The expression and subsequent purification of recombinant proteins are widely employed in biochemical studies. A powerful purification method involves the use of peptide affinity tags, which are fused to the protein of interest and used to expedite protein purification via affinity chromatography.1,2 A widely employed method utilizes immobilized metal-affinity chromatography (IMAC) to purify recombinant proteins containing a short affinity tag consisting of polyhistidine residues. IMAC is based on the interactions between a transition metal ion (Co2+, Ni2+, Cu2+, Zn2+) immobilized on a matrix and specific amino acid side chains. Histidine is the amino acid that exhibits the strongest interaction with immobilized metal ion matrices, as electron donor groups on the histidine imidazole ring readily form coordination bonds with the immobilized transition metal. Peptides containing sequences of consecutive histidine residues are efficiently retained on IMAC column matrices. Following washing of the matrix material, peptides containing polyhistidine sequences can be easily eluted by either adjusting the pH of the column buffer or adding free imidazole to the column buffer.3
IMAC is a versatile method that can be utilized to rapidly purify polyhistidine affinity-tagged proteins, resulting in 100-fold enrichments in a single purification step.4 Affinity-tagged protein purities can be achieved at up to 95% purity by IMAC in high yield.5,6 Purification using polyhistidine tags has been carried out successfully using a number of expression systems, including Escherichia coli,7 Saccharomyces cerevisiae,8 mammalian cells,9 and baculovirus-infected insect cells.10 However, this purification method may not be sufficient for tagged proteins expressed at low levels that require significantly greater than 100-fold enrichment or for the preparation of highly homogeneous protein samples. In such cases, either the use of a different affinity tag or the use of the polyhistidine tag in conjunction with additional purification techniques should be considered.
General Considerations
Incorporation of the Polyhistidine Affinity Tag
Affinity tags consisting of six polyhistidine residues are commonly used in IMAC. Whereas tags of six histidine residues are generally long enough to yield high-affinity interactions with the matrix, both shorter and longer affinity tags have been used successfully. In some cases the use of longer polyhistidine tags has resulted in increased purity due to the ability to use more stringent washing steps.11 Still, it is advisable to use the smallest number of histidine residues as required for efficient purification to minimize possible perturbation of protein function. In general, a six histidine tag is an appropriate choice for the first trial when adding a novel polyhistidine tag to a protein.
Polyhistidine affinity tags are commonly placed on either the N or the C terminus of recombinant proteins. Optimal placement of the tag is protein specific. A potential problem is inaccessibility of the protein tag to the immobilized metal due to occlusion of the tag in the folded protein. Moving the affinity tag to the opposite terminus of the protein or carrying out the purification under denaturing conditions often resolves this problem. In principle, it is possible that the affinity tag may interfere with protein activity, although the relatively small size and charge of the polyhistidine affinity tag ensure that protein activity is rarely affected. Thus, the affinity tag usually does not need to be removed following protein purification.12 If necessary, the affinity tag can be removed by use of a protease cleavage site inserted between the tag and the protein.13
Polyhistidine affinity tags are small enough to be incorporated easily into any expression vector. These tags can be added onto target genes by site-directed mutagenesis or by polymerase chain reaction methods. DNA fragments coding for the polyhistidine affinity tag can also be created from synthetic oligonucleotides and cloned into an appropriate location in the desired plasmid.14 Alternatively, there are a wide variety of commercially available cloning vectors for the generation and expression of polyhistidine-tagged recombinant proteins in different expression systems. Vectors for secreted polyhistidine affinity tagged proteins have also been developed for E. coli.15
Affinity Matrices
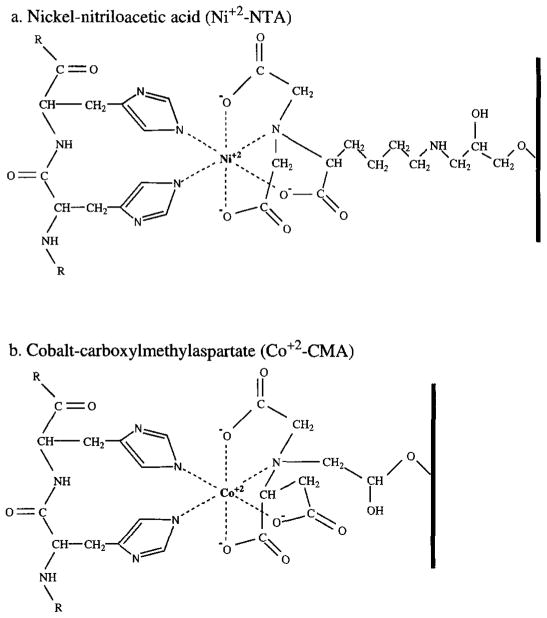
A variety of immobilized metal matrices are available for use in IMAC. Initial IMAC reports used iminodiacetic acid (IDA) as a matrix to chelate transition metals through three coordination sites.16 A problem with the use of IDA matrices is that the metal ion is only weakly bound to such a three-coordinate matrix. Metal leaching from the matrix during purification causes lowered yields and impure products.12 More recently, purification of polyhistidine affinity-tagged proteins has been facilitated by the development of the commercially available matrices nickel-nitrilotriacetic acid (Ni2+-NTA)17 and Co2+–carboxylmethylaspartate (Co2+-CMA),18 which are coupled to a solid support resin. These matrices securely coordinate metal ions through four coordination sites while leaving two of the transition metal coordination sites exposed to interact with histidine residues in the affinity tag. Molecular models of these interactions are shown in Fig. 1. Much of the versatility of IMAC stems from the ability of these resins to tolerate a wide range of conditions, including the presence of protein denaturants and detergents. The stability of metal binding in these resins also allows the resins to be regenerated and reused several times. The Ni2+–NTA matrix (available from Qiagen) has a binding capacity of 5–10 mg protein/ml of matrix resin and a high binding affinity (Kd = 10−13 M) for the six residue polyhistidine tag at pH 8.0.4 The Co2+–CMA matrix (Talon resin, available from Clontech) has a somewhat lower affinity for the polyhistidine affinity tag than the Ni2+–NTA resin, resulting in elution of the tagged proteins under milder conditions. The Co2+–CMA also has been reported to exhibit less nonspecific protein binding than the Ni2+–NTA resin, resulting in higher elution product purity.19 The binding capacity of the Co2+–CMA resin is also about 5–10 mg of protein/ml of resin.
Fig. 1.
Models of the interactions between the polyhistidine affinity tag and two immobilized metal affinity chromatography matrices, (a) The nickel–nitrilotriacetic acid matrix (Ni+2–NTA) [from J. Crowe, H. Döbeli, R. Gentz, E. Hochuli, D. Stüber, and K. Henco, Methods Mol. Biol. 31, 371 (1994)]. (b) The cobalt–carboxylmethylaspartate matrix (Co2+–CMA) (from G. Tchaga, Clontech, personal communication). In both cases, the metal ion exhibits octahedral coordination by four matrix ligands and two histidine side chains, the latter provided by the polyhistidine affinity tag.
Purification under Native and Denaturing Conditions
Purification using the polyhistidine tag can be performed under either native or denaturing conditions by IMAC. The use of mild buffer conditions and imidazole as the elutant often yields biologically active purification products. Proteins that remain soluble in the cytoplasm, or that are secreted, usually can be purified using these native conditions. However, purification under native conditions may be hindered if the target protein is insoluble, aggregates in inclusion bodies, or possesses a tertiary structure that occludes the polyhistidine affinity tag. In such cases, proteins can be purified by the use of denaturing conditions such as 6 M guanidinium hydrochloride or 8 M urea during the purification process. Interaction of the resin with the polyhistidine tag does not require a specific conformation of the peptide tag, which makes effective purification with the use of denaturing conditions possible. Purification under denaturing conditions can depress the activity of phosphatases and proteolytic enzymes.6 The use of urea as a denaturant is often preferable as 6 M guanidinium hydrochloride precipitates in the presence of SDS, interfering with subsequent SDS–PAGE analysis. Proteins purified under denaturing conditions can then be refolded into their active states by dialyzing away the denaturants.20 In some cases, proteins can be refolded while bound to the resin.21
Purification of Membrane Proteins
Polyhistidine-tagged membrane proteins can be purified by IMAC using detergent-containing buffers to solubilize the proteins during the chromatographic process.22,23 IMAC of membrane proteins has been carried out successfully in a variety of ionic and nonionic detergents. It is difficult to predict which detergent will be most suitable for IMAC in a given membrane protein system.11 Although caution should be used, the Ni2+–NTA and Co2+–CMA matrices are generally able to tolerate limited amounts of nonionic and ionic detergents. Following IMAC, it is possible to restore the activity of purified polyhistidine-tagged membrane proteins by reconstitution into membrane vesicles.14
Nonspecific Binding
A problem with the use of polyhistidine affinity tags is nonspecific binding of untagged proteins. Although histidine occurs relatively infrequently (2% of all protein residues are histidine), some cellular proteins contain two or more adjacent histidine residues.4 These proteins have an affinity for the IMAC matrix and may coelute with the protein of interest, resulting in significant contamination of the final product. This problem is generally more pronounced in systems other than E. coli. Mammalian systems, for example, have a higher natural abundance of proteins containing consecutive histidine residues.12 Disulfide bond formation between the protein of interest and other proteins can also lead to contamination. The use of 10 mM 2-mercaptoethanol in the loading, wash, and elution buffers generally eliminates this potential problem. Nonspecific hydrophobic interactions can also cause some copurification with the desired protein. Including low levels (up to 1%) of the nonionic detergent Triton X-100 or Tween 20 in the protein buffers can reduce these interactions without substantially affecting the binding of the tagged protein to the Ni2+-NTA or the Co2+–CMA matrices. The addition of salt (up to 500 mM NaCl), glycerol (up to 20%), or low levels of ethanol (up to 20%) can also reduce nonspecific hydrophobic protein interactions with these matrices. Optimum levels of these buffer components should be determined experimentally for individual proteins.
Purification Procedure
Design of Protein Binding, Washing, and Elution Steps
Binding of the polyhistidine-tagged proteins can be performed using either a column or a batch procedure. Cell lysis should be done in buffered solution adjusted to pH 8.0. When the column procedure is utilized, the resin is packed into a column and the cell lysate is slowly loaded (3 to 4 column volumes per hour) onto the column. The batch procedure involves incubating the affinity matrix resin in the cell lysate solution and then packing the resin into a column. During incubation at 4°, the resin can be suspended in the cell lysate solution by shaking or stirring. Use of the batch procedure often results in more efficient binding of the tagged protein. With either method, the use of the minimum amount of resin needed to bind the tagged protein is recommended. The tagged protein usually has a higher binding affinity than other proteins that bind nonspecifically to the resin. Thus, when the minimum amount of resin is used, the tagged protein will fill most of the available binding sites, reducing the number of nonspecific proteins that bind. Sodium chloride (up to 500 mM) and low levels of imidazole (up to 20 mM) can also be included in the binding buffer to reduce the number of proteins that bind nonspecifically to the resin. Most protease inhibitors, with the exception of metal-chelating agents such as EDTA, can be included in all buffers. There are commercially available protease inhibitor cocktails specifically designed for use in IMAC purifications (Sigma).
Following binding of the tagged protein, the column can be washed to remove nonspecific proteins that bind weakly to the column. If desired, the inclusion of imidazole (10–50 mM for Ni2+–NTA, 10 mM for Co2+–CMA) in the wash buffer will increase the stringency of the wash and elute nonspecifically bound proteins more effectively. Alternatively, a wash buffer with a pH lower than that of the binding buffer (pH 6.3 for Ni2+–NTA, pH 7.0 for Co2+–CMA) can be employed to remove nonspecifically bound proteins. Agents such as detergents, 2-mercaptoethanol, and sodium chloride are often included in the wash and binding buffer to reduce nonspecific protein binding.
Three different methods can be used to elute the tagged protein of interest. Lowering the pH (to 5.3–4.5 for Ni2+–NTA, 6.0 for Co2+–CMA) protonates the imidazole nitrogen atom of the histidine residue (pKa 6.0) and disrupts the coordination bond between the histidine and the transition metal. The histidine analog imidazole can also be used to competitively elute the bound polyhistidine residues (concentrations of 100 mM or higher for Ni2+–NTA, 50 mM or higher for Co2+–CMA). If the tagged protein forms oligomers, more stringent conditions, such as lower pH or higher concentrations of imidazole, may be required to elute the protein. While both of these elution methods are effective, the use of imidazole is often preferable as exposure to low pH may damage the protein of interest. Note, however, that heating a sample that contains imidazole to boiling prior to SDS–PAGE can cause acid-labile bonds to hydrolyze. Instead, it is recommended to heat the sample to no more than 37° for 5 min in the SDS loading buffer prior to analysis.12 Including chelating agents, such as EDTA (100 mM) in the elution buffer, can facilitate maximal elution of proteins from the resin. This treatment will strip the metal atoms away from the matrix, resulting in contamination of the elute. The presence of the chelating agent or the metal in the eluate may interfere with enzyme activity. Moreover, the matrix cannot be reused following the use of a chelating agent without recharging.
Protocol: Native Purification of a Soluble Polyhistidine-Tagged Protein
The following protocol is one that is designed for the purification of the soluble ERK2 protein tagged with six N-terminal histidine residues from E. coli utilizing a Ni2+–NTA resin under nondenaturing conditions (see Fig. 2).24 While this protocol may need minor optimization for other polyhistidine-tagged proteins, it should serve as a good starting point for the purification of soluble proteins in the native state. Urea or guanidinium hydrochloride can be added to all buffers to perform this procedure under denaturing conditions. Additional agents, including detergents, 2-mercaptoethanol, and various protease inhibitors, can also be added to the buffers to prevent nonspecific binding or proteolysis.
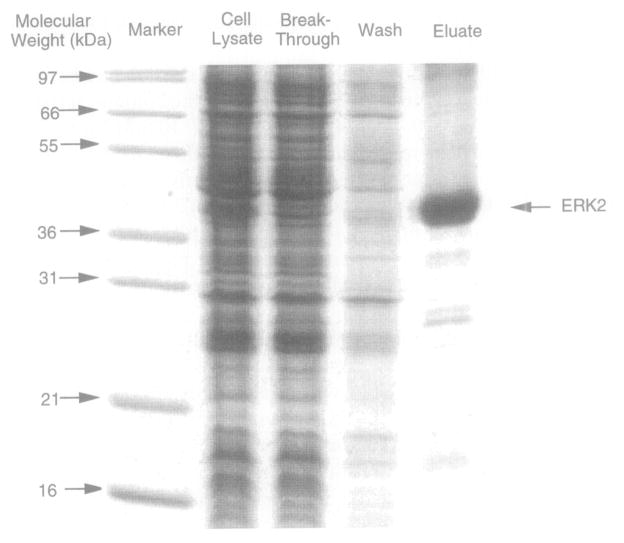
Fig. 2.
SDS–PAGE analysis of a representative polyhistidine-tagged protein purification using a nickel-nitrilotriacetic acid (Ni2+–NTA) matrix. The 42-kDa MAP-kinase protein ERK2 was affinity tagged at its N terminus with six histidines [D. J. Robbins, E. Zhen, H. Owaki, C. A. Vanderbuilt, D. Ebert, T. D. Geppert, and M. H. Cobb, J. Biol. Chem. 268, 5097 (1993)], and was isolated using the procedure detailed in the text. Shown are equal volumes of the cell lysate, the breakthrough material that failed to bind to the resin during the batch step, the wash material obtained after loading the resin into the column, and the eluate from the column. The 15% Laemmli gel was visualized by Coomassie staining.
Lyse cells expressing the tagged protein by sonication on ice in loading buffer (300 mM NaCl, 50 mM NaH2PO4, pH to 8.0 with NaOH). Approximately 3–5 ml of loading buffer should be used per gram (wet weight) of cells. Keep the lysate as cold as possible to minimize possible proteolysis.
Immediately centrifuge lysate by spinning at 30,000g for 30 min at 4°.
Add 50% Ni2+–NTA slurry (Qiagen) preequilibrated in ice-cold loading buffer to the supernatant. Add a sufficient amount of 50% Ni2+–NTA slurry to bind the polyhistidine-tagged protein (5–10 mg/ml resin). Stir or rotate at 4° for 1 hr.
Load the resin onto a column. Wash the resin with 20 column volumes of loading buffer at 4°.
Wash the resin with 20 column volumes of wash buffer at 4° (same as loading buffer but also containing 10 mM imidazole, pH to 8.0 with HCl).
Elute with a 20 column volume gradient of 10 to 250 mM imidazole in loading buffer (pH to 8.0 with HCl). Collect 1-ml fractions and assay for fractions containing the desired protein using SDS–PAGE. Pool the desired fractions.
Comparison of IMAC to Other Affinity Chromatographic Techniques
Because a wide number of other affinity tag systems are available, it is useful to consider the advantages and disadvantages of the polyhistidine affinity tag and IMAC. The nonspecific binding of proteins to the IMAC column is a major disadvantage, especially when the tagged protein is not expressed at high levels. In some cases, the use of other affinity tag systems results in higher degrees of enrichment of the affinity-tagged protein than can be attained when polyhistidine affinity tags are employed. For example, biotinylation-accepting domain affinity tags have been shown to provide protein samples of higher yield and purity than polyhistidine affinity tags.25,26
Still, the polyhistidine affinity tag also possesses several important advantages. This tag is easily added to the protein of interest and, for IMAC purification of highly expressed proteins, can readily provide purities of up to 95% with 90% recovery of the tagged protein in a single purification step. The relatively small size and charge of the histidine affinity tag ensure that it rarely affects protein function. Polyhistidine-tagged proteins can be eluted under mild conditions from the IMAC resin, which allows them to retain their biological activity. Purification can be carried out readily under denaturing conditions that allow for the purification of insoluble proteins and proteins in which the affinity tag is occluded by the native tertiary structure of the protein. The IMAC affinity matrix resin is not affected by protease or nuclease activities in the extract; thus unlike many other biological affinity procedures, IMAC can be used effectively as an initial purification step with crude cell lysates. Following IMAC, proteins can often be readily purified further using other chromatographic methods. The immobilization of polyhistidine-tagged proteins on IMAC matrices can also be used for protein–protein interaction studies.27 Overall, the use of IMAC to purify tagged proteins provides a rapid and inexpensive purification method in comparison to other affinity protein purification methods.
The advantages of the polyhistidine affinity tag can be combined with the advantages of other affinity tags through the use of multiaffinity fusion systems wherein two or more affinity tags are attached to the same protein. These multifusion systems provide great flexibility during the purification process by allowing for protein immobilization and elution to be performed under a variety of conditions.1,28 In addition, the use of multiaffinity tag purification procedures can result in products of higher purity than can be achieved using each individual affinity domain alone.29 A number of affinity tag domains have been coupled successfully with the polyhistidine tag on recombinant proteins, including the GST affinity tag,30 a modified S-peptide of ribonuclease A,29 and both the albumin-binding protein fusion domain and a biotinylation accepting domain.28
Acknowledgments
The authors thank Natalie Ahn and Melanie Cobb for providing the plasmid NpT7-5, which expresses six histidine-tagged ERK2. The authors thank Irene Ota, Eric Nalefski, Mark Benson, and Julie Poelchau for providing valuable comments regarding the manuscript. We also gratefully acknowledge NIH Grants GM 40731 and GM 48203.
References
- 1.Nilsson J, Ståhl S, Lundeberg J, Uhlén M, Nygren PÅ. Protein Expr Purif. 1997;11:1. doi: 10.1006/prep.1997.0767. [DOI] [PubMed] [Google Scholar]
- 2.Jarvik JW, Telmer CA. Annu Rev Genet. 1998;32:601. doi: 10.1146/annurev.genet.32.1.601. [DOI] [PubMed] [Google Scholar]
- 3.Porath J. Protein Expr Purif. 1992;3:263. doi: 10.1016/1046-5928(92)90001-d. [DOI] [PubMed] [Google Scholar]
- 4.Schmitt J, Hess H, Stunnenberg HG. Mol Biol Rep. 1993;18:223. doi: 10.1007/BF01674434. [DOI] [PubMed] [Google Scholar]
- 5.Hochuli E, Bannwarth W, Döbeli H, Gentz R, Stüber D. Bio/Technology. 1988;6:1321. [Google Scholar]
- 6.Janknecht R, de Martynoff G, Lou J, Hipskind RA, Nordheim A, Stunnenberg HG. Proc Natl Acad Sci USA. 1991;88:8972. doi: 10.1073/pnas.88.20.8972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Van Dyke MW, Sirito M, Sawadogo M. Gene. 1992;111:99. doi: 10.1016/0378-1119(92)90608-r. [DOI] [PubMed] [Google Scholar]
- 8.Kaslow DC, Shiloach J. Bio/Technology. 1994;12:494. doi: 10.1038/nbt0594-494. [DOI] [PubMed] [Google Scholar]
- 9.Janknecht R, Nordheim A. Gene. 1992;121:321. doi: 10.1016/0378-1119(92)90137-e. [DOI] [PubMed] [Google Scholar]
- 10.Kuusinen A, Arvola M, Oker-Blom C, Keinänen K. Eur J Biochem. 1995;233:720. doi: 10.1111/j.1432-1033.1995.720_3.x. [DOI] [PubMed] [Google Scholar]
- 11.Grisshammer R, Tucker J. Protein Expr Purif. 1997;11:53. doi: 10.1006/prep.1997.0766. [DOI] [PubMed] [Google Scholar]
- 12.Crowe J, Döbeli H, Gentz R, Hochuli E, Stüber D, Henco K. Methods Mol Biol. 1994;31:371. doi: 10.1385/0-89603-258-2:371. [DOI] [PubMed] [Google Scholar]
- 13.Nikolov DB, Hu SH, Lin J, Gasch A, Hoffmann A, Horikoshi M, Chua NH, Roeder RG, Burley SK. Nature. 1992;360:40. doi: 10.1038/360040a0. [DOI] [PubMed] [Google Scholar]
- 14.David NE, Gee M, Andersen B, Naider F, Thorner J, Stevens RC. J Biol, Chem. 1997;272:15553. doi: 10.1074/jbc.272.24.15553. [DOI] [PubMed] [Google Scholar]
- 15.Skerra A. Gene. 1994;141:79. doi: 10.1016/0378-1119(94)90131-7. [DOI] [PubMed] [Google Scholar]
- 16.Porath J, Carlsson J, Olsson I, Belfrage G. Nature. 1975;258:598. doi: 10.1038/258598a0. [DOI] [PubMed] [Google Scholar]
- 17.Hochuli E, Döbeli H, Schacher A. J Chromatogr. 1987;411:177. doi: 10.1016/s0021-9673(00)93969-4. [DOI] [PubMed] [Google Scholar]
- 18.Chaga G, Hopp J, Nelson P. Biotechnol Appl Biochem. 1999;29:19. [PubMed] [Google Scholar]
- 19.Yang T-T, Nelson PS, Bush GL, Meyer DI, Kain SR. Am Biotechnol Lab I. 1997;12 [Google Scholar]
- 20.Wingfield PT. In: Current Protocols in Protein Science. Coligan JE, Dunn BM, Ploegh HL, DW, Wingfield PT, editors. Wiley; New York: 1995. p. 6.1.1. [Google Scholar]
- 21.Sinha D, Bakhshi M, Vora R. Biotechniques. 1994;17:509. [PubMed] [Google Scholar]
- 22.Flachmann R, Kühlbrandt W. Proc Natl Acad Sci USA. 1996;93:14966. doi: 10.1073/pnas.93.25.14966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Janssen JJ, Bovee-Geurts PH, Merkx M, DeGrip WJ. J Biol Chem. 1995;270:11222. doi: 10.1074/jbc.270.19.11222. [DOI] [PubMed] [Google Scholar]
- 24.Robbins DJ, Zhen E, Owaki H, Vanderbilt CA, Ebert D, Geppert TD, Cobb MH. J Biol Chem. 1993;268:5097. [PubMed] [Google Scholar]
- 25.Tucker J, Grisshammer R. Biochem J. 1996;317:891. doi: 10.1042/bj3170891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Pouny Y, Weitzman C, Kaback HR. Biochemistry. 1998;37:15713. doi: 10.1021/bi981519z. [DOI] [PubMed] [Google Scholar]
- 27.Lu T, Van Dyke M, Sawadogo M. Anal Biochem. 1993;213:318. doi: 10.1006/abio.1993.1427. [DOI] [PubMed] [Google Scholar]
- 28.Nilsson J, Larsson M, Ståhl S, Nygren PÅ, Uhlén M. J Mol Recognit. 1996;9:585. doi: 10.1002/(sici)1099-1352(199634/12)9:5/6<585::aid-jmr306>3.0.co;2-z. [DOI] [PubMed] [Google Scholar]
- 29.Kim JS, Raines RT. Anal Biochem. 1994;219:165. doi: 10.1006/abio.1994.1251. [DOI] [PubMed] [Google Scholar]
- 30.Panagiotidis CA, Silverstein SJ. Gene. 1995;164:45. doi: 10.1016/0378-1119(95)00417-5. [DOI] [PubMed] [Google Scholar]