Plant development depends on the balance of stem cell renewal and differentiation in several stem cell niches that is achieved by yet unknown regulatory mechanisms. Using inducible RNA interference-mediated downregulation, Arabidopsis RETINOBLASTOMA-RELATED protein is shown to regulate the proliferation, maintenance, and differentiation of stem cells and organ production from stem cell niches.
Abstract
Several genes involved in the regulation of postembryonic organ initiation and growth have been identified. However, it remains largely unclear how developmental cues connect to the cell cycle. RETINOBLASTOMA RELATED (RBR) is a plant homolog of the tumor suppressor Retinoblastoma (pRb), which is a key regulator of the cell cycle. Using inducible RNA interference (RNAi) against Arabidopsis thaliana RBR (RBRi), we reduced RBR expression levels at different stages of plant development. Conditional reduction or loss of RBR function disrupted cell division patterns, promoted context-dependent cell proliferation, and negatively influenced establishment of cell differentiation. Several lineages of toti- and pluripotent cells, including shoot apical meristem stem cells, meristemoid mother cells, and procambial cells, failed to produce appropriately differentiated cells. Meristem activity was altered, leading to a disruption of the CLAVATA-WUSCHEL feedback loop and inhibition of lateral organ formation. Release of RBR from RNAi downregulation restored meristem activity. Gene profiling analyses soon after RBRi induction revealed that a change in RBR homeostasis is perceived as a stress, even before genes regulated by RBR-E2F become deregulated. The results establish RBR as a key cell cycle regulator required for coordination of cell division, differentiation, and cell homeostasis.
INTRODUCTION
Retinoblastoma (RB) was the first tumor suppressor gene identified in animals (Friend et al., 1986) and later was associated with cell cycle regulation, thus establishing pRb as a key regulator of cell proliferation. In animals, three members of the RB family of proteins (pRb, p107, and p130), also referred to as pocket proteins (Mulligan and Jacks, 1998), negatively regulate the G1-to-S phase transition during the cell cycle. It is now widely accepted that mitogenic signals activate several cyclin-dependent kinase (CDK)-cyclin complexes during progression through G1 to phosphorylate pRb, thereby releasing it from interactions with E2F/DP transcription factor complexes to facilitate S-phase entry (Weinberg, 1995). Subsequently, pRb was also found to regulate tissue-specific transcription factors that regulate cell differentiation, such as MyoD, which activates muscle-specific genes (De Falco et al., 2006). pRb also regulates germ cell proliferation, differentiation, and survival (Toppari et al., 2003) and is required for osteoblast, keratinocyte, and macrophage differentiation (Nead et al., 1998; Liu et al., 1999; Thomas et al., 2003). Together with the tumor suppressor p53, pRb regulates adipocyte differentiation and function (Hallenborg et al., 2009). The requirement for RB in cell differentiation therefore extends beyond regulation of cell cycle entry. For example, the loss of RB function in cardioblasts affects cell differentiation by modulating the activity of cardiogenic factors (Papadimou et al., 2005).
RB-related genes, termed RETINOBLASTOMA-RELATED (RBR; Ach et al., 1997), are also found in monocots (Grafi et al., 1996; Xie et al., 1996; Ach et al., 1997) and dicots (Nakagami et al., 1999; Kong et al., 2000) and even in unicellular algae (Umen and Goodenough, 2001). Animal pRb and plant RBR are similar in sequence and predicted protein structure (Kong et al., 2000) and, thus, likely share similar functions (Flemington et al., 1993; Ramirez-Parra et al., 2003; Gruissem, 2007). For example, loss of pRb function in mouse is embryo-lethal because embryos fail to develop properly (Jacks et al., 1992; Lee et al., 1992). In Arabidopsis thaliana, loss of RBR function is gametophyte-lethal because mitotically derived cells from the megaspore fail to differentiate into a functional female gametophyte (Ebel et al., 2004; Johnston et al., 2008; Johnston and Gruissem, 2009). Although all adult cells retain pRb in animals and RBR in plants (Savatier et al., 1994; Wildwater et al., 2005), except for their role in tumorgenesis and specific cell types, it has been difficult to understand the postembryonic functions of these proteins.
We investigated the role of Arabidopsis RBR in cell division and differentiation during development. Postembryonic plant development involves specific meristem stem cell niches in which cell division maintains a stem cell population and produces cells that continue to divide and differentiate (Brand et al., 2000; Tucker and Laux, 2007). Like tumor cells, stem cells are thought to proliferate indefinitely through cellular self-renewal capacity. Embryonic stem cells in animals have a very short G1 phase and a prolonged S phase, which is most likely regulated by pRb and has been proposed to maintain stemness during development (Stead et al., 2002; White and Dalton, 2005). Similarly, targeted RBR downregulation in the root apex alters root stem cell proliferation (Wildwater et al., 2005), suggesting that RBR has a function in stem cell maintenance as well.
Because rbr mutants are gametophytic lethal (Ebel et al., 2004), understanding postembryonic functions of RBR is only possible using conditional mutants. Previously, virus-induced gene silencing (VIGS) using Tobacco rattle virus (Park et al., 2005) or Tomato golden mosaic virus (Jordan et al., 2007) in Nicotiana benthamiana has been used to downregulate RBR in leaves. Similarly, conditional expression of viral proteins that bind RBR to interfere with RBR function has been reported (Desvoyes et al., 2006; Lageix et al., 2007). While these approaches provided insights into the function of RBR in cell proliferation, they are either spatially restricted or may obscure other RBR functions in development and differentiation. For example, VIGS is active in mature plants with little effect on tissue specificity and timing of silencing, thus making it difficult to observe acute effects of loss of RBR function. Similarly, expression of viral proteins may only restrict RBR regulation of cell proliferation but not other functions. In addition, viral RBR binding proteins, such as geminivirus AL or AC, are multifunctional (Orozco et al., 2000), and the role of the RepA/RBR complex in the regulation of transcription of E2F site–containing promoters is not well understood (Munoz-Martin et al., 2003). Likewise, the nanovirus Clink protein binds RBR but also SKP1, which functions in the proteolytic degradation pathway (Aoyama and Chua, 1997; Aronson et al., 2000).
To establish conditional mutants that only affect RBR function, we used a chemically inducible RBRi approach to rapidly downregulate RBR expression. Our results show that conditional downregulation and loss of RBR function uncouples division and differentiation of meristemoid cells in leaves and disrupts the appropriate division and maintenance of meristem stem cells. Thus, RBR has a critical function in the homeostasis of stem cells and organ production in every stem cell niche. Interestingly, gene expression profiling shows that acute downregulation of RBR function triggers an immediate stress response, while other known RBR targets respond only later and after protein levels are significantly reduced. In animals, pRb is required for blood cell differentiation under stress conditions (Spike et al., 2004), and in both animals and plants abiotic and biotic stress affects cell cycle regulation (Shackelford et al., 2000; Ma and Bohnert, 2007; Macleod, 2008; Klimova et al., 2009). Thus, in addition to the control of cell division and differentiation during development, RBR may also function in the early stress response to adjust cell proliferation to prevailing conditions.
RESULTS
Construction of Arabidopsis Lines with Inducible RNA Interference against RBR
We tested several induction systems for conditional downregulation of RBR expression. While cre/lox-mediated excision of a genomic RBR fragment produced sectors with mutant phenotypes in root meristems (Wildwater et al., 2005) and disturbed cell proliferation in leaves (see Supplemental Figure 1 online), we found that the central zone of the shoot apical meristem was recalcitrant to loss of lox alleles and RBR function. We then established conditional RBR mutant lines using the dexamethasone (DEX)-inducible GVG transcription factor system (Aoyama and Chua, 1997) but found that the DEX system by itself may produce phenotypes that depend on the expression level of GVG, as was previously reported by others (Kang et al., 1999; Andersen et al., 2003; Amirsadeghi et al., 2007). Although RBR protein levels could be effectively manipulated in DEX-inducible plants (data not shown), the GVG-induced phenotypes may obscure or amplify phenotypes produced by deregulation of RBR expression. We therefore constructed a new set of Arabidopsis conditional RBR mutant lines using the β-estradiol–inducible expression system, which we and others found to confer tight regulation of the XVE chimeric transcription factor (Brand et al., 2006).
A DNA hairpin (RBRhp) was constructed to target RBRi against the first six exons of RBR (see Supplemental Figure 2 online). RBRhp was cloned under the control of the OLexA promoter, which can be activated by the constitutively expressed XVE chimeric transcription factor after β-estradiol–dependent translocation to the nucleus (Brand et al., 2006). Pro35S:XVE and OLexA:RBRhp were independently transformed into Arabidopsis. Screening of several independent transgenic lines confirmed that the expression cassettes alone did not cause developmental phenotypes when plants were treated with β-estradiol. Control experiments using homozygous Pro35S:XVE; OLexA:GUS (β-glucuronidase [GUS]) lines showed that β-estradiol was taken up quickly and efficiently, resulting in GUS expression only after β-estradiol treatments (see Supplemental Figure 3 online). We cannot exclude, however, that β-estradiol–dependent XVE translocation to the nucleus may have different kinetics in different cell types. Homozygous Pro35S:XVE driver and OLexA:RBRhp target lines were then crossed, and F3 progenies were germinated on β-estradiol–containing Murashige and Skoog (MS) plates and screened to isolate inducible lines homozygous for the Pro35S:XVE; OLexA:RBRhp binary system, designated RBRi.
Quantitative RT-PCR and protein blot analyses confirmed that the RBRi system was inactive in the absence of β-estradiol but responded rapidly to induction. To investigate the effects of RBRi induction, protein and transcript quantification was performed on seedlings and young leaves, since we could detect the highest ProRBR:GUS reporter activities and RBR protein concentration in proliferating tissues (see Supplemental Figure 4 online). Twelve hours after induction by spraying 5 μM β-estradiol solution on 10-d-old seedlings, RBR mRNA (Figure 1A) and protein levels in young leaves were decreased by >50%, and RBR protein was no longer detectable 36 h after induction (Figure 1B). Similar results were obtained with 5-d-old whole seedlings germinated on plates containing β-estradiol (Figure 1C). The inducible, rapid, and effective downregulation of RBR transcript and protein using our RBRi system therefore provides a spatially and developmentally more tightly controlled conditional mechanism to specifically interfere with RBR function than virus-induced RBR silencing systems (Park et al., 2005; Jordan et al., 2007).
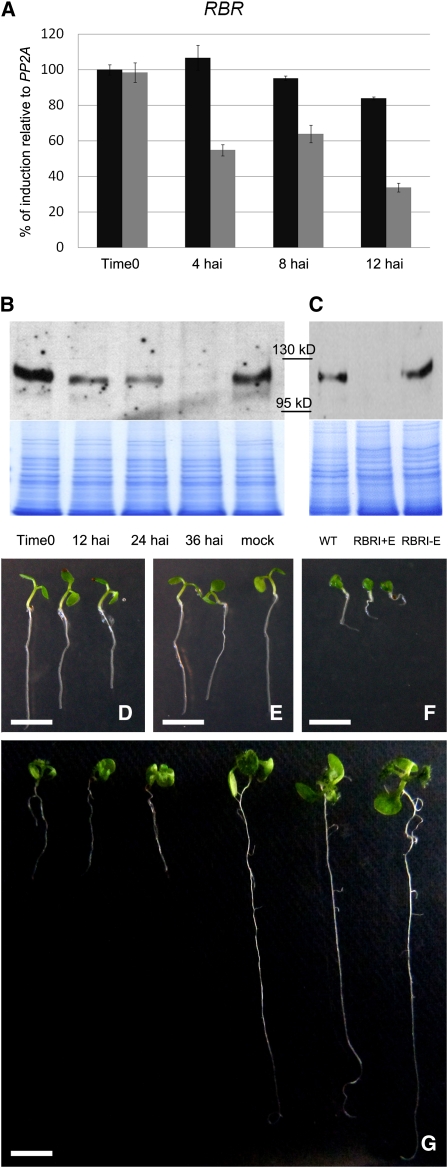
Figure 1.
Quantification of RBR Downregulation and Phenotypic Alterations in β-Estradiol–Induced RBRi Seedlings.
(A) RBR mRNA quickly decreases after RBRi induction. Twelve hours after spraying 10-d-old RBRi seedlings with β-estradiol, RBR mRNA in the first two young leaves was reduced to <40% of its initial level (gray columns). Black columns: untreated RBRi seedlings (n = 50 for each time point/genotype treatment). The error bars represent the se from three biological replicates.
(B) RBR protein levels in young leaves of 10-d-old seedlings were detected using an antibody against the N-terminal 374 amino acids of RBR. Thirty-six hours after β-estradiol spraying, RBR is barely visible on protein gels.
(C) Five days after germination on β-estradiol–containing plates (RBRi+E), RBR is not longer detectable compared with wild-type (WT) or uninduced RBRi seedlings (RBRi-E). The stained gel is shown as a quantitative control.
(D) Five-day-old wild-type seedlings germinated on β-estradiol–containing MS plates.
(E) Five-day-old RBRi seedlings germinated on β-estradiol–free MS plates.
(F) Five-day-old RBRi seedlings germinated on β-estradiol–containing MS plates.
(G) Wild-type and RBRi seedlings were germinated on MS medium plus β-estradiol and moved to β-estradiol–free MS plates 5 d after germination. One week after recovery, development of RBRi-induced seedlings remained delayed (left three plants) compared with wild-type seedlings of the same age (right three plants). Organ production in both shoot and root apices is strongly retarded in RBRi seedlings.
Bars = 0.5 cm.
Conditional Decrease of RBR Expression Causes Arrest of Plant Development
To analyze the physiological and developmental consequences of loss of RBR function, we induced RBRi expression at different times during development between the seedling stage and flowering. When germinated in the absence of β-estradiol, RBRi seedlings were indistinguishable from wild-type seedlings (Figures 1D and 1E). After 3 d of germination on β-estradiol–containing medium, RBRi seedling development was delayed and after 5 d, development was impaired. By 5 d, cotyledons had not expanded, hypocotyls had not elongated, new leaves failed to initiate, and root elongation was reduced (Figure 1F). The same seedlings resumed leaf production 1 week after transfer to β-estradiol–free plates and recovery of RBR protein levels (see Supplemental Figure 5 online), although development continued to be delayed compared with the wild type (Figure 1G). These results show that temporary loss of RBR function does not cause irreversible changes in development, although we cannot exclude that individual cells or cell types may have lost differentiation capacity (see below). To investigate the effects of RBR downregulation on organ production at different stages of plant development, we treated 12-d-old RBRi plants with 5 μM β-estradiol for seven consecutive days. By the end of the treatment, plants had expanded cotyledons and had developed the first two leaves, which were 40 to 60% (n = 12) smaller than those of the wild type after the treatment (Figures 2A and 2B). After RNA interference (RNAi) induction, however, RBRi plants only developed three to five additional very small leaves with a bumpy and strongly downward-curled morphology (Figures 2B and 2C). Differences between abaxial and adaxial cell division and/or expansion patterns were reported to cause alterations in the morphology of the leaf, reflected by up- or downward curling (Talbert et al., 1995; Berna et al., 1999; Dewitte et al., 2003). We measured leaf cell numbers in wild-type and RBRi adaxial and abaxial sides through quantification of scanning electron microscopy images (see below). The results showed an approximately two- and fourfold increase in cell number on the adaxial and abaxial sides, respectively, in RBRi plants when compared with the wild type (see Supplemental Figure 6 online). Because mutant leaves are smaller than the wild type but have more cells, we concluded that the curled phenotype was due to a reduction in cell expansion/cell differentiation that particularly affects the abaxial side. Together, these results show that the arrest of RBRi seedling/plant growth in the presence of β-estradiol is caused by loss of RBR function and not by treatment with β-estradiol, which does not affect development of wild-type Arabidopsis, even at a concentration of 25 μM (Moore et al., 2006).
Figure 2.
RBRi Leaf Mutant Phenotypes.
(A) Both 19-d-old wild-type (left) and RBRi (right) plants were sprayed with β-estradiol for five consecutive days beginning at day 14. The RBRi plant is smaller and leaf production is arrested.
(B) Top row: cotyledon and leaf morphogenesis of the 19-d-old wild-type plant. Bottom row: RBRi cotyledons and leaves. Leaf production was arrested in RBRi-induced plants, and leaves produced during the β-estradiol treatment were strongly delayed in development. Bar = 0.5 cm.
(C) A wild-type leaf number 6 on the left, compared with RBRi leaves number 5 and 6 from 19-d-old plants. RBRi leaves are small with a strong downward curl.
Downregulation of RBR Disrupts Stem Cell Maintenance and Organ Primordia Initiation in the Shoot Apical Meristem
The arrest of organ production prompted us to analyze the morphology of the RBRi shoot apical meristem (SAM). Thin sections from uninduced 3-d-old RBRi seedlings were indistinguishable from the wild type, and L1, L2, and L3 cell layers were clearly visible (Figures 3A and 3B). Four days after induction (DAI), cells of the L2 layer became detached from the L1 layer of RBRi SAMs (Figures 3C and 3D). Five DAI, L2 cells had lost their strict anticlinal division pattern and detachment from L1 layer cells was clearly visible (Figures 3E and 3F). Seven DAI, the L1 layer could still be recognized and had no apparent periclinal cell divisions, while undirected cell divisions in the internal SAM region had abolished the L2 and L3 layer organization (Figures 3G and 3H). At this stage, the detachment area we previously observed had disappeared, most likely because cell proliferation had increased (see Supplemental Figure 7 online). As a consequence of the disordered stem cell divisions, the apical dome became flatter, which resulted in a significant reduction in the typical height-to-width ratio of an active wild-type SAM.
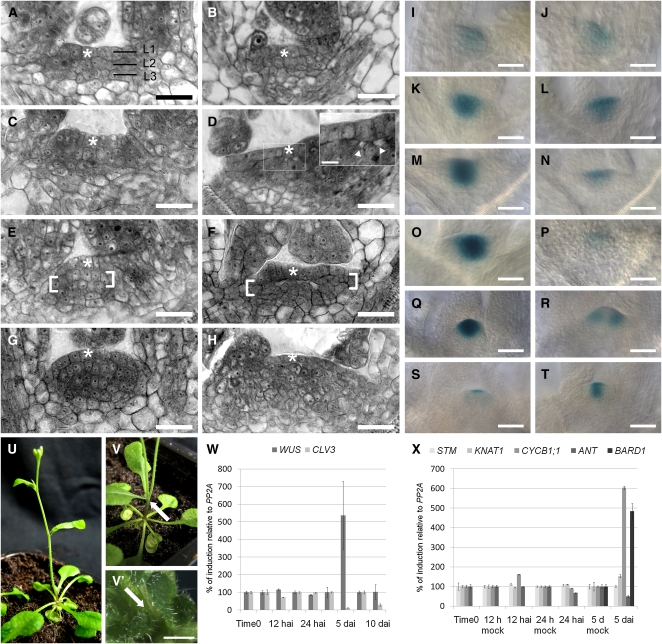
Figure 3.
SAM Morphological and Genetic Alterations in β-Estradiol–Treated RBRi Seedlings.
(A), (C), (E), and (G) Resin-embedded, toluidine blue–stained sections of wild-type SAMs from 3-, 4-, 5-, and 7-d-old seedlings, respectively. The apex acquires the dome shape morphology typical of the Arabidopsis SAM. Asterisks indicate the L1 layer.
(B), (D), (F), and (H) Resin-embedded, toluidine blue–stained sections of SAMs from 3-, 4-, 5-, and 7-d-old RBRi plants, respectively, germinated on β-estradiol. Beginning at day 4, morphological changes became visible in mutant SAMs. In the inset in (D), magnification of the boxed area, white arrowheads point to the beginning detachment of L2 from L1. On day 5 (F), the L2 layer became detached from the L1 layer, and disorganized cell divisions were observed in L2 (between square brackets, compare [E] and [F]). On day 7 (H), the L1 layer was still clearly visible, but cell division in L2 and L3 had become disorganized, resulting in the loss of the typical L2 anatomy. Mutant SAMs became flattened by day 5 (F) and afterwards expanded laterally and vertically, possibly because of cell overproliferation in L2 and L3.
(I), (K), (M), (O), and (Q) ProCLV3:GUS staining in untreated 3-, 4-, 5-, 8-, and 15-d-old RBRi; ProCLV3:GUS seedlings. The stronger GUS staining (3 h at 37°C) was necessary to compare the β-estradiol untreated and weakly staining treated RBRi seedlings.
(J), (L), (N), and (P) ProCLV3:GUS staining in 3-, 4-, 5-, and 8-d-old RBRi; ProCLV3:GUS seedlings germinated on β-estradiol.
(R) Thirty percent (n = 60) of the 15-d-old recovered seedlings germinated on β-estradiol developed twin ProCLV3:GUS sectors 1 week after transfer to β-estradiol–free soil.
(S) In rare cases, the ProCLV3:GUS signal remained confined to the L1 layer and no organ production was observed even 1 week after recovery from a 5-d-long β-estradiol treatment.
(T) Most of the seedlings recovered and showed a wild-type-like, although weaker, ProCLV3:GUS staining pattern.
(U), (V), and (V') Thirty percent (n = 60) of the recovered RBRi seedlings produced twin inflorescence stems originating at the first node ([V], arrow points at the bifurcation), or from the basal rosette ([V'], arrow pointing in between the two emerging young inflorescences), while the majority of the recovered RBRi plants had a single inflorescence stem (U).
(W) CLV3 and WUS expression in Arabidopsis RBRi shoot apices quantified by real-time PCR. In 5-d-old seedlings germinated on β-estradiol (n = 50 for each time point/treatment/analyzed gene expression), CLV3 is strongly downregulated, as confirmed by ProCLV3:GUS staining. The CLV3-WUS loop appears to be intact 5 d after RBRi induction because WUS became temporarily upregulated. Ten days after continuous induction, CLV3 expression levels was still reduced, while WUS mRNA levels had returned to nearly wild-type levels. Seedlings treated with β-estradiol for 5 d were arrested in organ production (see Figure 4). The error bars represent the se from two biological replicas and two technical replicas.
(X) Expression of meristem marker genes in Arabidopsis RBRi shoot apices quantified by real-time PCR. STM and KNAT1 RNA levels fluctuated only slightly at 12 and 24 h after RBR downregulation but remained at constant levels 5 d after germination on β-estradiol (n = 50 for each time point/treatment/analyzed gene expression). Consistent with the decreased organ production we observed in RBRi seedlings after β-estradiol treatment (Figure 4), ANT expression levels were downregulated 5 d after RBRi induction. Five days after germination on β-estradiol, CYCB1;1 was upregulated in the SAM, as we observed in leaves (see Figure 5). BARD1 (reported to limit WUS expression; Han et al., 2008), is strongly upregulated 5 d after induction. The error bars represent the se from two biological replicas and two technical replicas.
Bars = 10 μm in (A) to (H), 2.5 μm in the (D) inset, 20 μm in (I) to (V), and 2.5 mm in (V').
To understand the loss of ordered cell divisions in the RBRi SAM, we crossed a line expressing a reporter gene for stem cell identity, ProCLV3:GUS (Brand et al., 2000), with our RBRi lines. Three days after germination in the presence or absence of β-estradiol, the GUS signal observed in induced and uninduced RBRi; ProCLV3:GUS lines was comparable (Figures 3I and 3J). Four days after germination on β-estradiol, however, GUS expression in the L2 and L3 layers of the central zone was significantly reduced (Figures 3K and 3L) and had nearly ceased after 5 d (Figures 3M and 3N). Interestingly, the ProCLV3:GUS signal was still detectable in RBRi plants 8 d after continuous β-estradiol treatment but was confined to the L1 layer, which showed the least disturbance in cell organization (Figures 3O and 3P). Similar to observations of deletion of SAM sections by laser ablation (Reinhardt et al., 2003), the disorganized RBRi SAM can restore the CLV3 expression domain and leaf initiation after withdrawal of β-estradiol. Five days after recovery, the ProCLV3:GUS domain was reestablished in most of the RBRi SAMs (Figures 3Q to 3T). In ∼30% of the cases (n = 60), a split stem cell domain developed (Figure 3R), which produced double inflorescences (Figures 3U and 3V).
Analysis of CLV3 mRNA levels in 5-d-old RBRi seedlings treated with β-estradiol confirmed that CLV3 expression was reduced (Figure 3W). The reduction of CLV3 expression in the sub-L1 central zone may therefore explain the temporary increase of WUSCHEL (WUS) expression in 5-d-old treated seedlings (Figure 3W). Ten days after induction, WUS mRNA accumulation had returned to wild-type levels, although the CLV3 mRNA was still low. The results suggest that continuous RBR downregulation alters meristem organization to such an extent that stem cell maintenance is disrupted, although cell division is still supported, as revealed by the increased CYCB1;1 expression levels detected via quantitative PCR in shoot apices (Figure 3X). Interestingly, no significant changes were observed in the expression of WUS, SHOOTMERISTEMLESS (STM), and KNOTTED-LIKE1 (KNAT1) during the acute reduction phase of RBR (Figures 3W and 3X), suggesting that RBR does not directly regulate transcription of these genes. Rather, the loss of organized SAM cell division activity after downregulation of RBR expression might be responsible for the disruption of the CLV-WUS regulatory feedback loop. Alternatively, the detachment of L2 cells from the L1 layer may have disrupted cell–cell signaling that is critical for maintenance of the CLV-WUS regulatory feedback loop.
Disorganization of the SAM induced by RBR downregulation strongly affected organ primordia formation. In 5-d-old seedlings germinated on β-estradiol containing medium, the one or two detectable organ primordia were strongly delayed in development compared with the wild type (Figure 4). AINTEGUMENTA (ANT) was investigated for its known upregulation early during organ primordia initiation (Nole-Wilson and Krizek, 2006). Consistent with the decreased organ production we observed in RBRi seedlings after β-estradiol treatment, ANT expression was reduced 5 d after RBRi induction (Figure 3X). Together, our results suggest that RBR is required to maintain the SAM stem cell domain, most likely by organizing appropriate temporal and spatial cell divisions.
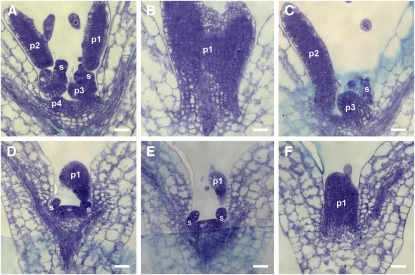
Figure 4.
Arrest of Organ Primordia Initiation in RBRi Seedlings Germinated on β-Estradiol.
(A) to (C) Five-day-old RBRi seedlings grown on β-estradiol–free medium. (B) and (C) are contiguous sections of (A). Four organ primordia were identified.
(D) to (F) Five-day-old RBRi seedlings grown on β-estradiol–containing medium. (E) and (F) are contiguous sections of (D). One organ primordium was identified.
p1 to p4, organ primordia; s, stipules. Bars = 20 μm.
RBR Regulates Meristemoid Proliferation and Subsequent Stomata Differentiation
Since RBR is thought to regulate the G1/S transition (Inze, 2005), we asked if the cell proliferation in RBRi leaves was similar to that observed in Arabidopsis leaves expressing the geminivirus RepA protein (Desvoyes et al., 2006) or with increased expression of E2Fa/DPa (De Veylder et al., 2002), both proteins or complexes of which interact with RBR (Xie et al., 1995). Two weeks after germination, epidermal cells in the proximal part of the second pair of leaves were still mitotically active, as indicated by ProCYCB1;1:CYCB1;1-GUS expression (Figures 5A and 5C). In the distal part of wild-type leaves, cell division had ceased (Figure 5C') and cells were expanding. When the 2-week-old RBRi plants had been treated with β-estradiol for 5 d, ProCYCB1;1:CYCB1;1-GUS expression was visible on the whole leaf blade in a punctuate pattern (Figures 5B, 5D, and 5D'). Thus, RBR loss of function can reactivate or continue mitotic activity in at least a restricted number of cells.
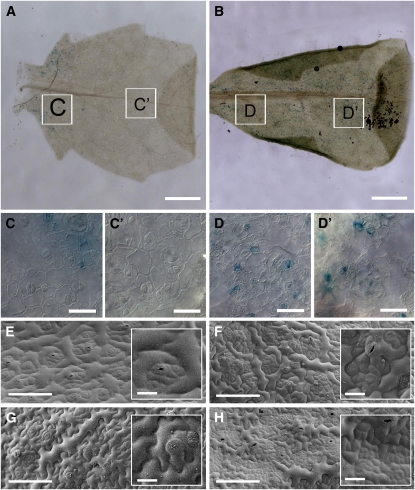
Figure 5.
Epidermal Pattern Variations in RBRi Leaves.
(A) to (D) Comparison of β-estradiol–treated CYCB1;1:GUS patterns in wild-type and RBRi leaves 2 weeks after germination. The GUS signal is visible on the proximal halves of the 2nd pair of both wild-type (C) and RBRi (D) leaves. Notice the absence of GUS signal in the distal half of wild-type leaves (C') but not in RBRi leaves (D').
(E) and (G) Scanning electron microscopy images of adaxial (E) and abaxial (G) sides of wild-type leaves from 15-d-old plants.
(F) and (H) Scanning electron microscopy pictures of adaxial (F) and abaxial (H) sides of RBRi leaves from 15-d-old plants sprayed with β-estradiol for five consecutive days.
Bars = 1 mm in (A) and (B), 20 μm in (C) to (D'), 100 μm in (E) to (H), and 20 μm in insets in (E) to (H).
We did not detect a significant increase in the number of mesophyll cells of RBRi leaves number 3 and 4 after 5 d of β-estradiol treatment (average number of mesophyll cells on proximal, middle, and distal leaf sides was 82 ± 25 in the wild type and 87 ± 33 in induced RBRi plants on a total analyzed area of 975,874 μm2), but we observed strong cell overproliferation in restricted areas on both adaxial and abaxial sides of the proximal leaf region. This increased but spatially restricted cell division activity resulted in large islands of small, brick-like cells (cf. Figures 5E and 5G to 5F and 5H). Increased cell proliferation, albeit to a lesser extent, occurred also in the middle-distal regions of RBRi leaves (see Supplemental Figure 8 online).
Since we observed a higher number of proliferating cell patches on the abaxial leaf side where more stomates are formed (Ferris et al., 2002), we asked if the overproliferating cells originated from meristemoid mother cells (MMCs) that give rise to the stomate cell lineage. To address this question, we crossed RBRi plants with the ProTOO-MANY-MOUTHS (ProTMM):TMM-GFP (green fluorescent protein) reporter construct, which is expressed throughout the stomata lineage. The first asymmetric cell division of MMCs gives rise to a small meristemoid and a larger neighboring cell (Nadeau and Sack, 2002). The meristemoid maintains stem cell–like activity and undergoes additional asymmetric cell divisions, which after each round produces a meristemoid and a large neighbor cell. The meristemoid eventually differentiates into a guard mother cell (GMC), which undergoes a single symmetrical cell division to generate a pair of guard cells (Nadeau and Sack, 2002).
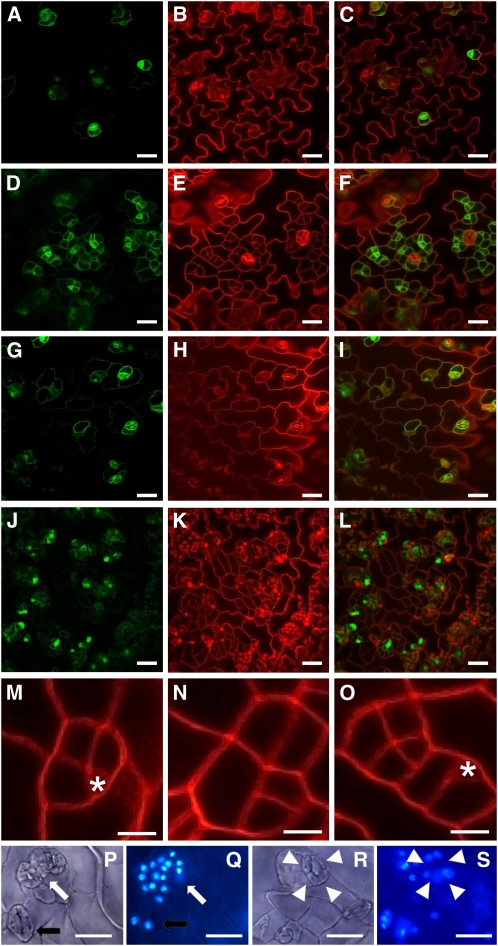
When ProTMM:TMM-GFP seedlings were treated with β-estradiol, GFP activity was detected only in guards cells and a neighboring cell as expected (Figures 6A to 6C). Three days after induction of 10-d-old RBRi seedlings, however, the overproliferating brick-like cells were all positive for TMM, which confirmed that they were derived from MMCs (Figures 6D to 6F). Two weeks after withdrawal of β-estradiol treatment, these overproliferated cells had expanded slightly, but retained their isomorphic shape (Figures 6G to 6L). Moreover, MMC divisions were mostly symmetric (Figures 6M to 6O), and the resulting cells had lost their potential to differentiate into mature stomata. These results suggest that RBR is required early to regulate the polarity of dividing MMCs, which is a prerequisite for patterning of the stomate lineage and epidermal cells (Geisler et al., 2000; Carlsbecker and Helariutta, 2005). Interestingly, genes required early during stomate development, such as STOMATAL DENSITY AND DISTRIBUTION1 (SDD1) (Von Groll et al., 2002) and TMM itself, were upregulated in leaves of β-estradiol–treated RBRi seedlings (see Supplemental Figure 9A online). Similarly, SPEECHLESS (SPCH), which was recently identified as a target of the MAPK cascade that modulates cell proliferation regulation (Lampard et al., 2008), was also upregulated. By contrast, genes required later during stomate differentiation, such as FAMA and MUTE (Bergmann and Sack, 2007), were not affected in their expression levels (see Supplemental Figures 9A to 9G online). RBR may also be involved in the regulation of the final cell division during stomate development, because stomates in leaves of β-estradiol–treated seedlings often contained four guard cells instead of two (Figures 6P to 6S).
Figure 6.
Confocal Laser Scanning Microscopy of Leaf Surfaces of Wild-Type and RBRi Plants.
Green channel, ProTMM:TMM-GFP; red channel, propidium iodide.
(A) to (C) In 13-d-old wild-type leaves, ProTMM:TMM-GFP expression was confined to stomates and one or two nearby pavement cells.
(D) to (F) In 13-d-old RBRi leaves from plants sprayed with β-estradiol for three consecutive days, all small proliferating cells expressed ProTMM:TMM-GFP.
(G) to (I) Wild-type leaf surface 2 weeks after β-estradiol treatment was discontinued.
(J) to (L) Two weeks after β-estradiol treatment was discontinued in RBRi plants, the small proliferating cells had not differentiated into stomata. The cells remained smaller and appeared to be less differentiated compared with the typical puzzle-shaped epidermal cells of wild-type or uninduced plants.
(M) to (O) In the wild type (M), asymmetric cell divisions of MMCs gave rise to epidermal spacer cells and founders of stomata (asterisks). In RBRi plants treated with β-estradiol ([N] and [O]), cell division was symmetric and disorganized.
(P) to (S) Differential interference contrast microscope ([P] and [R]) and 4',6-diamidino-2-phenylindole (DAPI) fluorescence ([Q] and [S]) images of RBRi leaves from 15-d-old plants treated with β-estradiol for five consecutive days. Clusters ([P] and [Q], white arrow) or single ([R] and [S], white arrowheads) four-celled stomates were observed in addition to wild-type-like two-celled stomates (black arrows).
Bars = 30 μm in (A) to (L), 20 μm in (M) to (O), and 20 μm in (P) to (S).
Reducing RBR Function Disrupts Procambial Cell File Maintenance
Based on the loss of RBR function on MMC proliferation and differentiation, we asked if other meristematic (or stem) cells that are required for tissue formation were similarly affected. Divisions of procambial cells in the vascular bundle produce xylem and phloem cells (Carlsbecker and Helariutta, 2005), and the procambial cells in leaves of 3-week-old RBRi seedlings sprayed with β-estradiol for five consecutive days had also strongly proliferated (Figures 7A to 7H'). The procambium typically consists of two or three organized cell files, which in leaves of β-estradiol–treated RBRi seedlings had greatly increased in number and become disorganized (Figures 7A to 7D). We sectioned RBRi and wild-type inflorescence stems from 5-week-old plants, induced for 9 d, between the first and the second internode. A strong proliferation of fascicular procambial cells was detected in induced plants only (Figures 7E and 7H'). Thus, reducing RBR function in developing organs or tissues is context dependent and appears to affect most strongly meristematic (or stem) cells that require RBR for regulation of proliferation and subsequent differentiation.
Figure 7.
Context-Dependent Overproliferation or Arrest of Meristematic Cells in RBRi Plants.
(A) to (H), (M), and (N) Transversal sections of resin-embedded tissues stained with Toluidine blue.
(A) and (B) Vascular bundles from the 4th leaf of 3-week-old RBRi plants treated with β-estradiol for 5 d (B) appear more convex than those from the wild type (A).
(C) and (D) Comparison of wild-type (C) and RBRi (D) vasculature shows that the procambium in RBRi plants treated with β-estradiol developed five to six cell layers (asterisks) instead of the two to three in wild-type plants.
(E) and (F) Stem lateral sections of 5-week-old wild-type (E) and RBRi plants treated with β-estradiol for nine consecutive days (F).
(G) and (H') A higher magnification of the vasculature meristem showed that the fascicular procambium (fc and highlighted in orange in [G'] and [H']) between the fascicular phloem (fp) and the fascicular xylem (fx) is thicker in RBRi plants treated with β-estradiol ([H] and [H']) compared with the wild type ([G] and [G']).
(I) and (J) RBRi induction impairs inflorescence elongation and affects cauline leaves. Three-week-old plants were sprayed with β-estradiol for five consecutive days. Wild-type plants (left) are taller and cauline laves are not bent downwards when compared with RBRi plants (right).
(K) Inflorescence meristem activity was arrested when 3-week-old flowering plants were subjected to 2 weeks of consecutive β-estradiol treatment. Wild-type plants (left) still produced flowers and lateral branches, whereas RBRi plants did not (right).
(L) and (M) Wild-type apex (asterisk) with two emerging lateral buds compared with arrested RBRi inflorescences from 5-week-old plants, treated with β-estradiol for fifteen consecutive days when 3 weeks old. Meristem organization was lost, and organ production was arrested.
(N) and (O) Top view of wild-type (left) and RBRi (right) inflorescence apices treated as in (F). The last organ produced by the RBRi inflorescence may result in a pin-like structure.
(P) and (Q) Thirty-day-old wild-type and RBRi plants sprayed with β-estradiol for two consecutive weeks. RBRi induction in flowering plants induces sterility.
(R) and (T) Five-week-old wild-type Arabidopsis inflorescences and flower, respectively.
(S) and (U) Five-week-old RBRi inflorescence, 10 d after recovery from a 5-d treatment with β-estradiol. Phyllotaxis is disturbed (asterisks), as multiple siliques may emerge from the same node, and internode length is altered. In 5% of the cases, flowers with increased numbers of petals and stamens (U) were observed.
Bars = 100 μm in (A) and (B), 20 μm in (C) and (D), 0.5 mm in (E) and (F), 100 μm (G) to (H'), and 100 μm in (L) and (M).
RBR Is Required for Inflorescence and Floral Meristem Maintenance
To determine if RBR is required for meristem maintenance and organ production at later stages of development, we induced RBRi expression in 3-week-old plants at the time of bolting. Five days after continuous RBRi expression, stem elongation was reduced and, similarly to RBRi rosette leaves, cauline leaves became strongly curled (Figures 7I and 7J). After an additional 10 d of continuous RBRi induction, the inflorescence stem had stopped growing (Figure 7K). Sections of the terminated inflorescence apex revealed a complete loss of the meristem structure (Figures 7L and 7M). Instead of an active dome-shaped meristem containing cytoplasmic dense cells, the mutant inflorescence apices had vacuolated cells. The last organ produced by the inflorescence meristem might not fully develop and terminated in a pin-like structure (Figures 7N and 7O). Fifty percent of the ovules produced by RBR loss-of-function mutants were aborted because RBR is required during gametophyte development (Ebel et al., 2004). We tested if RBR downregulation induced early during inflorescence development would also affect plant fertility. RBRi plants were sprayed with β-estradiol for two consecutive weeks beginning shortly before bolting (18 d after germination). In 30% of the plants (n = 12), no inflorescence stem elongated from the mutant basal rosette. Bolting plants remained stunted, lateral inflorescences initiated but arrested, and 70% of the siliques (6.8 per plant on average, n = 12) did not elongate further than 0.5 cm (Figures 7P and 7Q; see Supplemental Figure 10 online). These short siliques contained 100% aborted ovules (see Supplemental Figure 10D online). By contrast, in β-estradiol–treated wild-type plants (n = 12), only 16% of the siliques of the main inflorescence were shorter than 0.5 cm and 62% were longer than 1 cm (on a total number of 18.8 per plant), containing on average 48 seeds per silique.
When RBRi plants were continuously treated with β-estradiol for only 5 d after bolting followed by 10 d of recovery, the inflorescence meristem did not arrest organ production but showed phyllotactic alterations and occasionally flowers with supernumerary organs (Figures 7R to 7U). Internodal spacing varied (average = 0.6 cm) and ranged from 0 cm (i.e., changing from spiral to decussate floral pattern) to 1.1 cm, which is the average length in wild-type plants. Flowers with supernumerary petals and stamen were observed in 5% of the analyzed plants (n = 40). Together, these results suggest that RBR function is necessary during inflorescence and flower development to organize appropriate meristem cell divisions required for phyllotactic patterning and floral organ initiation.
Reducing RBR Function Does Not Accelerate Development-Dependent DNA Endoreduplication
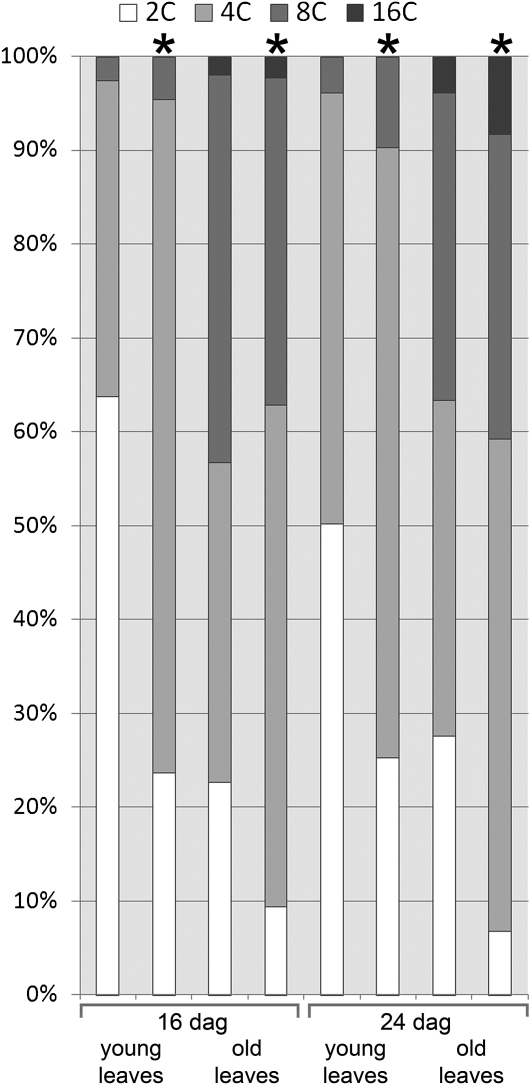
If RBR regulates only the G1–S transition, then loss of RBR function could be expected to deregulate cell division activity, but not affect DNA endoreduplication and nuclear ploidy. Alternatively, RBR may also regulate DNA endoreduplication, as was previously reported for Arabidopsis lines expressing the RepA protein, which binds RBR (Desvoyes et al., 2006). Strongly increased DNA ploidy levels in Arabidopsis lines ectopically expressing E2Fa and DPa are consistent with this possibility (Desvoyes et al., 2006). To investigate the effect of loss of RBR function on DNA ploidy, we treated 12-d-old RBRi plants with β-estradiol for 4 d and 16-d-old RBRi plants for 8 d. Mature leaves numbers 3 and 4 and young leaves numbers 5 and 6 (16-d-old plants) or 7 and 8 (24-d-old plants) from RBRi and control plants were analyzed for nuclear DNA ploidy. The results in Figure 8 and Supplemental Figure 11 online show that reducing RBR function did not cause a significant increase in leaf nuclear DNA ploidy. Both young and older β-estradiol–treated RBRi leaves had a higher number of 4C nuclei and only a small increase in the number of 8C in comparison to β-estradiol–treated wild-type plants. We did not observe nuclei with more than 16C DNA content or nuclei with intermediate DNA content. Thus, interference with RBR function by viral proteins (Desvoyes et al., 2006), and direct reduction of RBR protein levels has different effects on endoreduplication.
Figure 8.
Nuclear DNA Ploidy Analysis.
Leaf numbers 3 and 4 (old) and 5 to 8 (young) were collected from 16- and 24-d-old plants, respectively. The DNA content of purified nuclei was analyzed by flow cytometry. Asterisk indicates DNA content of nuclei from RBRi plants treated with β-estradiol. No asterisk indicates the DNA content from uninduced RBRi plants. dag, days after germination.
Transcriptome Analysis of Disrupting RBR Homeostasis Reveals a Stress Response and Perturbation of Cell Wall Biosynthesis
The rapid disappearance of RBR in RBRi lines after β-estradiol treatment (Figure 1B) suggested that the protein has a short half-life. This allowed us to investigate acute transcriptional responses to the disturbance of RBR homeostasis. We compared the gene expression profiles of the first two expanding leaves of 10-d-old RBRi and Pro35S:XVE plants at 3, 6, 12, and 24 h after induction (HAI) after β-estradiol treatment (see Supplemental Data Set 1 and Supplemental Tables 1 and 2 online). Many early upregulated genes were associated with different types of stress responses (i.e., common stress cluster 12; Ma and Bohnert, 2007; Ma et al., 2007) and particularly included transcription factors involved in the ethylene response, like ETHYLEN RESPONSE FACTOR2 (ERF2) and ERF5, and general stress responses, like RESPONSIVE TO DESICCATION26 (RD26) (Fujita et al., 2004), MYB family transcription factor (TF), SALT TOLERANCE ZINC FINGER (STZ/ZAT10) (Mittler et al., 2006), WRKY33 (Zheng et al., 2006), WRKY40 (Xu et al., 2006), and genes found in the ATTED (Obayashi et al., 2009) coexpression clusters of these factors (see Supplemental Figures 12 and 13 and Supplemental Table 3 online). Thus, it appears that early perturbation of RBR homeostasis is perceived by the leaf as a stress signal, which triggers the upregulation of stress response gene expression. Interestingly, we found that of 574 genes that were upregulated in plants overexpressing E2Fa and DPa (Naouar et al., 2009) in two different experiments, 11 and 16 were also upregulated 3 and 6 h, respectively, after induction in RBRi plants. This includes STZ/ZAT10, as well as other stress-regulated genes, a number of Zn-finger proteins, and biotic stress–related genes. These results, although having only a small significance (hypergeometrical distribution P values of 2.2E-03 and 5.6E-03, respectively), might suggest that at least part of the early response of stress response genes to perturbation of RBR homeostasis involves the RBR/E2F/DP regulatory pathway.
Another group of genes that was rapidly downregulated after β-estradiol treatment of RBRi plants includes members of the ARABINOGALACTAN protein family (AGP12, AGP17, and AGP18) (see Supplemental Figure 14 online). Genes involved in cell wall biogenesis were also deregulated in plants overexpressing E2Fa or E2Fa/DPa (Vlieghe et al., 2003; De Jager et al., 2009). Although the precise functions of AGP proteins are unknown, they appear to be necessary for cell adhesion and signal transduction processes (Gao and Showalter, 1999). The downregulation of AGP genes may explain the loss of cell adhesion between the L1 and L2 layers of the meristem in RBRi seedlings (Figures 3E and 3F).
Expression of cyclins (CYCA3;1 and CYCA3;2) and cyclin-dependent kinases (CDKB1;2 and CDKB2;1) began to increase after RBR downregulation. CYCA3;1/2 mRNAs peak in S phase, while CDKB1;2 mRNA appears in early G2 phase, and CDKB2;1 is a mitosis-specific gene (Menges et al., 2005). Upregulation of G2/M-specific genes reflects the stomata lineage proliferation we described in young leaves of RBRi-induced plants (Figures 5E to 5H). Additionally, several genes encoding proteins required for DNA replication-associated chromatin assembly, many of which contain E2F binding sites (Ramirez-Parra and Gutierrez, 2007), were upregulated 12 and 24 HAI (see Supplemental Figure 15 online). The upregulation of these genes is consistent with the increased cell proliferation due to RBR downregulation.
To identify potential cis-acting promoter motives in the set of deregulated genes, we analyzed the occurrence of 96 TF binding sites from the AGRIS website (http://Arabidopsis.med.ohio-state.edu/AtcisDB/bindingsites.html). Of the more specific TF sites, E2F binding sites are overrepresented in the group of genes upregulated 12 HAI. A variety of motifs have been used in analyses of E2F binding sites (Ishida et al., 2001; Kel et al., 2001; Stanelle et al., 2002; Vandepoele et al., 2005). We found that the stringently defined site TTTCCCGCC within the 5′ untranslated region and/or the first 500 bp of the promoter was the best predictor for involvement of RBR in gene-specific regulation. Sixteen (11.3%) and 27 (19.1%) of the 141 genes in this category are upregulated 12 and 24 HAI, respectively, whereas none are upregulated earlier, and only 4 (2.8%) were downregulated at a single time point. The only other motif overrepresented in upregulated genes in RBRi was the octamer motif CGCGGATC: 17 of 106 genes with this motif are upregulated at least at one time point, mostly at 12 and 24 HAI. Seven of the 10 histone genes containing this motif are upregulated above the twofold threshold.
DISCUSSION
The construction of an inducible RBRi system allowed us to reduce RBR levels rapidly and transiently, in the absence of confounding effects that may be associated with downregulation of RBR expression by VIGS (Park et al., 2005; Jordan et al., 2007) or expression of plant virus proteins that interact with RBR (Desvoyes et al., 2006; Lageix et al., 2007). Arabidopsis can recover from transient downregulation of RBR by reconstituting meristems and resuming growth (Figure 1), although certain cell populations that developed during the loss of RBR function may have been irreversibly changed in their differentiation status, causing transient alterations in morphology and anatomy (Figures 4 and 5). Prolonged downregulation of RBR resulted in arrest of plant growth, however, suggesting that RBR homeostasis ultimately is critical for meristem maintenance and sustained cell division. Earlier studies using targeted RBR-RNAi and Cre recombinase-mediated excision of an RBRlox allele indicated already that RBR is required to maintain the balance between stem cells and differentiating cells during early root development (Wildwater et al., 2005), although these experiments did not investigate very early or later effects of loss of RBR function. Invariably, all experiments of downregulating RBR during postembryonic plant development reported so far did not result in developmental growth arrest or abortion of organ production, as we observed in our experiments.
RBR Homeostasis Is Critical for Meristem Integrity and Function
Postembryonic organ development originates from totipotent stem cells that are maintained by continuous division to produce cells committed to differentiation into specific cell types. Prior to reaching their terminal differentiated state, these cells may undergo several more rounds of cell divisions before they withdraw from the cell cycle. Temporal and spatial regulation of cell division is therefore critical for coordinated organ development, and the underlying regulatory mechanisms are beginning to emerge. Retinoblastoma(-like) proteins have been implicated in regulatory decisions, most prominently in the regulation of the G1/S transition via E2F/DP transcription factors, which regulate the expression of cell cycle genes (De Veylder et al., 2007; Gruissem, 2007; van den Heuvel and Dyson, 2008), and in loading of DNA replication complexes (Bosco et al., 2001; Mukherjee et al., 2009). RB/E2F complexes have also been found to be associated with developmentally regulated promoters in Drosophila melanogaster, mouse, and Caenorhabditis elegans, often in combination with other transcription factors and chromatin modifiers (Korenjak and Brehm, 2005; Buttitta and Edgar, 2007; van den Heuvel and Dyson, 2008). In addition, RB has been implicated in E2F-independent regulatory mechanisms, including direct interaction with subunits of the anaphase-promoting complex during cell cycle exit (Binne et al., 2007).
Downregulation of RBR by induced RNAi expression resulted in arrest of leaf primordia formation and further shoot development. The observed defects in leaf morphology after RNAi induction suggest that leaf primordia that were initiated prior or during loss of RBR function could still continue to grow for a restricted period, although normal development was disrupted. Earlier reports have shown that RBR homeostasis is critical for regulation of cell differentiation in Arabidopsis root and tobacco (Nicotiana tabacum) shoot meristems, although the underlying mechanism was not well understood (Wildwater et al., 2005; Wyrzykowska et al., 2006). The disorganization of cells soon after RBR downregulation (Figure 3) suggests that RBR is required to maintain the typical anticlinal and periclinal division pattern in the L2 and L3 cell layers. Strikingly, loss of RBR function weakens cell–cell contacts between L1 and L2 cell layers, which may disrupt the temporal and spatial regulation of symplastic SAM domains and trafficking of molecules that regulate cell division pattern in L2 and L3 (Gisel et al., 1999). Cell ablation studies have shown that the L1 layer maintains meristem integrity, suggesting that L1 regulates stemness of the central SAM zone. L1 also integrates spatial auxin transport and signaling, which is required for primordia formation (Reinhardt et al., 2003). Weakening of L1-L2 cell–cell contact is consistent with the upregulation of PECTASE LYASE FAMILY PROTEIN2 (PLA2), which is involved in cell separation during lateral root emergence (Laskowski et al., 2006) and contains an E2F binding site in the promoter. One additional pectin esterase gene containing a canonical E2F site (TTTCCCGC) in its regulatory region showed a positive expression trend after RBRi induction. Eleven of 27 genes encoding arabinogalactan proteins (AGP) also became deregulated after downregulation of RBR, several of them shortly after RNAi induction. AGPs are necessary for cell adhesion and signal transduction processes (Gao and Showalter, 1999) and might contribute to cell–cell signaling between the SAM layers. In situ expression experiments of the deregulated genes will be required to better understand the role of L1-L2 signaling in SAM organization and function.
The weakening of L1-L2 cell–cell interactions could explain the change in CLV3 expression, which becomes confined to L1, and the subsequent transient increase in WUS expression (Figure 3W). At present, our results cannot distinguish if downregulation of RBR or the resulting loss of cell–cell contact between L1 and L2 disrupts the CLV-WUS feedback loop, which maintains the SAM stem cell niche that contributes to meristem activity and primordia formation (Brand et al., 2000, 2002; Rojo et al., 2002; Tucker and Laux, 2007). Loss of CLV3 activity leads to gradual conversion of bordering cells in the SAM peripheral zone to cells with central zone characteristics (Reddy and Meyerowitz, 2005), suggesting that CLV3 is involved in a signaling pathway required for the switch from stemness to differentiated cells. Similarly, RBR downregulation in the root meristem results in the expansion of the stem cell domain (Wildwater et al., 2005). However, loss of CLV3 function alone does not result in a disorganized cell patterning or loss of primordia initiation (Brand et al., 2000), indicating that RBR homeostasis is required for correct spatial and temporal maintenance of stem cell division or entry into the differentiation pathway leading to primordia formation. Recent microdissections of shoot meristem functional domains have identified genes that are preferentially expressed in the stem cell niche and leaf primordia (Brooks et al., 2009; Yadav et al., 2009). Many of these genes encode proteins known to interact with RB in animals (Morris and Dyson, 2001), including D-type cyclins and proteins involved in chromatin remodeling and DNA repair. Further experiments will be necessary to establish which of these interactions, when disrupted by progressive reduction of RBR levels, triggers unscheduled cell divisions and loss of meristem organization.
Interestingly, CLV3 expression and meristem function could be restored if RBR was allowed to accumulate again by withdrawing β-estradiol. It is possible that CLV3 expression in L1, which persists during RBR downregulation, is sufficient to reestablish the meristem by resetting the CLV/WUS feedback loop, which is known to be robust and flexible (Muller et al., 2006). About 30% of the arrested meristems recovered a twin stem cell domain, similar to meristems in which the central zone was ablated (Reinhardt et al., 2003). Modeling has shown that a L1-derived signal (e.g., CLV3) together with a central WUS-inducing signal may be sufficient to generate a layered meristem structure and to establish a twin stem cell domain after inactivation of the SAM central region (Jonsson et al., 2005). Together, our results show that loss of meristem signaling or cell division polarity caused by transient RBR reduction is reversible and therefore directly linked to RBR expression. When low RBR levels persisted, however, meristem activity could not recover and WUS expression was reduced again to normal levels, although CLV3 expression remained low (Figure 3). Thus, WUS expression might be repressed by a CLV-independent mechanism, such as BREAST CANCER ASSOCIATED RING1 (BARD1) (Han et al., 2008), which is strongly upregulated in induced RBRi seedlings (Figure 3). A detailed temporal and spatial analysis of WUS and other genes involved in SAM organization will be required to better understand their functions during RBR downregulation.
RBR Promotes Entry into Cell Differentiation
The function of RBR is not restricted to shoot and root apical meristems but also extends to stem cell–like populations in leaves and stems. Earlier attempts of reducing RBR function using VIGS or by expression of plant virus proteins that interact with RBR produced plants with overproliferating cells in leaves (Park et al., 2005; Desvoyes et al., 2006; Lageix et al., 2007), although the origins of these cell populations were not known. Overproliferating cells that we observed in leaves of RBRi plants were restricted to the leaf epidermis and expressed the TMM marker (Figures 5 and 6), suggesting that they originate from the stomate lineage. Stomate development involves a number of asymmetric cell divisions, beginning with the protodermal cell and proceeding through a MMC to the meristemoid, which may divide asymmetrically several times. Each division produces a new meristemoid and a stomate lineage ground cell. Finally the meristemoid differentiates into a GMC, which divides once to form the two guard cells (Bergmann and Sack, 2007). The cell divisions in Arabidopsis stomate development are at least partially regulated by epidermal patterning factors (EPFs), which interact with membrane receptors and associated kinases, such as TMM and ERECTA (ER) (-like) (Serna, 2009a). EPF2 is expressed early in the lineage and regulates the number of cells entering the stomata lineage (Hara et al., 2009; Hunt and Gray, 2009), most likely in concert with the basic-helix-loop-helix (bHLH) transcription factor SPCH (Lampard et al., 2008). The bHLH protein MUTE terminates the division program in late meristemoids and triggers differentiation into GMCs, which requires FAMA for the final division into mature guard cells (Serna, 2007). EPF2, TMM, SPCH, and the Ser-type endopeptidase SDD1 were upregulated in RBRi plants, while MUTE and FAMA were not (see Supplemental Figure 9 online), indicating that RBR is required for exit from the meristemoid division program. Since MUTE expression marks the transition of the stem cell–like meristemoids to guard cell morphogenesis (Pillitteri et al., 2007), we suggest that the proliferating TMM-expressing cells in the RBRi leaf epidermis maintain their stem cell character. Unlike cells in the SAM, however, these cells are unable to enter the stomatal differentiation program after reestablishing RBR levels, but either expand or remain small and triangular with high TMM expression. At present, we cannot distinguish if loss of RBR function increases the number of cells entering the stomatal lineage or results in proliferation of protodermal cells or MMCs, which are not yet committed to guard cell morphogenesis. The failure to undergo asymmetric divisions alone is unlikely the cause for this lack of commitment because cells in the breaking of asymmetry in the stomatal lineage1 mutant that are produced by incorrect symmetric divisions express MUTE and can form normal stomates (Dong et al., 2009). It is possible, therefore, that the TMM-expressing cells in the RBRi leaf represent protodermal or postprotodermal cells, in which restrictive signaling through TMM/ER/EPF2 cannot arrest cell cycle activity in the absence of RBR. During muscle cell differentiation, pRB promotes cell cycle withdrawal together with MyoD, a bHLH transcription factor. MyoD and pRb together also maintain durable cell cycle arrest by modulating cyclin-dependent transcription and affect transcriptional differentiation programs (De Falco et al., 2006; Serna, 2009b). It will be interesting to examine if SPCH plays a similar role in guard cell morphogenesis.
RBR and Cell Cycle Regulation
Depending on the developmental stage, RBR downregulation can at least transiently increase cell division activity in subpopulations of plant cells. Cells that had not exited the cell cycle when RBR was reduced continued to divide and lost their developmental constraints. In addition to cells in the SAM and leaf epidermis, procambial cells also proliferated (Figures 7A to 7H'), while we did not observe additional divisions of differentiated leaf mesophyll or root cells. These results suggest that at least two cell cycle regulation states exist in plants, in which RBR downregulation either induces continuous cycling and cytokinesis or cell cycle arrest. Most leaf cells have a 4C DNA content after RBR downregulation. Since in a population of rapidly dividing cells, ∼65% of the cells have a 2C content and only 35% have a 4C DNA content (Hirano et al., 2008), it appears unlikely that the observed high number of cells with a 4C DNA content and the low number of cells with a 2C content are due to a larger number of dividing cells in the RBR-downregulated leaves. Alternatively, nondividing cells may also be induced to bypass the G1/S cell cycle check point. However, the strongly increased number of leaf cells with a 4C DNA content in the absence of RBR suggests that most cells, either from the pool of cycling cells or from cells bypassing G1/S, were subsequently arrested or at least significantly delayed in G2/M (Figure 8).
In plant cells, the block of M phase progression is usually associated with DNA endoreduplication. The inactivation of RBR function by viral proteins or increased E2F/DP expression results in strongly increased DNA endoreduplication (Desvoyes et al., 2006; Jordan et al., 2007), while RBR downregulation by VIGS in tobacco leaves (Park et al., 2005), RNAi in Arabidopsis cell cultures (Hirano et al., 2008), and RNAi in Arabidopsis plants resulted in a similar increase in cells with a 4C DNA content. It is therefore possible that binding of plant viral proteins interferes with a function of RBR in the development-dependent regulation of DNA endoreduplication. This would explain why cells do not enter the DNA endoreduplication pathway when RBR function is strongly reduced or lost after VIGS- or RNAi-mediated downregulation of RBR expression.
In analogy to the better understood interactions between animal viruses and pRB (Felsani et al., 2006), it is likely that expression of plant viral proteins, which can interact with RBR (Kong and Hanley-Bowdoin, 2002), may interfere with RBR functions specific for cell cycle regulation or regulation of DNA replication. On the other hand, E2F/DP overexpression was associated with a strong increase in RBR expression (De Veylder et al., 2002) and might lead to enhanced E2F/DP-independent RBR functions.
While the established E2F/DP/RBR checkpoint supports the regulatory role of RBR at the G1/S transition, it does not explain why reduction of RBR levels results in the observed G2/M delay or arrest. We note that besides genes involved in DNA replication and cell cycle progression, WEE1 kinase is also upregulated soon after RBR downregulation. WEE1 kinase is required (but not sufficient) for inhibiting the cell cycle after activation of DNA integrity checkpoints (De Schutter et al., 2007).
Animal cells with a disturbed cell cycle due to pRb downregulation, unscheduled oncogene activity or expression of E2F factors have been shown to suffer DNA damage and to arrest in a senescence state due to DNA damage response check points (Bartkova et al., 2006; Di Micco et al., 2006; Pickering and Kowalik, 2006; Tort et al., 2006). It remains to be investigated whether RBR loss of function in plants leads to DNA damage or only signals to a DNA damage response.
Gene Expression Regulation by RBR
Retinoblastoma proteins function as transcriptional repressors in association with E2F transcription factors. Arabidopsis E2F target genes have been identified by the presence of a conserved E2F DNA sequence binding motif (Vandepoele et al., 2005; Naouar et al., 2009). In addition, pRB proteins are associated with several chromatin modifying complexes whose targets are less well defined (Buttitta and Edgar, 2007; van den Heuvel and Dyson, 2008) but include genes involved in developmental regulation (Korenjak and Brehm, 2005). The immediate consequences of acute downregulation of RBR at the transcriptional level, however, are not well understood. We found that typical E2F target genes were already upregulated 12 h after β-estradiol treatment (see Supplemental Data Set 1 and Supplemental Table 3 online), when RBR was reduced to <50% of levels prior to induction. Gene induction by relatively modest changes in RBR levels suggests that the RBR protein pool is not excessive but finely tuned with the needs of gene expression regulation, thereby allowing a fast and sensitive cellular response to regulation of RBR activity. The stoichiometry of RBR and E2F is likely established by E2F regulation of RBR transcription because the RBR promoter contains E2F binding consensus sequences, and RBR mRNA levels are increased in plants overexpressing E2F and DP (De Veylder et al., 2002; Naouar et al., 2009).
E2F binding sites of the type TTTCCCGCC (http://Arabidopsis.med.ohio-state.edu/AtcisDB/bindingsites.html) are present within 500 bp upstream of the start codon in ∼8% of the genes upregulated after β-estradiol treatment. The octamer CGCGGATC is another overrepresented motif in this group of upregulated genes. The motif is present in some genes that also contain classical E2F sites but also in a group of histone genes. Expression of histone genes needs to be coordinated with DNA replication, but how this is achieved in plants remains unknown.
Before the onset of E2F target gene transcription, RBR levels are reduced only slightly. Nevertheless, at 3 and 6 HAI, already several hundred genes become deregulated, including many transcription factor genes. For a large proportion of the upregulated genes, the response is transient and expression levels normalize or even decrease below wild-type levels at 24 HAI. Prominent among the induced genes are factors involved in the ethylene response, such as ERF2 and ERF5, in general stress responses, such as RD26 ((Fujita et al., 2004), ZAT10 (Mittler et al., 2006), WRKY33(Zheng et al., 2006), and WRKY40 (Xu et al., 2006), and in responses to reactive oxygen, different types of biotic and abiotic stresses and plant hormones. Furthermore, many of the genes in the ATTED coexpression clusters of these genes are coordinately upregulated (see Supplemental Figure 14 and Supplemental Table 3 online), suggesting that plants either experience stress by RBR downregulation or that a general stress signaling pathway is attenuated by RBR and becomes deregulated by small changes in RBR activity. Links between cell cycle responses and different stresses have been made for plants (Ma et al., 2009) and animal cells (Burhans and Heintz, 2009; Mavrogonatou and Kletsas, 2009), but the mechanisms underlying the crosstalk between cell cycle and stresses remain unknown.
Together, conditional downregulation of RBR expression during postembryonic plant development has revealed several new regulatory controls in which RBR function is required. Most central to these controls appears to be the coordination of cell cycle activity and differentiation in meristem tissues and pluripotent cell lineages that support organized plant development and growth. The phenotypic changes and transcriptional responses to RBR downregulation show that RBR in plants, similar to pRb in animals, occupies a critical node in a complex regulatory network. It will be interesting to establish how network components differ between animals and plants and which part of the plant network responds most acutely to changes in RBR homeostasis.
METHODS
Growth Conditions
Arabidopsis thaliana plants were grown on soil or 0.5× MS agar without sucrose under a 16-h-light/8-h-dark regime (long-day conditions) at 22°C.
DNA Manipulation, Plant Transformation, and RBRi Induction
Construction of Arabidopsis lines with conditional downregulation of RBR (RBRi). The RBR hairpin (RBRhp) was constructed by cloning the first five RBR exons and introns, plus a segment of the 6th exon, upstream of the corresponding inverted cDNA sequence using the native EcoRI site at position +1131. The 5′-end restriction site BglII and the 3′-end restriction site XhoI were used to subclone the RBRhp into the Gateway Entry vector pENTR2B. LR recombination was used to transfer the RBRhp into pMDC221, a Gateway-compatible, β-estradiol–inducible plant transformation vector developed by Mark Curtis and his team at the University of Zurich (Brand et al., 2006). The Pro35S:XVE driver vector was kindly provided by Mark Curtis (Brand et al., 2006). The RBR hairpin (RBRhp) DNA consisted of 1132 bp of the genomic sequence of the Arabidopsis RBR gene beginning from the start codon of RBR to the first EcoRI site plus, in antisense orientation, the corresponding region from an RBR cDNA clone (i.e., without the introns) as a 504-bp EcoRI-NcoI fragment. The two regions were assembled in a linker, providing a BglII site upstream (5′-AGATCTCTCTCTGTACAAGCTTCCATG-3′; BglII site underlined and RBR start codon in bold) and a SalI site downstream (5′-CCATGGGCCCCTTTTCGAACTGCGGATGCGACCAGTTGGCCATGTTTTTCCTCTAAGCTTGTACAGAGAGAGATCCCCAAGGGGTCGAC-3′;NcoI at end of hairpin and SalI sites underlined) of the cassette. These sites were used to excise the cassette and insert it into the compatible BamHI-XhoI sites of the Gateway-compatible pENTR-2B vector from Invitrogen. The Gateway LR Reaction kit (Invitrogen) was used to transfer the RBRhp cassette into the plant expression vector pMDC221 from Brand et al. (2006).
ProRBR:GUS was constructed by inserting a bidirectional reporter cassette between the HindIII-XbaI sites of the plasmid pK7WG2 (Karimi et al., 2002). The cassette consists of a 1152-bp-long fragment from a cloned region of the Arabidopsis genome spanning the intergenic region between the divergently transcribed genes AT3G12290 and RBR from start codon to start codon fused to the coding region of the GUS gene (followed by a WDV polyadenylation signal) for monitoring RBR promoter activity and to the coding region of the GFP gene (followed by a ACMV polyadenylation signal) for monitoring the eventual transcriptional activity in the opposite direction (corresponding to AT3G12290). The bidirectional promoter fragment was generated by PCR using Pwo polymerase, and PstI and KpnI sites were incorporated at the ends for directional fusion with the reporter genes. Sequences of the primers used for DNA cloning are listed in the Supplemental Methods online. The polyadenylation signals are described by Bieri et al. (2002), and all DNA fragments were assembled by standard restriction fragment ligation.
Sequences of the primers used for DNA cloning are listed in the Supplemental Methods online. The resulting constructs were transformed into Arabidopsis wild-type plants (accession Columbia) by a floral dip method (Clough and Bent, 1998). Transformants were selfed and the F2 progeny was screened for segregants with single transgene insertions and wild-type growth and development. Additional tests were performed to check for any side effects due to β-estradiol treatment or to induce overexpression of the XVE chimeric transcription factor. All of these tests had negative results. Homozygous Pro35S:XVE driver and RBRhp target lines were crossed and F3 progenies were screened on β-estradiol–containing MS plates to isolate inducible lines homozygous for the binary system. Six lines out of a total of 104 were found to be fully responsive to induction. Lines containing the complete system were named RBRi. A 5 μM β-estradiol solution in ethanol 70% (as suggested in Brand et al., 2006) was used in all inductions. To avoid potential problems with systemic β-estradiol transport, induction experiments with seedlings were performed by germination on 5 μM β-estradiol MS plates plus an additional direct supply of 5 μM β-estradiol solution to the examined plant tissues. β-Estradiol was poorly absorbed by siliques, as reported by Brand et al. (2006). Additional tests using Arabidopsis lines expressing a β-estradiol–inducible GUS construct confirmed that β-estradiol does not penetrate well into carpels. When flowering RBRi plants were induced, 0.005% of the surfactant Silwet was added to a 10 μm estradiol solution to improve tissue penetration of the inducer.
Expression Profiling
Seedlings of Arabidopsis (accession Columbia; lines Pro35S:XVE,RBRi) were grown on soil for 10 d without β-estradiol in growth chambers at 22°C under long-day photoperiods (16 h light/8 h darkness). After β-estradiol induction (time 0), the first pair of leaves was collected at 3, 6, 12, and 24 HAI and 5 DAI. RNA was extracted from leaves dissected from at least 40 seedlings. Three biological replicates were analyzed for time points 12 and 24 HAI. Two biological replicates were analyzed for time points 3 and 6 HAI.
Affymetrix Arabidopsis ATH1 GeneChips were used in the experiment (Affymetrix). Analysis was based on annotations compiled by The Arabidopsis Information Resource (http://www.Arabidopsis.org, version 2009-06-19). The arrays were scanned using an Affymetrix 3000 7G confocal scanner. Labeling of samples, hybridizations, and measurements was performed as described (Hennig et al., 2004).
Signal values were derived from the Affymetrix *.cel files using the GCRMA algorithm (Wu et al., 2004). All data processing was performed using the statistic language R (version 2.6.2) (R Development Core Team, 2009). Quality control was done using the affyQCReport package in R. Differentially expressed genes were identified using the limma package in R (Smyth, 2004). Multiple testing correction was done using the q-value method (Storey and Tibshirani, 2003). Due to relatively high variability between replicas, application of q-values below 0.1 as cutoff for significance led to exclusion of many genes that might be deregulated. To detect genes that might show a trend of gene expression changes already soon after RBR downregulation, and particularly to asses genes reported to be targets of RBR (e.g., E2F/DP-regulated genes), the signal log ratios (SLR) of all crosswise comparisons were determined between replicas (four for the two replicas of 3 and 6 h and nine for the three replicas of 12 and 24 h). The frequency of SLRs above 0.8 or below −0.8 was used as a measure of significance of differential expression and is indicated in Supplemental Data Set 1 online.
Data sets for comparison of expression data were derived from published analyses and websites (see descriptions and explanations in Supplemental Data Set 1 online in the Excel tab labeled “legend”) and converted to feature labels (tags) of the Arabidopsis genes present on the ATH1 GeneChip.
Antibody Production
For RBR antibody production, the N-terminal domain of RBR (encoding the first 374 amino acids) was cloned into the expression vector pQE31, which provides an N-terminal 6X histidine tag, and transformed into Escherichia coli M15Rep4 cells. Subsequently, primary transformants were screened for high expression of the protein with a colony blot procedure according to instructions from the Qiagen expressionist handbook (http://www1.qiagen.com/literature/handbooks/PDF/Protein/Expression/QXP_QIAexpressionist/1024473_QXPHB_0603.pdf). Induction and purification were done under native conditions as described in the Qiagen expressionist handbook. In short, a preculture was grown overnight in Luria-Bertani medium at 30°C in 200 mg/L ampicillin and 50 mg/L kanamycin. The next day, the culture was transferred and diluted 1:100 in SB (35 g tryptone, 20 g yeast, and 5 g NaCl/L) without antibiotics at 28°C until an OD of 0.4 was reached. Then the culture was shifted to 18°C, adapted for 1 h, and induced with 500 μM isopropyl β-d-1-thiogalactopyranoside. The cells were harvested (centrifugation for 15 min at 10,000 rpm with a SLA 3000 rotor) when they reached an OD of 1.5. For the subsequent steps, the cells were kept at 4°C. For cell lysis, the pellet was resuspended in 2.5 mL resuspension buffer (50 mM Na-phosphate, 300 mM NaCl, and 10 mM imidazole) per 100 mL culture and subsequently centrifuged at 12,000 rpm (SS34 rotor) for 5 min. The supernatant was discarded and the pellet resuspended in 2.5 mL lysis buffer (50 mM Na-phosphate, 300 mM NaCl, 10 mM imidazole, and 4 μg lysozyme) per 100 mL of cell culture. After an incubation time of 1 h, the extract was centrifuged at 20,000 rpm (SS34) for 1 h.
For antibody purification, 200 μL of Talon resin beads per 100 mL of culture were used. All the following steps were performed according to the Qiagen expressionist handbook with the following exceptions: instead of sodium phosphate buffer, we used 0.1 M MOPS buffer, pH 7.7, at 4°C for elution. Protein was dialyzed in MOPS buffer and used for immunization of rabbits (three times) or coupled to the affi-gel15 (Bio-Rad) resin according to manufacturer's protocol. Subsequently, 5 mL of immunized serum was diluted 10 times in TBS and passed over a column (N-terminal RBR coupled to 1 mL affi-gel15) five times. After washing, antibodies were eluted 10 times with 500 μL of 0.1 M glycine, pH 2.0, and collected in Eppendorf reaction tubes containing 42 μL 1 M Tris. Purity of antibodies was confirmed on dot and protein gel blots.
RNA and Protein Analysis
Total RNA was extracted from apices or leaves using TRIzol reagent (Invitrogen) following the manufacturer's instructions. The RNA was treated with 5 units of DNase I (Promega) for 30 min at 37°C, precipitated with 1/10 v/v of 3 M NaAc, pH 5.2, and 2 volumes of 100% ethanol. The RNA pellet was washed twice with 75% ethanol and dissolved in diethylpyrocarbonate-treated water. Two micrograms of total RNA was used to perform the reverse transcription reaction using SuperScript II (Invitrogen). Four microliters of a 1/10 dilution of the synthesized cDNA was used for PCR reactions. Real-time PCRs were performed in an optical 96-well plate with an ABI PRISM 7500 Fast sequence detection system (Applied Biosystems), using Fast SYBR Green to monitor double-stranded DNA synthesis. Reactions contained 12.5 μL 2× SYBR Green Master Mix reagent (Applied Biosystems), 4 μL of a 1/10 dilution of the synthesized cDNA, and 200 nmol of each gene-specific primer in a final volume of 20 μL. The following standard thermal profile was used for all PCRs: 95°C for 20 s; 40 cycles of 95°C for 3 s and 60°C for 30 s. Data were analyzed using the SDS 2.0 software (Applied Biosystems). CT values of the genes of interest were normalized to the CT value of SERINE/THREONINE PROTEIN PHOSPHATASE 2A (PP2A). Sequences of the primers used for PCR analyses are listed in the Supplemental Methods online.
Protein extracts were prepared from Arabidopsis by grinding shock-frozen tissue. Subsequently, extraction buffer (7 M urea, 2 M thiourea, 10% [v/v] isopropanol, 5% [v/v] glycerol, 2% [v/v] pharmalyte, 50 mM DTT, and 1× Complete protease inhibitor cocktail [Roche]) was added. Homogenates were centrifuged twice for 20 min at room temperature. Protein concentration was equilibrated (using a simple Bradford method with the Roth-Nanoquant solution, according to the manufacturer's protocol) until all samples contained the same concentration. Laemmli buffer and 100 μg of protein were added to each lane of an 8% SDS polyacrylamide gel. For each protein blot, a second gel was prepared as loading control, which was subsequently stained with Coomassie Brilliant Blue according to standard procedures (Neuhoff et al., 1988). Blotting was performed semidry onto nitrocellulose in 20% (v/v) methanol, 0.29% (w/v) glycine, 0.58% (w/v) Tris base, and 0.04% (w/v) SDS at 0.2 V/cm2 for 2 h. The membrane was incubated overnight at 4°C in TBST (150 mM NaCl, 50 mM Tris, pH 7.5, and 0.1% Tween 20) with 5% (w/v) dry milk powder. Blots were subsequently incubated for 3 h with a 1:400 dilution of α-RBR antibody in TBST plus milk. After three washes in TBST for 10 min per wash, secondary α-rabbit antibody, diluted 1:5000 in TBST plus milk, was added and the blot was incubated for another 2 h. After four final washes, chemiluminescent detection was performed with the ECL enhancer kit from Bio-Rad, according to the manufacturer's instructions.
Ploidy Analysis
For ploidy analysis, leaves were chopped in small pieces, incubated in 400 μL of nuclear extraction buffer (Partec) for 30 min on ice, filtered through a 30-μm mesh, mixed with 1 mL nuclear staining buffer (Partec), and incubated on ice for a further 10 min. The nuclei containing solution was then analyzed with a Partec ploidy analyzer.
Histological Analysis, GUS Staining, DAPI Staining, and Microscopy
GUS staining was performed as described by Brand et al. (2002). For histological analysis, samples were fixed in 4% formaldehyde, 50% ethanol, and 10% acetic acid overnight at 4°C. Samples were dehydrated in a progressive series of ethanol dilutions (50, 80, and 100%) for 1 hour each and embedded in Technovit (Kulzer) according to the manufacturer's instructions. Four-micrometer sections were prepared using a Leica RM2145 microtome and stained in 0.05% Toluidine blue. For DAPI staining, leaf tissue was fixed in 70% (v/v) ethanol overnight, incubated in a water solution containing 5 μg/mL DAPI for 20 min, and washed for 15 min in 70% (v/v) ethanol. Sample observation was performed with a Zeiss microscope using differential interference contrast optics (Zeiss). Images were recorded with an Axiocam HRC CCD camera (Zeiss). For scanning electron microscopy, plant material was treated as described previously (Kwiatkowska, 2004). A Leica SP1-2 was used for confocal microscopy, with settings recommended by the manufacturer.
Accession Numbers
Sequence data from this article can be found in the Arabidopsis Genome Initiative or GenBank/EMBL databases under the following accession numbers: WEE1, AT1G02970; tetrahydrofolate dehydrogenase/cyclohydrolase, putative, AT3G12290; BARD1, AT1G04020; PP2A, AT1G13320; STZ/ZAT10, AT1G27730; CYCA3;2, AT1G47210; STM, AT1G62360; PLA2, AT1G67750; myb family transcription factor, AT1G68670; CDKB2;1, AT1G76540; WRKY40, AT1G80840; WUS, AT2G17950; AGP17, AT2G23130; CLV3, AT2G27250; E2FA, AT2G36010; WRKY33, AT2G38470; CDKB1;2, AT2G38620; pectinesterase family protein, AT3G05620; MUTE, AT3G06120; RBR, AT3G12280; AGP12, AT3G13520; KNAT1, AT4G08150; RD26, AT4G27410; AGP18, AT4G37450; ANT, AT4G37750; DPA, AT5G02470; CYCA3;1, AT5G43080; ERF2, AT5G47220; SPCH, AT5G53210; ERF5, AT5G61600; SDD1, AT1G04110; TMM, AT1G80080; FAMA, AT3G24140; and EPF1, AT2G20875.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure 1. Altered Leaf Development in Seedlings Induced for cre/lox-Mediated Excision of RBR.
Supplemental Figure 2. Design of the β-Estradiol–Inducible RNAi against RBR (RBRi).
Supplemental Figure 3. Induction Test of Pro35S:XVE, olexA:GUS Plants by β-Estradiol Spraying or Supply in MS Plates.
Supplemental Figure 4. ProRBR:GUS Expression Patterns and RBR Protein Levels during Arabidopsis Development.
Supplemental Figure 5. RBR Protein Recovery after Estradiol Withdrawal.
Supplemental Figure 6. Cell Number Quantification on Adaxial and Abaxial Sides of Wild-Type and RBRi β-Estradiol–Treated Seedlings.
Supplemental Figure 7. Cell Number Quantification in Wild-Type and β-Estradiol–Treated RBRi Shoot Apical Meristems (SAMs).
Supplemental Figure 8. Quantification of Not-Puzzle-Shaped, Brick-Like Pavement Cells on Abaxial Sides of Wild-Type and β-Estradiol–Treated RBRi Leaves.
Supplemental Figure 9. Quantitative PCR Analysis of Genes Involved in Stomate Development in Wild-Type and β-Estradiol–Treated RBRi Leaves.
Supplemental Figure 10. Silique Elongation and Fertility in 30-d-Old Wild-Type and RBRi Plants Treated with β-Estradiol for Two Consecutive Weeks.
Supplemental Figure 11. Nuclear DNA Ploidy Analysis in Wild-Type and β-Estradiol–Treated RBRi Leaves.
Supplemental Figure 12. Exemplary Coexpression Clusters of Genes Deregulated in RBRi Plants after β-Estradiol Treatment.
Supplemental Figure 13. Quantitative PCR Analysis of WRKY33 and WRKY40, Two Transcription Factors Involved in Stress Response, in Wild-Type and RBRi-Induced Plants.
Supplemental Figure 14. Quantitative PCR Analysis of AGP12, 17, and 18 Expression Levels in Wild-Type and RBRi β-Estradiol–Treated Plants.
Supplemental Figure 15. Selection of DNA Replication–Related Genes Upregulated 12 (Light Gray) and 24 (Dark Gray) h after β-Estradiol Treatment.
Supplemental Table 1. Coexpression Analyses and Validation via Hypergeometrical Distribution of Genes Upregulated at 3, 6, 12, and 24 h after β-Estradiol Treatment of RBRi Seedlings.
Supplemental Table 2. Coexpression Analyses and Validation via Hypergeometrical Distribution of Genes Downregulated at 3, 6, 12, and 24 h after β-Estradiol Treatment of RBRi Seedlings.
Supplemental Table 3. Coexpression Analyses and Validation via Hypergeometrical Distribution between Genes Present in Several Stress-Related ATTED Clusters or E2F/DP Overexpression Experiments and Genes Deregulated at 3, 6, 12, and 24 h after β-Estradiol Treatment of RBRi Seedlings.
Supplemental Data Set 1. Average SLR Data from Gene Expression Analysis 3, 6, 12, and 24 h after β-Estradiol Treatment of Arabidopsis thaliana Plants Expressing an Inducible RBR RNAi Construct.
Supplemental Methods. Primer Sequences.
Supplemental References.
Acknowledgments
We thank Rüdiger Simon (Institut für Genetik, Heinrich Heine Universität Düsseldorf) for providing the CLV3:GUS reporter, Lieven De Veylder (Plant System Biology, VIB/Universiteit Gent) for CYCB1;1:GUS, Dominique Bergmann (Stanford University, CA) for pMUTE:MUTE-GFP and Fred Sacks (Department of Botany, The University of British Columbia) for the pTMM:TMM-GFP reporter lines, Vivien Exner (ETH Zurich) for support with ploidy analysis, and Sylvain Bischof (ETH Zurich) for comments on the manuscript. We thank Peter Wägli (Electron Microscopy Center, ETH Zurich) for scanning electron microscopy analyses, Catharine Aquino Fournier (Functional Genomics Center Zurich) for microarray processing, and Matthias Hirsch-Hoffmann (ETH Zurich) for bioinformatics support. This work was supported by a grant from the Swiss National Science Foundation to W.G.
References
- Ach R.A., Durfee T., Miller A.B., Taranto P., Hanley-Bowdoin L., Zambryski P.C., Gruissem W. (1997). RRB1 and RRB2 encode maize retinoblastoma-related proteins that interact with a plant D-type cyclin and geminivirus replication protein. Mol. Cell. Biol. 17: 5077–5086 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amirsadeghi S., McDonald A.E., Vanlerberghe G.C. (2007). A glucocorticoid-inducible gene expression system can cause growth defects in tobacco. Planta 226: 453–463 [DOI] [PubMed] [Google Scholar]
- Andersen S.U., Cvitanich C., Hougaard B.K., Roussis A., Gronlund M., Jensen D.B., Frokjaer L.A., Jensen E.O. (2003). The glucocorticoid-inducible GVG system causes severe growth defects in both root and shoot of the model legume Lotus japonicus. Mol. Plant Microbe Interact. 16: 1069–1076 [DOI] [PubMed] [Google Scholar]
- Aoyama T., Chua N.H. (1997). A glucocorticoid-mediated transcriptional induction system in transgenic plants. Plant J. 11: 605–612 [DOI] [PubMed] [Google Scholar]
- Aronson M.N., Meyer A.D., Gyorgyey J., Katul L., Vetten H.J., Gronenborn B., Timchenko T. (2000). Clink, a nanovirus-encoded protein, binds both pRB and SKP1. J. Virol. 74: 2967–2972 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartkova J., et al. (2006). Oncogene-induced senescence is part of the tumorigenesis barrier imposed by DNA damage checkpoints. Nature 444: 633–637 [DOI] [PubMed] [Google Scholar]
- Bergmann D.C., Sack F.D. (2007). Stomatal development. Annu. Rev. Plant Biol. 58: 163–181 [DOI] [PubMed] [Google Scholar]
- Berna G., Robles P., Micol J.L. (1999). A mutational analysis of leaf morphogenesis in Arabidopsis thaliana. Genetics 152: 729–742 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bieri S., Potrykus I., Fütterer J. (2002). Geminivirus sequences as bidirectional transcription termination/polyadenylation signals for economic construction of stably expressed transgenes. Mol. Breed. 10: 107–117 [Google Scholar]
- Binne U.K., Classon M.K., Dick F.A., Wei W., Rape M., Kaelin W.G., Jr, Naar A.M., Dyson N.J. (2007). Retinoblastoma protein and anaphase-promoting complex physically interact and functionally cooperate during cell-cycle exit. Nat. Cell Biol. 9: 225–232 [DOI] [PubMed] [Google Scholar]
- Bosco G., Du W., Orr-Weaver T.L. (2001). DNA replication control through interaction of E2F-RB and the origin recognition complex. Nat. Cell Biol. 3: 289–295 [DOI] [PubMed] [Google Scholar]
- Brand L., Horler M., Nuesch E., Vassalli S., Barrell P., Yang W., Jefferson R.A., Grossniklaus U., Curtis M.D. (2006). A versatile and reliable two-component system for tissue-specific gene induction in Arabidopsis. Plant Physiol. 141: 1194–1204 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brand U., Fletcher J.C., Hobe M., Meyerowitz E.M., Simon R. (2000). Dependence of stem cell fate in Arabidopsis on a feedback loop regulated by CLV3 activity. Science 289: 617–619 [DOI] [PubMed] [Google Scholar]
- Brand U., Grunewald M., Hobe M., Simon R. (2002). Regulation of CLV3 expression by two homeobox genes in Arabidopsis. Plant Physiol. 129: 565–575 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brooks L., III, et al. (2009). Microdissection of shoot meristem functional domains. PLoS Genet. 5: e1000476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burhans W.C., Heintz N.H. (2009). The cell cycle is a redox cycle: Linking phase-specific targets to cell fate. Free Radic. Biol. Med. 47: 1282–1293 [DOI] [PubMed] [Google Scholar]
- Buttitta L.A., Edgar B.A. (2007). Mechanisms controlling cell cycle exit upon terminal differentiation. Curr. Opin. Cell Biol. 19: 697–704 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carlsbecker A., Helariutta Y. (2005). Phloem and xylem specification: Pieces of the puzzle emerge. Curr. Opin. Plant Biol. 8: 512–517 [DOI] [PubMed] [Google Scholar]
- Clough S.J., Bent A.F. (1998). Floral dip: A simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 16: 735–743 [DOI] [PubMed] [Google Scholar]
- De Falco G., Comes F., Simone C. (2006). pRb: Master of differentiation. Coupling irreversible cell cycle withdrawal with induction of muscle-specific transcription. Oncogene 25: 5244–5249 [DOI] [PubMed] [Google Scholar]
- De Jager S.M., Scofield S., Huntley R.P., Robinson A.S., den Boer B.G., Murray J.A. (2009). Dissecting regulatory pathways of G1/S control in Arabidopsis: Common and distinct targets of CYCD3;1, E2Fa and E2Fc. Plant Mol. Biol. 71: 345–365 [DOI] [PubMed] [Google Scholar]
- De Schutter K., Joubes J., Cools T., Verkest A., Corellou F., Babiychuk E., Van Der Schueren E., Beeckman T., Kushnir S., Inze D., De Veylder L. (2007). Arabidopsis WEE1 kinase controls cell cycle arrest in response to activation of the DNA integrity checkpoint. Plant Cell 19: 211–225 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Desvoyes B., Ramirez-Parra E., Xie Q., Chua N.H., Gutierrez C. (2006). Cell type-specific role of the retinoblastoma/E2F pathway during Arabidopsis leaf development. Plant Physiol. 140: 67–80 [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Veylder L., Beeckman T., Beemster G.T., de Almeida Engler J., Ormenese S., Maes S., Naudts M., Van Der Schueren E., Jacqmard A., Engler G., Inze D. (2002). Control of proliferation, endoreduplication and differentiation by the Arabidopsis E2Fa-DPa transcription factor. EMBO J. 21: 1360–1368 [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Veylder L., Beeckman T., Inze D. (2007). The ins and outs of the plant cell cycle. Nat. Rev. Mol. Cell Biol. 8: 655–665 [DOI] [PubMed] [Google Scholar]
- Dewitte W., Riou-Khamlichi C., Scofield S., Healy J.M., Jacqmard A., Kilby N.J., Murray J.A. (2003). Altered cell cycle distribution, hyperplasia, and inhibited differentiation in Arabidopsis caused by the D-type cyclin CYCD3. Plant Cell 15: 79–92 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Di Micco R., et al. (2006). Oncogene-induced senescence is a DNA damage response triggered by DNA hyper-replication. Nature 444: 638–642 [DOI] [PubMed] [Google Scholar]
- Dong J., MacAlister C.A., Bergmann D.C. (2009). BASL controls asymmetric cell division in Arabidopsis. Cell 137: 1320–1330 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ebel C., Mariconti L., Gruissem W. (2004). Plant retinoblastoma homologues control nuclear proliferation in the female gametophyte. Nature 429: 776–780 [DOI] [PubMed] [Google Scholar]
- Felsani A., Mileo A.M., Paggi M.G. (2006). Retinoblastoma family proteins as key targets of the small DNA virus oncoproteins. Oncogene 25: 5277–5285 [DOI] [PubMed] [Google Scholar]
- Ferris R., Long L., Bunn S.M., Robinson K.M., Bradshaw H.D., Rae A.M., Taylor G. (2002). Leaf stomatal and epidermal cell development: Identification of putative quantitative trait loci in relation to elevated carbon dioxide concentration in poplar. Tree Physiol. 22: 633–640 [DOI] [PubMed] [Google Scholar]
- Flemington E.K., Speck S.H., Kaelin W.G., Jr (1993). E2F-1-mediated transactivation is inhibited by complex formation with the retinoblastoma susceptibility gene product. Proc. Natl. Acad. Sci. USA 90: 6914–6918 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friend S.H., Bernards R., Rogelj S., Weinberg R.A., Rapaport J.M., Albert D.M., Dryja T.P. (1986). A human DNA segment with properties of the gene that predisposes to retinoblastoma and osteosarcoma. Nature 323: 643–646 [DOI] [PubMed] [Google Scholar]
- Fujita M., Fujita Y., Maruyama K., Seki M., Hiratsu K., Ohme-Takagi M., Tran L.S., Yamaguchi-Shinozaki K., Shinozaki K. (2004). A dehydration-induced NAC protein, RD26, is involved in a novel ABA-dependent stress-signaling pathway. Plant J. 39: 863–876 [DOI] [PubMed] [Google Scholar]
- Gao M., Showalter A.M. (1999). Yariv reagent treatment induces programmed cell death in Arabidopsis cell cultures and implicates arabinogalactan protein involvement. Plant J. 19: 321–331 [DOI] [PubMed] [Google Scholar]
- Geisler M., Nadeau J., Sack F.D. (2000). Oriented asymmetric divisions that generate the stomatal spacing pattern in Arabidopsis are disrupted by the too many mouths mutation. Plant Cell 12: 2075–2086 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gisel A., Barella S., Hempel F.D., Zambryski P.C. (1999). Temporal and spatial regulation of symplastic trafficking during development in Arabidopsis thaliana apices. Development 126: 1879–1889 [DOI] [PubMed] [Google Scholar]
- Grafi G., Burnett R.J., Helentjaris T., Larkins B.A., DeCaprio J.A., Sellers W.R., Kaelin W.G., Jr (1996). A maize cDNA encoding a member of the retinoblastoma protein family: Involvement in endoreduplication. Proc. Natl. Acad. Sci. USA 93: 8962–8967 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gruissem W. (2007). Function of the Retinoblastoma-Related Protein in Plants. (Oxford, UK: Blackwell Scientific; ). [Google Scholar]
- Hallenborg P., Feddersen S., Madsen L., Kristiansen K. (2009). The tumor suppressors pRB and p53 as regulators of adipocyte differentiation and function. Expert Opin. Ther. Targets 13: 235–246 [DOI] [PubMed] [Google Scholar]
- Han P., Li Q., Zhu Y.X. (2008). Mutation of Arabidopsis BARD1 causes meristem defects by failing to confine WUSCHEL expression to the organizing center. Plant Cell 20: 1482–1493 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hara K., Yokoo T., Kajita R., Onishi T., Yahata S., Peterson K.M., Torii K.U., Kakimoto T. (2009). Epidermal cell density is autoregulated via a secretory peptide, EPIDERMAL PATTERNING FACTOR 2 in Arabidopsis leaves. Plant Cell Physiol. 50: 1019–1031 [DOI] [PubMed] [Google Scholar]
- Hennig L., Gruissem W., Grossniklaus U., Kohler C. (2004). Transcriptional programs of early reproductive stages in Arabidopsis. Plant Physiol. 135: 1765–1775 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirano H., Harashima H., Shinmyo A., Sekine M. (2008). Arabidopsis RETINOBLASTOMA-RELATED PROTEIN 1 is involved in G1 phase cell cycle arrest caused by sucrose starvation. Plant Mol. Biol. 66: 259–275 [DOI] [PubMed] [Google Scholar]
- Hunt L., Gray J.E. (2009). The signaling peptide EPF2 controls asymmetric cell divisions during stomatal development. Curr. Biol. 19: 864–869 [DOI] [PubMed] [Google Scholar]
- Inze D. (2005). Green light for the cell cycle. EMBO J. 24: 657–662 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishida S., Huang E., Zuzan H., Spang R., Leone G., West M., Nevins J.R. (2001). Role for E2F in control of both DNA replication and mitotic functions as revealed from DNA microarray analysis. Mol. Cell. Biol. 21: 4684–4699 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacks T., Fazeli A., Schmitt E.M., Bronson R.T., Goodell M.A., Weinberg R.A. (1992). Effects of an Rb mutation in the mouse. Nature 359: 295–300 [DOI] [PubMed] [Google Scholar]
- Johnston A.J., Gruissem W. (2009). Gametophyte differentiation and imprinting control in plants: Crosstalk between RBR and chromatin. Commun. Integr. Biol. 2: 144–146 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnston A.J., Matveeva E., Kirioukhova O., Grossniklaus U., Gruissem W. (2008). A dynamic reciprocal RBR-PRC2 regulatory circuit controls Arabidopsis gametophyte development. Curr. Biol. 18: 1680–1686 [DOI] [PubMed] [Google Scholar]
- Jonsson H., Heisler M., Reddy G.V., Agrawal V., Gor V., Shapiro B.E., Mjolsness E., Meyerowitz E.M. (2005). Modeling the organization of the WUSCHEL expression domain in the shoot apical meristem. Bioinformatics 21 (Suppl. 1): i232–i240 [DOI] [PubMed] [Google Scholar]
- Jordan C.V., Shen W., Hanley-Bowdoin L.K., Robertson D.N. (2007). Geminivirus-induced gene silencing of the tobacco retinoblastoma-related gene results in cell death and altered development. Plant Mol. Biol. 65: 163–175 [DOI] [PubMed] [Google Scholar]
- Kang H.G., Fang Y., Singh K.B. (1999). A glucocorticoid-inducible transcription system causes severe growth defects in Arabidopsis and induces defense-related genes. Plant J. 20: 127–133 [DOI] [PubMed] [Google Scholar]
- Karimi M., Inze D., Depicker A. (2002). GATEWAY vectors for Agrobacterium-mediated plant transformation. Trends Plant Sci. 7: 193–195 [DOI] [PubMed] [Google Scholar]
- Kel A.E., Kel-Margoulis O.V., Farnham P.J., Bartley S.M., Wingender E., Zhang M.Q. (2001). Computer-assisted identification of cell cycle-related genes: New targets for E2F transcription factors. J. Mol. Biol. 309: 99–120 [DOI] [PubMed] [Google Scholar]
- Klimova T.A., Bell E.L., Shroff E.H., Weinberg F.D., Snyder C.M., Dimri G.P., Schumacker P.T., Budinger G.R., Chandel N.S. (2009). Hyperoxia-induced premature senescence requires p53 and pRb, but not mitochondrial matrix ROS. FASEB J. 23: 783–794 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kong L.J., Hanley-Bowdoin L. (2002). A geminivirus replication protein interacts with a protein kinase and a motor protein that display different expression patterns during plant development and infection. Plant Cell 14: 1817–1832 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kong L.J., Orozco B.M., Roe J.L., Nagar S., Ou S., Feiler H.S., Durfee T., Miller A.B., Gruissem W., Robertson D., Hanley-Bowdoin L. (2000). A geminivirus replication protein interacts with the retinoblastoma protein through a novel domain to determine symptoms and tissue specificity of infection in plants. EMBO J. 19: 3485–3495 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korenjak M., Brehm A. (2005). E2F-Rb complexes regulating transcription of genes important for differentiation and development. Curr. Opin. Genet. Dev. 15: 520–527 [DOI] [PubMed] [Google Scholar]
- Kwiatkowska D. (2004). Surface growth at the reproductive shoot apex of Arabidopsis thaliana pin-formed 1 and wild type. J. Exp. Bot. 399: 1021–1032 [DOI] [PubMed] [Google Scholar]
- Lageix S., Catrice O., Deragon J.M., Gronenborn B., Pelissier T., Ramirez B.C. (2007). The nanovirus-encoded Clink protein affects plant cell cycle regulation through interaction with the retinoblastoma-related protein. J. Virol. 81: 4177–4185 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lampard G.R., Macalister C.A., Bergmann D.C. (2008). Arabidopsis stomatal initiation is controlled by MAPK-mediated regulation of the bHLH SPEECHLESS. Science 322: 1113–1116 [DOI] [PubMed] [Google Scholar]
- Laskowski M., Biller S., Stanley K., Kajstura T., Prusty R. (2006). Expression profiling of auxin-treated Arabidopsis roots: Toward a molecular analysis of lateral root emergence. Plant Cell Physiol. 47: 788–792 [DOI] [PubMed] [Google Scholar]
- Lee E.Y., Chang C.Y., Hu N., Wang Y.C., Lai C.C., Herrup K., Lee W.H., Bradley A. (1992). Mice deficient for Rb are nonviable and show defects in neurogenesis and haematopoiesis. Nature 359: 288–294 [DOI] [PubMed] [Google Scholar]
- Liu Q., VanHoy R.W., Zhou J.H., Dantzer R., Freund G.G., Kelley K.W. (1999). Elevated cyclin E levels, inactive retinoblastoma protein, and suppression of the p27(KIP1) inhibitor characterize early development of promyeloid cells into macrophages. Mol. Cell. Biol. 19: 6229–6239 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma Q., Dai X., Xu Y., Guo J., Liu Y., Chen N., Xiao J., Zhang D., Xu Z., Zhang X., Chong K. (2009). Enhanced tolerance to chilling stress in OsMYB3R–2 transgenic rice is mediated by alteration in cell cycle and ectopic expression of stress genes. Plant Physiol. 150: 244–256 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma S., Bohnert H.J. (2007). Integration of Arabidopsis thaliana stress-related transcript profiles, promoter structures, and cell-specific expression. Genome Biol. 8: R49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma S., Gong Q., Bohnert H.J. (2007). An Arabidopsis gene network based on the graphical Gaussian model. Genome Res. 17: 1614–1625 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Macleod K.F. (2008). The role of the RB tumour suppressor pathway in oxidative stress responses in the haematopoietic system. Nat. Rev. Cancer 8: 769–781 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mavrogonatou E., Kletsas D. (2009). High osmolality activates the G1 and G2 cell cycle checkpoints and affects the DNA integrity of nucleus pulposus intervertebral disc cells triggering an enhanced DNA repair response. DNA Repair (Amst.) 8: 930–943 [DOI] [PubMed] [Google Scholar]
- Menges M., de Jager S.M., Gruissem W., Murray J.A. (2005). Global analysis of the core cell cycle regulators of Arabidopsis identifies novel genes, reveals multiple and highly specific profiles of expression and provides a coherent model for plant cell cycle control. Plant J. 41: 546–566 [DOI] [PubMed] [Google Scholar]
- Mittler R., Kim Y., Song L., Coutu J., Coutu A., Ciftci-Yilmaz S., Lee H., Stevenson B., Zhu J.K. (2006). Gain- and loss-of-function mutations in Zat10 enhance the tolerance of plants to abiotic stress. FEBS Lett. 580: 6537–6542 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore I., Samalova M., Kurup S. (2006). Transactivated and chemically inducible gene expression in plants. Plant J. 45: 651–683 [DOI] [PubMed] [Google Scholar]
- Morris E.J., Dyson N.J. (2001). Retinoblastoma protein partners. Adv. Cancer Res. 82: 1–54 [DOI] [PubMed] [Google Scholar]
- Mukherjee P., Cao T.V., Winter S.L., Alexandrow M.G. (2009). Mammalian MCM loading in late-G(1) coincides with Rb hyperphosphorylation and the transition to post-transcriptional control of progression into S-phase. PLoS One 4: e5462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muller R., Borghi L., Kwiatkowska D., Laufs P., Simon R. (2006). Dynamic and compensatory responses of Arabidopsis shoot and floral meristems to CLV3 signaling. Plant Cell 18: 1188–1198 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mulligan G., Jacks T. (1998). The retinoblastoma gene family: Cousins with overlapping interests. Trends Genet. 14: 223–229 [DOI] [PubMed] [Google Scholar]
- Munoz-Martin A., Collin S., Herreros E., Mullineaux P.M., Fernandez-Lobato M., Fenoll C. (2003). Regulation of MSV and WDV virion-sense promoters by WDV nonstructural proteins: A role for their retinoblastoma protein-binding motifs. Virology 306: 313–323 [DOI] [PubMed] [Google Scholar]
- Nadeau J.A., Sack F.D. (2002). Control of stomatal distribution on the Arabidopsis leaf surface. Science 296: 1697–1700 [DOI] [PubMed] [Google Scholar]
- Nakagami H., Sekine M., Murakami H., Shinmyo A. (1999). Tobacco retinoblastoma-related protein phosphorylated by a distinct cyclin-dependent kinase complex with Cdc2/cyclin D in vitro. Plant J. 18: 243–252 [DOI] [PubMed] [Google Scholar]
- Naouar N., Vandepoele K., Lammens T., Casneuf T., Zeller G., van Hummelen P., Weigel D., Ratsch G., Inze D., Kuiper M., De Veylder L., Vuylsteke M. (2009). Quantitative RNA expression analysis with Affymetrix Tiling 1.0R arrays identifies new E2F target genes. Plant J. 57: 184–194 [DOI] [PubMed] [Google Scholar]
- Nead M.A., Baglia L.A., Antinore M.J., Ludlow J.W., McCance D.J. (1998). Rb binds c-Jun and activates transcription. EMBO J. 17: 2342–2352 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neuhoff V., Arold N., Taube D., Ehrhardt W. (1988). Improved staining of proteins in polyacrylamide gels including isoelectric focusing gels with clear background at nanogram sensitivity using Coomassie Brilliant Blue G-250 and R-250. Electrophoresis 9: 255–262 [DOI] [PubMed] [Google Scholar]
- Nole-Wilson S., Krizek B.A. (2006). AINTEGUMENTA contributes to organ polarity and regulates growth of lateral organs in combination with YABBY genes. Plant Physiol. 141: 977–987 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Obayashi T., Hayashi S., Saeki M., Ohta H., Kinoshita K. (2009). ATTED-II provides coexpressed gene networks for Arabidopsis. Nucleic Acids Res. 37: D987–D991 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orozco B.M., Kong L.J., Batts L.A., Elledge S., Hanley-Bowdoin L. (2000). The multifunctional character of a geminivirus replication protein is reflected by its complex oligomerization properties. J. Biol. Chem. 275: 6114–6122 [DOI] [PubMed] [Google Scholar]
- Papadimou E., Menard C., Grey C., Puceat M. (2005). Interplay between the retinoblastoma protein and LEK1 specifies stem cells toward the cardiac lineage. EMBO J. 24: 1750–1761 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park J.A., Ahn J.W., Kim Y.K., Kim S.J., Kim J.K., Kim W.T., Pai H.S. (2005). Retinoblastoma protein regulates cell proliferation, differentiation, and endoreduplication in plants. Plant J. 42: 153–163 [DOI] [PubMed] [Google Scholar]
- Pickering M.T., Kowalik T.F. (2006). Rb inactivation leads to E2F1-mediated DNA double-strand break accumulation. Oncogene 25: 746–755 [DOI] [PubMed] [Google Scholar]
- Pillitteri L.J., Sloan D.B., Bogenschutz N.L., Torii K.U. (2007). Termination of asymmetric cell division and differentiation of stomata. Nature 445: 501–505 [DOI] [PubMed] [Google Scholar]
- Ramirez-Parra E., Frundt C., Gutierrez C. (2003). A genome-wide identification of E2F-regulated genes in Arabidopsis. Plant J. 33: 801–811 [DOI] [PubMed] [Google Scholar]
- Ramirez-Parra E., Gutierrez C. (2007). E2F regulates FASCIATA1, a chromatin assembly gene whose loss switches on the endocycle and activates gene expression by changing the epigenetic status. Plant Physiol. 144: 105–120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- R Development Core Team (2009). R: A Language and Environment for Statistical Computing. (Vienna, Austria: R Foundation for Statistical Computing; ). [Google Scholar]
- Reddy G.V., Meyerowitz E.M. (2005). Stem-cell homeostasis and growth dynamics can be uncoupled in the Arabidopsis shoot apex. Science 310: 663–667 [DOI] [PubMed] [Google Scholar]
- Reinhardt D., Frenz M., Mandel T., Kuhlemeier C. (2003). Microsurgical and laser ablation analysis of interactions between the zones and layers of the tomato shoot apical meristem. Development 130: 4073–4083 [DOI] [PubMed] [Google Scholar]
- Rojo E., Sharma V.K., Kovaleva V., Raikhel N.V., Fletcher J.C. (2002). CLV3 is localized to the extracellular space, where it activates the Arabidopsis CLAVATA stem cell signaling pathway. Plant Cell 14: 969–977 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Savatier P., Huang S., Szekely L., Wiman K.G., Samarut J. (1994). Contrasting patterns of retinoblastoma protein expression in mouse embryonic stem cells and embryonic fibroblasts. Oncogene 9: 809–818 [PubMed] [Google Scholar]
- Serna L. (2007). bHLH proteins know when to make a stoma. Trends Plant Sci. 12: 483–485 [DOI] [PubMed] [Google Scholar]
- Serna L. (2009a). Cell fate transitions during stomatal development. Bioessays 31: 865–873 [DOI] [PubMed] [Google Scholar]
- Serna L. (2009b). Emerging parallels between stomatal and muscle cell lineages. Plant Physiol. 149: 1625–1631 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shackelford R.E., Kaufmann W.K., Paules R.S. (2000). Oxidative stress and cell cycle checkpoint function. Free Radic. Biol. Med. 28: 1387–1404 [DOI] [PubMed] [Google Scholar]
- Smyth G.K. (2004). Linear models and empirical bayes methods for assessing differential expression in microarray experiments. Stat. Appl. Genet. Mol. Biol. 3: Article3. [DOI] [PubMed] [Google Scholar]
- Spike B.T., Dirlam A., Dibling B.C., Marvin J., Williams B.O., Jacks T., Macleod K.F. (2004). The Rb tumor suppressor is required for stress erythropoiesis. EMBO J. 23: 4319–4329 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stanelle J., Stiewe T., Theseling C.C., Peter M., Putzer B.M. (2002). Gene expression changes in response to E2F1 activation. Nucleic Acids Res. 30: 1859–1867 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stead E., White J., Faast R., Conn S., Goldstone S., Rathjen J., Dhingra U., Rathjen P., Walker D., Dalton S. (2002). Pluripotent cell division cycles are driven by ectopic Cdk2, cyclin A/E and E2F activities. Oncogene 21: 8320–8333 [DOI] [PubMed] [Google Scholar]
- Storey J.D., Tibshirani R. (2003). Statistical significance for genomewide studies. Proc. Natl. Acad. Sci. USA 100: 9440–9445 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Talbert P.B., Adler H.T., Parks D.W., Comai L. (1995). The REVOLUTA gene is necessary for apical meristem development and for limiting cell divisions in the leaves and stems of Arabidopsis thaliana. Development 121: 2723–2735 [DOI] [PubMed] [Google Scholar]
- Thomas D.M., Yang H.S., Alexander K., Hinds P.W. (2003). Role of the retinoblastoma protein in differentiation and senescence. Cancer Biol. Ther. 2: 124–130 [PubMed] [Google Scholar]
- Toppari J., Suominenf J.S., Yan W. (2003). The role of retinoblastoma protein family in the control of germ cell proliferation, differentiation and survival. APMIS 111: 245–251 [DOI] [PubMed] [Google Scholar]
- Tort F., Bartkova J., Sehested M., Orntoft T., Lukas J., Bartek J. (2006). Retinoblastoma pathway defects show differential ability to activate the constitutive DNA damage response in human tumorigenesis. Cancer Res. 66: 10258–10263 [DOI] [PubMed] [Google Scholar]
- Tucker M.R., Laux T. (2007). Connecting the paths in plant stem cell regulation. Trends Cell Biol. 17: 403–410 [DOI] [PubMed] [Google Scholar]
- Umen J.G., Goodenough U.W. (2001). Control of cell division by a retinoblastoma protein homolog in Chlamydomonas. Genes Dev. 15: 1652–1661 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van den Heuvel S., Dyson N.J. (2008). Conserved functions of the pRB and E2F families. Nat. Rev. Mol. Cell Biol. 9: 713–724 [DOI] [PubMed] [Google Scholar]
- Vandepoele K., Vlieghe K., Florquin K., Hennig L., Beemster G.T., Gruissem W., Van de Peer Y., Inze D., De Veylder L. (2005). Genome-wide identification of potential plant E2F target genes. Plant Physiol. 139: 316–328 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vlieghe K., Vuylsteke M., Florquin K., Rombauts S., Maes S., Ormenese S., Van Hummelen P., Van de Peer Y., Inze D., De Veylder L. (2003). Microarray analysis of E2Fa-DPa-overexpressing plants uncovers a cross-talking genetic network between DNA replication and nitrogen assimilation. J. Cell Sci. 116: 4249–4259 [DOI] [PubMed] [Google Scholar]
- Von Groll U., Berger D., Altmann T. (2002). The subtilisin-like serine protease SDD1 mediates cell-to-cell signaling during Arabidopsis stomatal development. Plant Cell 14: 1527–1539 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weinberg R.A. (1995). The retinoblastoma protein and cell cycle control. Cell 81: 323–330 [DOI] [PubMed] [Google Scholar]
- White J., Dalton S. (2005). Cell cycle control of embryonic stem cells. Stem Cell Rev. 1: 131–138 [DOI] [PubMed] [Google Scholar]
- Wildwater M., Campilho A., Perez-Perez J.M., Heidstra R., Blilou I., Korthout H., Chatterjee J., Mariconti L., Gruissem W., Scheres B. (2005). The RETINOBLASTOMA-RELATED gene regulates stem cell maintenance in Arabidopsis roots. Cell 123: 1337–1349 [DOI] [PubMed] [Google Scholar]
- Wu Z., Irizarry R., Gentleman R., Murillo F., Spencer F. (2004). A Model Based Background Adjustment for Oligonucleotide Expression Arrays. (Baltimore, MD: Johns Hopkins University; ). [Google Scholar]
- Wyrzykowska J., Schorderet M., Pien S., Gruissem W., Fleming A.J. (2006). Induction of differentiation in the shoot apical meristem by transient overexpression of a retinoblastoma-related protein. Plant Physiol. 141: 1338–1348 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie Q., Sanz-Burgos A.P., Hannon G.J., Gutierrez C. (1996). Plant cells contain a novel member of the retinoblastoma family of growth regulatory proteins. EMBO J. 15: 4900–4908 [PMC free article] [PubMed] [Google Scholar]
- Xie Q., Suarez-Lopez P., Gutierrez C. (1995). Identification and analysis of a retinoblastoma binding motif in the replication protein of a plant DNA virus: Requirement for efficient viral DNA replication. EMBO J. 14: 4073–4082 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu X., Chen C., Fan B., Chen Z. (2006). Physical and functional interactions between pathogen-induced Arabidopsis WRKY18, WRKY40, and WRKY60 transcription factors. Plant Cell 18: 1310–1326 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yadav R.K., Girke T., Pasala S., Xie M., Reddy G.V. (2009). Gene expression map of the Arabidopsis shoot apical meristem stem cell niche. Proc. Natl. Acad. Sci. USA 106: 4941–4946 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng Z., Qamar S.A., Chen Z., Mengiste T. (2006). Arabidopsis WRKY33 transcription factor is required for resistance to necrotrophic fungal pathogens. Plant J. 48: 592–605 [DOI] [PubMed] [Google Scholar]