Abstract
Ebola virus (EBOV), a negative-sense RNA virus in the family Filoviridae, is known to cause severe hemorrhagic fever in humans and other primates. Infection with EBOV causes a high mortality rate and currently there is no FDA-licensed vaccine or therapeutic treatment available. Recently, heat-shock protein 90 (Hsp90), a molecular chaperone, was shown to be an important host factor for the replication of several negative-strand viruses. We tested the effect of several different Hsp90 inhibitors including geldanamycin, radicicol, and 17-allylamino-17-demethoxygeldanamycin (17-AAG; a geldanamycin analog) on the replication of Zaire EBOV. Our results showed that inhibition of Hsp90 significantly reduced the replication of EBOV. Classic Hsp90 inhibitors reduced viral replication with an effective concentration at 50% (EC50) in the high nanomolar to low micromolar range, while drugs from a new class of Hsp90 inhibitors showed markedly more potent inhibition. These compounds blocked EBOV replication with an EC50 in the low nanomolar range and showed significant potency in blocking replication in primary human monocytes. These results validated that Hsp90 is an important host factor for the replication of filoviruses and suggest that Hsp90 inhibitors may be therapeutically effective in treating EBOV infection.
Keywords: Ebola virus, Hsp90, therapeutic
1. INTRODUCTION
Ebola virus (EBOV) is the causative agent of Ebola hemorrhagic fever, an emerging viral disease that causes severe hemorrhagic fever in humans and non-human primates (NHPs). Outbreaks occur in Africa and mortality rates range from 25–90%. Development of vaccines and therapeutics are vital due to increasingly frequent outbreaks and the placement of EBOV as a category A agent by the Centers for Disease Control and Prevention; however, no effective countermeasures exist to treat this deadly viral disease.
EBOV is a filamentous, enveloped, nonsegmented, negative-sense RNA virus in the family Filoviridae. The genome is 18.9 kb in length and encodes seven structural proteins and one non-structural protein in the following order within the genome: 3’ non-coding region (leader), nucleoprotein (NP), virion protein 35 (VP35), VP40, glycoprotein (sGP and GP), VP30, VP24, RNA-dependent RNA-polymerase (RDRP; L) protein, and a 5’ non-coding region (trailer) (Sanchez, 2001). The RDRP shares significant sequence homology to RDRPs from other nonsegmented, negative-strand RNA viruses and is required for both viral transcription and replication of the viral genome (Whelan, Barr, and Wertz, 2004). A common theme of these RDRPs is that they require cooperating viral and host proteins to accomplish replication and/or transcription. Some of the important host factors required for RDRP function have been identified for other negative-strand RNA viruses but host factors required for EBOV RDRP function have not been described to date (Das et al., 1998; Gupta, Shaji, and Banerjee, 2003; Shen and Masters, 2001; Strauss and Strauss, 1999; Whelan, Barr, and Wertz, 2004).
Heat-shock protein 90 (Hsp90) is a molecular chaperone that guides the folding, intracellular disposition, and proteolytic turnover of many key regulators of cell growth and differentiation. Hsp90 has a specific set of client proteins in vivo such as steroid receptors, transcription factors, protein kinases, and oncogenes (Pratt and Toft, 2003). Inhibitors of Hsp90 have proven effective at driving cancer cells into apoptosis by preventing the proper folding of oncogenes required for promoting cancer cell growth. Because of this, several Hsp90 inhibitors are now in phase I and II clinical trials (Goetz et al., 2005; Whitesell and Lindquist, 2005).
Recently, Hsp90 was shown to be an important host factor for the replication of negative-strand viruses (Connor et al., 2007). In addition, the inhibition of Hsp90 has been shown to block vaccinia virus replication by interaction with the viral core protein 4a in the cytoplasm (Hung, Chung, and Chang, 2002). In the hepatitis C virus life cycle, Hsp90 is needed for proper cleavage of newly synthesized hepatitis C NSP2/3 protein (Ujino et al., 2009; Waxman et al., 2001) and activity of hepatitis B reverse transcriptase (Hu and Seeger, 1996; Hu, Toft, and Seeger, 1997; Stahl et al., 2007). In polio virus, Hsp90 is required for proper folding of the viral capsid protein and Hsp90 inhibitors showed antiviral activity (Geller et al., 2007).
Hsp90 has been shown to control viral polymerase function for several viruses. For influenza virus, Hsp90 binds to the PB2 subunit of the RNA polymerase and stimulates its activity (Momose et al., 2002). In herpes viruses, blocking Hsp90 significantly inhibits viral replication presumably due to improper localization of the viral polymerase (Burch and Weller, 2005; Li et al., 2004). In flock house virus, Hsp90 activity has proved to be important for stability and localization of the RNA polymerase (Kampmueller and Miller, 2005). Recently, it was reported that Hsp90 inhibitors impaired the replication of several prototype negative-strand RNA viruses [ (vesicular stomatitis virus), Paramyxovirus (SV5, HPIV-2 & 3, SV41), and a bunyavirus (La Crosse)] by destabilization of the L protein of the viral RDRP (Connor et al., 2007). It is thought that viruses have evolved to require the use of Hsp90 for proper folding of their RDRPs (Connor et al., 2007). Therefore, inhibitors of Hsp90 activity could act as broad-range antiviral agents and we speculated that this hypothesis would hold true for filoviruses; therefore, we evaluated the effect of several Hsp90 inhibitors on EBOV replication. Some were natural product inhibitors while others were synthetic inhibitors. The entire panel of Hsp90 inhibitors represented three different chemical structure classes with varying potencies. Our results showed that Hsp90 inhibitors significantly inhibited the replication of EBOV, suggesting their use as a potential therapeutic. There were differences in the inhibition of EBOV replication by different Hsp90 inhibitors, indicating that some classes of Hsp90 inhibitor may be superior choices for antiviral agents. The results of this study will aid in the design of more effective therapeutics to treat EBOV infection.
2. MATERIALS AND METHODS
2.1. Viruses
EBOV species Zaire was originally isolated in 1976 from a human patient (Johnson et al., 1977)and passaged twice in Vero E6 cells before use. The EBOV-green fluorescent protein (GFP) virus was derived by reverse genetics to generate a full-lenth cDNA clone inserted with the reporter gene eGFP (Towner et al., 2005).
2.2. Compounds
Geldanamycin, radicicol, and 17AAG were obtained from Invivogen (San Diego, CA) or from LC labs (Woburn, MA) and were re-suspended in DMSO. Compounds AV-1, 2, 3, and 81 were obtained from Serenex (now Pfizer; New York, NY) and were re-suspended in DMSO.
2.3. EBOV-GFP Microtiter Plate Assay
Compounds were screened using Vero cells (American Type Culture Collection, Manassa, VA) in 96-well plates. Compounds were diluted either threefold (1 µM to 0.5 nM) or twofold (50 µM to 0.4 µM) and added to the 96-well plates. One cohort allowed 3-h incubation with compounds before infection. Plates were infected at a low multiplicity of infection (MOI) with EBOV-GFP and incubated at 37°C. Plates were read at Ex 485, Em 515, cutoff 495 at 16, 24, 40, 48 h post infection (PI). Plates were then stained with crystal violet and read by spectrophotometer to evaluate cytotoxicity.
2.4. Yield-Reduction Assays
The effectiveness of the compounds was evaluated by virus yield-reduction assay using either Vero cells or primary human monocytes in 6-well plates. The cells were maintained in Modified Eagle’s medium (MEM) with 10% fetal bovine serum (FBS), and 1X GlutaMax (Invitrogen, Carlsbad, CA). Medium was removed from cells infected with ZEBOV at an MOI of 0.1 in 200 µl of medium (5% MEM, no antibiotics) that contained the following drug concentrations: 12.5 µM, 1 µM, 37 nM, 0.5 nM. Plates were incubated 1 h at 37°C with rocking every 15 min. Medium containing virus was removed and plates were washed 3 times with medium. After washing, 3 ml of medium containing the drug concentrations above was added and plates were incubated at 37°C with the following controls: virus, no drug; no virus, no drug; drug only, no virus. At 0, 24, 48, 96 h PI, 250 µl of medium was collected for titration by standard plaque assay on Vero cells or analyzed by real-time RT-PCR.
2.5. Plaque Assays
Plaque assays were completed using 90–100% confluent Vero cells in 6-well plates. Samples for titration were serially diluted 10-fold and 200 µL was added to each well. Plates were incubated for 1 h at 37°C with rocking every 15 min. A primary overlay containing 1X EBME, 5% FBS, and 0.5% agarose was added to each well. Plates were incubated at 37°C for 6 days followed by a secondary overlay, which was identical to the primary overlay with the addition of 5% neutral red. Plaque forming units (PFU) were counted on day 7 PI.
2.6. Real-time RT-PCR
The real-time RT-PCR assay used was previously published (Weidmann, Muhlberger, and Hufert, 2004). This assay was designed to detect the nucleoprotein gene of EBOV. PFU equivalents (PFUe) were determined using a known virus concentration (determined by plaque assay) whose RNA was extracted and was 10-fold serially diluted. In our hands the sensitivity of the assay was 0.04 PFUe.
2.7. Statistics
SAS version 9.1.3 (SAS Institute Inc., Cary, NC) was used to determine repeated measures ANOVA of viral replication samples between controls and treatment groups over time with step-down Sidak adjustment for multiple pairwise comparisons at each time point.
3. RESULTS
To test the hypothesis that Hsp90 is an important host factor for the replication of EBOV, we investigated the effect of several different Hsp90 inhibitors on the replication of EBOV in a virus-permissive cell line. We initially tested the effect of increasing concentrations of three different Hsp90 inhibitors: geldanamycin, 17-AAG, and radicicol. These compounds have an extensive history of use for the dissection of Hsp90 functions (Richter and Buchner, 2001), and represent two separate chemical classes (Taldone, Sun, and Chiosis, 2009). Geldanamycin is a benzoquinone ansamycin and 17-AAG is a geldanamycin derivative that is currently in Phase II clinical trials as an anticancer agent (Goetz et al., 2005; Whitesell and Lindquist, 2005). Radicicol is a natural product monorden and was the most potent Hsp90 inhibitor defined at the beginning of our studies (Clevenger and Blagg, 2004; Delmotte and Delmotte-Plaque, 1953).
Viral replication in the presence and absence of added drug was monitored using an EBOV that was engineered to express EGFP from an independent ORF (EBOV-GFP). This virus allowed the tracking of EGFP fluorescence as a robust indicator of viral replication (Towner et al., 2005). With this virus, the expression of GFP in Vero E6 cells that were treated with geldanamycin, 17-AAG, or radicicol was evaluated. Cells were mock-treated or treated with increasing concentrations of drug, from 0.5 nM to 50 µM and were then infected with virus 30 min post drug treatment. Expression of EGFP was determined by fluorescence measurements at 16, 24, 40, and 48-h PI. The effect of the drugs on inhibiting viral replication was then plotted as a percent reduction of GFP fluorescence, with a 90 % reduction representing a 10-fold drop in fluorescence expression (Fig 1).
Figure 1.
Effect of Hsp90 inhibitors geldanamycin, 17-AAG, and radidicol on the replication of EBOV-GFP. The dose response curves of geldanamycin (closed circles) 17-AAG (closed squares) and radicicol (closed triangles) at A) 16-h PI, B) 24-h PI, C) 40-h PI, and D) 48-h PI are plotted as percent decrease of a fluorescent signal compared to dimethylsulfoxide-treated control infection. Results show a decrease in the percent GFP reduction at increasing concentrations of compound.
All three Hsp90 inhibitors showed some inhibition of viral replication at each time point analyzed. The level of inhibition varied, but for all compounds, the greatest reduction in GFP was observed at 24 h PI (Figure 1B). Pre-incubation with compounds slightly improved efficacy of inhibition, but was not significant (data not shown). At 16-h PI, all compounds showed a maximum fluorescence reduction of approximately 60%, with the EC50 ranging from 43.8 nM with radicicol to 394.5 nM with 17-AAG. At all time points, radicicol was the most potent inhibitor of viral replication, reaching a maximum reduction of fluorescence of approximately 86% at 24-h PI (12.5 µM) with an EC50 of approximately 86.8 nM. Geldanamycin and 17-AAG both showed a weaker reduction in viral replication (maximum of 85 and 80% reduction, respectively) and lower potency (EC50 in the micromolar range) than was seen for radicicol (see Table 1).
Table 1.
EC50 values of geldanamycin, 17-AAG, and radicicol in nM ± standard deviation. Table also shows an approximate specificity index (SI50=CC50/EC50) calculated from values in Figure 1 and 2A
| Compounds | |||
|---|---|---|---|
| Time PI | Geldanamycin | 17-AAG | Radicicol |
| 16 | 378 ± 1.2 | 394.5 ± 1.5 | 43.8 ± 1.1 |
| 24 | 584.6 ± 1.1 | 2686 ± 1.9 | 86.75 ± 1.2 |
| 40 | 1149 ± 1.1 | 4210 ± 1.2 | 327.7 ± 1.3 |
| 48 | 1554 ± 1.1 | 5352 ± 1.2 | 1707 ± 1.7 |
| CC50 at 48hpi | 50µM | >50µM | >50µM |
| SI50 at 48hpi | ~25 | >10 | >25 |
During cytotoxicity testing of the Hsp90 inhibitors, all three drugs showed minimal toxicity at concentrations below 5 µM (Fig 2A). This is consistent with other reports of minimal toxicity of these compounds at or below this concentration (Kampmueller and Miller, 2005) However, all three compounds had significant effects on cellular homeostasis at high concentrations. At 25- and 50-µM levels of drug cell death rates of between 30 and 60% were observed, suggesting CC50 concentrations between 50 and 100µM and SI50 ratios of between 10 and 25 (see Table 1) When the effect of each Hsp90 inhibitor a minimally cytotoxic concentration was plotted as a function of time (inhibition at 1.6 µM, Fig2B), it is clear that while each compound blocked viral replication, this effect was not dramatic with any of the “classic” Hsp90 inhibitors. To determine if the results observed with the EBOV-GFP assays correlated with viral replication assays, we analyzed the same compounds by yield-reduction assays and determined viral titers by both real-time RT-PCR and plaque assays (Fig 3). For all three Hsp90 inhibitors, we observed a reduction in viral replication where the greatest reduction in viral replication was at 24- and 48-h PI for each compound. For geldanamycin, we observed a 1.9 log10 PFU/ml (0.5 log10 PFUe/ml) and 0.9 log10 PFU/ml (0.2 log10 PFUe/ml) reduction in virus at 24- and 48-h PI, respectively, at 12.5 µM (Fig 3A–B). For 17-AAG, we observed a 1.1 log10 PFU/ml (0.2 log10 PFUe/ml) reduction in virus at 24- and 48-h PI at similar concentrations of drug (Fig 3C–D). For radicicol, we observed a 1.3 log10 PFU/ml (1 log10 PFUe/ml) and 1 log10 PFU/ml (1.5 log10 PFUe/ml) reduction in virus at 24- and 48-h PI, respectively (Fig 3E–F). The viral yield reductions were statistically significant for geldanamycin (p=0.0205), 17AAG (p=0.0345), and radicicol (p=0.0100). These results showed good correlation between the microtiter plate assay and the yield reduction assay as analyzed b y plaque assay. Interestingly, when viral yield was tested by real-time RT-PCR, a different answer for viral yield reduction was obtained. For geldanamycin and 17-AAG, there appeared to be no change in viral output as measured by genome equivalents, regardless of the concentration of inhibitor used. Similarly, only the 12.5-µM concentration of radicicol showed a significant reduction in genome equivalents produced during infection. Thus, the real-time RT-PCR assay suggests that there is not a marked difference in viral replication after Hsp90 treatment. The plaque assays clearly showed viral titer reductions suggesting that these inhibitors may cause the budding of replication-defective viral particles. Alternatively, the medium could still contain the compound which could interfere with the results of the plaque assay.
Figure 2.
Toxicity and time-course of geldanamycin, 17-AAG, and radidicol during EBOV-GFP infection. A) Cell toxicity was determined by evaluating the viable cells by crystal violet stain. Percentage of surviving cells 48 h after drug treatment is plotted. B) % reduction of viral replication at 1.6 µM was plotted over the 48-h time-course.
Figure 3.
Compounds tested at 12.5 µM, 1 µM, 37 nM, and 1 nM by yield-reduction assay and supernatant analyzed by plaque assay (A, C, and E) or real-time RT-PCR (B, D, and F). PFU=plaque-forming units; PFUe=plaque-forming unit equivalents
These data showed that Hsp90 has some ability to reduce the replication of EBOV, but also highlighted that these inhibitors showed different abilities to block viral replication. The varying efficacies of these compounds led us to determine if chemically dissimilar inhibitors of Hsp90 activity showed increased effectiveness as viral replication inhibitors. Recently, a novel class of benzamide compounds have been identified as Hsp90 inhibitors(Barta et al., 2008). We determined the antiviral activity of four of these compounds (AV-1, AV-2, AV-3, and AV-81) in Vero cells and primary human monocytes.
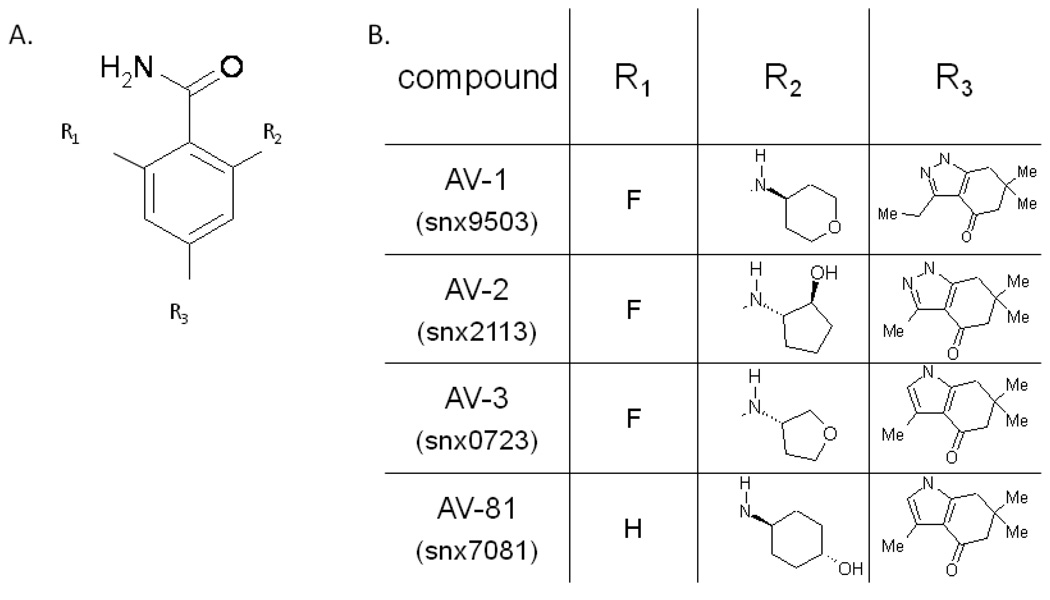
These compounds, whose structure is shown in Figure 4, represented a lead compound (AV-81, also published as SNX 7081(Huang et al., 2009; Rice et al., 2008) and three derivatives (AV-1 and A-V2 have not been published with specific compound names (Huang et al., 2009), AV-3 has been published as SNX 7023 (Putcha et al., 2009). They all contain the same core structure (see 4A) with various side-groups illustrated in the table shown in figure 4B.
Figure 4.
Structure of the Serenex compounds AV-1-3 and AV-81. A) Core structure and B) side-groups.
Similar to the experiments described above, when Vero cells were treated with increasing concentrations of either AV-1, AV-2, AV-3, or AV-81, and then infected with EBOV-EGFP, there was an inhibition of viral replication (Fig 4). All four compounds showed a significant ability to reduce viral gene expression, as measured by GFP production with a maximum of 60% reduction in fluorescence at 16-h PI and 80% reduction at 40- and 48-h PI for all compounds. All three were more potent than radicicol, geldanamycin, and 17AAG. Of the three compounds tested. AV-3 was the most potent, with an EC50 of 18.7 nM at 24-h PI and 27.3 and 27.7 at 40 and 48-h PI, respectively (Table 2). AV-2 and AV-1 were similarly potent, with EC50 within the range of 49.4 nM to 59.3 nM at 24- 48-h PI, which were well below the concentrations at which cytotoxicity was seen. AV-81 was the least potent with an EC50 ranging from 260 to 515.9 nM at 16- to 48-h PI. Similar GFP-expression assays were not possible in primary monocytes, because these cells are not uniformly adherent, but yield-reduction assays were done to analyze viral replication.
Table 2.
EC50 values of AV-1-3, and AV-81 in nM ± standard deviation
| Compounds | ||||
|---|---|---|---|---|
| Time PI | AV-1 | AV-2 | AV-3 | AV-81 |
| 16 | N/A | 452.8 ± 1.8 | N/A | 260 ± 1.7 |
| 24 | 49.4 ± 1.1 | 54.5 ± 1.1 | 18.7 ± 1.1 | 394 ± 1.1 |
| 40 | 54.7 ± 1.1 | 58.3 ± 1.1 | 27.3 ± 1.1 | 515.9 ± 1.1 |
| 48 | 55.2 ± 1.1 | 59.3 ± 1.1 | 27.7 ± 1.1 | 509.4 ± 1.1 |
Yield-reduction assays from Vero cells were determined at two concentrations of drug, one relatively low (37 nM) and the other high (12.5 µM). These assays showed only a limited reduction in viral titer (Fig 6A). However, yield-reduction assays using human monocytes showed a much more dramatic effect. In monocytes yield-reduction assays from cells treated with either 37 nm or 12.5 µM of drug showed significant decreases in viral replication (Fig 6B) over all time points. At 48-h PI, AV-1, AV-2, and AV-3 all showed ~0.5 log10 PFUe/ml reductions in EBOV production when used at 37 nM. At high drug concentrations, there was little evidence of replication in any drug-treated monocyte. Similar to the results of the microtiter plate assay, AV-81 was the least effective compound evaluated. When plaque reduction asays for monocytes were extended to 96-h PI, a more significant reduction in viral titers was seen in monocytes, showing that these compounds are highly effective at blocking viral replication over a significant period of time. No drug-related toxicities were noted in these assays at concentrations of drug up to 50µM. Above this concentration, toxicities were seen that were associated with DMSO, preventing an accurate calculation of CC50 and SI50. These compounds appear to be much more specific and potent than the “classic” Hsp90 inhibitors.
Figure 6.
Compounds were tested at 12.5 µM and 37 nM by yield-reduction assay in A) Vero or B) monocytes and supernatant analyzed by real-time RT-PCR.
4. DISCUSSION
The development of therapeutics to treat viral infections has generally targeted viral proteins. Recently the focus has shifted to targeting host factors that are co-opted for use in the life cycle of the virus. For example, Hsp90 chaperones, which constitute the most abundant folding enzymes in the cytosol, have been shown to play extensive roles for multiple viruses. Often, these proteins serve as host factors by binding viral proteins, preventing proper folding and promoting their degradation (Connor et al., 2007; Nakagawa et al., 2007). In addition to stability, Hsp90 chaperones are necessary for the intracellular transport of multiple viral proteins. This behavior is exemplified by the reverse transcriptase from hepatitis B and the polymerase PB2 from influenza virus (Hu et al., 2004; Naito et al., 2007).
Several studies have demonstrated that Hsp90 chaperones are promising targets for host-directed antiviral development. Several small-molecule Hsp90 inhibitors, including geldanamycin and its derivatives, originally characterized as anti-cancer therapeutics, reduce viral titers in cell culture models of vaccinia virus, influenza virus, vesicular stomatitis virus, several paramyxoviruses, La Crosse virus, and hepatitis C virus (Banerji, 2009; Chase et al., 2008; Connor et al., 2007; Hung, Chung, and Chang, 2002; Nakagawa et al., 2007). The use of Hsp90 inhibitors for antiviral development suggest that these inhibitors disrupt the function of the viral polymerase. This is in contrast to the use of Hsp90 inhibitors as an anti-cancer thereapeutic, which drive cancer cells into apoptosis. In the case of hepatitis C virus, combination therapy of polyethylene glycol-conjugated interferon (PEG-IFN) plus the Hsp90 inhibitor 17-dimethylaminoethylamino-demethoxygeldanamycin showed enhanced inhibition of viral replication in the livers of infected mice, compared to PEG-IFN alone (Nakagawa et al., 2007).
Here we show that Hsp90 inhibitors can also be used as inhibitors of EBOV replication. All compounds appeared to be effective as evaluated using our microtiter plate assay screen. Radicicol and the flouoridated benzamides (AV-1-3) were most effective and showed over 85% reduction in our microtiter plate screening assay. In Vero cells, the detection of inhibitory activity in the microtiter plate assay screen correlated with a significant reduction in viral titer. Our analysis showed that commercially available Hsp90 inhibitors such as geldanamycin, 17-AAG, and radicicol were less potent inhibitors of EBOV than the benzamide analogs AV-1-3 (Table 1 and 2). This difference in potency of inhibition has been observed not only for Ebola virus but also for other negative-strand RNA viruses (J. Connor, unpublished observations)
These results suggest that Hsp90 inhibitors with different structures will have varying efficacies against different viral pathogens. This could be due to inherent potencies of inhibition by the different compounds or to these compounds targeting different Hsp90 holoenzymes. Additionally, different cell lines may have different levels of Hsp90. The identity of the proteins within the complex are known to alter the ATP-binding pocket of Hsp90 (Pearl and Prodromou, 2006), where all of the drugs that we tested bound Hsp90. Our results point to the fluoridated benzamide compounds tested here as promising compounds for the blockage of the pertinent EBOV-required Hsp90 complex. Each of the benzamide compounds has very strong antiviral activity in primary human monocytes. Monocytes are known to be an important early target for EBOV replication (Geisbert et al., 2003), thus the significant antiviral effect of these compounds in these cells suggests that they might be particularly effective inhibitors of viral replication in vivo.
The mechanism of action of these Hsp90 inhibitors on EBOV remains to be determined. It is possible that these inhibitors cause a degradation in the viral polymerase as was observed for other negative-strand RNA viruses (Connor et al., 2007). However, given that our results using geldanamycin and 17-AAG showed that treated cells showed lower levels of EGFP production and lower infectious viral yields, but similar budded viral genomes to untreated cells, Hsp90 inhibition may also be important for the proper budding of infectious EBOV.
The role of Hsp90 for the replication of several viruses is an important topic of research with regard to therapeutic development. Because no effective countermeasures currently exist for EBOV, our results offer some insight into possible future development of potential therapeutics to treat this deadly disease. Our results with the benzamide Hsp90 inhibitors suggest that these compounds are a promising class of inhibitors with antiviral activity against EBOV. Future work in our laboratory will further investigate the role that Hsp90 plays on EBOV replication as well as other negative strand RNA viruses.
Figure 5.
Effect of novel Hsp90 inhibitors geldanamycin, 17-AAG, and radidicol on the replication of EBOV-GFP. The dose response curves of AV-1, AV-2, AV-3, and AV-81 at A) 16-h PI, B) 24-h PI, C) 40-h PI, and D) 48-h PI.
Acknowledgements
We thank Josh Johnson and Calli Lear for technical assistance and Diana Fisher for statistical analysis support. The research described herein was sponsored by DTRA; project number 02-4-4J-081, NIH Grant AI064606 and a subcontract from the NERCE/BEID U54 AI057159 awarded to JHC, who is a Peter T. Paul Career Development Professor. Opinions, interpretations, conclusions, and recommendations are those of the author and are not necessarily endorsed by the U.S. Army.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- Banerji U. Heat shock protein 90 as a drug target: some like it hot. Clin Cancer Res. 2009;15(1):9–14. doi: 10.1158/1078-0432.CCR-08-0132. [DOI] [PubMed] [Google Scholar]
- Barta TE, Veal JM, Rice JW, Partridge JM, Fadden RP, Ma W, Jenks M, Geng L, Hanson GJ, Huang KH, Barabasz AF, Foley BE, Otto J, Hall SE. Discovery of benzamide tetrahydro-4H-carbazol-4-ones as novel small molecule inhibitors of Hsp90. Bioorg Med Chem Lett. 2008;18(12):3517–3521. doi: 10.1016/j.bmcl.2008.05.023. [DOI] [PubMed] [Google Scholar]
- Burch AD, Weller SK. Herpes simplex virus type 1 DNA polymerase requires the mammalian chaperone hsp90 for proper localization to the nucleus. J Virol. 2005;79(16):10740–10749. doi: 10.1128/JVI.79.16.10740-10749.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chase G, Deng T, Fodor E, Leung BW, Mayer D, Schwemmle M, Brownlee G. Hsp90 inhibitors reduce influenza virus replication in cell culture. Virology. 2008;377(2):431–439. doi: 10.1016/j.virol.2008.04.040. [DOI] [PubMed] [Google Scholar]
- Clevenger RC, Blagg BS. Design, synthesis, and evaluation of a radicicol and geldanamycin chimera, radamide. Org Lett. 2004;6(24):4459–4462. doi: 10.1021/ol048266o. [DOI] [PubMed] [Google Scholar]
- Connor JH, McKenzie MO, Parks GD, Lyles DS. Antiviral activity and RNA polymerase degradation following Hsp90 inhibition in a range of negative strand viruses. Virology. 2007;362(1):109–119. doi: 10.1016/j.virol.2006.12.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Das T, Mathur M, Gupta AK, Janssen GM, Banerjee AK. RNA polymerase of vesicular stomatitis virus specifically associates with translation elongation factor-1 alphabetagamma for its activity. Proc Natl Acad Sci U S A. 1998;95(4):1449–1454. doi: 10.1073/pnas.95.4.1449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delmotte P, Delmotte-Plaque J. A new antifungal substance of fungal origin. Nature. 1953;171(4347):344. doi: 10.1038/171344a0. [DOI] [PubMed] [Google Scholar]
- Geisbert TW, Hensley LE, Larsen T, Young HA, Reed DS, Geisbert JB, Scott DP, Kagan E, Jahrling PB, Davis KJ. Pathogenesis of Ebola hemorrhagic fever in cynomolgus macaques: evidence that dendritic cells are early and sustained targets of infection. Am J Pathol. 2003;163(6):2347–2370. doi: 10.1016/S0002-9440(10)63591-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geller R, Vignuzzi M, Andino R, Frydman J. Evolutionary constraints on chaperone-mediated folding provide an antiviral approach refractory to development of drug resistance. Genes Dev. 2007;21(2):195–205. doi: 10.1101/gad.1505307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goetz MP, Toft D, Reid J, Ames M, Stensgard B, Safgren S, Adjei AA, Sloan J, Atherton P, Vasile V, Salazaar S, Adjei A, Croghan G, Erlichman C. Phase I trial of 17-allylamino-17-demethoxygeldanamycin in patients with advanced cancer. J Clin Oncol. 2005;23(6):1078–1087. doi: 10.1200/JCO.2005.09.119. [DOI] [PubMed] [Google Scholar]
- Gupta AK, Shaji D, Banerjee AK. Identification of a novel tripartite complex involved in replication of vesicular stomatitis virus genome RNA. J Virol. 2003;77(1):732–738. doi: 10.1128/JVI.77.1.732-738.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu J, Flores D, Toft D, Wang X, Nguyen D. Requirement of heat shock protein 90 for human hepatitis B virus reverse transcriptase function. J Virol. 2004;78(23):13122–13131. doi: 10.1128/JVI.78.23.13122-13131.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu J, Seeger C. Hsp90 is required for the activity of a hepatitis B virus reverse transcriptase. Proc Natl Acad Sci U S A. 1996;93(3):1060–1064. doi: 10.1073/pnas.93.3.1060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu J, Toft DO, Seeger C. Hepadnavirus assembly and reverse transcription require a multi-component chaperone complex which is incorporated into nucleocapsids. Embo J. 1997;16(1):59–68. doi: 10.1093/emboj/16.1.59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang KH, Veal JM, Fadden RP, Rice JW, Eaves J, Strachan JP, Barabasz AF, Foley BE, Barta TE, Ma W, Silinski MA, Hu M, Partridge JM, Scott A, DuBois LG, Freed T, Steed PM, Ommen AJ, Smith ED, Hughes PF, Woodward AR, Hanson GJ, McCall WS, Markworth CJ, Hinkley L, Jenks M, Geng L, Lewis M, Otto J, Pronk B, Verleysen K, Hall SE. Discovery of novel 2-aminobenzamide inhibitors of heat shock protein 90 as potent, selective and orally active antitumor agents. J Med Chem. 2009;52(14):4288–4305. doi: 10.1021/jm900230j. [DOI] [PubMed] [Google Scholar]
- Hung JJ, Chung CS, Chang W. Molecular chaperone Hsp90 is important for vaccinia virus growth in cells. J Virol. 2002;76(3):1379–1390. doi: 10.1128/JVI.76.3.1379-1390.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson KM, Lange JV, Webb PA, Murphy FA. Isolation and partial characterisation of a new virus causing acute haemorrhagic fever in Zaire. Lancet. 1977;1(8011):569–571. doi: 10.1016/s0140-6736(77)92000-1. [DOI] [PubMed] [Google Scholar]
- Kampmueller KM, Miller DJ. The cellular chaperone heat shock protein 90 facilitates Flock House virus RNA replication in Drosophila cells. J Virol. 2005;79(11):6827–6837. doi: 10.1128/JVI.79.11.6827-6837.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li YH, Tao PZ, Liu YZ, Jiang JD. Geldanamycin, a ligand of heat shock protein 90, inhibits the replication of herpes simplex virus type 1 in vitro. Antimicrob Agents Chemother. 2004;48(3):867–872. doi: 10.1128/AAC.48.3.867-872.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Momose F, Naito T, Yano K, Sugimoto S, Morikawa Y, Nagata K. Identification of Hsp90 as a stimulatory host factor involved in influenza virus RNA synthesis. J Biol Chem. 2002;277(47):45306–45314. doi: 10.1074/jbc.M206822200. [DOI] [PubMed] [Google Scholar]
- Naito T, Momose F, Kawaguchi A, Nagata K. Involvement of Hsp90 in assembly and nuclear import of influenza virus RNA polymerase subunits. J Virol. 2007;81(3):1339–1349. doi: 10.1128/JVI.01917-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakagawa S, Umehara T, Matsuda C, Kuge S, Sudoh M, Kohara M. Hsp90 inhibitors suppress HCV replication in replicon cells and humanized liver mice. Biochem Biophys Res Commun. 2007;353(4):882–888. doi: 10.1016/j.bbrc.2006.12.117. [DOI] [PubMed] [Google Scholar]
- Pearl LH, Prodromou C. Structure and mechanism of the Hsp90 molecular chaperone machinery. Annu Rev Biochem. 2006;75:271–294. doi: 10.1146/annurev.biochem.75.103004.142738. [DOI] [PubMed] [Google Scholar]
- Pratt WB, Toft DO. Regulation of signaling protein function and trafficking by the hsp90/hsp70-based chaperone machinery. Exp Biol Med (Maywood) 2003;228(2):111–133. doi: 10.1177/153537020322800201. [DOI] [PubMed] [Google Scholar]
- Putcha P, Danzer KM, Kranich L, Scott A, Silinski M, Mabbett S, Hicks CD, Veal JM, Steed PM, Hyman BT, McLean PJ. Brain permeable small molecule inhibitors of Hsp90 prevent alpha-synuclein oligomer formation and rescue alpha synuclein-induced toxicity. J Pharmacol Exp Ther. 2009 doi: 10.1124/jpet.109.158436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rice JW, Veal JM, Fadden RP, Barabasz AF, Partridge JM, Barta TE, Dubois LG, Huang KH, Mabbett SR, Silinski MA, Steed PM, Hall SE. Small molecule inhibitors of Hsp90 potently affect inflammatory disease pathways and exhibit activity in models of rheumatoid arthritis. Arthritis Rheum. 2008;58(12):3765–3775. doi: 10.1002/art.24047. [DOI] [PubMed] [Google Scholar]
- Richter K, Buchner J. Hsp90: chaperoning signal transduction. J Cell Physiol. 2001;188(3):281–290. doi: 10.1002/jcp.1131. [DOI] [PubMed] [Google Scholar]
- Sanchez A, Khan AS, Zaki SR, Nabel GJ, Ksiazek T, Peters CJ. Filoviridae: Marburg and Ebola Viruses. In: Knipe DM, Howley PM, editors. Fields Virology. Vol. 1. New York: Lippincott Williams & Wilkins; 2001. pp. 1279–1304. [Google Scholar]
- Shen X, Masters PS. Evaluation of the role of heterogeneous nuclear ribonucleoprotein A1 as a host factor in murine coronavirus discontinuous transcription and genome replication. Proc Natl Acad Sci U S A. 2001;98(5):2717–2722. doi: 10.1073/pnas.031424298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stahl M, Retzlaff M, Nassal M, Beck J. Chaperone activation of the hepadnaviral reverse transcriptase for template RNA binding is established by the Hsp70 and stimulated by the Hsp90 system. Nucleic Acids Res. 2007;35(18):6124–6136. doi: 10.1093/nar/gkm628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strauss JH, Strauss EG. Viral RNA replication. With a little help from the host. Science. 1999;283(5403):802–804. doi: 10.1126/science.283.5403.802. [DOI] [PubMed] [Google Scholar]
- Taldone T, Sun W, Chiosis G. Discovery and development of heat shock protein 90 inhibitors. Bioorg Med Chem. 2009;17(6):2225–2235. doi: 10.1016/j.bmc.2008.10.087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Towner JS, Paragas J, Dover JE, Gupta M, Goldsmith CS, Huggins JW, Nichol ST. Generation of eGFP expressing recombinant Zaire ebolavirus for analysis of early pathogenesis events and high-throughput antiviral drug screening. Virology. 2005;332(1):20–27. doi: 10.1016/j.virol.2004.10.048. [DOI] [PubMed] [Google Scholar]
- Ujino S, Yamaguchi S, Shimotohno K, Takaku H. Heat-shock protein 90 is essential for stabilization of the hepatitis C virus nonstructural protein NS3. J Biol Chem. 2009;284(11):6841–6846. doi: 10.1074/jbc.M806452200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waxman L, Whitney M, Pollok BA, Kuo LC, Darke PL. Host cell factor requirement for hepatitis C virus enzyme maturation. Proc Natl Acad Sci U S A. 2001;98(24):13931–13935. doi: 10.1073/pnas.241510898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weidmann M, Muhlberger E, Hufert FT. Rapid detection protocol for filoviruses. J Clin Virol. 2004;30(1):94–99. doi: 10.1016/j.jcv.2003.09.004. [DOI] [PubMed] [Google Scholar]
- Whelan SP, Barr JN, Wertz GW. Transcription and replication of nonsegmented negative-strand RNA viruses. Curr Top Microbiol Immunol. 2004;283:61–119. doi: 10.1007/978-3-662-06099-5_3. [DOI] [PubMed] [Google Scholar]
- Whitesell L, Lindquist SL. HSP90 and the chaperoning of cancer. Nat Rev Cancer. 2005;5(10):761–772. doi: 10.1038/nrc1716. [DOI] [PubMed] [Google Scholar]