Abstract
Division of spermatogonial stem cells1 produces daughter cells that either maintain their stem cell identity or undergo differentiation to form mature sperm. The Sertoli cell, the only somatic cell within seminiferous tubules, provides the stem cell niche through physical support and expression of surface proteins and soluble factors2,3. Here we show that the Ets related molecule4 (ERM) is expressed exclusively within Sertoli cells in the testis and is required for spermatogonial stem cell self-renewal. Mice with targeted disruption of ERM have a loss of maintenance of spermatogonial stem cell self-renewal without a block in normal spermatogenic differentiation and thus have progressive germ-cell depletion and a Sertoli-cell-only syndrome. Microarray analysis of primary Sertoli cells from ERM-deficient mice showed alterations in secreted factors known to regulate the haematopoietic stem cell niche. These results identify a new function for the Ets family transcription factors in spermatogenesis and provide an example of transcriptional control of a vertebrate stem cell niche.
Ets family transcription factors share a unique Ets DNA-binding domain and participate in a variety of developmental processes5. ERM4,6 belongs to a subfamily of Ets factors that also includes Pea3 and ER81 (ref. 5). Pea3 and ER81 are important for normal neuronal development7,8. ERM is expressed in several tissues including brain, lung and testis6. To study the function of ERM in vivo, we generated mice with an inactivated ERM allele (ERM−) (Supplementary Fig. 1). Heterozygous ERM+/− mice are normal, and interbreeding heterozygous ERM+/− mice yielded a mendelian (1:2:1) distribution of ERM−/−, ERM+/− and ERM+/+ in viable offspring (60:147:77, respectively), indicating that ERM is not critical for embryonic development or viability. The most marked phenotype in ERM−/− mice is the disruption of spermatogenesis, whereas other organs did not reveal obvious anatomical defects (Supplementary Fig. 2).
Spermatogenesis is a cyclic process involving the differentiation of spermatogonial stem cells, meiotic cell division and the formation of haploid spermatids. Sertoli cells are the only somatic cells within seminiferous tubules and provide the immediate environment for developing germ cells2. A balance between spermatogonial stem cell self-renewal and differentiation in the adult testis is essential for the maintenance of cyclic waves of spermatogenesis and fertility. Although ERM+/− males were fertile, adult ERM−/− males were sterile (n = 12). ERM−/− males had a significantly decreased testicular size (Fig. 1a). At 4 weeks of age, seminiferous tubules of wild-type and ERM−/− mice seemed similar, with multiple layers of germ cells indicating the normal initiation of spermatogenesis (Fig. 1b, c). By 6 weeks many ERM−/− tubules underwent progressive germ-cell maturation and depletion (Fig. 1d) and by 10 weeks most tubules were devoid of any germ cells (Fig. 1e), containing only morphologically normal Sertoli cells at the basement membrane, which expressed GATA-1 (Fig. 1f), a marker of mature Sertoli cells.
Figure 1. Spermatogonial depletion and Sertoli-cell-only syndrome in ERM−/− mice.
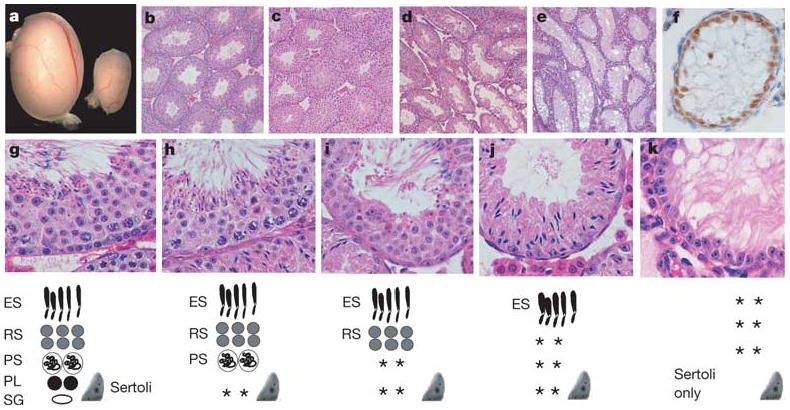
a, Ten-week wild-type (left) and ERM−/− (right) testes. b, Wild-type seminiferous tubules. c–e, Progressive germ-cell loss in ERM−/− testis at 4 weeks (c), 6 weeks (d) and 10 weeks (e). f, Persistent Sertoli cells in ERM−/− tubules indicated by GATA-1 expression at 10 weeks. g–k, Normal spermatogenesis with spermatogonial depletion. Histology of wild-type (g) or ERM−/− (h–k) testes at 6 weeks of age. Diagrams of seminiferous epithelium are shown at the bottom: ES, elongated spermatid; RS, round spermatid; PS, pachytene spermatocyte; PL, preleptotene spermatocyte; Sg, spermatogonium; Sertoli, Sertoli cell; asterisks, missing cell populations.
Histological analysis indicates that the primary defect in ERM−/− testis is the depletion of spermatogonia rather than a developmental block in spermatogenic differentiation. First, at 4 weeks or earlier, ERM−/− seminiferous tubules showed normal spermatogenic differentiation (Fig. 1c), indicating that ERM−/− primordial germ cells and embryonic gonocytes could give rise to spermatogonial stem cells and that a normal wave of spermatogenesis had occurred. However, by 6 weeks (Fig. 1g–k), an array of tubules showed either a selective loss of spermatogonia only or a combined loss of preleptotene and pachytene spermatocytes (Fig. 1h, i). Many tubules contained only elongated spermatids and Sertoli cells, with depletion of all precursor germ cells (Fig. 1j), whereas others were devoid of germ cells altogether (Fig. 1k). These data indicate that ERM deficiency is permissive of normal spermatogenic differentiation but causes germ-cell depletion through an initial loss of spermatogonial stem cells.
To assess the depletion of spermatogonia at the molecular level, we used microarray analysis to compare gene expression between wild-type and ERM−/− testis at 4 weeks of age, when the testes of the two phenotypes have nearly identical histological appearances. Of 50 genes whose expression was reduced to less than one-third in ERM−/− testis compared with the wild type, the greatest reduction was observed for spermatogonia-specific genes (Fig. 2a and Supplementary Table 1). For example, the spermatogonia marker Stra8 (ref. 9) was reduced 19-fold, and other spermatogonia-selective transcripts, including RNA-binding-motif protein (Rbm), Dazl, lymphoid-specific helicase (Lsh) and cellular retinoic-acid-binding protein (CRABP) were reduced 7-fold, 3.5-fold, 9-fold and 14-fold, respectively, in ERM−/− testis. By contrast, several markers of more mature germ cells were unchanged in expression, including the spermatid markers10 protamine 1 (Prm1) and Prm2, and transition proteins 1 and 2. Microarray results were confirmed by reverse-transcriptase-mediated polymerase chain reaction (RT–PCR) analysis in three distinct pairs of 4-week-old wild-type and ERM−/− littermates (Fig. 2a). Thus, at 4 weeks, when ERM−/− and wild-type testes are very similar in histological appearance, spermatogonia-specific genes were already greatly underrepresented in ERM−/− testes, again indicating that the defect is the loss of maintenance of spermatogonia. Finally, the loss of spermatogonial stems cells in ERM−/− mice was indicated by the absence of Plzf11 expression in ERM−/− testis in comparison with wild-type testis (Fig. 2b, c). The rare cells with morphological similarity to spermatogonia that remained in ERM−/− testis at 6 weeks of age were negative for Plzf expression, indicating that these are committed spermatogonia rather than undifferentiated spermatogonia.
Figure 2. Selective reduction of spermatogonia-specific genes in ERM−/− testes.
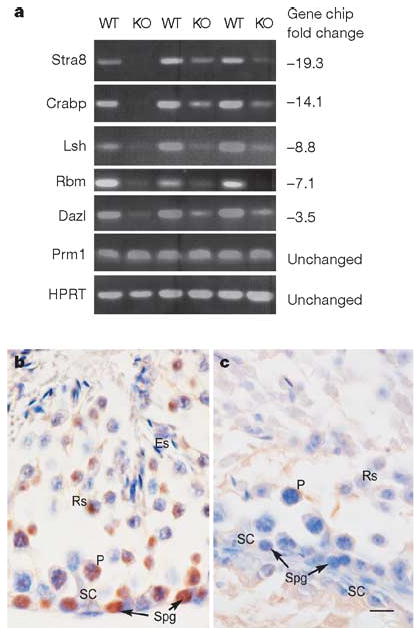
a, RT–PCR analyses of three independent pairs of wild-type (WT) and ERM−/− (KO) male littermates are shown for the indicated genes. Numbers at the right represent the relative fold reduction measured by microarray analysis (Supplementary Table 1). HPRT, hypoxanthine–guanine phosphoribosyltransferase. b, c, Expression of Plzf in 6-week-old wild-type (b) and ERM−/− (c) testes by immunohistochemistry. Cells indicated are as follows: ES, elongated spermatid; RS, round spermatid; PS, pachytene spermatocyte; Spg, spermatogonium; Sc, Sertoli cell. Scale bar, 10 μm.
Failure to maintain spermatogonia in ERM−/− testis could result from either an intrinsic requirement for ERM in germ cells or cell-extrinsic requirement for ERM in Sertoli or other cells. We first analysed ERM expression in c-kitW/W-v testes (Supplementary Fig. 3a), which have a Sertoli-cell-only phenotype12. ERM expression was increased in germ-cell-free c-kitW/W-v testis relative to wild-type testis, indicating that it is expressed by somatic cells. In addition, ERM was expressed in isolated Sertoli cells, but not in isolated spermatogonia, pachytene spermatocytes or round spermatids (Supplementary Fig. 3b), whereas Stra8 was expressed exclusively in spermatogonia, as expected. Further, we used several additional methods to show that ERM is exclusively expressed within Sertoli cell in the testis. First, by in situ hybridization, ERM messenger RNA was localized to the periphery of seminiferous tubules in wild-type testis but was absent centrally (Fig. 3a). The non-functional ERM− mRNA transcript was detected in the Sertoli-cell-only ERM−/− testis at 10 weeks (Fig. 3b), indicating direct expression by Sertoli cells. Second, we examined ERM expression histologically with an IRES-LacZ reporter cassette targeted to the ERM locus (N. Kurpios, S. Arber, J.A. Hassell, unpublished observations). ERM expression in ERM+/IRES-LacZ testis was found exclusively in Sertoli cells, was first detectable between 3 and 4 weeks of age and persisted throughout adulthood (Fig. 3c and Supplementary Fig. 4). This onset of ERM expression precedes the timing of spermatogonial loss, which is consistent with a requirement for ERM in the adult stem cell niche in the testis. Third, a fusion protein of ERM and green fluorescent protein (GFP) was localized to the nucleus of TM4 Sertoli cells (Fig. 3d). Last, we generated an ERM-specific monoclonal antibody, 3H7 (Supplementary Fig. 5), which identified ERM protein expression to be present exclusively within Sertoli cell nuclei of wild-type testis (Fig. 3e) and to be undetectable in ERM−/− testis (Fig. 3f). Thus, ERM expression in testis is specific to Sertoli cells, indicating that the loss of spermatogonia in ERM−/− testes might not be due to a cell-intrinsic defect of spermatogonia but rather to alterations in the microenvironment provided by Sertoli cells.
Figure 3. ERM expression in testis is restricted to Sertoli cells.
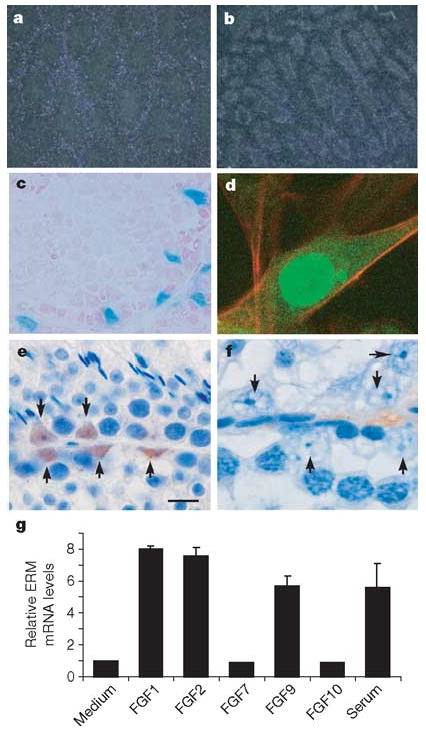
a, b, In situ hybridization for ERM mRNA in 10-week wild-type (a) and ERM−/− (b) testes. c, Testes from 6-week ERMIRES-LacZ heterozygous males were stained for LacZ expression. d, TM4 Sertoli cells were infected with ERM-GFP-RV retrovirus and analysed by confocal microscopy. e, f, Immunolocalization of ERM protein in adult wild-type (e) and ERM−/− (f) testis with anti-ERM monoclonal antibody. Arrows indicate nuclei of Sertoli cells in wild-type (e) and ERM−/− (f) testes. g, ERM mRNA expression in TM4 cells after treatment with various FGFs. Data are fold induction (means + s.d. for two independent measurements) compared with ERM expression in medium.
Signalling by fibroblast growth factor (FGF) has been reported to regulate the expression of ERM in Zebrafish13, and FGF9 deficiency in mice causes a possible defect in Sertoli cell differentiation14, prompting us to examine the effect of FGF on ERM expression in Sertoli cells (Fig. 3g). ERM mRNA was induced by FGF1, FGF2 and FGF9, but not by FGF7 and FGF10, in the murine Sertoli cell line TM4 (Fig. 3g). These results indicate a potential activity of FGF receptor signalling in regulating ERM expression and Sertoli cell function in vivo.
Spermatogonial depletion could result from alterations in spermatogonial proliferation or apoptosis. We therefore examined cell proliferation in ERM−/− testes at 3 and 4 weeks by labelling in vivo with bromodeoxyuridine (BrdU) (Fig. 4a–d). At 3 weeks, before the loss of germ cells, BrdU incorporation by spermatogonia was normal in ERM−/− testis (Fig. 4a, b). However, at 4 weeks, BrdU incorporation was almost absent in ERM−/− testis, compared with labelling in wild-type (Fig. 4c, d). To determine whether this loss was due to increased apoptosis, we used TdT-mediated dUTP nick end labelling (TUNEL), which showed no alteration in TUNEL staining in ERM−/− testes at 4 or 6 weeks (Fig. 4e, f, and Supplementary Table 2). Further, expression of proapoptotic and antiapoptotic genes in the Bcl-2 family also showed no difference between ERM−/− and wild-type testes (Fig. 4g). These results indicate that the defect was caused by decreased self-renewal of spermatogonial stem cells. Finally, serum hormone levels were measured to determine whether the pituitary–testis axis had contributed to the phenotype. We found no significant difference in serum levels of testosterone or follicle-stimulating hormone between wild-type and ERM−/− mice (Supplementary Table 3). Together, these results indicate that ERM expression by Sertoli cells might be required for spermatogonial stem cell self-renewal, and that loss of spermatogonial stem cells in ERM−/− testis is due to a defect in the stem cell niche provided by ERM-expressing Sertoli cells rather than an endocrine disorder.
Figure 4. Failure of stem cell self-renewal causes spermatogonial depletion.
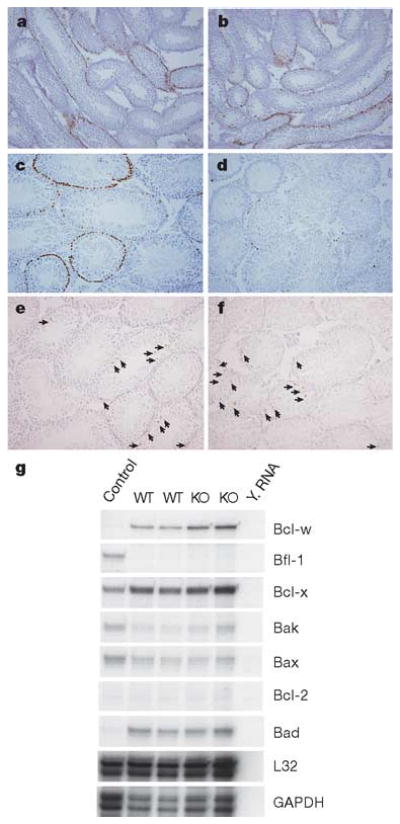
a–d, Loss of proliferating spermatogonia in ERM−/− testis, demonstrated by BrdU labelling in vivo of wild-type (a, c) and ERM−/− (b, d) testes at 3 weeks (a, b) and 4 weeks (c, d). e, f, Comparable levels of spermatogonial apoptosis, determined by TUNEL staining, in wild-type (e) and ERM−/− (f) testes at 4 weeks of age. Arrows indicate apoptotic cells. g, Expression of apoptosis-related genes was unchanged in ERM−/− testes as indicated by RNase protection assay of total RNA from 4-week-old wild-type (WT) and ERM−/− (KO) testes. GAPDH, glyceraldehyde3phosphate dehydrogenase. Y. RNA, Yeast tRNA.
Several somatic signalling pathways regulating spermatogonial stem cell self-renewal have recently been discovered3,15,16. Mutations in two genes, promyelocytic zinc-finger (Plzf)11,17 and glial-cell-derived neurotrophic factor (GDNF)3 result in a defect in mammalian spermatogonial stem cell self-renewal. By comparison, the self-renewal defect in ERM−/− testes is more severe and differs mechanistically from that caused by mutations in Plzf or GDNF. First, Plzf is a transcription factor expressed by spermatogonia11,17, not by Sertoli cells, and the loss of Plzf causes a spermatogonia-intrinsic defect. Loss of spermatogonia in Plzf-deficient mice is not complete even at 8 months11,17, indicating that spermatogonial self-renewal can occur for extended periods in the absence of Plzf, in contrast to more rapid and complete loss of spermatogonia in ERM−/− testes. Second, GDNF3, which is secreted by Sertoli cells, was implicated in spermatogonial self renewal because GDNF+/− mice show gradual spermatogonial depletion3. GDNF−/− testes have not been analysed because of embryonic lethality.
To test whether ERM regulates GDNF, we performed a microarray analysis of purified wild-type and ERM−/− primary Sertoli cells (Supplementary Fig. 6 and Supplementary Table 4). GDNF expression was unchanged in ERM−/− testis in primary Sertoli cells from 4-week-old wild-type and ERM−/− testes, indicating that it is not a target of ERM. In contrast, several other genes were greatly reduced in ERM−/− Sertoli cells. Genes with the greatest reduction included the chemokines, CXCL-12 (SDF-1), CXCL5 (LIX) and CCL7 (MCP-3), reduced 9-fold, 10-fold and 25-fold, respectively (Supplementary Fig. 6 and Supplementary Table 4). SDF-1 and CXCL5 have been implicated in regulating the stem cell niche in other systems18,19. SDF-1 is involved in haematopoietic stem cell (HSC) migration, retention and self-renewal18 and is required for the migration of primordial germ cells towards the genital ridge20. CXCL5 was recently implicated in HSC maintenance19. Conceivably these chemokines are niche signalling molecules regulating spermatogonial stem cell self-renewal, but lethality caused by deficiencies of SDF-1 and its receptor CXCR4 prevent an immediate analysis of their role in adult spermatogenesis21,22. Matrix metalloproteinase-12 (MMP-12) showed a tenfold reduction in ERM−/− Sertoli cells. Interestingly, MMP-9, a related MMP family member, is involved in the recruitment of HSCs to the bone marrow niche23.
Interactions with niche cells are crucial for maintaining stem cell character, and several molecules produced by niche cells can regulate the capacity of the niche to support stem cell self-renewal24. As an example of the transcriptional control of a stem cell niche, ERM provides the opportunity to explore how the capacity of the stem cell niche to maintain spermatogonial self-renewal is transcriptionally coordinated.
Methods
Generation of ERM mutant mice
ERM exons 2–5 encoding the initiation codon and transcriptional activation domain were deleted (Supplementary Fig. 1a). The targeting vector was constructed in pLNTK by using a 1.6-kilobase (kb) genomic fragment (left arm) upstream of the mouse ERM exon 2, and a 4-kb genomic fragment (right arm) downstream of exon 5. The left arm was generated by PCR from genomic DNA with the use of the oligonucleotides left arm forward (f), 5′-TTTTGTCGACGCGGCCGCTTTTGGAATCTCTTAGGG AAGTTT-3′ (SalI tailed), and left arm reverse (r), 5′-CCC CTCGAG TTTCCCTCTTGCCTGTGTAGCCA-3′ (XhoI tailed). The 1.6-kb PCR fragment was digested with XhoI and SalI and ligated into the XhoI site of pLNTK vector. The right arm was generated by PCR with the use of the oligonucleotides right arm forward (f), 5′-AAAACTCGAGATACAAAGGATTGCAAAGGCT-3′ (XhoI tailed), and right arm reverse (r), 5′-GGGACTCGAGTTCTGAAATTG TTTGGCCTTGGA-3′ (XhoI tailed), digested with XhoI and ligated into the SalI site of targeting vector. The targeting vector DNA was electroporated into MC50 embryonic stem cells (a gift from R. Schreiber). Positive clones were identified by Southern blot analysis with 5′ and 3′ probes (Supplementary Fig. 1a). In vitro Cre-mediated neo excision was performed on two distinct recombinant clones, 1CD3 and 1CC5, generating neo-deleted clones E7 and A7, respectively. Blastocyst injection was performed for all four clones and each generated germline transmission of the targeted ERM allele. Male chimaeras were crossed with 129SvEv females to establish ERM mutants on the 129SvEv genetic background. Homozygous mice were obtained by intercrossing heterozygous siblings. The phenotypes for all four lines were indistinguishable grossly and microscopically. For the results shown in this study, the E7 neo-deleted strain was used.
In situ hybridization
A 345-base-pair fragment of the ERM cDNA was obtained by RT–PCR with the use of the oligonucleotides ERM-345(f), 5′-CCGAGTT GTCGTCCTGTAG-3′, and ERM-345(r), 5′-ACTGGCTTTCAGGCATCATC-3′, and cloned into pGEM-Teasy vector used for the synthesis of anti-sense and sense probes. Cryostat sections were hybridized with 35S-labelled antisense RNA (cRNA) probe.
Generation of ERM-specific monoclonal antibody, and histology
ERM region encoded by exons 7 and 8, lacking homology to Pea3 and ER81, was amplified by RT–PCR with the use of the primers 5′-GGAATTCCATATGTGTGCCTA CGATAGGAAGCCTCCC-3′ and CGGGATCCTTATCTCTGTTCTGATGGA TACTGG-3′ and cloned into NdeI/BamHI sites of pET28a (Novagen). His-tagged ERM recombinant protein (12 kDa) was induced with 1 mM isopropyl β-d-thiogalactoside in Escherichia coli BL21 (Invitrogen) and purified by Ni2+-nitrilotriacetate and size-exclusion chromatography. Hybridomas were generated from immunized hamsters and screened by ELISA against purified ERM protein. The hybridoma 3H7 monoclonal antibody (mAb) was used as supernatant for immunohistochemistry.
Immunohistochemistry was performed on sections fixed in 10% formalin. mAb 3H7 was used with goat anti-hamster biotinylated secondary antibody at 1:1000 dilution. Anti-GATA-1 rat mAb (Santa Cruz) was used at 1:100 dilution. Anti-Plzf antibody (Calbiochem) was used at 1:1000 dilution. Vectastain ABC kit and DAB substrate kit (Vector Laboratories) were used for immunohistochemistry. Sections were counterstained with haematoxylin. Analysis of the ERM–LacZ cassette used frozen sections stained overnight with 5-bromo-4-chloro-3-indolyl-β-Dgalactoside staining buffer at 37 °C, counterstained with nuclear Fast red.
An ERM-GFP fusion protein was created by deletion of the IRES from the plasmid ERM-RV25 by Quick change mutagenesis (Strategene) with the oligonucleotides ERM-GFP top (5′-CTTCGCTTACGTGAGCAAGGGCGAGGAGC-3′ and ERM-GFP bot (5′-CCTTGCTCACGTAAGCGAAGCCTTCGGTGTA-3′), to produce ERM-GFP-RV. TM4 cells were infected with ERM-GFP-RV or GFP-RV and purified by cell sorting. Cellular localization of ERM-GFP fusion protein was captured by confocal microscopy.
TM4 cells were maintained in serum-free medium for 24 h and treated with medium alone or 1 nM FGF1, FGF2, FGF7, FGF9 and FGF10 (PeproTech) or 10% fetal calf serum. RNA was harvested after 3 h and real-time RT–PCR was performed for ERM and glyceraldehyde3phosphate dehydrogenase using an ABI Prism 7700 Sequence Detector (Applied Biosystems). ERM-specific primers used for this RT–PCR were 5′-CAGGAGCCCCGAGATTACTG-3′ and 5′-CCGCCTCTCATGTAGGATGAC-3′. Data are represented as fold expression of normalized ERM expression over the medium control.
Cell proliferation and apoptosis assays
Mice were injected intraperitoneally with BrdU (Sigma) at a concentration of 50 mg kg−1. After 4 h, testes and small intestine were isolated and fixed in 10% buffered formalin. Paraffin-embedded sections were processed with the BrdU in-situ detection kit (BD Biosciences). Germ-cell apoptosis was analysed by TUNEL labelling with Apoptag in situ apoptosis detection kit (Intergen) in accordance with the manufacturer's protocol. Expression of proapoptotic and antiapoptotic genes in 4-week-old wild-type and ERM−/− testes was quantified by the mAPO-2 Multi-probe RNase protection assay kit (BD Biosciences) in accordance with the manufacturer's protocol.
Microarray analysis of wild-type and ERM−/− testes and primary Sertoli cells
Wild-type and ERM−/− testes (n = 4) were isolated at 4 weeks of age, and total RNA was extracted separately with RNeasy kit (Qiagen). RNA (10 μg) was pooled, and biotinylated cRNA target was independently generated from each pool. Each cRNA was hybridized to an Affymetrix U74Av2 Murine Genome Array. Microarray analysis of primary Sertoli cells isolated26 from wild-type or ERM−/− mice was carried out similarly.
RT–PCR analysis
We used semiquantitative PCR for confirmation of Affymetrix gene array results in Fig. 2a and Supplementary Fig. 5. RT–PCR amplifications were titrated within the linear range for each primer pair. Primers used are listed in Supplementary Table 5.
Supplementary Material
Acknowledgments
We thank M. White for blastocyst microinjections; X. Cheng for helpful discussion; B. Sleckman for targeting vector; D. Ornitz, M. Griswold and K. Sheehan for reagents and antibody production; and T. Jessell for help with ERMIRES-LacZ mice. J.A.H. acknowledges funding from the Canadian Breast Cancer Research Alliance, the Canadian Institutes of Health Research and the DOD Breast Cancer Research Program. N.A.K. was supported by a DOD Breast Cancer Research Program Scholarship. K.M.M. is an investigator of the Howard Hughes Medical Institute.
Footnotes
Supplementary Information is linked to the online version of the paper at www.nature.com/nature.
The authors declare no competing financial interests.
References
- 1.Kubota H, Avarbock MR, Brinster RL. Spermatogonial stem cells share some, but not all, phenotypic and functional characteristics with other stem cells. Proc Natl Acad Sci USA. 2003;100:6487–6492. doi: 10.1073/pnas.0631767100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Griswold MD. The central role of Sertoli cells in spermatogenesis. Semin Cell Dev Biol. 1998;9:411–416. doi: 10.1006/scdb.1998.0203. [DOI] [PubMed] [Google Scholar]
- 3.Meng X, et al. Regulation of cell fate decision of undifferentiated spermatogonia by GDNF. Science. 2000;287:1489–1493. doi: 10.1126/science.287.5457.1489. [DOI] [PubMed] [Google Scholar]
- 4.Monte D, Baert JL, Defossez PA, de Launoit Y, Stehelin D. Molecular cloning and characterization of human ERM, a new member of the Ets family closely related to mouse PEA3 and ER81 transcription factors. Oncogene. 1994;9:1397–1406. [PubMed] [Google Scholar]
- 5.Seth A, et al. The ets gene family. Cell Growth Differ. 1992;3:327–334. [PubMed] [Google Scholar]
- 6.Chotteau-Lelievre A, Desbiens X, Pelczar H, Defossez PA, de Launoit Y. Differential expression patterns of the PEA3 group transcription factors through murine embryonic development. Oncogene. 1997;15:937–952. doi: 10.1038/sj.onc.1201261. [DOI] [PubMed] [Google Scholar]
- 7.Arber S, Ladle DR, Lin JH, Frank E, Jessell TM. ETS gene Er81 controls the formation of functional connections between group Ia sensory afferents and motor neurons. Cell. 2000;101:485–498. doi: 10.1016/s0092-8674(00)80859-4. [DOI] [PubMed] [Google Scholar]
- 8.Livet J, et al. ETS gene Pea3 controls the central position and terminal arborization of specific motor neuron pools. Neuron. 2002;35:877–892. doi: 10.1016/s0896-6273(02)00863-2. [DOI] [PubMed] [Google Scholar]
- 9.Oulad-Abdelghani M, et al. Characterization of a premeiotic germ cell-specific cytoplasmic protein encoded by Stra8, a novel retinoic acid-responsive gene. J Cell Biol. 1996;135:469–477. doi: 10.1083/jcb.135.2.469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sassone-Corsi P. Unique chromatin remodeling and transcriptional regulation in spermatogenesis. Science. 2002;296:2176–2178. doi: 10.1126/science.1070963. [DOI] [PubMed] [Google Scholar]
- 11.Buaas FW, et al. Plzf is required in adult male germ cells for stem cell self-renewal. Nature Genet. 2004;36:647–652. doi: 10.1038/ng1366. [DOI] [PubMed] [Google Scholar]
- 12.De Franca LR, et al. Sertoli cells in testes containing or lacking germ cells: a comparative study of paracrine effects using the W (c-kit) gene mutant mouse model. Anat Rec. 1994;240:225–232. doi: 10.1002/ar.1092400209. [DOI] [PubMed] [Google Scholar]
- 13.Raible F, Brand M. Tight transcriptional control of the ETS domain factors Erm and Pea3 by Fgf signalling during early zebrafish development. Mech Dev. 2001;107:105–117. doi: 10.1016/s0925-4773(01)00456-7. [DOI] [PubMed] [Google Scholar]
- 14.Colvin JS, Green RP, Schmahl J, Capel B, Ornitz DM. Male-to-female sex reversal in mice lacking fibroblast growth factor 9. Cell. 2001;104:875–889. doi: 10.1016/s0092-8674(01)00284-7. [DOI] [PubMed] [Google Scholar]
- 15.Kiger AA, White-Cooper H, Fuller MT. Somatic support cells restrict germline stem cell self-renewal and promote differentiation. Nature. 2000;407:750–754. doi: 10.1038/35037606. [DOI] [PubMed] [Google Scholar]
- 16.Tran J, Brenner TJ, DiNardo S. Somatic control over the germline stem cell lineage during Drosophila spermatogenesis. Nature. 2000;407:754–757. doi: 10.1038/35037613. [DOI] [PubMed] [Google Scholar]
- 17.Costoya JA, et al. Essential role of Plzf in maintenance of spermatogonial stem cells. Nature Genet. 2004;36:653–659. doi: 10.1038/ng1367. [DOI] [PubMed] [Google Scholar]
- 18.Christensen JL, Wright DE, Wagers AJ, Weissman IL. Circulation and chemotaxis of fetal hematopoietic stem cells. PLoS Biol. 2004;2:E75. doi: 10.1371/journal.pbio.0020075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Choong ML, Yong YP, Tan AC, Luo B, Lodish HF. LIX: a chemokine with a role in hematopoietic stem cells maintenance. Cytokine. 2004;25:239–245. doi: 10.1016/j.cyto.2003.11.002. [DOI] [PubMed] [Google Scholar]
- 20.Molyneaux KA, et al. The chemokine SDF1/CXCL12 and its receptor CXCR4 regulate mouse germ cell migration and survival. Development. 2003;130:4279–4286. doi: 10.1242/dev.00640. [DOI] [PubMed] [Google Scholar]
- 21.Nagasawa T, et al. Defects of B-cell lymphopoiesis and bone-marrow myelopoiesis in mice lacking the CXC chemokine PBSF/SDF-1. Nature. 1996;382:635–638. doi: 10.1038/382635a0. [DOI] [PubMed] [Google Scholar]
- 22.Tachibana K, et al. The chemokine receptor CXCR4 is essential for vascularization of the gastrointestinal tract. Nature. 1998;393:591–594. doi: 10.1038/31261. [DOI] [PubMed] [Google Scholar]
- 23.Heissig B, et al. Recruitment of stem and progenitor cells from the bone marrow niche requires MMP-9 mediated release of kit-ligand. Cell. 2002;109:625–637. doi: 10.1016/s0092-8674(02)00754-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Fuchs E, Tumbar T, Guasch G. Socializing with the neighbors: stem cells and their niche. Cell. 2004;116:769–778. doi: 10.1016/s0092-8674(04)00255-7. [DOI] [PubMed] [Google Scholar]
- 25.Ouyang W, et al. The Ets transcription factor ERM is Th1-specific and induced by IL-12 through a Stat4-dependent pathway. Proc Natl Acad Sci USA. 1999;96:3888–3893. doi: 10.1073/pnas.96.7.3888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Karl AF, Griswold MD. Sertoli cells of the testis: preparation of cell cultures and effects of retinoids. Methods Enzymol. 1990;190:71–75. doi: 10.1016/0076-6879(90)90010-x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


