Abstract
Rev1, a Y-family DNA polymerase, contributes to spontaneous and DNA damage-induced mutagenic events. In this paper, we have employed pre-steady state kinetic methodology to establish a kinetic basis for nucleotide selection by human Rev1, a unique nucleotidyl transferase that uses a protein template-directed mechanism to preferentially instruct dCTP incorporation. This work demonstrated that the high incorporation efficiency of dCTP is dependent on both substrates: an incoming dCTP and a templating base dG. The extremely low base substitution fidelity of human Rev1 (100 to 10-5) was due to the preferred misincorporation of dCTP with templating bases dA, dT, and dC over correct dNTPs. Using non-natural nucleotide analogs, we showed that hydrogen bonding interactions between residue R357 of human Rev1 and an incoming dNTP are not essential for DNA synthesis. Lastly, human Rev1 discriminates between ribonucleotides and deoxyribonucleotides mainly by reducing the rate of incorporation, and the sugar selectivity of human Rev1 is sensitive to both the size and orientation of the 2′-substituent of a ribonucleotide.
The human genome encodes at least 16 DNA polymerases (Pol) that are involved in replicating and maintaining the integrity of genomic DNA. Human DNA polymerases are classified into four families: A, B, X, and Y. Y-family DNA polymerases are involved in DNA damage tolerance pathways, whereby a Y-family enzyme rescues stalled DNA replication at sites of DNA damage. Humans have four known Y-family members: Pol η, Pol ι, Pol κ, and Rev1. Rev1 is found in the genome of all eukaryotes (1) and is capable of functioning in both catalytic and structural roles. Composed of 1,251 amino acids (2), human Rev1 (hRev1) is organized into a central catalytic domain that is flanked by an N-terminal BRCT domain and a C-terminus with two ubiquitin-binding motifs and a domain for polymerase interactions (3). As a scaffold protein, Rev1 interacts with proliferating cell nuclear antigen (PCNA) (4-7), ubiquitinated proteins (5, 6), and DNA polymerases η, κ, ι, and ζ (8-15). These findings support a model, whereby Rev1 is involved in polymerase switching at sites of DNA damage (16-18). In regards to enzymatic activity, hRev1 preferentially inserts dCTP opposite a templating base dG (2, 19-22), however, unlike other human DNA polymerases, this incorporation event proceeds in a protein template-directed manner rather than a DNA template-dependent manner with Watson-Crick base pairing (23). Instead, the incoming dCTP hydrogen bonds with R357, and the extrahelical template base dG is accommodated in a hydrophobic pocket while L358 rests in the conventional location of a templating base (Figure 1) (23).
Figure 1. Active site of hRev1.

Important active site residues that interact with an incoming dCTP or the templating base dG are shown (PDB 3GQC). The dashed lines represent hydrogen bonds, and the four magnesium ions are shown as gray spheres.
Rev1 and Polζ are responsible for the majority of spontaneous and DNA damage-induced mutagenic events in yeast; early studies reveal similar findings in mammalian cell lines (24-26). In human tissues, the rev1 gene is ubiquitously expressed, but the highest level of expression is in human testis and ovary based on RT-PCR results (2, 8, 19). Furthermore, hRev1 has been observed at replication foci during both G1 and S phases following UV-irradiation (27). However, it has also been reported that the protein levels of hRev1 are unaffected by UV irradiation or cell cycle progression (28). In addition to a role in translesion synthesis, Rev1 has been implicated in somatic hypermutation, and current data suggests the catalytic domain participates in the generation of C to G transversions (29, 30). To better understand the enzymatic function of hRev1, we have performed pre-steady state kinetic analysis on a truncated version of hRev1. Our studies established a kinetic basis for nucleotide selection by hRev1.
Experimental Procedures
Materials
These chemicals were purchased from the following companies: [γ-32P]ATP, MP Biomedicals; deoxyribonucleotide 5′-triphosphates, GE Healthcare; ribonucleotide 5′-triphosphates, MBI Fermentas; 2′-aracytidine-5′-triphosphate (araCTP), 2′-deoxy-2′,2′-difluorocytidine-5′-triphosphate (GemCTP), 2′-fluoro-2′-deoxycytidine-5′-triphosphate (2′-F-CTP), 2′-O-methylcytidine-5′-triphosphate (2′-OCH3-CTP), and 5-nitroindole 5′-triphosphate (dNITP), TriLink Biotechnologies; Bio-Spin 6 columns, Bio-Rad Laboratories; OptiKinase™, USB Corporation; synthetic oligodeoxyribonucleotides 21-mer, 5′-phosphorylated 19-mer, and 41-mers, Integrated DNA Technologies. Pyrene 5′-triphosphate (dPTP) was a generous gift from Dr. John-Stephen Taylor (Washington University at St. Louis).
Expression and purification of hRev1
The expression plasmid pBAD-REV1S, a generous gift from K. Kamiya at Hiroshima University, encoded a truncated version of human Rev1 (341-829) (31). The expression and purification of truncated human Rev1 was performed as previously described (19).
DNA substrates
Commercially synthesized oligomers in Table 1 were purified using polyacrylamide gel electrophoresis (32, 33). The 21-mer primer was radiolabeled with [γ-32P]ATP and OptiKinase™ according to the manufacturer's protocol, and the unreacted [γ-32P]ATP was subsequently removed via a Bio-Spin 6 column. The primer-template DNA substrates (32) and single-nucleotide gap DNA substrate (33) were annealed as described previously.
Table 1.
Sequences of the D-DNA substratesa
| D-G | 5′–CGCAGCCGTCCAACCAACTCA–3′ |
| 3′–GCGTCGGCAGGTTGGTTGAGTGTCAGCTAGGTTACGGCAGG–5′ | |
| D-A | 5′–CGCAGCCGTCCAACCAACTCA–3′ |
| 3′–GCGTCGGCAGGTTGGTTGAGTATCAGCTAGGTTACGGCAGG–5′ | |
| D-T | 5′–CGCAGCCGTCCAACCAACTCA–3′ |
| 3′–GCGTCGGCAGGTTGGTTGAGTTTCAGCTAGGTTACGGCAGG–5′ | |
| D-C | 5′–CGCAGCCGTCCAACCAACTCA–3′ |
| 3′–GCGTCGGCAGGTTGGTTGAGTCTCAGCTAGGTTACGGCAGG–5′ | |
| D-G Gap | 5′–CGCAGCCGTCCAACCAACTCA AGTCGATCCAATGCCGTCC–3′ |
| 3′–GCGTCGGCAGGTTGGTTGAGTGTCAGCTAGGTTACGGCAGG–5′ |
Each DNA substrate is composed of a 5′-radiolabeled 21-mer and a 41-mer template which has the unique template bases in bold. D-G Gap has a 5′-phosphorylated 19-mer.
Measurement of the kp and Kd for single nucleotide incorporation
Kinetic assays were completed using buffer R (50 mM HEPES, pH 7.5 at 37 °C, 5 mM MgCl2, 50 mM NaCl, 0.1 mM EDTA, 5 mM DTT, 10% glycerol, and 0.1 mg/ml BSA). All kinetic experiments described herein were performed at 37 °C, and the reported concentrations were final after mixing all of the components. A pre-incubated solution containing hRev1 (120 nM) and 5′-[32P]-radiolabeled DNA substrate (30 nM) was mixed with increasing concentrations (0.02-800 μM) of nucleotide in buffer R at 37 °C. Aliquots of the reaction mixtures were quenched at various times using 0.37 M EDTA. A rapid chemical-quench flow apparatus (KinTek) was utilized for fast nucleotide incorporations. Reaction products were resolved using sequencing gel electrophoresis (17% acrylamide, 8 M urea) and quantitated with a Typhoon TRIO (GE Healthcare). The time course of product formation at each nucleotide concentration was fit to a single-exponential equation (Eq. 1) using a nonlinear regression program, KaleidaGraph (Synergy Software), to yield an observed rate constant of nucleotide incorporation (kobs). The kobs values were then plotted as a function of nucleotide concentration and fit using the hyperbolic equation (Eq. 2) which resolved the kp and Kd values for nucleotide incorporation catalyzed by hRev1.
| Eq. 1 |
| Eq. 2 |
Results
Kinetic basis of dNTP selection
Transient state kinetic methods were employed to measure the substrate specificity and polymerase fidelity of a truncated form of hRev1. A pre-incubated solution of hRev1 (120 nM) and 5′-[32P]-labeled D-G DNA (30 nM) was mixed with increasing concentrations of dCTP•Mg2+ (see Experimental Procedures). These single-turnover conditions in which hRev1 is in molar excess over DNA permits the direct observation of the DNA substrate being converted to the extended DNA product in a single pass through the enzymatic pathway (34). The extended DNA product was quantitated, plotted (Figure 2), and fit to the appropriate equations (Equations 1 or 2) that resolved a maximum rate of nucleotide incorporation (kp) of 22.4 ± 0.9 s-1 and an equilibrium dissociation constant (Kd) of 2.2 ± 0.3 μM (Table 2). Notably, Tsai and Johnson report that nucleotide binding to T7 DNA polymerase, an A-family enzyme, induces several conformational changes preceding the incorporation step, thereby arguing that the measured Kd value under single-turnover reaction conditions is not a true equilibrium dissociation constant (35). Since there is no published evidence to support the existence of such conformational changes for the protein template-directed hRev1, we assume the Kd values measured in this paper reflect the true nucleotide binding affinity (1/Kd). To examine how efficient hRev1 incorporates dCTP opposite other templating bases, we performed similar single-turnover assays using DNA substrates with dA (D-A), dC (D-C), and dT (D-T) as the template base (Table 2). The substrate specificity constants (kp/Kd), efficiency ratio, and fidelity were calculated. The ground-state binding affinity dropped 4- to 55-fold while the rate for dCTP incorporation was reduced by 7- to 12-fold when the templating base was not dG. Overall, the catalytic efficiency was up to 360-fold greater when dCTP was inserted into D-G. The preferential order of dCTP incorporation opposite the four template bases was dG ≫ dA > dT ≈ dC.
Figure 2. Concentration dependence on the pre-steady state rate constant of deoxycytidyl transferase catalyzed by hRev1.
(A) A pre-incubated solution of hRev1 (120 nM) and 5′-[32P]-labeled D-6T (30 nM) was rapidly mixed with increasing concentrations of dCTP •Mg2+ (0.2 μM, ●; 0.5 μM, ○; 1 μM, ■; 2 μM, □; 5 μM, ▲; 10 μM, △; and 25 μM, ◆) for various time intervals. The solid lines are the best fits to a single-exponential equation which determined the observed rate constant, kobs. (B) The kobs values were plotted as a function of dCTP concentration. The data (●) were then fit to a hyperbolic equation, yielding a kp of 22.4 ± 0.9 s-1 and a Kd of 2.2 ± 0.3 μM.
Table 2.
Kinetic parameters for nucleotide incorporation into D-DNA catalyzed by hRev1 at 37 °C.
| dNTP | kp (s-1) | Kd (μM) | kp/Kd (μM-1s-1) | Efficiency Ratioa | Fidelityb |
|---|---|---|---|---|---|
| Template dG (D-G) | |||||
| dCTP | 22.4 ± 0.9 | 2.2 ± 0.3 | 10 | ||
| dATP | 0.050 ± 0.004 | 70 ± 20 | 7.1 × 10-4 | 1.4 × 104 | 7.0 × 10-5 |
| dGTP | 6.3 ± 0.3 | 90 ± 10 | 7.0 × 10-2 | 1.5 × 102 | 6.8 × 10-3 |
| dTTP | 0.88 ± 0.06 | 22 ± 7 | 4.0 × 10-2 | 2.5 × 102 | 3.9 × 10-3 |
| Template dA (D-A) | |||||
| dTTP | 0.092 ± 0.007 | 44 ± 6 | 2.1 × 10-3 | 4.9 × 103 | |
| dCTP | 1.87 ± 0.05 | 9.5 ± 0.8 | 2.0 × 10-1 | 5.2 × 101 | 9.9 × 10-1 |
| Template dT (D-T) | |||||
| dATP | 0.00235 ± 0.00008 | 2.7 ± 0.4 | 8.7 × 10-4 | 1.2 × 104 | |
| dCTP | 1.93 ± 0.05 | 35 ± 2 | 5.5 × 10-2 | 1.8 × 102 | 9.8 × 10-1 |
| Template dC (D-C) | |||||
| dGTP | 0.0073 ± 0.0010 | 30 ± 10 | 2.4 × 10-4 | 4.2 × 104 | |
| dCTP | 3.4 ± 0.2 | 120 ± 10 | 2.8 × 10-2 | 3.6 × 102 | 9.9 × 10-1 |
| Template dG (D-G Gap) | |||||
| dCTP | 11 ± 1 | 8 ± 2 | 1.4 | 7.4 | |
Calculated as (kp/Kd)dCTP:D-G/(kp/Kd)dNTP:dN.
Calculated as (kp/Kd)incorrect/[(kp/Kd)correct + (kp/Kd)incorrect].
Next, we measured the catalytic efficiency of nucleotide incorporation for the three remaining Watson-Crick base pair combinations under single-turnover conditions and the kinetic data are listed in Table 2. Compared to dCTP:dG, the catalytic efficiency of hRev1 dropped 4,900-, 12,000- and 42,000-fold for dTTP:dA, dATP:dT, and dGTP:dC, respectively. Despite a change in the identity of an incoming dNTP, the template preference remained the same based on the substrate specificity constant as observed with dCTP. The binding affinity remained high for dATP but was ∼14- and 20-fold weaker for dGTP and dTTP. Furthermore, the rate of dCTP incorporation into D-A, D-T, and D-C DNA was up to 820-fold faster than the canonical dNTP, therefore, the strong dCTP preference by hRev1 with templating bases dA, dT, and dC leads to an extremely low fidelity of ∼1 (Table 2). Please note, enzyme fidelity is calculated using the standard kinetic equation, (kp/Kd)incorrect/[(kp/Kd)correct + (kp/Kd)incorrect]. When fidelity approaches a value of 1, this indicates that a misincorporation is favored over the canonical Watson-Crick base pair and that a correct incorporation is not likely to occur. Therefore, to better understand the frequency of a correct incorporation catalyzed by hRev1, the following equation was used: (kp/Kd)correct/(kp/Kd)dCTP:dN. Here, the frequency of a correct incorporation is calculated to be 1.1 × 10-2, 1.6 × 10-2, and 8.6 × 10-3 for dTTP:dA, dATP:dT, and dGTP:dC, respectively. These values translate into approximately one correct incorporation (dTTP, dATP, or dGTP) per 100 dCTP misincorporations.
Since hRev1 displayed greater catalytic efficiency when dG is the template base, we determined the substrate specificity constant for the incorporation of the other dNTPs into D-G DNA (Table 2). The efficiency to form base pairs dATP:dG, dGTP:dG, and dTTP:dG was 1-, 290-, and 20-fold greater than dATP:dT, dGTP:dC, and dTTP:dA, respectively. Surprisingly, relative to the other template bases, the rate of nucleotide incorporation was up to 860-fold faster when the substrate had dG positioned as the template base. Meanwhile, the Kd value was at least 10-fold higher for non-dCTP addition into D-G DNA. The fidelity of hRev1 inserting dNTPs opposite dG ranged from 10-3 to 10-5.
It has been shown that hRev1 may participate in cellular processes that involve gapped DNA (36). Determining the pre-steady state kinetic parameters for dCTP incorporation into a single-nucleotide gapped DNA substrate (D-G Gap) revealed that hRev1 is 7-fold more efficient with the primer-template D-G DNA substrate (Table 2). This modest effect can be attributed to a 2-fold slower rate and a 4-fold weaker binding affinity for dCTP incorporation.
Importance of hydrogen bonding and base stacking
Crystallographic studies have shown that hRev1 utilizes a protein template-directed mechanism to instruct dCTP incorporation through hydrogen bonding between cytosine and residue R357 of hRev1 (Figure 1) (23). To evaluate the roles of hydrogen bonding, base stacking, and base size during DNA synthesis, we have measured the catalytic efficiency of hRev1 incorporating two non-natural nucleotide analogs into D-G DNA (Figure 3A and Table 3). Both dPTP and dNITP lack the ability to form strong hydrogen bonds, possess greater base stacking energy, and are physically larger than dCTP (37). hRev1 can incorporate both analogs, although, the incorporation efficiency drops by 3,500- and 11,000-fold for dNITP and dPTP, respectively. Both analogs are incorporated with significantly reduced rates (at least 490-fold) and modestly weakened binding affinities (at least 7-fold). These data suggested that hydrogen bonding is not essential for catalysis, but it does enhance the rate and binding affinity for dCTP incorporation.
Figure 3. Chemical structures of nucleotide analogs.
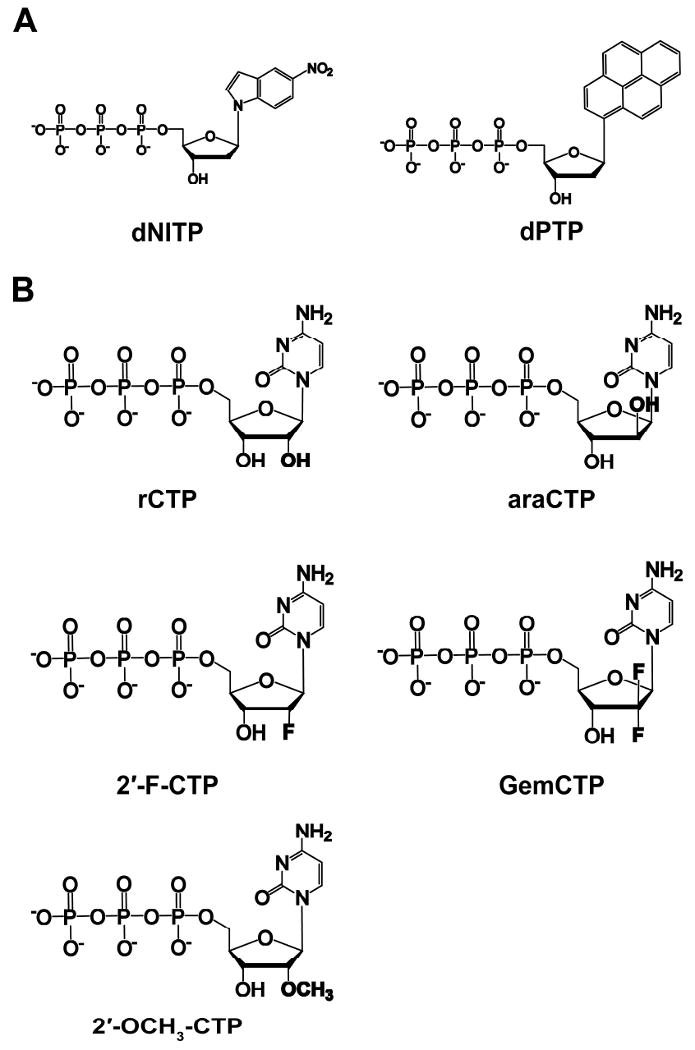
(A) non-natural nucleotide analogs and (B) CTP analogs used in this work.
Table 3.
Kinetic parameters for non-natural nucleotide analog incorporation into D-G DNA catalyzed by hRev1 at 37 °C.
| dNTP | kp (s-1) | Kd (μM) | kp/Kd (μM-1s-1) | Efficiency Ratioa |
|---|---|---|---|---|
| dCTP | 22.4 ± 0.9 | 2.2 ± 0.3 | 10 | |
| dATP | 0.050 ± 0.004 | 70 ± 20 | 7.1 × 10-4 | 1.4 × 104 |
| dNITP | 0.0457 ± 0.0006 | 15.8 ± 0.6 | 2.9 × 10-3 | 3.5 × 103 |
| dPTP | 0.0228 ± 0.0008 | 25 ± 3 | 9.1 × 10-4 | 1.1 × 104 |
Calculated as (kp/Kd)dCTP/(kp/Kd)dNTP.
Kinetic basis of ribonucleotide selection
The concentration of cellular dNTP pools fluctuate during the cell cycle, and the levels are 10- to 200-fold less than the ribonucleotide (rNTP) pools which remain relatively high and constant (38, 39). Since hRev1 has been shown to be present outside of S phase (28), we have evaluated the sugar selectivity of hRev1 by measuring the substrate specificity constant for various CTP analogs (Figure 3B and Table 4). hRev1 discriminates between dCTP and rCTP by 280-fold, and this is mostly due to a 230-fold rate decrease. To better understand how size and orientation affect the degree of sugar selectivity, we have used araCTP (an anti-cancer drug that is a steric isomer of rCTP with the 2′-OH pointed above the ribose ring), 2′-F-CTP (the 2′-F group is smaller than the 2′-OH), GemCTP (an anti-cancer drug with two fluorines at the 2′ position), and 2′-OCH3-CTP (the 2′-OCH3 group is larger than the 2′-OH). Orientation and reduced size of the 2′ group are important factors because the efficiency of hRev1 incorporating araCTP and 2′-F-CTP was similar to dCTP. In contrast, the increased volume of the 2′-methoxy group enhanced the magnitude of discrimination to 6,700. Surprisingly, most of the sugar selection was kp driven for hRev1. The one exception is for GemCTP where the Kd value increased by 13-fold.
Table 4.
Kinetic parameters for CTP analog incorporation into D-G DNA catalyzed by hRev1 at 37 °C.
| NTP | kp (s-1) | Kd (μM) | kp/Kd (μM-1s-1) | Sugar Selectivitya |
|---|---|---|---|---|
| dCTP | 22.4 ± 0.9 | 2.2 ± 0.3 | 10 | |
| rCTP | 0.098 ± 0.002 | 2.7 ± 0.2 | 3.6 × 10-2 | 280 |
| araCTP | 6.3 ± 0.5 | 4 ± 1 | 1.6 | 6 |
| 2′-F-CTP | 19.2 ± 0.5 | 3.5 ± 0.4 | 5.5 | 2 |
| GemCTP | 6.8 ± 0.4 | 29 ± 6 | 2.3 × 10-1 | 43 |
| 2′-OCH3-CTP | 0.0122 ± 0.0006 | 8 ± 1 | 1.5 × 10-3 | 6,700 |
Calculated as (kp/Kd)dCTP/(kp/Kd)analog.
Discussion
Comparison of base substitution fidelity
As a dCTP transferase, Rev1 is a DNA polymerase with extremely low fidelity due to the preference to form dCTP:dN base pairs over canonical Watson-Crick base pairs dTTP:dA, dATP:dT, and dGTP:dC. Using pre-steady state kinetic methods, we have established a base substitution fidelity of 100 to 10-5 for truncated hRev1 synthesizing undamaged DNA (Table 2). This fidelity range is similar to other human Y-family DNA polymerases (40) and a fidelity range of 100 to 10-4 that was estimated for full-length hRev1 under semi-steady-state kinetic conditions by Zhang et al. (22). In their studies, Zhang et al. used too much full-length hRev1 (14 fmol) in the reactions with 50 fmol of DNA and various dNTPs at 30 °C (22), possibly due to the lack of quantifiable reaction products during non-dCTP incorporations. Thus, their semi-steady-state kinetic parameters cannot be used to kinetically describe nucleotide incorporation catalyzed by hRev1. In this paper, we employed pre-steady state kinetic methods to investigate the kinetic basis for nucleotide selection and enzyme fidelity for hRev1. Our kinetic data revealed that hRev1 discriminates at both the nucleotide binding (Kd) and incorporation (kp) steps. Overall, hRev1 prefers dCTP:dG with a 20-fold tighter binding affinity and 14-fold faster rate of incorporation (on average) with undamaged DNA relative to the other tested dNTP:dN base pair combinations (Table 2).
Pre-steady state kinetic analyses have been completed with a truncated form of yeast Rev1 (yRev1, 1-746) (41). In stark contrast, yRev1 selects incoming nucleotides mostly at the nucleotide binding step (Kd). The catalytic efficiency for dCTP:dG is 660-fold greater for the human enzyme, and this effect is governed by a ∼1,900-fold faster rate of dCTP incorporation catalyzed by hRev1 at 37 °C (22.4 s-1) versus yRev1 at 22 °C (0.012 s-1), although, hRev1 (2.2 μM) binds dCTP with a 3-fold weaker affinity than yRev1 (0.78 μM) (41). Interestingly, significant kinetic differences have been observed for human and yeast Polη at varying reaction temperatures, too (42). Thus, it is important to exercise caution when extending conclusions about DNA polymerase homologs derived from different organisms (14, 43).
Effect of DNA substrate on the catalytic efficiency of hRev1
Translesion DNA synthesis has been proposed to proceed through a polymerase-switching or gap-filling model (44). Also, Rev1 has been shown to be important during UV-induced post replicative gap-filling processes that likely occur outside of S phase (36, 44). Although the incorporation efficiency dropped by ∼7-fold from non-gapped to gapped DNA, hRev1 is capable of accommodating a single-nucleotide gap DNA substrate despite lacking the signature helix-hairpin-helix (HhH) motif that Polβ and Polλ, two X-family DNA polymerases specialized for gap-filling DNA synthesis, use to bind the downstream strand. Moreover, the gap-filling efficiency of 1.4 μM-1s-1 for hRev1 is close or similar to the values measured for rat Polβ (6.6 μM-1s-1) and human Polλ (1.8 μM-1s-1) (Table 2) (45, 46). More studies are needed to evaluate whether hRev1 plays a role in gap-filling DNA synthesis in vivo.
Kinetic basis for nucleotide selection
Watson-Crick hydrogen bond formation between the template base and incoming dNTP has been shown to play an important role in nucleotide selection by many DNA polymerases including T7 DNA polymerase (47). However, hRev1 does not use this DNA template-dependent mechanism to select incoming dNTPs. Instead, it uses the protein template-directed mechanism while the templating base dG is evicted from the active site by L358 so that it fits into a hydrophobic pocket surrounded by F525, K770, and H774 (Figure 1). To probe whether hydrogen bonds between cytosine and R357 are essential for catalysis by hRev1, we examined if hRev1 could incorporate dNITP and dPTP which are unable to form hydrogen bonds. Although efficiency was reduced dramatically (Table 3), these non-natural nucleotide analogs were incorporated into DNA by hRev1. These results suggested that hydrogen bonds formed between the incoming dNTP and R357 are important, but not absolutely essential for efficient nucleotide incorporation catalyzed by hRev1 and that an oversized nucleobase with strong base stacking energy can be accommodated. To better understand the role of hydrogen bonds, additional studies need to be performed using isosteric, non-hydrogen bonding dCTP analogs.
Previously, Howell et al. (41) proposed possible interactions (i.e. hydrogen bonds and base conformations) for the four dNTP:Arg combinations based on the X-ray crystal structures of yRev1•DNA•dCTP (48) and E. coli MutM DNA glycosylase•DNA (49). Interestingly, the number of hydrogen bonds correlates with the substrate specificity of dNTP incorporation into DNA with dG as the template for both yRev1 and hRev1: dCTP (2 hydrogen bonds) > dGTP (2 hydrogen bonds if dGTP adopts a syn conformation) ≈ dTTP (1 hydrogen bond) > dATP (0 hydrogen bonds) (41, 50). However, the identity of the template base also contributes to catalytic efficiency since dCTP misincorporation is less efficient for hRev1 (Table 2). Thus, optimal catalytic activity (kp/Kd) of hRev1 depends on both substrates: an incoming dCTP and the template base dG.
Kinetic basis for ribonucleotide exclusion
Most DNA polymerases prevent ribonucleotide incorporation via a steric clash between the 2′-OH group of an incoming rNTP and a protein backbone segment (51) or bulky side chain residue of the enzyme (52-56). This mechanism usually yields sugar selectivity values greater than 1,000-fold (51, 52, 54-57). hRev1 discriminates between dCTP and rCTP by 280-fold, a value that is relatively low compared to other DNA polymerases (Table 4). Like other DNA polymerases, hRev1 possesses a putative steric gate residue F428 but its benzene ring almost parallels and stacks to the ribose ring (Figure 1) (23). Thus, it is unclear how hRev1 discriminates against rNTPs. In general, the kinetic basis for rNTP discrimination by most DNA polymerases is via weakened binding and slowed incorporation of rNTPs. Using CTP analogs, we showed that the mechanism of ribonucleotide selection employed by hRev1 is influenced by both the size and orientation of the 2′ group (Table 5). With varying sizes of the 2′ substituent, the Kd values for 2′-F-CTP, rCTP, araCTP, and 2′-OCH3-CTP were not affected significantly. This is probably due to the favorable hydrogen bonding interactions between residue R357 of hREV1 and the cytosine base which compensated for the steric effect of the 2′ substituent. However, the binding of GemCTP to hRev1•D-G DNA was perturbed the most with a 13-fold lower affinity than dCTP. The geminal difluoro group of GemCTP has more electronegativity than the deoxyribose of dCTP, and an embedded GemCMP residue in duplex DNA adopts a C3′-endo pucker (58). These may affect how GemCTP was positioned in the active site and how it interacted with R357 and F428 of hRev1, leading to the lower affinity. Interestingly, a similar conclusion has been drawn for the human mitochondrial DNA polymerase γ incorporating GemCTP (59). In comparison, Table 4 shows that the kp variation is much larger than the Kd range for the CTP analogs. If the ribose 2′ substituent either has a small size (e.g. 2′-F in both 2′-F-CTP and GemCTP) or is orientated above the ribose ring (e.g. 2′-OH in araCTP), it has a small impact on the kp value. Contrary to these trends, the kp values for rCTP and 2′-OCH3-CTP are 200- to 2,000-fold lower than that of dCTP. Together, these results suggested that, inconsistent with the general kinetic trends observed with other DNA polymerases (see above), the steric clash of the 2′-OH of the incoming rCTP with F428 of hRev1 mostly impacts the incorporation step (kp) rather than the ground-state binding step (Kd). In addition to the major contribution of the templating base dG to the high dCTP incorporation efficiency (see above discussion), our kinetic data further dissect the contribution of each chemical moiety toward the high efficiency of dCTP incorporation catalyzed by hRev1: the ribose 2′-H of dCTP significantly contributes to the fast kp while the cytosine of dCTP contributes to the low Kd for dCTP binding. We are currently elucidating the kinetic mechanism of dCTP incorporation in order to mechanistically understand how these chemical moieties of dCTP influence its kp and Kd.
Acknowledgments
We thank Dr. John-Stephen Taylor for providing us dPTP and Dr. Kenji Kamiya for providing the expression plasmid pBAD-REV1S.
Abbreviations
- 2′-F-CTP
2′-fluoro-2′-deoxycytidine-5′-triphosphate
- 2′-OCH3-CTP
2′-O-methylcytidine-5′-triphosphate
- araCTP
2′-aracytidine-5′-triphosphate
- BSA
bovine serum albumin
- dNITP
5-nitroindole 5′-triphosphate
- dNTP
2′-deoxyribonucleotide 5′-triphosphate
- dPTP
pyrene 5′-triphosphate
- DTT
dithiothreitol
- EDTA
ethylenediaminetetraacetic acid
- GemCTP
2′-deoxy-2′,2′-difluorocytidine-5′-triphosphate
- HhH
helix-hairpin-helix
- hRev1
human Rev1
- PCNA
proliferating cell nuclear antigen
- Pol
DNA polymerase
- Polη
DNA polymerase eta
- Polι
DNA polymerase iota
- Polκ
DNA polymerase kappa
- rNTP
ribonucleotide-5′-triphosphate
- yRev1
yeast Rev1
Footnotes
This work was supported by the National Institutes of Health Grants GM079403 and ES009127 to Z.S. J.A.B was supported by an American Heart Association Pre-doctoral Fellowship (Grant 0815382D) and an International P.E.O. Scholar Award. J.D.F. was supported by a Post-doctoral Fellowship from a Pulmonary National Institutes of Health Training Grant 5T32HL007946 (PI: Mark D. Wewers).
References
- 1.Burgers PM, Koonin EV, Bruford E, Blanco L, Burtis KC, Christman MF, Copeland WC, Friedberg EC, Hanaoka F, Hinkle DC, Lawrence CW, Nakanishi M, Ohmori H, Prakash L, Prakash S, Reynaud CA, Sugino A, Todo T, Wang Z, Weill JC, Woodgate R. Eukaryotic DNA polymerases: proposal for a revised nomenclature. J Biol Chem. 2001;276:43487–43490. doi: 10.1074/jbc.R100056200. [DOI] [PubMed] [Google Scholar]
- 2.Lin W, Xin H, Zhang Y, Wu X, Yuan F, Wang Z. The human REV1 gene codes for a DNA template-dependent dCMP transferase. Nucleic Acids Res. 1999;27:4468–4475. doi: 10.1093/nar/27.22.4468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Yang W, Woodgate R. What a difference a decade makes: insights into translesion DNA synthesis. Proc Natl Acad Sci U S A. 2007;104:15591–15598. doi: 10.1073/pnas.0704219104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ross AL, Simpson LJ, Sale JE. Vertebrate DNA damage tolerance requires the C-terminus but not BRCT or transferase domains of REV1. Nucleic Acids Res. 2005;33:1280–1289. doi: 10.1093/nar/gki279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Guo C, Sonoda E, Tang TS, Parker JL, Bielen AB, Takeda S, Ulrich HD, Friedberg EC. REV1 protein interacts with PCNA: significance of the REV1 BRCT domain in vitro and in vivo. Mol Cell. 2006;23:265–271. doi: 10.1016/j.molcel.2006.05.038. [DOI] [PubMed] [Google Scholar]
- 6.Guo C, Tang TS, Bienko M, Parker JL, Bielen AB, Sonoda E, Takeda S, Ulrich HD, Dikic I, Friedberg EC. Ubiquitin-binding motifs in REV1 protein are required for its role in the tolerance of DNA damage. Mol Cell Biol. 2006;26:8892–8900. doi: 10.1128/MCB.01118-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wood A, Garg P, Burgers PM. A ubiquitin-binding motif in the translesion DNA polymerase Rev1 mediates its essential functional interaction with ubiquitinated proliferating cell nuclear antigen in response to DNA damage. J Biol Chem. 2007;282:20256–20263. doi: 10.1074/jbc.M702366200. [DOI] [PubMed] [Google Scholar]
- 8.Murakumo Y, Ogura Y, Ishii H, Numata S, Ichihara M, Croce CM, Fishel R, Takahashi M. Interactions in the error-prone postreplication repair proteins hREV1, hREV3, and hREV7. J Biol Chem. 2001;276:35644–35651. doi: 10.1074/jbc.M102051200. [DOI] [PubMed] [Google Scholar]
- 9.Guo C, Fischhaber PL, Luk-Paszyc MJ, Masuda Y, Zhou J, Kamiya K, Kisker C, Friedberg EC. Mouse Rev1 protein interacts with multiple DNA polymerases involved in translesion DNA synthesis. Embo J. 2003;22:6621–6630. doi: 10.1093/emboj/cdg626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Masuda Y, Ohmae M, Masuda K, Kamiya K. Structure and enzymatic properties of a stable complex of the human REV1 and REV7 proteins. J Biol Chem. 2003;278:12356–12360. doi: 10.1074/jbc.M211765200. [DOI] [PubMed] [Google Scholar]
- 11.Tissier A, Kannouche P, Reck MP, Lehmann AR, Fuchs RPP, Cordonnier A. Co-localization in replication foci and interaction of human Y-family members, DNA polymerase pol eta and REV1 protein. DNA Repair. 2004;3:1503–1514. doi: 10.1016/j.dnarep.2004.06.015. [DOI] [PubMed] [Google Scholar]
- 12.Ohashi E, Murakumo Y, Kanjo N, Akagi J, Masutani C, Hanaoka F, Ohmori H. Interaction of hREV1 with three human Y-family DNA polymerases. Genes Cells. 2004;9:523–531. doi: 10.1111/j.1356-9597.2004.00747.x. [DOI] [PubMed] [Google Scholar]
- 13.Yuasa MS, Masutani C, Hirano A, Cohn MA, Yamaizumi M, Nakatani Y, Hanaoka F. A human DNA polymerase eta complex containing Rad18, Rad6 and Rev1; proteomic analysis and targeting of the complex to the chromatin-bound fraction of cells undergoing replication fork arrest. Genes Cells. 2006;11:731–744. doi: 10.1111/j.1365-2443.2006.00974.x. [DOI] [PubMed] [Google Scholar]
- 14.Kosarek JN, Woodruff RV, Rivera-Begeman A, Guo C, D'Souza S, Koonin EV, Walker GC, Friedberg EC. Comparative analysis of in vivo interactions between Rev1 protein and other Y-family DNA polymerases in animals and yeasts. DNA Repair (Amst) 2008;7:439–451. doi: 10.1016/j.dnarep.2007.11.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ohashi E, Hanafusa T, Kamei K, Song I, Tomida J, Hashimoto H, Vaziri C, Ohmori H. Identification of a novel REV1-interacting motif necessary for DNA polymerase kappa function. Genes Cells. 2009;14:101–111. doi: 10.1111/j.1365-2443.2008.01255.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kannouche P, Stary A. Xeroderma pigmentosum variant and error-prone DNA polymerases. Biochimie. 2003;85:1123–1132. doi: 10.1016/j.biochi.2003.10.009. [DOI] [PubMed] [Google Scholar]
- 17.Friedberg EC, Lehmann AR, Fuchs RP. Trading places: how do DNA polymerases switch during translesion DNA synthesis? Mol Cell. 2005;18:499–505. doi: 10.1016/j.molcel.2005.03.032. [DOI] [PubMed] [Google Scholar]
- 18.Lehmann AR, Niimi A, Ogi T, Brown S, Sabbioneda S, Wing JF, Kannouche PL, Green CM. Translesion synthesis: Y-family polymerases and the polymerase switch. DNA Repair (Amst) 2007;6:891–899. doi: 10.1016/j.dnarep.2007.02.003. [DOI] [PubMed] [Google Scholar]
- 19.Masuda Y, Takahashi M, Tsunekuni N, Minami T, Sumii M, Miyagawa K, Kamiya K. Deoxycytidyl transferase activity of the human REV1 protein is closely associated with the conserved polymerase domain. J Biol Chem. 2001;276:15051–15058. doi: 10.1074/jbc.M008082200. [DOI] [PubMed] [Google Scholar]
- 20.Masuda Y, Kamiya K. Biochemical properties of the human REV1 protein. FEBS Lett. 2002;520:88–92. doi: 10.1016/s0014-5793(02)02773-4. [DOI] [PubMed] [Google Scholar]
- 21.Choi JY, Guengerich FP. Kinetic analysis of translesion synthesis opposite bulky N2- and O6-alkylguanine DNA adducts by human DNA polymerase REV1. J Biol Chem. 2008;283:23645–23655. doi: 10.1074/jbc.M801686200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zhang Y, Wu X, Rechkoblit O, Geacintov NE, Taylor JS, Wang Z. Response of human REV1 to different DNA damage: preferential dCMP insertion opposite the lesion. Nucleic Acids Res. 2002;30:1630–1638. doi: 10.1093/nar/30.7.1630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Swan MK, Johnson RE, Prakash L, Prakash S, Aggarwal AK. Structure of the human Rev1-DNA-dNTP ternary complex. J Mol Biol. 2009;390:699–709. doi: 10.1016/j.jmb.2009.05.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lawrence CW. Cellular roles of DNA polymerase zeta and Rev1 protein. DNA Repair (Amst) 2002;1:425–435. doi: 10.1016/s1568-7864(02)00038-1. [DOI] [PubMed] [Google Scholar]
- 25.Clark DR, Zacharias W, Panaitescu L, McGregor WG. Ribozyme-mediated REV1 inhibition reduces the frequency of UV-induced mutations in the human HPRT gene. Nucleic Acids Res. 2003;31:4981–4988. doi: 10.1093/nar/gkg725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Simpson LJ, Sale JE. Rev1 is essential for DNA damage tolerance and non-templated immunoglobulin gene mutation in a vertebrate cell line. Embo J. 2003;22:1654–1664. doi: 10.1093/emboj/cdg161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Murakumo Y, Mizutani S, Yamaguchi M, Ichihara M, Takahashi M. Analyses of ultraviolet-induced focus formation of hREV1 protein. Genes Cells. 2006;11:193–205. doi: 10.1111/j.1365-2443.2006.00938.x. [DOI] [PubMed] [Google Scholar]
- 28.Akagi J, Masutani C, Kataoka Y, Kan T, Ohashi E, Mori T, Ohmori H, Hanaoka F. Interaction with DNA polymerase eta is required for nuclear accumulation of REV1 and suppression of spontaneous mutations in human cells. DNA Repair (Amst) 2009;8:585–599. doi: 10.1016/j.dnarep.2008.12.006. [DOI] [PubMed] [Google Scholar]
- 29.Ross AL, Sale JE. The catalytic activity of REV1 is employed during immunoglobulin gene diversification in DT40. Mol Immunol. 2006;43:1587–1594. doi: 10.1016/j.molimm.2005.09.017. [DOI] [PubMed] [Google Scholar]
- 30.Jansen JG, Langerak P, Tsaalbi-Shtylik A, van den Berk P, Jacobs H, de Wind N. Strand-biased defect in C/G transversions in hypermutating immunoglobulin genes in Rev1-deficient mice. J Exp Med. 2006;203:319–323. doi: 10.1084/jem.20052227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Masuda Y, Kamiya K. Role of single-stranded DNA in targeting REV1 to primer termini. J Biol Chem. 2006;281:24314–24321. doi: 10.1074/jbc.M602967200. [DOI] [PubMed] [Google Scholar]
- 32.Fiala KA, Suo Z. Pre-steady-state kinetic studies of the fidelity of Sulfolobus solfataricus P2 DNA polymerase IV. Biochemistry. 2004;43:2106–2115. doi: 10.1021/bi0357457. [DOI] [PubMed] [Google Scholar]
- 33.Fiala KA, Abdel-Gawad W, Suo Z. Pre-steady-state kinetic studies of the fidelity and mechanism of polymerization catalyzed by truncated human DNA polymerase lambda. Biochemistry. 2004;43:6751–6762. doi: 10.1021/bi049975c. [DOI] [PubMed] [Google Scholar]
- 34.Johnson KA. Transient-state kinetic analysis of enzyme reaction pathways. Enzymes. 1992;20:1–61. [Google Scholar]
- 35.Tsai YC, Johnson KA. A new paradigm for DNA polymerase specificity. Biochemistry. 2006;45:9675–9687. doi: 10.1021/bi060993z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Jansen JG, Tsaalbi-Shtylik A, Hendriks G, Gali H, Hendel A, Johansson F, Erixon K, Livneh Z, Mullenders LH, Haracska L, de Wind N. Separate domains of Rev1 mediate two modes of DNA damage bypass in mammalian cells. Mol Cell Biol. 2009;29:3113–3123. doi: 10.1128/MCB.00071-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Guckian KM, Schweitzer BA, Ren RXF, Sheils CJ, Tahmassebi DC, Kool ET. Factors contributing to aromatic stacking in water: evaluation in the context of DNA. J Am Chem Soc. 2000;122:2213–2222. doi: 10.1021/ja9934854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kornberg A, Baker TA. DNA replication. 2nd. W.H. Freeman; New York: 1992. [Google Scholar]
- 39.Traut TW. Physiological concentrations of purines and pyrimidines. Mol Cell Biochem. 1994;140:1–22. doi: 10.1007/BF00928361. [DOI] [PubMed] [Google Scholar]
- 40.McCulloch SD, Kunkel TA. The fidelity of DNA synthesis by eukaryotic replicative and translesion synthesis polymerases. Cell Res. 2008;18:148–161. doi: 10.1038/cr.2008.4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Howell CA, Prakash S, Washington MT. Pre-steady-state kinetic studies of protein-template-directed nucleotide incorporation by the yeast Rev1 protein. Biochemistry. 2007;46:13451–13459. doi: 10.1021/bi701429v. [DOI] [PubMed] [Google Scholar]
- 42.Washington MT, Johnson RE, Prakash L, Prakash S. The mechanism of nucleotide incorporation by human DNA polymerase eta differs from that of the yeast enzyme. Mol Cell Biol. 2003;23:8316–8322. doi: 10.1128/MCB.23.22.8316-8322.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Poltoratsky V, Horton JK, Prasad R, Wilson SH. REV1 mediated mutagenesis in base excision repair deficient mouse fibroblast. DNA Repair (Amst) 2005;4:1182–1188. doi: 10.1016/j.dnarep.2005.05.002. [DOI] [PubMed] [Google Scholar]
- 44.Waters LS, Minesinger BK, Wiltrout ME, D'Souza S, Woodruff RV, Walker GC. Eukaryotic translesion polymerases and their roles and regulation in DNA damage tolerance. Microbiol Mol Biol Rev. 2009;73:134–154. doi: 10.1128/MMBR.00034-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ahn J, Kraynov VS, Zhong X, Werneburg BG, Tsai MD. DNA polymerase beta: effects of gapped DNA substrates on dNTP specificity, fidelity, processivity and conformational changes. Biochem J. 1998;331(Pt 1):79–87. doi: 10.1042/bj3310079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Fiala KA, Duym WW, Zhang J, Suo Z. Up-regulation of the fidelity of human DNA polymerase lambda by its non-enzymatic proline-rich domain. J Biol Chem. 2006;281:19038–19044. doi: 10.1074/jbc.M601178200. [DOI] [PubMed] [Google Scholar]
- 47.Kool ET, Sintim HO. The difluorotoluene debate--a decade later. Chem Commun (Camb) 2006:3665–3675. doi: 10.1039/b605414e. [DOI] [PubMed] [Google Scholar]
- 48.Nair DT, Johnson RE, Prakash L, Prakash S, Aggarwal AK. Rev1 employs a novel mechanism of DNA synthesis using a protein template. Science. 2005;309:2219–2222. doi: 10.1126/science.1116336. [DOI] [PubMed] [Google Scholar]
- 49.Fromme JC, Verdine GL. Structural insights into lesion recognition and repair by the bacterial 8-oxoguanine DNA glycosylase MutM. Nat Struct Biol. 2002;9:544–552. doi: 10.1038/nsb809. [DOI] [PubMed] [Google Scholar]
- 50.Haracska L, Prakash S, Prakash L. Yeast Rev1 protein is a G template-specific DNA polymerase. J Biol Chem. 2002;277:15546–15551. doi: 10.1074/jbc.M112146200. [DOI] [PubMed] [Google Scholar]
- 51.Brown JA, Fiala KA, Fowler JD, Sherrer SM, Newmister SA, Duym WW, Suo Z. A Novel Mechanism of Sugar Selection Utilized by a Human X-Family DNA Polymerase. J Mol Biol. 2010;395:282–290. doi: 10.1016/j.jmb.2009.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Astatke M, Ng K, Grindley ND, Joyce CM. A single side chain prevents Escherichia coli DNA polymerase I (Klenow fragment) from incorporating ribonucleotides. Proc Natl Acad Sci U S A. 1998;95:3402–3407. doi: 10.1073/pnas.95.7.3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Gardner AF, Jack WE. Determinants of nucleotide sugar recognition in an archaeon DNA polymerase. Nucleic Acids Res. 1999;27:2545–2553. doi: 10.1093/nar/27.12.2545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Bonnin A, Lazaro JM, Blanco L, Salas M. A single tyrosine prevents insertion of ribonucleotides in the eukaryotic-type phi29 DNA polymerase. J Mol Biol. 1999;290:241–251. doi: 10.1006/jmbi.1999.2900. [DOI] [PubMed] [Google Scholar]
- 55.Yang G, Franklin M, Li J, Lin TC, Konigsberg W. A conserved Tyr residue is required for sugar selectivity in a Pol alpha DNA polymerase. Biochemistry. 2002;41:10256–10261. doi: 10.1021/bi0202171. [DOI] [PubMed] [Google Scholar]
- 56.DeLucia AM, Grindley ND, Joyce CM. An error-prone family Y DNA polymerase (DinB homolog from Sulfolobus solfataricus) uses a ‘steric gate’ residue for discrimination against ribonucleotides. Nucleic Acids Res. 2003;31:4129–4137. doi: 10.1093/nar/gkg417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Nick McElhinny SA, Ramsden DA. Polymerase mu is a DNA-directed DNA/RNA polymerase. Mol Cell Biol. 2003;23:2309–2315. doi: 10.1128/MCB.23.7.2309-2315.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Konerding D, James TL, Trump E, Soto AM, Marky LA, Gmeiner WH. NMR structure of a gemcitabine-substituted model Okazaki fragment. Biochemistry. 2002;41:839–846. doi: 10.1021/bi015678l. [DOI] [PubMed] [Google Scholar]
- 59.Fowler JD, Brown JA, Johnson KA, Suo Z. Kinetic investigation of the inhibitory effect of gemcitabine on DNA polymerization catalyzed by human mitochondrial DNA polymerase. J Biol Chem. 2008;283:15339–15348. doi: 10.1074/jbc.M800310200. [DOI] [PMC free article] [PubMed] [Google Scholar]



