Abstract
Defects in pituitary gland organogenesis are sometimes associated with congenital anomalies that affect head development. Lesions in transcription factors and signaling pathways explain some of these developmental syndromes. Basic research studies, including the characterization of genetically engineered mice, provide a mechanistic framework for understanding how mutations create the clinical characteristics observed in patients. Defects in BMP, WNT, Notch, and FGF signaling pathways affect induction and growth of the pituitary primordium and other organ systems partly by altering the balance between signaling pathways. The PITX and LHX transcription factor families influence pituitary and head development and are clinically relevant. A few later-acting transcription factors have pituitary-specific effects, including PROP1, POU1F1 (PIT1), and TPIT (TBX19), while others, such as NeuroD1 and NR5A1 (SF1), are syndromic, influencing development of other endocrine organs. We conducted a survey of genes transcribed in developing mouse pituitary to find candidates for cases of pituitary hormone deficiency of unknown etiology. We identified numerous transcription factors that are members of gene families with roles in syndromic or nonsyndromic pituitary hormone deficiency. This collection is a rich source for future basic and clinical studies.
Keywords: bHLH, beta helix-loop-helix, HMG, high mobility group, T-box, forkhead, hypopituitarism
Introduction
Human growth insufficiency
Height of 2 or more standard deviations (SD) below the mean for age and sex is defined as short stature. Metabolic or endocrine disorders usually cause proportionate short stature, while skeletal defects often cause disproportionate short stature [1-4]. The sitting height to standing height ratio can be used to distinguish proportionate and disproportionate short stature in cases where the distinction is not immediately obvious. Skeletal and hypothalamic-pituitary axis-based growth insufficiencies occur with similar frequencies. Genetic causes of growth hormone deficiency (GHD) are thought to occur in approximately 1/4,000 to 1/10,000 births [5,6]. These can be syndromic, including pituitary and head defects as well as defects in the development of other organs, or non-syndromic, with pituitary gland-specific effects (Table 1). Because the pituitary gland is critical for the development and function of many other organs, all defects in pituitary organogenesis cause secondary effects on target organs. Syndromic pituitary deficiencies include effects on non-pituitary tissues that are not within the expectations for secondary effects on target organs. Genetic defects in the GH gene itself and mutations in the growth hormone releasing hormone receptor (GHRHR) cause isolated GHD (IGHD), but most cases of IGHD are idiopathic (reviewed in: [7]). Patients with mutations in the growth hormone receptor gene (Laron dwarfism, a growth hormone insensitivity syndrome) have a clinical presentation similar to patients with IGHD, but they have elevated levels of GH and are treated with insulin like growth factor because they are unable to respond to GH therapy (Reviewed in: [8]).
Table 1.
Variety of transcription factor defects affect pituitary function
| Gene | DNA binding motif | Clinical features, mouse phenotypes |
|---|---|---|
| Syndromic: affecting pituitary development and other head structures | ||
| PITX2 | Paired/bicoid homeo | Rieger syndrome: eyes, teeth, umbilical defects Rarely, isolated GH deficiency, haploinsufficient in humans but not obviously so in mice |
| OTX2 | POU homeo | Anophthalmia, microphthalmia, hypopituitarism |
| LHX3 | LIM homeo | GH, TSH, PRL, LH, FSH, ACTH, variable including rigid cervical spine, sensorineural deafness |
| LHX4 | LIM homeo | GH, TSH, PRL, LH, FSH, ACTH, cerebellar and skull defects |
| SOX2 | HMG box | Hypogonadotrophic hypogonadism, rare isolated GH deficiency |
| SOX3 | HMG box | MPHD, mental retardation |
| HESX1 | Paired homeo | Variable including septo-optic dysplasia and severe or mild pituitary hypoplasia or aplasia; GH, TSH, PRL, LH, FSH, ACTH, or IGHD |
| GLI2 | Kruppel family | Holoprosencephaly, cleft lip, central incisor, hypopituitarism |
| Nonsyndromic: affecting pituitary development | ||
| PROP1 | Paired homeo | Progressive hypopituitarism, GH, TSH, PRL, LH, FSH, ACTH |
| POU1F1 | POU homeo | GH, TSH, PRL |
| TPIT | T box | ACTH |
| OTX1 | POU homeo | No human mutations described, mice have delayed growth, puberty |
| Syndromic: affecting pituitary development and other peripheral organs | ||
| NR5A1 | Nuclear receptor | LH, FSH, 46, XY disorder of sexual development, hypogonadism, premature ovarian failure, adrenal failure |
The availability of recombinant growth hormone has generally led to effective correction of growth insufficiency in children with multiple or IGHD due to pituitary developmental defects, but it is not always efficacious for some idiopathic short stature patients or for other problems associated with syndromic pituitary hormone deficiency (i.e. septo-optic dysplasia and other severe craniofacial abnormalities) [9-13]. Thus, treatment of children with pituitary hormone deficiency can be challenging, as well as expensive.
Are mouse studies informative for clinical endocrinologists?
Studies in genetically engineered and mutant mice have advanced the understanding of the mechanisms underlying pituitary organogenesis defects that lead to short stature [14,15]. In many cases, genes discovered in the mouse led to the discovery of lesions in human patients and have revealed the mechanism of action and genetic hierarchy of control of pituitary cell specification and growth [16]. For example, the discovery of the etiology of the Snell and Ames dwarf mutations (Pou1f1 and Prop1, respectively), and characterization of the phenotypes of genetically engineered mice with mutations in Tpit (officially Tbx19), Hesx1, Lhx3 and Lhx4, paved the way for identification of the mutated human genes. Some genes necessary for normal growth in mice, i.e. Aes, have not yet been reported to have lesions in human patients, but there can be a considerable lag between discovery in mice and identification of rare human patients [17,18].
The ability of mouse mutants to predict the correct human patient characteristics for screening is remarkable, as evidenced by Pou1f1, Prop1, Tpit, Hesx1, Lhx3 and Lhx4, although the correspondence is imperfect. For example, LHX4 mutations cause similar hormone deficiencies in humans and mice, and while the mouse mutations are recessive and cause perinatal lethality, the human mutations are haploinsufficient and viable [19-23]. PROP1 mutations are another example of an imperfect correspondence between the mouse and human features. In mice, lesions in Prop1 cause pituitary hypoplasia and congenital pituitary hormone deficiency, including GH, TSH, PRL, and gonadotropin deficiencies [24,25]. Both male and female mutant mice go through puberty and become fertile with GH, thyroid hormone and PRL supplements, suggesting that the gonadotropin deficiency is secondary to the lack of POU1F1 [26,27]. In contrast, humans have variable pituitary size and progressive hormone deficiency, usually with failure to undergo puberty and the additional involvement of evolving ACTH deficiency, which can be fatal if untreated [28-31]. The missense mutation (S83P) in the spontaneous Ames dwarf mutant, Prop1df/df, minimally transactivates an artificial paired homeodomain binding site in cell transfection assays, while the most common human PROP1 mutation creates a frame shift and likely complete loss of function [32,33]. This does not account for the differences in pituitary dysfunction between the species, as mice homozygous for genetically engineered Prop1 loss of function alleles have features similar to the missense mutation, and the S83P mutant appears to have no activity in culture on the Pou1f1 early enhancer, which is considered a bona fide target [32,34,35].
Dissimilarities in the features that characterize mouse mutants and human patients may be attributable to differences in the effect of the mutations (i.e. partial vs. complete loss of function), species differences in temporal or spatial expression, overlapping gene function amongst gene family members, and/or “genome variation.” Genome variation means different phenotypic manifestations of the same genetic defect due to the influence of other genes in the genome that have functional variant alleles segregating in the population. Analysis of mutant mice on different strain backgrounds can be exploited to uncover the influence of these modifier genes that magnify or minimize the manifestations of reduced function in other genes [36]. For example, genetic background has a profound effect on the survival of Prop1 mutant mice, ranging from neonatal lethal, juvenile lethal to completely viable [34]. The utility of genetically engineered mice and well defined inbred strains may make it possible to tease out the genetic risk factors that could cause some human mutations to have mild effects in some individuals and more severe ones in other patients with the same mutation [36,37].
Cell-cell signaling plays a critical role in pituitary organogenesis
Classic embryology experiments involving tissue transplantation and recombination reveal that diffusible molecules produced by the neural tissue located dorsal to Rathke’s pouch, the primordia for the intermediate and anterior lobes of the pituitary gland, are essential for pouch induction and growth [38-43]. Subsequently, members of the WNT, BMP, FGF, Notch, and hedgehog pathways were discovered to have profound effects on pituitary development [35,41,43-56]. Some essential signaling molecules are expressed in the infundibulum, but there are some in the mesenchyme surrounding the pituitary, i.e. Tgfbi, and some in the pouch itself [57]. This suggests that the regulation of pituitary development by signaling molecules is complex.
Expression of noggin, an antagonist of BMP signaling, TCF7L2, an effector of canonical WNT signaling, and WNT5A, typically acting in the non-canonical pathway, are critical for maintaining the balance of signaling pathways necessary for normal pituitary growth and morphology [17,44-47]. For example, excessive BMP signaling in noggin mutant mice is associated with reduced FGF10 expression, alteration in the SHH signaling domain, and multiple invaginations of Rathke’s pouch [46]. TCF7L2 deficient mice exhibit expansion of the FGF10 and BMP signaling domains and an abnormally large Rathke’s pouch and subsequently oversized anterior lobe [47]. Finally, WNT5A deficient mice also have expanded FGF and BMP signaling domains, and the pouch is dysmorphic but not markedly oversized [44]. In each of these cases, disruption of one signaling pathway has pleiotropic effects on other signaling pathways. This paradigm is emerging as a common theme for signaling pathway function in pituitary development.
Popular models suggest that signaling molecules influence the spatial patterns of pituitary transcription factor expression, leading to the emergence of specialized cell types that produce pituitary hormones, yet there is also compelling evidence that alterations in signaling pathways affect the morphology and size of the organ more than cell specification [17,41,44-47,56]. The noggin, WNT5A, and TCF7L2 mutants are each able to generate the 5 major hormone-producing cells of the anterior lobe despite variations in size and shape of the organ. Rizzoti et al. recently reviewed the effects of various genetic lesions on pituitary growth and shape [58].
The developing pituitary transcriptome contains many members of the BMP, FGF, WNT, Notch and SHH signaling pathways [57]. Using Genomatix software we identified 61 additional genes in these pathways that are expressed at a time when they could influence pituitary development. 17 genes may be involved in cross talk between the pathways. Gene ontology terms revealed an additional 72 genes that could contribute to cell signaling in the developing pituitary gland. RT-PCR surveys of WNT genes expressed in and around the developing organ have identified many different candidates for regulation of β-catenin activity, but little is known about the functional significance of many of these genes [35,44]. Several pituitary transcription factors are regulated by β-catenin, including the EGR1, NR5A1 complex, PITX2, and the HESX1, PROP1 complex [35,59-62]. Because β-catenin is regulated by G-protein coupled receptors, some of the pituitary transcription factors that respond to β-catenin could be independent of WNT molecules themselves, which is an area for future study [63]. Many of the signaling pathways involved in pituitary development play important roles in ontogeny of other organs, leading to lethality in mice homozygous for complete loss of function alleles. Thus, it seems unlikely, but not impossible, that genes in these pathways will be responsible for hypopituitarism in humans.
Transcription factor regulation of pituitary development
Many transcription factors play important roles in pituitary development and hormone production (Table 1). The early-acting genes are not pituitary specific, and lesions in these genes cause defects in development of craniofacial or other structures. Some of these are homeobox genes with overlapping functions and multiple roles during ontogeny, i.e. Pitx1 and Pitx2, Lhx3 and Lhx4 [19,64-69]. Defects in some of these genes cause apoptosis, reduced cell proliferation, or both, which ultimately results in pituitary hypoplasia. The functions of genes like Nr5a1, Pitx2, and Gata2, a downstream target of Pitx2, with broad expression patterns and roles in the pituitary as well as other critical organs, can be dissected by tissue specific disruption in mice [70,71]. Such studies reveal roles for Gata2 in thyrotropin and gonadotropin production, and implicate Gata3 as a gene with potential for compensatory activity [72,73].
Prop1 and Pou1f1 are examples of homeodomain transcription factors critical for pituitary development, specifically. Mutations in the human ortholog of Prop1 are the most common known cause of multiple pituitary hormone deficiency in humans [16,33,74,75]. There are dramatic differences in the effects of Prop1 and Pou1f1 mutations on fetal and neonatal pituitary development in mice. Prop1 mutants have poor pituitary vascularization and dysmorphology that appears to result, in part, from the failure of progenitors to migrate away from the proliferative zone and undergo differentiation [76,77]. The defect may result from failure to undergo epithelial to mesenchymal transition, as Prop1 is required for normal N-cadherin expression, and changes in cadherin gene expression are typically associated with epithelial to mesenchymal transition [78-80]. In contrast, there are no obvious effects on pituitary vascularization or morphology in Pou1f1 mouse mutants.
Pou1f1 is generally accepted as a direct downstream target of Prop1. This is based on the ability of PROP1 to transactivate a DNA fragment of Pou1f1 that contains the early enhancer in cell culture and the occupancy of PROP1 at that site by chromatin immunoprecipitation in extracts of microdissected embryonic pituitary glands at e12.5 and e13.5 [32,35]. Careful review of the evidence suggests that the story may be more complicated. First, there is a profound temporal delay (approximately 4 days) between activation of Prop1 and Pou1f1 expression in mice, which is unusual for a direct downstream target [32]. Second, human newborns with loss of function alleles in PROP1 have low but biologically significant levels of TSH, GH and PRL initially, suggesting that PROP1 is not required for initial expression of POU1F1 in humans [30]. Similarly, mice with Prop1 mutations express limited amounts of Pou1f1 and its targets Tshb, Gh, and Prl [81,82]. More work needs to be done to reconcile these apparently conflicting observations and clarify the role of PROP1 in humans and mice.
We hypothesize that the role of PROP1 is to generate precursor cells that are capable of becoming hormone-producing cells of the anterior lobe and promote the transition from proliferation to differentiation. It may also play a role in regulating the accessibility of the POU1F1 regulatory elements. POU1F1 is activated in some of the precursor cells to promote differentiation into somatotrophs, thyrotrophs and lactotrophs and to expand the proliferation of that lineage after birth. If this hypothesis is true, the progressive hormone deficiency in humans with PROP1 mutations could arise by depletion of the progenitor pool, and the more severe, congenital hormone deficiency in Prop1 mutant mice could be due to a stronger and/or earlier requirement of Prop1 for establishing the precursor pool in mice than humans. Investigation of genes expressed in the developing pituitary gland between peak Prop1 and Pou1f1 expression may uncover direct targets of Prop1 that are intermediates between Prop1 and Pou1f1. Neurod4 (also known as Math3) is a candidate for an intermediate, as it is activated at e13.5 before Pou1f1 is generally detected, although maintenance of Neurod4 expression requires Pou1f1 [53]. Novel genes expressed at these early developmental times will be candidates to explain pituitary deficiency diseases of unknown etiology.
Several of the known pituitary transcription factors were discovered using the approach of defining the key cis-acting sequences in hormone genes and the trans-acting factors that bind to them [83-88]. Additional advances could be made by pursuing this strategy more extensively and/or by identifying the regulatory sequences for some of the early-acting pituitary specific transcription factors and their binding factors, as well as the downstream targets of key transcription factors. For example, we used comparative genomics and bioinformatics to identify regulatory sequences in Prop1, and confirmed their relevance in cell culture and transgenic mice [89]. A highly conserved intragenic enhancer that affects spatial expression of a Prop1 transgene is a target of Notch signaling [53,89]. This suggests that screening for PROP1 mutations in human patients should include a scan of the intronic enhancer that controls spatial expression of the gene in mice [90].
Another approach is to identify gene expression differences in the pituitary glands of normal and mutant mice to identify potential downstream targets of Prop1 and Pou1f1 [57,91-94]. This gene discovery approach has revealed new members of transcription factor families that are exciting candidates for regulating pituitary development and the basis of human hormone deficiency disease. Here we report the discovery of transcription factors expressed in the developing mouse pituitary gland that are members of several important gene families including basic helix-loop-helix, high mobility group, and T-box. These genes are intriguing candidates for future functional studies and evaluation in human patients. In addition, we present a summary of the clinical features associated with hypopituitarism caused by known transcription factors, with the purpose of streamlining molecular diagnostic studies.
Results and Discussion
Prioritizing genes for molecular studies in human patients
There are approximately a dozen different transcription factor genes that are mutated in cases of short stature and/or pituitary gland dysfunction (Table 1). These are classified based on the type of pituitary defect that they produce as well as any other clinical features. Several of these genes are expressed in the developing hypothalamus and are likely to affect anterior pituitary gland development by disrupting the normal balance of signaling pathways and inductive factors produced by the hypothalamus. For example, GLI2, SOX2, SOX3, and TCF7L2 are primarily expressed in the neural ectoderm [17,47,95-100]. Some of the pituitary transcription factor genes are large and can pose difficulties for DNA sequence analysis because of high GC content. Thus, it is useful to predict which of the many genes are most likely to be mutated given a set of clinical characteristics.
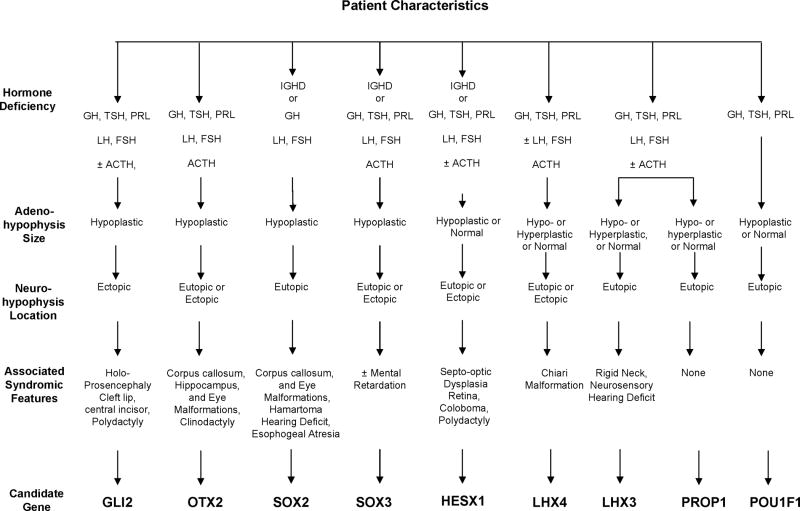
The primary sources of diagnostic information for a clinical endocrinologist are the family history, evidence of growth retardation based on the longitudinal height and weight curve of population matched controls, basal and stimulated levels of circulating hormones, and imaging to analyze the shape of the sella turcica, the size and position of the anterior pituitary, the pituitary stalk and the posterior pituitary. Non-pituitary related syndromic features such as Chiari malformation, craniofacial abnormalities, limited neck rotation, eye abnormalities, and hearing deficits can suggest a causative gene that could account for hypopituitarism and the syndromic features (Fig. 1). Congenital hypopituitarism associated to dysmorphic features are generally linked to early-expressed genes during pituitary development as GLI2, SOX2, SOX3, HESX1, LHX3, LHX4, whereas non-syndromic hypopituitarism is generally due to defects in later-expressed, pituitary-specific genes such as PROP1 and POU1F1 (Fig. 1). There can be a large spectrum of clinical features associated with any one gene. For example, HESX1 mutations can cause multiple pituitary hormone deficiencies or occasionally, isolated GH deficiency [101,102]. Obviously, not all patients with mutations in the same gene have identical clinical findings, but there are trends that suggest prioritization of the molecular diagnostic tests. Distinguishing between hypothalamic and pituitary origins of hormone deficiency is useful in planning the strategy for genetic analysis, although this is not without controversy [7,103]. There is a lack of data in the literature correlating TRH stimulation pattern in patients with known genetic defects associated to hypothalamic or pituitary cells differentiation. In a study of 43 patients with MPHD, 26% were compatible with pituitary hypopituitarism, and 70% had hormonal responses suggestive of hypothalamic hypopituitarism [104]. Ectopic posterior lobe was a characteristic of putative hypothalamic hypopituitarism.
Figure 1. A guide for planning genetic screening for hypopituitary patients based on clinical findings.
The patient characteristics identified by hormone screening, imaging studies, and analyses of syndromic features are itemized for each candidate gene based on currently known patient mutations. Note that for most genes there are variable hormone deficiencies, pituitary size and placement, and variable syndromic features.
Mutations in HESX1, LHX3, LHX4, GLI2, SOX2, and SOX3 are rare causes of congenital hypopituitarism, while PROP1 defects are a common cause of familial, congenital hypopituitarism [15,75,105,106]. Although PROP1 mutations have a variable effect the size of the gland, all cases described to date have an intact stalk, eutopic (normally placed) posterior pituitary, and hormone deficiencies that tend to worsen progressively [28-30,33]. Other genes that can be mutated in patients with MPHD, eutopic posterior lobe and normal pituitary stalk are POU1F1 and LHX3, although LHX3 mutations can present with cervical stiffness (Fig. 1). Despite the common occurrence of PROP1 mutations in multiple ethnic groups, and the rare mutations reported in other transcription factor genes, most patients with hypopituitarism have no identifiable genetic lesions [104]. Ectopic posterior pituitary and stalk disruptions are common features of patients with unknown etiology [101].
There have been impressive advances in identifying the molecular basis for pituitary hormone deficiency over the past 20 years. It is also appreciated that non-genetic traumatic events can cause hypopituitarism [104]. Yet birth trauma does not appear to be an obvious contributor to the majority of cases of hypopituitarism without a molecular diagnosis. Thus, additional genes are likely to be involved, and alternative approaches must be taken to identify these genes.
Gene discovery to identify new candidate genes
In an effort to identify novel candidate genes for hypopituitarism, we undertook a project to discover genes expressed during a critical period of mouse pituitary gland development [57,93]. A brief description of library preparation and analysis follows. We dissected pituitary glands from normal mice at e12.5 (CD1 strain) and from DF/B-Prop1df/df mutants and their wild type littermates at e14.5 and genotyped individual fetuses using previously described protocols [92]. Appropriate samples were pooled based on genotype, total RNA isolated, and cDNA prepared using the cap-trapper method to enhance the representation of full-length clones and normalization to enrich for rare sequences [93,107]. Two subtracted libraries were prepared to enhance the representation of Prop1 targets [57]. One subtracted library was enriched for genes activated at e14.5 but not e12.5, and the other was enriched for genes expressed in normal pituitaries but not in Prop1 mutants at e14.5. Single pass DNA sequencing was conducted on 56,716 clones using an algorithm to maximize gene discovery and minimize re-sequencing cDNAs representing the same genes. The cDNA sequences were analyzed by comparison with publicly available databases using an expect value of 10-5 to favor putative identifications that could be validated by more complete sequence analysis. We placed the matches of single pass sequence with public database entries into a local database that is searchable by gene name, RefSeq ID, Unigene ID, gene ontology terms (GO), and DNA sequence and identified 12,009 different expressed genes. To obtain database access contact Drs. Camper or Lyons at scamper@umich.edu or boblyons@umich.edu.
Analysis of the database revealed numerous homeobox genes not previously known to be expressed in the pituitary gland, members of signaling pathways, and genes involved in cell proliferation, apoptosis, cell migration, and cell adhesion [57,94]. Here we highlight transcription factors identified by this approach that are members of the PITX, and LHX gene families, as well as the forkhead group, families of basic helix-loop-helix (bHLH), high mobility group (HMG), and the T-box family. Members of each of these families have been implicated in pituitary development and function, and there are several precedents for overlapping and critical functions of related genes within a single family for pituitary development [19,65].
Initial validation studies reveal that the single pass sequences obtained from the 3’ end are not as reliable as those from the 5’ end, probably due to the difficulty of accurate amplification through stretches of polyadenine. In rare cases we identified chimeric clones or unexplained differences between the single pass sequence and the validation by complete sequencing, but in most cases the predictions are accurate [57]. Another important type of verification is demonstration that the cDNA is expressed in the pituitary gland because the embryonic pituitary dissection includes some surrounding mesenchyme, oral ectoderm, and neural ectoderm. Complete sequencing and expression studies have not yet been done on all of the genes in the basic helix-loop-helix (bHLH) and high mobility group (HMG) families reported here or for another study based on gene ontology, biological process terms [94]. Thus, this report represents a foundation for comprehensive validation and future studies.
The PITX gene family
The PITX gene family illustrates how multiple members within the same gene family can have unique and overlapping functions in organ development. PITX2 mutations are one cause of Rieger syndrome, a genetically heterogeneous, dominant disorder characterized by defects in development of the eyes, teeth, and umbilical cord [108]. Some individuals have additional abnormalities including heart defects, and rarely, growth hormone insufficiency. Molecular studies show that missense mutations can cause loss of function, gain of function, or dominant negative effects, but loss of function mutations appear to be the most common, consistent with a haploinsufficiency disorder [109].
Mice heterozygous for loss of function mutations typically have no obvious abnormalities, and eye and tooth defects occur with extremely low penetrance. Multiple genetic backgrounds studied do not increase the penetrance (Camper and Gage, unpublished observations). This suggests that reduced levels of PITX2 are better tolerated in mice than humans. Homozygous mutants die at approximately e12.5 due to severe heart and abdominal body wall defects [67]. Multiple organs are severely affected, including the eyes, teeth, and lungs, which exhibit isomerization. The pituitary gland exhibits hypoplasia, partly caused by enhanced cell death at the ventral aspect that is suggestive of defective sonic hedgehog and/or FGF signaling [65].
Mice homozygous for a partial loss of function allele, Pitx2neo, have milder pituitary defects, with little reduction in organ size, but reduction in the Pou1f1 lineage, resulting in reduced expression of both GH and TSH [66]. The reduced differentiation of this lineage could explain the reduction in GH secretion that characterizes some patients with PITX2 lesions. Homozygotes for this hypomorphic allele lack gonadotrophs, as there is virtually no detectable Nr5a1, Lhb or Fshb expression. Gata2 expression is profoundly reduced as well. There are no reports of hypogonadism in Rieger patients, but it is possible that feedback regulation from the ovaries and testes normalizes gonadotropin production after birth. Indeed, there are some animal models that exhibit transient hypogonadism and delayed puberty but eventually become fertile [110-112].
No PITX1 mutations have been described in humans to date. Pitx1 loss of function is recessive lethal in mice [113,114]. Mutants have multiple defects including reduction in the mandible and hind limbs, but the pituitary is mildly affected, exhibiting a reduction in the number of gonadotropes at birth. Despite the fact that Pitx1 is dispensable for development of a normally sized pituitary gland and differentiation of the major cell types of the anterior lobe at birth, Pitx1 and Pitx2 have overlapping functions early in pituitary development and are required for activation of Lhx3 [65,66,115].
The LHX family
Several LIM homeodomain transcription factors are expressed in the pituitary gland: Lhx2, Lhx3, Lhx4 and Isl1. The first to be implicated in the pituitary gland was Lhx2, which was identified by cloning the transcription factor that bound to a critical cis-acting sequence in the gene that encodes the alpha subunit of the pituitary glycoprotein hormones, Cga, in the gonadotrope- and thyrotrope-like cell lines, T3-1 and TSH, respectively [116]. Additional studies demonstrated that LHX2 interacts with a LIM domain binding protein (LBD1) to regulate Cga expression, and the single-stranded DNA binding protein, SSBP3, regulates the abundance of this complex [117]. We identified Lhx2 expression in a cDNA library made by subtracting e12.5 Rathke’s pouch cDNA from e14.5, suggesting that Lhx2 could have a role in pituitary development [57]. To explore the spatial expression of Lhx2 in early pituitary development we carried out immunohistochemical staining (Figure 2). The majority of the stain appears in the neural ectoderm, dorsal to the prospective posterior lobe and signaling center for BMP and FGF. Staining is also readily detectable in the caudal hindbrain, but little or no staining is detected in the developing pituitary gland at these stages. These immunohistochemistry results are consistent with the failure to detect Lhx2 transcripts in Rathke’s pouch or its derivatives at e10.5 and e14.5 [118-120]. This suggests that LHX2 regulation of Cga expression initiates later in development or requires amounts of the protein that are difficult to detect. In some tissues Lhx2 is expressed after stem cells become committed progenitors but before terminal differentiation occurs [121]. If Lhx2 plays a role in pituitary gland, it might explain our difficulties in detecting immunoreactivity. Lhx2 mutants die during gestation from defects in erythropoiesis and exhibit anophthalmia and brain defects [122]. Additional studies are necessary to define the role of Lhx2 in pituitary gland development.
Figure 2. LHX2 expression in the neural ectoderm.
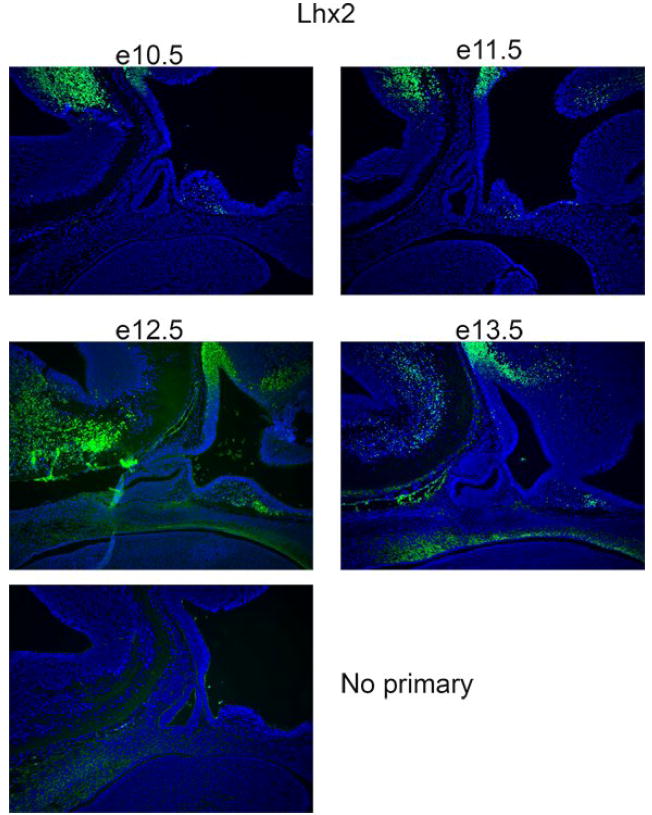
LHX2 immunoreactivity (green) is detected in sagittal sections of developing mice at e10.5 through e13.5. DAPI (blue) counterstain reveals nuclei of individual cells.
LHX4 is a LIM homeodomain transcription factor involved in pituitary organogenesis, and crucial for the genesis and development of Rathke’s pouch. As a LIM homeodomain transcription factor, LHX4 shows significant structural similarity with LHX3, suggesting a possible overlap between each factor during Rathke’s pouch formation [123]. This point is suggested by the observation of similar activities of LHX4 and LHX3 in assays using pituitary hormone promoter genes, and by the role of each factor in ventral motor neuron differentiation [124].
LHX4 mutations produce a wide intra- and inter-family range of phenotypes in humans, both in terms of hypopituitarism and of pituitary/cerebral MRI images of the morphology. The gland may exhibit hypo- or hyperplasia, variable ectopic posterior lobe, and assorted intracranial abnormalities including Chiari syndrome and corpus callosum hypoplasia, and poorly developed sella turcica. To date, 5 heterozygous mutations, including 1 intronic lesion, are reported, suggesting that the mechanism underlying the functional defect is haplo-insufficiency [20,21,125,126].
LHX3 mutations generally lead to MPHD with variablity in corticotrope axis function, abnormal neck rotation, mild to severe hearing impairments, and/or mental retardation. The pituitary can either be hypo- or hyperplastic, or even associated with a microadenoma. Only 9 LHX3 mutations have been reported, and all are inherited in an autosomal recessive manner [22,127-130].
Mice homozygous for an Lhx4 disruption induced by homologous recombination (Lhx4-/-) have an abnormal pituitary phenotype and die soon after birth from lung defects, whereas heterozygous animals (Lhx4+/-) seem unaffected [19,131]. It is possible that homozygous LHX4 mutation causes lethality in humans. It is noteworthy that mice heterozygous for Lhx4 loss of function do not appear affected, just as PITX2 haploinsufficiency is evident in humans but not mice. This species difference in the tolerance for reduced LHX4 and PITX2 levels illustrates a limitation of the comparison between human and mice, though the accessibility of tissues in mice throughout development is invaluable for understanding the mechanisms that underlie human pituitary developmental defects.
Mice homozygous for Lhx3 disruption (Lhx3-/-) exhibit a severe phenotype with death within 24 hours after birth [64]. In contrast, Lhx3+/- mice appear normal. Embryonic Lhx3-/- mice show normal rudimentary Rathke’s pouch formation but lack further pituitary development from e10.5 onward and undergo apoptosis [69,132]. Dorsal-ventral patterning is modified with dorsal location of some progenitors normally located in the ventral aspect of the gland [69]. At birth, Lhx3-/- mice lack the anterior and intermediate lobes of pituitary gland.
Lhx3 and Lhx4 are expressed throughout the pouch at e9.5 [19]. At e12.5 Lhx3 continues to be expressed throughout the pouch in a gradient with higher protein levels at the dorsal aspect of the pouch [131], while Lhx4 expression becomes restricted to the developing anterior lobe. At e15.5, Lhx4 decreases, while Lhx3 continues to be expressed at high levels. The overlap in gene expression suggests the possibility of functional overlap, which is borne out by analysis of double mutant mice.
Lhx3-/- and Lhx4-/- single mutants form a definitive pouch [19,64]. The pouch fails to expand in Lhx3 mutants due to increased apoptosis resulting in severe pituitary hypoplasia [69,132]. Lhx3 nulls exhibit reduced ACTH immunostaining and deficiency of all other hormones normally produced in the anterior lobe. The Lhx4-/- phenotype is less severe. Specification of five hormone-producing cell types occurs, but expansion of these lineages is greatly reduced, and increased apoptosis is evident [131]. One wild type Lhx3 or Lhx4 allele is sufficient for formation of a definitive pouch, as evidenced by analysis of Lhx3-/-, Lhx4+/- and Lhx3+/-, Lhx4-/- mutants. Loss of all alleles for Lhx3 and Lhx4 results in formation of a rudimentary pouch, which fails to grow into a definitive pouch and remains under the sphenoid cartilage [19]. The fact that a definitive pouch is formed in the absence of Lhx3, but only a rudimentary pouch is formed when both Lhx3 and Lhx4 are deleted, suggests that Lhx4 can substitute for the function of Lhx3 to support the formation of a definitive pouch. The fact that deletion of Lhx3 alone results in loss of most of the anterior lobe cell types suggests that Lhx4 can not substitute for the function of Lhx3 to activate a pituitary-specific transcription program [133].
Isl1 expression is detectable throughout Rathke’s pouch at e9.5 and by e12.5 it is restricted to the ventral, differentiating cells that express Cga and Foxl2 [69,131]. Lhx3 and Lhx4 mutants have different effects on Isl1 expression in the pituitary gland, causing a gain and loss of expression, respectively [69]. Isl1 is implicated as a lineage specific transcription factor in cell fate choice in progenitors of the retina, heart, forebrain, and motor neurons [134-138]. Isl1 deficiency causes an arrest in pituitary gland development at an early stage, and the embryos die at e11.5 [134]. Thus, it is an essential regulator of the early steps, but its role, if any, in later stages is unknown.
Forkhead factors are essential for diverse developmental processes
There are several common functional themes among forkhead factors. First, they are responsible for numerous autosomal dominant human developmental disorders. For example, mutations in four different forkhead genes affect ocular development. Mutations in FOXC1 result in Axenfeld-Rieger anomaly that is characterized by facial, teeth and eye malformations [139-142]. FOXC2 mutations result in lymphedema and distichiasis, a double row of eyelashes [143,144]. Mutations in FOXE3 result in malformations in the anterior segment of the eye referred to as Peter’s anomaly [145,146]. FOXL2 mutations cause eyelid malformations and premature ovarian failure [147,148]. Mutations in the orthologous mouse genes cause similar phenotypes [149-161].
Secondly, many forkhead proteins are important for cell cycle regulation and act as tumor suppressors. Overexpression of the forkhead transcription factors FoxO3a, AFX, or FoxO1a cause growth suppression in a number of cell lines, including a Ras-transformed cell line and a cell line lacking a known tumor suppressor [162]. Amplification of the forkhead gene, FoxA1, occurs in lung tumors and esophageal adenocarcinomas implicating this gene in tumorigenesis [163].
Finally, approximately half of the known null mutations in forkhead genes result in death before or shortly after birth [164]. This raises three important points: 1) this family of genes is very important for normal development, 2) members of this family generally do not exhibit functional redundancy, 3) mouse models of forkhead gene deficiency are good predictors of the human phenotype. Null mouse models have been described for approximately 31 forkhead genes so far and all except for Foxo4 result in an abnormal phenotype. Moreover, a number of these knockouts involved members of the same subfamily. For example, Foxa1 knockout mice die postnatally with severe growth retardation [165], and Foxa2 knockout mice do not develop beyond embryonic day 8.5 (e8.5) and lack the node, notochord and foregut [166-169]. These data suggest that FOXA1 and FOXA2 cannot compensate for each other, in contrast to Foxc1 and Foxc2. Loss of Foxc1 results in multiple abnormalities including hydrocephalus, skeletal, ocular, renal and cardiovascular defects, and Foxc2 deficiency causes skeletal, cardiovascular and ocular defects, indicating that each gene is required independently for development of several organ systems including the skeleton. Loss of both Foxc1 and Foxc2 disrupts somitogenesis, revealing an early overlapping function.
Forkhead factors in pituitary development
Foxl2 (Pfrk) is expressed in the prospective anterior lobe of the developing pituitary gland starting at e11.5 and continuing into adulthood in gonadotrope and thyrotrope cells of the anterior pituitary [56,170]. In addition, FOXL2 is part of a transcription complex that binds the gonadotropin-releasing hormone receptor gene in gonadotrope cells [171]. Finally, Foxl2 stimulates expression of Cga (αGSU) in cell culture studies and when overexpressed in transgenic mice [170].
Human patients heterozygous for FOXL2 mutations have dominant blepharophimosis ptosis epicanthus inversus syndrome (BPES, eyelid abnormalities) and premature ovarian failure [172]. Mice homozygous for loss of function alleles are mostly nonviable, but those that survive have craniofacial and ovarian abnormalities [160]. While the effects of this deficiency on the pituitaries of most mutants has not been reported, Foxl2 transgene expression is sufficient to drive expression of Cga at ectopic sites within the pituitary primordium, and it has permanent expression in thyrotropes and gonadotropes, suggesting a role in gonadotrope differentiation and function [170]. Consistent with a role in gonadotrope differentiation, FOXL2 regulates the expression of Gnrhr, Fshb, and follistatin in gonadotropes [171,173,174]. During pituitary development, FOXL2 protein is localized to quiescent cells, suggesting that FOXL2 may be important for inhibiting cell proliferation [170]. This idea is supported by the discovery that mutations in FOXL2 are associated with aggressive ovarian granulosa cell tumors in children [175].
Several other forkhead genes are expressed in the pituitary gland: Foxe1, Foxf1, Foxa1, and Foxd1. Foxe1 expression is first detected at e9.5 in the ectoderm that will give rise to the anterior pituitary [176]. Foxf1 is expressed in the mesenchyme of the developing pituitary gland by e9.5 [177]. FOXA1 (a.k.a. HNF-3α) represses growth hormone expression in mouse and human pituitary tissue by binding to the P sequence element C of the human GH gene [178]. FOXD1 (or brain factor 2, BF2) is expressed in the diencephalon, retina, and kidney. Mutations in Foxd1 affect the retina, optic chiasm and kidney [179]. The kidneys are small with decreased ureteric branching, and the mice die within 24 hours after birth due to renal failure. Ectopic BMP signaling is thought to be responsible for the dysmorphology and loss of kidney function [180]. Foxd1 expression is detectable in the pituitary gland after birth, and during development it is evident in the diencephalon and the mesenchyme surrounding the pituitary at e10.5. Interestingly, these regions are essential for the production of BMPs, which are required for normal pituitary development, and thus FOXD1 regulation of BMP production in these tissues likely contributes to normal pituitary development.
Basic helix-loop-helix family is highly represented in the developing pituitary gland
Members of the basic helix-loop-helix (bHLH) family of transcriptions factors are found in all eukaryotes, and they bind DNA in complexes of homo- or heterodimers through a conserved helix-loop-helix domain [181]. These transcription factors play diverse roles in many developmental pathways and tissues. Myod, Myog, and Myf5 are bHLH proteins that are instrumental in the differentiation of skeletal muscle (Reviewed in [182]), while Ascl1, Neurog1, and Neurod1 participate in neuronal differentiation [183-189]. Several members of the bHLH family are known to be expressed in pituitary development: Ascl1, Neurod1, Neurod4, and Hes1. Roles of these genes in corticotrope, somatotrope, and melanotrope development will be reviewed below.
Neurod1 (also known as BETA2, BHF-1, bHLHa3, NeuroD) is expressed in the embryonic pituitary at e12.5, where it precedes the appearance of POMC in the corticotrope lineage, and it is down regulated at e15.5 [190,191]. Neurod1 has a role in corticotrope development and function, and recent evidence suggests that it may affect gene expression in gonadotropes as well [192]. Loss of Neurod1 delays the terminal differentiation of corticotropes from e12.5 to e16.5, indicating that it is a critical factor for promoting corticotrope differentiation, although it is not required [86,193]. NEUROD1 and TPIT (a T box gene officially named TBX19, discussed below) both bind to the POMC promoter to drive expression. However, neither Neurod1 nor Tpit is required for POMC expression, nor does loss of one prevent the binding of the other to the POMC promoter, suggesting that these transcription factors act independently of each other to drive POMC expression [194,195].
Neurod4 (also known as Math3, Atoh3, bHLHa4) is a downstream target of Pou1f1 that is necessary for maintenance of the somatotrope cells [53].
The bHLH factor, Hes1, is necessary for POMC expression in melanotropes in the intermediate lobe [49]. The Hes transcription factors transduce signals from the Notch signaling pathway, along with the Hey class of bHLH transcription factors. Loss of Hes1 results in a cell fate switch such that intermediate lobe cells differentiate as GH hormone secreting somatotropes instead of POMC expressing melanotropes [49]. Hes1 plays an additional role in maintaining anterior lobe precursor cells such that they do not differentiate prematurely [52,53,78]. Recent studies demonstrate that this is accomplished through control of cell cycle regulators [196].
The expression patterns of bHLH transcriptions factors Hey1 and Hes6 suggest possible roles in the developing pituitary gland, but more studies are necessary to assess their functional significance. The Hey1 expression domain overlaps with Hes1 in presumptive precursor cells in the pouch, suggesting that these genes may have overlapping functions in regulating the progression of cells from proliferation to differentiation [49,50]. In contrast, Hes6 is expressed in the differentiating cells of the anterior lobe, positioning it for maintaining quiescence and/or cell specification [48]. The contributions of these bHLH transcription factors to pituitary gland development are still speculative.
We searched our cDNA encyclopedia for members of the bHLH family and identified 33 genes including expected factors such as Neurod1, Hes6 and Hey1 as well as additional bHLH family members whose embryonic pituitary expression was not previously known (Table 2). Among these new bHLH family members are the Id genes, which act downstream of BMP signaling [57]. Identification of Id3 in the library allowed for its use as a reporter of BMP signaling in the ventral diencephalon overlying Rathke’s pouch [46].
Table 2.
Developing pituitary transcriptome contains many transcription factors of the bHLH, HMG and Tbox families
| bHLH = basic helix-loop-helix | |||
|---|---|---|---|
| Gene symbol | Full gene name | Gene symbol | Full gene name |
| Ahr | aryl-hydrocarbon receptor | Mlx | MAX-like protein X |
| Arnt | aryl hydrocarbon receptor nuclear translocator | Mnt | max binding protein |
| Arntl | aryl hydrocarbon receptor nuclear | Msc | musculin |
| Ascl1 | translocator-like achaete-scute complex homolog-like 1 (Mash1) | Mxi1 | Max interacting protein 1 |
| Bhlhb9 | basic helix-loop-helix domain containing, class B9 | Mycl1 | v-myc myelocytomatosis viral oncogene homolog 1, lung carcinoma derived |
| Ebf2 | early B-cell factor 2 | Mycn | v-myc myelocytomatosis viral related oncogene, neuroblastoma derived |
| Ebf4 | early B-cell factor 4 | Neurod1 | neurogenic differentiation 1 |
| Hes6 | hairy and enhancer of split 6 | Npas3 | neuronal PAS domain protein 3, transcript variant 2 |
| Hey1 | hairy/enhancer-of-split related with YRPW motif 1 | Srebf1 | sterol regulatory element binding factor 1 |
| Hif1a | hypoxia inducible factor 1, alpha subunit | Tcf23 | transcription factor 23 |
| Hif3a | hypoxia inducible factor 3, alpha subunit | Tcf25 | transcription factor 25 |
| Id1 | inhibitor of DNA binding 1 | Tcf4 | transcription factor 4 |
| Id2 | inhibitor of DNA binding 2 | Tcfe2a | transcription factor E2a |
| Id3 | inhibitor of DNA binding 3 | Tcfe3 | transcription factor E3 |
| Id4 | inhibitor of DNA binding 4 | Tcfeb | transcription factor EB |
| Max | Max protein | Usf2 | upstream transcription factor 2 |
| Mesp2 | mesoderm posterior 2 | ||
| HMG = High mobility group | |||
| Bbx | bobby sox homolog | Nsbp1 | nucleosome binding protein 1 |
| Cic | capicua homolog | Sox2 | SRY-box containing gene 2 |
| Hmga1 | high mobility group AT-hook 1 | Sox9 | SRY-box containing gene 9 |
| Hmga2 | high mobility group AT-hook 2 | Sox11 | SRY-box containing gene 11 |
| Hmgb1 | high mobility group box 1 | Sox12 | SRY-box containing gene 12 |
| Hmgb2 | high mobility group box 2 | Sox30 | SRY-box containing gene 30 |
| Hmgb3 | high mobility group box 3 | Ssrp1 | structure specific recognition protein 1 |
| Hmgn1 | high mobility group nucleosomal binding domain 1 | Taf1 | TAF1 RNA polymerase II, TATA box binding protein (TBP)- associated factor |
| Hmgn3 | high mobility group nucleosomal binding domain 3 | Tfam | transcription factor A, mitochondrial |
| Mll3 | myeloid/lymphoid or mixed-lineage leukemia 3 | ||
| T-box | |||
| Tbx19 | T-box19 (Tpit) | Tbx3 | T-box 3 |
| Tbx2 | T-box 2 | ||
Interestingly, the bHLH member, aryl-hydrocarbon receptor (Ahr), and the aryl-hydrocarbon interacting protein (Aip, a tetratrico peptide repeat containing protein) are contained in the developmental library. AIP is associated with increased risk of pituitary adenomas that secrete GH in some populations; and the molecular mechanism appears to be loss of tumor suppression [197,198]. The overall contribution of mutations in AIP to sporadic adenoma risk world wide appears to be low [199], but the presence of this Ahr, Aip complex during the development of the pituitary suggests that an early developmental mechanism for growth regulation may be recapitulated during adenoma formation.
High mobility group genes expressed in the developing pituitary
The high mobility group or HMG class of DNA binding proteins bind DNA through a conserved domain that consists of three alpha helices arranged in an “L-shape.” These proteins are known to bend the DNA to which they are bound [200]. The Sox genes, which are related to SRY, the mammalian male sex determination gene, are members of the HMG family, and they have received increased attention recently because of their role in stem cell maintenance. Sox2 is expressed in embryonic stem cells and stem cells from a variety of tissues, including a potential stem cell population in the pituitary [201,202]. The presence of a pituitary stem cell population that gives rise to all the cell types of the anterior lobe is an exciting development as it has been a proposed mechanism for explaining the plasticity of the pituitary gland, which can change its cellular make up in response to changing physiological demands.
Sox2 and Sox3 are also critical factors in embryonic pituitary development. Both are expressed in the ventral diencephalon overlying Rathke’s pouch [100,202,203]. Sox3 homozygous null mice have disruptions in the patterning of the ventral diencephalon such that both Bmp4 and Fgf8 expression domains, which are critical for Rathke’s pouch induction and proliferation, are expanded, resulting in a dysmorphic Rathke’s pouch. Sox3 null mice also have hypopituitarism with decreased levels of GH, LH, FSH, and TSH [97]. Sox2 heterozygous mice have a dysmorphic Rathke’s pouch, which likely results from a similar mechanism as Sox3 in the ventral diencephalon. Unlike Sox3, Sox2 is also expressed in Rathke’s pouch so that the hypopituitarism observed in Sox2 heterozygous mice may result from a direct affect of Sox2 in the anterior lobe [106]. Both SOX2 and SOX3 are associated with human disorders; SOX3 causes X-linked panhypopituitarism [100], while SOX2 mutations cause anterior pituitary hypoplasia and hypogonadotropic hypogonadism [203].
NUPR1 (p8) is a high mobility group protein that was discovered as a differentially expressed gene in cell lines representing different stages of gonadotrope development [204]. It is expressed during late gestation in mice, concomitant with the activation of the gonadotropin beta subunit genes. It is essential for timely activation of gonadotropin expression [205]. In addition, it is implicated in pituitary tumorigenesis [206,207].
Given the importance of HMG genes in pituitary development we screened our developmental library for expected and novel HMG family members expressed in the pituitary (Table 2). We identified 19 different HMG genes in our libraries. Five members of the Sox group were identified, including Sox2. Mixed-lineage leukemia 3 (Mll3) is in this set. Myeloid leukemias have deletions in MLL3 [208]. Further analysis of these HMG genes will enrich our understanding of pituitary gland development, and perhaps adenoma formation.
Several T-box genes are expressed in the developing pituitary gland
The T-box genes are a family of transcription factors that bind DNA through the ~200 amino acid T-box domain. They are highly conserved, found in all metazoans and every known vertebrate genome. T-box genes can act as transcriptional regulators, either as activators or repressors, in a context-dependent manner. T-box genes are involved in early embryogenesis, extra-embryonic tissue survival, cell fate decisions, embryonic patterning and organogenesis (reviewed in [209]). The first T-box gene to be identified in the pituitary was Tbx19 (Tpit) [86]. In the absence of TPIT, the POMC lineages, including intermediate lobe melanotropes and anterior lobe corticotropes, fail to differentiate fully [210]. In addition, TPIT inactivation results in a cell fate change, permitting prospective melanotropes to differentiate into gonadotropes and Pou1f1-independent thyrotropes. The phenotype of mutant mice predicted the clinical characteristics of human patients with TPIT mutations [195].
We identified Tpit, Tbx2 and Tbx3 as genes expressed in the developing pituitary gland between e12.5 and e14.5 using our cDNA encyclopedia. Tbx2 and Tbx3 expression was detected by RT-PCR in cDNA prepared from dissected pituitary glands at e12.5, e14.5, e18.5, and in Prop1df/df cDNA at e14.5. Localization of Tbx2 and Tbx3 transcripts by in situ hybridization suggests that there is little or no overlap of either gene expression pattern with that of Tpit (Fig. 3) and [211]. Both genes are strongly expressed in the developing ventral diencephalon and the posterior lobe of the pituitary gland. Prominent expression of Tbx3 overlaps the area of the neural ectoderm where factors that induce pituitary growth, BMP and FGF, are expressed. There are no obvious differences in Tbx3 expression in normal and Prop1df/df mice (Fig. 3). Tbx3 is expressed in the rostral tip, where Pou1f1- independent thyrotropes are located [211]. These transcriptional repressors, Tbx2 and Tbx3, might have functional overlap in regulating posterior lobe development because the expression patterns overlap, but neither gene has overlapping expression with Tpit. The function of these genes in pituitary development could best be determined using organ specific inducible loss of function models because of the embryonic lethality and ancillary organ defects characteristic of mice homozygous for systemic null alleles [212,213].
Figure 3. Tbx3 expression in the ventral diencephalon and prospective posterior lobe of the pituitary gland during development.
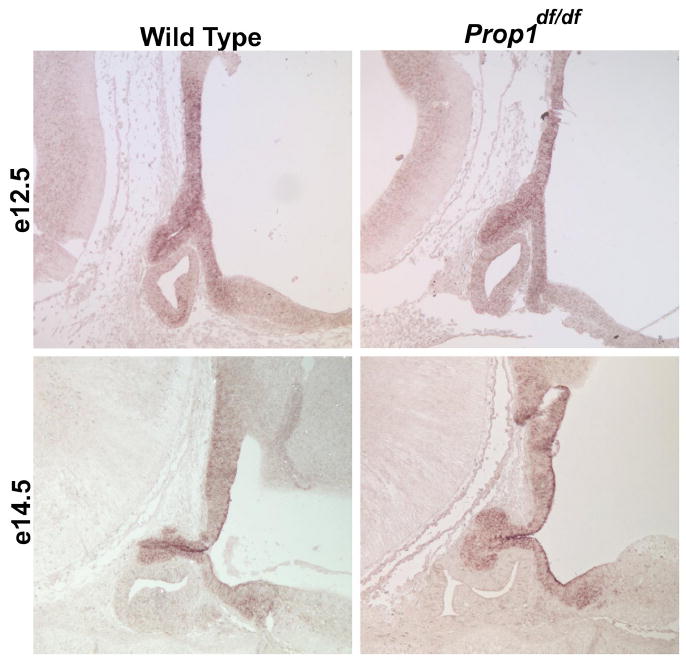
Tbx3 transcripts are readily detectable in mid-sagittal sections of developing normal and Prop1df/df mouse embryos at e12.5 and e14.5 by in situ hybridization.
Several other T-box family members are expressed in and around the developing pituitary gland, but they have not been studied extensively and were not detected in our cDNA encyclopedia. These include Tbx15, Tbx18, and the T-box brain gene 1, Tbr1 (www.genepaint.org). While Tbx18 and Tbr1 do not display overlapping expression patterns with Tbx2, 3, or Tpit, they appear to be expressed in a subset of anterior lobe cells at e14.5, consistent with expression of Pou1f1 in that region.
Future directions
The developing pituitary cDNA libraries we made and analyzed reveal that the transcriptome has great depth at the time the cells are differentiating and the organ is undergoing substantial growth. There are already several compelling precedents for functional overlap of transcription factors within a particular gene family, i.e. PITX and LHX families. Thus, the discovery of many members of the Forkhead, HMG, bHLH and Tbox families, suggested that there may be genes with essential functions that overlap important transcription factors in these families like Foxl2, Nupr1, NeuroD1, Hes1, Sox2 and Sox3, and Tpit. Expression studies suggest that Tbx2, 3 have distinct functions from Tpit, and Lhx2 may act differently than the Lhx3, 4 genes. The sheer number of genes in these families that are expressed at a time when they could have an important impact suggests that the ideal strategy for identifying the genes with essential functions is a high throughput screen. Given the strong parallels between the function of orthologous developmental regulators in fish and mammalian pituitary gland, it is possible that zebrafish could provide the basis for such a screen (reviewed in: [214]). Alternatively, embryonic stem cells have recently been coaxed to differentiate into pituitary hormone producing cells, suggesting embryonic stem cells might be adaptable for screening studies [215,216]. Success with such a high throughput screening approach would be invaluable for nominating candidates for human mutation screening in cases of hypopituitarism of unknown etiology.
Acknowledgments
NIH R37HD030428, R01HD034283 (SAC); University of Michigan Center for Computational Medicine and Biology, Clinical Translational Science Award (SAC), Reproductive Sciences Training Grant (NIH T32 HD07048 (SWD & BSE), NIH NRSA F32-HD046300 (BSE), Endocrine Society, International Scholar’s Program (LC), Novo-Nordisk and University of Michigan Center for Genetics in Health and Medicine (FC).
Footnotes
Note added in proof: Recent analysis of Lhx2 by in situ hybridization shows a broader range of expression in the ventral diencephalon than we report by immunohistochemistry. This report also reveals a direct role for LHX2 in the development of the posterior lobe of the pituitary gland. It is required in the infundibulum to restrict growth factor expression and suppress proliferation, and it is necessary for vasopressin expression in the posterior lobe. LHX2 deficiency causes dysmorphology of the intermediate and anterior lobes indirectly because no expression is detected there. LHX2 is not required for specification of the hormone producing cells of the anterior lobe. (Zhao Y, Mailloux CM, Hermesz E, Palkóvits M, Westphal H. A role of the LIM-homeobox gene Lhx2 in the regulation of pituitary development. Dev Biol. 2009 Nov 6. [Epub ahead of print]
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Weedon MN, Frayling TM. Reaching new heights: insights into the genetics of human stature. Trends Genet. 2008;24:595–603. doi: 10.1016/j.tig.2008.09.006. [DOI] [PubMed] [Google Scholar]
- 2.Rimoin DL, Cohn D, Krakow D, Wilcox W, Lachman RS, Alanay Y. The skeletal dysplasias: clinical-molecular correlations. Ann N Y Acad Sci. 2007;1117:302–9. doi: 10.1196/annals.1402.072. [DOI] [PubMed] [Google Scholar]
- 3.Cha KB, Karolyi IJ, Hunt A, Wenglikowski AM, Wilkinson JE, Dolan DF, Dootz G, Finnegan AA, Seasholtz AF, Hankenson KD, Siracusa LD, Camper SA. Skeletal dysplasia and male infertility locus on mouse chromosome 9. Genomics. 2004;83:951–60. doi: 10.1016/j.ygeno.2003.12.020. [DOI] [PubMed] [Google Scholar]
- 4.Jorge AA, Souza SC, Nishi MY, Billerbeck AE, Liborio DC, Kim CA, Arnhold IJ, Mendonca BB. SHOX mutations in idiopathic short stature and Leri-Weill dyschondrosteosis: frequency and phenotypic variability. Clin Endocrinol (Oxf) 2007;66:130–5. doi: 10.1111/j.1365-2265.2006.02698.x. [DOI] [PubMed] [Google Scholar]
- 5.Procter AM, Phillips JA, 3rd, Cooper DN. The molecular genetics of growth hormone deficiency. Hum Genet. 1998;103:255–72. doi: 10.1007/s004390050815. [DOI] [PubMed] [Google Scholar]
- 6.Patel L, McNally RJ, Harrison E, Lloyd IC, Clayton PE. Geographical distribution of optic nerve hypoplasia and septo-optic dysplasia in Northwest England. J Pediatr. 2006;148:85–8. doi: 10.1016/j.jpeds.2005.07.031. [DOI] [PubMed] [Google Scholar]
- 7.Hernandez LM, Lee PD, Camacho-Hubner C. Isolated growth hormone deficiency. Pituitary. 2007;10:351–7. doi: 10.1007/s11102-007-0073-3. [DOI] [PubMed] [Google Scholar]
- 8.Savage MO, Attie KM, David A, Metherell LA, Clark AJ, Camacho-Hubner C. Endocrine assessment, molecular characterization and treatment of growth hormone insensitivity disorders. Nat Clin Pract Endocrinol Metab. 2006;2:395–407. doi: 10.1038/ncpendmet0195. [DOI] [PubMed] [Google Scholar]
- 9.Bryant J, Baxter L, Cave CB, Milne R. Recombinant growth hormone for idiopathic short stature in children and adolescents. Cochrane Database Syst Rev. 2007:CD004440. doi: 10.1002/14651858.CD004440. [DOI] [PubMed] [Google Scholar]
- 10.Hintz RL. Growth hormone treatment of idiopathic short stature: clinical studies. Growth Horm IGF Res. 2005;15(Suppl A):S6–8. doi: 10.1016/j.ghir.2005.06.003. [DOI] [PubMed] [Google Scholar]
- 11.Zucchini S. Growth hormone use in the treatment of idiopathic short stature. Curr Opin Investig Drugs. 2008;9:396–401. [PubMed] [Google Scholar]
- 12.Zucchini S, Wasniewska M, Cisternino M, Salerno M, Iughetti L, Maghnie M, Street ME, Caruso-Nicoletti M, Cianfarani S. Adult height in children with short stature and idiopathic delayed puberty after different management. Eur J Pediatr. 2008;167:677–81. doi: 10.1007/s00431-007-0576-y. [DOI] [PubMed] [Google Scholar]
- 13.Cohen P, Rogol AD, Deal CL, Saenger P, Reiter EO, Ross JL, Chernausek SD, Savage MO, Wit JM. Consensus statement on the diagnosis and treatment of children with idiopathic short stature: a summary of the Growth Hormone Research Society, the Lawson Wilkins Pediatric Endocrine Society, and the European Society for Paediatric Endocrinology Workshop. J Clin Endocrinol Metab. 2008;93:4210–7. doi: 10.1210/jc.2008-0509. [DOI] [PubMed] [Google Scholar]
- 14.Zhu X, Gleiberman AS, Rosenfeld MG. Molecular physiology of pituitary development: signaling and transcriptional networks. Physiol Rev. 2007;87:933–63. doi: 10.1152/physrev.00006.2006. [DOI] [PubMed] [Google Scholar]
- 15.Kelberman D, Dattani MT. Hypothalamic and pituitary development: novel insights into the aetiology. Eur J Endocrinol. 2007;157(Suppl 1):S3–14. doi: 10.1530/EJE-07-0156. [DOI] [PubMed] [Google Scholar]
- 16.Kelberman D, Dattani MT. Role of Transcription Factors in Midline Central Nervous System and Pituitary Defects. Endocr Dev. 2009;14:67–82. doi: 10.1159/000207478. [DOI] [PubMed] [Google Scholar]
- 17.Brinkmeier ML, Potok MA, Cha KB, Gridley T, Stifani S, Meeldijk J, Clevers H, Camper SA. TCF and Groucho-related genes influence pituitary growth and development. Mol Endocrinol. 2003;17:2152–61. doi: 10.1210/me.2003-0225. [DOI] [PubMed] [Google Scholar]
- 18.Wang W, Wang YG, Reginato AM, Glotzer DJ, Fukai N, Plotkina S, Karsenty G, Olsen BR. Groucho homologue Grg5 interacts with the transcription factor Runx2-Cbfa1 and modulates its activity during postnatal growth in mice. Dev Biol. 2004;270:364–81. doi: 10.1016/j.ydbio.2004.03.003. [DOI] [PubMed] [Google Scholar]
- 19.Sheng HZ, Moriyama K, Yamashita T, Li H, Potter SS, Mahon KA, Westphal H. Multistep control of pituitary organogenesis. Science. 1997;278:1809–12. doi: 10.1126/science.278.5344.1809. [DOI] [PubMed] [Google Scholar]
- 20.Castinetti F, Saveanu A, Reynaud R, Quentien MH, Buffin A, Brauner R, Kaffel N, Albarel F, Guedj AM, El Kholy M, Amin M, Enjalbert A, Barlier A, Brue T. A novel dysfunctional LHX4 mutation with high phenotypical variability in patients with hypopituitarism. J Clin Endocrinol Metab. 2008;93:2790–9. doi: 10.1210/jc.2007-2389. [DOI] [PubMed] [Google Scholar]
- 21.Pfaeffle RW, Hunter CS, Savage JJ, Duran-Prado M, Mullen RD, Neeb ZP, Eiholzer U, Hesse V, Haddad NG, Stobbe HM, Blum WF, Weigel JF, Rhodes SJ. Three novel missense mutations within the LHX4 gene are associated with variable pituitary hormone deficiencies. J Clin Endocrinol Metab. 2008;93:1062–71. doi: 10.1210/jc.2007-1525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Rajab A, Kelberman D, de Castro SC, Biebermann H, Shaikh H, Pearce K, Hall CM, Shaikh G, Gerrelli D, Grueters A, Krude H, Dattani MT. Novel mutations in LHX3 are associated with hypopituitarism and sensorineural hearing loss. Hum Mol Genet. 2008;17:2150–9. doi: 10.1093/hmg/ddn114. [DOI] [PubMed] [Google Scholar]
- 23.Kristrom B, Zdunek AM, Rydh A, Jonsson H, Sehlin P, Escher SA. A novel mutation in the LIM homeobox 3 gene is responsible for combined pituitary hormone deficiency, hearing impairment, and vertebral malformations. J Clin Endocrinol Metab. 2009;94:1154–61. doi: 10.1210/jc.2008-0325. [DOI] [PubMed] [Google Scholar]
- 24.Bartke A, Goldman BD, Bex F, Dalterio S. Effects of prolactin (PRL) on pituitary and testicular function in mice with hereditary PRL deficiency. Endocrinology. 1977;101:1760–6. doi: 10.1210/endo-101-6-1760. [DOI] [PubMed] [Google Scholar]
- 25.Tang K, Bartke A, Gardiner CS, Wagner TE, Yun JS. Gonadotropin secretion, synthesis, and gene expression in human growth hormone transgenic mice and in Ames dwarf mice. Endocrinology. 1993;132:2518–24. doi: 10.1210/endo.132.6.8504754. [DOI] [PubMed] [Google Scholar]
- 26.Buckwalter MS, Katz RW, Camper SA. Localization of the panhypopituitary dwarf mutation (df) on mouse chromosome 11 in an intersubspecific backcross. Genomics. 1991;10:515–26. doi: 10.1016/0888-7543(91)90430-m. [DOI] [PubMed] [Google Scholar]
- 27.Soares MJ, Bartke A, Colosi P, Talamantes F. Identification of a placental lactogen in pregnant Snell and Ames dwarf mice. Proc Soc Exp Biol Med. 1984;175:106–8. doi: 10.3181/00379727-175-41775. [DOI] [PubMed] [Google Scholar]
- 28.Pernasetti F, Toledo SP, Vasilyev VV, Hayashida CY, Cogan JD, Ferrari C, Lourenco DM, Mellon PL. Impaired adrenocorticotropin-adrenal axis in combined pituitary hormone deficiency caused by a two-base pair deletion (301-302delAG) in the prophet of Pit-1 gene. J Clin Endocrinol Metab. 2000;85:390–7. doi: 10.1210/jcem.85.1.6324. [DOI] [PubMed] [Google Scholar]
- 29.Reynaud R, Gueydan M, Saveanu A, Vallette-Kasic S, Enjalbert A, Brue T, Barlier A. Genetic screening of combined pituitary hormone deficiency: experience in 195 patients. J Clin Endocrinol Metab. 2006;91:3329–36. doi: 10.1210/jc.2005-2173. [DOI] [PubMed] [Google Scholar]
- 30.Bottner A, Keller E, Kratzsch J, Stobbe H, Weigel JF, Keller A, Hirsch W, Kiess W, Blum WF, Pfaffle RW. PROP1 mutations cause progressive deterioration of anterior pituitary function including adrenal insufficiency: a longitudinal analysis. J Clin Endocrinol Metab. 2004;89:5256–65. doi: 10.1210/jc.2004-0661. [DOI] [PubMed] [Google Scholar]
- 31.Riepe FG, Partsch CJ, Blankenstein O, Monig H, Pfaffle RW, Sippell WG. Longitudinal imaging reveals pituitary enlargement preceding hypoplasia in two brothers with combined pituitary hormone deficiency attributable to PROP1 mutation. J Clin Endocrinol Metab. 2001;86:4353–7. doi: 10.1210/jcem.86.9.7828. [DOI] [PubMed] [Google Scholar]
- 32.Sornson MW, Wu W, Dasen JS, Flynn SE, Norman DJ, O’Connell SM, Gukovsky I, Carriere C, Ryan AK, Miller AP, Zuo L, Gleiberman AS, Andersen B, Beamer WG, Rosenfeld MG. Pituitary lineage determination by the Prophet of Pit-1 homeodomain factor defective in Ames dwarfism. Nature. 1996;384:327–33. doi: 10.1038/384327a0. [DOI] [PubMed] [Google Scholar]
- 33.Deladoey J, Fluck C, Buyukgebiz A, Kuhlmann BV, Eble A, Hindmarsh PC, Wu W, Mullis PE. “Hot spot” in the PROP1 gene responsible for combined pituitary hormone deficiency. J Clin Endocrinol Metab. 1999;84:1645–50. doi: 10.1210/jcem.84.5.5681. [DOI] [PubMed] [Google Scholar]
- 34.Nasonkin IO, Ward RD, Raetzman LT, Seasholtz AF, Saunders TL, Gillespie PJ, Camper SA. Pituitary hypoplasia and respiratory distress syndrome in Prop1 knockout mice. Hum Mol Genet. 2004;13:2727–35. doi: 10.1093/hmg/ddh311. [DOI] [PubMed] [Google Scholar]
- 35.Olson LE, Tollkuhn J, Scafoglio C, Krones A, Zhang J, Ohgi KA, Wu W, Taketo MM, Kemler R, Grosschedl R, Rose D, Li X, Rosenfeld MG. Homeodomain-mediated beta-catenin-dependent switching events dictate cell-lineage determination. Cell. 2006;125:593–605. doi: 10.1016/j.cell.2006.02.046. [DOI] [PubMed] [Google Scholar]
- 36.Nadeau JH. Modifier genes and protective alleles in humans and mice. Curr Opin Genet Dev. 2003;13:290–5. doi: 10.1016/s0959-437x(03)00061-3. [DOI] [PubMed] [Google Scholar]
- 37.Badano JL, Katsanis N. Beyond Mendel: an evolving view of human genetic disease transmission. Nat Rev Genet. 2002;3:779–89. doi: 10.1038/nrg910. [DOI] [PubMed] [Google Scholar]
- 38.Couly GF, Le Douarin NM. Mapping of the early neural primordium in quail-chick chimeras. I. Developmental relationships between placodes, facial ectoderm, and prosencephalon. Dev Biol. 1985;110:422–39. doi: 10.1016/0012-1606(85)90101-0. [DOI] [PubMed] [Google Scholar]
- 39.ElAmraoui A, Dubois PM. Experimental evidence for the early commitment of the presumptive adenohypophysis. Neuroendocrinology. 1993;58:609–615. doi: 10.1159/000126599. [DOI] [PubMed] [Google Scholar]
- 40.Hermesz E, Williams-Simons L, Mahon KA. A novel inducible element, activated by contact with Rathke’s pouch, is present in the regulatory region of the Rpx/Hesx1 homeobox gene. Dev Biol. 2003;260:68–78. doi: 10.1016/s0012-1606(03)00218-5. [DOI] [PubMed] [Google Scholar]
- 41.Ericson J, Norlin S, Jessell TM, Edlund T. Integrated FGF and BMP signaling controls the progression of progenitor cell differentiation and the emergence of pattern in the embryonic anterior pituitary. Development. 1998;125:1005–15. doi: 10.1242/dev.125.6.1005. [DOI] [PubMed] [Google Scholar]
- 42.Gleiberman AS, Fedtsova NG, Rosenfeld MG. Tissue interactions in the induction of anterior pituitary: role of the ventral diencephalon, mesenchyme, and notochord. Dev Biol. 1999;213:340–53. doi: 10.1006/dbio.1999.9386. [DOI] [PubMed] [Google Scholar]
- 43.Takuma N, Sheng HZ, Furuta Y, Ward JM, Sharma K, Hogan BL, Pfaff SL, Westphal H, Kimura S, Mahon KA. Formation of Rathke’s pouch requires dual induction from the diencephalon. Development. 1998;125:4835–40. doi: 10.1242/dev.125.23.4835. [DOI] [PubMed] [Google Scholar]
- 44.Potok MA, Cha KB, Hunt A, Brinkmeier ML, Leitges M, Kispert A, Camper SA. WNT signaling affects gene expression in the ventral diencephalon and pituitary gland growth. Dev Dyn. 2008;237:1006–20. doi: 10.1002/dvdy.21511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Cha KB, Douglas KR, Potok MA, Liang H, Jones SN, Camper SA. WNT5A signaling affects pituitary gland shape. Mech Dev. 2004;121:183–94. doi: 10.1016/j.mod.2003.12.002. [DOI] [PubMed] [Google Scholar]
- 46.Davis SW, Camper SA. Noggin regulates Bmp4 activity during pituitary induction. Dev Biol. 2007;305:145–60. doi: 10.1016/j.ydbio.2007.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Brinkmeier ML, Potok MA, Davis SW, Camper SA. TCF4 deficiency expands ventral diencephalon signaling and increases induction of pituitary progenitors. Dev Biol. 2007;311:396–407. doi: 10.1016/j.ydbio.2007.08.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Raetzman LT, Ross SA, Cook S, Dunwoodie SL, Camper SA, Thomas PQ. Developmental regulation of Notch signaling genes in the embryonic pituitary: Prop1 deficiency affects Notch2 expression. Dev Biol. 2004;265:329–40. doi: 10.1016/j.ydbio.2003.09.033. [DOI] [PubMed] [Google Scholar]
- 49.Raetzman LT, Cai JX, Camper SA. Hes1 is required for pituitary growth and melanotrope specification. Dev Biol. 2007;304:455–66. doi: 10.1016/j.ydbio.2006.11.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Raetzman LT, Wheeler BS, Ross SA, Thomas PQ, Camper SA. Persistent expression of Notch2 delays gonadotrope differentiation. Mol Endocrinol. 2006;20:2898–908. doi: 10.1210/me.2005-0394. [DOI] [PubMed] [Google Scholar]
- 51.Treier M, O’Connell S, Gleiberman A, Price J, Szeto DP, Burgess R, Chuang PT, McMahon AP, Rosenfeld MG. Hedgehog signaling is required for pituitary gland development. Development. 2001;128:377–86. doi: 10.1242/dev.128.3.377. [DOI] [PubMed] [Google Scholar]
- 52.Kita A, Imayoshi I, Hojo M, Kitagawa M, Kokubu H, Ohsawa R, Ohtsuka T, Kageyama R, Hashimoto N. Hes1 and Hes5 control the progenitor pool, intermediate lobe specification, and posterior lobe formation in the pituitary development. Mol Endocrinol. 2007;21:1458–66. doi: 10.1210/me.2007-0039. [DOI] [PubMed] [Google Scholar]
- 53.Zhu X, Zhang J, Tollkuhn J, Ohsawa R, Bresnick EH, Guillemot F, Kageyama R, Rosenfeld MG. Sustained Notch signaling in progenitors is required for sequential emergence of distinct cell lineages during organogenesis. Genes Dev. 2006;20:2739–53. doi: 10.1101/gad.1444706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Ezzat S, Zheng L, Zhu XF, Wu GE, Asa SL. Targeted expression of a human pituitary tumor-derived isoform of FGF receptor-4 recapitulates pituitary tumorigenesis. J Clin Invest. 2002;109:69–78. doi: 10.1172/JCI14036. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 55.Ohuchi H, Hori Y, Yamasaki M, Harada H, Sekine K, Kato S, Itoh N. FGF10 acts as a major ligand for FGF receptor 2 IIIb in mouse multi-organ development. Biochem Biophys Res Commun. 2000;277:643–9. doi: 10.1006/bbrc.2000.3721. [DOI] [PubMed] [Google Scholar]
- 56.Treier M, Gleiberman AS, O’Connell SM, Szeto DP, McMahon JA, McMahon AP, Rosenfeld MG. Multistep signaling requirements for pituitary organogenesis in vivo. Genes Dev. 1998;12:1691–704. doi: 10.1101/gad.12.11.1691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Brinkmeier ML, Davis SW, Carninci P, MacDonald JW, Kawai J, Ghosh D, Hayashizaki Y, Lyons RH, Camper SA. Discovery of transcriptional regulators and signaling pathways in the developing pituitary gland by bioinformatic and genomic approaches. Genomics. 2009;93:449–60. doi: 10.1016/j.ygeno.2008.11.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Rizzoti K, Lovell-Badge R. Early development of the pituitary gland: induction and shaping of Rathke’s pouch. Rev Endocr Metab Disord. 2005;6:161–72. doi: 10.1007/s11154-005-3047-7. [DOI] [PubMed] [Google Scholar]
- 59.Salisbury TB, Binder AK, Grammer JC, Nilson JH. GnRH-regulated expression of Jun and JUN target genes in gonadotropes requires a functional interaction between TCF/LEF family members and beta-catenin. Mol Endocrinol. 2009;23:402–11. doi: 10.1210/me.2008-0370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Salisbury TB, Binder AK, Grammer JC, Nilson JH. Maximal activity of the luteinizing hormone beta-subunit gene requires beta-catenin. Mol Endocrinol. 2007;21:963–71. doi: 10.1210/me.2006-0383. [DOI] [PubMed] [Google Scholar]
- 61.Garcia-Lavandeira M, Quereda V, Flores I, Saez C, Diaz-Rodriguez E, Japon MA, Ryan AK, Blasco MA, Dieguez C, Malumbres M, Alvarez CV. A GRFa2/Prop1/stem (GPS) cell niche in the pituitary. PLoS ONE. 2009;4:e4815. doi: 10.1371/journal.pone.0004815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Kioussi C, Briata P, Baek SH, Rose DW, Hamblet NS, Herman T, Ohgi KA, Lin C, Gleiberman A, Wang J, Brault V, Ruiz-Lozano P, Nguyen HD, Kemler R, Glass CK, Wynshaw-Boris A, Rosenfeld MG. Identification of a Wnt/Dvl/beta-Catenin --> Pitx2 pathway mediating cell-type-specific proliferation during development. Cell. 2002;111:673–85. doi: 10.1016/s0092-8674(02)01084-x. [DOI] [PubMed] [Google Scholar]
- 63.Gardner S, Maudsley S, Millar RP, Pawson AJ. Nuclear stabilization of beta-catenin and inactivation of glycogen synthase kinase-3beta by gonadotropin-releasing hormone: targeting Wnt signaling in the pituitary gonadotrope. Mol Endocrinol. 2007;21:3028–38. doi: 10.1210/me.2007-0268. [DOI] [PubMed] [Google Scholar]
- 64.Sheng HZ, Zhadanov AB, Mosinger B, Jr, Fujii T, Bertuzzi S, Grinberg A, Lee EJ, Huang SP, Mahon KA, Westphal H. Specification of pituitary cell lineages by the LIM homeobox gene Lhx3. Science. 1996;272:1004–7. doi: 10.1126/science.272.5264.1004. [DOI] [PubMed] [Google Scholar]
- 65.Charles MA, Suh H, Hjalt TA, Drouin J, Camper SA, Gage PJ. PITX genes are required for cell survival and Lhx3 activation. Mol Endocrinol. 2005;19:1893–903. doi: 10.1210/me.2005-0052. [DOI] [PubMed] [Google Scholar]
- 66.Suh H, Gage PJ, Drouin J, Camper SA. Pitx2 is required at multiple stages of pituitary organogenesis: pituitary primordium formation and cell specification. Development. 2002;129:329–37. doi: 10.1242/dev.129.2.329. [DOI] [PubMed] [Google Scholar]
- 67.Gage PJ, Suh H, Camper SA. Dosage requirement of Pitx2 for development of multiple organs. Development. 1999;126:4643–51. doi: 10.1242/dev.126.20.4643. [DOI] [PubMed] [Google Scholar]
- 68.Gage PJ, Suh H, Camper SA. The bicoid-related Pitx gene family in development. Mamm Genome. 1999;10:197–200. doi: 10.1007/s003359900970. [DOI] [PubMed] [Google Scholar]
- 69.Ellsworth BS, Butts DL, Camper SA. Mechanisms underlying pituitary hypoplasia and failed cell specification in Lhx3-deficient mice. Dev Biol. 2008;313:118–29. doi: 10.1016/j.ydbio.2007.10.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Zhao L, Bakke M, Krimkevich Y, Cushman LJ, Parlow AF, Camper SA, Parker KL. Hypomorphic phenotype in mice with pituitary-specific knockout of steroidogenic factor 1. Genesis. 2001;30:65–9. doi: 10.1002/gene.1034. [DOI] [PubMed] [Google Scholar]
- 71.Zhao L, Bakke M, Krimkevich Y, Cushman LJ, Parlow AF, Camper SA, Parker KL. Steroidogenic factor 1 (SF1) is essential for pituitary gonadotrope function. Development. 2001;128:147–54. doi: 10.1242/dev.128.2.147. [DOI] [PubMed] [Google Scholar]
- 72.Charles MA, Saunders TL, Wood WM, Owens K, Parlow AF, Camper SA, Ridgway EC, Gordon DF. Pituitary-specific Gata2 knockout: effects on gonadotrope and thyrotrope function. Mol Endocrinol. 2006;20:1366–77. doi: 10.1210/me.2005-0378. [DOI] [PubMed] [Google Scholar]
- 73.Charles MA, Mortensen AH, Potok MA, Camper SA. Pitx2 deletion in pituitary gonadotropes is compatible with gonadal development, puberty, and fertility. Genesis. 2008;46:507–14. doi: 10.1002/dvg.20398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Mody S, Brown MR, Parks JS. The spectrum of hypopituitarism caused by PROP1 mutations. Best Pract Res Clin Endocrinol Metab. 2002;16:421–31. doi: 10.1053/beem.2002.0218. [DOI] [PubMed] [Google Scholar]
- 75.Cogan JD, Wu W, Phillips JA, 3rd, Arnhold IJ, Agapito A, Fofanova OV, Osorio MG, Bircan I, Moreno A, Mendonca BB. The PROP1 2-base pair deletion is a common cause of combined pituitary hormone deficiency. J Clin Endocrinol Metab. 1998;83:3346–9. doi: 10.1210/jcem.83.9.5142. [DOI] [PubMed] [Google Scholar]
- 76.Ward RD, Stone BM, Raetzman LT, Camper SA. Cell proliferation and vascularization in mouse models of pituitary hormone deficiency. Mol Endocrinol. 2006;20:1378–90. doi: 10.1210/me.2005-0409. [DOI] [PubMed] [Google Scholar]
- 77.Ward RD, Raetzman LT, Suh H, Stone BM, Nasonkin IO, Camper SA. Role of PROP1 in pituitary gland growth. Mol Endocrinol. 2005;19:698–710. doi: 10.1210/me.2004-0341. [DOI] [PubMed] [Google Scholar]
- 78.Himes AD, Raetzman LT. Premature differentiation and aberrant movement of pituitary cells lacking both Hes1 and Prop1. Dev Biol. 2009;325:151–61. doi: 10.1016/j.ydbio.2008.10.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Kikuchi M, Yatabe M, Fujiwara K, Takigami S, Sakamoto A, Soji T, Yashiro T. Distinctive localization of N- and E-cadherins in rat anterior pituitary gland. Anat Rec A Discov Mol Cell Evol Biol. 2006;288:1183–9. doi: 10.1002/ar.a.20384. [DOI] [PubMed] [Google Scholar]
- 80.Kikuchi M, Yatabe M, Kouki T, Fujiwara K, Takigami S, Sakamoto A, Yashiro T. Changes in E- and N-cadherin expression in developing rat adenohypophysis. Anat Rec (Hoboken) 2007;290:486–90. doi: 10.1002/ar.20516. [DOI] [PubMed] [Google Scholar]
- 81.Gage PJ, Lossie AC, Scarlett LM, Lloyd RV, Camper SA. Ames dwarf mice exhibit somatotrope commitment but lack growth hormone-releasing factor response. Endocrinology. 1995;136:1161–7. doi: 10.1210/endo.136.3.7867569. [DOI] [PubMed] [Google Scholar]
- 82.Gage PJ, Roller ML, Saunders TL, Scarlett LM, Camper SA. Anterior pituitary cells defective in the cell-autonomous factor, df, undergo cell lineage specification but not expansion. Development. 1996;122:151–60. doi: 10.1242/dev.122.1.151. [DOI] [PubMed] [Google Scholar]
- 83.Ingraham HA, Chen RP, Mangalam HJ, Elsholtz HP, Flynn SE, Lin CR, Simmons DM, Swanson L, Rosenfeld MG. A tissue-specific transcription factor containing a homeodomain specifies a pituitary phenotype. Cell. 1988;55:519–29. doi: 10.1016/0092-8674(88)90038-4. [DOI] [PubMed] [Google Scholar]
- 84.Bodner M, Castrillo JL, Theill LE, Deerinck T, Ellisman M, Karin M. The pituitary-specific transcription factor GHF-1 is a homeobox-containing protein. Cell. 1988;55:505–18. doi: 10.1016/0092-8674(88)90037-2. [DOI] [PubMed] [Google Scholar]
- 85.Lamonerie T, Tremblay JJ, Lanctot C, Therrien M, Gauthier Y, Drouin J. Ptx1, a bicoid-related homeo box transcription factor involved in transcription of the pro-opiomelanocortin gene. Genes Dev. 1996;10:1284–95. doi: 10.1101/gad.10.10.1284. [DOI] [PubMed] [Google Scholar]
- 86.Lamolet B, Pulichino AM, Lamonerie T, Gauthier Y, Brue T, Enjalbert A, Drouin J. A pituitary cell-restricted T box factor, Tpit, activates POMC transcription in cooperation with Pitx homeoproteins. Cell. 2001;104:849–59. doi: 10.1016/s0092-8674(01)00282-3. [DOI] [PubMed] [Google Scholar]
- 87.Gordon DF, Woodmansee WW, Black JN, Dowding JM, Bendrick-Peart J, Wood WM, Ridgway EC. Domains of Pit-1 required for transcriptional synergy with GATA-2 on the TSH beta gene. Mol Cell Endocrinol. 2002;196:53–66. doi: 10.1016/s0303-7207(02)00223-x. [DOI] [PubMed] [Google Scholar]
- 88.Gordon DF, Lewis SR, Haugen BR, James RA, McDermott MT, Wood WM, Ridgway EC. Pit-1 and GATA-2 interact and functionally cooperate to activate the thyrotropin beta-subunit promoter. J Biol Chem. 1997;272:24339–47. doi: 10.1074/jbc.272.39.24339. [DOI] [PubMed] [Google Scholar]
- 89.Ward RD, Davis SW, Cho M, Esposito C, Lyons RH, Cheng JF, Rubin EM, Rhodes SJ, Raetzman LT, Smith TP, Camper SA. Comparative genomics reveals functional transcriptional control sequences in the Prop1 gene. Mamm Genome. 2007 doi: 10.1007/s00335-007-9008-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Carvalho LR, Ward RD, Davis SW, Nishi MY, Cogan JD, Donohoue PA, Rhodes S, Walvoord EC, Lyons RH, Phillips JA, Arnhold IJP, Mendonca BB, Camper SA. Analysis of PROP1 transcriptional regulatory sequences in patients with hypopituitarism. 89th Endocrine Society Annual Meeting; Toronto. 2007. [Google Scholar]
- 91.Douglas KR, Brinkmeier ML, Kennell JA, Eswara P, Harrison TA, Patrianakos AI, Sprecher BS, Potok MA, Lyons RH, Jr, MacDougald OA, Camper SA. Identification of members of the Wnt signaling pathway in the embryonic pituitary gland. Mamm Genome. 2001;12:843–51. doi: 10.1007/s00335-001-2076-0. [DOI] [PubMed] [Google Scholar]
- 92.Douglas KR, Camper SA. Partial transcriptome of the developing pituitary gland. Genomics. 2000;70:335–46. doi: 10.1006/geno.2000.6400. [DOI] [PubMed] [Google Scholar]
- 93.Carninci P, Waki K, Shiraki T, Konno H, Shibata K, Itoh M, Aizawa K, Arakawa T, Ishii Y, Sasaki D, Bono H, Kondo S, Sugahara Y, Saito R, Osato N, Fukuda S, Sato K, Watahiki A, Hirozane-Kishikawa T, Nakamura M, Shibata Y, Yasunishi A, Kikuchi N, Yoshiki A, Kusakabe M, Gustincich S, Beisel K, Pavan W, Aidinis V, Nakagawara A, Held WA, Iwata H, Kono T, Nakauchi H, Lyons P, Wells C, Hume DA, Fagiolini M, Hensch TK, Brinkmeier M, Camper S, Hirota J, Mombaerts P, Muramatsu M, Okazaki Y, Kawai J, Hayashizaki Y. Targeting a complex transcriptome: the construction of the mouse full-length cDNA encyclopedia. Genome Res. 2003;13:1273–89. doi: 10.1101/gr.1119703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Davis SW, Potok MA, Brinkmeier ML, Carninci P, Lyons RH, MacDonald JW, Fleming MT, Mortensen AH, Egashira N, Ghosh D, Steel KP, Osamura RY, Hayashizaki Y, Camper SA. Genetics, gene expression and bioinformatics of the pituitary gland. Horm Res. 2009;71(Suppl 2):101–15. doi: 10.1159/000192447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Gustavsson P, Schoumans J, Staaf J, Jonsson G, Carlsson F, Kristoffersson U, Borg A, Nordenskjold M, Dahl N. Hemizygosity for chromosome 2q14.2-q22.1 spanning the GLI2 and PROC genes associated with growth hormone deficiency, polydactyly, deep vein thrombosis and urogenital abnormalities. Clin Genet. 2006;69:441–3. doi: 10.1111/j.1399-0004.2006.00601.x. [DOI] [PubMed] [Google Scholar]
- 96.Tziaferi V, Kelberman D, Dattani MT. The role of SOX2 in hypogonadotropic hypogonadism. Sex Dev. 2008;2:194–9. doi: 10.1159/000152035. [DOI] [PubMed] [Google Scholar]
- 97.Woods KS, Cundall M, Turton J, Rizotti K, Mehta A, Palmer R, Wong J, Chong WK, Al-Zyoud M, El-Ali M, Otonkoski T, Martinez-Barbera JP, Thomas PQ, Robinson IC, Lovell-Badge R, Woodward KJ, Dattani MT. Over- and underdosage of SOX3 is associated with infundibular hypoplasia and hypopituitarism. Am J Hum Genet. 2005;76:833–49. doi: 10.1086/430134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Weiss J, Meeks JJ, Hurley L, Raverot G, Frassetto A, Jameson JL. Sox3 is required for gonadal function, but not sex determination, in males and females. Mol Cell Biol. 2003;23:8084–91. doi: 10.1128/MCB.23.22.8084-8091.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Laumonnier F, Ronce N, Hamel BC, Thomas P, Lespinasse J, Raynaud M, Paringaux C, Van Bokhoven H, Kalscheuer V, Fryns JP, Chelly J, Moraine C, Briault S. Transcription factor SOX3 is involved in X-linked mental retardation with growth hormone deficiency. Am J Hum Genet. 2002;71:1450–5. doi: 10.1086/344661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Rizzoti K, Brunelli S, Carmignac D, Thomas PQ, Robinson IC, Lovell-Badge R. SOX3 is required during the formation of the hypothalamo-pituitary axis. Nat Genet. 2004;36:247–55. doi: 10.1038/ng1309. [DOI] [PubMed] [Google Scholar]
- 101.Dattani MT, Martinez-Barbera JP, Thomas PQ, Brickman JM, Gupta R, Martensson IL, Toresson H, Fox M, Wales JK, Hindmarsh PC, Krauss S, Beddington RS, Robinson IC. Mutations in the homeobox gene HESX1/Hesx1 associated with septo-optic dysplasia in human and mouse. Nat Genet. 1998;19:125–33. doi: 10.1038/477. [DOI] [PubMed] [Google Scholar]
- 102.Thomas PQ, Dattani MT, Brickman JM, McNay D, Warne G, Zacharin M, Cameron F, Hurst J, Woods K, Dunger D, Stanhope R, Forrest S, Robinson IC, Beddington RS. Heterozygous HESX1 mutations associated with isolated congenital pituitary hypoplasia and septo-optic dysplasia. Hum Mol Genet. 2001;10:39–45. doi: 10.1093/hmg/10.1.39. [DOI] [PubMed] [Google Scholar]
- 103.Mehta A, Hindmarsh PC, Stanhope RG, Brain CE, Preece MA, Dattani MT. Is the thyrotropin-releasing hormone test necessary in the diagnosis of central hypothyroidism in children. J Clin Endocrinol Metab. 2003;88:5696–703. doi: 10.1210/jc.2003-030943. [DOI] [PubMed] [Google Scholar]
- 104.Osorio MG, Marui S, Jorge AA, Latronico AC, Lo LS, Leite CC, Estefan V, Mendonca BB, Arnhold IJ. Pituitary magnetic resonance imaging and function in patients with growth hormone deficiency with and without mutations in GHRH-R, GH-1, or PROP-1 genes. J Clin Endocrinol Metab. 2002;87:5076–84. doi: 10.1210/jc.2001-011936. [DOI] [PubMed] [Google Scholar]
- 105.McNay DE, Turton JP, Kelberman D, Woods KS, Brauner R, Papadimitriou A, Keller E, Keller A, Haufs N, Krude H, Shalet SM, Dattani MT. HESX1 mutations are an uncommon cause of septooptic dysplasia and hypopituitarism. J Clin Endocrinol Metab. 2007;92:691–7. doi: 10.1210/jc.2006-1609. [DOI] [PubMed] [Google Scholar]
- 106.Kelberman D, Dattani MT. The role of transcription factors implicated in anterior pituitary development in the aetiology of congenital hypopituitarism. Ann Med. 2006;38:560–77. doi: 10.1080/07853890600994963. [DOI] [PubMed] [Google Scholar]
- 107.Carninci P, Shibata Y, Hayatsu N, Sugahara Y, Shibata K, Itoh M, Konno H, Okazaki Y, Muramatsu M, Hayashizaki Y. Normalization and subtraction of cap-trapper-selected cDNAs to prepare full-length cDNA libraries for rapid discovery of new genes. Genome Res. 2000;10:1617–30. doi: 10.1101/gr.145100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Semina EV, Reiter R, Leysens NJ, Alward WL, Small KW, Datson NA, Siegel-Bartelt J, Bierke-Nelson D, Bitoun P, Zabel BU, Carey JC, Murray JC. Cloning and characterization of a novel bicoid-related homeobox transcription factor gene, RIEG, involved in Rieger syndrome. Nat Genet. 1996;14:392–9. doi: 10.1038/ng1296-392. [DOI] [PubMed] [Google Scholar]
- 109.Suh H, Martin DM, Charles MA, Nasonkin IO, Gage PJ, Camper SA. Role of PITX2 in the pituitary gland. In: Amendt BA, editor. The molecular mechanism of Axenfeld-Rieger syndrome. Springer; 2005. pp. 54–64. [Google Scholar]
- 110.Acampora D, Mazan S, Tuorto F, Avantaggiato V, Tremblay JJ, Lazzaro D, di Carlo A, Mariano A, Macchia PE, Corte G, Macchia V, Drouin J, Brulet P, Simeone A. Transient dwarfism and hypogonadism in mice lacking Otx1 reveal prepubescent stage-specific control of pituitary levels of GH, FSH and LH. Development. 1998;125:1229–39. doi: 10.1242/dev.125.7.1229. [DOI] [PubMed] [Google Scholar]
- 111.Cushman LJ, Watkins-Chow DE, Brinkmeier ML, Raetzman LT, Radak AL, Lloyd RV, Camper SA. Persistent Prop1 expression delays gonadotrope differentiation and enhances pituitary tumor susceptibility. Hum Mol Genet. 2001;10:1141–53. doi: 10.1093/hmg/10.11.1141. [DOI] [PubMed] [Google Scholar]
- 112.Vesper AH, Raetzman LT, Camper SA. Role of prophet of Pit1 (PROP1) in gonadotrope differentiation and puberty. Endocrinology. 2006;147:1654–63. doi: 10.1210/en.2005-1080. [DOI] [PubMed] [Google Scholar]
- 113.Lanctot C, Moreau A, Chamberland M, Tremblay ML, Drouin J. Hindlimb patterning and mandible development require the Ptx1 gene. Development. 1999;126:1805–10. doi: 10.1242/dev.126.9.1805. [DOI] [PubMed] [Google Scholar]
- 114.Szeto DP, Rodriguez-Esteban C, Ryan AK, O’Connell SM, Liu F, Kioussi C, Gleiberman AS, Izpisua-Belmonte JC, Rosenfeld MG. Role of the Bicoid-related homeodomain factor Pitx1 in specifying hindlimb morphogenesis and pituitary development. Genes Dev. 1999;13:484–94. doi: 10.1101/gad.13.4.484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Tremblay JJ, Lanctot C, Drouin J. The pan-pituitary activator of transcription, Ptx1 (pituitary homeobox 1), acts in synergy with SF-1 and Pit1 and is an upstream regulator of the Lim-homeodomain gene Lim3/Lhx3. Mol Endocrinol. 1998;12:428–41. doi: 10.1210/mend.12.3.0073. [DOI] [PubMed] [Google Scholar]
- 116.Roberson MS, Schoderbek WE, Tremml G, Maurer RA. Activation of the glycoprotein hormone alpha-subunit promoter by a LIM-homeodomain transcription factor. Mol Cell Biol. 1994;14:2985–93. doi: 10.1128/mcb.14.5.2985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Cai Y, Xu Z, Nagarajan L, Brandt SJ. Single-stranded DNA-binding proteins regulate the abundance and function of the LIM-homeodomain transcription factor LHX2 in pituitary cells. Biochem Biophys Res Commun. 2008;373:303–8. doi: 10.1016/j.bbrc.2008.06.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Lu CH, Rincon-Limas DE, Botas J. Conserved overlapping and reciprocal expression of msh/Msx1 and apterous/Lhx2 in Drosophila and mice. Mech Dev. 2000;99:177–81. doi: 10.1016/s0925-4773(00)00479-2. [DOI] [PubMed] [Google Scholar]
- 119.Visel A, Thaller C, Eichele G. GenePaint.org: an atlas of gene expression patterns in the mouse embryo. Nucleic Acids Res. 2004;32:D552–6. doi: 10.1093/nar/gkh029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Visel A, Carson J, Oldekamp J, Warnecke M, Jakubcakova V, Zhou X, Shaw CA, Alvarez-Bolado G, Eichele G. Regulatory pathway analysis by high-throughput in situ hybridization. PLoS Genet. 2007;3:1867–83. doi: 10.1371/journal.pgen.0030178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Rhee H, Polak L, Fuchs E. Lhx2 maintains stem cell character in hair follicles. Science. 2006;312:1946–9. doi: 10.1126/science.1128004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Porter FD, Drago J, Xu Y, Cheema SS, Wassif C, Huang SP, Lee E, Grinberg A, Massalas JS, Bodine D, Alt F, Westphal H. Lhx2, a LIM homeobox gene, is required for eye, forebrain, and definitive erythrocyte development. Development. 1997;124:2935–44. doi: 10.1242/dev.124.15.2935. [DOI] [PubMed] [Google Scholar]
- 123.Mullen RD, Colvin SC, Hunter CS, Savage JJ, Walvoord EC, Bhangoo AP, Ten S, Weigel J, Pfaffle RW, Rhodes SJ. Roles of the LHX3 and LHX4 LIM-homeodomain factors in pituitary development. Mol Cell Endocrinol. 2007;265-266:190–5. doi: 10.1016/j.mce.2006.12.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Sharma K, Sheng HZ, Lettieri K, Li H, Karavanov A, Potter S, Westphal H, Pfaff SL. LIM homeodomain factors Lhx3 and Lhx4 assign subtype identities for motor neurons. Cell. 1998;95:817–28. doi: 10.1016/s0092-8674(00)81704-3. [DOI] [PubMed] [Google Scholar]
- 125.Machinis K, Amselem S. Functional relationship between LHX4 and POU1F1 in light of the LHX4 mutation identified in patients with pituitary defects. J Clin Endocrinol Metab. 2005;90:5456–62. doi: 10.1210/jc.2004-2332. [DOI] [PubMed] [Google Scholar]
- 126.Machinis K, Pantel J, Netchine I, Leger J, Camand OJ, Sobrier ML, Dastot-Le Moal F, Duquesnoy P, Abitbol M, Czernichow P, Amselem S. Syndromic short stature in patients with a germline mutation in the LIM homeobox LHX4. Am J Hum Genet. 2001;69:961–8. doi: 10.1086/323764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Sobrier ML, Attie-Bitach T, Netchine I, Encha-Razavi F, Vekemans M, Amselem S. Pathophysiology of syndromic combined pituitary hormone deficiency due to a LHX3 defect in light of LHX3 and LHX4 expression during early human development. Gene Expr Patterns. 2004;5:279–84. doi: 10.1016/j.modgep.2004.07.003. [DOI] [PubMed] [Google Scholar]
- 128.Netchine I, Sobrier ML, Krude H, Schnabel D, Maghnie M, Marcos E, Duriez B, Cacheux V, Moers A, Goossens M, Gruters A, Amselem S. Mutations in LHX3 result in a new syndrome revealed by combined pituitary hormone deficiency. Nat Genet. 2000;25:182–6. doi: 10.1038/76041. [DOI] [PubMed] [Google Scholar]
- 129.Bhangoo AP, Hunter CS, Savage JJ, Anhalt H, Pavlakis S, Walvoord EC, Ten S, Rhodes SJ. Clinical case seminar: a novel LHX3 mutation presenting as combined pituitary hormonal deficiency. J Clin Endocrinol Metab. 2006;91:747–53. doi: 10.1210/jc.2005-2360. [DOI] [PubMed] [Google Scholar]
- 130.Pfaeffle RW, Savage JJ, Hunter CS, Palme C, Ahlmann M, Kumar P, Bellone J, Schoenau E, Korsch E, Bramswig JH, Stobbe HM, Blum WF, Rhodes SJ. Four novel mutations of the LHX3 gene cause combined pituitary hormone deficiencies with or without limited neck rotation. J Clin Endocrinol Metab. 2007;92:1909–19. doi: 10.1210/jc.2006-2177. [DOI] [PubMed] [Google Scholar]
- 131.Raetzman LT, Ward R, Camper SA. Lhx4 and Prop1 are required for cell survival and expansion of the pituitary primordia. Development. 2002;129:4229–39. doi: 10.1242/dev.129.18.4229. [DOI] [PubMed] [Google Scholar]
- 132.Zhao Y, Morales DC, Hermesz E, Lee WK, Pfaff SL, Westphal H. Reduced expression of the LIM-homeobox gene Lhx3 impairs growth and differentiation of Rathke’s pouch and increases cell apoptosis during mouse pituitary development. Mech Dev. 2006;123:605–13. doi: 10.1016/j.mod.2006.06.005. [DOI] [PubMed] [Google Scholar]
- 133.Sheng HZ, Westphal H. Early steps in pituitary organogenesis. Trends Genet. 1999;15:236–40. doi: 10.1016/s0168-9525(99)01742-4. [DOI] [PubMed] [Google Scholar]
- 134.Cai CL, Liang X, Shi Y, Chu PH, Pfaff SL, Chen J, Evans S. Isl1 identifies a cardiac progenitor population that proliferates prior to differentiation and contributes a majority of cells to the heart. Dev Cell. 2003;5:877–89. doi: 10.1016/s1534-5807(03)00363-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Elshatory Y, Gan L. The LIM-homeobox gene Islet-1 is required for the development of restricted forebrain cholinergic neurons. J Neurosci. 2008;28:3291–7. doi: 10.1523/JNEUROSCI.5730-07.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Elshatory Y, Everhart D, Deng M, Xie X, Barlow RB, Gan L. Islet-1 controls the differentiation of retinal bipolar and cholinergic amacrine cells. J Neurosci. 2007;27:12707–20. doi: 10.1523/JNEUROSCI.3951-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Pan L, Deng M, Xie X, Gan L. ISL1 and BRN3B co-regulate the differentiation of murine retinal ganglion cells. Development. 2008;135:1981–90. doi: 10.1242/dev.010751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Pfaff SL, Mendelsohn M, Stewart CL, Edlund T, Jessell TM. Requirement for LIM homeobox gene Isl1 in motor neuron generation reveals a motor neuron-dependent step in interneuron differentiation. Cell. 1996;84:309–20. doi: 10.1016/s0092-8674(00)80985-x. [DOI] [PubMed] [Google Scholar]
- 139.Nishimura DY, Searby CC, Alward WL, Walton D, Craig JE, Mackey DA, Kawase K, Kanis AB, Patil SR, Stone EM, Sheffield VC. A spectrum of FOXC1 mutations suggests gene dosage as a mechanism for developmental defects of the anterior chamber of the eye. Am J Hum Genet. 2001;68:364–72. doi: 10.1086/318183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Mirzayans F, Gould DB, Heon E, Billingsley GD, Cheung JC, Mears AJ, Walter MA. Axenfeld-Rieger syndrome resulting from mutation of the FKHL7 gene on chromosome 6p25. Eur J Hum Genet. 2000;8:71–4. doi: 10.1038/sj.ejhg.5200354. [DOI] [PubMed] [Google Scholar]
- 141.Mears AJ, Jordan T, Mirzayans F, Dubois S, Kume T, Parlee M, Ritch R, Koop B, Kuo WL, Collins C, Marshall J, Gould DB, Pearce W, Carlsson P, Enerback S, Morissette J, Bhattacharya S, Hogan B, Raymond V, Walter MA. Mutations of the forkhead/winged-helix gene, FKHL7, in patients with Axenfeld-Rieger anomaly. Am J Hum Genet. 1998;63:1316–28. doi: 10.1086/302109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Lehmann OJ, Ebenezer ND, Jordan T, Fox M, Ocaka L, Payne A, Leroy BP, Clark BJ, Hitchings RA, Povey S, Khaw PT, Bhattacharya SS. Chromosomal duplication involving the forkhead transcription factor gene FOXC1 causes iris hypoplasia and glaucoma. Am J Hum Genet. 2000;67:1129–35. doi: 10.1016/s0002-9297(07)62943-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Fang J, Dagenais SL, Erickson RP, Arlt MF, Glynn MW, Gorski JL, Seaver LH, Glover TW. Mutations in FOXC2 (MFH-1), a forkhead family transcription factor, are responsible for the hereditary lymphedema-distichiasis syndrome. Am J Hum Genet. 2000;67:1382–8. doi: 10.1086/316915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Finegold DN, Kimak MA, Lawrence EC, Levinson KL, Cherniske EM, Pober BR, Dunlap JW, Ferrell RE. Truncating mutations in FOXC2 cause multiple lymphedema syndromes. Hum Mol Genet. 2001;10:1185–9. doi: 10.1093/hmg/10.11.1185. [DOI] [PubMed] [Google Scholar]
- 145.Semina EV, Brownell I, Mintz-Hittner HA, Murray JC, Jamrich M. Mutations in the human forkhead transcription factor FOXE3 associated with anterior segment ocular dysgenesis and cataracts. Hum Mol Genet. 2001;10:231–6. doi: 10.1093/hmg/10.3.231. [DOI] [PubMed] [Google Scholar]
- 146.Ormestad M, Blixt A, Churchill A, Martinsson T, Enerback S, Carlsson P. Foxe3 haploinsufficiency in mice: a model for Peters’ anomaly. Invest Ophthalmol Vis Sci. 2002;43:1350–7. [PubMed] [Google Scholar]
- 147.Crisponi L, Deiana M, Loi A, Chiappe F, Uda M, Amati P, Bisceglia L, Zelante L, Nagaraja R, Porcu S, Ristaldi MS, Marzella R, Rocchi M, Nicolino M, Lienhardt-Roussie A, Nivelon A, Verloes A, Schlessinger D, Gasparini P, Bonneau D, Cao A, Pilia G. The putative forkhead transcription factor FOXL2 is mutated in blepharophimosis/ptosis/epicanthus inversus syndrome. Nat Genet. 2001;27:159–66. doi: 10.1038/84781. [DOI] [PubMed] [Google Scholar]
- 148.De Baere E, Dixon MJ, Small KW, Jabs EW, Leroy BP, Devriendt K, Gillerot Y, Mortier G, Meire F, Van Maldergem L, Courtens W, Hjalgrim H, Huang S, Liebaers I, Van Regemorter N, Touraine P, Praphanphoj V, Verloes A, Udar N, Yellore V, Chalukya M, Yelchits S, De Paepe A, Kuttenn F, Fellous M, Veitia R, Messiaen L. Spectrum of FOXL2 gene mutations in blepharophimosis-ptosis-epicanthus inversus (BPES) families demonstrates a genotype--phenotype correlation. Hum Mol Genet. 2001;10:1591–600. doi: 10.1093/hmg/10.15.1591. [DOI] [PubMed] [Google Scholar]
- 149.Kume T, Deng K, Hogan BL. Murine forkhead/winged helix genes Foxc1 (Mf1) and Foxc2 (Mfh1) are required for the early organogenesis of the kidney and urinary tract. Development. 2000;127:1387–95. doi: 10.1242/dev.127.7.1387. [DOI] [PubMed] [Google Scholar]
- 150.Kume T, Deng KY, Winfrey V, Gould DB, Walter MA, Hogan BL. The forkhead/winged helix gene Mf1 is disrupted in the pleiotropic mouse mutation congenital hydrocephalus. Cell. 1998;93:985–96. doi: 10.1016/s0092-8674(00)81204-0. [DOI] [PubMed] [Google Scholar]
- 151.Hong HK, Lass JH, Chakravarti A. Pleiotropic skeletal and ocular phenotypes of the mouse mutation congenital hydrocephalus (ch/Mf1) arise from a winged helix/forkhead transcriptionfactor gene. Hum Mol Genet. 1999;8:625–37. doi: 10.1093/hmg/8.4.625. [DOI] [PubMed] [Google Scholar]
- 152.Kidson SH, Kume T, Deng K, Winfrey V, Hogan BL. The forkhead/winged-helix gene, Mf1, is necessary for the normal development of the cornea and formation of the anterior chamber in the mouse eye. Dev Biol. 1999;211:306–22. doi: 10.1006/dbio.1999.9314. [DOI] [PubMed] [Google Scholar]
- 153.Winnier GE, Kume T, Deng K, Rogers R, Bundy J, Raines C, Walter MA, Hogan BL, Conway SJ. Roles for the winged helix transcription factors MF1 and MFH1 in cardiovascular development revealed by nonallelic noncomplementation of null alleles. Dev Biol. 1999;213:418–31. doi: 10.1006/dbio.1999.9382. [DOI] [PubMed] [Google Scholar]
- 154.Iida K, Koseki H, Kakinuma H, Kato N, Mizutani-Koseki Y, Ohuchi H, Yoshioka H, Noji S, Kawamura K, Kataoka Y, Ueno F, Taniguchi M, Yoshida N, Sugiyama T, Miura N. Essential roles of the winged helix transcription factor MFH-1 in aortic arch patterning and skeletogenesis. Development. 1997;124:4627–38. doi: 10.1242/dev.124.22.4627. [DOI] [PubMed] [Google Scholar]
- 155.Smith RS, Zabaleta A, Kume T, Savinova OV, Kidson SH, Martin JE, Nishimura DY, Alward WL, Hogan BL, John SW. Haploinsufficiency of the transcription factors FOXC1 and FOXC2 results in aberrant ocular development. Hum Mol Genet. 2000;9:1021–32. doi: 10.1093/hmg/9.7.1021. [DOI] [PubMed] [Google Scholar]
- 156.Winnier GE, Hargett L, Hogan BL. The winged helix transcription factor MFH1 is required for proliferation and patterning of paraxial mesoderm in the mouse embryo. Genes Dev. 1997;11:926–40. doi: 10.1101/gad.11.7.926. [DOI] [PubMed] [Google Scholar]
- 157.Blixt A, Mahlapuu M, Aitola M, Pelto-Huikko M, Enerback S, Carlsson P. A forkhead gene, FoxE3, is essential for lens epithelial proliferation and closure of the lens vesicle. Genes Dev. 2000;14:245–54. [PMC free article] [PubMed] [Google Scholar]
- 158.Brownell I, Dirksen M, Jamrich M. Forkhead Foxe3 maps to the dysgenetic lens locus and is critical in lens development and differentiation. Genesis. 2000;27:81–93. doi: 10.1002/1526-968x(200006)27:2<81::aid-gene50>3.0.co;2-n. [DOI] [PubMed] [Google Scholar]
- 159.Ottolenghi C, Omari S, Garcia-Ortiz JE, Uda M, Crisponi L, Forabosco A, Pilia G, Schlessinger D. Foxl2 is required for commitment to ovary differentiation. Hum Mol Genet. 2005;14:2053–62. doi: 10.1093/hmg/ddi210. [DOI] [PubMed] [Google Scholar]
- 160.Schmidt D, Ovitt CE, Anlag K, Fehsenfeld S, Gredsted L, Treier AC, Treier M. The murine winged-helix transcription factor Foxl2 is required for granulosa cell differentiation and ovary maintenance. Development. 2004;131:933–42. doi: 10.1242/dev.00969. [DOI] [PubMed] [Google Scholar]
- 161.Uda M, Ottolenghi C, Crisponi L, Garcia JE, Deiana M, Kimber W, Forabosco A, Cao A, Schlessinger D, Pilia G. Foxl2 disruption causes mouse ovarian failure by pervasive blockage of follicle development. Hum Mol Genet. 2004;13:1171–81. doi: 10.1093/hmg/ddh124. [DOI] [PubMed] [Google Scholar]
- 162.Medema RH, Kops GJ, Bos JL, Burgering BM. AFX-like Forkhead transcription factors mediate cell-cycle regulation by Ras and PKB through p27kip1. Nature. 2000;404:782–7. doi: 10.1038/35008115. [DOI] [PubMed] [Google Scholar]
- 163.Lin L, Miller CT, Contreras JI, Prescott MS, Dagenais SL, Wu R, Yee J, Orringer MB, Misek DE, Hanash SM, Glover TW, Beer DG. The hepatocyte nuclear factor 3 alpha gene, HNF3alpha (FOXA1), on chromosome band 14q13 is amplified and overexpressed in esophageal and lung adenocarcinomas. Cancer Res. 2002;62:5273–9. [PubMed] [Google Scholar]
- 164.Carlsson P, Mahlapuu M. Forkhead transcription factors: key players in development and metabolism. Dev Biol. 2002;250:1–23. doi: 10.1006/dbio.2002.0780. [DOI] [PubMed] [Google Scholar]
- 165.Kaestner KH, Katz J, Liu Y, Drucker DJ, Schutz G. Inactivation of the winged helix transcription factor HNF3alpha affects glucose homeostasis and islet glucagon gene expression in vivo. Genes Dev. 1999;13:495–504. doi: 10.1101/gad.13.4.495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Ang SL, Rossant J. HNF-3 beta is essential for node and notochord formation in mouse development. Cell. 1994;78:561–74. doi: 10.1016/0092-8674(94)90522-3. [DOI] [PubMed] [Google Scholar]
- 167.Weinstein DC, Ruiz i Altaba A, Chen WS, Hoodless P, Prezioso VR, Jessell TM, Darnell JE., Jr The winged-helix transcription factor HNF-3 beta is required for notochord development in the mouse embryo. Cell. 1994;78:575–88. doi: 10.1016/0092-8674(94)90523-1. [DOI] [PubMed] [Google Scholar]
- 168.Sund NJ, Vatamaniuk MZ, Casey M, Ang SL, Magnuson MA, Stoffers DA, Matschinsky FM, Kaestner KH. Tissue-specific deletion of Foxa2 in pancreatic beta cells results in hyperinsulinemic hypoglycemia. Genes Dev. 2001;15:1706–15. doi: 10.1101/gad.901601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Sund NJ, Ang SL, Sackett SD, Shen W, Daigle N, Magnuson MA, Kaestner KH. Hepatocyte nuclear factor 3beta (Foxa2) is dispensable for maintaining the differentiated state of the adult hepatocyte. Mol Cell Biol. 2000;20:5175–83. doi: 10.1128/mcb.20.14.5175-5183.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Ellsworth BS, Egashira N, Haller JL, Butts DL, Cocquet J, Clay CM, Osamura RY, Camper SA. FOXL2 in the pituitary: molecular, genetic, and developmental analysis. Mol Endocrinol. 2006;20:2796–805. doi: 10.1210/me.2005-0303. [DOI] [PubMed] [Google Scholar]
- 171.Ellsworth BS, Burns AT, Escudero KW, Duval DL, Nelson SE, Clay CM. The gonadotropin releasing hormone (GnRH) receptor activating sequence (GRAS) is a composite regulatory element that interacts with multiple classes of transcription factors including Smads, AP-1 and a forkhead DNA binding protein. Mol Cell Endocrinol. 2003;206:93–111. doi: 10.1016/s0303-7207(03)00235-1. [DOI] [PubMed] [Google Scholar]
- 172.Loffler KA, Zarkower D, Koopman P. Etiology of ovarian failure in blepharophimosis ptosis epicanthus inversus syndrome: FOXL2 is a conserved, early-acting gene in vertebrate ovarian development. Endocrinology. 2003;144:3237–43. doi: 10.1210/en.2002-0095. [DOI] [PubMed] [Google Scholar]
- 173.Blount AL, Schmidt K, Justice NJ, Vale WW, Fischer WH, Bilezikjian LM. FoxL2 and Smad3 coordinately regulate follistatin gene transcription. J Biol Chem. 2009;284:7631–45. doi: 10.1074/jbc.M806676200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Lamba P, Fortin J, Tran S, Wang Y, Bernard DJ. A novel role for the forkhead transcription factor FOXL2 in activin A-regulated follicle-stimulating hormone beta subunit transcription. Mol Endocrinol. 2009;23:1001–13. doi: 10.1210/me.2008-0324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Kalfa N, Philibert P, Patte C, Ecochard A, Duvillard P, Baldet P, Jaubert F, Fellous M, Sultan C. Extinction of FOXL2 expression in aggressive ovarian granulosa cell tumors in children. Fertil Steril. 2007;87:896–901. doi: 10.1016/j.fertnstert.2006.11.016. [DOI] [PubMed] [Google Scholar]
- 176.Zannini M, Avantaggiato V, Biffali E, Arnone MI, Sato K, Pischetola M, Taylor BA, Phillips SJ, Simeone A, Di Lauro R. TTF-2, a new forkhead protein, shows a temporal expression in the developing thyroid which is consistent with a role in controlling the onset of differentiation. EMBO J. 1997;16:3185–97. doi: 10.1093/emboj/16.11.3185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.Kalinichenko VV, Gusarova GA, Shin B, Costa RH. The forkhead box F1 transcription factor is expressed in brain and head mesenchyme during mouse embryonic development. Gene Expr Patterns. 2003;3:153–8. doi: 10.1016/s1567-133x(03)00010-3. [DOI] [PubMed] [Google Scholar]
- 178.Norquay LD, Yang X, Jin Y, Detillieux KA, Cattini PA. Hepatocyte nuclear factor-3alpha binding at P sequences of the human growth hormone locus is associated with pituitary repressor function. Mol Endocrinol. 2006;20:598–607. doi: 10.1210/me.2005-0221. [DOI] [PubMed] [Google Scholar]
- 179.Herrera E, Marcus R, Li S, Williams SE, Erskine L, Lai E, Mason C. Foxd1 is required for proper formation of the optic chiasm. Development. 2004;131:5727–39. doi: 10.1242/dev.01431. [DOI] [PubMed] [Google Scholar]
- 180.Hatini V, Huh SO, Herzlinger D, Soares VC, Lai E. Essential role of stromal mesenchyme in kidney morphogenesis revealed by targeted disruption of Winged Helix transcription factor BF-2. Genes Dev. 1996;10:1467–78. doi: 10.1101/gad.10.12.1467. [DOI] [PubMed] [Google Scholar]
- 181.Murre C, McCaw PS, Vaessin H, Caudy M, Jan LY, Jan YN, Cabrera CV, Buskin JN, Hauschka SD, Lassar AB, et al. Interactions between heterologous helix-loop-helix proteins generate complexes that bind specifically to a common DNA sequence. Cell. 1989;58:537–44. doi: 10.1016/0092-8674(89)90434-0. [DOI] [PubMed] [Google Scholar]
- 182.Arnold HH, Braun T. Targeted inactivation of myogenic factor genes reveals their role during mouse myogenesis: a review. Int J Dev Biol. 1996;40:345–53. [PubMed] [Google Scholar]
- 183.Chu K, Nemoz-Gaillard E, Tsai MJ. BETA2 and pancreatic islet development. Recent Prog Horm Res. 2001;56:23–46. doi: 10.1210/rp.56.1.23. [DOI] [PubMed] [Google Scholar]
- 184.Cho JH, Tsai MJ. The role of BETA2/NeuroD1 in the development of the nervous system. Mol Neurobiol. 2004;30:35–47. doi: 10.1385/MN:30:1:035. [DOI] [PubMed] [Google Scholar]
- 185.Henke RM, Meredith DM, Borromeo MD, Savage TK, Johnson JE. Ascl1 and Neurog2 form novel complexes and regulate Delta-like3 (Dll3) expression in the neural tube. Dev Biol. 2009;328:529–40. doi: 10.1016/j.ydbio.2009.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 186.Fratticci A, Grieco FA, Spilioti C, Giangaspero F, Ventura L, Esposito V, Piccirilli M, Santoro A, Gulino A, Cantore G, Alesse E, Jaffrain-Rea ML. Differential expression of neurogenins and NeuroD1 in human pituitary tumours. J Endocrinol. 2007;194:475–84. doi: 10.1677/JOE-07-0020. [DOI] [PubMed] [Google Scholar]
- 187.Cai L, Morrow EM, Cepko CL. Misexpression of basic helix-loop-helix genes in the murine cerebral cortex affects cell fate choices and neuronal survival. Development. 2000;127:3021–30. doi: 10.1242/dev.127.14.3021. [DOI] [PubMed] [Google Scholar]
- 188.Cau E, Gradwohl G, Fode C, Guillemot F. Mash1 activates a cascade of bHLH regulators in olfactory neuron progenitors. Development. 1997;124:1611–21. doi: 10.1242/dev.124.8.1611. [DOI] [PubMed] [Google Scholar]
- 189.Mizuguchi R, Kriks S, Cordes R, Gossler A, Ma Q, Goulding M. Ascl1 and Gsh1/2 control inhibitory and excitatory cell fate in spinal sensory interneurons. Nat Neurosci. 2006;9:770–8. doi: 10.1038/nn1706. [DOI] [PubMed] [Google Scholar]
- 190.Poulin G, Lebel M, Chamberland M, Paradis FW, Drouin J. Specific protein-protein interaction between basic helix-loop-helix transcription factors and homeoproteins of the Pitx family. Mol Cell Biol. 2000;20:4826–37. doi: 10.1128/mcb.20.13.4826-4837.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 191.Poulin G, Turgeon B, Drouin J. NeuroD1/beta2 contributes to cell-specific transcription of the proopiomelanocortin gene. Mol Cell Biol. 1997;17:6673–82. doi: 10.1128/mcb.17.11.6673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 192.Cherrington BD, Bailey JS, Diaz AL, Mellon PL. NeuroD1 and Mash1 temporally regulate GnRH receptor gene expression in immortalized mouse gonadotrope cells. Mol Cell Endocrinol. 2008;295:106–14. doi: 10.1016/j.mce.2008.07.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 193.Lavoie PL, Budry L, Balsalobre A, Drouin J. Developmental dependence on NurRE and EboxNeuro for expression of pituitary proopiomelanocortin. Mol Endocrinol. 2008;22:1647–57. doi: 10.1210/me.2007-0567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 194.Lamolet B, Poulin G, Chu K, Guillemot F, Tsai MJ, Drouin J. Tpit-independent function of NeuroD1(BETA2) in pituitary corticotroph differentiation. Mol Endocrinol. 2004;18:995–1003. doi: 10.1210/me.2003-0127. [DOI] [PubMed] [Google Scholar]
- 195.Pulichino AM, Vallette-Kasic S, Couture C, Gauthier Y, Brue T, David M, Malpuech G, Deal C, Van Vliet G, De Vroede M, Riepe FG, Partsch CJ, Sippell WG, Berberoglu M, Atasay B, Drouin J. Human and mouse TPIT gene mutations cause early onset pituitary ACTH deficiency. Genes Dev. 2003;17:711–6. doi: 10.1101/gad.1065603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 196.Monahan P, Rybak S, Raetzman LT. The Notch Target gene Hes1 regulates cell cycle inhibitor expression in the developing pituitary. Endocrinology. 2009 doi: 10.1210/en.2009-0206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 197.Vierimaa O, Georgitsi M, Lehtonen R, Vahteristo P, Kokko A, Raitila A, Tuppurainen K, Ebeling TM, Salmela PI, Paschke R, Gundogdu S, De Menis E, Makinen MJ, Launonen V, Karhu A, Aaltonen LA. Pituitary adenoma predisposition caused by germline mutations in the AIP gene. Science. 2006;312:1228–30. doi: 10.1126/science.1126100. [DOI] [PubMed] [Google Scholar]
- 198.Leontiou CA, Gueorguiev M, van der Spuy J, Quinton R, Lolli F, Hassan S, Chahal HS, Igreja SC, Jordan S, Rowe J, Stolbrink M, Christian HC, Wray J, Bishop-Bailey D, Berney DM, Wass JA, Popovic V, Ribeiro-Oliveira A, Jr, Gadelha MR, Monson JP, Akker SA, Davis JR, Clayton RN, Yoshimoto K, Iwata T, Matsuno A, Eguchi K, Musat M, Flanagan D, Peters G, Bolger GB, Chapple JP, Frohman LA, Grossman AB, Korbonits M. The role of the aryl hydrocarbon receptor-interacting protein gene in familial and sporadic pituitary adenomas. J Clin Endocrinol Metab. 2008;93:2390–401. doi: 10.1210/jc.2007-2611. [DOI] [PubMed] [Google Scholar]
- 199.Daly AF, Vanbellinghen JF, Khoo SK, Jaffrain-Rea ML, Naves LA, Guitelman MA, Murat A, Emy P, Gimenez-Roqueplo AP, Tamburrano G, Raverot G, Barlier A, De Herder W, Penfornis A, Ciccarelli E, Estour B, Lecomte P, Gatta B, Chabre O, Sabate MI, Bertagna X, Garcia Basavilbaso N, Stalldecker G, Colao A, Ferolla P, Wemeau JL, Caron P, Sadoul JL, Oneto A, Archambeaud F, Calender A, Sinilnikova O, Montanana CF, Cavagnini F, Hana V, Solano A, Delettieres D, Luccio-Camelo DC, Basso A, Rohmer V, Brue T, Bours V, Teh BT, Beckers A. Aryl hydrocarbon receptor-interacting protein gene mutations in familial isolated pituitary adenomas: analysis in 73 families. J Clin Endocrinol Metab. 2007;92:1891–6. doi: 10.1210/jc.2006-2513. [DOI] [PubMed] [Google Scholar]
- 200.Thomas JO. HMG1 and 2: architectural DNA-binding proteins. Biochem Soc Trans. 2001;29:395–401. doi: 10.1042/bst0290395. [DOI] [PubMed] [Google Scholar]
- 201.Masui S, Nakatake Y, Toyooka Y, Shimosato D, Yagi R, Takahashi K, Okochi H, Okuda A, Matoba R, Sharov AA, Ko MS, Niwa H. Pluripotency governed by Sox2 via regulation of Oct3/4 expression in mouse embryonic stem cells. Nat Cell Biol. 2007;9:625–35. doi: 10.1038/ncb1589. [DOI] [PubMed] [Google Scholar]
- 202.Fauquier T, Rizzoti K, Dattani M, Lovell-Badge R, Robinson IC. SOX2-expressing progenitor cells generate all of the major cell types in the adult mouse pituitary gland. Proc Natl Acad Sci U S A. 2008;105:2907–12. doi: 10.1073/pnas.0707886105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 203.Kelberman D, Rizzoti K, Avilion A, Bitner-Glindzicz M, Cianfarani S, Collins J, Chong WK, Kirk JM, Achermann JC, Ross R, Carmignac D, Lovell-Badge R, Robinson IC, Dattani MT. Mutations within Sox2/SOX2 are associated with abnormalities in the hypothalamo-pituitary-gonadal axis in mice and humans. J Clin Invest. 2006;116:2442–55. doi: 10.1172/JCI28658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 204.Quirk CC, Seachrist DD, Nilson JH. Embryonic expression of the luteinizing hormone beta gene appears to be coupled to the transient appearance of p8, a high mobility group-related transcription factor. J Biol Chem. 2003;278:1680–5. doi: 10.1074/jbc.M209906200. [DOI] [PubMed] [Google Scholar]
- 205.Million Passe CM, White CR, King MW, Quirk PL, Iovanna JL, Quirk CC. Loss of the protein NUPR1 (p8) leads to delayed LHB expression, delayed ovarian maturation, and testicular development of a sertoli-cell-only syndrome-like phenotype in mice. Biol Reprod. 2008;79:598–607. doi: 10.1095/biolreprod.108.068304. [DOI] [PubMed] [Google Scholar]
- 206.Brannon KM, Million Passe CM, White CR, Bade NA, King MW, Quirk CC. Expression of the high mobility group A family member p8 is essential to maintaining tumorigenic potential by promoting cell cycle dysregulation in LbetaT2 cells. Cancer Lett. 2007;254:146–55. doi: 10.1016/j.canlet.2007.03.011. [DOI] [PubMed] [Google Scholar]
- 207.Mohammad HP, Seachrist DD, Quirk CC, Nilson JH. Reexpression of p8 contributes to tumorigenic properties of pituitary cells and appears in a subset of prolactinomas in transgenic mice that hypersecrete luteinizing hormone. Mol Endocrinol. 2004;18:2583–93. doi: 10.1210/me.2004-0163. [DOI] [PubMed] [Google Scholar]
- 208.Ruault M, Brun ME, Ventura M, Roizes G, De Sario A. MLL3, a new human member of the TRX/MLL gene family, maps to 7q36, a chromosome region frequently deleted in myeloid leukaemia. Gene. 2002;284:73–81. doi: 10.1016/s0378-1119(02)00392-x. [DOI] [PubMed] [Google Scholar]
- 209.Naiche LA, Harrelson Z, Kelly RG, Papaioannou VE. T-box genes in vertebrate development. Annu Rev Genet. 2005;39:219–39. doi: 10.1146/annurev.genet.39.073003.105925. [DOI] [PubMed] [Google Scholar]
- 210.Pulichino AM, Vallette-Kasic S, Tsai JP, Couture C, Gauthier Y, Drouin J. Tpit determines alternate fates during pituitary cell differentiation. Genes Dev. 2003;17:738–47. doi: 10.1101/gad.1065703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 211.Pontecorvi M, Goding CR, Richardson WD, Kessaris N. Expression of Tbx2 and Tbx3 in the developing hypothalamic-pituitary axis. Gene Expr Patterns. 2008;8:411–7. doi: 10.1016/j.gep.2008.04.006. [DOI] [PubMed] [Google Scholar]
- 212.Harrelson Z, Kelly RG, Goldin SN, Gibson-Brown JJ, Bollag RJ, Silver LM, Papaioannou VE. Tbx2 is essential for patterning the atrioventricular canal and for morphogenesis of the outflow tract during heart development. Development. 2004;131:5041–52. doi: 10.1242/dev.01378. [DOI] [PubMed] [Google Scholar]
- 213.Davenport TG, Jerome-Majewska LA, Papaioannou VE. Mammary gland, limb and yolk sac defects in mice lacking Tbx3, the gene mutated in human ulnar mammary syndrome. Development. 2003;130:2263–73. doi: 10.1242/dev.00431. [DOI] [PubMed] [Google Scholar]
- 214.Pogoda HM, Hammerschmidt M. Molecular genetics of pituitary development in zebrafish. Semin Cell Dev Biol. 2007;18:543–58. doi: 10.1016/j.semcdb.2007.04.004. [DOI] [PubMed] [Google Scholar]
- 215.Wagner J, Lepore D, Thomas P. Differentiation of mouse embryonic stem cells into growth hormone and prolactin expressing cells in vitro. Mol Cell Endocrinol. 2007;273:68–74. doi: 10.1016/j.mce.2007.05.007. [DOI] [PubMed] [Google Scholar]
- 216.Zhao X, Teng R, Asanuma K, Okouchi Y, Johkura K, Ogiwara N, Sasaki K. Differentiation of mouse embryonic stem cells into gonadotrope-like cells in vitro. J Soc Gynecol Investig. 2005;12:257–62. doi: 10.1016/j.jsgi.2005.01.004. [DOI] [PubMed] [Google Scholar]