DNA strand. The model shows that the DNA binding groove of topoisomerase V is wide enough to bind DNA and that the movement of linker helix and (HhH)2 domains are required to accommodate the DNA. The electrostatic potential was calculated with a dielectric constant of 80 for solvent and 2 for protein. The surface is colored with a blue to red gradient from +10 to −10 KbT/ec.
Phosphate ions bind in the groove near the topoisomerase active site
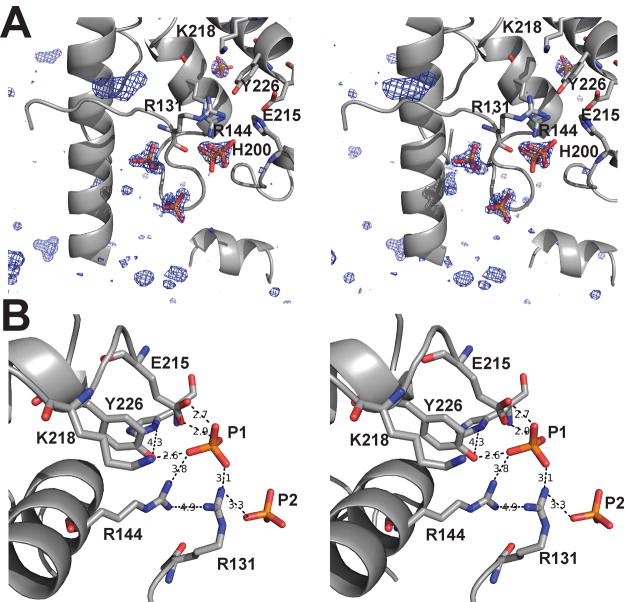
An interesting observation stemming from the Form II and Form III Topo-44 structures is the presence of phosphate ions near the positively charged DNA binding groove. All three Topo-44 crystal forms were crystallized in the presence of phosphate-citrate buffer, but only Form II and Form III structures showed phosphate ions bound to the protein, which were assigned based on electron density consistent with a tetrahedral phosphate ion (Figure 4A). Form II and Form III crystals include 1–1.2 M guanidium hydrochloride in the crystallization solution. The high resolution Form III structure shows clear density for three guanidium ions bound to the protein, two very well ordered and one with weak density. The presence of guanidium hydrochloride in the crystals appears to trigger a conformational change allowing the binding of phosphate ions to the protein. It is interesting to note that Form I crystals did not show any bound phosphate albeit its presence in the crystallization condition. This could be due to the absence of guanidium hydrochloride to trigger the binding of phosphate ions as observed in Form II and Form III structures. There are three phosphate ions in the B subunit and none in the A subunit of the Form II Topo-44 structure. Two of the phosphates are in the topoisomerase active site and one of them forms close contacts with the putative active site residues in the topoisomerase domain (Figure 4B). Form III crystal has seven phosphate ions, three in each subunit and one between both the subunits. In the Form III structure, the phosphate ion near the active site Tyr226 is absent, but it shows several new locations for phosphate ions, especially in the positively charged groove containing the topoisomerase active site (Figure 5A). An overlay of the A and B subunits of the Topo-44 Form III structure with the B subunit of Topo-44 Form II structure shows eight unique phosphate ions (Figure 5A). It clearly shows that there are more phosphate ions bound in the positively charged groove compared to other regions of the protein.
Figure 4. Phosphate ions present near the active site of the Topo-44 structure.
A) Stereo view of a Form III difference electron density map calculated with a model not including the phosphates. The electron density is contoured at 3.7σ and shows the tetrahedral shape of the phosphate ions. The active site residues are shown in stick. B) Stereo view of the interaction of the phosphate ions with the putative active site residues. The B subunit of Form II structure was superimposed onto the B subunit of Form III structure and the phosphates ions from both structures are shown together with the Form II B subunit protein backbone. The interactions made by the phosphate ion with the active site residues and the corresponding distances in Å are represented as black dotted lines.
Figure 5. Representation of the unique phosphate ions from Form II and Form III Topo-44 structures.
A) An overlay of the A (magenta) and B (brown) subunits of the Form III structure and B (blue) subunit of Form II Topo-44 structures. The positions of eight unique phosphate ions (orange spheres) are shown. Note that most phosphate ions are found along the DNA binding groove of the topoisomerase domain. B) The phosphate ions in the DNA binding groove are separated by distances of around 7 Å. The protein backbone is that of the B subunit of Form III structure. The active site residues are represented as sticks and distances in Å between adjacent phosphate ions are shown as black dotted lines.
Taking into account all structures, there are five unique phosphate ion binding sites in the putative DNA binding groove and an additional one near its end and close to the start of the linker helix. Several pairs of phosphates in the groove are separated by a distance of around 7 Å (Figure 5B), which would be consistent with the phosphate-phosphate distance in adjacent nucleotides in a DNA double helix. One of the phosphates (P1) is found near the active site tyrosine and is coordinated by Tyr226 and Arg131, two residues that have been implicated in cleavage and religation of the DNA (Taneja et al., 2006), and by Glu215, whose charge may be important for interactions with DNA (R.R. and A.M., unpublished observations). The side chains of the tyrosine and the glutamate residues are in contact with Arg144 and His200, the other putative active site residues, and these interactions may help to orient them for the catalytic reaction. Adjacent to P1, there is a second phosphate (P2) at a distance of 7.5 Å which is trapped between the topoisomerase domain and an HhH motif. P2 is coordinated by Arg131, an active site residue, in addition to Arg108 from the topoisomerase domain and Arg293 and serines 322 and 324 from the second HhH motif (Figure 6C). Three more phosphates are found in the groove (P3, P4, and P5) coordinated mainly by positively charged residues, such as Arg37, Lys47, Arg108, Lys134, and Arg135 from the topoisomerase domain and also residues from the linker helix such as Tyr289 and Arg293. In general, some of the side chains can contact more than one phosphate. The distance between P3 and P4 and P4 and P5 is 6.8 Å and 6.5 Å respectively. A final phosphate (P6) is located at the start of the linker helix and on the edge of the groove (Figure 5A).
Discussion
Topoisomerase V is active at very high temperatures (122°C) and high salt concentrations. DNA relaxation assays with various topoisomerase V fragments (Topo-44 and Topo-61) show that a temperature above 60° C is required for optimal activity, although longer fragments of topoisomerase V can relax DNA at lower temperatures (Taneja et al., 2007). Topo-44 was first identified by limited proteolytic digestion of the full length topoisomerase V protein (Belova et al., 2002) at 80°C. In contrast, Topo-61 is the shortest fragment showing topoisomerase activity when the proteolytic reaction is performed at 37°C (Belova et al., 2002). The N-terminal Topo-31 fragment, which contains neither HhH motifs nor the linker helix, was identified as the smallest region spanning the topoisomerase domain from the crystal structure of Topo-61 fragment (Taneja et al., 2006) and it was suggested that it could represent the minimal domain capable of relaxing DNA. Relaxation experiments with this minimal domain show that this is indeed the case, although the activity is not as robust as with longer fragments. As expected, Topo-31 does not require magnesium for activity, but magnesium enhances it, as is the case for type IB topoisomerases, which also uses a swiveling mechanism for DNA relaxation (Stewart et al., 1996). The optimal pH for activity for the Topo-31 and the Topo-44 fragments is around 5. This pH dependence is not observed for the Topo-78 fragment. The DNA relaxation by shorter fragments of topoisomerase V at pH 5 could point to the involvement of some ionizable side chains in the relaxation activity. It could also be simply due to the effects of various side chains on DNA binding. Further experiments with different active site mutations in both longer and shorter fragments of topoisomerase V will be required to probe the pH dependence of the relaxation reaction by shorter topoisomerase V fragments.
Gel shift experiments show that Topo-44 and also longer fragments (Topo-78) can bind double stranded DNA. Surprisingly, Topo-31 does not show DNA binding activity in these assays even though it is still capable of relaxing DNA. It appears that the presence of the (HhH)2 domains stabilizes the DNA/protein complex. One possibility is that the (HhH)2 domains could play a similar role to the cap domain present in type IB enzymes, which helps to encircle the DNA during the swiveling reaction (Redinbo et al., 1998). In addition, both short fragments of topoisomerase V do not bind single stranded DNA, whereas Topo-78 can form a stable complex with single stranded DNA (data not shown). (HhH)2 domains binding to single stranded DNA has been observed before. For instance, the N-terminal 8 kDa of mammalian polymerase β, which contains a single HhH motif, binds to single stranded DNA through both helices (Kumar et al., 1990; Liu et al., 1994). The exact mode of single stranded DNA binding by Topo-78 or the possible role in relaxation or repair activities is not yet clear.
The structure of Topo-61 showed that the topoisomerase active site of topoisomerase V is buried by one of the (HhH)2 domains suggesting that conformational changes are essential for the protein to bind DNA. The present structures of Topo-44 reinforce this observation and show that the (HhH)2 domains can change their position relative to the topoisomerase domain and that this change is mediated by the movement of the linker helix. The (HhH)2 domains act as rigid individual units, as evidenced by the fact that in different structures they show the same structure and relative orientation of the two HhH motifs. The topoisomerase domain also appears to be rigid showing the same structure even in the total absence of the rest of the protein. The linker helix (residues 269-295), which is a long bent helix, serves as a hinge for the movement of the (HhH)2 domains away from the rigid topoisomerase domain, possibly by responding to interactions with double stranded DNA. This movement has to be quite large. The Topo-44 structures in the absence of DNA capture the regions that move, but do not show the full extent of the movement or indicate the way the HhH motifs interact with DNA.
As mentioned before, topoisomerase V binds double stranded DNA and has a groove wide enough to accommodate double strand DNA (Figure 6C). The presence of an HTH domain normally associated with DNA binding, the positively charged nature of it, and several phosphates bound along it suggest that this groove could be involved in DNA binding. In addition, the active site is found in this groove and some residues form part of the HTH domain. Previously, DNA was modeled bound to the topoisomerase domain (Taneja et al., 2006) based on the structures of HTH domains in complex with DNA but there was no evidence to support it. Using the phosphates present in the groove in the current structures, it is possible to refine this model. A superposition of the B subunit of Form II and the A and B subunits of Form III Topo-44 structures shows five different phosphate ions in the positively charged groove which are separated by a distance of around 7 Å, consistent with the distance of consecutive phosphates in B DNA of ~6.4 Å. A sixth phosphate ion is found outside the groove near the linker helix. A double stranded DNA molecule was modeled into the groove based on the positions of the phosphate ions (Figure 6). Only five out of the six phosphates could be placed on the DNA molecule, as one of them was inconsistent with a double stranded molecule. Phosphate ions P3, P4, and P5 would correspond to three adjacent phosphates in one DNA strand, while P1, located near the active site, would belong to the opposite strand. A final phosphate (P6) is away from the groove and near the linker helix (Figure 6A, 6B). The fit to the four inner phosphates is excellent and the DNA can be accommodated in the groove of the Topo-31 structure without the need for any major rearrangements of the protein backbone. The fifth phosphate (P6) does not fit as well and a better fit would require movement of either the protein or the DNA, but the change would be relatively modest. Several side chains would need to move, but these changes would also be minor. The major change needed to accommodate the DNA in the structures with the (HhH)2 domains present is the movement of the (HhH)2 domains away from the topoisomerase domain (Figure 6B). The movement of (HhH)2 domains should be feasible as is evident from the Topo-44 structures showing different orientations of the (HhH)2 domains. The location of the (HhH)2 domains after DNA binding is not evident, but one possibility is that they would help enclose the DNA to form a clamp around it, similar to the arrangement in type IB enzymes.
In the model of the topoisomerase domain in complex with DNA, the active site residues are in close contact with the backbone of DNA. The catalytic Tyr226 is pointing towards the phosphate of the DNA backbone; Arg131 and Arg144 are positioned to stabilize the protein-DNA covalent complex. Surprisingly Glu215 also appears to interact directly with the DNA phosphate backbone. The other active site residues like His200 and Lys 218 are also near the DNA. The active site is located near the end of the groove, where it widens. At this end, the DNA fits loosely in the groove, which is spacious to accommodate the movement of the strands. The ‘constrained swiveling’ mechanism employed by type IB and IC enzymes necessitates rotation of one strand about the other after forming the covalent protein-DNA intermediate. The position of the active site at the wider end of the putative DNA binding groove would facilitate the rotation of the DNA strand at this end, while holding the rest of the DNA in place through extensive interactions along the groove.
Even though type IB and IC enzymes have a similar overall mechanism of action, the structures of fragments of topoisomerase V suggest many differences. Type IB enzymes have two domains which come together to form a C-shaped clamp around the DNA (Perry et al., 2006; Redinbo et al., 1998; Stewart et al., 1998) The protein has an open stage where these domains are separate and this helps in the entry and release of the DNA from the protein active site. A wide DNA binding cavity is not observed in the topoisomerase V structures. Instead, the structures show a positively charged groove which is always present in the protein and does not require domain rearrangements to form. DNA can access this groove after a conformational change involving the movement of the (HhH)2 domains exposing the active site. The (HhH)2 domains could help enclose DNA during the swiveling of the DNA, forming a similar enclosure to the one observed for type IB enzymes. It is not known whether all HhH motifs contact DNA simultaneously, but this appears unlikely without a major rearrangement of the motifs. It is likely that only some of the HhH motifs contact DNA at any given time or that some of the motifs do not have the capacity to bind DNA. Finally, similar to type IB enzymes (Cheng and Shuman, 1998), the putative domain enclosing the DNA is dispensable for activity, although it enhances the relaxation activity markedly. Thus, it is likely that type IB and IC enzymes have several overall similarities in the way that they interact with DNA, but the atomic details are markedly different.
There are still many details of the atomic mechanism of type IC topoisomerases that need to be understood. The present functional and structural studies provide new information about topoisomerase V including the observations that the Topo-31 is the minimal fragment capable of DNA relaxation, the (HhH)2 domains enhance binding of the protein to DNA, the changes in relative orientation of the domains is mediated by the linker helix, and several phosphate ions bind in a positively charged groove. Furthermore, the position of the phosphate ions in the groove helped in the placement of DNA in the topoisomerase domain and this provides an initial model of how topoisomerase V interacts with DNA. Thus the present study helps to establish the role of different domains more clearly, to illustrate a mechanism to drive the conformational changes needed for activity, and to suggest a possible manner of binding DNA. Additional work on structures of protein/DNA complexes and intermediates in the swiveling reaction are needed to understand the way this new type of topoisomerases interacts with DNA to perform a complex reaction.
Experimental Procedures
Protein purification
The N-terminal 31 kDa (Topo-31: residues 1-269), and 44 kDa (Topo-44: residues 1 to 380) fragments of topoisomerase V protein were cloned into the pET15b plasmid and transformed into Escherichia coli BL21 Rosetta (DE3) cells. The N-terminal 78 kDa (Topo-78: residues 1 to 685) fragment of topoisomerase V protein was cloned into the pET14b plasmid (Belova et al., 2002) and transformed into Escherichia coli BL21(DE3) cells. For protein production, cells were grown at 37° C in LB medium containing 100 μg/ml ampicillin and 100 μg/ml chloramphenicol for Rosetta cells and LB medium with 100 μg/ml ampicillin for BL21(DE3) cells to an optical density (OD600) of 0.6. The cells were then cooled down on ice, followed by the addition of isopropyl β-D-1-thiogalactopyranoside (IPTG) to a final 0.5 mM concentration, and grown overnight at 16° C. Cells were harvested and resuspended in 50 mM Tris pH 8, 500 mM NaCl, 0.5 mM EDTA, 1 mM DTT, flash frozen in liquid nitrogen and stored at −80° C. After thawing the pellet, pepstatin (1μg/ml), benzamidine (1mM), PMSF (1mM), and Brij 58 (0.1%) were added to the cells and the protein was purified as described earlier (Taneja et al., 2006) The protein was further purified by anion exchange and gel filtration chromatography. Pure protein was concentrated and stored in 50 mM Tris pH 8, 250 mM NaCl, and 1 mM DTT. The seleno-methionine substituted Topo-44 was prepared from cells grown in a minimal medium supplemented with nutrients and salts (Doublie, 1997); protein purification followed the same procedure as for the native protein except that 5mM DTT was used in all the purification steps and for storage.
Relaxation assays
Relaxation assays with the different topoisomerase V fragments were carried out at pH values ranging from 4 to 10. The pH of the buffers was adjusted at 65 °C to account for the change in pH at higher temperature. The different buffers used were: sodium acetate for pH 4 and 5, MES for pH 6, HEPES for pH 7, TRIS for pH 8, CHES for pH 9, and CAPS for pH 10. Topoisomerase activity assays were performed by incubating varying amounts of protein (Topo-31, Topo-44 or Topo-78) with 0.2 μg negatively supercoiled pUC19 DNA in 50 mM of the required buffer, 30 mM NaCl, 0.2 mM or 5 mM EDTA or 1 mM MgCl2. The reactions were carried out at 65 °C for 15 min and terminated by cooling and addition of SDS to a final 1% concentration. The products were resolved on a 1% agarose gel and visualized by ethidium bromide staining.
Electrophoretic Mobility Shift Assay
For Electrophoretic Mobility Shift Assay (EMSA), 4 μM of a 39mer double stranded DNA oligonucleotide (5′ GCGACGCGAGGCTGGATGGCCTTCCCCATTATGATTCTT3′) was incubated with different concentrations of topoisomerase V fragments in 50 mM sodium acetate pH 5, 30 mM NaCl, 1 mM MgCl2 at 65 °C for 30 minutes. Glycerol was added to the reaction mixture to a final concentration of 8% and the products were separated on a 4 % acrylamide native gel. The gel was stained with ethidium bromide to detect the DNA. When a stable protein-DNA complex was formed, there was an upward shift in the band indicating a higher molecular weight complex.
Crystallization
Topo-31 crystals were grown using the sitting drop vapor diffusion method equilibrated against, 23% PEG 6000, 0.1 M Na citrate pH 5.5, at 22°C. For data collection, the Topo-31 crystals were cryo-protected by adding glycerol to the mother liquor to a final 20% concentration. Topo-44 was crystallized by the hanging drop vapor diffusion method under three different crystallization conditions (Forms I, II, and III). Crystal Form I grew under 0.1 M phosphate citrate pH 5, 0.2 M NaCl, 15% PEG 3350 and 8% dioxane. The crystals were cryo-protected by increasing the PEG concentration to 30%. Form II crystals grew under 0.1 M phosphate citrate pH 5, 0.2 M NaCl, 16% PEG 8000 and 1M guanidium hydrochloride. For cryo-protection, they were transferred to a solution with 1.5X reservoir solution and 20% 2,3 butanediol or 20% DMSO for 10 seconds and immediately flash frozen under liquid nitrogen. Form III crystals grew under 0.1 M phosphate citrate pH 5.5, 0.15 M sodium sulfate, 0.01 M MgCl2, 1 M guanidium hydrochloride, and 28 % PEG 3350. The crystals were grown at 30°C and were cryo-protected by increasing the PEG concentration to 40%. Further details of crystallization are presented in the Supplementary Information.
Data collection and structure determination
Diffraction data were collected at the Dupont Northwestern Dow and Life Science Collaborative Access Team stations (DND and LS CAT) at the Advanced Photon Source in Argonne National Laboratory. Data collection and refinement statistics are shown in Table I. All data were processed and integrated using XDS (Kabsch, 1993) and scaled with SCALA (Collaborative-Computational-Project-4, 1994). Data on the Topo-31 crystals were collected to 2.4 Å resolution. The structure was solved by Molecular Replacement (McCoy et al., 2007) using the topoisomerase domain from the Topo-61 structure (residues 1-266) (Taneja et al., 2006) as the search model. It was refined with refmac5 (Murshudov et al., 1997) and Phenix (Afonine et al., 2005) to a final Rwork of 20.0 % and Rfree of 24.8 %. Topo-44 Form I crystals diffract to 1.8 Å. The structure of Form I crystals was solved by Molecular Replacement (McCoy et al., 2007) using the topoisomerase domain from the Topo-61 structure as the search model. Model rebuilding was performed using coot (Emsley and Cowtan, 2004), and refinement using refmac5 (Murshudov et al., 1997). The final Rwork and Rfree are 17.5 % and 22.0 % respectively.
Table 1.
Data collection and refinement statistics
| Topo-31 | Topo-44 Form I | Topo-44 Form II | Topo-44 Form III | |
|---|---|---|---|---|
| Data Collection | ||||
| Space group | C2221 | C121 | P41212 | P212121 |
| Cell dimensions | a=106.7 Å, b=119.4 Å, c=63.7 Å | a=104.2 Å, b=47.7 Å, c=81.2 Å (β=112.48) | a=b=70.1 Å, c=349.6 Å | a=63.6 Å, b=80.1 Å, c=137.2 Å |
| Resolution (Å)a | 79.56 – 2.4 (2.53 – 2.4) | 75.05 – 1.82 (1.91 – 1.82) | 29.5- 2.6 (2.72-2.6) | 28.9-1.4 (1.46-1.4) |
| Number of observed reflections | 78,729 (11,538 | 134,411 (13,220) | 227,408 (19,917) | 1,157,917 (126,319) |
| Number of unique reflections | 16,259 (2,346) | 32,998 (4,301) | 28,151 (3,331) | 136,662 (15,986) |
| Completeness (%) | 99.8 (99.8) | 98.3 (88.6) | 99.9 (100.0) | 98.8 (95.5) |
| Multiplicity | 4.8 (4.9) | 4.1 (3.1) | 8.1 (6.0) | 8.5 (7.9) |
| Rmerge (%)b | 4.7 (71.1) | 4.0 (16.3) | 7.4 (52.2) | 4.5 (37.9) |
| Rmeas (%)c | 5.3 (79.6) | 4.6 (19.4) | 7.9 (57.2) | 4.8 (40.5) |
| ≪I>/σ(<I>)>d | 20.5 (2.5) | 23.0 (6.8) | 19 (3.2) | 27.5 (5.3) |
| Refinement | ||||
| Resolution (Å) | 79.56 - 2.4 (2.46 - 2.4) | 28.06 -1.82 (1.87 – 1.82) | 29.14 – 2.6 (2.67 – 2.6) | 28.9 - 1.4 (1.44 - 1.4) |
| Number of reflections working/test | 15,419/821 | 31,317/1,673 | 26,710/1,438 | 129,802/6,859 |
| Rwork (%)e | 20.0(24.3) | 17.5 (17.9) | 24.1(36.6) | 16.5 (19.3) |
| Rfree(%)f | 24.8 (31.1) | 22.0 (24.8) | 28.9 (45.1) | 18.4 (22.1) |
| Protein residues/atomsg | 269/2,203 | 376/3212 | 727/5,970 | 738/7,511 |
| Atoms in alternate conformations | 0 | 258 (20 protein residues) | 8 (1 protein residue) | 2846 (157 protein residues) |
| Water molecules | 29 | 238 | 30 | 573 |
| Other atoms | - | - | 3 PO4 | 7 PO4, 3 Gmh, 3 Mg++, 2 Cl− |
| B-factor (Å2) | ||||
| Protein atoms (chain) | 68.4 | 22.8 | A:53.8; B:58.2 | A:13.4; B:14.9 |
| Water molecules | 59.1 | 29.3 | 40.0 | 23.7 |
| r.m.s. deviations | ||||
| bond lengths (Å) | 0.015 | 0.006 | 0.01 | 0.009 |
| bond angles (°) | 1.42 | 0.920 | 1.2 | 1.2 |
| Ramachandran ploti | ||||
| Favored regions (%) | 94.3 | 98.9 | 96.2 | 98.5 |
| Outliers (%) | 0.0 | 0.0 | 0.3 | 0 |
Numbers in parenthesis correspond to highest resolution shell.
Rmerge= Σ|I − <I>|/ΣI, where I is the observed intensity and <I> the average intensity obtained from multiple measurements.
Rmeas as described in Diederichs and Karplus (Diederichs and Karplus, 1997).
≪I>/σ(<I>)> = Mean Ih over the standard deviation of the mean Ih averaged over all reflections in a resolution shell.
Rwork= Σ ||Fo| − |Fc||/Σ|Fo|, where |Fo| is the observed structure factor amplitude and |Fc| the calculated structure factor amplitude.
Rfree: Rfactor based on 5% of the data excluded from refinement.
Total number of protein atoms, including those in alternate conformations.
Gm: guanidinum ion.
As reported by Molprobity (Davis et al., 2004).
For Topo-44 Form II and Form III crystals, seleno-methionine derivatized crystals were used for single-wavelength anomalous dispersion (SAD) phasing. AutoSharp (Vonrhein et al., 2007) was used for locating the selenium atoms; model building was done using coot (Emsley and Cowtan, 2004), and refinement was carried out using refmac5 (Murshudov et al., 1997) Three phosphate ions were noticed in the Form II structure; two of which present in the topoisomerase active site and are separated by a distance of ~7.5 Å. The structure was refined to a final Rwork of 24.1 % and Rfree of 28.9 %. Topo-44 Form III crystals diffracted to 1.4 Å. The final Rwork and Rfree are 16.5 % and 18.4%, respectively. An interesting observation is the presence of both phosphate and guanidium ions in the Form III Topo-44 structure. The linker helix and part of the first HhH motif of the B monomer show alternate conformations and were built as two separate chains with occupancy of 0.5 each. Further details on data collection and structure determination are given in the Supplementary Information.
All figures were made with Pymol (DeLano, 2002) and the electrostatic surfaces were calculated with APBS (Baker et al., 2001).
Supplementary Material
Acknowledgments
We acknowledge staff and instrumentation support from the Keck Biophysics Facility and the Center for Structural Biology at Northwestern University, and DND and LS-CAT at the Advanced Photon Source (APS) at Argonne National Laboratory. Support from the R.H. Lurie Comprehensive Cancer Center of Northwestern University to the Structural Biology Facility is also acknowledged. DND-CAT is supported by Dupont, DOW and the NSF. LS-CAT was supported by the Michigan Economic Development Corporation and the Michigan Technology Tri-Corridor. Use of the APS is supported by the Department of Energy (DOE). Research was supported by NIH grant GM51350 (to AM).
Footnotes
Protein data bank accession codes
The final structure factors and coordinates of Topo-31, Topo-44 Form I, Form II, and Form III have been deposited in the Protein Data Bank with accession codes 3M7G, 3M7D, 3M6K, and 3M6Z respectively.
Supplementary data are available at Structure Journal Online.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Afonine PV, Grosse-Kunstleve RW, Adams PD. A robust bulk-solvent correction and anisotropic scaling procedure. Acta Crystallogr D Biol Crystallogr. 2005;61:850–855. doi: 10.1107/S0907444905007894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baker NA, Sept D, Joseph S, Holst MJ, McCammon JA. Electrostatics of nanosystems: application to microtubules and the ribosome. Proc Natl Acad Sci U S A. 2001;98:10037–10041. doi: 10.1073/pnas.181342398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baker NM, Rajan R, Mondragon A. Structural studies of type I topoisomerases. Nucleic Acids Res. 2009;37:693–701. doi: 10.1093/nar/gkn1009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belova GI, Prasad R, Kozyavkin SA, Lake JA, Wilson SH, Slesarev AI. A type IB topoisomerase with DNA repair activities. Proc Natl Acad Sci U S A. 2001;98:6015–6020. doi: 10.1073/pnas.111040498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belova GI, Prasad R, Nazimov IV, Wilson SH, Slesarev AI. The domain organization and properties of individual domains of DNA topoisomerase V, a type 1B topoisomerase with DNA repair activities. J Biol Chem. 2002;277:4959–4965. doi: 10.1074/jbc.M110131200. [DOI] [PubMed] [Google Scholar]
- Champoux JJ. DNA Topoisomerases: Structure, Function, and Mechanism. Annu Rev Biochem. 2001;70:369–413. doi: 10.1146/annurev.biochem.70.1.369. [DOI] [PubMed] [Google Scholar]
- Cheng C, Shuman S. A catalytic domain of eukaryotic DNA topoisomerase I. J Biol Chem. 1998;273:11589–11595. doi: 10.1074/jbc.273.19.11589. [DOI] [PubMed] [Google Scholar]
- Collaborative-Computational-Project-4. The CCP4 suite: programs for protein crystallography. Acta Crystallogr D. 1994;50:760–763. doi: 10.1107/S0907444994003112. [DOI] [PubMed] [Google Scholar]
- Davis IW, Murray LW, Richardson JS, Richardson DC. MOLPROBITY: structure validation and all-atom contact analysis for nucleic acids and their complexes. Nucleic Acids Res. 2004;32:W615–619. doi: 10.1093/nar/gkh398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeLano WL. The PyMol Molecular Graphics System. San Carlos, CA: DeLano Scientific; 2002. [Google Scholar]
- Diederichs K, Karplus PA. Improved R-factors for diffraction data analysis in macromolecular crystallography. Nat Struct Biol. 1997;4:269–275. doi: 10.1038/nsb0497-269. [DOI] [PubMed] [Google Scholar]
- Doublie S. Preparation of selenomethionyl proteins for phase determination. Methods Enzymol. 1997;276:523–530. [PubMed] [Google Scholar]
- Emsley P, Cowtan K. Coot: model-building tools for molecular graphics. Acta Crystallogr D Biol Crystallogr. 2004;60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- Forterre P. DNA topoisomerase V: a new fold of mysterious origin. Trends Biotechnol. 2006;24:245–247. doi: 10.1016/j.tibtech.2006.04.006. [DOI] [PubMed] [Google Scholar]
- Forterre P, Gribaldo S, Gadelle D, Serre MC. Origin and evolution of DNA topoisomerases. Biochimie. 2007;89:427–446. doi: 10.1016/j.biochi.2006.12.009. [DOI] [PubMed] [Google Scholar]
- Gellert M. DNA Topoisomerases. Annu Rev Biochem. 1981;50:879–910. doi: 10.1146/annurev.bi.50.070181.004311. [DOI] [PubMed] [Google Scholar]
- Huber R, Kurr M, Jannasch HW, Stetter KO. A novel group of abyssal methanogenic archaebacteria (Methanopyrus) growing at 110 °C. Nature. 1989;342:833–834. [Google Scholar]
- Kabsch W. Automatic processing of rotation diffraction data from crystals of initially unknown symmetry and cell constants. J Appl Crystallogr. 1993;26:795–800. [Google Scholar]
- Koster DA, Croquette V, Dekker C, Shuman S, Dekker NH. Friction and torque govern the relaxation of DNA supercoils by eukaryotic topoisomerase IB. Nature. 2005;434:671–674. doi: 10.1038/nature03395. [DOI] [PubMed] [Google Scholar]
- Kumar A, Widen SG, Williams KR, Kedar P, Karpel RL, Wilson SH. Studies of the domain structure of mammalian DNA polymerase beta. Identification of a discrete template binding domain. J Biol Chem. 1990;265:2124–2131. [PubMed] [Google Scholar]
- Liu D, DeRose EF, Prasad R, Wilson SH, Mullen GP. Assignments of 1H, 15N, and 13C resonances for the backbone and side chains of the N-terminal domain of DNA polymerase beta. Determination of the secondary structure and tertiary contacts. Biochemistry. 1994;33:9537–9545. doi: 10.1021/bi00198a020. [DOI] [PubMed] [Google Scholar]
- Maxwell A. DNA gyrase as a drug target. Biochem Soc Trans. 1999;27:48–53. doi: 10.1042/bst0270048. [DOI] [PubMed] [Google Scholar]
- McCoy AJ, Grosse-Kunstleve RW, Adams PD, Winn MD, Storoni LC, Read RJ. Phaser crystallographic software. J Appl Crystallogr. 2007;40:658–674. doi: 10.1107/S0021889807021206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murshudov GN, Vagin AA, Dodson EJ. Refinement of macromolecular structures by the maximum-likelihood method. Acta Crystallogr D. 1997;53:240–255. doi: 10.1107/S0907444996012255. [DOI] [PubMed] [Google Scholar]
- Perry K, Hwang Y, Bushman FD, Van Duyne GD. Structural basis for specificity in the poxvirus topoisomerase. Mol Cell. 2006;23:343–354. doi: 10.1016/j.molcel.2006.06.015. [DOI] [PubMed] [Google Scholar]
- Pommier Y. Diversity of DNA topoisomerases I and inhibitors. Biochimie. 1998;80:255–270. doi: 10.1016/s0300-9084(98)80008-4. [DOI] [PubMed] [Google Scholar]
- Redinbo MR, Stewart L, Kuhn P, Champoux JJ, Hol WG. Crystal structures of human topoisomerase I in covalent and noncovalent complexes with DNA. Science. 1998;279:1504–1513. doi: 10.1126/science.279.5356.1504. [DOI] [PubMed] [Google Scholar]
- Rothenberg ML. Topoisomerase I inhibitors: review and update. Ann Oncol. 1997;8:837–855. doi: 10.1023/a:1008270717294. [DOI] [PubMed] [Google Scholar]
- Schoeffler AJ, Berger JM. DNA topoisomerases: harnessing and constraining energy to govern chromosome topology. Q Rev Biophys. 2008;41:41–101. doi: 10.1017/S003358350800468X. [DOI] [PubMed] [Google Scholar]
- Slesarev AI, Stetter KO, Lake JA, Gellert M, Krah R, Kozyavkin SA. DNA topoisomerase V is a relative of eukaryotic topoisomerase I from a hyperthermophilic prokaryote. Nature. 1993;364:735–737. doi: 10.1038/364735a0. [DOI] [PubMed] [Google Scholar]
- Stewart L, Ireton GC, Parker LH, Madden KR, Champoux JJ. Biochemical and biophysical analyses of recombinant forms of human topoisomerase I. J Biol Chem. 1996;271:7593–7601. doi: 10.1074/jbc.271.13.7593. [DOI] [PubMed] [Google Scholar]
- Stewart L, Redinbo MR, Qiu X, Hol WG, Champoux JJ. A model for the mechanism of human topoisomerase I. Science. 1998;279:1534–1541. doi: 10.1126/science.279.5356.1534. [DOI] [PubMed] [Google Scholar]
- Taneja B, Patel A, Slesarev A, Mondragon A. Structure of the N-terminal fragment of topoisomerase V reveals a new family of topoisomerases. EMBO J. 2006;25:398–408. doi: 10.1038/sj.emboj.7600922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taneja B, Schnurr B, Slesarev A, Marko JF, Mondragon A. Topoisomerase V relaxes supercoiled DNA by a constrained swiveling mechanism. Proc Natl Acad Sci U S A. 2007;104:14670–14675. doi: 10.1073/pnas.0701989104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vonrhein C, Blanc E, Roversi P, Bricogne G. Automated structure solution with autoSHARP. Methods Mol Biol. 2007;364:215–230. doi: 10.1385/1-59745-266-1:215. [DOI] [PubMed] [Google Scholar]
- Wang HK, Morris-Natschke SL, Lee KH. Recent advances in the discovery and development of topoisomerase inhibitors as antitumor agents. Med Res Rev. 1997;17:367–425. doi: 10.1002/(sici)1098-1128(199707)17:4<367::aid-med3>3.0.co;2-u. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.