SUMMARY
Intracellular iron homeostasis is critical for survival and proliferation. Lipocalin 24p3 is an iron trafficking protein that binds iron through association with a bacterial siderophore, such as enterobactin, or a postulated mammalian siderophore. Here we show that the iron-binding moiety of the 24p3-associated mammalian siderophore is 2,5-dihydroxybenzoic acid (2,5-DHBA), which is similar to 2,3-DHBA, the iron-binding component of enterobactin. We find that the murine enzyme responsible for 2,5-DHBA synthesis is the homologue of bacterial EntA, which catalyzes 2,3-DHBA production during enterobactin biosynthesis. RNA interference-mediated knockdown of the murine homologue of EntA results in siderophore depletion. Mammalian cells lacking the siderophore accumulate abnormally high amounts of cytoplasmic iron, resulting in elevated levels of reactive oxygen species, whereas the mitochondria are iron deficient. Siderophore-depleted mammalian cells and zebrafish embryos fail to synthesize heme, an iron-dependent mitochondrial process. Our results reveal features of intracellular iron homeostasis that are conserved from bacteria through humans.
INTRODUCTION
The acquisition of iron by cells is critical for survival, growth and differentiation (Andrews, 2008). Cells require a constant source of iron, which serves as a functional component of hemoproteins and iron-sulfur (Fe-S) cluster-containing proteins. Almost all mammalian cells and tissues are thought to acquire iron by receptor-mediated endocytosis of iron-loaded transferrin. Following uptake, intracellular iron levels must be tightly regulated, as excess free iron can catalyze the production of toxic reactive oxygen species (ROS; reviewed in Emerit et al., 2001).
In addition to its cytoplasmic function, iron also has a critical role in mitochondria, where heme and various Fe-S cluster-containing proteins are synthesized (Rouault and Tong, 2005). Mitochondrial iron levels must be tightly regulated; an inadequate supply of iron can impair the metabolic and respiratory activities of the organelle, whereas excess mitochondrial iron promotes the generation of ROS, which are produced as a side reaction of mitochondrial electron transport (Foury and Cazzalini, 1997).
Iron is ultimately delivered to proteins or transporters by metallochaperones, such as the mitochondrial iron chaperone frataxin (Bulteau et al., 2004) and the cytosolic iron chaperone PCBP1 (Shi et al., 2008). However, an unresolved question is the form in which iron, which has low solubility in aqueous solution, is trafficked from the transferrin-containing endosome to other cellular sites, such as mitochondria. Although most cytoplasmic iron is tightly bound to proteins, it is generally believed that a fraction of cytoplasmic iron is complexed with a low molecular weight carrier to form the “free” (also called labile or chelatable) iron pool (Breuer et al., 2008). In support of this idea, several early studies suggested that mammalian cells and tissues contain low molecular weight siderophores that bind iron with high affinity (Fernandez-Pol, 1978; Jones et al., 1980).
Recently, new evidence for the existence of a mammalian siderophore has emerged from studies of the lipocalin 24p3 (also known as lipocalin-2 (Lcn2)). Lipocalins are a family of secreted proteins that can bind small molecular weight ligands and have diverse molecular recognition properties and functions (Akerstrom et al., 2000). 24p3 has been found to be involved in multiple biological processes including apoptosis (Devireddy et al., 2005), innate immunity (Flo et al., 2004) and renal development (Yang et al., 2002).
Several groups have shown that 24p3 can bind iron and deliver it to or remove it from cells (Devireddy et al., 2005; Goetz et al., 2002; Yang et al., 2002). The iron-status of 24p3 is the critical determinant of its activity. Iron-containing 24p3 (holo-24p3) binds to a cell surface receptor called 24p3R, is internalized, and releases its bound iron thereby increasing intracellular iron concentration (Devireddy et al., 2005). By contrast, iron-lacking 24p3 (apo-24p3) binds to 24p3R, is internalized, chelates iron and transfers it to the extracellular medium thereby reducing intracellular iron concentration.
24p3 does not have an intrinsic ability to interact with iron, but rather binds iron through an associated, small molecular weight, iron-chelating compound (Devireddy et al., 2005; Goetz et al., 2002; Yang et al., 2002). Significantly, the classic bacterial siderophore, enterobactin, can bind 24p3 and function as the iron-chelating moiety (Yang et al., 2002). Interestingly, apo-24p3 produced in enterobactin-deficient bacteria acquires iron-binding ability following delivery to, and internalization in, mammalian cells (Devireddy et al., 2005). These results indicate that mammalian cells contain a siderophore, which may have structural and functional similarities to bacterial enterobactin.
Here we characterize the iron-binding moiety of the 24p3-associated mammalian siderophore and identify one of the cellular enzymes involved in its synthesis. This information has enabled us to deplete the siderophore from mammalian cells and zebrafish embryos and, for the first time, study the role of the siderophore in intracellular iron homeostasis.
RESULTS
2,5-DHBA is the Iron-Binding Moiety of the 24p3-Associated Mammalian Siderophore
We have previously shown that FL5.12 cells, a murine interleukin-3 (IL-3)-dependent pro-B lymphocytic cell line, harbor an intracellular co-factor that enables 24p3 to bind iron (Devireddy et al., 2005). To gain insight into the composition of this cofactor we derived an FL5.12 cell line stably expressing an ecdysone-inducible, epitope-tagged derivative of 24p3 (FL5.12/Ec-24p3 cells). Figure 1A shows that treatment of FL5.12/Ec-24p3 cells with the ecdysone analog ponasterone A resulted in the accumulation of 24p3 in the conditioned medium (CM). To test whether the induced 24p3 was able to bind iron, radiolabeled iron (55FeCl3) was added to the CM, 24p3 was immunoprecipitated, and 24p3 iron binding assessed by measuring radioactivity in the 24p3 immunoprecipitate. Figure 1B shows that 24p3 present in the CM from ponasterone A-treated FL5.12/Ec-24p3 cells bound to 55Fe, indicating, as expected, that the 24p3 was siderophore-associated.
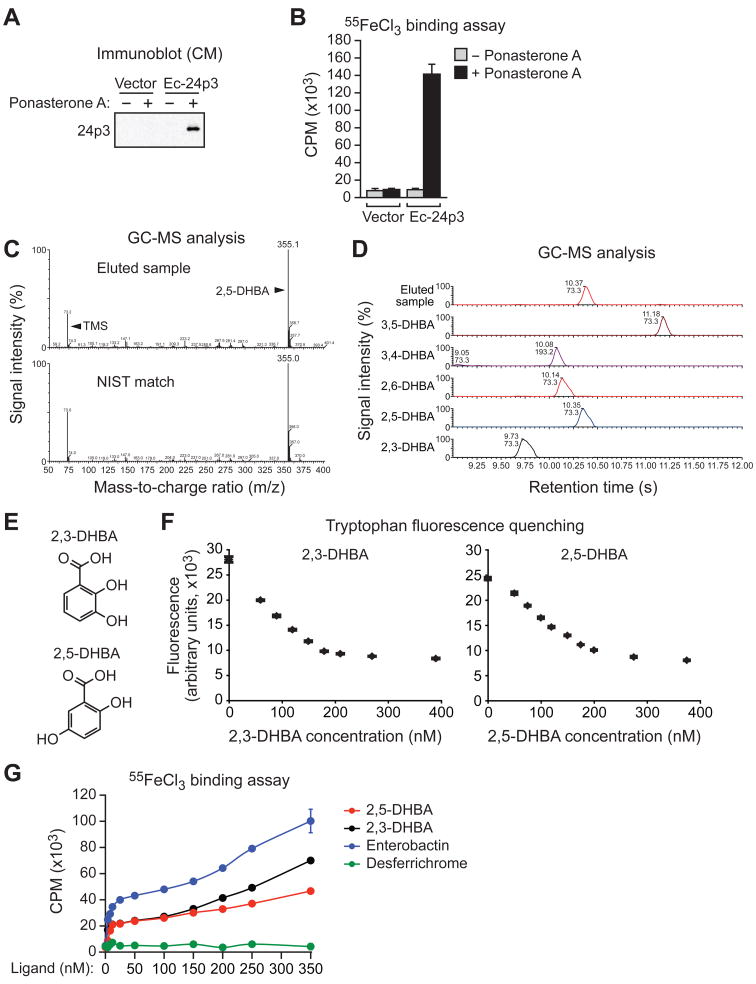
Figure 1. 2,5-DHBA is the Iron-Binding Moiety of the 24p3-Associated Mammalian Siderophore.
(A) Immunoblot analysis showing 24p3 in the CM of FL5.12 cells expressing either vector or Ec-24p3 in the presence or absence of ponasterone A.
(B) Iron binding assay. 24p3 was immunoprecipitated from the CM of cells described in (A) and analyzed for the presence of 55Fe. CPM, counts per minute. Error bars indicate SD.
(C) (Top) GC-MS analysis on the TMS-derivatized, small molecule flow-through fraction eluted from immunopurified 24p3. (Bottom) Matching spectrum from the NIST database.
(D) GC-MS analysis on the eluted sample from immunopurified 24p3 and a set of DHBA standards.
(E) Chemical structures of 2,3-DHBA and 2,5-DHBA.
(F) Tryptophan fluorescence quenching assay. 2,3-DHBA (left) or 2,5-DHBA (right) was added to apo-24p3 at increasing concentrations, and tryptophan fluorescence was monitored. Error bars indicate SD.
(G) Iron binding assay. Purified apo-24p3 was incubated with 55FeCl3 and increasing concentrations of either 2,3-DHBA, 2,5-DHBA, iron-free enterobactin or desferrichrome, and radioactivity was measured in the 24p3 immunoprecipitate. Error bars indicate SD. See also Figure S1.
To identify the 24p3-associated siderophore, 24p3 was immunopurified from the CM of ponasterone A-treated FL5.12/Ec-24p3 cells, the putative 24p3-siderophore complex was eluted with acetic acid, and protein was then separated from small molecules by size exclusion chromatography. The small molecule flow-through fraction was derivatized with trimethylsilate (TMS) and subjected to gas chromatography-mass spectrometry (GC-MS). As shown in Figure 1C, a single prominent GC peak was detected at 355 m/z (mass-to-charge ratio), and a search of the National Institute of Standards and Technology (NIST) database of known MS spectra yielded a match to tri-TMS-2,5-dihydroxybenzoic acid (tri-TMS-2,5-DHBA), which was confirmed using a set of DHBA standards (Figure 1D). GC-MS analysis under milder conditions [N,O-Bis(trimethylsilyl)trifluoroacetamide (BSTFA)-pyridine] did not yield a peak for tri-TMS-2,5-DHBA whereas free 2,5-DHBA was fully derivatized (data not shown) indicating that the 24p3-associated 2,5-DHBA is a component of a larger molecule. Notably, 2,5-DHBA could also be detected and quantified by TMS derivatization and GC-MS analysis of cytoplasmic extracts from FL5.12 cells cultured in the presence of IL-3 (Figure S1A), which lack 24p3 (Devireddy et al., 2001).
2,5-DHBA is related to 2,3-DHBA (Figure 1E), the iron-binding moiety of bacterial enterobactin (reviewed in Raymond et al., 2003). We performed several experiments to determine whether, like 2,3-DHBA, 2,5-DHBA could also chelate iron and bind 24p3. The iron-binding pocket of 24p3 has a single tryptophan residue and the occupation of this site by enterobactin or 2,3-DHBA quenches its intrinsic fluorescence (Goetz et al., 2002). Figure 1F (left panel) shows, consistent with previous results, that addition of 2,3-DHBA quenched the intrinsic fluorescence emitted by tryptophan. Addition of 2,5-DHBA also quenched tryptophan fluorescence (Figure 1F, right panel), indicating that 2,5-DHBA bound to 24p3. 2,5-DHBA and 2,3-DHBA bound 24p3 with roughly comparable affinities of 12 and 8 nM, respectively, as calculated by non-linear regression analysis. As expected, desferrichrome, a structurally unrelated siderophore that does not bind to 24p3 (Flo et al., 2004; Goetz et al., 2002), had no effect on tryptophan fluorescence (Figure S1B).
We next assessed the ability of 2,5-DHBA to confer iron binding to 24p3. Purified apo-24p3 was incubated with 55FeCl3 and increasing concentrations of 2,5-DHBA or, as positive controls, 2,3-DHBA or enterobactin, and following immunoprecipitation of 24p3, radioactivity was measured in the 24p3 immunoprecipitate. Figure 1G shows that addition of 2,3-DHBA, enterobactin or 2,5-DHBA to apo-24p3 conferred the ability to bind iron, whereas desferrichrome did not. The greater level of iron binding observed with enterobactin compared to DHBA is most likely due to the higher affinity of enterobactin for 24p3 (Goetz et al., 2002). Order of addition experiments confirmed that 2,5-DHBA could stably bind to 24p3 in the absence of iron (Figure S1C).
Identification of a Murine Homologue of Bacterial EntA that Mediates 2,5-DHBA Synthesis
In bacteria, synthesis of 2,3-DHBA from its precursor, 2,3-dihydro-2,3-dihydroxybenzoic acid (2,3-diDHBA), is catalyzed by the EntA protein (Liu et al., 1989). The finding that the mammalian siderophore had structural similarities to enterobactin prompted us to perform database searches for proteins homologous to bacterial EntA. As shown in Figure 2A, these searches revealed putative human, mouse, zebrafish and yeast homologues of EntA. The human homologue was identified as a short chain dehydrogenase/reductase family member originally called DHRS6 and now referred to as BDH2 (Guo et al., 2006). RT-PCR analysis indicates that the murine Bdh2 gene was widely expressed in mouse tissues (Figure S2A).
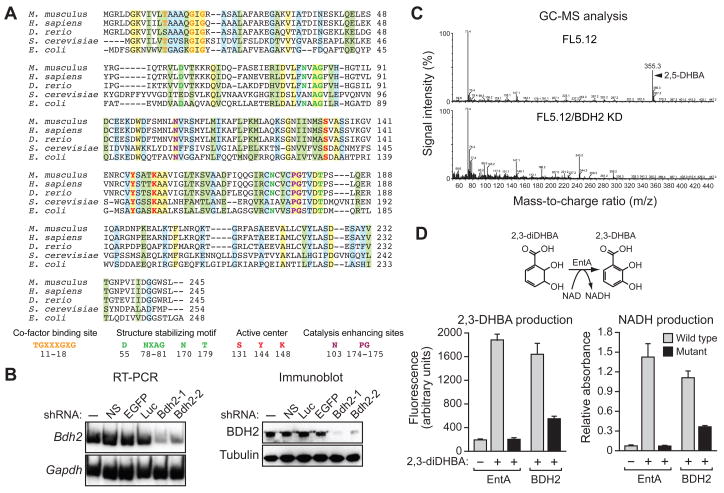
Figure 2. Identification of a Murine Homologue of Bacterial EntA that Mediates 2,5-DHBA Synthesis.
(A) Multiple sequence alignment of EntA/BDH2 homologues. Identical residues between all five proteins are indicated in yellow; conserved residues in green, and semi-conserved residues in blue.
(B) RT-PCR (left) and immunoblot (right) analysis monitoring murine Bdh2 expression in FL5.12 cells treated with one of two unrelated shRNAs targeting Bdh2 or a control non-silencing (NS), enhanced green fluorescent protein (EGFP) or luciferase (Luc) shRNA.
(C) GC-MS analysis of the eluted 24p3 sample prepared from parental FL5.12 cells or FL5.12 cells expressing a Bdh2 shRNA.
(D) Wild-type and mutant bacterial EntA and mouse BDH2 proteins were analyzed for their ability to convert 2,3-diDHBA to 2,3-DHBA (left) or NAD to NADH (right). Error bars indicate SD. Mutant EntA proteins were expressed at comparable levels to the wild-type proteins (Figure S2C). See also Figure S2.
To determine whether BDH2 was in fact involved in synthesis of 24p3-associated 2,5-DHBA, we derived two unrelated shRNAs directed against murine Bdh2 (Bdh2-1 and Bdh2-2). The RT-PCR and immunoblot results of Figure 2B show that both shRNAs knocked down Bdh2 expression. Bdh2-1 was more efficient than Bdh2-2 at knocking down Bdh2 expression, which is relevant to results of some of the functional experiments presented below. We then repeated the GC-MS analysis of Figure 1C in BDH2 knockdown (KD) cells. Figure 2C shows that in contrast to parental FL5.12 cells, in BDH2 KD cells there was no detectable 24p3-associated 2,5-DHBA.
Finally, we tested whether BDH2 was a functional homologue of bacterial EntA by asking whether it could catalyze an equivalent enzymatic reaction. The NAD+-dependent oxidation of 2,3-diDHBA to 2,3-DHBA can be monitored by ultraviolet (UV) fluorescence (Liu et al., 1989). Figure 2D (left panel) shows, as expected, that E. coli EntA efficiently catalyzed the production of 2,3-DHBA from 2,3-diDHBA, whereas this reaction was not catalyzed by an EntA derivative containing a point mutation (Tyr144 to Phe) within the predicted active site (Filling et al., 2002). Significantly, 2,3-DHBA production was also catalyzed by recombinant BDH2 but not by a mutant BDH2 derivative (Tyr147 to Phe). As expected, in these same reactions wild-type EntA and BDH2 proteins, but not their mutant derivatives, catalyzed the conversion of NAD to NADH (Figure 2D, right panel). Collectively, these results indicate BDH2 is a homologue of bacterial EntA.
BDH2 is Required for 24p3-Mediated Iron Transport and Apoptosis
The finding that following BDH2 knockdown there was no detectable 24p3-associated 2,5-DHBA suggested that 24p3 would be unable to bind iron in BDH2 KD cells. To test this prediction, we analyzed 24p3 iron-binding activity in cells expressing a Bdh2 shRNA. Figure 3A shows that 24p3 present in the CM from IL-3-deprived FL5.12 cells or ponasterone A-treated FL5.12/Ec-24p3 cells bound to 55Fe, whereas 24p3 iron-binding activity was markedly decreased in BDH2-depleted cells despite near equivalent amounts of 24p3 in the CM.
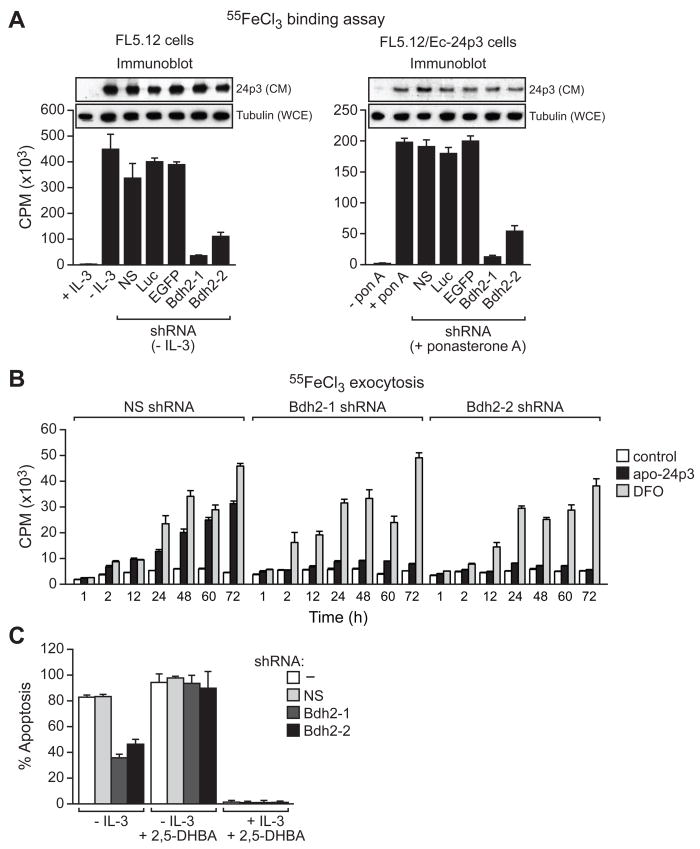
Figure 3. BDH2 is Required for 24p3-Mediated Iron Transport and Apoptosis.
(A) Iron binding assay. (Left) FL5.12 cells were treated in the presence or absence of IL-3, and with a Bdh2 or control shRNA, and the iron-binding ability of 24p3 in the CM was monitored as described in Figure 1B. Inset, immunoblot analysis monitoring 24p3 levels in the CM and tubulin levels in the whole cell extract (WCE). (Right) FL5.12/Ec-24p3 cells were treated in the presence or absence of ponasterone A. Error bars indicate SD.
(B) Iron exocytosis assay. 55FeCl3-loaded FL5.12 cells were either untreated (control) or treated with apo-24p3 or DFO, and the presence of 55Fe in the medium was quantified.
(C) Apoptosis assay. FL5.12 cells cultured in the presence or absence of IL-3, with or without 2,5-DHBA, and expressing a NS or Bdh2 shRNA were analyzed for apoptosis by annexin V-FITC staining.
We next performed a series of experiments to analyze the ability of 24p3 to mediate iron transport in BDH2-depleted cells. Following receptor-mediated internalization, apo-24p3 chelates intracellular iron and exits the cell via the recycling endosome thereby transferring iron to the extracellular medium. The resultant decrease in intracellular iron concentration induces apoptosis (Devireddy et al., 2005). We predicted that in the absence of a functional siderophore, apo-24p3 would be unable to deplete intracellular iron and therefore fail to induce apoptosis.
As a first test of this prediction we analyzed the ability of apo-24p3 to transfer iron to the extracellular medium in BDH2 KD cells. FL5.12 cells were loaded with radiolabeled iron by pre-incubation with 55FeCl3 followed by re-culturing in fresh medium lacking 55Fe. 55Fe-loaded cells were treated with apo-24p3 or, as a control, the membrane-permeable iron chelator desferrioxamine (DFO), and the accumulation of 55Fe in the medium was determined by liquid scintillation counting. Figure 3B shows that addition of apo-24p3 or DFO to control non-silencing shRNA-treated cells led to the time-dependent accumulation of 55Fe in the medium. By contrast, in BDH2 KD cells, DFO but not apo-24p3 was able to transfer 55Fe to the extracellular medium.
We next analyzed the ability of 24p3 to induce apoptosis. We have previously shown that in FL5.12 cells, IL-3 deprivation induces 24p3 expression resulting in apoptosis (Devireddy et al., 2001). Figure 3C shows that in BDH2 KD FL5.12 cells the level of apoptosis resulting from IL-3 deprivation was markedly reduced compared to cells treated with a non-silencing shRNA. Remarkably, addition of 2,5-DHBA restored the ability of BDH2 KD FL5.12 cells to induce apoptosis following IL-3 deprivation. Notably, in FL5.12 cells cultured in the presence of IL-3, and which therefore do not express 24p3, addition of 2,5-DHBA did not promote apoptosis. Collectively, these results indicate that BDH2 is required for synthesis of the 24p3-associated mammalian siderophore and thus for 24p3-mediated iron binding and trafficking activities.
BDH2 is Required to Maintain Normal Cytoplasmic Iron Levels
Next, we performed a series of experiments to determine the role of the siderophore in intracellular iron homeostasis. First, we used a colorimetric assay to measure cytoplasmic iron concentration in cells containing or lacking the siderophore. The results of Figure 4A show that total cytoplasmic iron levels were markedly elevated in BDH2 KD cells. Second, we measured free iron concentration using calcein, a fluorescent cell permeable dye whose emission is quenched by iron binding (Kakhlon and Cabantchik, 2002). Figure 4B shows that BDH2 KD cells contained substantially higher free iron levels, as evidenced by a decrease in fluorescence intensity. Similar results were obtained in murine mIMCD kidney cells (Figure S3A–C).
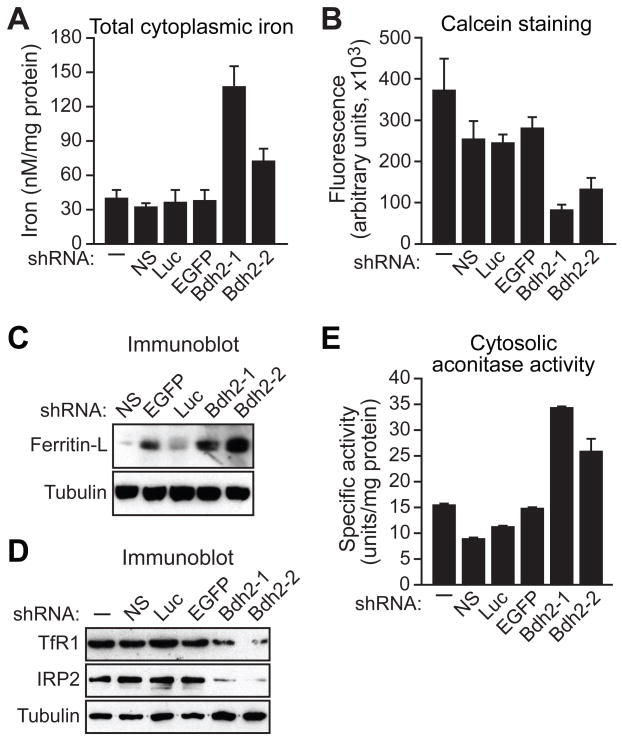
Figure 4. BDH2 is Required to Maintain Normal Cytoplasmic Iron Levels.
(A–E) FL5.12 cells expressing a Bdh2 or control shRNA were analyzed for total cytoplasmic iron levels by colorimetric analysis (A), for free cytoplasmic iron concentration by fluorescence calcein assay (B), for ferritin-L (C) and Tfr1 and IRP2 (D) levels by immunoblot analysis, and for cytosolic aconitase activity (E). Error bars indicate SD. See also Figures S3A–D and S4.
Intracellular iron levels were also assessed by immunoblotting for two iron-regulated proteins, ferritin-L and transferrin receptor 1 (TfR1). In BDH2 KD cells ferritin-L levels were substantially elevated (Figures 4C and S3D), whereas TfR1 levels were decreased (Figure 4D), both of which are indicative of increased intracellular iron concentration.
We also analyzed the iron-regulatory proteins IRP1 and IRP2. Upon elevated iron concentration, IRP2 undergoes rapid proteasomal degradation (Iwai et al., 1995). In BDH2 KD cells IRP2 levels were decreased, indicative of increased iron concentration (Figure 4D). IRP1 contains an Fe-S cluster that, upon elevated iron concentration, undergoes a conformational switch converting the RNA binding protein to a cytosolic aconitase (Rouault et al., 1991). We found that BDH2 KD cells had increased cytosolic aconitase activity, as expected if cytoplasmic iron levels are elevated (Figure 4E and see also Figure S4). Thus, based upon multiple, independent lines of evidence, cells lacking a functional siderophore accumulate abnormally high levels of cytoplasmic iron.
The Siderophore Protects Cells from Oxidative Stress
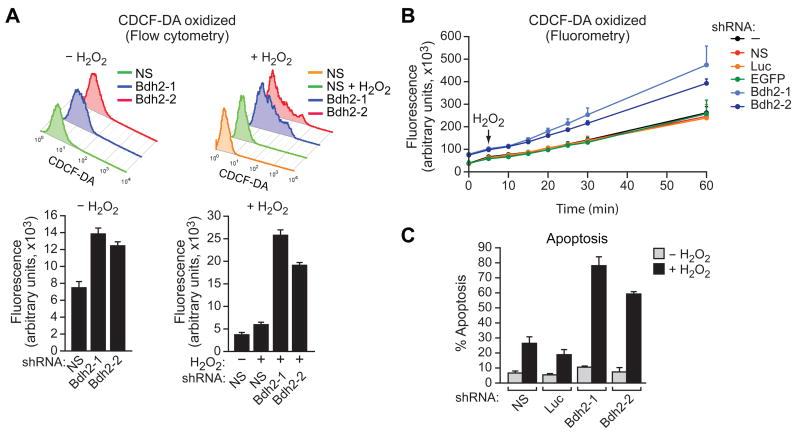
Free iron can generate ROS, prompting us to measure ROS levels in siderophore-depleted cells. In these experiments, we imposed an oxidative stress by addition of hydrogen peroxide. In the first set of experiments, the accumulation of ROS was measured by preloading cells with the non-fluorescent probe 2′-7′ carboxy-dichlorodihydrofluorescein-diacetate (CDDHCF-DA), which is rapidly converted to the highly fluorescent compound 2′,7′-carboxy-dichlorofluorescein-diacetate (CDCF-DA) by ROS. The results, assayed by either flow cytometry (Figure 5A) or fluorometry (Figures 5B and S3E), show that in BDH2 KD cells there was higher ROS production as evidenced by increased CDCF-DA fluorescence. Significantly, basal ROS levels in the absence of hydrogen peroxide were also substantially higher in BDH2 KD cells (Figure 5A).
Figure 5. The Siderophore Protects Cells from Oxidative Stress.
(A) FL5.12 cells expressing a NS or Bdh2 shRNA and treated in the presence or absence of hydrogen peroxide (H2O2) were analyzed for ROS by monitoring CDCF-DA fluorescence using flow cytometry. Error bars indicate SD.
(B) FL5.12 cells expressing a control or Bdh2 shRNA were analyzed for ROS by monitoring CDCF-DA fluorescence using fluorometry. Error bars indicate SD.
(C) FL5.12 cells expressing a control or Bdh2 shRNA were treated in the presence or absence of H2O2 and apoptosis was measured by annexin V-FITC staining. Error bars indicate SD. See also Figure S3E.
Increased ROS can lead to apoptosis (reviewed in Simon et al., 2000). Therefore, we predicted siderophore-depletion would result in increased apoptosis following oxidative stress. To test this idea, cells were treated with hydrogen peroxide and apoptosis was measured by annexin V-FITC staining. The results of Figure 5C show that siderophore-depleted cells were markedly more sensitive to oxidative stress-induced apoptosis, consistent with their elevated cytoplasmic iron levels.
BDH2 is Required for Mitochondrial Iron Homeostasis
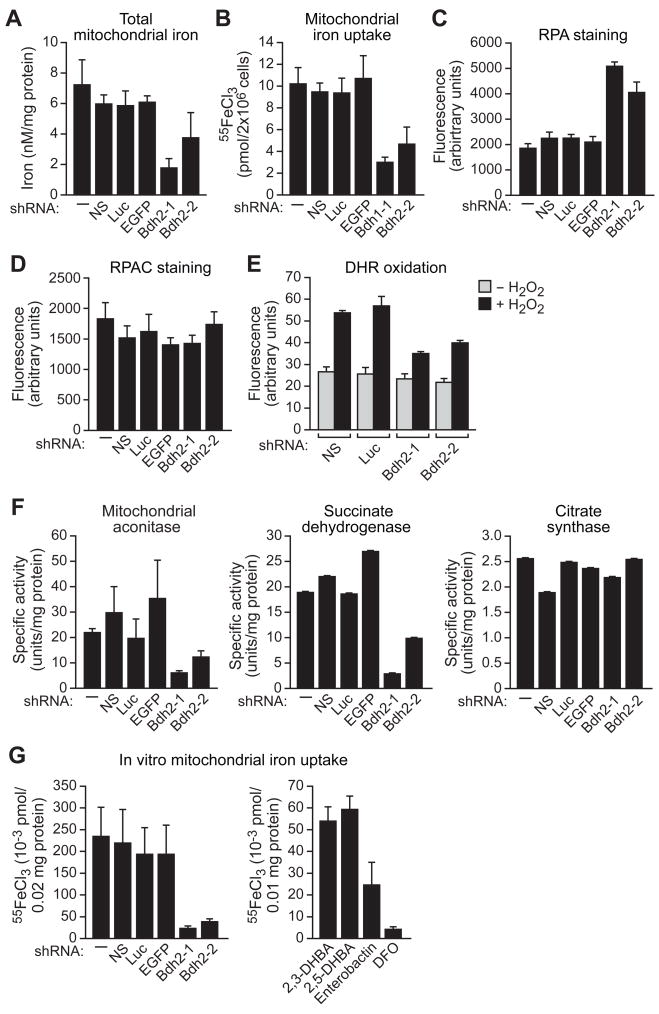
We next performed a series of experiments to assess mitochondrial iron levels in siderophore-depleted cells. Unexpectedly, total mitochondrial iron levels were significantly reduced in BDH2 KD cells (Figures 6A and S3F). To confirm this result, we analyzed incorporation of radioactive iron into mitochondria. In brief, iron-deprived cells were labeled with 55FeCl3, mitochondria were isolated (Figure S5), and the amount of incorporated 55Fe quantified. Figure 6B shows that mitochondrial radioactive iron incorporation was significantly reduced following BDH2 knockdown.
Figure 6. BDH2 is Required for Mitochondrial Iron Homeostasis.
(A–F) FL5.12 cells expressing a Bdh2 or control shRNA were analyzed for mitochondrial iron levels by colorimetric analysis (A), for mitochondrial iron uptake (B), for RPA (C) and RPAC (D) fluorescence by fluorometry, for DHR 123 oxidation in the presence or absence of H2O2 by flow cytometry (E), and for activities of mitochondrial aconitase, succinate dehydrogenase and citrate synthase (F).
(G) In vitro mitochondrial iron import. (Left) Proteolytically treated cytosolic extracts from control or Bdh2 shRNA-treated FL5.12 cells were added to purified mitochondria, and iron import was determined by monitoring mitochondrial incorporation of 55FeCl3. (Right) Purified mitochondria were treated with 2,3-DHBA, 2,5-DHBA, enterobactin or DFO, and iron import was monitored. See also Figures S3F-J and S5.
We next asked whether mitochondrial free iron was also altered in siderophore-depleted cells. The fluorescent mitochondrial iron indicator rhodamine B-[(1,10-phenanthrolin-5-yl)aminocarbonyl]benzyl ester (RPA) contains a rhodamine moiety for mitochondrial targeting and a 1,10-phenanthroline moiety for iron chelation; iron binding quenches the fluorescence of RPA (Rauen et al., 2007). Following RPA addition, fluorescence was significantly higher in BDH2 KD cells compared to control cells (Figures 6C and S3G), indicative of reduced mitochondrial iron levels. A control experiment using a structurally similar compound, rhodamine B-[(phenanthren-9-yl)aminocarbonyl]benzyl (RPAC), which contains the same fluorophore and linker as RPA but lacks iron-chelating properties and thus its fluorescence is not responsive to iron (Rauen et al., 2007), confirmed that the results of Figure 6C are explained by differences in mitochondrial iron levels and not by an inability of RPA to enter into the mitochondria of BDH2 KD cells (Figure 6D, and see also Figure S3H).
The lower concentration of mitochondrial free iron in BDH2 KD cells should result in reduced levels of mitochondrial ROS. To test this prediction, we measured mitochondrial ROS by incubating cells with the redox-sensitive probe dihydrorhodamine (DHR 123), which is converted by oxidation to the fluorescent compound rhodamine 123. Following DHR 123 loading and hydrogen peroxide addition, fluorescence intensity in BDH2 KD cells was significantly reduced, indicative of decreased mitochondrial ROS (Figures 6E and S3I).
As a final confirmation of altered mitochondrial iron levels, we measured the activity of several iron-dependent mitochondrial enzymes. Figure 6F shows that the activities of the Fe-S cluster-containing enzymes, mitochondrial aconitase and succinate dehydrogenase, were substantially reduced in BDH2 KD cells (see also Figure S3J). By contrast, citrate synthase, which does not contain an Fe-S cluster and whose function is not iron dependent, was comparably active in control and BDH2 KD cells.
The results described above raised the possibility that iron is imported into mitochondria as an iron-siderophore complex. To provide additional support for this conclusion we performed a series of in vitro mitochondrial iron import experiments. Proteolytically treated cytosolic extracts from control or BDH2 KD cells were added to purified mitochondria, and iron import was determined by monitoring mitochondrial incorporation of 55FeCl3. The results of Figure 6G (left panel) show that all control cytosolic extracts supported substantial levels of mitochondrial iron import. Significantly, mitochondrial iron import was markedly reduced in reaction mixtures containing cytosolic extracts from BDH2 KD cells. In a second experiment, we found that addition of 2,5-DHBA, 2,3-DHBA and, to a lesser extent, enterobactin enabled purified mitochondria to import radioactive iron (Figure 6G, right panel). By contrast, DFO failed to support mitochondrial iron import.
Requirement of Vertebrate EntA Homologues for Heme Synthesis
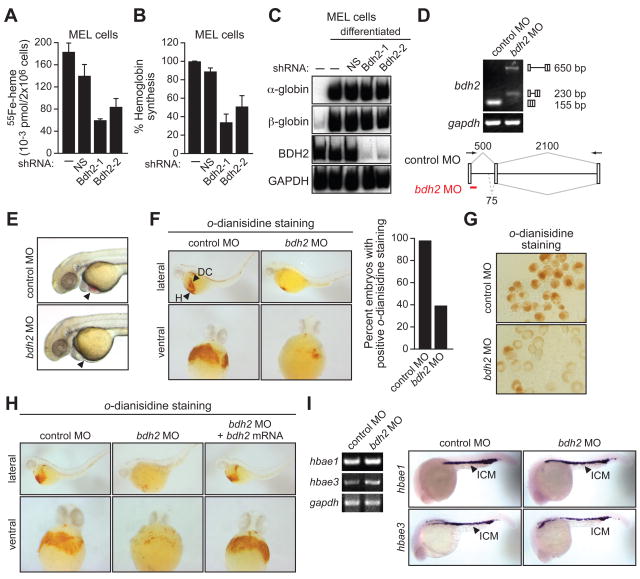
Synthesis of heme occurs in the mitochondria and insufficient mitochondrial iron is known to decrease heme synthesis, resulting in lower hemoglobin levels (reviewed in Napier et al., 2005). To determine the role of BDH2 in synthesis of heme and hemoglobin we performed an initial series of experiment in mouse erythroleukemia (MEL) cells. Untreated MEL cells or MEL cells expressing a control or BDH2 shRNA were differentiated into erythroid cells by addition of dimethyl sulfoxide and assayed for the production of heme and hemoglobin. We found that BDH2 knockdown resulted in decreased levels of both heme (Figure 7A and see Figure S4) and hemoglobin (Figure 7B). The RT-PCR results of Figure 7C show that expression of α- and β-globin was normal in BDH2 KD cells and is thus not responsible for the decreased hemoglobin levels.
Figure 7. Requirement of Vertebrate EntA Homologues for Heme Synthesis.
(A) MEL cells expressing a NS or Bdh2 shRNA were differentiated. Following labeling with 55Fe, cells were lysed and heme was extracted and analyzed for 55Fe incorporation. Error bars indicate SD.
(B) MEL cells were differentiated as described in (A) and following cell lysis, hemoglobin levels were determined by benzidine reaction. Hemoglobin content was expressed relative to that in untreated MEL cells cultured in the absence of DMSO. Error bars indicate SD.
(C) MEL cells were differentiated as described in (A) and α- and β-globin mRNA levels monitored by RT-PCR. The levels of Bdh2 and Gapdh were monitored as controls.
(D) RT-PCR analysis. The bdh2 MO was injected into one-cell stage embryos and 24 hr later, RNA was extracted and analyzed by RT-PCR. Treatment with the bdh2 MO resulted in retention of the entire intron (upper band) as well as activation of a cryptic splice site that resulted in partial intron retention (lower band).
(E) Phenotypic analysis of control MO- and bdh2 MO-injected embryos at 72 hpf. Bdh2-depleted embryos lack hemoglobinized erythrocytes, as evidenced by the absence of red color (arrow).
(F) Whole-mount o-dianisidine staining of control MO and bdh2 MO-injected embryos at 72 hpf. Shown are lateral (top) and ventral (bottom) views of the anterior region of embryos. Control MO-injected embryos show positive o-dianisidine staining at the duct of Cuvier (DC) and the heart (H), which is markedly reduced in bdh2 MO-injected embryos. (Right) Quantification of the percent embryos with normal o-dianisidine staining.
(G) Blood cells were isolated from control or bdh2 MO-injected embryos and analyzed for hemoglobinization by o-dianisidine staining.
(H) Whole-mount o-dianisidine staining of embryos (72 hpf) injected with a control MO or bdh2 MO, or bdh2 MO-treated embryos co-injected with a bdh2 mRNA.
(I) RT-PCR (left) and in situ hybridization analysis (right) for hbae1 and hbae3 expression in control and bdh2 MO-injected embryos. See also Figures S4 and S6.
To confirm and extend these cell culture results we analyzed the role of the siderophore in zebrafish embryos. As stated above, we found that zebrafish also contained an EntA homologue (see Figure 2A). Whole-mount in situ hybridization analysis revealed that bdh2 mRNA was widely distributed throughout the zebrafish embryo (Figure S6A). To ablate Bdh2 function in zebrafish embryos, we designed an antisense morpholino oligonucleotide (MO) to induce an aberrant splicing event, which was confirmed by RT-PCR analysis (Figure 7D). The bdh2 MO was injected into wild-type one-cell stage fertilized embryos in parallel with a randomized control MO. Figure 7E shows that bdh2 MO-injected embryos developed normally and were morphologically indistinguishable from control MO-injected embryos. Notably, however, Bdh2-depleted embryos had hypochromic blood (i.e., lacking red color), suggesting an absence of hemoglobinized erythrocytes. Identical results were obtained with a second, unrelated bdh2 MO (Figure S6B).
To determine the basis for the hypochromia, we analyzed hemoglobin content by whole embryo staining with o-dianisidine, a heme-specific stain (Iuchi and Yamamoto, 1983). Control MO-injected embryos displayed intense o-dianisidine staining at the duct of Cuvier and the heart, indicative of normal hemoglobinized blood flow over the yolk sac, whereas Bdh2-depleted embryos exhibited a marked reduction in o-dianisidine-staining (Figures 7F and S4B). Reduced o-dianisidine staining was also evident in blood cells isolated from Bdh2-depleted embryos (Figure 7G). Injection of a bdh2 mRNA bearing a mutation that prevented hybridization with the bdh2 MO rescued the normal pattern of o-dianisidine staining in Bdh2-depleted embryos (Figure 7H). Notably, in Bdh2-depleted embryos the globin genes hbae1 and hbae3 were expressed normally (Figure 7I), indicating that the decreased hemoglobin levels are not due to reduced globin gene expression.
DISCUSSION
In this study, we have confirmed the existence of a mammalian siderophore and provided insight into its structure, biosynthesis and function. We have identified a mammalian homologue to EntA, a bacterial protein that has a critical role in synthesis of the bacterial siderophore enterobactin. RNA interference-mediated knockdown of BDH2 depletes cells of the siderophore and results in profound dysregulation of cytoplasmic and mitochondrial iron levels (summarized in Figure S7). The majority of our experiments were performed in hematopoietic FL5.12 cells but BDH2 is broadly expressed (Figure S2A) and we have obtained similar results in other mammalian cell types (Figures 7A–C and S3). Finally, we have also identified putative zebrafish and yeast homologues (see Figure 2A) with functional similarities to mammalian ENTA (Figures 7D–I and S4, respectively). Collectively, our results reveal features of intracellular iron homeostasis that are conserved from bacteria through humans.
Although we have identified the iron-binding moiety of the mammalian siderophore, the composition and structure of the intact siderophore remain to be determined. Notably, in addition to EntA, we have identified putative mammalian homologues of EntE and EntF (Figure S2B), two proteins required for the final stage of enterobactin synthesis. These results suggest that there are likely to be additional similarities between the biosynthetic pathways of the mammalian siderophore and bacterial enterobactin.
Role of the Mammalian Siderophore in Cytoplasmic Iron Homeostasis
Although the vast majority of intracellular iron is safely bound to proteins, a small portion of intracellular iron (0.2–3%) is bound to low molecular weight ligands and forms a transit pool for intracellular trafficking and synthesis of iron-containing proteins. A number of putative iron-binding ligands have been identified, including peptides, proteins and nucleotides (Breuer et al., 2008). However, the function of these putative iron-binding ligands in iron trafficking and homeostasis has not been established.
We have presented several lines of evidence that demonstrate the iron-siderophore complex is a critical functional component of the intracellular free iron pool. First, the ability of intracellularly-delivered apo-24p3 to substantially deplete cells of iron indicates that much of the cytoplasmic free iron is bound to the siderophore (Devireddy et al., 2005). Second, depletion of the siderophore profoundly perturbs intracellular iron homeostasis, resulting in markedly increased cytoplasmic iron levels. The increased cytoplasmic iron levels in siderophore-depleted cells is due, at least in part, to decreased transport of iron from the cytoplasm to the mitochondria (see below).
We have found that siderophore-depleted cells contain higher levels of ROS and are substantially more sensitive to oxidative stress. Previous studies have shown that chelated iron is much less reactive than free iron in generating ROS (Blat et al., 2008; Lukinova et al., 2009). Thus, we propose that a major function of the siderophore is to protect cells from oxidative stress while providing a source of free iron.
Role of the Siderophore in Mitochondrial Iron Homeostasis
Several lines of evidence indicate that the siderophore we have identified here plays an important role in mitochondrial iron import. First, we have found that despite markedly elevated cytoplasmic iron levels, the mitochondria of siderophore-depleted cells are iron deficient. Second, our in vitro mitochondrial import experiments provide evidence that iron is imported into mitochondria as an iron-siderophore complex (see Figure 6G). Previous studies have identified two proteins involved in mitochondrial iron import, mitoferrin 1 (Mfrn1) and mitoferrin 2 (Mfrn2), which are homologous members of the mitochondrial solute carrier family (Paradkar et al., 2009; Shaw et al., 2006). What, then, is the relationship between the siderophore- and Mfrn-mediated iron import pathways? One possibility is that there are two independent mitochondrial import pathways, one involving Mfrns and a second involving the siderophore. Alternatively, a parsimonious explanation for these collective results is that the iron-siderophore complex, rather than free iron, is the form in which iron is transported by Mfrns.
EXPERIMENTAL PROCEDURES
Cell Lines
FL5.12/Ec-24p3 cells were generated as previously described (Devireddy et al., 2001). Murine erythroleukemia (MEL) cells were maintained in DMEM supplemented with 10% FBS and differentiated by incubation in 1.5% dimethyl sulfoxide.
24p3-Iron Binding Assays
CM from ponasterone A-treated FL5.12/Ec-24p3 cells or IL-3-deprived FL5.12 cells was incubated with 5 mCi 55FeCl3 (Perkin-Elmer) overnight at 4°C. 24p3 was immunoprecipitated from the CM of FL5.12/Ec-24p3 cells using an anti-HA antibody (Santa Cruz Biotechnology) or from the CM of IL-3-deprived FL5.12 cells using a rabbit polyclonal anti-24p3 antibody raised against full-length recombinant 24p3-derived from E. coli strain BL21. The immunoprecipitate was analyzed for the presence of 55Fe by liquid scintillation counting.
For the in vitro iron binding assays of Figure 1G, purified apo-24p3 (Goetz et al., 2002) was incubated with 500 nM 55FeCl3 and increasing concentrations of either 2,3-DHBA, 2,5-DHBA, iron-free enterobactin (EMC Microcollections) or desferrichrome (EMC Microcollections). Following immunoprecipitation of 24p3, radioactivity was measured in the 24p3 immunoprecipitate.
Gas Chromatography-Mass Spectrometry
The immunopurified 24p3-siderophore complex was incubated with 1% acetic acid for 2 hr at room temperature, and passed through a Centricon YM-10 column (Millipore). The small molecule flow-through fraction was derivatized with BSTFA-pyridine (N,O-bis[Trimethylsilyl]trifluoroacetamide; Pierce Biotechnology), 1:1 at 80°C for 1 hr and subjected to GC-MS. The NIST library search was performed against 191,436 MS spectra (www.nist.gov/srd/nist1a.htm). DHBA standards were obtained from Sigma Chemical Company. GC-MS analysis was also performed under milder conditions (BSTFA-pyridine, 1:1 at 40°C for 15 min).
Tryptophan Fluorescence Quenching Assay
Briefly, 120 nM apo-24p3 was incubated with increasing amounts of 2,3- or 2,5-DHBA and the fluorescence intensity was determined at 280 nm (excitation)/340 nm (emission) as previously described (Goetz et al., 2002).
EntA Homologue Identification
The E. coli EntA protein (NP_415128) was used to perform BLAST searches against non-redundant protein sequence databases to identify homologues in Mus musculus (NP_081484.1; also known as Bdh2/Dhrs6), Homo sapiens (NP_064524.3; BDH2/DHRS6), Danio rerio (NP_001017809.1; zgc:110323) and Saccharomyces cerevisiae (NP_012301.1; YIR035C). The amino acid sequences were aligned using ClustalW2 sequence analysis software.
RNA Interference
Non-silencing (RS 1707), luciferase (RHS 1705) and EGFP (RHS 1706) shRNAs were purchased from Open Biosystems, and Bdh2 shRNAs were synthesized (Table S1). Naïve FL5.12 or FL5.12/Ec-24p3 cells were transduced with retroviruses expressing shRNAs and selected for puromycin resistance (2 μg/ml). Knockdown efficiency was assessed by RT-PCR (see Table S1 for primer sequences) or by immunoblot analysis using a rabbit polyclonal antibody raised against recombinant E. coli EntA protein or a monoclonal anti-α-tubulin antibody (Sigma).
2,3-diDHBA Oxidation Assay
Site-directed mutagenesis to create the Y144F/Y147F mutant was performed using a kit from Stratagene. Wild-type and mutant EntA proteins were purified as previously described (Sundlov et al., 2006) and assayed for enzymatic activity as previously described (Liu et al., 1989). Conversion of 2,3-diDHBA to 2,3-DHBA was monitored at 306 nm (excitation)/437 nm (emission), and the conversion of NAD to NADH was measured at 340 nm.
Iron Exocytosis Assays
FL5.12 cells expressing a non-silencing or Bdh2 shRNA were deprived of iron by culturing in serum-free MEM supplemented with IL-3 for 12 hr, and then incubated with 100 μCi 55FeCl3 in serum-containing MEM supplemented with IL-3 for 24 hr. Labeled cells were washed, placed in fresh growth medium and treated with 10 μg GST or apo-24p3 or 100 μM DFO (Calbiochem), and the accumulation of 55Fe in the medium was determined by liquid scintillation counting.
Assessment of Intracellular Iron Levels
For determination of total cytoplasmic iron, mitochondrial-depleted cytoplasmic extracts were analyzed using Iron Color Reagent (Sigma). Iron concentrations in the samples were calculated from a standard curve plotted from iron standards (Sigma). Free iron was measured using the iron-sensitive probes Calcein Green (CALG; Molecular Probes) or Blue (B-AM; Molecular Probes), and RPA or RPAC (Axxora), for cytosolic and mitochondrial compartments, respectively, according to the manufacturer’s instructions. For immunoblot analysis, blots were probed with antibodies against Ferritin-L (Santa Cruz Biotechnology), TfR-1 (Zymed), IRP-1 (gift from Betty Leibold) or IRP-2 (Novus Biologicals). Blots were stripped and reprobed with an α-Tubulin antibody (Santa Cruz Biotechnology).
Total mitochondrial iron was determined by colorimetric assay as described above. To measure mitochondrial incorporation of radioactive iron, cells were labeled with 55FeCl3 as described above, mitochondria from labeled cells were isolated (see Supplementary Experimental Procedures), and the amount of incorporated 55Fe was determined by liquid scintillation. To assess mitochondrial free iron, cells were treated with 1 μM RPA or RPAC, and fluorescence was measured at 544 nm (excitation)/590 nm (emission).
Measurement of ROS Species
The redox-mediated conversion of non-fluorescent CDDHCF-DA (25 μM) to the fluorescent CDCF-DA was measured either by flow cytometry or using a fluorescence plate reader [488 nm (excitation)/517 nm (emission)]. Mitochondrial ROS formation was determined by analyzing oxidation of DHR 123 (50 mM) to rhodamine 123 by fluorometry [485 nm (excitation)/535 nm (emission)] or flow cytometry. Cytosolic and mitochondrial ROS were also determined following treatment with 50 μM H2O2 for 24 hr. Apoptosis was measured by annexin V-FITC staining 24 hr after treatment with 100 μM H2O2.
Enzymatic Activity Assays
Cytosolic aconitase activity was measured as previously described (Gardner et al., 1994); one unit was defined as the amount of enzyme necessary to produce 1 mM NADPH per minute (e340 = 3.13 mM−1 cm−1; extinction coefficient adjusted for the path length of the solution in the well). Mitochondrial aconitase was determined as previously described (Campanella et al., 2009) using an extinction coefficient of 1.8 mM−1 cm−1. Citrate synthase activity was assayed as previously described (Powell and Jackson, 2003) using an extinction coefficient of 6.8 mM−1 cm−1. Succinate dehydrogenase was assayed as previously described (Campanella et al., 2009) using an extinction coefficient of 9.65 mM−1 cm−1.
In vitro Mitochondrial Iron Import Assays
Cytosolic extracts (50 μg) from control or BDH2 KD FL5.12 cells were treated with 1 unit of carboxypeptidase immobilized onto agarose beads (Thermo Scientific) overnight at 4°C, and the beads removed by centrifugation. The treated extracts were then incubated with 100 μM 55FeCl3 for 1 hr at 37°C. Extracts were then added to purified mitochondria (0.02 mg) isolated from naïve cells and incubated for 1 hr at 37°C, and the amount of 55Fe in lysed mitochondria was determined by liquid scintillation. For control experiments, 1 μM 2,3-DHBA, 2,5-DHBA, DFO or iron-free enterobactin was incubated with 1 μM 55FeCl3 (or 3 μM for enterobactin) overnight at 4°C, followed by addition of mitochondria (0.01 mg), incubation for 1 hr at 37°C and determination of 55Fe in lysed mitochondria.
Heme and Hemoglobin Quantification
Heme measurements were performed as described previously (Shaw et al., 2006). Briefly, differentiated MELs were washed with 1x PBS, placed in MEM supplemented with 0.5% FBS for 2 hr at 37°C, and then in DMEM supplemented with 5% dialyzed FBS and 55Fe-Transferrin with 1.2 mM ∂-aminolevulinic acid for 8 hr at 37°C. Cells were then washed with 1x PBS and lysed in RIPA lysis buffer. Hemoproteins were denatured by adding 0.1 N HCl, and heme was extracted by adding an equal amount of cyclohexanone. The CPM in the organic phase was then determined by liquid scintillation counting. For hemoglobin quantification, differentiated MELs were washed with 1x PBS, and lysed with 1.5% octyl glucoside for 30 min on ice. The lysates were clarified by centrifugation at 13,000 rpm for 10 min at 4°C. For hemoglobin determination, a volume corresponding to 50 μg of protein was added to 100 μl freshly prepared benzidine (0.5 mg/ml in 10% acetic acid) and 10 μl of 30% H2O2. After 10 min, optical density was determined at 604 nm in a plate reader (Perkin Elmer). RT-PCR analysis was performed using primers listed in Table S1.
Zebrafish Experiments
Whole-mount in situ hybridization was performed as previously described (Hart et al., 2007). Riboprobes were prepared from full-length zebrafish cDNAs cloned into the pGEM-T TA cloning vector (Promega) (see Table S1 for primer sequences). Staining for o-dianisidine was performed as previously described (Detrich et al., 1995).
MOs (Table S1) were injected into 1–2 cell stage wild-type embryos at a range of concentrations and their effect on morphology was assessed up to 72 hr post-injection. For rescue experiments, full-length human BDH2 cDNA was amplified from p11-DHRS6 (provided by Upo Opperman), fused in-frame at the C-terminus with myc, and inserted into pCS2+ (provided by David Turner). Capped sense mRNA was synthesized using the mMESSAGE mMACHINE kit (Ambion) and injected immediately following MO injections into the same embryos. For RT-PCR analysis, total RNA was prepared using Trizol reagent (Invitrogen) and RT-PCR was performed using primers listed in Table S1.
Circulating blood cells were isolated from zebrafish by tail amputation as previously described (Brownlie et al., 1998). Cells were cytospun onto glass slides and fixed for 5 min in 12% glacial acetic acid containing 0.4% benzidine (Sigma). H2O2 was added to a final concentration of 0.3% and cells were incubated at room temperature for 10–15 min and photographed immediately.
Statistical Analysis
Results were analyzed using either ANOVA or Student’s t-test, and for all graphs presented P<0.05.
Research Highlights
The iron-binding moiety of the mammalian siderophore is 2,5-DHBA
The mouse enzyme responsible for 2,5-DHBA synthesis is homologous to bacterial EntA
Mammalian cells lacking the siderophore accumulate high amounts of cytoplasmic iron
Iron is imported into mitochondria as an iron-siderophore complex
Supplementary Material
Acknowledgments
We thank Andrew Gulick, Michael Mueller, Udo Oppermann, Betty Leibold, Kostas Pantopoulos, Barry Paw, Arthur Skoultchi, David Plas, Sonia Levi, Jerry Kaplan, Maurice Burg and David Turner for providing reagents; Andy Dancis, Heidi O’Neill, Yavuz Oktay, Stacy Dalheimer, Jon Donze, David Yao, Wei-Dong Chen, Marvin Neiman, Kiran Velpula and Jun Liu for assistance with various experimental procedures; Jim and Barbara Evans from the UMMS Proteomics and Mass Spectrometry Facility for GC-MS analysis; and Sara Evans for editorial assistance. This work is supported by NIH R01 DK081395-01 and Case Western Reserve University start up funds to L.R.D. and NIH R01 CA115817-04 to M.R.G. L.R.D. is also a recipient of career developmental awards from March of Dimes and American Society of Hematology. M.R.G. is an Investigator of the Howard Hughes Medical Institute.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Akerstrom B, Flower DR, Salier JP. Lipocalins: unity in diversity. Biochim Biophys Acta. 2000;1482:1–8. doi: 10.1016/s0167-4838(00)00137-0. [DOI] [PubMed] [Google Scholar]
- Andrews NC. Forging a field: the golden age of iron biology. Blood. 2008;112:219–230. doi: 10.1182/blood-2007-12-077388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blat D, Weiner L, Youdim MB, Fridkin M. A novel iron-chelating derivative of the neuroprotective peptide NAPVSIPQ shows superior antioxidant and antineurodegenerative capabilities. J Med Chem. 2008;51:126–134. doi: 10.1021/jm070800l. [DOI] [PubMed] [Google Scholar]
- Breuer W, Shvartsman M, Cabantchik ZI. Intracellular labile iron. Int J Biochem Cell Biol. 2008;40:350–354. doi: 10.1016/j.biocel.2007.03.010. [DOI] [PubMed] [Google Scholar]
- Brownlie A, Donovan A, Pratt SJ, Paw BH, Oates AC, Brugnara C, Witkowska HE, Sassa S, Zon LI. Positional cloning of the zebrafish sauternes gene: a model for congenital sideroblastic anaemia. Nat Genet. 1998;20:244–250. doi: 10.1038/3049. [DOI] [PubMed] [Google Scholar]
- Bulteau AL, O’Neill HA, Kennedy MC, Ikeda-Saito M, Isaya G, Szweda LI. Frataxin acts as an iron chaperone protein to modulate mitochondrial aconitase activity. Science. 2004;305:242–245. doi: 10.1126/science.1098991. [DOI] [PubMed] [Google Scholar]
- Campanella A, Rovelli E, Santambrogio P, Cozzi A, Taroni F, Levi S. Mitochondrial ferritin limits oxidative damage regulating mitochondrial iron availability: hypothesis for a protective role in Friedreich ataxia. Hum Mol Genet. 2009;18:1–11. doi: 10.1093/hmg/ddn308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Detrich HW, 3rd, Kieran MW, Chan FY, Barone LM, Yee K, Rundstadler JA, Pratt S, Ransom D, Zon LI. Intraembryonic hematopoietic cell migration during vertebrate development. Proc Natl Acad Sci USA. 1995;92:10713–10717. doi: 10.1073/pnas.92.23.10713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Devireddy LR, Gazin C, Zhu X, Green MR. A cell-surface receptor for lipocalin 24p3 selectively mediates apoptosis and iron uptake. Cell. 2005;123:1293–1305. doi: 10.1016/j.cell.2005.10.027. [DOI] [PubMed] [Google Scholar]
- Devireddy LR, Teodoro JG, Richard FA, Green MR. Induction of apoptosis by a secreted lipocalin that is transcriptionally regulated by IL-3 deprivation. Science. 2001;293:829–834. doi: 10.1126/science.1061075. [DOI] [PubMed] [Google Scholar]
- Emerit J, Beaumont C, Trivin F. Iron metabolism, free radicals, and oxidative injury. Biomed Pharmacother. 2001;55:333–339. doi: 10.1016/s0753-3322(01)00068-3. [DOI] [PubMed] [Google Scholar]
- Fernandez-Pol JA. Isolation and characterization of a siderophore-like growth factor from mutants of SV40-transformed cells adapted to picolinic acid. Cell. 1978;14:489–499. doi: 10.1016/0092-8674(78)90235-0. [DOI] [PubMed] [Google Scholar]
- Filling C, Berndt KD, Benach J, Knapp S, Prozorovski T, Nordling E, Ladenstein R, Jornvall H, Oppermann U. Critical residues for structure and catalysis in short-chain dehydrogenases/reductases. J Biol Chem. 2002;277:25677–25684. doi: 10.1074/jbc.M202160200. [DOI] [PubMed] [Google Scholar]
- Flo TH, Smith KD, Sato S, Rodriguez DJ, Holmes MA, Strong RK, Akira S, Aderem A. Lipocalin 2 mediates an innate immune response to bacterial infection by sequestrating iron. Nature. 2004;432:917–921. doi: 10.1038/nature03104. [DOI] [PubMed] [Google Scholar]
- Foury F, Cazzalini O. Deletion of the yeast homologue of the human gene associated with Friedreich’s ataxia elicits iron accumulation in mitochondria. FEBS Lett. 1997;411:373–377. doi: 10.1016/s0014-5793(97)00734-5. [DOI] [PubMed] [Google Scholar]
- Gardner PR, Nguyen DD, White CW. Aconitase is a sensitive and critical target of oxygen poisoning in cultured mammalian cells and in rat lungs. Proc Natl Acad Sci USA. 1994;91:12248–12252. doi: 10.1073/pnas.91.25.12248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goetz DH, Holmes MA, Borregaard N, Bluhm ME, Raymond KN, Strong RK. The neutrophil lipocalin NGAL is a bacteriostatic agent that interferes with siderophore-mediated iron acquisition. Mol Cell. 2002;10:1033–1043. doi: 10.1016/s1097-2765(02)00708-6. [DOI] [PubMed] [Google Scholar]
- Guo K, Lukacik P, Papagrigoriou E, Meier M, Lee WH, Adamski J, Oppermann U. Characterization of human DHRS6, an orphan short chain dehydrogenase/reductase enzyme: a novel, cytosolic type 2 R-beta-hydroxybutyrate dehydrogenase. J Biol Chem. 2006;281:10291–10297. doi: 10.1074/jbc.M511346200. [DOI] [PubMed] [Google Scholar]
- Hart DO, Raha T, Lawson ND, Green MR. Initiation of zebrafish haematopoiesis by the TATA-box-binding protein-related factor Trf3. Nature. 2007;450:1082–1085. doi: 10.1038/nature06349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iuchi I, Yamamoto M. Erythropoiesis in the developing rainbow trout, Salmo gairdneri irideus: histochemical and immunochemical detection of erythropoietic organs. J Exp Zool. 1983;226:409–417. doi: 10.1002/jez.1402260311. [DOI] [PubMed] [Google Scholar]
- Iwai K, Klausner RD, Rouault TA. Requirements for iron-regulated degradation of the RNA binding protein, iron regulatory protein 2. EMBO J. 1995;14:5350–5357. doi: 10.1002/j.1460-2075.1995.tb00219.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones RL, Peterson CM, Grady RW, Cerami A. Low molecular weight iron-binding factor from mammalian tissue that potentiates bacterial growth. J Exp Med. 1980;151:418–428. doi: 10.1084/jem.151.2.418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kakhlon O, Cabantchik ZI. The labile iron pool: characterization, measurement, and participation in cellular processes(1) Free Radic Biol Med. 2002;33:1037–1046. doi: 10.1016/s0891-5849(02)01006-7. [DOI] [PubMed] [Google Scholar]
- Liu J, Duncan K, Walsh CT. Nucleotide sequence of a cluster of Escherichia coli enterobactin biosynthesis genes: identification of entA and purification of its product 2,3-dihydro-2,3-dihydroxybenzoate dehydrogenase. J Bacteriol. 1989;171:791–798. doi: 10.1128/jb.171.2.791-798.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lukinova N, Iacovelli J, Dentchev T, Wolkow N, Hunter A, Amado D, Ying GS, Sparrow JR, Dunaief JL. Iron chelation protects the retinal pigment epithelial cell line ARPE-19 against cell death triggered by diverse stimuli. Invest Ophthalmol Vis Sci. 2009;50:1440–1447. doi: 10.1167/iovs.08-2545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Napier I, Ponka P, Richardson DR. Iron trafficking in the mitochondrion: novel pathways revealed by disease. Blood. 2005;105:1867–1874. doi: 10.1182/blood-2004-10-3856. [DOI] [PubMed] [Google Scholar]
- Paradkar PN, Zumbrennen KB, Paw BH, Ward DM, Kaplan J. Regulation of mitochondrial iron import through differential turnover of mitoferrin 1 and mitoferrin 2. Mol Cell Biol. 2009;29:1007–1016. doi: 10.1128/MCB.01685-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Powell CS, Jackson RM. Mitochondrial complex I, aconitase, and succinate dehydrogenase during hypoxia-reoxygenation: modulation of enzyme activities by MnSOD. Am J Physiol Lung Cell Mol Physiol. 2003;285:L189–198. doi: 10.1152/ajplung.00253.2002. [DOI] [PubMed] [Google Scholar]
- Rauen U, Springer A, Weisheit D, Petrat F, Korth HG, de Groot H, Sustmann R. Assessment of chelatable mitochondrial iron by using mitochondrion-selective fluorescent iron indicators with different iron-binding affinities. Chembiochem. 2007;8:341–352. doi: 10.1002/cbic.200600311. [DOI] [PubMed] [Google Scholar]
- Raymond KN, Dertz EA, Kim SS. Enterobactin: an archetype for microbial iron transport. Proc Natl Acad Sci USA. 2003;100:3584–3588. doi: 10.1073/pnas.0630018100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rouault TA, Stout CD, Kaptain S, Harford JB, Klausner RD. Structural relationship between an iron-regulated RNA-binding protein (IRE-BP) and aconitase: functional implications. Cell. 1991;64:881–883. doi: 10.1016/0092-8674(91)90312-m. [DOI] [PubMed] [Google Scholar]
- Rouault TA, Tong WH. Iron-sulphur cluster biogenesis and mitochondrial iron homeostasis. Nat Rev Mol Cell Biol. 2005;6:345–351. doi: 10.1038/nrm1620. [DOI] [PubMed] [Google Scholar]
- Shaw GC, Cope JJ, Li L, Corson K, Hersey C, Ackermann GE, Gwynn B, Lambert AJ, Wingert RA, Traver D, et al. Mitoferrin is essential for erythroid iron assimilation. Nature. 2006;440:96–100. doi: 10.1038/nature04512. [DOI] [PubMed] [Google Scholar]
- Shi H, Bencze KZ, Stemmler TL, Philpott CC. A cytosolic iron chaperone that delivers iron to ferritin. Science. 2008;320:1207–1210. doi: 10.1126/science.1157643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simon HU, Haj-Yehia A, Levi-Schaffer F. Role of reactive oxygen species (ROS) in apoptosis induction. Apoptosis. 2000;5:415–418. doi: 10.1023/a:1009616228304. [DOI] [PubMed] [Google Scholar]
- Sundlov JA, Garringer JA, Carney JM, Reger AS, Drake EJ, Duax WL, Gulick AM. Determination of the crystal structure of EntA, a 2,3-dihydro-2,3-dihydroxybenzoic acid dehydrogenase from Escherichia coli. Acta Crystallogr D Biol Crystallogr. 2006;62:734–740. doi: 10.1107/S0907444906015824. [DOI] [PubMed] [Google Scholar]
- Yang J, Goetz D, Li JY, Wang W, Mori K, Setlik D, Du T, Erdjument-Bromage H, Tempst P, Strong R, et al. An iron delivery pathway mediated by a lipocalin. Mol Cell. 2002;10:1045–1056. doi: 10.1016/s1097-2765(02)00710-4. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.