Human meibomian gland epithelial cells were immortalized, and a defined culture system was established for the maintenance of the cells. These cultures have been, are, and will be, extremely useful for identifying factors that regulate meibomian gland activity.
Abstract
Purpose.
Meibomian gland epithelial cells are essential in maintaining the health and integrity of the ocular surface. However, very little is known about their physiological regulation. In this study, the cellular control mechanisms were explored, first to establish a defined culture system for the maintenance of primary epithelial cells from human meibomian glands and, second, to immortalize these cells, thereby developing a preclinical model that could be used to identify factors that regulate cell activity.
Methods.
Human meibomian glands were removed from lid segments after surgery, enzymatically digested, and dissociated. Isolated epithelial cells were cultured in media with or without serum and/or 3T3 feeder layers. To attempt immortalization, the cells were exposed to retroviral human telomerase reverse transcriptase (hTERT) and/or SV40 large T antigen cDNA vectors, and antibiotic-resistant cells were selected, expanded, and subcultured. Analyses for possible biomarkers, cell proliferation and differentiation, lipid-related enzyme gene expression, and the cellular response to androgen were performed with biochemical, histologic, and molecular biological techniques.
Results.
It was possible to isolate viable human meibomian gland epithelial cells and to culture them in serum-free medium. These cells proliferated, survived through at least the fifth passage, and contained neutral lipids. Infection with hTERT immortalized these cells, which accumulated neutral lipids during differentiation, expressed multiple genes for lipogenic enzymes, responded to androgen, and continued to proliferate.
Conclusions.
The results show that human meibomian gland epithelial cells may be isolated, cultured, and immortalized.
Meibomian glands are essential in maintaining the health and integrity of the ocular surface.1–6 These glands actively synthesize lipids and secrete them at the upper and lower eyelid margins just anterior to the mucocutaneous junctions. These glandular lipids then spread onto the tear film and promote the stability and prevent the evaporation of this film.1–6 Meibomian gland lipids may also help preserve visual acuity, provide lubrication during blinking, interfere with bacterial colonization, and prevent tear overflow.1–6 Meibomian gland dysfunction (MGD), in turn, leads to tear film instability and evaporation1–6 and is thought to be the major cause of dry eye syndrome throughout the world.7
However, aside from its regulation by sex steroids8–13 or the negative impact of retinoic acid,14 almost nothing is known about the physiological control of the meibomian gland in health or disease. This dearth of knowledge is somewhat surprising, given that the meibomian gland is a large sebaceous gland, and numerous articles have been published about the neural, hormonal, and secretagogue modulation of nonocular sebaceous gland tissue.15–24 These reports have shown that the nature of sebaceous gland regulation may vary significantly depending on the type of gland and its skin location. And of particular importance, much of this knowledge of sebaceous gland control has been generated by research with primary, and especially, immortalized sebaceous gland epithelial cells.25–28
We seek to advance our understanding of the regulation of meibomian gland function and the mechanisms underlying MGD. We also seek to translate this knowledge into the development of novel and unique therapeutic strategies to treat MGD and evaporative dry eye. Toward that end, we had a twofold purpose in this study: first, to establish a defined culture system for the maintenance of primary epithelial cells from human meibomian glands and, second, to immortalize these meibomian gland epithelial cells, thereby developing cell cultures that could be useful for identifying factors that regulate meibomian gland epithelial cell activity.
Materials and Methods
Cellular Procedures
Isolation and Initial Culture of Primary Human Meibomian Gland Epithelial Cells.
Human eyelid tissues were obtained within 24 hours after eyelid surgeries (five women, four men; age range, 32–85 years). These tissues were used for evaluating primary cell culture conditions, generating cells for immortalization, and conducting immunohistochemical studies. The use of human tissues was approved by the Institutional Review Boards of Massachusetts Eye and Ear Infirmary and Schepens Eye Research Institute and adhered to the tenets of the Declaration of Helsinki. Tarsal plates were isolated from eyelid tissues under a dissecting microscope (Bausch & Lomb, Rochester, NY) by removing skin, SC tissue, muscle, and palpebral conjunctiva. The tarsal plates, which contained two to five meibomian glands, were further digested with 0.25% collagenase A (Roche Diagnostics, GmbH, Mannheim, Germany) and 0.6 U/mL dispase II (Roche Applied Science, Indianapolis, IN) at 37°C for 3 hours. Single glands were then isolated under a dissecting microscope (Bausch & Lomb). Epithelial cells were dissociated into a single-cell suspension by 0.05% trypsin and 0.05% EDTA (Invitrogen-Gibco, Grand Island, NY) treatment for 5 minutes and cultured in keratinocyte serum-free medium (SFM) (Invitrogen-Gibco) for 10 to 12 days before subculture. The fibroblasts were removed with a rubber policeman 6 to 7 days after seeding.
Preparation of 3T3 Fibroblasts.
3T3 fibroblasts (a gift from Andrius Kazlauskas, Schepens Eye Research Institute, Boston, MA) at 80% confluence were incubated with mitomycin C (4 μg/mL; Roche Diagnostics, GmbH) for 2 hours at 37°C under 5% CO2/95% air, trypsinized, and plated onto cell culture dishes (BD Biosciences, Lincoln Park, NJ) at a density of 2.2 × 104 cells/cm2. These feeder cells were used 4 to 24 hours after plating.
Incubation of Primary Human Meibomian Gland Epithelial Cells in Various Culture Conditions.
Primary human meibomian gland epithelial cells were cultured at passage 2 under three conditions: SFM, SFM in the presence of a mitomycin C-treated 3T3 feeder layer, and serum-containing medium with the mitomycin C-treated 3T3 feeder layer. The serum-containing medium consisted of an equal volume of Dulbecco's modified Eagle's medium (DMEM) and Ham's F12, supplemented with 10% fetal bovine serum (FBS; Invitrogen-Gibco), 5 μg/mL insulin, 2.5 μg/mL epidermal growth factor (EGF), 8.4 ng/mL cholera toxin A subunit (Sigma-Aldrich, St. Louis, MO), 0.5 μg/mL hydrocortisone, 50 μg/mL gentamicin, and 1.25 μg/mL amphotericin B (Invitrogen-Gibco). Cultures were incubated at 37°C with 5% CO2/95% air. Media were changed every 2 days. On reaching 70% to 80% confluence, the feeder layer was removed, and the epithelial cells were subcultured to the next passage.
Clonal Analysis.
Human meibomian gland epithelial cells were seeded in SFM at a plating density of 1000 cells per 60-mm dish and cultured for 5 days. The cells were fixed with 4% paraformaldehyde and stained with 1% rhodamine B (Sigma-Aldrich). The total number of colonies that consisted of four or more cells was counted under a dissecting microscope. Colony-forming efficiency equaled the (number of colonies/number of cells seeded) × 100%. Each experiment was performed in triplicate.
Cell Proliferation Assay.
Cell proliferation was assessed by measuring the incorporation of 5-bromo-2-deoxyuridine (BrdU) into the newly synthesized DNA of replicating cells (Cell Proliferation Biotrak ELISA System; Amersham Biosciences, Piscataway, NJ). Meibomian gland epithelial cell cultures were plated at a seeding density of 5000 cells/well in 24-well dishes and allowed to grow for 5 days in the above three culture conditions. Then, BrdU (10 μM) was added 4 hours before the termination of the incubation. The detection of BrdU was performed according to the manufacturer's instructions. Statistical analysis was performed by using ANOVA and the Fisher PLSD multiple comparisons test.
Retroviral Immortalization of Human Meibomian Gland Epithelial Cells.
Retroviral vectors were used to introduce exogenous genes into human meibomian gland epithelial cells. In brief, bacteria containing pBABE-puro-hTERT plasmid29 (plasmid 1771, Addgene, Cambridge, MA) or pBABE-neo-large T cDNA plasmid30 (Addgene plasmid 1780, Cambridge, MA) were grown overnight at 37°C in LB broth. Plasmid DNA was extracted with a kit (Plasmid Maxi Kit; Qiagen, Valencia, CA). Plasmid DNA (25 μg) or transfection reagent (156 μL; Lipofectamine; Invitrogen-Gibco) was each mixed individually with 1.8 mL of reduced-serum medium (OptiMem; Invitrogen-Gibco). The two were then mixed and kept at room temperature for 45 minutes to allow complexing of the DNA with the transfection reagent. The mixture was added slowly to 293GPG viral packaging cells (a gift from Andrius Kazlauskas), which had been cultured on 15-cm culture dishes in DMEM containing 10% heat-inactivated FBS, 50 U/mL penicillin-streptomycin, 1 μg/mL tetracycline, 2 μg/mL puromycin (Sigma-Aldrich), and 3 mg/mL geneticin (G418; Sigma-Aldrich). After a 7-hour incubation period, additional DMEM containing 10% heat-inactivated FBS and 50 U/mL penicillin-streptomycin was added. Medium containing virus was collected from days 2 to 6 after transfection and spun at 1500 rpm for 10 minutes in a centrifuge (GPK; Beckman, Fullerton, CA). The virus pellet was collected by centrifuging the supernatant at 25,000g for 90 minutes at 4°C. The virus pellet was resuspended in sterile medium which contained 50 mM Tris (pH 7.8), 130 mM NaCl, and 1 mM EDTA. The virus solution was stored at −80°C until use.
First-passaged human meibomian gland epithelial cells from a 58-year-old man were used for the immortalization. The cells were seeded on a six-well culture plate at 2 × 105 cells/well in SFM. When the cells grew to 60% and 70% confluence, the medium was changed to 2 mL SFM containing 8 μg/mL polybrene (Millipore, Billerica, MA). The concentrated virus solution containing pBABE-puro-hTERT or pBABE-neo-large T cDNA, or both, was added to the well and incubated at 37°C in 5% CO2/95% air for 24 hours.
The cells were subcultured into 10-cm culture dishes and incubated with SFM containing selective antibiotics: puromycin (2 μg/mL) for cells infected with pBABE-puro-hTERT alone or combined with pBABE-neo-large T cDNA, and neomycin (2 mg/mL) for cells infected only with pBABE-neo-large T cDNA-infected cells. The cells were selected for 7 to 14 days. Drug-resistant cells were expanded and subcultured.
Androgen Treatment of Immortalized Human Meibomian Gland and Conjunctival Epithelial Cells.
Immortalized meibomian gland (passage 11) and conjunctival (passage 26; generous gift from Ilene Gipson, Schepens Eye Research Institute, Boston, MA) epithelial cells (3 × 105/well; n = 3 wells/treatment group) were cultured to 80% confluence, then treated with either the ethanol vehicle or 10 nM DHT (Steraloids, Wilton, NH) for 3 or 4 days, respectively, as previously described (Khandelwal P, et al. IOVS 2009;50:ARVO E-Abstract 4266).
Growth Kinetics.
Immortalized human meibomian gland epithelial cells were seeded in SFM in six-well culture plates at a density of 3.76 × 104 cells per well and cultured for 1, 3, 5, 7, and 10 days. Total cells at each time point were counted with a hemocytometer.
General Procedures
Karyotype Analysis.
Karyotype analysis of the immortalized human meibomian gland epithelial cells was performed at the Center for Human Genetics, Boston University School of Medicine (Boston, MA). The cells were treated with colcemid, lysed, and fixed in methanol-acetic acid. Chromosomes were stained with Wright solution, and 15 cells in metaphase were analyzed.
Nile Red Staining.
Neutral lipid expression during differentiation was determined by staining the cells with the lipophilic dye Nile red (Sigma-Aldrich). The hTERT-immortalized human meibomian gland epithelial cells were cultured on chamber slides in SFM to 70% to 80% confluence and then changed to three types of media: SFM, SFM with 1.2 mM calcium, and serum-containing medium which consisted of an equal volume of DMEM and Ham's F12, supplemented with 10% FBS, and 10 ng/mL EGF. The cells were cultured for an additional 1, 3, 5, 7, and 14 days, then fixed with 4% paraformaldehyde for 10 minutes and stained with Nile Red for 15 minutes at room temperature. After extensive washing, the cells were observed with a confocal microscope (TCS SP2; Leica, Bannockburn, IL) using a 580 ± 30-nm band-pass filter, which detects neutral lipid and phospholipids.31
Oil Red O Staining.
To detect neutral lipids, the cells were cultured in the above three media conditions for 5 days and fixed with 10% neutral buffered formalin for 15 minutes. The samples were then stained with 0.3% Oil red O (Fisher Biotec, Wembley, WA, Australia) in 60% isopropanol for 15 minutes, rinsed with 60% isopropanol, and observed under a phase-contrast microscope (Eclipse TS100; Nikon, Avon, MA).
Biochemical Methods
To evaluate the lipid profile in hTERT-immortalized human meibomian gland epithelial cells, the lipids were extracted by the method of Folch et al.32 Total lipids were analyzed by an adaptation of a protocol devised by Christie,33 and phospholipids were characterized as previously described.34 For fatty acid analysis, a lipid extract aliquot was mixed with an internal standard (FIM-FAME-7; Matreya, Pleasant Gap, PA) and subjected to base-catalyzed methanolysis to free the ester-bound fatty acids and convert them to their methyl esters (FAME). The FAMEs were separated by capillary gas chromatography and ionized by ammonia chemical ionization, and their mass spectra were acquired (Quattro-II triple quadrupole GC-MS/MS instrument; Water, Milford, MA). Fatty acids were identified by their molecular masses as shown in the spectra. Peak area ratios of each FAME peak to the internal standard were compared with those ratios in an external standard mix to calculate the concentration of each fatty acid.
Histologic Techniques
Immunocytochemistry.
Meibomian gland epithelial cells cultured to 70% and 80% confluence on chamber slides were fixed with 4% paraformaldehyde for 10 minutes at room temperature. After blocking with 3% bovine serum albumin (BSA)/0.3% Triton X-100/phosphate buffer solution (PBS) for 30 minutes at room temperature, the cells were incubated for 2 hours at room temperature with a mouse monoclonal IgG antibody to SV40 large T antigen (Calbiochem Inc., Temecula, CA) in 1% BSA/PBS at a dilution of 1:100. After they were washed with PBS, the slides were incubated with FITC-conjugated rabbit anti-mouse IgG (Chemicon) for 1 hour at room temperature and mounted (Vectashield antifade medium; Vector Laboratories, Burlingame, CA). For negative control experiments, the primary antibody was omitted. The slides were examined with a fluorescence microscope (Eclipse E800; Nikon, Tokyo, Japan).
Immunohistochemistry.
Human lid, corneal, and conjunctival tissues (gifts from Ilene Gipson, Schepens Eye Research Institute) were embedded in OCT compound and frozen at −80°C until they were used. Sections (6 μm) were cut and placed on microscope slides (Superfrost Plus; VWR International, West Chester, PA). They were fixed with 10% neutral buffered formalin for 10 minutes, blocked with 3% BSA/PBS (pH 7.4) for 30 minutes, and incubated with a 1:10 dilution of goat anti-human kallikrein 6 IgG antibody (R&D Systems, Inc. Minneapolis, MN) in 1% BSA/PBS for 2 hours at room temperature. After they were washed with PBS, the slides were incubated with FITC-conjugated donkey anti-goat IgG (1:100 dilution in 1% BSA/PBS; Chemicon) for 1 hour at room temperature. The slides were mounted with mounting medium with 4′,6-diamidino-2-phenylindole (Vectashield; Vector Laboratories) as a counterstain. For negative controls, either the primary antibody was preincubated with recombinant human kallikrein 6 (1.5 μg; R&D Systems, Inc.) or it was omitted. The slides were examined by fluorescence microscope (Eclipse E800; Nikon).
Molecular Biological Procedures
Real-time PCR Detection of Telomerase Activity.
Telomerase activity was detected by quantitative telomerase detection (Allied Biotech Inc., Ijamsville, MD), according to the manufacturer's instructions. Briefly, primary and hTERT-immortalized meibomian gland epithelial cells were grown to 80% confluence, trypsinized, centrifuged, and washed. The cell pellets were stored at −80°C until use. The pellets were resuspended in 200 μL lysis buffer, incubated on ice for 30 minutes, and spun at 12,000g for 30 minutes at 4°C. The supernatants (i.e., cell extracts) were transferred to fresh tubes and the protein concentrations were determined with a BCA protein assay kit (Pierce, Rockford, lL). Protein concentrations for samples were adjusted to 500 ng/μL. In negative control experiments, aliquots of cell extracts were incubated at 85°C for 10 minutes to inactivate enzyme activity. Dilutions of an oligonucleotide with an identical sequence as telomere primers were used to generate a standard curve. The real-time PCR reaction system was composed of 12.5 μL premix, 1.0 μL cell extracts, heat-inactivated extracts or template controls, and 11.5 μL water. Real-time PCR reactions, which were performed on a sequence-detection system (model 7900; Applied Biosystems Inc. Foster City, CA), were 25°C for 20 minutes and 95°C for 10 minutes, followed by three-step cycling for 40 cycles: 95°C for 30 seconds, 60°C for 30 seconds, and 72°C for 30 seconds. Statistical analysis was performed by using ANOVA and the Fisher PLSD multiple comparisons test.
Gene Expression Analyses.
Total RNA was extracted (Trizol; Invitrogen, Carlsbad, CA) from human meibomian glands (n = 12 samples), as well as from primary human meibomian gland epithelial cells (n = 12 samples), hTERT-immortalized human meibomian gland epithelial cells (n = 30 samples) and immortalized human conjunctival epithelial cells (n = 6 samples) that had been cultured in a variety of conditions, including exposure to exogenous calcium, FBS, EGF, bovine pituitary extract (BPE), or DHT (i.e., Liu S, et al. IOVS 2008;49:ARVO E-Abstract 88; Liu S, et al. IOVS 2009;50:ARVO E-Abstract 3669; Khandelwal P, et al. IOVS 2009;50:ARVO E-Abstract 4266; Khandelwal P, et al. IOVS 2010;51:ARVO E-Abstract 4158). Total RNA samples were further purified (RNeasy MinElute Cleanup Kit; Qiagen Inc.) and run on a bioanalyzer (RNA 6000 Nano LabChip. with a model 2100 Bioanalyzer; Agilent Technologies, Palo Alto, CA) to verify RNA integrity.
The RNA (100 ng) samples were processed by Asuragen (Austin, TX) for the quantitation of mRNA levels by using gene expression profiling (HumanHT-12 v3 Expression BeadChips; Illumina San Diego, CA), as reported elsewhere (Liu S, et al., manuscripts in preparation; Khandelwal P, et al., manuscripts in preparation). These chips target more than 25,000 annotated genes with more than 48,000 probes derived from National Center for Biotechnology Information (NCBI) reference sequences and the UniGene databases (provided in the public domain by the NCBI, Bethesda, MD, http://www.ncbi.nlm.nih.gov/UniGene). Data were processed (BeadStudio software v3; Illumina) using background subtraction and cubic spline normalization. Normalized hybridization intensity values were adjusted by adding a constant such that the lowest intensity value for any sample equaled 16.35
Normalized data were evaluated with a comprehensive program (GeneSifter.Net software; Geospiza, Seattle, WA) that also produced gene ontology and z-score reports. These ontologies encompassed biological processes and molecular functions and were organized according to the guidelines of the Gene Ontology Consortium (http://www.geneontology.org/GO.downloads.ontology.shtml).36
The gene expression of the various human meibomian gland samples was compared to the gene expression profile of human conjunctival and corneal epithelia, as shown in the Gene Expression Omnibus Series accession number GSE5543,37 as well as to genes expressed in sebaceous glands and immortalized human sebaceous gland epithelial cells (GSE10432).26–28,38
Results
Culture of Primary Human Meibomian Gland Epithelial Cells
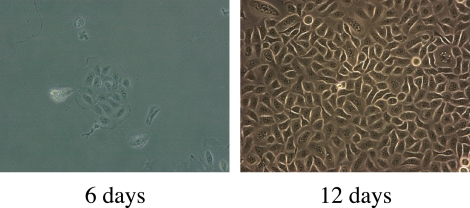
Single attached primary human meibomian gland epithelial cells divided to form four-cell colonies by days 3 to 4 after seeding and continued to proliferate to form 16- to 32-cell colonies by day 6 (Fig. 1). Each colony consisted of a collection of small, ovoid or polygonal cells. Confluent colonies at day 12 had cobblestone morphology (Fig. 1).
Figure 1.
Appearance of primary human meibomian gland epithelial cells after culture in SFM for 6 and 12 days. Magnification, ×200.
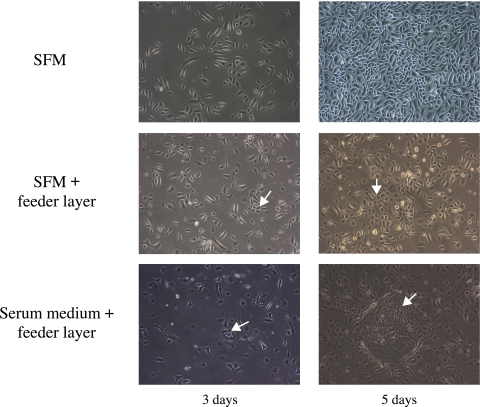
Early passages of meibomian gland epithelial cells cultured in SFM continued to proliferate rapidly and could be passaged at least five times (Fig. 2). Human meibomian gland epithelial cells cultured in SFM in the presence of the 3T3 feeder layer formed four- to six-cell colonies by day 3. Epithelial cells continued to proliferate and distributed among the 3T3 cells (Fig. 2). Human meibomian gland epithelial cells cultured in serum-containing medium in the presence of the feeder layer formed four- to six-cell colonies by day 3. The colonies grew in size over time. Cells in these colonies were small and tightly arranged, whereas 3T3 cells around the colonies formed a distinct clonal margin (Fig. 2).
Figure 2.
Appearance of primary human meibomian gland epithelial cells (passage 2) after culturing in SFM, SFM in the presence of a 3T3 feeder layer, and serum-containing medium in the presence of a 3T3 feeder layer for 3 and 5 days. Arrows: the meibomian gland epithelial cell colonies. Magnification, ×200.
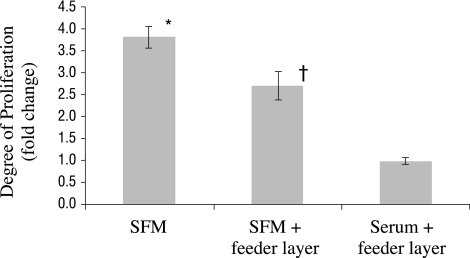
There was a significant (P < 0.05) increase in the proliferation of primary human meibomian gland epithelial cells when cultured in SFM, compared with incubation on a feeder layer in the presence or absence of serum (Fig. 3). Cell proliferation in SFM with the feeder layer, in turn, was significantly (P < 0.05) greater than that in serum-containing medium with the feeder layer (Fig. 3).
Figure 3.
Proliferation of primary human meibomian gland epithelial cells (passage 2; n = 3 wells/condition) in different culture conditions. *Significantly (P < 0.05) greater proliferation than when cultured on a feeder layer in the presence or absence of serum; †significantly (P < 0.05) greater proliferation than when cultured in serum-containing medium on a feeder layer.
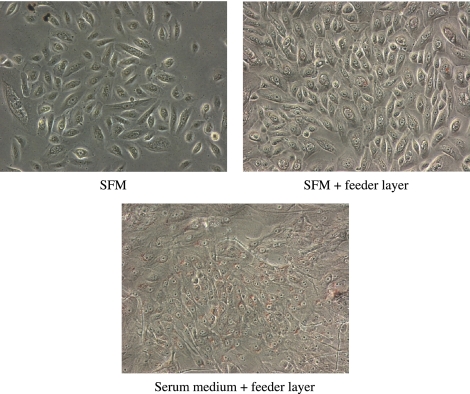
Neutral lipid droplets in primary human meibomian gland epithelial cells were detected by Oil red O staining. Cells cultured in serum-containing medium with the feeder layer appeared to have more pronounced staining when compared with cells grown in SFM or SFM with the feeder layer. Cells cultured in SFM showed minimal Oil red O staining (Fig. 4).
Figure 4.
Accumulation of neutral lipid droplets in primary human meibomian gland epithelial cells (passage 2). The cells were cultured for 5 days in SFM, or on feeder layers in the presence or absence of serum and stained with Oil red O. The cells cultured in serum-containing medium with the feeder layer appeared to have more pronounced staining than did the cells grown in SFM with or without the feeder layer. Magnification, ×200.
Immortalization of Human Meibomian Gland Epithelial Cells
To attempt immortalization, we exposed human meibomian gland epithelial cells to retroviral hTERT and/or SV40 large T antigen vectors and selected antibiotic-resistant cells.
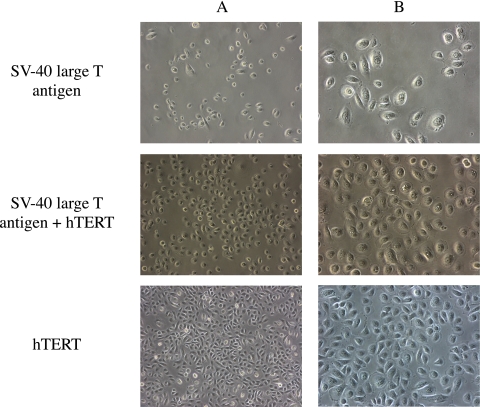
Only one cell colony infected with SV40 large T antigen retroviral vector survived antibiotic selection. The cells in the colony were serially subcultured (Fig. 5). After 4.5 months, they had reached passage 13, but then stopped proliferating.
Figure 5.
Appearance of human meibomian gland epithelial cells after infection with SV40 large T antigen (passage 9), SV40 large T antigen and hTERT (passage 16), or hTERT (passage 23). Magnification: (A) ×100; (B) ×200.
Human meibomian gland epithelial cells infected with both SV40 large T antigen and hTERT retroviral vectors kept proliferating and grew beyond passage 25 during a 6-month culture period (Fig. 5).
Human meibomian gland epithelial cells infected with the hTERT retroviral vector survived antibiotic selection and proliferated. Cell division seemed to slow at passage 5 to 6. However, small, rapidly dividing cells appeared at passage 6 and continued to proliferate beyond passage 70 over a 15-month time span (Fig. 5). Under phase-contrast microscopy, these cells show typical polygonal epithelial cell morphology and form a cobblestone-like cell sheet when confluent. This epithelial-cell–like appearance was maintained at every passage.
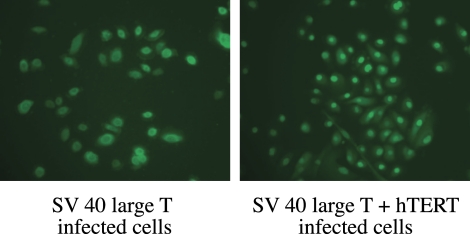
Immunofluorescent staining for SV40 large T antigen demonstrated nuclear expression in SV40-infected large T cells, as well as in large T cells infected with SV40 antigen and hTERT (Fig. 6).
Figure 6.
Immunofluorescent staining of SV40 large T antigen in nuclei of cells infected with SV40 large T antigen or with SV40 large T antigen and hTERT. Magnification: (left) ×400; (right) ×200.
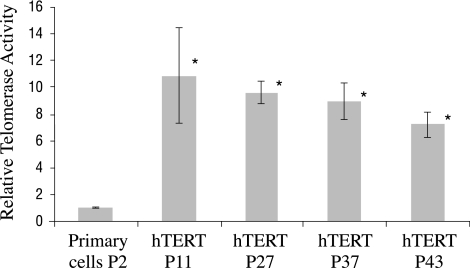
Quantitative real-time PCR analysis showed that telomerase activity in hTERT-immortalized cells (n = 3 wells/passage) at passages 11, 27, 37, and 43 was significantly (P < 0.05) higher than that of the primary cultured cells at passage 2 (Fig. 7).
Figure 7.
Real-time PCR quantitative telomerase activity. *Telomerase activity in hTERT immortalized cells (n = 3 wells/passage) at passages 11, 27, 37, and 43 was significantly higher than that of the primary cultured cells at passage 2 (P < 0.05).
Cellular Growth Kinetics and Colony-Forming Efficiency
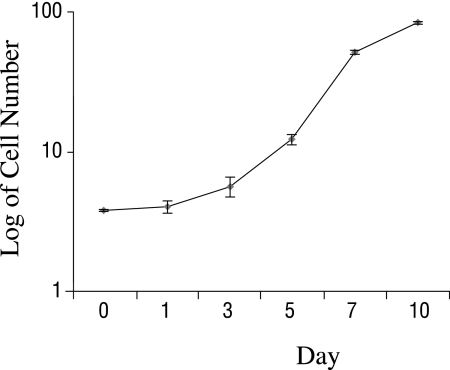
Measurements of cellular growth kinetics were obtained on hTERT-immortalized cells (n = 3 wells) at passage 16. By 24 hours after plating, the number of cells increased by 6%, from 3.76 × 104 cells/well to 3.99 ± 0.38 × 104 cells/well. From 24 to 72 hours, the cell number rose by 40%, to 5.58 ± 0.88 × 104 cells/well. From 72 to 120 to 168 hours, cells underwent successive 1.12 (to 12.11 ± 1.01 × 104 cells/well) and 2.09 (to 51.64 ± 1.14 × 104cells/well) population doublings, respectively. The average population-doubling time during the log growth phase was 27.39 hours. By 168 hours, the cells had reached 90% confluence. Cells reached confluence by 240 hours (day 10) after plating (Fig. 8).
Figure 8.
Kinetics of growth of hTERT-immortalized human meibomian gland epithelial cells. During the log growth phase, the average population-doubling time was 27.39 hours. Cells reached 90% confluence by 168 hours and 100% confluence by 240 hours after plating.
The colony-forming efficiency (n = 3 wells/group) of primary human meibomian gland epithelial cells at passage 2 equaled 11.20% ± 1.39%, whereas that of hTERT-immortalized cells at passage 17 was significantly (P < 0.05) greater at 24.78% ± 1.62%.
Karyotypes of Immortalized Human Meibomian Gland Epithelial Cells
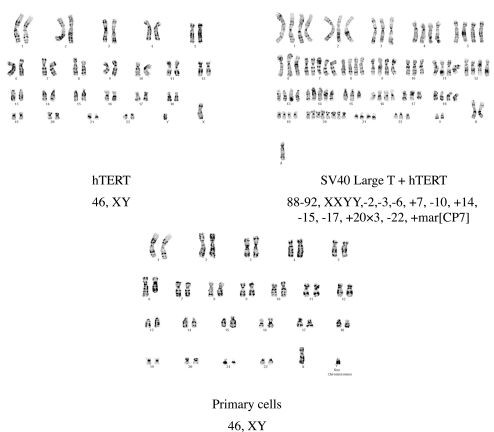
The karyotype of hTERT-immortalized human meibomian gland epithelial cells at passage 23 was 46, XY. This represented a normal male karyotype and was identical with that of the “male” parent primary cells. No evidence of any chromosome abnormality was observed (Fig. 9).
Figure 9.
Karyotype analyses of primary (passage 2) human meibomian gland epithelial cells, as well as cells immortalized with hTERT or SV40 large T and hTERT. The 46, XY karyotype of the hTERT-immortalized human meibomian gland epithelial cells at passage 23 was normal and identical with that of the male parent primary cells. The karyotype of human meibomian gland epithelial cells immortalized with both SV40 large T antigen and hTERT was abnormal.
At later passages, the karyotype of hTERT-immortalized human meibomian gland epithelial cells underwent some alterations. Cells at passage 35 revealed the presence of three subpopulations: 46, XY; 47, XY, +5; 48, XY, +5, +20. Cells at passage 46 showed a male karyotype with trisomy 5 and trisomy 20. Cells at passage 59 showed additional changes at chromosomes 4 and 1.
The karyotype of human meibomian gland epithelial cells immortalized with both SV40 large T antigen and hTERT was quite abnormal. These cells at passage 23 showed tetraploidy and hypotetraploidy, 88-92, XXYY. Moreover, these cells had extra chromosomes 7 and 14, three further copies of chromosome 20, and loss of one of the chromosomes 2, 3, 6, 10, 15, 17, and 22. A marker chromosome of unknown origin was also detected (Fig. 9).
Neutral Lipid Accumulation during Differentiation of hTERT-Immortalized Human Meibomian Gland Epithelial Cells
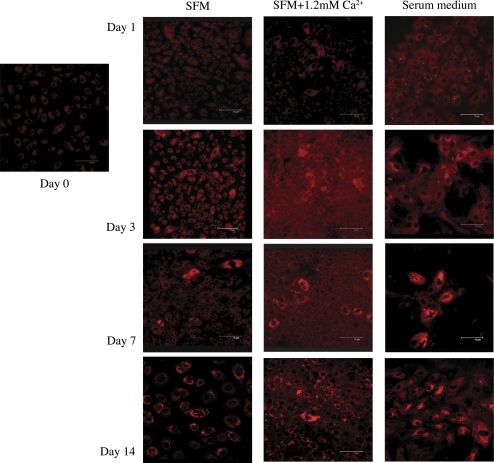
Neutral lipids accumulate in sebaceous gland epithelial cells during differentiation,39,40 which, in turn, may be induced in various cell types by exposure to calcium or serum.41–43 Given this background, we sought to determine whether hTERT-immortalized human meibomian gland epithelial cells accumulate intracellular neutral lipids during differentiation. Cells were cultured in SFM, SFM with 1.2 mM calcium, or serum-containing medium for increasing lengths of time and then stained with Nile red.
Our microscopy results show minimal staining of cells when subconfluent and then an apparent, time-dependent increase in granular lipid staining (Fig. 10). Cultures containing serum appeared to express lipids more rapidly (e.g., by 24 hours), but ultimately all culture conditions led to increased staining. These findings indicate that the hTERT-immortalized cells differentiate in serum-free, as well as in serum-containing, media.
Figure 10.
Nile red staining of hTERT-immortalized human meibomian gland epithelial cells (passage 31) cultured in SFM, SFM, with 1.2 mM calcium or serum-containing medium. Cells showed minimal granular lipid staining when subconfluent at day 0. This cellular staining underwent an apparent time-dependent increase in all culture conditions. Cell cultures containing serum appeared to express lipids more rapidly (e.g., by 24 hours). Magnification, 400×.
Preliminary analyses of the lipid profile of hTERT-immortalized human meibomian gland epithelial cells (1.5 × 106 cells; passage 41),that had been cultured in SFM indicated the presence of wax esters; cholesterol esters; tri-, di-, and monoglycerides; cholesterol; phosphocholine; sphingolipids; and oleic, palmitic, palmitoleic, and stearic fatty acids, among other species.
Comparison of the Gene Expression Profile in hTERT-Immortalized Meibomian Gland Epithelial Cells to Those of Sebaceous Gland Epithelial Cells, Primary Human Meibomian Gland Epithelial Cells, and Human Meibomian Gland Tissues
To determine whether hTERT-immortalized human meibomian gland epithelial cells express genes common to other sebaceous gland epithelial cells, we scanned meibomian samples for the expression of lipid-, keratin-, and protease-related genes known to be transcribed in sebocytes, as reported in Gene Expression Omnibus Series accession number GSE10432 (http://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE10432).27 We also examined whether these selected genes were expressed in primary human meibomian gland epithelial cells and in human meibomian glands.
As shown in Table 1, the genes encoding acyl-coenzyme A dehydrogenase, farnesyl diphosphate synthase, farnesyl-diphosphate, farnesyltransferase 1, farnesyltransferase, CAAX box α, farnesyltransferase, CAAX box β, fatty acid desaturase 1, fatty acid synthase, kallikrein-related peptidase 6, kallikrein-related peptidase 6, keratin 7, keratin 13, squalene epoxidase, stearoyl-CoA desaturase, and sterol regulatory element binding transcription factor 1 were expressed in all sebaceous gland samples. Similarly, these genes were expressed in almost all meibomian gland cell and tissue samples.
Table 1.
Examples of Lipid-, Keratin- and Protease-Related Genes Expressed in the Various Tissues Studied
| Gene ID | Gene | Immortalized SGE Cells | hTERT-Immortalized MGE Cells | Primary Human MGE Cells | Human Meibomian Gland |
|---|---|---|---|---|---|
| ACADM | Acyl-coenzyme A dehydrogenase | 100 | 100 | 100 | 100 |
| FDPS | Farnesyl diphosphate synthase | 100 | 100 | 100 | 100 |
| FDFT1 | Farnesyl-diphosphate farnesyltransferase 1 | 100 | 100 | 100 | 100 |
| FNTA | Farnesyltransferase, CAAX box, α | 100 | 100 | 100 | 100 |
| FNTB | Farnesyltransferase, CAAX box, β | 100 | 100 | 100 | 100 |
| FADS1 | Fatty acid desaturase 1 | 100 | 100 | 75 | 100 |
| FASN | Fatty acid synthase | 100 | 100 | 100 | 100 |
| KLK6 | Kallikrein-related peptidase 6, Variant A | 100 | 93 | 100 | 100 |
| KLK6 | Kallikrein-related peptidase 6, Variant B | 100 | 100 | 100 | 92 |
| KRT7 | Keratin 7 | 100 | 100 | 100 | 100 |
| KRT13 | Keratin 13 | 100 | 100 | 100 | 100 |
| SQLE | Squalene epoxidase | 100 | 100 | 100 | 100 |
| SCD | Stearoyl-CoA desaturase | 100 | 100 | 100 | 100 |
| SREBF1 | Sterol regulatory element binding transcription factor 1 | 100 | 100 | 100 | 100 |
Data are the percentage of samples of immortalized human sebaceous gland epithelial (SGE) cells (n = 6) primary (n = 12) and hTERT-immortalized human meibomian gland epithelial (MGE) cells (n = 30), and human meibomian glands (n = 12) that contained the designated genes. The data for immortalized sebocytes are reported in Gene Expression Omnibus Series accession number GSE10432 (NCBI, Bethesda, MD, http://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE10432).27 Analogous gene expression findings are published for ACADM, FNTA, FNTB, FADS1, FASN, KRT7, KRT13, and SCD in immortalized sebocytes and/or sebaceous glands.26,28,38 The primary and immortalized meibomian gland epithelial cells were cultured under a variety of conditions, including exposure to exogenous calcium, FBS, EGF, BPE, or DHT (i.e., Liu S, et al. IOVS:2008;49:ARVO E- Abstract 88; Liu S, et al. IOVS 2009;40:ARVO E-abstract 3669; Khandelwal P, et al. IOVS 2009;50:ARVO E-abstract 4266; Khandelwal P, et al. IOVS 2010;51:ARVO E-abstract 4158).
Influence of Androgen Treatment on Lipid-Related Gene Expression in hTERT-Immortalized Human Meibomian Gland Epithelial Cells
Nonocular sebaceous gland epithelial cells are known to respond to androgens by increasing the transcription of lipid-related genes and producing proteins that augment both the synthesis and secretion of lipids.44,45 To determine whether hTERT-immortalized human meibomian gland epithelial cells respond in a similar manner, we analyzed the effect of DHT on the cellular expression of lipid-related gene ontologies. For comparison, we also evaluated the influence of androgen exposure on immortalized human conjunctival epithelial cells. These conjunctival epithelial cells, because they are not of sebaceous gland origin, should not respond to androgen treatment with an upregulation of numerous genes associated with lipid pathways. Meibomian gland (passage 11) and conjunctival (passage 26) epithelial cells (3 × 105/well; n = 3 wells/treatment group) were cultured to 80% confluence, then treated with either vehicle or 10 nM DHT. After optimal time courses (conjunctival cells, 4 days; meibomian cells, 3 days), as previously demonstrated (Khandelwal P, et al. IOVS 2009;50:ARVO E-Abstract 4266), the cells were processed for analysis (Illumina BeadChips and Geospiza software).
Our results show that DHT has a significant impact on the expression of numerous genes related to lipid metabolic pathways in hTERT-immortalized human meibomian gland epithelial cells. As shown in Table 2, DHT stimulated 25 different ontologies (with ≥5 genes) involved with lipid biosynthesis, homeostasis, transport, and binding, as well as with cholesterol, fatty acid, phospholipid, and steroid dynamics. Twelve of these ontologies containing multiple genes had z-scores greater than 3.0. Androgen exposure also significantly increased the expression of ontologies (<5 genes) associated with fatty acid elongation and phospholipid dephosphorylation, and with the activities of fatty-acyl-CoA synthase, 7-dehydrocholesterol reductase, C-3 sterol dehydrogenase, lathosterol oxidase, malate dehydrogenase, C-5 sterol desaturase, 3-α-and 20-α-hydroxysteroid dehydrogenase, and ATP citrate synthase (data not shown).
Table 2.
Influence of DHT on the Expression of Lipid-Related Gene Ontologies in hTERT-Immortalized Human Meibomian Gland Epithelial Cells
| Ontologies | DHT Genes ↑ | Plac Genes ↑ | DHT z-score | Plac z-score |
|---|---|---|---|---|
| Lipid metabolic process | 87 | 43 | 5.08 | −1.94 |
| Cellular lipid metabolic process | 71 | 32 | 4.76 | −2.09 |
| Lipid binding | 45 | 22 | 2.93 | −1.8 |
| Lipid biosynthetic process | 34 | 16 | 3.03 | −1.43 |
| Steroid metabolic process | 27 | 11 | 4 | −0.88 |
| Fatty acid metabolic process | 28 | 6 | 4.25 | −2.28 |
| Lipid transport | 16 | 7 | 2.44 | −0.9 |
| Phospholipid binding | 16 | 6 | 2.02 | −1.45 |
| Sterol metabolic process | 15 | 6 | 3.42 | −0.42 |
| Cholesterol metabolic process | 13 | 6 | 3.01 | −0.17 |
| Steroid biosynthetic process | 12 | 5 | 2.63 | −0.55 |
| Fatty acid biosynthetic process | 12 | 3 | 2.86 | −1.26 |
| Sterol biosynthetic process | 9 | 3 | 4.08 | 0.1 |
| Protein amino acid lipidation | 9 | 3 | 3.17 | −0.35 |
| Lipid modification | 9 | 2 | 2.11 | −1.4 |
| Cholesterol biosynthetic process | 7 | 3 | 3.66 | 0.59 |
| Acetyl-CoA catabolic process | 5 | 4 | 2.81 | 1.82 |
| Acetyl-CoA metabolic process | 5 | 4 | 2.48 | 1.54 |
| Steroid dehydrogenase activity | 6 | 2 | 3.15 | 0.02 |
| Fatty acid oxidation | 7 | 1 | 2.57 | −1.22 |
| Lipid oxidation | 7 | 1 | 2.57 | −1.22 |
| Lipid homeostasis | 6 | 2 | 2.23 | −0.47 |
| Fatty acid binding | 6 | 1 | 3.05 | −0.76 |
| Steroid dehydrogenase activity, acting on the CH-OH group of donors, NAD or NADP as acceptor | 5 | 2 | 2.7 | 0.2 |
Lipid-related gene ontologies, with some of the highest and lowest z-scores, were selected after the analysis of log-transformed microarray data. A z-score is the statistical rating of the relative expression of gene ontologies, and depicts how much each ontology is over- or underrepresented in a given gene list.46 Positive z-scores reflect gene ontology terms with a greater number of genes meeting the criterion than is expected by chance, whereas negative z-scores represent gene ontology terms with fewer genes meeting the criterion than expected by chance. A z-score close to 0 indicates that the number of genes meeting the criterion is approximately the expected number.46 z-scores with values >2.0 or <−2.0 are quite significant. Biological process and molecular function ontologies with such z-scores and with ≥6 genes are highlighted in bold. DHT Genes ↑, number of genes upregulated in DHT-treated cells, as compared with those of the placebo group; Plac Genes ↑, number of genes upregulated in placebo (i.e., ethanol vehicle)-treated cells, relative to those of the DHT group; z-score, specific score for the upregulated genes in the placebo- and hormone-treated cells.
In contrast, DHT treatment had a relatively minor effect on lipid-related ontologies in immortalized human conjunctival epithelial cells. Androgen action upregulated several ontologies associated with lipid metabolism, but none of these effects had a z-score higher than 3.0. Androgen administration also downregulated several lipid-linked ontologies (Table 3).
Table 3.
Impact of DHT on the Expression of Lipid-Related Gene Ontologies in Immortalized Human Conjunctival Epithelial Cells
| Ontologies | DHT Genes ↑ | Plac Genes ↑ | DHT z-score | Plac z-score |
|---|---|---|---|---|
| Lipid metabolic process | 64 | 64 | 2.11 | 0.21 |
| Lipid binding | 40 | 34 | 2.25 | −0.11 |
| Phospholipid binding | 16 | 13 | 2.2 | 0.46 |
| Glycerophospholipid biosynthetic process | 8 | 10 | 2.27 | 2.57 |
| Regulation of lipid metabolic process | 10 | 6 | 2.56 | 0.07 |
| Sterol biosynthetic process | 7 | 8 | 2.95 | 2.89 |
| Lipid homeostasis | 6 | 3 | 2.37 | −0.02 |
| Sterol homeostasis | 5 | 3 | 2.32 | 0.42 |
| Phospholipase activity | 4 | 1 | −0.47 | −2.15 |
| Sterol metabolic process | 8 | 14 | 0.73 | 2.32 |
| Protein amino acid lipidation | 4 | 10 | 0.45 | 3.11 |
| Acetyl-CoA catabolic process | 1 | 5 | −0.43 | 2.36 |
| Fatty acid beta-oxidation | 1 | 5 | −0.52 | 2.15 |
| Acetyl-CoA metabolic process | 1 | 5 | −0.56 | 2.05 |
Lipid-related gene ontologies with the highest and lowest z-scores were selected after the analysis of log-transformed microarray data. Terms are explained in the legend to Table 2.
Overall, our findings demonstrate that the effect of DHT on hTERT-immortalized human meibomian gland epithelial cells is quite distinct and is not duplicated in immortalized human conjunctival epithelial cells.
Identification of a Biomarker for Human Meibomian Gland Epithelial Cells
To determine whether we could identify a unique biomarker for human meibomian gland epithelial cells, we compared the gene expression profile of normal human meibomian gland tissues with those of human conjunctival and corneal epithelia (Gene Expression Omnibus Series accession number GSE5543).37 Through this process, we identified several genes that were highly expressed in human meibomian gland tissue, but minimally expressed in human conjunctiva and cornea. One of these representative genes was kallikrein-related peptidase 6 (KLK6), transcript variant B.
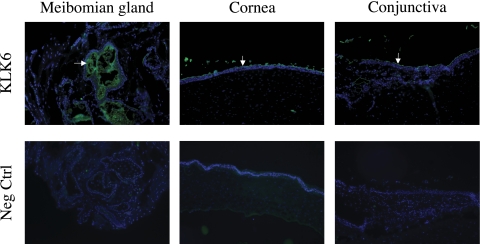
Given this information, we assessed whether parallel differences exist in the expression of KLK6 protein in meibomian gland, conjunctival, and corneal tissues. Our immunohistochemical analyses showed intense staining of KLK6 protein in the superficial layers of meibomian gland epithelia (Fig. 11). However, similar intense staining was also identified in the superficial layer of human conjunctival and corneal epithelia (Fig. 11). These findings demonstrate that KLK6 protein is not a specific marker for human meibomian gland epithelial cells.
Figure 11.
Immunofluorescent staining of KLK6 protein in the human meibomian gland, cornea, and conjunctiva. The KLK6 was identified in the superficial epithelia of all three tissues, indicating that this protein is not a specific marker for human meibomian gland epithelial cells. For negative control sections, the primary antibody was preincubated with human KLK6. Magnification, ×100.
Discussion
Our studies demonstrate that it is possible to isolate viable human meibomian gland epithelial cells and to culture them in serum-free medium. These cells proliferate, survive through at least the fifth passage, and contain neutral lipids. Our results also show that infection with hTERT immortalized these cells, which accumulated neutral lipids during differentiation, expressed multiple genes for lipogenic enzymes, responded to androgen exposure, and continued to proliferate. Thus, we have created an immortalized human meibomian gland epithelial cell line that may be used to evaluate the physiological regulation of these cells.
Primary human meibomian gland epithelial cells were cultured in both serum-containing and serum-free media. When human meibomian gland epithelial cells were cultured in serum-containing media on 3T3 feeder layers, they formed colonies and accumulated neutral lipids in a manner similar to sebaceous gland epithelial cells from human skin.39,40 Primary meibomian gland epithelial cell cultures could also be generated in SFM and without the 3T3 fibroblast layer. Fujie et al.47 showed that dispersed primary cultures of human skin sebocytes could be passaged six times, whereas explant cultures ceased growing after passage 3. We found that dispersed primary cultures of human meibomian gland epithelial cells, when grown in SFM, could be passaged five times. We also observed that when meibomian gland epithelial cells underwent rapid mitosis, there was minimal intracellular lipid staining. After cell proliferation decelerated, cells started to accumulate lipids and show pronounced Oil red O and Nile red staining. This phenomenon has also been noted in other sebaceous gland epithelial cell cultures.39
However, the use of primary cultures of human meibomian gland epithelial cells for medical research is hampered by their finite proliferative capacity, as well as by the very limited source amount of meibomian gland tissue available from small lid segments. These difficulties create a need for an extended lifespan or “immortalized” cell line, which retains the ability to proliferate indefinitely in culture. There is substantial evidence that human somatic cells senesce in a two-stage manner: mortality stage 1 and mortality stage 2.48–51 Mortality stage 1 (M1), also called replicative senescence, is attributed to pRb- and/or p53-mediated cell cycle arrest. Viral oncogenes that can inactivate both p53 and p16Ink4a/pRB checkpoint pathways, such as SV40 large T antigen and the E6/E7 proteins of the HPV 16 virus, can overcome M1 and provide cells with an extended lifespan.48,52,53 Human corneal epithelial cells and human skin sebaceous gland epithelial cells have been successfully immortalized by SV40 large T antigen.26,54 Mortality stage 2 (M2), also called crisis, is due to shortening of telomeres. In normal human somatic cells, each cell division is associated with the loss of 50 to 200 bp of telomeric DNA. At a critical telomere length, cells eventually enter mortality stage 2 and die.48–51 Telomeres are maintained by telomerase, a ribonucleoprotein enzyme normally silent in somatic cells.55 hTERT contains the catalytic subunit for telomerase. Introduction of hTERT into human cells leads to the activation of telomerase, preventing telomere erosion and subsequent telomere-dependent senescence.56–58 Previous reports of hTERT infection in human epithelial cells suggest that a knockdown of p16INK4 and/or p53 tumor suppressor pathways is required for cellular immortalization.49,59,60 However, many cell types including human corneal epithelial cells, esophageal epithelial cells, fibroblasts, retinal pigmented epithelial cells, and endothelial cells have been successfully immortalized by the use of hTERT alone.56,58,61,62
Research has shown that human cell lines developed by transformation with viral oncoproteins including adenovirus E1A, the SV40 large T antigen, and HPV16-E6/E7 were genomically unstable and displayed cellular properties that differed from their normal counterparts.26,54,63,64 Human cell lines immortalized with hTERT exhibit genetic stability, normal contact inhibition, and maintain the capacity to differentiate.58,62 In our study, we demonstrated that human meibomian gland epithelial cells infected with SV40 large T alone had an expanded lifespan. However, the cells were not immortalized and eventually entered M2 because of shortening of telomeres. Meibomian gland epithelial cells were immortalized by introduction of both SV40 large T and hTERT, but, considerable variations in chromosome numbers and the presence of marked chromosomal aberrations were found in those cells.
We immortalized human meibomian gland epithelial cells with hTERT alone. The hTERT-immortalized human meibomian gland epithelial cells maintained a polygonal epithelial cell appearance. Their morphology was similar to that of human skin sebocytes cultured in serum-free medium.47 Their colony-forming efficiency was higher than that of the primary cells at passage 2, indicating that the immortalized cells maintained a high colony growth ability. Their population-doubling time was much shorter than that of the human sebaceous gland cell line (SZ 95), which was reported as 52.4 ± 1.6 hours.26 The SZ95 cells were generated by SV40 large T antigen transfection and had numerous chromosome aberrations, a highly abnormal hyperdiploid-aneuploid karyotype, and structural anomalies. Whether the difference in population-doubling times of immortalized meibocytes and sebocytes is attributable to these chromosome, karyotype, and structural disparities should be further investigated.
The hTERT-immortalized human meibomian gland epithelial cells maintained normal karyotype for more than 23 passages. Even though the epithelial-cell–like appearance and the ability to accumulate lipids were maintained in cells of later passages, we found duplication of chromosome 5 and 20 in passage 35 and beyond. The same gain in chromosomes was also noted in other epithelial cells immortalized by hTERT.65–67 Investigators have suggested that this increased number of chromosomes may contribute to cellular immortalization.65,66 However, the chromosomal gain could also be the result of mitotic nondisjunction due to integration of the proviral cDNA into the cell's genome after retroviral transfection.65
Our microscopy results indicate that the hTERT-immortalized human meibomian gland epithelial cells differentiate in both serum-containing and serum-free media. To explain these findings, we initially hypothesized that differentiation in SFM (i.e., control) might simply be due to the cells' achieving confluence. However, a more likely explanation is that this differentiation response in SFM is induced by EGF and BPE. Invitrogen recommends including these supplements in SFM when formulating their keratinocyte growth medium in serum-free conditions. These growth factors, in turn, are known to stimulate the differentiation of human sebaceous and rabbit meibomian gland epithelial cells.68,69 Consequently, use of SFM alone would be expected to promote cellular differentiation.
The hTERT-immortalized meibomian gland epithelial cells express numerous lipid-, keratin- and protease-related genes that are common to other sebaceous gland epithelial cells, as well as to primary human meibomian gland epithelial cells and human meibomian glands. Also like sebocytes,70–72 the hTERT-immortalized meibomian gland epithelial cells respond to DHT treatment with a significant increase in the expression of genes related to lipid metabolic pathways. This hormone response is similar to the androgen influence on meibomian glands in vivo,11,13,73,74 wherein testosterone upregulates many genes related to lipogenic, steroidogenic, and cholesterogenic pathways. In contrast, this hTERT-immortalized meibomian gland epithelial cell response to androgen exposure is not duplicated in immortalized human conjunctival epithelial cells. These combined findings demonstrate that our hTERT-immortalized epithelial cells are not only meibomian gland in origin, but also respond in a manner analogous to meibomian gland epithelial cells in vivo.
We attempted to identify a unique biomarker for human meibomian gland epithelial cells. Our comparison of the gene expression profiles of normal human meibomian gland tissues with those of human conjunctival and corneal epithelia indicated that KLK6 may be such a marker. KLK6 is a member of the tissue kallikrein gene family. This family consists of genes encoding 15 different secreted serine proteases, all of which are localized in a cluster on chromosome 19.75 The KLKs are expressed in many tissues, including steroid hormone-producing or -dependent tissues, such as the prostate, breast, ovary, and testis, and have diverse physiological functions.76 However, our analysis of KLK6 protein expression showed that KLK6 was present, not only in meibomian gland tissue, but also in the superficial layers of conjunctival and corneal epithelia. The KLK6 protein has also been shown to be expressed in stratum corneum and stratum granulosum in normal skin.77 Therefore, our results demonstrate that KLK6 is not a specific biomarker for meibomian gland. One explanation for the low KLK6 gene expression level in human conjunctiva and cornea samples is that the superficial epithelial layers of these tissues may have sloughed off during their processing and/or preservation. Consequently, this possible source of KLK6 mRNA would not have been available for the microarray experiments.37
In conclusion, we have demonstrated that it is possible to culture and immortalize responsive human meibomian gland epithelial cells. We believe that these cells will be invaluable in helping to identify factors that regulate meibomian gland epithelial cell activity. We also believe that these cells will serve as an excellent preclinical model for the development of novel and unique therapeutic strategies to treat MGD.
Acknowledgments
The authors thank Stephen M. Richards, Hetian Lei, Kristine Lo, Aaron Fay, Ilene Gipson, and Andrius Kazlauskas (Boston, MA) for their technical and clinical assistance; Robert A. Weinberg (Cambridge, MA) for the plasmid vectors; and James Evans and Barbara Evans (Worcester, MA) for their assistance with mass spectrometry.
Footnotes
Supported by Grant EY05612 from the National Institutes of Health and by Alcon Research, Ltd.
Disclosure: S. Liu, None; M.P. Hatton, None; P. Khandelwal, None; D.A. Sullivan, None
References
- 1.McCulley JP, Shine WE. Meibomian gland function and the tear lipid layer. Ocul Surf 2003;1:97–106 [DOI] [PubMed] [Google Scholar]
- 2.Foulks GN, Bron AJ. Meibomian gland dysfunction: a clinical scheme for description, diagnosis, classification, and grading. Ocul Surf 2003;1:107–126 [DOI] [PubMed] [Google Scholar]
- 3.Bron AJ, Tiffany JM, Gouveia SM, Yokoi N, Voon LW. Functional aspects of the tear film lipid layer. Exp Eye Res 2004;78:347–360 [DOI] [PubMed] [Google Scholar]
- 4.Bron AJ, Sci FM, Tiffany JM. The contribution of meibomian disease to dry eye. Ocul Surf 2004;2:149–165 [DOI] [PubMed] [Google Scholar]
- 5.Driver PJ, Lemp MA. Meibomian gland dysfunction. Surv Ophthalmol 1996;40:343–367 [DOI] [PubMed] [Google Scholar]
- 6.Tiffany J. Physiological functions of the meibomian glands. Prog Retin Eye Res 1995;14:47–74 [Google Scholar]
- 7.Shimazaki J, Sakata M, Tsubota K. Ocular surface changes and discomfort in patients with meibomian gland dysfunction. Arch Ophthalmol 1995;113:1266–1270 [DOI] [PubMed] [Google Scholar]
- 8.Sullivan DA, Sullivan BD, Ullman MD, et al. Androgen influence on the meibomian gland. Invest Ophthalmol Vis Sci 2000;41:3732–3742 [PubMed] [Google Scholar]
- 9.Sullivan BD, Evans JE, Cermak JM, Krenzer KL, Dana MR, Sullivan DA. Complete androgen insensitivity syndrome: effect on human meibomian gland secretions. Arch Ophthalmol 2002;120:1689–1699 [DOI] [PubMed] [Google Scholar]
- 10.Cermak JM, Krenzer KL, Sullivan RM, Dana MR, Sullivan DA. Is complete androgen insensitivity syndrome associated with alterations in the meibomian gland and ocular surface? Cornea 2003;22:516–521 [DOI] [PubMed] [Google Scholar]
- 11.Schirra F, Suzuki T, Richards SM, et al. Androgen control of gene expression in the mouse meibomian gland. Invest Ophthalmol Vis Sci 2005;46:3666–3675 [DOI] [PubMed] [Google Scholar]
- 12.Suzuki T, Schirra F, Richards SM, Jensen RV, Sullivan DA. Estrogen and progesterone control of gene expression in the mouse meibomian gland. Invest Ophthalmol Vis Sci 2008;49:1797–1808 [DOI] [PubMed] [Google Scholar]
- 13.Sullivan DA, Jensen RV, Suzuki T, Richards SM. Do sex steroids exert sex-specific and/or opposite effects on gene expression in lacrimal and meibomian glands? Mol Vis 2009;15:1553–1572 [PMC free article] [PubMed] [Google Scholar]
- 14.Kremer I, Gaton DD, David M, Gaton E, Shapiro A. Toxic effects of systemic retinoids on meibomian glands. Ophthalmic Res 1994;26:124–128 [DOI] [PubMed] [Google Scholar]
- 15.Zouboulis CC. Acne and sebaceous gland function. Clin Dermatol 2004;22:360–366 [DOI] [PubMed] [Google Scholar]
- 16.Thody AJ, Shuster S. Control of sebaceous gland function in the rat by alpha-melanocyte-stimulating hormone. J Endocrinol 1975;64:503–510 [PubMed] [Google Scholar]
- 17.Deplewski D, Rosenfield RL. Growth hormone and insulin-like growth factors have different effects on sebaceous cell growth and differentiation. Endocrinology 1999;140:4089–4094 [DOI] [PubMed] [Google Scholar]
- 18.Cabeza M, Miranda R, Arias E, Diaz de Leon L. Inhibitory effect of propranolol on lipid synthesis in gonadectomized male hamster flank organs. Arch Med Res 1998;29:291–295 [PubMed] [Google Scholar]
- 19.Chen W, Kelly MA, Opitz-Araya X, Thomas RE, Low MJ, Cone RD. Exocrine gland dysfunction in MC5-R-deficient mice: evidence for coordinated regulation of exocrine gland function by melanocortin peptides. Cell 1997;91:789–798 [DOI] [PubMed] [Google Scholar]
- 20.Zouboulis CC, Bohm M. Neuroendocrine regulation of sebocytes: a pathogenetic link between stress and acne. Exp Dermatol 2004;13(suppl 4):31–35 [DOI] [PubMed] [Google Scholar]
- 21.Zouboulis CC, Seltmann H, Hiroi N, et al. Corticotropin-releasing hormone: an autocrine hormone that promotes lipogenesis in human sebocytes. Proc Natl Acad Sci U S A 2002;99:7148–7153 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zouboulis CC, Chen WC, Thornton MJ, Qin K, Rosenfield R. Sexual hormones in human skin. Horm Metab Res 2007;39:85–95 [DOI] [PubMed] [Google Scholar]
- 23.Zouboulis CC. Human skin: an independent peripheral endocrine organ. Horm Res 2000;54:230–242 [DOI] [PubMed] [Google Scholar]
- 24.Zouboulis CC, Baron JM, Bohm M, et al. Frontiers in sebaceous gland biology and pathology. Exp Dermatol 2008;17:542–551 [DOI] [PubMed] [Google Scholar]
- 25.Zouboulis CC, Schagen S, Alestas T. The sebocyte culture: a model to study the pathophysiology of the sebaceous gland in sebostasis, seborrhoea and acne. Arch Dermatol Res 2008;300:397–413 [DOI] [PubMed] [Google Scholar]
- 26.Zouboulis CC, Seltmann H, Neitzel H, Orfanos CE. Establishment and characterization of an immortalized human sebaceous gland cell line (SZ95). J Invest Dermatol 1999;113:1011–1020 [DOI] [PubMed] [Google Scholar]
- 27.Nelson AM, Zhao W, Gilliland KL, Zaenglein AL, Liu W, Thiboutot DM. Neutrophil gelatinase-associated lipocalin mediates 13-cis retinoic acid-induced apoptosis of human sebaceous gland cells. J Clin Invest 2008;118:1468–1478 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Harrison WJ, Bull JJ, Seltmann H, Zouboulis CC, Philpott MP. Expression of lipogenic factors galectin-12, resistin, SREBP-1, and SCD in human sebaceous glands and cultured sebocytes. J Invest Dermatol 2007;127:1309–1317 [DOI] [PubMed] [Google Scholar]
- 29.Counter CM, Hahn WC, Wei W, et al. Dissociation among in vitro telomerase activity, telomere maintenance, and cellular immortalization. Proc Natl Acad Sci U S A 1998;95:14723–14728 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hahn WC, Dessain SK, Brooks MW, et al. Enumeration of the simian virus 40 early region elements necessary for human cell transformation. Mol Cell Biol 2002;22:2111–2123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hong I, Lee MH, Na TY, Zouboulis CC, Lee MO. LXRalpha enhances lipid synthesis in SZ95 sebocytes. J Invest Dermatol 2008;128:1266–1272 [DOI] [PubMed] [Google Scholar]
- 32.Folch J, Lees M, Sloane Stanley GH. A simple method for the isolation and purification of total lipides from animal tissues. J Biol Chem 1957;226:497–509 [PubMed] [Google Scholar]
- 33.Christie WW. Rapid separation and quantification of lipid classes by high performance liquid chromatography and mass (light-scattering) detection. J Lipid Res 1985;26:507–512 [PubMed] [Google Scholar]
- 34.Brugger B, Erben G, Sandhoff R, Wieland FT, Lehmann WD. Quantitative analysis of biological membrane lipids at the low picomole level by nano-electrospray ionization tandem mass spectrometry. Proc Natl Acad Sci U S A 1997;94:2339–2344 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Shi L, Reid LH, Jones WD, et al. The MicroArray Quality Control (MAQC) project shows inter- and intraplatform reproducibility of gene expression measurements. Nat Biotechnol 2006;24:1151–1161 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ashburner M, Ball CA, Blake JA, et al. Gene ontology: tool for the unification of biology: The Gene Ontology Consortium. Nat Genet 2000;25:25–29 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Turner HC, Budak MT, Akinci MA, Wolosin JM. Comparative analysis of human conjunctival and corneal epithelial gene expression with oligonucleotide microarrays. Invest Ophthalmol Vis Sci 2007;48:2050–2061 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Thiboutot D, Jabara S, McAllister JM, et al. Human skin is a steroidogenic tissue: steroidogenic enzymes and cofactors are expressed in epidermis, normal sebocytes, and an immortalized sebocyte cell line (SEB-1). J Invest Dermatol 2003;120:905–914 [DOI] [PubMed] [Google Scholar]
- 39.Ito A, Sakiguchi T, Kitamura K, Akamatsu H, Horio T. Establishment of a tissue culture system for hamster sebaceous gland cells. Dermatology 1998;197:238–244 [DOI] [PubMed] [Google Scholar]
- 40.Xia LQ, Zouboulis C, Detmar M, Mayer-da-Silva A, Stadler R, Orfanos CE. Isolation of human sebaceous glands and cultivation of sebaceous gland-derived cells as an in vitro model. J Invest Dermatol 1989;93:315–321 [PubMed] [Google Scholar]
- 41.Stanley JR, Yuspa SH. Specific epidermal protein markers are modulated during calcium-induced terminal differentiation. J Cell Biol 1983;96:1809–1814 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Hennings H, Holbrook KA. Calcium regulation of cell-cell contact and differentiation of epidermal cells in culture: an ultrastructural study. Exp Cell Res 1983;143:127–142 [DOI] [PubMed] [Google Scholar]
- 43.Eisinger M, Lee JS, Hefton JM, Darzynkiewicz Z, Chiao JW, de Harven E. Human epidermal cell cultures: growth and differentiation in the absence of differentiation in the absence of dermal components or medium supplements. Proc Natl Acad Sci U S A 1979;76:5340–5344 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Puy LA, Turgeon C, Gagne D, Labrie Y, et al. Localization and regulation of expression of the FAR-17A gene in the hamster flank organs. J Invest Dermatol 1996;107:44–50 [DOI] [PubMed] [Google Scholar]
- 45.Rosignoli C, Nicolas JC, Jomard A, Michel S. Involvement of the SREBP pathway in the mode of action of androgens in sebaceous glands in vivo. Exp Dermatol 2003;12:480–489 [DOI] [PubMed] [Google Scholar]
- 46.Doniger SW, Salomonis N, Dahlquist KD, Vranizan K, Lawlor SC, Conklin BR. MAPPFinder: using Gene Ontology and GenMAPP to create a global gene-expression profile from microarray data. Genome Biol 2003;4:R7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Fujie T, Shikiji T, Uchida N, Urano Y, Nagae H, Arase S. Culture of cells derived from the human sebaceous gland under serum-free conditions without a biological feeder layer or specific matrices. Arch Dermatol Res 1996;288:703–708 [DOI] [PubMed] [Google Scholar]
- 48.Wright WE, Pereira-Smith OM, Shay JW. Reversible cellular senescence: implications for immortalization of normal human diploid fibroblasts. Mol Cell Biol 1989;9:3088–3092 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kiyono T, Foster SA, Koop JI, McDougall JK, Galloway DA, Klingelhutz AJ. Both Rb/p16INK4a inactivation and telomerase activity are required to immortalize human epithelial cells. Nature 1998;396:84–88 [DOI] [PubMed] [Google Scholar]
- 50.Farwell DG, Shera KA, Koop JI, et al. Genetic and epigenetic changes in human epithelial cells immortalized by telomerase. Am J Pathol 2000;156:1537–1547 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Shay JW, Wright WE. Senescence and immortalization: role of telomeres and telomerase. Carcinogenesis 2005;26:867–874 [DOI] [PubMed] [Google Scholar]
- 52.Lee KM, Choi KH, Ouellette MM. Use of exogenous hTERT to immortalize primary human cells. Cytotechnology 2004;45:33–38 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Ahuja D, Saenz-Robles MT, Pipas JM. SV40 large T antigen targets multiple cellular pathways to elicit cellular transformation. Oncogene 2005;24:7729–7745 [DOI] [PubMed] [Google Scholar]
- 54.Araki-Sasaki K, Ohashi Y, Sasabe T, et al. An SV40-immortalized human corneal epithelial cell line and its characterization. Invest Ophthalmol Vis Sci 1995;36:614–621 [PubMed] [Google Scholar]
- 55.Weng NP, Hathcock KS, Hodes RJ. Regulation of telomere length and telomerase in T and B cells: a mechanism for maintaining replicative potential. Immunity 1998;9:151–157 [DOI] [PubMed] [Google Scholar]
- 56.Bodnar AG, Ouellette M, Frolkis M, et al. Extension of life-span by introduction of telomerase into normal human cells. Science 1998;279:349–352 [DOI] [PubMed] [Google Scholar]
- 57.Vaziri H, Benchimol S. Reconstitution of telomerase activity in normal human cells leads to elongation of telomeres and extended replicative life span. Curr Biol 1998;8:279–282 [DOI] [PubMed] [Google Scholar]
- 58.Robertson DM, Li L, Fisher S, et al. Characterization of growth and differentiation in a telomerase-immortalized human corneal epithelial cell line. Invest Ophthalmol Vis Sci 2005;46:470–478 [DOI] [PubMed] [Google Scholar]
- 59.Weinberg RA. Telomeres: bumps on the road to immortality. Nature 1998;396:23–24 [DOI] [PubMed] [Google Scholar]
- 60.Rheinwald JG, Hahn WC, Ramsey MR, et al. A two-stage, p16(INK4A)- and p53-dependent keratinocyte senescence mechanism that limits replicative potential independent of telomere status. Mol Cell Biol 2002;22:5157–5172 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Yang J, Chang E, Cherry AM, et al. Human endothelial cell life extension by telomerase expression. J Biol Chem 1999;274:26141–26148 [DOI] [PubMed] [Google Scholar]
- 62.Morales CP, Gandia KG, Ramirez RD, Wright WE, Shay JW, Spechler SJ. Characterisation of telomerase immortalised normal human oesophageal squamous cells. Gut 2003;52:327–333 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Kahn CR, Young E, Lee IH, Rhim JS. Human corneal epithelial primary cultures and cell lines with extended life span: in vitro model for ocular studies. Invest Ophthalmol Vis Sci 1993;34:3429–3441 [PubMed] [Google Scholar]
- 64.Mohan RR, Possin DE, Mohan RR, Sinha S, Wilson SE. Development of genetically engineered tet HPV16–E6/E7 transduced human corneal epithelial clones having tight regulation of proliferation and normal differentiation. Exp Eye Res 2003;77:395–407 [DOI] [PubMed] [Google Scholar]
- 65.Ramirez RD, Sheridan S, Girard L, et al. Immortalization of human bronchial epithelial cells in the absence of viral oncoproteins. Cancer Res 2004;64:9027–9034 [DOI] [PubMed] [Google Scholar]
- 66.Gu Y, Li H, Miki J, et al. Phenotypic characterization of telomerase-immortalized primary non-malignant and malignant tumor-derived human prostate epithelial cell lines. Exp Cell Res 2006;312:831–843 [DOI] [PubMed] [Google Scholar]
- 67.Troester MA, Hoadley KA, Sorlie T, et al. Cell-type-specific responses to chemotherapeutics in breast cancer. Cancer Res 2004;64:4218–4226 [DOI] [PubMed] [Google Scholar]
- 68.Zhang L, Anthonavage M, Huang Q, Li WH, Eisinger M. Proopiomelanocortin peptides and sebogenesis. Ann N Y Acad Sci 2003;994:154–161 [DOI] [PubMed] [Google Scholar]
- 69.Maskin SL, Tseng SC. Clonal growth and differentiation of rabbit meibomian gland epithelium in serum-free culture: differential modulation by EGF and FGF. Invest Ophthalmol Vis Sci 1992;33:205–217 [PubMed] [Google Scholar]
- 70.Hall DW, Van den Hoven WE, Noordzij-Kamermans NJ, Jaitly KD. Hormonal control of hamster ear sebaceous gland lipogenesis. Arch Dermatol Res 1983;275:1–7 [DOI] [PubMed] [Google Scholar]
- 71.Akimoto N, Sato T, Iwata C, et al. Expression of perilipin A on the surface of lipid droplets increases along with the differentiation of hamster sebocytes in vivo and in vitro. J Invest Dermatol 2005;124:1127–1133 [DOI] [PubMed] [Google Scholar]
- 72.Kurokawa I, Danby FW, Ju Q, et al. New developments in our understanding of acne pathogenesis and treatment. Exp Dermatol 2009;18:821–832 [DOI] [PubMed] [Google Scholar]
- 73.Schirra F, Richards SM, Liu M, Suzuki T, Yamagami H, Sullivan DA. Androgen regulation of lipogenic pathways in the mouse meibomian gland. Exp Eye Res 2006;83:291–296 [DOI] [PubMed] [Google Scholar]
- 74.Schirra F, Richards SM, Sullivan DA. Androgen influence on cholesterogenic enzyme mRNA levels in the mouse meibomian gland. Curr Eye Res 2007;32:393–398 [DOI] [PubMed] [Google Scholar]
- 75.Yousef GM, Diamandis EP. The new human tissue kallikrein gene family: structure, function, and association to disease. Endocr Rev 2001;22:184–204 [DOI] [PubMed] [Google Scholar]
- 76.Pampalakis G, Sotiropoulou G. Tissue kallikrein proteolytic cascade pathways in normal physiology and cancer. Biochim Biophys Acta 2007;1776:22–31 [DOI] [PubMed] [Google Scholar]
- 77.Komatsu N, Saijoh K, Toyama T, et al. Multiple tissue kallikrein mRNA and protein expression in normal skin and skin diseases. Br J Dermatol 2005;153:274–281 [DOI] [PubMed] [Google Scholar]