Abstract
Krüppel-like factor 6 (Klf6; copeb in zebrafish) is a zinc-finger transcription factor and tumor suppressor gene. Klf6−/− mice have defects in hematopoiesis and angiogenesis and do not form a liver. However, the vascular abnormalities in Klf6−/− mice obfuscate its role in liver development since these two processes are linked in mammals. We utilized zebrafish and mouse ES cells to investigate the role of copeb in endoderm specification and hepatogenesis separate from its function in angiogenesis. During zebrafish development, copeb expression is enriched in digestive organs. Morpholino knockdown of copeb blocks expansion of the liver, pancreas and intestine, but does not affect their specification, differentiation or the vascularization of the liver. Decreased hepatocyte proliferation in copeb morphants is accompanied by upregulation of the cell cycle inhibitor, cdkn1a, a Copeb transcriptional target. A cell autonomous role for Klf6 in endoderm and hepatic development was investigated by manipulating Klf6 expression in mouse ES cells driven to differentiate along the hepatic lineage. Expression of the endoderm markers Hnf3β, Gata4, Sox17, and CxCr4 is not induced in Klf6−/− cells but is upregulated in ES cells over-expressing Klf6. Collectively, these findings indicate that copeb/Klf6 is essential for the development of endoderm-derived organs.
Keywords: hepatogenesis, copeb/klf6, zebrafish, ES cells, endoderm
Introduction
Hepatogenesis occurs in three phases: patterning of the endoderm to give rise to liver primordium, differentiation of hepatoblasts and the outgrowth and morphogenesis of the hepatic bud to form a functional liver with distinct lobes. While numerous studies have identified genes required for the first two phases, much less is known about the genes required for hepatic outgrowth and morphogenesis (Chu and Sadler, 2009; Zaret, 2002; Zhao and Duncan, 2005). The proliferation of hepatoblasts during this phase is tightly regulated, with evidence from mouse models suggesting that both growth stimulatory and inhibitory signals are involved; however, few experiments have tested this hypothesis directly. It is not yet clear whether hepatoblast proliferation is regulated in a tissue specific manner, or if the signals that control organ expansion are universal.
Klf6 belongs to the Sp1/Krüppel-like family of transcription factors, and is a potent growth regulator and tumor suppressor gene inactivated in several human cancers (Cho et al., 2005; DiFeo et al., 2008; Kremer-Tal et al., 2004; Narla et al., 2005a; Narla et al., 2001; Reeves et al., 2004). KLF6 is highly conserved, with homologs expressed in a number of model organisms, including zebrafish, (Oates et al., 2001) where it is termed the core promoter element binding protein (copeb). The KLF family includes at least 18 members, all of which share a highly conserved C-terminal C2H2 zinc finger DNA binding domain, whereas the N-terminal activation domain is distinct with each Krüppel-like factor (Bieker, 2001). KLFs regulate diverse processes including embryonic development, cell differentiation, proliferation and apoptosis (Suske et al., 2005).
All members of the Klf family are important developmental regulators; each knockout model to date has displayed profound developmental defects. Mouse models have revealed roles for Klfs in varied developmental events including β-globin synthesis during erythropoiesis (Klf1) (Nuez et al., 1995; Perkins et al., 1995), blood vessel stability (Klf2) (Kuo et al., 1997), and epithelial barrier integrity (Klf4) (Segre et al., 1999). Recent studies in zebrafish have shown that klf4 is essential for embryonic development, for formation of meso-endodermally derived structures and for embryonic erythropoiesis (Gardiner et al., 2005; Gardiner et al., 2007). Additionally, a study investigating retinal nerve regeneration in the adult zebrafish retina identified copeb and klf7 as upregulated following injury and essential for axon outgrowth (Veldman et al., 2007). Interestingly, several genes required for nerve regeneration in the latter study are also required for zebrafish eye development, further supporting a role for Klf6 in developmentally regulated growth. Thus, it is clear that Klf transcription factors play specific and critical roles during development that correspond to their sites of expression in the embryo.
Our previous studies in mice have shown that Klf6 is essential for embryogenesis and suggest a role in liver development. Klf6 knockout mice do not survive past 12.5 days of development, with markedly reduced hematopoiesis and disorganized vascularization (Matsumoto et al., 2006). Assessment of the hematopoietic potential of Klf6−/− embryonic stem (ES) cells further supported the role of KLF6 in hematopoiesis and vasculogenesis, demonstrated by significantly reduced expression of the endothelial marker FLK1 and impaired differentiation into primitive erythrocytes and macrophages in a hematopoietic colony formation assay (Matsumoto et al., 2006). These defects result in profound anemia of the Klf6 knockout embryos.
Although Klf6−/− embryos do not appear to have a liver (Matsumoto et al., 2006), direct assessment of liver formation in this model is difficult, since hepatogenesis in mammals is dependent upon vascular and hematopoietic development (Matsumoto et al., 2001; Nikolova and Lammert, 2003). Therefore, it is unclear whether the liver defect in these mice results directly from the loss of Klf6, or whether this effect is secondary to the abnormal endothelial cell development and/or expansion. Two studies indicate that Klf6/Copeb is required for growth: copeb knock-down impedes growth of zebrafish axons following injury (Veldman et al., 2007) and depleting Klf6 in ES cells hinders proliferation (Matsumoto et al., 2006). These data lead to the intriguing possibility that Klf6/Copeb function changes with context; it may promote cell division during development or regeneration yet play an opposite role in tissue homeostasis tissues in adults, where it functions as a tumor suppressor. A role for Klf6 in liver growth is indicated by our finding that its over-expression in hepatocytes results in post-natal liver hypoplasia (Narla et al., 2007). Thus, although it is clear that Klf6 is a growth suppressor, alternative splicing (Narla et al., 2005a; Yea et al., 2008) and interaction with different co-factors may change its function. Since Klf6 regulates cell division and is implicated in both fibrosis (Ratziu et al., 1998) and cancer (Kremer-Tal et al., 2004; Yea et al., 2008) in the adult liver, we hypothesize that it may control cell proliferation during hepatic outgrowth in embryos.
Several features of the zebrafish make it an excellent alternative model to explore the role of copeb in liver development (reviewed in Chu and Sadler, 2009): (i) liver budding, morphogenesis and hepatocyte differentiation in zebrafish embryo do not require endothelial cells (Field et al., 2003b), (ii) embryonic hematopoiesis in zebrafish does not take place in the liver, as it does in mammals and (iii) zebrafish embryos receive enough oxygen through diffusion to allow for relatively normal development in the absence of vasculature for several days. iv) Zebrafish have proven useful for identification of new genes that are required for late stages of hepatic development, including morphogenesis and outgrowth (Chen et al., 2005; Chu and Sadler, 2009; Farooq et al., 2008; Mayer and Fishman, 2003; Noel et al., 2008; Sadler et al., 2007). Therefore, although some aspects of hepatic development, including hepatocyte polarization (Sakaguchi et al., 2008) and possibly hepatic outgrowth (Korzh et al., 2008) may require vascularization, most of hepatogenesis does not require hematopoiesis or vasculogenesis. Thus, even if copeb is required for angiogenesis in zebrafish, we will be able to assess an independent function in the development of the liver.
The early stages of hepatogenesis are relatively well studied, and the main players in hepatic patterning appear to be conserved between zebrafish and mammals (Chu and Sadler, 2009; Field et al., 2003b; Zaret, 2002). In zebrafish, the alimentary canal and its associated organs, the liver, gallbladder, and pancreas all emerge from the gut primordium (Ober et al., 2003). Patterning studies illustrate that liver precursors lie within the foregut endoderm, and aggregate shortly after 24 hours post-fertilization (hpf) to form a hepatic bud as marked by the expression of hhex and prox1, which are essential for hepatogenesis in both organisms (Bort et al., 2006; Sosa-Pineda et al., 2000; Wallace et al., 2001). In contrast to knowledge about liver specification, there is only a superficial understanding of the genes that govern outgrowth and the morphogenic process that results in the formation of multiple liver lobes. Hepatic outgrowth in all species is characterized by cell proliferation. Hepatic outgrowth requires both mitogenic signals (Shin et al., 2007; Zaret, 2002) and active suppression of hepatocyte apoptosis (Li et al., 1999; Rosenfeld et al., 2000; Rudolph et al., 2000). In zebrafish, the liver bud expands from the left, crosses the midline ventrally and forms a smaller right lobe (Field et al., 2003b) and mutants that are defective in hepatic outgrowth demonstrate both decreased hepatocyte proliferation and increased apoptosis. To investigate whether Klf6/copeb plays a direct role in liver development we utilized zebrafish and mouse ES cells in which Klf6/copeb levels were manipulated. Collectively, these data indicate that Klf6/copeb is essential for endoderm development and hepatic outgrowth.
Materials and Methods
Zebrafish maintenance, morpholino and RNA injection
Wild-type (TAB5 and TAB 14) zebrafish were maintained at 28°C under standard laboratory conditions. Embryos collected after natural matings were maintained in egg water (0.6g/L Crystal Sea Marine Mix, 0.1mg/L methylene blue). The Tg(fli1a:EGFP) line was provided by J. Torres Vazquez. The LiPan Tg(fabp10:ds-Red;ela3l:EGFP;ins:RFP) line was provided by D. Stainier and described by Korzh et al. (2008). All experiments were conducted in accordance with the guidelines approved by the committee on animal care at Mount Sinai School of Medicine.
Morpholino oligonucleotides (MO) for copeb ATG (5′-tgcacattggtagaacatccattgc-’3), copeb splice donor (5′-tcactttaaaaatcagttacctgtt-3′) and a control (5′-acctggctaaaagccgaaatggcgc-3′) morpholino were purchased from Gene Tools, LLC (Philomath, OR). Morpholinos were diluted in sterile water to 0.25 mM and 0.50 mM, and approximately 6-8 nl of each MO were injected into 1-2 cell stage embryos using a Narishege IM-300 microinjector resulting in an estimated final internal concentration of 2.0 μM and 4.0 μM, respectively. Injection volume was estimated by determining the average number of injections needed to completely expel 1 ul of morpholino from the injection needle.
copeb mRNA was generated using a degenerate 5′ primer in order to avoid interaction with the copeb MO. It was prepared by first creating cDNA enriched for copeb by reverse transcription of total RNA from 5 dpf WT embryos primed with a copeb specific primer (copeb-5-cDNA: 5′-CACTTTCATTCAGGACCGAGTTT-3′). The copeb coding region was PCR amplified using a 5′ degenerate primer that retains the amino acid sequence, but incorporates 9 mis-matched bases with the copeb morpholino (copeb-dg5; 5′-ATGGACGTCCTCCCGATGTGTAGCATTTTTCAGGAACTTCAGATC-3′) and a 3′ reverse primer (copeb-3: 5′-TCAGAGGTGCCTCTTCATGTGC-3′). The PCR product was cloned into the pCR8 vector using Topo cloning (Invitrogen) and sequenced in full to confirm that the mis-matched bases were the only changes. LR recombination was carried out with the NR221 vector (Villefranc et al., 2007) using Gateway Cloning (Invitrogen). Finally, mRNA encoding copeb was prepared using mMESSAGE mMACHINE SP6 Kit (Ambion; AM1340) following the manufacturer’s protocol. Approximately 8 nl of mRNA was injected into at least 150 embryos from 4 clutches at a concentration of 200ng/ul either alone or in combination with 0.25mM copeb morpholino.
Whole mount in-situ hybridization (WISH)
The probes for gata6, hex, and prox1 were provided by the Peng laboratory, cmcl2 (Holtzinger and Evans, 2005) was provided by the Evans’ laboratory, trypsin (Biemar et al., 2001) and intestinal fatty acid binding protein (ifapb) (Mudumana et al., 2004) was provided by the Zon laboratory. fabp10 and insulin probes were previously described (Sadler et al., 2007). The copeb probe covers a 941 base pair region spanning position 487 to position 1427 according to the sequence in accession number NM_201461. This sequence has minimal similarity with other members of the Klf6 family, and shares only 31% identity with the most closely related klf family member in zebrafish, si:dkeyp-12a9.2 (also called “similar to Kruppel-like factor 6”) and therefore should be specific to copeb when high-stringency washes are employed. The probe-producing plasmid was prepared by first using PCR to amplify a 1,105 bp fragment from a pooled 1-5 dpf cDNA sample using the primers: copeb-p3-forward: 5′-CACTTTCATTCAGGACCGAGTTT-3′, copeb-p5- reverse: 5′-TGGAGATAAGGGCGATGGG-3′. The product was then digested with EcoRI to generate a 941 base pair fragment that was ligated into pre-digested PGEM T-easy vector (Promega). The correct clone was identified by sequence analysis. DIG labeled sense and antisense RNA probes were synthesized using T7 and Sp6 RNA polymerase, respectively according to standard protocols.
Embryos were fixed in 4% paraformaldehyde (PFA) in phosphate buffer saline (PBS) at 4°C overnight, dehydrated in a graded series of methanol/PBS and stored in 100% methanol at −20 °C. They were re-hydrated from methanol to PBS with 0.1% Tween (PBST) in a graded methanol series. Embryos older than 24 hpf were bleached with solution containing 3.25% hydrogen peroxide, 2.5% formamide, and 0.1 X SCC. The embryos were permeabilized using Proteinase K (10 μg/ml, Roche), refixed in 4% PFA, and prehybridized in 50% formamide/5X SSC/0.1% Tween/20.5 mg/ml torula RNA/50ug/ml heparin for 2 hours. Probes were added at 0.5-2 ng/ul and incubated overnight followed by two 20 minute washes each in 2XSSCT/50%Formamide, 2XSSCT/25%Formamide, 20 minutes in 2X SCCT, and three 30 minute washes in 0.2X SCCT. All steps were carried out at 68°C. The embryos were blocked for 2 hours at room temperature in 5% lamb serum/PBST and incubated with anti-DIG antibody conjugated to alkaline phosphatase (1:2000; Roche) overnight at 4 °C. Embryos were washed with two 10 minute washes followed by three 30 minute washes in 0.5% lamb serum/PBST, and three 10 minute washes with alkaline phosphatase (AP) buffer (100 mM Tris pH 9.5/100 mM NaCl/0.1% Tween). AP development was carried out with nitro blue tetrazolium and 5-bromo-4-chloro-3-indolil phosphate in AP buffer until specific staining was observed. AP development was stopped by washing embryos with PBST and then were fixed with 4% PFA for 30 minutes and stored in 80% glycerol. Photographs were taken with Nikon Digital Sight DS-5M color camera using a Nikon SMZ1500 stereoscope.
Histology, immunostaining, acridine orange staining and TUNEL staining
PFA fixed embryos were processed for paraffin sectioning and Hematoxylin and Eosin (H&E) staining as described (Sadler et al., 2005). Proliferating cell nuclear antigen (PCNA; Sigma) immunohistochemistry was carried out as described (Sadler et al., 2007) and the number of PCNA positive nuclei was counted in the entire liver for 2 control fish and 3 morphants.
For whole mount immunofluorescence, PFA fixed embryos were washed with PBST, then with PBS containing 0.2% saponin (PBS-S), and blocked with 10% lamb serum in PBS-S for 30 minutes. Rabbit Anti-phospho-histone H3 (Santa Cruz Biotech) was diluted in blocking buffer (1:500) and incubated overnight at 4°C. Embryos were washed with PBS-S and incubated with 1:1000 goat anti-Rabbit Alexa 488 (Invitrogen) for 2 hours, washed with PBS-S and stored in 80% glycerol. Staining was visualized under fluorescence on a Nikon SMZ1500 stereoscope and images were taken with Nikon Digital Sight DS-5M black and white camera. At least 20 fish per sample per time point were observed for each experiment, and each experiment was repeated on 2 clutches.
Acridine orange was added to the water of live embryos at a final concentration of 5 μg/ml for 30 minutes. At least 40 embryos for each time point were observed, and representative embryos were imaged.
Terminal deoxynucleotidyl transferase dUTP nick end labeling (TUNEL) staining was carried out on cryosections using the Roche In Situ Cell Death Detection Kit with Fluorescein according to the manufactures protocol. Following TUNEL labeling, sections were stained with 1:200 dilution of CY3-Streptavidin (Sigma) in PBST for 2 hours. Coverslips were mounted using Vectashield Mounting Media with DAPI (Vector Labs). Images were acquired on a Leica DM6000. ES cell culture
Klf6+/+ and Klf6−/− mouse ES cells were generated using previously described methods (Matsumoto et al., 2006). The cells were maintained in serum-free media and differentiated using the serum-free protocol for hepatic differentiation as described by Gouon-Evans et al. (2006) with the following modifications. After inducing endoderm formation with 50 ng/ml activin A at day 2, instead of flow cytometry analysis, real-time quantitative PCR (qPCR) was used to examine when endoderm markers are highly expressed. Doxycycline-inducible Klf6 ES cells were generated using methods previously described (Matsumoto et al., 2006). To induce Klf6 expression, 2 μg/ml doxcycline (Sigma) was added to the culture medium at day 1 or day 2.
RNA extraction and real-time quantitative PCR (qPCR)
Total RNA was extracted from zebrafish embryos or ES cells using an RNeasy kit (Qiagen). cDNAs were synthesized from 0.5 μg total RNA using Quanta Bio superscript IV. Primer efficacy was validated through serial dilutions of cDNA samples. qPCR was performed in triplicate using Roche Sybergreen on the Roche LightCycler. The primer sequences used are listed in Table S1. Ct values were normalized against ribosomal protein P0 (rppo) in zebrafish or Gapdh in ES cells, and expression was calculated by the ΔCt method. Melting curves for each gene were analyzed to confirm homogeneity of the DNA product. The copeb product was sequenced to confirm specificity.
Results
copeb expression is enriched in digestive organs
Copeb/Klf6 protein sequences are highly conserved between zebrafish and mammals: human and zebrafish proteins share 75% identity overall (74% identity between zebrafish and mouse), and 100% identity in the zinc finger and DNA binding domains (Fig S1 a). The copeb gene is located on chromosome 24 and appears to be the closest related homolog of mammalian KLF6 (Fig. S1 b). Another homologous gene, si:dkeyp-12a9.2 (also called “similar to Kruppel-like factor 6”) is located on chromosome 2 and has recently been predicted in zebrafish based on genome sequence analysis. The protein encoded by si:dkeyp-12a9.2 is 53% identical to human KLF6 and 64% identical to zebrafish Copeb. The highest identity is found in the 200 amino acids at the N-terminus and the first 52 amino acids, but is highly divergent in the middle of the protein (Fig. S1 a). The existence of these two distinct genes may reflect the genome duplication that occurred during zebrafish evolution. Both genes are expressed in 5 dpf zebrafish larvae (Fig. 1 a and not shown), raising the possibility that they both function in the embryo. When designing morpholinos, probes and primers, we took efforts to assure specificity for copeb.
Figure 1. copeb expression is enriched in endoderm organs during zebrafish development.
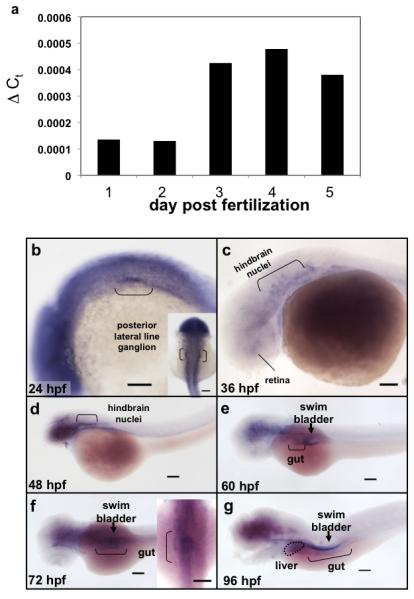
(a) Total RNA was extracted from 10 embryos taken from the same clutch on 1-5 dpf was assessed for copeb RNA expression by qPCR relative to rppo by the ΔCt method. (b-g) copeb expression pattern in developing zebrafish embryos examined by WISH at 24 hpf, 36 hpf (c), 48 hpf (d), 60 hpf (e) 72 hpf (f) and 96 hpf (g). Structures previously shown to express copeb in the early embryo are labeled in b-d. Endoderm derived structures, including the gut (brackets), liver (white arrow) and swim bladder (black arrowhead) are labeled in e-g. Other structures expressing copeb are labeled. Scale bar = 100 microns.
To define the temporal and spatial expression of copeb during zebrafish development, we used qPCR to quantify copeb mRNA and WISH to examine its expression pattern. Using primers that only amplified copeb, as confirmed by sequence analysis of the amplified product (not shown), qPCR analysis revealed that copeb was highly expressed in oocytes (60-fold higher than in embryos at 1 dpf; not shown), indicating that copeb is maternally provided, similar to many embryonic essential genes. copeb expression increased moderately between 2-3 dpf, and remained constant through 5 dpf (Fig. 1 a).
A genome-wide gene expression study in zebrafish identified copeb in the central nervous system, posterior lateral line ganglion and metencephalic blood vessels (Thisse et al., 2001). Our data confirm and expand upon these observations. A probe designed to be specific for copeb was used in WISH with stringent hybridization and wash conditions to minimize detection of other klf family members. Low level, ubiquitous expression was detected in 24-48 hpf embryos with higher levels in the posterior lateral line ganglion, hindbrain nuclei and eye (Fig. 1 b-d). The widespread expression of copeb was restricted to the brain and endodermal organs after 60 hpf, during which copeb was seen in the developing gut and the endodermally derived swim bladder primordium (Fig. 1 e-g). In 72 and 96 hpf embryos, copeb was enriched in the gut and at 72 hours was found at low levels in the liver bud (Fig. 1 f). By 96 hpf, copeb expression was clearly observed in the liver, gut and swim bladder (Fig. 1 g). Moderate expression was detected in the pancreas after 72 hpf (not shown). These data are consistent with the hypothesis that copeb functions in the developing brain and digestive organs.
copeb morphants exhibit vascular and hepatic defects
To elucidate the function of copeb in development, morpholino oligomers were used to target either the splicing of exon 2 to exon 3 (Fig. S3) or to the initiator ATG of the copeb coding sequence (accession number NM_201461) to block translation of both maternal and zygotic transcripts. There is no sequence similarity between the splice blocking morpholino and si:dkeyp-12a9.2 (accession number XM_001920617) and the ATG-targeting morpholino has 6 mis-matches with si:dkeyp-12a9.2. Therefore, both morpholinos are predicted to be specific for copeb. A morpholino that is not known to be homologous to any zebrafish transcript was used at the same concentration as the copeb morpholinos in order to control for injection or morpholino-induced toxicity.
To select the maximal tolerable morpholino concentration, embryos were injected with 6-8 nanoliters of morpholino concentrations ranging from 0.1 mM to 1.0 mM to achieve an approximate concentration of 1.0 μM to 8.0 μM in the embryos, and scored for mortality and developmental abnormalities. For both morpholinos, 1.0 μM had no effect, while high mortality was observed using 8.0 μM (not shown). Nearly 70% of embryos injected with 2.0 μM and 4.0 μM of the ATG targeting morpholino survived to 1 dpf, with no significant mortality in the number of live embryos from 1 to 4 dpf (Fig. S2).
PCR analysis was used to establish the efficacy of the splice blocking morpholino. cDNA prepared from phenotypically abnormal morphants and from their uninjected siblings on 1-4 dpf was amplified using primers that flank the splice site at the boundary of exon 2 and intron 2 (Fig. S3 a). Sequence analysis of the shorter product amplified in morphants revealed a 49 bp deletion that removed bases 630-678 of the coding sequence due to splicing to an alternative splice site in exon 2 (Fig. S3 b). This deletion causes a frame shift that is predicted to result in premature termination of the protein (Fig. S3 d). While the truncated protein may retain the functional properties of the N-terminus, the zinc finger domain is eliminated and therefore the ability to function in transcriptional regulation is likely to be lost. Moreover, since splicing of human KLF6 alters the C-terminus and creates a dominant negative (Narla et al., 2005b), the remaining portion of Copeb in the splice-blocked morphants may interfere with any remaining full length protein, thus creating 2 means by which this morpholino could block Copeb.
To examine the effect of copeb knock-down on development, we assessed the gross morphology of embryos injected with 2.0 μM of exon 2 splice donor morpholino (splice donor MO; Fig. 2 a). Most copeb morphants were only moderately abnormal by 24 dpf, with slightly smaller eyes and head. After 48 hpf, these defects are more pronounced, and pericardial edema developed in nearly all morphants. To examine the requirement for copeb in liver development we assessed liver size in copeb splice donor morphants using the transgenic line Tg(fabp10:RFP) that expresses RFP only in hepatocytes starting at 72 hpf (Korzh et al., 2008). Interestingly, copeb morphants have small livers at 72 and 96 hpf, as representative images show in Figure 2a. A similar phenotype was observed in embryos injected with the ATG-targeting morpholino (Fig. 2b and see below).
Figure 2. copeb is required for embryonic development.
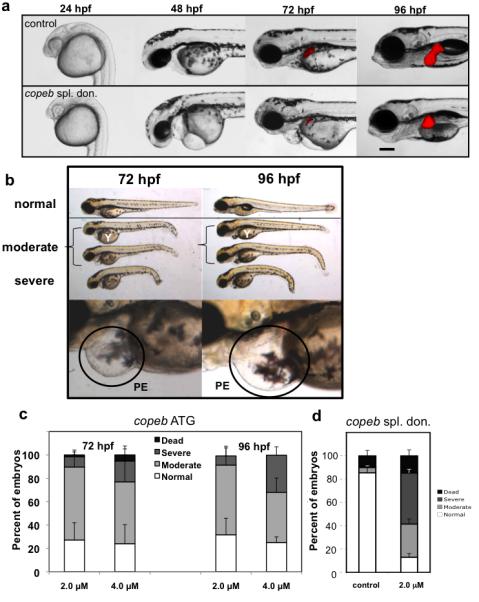
Live copeb splice donor morphants and copeb ATG morphants were examined on 1- 4 dpf. (a) Live images of Tg(fabp10-RFP) embryos showed copeb slice donor morphants exhibit pericardial, yolk sac and cephalic edema at 48 and 72 hpf and a small for size liver. All images are taken at the same magnification; scale bar =100 μm. (b) Live images of embryos showed a range of phenotypes of copeb ATG morpholino-injected embryos: normal (resembling embryos injected with the control morpholino (top), moderately affected embryos with pericardial edema, unconsumed yolk (Y) and blood congestion (middle 2 embryos) and severely affected, runted embryos (bottom). Bottom panels show the pericardial edema (PE; black circle) in morphants. These characteristics were observed in at least 10 clutches of copeb ATG morphants. (c) Quantification of the percent of embryos displaying the morphant phenotype at 72 and 96 hpf demonstrate that the phenotype becomes more severe as a function of morpholino concentration and time. Values represent the average of four clutches of at least 100 embryos were scored for each clutch on each day. Error bars indicate the standard deviation of the mean. (d) copeb splice donor morphants were quantified by phenotype and compared to uninjected controls at 96 hpf. Data is the average of at least 50 embryos per group in 3 clutches. Error bars indicate the standard deviation of the mean.
By 60 hpf, a few ATG morphants developed a mild phenotype including slightly under consumed yolk and smaller head and eyes (data not shown) and by 3 dpf, morphants were small overall, developed pericardial edema, blood congestion, under consumption of yolk, as well as micropthalmia and micrognathia (Fig. 2b). The aforementioned phenotype is consistent with the previously reported role for copeb in development of the retinal ganglion cell regeneration (Veldman et al., 2007). The liver also appeared small in the ATG morphants at 72 hpf. The phenotype increased in severity on 4 dpf (Fig. 2b-c). These traits were used to categorize morphants into three groups: normal (resembled control embryos), moderate (had two or more of the above mentioned characteristics), and severe (grossly runted/underdeveloped). Figure 2c illustrates that higher morpholino concentration resulted in a higher percentage of severely affected embryos (8.7% for 2 μM vs. 17.8% for 4 μM at 3 dpf) and that the phenotype progressed in severity, with 31.9% of the 4 μM morphants at 4 dpf scored as severely affected (Fig. 2c). A similar distribution of affected embryos was observed at 4 dpf in the splice blocked morphants (Fig. 2d). We also observed a smaller liver and underdeveloped gut in the moderately affected 4 dpf morphants (see below), whereas the severely affected embryos were significantly developmentally retarded, precluding assessment of the liver and gut. Except where otherwise noted, all experiments were carried out using on moderately affected morphants generated from the ATG morpholino, which targets both the maternal and the zygotic copeb.
We do not attribute the copeb morphant phenotype to off-target effects, since none of the embryos injected with the same or 2 times higher concentrations of the control morpholino exhibited any of these phenotypes in any experiments. Moreover, very similar phenotypes are achieved using two different morpholinos. Using a p53 morpholino is a common control to eliminate non-specific morpholino toxicity (Robu et al., 2007). However, given that loss of KLF6 can directly lead to p53 upregulation, (Sirach et al., 2007), using a p53 morpholino as a control may confound analysis of the copeb morphants. Since we do not observe the massive brain necrosis that characterizes non-specific morpholino toxicity, we believe that the phenotype observed in copeb morphants is specific. While we observed the inherent variability between clutches that is characteristic of these experiments, both the survival and the number of affected larvae on 4 dpf improved by co-injecting 2 μM copeb morpholino with mRNA encoding zebrafish copeb (Fig. S4a-b). Importantly, the most intriguing phenotype in the morphants – reduced size of the liver and pancreas (see below) - were rescued by co-injecting 2 μM copeb ATG morpholino with mRNA encoding zebrafish copeb (Fig. S4c).
Defective vasculogenesis and hematopoiesis are the most striking phenotypes of the Klf6−/− mouse embryos (Matsumoto et al., 2006). Consistent with this finding, up to 30% of the copeb morphants had no circulating blood on 2 dpf (data not shown), however, all morphants had circulating blood by 3 dpf. We separated the bloodless morphants and followed their development compared to their siblings with blood, and did not find any difference in severity of phenotype later in development. If copeb plays a role in vasculogenesis and hematopoiesis in zebrafish as it does in mice, then markers of a number of blood cell lineages should be down regulated in copeb morphants. qPCR was used to compare the expression of markers of endothelial cells (kinase insert domain receptor; kdr), erythrocytes (hemoglobin beta embryonic-2; hbbe2), macrophages (lympocte cytosolic plastin-1; lcp1) and granulocytes (myeoloid-specific peroxidase: mpx) in 4 dpf copeb morphant and control embryos. We found significantly decreased expression of all these markers, but there was no effect in the expression of kidney marker nephrosis 1 (nphs1L; Fig. S5 a). The effect of copeb knockdown on vasculogeneis was assessed by injecting the copeb morpholino into embryos from the Tg(fli1a:EGFP) transgenic line that expresses GFP in endothelial cells. Although the liver was smaller in copeb morphants, the vascular network was intact (Fig S5 b-e). These findings suggest that while copeb is required for some aspects of hematopoiesis in mice and zebrafish, 4 dpf morphants have normal hepatic sinusoids (Fig. 3 l) and vascular network (Fig. S5 b-e). Thus, either copeb is dispensable for development of hepatic vasculature in zebrafish or the incomplete knock-down achieved with the ATG morpholino may allow for vasculogenesis to occur. Regardless, these data indicate that the phenotype of copeb is not secondary to defect in the vasculature.
Figure 3. copeb is required for achieving proper liver size but is dispensable for hepatic patterning.
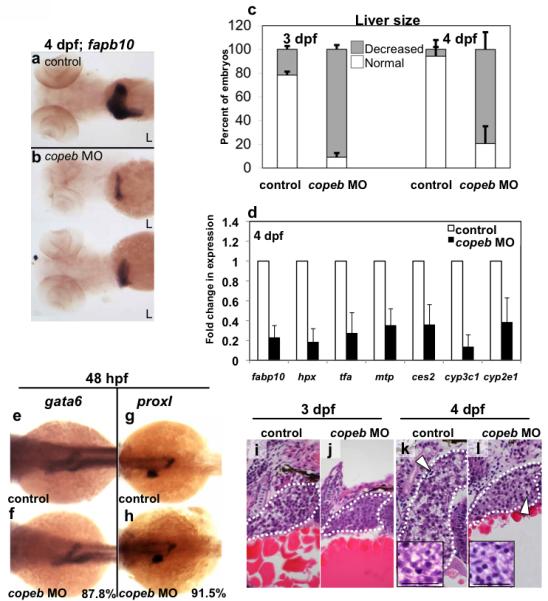
(a-b) fabp10 WISH reveals that the small liver in 4 dpf copeb morphants extends across the midline. Two representative morphants are shown. L indicates the left side of the embryo, where the liver bud forms. (c) The number of embryos with a liver size that was less than 50% of normal was scored in 3 clutches. The total n=75 for morphants and for controls at 3 dpf n=71 for each at 4 dpf. p<0.01 when comparing morphants to controls for both time points. (d) mRNA expression of genes implicated in hepatocyte function were determined by qPCR in control embryos compared to copeb morphants at 4 dpf. Expression levels were normalized to control embryos, the experiments were repeated in 3 independent clutches using 30 embryos per sample. Errors bars show standard deviation. All genes are significantly down-regulated (p<0.05). (e-h) WISH analysis of gata6 (e,f) and prox1 (g,h) in control embryos (e,g) and copeb morphants (f,h) at 48 hpf. All control embryos had the same staining pattern. The percent of copeb morphants with staining that is indistinguishable from control embryos, as shown in the representative image, is indicated in the lower right corner of f and g (gata6: n=41, prox1: n=70). (i-l) Serial sections of control embryos and copeb morphants on 3 and 4 dpf were stained with H&E and the largest section through the liver (outlined) was imaged, illustrating the reduced size of the liver. Arrows point to sinusoids containing nucleated endothelial cells. Hepatocyte morphology is the same in 4 dpf control and copeb morphants (inset; scale bar = 40 μM).
copeb is required for hepatic outgrowth
The wild-type zebrafish liver bud appears at approximately 44 hpf (Chu and Sadler, 2009; Ober et al., 2003; Wallace and Pack, 2003), and between 3 and 5 dpf undergoes rapid expansion during hepatic outgrowth to form the left and right liver lobes (Chu and Sadler, 2009; Field et al., 2003b; Pack et al., 1996). Embryos from the LiPan transgenic line Tg(fabp10:RFP;ela3l:GFP) which expresses RFP in hepatocytes and GFP in the exocrine pancreas (Korzh et al., 2008) were injected with the copeb morpholino which resulted in a small liver in over 64% of copeb morphants on 4 dpf (Fig. 2 a and Fig. S4 c). To expand on this observation, liver size was assessed by WISH using the specific hepatocyte marker, fatty acid binding protein 10 (fabp10) which revealed a small liver in all morphants (Fig. 3a-c). Importantly, the small liver phenotype was rescued by co-injecting the ATG morpholino with copeb mRNA (Fig. S4c-d). Consistent with our finding of small liver size, qPCR analysis showed markedly decreased expression of the liver-specific markers fabp10, hemopexin (hpx), and transferrin-a (tfa) as well as liver-expressed genes ces2, cytochrome P450, subfamily c, polypeptide 1 (cyp3c1), cyp2e1, microsomal triglyceride transfer protein (mtp) in copeb morphants (Fig. 3 d), whereas the expression of other genes, such as nephrosis 1 (Fig. S5) and insulin (Fig. 6 c) remained unchanged. While most morphants failed to undergo expansion of the liver bud to form the right liver lobe, their undersized left liver lobe achieved its normal crescent shape. This is notably different from the other zebrafish mutants that have hepatogenesis defects resulting in a small liver, where the liver consists of only a spherical left lobe (Farooq et al., 2008; Mayer and Fishman, 2003; Noel et al., 2008; Sadler et al., 2007). These data indicate that copeb is required for the proper formation of the liver, however, it also suggests that some aspects of hepatic morphogenesis are unaffected in copeb morphants.
Figure 6. copeb is required for expansion of the exocrine pancreas and intestine.
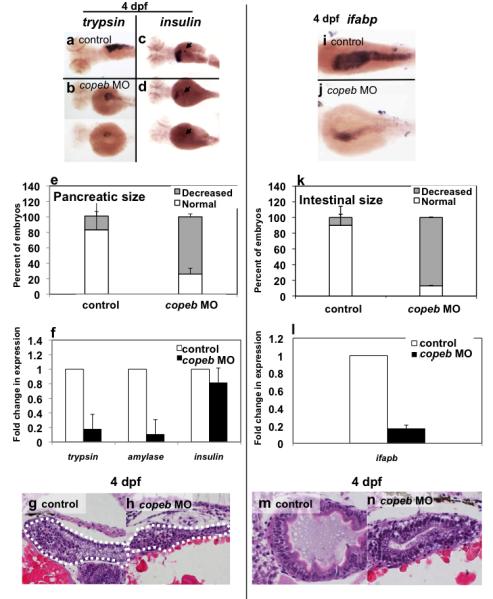
Pancreatic development was assessed by WISH for trypsin (a,b) and insulin (arrowhead in c,d) in 4 dpf uninjected controls (a,c) and copeb ATG morphants (b,d). The fabp10 probe was included with the insulin probe as a control. Two morphants are shown to indicate the range of phenotype. (e) The size of the exocrine pancreas was scored as either normal, indicating a wide pancreatic head and long tail, or reduced, indicating that the trypsin stained tissue was less than ½ what is typically observed in control fish; 73.9% of the copeb morphants had reduced pancreas size versus 17.7% in controls (n=50, p<0.01). (f) Expression of markers of the exocrine pancreas (trypsin and amylase) and endocrine pancreas (insulin) were analyzed by qPCR, with rppo as a reference in 4 dpf morphant and control larvae. The average fold change 3 clutches of morphants were determined by normalizing to the average expression in uninjected controls. (g,h) H&E stained sagittal sections of 4 dpf embryos through the widest part of the pancreas (outlined) illustrate a well formed exocrine pancreas in control embryos and a significantly smaller exocrine pancreas in copeb morphants. (i,j) WISH using ifabp as a marker of gut epithelial cells on 4 dpf embryos revealed slight staining of only the intestinal bulb in copeb morphants. (k) 4 dpf uninjected controls and copeb morphants processed by WISH for ifabp. Controls and morphants from 3 clutches (n=50 for each) were scored as normal if they had staining in a thick and curved intestinal bulb as well as staining in the distal intestine, or as decreased if the staining was confined to the intestinal bulb and if the intestine was thin. Decreased gut ifabp staining was observed in 10.0% of the control versus 87.1% of the morphants (p<0.01). (l) ifabp mRNA level in 4 dpf embryos using qPCR with rppo as a reference was averaged in 3 clutches of copeb morphants and normalized to the average expression in controls. (m,n) H&E staining of 4 dpf embryos shows well-differentiated intestinal epithelial cells copeb morphants, however the intestinal villi do not form.
The small liver phenotype may result from either a failure of the liver bud to expand or to undergo proper patterning or differentiation. To distinguish between these possibilities, we examined whether the expression of genes involved in hepatic patterning and specification was disrupted as a result of copeb knockdown. Expression of gata6 (Fig. 3 e,f), which regulates endoderm development (Field et al., 2003b; Zhao et al., 2005) and prox1 (Fig. 3 g,h), which controls hepatic and pancreatic patterning (Sosa-Pineda et al., 2000), were identical in morphant and control embryos at 48 hpf. This suggests that liver specification and budding do not depend on copeb.
Although liver size was significantly reduced in morphant embryos, the morphant liver clearly expresses a number of other genes directing hepatocyte function, indicating that hepatocyte differentiation occurs in copeb morphants. Furthermore, histological analysis of morphants on 3 dpf (Fig. 3 i,j) and 4 dpf (Fig. 3 k,l) demonstrated normal cellular appearance, organization and bile duct formation. Importantly, the sinusoids of copeb morphants contained erythrocytes (Fig. 3 l), which indicate that the reduction of hematopoietic markers caused by loss of copeb (Fig. S5) does not completely block formation of blood cells. Moreover, direct visualization of the hepatic vasculature using the Tg(fli1:EGFP) transgenic line demonstrates that copeb morphants have well developed blood vessels in the liver on 4 dpf (Fig. S5 b-e).
These data suggest that copeb is not required for hepatic patterning or differentiation. The expansion of the liver bud begins on early 3 dpf (Chu and Sadler, 2009). The finding that copeb morphants have normal hepatic patterning but a small liver at 72 hpf suggests that reduced liver size in copeb morphants is attributed to a defect in hepatic outgrowth. Both mitogenic and anti-apoptotic signals are required for hepatic outgrowth in mammals (Rosenfeld et al., 2000; Rudolph et al., 2000; Tanaka et al., 1999) and in zebrafish (Sadler et al., 2007). There was no evidence of increased apoptosis in the liver based on histological evaluation of serial sections through the morphant livers (not shown). Moreover, although morphants have a modest increase in cell death in the brain, there is no increase in cell death in the liver on 3 and 4 dpf as measured by acridine orange staining (Fig. 4 a) and TUNEL staining (Fig. 4 b). This indicates that cell death does not account for the small liver phenotype.
Figure 4. Decreased endoderm-derived organ size in copeb morphants is not due to increased cell death.
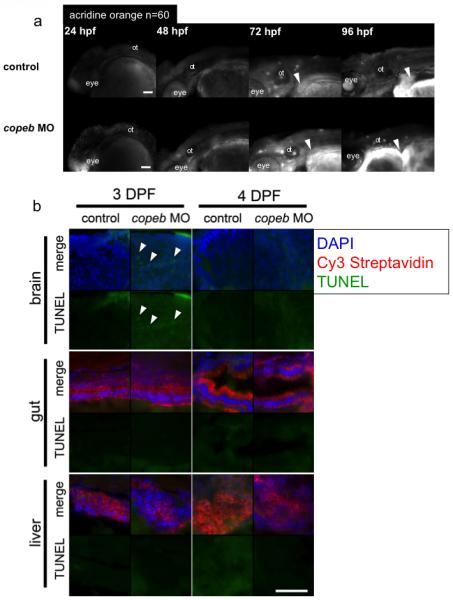
(a) Cell death was examined in live copeb ATG morphants and control embryos at 24, 48, 72 and 96 hpf by acridine orange. There was increased labeling of the head and eye in copeb morphants at 24 hpf but virtually no staining in the control and morphants at any other time. White arrowheads point to the liver, which does not have any acridine positive cells in either morphants or controls. n=60 for each sample at each time point. ot=otolith. (b) No increase in apoptosis is observed in endoderm-derived organs in 3 and 4 dpf copeb ATG morphants compared to controls. Sections of embryos on 3 and 4 dpf were stained with Cy3-streaptavidin to label the liver and gut epithelium, and were processed for TUNEL using fluorescein. TUNEL positive cells (white arrow heads) are observed only in very few cells in 3 dpf sections in the brain of copeb ATG morphants. Blue = DAPI, Red = Cy3 Strepavidin, Green = TUNEL, scale bar = 50mM.
Two methods were used to determine whether copeb depletion causes cell proliferation defects. First, global changes in cell division were assessed by whole mount staining for phosphorylated Histone-H3 (pHist-H3), which marks cells in the G2/M phase of the cell cycle. While we did observe modest increase in dying cells in the brain on 1 dpf (Fig. 4a), pHist-H3 staining of morphants was indistinguishable from control embryos on days 1-3 of development (Fig. 5 a), indicating that cell proliferation in the entire embryo is not dependent on copeb. Next, we examined cell proliferation in hepatocytes using PCNA immunostaining on serial sections of the liver from control and morphant larvae at 4 dpf, the stage of hepatic outgrowth in which proliferation is most robust. Nearly all the cells in zebrafish larval liver are hepatocytes (Chu and Sadler, 2009) which are clearly identifiable by their size, shape and appearance of their nuclei. All hepatocyte nuclei in serial sections through multiple embryos were scored for PCNA immunostaining as a marker for cells in S and G2 of the cell cycle (Fig. 5 b,c). Nearly 50% of control hepatocytes were PCNA positive in 4 dpf larvae whereas only ~15% of copeb morphant hepatocytes are PCNA positive (Fig. 4 c-d). These data indicate that copeb is not required for suppressing apoptosis, but is required for hepatocyte proliferation during hepatic outgrowth.
Figure 5. Decreased endoderm-derived organ size in copeb morphants is due to decreased proliferation.
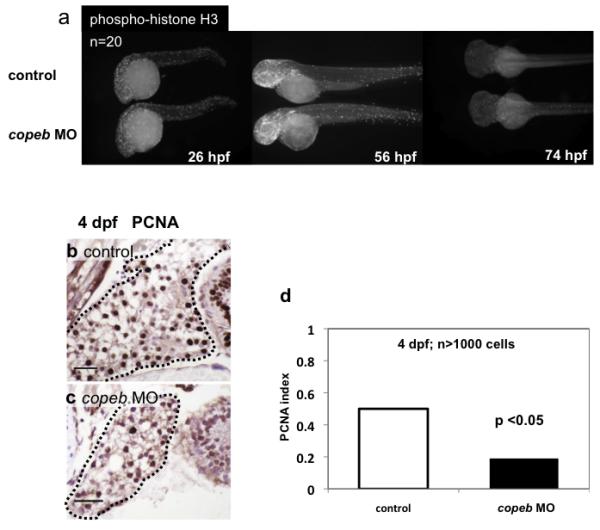
(a) Proliferation in whole embryos was examined at 26, 56, and 74 hpf by immunofluorescence using an antibody against phosphorylated Histone H3 (p-histone H3) to identify cells in the G2/M phase of the cell cycle. High levels of proliferation are found in the head, eyes and tails of both morphants and controls; n=20 for each sample at each time point. (b-d) Hepatocyte proliferation was assessed by nuclear PCNA staining of serial sections of the liver (outlined) in control embryos (b) and copeb morphants (c) on 4 dpf and was quantified in (d). scale bar = 20 microns. At least 1000 hepatocytes in at least 2 control and 3 morphant embryos were scored for each sample.
copeb is required for growth of the pancreas and intestine
To determine whether copeb is also required for the development of other endodermally derived organs, we examined the pancreas and intestine in copeb splice blocking and ATG morphants in the LiPan line (Fig. S4 c) and via WISH in ATG morphants. While there was no difference in early endodermal patterning in ATG morphants, revealed by gata6 staining (Fig. 3 e,f), the exocrine pancreas was significantly smaller in all 4 dpf morphants (Fig. S4c). Thus, it is possible that, as in the liver, copeb knockdown may result in a failure to undergo maturation or expansion of other endodermal organs.
The zebrafish pancreas originates from two anlagen, the first (posterior) is initiated at approximately 24 hpf and the second (anterior) at around 40 hpf. These two buds merge at 52 hpf to form the morphologically identifiable pancreas (Field et al., 2003a). We used four means to evaluate whether pancreatic development is affected by copeb depletion. First, the pancreas in live tg(ela3l:EGFP) fish (Fig. S4 c) was smaller in nearly all morphants (Fig. S4 c). Second, WISH (Fig. 6 a-e) showed that 73.9% of copeb morphants have a small pancreas based on WISH, compared to 17.7% of control embryos (n=50, p<0.01). This degree of variation in controls reflects the natural variation that is inherent when examining fish across multiple clutches. Third, qPCR analysis showed that genes expressed specifically by pancreatic acinar cells were significantly reduced in 4 dpf morphants compared to control embryos (Fig. 6 f). Finally, histological sections revealed a small exocrine pancreas with differentiated acinar cells (Fig. 6 g,h). In contrast, the expression of insulin, a marker of the endocrine pancreas, was unaffected (Fig. 6 c,d). This suggests that exocrine pancreas maturation or outgrowth requires copeb.
Using a similar approach, we found that copeb also plays a role in intestinal development. In the zebrafish, the endoderm-intestine transition occurs at approximately 60 hpf and is marked by the formation of columnar epithelium with highly organized brush border microvilli and by expression of gut-specific proteins such as intestine fatty acid-binding protein (ifabp) and alkaline phosphatase (Ng et al., 2005; Wallace and Pack, 2003). Figure 6 i-k shows that in 3 independent clutches of 4 dpf embryos, a 87% of morphant embryos displayed a lower level of ifapb expression compared to only 10% of control embryos (total n=50 embryos per group, p<0.01). The finding that some control embryos have a reduction of ifabp staining reflects either minor inconsistencies in WISH staining due to technical reasons or the occasional developmentally abnormal embryo that is typically observed in any given clutch. Reduced ifabp expression was also confirmed by qPCR (Fig. 6l). Additionally, histological analysis indicated that although the intestinal columnar epithelium developed, the gut tube in copeb morphants was small, the intestinal bulb did not expand and villi were not observed (Fig. 6 m,n). In the liver, PCNA immunostaining on 3 and 4 dpf was markedly decreased in the gut of the copeb morphants (not shown) and there was no increase in apoptotic cells based on TUNEL staining (Fig. 4b). Based on these results, we conclude that copeb plays a role in regulating endoderm organ outgrowth. In contrast, the histological morphology of mesoderm-derived organs, such as kidney and muscle, as well as neural tissue, appeared normal (not shown). Taken together, these data indicate that copeb is required for liver, gut and pancreas expansion but not for the specification of these organs. Given that Klf6 is implicated in growth control, we hypothesize that it plays a role in proliferation of these tissues.
cdkn1a expression is up-regulated in copeb morphants
Klf6 is a tumor suppressor gene inactivated in many adult cancer tissues (Kremer-Tal et al., 2004; Narla et al., 2001; Reeves et al., 2004), and may also act as a growth promoter during development. This is supported by the finding that Klf6−/− ES cells proliferate 50% slower than their wild-type counterparts (Matsumoto et al., 2006) and that digestive organ proliferation was significantly reduced in copeb depleted zebrafish embryos. Klf6 functions as a growth-suppressor in part by the upregulation of p21 (Narla et al., 2001), the cyclin-dependent kinase inhibitor known as cdkn1a in zebrafish. cdkn1a expression levels were consistently up-regulated from 1-4 dpf in copeb morphants, and strikingly induced in 2 dpf morphants (Fig. 7a). Importantly, the increase occurs prior to the onset of the morphant phenotype, which presents the intriguing possibility that it contributes to the defect in proliferation observed later in the development of copeb morphants. While cdkn1a is expressed ubiquitously throughout development, the increase is best directly observed in copeb morphants in the hatching gland at 24 hpf (Fig. 7 b,c) by WISH. While cdkn1a may be increased in a tissue specific manner at other stages, the high level of ubiquitous expression makes this difficult to assess using WISH.
Figure 7. cdkl1a expression is increased in copeb morphants.
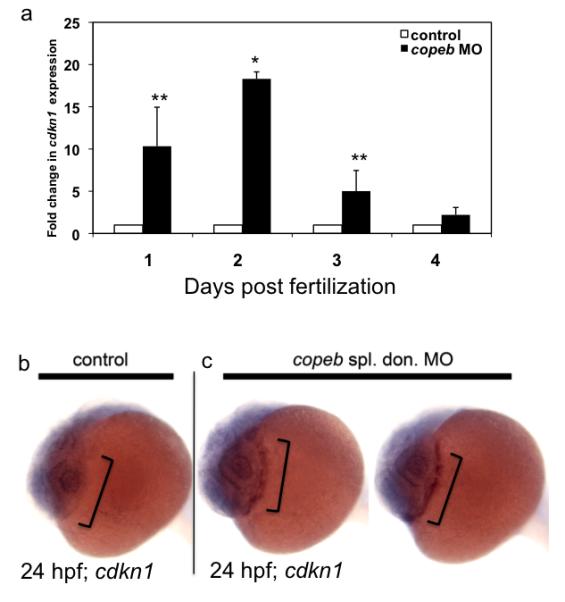
(a) cdkl1a was assessed by qPCR in control or copeb morphant embryos on 1-4 dpf. Levels were averaged from 3 experiments and normalized to the averages of controls, with errors bars showing standard deviation. * indicates p<0.001; ** p=0.07. (b) WISH for cdkl1a in 24 hpf controls and copeb splice donor morphants shows an increase in cdkI1a expression in the hatching gland (brackets) of morphants.
Klf6 is required for endoderm formation in mouse ES cells
Our zebrafish data indicates that copeb is expressed in the developing liver bud and that copeb is required for hepatic outgrowth, however, it is not clear whether this effect is due to a cell autonomous role of copeb in hepatocytes. To investigate whether copeb (which is referred to as ‘Klf6’ when discussing mouse samples) serves a necessary and sufficient, cell autonomous role for liver development in mammals, we previously developed a protocol to drive mouse ES cells into hepatocytes in vitro (Gouon-Evans et al., 2006). As outlined in Figure S6a, ES cells in serum-free media are first driven to form embryoid bodies (EBs). At day 2 of differentiation, activin A was added to the EB culture to induce endoderm formation. When endoderm markers are highly expressed, EBs are dissociated and re-aggregated with activin A, FGF and BMP4, which induces hepatic specification. Aggregates were plated on gelatin-coated dishes in media including cytokines and dexamethasone to allow expansion and maturation of hepatic cells.
This protocol was used to differentiate Klf6+/+ ES cells into the hepatic lineage. As expected, high levels of the early markers of hepatocyte differentiation were detected by qPCR, including alpha fetoprotein (Afp) (Fig. S6 c) and transferrin (Ttr) (Fig. S6 d) at day 8, as well as the late hepatic marker albumin (Alb) at day 12 (Fig. S6 e). The expression of Klf6 during hepatic differentiation undergoes a biphasic increase in Klf6 at day 2, prior to the addition of activin A, and again following hepatic specification from day 6 through 12 (Fig. S6 b).
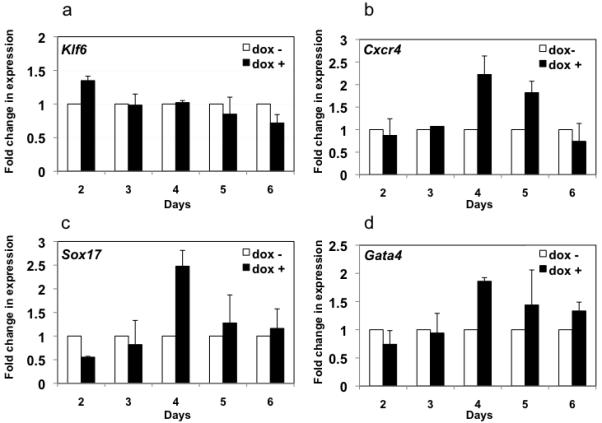
Given the role for copeb in zebrafish hepatogenesis, we explored whether Klf6 is required for hepatic differentiation in vitro by comparing the capacity of Klf6+/+ and Klf6−/− ES cells to undergo hepatic differentiation. As expected, Klf6 expression was not detected in Klf6−/− EBs (Fig. 8 a). While the Klf6+/+ ES cells were able to differentiate into endoderm and subsequently into the hepatic lineage, Klf6−/− ES cells did not form hepatic cells and did not express any of the hepatic specific markers (data not shown). We hypothesized that this was due to the failure to form endoderm, which was tested by analyzing expression of genes marking early and late stages of endoderm formation including Hnf3β, Cxcr4, Sox17 and Gata4. The expression of all of these genes was suppressed in the knock-out cells at all time points (Fig. 8 b-e). These data indicate that Klf6 is necessary for endoderm formation in vitro. To our surprise, a similar increase in p21 expression was not observed in Klf6−/− ES cells during differentiation (Fig. S6), although their rate of proliferation was decreased (not shown), suggesting that other factors may compensate the lack of Klf6 in regulating p21 expression in mouse ES cells.
Figure 8. Markers of definitive endoderm are not expressed in Klf6−/− ES cells.
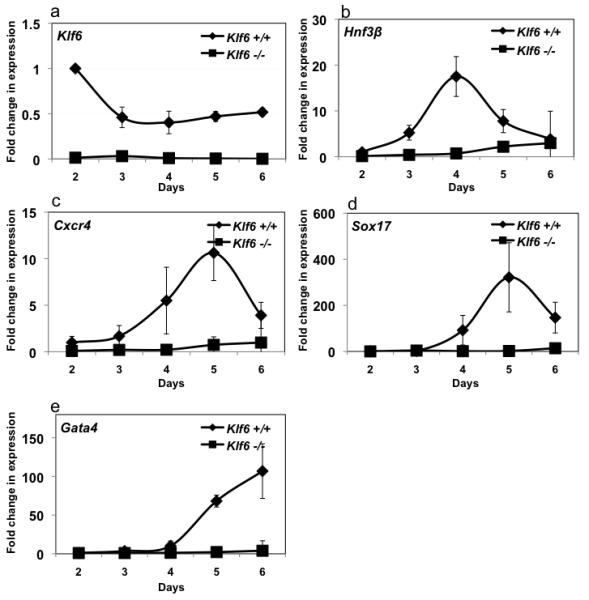
Klf6+/+ and Klf6−/− ES cells were differentiated along the endoderm linage and RNA samples were obtained each day following activin addition to the EBs (see Figure S3). qPCR was used to detect Klf6 (a), Hnf3b (b), Cxcr4 (c), Sox17 (d), Gata4 (e) and compared to Gapdh as a reference. Levels were normalized to the expression in Klf6+/+ ES cells on day 2 of the differentiation protocol. Errors bars show standard deviation from three independent experiments.
To investigate whether increasing Klf6 expression in wild-type ES cells is sufficient to drive endoderm differentiation, we created an ES cell line in which Klf6 expression increased by 40% with a single addition of doxycycline (Fig. 9 a). Indeed, induction of Klf6 on day 1 of the hepatocyte differentiation protocol (Fig S5), just prior to the addition of activin, caused the expression of endoderm maker genes Cxcr4, Sox17, and Gata4 to be increased during the endoderm induction phase (Fig. 8 b-d). When doxycycline was added at day 2, there was no effect on endodermal differentiation even though induction of Klf6 expression was comparable (data not shown). This suggests that KLF6 functions in temporal-specific manner at an early stage of endoderm formation in vitro.
Figure 9. Forced Klf6 expression enhances endoderm differentiation.
(a) Stable ES cells expressing Klf6 under a doxycyclin inducible promoter were cultured as EBs for 1 day, and then doxycycline was added, resulting in a 40% increase in Klf6 mRNA assessed by qPCR, using Gapdh as a reference (a). (b-d) Expression of Cxcr4, Sox17, Gata4 were detected in cells with and without doxycycline by qPCR and normalized to the expression level in cells without Klf6 induction (dox-). Errors bars show standard deviation from three independent experiments.
Discussion
Endoderm organogenesis includes stages of cell fate specification, bud formation, cell differentiation and organ expansion (Chu and Sadler, 2009; Zhao and Duncan, 2005). The successful identification of genes that control the early stages of hepatogenesis has not been matched by efforts to understand hepatic outgrowth. Outgrowth is an important developmental process, since genes that regulate hepatic outgrowth are also critical for liver regeneration in adults (Apte et al., 2007; Sadler et al., 2007; Tan et al., 2006; Tan et al., 2008), indicating that the response to hepatic injury involves some of the signals that are essential for hepatogenesis. Our data lead to the hypothesis that Klf6 is induced as an immediate-early gene in response to liver injury in hepatic mesenchymal cells (Ratziu et al., 1998) and is required for the outgrowth and specification of endoderm formation in mouse ES cells in vitro.
There is little known about the signals that are required to coordinate the outgrowth phase of hepatogenesis, although data from mice indicates that both suppression of apoptosis and mitogenic stimulation may be essential. Studies in mice are complicated both by the presence of hematopoiesis in the mammalian embryonic liver, and by the requirement for endothelial cells in mammalian liver development. Therefore, mouse mutants that affect either angiogenesis or blood development will indirectly impair liver development. Klf6 is one such example: our previous work has shown that yolk sac vascularization does not occur in Klf6−/− mouse embryos and that hematopoiesis is blocked in Klf6−/− embryos and ES cell differentiation cultures, resulting in death by day 12.5 of development (Matsumoto et al., 2006). Similarly, we report here that copeb is essential for embryogenesis in zebrafish, and the copeb morphant phenotype partially recapitulates the phenotype observed in the Klf6−/− mice with reduced hematopoiesis (Matsumoto et al., 2006). While hepatic outgrowth (Korzh et al., 2008) and hepatocyte polarization (Toyoshima et al., 2008) are partially dependent on endothelial cells, zebrafish differ from mammals (Nikolova and Lammert, 2003) as many aspects of zebrafish hepatogenesis are independent of hematopoiesis and endothelial cells. Although some copeb morphants do not have normal blood flow early in development, and markers of several hematopoietic lineages are decreased in copeb morphants, development of hepatic vasculature does not require copeb. Therefore, the lack of endothelial cells cannot fully explain the hepatic phenotype generated by depleting Copeb. Additionally, the small liver on 3 and 4 dpf in copeb morphants is not attributable to defects in patterning, endoderm development or hepatocyte proliferation. This distinguishes copeb from the prometheus mutant, which fails to undergo liver specification (Chu and Sadler, 2009; Ober et al., 2006). Instead, copeb is required for hepatic outgrowth as well as expansion of the exocrine pancreas and intestine. The expression pattern of copeb corresponds well with the phenotype displayed by copeb morphant embryos: it was enriched in endoderm-derived organs, including liver, pancreas, intestine, and swim-bladder at 60 hpf, by which time endoderm organ specification has already occurred (Chu and Sadler, 2009; Ober et al., 2003). This suggests that copeb is required for cell proliferation that drives expansion of endodermally derived organs, but loss of copeb does not induce cell death.
There are a few zebrafish models with a phenotype similar to the copeb morphants; however, some striking differences among these indicate that copeb is part of a different regulatory network. For example, in the nil per os mutant, underdevelopment of digestive organs results from arrested cell differentiation (Mayer and Fishman, 2003). This is different from the copeb morphants, where the liver, gut and pancreatic cells on 4 and 5 dpf appear to be differentiated based on morphological characteristics and that markers of terminal differentiation, albeit reduced, are expressed by the pancreatic and hepatic cells in copeb morphants. Hepatic outgrowth is blocked in zebrafish embryos with depletion of genes that play a role in chromatin remodeling including hdac (Farooq et al., 2008; Noel et al., 2008) and uhrf1 (Sadler et al., 2007); however there is a significant amount of apoptosis in the liver and gut of uhrf1 mutants (Sadler et al., 2007), which is not found in copeb morphants. It is interesting that liver morphogenesis is also affected in all of the previously described zebrafish models with defective hepatic outgrowth, which is visualized by the failure of the small left-lobe to elongate into the characteristic crescent shape and to cross the midline to form the right lobe. In contrast, while the liver size was drastically smaller in the copeb morphants, their livers displayed a normal shape and crossed the midline, thereby preserving both the left and right lobe, and suggesting that copeb functions primarily in organ expansion and that morphogenesis and proliferation are separate developmental events.
copeb morphants most resemble def mutants, which undergo normal endoderm formation but fail to expand the liver, gut and pancreas. The def gene regulates endoderm-derived organ outgrowth through selective up-regulation of 113p53, a truncated isoform of p53, which subsequently leads to the activation of p53-responsive genes such as mdm2 and cdkn1a (Chen et al., 2005). Similarly, cdkn1a expression is up-regulated in copeb morphants from 1 to 4 dpf. Intriguingly, in adult mammalian tissues, KLF6’s growth-suppressive properties are mediated in part through direct transcriptional activation of CDKN1A (Narla et al., 2007). Over expression of either Cdkn1a (Wu et al., 1996) or Klf6 (Narla et al., 2007) in hepatocytes causes marked decrease in cell proliferation and small for size livers. Indeed, this phenotype is reminiscent of what is observed in the copeb morphant zebrafish, but in contrast to the activity of KLF6 in ES cells (Matsumoto et al., 2006). To our surprise, although we observed a 50% decrease in proliferation of Klf6−/− ES cells (not shown), Cdkn1a expression is not significantly increased. While the mechanisms underlying these divergent effects of Copeb/KLF6 on Cdkn1a are unknown, we speculate that they result from different repertoires of co-activators or repressors between embryonic and adult cells; thus, Copeb/KLF6 can act as trans-activator in one context and trans-repressor in the other, a characteristic feature of other KLF family members. It is also possible that the increase in cdkn1a expression is not a direct result of knocking down copeb, but rather a secondary effect of a cascade of events initiated by copeb depletion. Finally, some copeb-dependent events occurring outside of the liver may affect hepatic outgrowth in zebrafish.
While copeb contributes to the final stages of organogenesis of the liver, gut and pancreas in vivo, it is necessary and sufficient for early endoderm formation in mouse ES cells. These findings demonstrate that, in addition to playing a role in development of the mesoderm, blood and endothelial cells (Matsumoto et al., 2006), Klf6/copeb also participates in endoderm induction. Interestingly, while mesoderm induction is only delayed in Klf6−/− ES cell (Matsumoto et al., 2006), endoderm induction is almost completely blocked. It is possible that the defects observed in germ layer formation in Klf6−/− (Matsumoto et al., 2006) mice may contribute to the defects we observe in both mesoderm and endoderm cell differentiation in vitro. This may be attributed to an important technical difference between these two studies: serum was included in the protocol for differentiating ES cells to mesoderm, whereas this study used serum free media for endoderm and hepatic differentiation. The elimination of serum may prevent activation of other members of the KLF family, which could compensate for the loss of Klf6.
Technical issues may also explain the discrepancy between our in vivo data, where copeb is required for late stages of organogenesis, and in vitro, where it is required for early endoderm formation. In zebrafish, morpholino injection causes transient and sometimes incomplete knock-down. The residual copeb mRNA that may be retained in morphants may allow for endoderm specification. Additionally, it is possible that Copeb protein is maternally provided in zebrafish, and that this supports early embryonic development even when translation of the copeb message is blocked with the morpholino. In contrast, mutant mouse ES cells are completely devoid of Klf6 mRNA and protein, and therefore the earliest stage in development at which it is required provides the most evident phenotype. The most likely explanation is that other members of the Klf family can compensate for the loss of copeb. The copeb morpholinos do not likely target si:dkeyp-12a9.2 or any other Klf family members and thus these could serve a redundant function with copeb in early endoderm development in zebrafish. This underscores the benefit of using complementary model systems to allow examination of the divergent roles played by the same gene under different conditions.
KLF6 is a tumor suppressor gene in humans, yet it acts as a growth promoter in zebrafish development and mouse ES cell differentiation. Similarly, divergent activities between developing and adult tissue are displayed by other tumor suppressors. For example, Smad4/Dpc4−/− embryos are embryonic lethal by E 7.5 and show reduced proliferation in vivo and in vitro (Sirard et al., 1998). Similarly, zebrafish bearing a mutation in the neurofibromatosis 2 (nf2a) gene and nf2a morphants do not survive embryogenesis and develop hepatic and biliary defects (Sadler et al., 2005). These findings form an emerging picture of how loss of tumor suppressors may lead to growth deregulation in adult cells. Thus, as recently reported for E2F1-3 (Chong et al., 2009), the same genes which contribute to developmentally regulated, tissue specific proliferation or cell survival in embryos play a different role in adults.
In summary, our findings support the role of Klf6/Copeb at several different stages of development. In early stages of ES cell differentiation, Klf6 promotes ES-cell proliferation and subsequently contributes to differentiation into endodermal cells. copeb is also essential for embryonic survival and for regulating digestive organ growth/expansion in zebrafish. These findings indicate that Klf6/copeb plays a role in liver formation that is independent of its contribution to endothelial cell development. In aggregate, our findings reinforce the importance of Klf6/copeb in liver development and growth, and underscore the value of complementary studies in zebrafish and ES cell models to understand hepatogenesis.
Supplementary Material
Figure S1. Mammalian and zebrafish KLF6/Copeb protein sequences are highly homologous. (a). Alignment of human, mouse and zebrafish KLF6/Copeb protein sequences compared to the similar to kruppel like factor 6 (official gene name si:dkeyp-12a9.2). There is 75% identity between the human and zebrafish KLF6/Copeb proteins. Only 54% identity is seen between zebrafish Copeb and the related protein encoded by si:dkeyp-12a9.2 (called “similar to klf6” in figure). * indicates identical amino acids and : indicates isoelectric amino acids. Blue boxes delineate the C2H2 zinc finger DNA binding domains. (b) Phylogram demonstrates that Copeb is closely related to mammalian KLF6.
Figure S2. copeb ATG morpholino injection induces early embryonic lethality in few embryos. The percent survival of embryos injected with either 2 or 4 μM copeb ATG morpholino was normalized to that observed in embryos from the same clutch that were either injected with a control morpholino or were uninjected. No death was observed in any clutches after 1 dpf. Values are averaged from over 300 embryos at each time point from at least 4 clutches. The error bars indicate the standard deviation. Similar results were obtained with the exon 2 splice donor blocking morpholino.
Figure S3. copeb-splice blocking morpholino creates a truncated protein due to mis-splicing of zygotic Copeb. (a) A copeb-splicing morpholino was designed to target the splicing donor site of copeb exon 2. (b) RT-PCR was performed on cDNA samples prepared from uninjected control and morphants on 1-4 dpf embryos with a pair of primers on exons 2 and 3, (copeb-e2-f and copeb-e3-r, respectively) as indicated by the arrows in (a). (c) The bands generated by the PCR from control and morphant cDNA shown in (c) were sequenced, revealing a 49 bp deletion in the morphant mRNA due to splicing to a cryptic splice donor site in exon 2 (black box). The corresponding base pair of the copeb coding sequence is labeled below the sequencing traces. (d) The product of the mis-spliced mRNA is predicted to be prematurely truncated due to a frame shift, eliminating the C2H2 zinc finger that is essential for Copeb function.
Figure S4. Restoration of copeb rescues the liver and pancreas phenotypes in copeb morphants. Embryos from Tg(fabp10:RFP;ela3l:EGFP) carriers were injected with 2 μM copeb ATG morpholino, copeb mRNA, a combination of the two or uninjected as controls and were scored for viability (a) and any developmental abnormality, termed “affected” (b); n is the total number of embryos scored in all experiments, which were carried out on at least 3 clutches. (c) Live images of representative 4 dpf larvae shows normal size pancreas in control larvae, marked by GFP expression, whereas the liver and pancreas is reduced in larvae injected with either the copeb splice donor morpholino or ATG morpholino. The liver and pancreas size is rescued by co-injecting the ATG morpholino with copeb mRNA. (d) 4 dpf larvae from Tg(fabp10:RFP;ela3l:EGFP) fish were injected with the copeb ATG morpholino, copeb RNA or both were scored for liver size based on RFP. Total n from 4 clutches is indicated below graph.
Figure S5. copeb morphants have decreased expression of blood cell genes but normal hepatic vasculature. (a) mRNA levels of marker genes for mesoderm-derived organ system such as blood and kidney were analyzed using QPCR in control embryos and copeb morphants at 4 dpf, and their levels in control embryos were set to be 1. Compared to control embryos, copeb morphants exhibited significantly lower expression of genes involved in vasculogenesis and hematopoiesis. However, expression of the kidney marker (npsh1l) was not significantly different between control and morphants. Errors bars show standard deviation from three independent experiments; * indicates p<0.05. (b) The copeb morpholino was injected into embryos expressing GFP in endothelial cells Tg(fli1a:EGFP) were imaged live on 4 dpf. The vascular network is similar in the liver in control (b-c) and copeb morphants (d-e). Scale bars = 500μm (b,d) and 100μm (c,e)
Figure S6. Klf6+/+ ES cells are able to differentiated in to the hepatic lineage using the serum-free hepatic differentiation protocol. (a) A schematic drawing showing the serum-free hepatic differentiation protocol. Klf6+/+ ES cells were pushed into the hepatic lineage using the serum-free protocol. Expression of marker genes for hepatic differentiation was analyzed in these cells using QPCR from day 0-12. mRNA levels at day 0 were set to be 1. Klf6 expression exhibited a biphasic increase at day 2 and day 8 (b). Expression of early liver marker Afp (c) and Ttr (d) could be detected at day 8, while expression of Albumin, a later hepatic marker, peaked at day 12 (e).
Figure S7. p21 expression does not require Klf6 in ES cell differentiation cultures. Klf6−/− ES cells on days 2-6 of differentiation were analyzed for p21 expression by qPCR. Expression was normalized to WT cells, averaged over 3 experiments and the error bars display the standard error.
Table S1. Primer sequences for qPCR
Acknowledgments
We are indebted to L. Loughlin and A. Mir for careful reading of the manuscript and to D. Howarth and J. Loke for technical assistance. Support was provided by the March of Dimes, Breast Cancer Alliance, American Gastroenterological Association and the NIH (R01DK080789-01A1 to KCS) and by the NIH (RO1 DK37340, RO1 DK56621 and P20 AA017067 to SLF). X. Zhao was supported by a Howard Hughes Medical Institute Research Training Fellowship for Medical Students.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Apte U, Zeng G, Thompson MD, Muller P, Micsenyi A, Cieply B, Kaestner KH, Monga SP. beta-Catenin is critical for early postnatal liver growth. Am J Physiol Gastrointest Liver Physiol. 2007;292:G1578–1585. doi: 10.1152/ajpgi.00359.2006. [DOI] [PubMed] [Google Scholar]
- Bieker JJ. Kruppel-like factors: three fingers in many pies. J Biol Chem. 2001;276:34355–34358. doi: 10.1074/jbc.R100043200. [DOI] [PubMed] [Google Scholar]
- Biemar F, Argenton F, Schmidtke R, Epperlein S, Peers B, Driever W. Pancreas development in zebrafish: early dispersed appearance of endocrine hormone expressing cells and their convergence to form the definitive islet. Dev Biol. 2001;230:189–203. doi: 10.1006/dbio.2000.0103. [DOI] [PubMed] [Google Scholar]
- Bort R, Signore M, Tremblay K, Barbera J.P. Martinez, Zaret KS. Hex homeobox gene controls the transition of the endoderm to a pseudostratified, cell emergent epithelium for liver bud development. Dev Biol. 2006;290:44–56. doi: 10.1016/j.ydbio.2005.11.006. [DOI] [PubMed] [Google Scholar]
- Chen J, Ruan H, Ng SM, Gao C, Soo HM, Wu W, Zhang Z, Wen Z, Lane DP, Peng J. Loss of function of def selectively up-regulates Delta113p53 expression to arrest expansion growth of digestive organs in zebrafish. Genes Dev. 2005;19:2900–2911. doi: 10.1101/gad.1366405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho YG, Kim CJ, Park CH, Yang YM, Kim SY, Nam SW, Lee SH, Yoo NJ, Lee JY, Park WS. Genetic alterations of the KLF6 gene in gastric cancer. Oncogene. 2005;24:4588–4590. doi: 10.1038/sj.onc.1208670. [DOI] [PubMed] [Google Scholar]
- Chong JL, Wenzel PL, Saenz-Robles MT, Nair V, Ferrey A, Hagan JP, Gomez YM, Sharma N, Chen HZ, Ouseph M, Wang SH, Trikha P, Culp B, Mezache L, Winton DJ, Sansom OJ, Chen D, Bremner R, Cantalupo PG, Robinson ML, Pipas JM, Leone G. E2f1-3 switch from activators in progenitor cells to repressors in differentiating cells. Nature. 2009;462:930–934. doi: 10.1038/nature08677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chu J, Sadler KC. New school in liver development: lessons from zebrafish. Hepatology. 2009;50:1656–1663. doi: 10.1002/hep.23157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DiFeo A, Feld L, Rodriguez E, Wang C, Beer DG, Martignetti JA, Narla G. A functional role for KLF6-SV1 in lung adenocarcinoma prognosis and chemotherapy response. Cancer Res. 2008;68:965–970. doi: 10.1158/0008-5472.CAN-07-2604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farooq M, Sulochana KN, Pan X, To J, Sheng D, Gong Z, Ge R. Histone deacetylase 3 (hdac3) is specifically required for liver development in zebrafish. Dev Biol. 2008 doi: 10.1016/j.ydbio.2008.02.034. [DOI] [PubMed] [Google Scholar]
- Field HA, Dong PD, Beis D, Stainier DY. Formation of the digestive system in zebrafish. II. Pancreas morphogenesis. Dev Biol. 2003a;261:197–208. doi: 10.1016/s0012-1606(03)00308-7. [DOI] [PubMed] [Google Scholar]
- Field HA, Ober EA, Roeser T, Stainier DY. Formation of the digestive system in zebrafish. I. Liver morphogenesis. Dev Biol. 2003b;253:279–290. doi: 10.1016/s0012-1606(02)00017-9. [DOI] [PubMed] [Google Scholar]
- Gardiner MR, Daggett DF, Zon LI, Perkins AC. Zebrafish KLF4 is essential for anterior mesendoderm/pre-polster differentiation and hatching. Dev Dyn. 2005;234:992–996. doi: 10.1002/dvdy.20571. [DOI] [PubMed] [Google Scholar]
- Gardiner MR, Gongora MM, Grimmond SM, Perkins AC. A global role for zebrafish klf4 in embryonic erythropoiesis. Mech Dev. 2007;124:762–774. doi: 10.1016/j.mod.2007.06.005. [DOI] [PubMed] [Google Scholar]
- Gouon-Evans V, Boussemart L, Gadue P, Nierhoff D, Koehler CI, Kubo A, Shafritz DA, Keller G. BMP-4 is required for hepatic specification of mouse embryonic stem cell-derived definitive endoderm. Nat Biotechnol. 2006;24:1402–1411. doi: 10.1038/nbt1258. [DOI] [PubMed] [Google Scholar]
- Holtzinger A, Evans T. Gata4 regulates the formation of multiple organs. Development. 2005;132:4005–4014. doi: 10.1242/dev.01978. [DOI] [PubMed] [Google Scholar]
- Korzh S, Pan X, Garcia-Lecea M, Winata CL, Wohland T, Korzh V, Gong Z. Requirement of vasculogenesis and blood circulation in late stages of liver growth in zebrafish. BMC Dev Biol. 2008;8:84. doi: 10.1186/1471-213X-8-84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kremer-Tal S, Reeves HL, Narla G, Thung SN, Schwartz M, Difeo A, Katz A, Bruix J, Bioulac-Sage P, Martignetti JA, Friedman SL. Frequent inactivation of the tumor suppressor Kruppel-like factor 6 (KLF6) in hepatocellular carcinoma. Hepatology. 2004;40:1047–1052. doi: 10.1002/hep.20460. [DOI] [PubMed] [Google Scholar]
- Kuo CT, Veselits ML, Barton KP, Lu MM, Clendenin C, Leiden JM. The LKLF transcription factor is required for normal tunica media formation and blood vessel stabilization during murine embryogenesis. Genes Dev. 1997;11:2996–3006. doi: 10.1101/gad.11.22.2996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Q, Van Antwerp D, Mercurio F, Lee KF, Verma IM. Severe liver degeneration in mice lacking the IkappaB kinase 2 gene. Science. 1999;284:321–325. doi: 10.1126/science.284.5412.321. [DOI] [PubMed] [Google Scholar]
- Matsumoto K, Yoshitomi H, Rossant J, Zaret KS. Liver Organogenesis Promoted by Endothelial Cells Prior to Vascular Function. Science. 2001;294:559–563. doi: 10.1126/science.1063889. [DOI] [PubMed] [Google Scholar]
- Matsumoto N, Kubo A, Liu H, Akita K, Laub F, Ramirez F, Keller G, Friedman SL. Developmental regulation of yolk sac hematopoiesis by Kruppel-like factor 6. Blood. 2006;107:1357–1365. doi: 10.1182/blood-2005-05-1916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayer AN, Fishman MC. Nil per os encodes a conserved RNA recognition motif protein required for morphogenesis and cytodifferentiation of digestive organs in zebrafish. Development. 2003;130:3917–3928. doi: 10.1242/dev.00600. [DOI] [PubMed] [Google Scholar]
- Mudumana SP, Wan H, Singh M, Korzh V, Gong Z. Expression analyses of zebrafish transferrin, ifabp, and elastaseB mRNAs as differentiation markers for the three major endodermal organs: liver, intestine, and exocrine pancreas. Dev Dyn. 2004;230:165–173. doi: 10.1002/dvdy.20032. [DOI] [PubMed] [Google Scholar]
- Narla G, Difeo A, Reeves HL, Schaid DJ, Hirshfeld J, Hod E, Katz A, Isaacs WB, Hebbring S, Komiya A, McDonnell SK, Wiley KE, Jacobsen SJ, Isaacs SD, Walsh PC, Zheng SL, Chang BL, Friedrichsen DM, Stanford JL, Ostrander EA, Chinnaiyan AM, Rubin MA, Xu J, Thibodeau SN, Friedman SL, Martignetti JA. A germline DNA polymorphism enhances alternative splicing of the KLF6 tumor suppressor gene and is associated with increased prostate cancer risk. Cancer Res. 2005a;65:1213–1222. doi: 10.1158/0008-5472.CAN-04-4249. [DOI] [PubMed] [Google Scholar]
- Narla G, DiFeo A, Yao S, Banno A, Hod E, Reeves HL, Qiao RF, Camacho-Vanegas O, Levine A, Kirschenbaum A, Chan AM, Friedman SL, Martignetti JA. Targeted inhibition of the KLF6 splice variant, KLF6 SV1, suppresses prostate cancer cell growth and spread. Cancer Res. 2005b;65:5761–5768. doi: 10.1158/0008-5472.CAN-05-0217. [DOI] [PubMed] [Google Scholar]
- Narla G, Heath KE, Reeves HL, Li D, Giono LE, Kimmelman AC, Glucksman MJ, Narla J, Eng FJ, Chan AM, Ferrari AC, Martignetti JA, Friedman SL. KLF6, a candidate tumor suppressor gene mutated in prostate cancer. Science. 2001;294:2563–2566. doi: 10.1126/science.1066326. [DOI] [PubMed] [Google Scholar]
- Narla G, Kremer-Tal S, Matsumoto N, Zhao X, Yao S, Kelley K, Tarocchi M, Friedman SL. In vivo regulation of p21 by the Kruppel-like factor 6 tumor-suppressor gene in mouse liver and human hepatocellular carcinoma. Oncogene. 2007;26:4428–4434. doi: 10.1038/sj.onc.1210223. [DOI] [PubMed] [Google Scholar]
- Ng AN, de Jong-Curtain TA, Mawdsley DJ, White SJ, Shin J, Appel B, Dong PD, Stainier DY, Heath JK. Formation of the digestive system in zebrafish: III. Intestinal epithelium morphogenesis. Dev Biol. 2005;286:114–135. doi: 10.1016/j.ydbio.2005.07.013. [DOI] [PubMed] [Google Scholar]
- Nikolova G, Lammert E. Interdependent development of blood vessels and organs. Cell Tissue Res. 2003;314:33–42. doi: 10.1007/s00441-003-0739-8. [DOI] [PubMed] [Google Scholar]
- Noel ES, Casal-Sueiro A, Busch-Nentwich E, Verkade H, Dong PD, Stemple DL, Ober EA. Organ-specific requirements for Hdac1 in liver and pancreas formation. Dev Biol. 2008;322:237–250. doi: 10.1016/j.ydbio.2008.06.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nuez B, Michalovich D, Bygrave A, Ploemacher R, Grosveld F. Defective haematopoiesis in fetal liver resulting from inactivation of the EKLF gene. Nature. 1995;375:316–318. doi: 10.1038/375316a0. [DOI] [PubMed] [Google Scholar]
- Oates AC, Pratt SJ, Vail B, Yan Y, Ho RK, Johnson SL, Postlethwait JH, Zon LI. The zebrafish klf gene family. Blood. 2001;98:1792–1801. doi: 10.1182/blood.v98.6.1792. [DOI] [PubMed] [Google Scholar]
- Ober EA, Field HA, Stainier DY. From endoderm formation to liver and pancreas development in zebrafish. Mech Dev. 2003;120:5–18. doi: 10.1016/s0925-4773(02)00327-1. [DOI] [PubMed] [Google Scholar]
- Ober EA, Verkade H, Field HA, Stainier DY. Mesodermal Wnt2b signalling positively regulates liver specification. Nature. 2006 doi: 10.1038/nature04888. [DOI] [PubMed] [Google Scholar]
- Pack M, Solnica-Krezel L, Malicki J, Neuhauss SC, Schier AF, Stemple DL, Driever W, Fishman MC. Mutations affecting development of zebrafish digestive organs. Development. 1996;123:321–328. doi: 10.1242/dev.123.1.321. [DOI] [PubMed] [Google Scholar]
- Perkins AC, Sharpe AH, Orkin SH. Lethal beta-thalassaemia in mice lacking the erythroid CACCC-transcription factor EKLF. Nature. 1995;375:318–322. doi: 10.1038/375318a0. [DOI] [PubMed] [Google Scholar]
- Ratziu V, Lalazar A, Wong L, Dang Q, Collins C, Shaulian E, Jensen S, Friedman SL. Zf9, a Kruppel-like transcription factor up-regulated in vivo during early hepatic fibrosis. Proc Natl Acad Sci U S A. 1998;95:9500–9505. doi: 10.1073/pnas.95.16.9500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reeves HL, Narla G, Ogunbiyi O, Haq AI, Katz A, Benzeno S, Hod E, Harpaz N, Goldberg S, Tal-Kremer S, Eng FJ, Arthur MJ, Martignetti JA, Friedman SL. Kruppel-like factor 6 (KLF6) is a tumor-suppressor gene frequently inactivated in colorectal cancer. Gastroenterology. 2004;126:1090–1103. doi: 10.1053/j.gastro.2004.01.005. [DOI] [PubMed] [Google Scholar]
- Robu ME, Larson JD, Nasevicius A, Beiraghi S, Brenner C, Farber SA, Ekker SC. p53 activation by knockdown technologies. PLoS Genet. 2007;3:e78. doi: 10.1371/journal.pgen.0030078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosenfeld ME, Prichard L, Shiojiri N, Fausto N. Prevention of hepatic apoptosis and embryonic lethality in RelA/TNFR-1 double knockout mice. Am J Pathol. 2000;156:997–1007. doi: 10.1016/S0002-9440(10)64967-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rudolph D, Yeh WC, Wakeham A, Rudolph B, Nallainathan D, Potter J, Elia AJ, Mak TW. Severe liver degeneration and lack of NF-kappaB activation in NEMO/IKKgamma-deficient mice. Genes Dev. 2000;14:854–862. [PMC free article] [PubMed] [Google Scholar]
- Sadler KC, Amsterdam A, Soroka C, Boyer J, Hopkins N. A genetic screen in zebrafish identifies the mutants vps18, nf2 and foie gras as models of liver disease. Development. 2005;132:3561–3572. doi: 10.1242/dev.01918. [DOI] [PubMed] [Google Scholar]
- Sadler KC, Krahn KN, Gaur NA, Ukomadu C. Liver growth in the embryo and during liver regeneration in zebrafish requires the cell cycle regulator, uhrf1. Proc Natl Acad Sci U S A. 2007;104:1570–1575. doi: 10.1073/pnas.0610774104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakaguchi TF, Sadler KC, Crosnier C, Stainier DY. Endothelial signals modulate hepatocyte apicobasal polarization in zebrafish. Curr Biol. 2008;18:1565–1571. doi: 10.1016/j.cub.2008.08.065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Segre JA, Bauer C, Fuchs E. Klf4 is a transcription factor required for establishing the barrier function of the skin. Nat Genet. 1999;22:356–360. doi: 10.1038/11926. [DOI] [PubMed] [Google Scholar]
- Shin D, Shin CH, Tucker J, Ober EA, Rentzsch F, Poss KD, Hammerschmidt M, Mullins MC, Stainier DY. Bmp and Fgf signaling are essential for liver specification in zebrafish. Development. 2007;134:2041–2050. doi: 10.1242/dev.000281. [DOI] [PubMed] [Google Scholar]
- Sirach E, Bureau C, Peron JM, Pradayrol L, Vinel JP, Buscail L, Cordelier P. KLF6 transcription factor protects hepatocellular carcinoma-derived cells from apoptosis. Cell Death Differ. 2007;14:1202–1210. doi: 10.1038/sj.cdd.4402114. [DOI] [PubMed] [Google Scholar]
- Sirard C, de la Pompa JL, Elia A, Itie A, Mirtsos C, Cheung A, Hahn S, Wakeham A, Schwartz L, Kern SE, Rossant J, Mak TW. The tumor suppressor gene Smad4/Dpc4 is required for gastrulation and later for anterior development of the mouse embryo. Genes Dev. 1998;12:107–119. doi: 10.1101/gad.12.1.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sosa-Pineda B, Wigle JT, Oliver G. Hepatocyte migration during liver development requires Prox1. Nat Genet. 2000;25:254–255. doi: 10.1038/76996. [DOI] [PubMed] [Google Scholar]
- Suske G, Bruford E, Philipsen S. Mammalian SP/KLF transcription factors: bring in the family. Genomics. 2005;85:551–556. doi: 10.1016/j.ygeno.2005.01.005. [DOI] [PubMed] [Google Scholar]
- Tan X, Behari J, Cieply B, Michalopoulos GK, Monga SP. Conditional deletion of beta-catenin reveals its role in liver growth and regeneration. Gastroenterology. 2006;131:1561–1572. doi: 10.1053/j.gastro.2006.08.042. [DOI] [PubMed] [Google Scholar]
- Tan X, Yuan Y, Zeng G, Apte U, Thompson MD, Cieply B, Stolz DB, Michalopoulos GK, Kaestner KH, Monga SP. Beta-catenin deletion in hepatoblasts disrupts hepatic morphogenesis and survival during mouse development. Hepatology. 2008;47:1667–1679. doi: 10.1002/hep.22225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka M, Fuentes ME, Yamaguchi K, Durnin MH, Dalrymple SA, Hardy KL, Goeddel DV. Embryonic lethality, liver degeneration, and impaired NF-kappa B activation in IKK-beta-deficient mice. Immunity. 1999;10:421–429. doi: 10.1016/s1074-7613(00)80042-4. [DOI] [PubMed] [Google Scholar]
- Thisse B, Pfumio S, Fürthauer M, B. L, Heyer V, Degrave A, Woehl R, Lux A, Steffan T, Charbonnier XQ, Thisse C. Expression of the zebrafish genome during embryogenesis. 2001 zfin.org online publication.
- Toyoshima Y, Monson C, Duan C, Wu Y, Gao C, Yakar S, Sadler KC, Leroith D. The Role of Insulin Receptor Signaling in Zebrafish Embryogenesis. Endocrinology. 2008;149:5996–6005. doi: 10.1210/en.2008-0329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Veldman MB, Bemben MA, Thompson RC, Goldman D. Gene expression analysis of zebrafish retinal ganglion cells during optic nerve regeneration identifies KLF6a and KLF7a as important regulators of axon regeneration. Dev Biol. 2007;312:596–612. doi: 10.1016/j.ydbio.2007.09.019. [DOI] [PubMed] [Google Scholar]
- Villefranc JA, Amigo J, Lawson ND. Gateway compatible vectors for analysis of gene function in the zebrafish. Dev Dyn. 2007;236:3077–3087. doi: 10.1002/dvdy.21354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wallace KN, Pack M. Unique and conserved aspects of gut development in zebrafish. Dev Biol. 2003;255:12–29. doi: 10.1016/s0012-1606(02)00034-9. [DOI] [PubMed] [Google Scholar]
- Wallace KN, Yusuff S, Sonntag JM, Chin AJ, Pack M. Zebrafish hhex regulates liver development and digestive organ chirality. Genesis. 2001;30:141–143. doi: 10.1002/gene.1050. [DOI] [PubMed] [Google Scholar]
- Wu H, Wade M, Krall L, Grisham J, Xiong Y, Van Dyke T. Targeted in vivo expression of the cyclin-dependent kinase inhibitor p21 halts hepatocyte cell-cycle progression, postnatal liver development and regeneration. Genes Dev. 1996;10:245–260. doi: 10.1101/gad.10.3.245. [DOI] [PubMed] [Google Scholar]
- Yea S, Narla G, Zhao X, Garg R, Tal-Kremer S, Hod E, Villanueva A, Loke J, Tarocchi M, Akita K, Shirasawa S, Sasazuki T, Martignetti JA, Llovet JM, Friedman SL. Ras promotes growth by alternative splicing-mediated inactivation of the KLF6 tumor suppressor in hepatocellular carcinoma. Gastroenterology. 2008;134:1521–1531. doi: 10.1053/j.gastro.2008.02.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zaret KS. Regulatory phases of early liver development: paradigms of organogenesis. Nat Rev Genet. 2002;3:499–512. doi: 10.1038/nrg837. [DOI] [PubMed] [Google Scholar]
- Zhao R, Duncan SA. Embryonic development of the liver. Hepatology. 2005;41:956–967. doi: 10.1002/hep.20691. [DOI] [PubMed] [Google Scholar]
- Zhao R, Watt AJ, Li J, Luebke-Wheeler J, Morrisey EE, Duncan SA. GATA6 is essential for embryonic development of the liver but dispensable for early heart formation. Mol Cell Biol. 2005;25:2622–2631. doi: 10.1128/MCB.25.7.2622-2631.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1. Mammalian and zebrafish KLF6/Copeb protein sequences are highly homologous. (a). Alignment of human, mouse and zebrafish KLF6/Copeb protein sequences compared to the similar to kruppel like factor 6 (official gene name si:dkeyp-12a9.2). There is 75% identity between the human and zebrafish KLF6/Copeb proteins. Only 54% identity is seen between zebrafish Copeb and the related protein encoded by si:dkeyp-12a9.2 (called “similar to klf6” in figure). * indicates identical amino acids and : indicates isoelectric amino acids. Blue boxes delineate the C2H2 zinc finger DNA binding domains. (b) Phylogram demonstrates that Copeb is closely related to mammalian KLF6.
Figure S2. copeb ATG morpholino injection induces early embryonic lethality in few embryos. The percent survival of embryos injected with either 2 or 4 μM copeb ATG morpholino was normalized to that observed in embryos from the same clutch that were either injected with a control morpholino or were uninjected. No death was observed in any clutches after 1 dpf. Values are averaged from over 300 embryos at each time point from at least 4 clutches. The error bars indicate the standard deviation. Similar results were obtained with the exon 2 splice donor blocking morpholino.
Figure S3. copeb-splice blocking morpholino creates a truncated protein due to mis-splicing of zygotic Copeb. (a) A copeb-splicing morpholino was designed to target the splicing donor site of copeb exon 2. (b) RT-PCR was performed on cDNA samples prepared from uninjected control and morphants on 1-4 dpf embryos with a pair of primers on exons 2 and 3, (copeb-e2-f and copeb-e3-r, respectively) as indicated by the arrows in (a). (c) The bands generated by the PCR from control and morphant cDNA shown in (c) were sequenced, revealing a 49 bp deletion in the morphant mRNA due to splicing to a cryptic splice donor site in exon 2 (black box). The corresponding base pair of the copeb coding sequence is labeled below the sequencing traces. (d) The product of the mis-spliced mRNA is predicted to be prematurely truncated due to a frame shift, eliminating the C2H2 zinc finger that is essential for Copeb function.
Figure S4. Restoration of copeb rescues the liver and pancreas phenotypes in copeb morphants. Embryos from Tg(fabp10:RFP;ela3l:EGFP) carriers were injected with 2 μM copeb ATG morpholino, copeb mRNA, a combination of the two or uninjected as controls and were scored for viability (a) and any developmental abnormality, termed “affected” (b); n is the total number of embryos scored in all experiments, which were carried out on at least 3 clutches. (c) Live images of representative 4 dpf larvae shows normal size pancreas in control larvae, marked by GFP expression, whereas the liver and pancreas is reduced in larvae injected with either the copeb splice donor morpholino or ATG morpholino. The liver and pancreas size is rescued by co-injecting the ATG morpholino with copeb mRNA. (d) 4 dpf larvae from Tg(fabp10:RFP;ela3l:EGFP) fish were injected with the copeb ATG morpholino, copeb RNA or both were scored for liver size based on RFP. Total n from 4 clutches is indicated below graph.
Figure S5. copeb morphants have decreased expression of blood cell genes but normal hepatic vasculature. (a) mRNA levels of marker genes for mesoderm-derived organ system such as blood and kidney were analyzed using QPCR in control embryos and copeb morphants at 4 dpf, and their levels in control embryos were set to be 1. Compared to control embryos, copeb morphants exhibited significantly lower expression of genes involved in vasculogenesis and hematopoiesis. However, expression of the kidney marker (npsh1l) was not significantly different between control and morphants. Errors bars show standard deviation from three independent experiments; * indicates p<0.05. (b) The copeb morpholino was injected into embryos expressing GFP in endothelial cells Tg(fli1a:EGFP) were imaged live on 4 dpf. The vascular network is similar in the liver in control (b-c) and copeb morphants (d-e). Scale bars = 500μm (b,d) and 100μm (c,e)
Figure S6. Klf6+/+ ES cells are able to differentiated in to the hepatic lineage using the serum-free hepatic differentiation protocol. (a) A schematic drawing showing the serum-free hepatic differentiation protocol. Klf6+/+ ES cells were pushed into the hepatic lineage using the serum-free protocol. Expression of marker genes for hepatic differentiation was analyzed in these cells using QPCR from day 0-12. mRNA levels at day 0 were set to be 1. Klf6 expression exhibited a biphasic increase at day 2 and day 8 (b). Expression of early liver marker Afp (c) and Ttr (d) could be detected at day 8, while expression of Albumin, a later hepatic marker, peaked at day 12 (e).
Figure S7. p21 expression does not require Klf6 in ES cell differentiation cultures. Klf6−/− ES cells on days 2-6 of differentiation were analyzed for p21 expression by qPCR. Expression was normalized to WT cells, averaged over 3 experiments and the error bars display the standard error.
Table S1. Primer sequences for qPCR


