Summary
Chromosome segregation in metazoans requires the alignment of sister-kinetochores onto the metaphase plate. During chromosome alignment, bioriented kinetochores move chromosomes by regulating the plus-end dynamics of the attached microtubules. The bundles of kinetochore-bound microtubules alternate between growth and shrinkage, leading to regular oscillations along the spindle axis. However, the molecular mechanisms that coordinate microtubule plus-end dynamics remain unknown. Here we show that CENP-H, a subunit of the CENP-A NAC/CAD kinetochore complex, is essential for this coordination, as kinetochores lacking CENP-H establish bioriented attachments, but fail to generate regular oscillations, due to an uncontrolled rate of microtubule plus-end turnover. These alterations lead to rapid erratic movements that disrupt metaphase plate organization. Moreover, we show that the abundance of the CENP-A NAC/CAD subunits CENP-H and CENP-I dynamically change on individual sister-kinetochores in vivo, as they preferentially bind the sister-kinetochore attached to growing microtubules, and that one other subunit, CENP-Q, binds microtubules in vitro. Thus, we propose that CENP-A NAC/CAD is a direct regulator of kinetochore-microtubule dynamics, which physically links centromeric DNA to microtubule plus-ends.
Introduction
Chromosome alignment is a multistep process, in which kinetochores establish stable bioriented attachments to the plus-end of spindle microtubules (MTs). The main players required for bioriented attachments are the KMN (Knl1-Mis12-Ndc80) kinetochore complex, which binds and stabilises end-on attached MTs1, and error-correction mechanisms based on the protein kinase Aurora B, which detaches improper kinetochore-MT (kMT) attachments by phosphorylating the KMN complex2. In mammalian cells, bioriented sister-kinetochore pairs on the metaphase plate undergo regular oscillations along the spindle axis driven by the coupled MT depolymerisation at the leading kinetochore and MT polymerisation at the trailing kinetochore3-5. Importantly, 25-30 MT plus-ends bind each kinetochore to form MT-bundles, known as kinetochore-fibres (k-fibres)6. Although k-fibres are embedded into kinetochores, the plus-ends of the individual MTs remain dynamic with a rate of tubulin turnover 20-fold slower than the plus-ends of free spindle-MTs7. The key to understanding how kinetochores generate directional movement and align chromosomes is to identify the proteins and molecular mechanisms that coordinate the dynamics of individual MT plus-ends within k-fibres.
Several MT-associated proteins (MAPs) accumulate onto end-on attached kinetochores and regulate kMT dynamics8, 9. These include the kinesins Kif18a and MCAK, which possess MT-depolymerising activity, and members of the CLASP and EB protein families. Kif18a and MCAK modulate the speed of kinetochore oscillations, but do not coordinate MT plus-end dynamics, as their depletion does not abrogate chromosome oscillations10, 11. EB1, which binds to growing plus-ends in vivo and in vitro, controls the assembly of entire plus-end binding protein-network12-14. Finally, CLASPs have been implicated in the regulation of kMT plus-end dynamics and the generation of poleward MT flux in various systems15-18. However, since these MAPs also associate with the plus-ends of free spindle-MTs and because free- and kinetochore-MT plus-ends have different tubulin turnovers, one has to assume that kinetochore-specific factors must coordinate kMT plus-end dynamics.
Consistent with this hypothesis, microinjection of antibodies against Hec1, a component of the MT-binding NDC80 complex, or introduction of a Hec1 mutant that cannot be phosphorylated by Aurora B, suppresses kMT tubulin turnover and disrupts chromosome alignment in Ptk1 cells19. However, this result reflects a gain of function of Hec1, since loss of Hec1 disrupts MT attachments20. Other potential candidates are the 15 proteins that associate with centromere-specific, CENP-A-containing nucleosomes, (CENP-H/I/K/L/M/N/O/P/Q/R/S/T/U/W/X) and form a protein network termed CENP-A NAC/CAD, ICEN or CCAN complex21-24. Loss of CENP-H, -I, -K or CENP-N, which disrupts the kinetochore localization of all of these proteins except CENP-T/W, does not affect the ability of kinetochores to bind MT plus-ends, but instead impairs chromosome congression23, 25-27. Moreover, depletion of CENP-O (also known as Mcm21R) disrupts kinetochore-localization of CENP-U/P/Q/R28 and destabilises k-fibres29, thus potentially linking CENP-A NAC/CAD to k-fibre regulation. Therefore, we investigated whether and how CENP-A NAC/CAD subunits contribute to the regulation of kMT plus-end dynamics.
Results
Loss of CENP-H disrupts sister-kinetochore oscillations and metaphase plate alignment
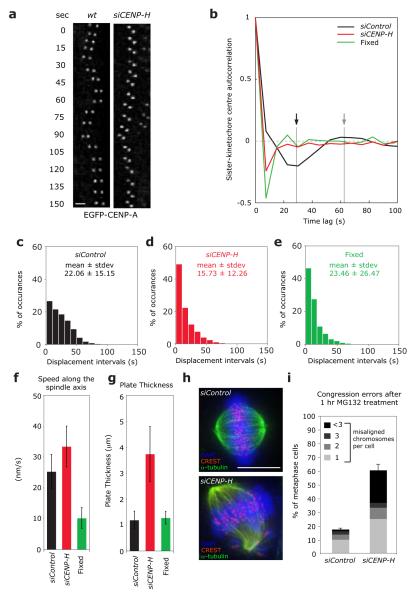
To investigate whether CENP-A NAC/CAD is involved in the control of kMT dynamics and kinetochore oscillations we depleted the CENP-H subunit, which is required for the loading of CENP-H/I/K/L/M/N/O/P/Q/R/S/X, but not CENP-T/W onto kinetochores (shown for CENP-H/I/K/L/O/P/N in Supplementary Information, Fig. S1, but see also22, 23, 25-27). Importantly in CENP-H-depleted cells CENP-C remains kinetochore-bound, confirming previous reports that CENP-C acts upstream of CENP-A NAC/CAD (Supplementary Information, Fig. 1; see25). To analyse sister-kinetochore oscillations, we used an EGFP-CENP-A HeLa cell line and a recently developed live-cell kinetochore-tracking assay, which measures sister-kinetochore dynamics on the metaphase plate30. To quantify the regularity of sister-kinetochore oscillations, we calculated the autocorrelation function of sister-kinetochore movements along the spindle axis. Sister-kinetochores in control-depleted cells behave as semi-regular oscillators (Fig. 1a, b), resulting in an autocorrelation curve in which the first negative side-lobe indicates the mean half-period, the second positive lobe indicates the mean full period of the oscillator, and the magnitude of the side-lobes reflects its regularity. In contrast, the autocorrelation curve of CENP-H-depleted or fixed cells was flat, indicating a lack of regular oscillations on the metaphase plate (Fig. 1a, b). In addition, in control-depleted cells the time intervals between directional switches were broadly distributed, whilst in CENP-H-depleted cells the distribution was identical to fixed cells, indicating random movements (Fig. 1c-e; kinetochores in fixed cells “move” randomly due to the inherent uncertainty in positional measurements). These sister-kinetochores were not immobile as their average speed was 40% higher than control-depleted cells and 5 times higher than fixed cells (Fig. 1f). CENP-H depletion also widened the average metaphase plate thickness (3.73 μm in siCENP-H-treated cells compared to 1.18 μm in control-depleted cells; Fig. 1g). These metaphase plate organization defects are the likely reason for the chromosome alignment defects in CENP-H-depleted cells (Fig. 1h, i)27, 31. We conclude that CENP-H loss abrogates regular chromosome oscillations and disorganizes metaphase plate alignment, suggesting a crucial role in the regulation of kMT plus-end dynamics.
Figure 1.
Loss of CENP-H abolishes sister-kinetochore oscillations and disrupts metaphase plate alignment. (a) Live cell stills of single kinetochore pair oscillations in wild-type or CENP-H-depleted EGFP-CENP-A HeLa cells. Scale bar = 2 μm (b) Autocorrelation function of translational sister-kinetochore movements along the spindle axis (kinetochore oscillation) of siControl (black line), siCENP-H (red line) or fixed cells (green line). The autocorrelation function was calculated by combining all aligned sister-kinetochore pairs for each condition (Supplementary Information, Table S2). The first negative lobe in control-depleted cells (black arrow) indicates the half-period of the mean oscillation period, while its depth indicates the oscillation regularity. The second positive lobe (gray arrow) indicates the full period. Note that a random motion always produces a negative value at the first lag30 (c-e) Histograms of the mean interval time between directional switches of the sister-kinetochore pairs along the spindle axis of siControl (black bars), siCENP-H (red bars) or fixed cells (green bars). For each condition, the mean values and SDs are indicated. (f) Average sister-kinetochore pair speed along the spindle axis of siControl (black bar), siCENP-H (red bar) or fixed cells (green bar). Number of independent experiments n = 3. Error bars represent SD. (g) Width of the metaphase plate of siControl (black bar), siCENP-H (red bar) or fixed cells (green bar). Number of independent experiments n = 3. Error bars represent SD. (h) Representative images of control or CENP-H-depleted cells arrested in metaphase for 1 hour with the proteasome inhibitor MG132 and stained with DAPI (DNA; blue), α-tubulin (MTs; green) and CREST antisera (kinetochores; red). (i) Percentage of cells with one, two, three, or more than three uncongressed chromosomes following a 1 hour treatment with MG132 calculated from images such as shown in (h). Chromosomes in metaphase cells were counted as unaligned if they were located outside the central 30% of the mitotic spindle. Number of independent experiments n = 3 with 20 cells each. Scale bar = 10 μm. Error bars represent SEM.
Loss of CENP-H hyperstabilises k-fibres and elevates plus-end tubulin turnover
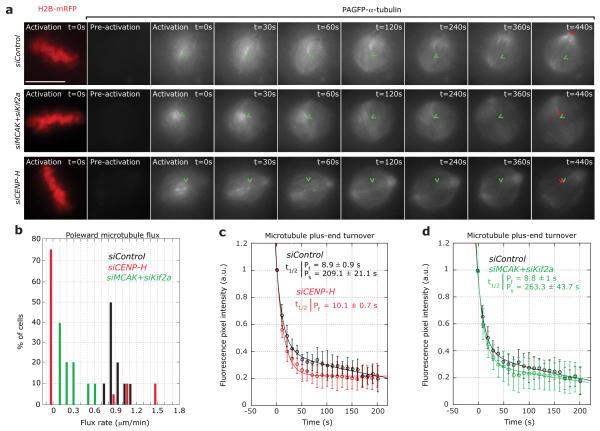
To test how CENP-H depletion affects kMT plus-end dynamics, we established a stable HeLa cell line expressing Histone-H2B-mRFP to identify metaphase cells, and photoactivatable (PA)GFP-α-tubulin to measure tubulin turnover at MT plus-ends. We generated fluorescent marks on the metaphase plate and monitored two parameters: (1) the movement of the marks towards the poles as a measure of poleward MT flux, which reflects MT treadmilling, and (2) the time-dependent decrease in fluorescence intensity of the activated marks, which provides a readout of tubulin turnover at MT plus-ends (expressed as the half-life (t1/2)). In control-depleted cells, the activated marks moved towards the spindle pole with a mean rate of 0.9 ± 0.3 μm/min, (Fig. 2a, b; Supplementary Information, Video S1). The rate of plus-end tubulin turnover was biphasic, reflecting the two previously reported MT populations7, 19: a fast population of free spindle-MTs (t1/2 = 8.9 ± 0.9 s, 75% of all MTs), and a second population with low turnover, which represents k-fibres (t1/2 = 209.1 ± 21.1 s, 25% of all MTs; Fig. 2a, c). In contrast, in CENP-H-depleted cells, MT flux was abrogated in 75% of the cells (Fig. 2a, b; Supplementary Information, Video S2). Moreover, the tubulin turnover at plus-ends was very rapid (Fig. 2a, c) and the data set fitted to a single exponential with a half-life similar to free spindle-MT plus-ends (t1/2 = 10.1 ± 0.7 s; Fig. 2c). As previously reported, co-depletion of the MT depolymerases Kif2a and MCAK did not affect plus-end tubulin turnover, even though MT flux was severely reduced, indicating that the high plus-end turnover rate in CENP-H-depleted cells was not due to reduced poleward MT flux (Fig. 2a, b, and d)32.
Figure 2.
Loss of CENP-H abrogates MT flux and abolishes control of kMT turnover (a) Successive frames every 30 seconds before and after photoactivation of stable PAGFP-α-tubulin/H2B-mRFP HeLa cells treated with siControl, siCENP-H or siMCAK+siKif2a RNAs. PAGFP-α-tubulin fluorescence was activated in a circular region near the chromosome mass in metaphase cells (detected by the H2B-mRFP signal). A H2B-mRFP (DNA) frame is shown for the first time point of the live-cell movie after activation. (b) Quantification of poleward MT flux rates in cells treated with siControl (black bars), siCENP-H (red bars) or siMCAK+siKif2a (green bars) RNAs. n = 20 cells each (c-e) Quantification of fluorescence intensity decay of the activated regions over time in siControl (black), siCENP-H (red) or siMCAK+siKif2a (green) treated cells. The lines through the data points were fitted to a double exponential equation of the type I = Pf.exp(−kf.t) + Ps.exp(−ks.t), which correspond to previously described slow and fast MT populations7, 19. Analysis of the siCENP-H fluorescence loss indicated that the data fitted to single exponential curve (R2 = 0.99). Indicated are the corresponding half-lives of the fast and slow MT populations. Green arrowheads mark the initial position of the photoactivated spot and the red arrowheads show the final position of the activated spot. Scale bar = 10 μm. Error bars represent SD.
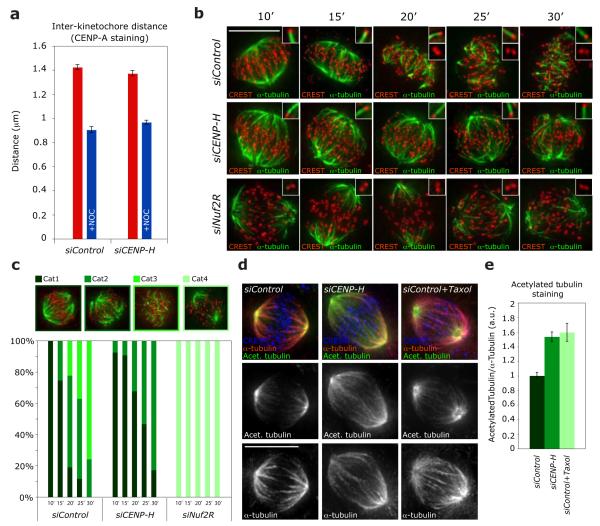
The presence of a single population of plus-ends, exhibiting a high turnover rate, could be explained: (1) either if CENP-H-depleted cells have a minimal number of MTs within k-fibres, which could explain the difficulty to coordinate directional movements, or (2) if CENP-H-depleted kinetochores cannot reduce the k-fibre plus-end turnover. To differentiate between those possibilities, we measured whether bioriented sister-kinetochores on the metaphase plate have normal inter-kinetochore distances, since a reduction in kMT numbers within a k-fibre decreases the inter-kinetochore distance33. We found that CENP-H- and control-depleted cells had the same inter-kinetochore distances and that this distance was reduced to the same extent when k-fibres were disrupted by the MT-depolymerising drug nocodazole (Fig. 3a). To visualise k-fibres we used a cold-stable assay34. A 10 min treatment with ice-cold medium depolymerised all non-kMTs in control-depleted cells, while longer treatments of up to 30 min progressively reduced k-fibres (Fig. 3b, c). Cells lacking the NDC80 complex component Nuf2R had no k-fibres after 10 min cold treatment, consistent with previous reports (Fig. 3b, c)20. In contrast, in CENP-H-depleted cells the k-fibre network was as intense as in control-depleted cells. Moreover, these k-fibres were more cold-resistant, as many long k-fibres persisted even after a 30 min cold treatment (Fig. 3b, c). This indicated that CENP-H-depleted cells have k-fibres with a normal number of MTs, and implied that their MT lattice is hyperstable, while their plus-ends have a high tubulin turnover. To analyse the turnover of the spindle-MT lattice, we measured the immunofluorescence ratio of acetylated α-tubulin to normal α-tubulin in mitotic spindles of siControl, siCENP-H or taxol treated cells. Tubulin acetylation is time-dependent and occurs preferentially on stable MTs, providing a readout of MT turnover35. Both treatment with taxol or CENP-H depletion increased the levels of tubulin acetylation by 55% when compared to control-depleted cells (Fig. 3d, e). We conclude that, in CENP-H-depleted cells, the k-fibre lattice is hyperstable, while the plus-ends are rapidly turned over.
Figure 3.
Loss of CENP-H increases kMT stability. (a) Average inter-kinetochore distances of fixed siControl or siCENP-H transfected EGFP-CENP-A cells treated with or without nocodazole. The quantification was based on the location of CENP-A signals in over 100 cells (>5 kinetochore pairs per cell) from 4 independent experiments without nocodazole and in 10 cells (>5 kinetochore pairs per cell) from 1 experiment with nocodazole. Error bars represent SEM. (b) Representative images of siControl, siCENP-H and siNuf2R transfected cells treated for 10, 15, 20, 25 or 30 min with ice-cold medium and stained with CREST antisera (kinetochores; red) and α-tubulin antibodies (MTs; green). Scale bar = 10 μm (c) Quantification of stable kMTs in cold-treated cells. Bar graph indicates the percentage of cells with all kMTs intact (Cat1), lacking few kMTs (Cat2), few short kMTs left (Cat3) or no kMTs (Cat4). (d) Representative images of cells treated with siControl, siCENP-H or siControl+Taxol RNAs and stained with acetylated-α-tubulin (stable MTs; green), α-tubulin (all MTs; red) and CREST antisera (kinetochores; blue). N = 50 cells Scale bar = 10 μm (e) Immunofluorescence quantification of the ratio of acetylated-α-tubulin to α-tubulin in mitotic spindles of cells after treatment with siControl, siCENP-H or siControl+Taxol from images such as shown in (d). n = 70 cells.
CENP-A NAC/CAD controls plus-end dynamics independently of Aurora B and differentially binds to kinetochores bound to growing or shrinking k-fibres
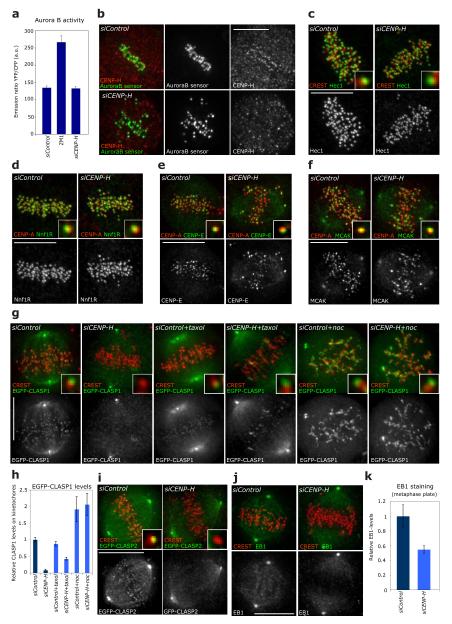
How does CENP-H control kMT plus-end turnover at the molecular level? One possibility is that the Aurora B/KMN network could be affected by loss of CENP-H. However, using a FRET-based reporter assay36, we find that CENP-H depletion does not influence the kinase activity of Aurora B (Fig. 4a, b). Moreover, multiple components of the KMN network, MCAK and CENP-E, all downstream targets of Aurora B, remained kinetochore-bound in the absence of CENP-H, indicating that CENP-H depletion affects kMT plus-end dynamics in an Aurora B- and KMN-independent manner (Fig. 4c-f, but also27, 37-40). In addition, CENP-H depletion did not impair the recruitment at kinetochores of p150Glued (dynein complex), CENP-F, the kinesin Kif18a, the spindle checkpoint proteins Bub1 and BubR1, the cohesion factor Sgo1 or Ska1, a component of the MT-binding Ska complex (Supplementary Information, Fig. S2)41. This indicated that CENP-H depletion does not act as a scaffold for kinetochore-bound MT-regulators.
Figure 4.
CENP-A NAC/CAD regulates kMT dynamics in an Aurora B/KMN independent manner (a) Graphs showing the Aurora B FRET-based sensor emission ratio of YFP/CFP for mock, ZM1 (Aurora B inhibitor) and siCENP-H-treated cells. An increase in emission ratio indicates dephosphorylation (lower Aurora B activity). Error bars represent SEM based on n = 3 independent experiments. (b) Representative images of siControl or siCENP-H-treated cells fixed after FRET analysis and stained with CENP-H antisera. (c-f) Representative images of siControl or siCENP-H RNA-treated cells stained with CREST or CENP-A (red) and Hec1 (c), Nnf1R (d), CENP-E (e) or MCAK (f) antisera (green). Insets show higher magnification views of a single kinetochore. (g) Representative images of HeLa cells stably expressing EGFP-CLASP1 treated with siControl or siCENP-H RNAs and either no drug, a 1μM taxol or a 1μg/ml nocodazole treatment. Cells were stained with CREST antisera (to mark kinetochores; red). Insets show higher magnification views of a single kinetochore. (h) Quantification of the EGFP-CLASP1 signal on kinetochores normalized with the signal at the spindle poles (see Supplementary Information, Fig. S3 for methodological details for indicated treatments) n > 25 kinetochores for each treatment. Error bars represent SEM based on n = 3 independent experiments. (i) Representative images of HeLa cells transiently expressing EGFP-CLASP2 treated with siControl, siCENP-H and stained with CREST antisera (to mark kinetochores; red). Insets show higher magnification views of a single kinetochore. (j) Representative images of siControl or siCENP-H RNA-treated cells stained with CREST (to mark kinetochores; red) and EB1 antisera (green). (k) Immunofluorescence quantification of EB1 levels on the metaphase plate region after treatment with siControl or siCENP-H RNAs from images such as shown in (j), n = 25 cells Error bars represent SEM based on n = 2 independent experiments. Scale bars = 10 μm.
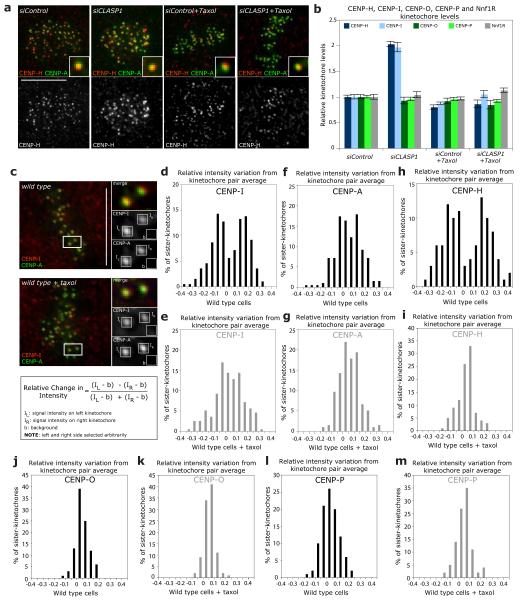
Another possibility is that CENP-A NAC/CAD provides a scaffold for the recruitment of MAPs to kinetochores. Quantitative immunofluorescence indicated that CENP-H loss abrogated the binding of CLASP1 and CLASP2 to kinetochores and led to a two-fold reduction in kinetochore-bound EB1 (Fig. 4g-k). This phenotype was specific, as depletion of the KMN components Nuf2R and Nnf1R (Mis12 complex) did not affect CLASP binding (Supplementary Information, Table S1). However, since blocking kMT plus-end turnover reduces the levels of CLASPs on kinetochores in Ptk1 cells19, we treated CENP-H-depleted cells either with taxol or with nocodazole to test whether the loss of CLASP on kinetochores reflects modified kMT dynamics. Such treatments restored the levels of kinetochore-bound CLASP1 to 42% and 103%, respectively, of control levels, indicating that CENP-H depletion affects CLASP levels at kinetochores because it changes MT dynamics (Fig. 4g, h).
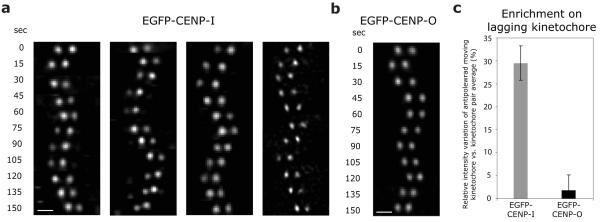
In the reciprocal experiments, CLASP1 depletion increased the levels of the CENP-A NAC/CAD components CENP-H and CENP-I on kinetochores by a factor of two, while leaving the levels of the CENP-A NAC/CAD components CENP-O and CENP-P or the levels of Nnf1R unaffected (Fig. 5a, b). Given that CLASPs regulate kMT dynamics15, this suggested that the stoichiometry of the CENP-A NAC/CAD network at kinetochores is modulated by changes in kMT dynamics. Consistent with this hypothesis, blocking kMT dynamics in siCLASP1-treated cells with taxol reduced the abundance of kinetochore-bound CENP-H and CENP-I to normal levels (Fig. 5a, b). We next determined whether CENP-H and CENP–I levels at kinetochores are also sensitive to the state of kMT dynamics in unperturbed cells. Metaphase kinetochore pairs have a leading sister-kinetochore bound to depolymerising kMTs, and a trailing sister-kinetochore bound to growing kMTs14. If CENP-I and CENP-H protein levels were sensitive to kMT dynamics then one would predict an asymmetry in CENP-I and CENP-H signal intensity between the two sisters. Using immunofluorescence we measured the CENP-I and CENP-H levels of each sister-kinetochore and calculated by how much the intensities on individual sister-kinetochores differed from the kinetochore pair average intensity (Fig. 5c). This revealed a bimodal distribution for both proteins, indicating the existence of two types of sister-kinetochore populations, one with CENP-I and CENP-H levels 15% smaller than the average, and the other one 15% higher than the average (Fig. 5d, h). The asymmetry in CENP-I and CENP-H levels was dependent on MT dynamics, as a taxol treatment yielded a unimodal distribution (Fig. 5e, i). In contrast, CENP-A, CENP-O and CENP-P had a unimodal distribution peaking at 0 in untreated or taxol treated cells, indicating that their levels are unaffected by changes in kMT dynamics (Fig. 5f, g, j-m). This implied that CENP-A NAC/CAD differentiates between kinetochores bound to growing or shrinking MTs, by incorporating more or less CENP-I/H in response to changes in MT dynamics. To test this model, we established stable cell lines expressing either EGFP-CENP-I or EGFP-CENP-O. Using live cell imaging, we found that the levels of CENP-I on oscillating kinetochore pairs were asymmetric, as the protein accumulates preferentially at anti-poleward moving kinetochores attached to growing MTs (30% higher levels; Fig. 6a, c; Supplementary Information, Video S3). In contrast, the levels of CENP-O were symmetric (Fig. 6b, c). We conclude that CENP-I and -H accumulate on the sister-kinetochores bound to growing k-fibres in a manner similar to EB1 or Kif18a10, 14, suggesting that CENP-A NAC/CAD is a direct regulator of kMT plus-ends.
Figure 5.
CENP-H and CENP-I differentially binds to kinetochores attached to growing kinetochore fibres. (a) Representative images of siControl, siCLASP1, siControl+Taxol or siCENP-H+Taxol cells stained with CENP-A antisera (to mark kinetochores; green) and CENP-H antisera (red). Insets show higher magnification views of a single kinetochore. (b) Immunofluorescence quantification of CENP-H, CENP-I, CENP-O, CENP-P and Nnf1R kinetochore levels in cells treated with siControl, siCENP-H, siControl+Taxol or siCENP-H+Taxol RNAs from images such as shown in (a). Error bars represent SEM based on n = 3 independent experiments (c-m) To calculate the asymmetry of the CENP-I (d, e), CENP-A (f, g), CENP-H (h, i), CENP-O (j, k) and CENP-P (I, m) intensities on sister-kinetochore pairs, we determined the intensities of the “left” sister-kinetochore (IL), the “right” sister-kinetochore (IR) and the background value (b) as schematized in (c). If one assumes that x is the average signal intensity of one pair, then the left sister-kinetochore will have an intensity of IL = x + Δ, while the right sister-kinetochore will have an intensity of IR= x - Δ (Δ being the relative positive or negative amount the two sister-kinetochores deviate from the average). By dividing the difference of the background-subtracted intensities over their sum one obtains Δ/x, which indicates by how much the intensity of the left kinetochore is larger or smaller than the average of the two sisters. We then plotted the distribution of Δ/x in cells treated with or without taxol as indicated. A bimodal distribution indicates the existence of two separate sister-kinetochore populations, while a unimodal distribution indicates the absence of protein level asymmetry. Note that CENP-I and CENP-A intensities were measured from the same sister-kinetochores (n = 200 kinetochore pairs for CENP-I and CENP-A; n = 100 kinetochore-pairs for CENP-H, CENP-O and CENP-P). Scale bars = 10 μm.
Figure 6.
CENP-I preferentially accumulates on the sister-kinetochore bound to growing MTs (a, b) Representative live cell imaging frames of single oscillating sister-kinetochore pairs of wild type EGFP-CENP-I (x4) or EGFP-CENP-O (x1) cells. (c) Enrichment of EGFP-CENP-I or EGFP-CENP-O on anti-poleward moving sister-kinetochores. The quantifications and calculations were done as in Figure 5c, but in live cells and defining that the “left” kinetochore is the sister-kinetochore that moves in antipoleward manner and is bound to growing MTs. Error bars represent SEM based on n = 4 (EGFP-CENP-I) and n = 3 (EGFP-CENP-O) independent experiments. Scale bars = 2 μm.
CENP-A NAC/CAD subunits interact directly with MTs
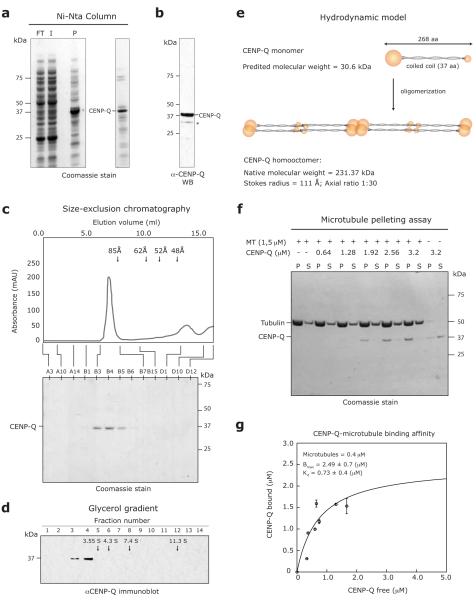
We next tested whether CENP-A NAC/CAD subunits can physically interact with MTs in vitro. Expression of individual CENP-A NAC/CAD subunits in E. coli was unsuccessful (data not shown). Previous studies have, however, demonstrated that co-expression of chicken CENP-P, CENP-Q, CENP-U and CENP-O can form a stable subcomplex28. We, therefore, attempted to express the human orthologues of these proteins in addition to CENP-H and CENP-S in E. coli from three separate expression vectors in which CENP-Q contained an amino-terminal His-tag. Elution from a nickel-NTA column, mass spectroscopy and immunoblotting analysis of eluting proteins revealed a fraction that contained CENP-Q, low amounts of CENP-P and other contaminating bands (Fig. 7a, b; data now shown). A second purification step by size-exclusion chromatography yielded a fraction with Stoke radius of 111 Å that only contained CENP-Q (Fig. 7c). Analysis by glycerol gradient centrifugation revealed a Svedberg coefficient of 2.34 S for CENP-Q (Fig. 7d). By combining this experimental S-value with the Stokes radius we calculated that soluble CENP-Q forms a homooctameric complex with a native molecular weight of 260 kDa and an axial ratio of around 30:1 (assuming the complex to be a prolate ellipsoid, Fig. 7e)42, 43. Using a MT-pelleting assay, we tested whether CENP-Q complex can bind MTs. In the absence of taxol-stabilised MTs, CENP-Q protein remained in the supernatant, whereas, in the presence of MTs, CENP-Q appeared in the pellet fractions in a concentration-dependent manner, indicating that CENP-Q binds MTs in vitro (Fig. 7f). We determined the stoichiometry of CENP-Q:MT binding by titration of CENP-Q using a constant MT concentration. Saturation of binding was at a ratio of ~6:1 with a fitted Kd of ~0.7 μM (Fig. 7g). This implies that one CENP-Q homooctamer binds to one tubulin heterodimer, indicating that CENP-Q binds the MT lattice and does not preferentially bind MT plus-ends. Since the MT-affinity of CENP-Q is better than the bona fide MT-binding NDC80 complex (3 mM1) and similar to that of the KMN network (Kd of ~0.5 μM), or the Ska complex (Kd of ~0.3 μM)1, 44, we conclude that CENP-Q, directly binds MTs in vitro.
Figure 7.
The CENP-A NAC/CAD subunit CENP-Q makes direct physical interactions with MTs. (a) SDS-PAGE and coomassie staining of flow-through (FT), input (I), pellet (P) and elution fractions from nickel-NTA chromatography. The band corresponding to CENP-Q is indicated based on mass spectroscopy analysis. (b) Immunoblot of the eluted fraction with anti-CENP-Q antisera. Asterix-marked band corresponds to degradation products of CENP-Q (c) Hydrodynamic analysis of CENP-Q on size exclusion chromatography. Arrows indicate the elution volumes of standards with known Stokes radius: Thyroglobulin (85 Å), Ferritin (62 Å), Catalase (52 Å) and Aldolase (48 Å). (d) Fractions from 5-40% glycerol-gradient analysis of CENP-Q. Arrows indicate the migration of standards with known S values: Catalase (11.3 S), Aldolase (7.4 S), BSA (4.3 S) and Ovalbumin (3.55 S). (e) Model for the predicted organization of CENP-Q based on the biochemical experiments. (f) MT-pelleting assay performed in BRB80 + 50 mM NaCl, 1 mM ADP using 1.5 μM taxol-stabilised pig-brain MTs and purified CENP-Q protein in different concentrations (0.64-3.2 μM) as indicated. Supernatant (S) and pellet (P) fractions are shown. (g) Quantification of CENP-Q:MTs binding affinity. Quantification was performed using 0.4 μM taxol-stabilised pig-brain MTs and purified CENP-Q protein (0.64-3.2 μM). The average of multiple samples were plotted. Error bars indicate SD.
Discussion
In this study, we investigated the potential contribution of the CENP-A NAC/CAD complex to the regulation of kMT dynamics and found that CENP-H is required for controlled kMT plus-end turnover and regular chromosome oscillations. Although sister-kinetochore oscillations have been under investigation for over 70 years4, their exact function remains unclear. Our CENP-H depletion data imply that kinetochore oscillations are essential to precisely find the spindle equator and establish an organized metaphase plate. Moreover, our data suggest that CENP-A NAC/CAD directly binds MTs. Therefore, we propose that CENP-A NAC/CAD provides a direct physical and functional link between centromeric nucleosomes and MT plus-end coordination that is required for precise chromosome alignment.
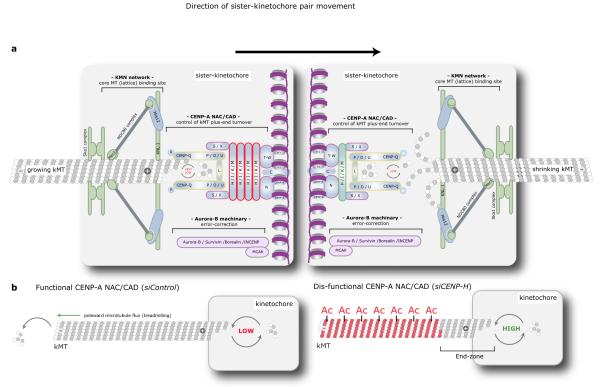
Kinetochores contain two conserved core complexes: KMN and CENP-A NAC/CAD, which are ubiquitous in metazoans, fungi, plants and protists45. Previous studies proposed that MT-attachment and regulation of kMT dynamics are both regulated by the KMN-Aurora B pathway, while CENP-A NAC/CAD was thought to act as structural scaffold within the kinetochore37, 46. Indeed, microinjection of Ndc80 antibodies or expression of a non-phosphorylatable Ndc80 mutants lead to hyperstable kMTs and low plus-end kMT turnover19, suggesting that MT plus-end turnover and kMT stability are linked. However, our data show that these two functions are separate since it is possible to have high plus-end kMT turnover that is equivalent to free spindle-MTs, without decreasing overall kMT stability. We propose an alternative model, in which the two essential mechanical functions of kinetochores are controlled in a separate manner: (1) lateral k-fibre binding and stabilisation by a machinery comprising KMN, chromosome passenger and perhaps the Ska complexes2, 44, 47; (2) regulation of kMT plus-end dynamics by CENP-A NAC/CAD (Fig. 8a). Kinetochores lacking CENP-H, and thus lacking all CENP-A NAC/CAD subunits, with the exception of Histone H3-binding CENP-T/W subunits, still recruit and stabilise kMTs through attachments to the MT lattice. However, these kMT plus-ends undergo fast tubulin turnover, resulting in rapid switches between growth and shrinkage, whilst leaving the distal lattice region stable (Fig. 8b). The high turnover rate explains the absence of poleward MT flux in 75% of the cells, as newly incorporated tubulin subunits are rapidly removed from the plus-ends, thus preventing MT treadmilling (we also observe an increase of MT nucleation at spindle poles, which explains why spindles do not shorten; unpublished observation, A.C.A., C.P.S, A.D.M and P.M). Our model is consistent with the fact that CENP-H depletion affects kMT plus-end dynamics independently of Aurora B and KMN. Finally, we propose that CENP-A NAC/CAD does not act as a scaffold, but is a bona fide direct regulator of kMT plus-ends. Indeed, recent high-resolution imaging of human kinetochore proteins shows that CENP-A NAC/CAD is located in the immediate vicinity of the kMT plus-ends48. Moreover, the CENP-A NAC/CAD components CENP-I/H preferentially accumulate on kinetochores bound to growing MTs - a differential behaviour only known for the direct MT regulators EB1 and Kif18a10, 14. Finally, our MT-pelleting experiments indicate that CENP-Q binds MTs in vitro with an affinity that is comparable to the Ska complex and better than the NDC80 complex. In the future, it will be crucial to investigate the molecular mechanisms that control the preferential binding of CENP-H/-I to antipoleward moving sister-kinetochores in such a dynamic manner, and to determine whether additional CENP-A NAC/CAD subunits bind MTs.
Figure 8.
Model for the function of the CENP-A NAC/CAD in controlling kMT dynamics (a) Schematic view of the human kinetochore-MT interface showing the outer KMN and Ska complexes as well as the inner CENP-A NAC/CAD complex. While the Ndc80/Nuf2 and KNL-1 subunits and Ska1/Ska2 subunits are essential for MT attachment, CENP-A NAC/CAD regulates the turnover of kMT plus-ends, by controlling dynamic instability. Importantly, there is a differential stoichiometry of subunits within the CENP-A NAC/CAD complex that is dependent on whether a sister-kinetochore is bound to growing or shrinking MTs. The sister-kinetochore bound to growing microtubules contains high levels of the CENP-H, -I, -K, -M complex (red), whereas the sister-kinetochore bound to shrinking MTs contain low levels of this complex (green). (b) Explanatory model for the lack of correlation between kMT stability and plus-end MT turnover. In control cells, kMT plus-ends exhibit low rates of dynamic instability allowing continual incorporation of new tubulin heterodimers. In contrast, in the absence of the CENP-A NAC/CAD complex, kinetochores still bind and stabilise kMTs through attachments to the MT lattice, but the dynamic instability at the plus-ends is dramatically increased. This results in a high turnover of the MT lattice proximal to the plus-ends (the “end-zone”), due to the elevated frequency of catastrophe and rescue events. Presumably these changes in kMTs dynamics prevent the establishment of regular kinetochore oscillations (see discussion for detail). Moreover, the MT lattice distal to the end-zone becomes hyperstable, causing an interruption of poleward MT flux, as MTs cannot undergo treadmilling any more.
CENP-H is essential for the generation of regular chromosome oscillations and metaphase plate organization, indicating that it is necessary for the coordination of individual MTs within k-fibres. We propose that this function relies on the ability of CENP-A NAC/CAD to control tubulin turnover at the kMT plus-ends. Previous studies found that the rate of tubulin turnover is tightly controlled during chromosome segregation, as it is 20 times lower compared to free spindle-MTs during metaphase7. We propose that kMT plus-ends coordination requires the damping of tubulin turnover. Only within this “kinetochore microenvironment”, provided by CENP-A NAC/CAD, can MT regulatory factors influence the behaviour of multiple kMTs in parallel and generate chromosome oscillations. Without such damping the multiple MT plus-ends may be too dynamic and the influence of regulatory factors is drowned in a sea of rapid growth and shrinkage events. Strikingly the stoichiometry of the CENP-A NAC/CAD network is not the same on sister-kinetochores bound to growing or shrinking k-fibres, as CENP-I/H preferentially bind to kinetochores attached to growing MTs, whilst two subunits of the CENP-O subcomplex28, CENP-P/O, do not show any preference (Fig. 8a). As we have previously found, the CENP-A NAC/CAD complex is regulated through an internal feedback loop, in which proteins of the CENP-O complex regulate the function of CENP-H/I27, we speculate that the CENP-A NAC/CAD differentially regulates MT turnover on both sister-kinetochores. In particular, the turnover might be lower at the sister-kinetochore bound to growing MTs to suppress a sudden burst of depolymerisation, which could disrupt regular oscillations. Such an asymmetric distribution could be part of a feedback mechanism that is predicted by modelling to be necessary for the emergence of chromosome oscillations49. Therefore, it will be key in the future to determine whether these changes only respond to evolving MT dynamics, or whether they play a more pro-active role in the regulation of kinetochore oscillations.
Material and Methods
Cell culture, siRNA transfection, and drug treatments
HeLa cells were grown in Dulbecco’s modified medium (DMEM) containing 10% FCS, 100 U/ml penicillin and 100 μg/ml streptomycin at 37°C with 5% CO2 in a humidified incubator. The stable HeLa EGFP-CENP-A and HeLa EGFP-CLASP1 cell lines were grown as described16, 30. To establish the stable HeLa cell line expressing Histone H2B-mRFP and photoactivatable GFP-α-tubulin, H2B-mRFP cells were transfected with a PAGFP-α-tubulin-IRES-puro vector using FuGENE6 (Roche) and positive cells selected in the presence of 500 ng/ml puromycin. The cell line was maintained in DMEM containing 10% FCS, 100 U/ml penicillin, 100 μg/ml streptomycin, 250 ng/ml puromycin and 500 μg/ml G418. To obtain the stable cell line EGFP-CENP-I and EGFP-CENP-O, HeLa cells were transfected with an EGFP-CENP-I-IRES-puro or EGFP-CENP-O-IRES-puro vector using FuGENE6 (Roche). Both cells lines were selected and grown in DMEM containing 10% FCS, 100 U/ml penicillin, 100 μg/ml streptomycin and 500 ng/ml puromycin. Live-cell imaging experiments were performed at 37°C in Lab-Tek II chambers (Thermo Fisher Scientific) with Leibovitz L-15 medium containing 10% FCS. Short RNAi oligonucleotides (Qiagen or Invitrogen; Control (LaminA)50, CENP-H (oligo 1 and 2)27, CENP-L27, CLASP129, MCAK51, CENP-P (5′-GAACCCTGGTAGGACTGCTTGGAAT-3′), EB152, Kif2a53, Nuf2R54, Nnf1R55) were transfected as described and analysed 48 or 72 hrs after transfection50. Note that the most critical CENP-H experiments were repeated with a second CENP-H siRNA oligonucleotide, which gave qualitatively the same results (Supplementary Information, Fig. S4). All siRNA treatments were validated by immunofluorescence or immunoblotting (Supplementary Information, Fig. S5). For drug treatments, HeLa cells were treated with 1 μM MG132 for 1hr, 1 μM taxol for 20 min, or 1 μg/ml nocodazole for 1 hr before fixation for immunofluorescence. Transient transfection of EGFP-CLASP256 was performed using FuGENE 6 (Roche).
Antibody production
Rabbit antibodies were raised against Rubredoxin-MCAK expressed in Escherichia coli and purified from inclusion bodies (NeoMPS). Peptide antibodies were raised in rabbits against (CENP-P) CRRACEDPPAPWEEKSRVQK, (CENP-L) CSRPEDYFIGATPLQKRLE and (CENP-Q) CKKNAQQLKRNPKRKKDNEE peptides (Pepceuticals). Rabbit anti-CENP-P antibodies were affinity-purified against the immunizing peptide using a Microlink Peptide Coupling Kit (Thermo Scientific).
Immunofluorescence microscopy
Cells were fixed at room temperature for 8 min in 20 mM Pipes (pH 6.8), 10 mM EGTA, 1 mM MgCl2, 0.2% Triton X-100, 4% formaldehyde. For cold-stable assays, cells were incubated on ice-cold medium 10-30min before fixation. The following primary antibodies were used: rabbit anti-MCAK (1:2000; this study), mouse anti-EB1 (1:500; BD biosciences), rabbit anti-CENP-H (1:4000)27, rabbit anti-CENP-I (1:1000)27, rabbit anti-CENP-O (1:500)55, rabbit anti-CENP-P (1:500; this study), rabbit anti-Nnf1R (1:1000)55, mouse anti-CENP-A (1:1000; Abcam), mouse anti-α-tubulin (1:10000; Sigma-Aldrich), CREST antisera (1:400; Antibodies Incorporated), rabbit anti-α-tubulin (1:1000; Abcam), mouse anti-acetylated-α-tubulin (1:1000; Sigma-Aldrich), rabbit anti-CENP-E (1:1500)54, rabbit CENP-N (1:2000)27, rabbit anti-CENP-K (1:1000)27, rabbit anti-CENP-L (1:1000; this study), mouse anti-CENP-C (1:1000; Abcam), rabbit anti-CENP-F (1:2000; Novus Biologicals), mouse anti-p150Glued (1:500; BD biosciences), rabbit anti-Kif18a (1:1000; Bethyl Laboratories), rabbit anti-Ska1 (1:500)57, rabbit anti-Bub1 (1:8000)58, mouse anti-BubR1 (1:1500; Abcam), rabbit anti-Sgo1 (1:500; Abcam). Cross-adsorbed secondary antibodies were used (Invitrogen). 3D image stacks of mitotic cells were acquired in 0.2 μm steps using a 60x or 100x oil NA 1.4 objective on an Olympus DeltaVision microscope (Applied Precision, LLC) equipped with a DAPI-FITC-Rhod/TRCY5 filter set (Chroma) and a CoolSNAP HQ camera (Roper Scientific). The 3D image stacks were deconvolved with SoftWorx (Applied Precision, LLC) and mounted in figures using Photoshop and Illustrator (Adobe). For quantitative measurements, signals were determined and quantified with SoftWorx as described in Supplementary Information, Figure S3.
Live still frames and live cell imaging
Fluorescence time-lapse imaging of the EGFP-CENP-I or EGFP-CENP-O cell lines was recorded with a 100x NA 1.35 objective on an Olympus DeltaVision microscope (Applied Precision, LLC) equipped with a CoolSNAP HQ2 camera (Roper Scientific) and a EGFP filter set (Chroma) at a temporal resolution of 15 s for a total time of 3 min. For each time point 20 z-sections 0.5 μm apart were acquired. Live still frames were assembled in Adobe Illustrator by manually tracking sister-kinetochore pairs, cropping them out and adjusting the intensity levels (including gamma function) in Adobe Photoshop in order to compensate for photobleaching. These treatments were always applied to the whole image, i.e. both sister-kinetochores in a pair were subjected to the same adjustments.
Photoactivation experiments
Photoactivation experiments were performed on bipolar metaphase spindles (identified by the H2B-mRFP signal) as described previously29 using a 50 ms pulse from a 406 nm laser (20%) on a DeltaVision RT microscope equipped with a quantifiable laser module (Applied Precision, LLC). Fluorescence images were captured every 10 s for 530 s using a 100x oil NA 1.4 objective and FITC filter set (Applied Precision, LLC). Fluorescence intensities were measured in a 20 × 20 pixel area using SoftWorx (Applied Precision, LLC) over the first 200 s of each movie. Background spindle signal was subtracted for each time frame by measuring the same pixel area on the opposite side of the photoactivated spindle. Cells that underwent anaphase in this period were discarded, assuring that all measurements were taken from cells in late prometaphase. The values were corrected for photobleaching by determining the percentage of fluorescence loss during the first 200 s in activated cells treated with 10 μM taxol. The average data was fitted to a double exponential curve I = Pf.exp(-kf.t) + Ps.exp(−ks.t) where “I” is the proportion of the initial fluorescence intensity, “P” is the proportion of fluorescence decay due to the fast (f) or slow (s) process, “k” is the rate constant for fluorescence decay of the fast (f) or slow (s) process. Curve fitting was performed using KaleidaGraph (Synergy Software). The initial parameters Pf, kf, Ps and ks were set as 1, 0.1, 0.2 and 3.3×10−3 respectively, based on previous results7. The turnover half time (tl/2) for each process was calculated as ln2/k for each fast and slow process.
Kinetochore tracking assay
For the kinetochore tracking experiments, fluorescence time-lapse imaging of an EGFP-CENP-A cell line was recorded with a 100x NA 1.35 objective on an Olympus DeltaVision microscope (Applied Precision, LLC) equipped with a CoolSNAP HQ2 camera (Roper Scientific) and a EGFP filter set (Chroma) at a temporal resolution of 7.5s and subjected to the kinetochore tracking assay analysis30. For each time-lapse movie, the position of the metaphase plate was estimated by fitting a plane to the calculated kinetochore positions. To characterize the dynamics of individual sister-kinetochore pairs located on the metaphase plate over time, we followed sister-kinetochore centre position along the normal to the metaphase plate (spindle axis). The autocorrelation function of sister-kinetochore movements along the spindle axis yielded the periodicity of sister-kinetochore oscillations. The displacement intervals correspond to the duration between consecutive directional switches of sister-kinetochore pairs. The average speed of sister-kinetochores along the spindle axis was calculated as the standard deviation of the distribution of all sister-kinetochore frame-to-frame movements. The metaphase plate thickness was calculated as the standard deviation of the distribution of aligned sister centre positions along the normal to the metaphase plate.
FRET-based sensor for Aurora B activity
HeLa cells were transfected first with 200 nM CENP-H siRNAs using Oligofectamine (Qiagen) and then 24 hrs later with centromere-targeted Aurora B phosphorylation sensor36 using FuGENE6 (Roche). Cells were imaged live 48 hrs after the sensor transfection, and mitotic cells randomly selected. Imaging and analysis were performed as previously described59. For treatment with the Aurora B kinase inhibitor ZM1, cells were treated with 2 μM for 1 hr before imaging. For immunofluorescence staining, cells were fixed at −20°C with pre-cooled methanol for 10 min. Images of fixed cells were acquired using the same spinning disk confocal as for the live cell imaging.
Immunoblotting
Whole-cell lysate preparation and immunoblotting were carried out as described55, using rabbit anti-CLASP1 (1:500)15, mouse anti-α-tubulin (1:10000; Sigma-Aldrich) and rabbit anti-CENP-Q (1:500; this study) as and anti-mouse or anti-rabbit HRP conjugated secondary antibodies (Amersham).
Protein purification, mass spectroscopy and MT-binding assays
Full-length CENP-A NAC/CAD genes were amplified from cDNAs (GENEART) and cloned into E. coli expression vectors (Novagen). The genes for CENP-Q and CENP-P were inserted into pRSFDuet-1 with an amino-terminal his-tag on CENP-Q. CENP-S and CENP-O genes were inserted into pCDFDuet-1 and CENP-H and CENP-U genes into pACYCDuet-1. The plasmids were co-transformed into BL21-CodonPlus-(DE3) E. coli and expression induced with 0.5mM IPTG, shaking overnight at 15 °C. Cell pellets were resuspended in 50 mM Tris-Cl, 100 mM NaCl, 30 mM imidazole and 5 mM β-mercaptoethanol and lysed with 0.1 mg/ml lysozyme for 1hr at 4°C. Extracts were clarified at 16000 rpm for 40 min at 4 °C. Clarified extracts were purified with a Ni-NTA column (Qiagen) and the proteins eluted with a 500 mM imidazole gradient. Proteins were analysed by SDS-PAGE and simply blue staining (Invitrogen). Proteins in each band were identified by trypsin digest and microcapillarity liquid chromatography-tandem mass spectrometry (MC-MS/MS) at the FingerPrints Proteomics Facility, University of Dundee. Hydrodynamic analysis of CENP-Q was performed as described60, except that we used a Superose 12 10/300 GL (GE Healthcare) for the size-exclusion chromatography and all the assays were done in 50 mM Tris-Cl, 100 mM NaCl, and 5 mM β-mercaptoethanol buffer. MT pelleting assays were performed as described previously61, except that we used BRB80 buffer (80 mM Pipes pH 6.8, 1 mM MgCl2 and 1 mM EGTA) containing 50 mM NaCl. The concentration of CENP-Q and tubulin in micromolar used in the MT pelleting assays was calculated as follows: [CENP-Q or tubulin] = Abs(280 nm)/ε, where the extinction coefficient (ε) for CENP-Q = 8.25×10^−3 and for tubulin = 0.10583. The concentrations were confirmed by Commasie Blue Staining and comparison with a BSA standard curve. To obtain the CENP-Q:MT binding affinity parameters, the amounts of CENP-Q bound to MT (corrected for any insoluble material found in the fraction lacking MTs) and not bound to MT were obtained by quantification of the MT pelleting assays using Quantity One software (Bio-Rad Laboratories). The average data obtained was fitted to a hyperbolic function y=(Bmax*x)/(Kd+x) where “y” is the amount of CENP-Q bound to MT and “x” is the amount of CENP-Q not bound to MT, obtained previously. Curve fitting was performed using KaleidaGraph (Synergy Software).
Supplementary Material
Acknowledgements
We thank the ETH Light Microscopy Centre and Yves Barral for microscopy support; Anna Akhmanova, Helder Maiato, Michel Steinmetz, Daniel Gerlich, Erich Nigg, Stephan Diekmann and Jennifer Lippincot-Schwarz for reagents, Jason Swedlow, Gaudenz Danuser for the joint development of the kinetochore tracking assay, Satyarebala Borusu for the EGFP-CENP-O cell line, Jennifer Winter for initial photoactivation observations, Itsaso Olasagasti for helping with the CENP-I intensities calculations and Kunyoshi Kaseda for helping analyzing the photoactivation experiments. The authors are grateful to Yves Barral, Monica Gotta, Helder Maiato, Jonathon Pines and members of the Barral, McAinsh and Meraldi labs for helpful discussions. Work in the McAinsh lab was supported by Marie Curie Cancer Care (A.D.M. and C.P.S.) and by a Fundação para a Ciência e Tecnologia fellowship (C.P.S.). A.C.A. is a member of the Life Science Zurich Graduate School in Molecular Life Sciences. Work in the Meraldi lab (A.C.A, R.H. and P.M.) was supported by a SNSF-Förderungprofessur and a EURYI award.
References
- 1.Cheeseman IM, Chappie JS, Wilson-Kubalek EM, Desai A. The conserved KMN network constitutes the core microtubule-binding site of the kinetochore. Cell. 2006;127:983–997. doi: 10.1016/j.cell.2006.09.039. [DOI] [PubMed] [Google Scholar]
- 2.Tanaka TU, Desai A. Kinetochore-microtubule interactions: the means to the end. Curr Opin Cell Biol. 2008;20:53–63. doi: 10.1016/j.ceb.2007.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Skibbens RV, Skeen VP, Salmon ED. Directional instability of kinetochore motility during chromosome congression and segregation in mitotic newt lung cells: a push-pull mechanism. J Cell Biol. 1993;122:859–875. doi: 10.1083/jcb.122.4.859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lewis WH. Changes of viscosity and cell activity. Science. 1939;89:400. [Google Scholar]
- 5.Hughes AF, Swann MM. Anaphase movements in the living cell. J. Exp. Biol. 1948;25:45–70. [Google Scholar]
- 6.Rieder CL. The structure of the cold-stable kinetochore fiber in metaphase PtK1 cells. Chromosoma. 1981;84:145–158. doi: 10.1007/BF00293368. [DOI] [PubMed] [Google Scholar]
- 7.Zhai Y, Kronebusch PJ, Borisy GG. Kinetochore microtubule dynamics and the metaphase-anaphase transition. J Cell Biol. 1995;131:721–734. doi: 10.1083/jcb.131.3.721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Akhmanova A, Steinmetz MO. Tracking the ends: a dynamic protein network controls the fate of microtubule tips. Nat Rev Mol Cell Biol. 2008;9:309–322. doi: 10.1038/nrm2369. [DOI] [PubMed] [Google Scholar]
- 9.Maiato H, DeLuca J, Salmon ED, Earnshaw WC. The dynamic kinetochore-microtubule interface. J Cell Sci. 2004;117:5461–5477. doi: 10.1242/jcs.01536. [DOI] [PubMed] [Google Scholar]
- 10.Stumpff J, von Dassow G, Wagenbach M, Asbury C, Wordeman L. The kinesin-8 motor Kif18A suppresses kinetochore movements to control mitotic chromosome alignment. Dev Cell. 2008;14:252–262. doi: 10.1016/j.devcel.2007.11.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wordeman L, Wagenbach M, von Dassow G. MCAK facilitates chromosome movement by promoting kinetochore microtubule turnover. J Cell Biol. 2007;179:869–879. doi: 10.1083/jcb.200707120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Honnappa S, et al. An EB1-binding motif acts as a microtubule tip localization signal. Cell. 2009;138:366–376. doi: 10.1016/j.cell.2009.04.065. [DOI] [PubMed] [Google Scholar]
- 13.Bieling P, et al. Reconstitution of a microtubule plus-end tracking system in vitro. Nature. 2007;450:1100–1105. doi: 10.1038/nature06386. [DOI] [PubMed] [Google Scholar]
- 14.Tirnauer JS, Canman JC, Salmon ED, Mitchison TJ. EB1 targets to kinetochores with attached, polymerizing microtubules. Mol Biol Cell. 2002;13:4308–4316. doi: 10.1091/mbc.E02-04-0236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Maiato H, et al. Human CLASP1 is an outer kinetochore component that regulates spindle microtubule dynamics. Cell. 2003;113:891–904. doi: 10.1016/s0092-8674(03)00465-3. [DOI] [PubMed] [Google Scholar]
- 16.Pereira AL, et al. Mammalian CLASP1 and CLASP2 cooperate to ensure mitotic fidelity by regulating spindle and kinetochore function. Mol Biol Cell. 2006;17:4526–4542. doi: 10.1091/mbc.E06-07-0579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hannak E, Heald R. Xorbit/CLASP links dynamic microtubules to chromosomes in the Xenopus meiotic spindle. J Cell Biol. 2006;172:19–25. doi: 10.1083/jcb.200508180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Maiato H, Khodjakov A, Rieder CL. Drosophila CLASP is required for the incorporation of microtubule subunits into fluxing kinetochore fibres. Nat Cell Biol. 2005;7:42–47. doi: 10.1038/ncb1207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.DeLuca JG, et al. Kinetochore microtubule dynamics and attachment stability are regulated by Hec1. Cell. 2006;127:969–982. doi: 10.1016/j.cell.2006.09.047. [DOI] [PubMed] [Google Scholar]
- 20.DeLuca J, Moree B, Hickey JM, Kilmartin JV, Salmon ED. hNuf2 inhibition blocks stable kinetochore-microtubule attachment and induces mitotic cell death in HeLa cells. Journal of Cell Biology. 2002;159:549–555. doi: 10.1083/jcb.200208159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Izuta H, et al. Comprehensive analysis of the ICEN (Interphase Centromere Complex) components enriched in the CENP-A chromatin of human cells. Genes Cells. 2006;11:673–684. doi: 10.1111/j.1365-2443.2006.00969.x. [DOI] [PubMed] [Google Scholar]
- 22.Foltz DR, et al. The human CENP-A centromeric nucleosome-associated complex. Nat Cell Biol. 2006;8:458–469. doi: 10.1038/ncb1397. [DOI] [PubMed] [Google Scholar]
- 23.Okada M, et al. The CENP-H-I complex is required for the efficient incorporation of newly synthesized CENP-A into centromeres. Nat Cell Biol. 2006 doi: 10.1038/ncb1396. [DOI] [PubMed] [Google Scholar]
- 24.Carroll CW, Silva MC, Godek KM, Jansen LE, Straight AF. Centromere assembly requires the direct recognition of CENP-A nucleosomes by CENP-N. Nat Cell Biol. 2009;11:896–902. doi: 10.1038/ncb1899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hori T, et al. CCAN makes multiple contacts with centromeric DNA to provide distinct pathways to the outer kinetochore. Cell. 2008;135:1039–1052. doi: 10.1016/j.cell.2008.10.019. [DOI] [PubMed] [Google Scholar]
- 26.Amano M, et al. The CENP-S complex is essential for the stable assembly of outer kinetochore structure. J Cell Biol. 2009;186:173–182. doi: 10.1083/jcb.200903100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.McClelland SE, et al. The CENP-A NAC/CAD kinetochore complex controls chromosome congression and spindle bipolarity. Embo J. 2007;26:5033–5047. doi: 10.1038/sj.emboj.7601927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hori T, Okada M, Maenaka K, Fukagawa T. CENP-O class proteins form a stable complex and are required for proper kinetochore function. Mol Biol Cell. 2008;19:843–854. doi: 10.1091/mbc.E07-06-0556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Toso A, et al. Kinetochore-generated pushing forces separate centrosomes during bipolar spindle assembly. J Cell Biol. 2009;184:365–372. doi: 10.1083/jcb.200809055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Jaqaman K, et al. Quantitative 4D live-cell imaging reveals regulation of kinetochore alignment within the metaphase plate by centromere stiffness and microtubule depolymerases. J. Cell Biol. 2010 doi: 10.1083/jcb.200909005. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Fukagawa T, et al. CENP-H, a constitutive centromere component, is required for centromere targeting of CENP-C in vertebrate cells. EMBO J. 2001;20:4603–4617. doi: 10.1093/emboj/20.16.4603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ganem NJ, Upton K, Compton DA. Efficient mitosis in human cells lacking poleward microtubule flux. Curr Biol. 2005;15:1827–1832. doi: 10.1016/j.cub.2005.08.065. [DOI] [PubMed] [Google Scholar]
- 33.Putkey FR, et al. Unstable kinetochore-microtubule capture and chromosomal instability following deletion of CENP-E. Dev Cell. 2002;3:351–365. doi: 10.1016/s1534-5807(02)00255-1. [DOI] [PubMed] [Google Scholar]
- 34.Salmon ED, Segall RR. Calcium-labile mitotic spindles isolated from sea urchin eggs (Lytechinus variegatus) J Cell Biol. 1980;86:355–365. doi: 10.1083/jcb.86.2.355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Westermann S, Weber K. Post-translational modifications regulate microtubule function. Nat Rev Mol Cell Biol. 2003;4:938–947. doi: 10.1038/nrm1260. [DOI] [PubMed] [Google Scholar]
- 36.Fuller BG, et al. Midzone activation of aurora B in anaphase produces an intracellular phosphorylation gradient. Nature. 2008;453:1132–1136. doi: 10.1038/nature06923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Cheeseman IM, Hori T, Fukagawa T, Desai A. KNL1 and the CENP-H/I/K complex coordinately direct kinetochore assembly in vertebrates. Mol Biol Cell. 2008;19:587–594. doi: 10.1091/mbc.E07-10-1051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Johnson VL, Scott MI, Holt SV, Hussein D, Taylor SS. Bub1 is required for kinetochore localization of BubR1, Cenp-E, Cenp-F and Mad2, and chromosome congression. J Cell Sci. 2004;117:1577–1589. doi: 10.1242/jcs.01006. [DOI] [PubMed] [Google Scholar]
- 39.Andrews PD, et al. Aurora B regulates MCAK at the mitotic centromere. Dev Cell. 2004;6:253–268. doi: 10.1016/s1534-5807(04)00025-5. [DOI] [PubMed] [Google Scholar]
- 40.Lan W, et al. Aurora B phosphorylates centromeric MCAK and regulates its localization and microtubule depolymerization activity. Curr Biol. 2004;14:273–286. doi: 10.1016/j.cub.2004.01.055. [DOI] [PubMed] [Google Scholar]
- 41.Santaguida S, Musacchio A. The life and miracles of kinetochores. EMBO J. 2009 doi: 10.1038/emboj.2009.173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Harding SE, Colfen H. Inversion formulae for ellipsoid of revolution macromolecular shape functions. Anal Biochem. 1995;228:131–142. doi: 10.1006/abio.1995.1324. [DOI] [PubMed] [Google Scholar]
- 43.Schuyler SC, Pellman D. Analysis of the size and shape of protein complexes from yeast. Methods Enzymol. 2002;351:150–168. doi: 10.1016/s0076-6879(02)51845-0. [DOI] [PubMed] [Google Scholar]
- 44.Welburn JP, et al. The human kinetochore Ska1 complex facilitates microtubule depolymerization-coupled motility. Dev Cell. 2009;16:374–385. doi: 10.1016/j.devcel.2009.01.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Meraldi P, McAinsh AD, Rheinbay E, Sorger PK. Phylogenetic and structural analysis of centromeric DNA and kinetochore proteins. Genome Biol. 2006;7:R23. doi: 10.1186/gb-2006-7-3-r23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Cheeseman IM, Desai A. Molecular architecture of the kinetochore-microtubule interface. Nat Rev Mol Cell Biol. 2008;9:33–46. doi: 10.1038/nrm2310. [DOI] [PubMed] [Google Scholar]
- 47.Gaitanos TN, et al. Stable kinetochore-microtubule interactions depend on the Ska complex and its new component Ska3/C13Orf3. Embo J. 2009;28:1442–1452. doi: 10.1038/emboj.2009.96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Wan X, et al. Protein architecture of the human kinetochore microtubule attachment site. Cell. 2009;137:672–684. doi: 10.1016/j.cell.2009.03.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Liu J, Desai A, Onuchic JN, Hwa T. An integrated mechanobiochemical feedback mechanism describes chromosome motility from prometaphase to anaphase in mitosis. Proc Natl Acad Sci U S A. 2008;105:13752–13757. doi: 10.1073/pnas.0807007105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Elbashir SM, et al. Duplexes of 21-nucleotide RNAs mediate RNA interference in cultured mammalian cells. Nature. 2001;411:494–498. doi: 10.1038/35078107. [DOI] [PubMed] [Google Scholar]
- 51.Cassimeris L, Morabito J. TOGp, the human homolog of XMAP215/Dis1, is required for centrosome integrity, spindle pole organization, and bipolar spindle assembly. Mol Biol Cell. 2004;15:1580–1590. doi: 10.1091/mbc.E03-07-0544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Draviam VM, Shapiro I, Aldridge B, Sorger PK. Misorientation and reduced stretching of aligned sister kinetochores promote chromosome missegregation in EB1- or APC-depleted cells. EMBO J. 2006;25:2814–2827. doi: 10.1038/sj.emboj.7601168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Ganem NJ, Compton DA. The KinI kinesin Kif2a is required for bipolar spindle assembly through a functional relationship with MCAK. J Cell Biol. 2004;166:473–478. doi: 10.1083/jcb.200404012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Meraldi P, Draviam VM, Sorger PK. Timing and checkpoints in the regulation of mitotic progression. Dev Cell. 2004;7:45–60. doi: 10.1016/j.devcel.2004.06.006. [DOI] [PubMed] [Google Scholar]
- 55.McAinsh AD, Meraldi P, Draviam VM, Toso A, Sorger PK. The human kinetochore proteins Nnf1R and Mcm21R are required for accurate chromosome segregation. Embo J. 2006;25:4033–4049. doi: 10.1038/sj.emboj.7601293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Akhmanova A, et al. Clasps are CLIP-115 and -170 associating proteins involved in the regional regulation of microtubule dynamics in motile fibroblasts. Cell. 2001;104:923–935. doi: 10.1016/s0092-8674(01)00288-4. [DOI] [PubMed] [Google Scholar]
- 57.Hanisch A, Sillje HH, Nigg EA. Timely anaphase onset requires a novel spindle and kinetochore complex comprising Ska1 and Ska2. Embo J. 2006;25:5504–5515. doi: 10.1038/sj.emboj.7601426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Klebig C, Korinth D, Meraldi P. Bub1 regulates chromosome segregation in a kinetochore-independent manner. J Cell Biol. 2009;185:841–858. doi: 10.1083/jcb.200902128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Liu D, Vader G, Vromans MJ, Lampson MA, Lens SM. Sensing chromosome bi-orientation by spatial separation of aurora B kinase from kinetochore substrates. Science. 2009;323:1350–1353. doi: 10.1126/science.1167000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.McClelland SE, McAinsh AD. Hydrodynamic analysis of human kinetochore complexes during mitosis. Methods Mol Biol. 2009;545:81–98. doi: 10.1007/978-1-60327-993-2_5. [DOI] [PubMed] [Google Scholar]
- 61.Braun M, Drummond DR, Cross RA, McAinsh AD. The kinesin-14 Klp2 organizes microtubules into parallel bundles by an ATP-dependent sorting mechanism. Nat Cell Biol. 2009;11:724–730. doi: 10.1038/ncb1878. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.