Abstract
The nuclear envelope is increasingly viewed from an electrophysiological perspective by researchers interested in signal transduction pathways that influence gene transcription and other processes in the nucleus. Here, we describe evidence for ion channels and transporters in the nuclear membranes and for possible ion gating by the nuclear pores. We argue that a systems-level understanding of cellular regulation is likely to require the assimilation of nuclear electrophysiology into molecular and biochemical signaling pathways.
Keywords: Ion channel, nuclear Ca+2 signaling, nuclear electrophysiology, nuclear membrane, nuclear pore
INTRODUCTION
The defining feature of eukaryotic cells is the nucleus—an organelle that houses the nuclear genome and the molecular machineries required for DNA replication and transcription. The nucleus is bounded by a nuclear envelope (NE) comprising two functionally distinct membranes: the inner nuclear membrane (INM), which interacts with nucleoskeletal proteins and peripheral chromatin, and the outer nuclear membrane (ONM), which is continuous with the endoplasmic reticulum (ER). The compartment between the INM and ONM, referred to as the perinuclear space, is contiguous with the lumen of the ER. The function and contents of the perinuclear space are unknown, but it likely serves as an intracellular store for calcium (Ca2+) and other inorganic ions. Extensions of the INM can infiltrate into the nuclear interior to form a nucleoplasmic reticulum. The INM and ONM are fused at the nuclear pore complexes (NPCs), which provide the only direct passageway between the nucleoplasm and cytoplasm (Figure 1A).
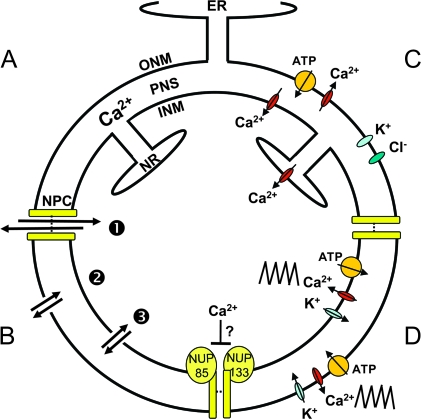
Figure 1.
Membranes, Ion Channels, and Transporters of the NE.
(A) Membrane structure of the nuclear envelope. ONM, outer nuclear membrane; INM, inner nuclear membrane; PNS, perinuclear space; ER, endoplasmic reticulum; NR, nucleoplasmic reticulum; NPC, nuclear pore complex (depicted as two yellow bars).
(B) Three routes of ion transport at the nuclear periphery (bidirectional black arrows): through the NPC (1), across the ONM (2), and across the INM (3).
(C) Reported ion channel activities in the NE of mammalian cells. IP3-regulated Ca2+ channels in the INM (including the NR) and ONM are presumed to release Ca2+ from the perinuclear space into the nucleoplasm and cytoplasm, respectively. A Ca2+-ATPase has been identified in the ONM. The direction of ion flow is unknown for K+ and Cl– channels.
(D) Nuclear membrane ion channels, transporters, and NUPs acting in the Nod factor signaling pathway. Related cation channels (DMI1, CASTOR, POLLUX, and SYM8) and two NPC proteins (NUP85 and NUP133; enlarged for emphasis) have been identified in genetic screens for nodulation-defective mutants in several species of leguminous plants. These proteins are needed for Ca2+ oscillations (zig-zag line) that occur on both sides of the NE during Nod factor signaling (Sieberer et al., 2009). CASTOR and POLLUX (blue ovals) may be K+ channels that regulate the membrane potential of the INM and/or ONM to trigger release of Ca2+ from the perinuclear space through as yet unidentified voltage-sensitive Ca2+ channels (red ovals) (Charpentier et al., 2008). An (unknown) Ca2+-ATPase (orange spheres) presumably pumps Ca2+ back into the perinuclear space. A hypothetical function of NUP85 and NUP133 in perinuclear Ca2+ oscillations is regulation of the Ca2+ permeability of NPCs (Downie and Oldroyd, 2008).
Despite the obvious structural and functional importance of the NE, certain aspects—including the electrophysiological properties of the extensive double membrane system and the NPCs—remain incompletely understood. There are several reasons for this knowledge gap. First, interest in the nuclear membranes per se has been overshadowed by research on abundant and chromatin-associated proteins of the NE and on regulated transport of macromolecules through the NPCs, which are usually regarded as large aqueous openings that are unable to impede ion flow. Although new findings have revealed the participation of some NE proteins in molecular signaling pathways (Worman, 2005; Dauer and Worman, 2009), comparatively little attention has been given to investigating ion channels and transporters in the nuclear membranes and ion gating by the NPCs. However, all other membranes in the cell, including those in the double membrane systems of mitochondria and chloroplasts (Xiong et al., 2006), contain ion channels and pumps that are essential for their physiological functions. The membranes of the NE are unlikely to be exceptions in this respect and their ability to impart an electrophysiological dimension to regulation in the nucleus should not be overlooked.
Another reason for an incomplete understanding of ‘nuclear electrophysiology’ (Bustamante, 2006) is that nuclear membranes, which constitute only a small fraction of total cellular membranes, are difficult to access and to purify. For example, in intact cells and isolated nuclei, the INM is inaccessible to patch clamp pipets and the perinuclear space is too small to be impaled by microelectrodes. The continuity of the ONM with the ER complicates the isolation of nuclear membranes from other cellular membranes and routine methods for cleanly separating the INM from the ONM are not yet available (Batrakou et al., 2009). These problems have hampered efforts to obtain homogeneous membrane preparations that are suitable for electrophysiological analyses in vitro.
Several recent reviews have described the growing list of functions ascribed to the NE and NPCs. These functions include not only macromolecular transport, but also roles in signal transduction, development, and cell cycle regulation (Meier and Brkljacic, 2009a, 2009b; Hetzer and Wente, 2009; Dauer and Worman, 2009; Batrakou et al., 2009). Here, we focus on aspects that are not considered in detail in most other reviews, namely ion channels and transporters in the nuclear membranes and ion permeability of NPCs. Despite areas of controversy and uncertainty, accumulating evidence suggests a pivotal role for nuclear ion channels and transporters in signaling pathways that influence processes in the nucleus, which is increasingly regarded as a ‘cell within a cell’ (Mazzanti et al., 2001; Bkaily et al., 2009; Rodrigues et al., 2009). After discussing evidence for nuclear membrane ion channels and transporters and for ion gating by the NPCs, we propose that a systems-level understanding of cellular physiology is likely to require the incorporation of nuclear electrophysiology into biochemical and molecular signaling pathways. We end by suggesting promising experimental approaches and directions for future research.
PROTEOMICS OF NEs AND NPCs
Plant NEs and NPCs are not as well characterized as their counterparts in yeast and animal cells (Xu and Meier, 2007; Fiserova et al., 2009). Therefore, we discuss NE and NPC proteins, with an emphasis on ion channels and transporters, in the context of what is known from non-plant systems. Descriptions of additional proteins in the NE and NPC can be found in recent reviews (Batrakou et al., 2009; Dauer and Worman, 2009; Hetzer and Wente, 2009)
NE Proteome in Mammalian Cells
Previous biochemical and immunological analyses identified a number of abundant and/or human disease-related proteins of the NE (Dauer and Worman, 2009), but recent high-throughput approaches have greatly expanded the NE proteome by revealing hundreds of proteins in the NE of mammalian cells (Schirmer et al., 2003; Schirmer and Gerace, 2005; Batrakou et al., 2009). A ‘subtractive’ proteomics approach that distinguished NE proteins from microsomal membrane proteins detected many different ion channels and transporters, including channels associated with Ca2+ signaling (described in more detail below) as well as Zn2+ transporters, Na2+/H+ exchangers, and other ion transporters (Schirmer et al., 2003; Batrakou et al., 2009). The NE proteome thus suggests an unanticipated diversity of ion channels and transporters that co-exist with the well known NPC-associated transport proteins (Batrakou et al., 2009). Whether these ion channels and transporters reside exclusively in the NE is not yet certain in most cases. Many are likely to be present in other cellular membranes given that ∼ 40% of organellar proteins have been found in multiple locations (Foster et al., 2006). Hence, in addition to nucleus-specific functions, the nuclear membranes may to some extent ‘recapitulate’ functions of other organelles (Batrakou et al., 2009). Whether the ion channels and transporters identified in the NE proteome are in the INM or ONM also remains to be determined. However, as described below, the proteomics data are supported by findings of ion channels in the INM and ONM by nuclear patch clamping and other techniques.
NPC Proteome in Yeast and Mammals
Early estimates from electron microscopic images suggested that NPCs, which are the most conspicuous structural feature of the NE, contain up to 100 distinct proteins (Batrakou et al., 2009). Recent proteomics analyses of yeast and mammalian NPCs, however, have reduced this number to approximately 30 different core proteins. The NPC proteins, termed nucleoporins (NUPs), can be divided into three classes (Lim et al., 2008; Hetzer and Wente, 2009; Batrakou et al., 2009). Transmembrane NUPs anchor the NPC to the double membrane system; structural NUPs comprise the inner framework of the NPC; and FG-NUPs, so-called because they contain repeats of phenylalanine (F) and glycine (G), contribute to the permeability properties of the NPC (Terry and Wente, 2009). At least eight copies of each NUP protein are assembled into a doughnut-shaped complex with eight-fold symmetry and an outer diameter of around 120 nm (Lim et al., 2008; Hetzer and Wente, 2009). The channel activities of NPCs are considered below.
Plant NEs and NPCs
Proteomics analyses of plant NEs and NPCs have not yet been carried out but current information is consistent with overall similarity to their counterparts in yeast and mammalian cells (Meier and Brkljacic, 2009a). Most of the NUPs identified in mammals are also present in plants (Xu and Meier, 2007; Meier and Brkljacic, 2009b) and plant NPCs display in electron micrographs the conserved three-dimensional architecture found in other eukaryotes (Fiserova et al., 2009; Meier and Brkljacic, 2009a). Although comprehensive information on the protein composition of the INM and ONM in plants is not yet available, several nuclear membrane ion channels have been identified in plants by nuclear patch clamping and through genetic screens (discussed further below).
ROUTES OF ION FLUXES AND TRANSPORT AT THE NUCLEAR PERIPHERY
Because of the unique structure of the NE, there are three possible routes for ion fluxes and transport at the perimeter of the nucleus (Figure 1B): (1) through the NPCs between the cytoplasm and the nucleoplasm; (2) across the ONM between the cytoplasm and perinuclear space; and (3) across the INM between the nucleoplasm and the perinuclear space. The presence of ion channels and transporters in the INM and ONM is generally accepted by researchers in the field whereas the ion permeability of the NPCs remains an area of controversy.
Channels of the NPC
Two types of channel have been associated with the NPC. A large central channel provides the major conduit for bidirectional transport between the nucleoplasm and the cytoplasm. The central channel has an average diameter of ∼9–10 nm (Bootman et al., 2009) but can expand to accommodate particles up to ∼39 nm in diameter (Panté and Kann, 2002). The conventional view is that the central channel permits passive diffusion of ions, small molecules and proteins (<30–40 kD) and facilitates selective transport of larger proteins and nucleoprotein complexes that carry a signal sequence (Hetzer and Wente, 2009; Meier and Brkljacic, 2009b). FG-NUPs contribute to the size-selectivity of the central channel, perhaps by forming a hydrogel-like meshwork (Frey et al., 2006; Mohr et al., 2009), although other models have also been proposed (Terry and Wente, 2009). Another factor reported to affect the size-selectivity of the central channel is Ca2+ depletion from the perinuclear space, which triggers conformational changes in the NPC that reduce the passive diffusion of intermediate-sized molecules (∼10–40 kD) (Stehno-Bittel et al., 1995; Wang and Clapham, 1999). Surrounding the central channel are eight smaller peripheral channels (with approximate diameters of 8 nm; Lim et al., 2008), which have been observed in three-dimensional reconstructions of electron microscopic data (Hinshaw et al., 1992). Although the functions of the peripheral channels are still unclear, they are proposed to provide routes for passive diffusion of ions and small molecules (Hinshaw et al., 1992; Shanin et al., 2001; Mazzanti et al., 2001; Kramer et al., 2007).
The NPC as an Ion Channel
The ion conductivity of the NPCs has been a contentious issue for decades, primarily because discrepant results are obtained depending on the experimental method and cell type used (Gerasimenko and Gerasimenko, 2004; Mazzanti et al., 2001; Bustamante, 2006). The notion of the NPC as a gated ion channel is difficult to reconcile with the large diameter of the central channel, which has a predicted single channel conductance of ∼1 nanosiemens (nS) (Mazzanti et al., 2001; Bootman et al., 2009). A channel of this size should allow free diffusion of small molecules and inorganic ions, which, in their hydrated form, are only several Å in diameter. Moreover, even if the central channel is partially blocked during macromolecular trafficking, it is assumed that the smaller peripheral channels of the NPC would still allow constitutive ion flow (Hinshaw et al., 1992; Kramer et al., 2007; Bootman et al., 2009).
This conventional view of NPC ion permeability, which derives largely from molecular flux data and ultrastructural studies, is challenged by results obtained using electrophysiological methods (Mazzanti et al., 2001; Bustamante, 2006). Early work with microelectrodes measured a high electrical resistance of NEs in certain cell types (e.g. Dipteran salivary glands) but not in others (most notably, oocytes of amphibians and marine invertebrates) (Loewenstein et al., 1966). These findings were later supported by patch clamp studies on isolated animal and plant nuclei, which demonstrated the existence of cation-selective channels with multiple conductances ranging from 50 pS to 1 nS (Mazzanti et al., 1990; Matzke et al., 1990, 1992; Mazzanti, 1998). The wide range of conductance values could reflect substates of a large conductance channel (Matzke et al., 1990) or multiple channels of the same type (Mazzanti et al., 2001). Patch clamp detection of ion channels in nuclei isolated from cells deficient in internal membranes ((mature avian erythrocytes (Matzke et al., 1990) and immature coconut endosperm (Matzke et al., 1992)) minimized the possibility that the nucleus-associated ion channel activity was due to contamination with other cellular membranes. The ability to obtain high-resistance gigaohm seals during patch clamping in the nucleus-attached mode, when dozens of NPCs may be encompassed by the patch pipette (Mazzanti et al., 2001; Taylor et al., 2009), suggested that the NPCs can exist in a closed state that restricts ion flow (Mazzanti, 1998; Mazzanti et al., 2001; Bustamante, 2006). Further nuclear patch clamping experiments supported the idea that NPCs possess ion channel activity and that the diameter of the channel can be modified by Ca2+, ATP, and other factors (Mazzanti, 1998; Mazzanti et al., 2001; Bustamante, 2006).
The possibility of ion impermeable states of the NPC has gained further support from findings on independent regulation of Ca2+ in the nuclear compartment. Although the permeability of the NPCs to Ca2+ remains controversial (Gerasimenko and Gerasimenko, 2004; Alonso et al., 2006; Mellström et al., 2008; Bootman et al., 2009; Laude and Simpson, 2009), experiments in plant and animal cells have indicated that Ca2+ in the nucleoplasm and cytoplasm exhibits distinct and independent signatures in response to a number of stimuli (Brière et al., 2006; Walter et al., 2007; Mazars et al., 2008; Kim et al., 2009; Bootman et al., 2009). One interpretation is that the NPCs can be gated for Ca2+ (and perhaps other inorganic ions) such that ion flow is blocked under certain circumstances or in specific cell types (Mazzanti, 1998; Mellström et al., 2008; Meier and Brkljacic, 2009b; Taylor et al., 2009). An alternative view holds that the NPCs are, at most, a diffusion barrier for Ca2+ and create a kinetic delay in equilibration of Ca2+ in the nuclear and cytoplasmic compartments (Bootman et al., 2009). This alternative interpretation, however, relies in part on results from experiments performed on amphibian oocytes (Danker et al., 1999), which were shown in early work to have NEs with an exceptionally low electrical resistance (Loewenstein et al., 1966).
The ion permeability of NPCs thus remains an open question. Hypothetical gating mechanisms by which the NPCs might regulate ion conductance invoke a double iris mechanism (Akey, 1990) that can adopt a spectrum of ion conductance states including completely closed (Bustamante, 2006). (Note that the iris-like structures proposed for the central channel by Akey (1990) differ from those suggested for the ‘nuclear basket’, a filamentous structure projecting into the nucleoplasm from the NPC (Stoffler et al., 1999; Bustamante, 2006).) Variations on the iris-diaphragm theme consider separate contributions by the central channel and peripheral channels, which may have distinct gating mechanisms, in regulating ion flow through the NPC (Mazzanti, 1998). Clearly, the elaborate architecture of the NPC endows it with complex transport properties that remain incompletely understood (Lim et al., 2008). To ultimately judge the full ion conductance characteristics of NPCs, further investigations using advanced electrophysiological tools and optical probes (Scanziani and Häusser, 2009) need to be carried out on nuclei from multiple cell types grown under various conditions.
Ion Channels in the INM and ONM
Irrespective of the ion permeability properties of NPCs, ion gradients can be generated and maintained across either the INM or ONM, with ions stored in and released from the perinuclear space (Figure 1A and 1B). In addition to their identification in proteomics analyses (see above), ion channels and transporters in the INM and ONM have been discovered using genetic, immunological, pharmacological, and electrophysiological approaches. The most comprehensive results concern Ca2+ ions and there is now convincing evidence, particularly from animal cells, for the presence of Ca2+ channels and transporters in the nuclear membranes.
Calcium
As a second messenger in signal transduction pathways, Ca2+ regulates a variety of processes in animal and plant cells. At rest, free Ca2+ is normally kept at a low concentration (∼100 nM) in the cytoplasm and nucleoplasm but can be released in response to various stimuli from intracellular stores, including the ER and perinuclear space, through intracellular Ca2+ release channels (Laude and Simpson, 2009). The concentration of Ca2+ in the perinuclear space is likely to be similar to that in the lumen of the ER, which is estimated to be 100–300 μM (Grygorczyk and Grygorczyk, 1998). Several recent reviews have focused on nuclear Ca2+ signaling in animal (Bootman et al., 2009) and plant cells (Mazars et al., 2008; McAinsh and Pittman, 2009). Nuclear Ca2+ can directly or indirectly regulate gene transcription (Kim et al., 2009; Mellström et al., 2008) and influence cell-cycle regulation and other nuclear processes (Bootman et al., 2009; Kim et al., 2009; Rodrigues et al., 2009). Although there are still unresolved questions about the initiation of Ca2+ signals within the nuclear compartment (Gerasimenko and Gerasimenko, 2004; Bootman et al., 2009; Rodrigues et al., 2009), the presence of the required channel and transport machinery in NEs argues for an autonomous Ca2+ signaling system in the nucleus (Mazars et al., 2008; Bootman et al., 2009; Rodrigues et al., 2009).
Major intracellular Ca2+ release channels in animal cells are inositol (1,4,5)-triphosphate receptors (IP3Rs) and the related ryanodine receptors (RyRs) (Gerasimenko and Gerasimenko, 2004; Bootman et al., 2009). IP3Rs have been detected by biochemical and immunological methods in both the INM and ONM of animal cells (Humbert et al., 1996; Bootman et al., 2009) (Figure 1C). Those in the INM enable the release of Ca2+ from perinuclear stores directly into the nucleoplasm (Humbert et al., 1996; Bootman et al., 2009; Taylor et al., 2009). Functional RyRs have been localized to the INM by immunological techniques (Gerasimenko and Gerasimenko, 2004). Ca2+-ATPases and inositol 1,3,4,5-tetrakisphosphate-operated Ca2+ channels, which replenish the Ca2+ store in the perinuclear space, have been detected in the ONM but not yet in the INM of animal cells (Humbert et al., 1996; Mazars et al., 2008; Bootman et al., 2009) (Figure 1C). IP3Rs and RyRs are also present in the nucleoplasmic reticulum, which allows sub-nuclear control of Ca2+ signaling (Echevarria et al., 2003; Marius et al., 2006; Bootman et al., 2009) (Figure 1C).
Nuclear and cytoplasmic Ca2+ can also be regulated independently in plant cells, but the channels involved have not yet been defined (Mazars et al., 2008; McAinsh and Pittman, 2009). There is considerable evidence that IP3 signaling influences many processes in plants, including stomatal guard cell closure, pollen tube elongation, and responses to various abiotic stresses (Krinke et al., 2007; Wheeler and Brownlee, 2008). Surprisingly, however, genes encoding recognizable homologs to animal IP3Rs and RyR channels have not been found in sequenced land plant genomes (Wheeler and Brownlee, 2008; McAinsh and Pittman, 2009; Ward et al., 2009). Identifying the genes involved in IP3 signaling in land plants is thus an important goal for the future (Krinke et al., 2007).
Interestingly, the genome of the unicellular green alga Chlamydomonas reinhardtii encodes an IP3R and several other Ca2+ channels found only in animals (Wheeler and Brownlee, 2008; Ward et al., 2009). Although their functions and sub-cellular locations are unknown, the presence of these animal-type Ca2+ channels in C. reinhardtii, which has been referred to as a ‘green animal’ (Ward et al., 2009), suggests that the channels were present in ancestral eukaryotes and eliminated during the evolution of land plants (Wheeler and Brownlee, 2008). Assessing the presence of various Ca2+ channels in other green algal species and the roles of IP3Rs in C. reinhardtii should lead to an improved understanding of the functions and evolution of Ca2+ signaling in plants (Wheeler and Brownlee, 2008).
Nuclear Membrane Channels for Other Ions
Nuclear patch clamping has detected different types of K+ channels of varying conductance in the NE of animal cells (Guihard et al., 2000; Mazzanti et al., 2001; Bkaily et al., 2009) (Figure 1C). In principle, patch clamping of intact nuclei should primarily detect channels in the ONM (Bkaily et al., 2009), although channels in the INM may also be accessible to the patch pipette when isolated nuclear membranes are incorporated into proteoliposomes (Matzke et al., 1990; Guihard et al., 2000). Membrane potentials created by K+ channels across the INM and ONM could regulate other nuclear membrane voltage-dependent ion channels, such as certain Ca+2 and chloride channels (Bkaily et al., 2009). Several chloride channels, which may participate in osmotic volume regulation of the nucleus, have been detected in the NE of rat liver nuclei by patch clamp analysis (Guihard et al., 2000; Mazzanti et al., 2001; Bkaily et al., 2009) (Figure 1C). Although the nuclear patch clamp results are basically compatible with the NE proteome reported for mammalian cells (Schirmer et al., 2003; Batrakou et al., 2009), additional work is required to match the ion channel activities detected by nuclear patch clamping with specific ion channels identified in the proteomics analysis.
Because many plant ion channel families have homologs in animals (Ward et al., 2009), some of the NE channels detected by nuclear patch clamping in animals may eventually be shown to have counterparts in plant cells. Nuclear patch clamping identified a cation-selective, Ca2+- and voltage-dependent channel in the NE of red beet (Grygorczyk and Grygorczyk, 1998). The NE channel was affected only by changes in nucleoplasmic Ca2+, not cytoplasmic Ca+2, and was therefore suggested to be specific for Ca2+-dependent processes in the nucleus. Further work is required to clarify the function and location of this channel in the NE. A somewhat similar channel was detected by patch clamping of ER membranes from red beet taproots (Brito-Argáez et al., 2008). Given the continuity between the ER and ONM, this channel is likely to also be in the ONM, but additional experiments are needed to verify this supposition.
NUCLEAR MEMBRANE ION CHANNELS AND PERINUCLEAR Ca2+ OSCILLATIONS IN ROOT NODULE DEVELOPMENT
The Rhizobium–legume endosymbiotic association illustrates beautifully the key role of nuclear membrane ion channels in a signal transduction pathway that reprograms gene expression and cell identity (Oldroyd and Downie, 2008; Crespi and Frugler, 2008). Nodulation (Nod) factors produced by rhizobial bacteria in the soil interact with the surface of root hair cells to initiate a cascade of signaling events, including perinuclear Ca2+ oscillations, which culminate in the expression of early nodulin genes through a Ca2+-regulated transcriptional pathway. The outcome is the development of a specialized organ, the root nodule, in which the bacteria fix nitrogen and utilize nutrients synthesized by the plant. Forward genetic screens for nodulation-defective mutants identified several related cation channels that are essential for perinuclear Ca2+ oscillations: DMI1 (DOESN'T MAKE INFECTIONS1) in Medicago truncatula, CASTOR and POLLUX in Lotus japonicus, and SYM8 (SYMBIOSIS8) in Pisum sativum (Oldroyd and Downie, 2008). Deposition in the NE has been demonstrated for DMI1 (Riely et al., 2007) and for CASTOR and POLLUX (Charpentier et al., 2008) but whether these nuclear ion channels are concentrated in the INM or ONM remains to be determined.
It is still not known how DMI1, CASTOR, POLLUX, and related proteins contribute to perinuclear Ca2+ oscillations. DMI1 does not seem to be a Ca2+ channel but experiments in yeast suggest that it may help to regulate Ca2+ release (Peiter et al., 2007). CASTOR and POLLUX are cation channels that appear to have a preference for K+ over Ca2+ (Charpentier et al., 2008). Perhaps CASTOR and POLLUX generate a K+ gradient across the INM and/or ONM, which triggers the opening of unidentified voltage-sensitive Ca2+ channels in the NE (Oldroyd and Downie, 2008; Charpentier et al., 2008) (Figure 1D). A recent study used a nuclear-localized cameleon, which is a genome-encoded, fluorescent resonance energy transfer (FRET)-based Ca2+ sensor (Allan et al., 1999; Nagai et al., 2004; Palmer and Tsien, 2006), to study the spatial distribution of perinuclear Ca2+ oscillations in root hair cells of M. truncatula. Intriguingly, Ca2+ oscillations were observed to occur on both sides of the NE (Sieberer et al., 2009) (Figure 1D). Further work is needed to unravel the individual contributions of Ca2+ oscillations in the nuclear and cytoplasmic compartments to Nod factor signaling, and the identities and locations of the ion channels involved.
In addition to nuclear membrane ion channels, two NPC proteins, NUP85 and NUP133, have also been identified in forward genetic screens for nodulation-defective mutants (Kanamori et al., 2006; Saito et al., 2007). The roles of NUP85 and NUP133 in Nod factor signaling are uncertain, but both proteins are necessary for perinuclear Ca2+ oscillations. One hypothesis is that they modulate the Ca2+ permeability of the NPCs (Figure 1D) but other possibilities, such as facilitating import of ion channels into the nucleus, have also been suggested (Kanamori et al., 2006; Saito et al., 2007; Oldroyd and Downie, 2008).
SYSTEMS BIOLOGY: INTEGRATING ELECTRICAL, MOLECULAR, AND BIOCHEMICAL SIGNALLING
The ultimate aim of systems biology is ‘to understand the dynamic networks of regulation and interactions that allows cells and organisms to live in a highly interactive environment’ (Latterich, 2005). Achieving this goal is likely to require knowledge of bioelectrical controls and how they interface with molecular and biochemical signaling pathways (Elson, 2007; McCaig et al., 2009; Blackiston et al., 2009). Examples are emerging in which nuclear electrophysiology is embedded within the broader context of cellular signaling. One of the most compelling cases is Nod factor signaling in legumes, which represents a paradigm for nuclear electrophysiological changes occurring within a signal transduction pathway that initiates at the plasma membrane and ends in the nucleus with changes in gene transcription.
The most important contributions of nuclear membrane ion channel and transporters to signaling and gene expression are likely to involve regulating free ion concentrations in the nucleoplasm and generating ion gradients across the INM and ONM. In addition to the well known ability of nuclear free Ca2+ to directly or indirectly regulate soluble transcription factors (Mellström et al., 2008; Kim et al., 2009), recent studies have forged a more direct functional link between ion channels and gene transcription by showing that some transcription factors act in molecular complexes containing voltage-gated Ca2+ channels (Kaczmarek, 2006; Barbado et al., 2009). Ion gradients across cellular membranes create electric fields that can regulate the activities of voltage-gated channels and transporters as well as membrane-bound enzymes and other proteins that possess voltage-sensing domains (Murata et al., 2005; Bezanilla, 2008). Regulation by membrane electric fields has been considered for plasma membrane proteins but the same principles can also be applied to nuclear membrane proteins (Olivotto et al., 1996; Matzke and Matzke, 1991, 1996). Further insight into how membrane potentials regulate cell homeostasis is likely to emerge from discoveries of new types of voltage sensor in membrane proteins (Bezanilla, 2008) and novel ways to couple the voltage sensors of ion channels to biochemical reactions (Kaczmarek, 2006).
Membrane electric fields can directly affect the genome by altering the charge transfer properties of DNA (Elson, 2007; Jakobsson and Stafström, 2009), which are important for signaling in repair of oxidative damage and transcription factor binding (Merino et al., 2008). Electrical continuity between the plasma membrane and nuclear membranes may be established by intracellular membranes and cytoskeletal elements, allowing signals at the cell surface to be rapidly conveyed to the nucleus (Matzke and Matzke, 1991; Shemer et al., 2008). Cellular-wide electric fields that can connect remote parts of the cell are being mapped using newly developed nanotechnological devices (Tyner et al., 2007). Electrical stimulation of plant cells can trigger important biological responses, including cell division, polarization, and morphogenesis, which implies an electrical influence on nuclear activities (Montané and Teissié, 1992; Peng and Jaffe, 1976; Carmen, 2006). These concepts and novel experimental approaches are likely to increase awareness of the electrical dimension of the NE and how it may be incorporated into the broader context of systems biology.
FUTURE EXPERIMENTAL DIRECTIONS
Nuclear electrophysiology in plants is a young field that offers many challenging problems for future research. One priority is determining the plant NE proteome to identify the full ion channel and transporter inventory of the INM and ONM. For these experiments, it will be important to obtain pure nuclear membrane preparations. A promising source for initial studies is the liquid endosperm of immature coconuts, which contains free-floating nuclei that have proven useful for patch clamp experiments (Matzke et al., 1992; Bustamante, 2006) as well as enzymatic and respiratory studies (Cutter et al., 1952a, 1952b, 1955; Wilson and Cutter, 1952). The sequence of the coconut genome, which will be needed for protein identification, may soon be available given the rapid pace of genome sequencing efforts (e.g. the oil palm genome has recently been completed (Schill, 2009)). The proteomics analysis should eventually be extended to other cell types, since findings in mammalian cells indicate that the NE proteome displays cell-type specificity (Batrakou et al., 2009). In addition, the protein composition of the INM and ONM may also vary because of intracellular protein trafficking that occurs in the physiological context of the cell. Transient association of channels, transporters, and regulators with the nuclear membranes may dramatically affect the permeability of the nucleus to ions and other molecules.
To study ion channel activity in nuclear membranes, it will be essential to develop methods for cleanly separating the INM from ONM, which can then be used individually for patch clamping and other electrophysiological analyses in vitro. Determining the ionic compositions and concentrations of distinct nuclear compartments will allow estimates of the direction of ion flow. Additional measurements of the nuclear electrical potential compared to the cytoplasm in different cell types may reveal cell-type-specific values (Loewenstein et al., 1966) similar to those found for plasma membrane potential (Levin, 2007).
In silico studies have identified a number of ion channels and transporters in Arabidopsis with a predicted nuclear localization signal (NLS), which suggests deposition in the INM (Matzke et al., 2001). Although the mechanisms by which proteins are localized to the INM are still under investigation (Lusk et al., 2007), around two-thirds of INM proteins in mammals have a predicted NLS (Zuleger et al., 2008) indicating the general importance of these sequences for INM targeting. However, proteins that lack a canonical NLS can also be localized to the INM. Nurim, for example, is tightly associated with the INM in mammalian cells despite the absence of either a recognizable NLS or a hydrophilic N-terminal domain found in many mammalian INM proteins (Hofmeister and O'Hare, 2005). The predicted NLS of DND1 (DEFENSE NO DEATH1), a cyclic nucleotide-gated cation channel important for pathogen responses in Arabidopsis, has been shown to target soluble green fluorescent protein (GFP) to the nucleus (Matzke et al., 2009). Further work is required to determine whether DND1 is indeed a nuclear ion channel. Signals specific for ONM proteins have not been identified, but ER localization signals (Cutler et al., 2000) should suffice for ONM targeting.
Nuclear localization of ion channels and transporters can be confirmed by targeting GFP-fusion proteins to the NE in transformed cells or by immuno-localization (Schirmer and Gerace, 2005). These approaches, however, are not perfect. For example, overexpression of fusion proteins can lead to accumulation in other membranes (Schirmer and Gerace, 2005). Overexpression of GFP-fusion proteins from the cauliflower mosaic virus 35S promoter may explain initial findings that CASTOR and POLLUX are present in plastids of root cells (Imaizumi-Anraku et al., 2005) whereas subsequent immuno-gold labeling and electron microscopy established their location in the NE (Charpentier et al., 2008). The latter is the method of choice for determining sub-nuclear locations of ion channels and transporter proteins if antibodies to the native protein are available or if GFP-fusion proteins are expressed from the endogenous promoter. However, immuno-gold labeling is not trivial, as fixation methods for electron microscopy do not always preserve the structure of nuclear membranes (Charpentier et al., 2008). Ion channels and transporters that have confirmed INM or ONM locations can be functionally analyzed in Arabidopsis using the extensive T-DNA insertion collections that are available, and requirements for NE localization investigated using site-directed mutagenesis to disrupt putative NLSs or other targeting signals.
Advances in nuclear electrophysiology can be expected from increased use of nuclear-targeted, genome-encoded sensors that can monitor changes in nuclear Ca2+ and INM or ONM electrical potential in real time in living plants (Figure 2A). As mentioned previously, nuclear-localized camelon has been used to demonstrate that Ca2+ oscillations occur on both sides of the NE during Nod factor signaling in M. truncatula root hair cells (Sieberer et al., 2009). A nuclear-localized cameleon line has been established in Arabidopsis (Figure 2B) and can be used, for example, to study nuclear Ca2+ changes in roots exposed to the beneficial fungus Piriformospora indica (Vadassery and Oelmüller, 2009). Given the success in identifying the nuclear ion channels DMI1, CASTOR, POLLUX, and SYM8 in forward genetic screens in legumes, it may be possible to design forward genetic screens using the Arabidopsis nuclear cameleon line to identify factors important for nuclear Ca2+ signaling in development and stress responses.
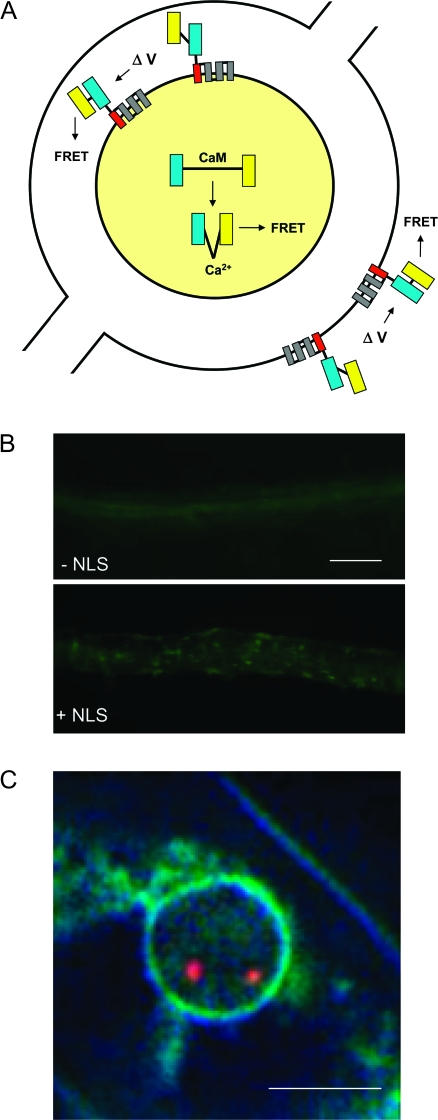
Figure 2.
Genome Encoded Sensors of Nuclear Ca2+ and Membrane Potential.
(A) Schematic drawing of nuclear-targeted, genome encoded, FRET-based sensors of free Ca2+ (cameleon) and membrane electrical potential (voltage-sensitive fluorescent protein: VSFP). Cameleon is a soluble protein (indicated by yellow nucleoplasm) containing a calmodulin (CaM) domain positioned between CFP (cyan fluorescent protein) (blue bar) and YFP (yellow fluorescent protein) (yellow bar). Binding of four Ca2+ ions to the calmodulin moiety induces a conformational change, bringing the CFP and YFP closer so that FRET can occur. The VSFP is an integral membrane protein containing four transmembrane domains (linked bars in INM and ONM). The voltage-sensitive fourth transmembrane helix (S4; red bar) is fused to CFP and YFP. When the membrane potential changes (ΔV), S4 rotates, aligning CFP and YFP so that FRET occurs. The concentric rings of the INM and ONM, with the intervening perinuclear space and continuity of the ONM to the ER, are shown. For simplicity, the NPCs are omitted.
(B) Roots of Arabidopsis plants transformed with cameleon lacking an NLS display diffuse fluorescence throughout the root (top). When joined to an NLS, cameleon is concentrated in nuclei (bottom). Nuclear localization was achieved by inserting three copies of the SV40 NLS between a 35S promoter and the cameleon coding sequence (A.J.M. Matzke, unpublished work). A nucleoplasmin-tagged cameleon has been localized to the nucleus in M. truncatula root hair cells (Sieberer et al., 2009). Bar indicates 100 μm.
(C) VSFP (blue–yellow fluorescence) accumulating at the NE in Arabidopsis root cells. VSFP1, the first prototype of VSFP (Sakai et al., 2001), has been expressed in transgenic Arabidopsis plants under the control of the 35S promoter and a plant transcriptional terminator. The VSFP1 localizes to the plasma membrane and most internal membranes, including (sporadically) the NE, as shown here. Targeting of VSFP to the INM and ONM is still under development, as is the adaptation for plants of second and third-generation VSFPs that have enhanced voltage sensitivity (Perron et al., 2009) (A.J.M. Matzke, unpublished results). The red dots represent transgene loci that are tagged with red fluorescent protein (Matzke et al., 2010), which will allow correlations between changes in nuclear Ca2+ and/or INM potential and transgene position in interphase nuclei. Bar indicates 5 μm.
It should eventually be possible to investigate changes in electrical potential across the INM and ONM using genome-encoded, voltage-sensitive fluorescent proteins (VSFP) (Sakai et al., 2001; Perron et al., 2009) provided the VSFP can be targeted preferentially to these membranes (Figure 2A and 2C). Both the nuclear-localized cameleon and VSFP lines can be used in conjunction with fluorescent-tagged transgenes (Matzke et al., 2010) to study correlations between changes in nuclear Ca2+, INM potential, and chromatin dynamics (Figure 2C).
CONCLUSIONS
The NE is arguably the most complex organellar membrane in the cell (Batrakou et al., 2009) and the one positioned to directly influence the nuclear genome in a variety of ways. Nuclear electrophysiology offers enormous opportunities for understanding signal transduction pathways that traverse the NE to modulate gene expression and other nuclear processes. By now, there is ample evidence for independent nuclear Ca2+ signaling; however, knowledge of the ion channel and transport capabilities of the NE is likely to grow as researchers focus on the nuclear membrane system and consider ions other than Ca2+ as second messengers in cellular signaling (Orlov and Hamet, 2006). Reassessing the ion permeability of NPCs seems appropriate given the enhanced regulatory capability inherent in establishing distinct ion environments in the nuclear and cytoplasmic compartments. In the future, we can expect that continued technological improvements, conceptual innovations, and the development of suitable experimental systems will accelerate research and expand our knowledge of the elusive electrophysiological aspects of the nucleus.
FUNDING
Work in the Matzke lab on nuclear membranes is supported in part by the Austrian Academy of Sciences and by a grant from the Austrian Fonds zur Förderung der wissenschaftlichen Forschung (grant no. P19572-B12).
Acknowledgments
We thank Jeff Harper for the cameleon construct 35S-YC3.60, Thomas Knöpfel for the VSFP1 construct, and Eric Schirmer for helpful correspondence. No conflict of interest declared.
References
- Akey CW. Visualization of transport-related configurations of the nuclear pore transporter. Biophys. J. 1990;58:341–355. doi: 10.1016/S0006-3495(90)82381-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allan GJ, Kwak JM, Chu SP, Llopis J, Tsien RY, Harper JF, Schroeder JI. Cameleon calcium indicator reports cytoplasmic calcium dynamics in Arabidopsis guard cells. Plant J. 1999;19:735–747. doi: 10.1046/j.1365-313x.1999.00574.x. [DOI] [PubMed] [Google Scholar]
- Alonso MT, Villalobos C, Chamero P, Alvarez J, García-Sancho J. Calcium microdomains in mitochondria and nucleus. Cell Calcium. 2006;40:513–525. doi: 10.1016/j.ceca.2006.08.013. [DOI] [PubMed] [Google Scholar]
- Barbado M, Fablet K, Ronjat M, De Waard M. Gene regulation by voltage-dependent calcium channels. Biochim. Biophys. Acta. 2009;1793:1096–1104. doi: 10.1016/j.bbamcr.2009.02.004. [DOI] [PubMed] [Google Scholar]
- Batrakou DG, Kerr ARW, Schirmer EC. Comparative proteomic analyses of the nuclear envelope and pore complex suggests a wide range of heretofore unexpected functions. J. Proteomics. 2009;72:56–70. doi: 10.1016/j.jprot.2008.09.004. [DOI] [PubMed] [Google Scholar]
- Bezanilla F. How membrane proteins sense voltage. Nat. Rev. Mol. Cell Biol. 2008;9:323–332. doi: 10.1038/nrm2376. [DOI] [PubMed] [Google Scholar]
- Bkaily G, Avedanian L, Jacques D. Nuclear membrane receptors and channels as targets for drug development in cardiovascular diseases. Can. J. Physiol. Pharmacol. 2009;87:108–119. doi: 10.1139/Y08-115. [DOI] [PubMed] [Google Scholar]
- Blackiston DJ, McLaughlin KA, Levin M. Bioelectric controls of cell proliferation. Cell Cycle. 2009;8:3527–3536. doi: 10.4161/cc.8.21.9888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bootman MD, Fearnley C, Smyrnias I, MacDonald F, Roderick HL. An update on nuclear calcium signalling. J. Cell Sci. 2009;122:2337–2350. doi: 10.1242/jcs.028100. [DOI] [PubMed] [Google Scholar]
- Brière C, Xiong TC, Mazars C, Ranjeva R. Autonomous regulation of free Ca2+ concentrations in isolated plant nuclei: a mathematical analysis. Cell Calcium. 2006;39:293–303. doi: 10.1016/j.ceca.2005.11.005. [DOI] [PubMed] [Google Scholar]
- Brito-Argáez L, Canto-Canché B, Hernández-Sotomayor SMT, Martínez-Estevez M, Pottosin II. Patch clamp characterization of a non-selective cation channel of ER membranes purified from Beta vulgaris taproots. Physiologia Plantarum. 2008;132:399–206. doi: 10.1111/j.1399-3054.2007.01031.x. [DOI] [PubMed] [Google Scholar]
- Bustamante JO. Current concepts in nuclear pore electrophysiology. Can. J. Physiol. Pharmacol. 2006;84:347–365. doi: 10.1139/y05-096. [DOI] [PubMed] [Google Scholar]
- Carmen GC. Electrical control of plant morphogenesis. In: Datta Gupta S, Ibaraki Y, editors. Plant Tissue Culture Engineering. Netherlands: Springer Verlag; 2006. pp. 397–415. [Google Scholar]
- Charpentier M, Bredemeier R, Wanner G, Takeda N, Schleiff E, Parniske M. Lotus japonicus CASTOR and POLLUX are ion channels essential for perinuclear calcium spiking in legume root endosymbiosis. Plant Cell. 2008;20:3467–3479. doi: 10.1105/tpc.108.063255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crespi M, Frugler F. De novo organ formation from differentiated cells: root nodule organogenesis. Sci. Signal. 2008;49 doi: 10.1126/scisignal.149re11. re11. [DOI] [PubMed] [Google Scholar]
- Cutler SR, Ehrhardt DW, Griffitts JS, Somerville CR. Random GFP::cDNA fusions enable visualization of subcellular structures in cells of Arabidopsis at high frequency. Proc. Natl Acad. Sci. U S A. 2000;97:3718–3723. doi: 10.1073/pnas.97.7.3718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cutter VM, Wilson KS, Dube JF. The endogenous oxygen uptake of tissues in the developing fruit of Cocos nucifera. Am. J. Botany. 1952a;39:51–56. [Google Scholar]
- Cutter VM, Wilson KS, Dube GR. The isolation of living nuclei from the endosperm of Cocos nucifera. Science. 1952b;115:58–59. doi: 10.1126/science.115.2977.58. [DOI] [PubMed] [Google Scholar]
- Cutter VM, Wilson KS, Freeman B. Nuclear behavior and cell formation in the developing endosperm of Cocos nucifera. Am. J. Botany. 1955;42:109–115. [Google Scholar]
- Danker T, Schillers H, Storck J, Shahin V, Krämer B, Wilhelm M, Oberleithner H. Nuclear hourglass technique: novel approach detects electrically open pores in Xenopus laevis oocyte. Proc. Natl Acad. Sci. U S A. 1999;96:13530–13535. doi: 10.1073/pnas.96.23.13530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dauer WT, Worman HJ. The nuclear envelope as a signalling node in development and disease. Dev. Cell. 2009;17:626–638. doi: 10.1016/j.devcel.2009.10.016. [DOI] [PubMed] [Google Scholar]
- Echevarria W, Leite MF, Guerra MT, Zipfel WR, Nathanson MH. Regulation of calcium signals in the nucleus by a nucleoplasmic reticulum. Nat. Cell Biol. 2003;5:440–446. doi: 10.1038/ncb980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elson E. Developmental control in animals and a biological role for DNA charge transfer. Prog. Biophys. Mol. Biol. 2007;95:1–15. doi: 10.1016/j.pbiomolbio.2006.07.001. [DOI] [PubMed] [Google Scholar]
- Fiserova J, Kiseleva E, Goldberg MW. Nuclear envelope and nuclear pore complex structure and organization in tobacco BY-2 cells. Plant J. 2009;59:243–255. doi: 10.1111/j.1365-313X.2009.03865.x. [DOI] [PubMed] [Google Scholar]
- Foster LJ, Hoog CL, Zhang Y, Zhang Y, Xie X, Mootha VK, Mann M. A mammalian organelle map by protein correlation profiling. Cell. 2006;125:187–199. doi: 10.1016/j.cell.2006.03.022. [DOI] [PubMed] [Google Scholar]
- Frey S, Richter RP, Görlich D. FG-rich repeats of nuclear pore proteins form a three dimensional meshwork with hydrogel-like properties. Science. 2006;314:815–817. doi: 10.1126/science.1132516. [DOI] [PubMed] [Google Scholar]
- Gerasimenko O, Gerasimenko J. New aspects of nuclear calcium signalling. J. Cell Sci. 2004;117:3087–3094. doi: 10.1242/jcs.01295. [DOI] [PubMed] [Google Scholar]
- Grygorczyk C, Grygorczyk R. A Ca2+- and voltage-dependent cation channel in the nuclear envelope of red beet. Biochim. Biophys. Acta. 1998;1375:117–130. doi: 10.1016/s0005-2736(98)00142-4. [DOI] [PubMed] [Google Scholar]
- Guihard G, Proteau S, Payet MD, Escande D, Rousseau E. Patch-clamp study of liver nuclear ionic channels reconstituted into giant proteoliposomes. FEBS Lett. 2000;476:234–239. doi: 10.1016/s0014-5793(00)01752-x. [DOI] [PubMed] [Google Scholar]
- Hetzer MW, Wente SR. Border control at the nucleus: biogenesis and organization of the nuclear membrane and pore complexes. Dev. Cell. 2009;17:606–616. doi: 10.1016/j.devcel.2009.10.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hinshaw JE, Carragher BO, Milligan RA. Architecture and design of the nuclear pore complex. Cell. 1992;69:1133–1141. doi: 10.1016/0092-8674(92)90635-p. [DOI] [PubMed] [Google Scholar]
- Hofmeister H, O'Hare P. Analysis of the localization and topology of nurim, a polytopic protein tightly associated with the inner nuclear membrane. J. Biol. Chem. 2005;280:2512–2521. doi: 10.1074/jbc.M410504200. [DOI] [PubMed] [Google Scholar]
- Humbert JP, Matter N, Artault J-C, Köppler P, Malviya AN. Inositol 1,4,5-triphosphate receptor is located to the inner nuclear membrane vindicating regulation of nuclear calcium signalling by inositol 1,4,5-triphosphate. J. Biol. Chem. 1996;271:478–485. doi: 10.1074/jbc.271.1.478. [DOI] [PubMed] [Google Scholar]
- Imaizumi-Ankaru H, et al. Plastid proteins crucial for symbiotic fungal and bacterial entry into plant roots. Nature. 2005;433:527–531. doi: 10.1038/nature03237. [DOI] [PubMed] [Google Scholar]
- Jakobsson M, Stafström S. Hole mobility and transport mechanisms in λ-DNA.J. Chem. Physics. 2009;131:155102–155111. doi: 10.1063/1.3244677. [DOI] [PubMed] [Google Scholar]
- Kaczmarek LK. Non-conducting functions of voltage-gated ion channels. Nat. Rev. Neurosci. 2006;7:761–771. doi: 10.1038/nrn1988. [DOI] [PubMed] [Google Scholar]
- Kanamori N, et al. A nucleoporin is required for induction of Ca2+ spiking in legume nodule and fungal symbiosis. Proc. Natl Acad. Sci. U S A. 2006;103:359–364. doi: 10.1073/pnas.0508883103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim MC, Chung WS, Yun DJ, Cho MJ. Calcium and calmodulin- mediated regulation of gene expression in plants. Mol. Plant. 2009;2:13–21. doi: 10.1093/mp/ssn091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kramer A, Ludwig Y, Shahin V, Oberleithner H. A pathway separate from the central channel through the nuclear pore complex for inorganic ions and small molecules. J. Biol. Chem. 2007;282:31437–31443. doi: 10.1074/jbc.M703720200. [DOI] [PubMed] [Google Scholar]
- Krinke O, Novotná Z, Valentová O, Martinec J. Inositol triphosphate receptor in higher plants: is it real? J. Exp. Bot. 2007;58:361–376. doi: 10.1093/jxb/erl220. [DOI] [PubMed] [Google Scholar]
- Latterich M. Molecular systems biology at the crossroads: to know less about more, or to know more about less? Proteome Science. 2005;3:8. doi: 10.1186/1477-5956-3-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laude AJ, Simpson AWM. Compartmentalized signalling: Ca2+ compartments, microdomains and the many facets of Ca2+ signalling. FEBS J. 2009;276:1800–1816. doi: 10.1111/j.1742-4658.2009.06927.x. [DOI] [PubMed] [Google Scholar]
- Levin M. Large-scale biophysics: ion flows and regeneration. Trends Cell Biol. 2007;17:261–270. doi: 10.1016/j.tcb.2007.04.007. [DOI] [PubMed] [Google Scholar]
- Lim RYH, Aebi U, Fahrenkrog B. Towards reconciling structure and function in the nuclear pore complex. Histochem. Cell Biol. 2008;129:105–116. doi: 10.1007/s00418-007-0371-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loewenstein WR, Kanno Y, Ito S. Permeability of nuclear membranes. Ann. N.Y. Acad. Sci. 1966;137:708–716. doi: 10.1111/j.1749-6632.1966.tb50192.x. [DOI] [PubMed] [Google Scholar]
- Lusk CP, Blobel G, King MC. Highway to the inner nuclear membrane: rules for the road. Nat. Rev. Mol. Cell Biol. 2007;8:414–420. doi: 10.1038/nrm2165. [DOI] [PubMed] [Google Scholar]
- Marius P, Guerra MT, Nathanson MH, Ehrlich BE, Leite MF. Calcium release from ryanodine receptors in the nucleoplasmic reticulum. Cell Calcium. 2006;39:65–73. doi: 10.1016/j.ceca.2005.09.010. [DOI] [PubMed] [Google Scholar]
- Matzke AJM, Matzke M. The electrical properties of the nuclear envelope, and their possible role in the regulation of eukaryotic gene expression. Bioelectrochem. Bioenerg. 1991;25:357–370. [Google Scholar]
- Matzke AJM, Behensky T, Weiger T, Matzke MA. A large conductance ion channel in the nuclear envelope of a higher plant cell. FEBS Lett. 1992;302:81–85. doi: 10.1016/0014-5793(92)80290-w. [DOI] [PubMed] [Google Scholar]
- Matzke AJM, Watanabe K, van der Winden J, Naumann U, Matzke M. High frequency, cell type-specific visualization of fluorescent-tagged genomic sites in interphase and mitotic cells of living Arabidopsis plants. Plant Methods. 2010;6:2. doi: 10.1186/1746-4811-6-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matzke AJM, Weiger T, Matzke MA. Detection of a large cation-selective channel in nuclear envelope of avian erythrocytes. FEBS Lett. 1990;271:161–164. doi: 10.1016/0014-5793(90)80397-2. [DOI] [PubMed] [Google Scholar]
- Matzke M, Aufsatz W, Gregor W, van der Winden J, Papp I, Matzke AJM. Ion transporters in the nucleus? Plant Physiol. 2001;127:10–13. doi: 10.1104/pp.127.1.10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matzke M, Weiger TM, Papp I, Matzke AJM. Nuclear membrane ion channels mediate root nodule development. Trends Plant Sci. 2009;14:296–298. doi: 10.1016/j.tplants.2009.03.009. [DOI] [PubMed] [Google Scholar]
- Matzke MA, Matzke AJM. Electric fields and the nuclear membrane. BioEssays. 1996;18:849–850. doi: 10.1002/bies.950181013. [DOI] [PubMed] [Google Scholar]
- Mazars C, Bourque S, Mithöfer A, Pugin A, Ranjeva R. Calcium homeostasis in plant cell nuclei. New Phytologist. 2008;181:261–274. doi: 10.1111/j.1469-8137.2008.02680.x. [DOI] [PubMed] [Google Scholar]
- Mazzanti M. Ion permeability of the nuclear envelope. News Physiol. Sci. 1998;13:44–50. doi: 10.1152/physiologyonline.1998.13.1.44. [DOI] [PubMed] [Google Scholar]
- Mazzanti M, Bustamante JO, Oberleithner H. Electrical dimension of the nuclear envelope. Physiol. Rev. 2001;81:1–19. doi: 10.1152/physrev.2001.81.1.1. [DOI] [PubMed] [Google Scholar]
- Mazzanti M, DeFelice LJ, Cohen J, Malter H. Ion channels in the nuclear envelope. Nature. 1990;343:764–767. doi: 10.1038/343764a0. [DOI] [PubMed] [Google Scholar]
- McAinsh MR, Pittman JK. Shaping the calcium signature: New Phytologist. 2009;181:275–294. doi: 10.1111/j.1469-8137.2008.02682.x. [DOI] [PubMed] [Google Scholar]
- McCaig CD, Song B, Rajnicek AM. Electrical dimensions in cell science. J. Cell Sci. 2009;122:4267–4276. doi: 10.1242/jcs.023564. [DOI] [PubMed] [Google Scholar]
- Meier I, Brkljacic J. Adding pieces to the puzzling plant nuclear envelope. Curr. Opin. Plant Biol. 2009a;12:1–8. doi: 10.1016/j.pbi.2009.09.016. [DOI] [PubMed] [Google Scholar]
- Meier I, Brkljacic J. The nuclear pore and plant development. Curr. Opin. Plant Biol. 2009b;12:87–96. doi: 10.1016/j.pbi.2008.09.001. [DOI] [PubMed] [Google Scholar]
- Mellström B, Savignac M, Gomez-Villafuertes R, Maranjo JR. Ca2+-operated transcriptional networks: molecular mechanisms and in vivo models. Physiol. Rev. 2008;88:421–449. doi: 10.1152/physrev.00041.2005. [DOI] [PubMed] [Google Scholar]
- Merino EJ, Boal AK, Barton JK. Biological contexts for DNA charge transport chemistry. Curr. Opin. Chem. Biol. 2008;12:229–237. doi: 10.1016/j.cbpa.2008.01.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mohr D, Frey S, Fischer T, Güttler T, Görlich D. Characterisation of the passive permeability barrier of nuclear pore complexes. EMBO J. 2009;28:2541–2553. doi: 10.1038/emboj.2009.200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Montané MH, Teissié J. Electrostimulation of plant protoplast division Part 1. experimental results. Bioelectrochem. Bioenerg. 1992;29:59–70. [Google Scholar]
- Murata Y, Iwasaki H, Sasaki M, Inaba K, Okamura Y. Phosphoinositide phosphatase activity coupled to an intrinsic voltage sensor. Nature. 2005;435:1239–1243. doi: 10.1038/nature03650. [DOI] [PubMed] [Google Scholar]
- Nagai T, Yamada S, Tominaga T, Ichikawa M, Miyawaki A. Expanded dynamic range of fluorescent indicators for Ca2+ by circularly permuted yellow fluorescent proteins. Proc. Natl Acad. Sci. U S A. 2004;101:10554–10559. doi: 10.1073/pnas.0400417101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oldroyd GED, Downie JA. Coordinating nodule morphogenesis with Rhizobial infection in legumes. Annu. Rev. Plant Biol. 2008;59:519–546. doi: 10.1146/annurev.arplant.59.032607.092839. [DOI] [PubMed] [Google Scholar]
- Olivotto M, Arcangeli A, Carlà M, Wanke R. Electric fields at the plasma membrane level: a neglected element in the mechanisms of cell signalling. BioEssays. 1996;18:495–504. doi: 10.1002/bies.950180612. [DOI] [PubMed] [Google Scholar]
- Orlov SN, Hamet P. Intracellular monovalent ions as second messengers. J. Membrane Biol. 2006;201:161–172. doi: 10.1007/s00232-006-0857-9. [DOI] [PubMed] [Google Scholar]
- Palmer AE, Tsien RY. Measuring calcium signalling using genetically targetable fluorescent indicators. Nat. Protocols. 2006;3:1057–1065. doi: 10.1038/nprot.2006.172. [DOI] [PubMed] [Google Scholar]
- Panté N, Kann M. Nuclear pore complex is able to transport macromolecules with diameters of ∼ 39 nm. Mol. Biol. Cell. 2002;13:425–434. doi: 10.1091/mbc.01-06-0308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peiter E, et al. The Medicago truncatula DMI1 protein modulates cytosolic calcium signalling. Plant Physiol. 2007;145:192–203. doi: 10.1104/pp.107.097261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng HB, Jaffe LF. Polarization of fucoid eggs by steady electrical fields. Dev. Biol. 1976;53:277–284. doi: 10.1016/0012-1606(76)90229-3. [DOI] [PubMed] [Google Scholar]
- Perron A, Mutoh H, Akermann W, Ghimire Gautam S, Dimitrov D, Iwamoto Y, Knöpfel T. Second and third generation voltage-sensitive fluorescent proteins for monitoring membrane potential. Frontiers in Molecular Neuroscience. 2009;2:1–8. doi: 10.3389/neuro.02.005.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riely BK, Lougnon G, Ané J-M, Cook DR. The symbiotic ion channel homolog DMI1 is localized in the nuclear membrane of Medicago truncatula roots. Plant. J. 2007;49:208–216. doi: 10.1111/j.1365-313X.2006.02957.x. [DOI] [PubMed] [Google Scholar]
- Rodrigues MA, Gomes DA, Nathanson MH, Leite MF. Nuclear calcium signalling: a cell within a cell. Brazilian Journal of Medical and Biological Research. 2009;42:17–20. doi: 10.1590/s0100-879x2008005000050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saito K, et al. NUCLEOPORIN85 is required for calcium spiking, fungal and bacterial symbioses, and seed production in Lotus japonicus. Plant Cell. 2007;19:610–624. doi: 10.1105/tpc.106.046938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakai R, Repunte-Canonigo V, Raj CD, Knöpfel T. Design and characterization of a DNA-encoded, voltage-sensitive fluorescent protein. Eur. J. Neurosci. 2001;13:2314–2318. doi: 10.1046/j.0953-816x.2001.01617.x. [DOI] [PubMed] [Google Scholar]
- Scanziani M, Häusser M. Electrophysiology in the age of light. Nature. 2009;461:930–939. doi: 10.1038/nature08540. [DOI] [PubMed] [Google Scholar]
- Schill SR. Consortium completes oil palm genome sequencing. Biodiesel Magazine. 2009 November 2009 issue. [Google Scholar]
- Schirmer EC, Gerace L. The nuclear membrane proteome: extending the envelope. Trends Biochem. Sci. 2005;30:551–558. doi: 10.1016/j.tibs.2005.08.003. [DOI] [PubMed] [Google Scholar]
- Schirmer EC, Florens L, Guan T, Yates JR, 3rd, Gerace L. Nuclear membrane proteins with potential disease links found by subtractive proteomics. Science. 2003;301:1380–1382. doi: 10.1126/science.1088176. [DOI] [PubMed] [Google Scholar]
- Shanin V, Danker T, Enss K, Ossig R, Oberleithner H. Evidence for Ca2+- and ATP-sensitive peripheral channels in nuclear pore complexes. FASEB J. 2001;15:1895–1901. doi: 10.1096/fj.00-0838com. [DOI] [PubMed] [Google Scholar]
- Shemer I, Brinne B, Tegner J, Grillner S. Electronic signals along intracellular membranes may interconnect dendritic spines and nucleus. PLoS Computational Biology. 2008;4 doi: 10.1371/journal.pcbi.1000036. e1000036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sieberer BJ, Chabaud M, Timmers AC, Monin A, Fournier J, Barker DG. A nuclear-targeted cameleon demonstrates intranuclear Ca2+ spiking in Medicago truncatula root hairs in response to rhizobial nodulation factors. Plant Physiol. 2009;151:1197–1206. doi: 10.1104/pp.109.142851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stehno-Bittel L, Perez-Terzic C, Clapham DE. Diffusion across the nuclear envelope inhibited by depletion of the nuclear Ca2+ store. Science. 1995;270:1835–1838. doi: 10.1126/science.270.5243.1835. [DOI] [PubMed] [Google Scholar]
- Stoffler D, Goldie KN, Feja B, Aebi U. Calcium-mediated structural changes of native nuclear pore complexes monitored by time-lapse atomic force microscopy. J. Mol. Biol. 1999;287:741–752. doi: 10.1006/jmbi.1999.2637. [DOI] [PubMed] [Google Scholar]
- Taylor CW, Rahman T, Tovey SC, Dedos SG, Taylor EJA, Velamakanni S. IP3 receptors: some lessons from DT40 cells. Immunological Reviews. 2009;231:23–44. doi: 10.1111/j.1600-065X.2009.00807.x. [DOI] [PubMed] [Google Scholar]
- Terry LJ, Wente SR. Flexible gates: dynamic topologies and functions for FG nucleoporins in nucleocytoplasmic transport. Eukaryotic Cell. 2009;8:1814–1827. doi: 10.1128/EC.00225-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tyner KM, Kopelman R, Philbert MA. ‘Nanosized voltmeter’ enables cellular-wide electric field mapping. Biophys. J. 2007;93:1163–1174. doi: 10.1529/biophysj.106.092452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vadassery J, Oelmüller R. Calcium signalling in pathogenic and beneficial plant microbe interactions. Plant Signaling and Behavior. 2009;4:1024–1027. doi: 10.4161/psb.4.11.9800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walter A, Mazars C, Maitrejean M, Hopke J, Ranjeva R, Boland W, Mithöfer A. Structural requirements of jasmonates and synthetic analogues as inducers of Ca2+ signals in the nucleus and the cytosol of plant cells. Angew. Chem. Int. 2007;46:4783–4785. doi: 10.1002/anie.200604989. [DOI] [PubMed] [Google Scholar]
- Wang H, Clapham DE. Conformational changes of the in situ nuclear pore complex. Biophys. J. 1999;77:241–247. doi: 10.1016/S0006-3495(99)76885-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ward JM, Mäser P, Schroeder JI. Plant ion channels: gene families, physiology, and functional genomics analyses. Annu. Rev. Physiol. 2009;71:59–82. doi: 10.1146/annurev.physiol.010908.163204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wheeler GL, Brownlee C. Ca2+ signalling in plants and green algae: changing channels. Trends Plant Sci. 2008;13:506–514. doi: 10.1016/j.tplants.2008.06.004. [DOI] [PubMed] [Google Scholar]
- Wilson KS, Cutter VM. The distribution of acid phosphatases during development of the fruit of Cocos nucifera. Am. J. Botany. 1952;39:57–58. [Google Scholar]
- Worman HJ. Inner nuclear membrane and signal transduction. J. Cell. Biochem. 2005;96:1185–1192. doi: 10.1002/jcb.20650. [DOI] [PubMed] [Google Scholar]
- Xiong T-C, Bourque S, Lecourieux D, Amelot N, Grat S, Brière C, Mazars C, Pugin A, Ranjeva R. Calcium signalling in plant cell organelles delimited by a double membrane. Biochim. Biophys. Acta. 2006;1763:1209–1215. doi: 10.1016/j.bbamcr.2006.09.024. [DOI] [PubMed] [Google Scholar]
- Xu XM, Meier I. The nuclear pore comes to the fore. Trends Plant Sci. 2007;13:20–27. doi: 10.1016/j.tplants.2007.12.001. [DOI] [PubMed] [Google Scholar]
- Zuleger N, Korfail N, Schirmer EC. Inner nuclear membrane protein transport is mediated by multiple mechanisms. Biochem. Soc. Trans. 2008;36:1373–1377. doi: 10.1042/BST0361373. [DOI] [PubMed] [Google Scholar]