This study identifies AMSH3 as a major deubiquitinating enzyme in Arabidopsis that hydrolyzes ubiquitin chains in vitro and in vivo. It finds that AMSH3 is essential for proper vacuole biogenesis, trafficking from the Golgi to the vacuole, and the vacuolar trafficking of endocytosed cargo.
Abstract
Ubiquitination, deubiquitination, and the formation of specific ubiquitin chain topologies have been implicated in various cellular processes. Little is known, however, about the role of ubiquitin in the development of cellular organelles. Here, we identify and characterize the deubiquitinating enzyme AMSH3 from Arabidopsis thaliana. AMSH3 hydrolyzes K48- and K63-linked ubiquitin chains in vitro and accumulates both ubiquitin chain types in vivo. amsh3 mutants fail to form a central lytic vacuole, accumulate autophagosomes, and mis-sort vacuolar protein cargo to the intercellular space. Furthermore, AMSH3 is required for efficient endocytosis of the styryl dye FM4-64 and the auxin efflux facilitator PIN2. We thus present evidence for a role of deubiquitination in intracellular trafficking and vacuole biogenesis.
INTRODUCTION
Ubiquitin conjugation and the formation of ubiquitin chains with different linkage topologies have been associated with different biological processes (Kerscher et al., 2006). Best understood is the role of K48-linked ubiquitin chains in targeting proteins for degradation by the 26S proteasome (Hershko and Ciechanover, 1998). However, in recent years a range of additional ubiquitin (chain) functions have been uncovered in the regulation of other cellular processes, including membrane trafficking, protein kinase activation, DNA repair, and chromatin dynamics (Acconcia et al., 2009; Chen and Sun, 2009). In yeast and animal cells, monoubiquitination or the formation of K63-linked ubiquitin chains initiates the endocytosis of proteins and thereby promotes the degradation of these proteins in the vacuole or lysosome (Mukhopadhyay and Riezman, 2007). Endocytosis is promoted by the endosomal sorting complexes required for transport (ESCRT) 0, I, II, and III. Several proteins of the ESCRT machinery have ubiquitin binding domains that provide the critical affinity for (poly-)ubiquitinated cargo.
Ubiquitination is a reversible process, and monoubiquitin and ubiquitin chains are hydrolyzed by deubiquitinating enzymes (DUBs). DUBs serve to counterbalance ubiquitination within a cell and contribute to the regulation of cellular processes and to the maintenance of free ubiquitin pools (Reyes-Turcu et al., 2009). Eukaryotic DUBs can be classified into five distinct families based on their active site and domain organization (Komander et al., 2009). We are particularly interested in metalloprotease DUBs with an MPN+/JAMM domain (MPR1, PAD1 N-terminal+/JAB1/MPN/MOV34; hitherto MPN+ domain) (Maytal-Kivity et al., 2002; Ambroggio et al., 2004; Clague and Urbe, 2006; Komander et al., 2009). To date, the best-studied MPN+ domain proteins are the evolutionarily conserved RPN11/Poh and CSN5/Jab1: RPN11/Poh1 hydrolyzes ubiquitin chains prior to protein degradation by the 26S proteasome, and CSN5/Jab1 removes the ubiquitin-related protein NEDD8 from the cullin subunit of E3 ligases (Cope et al., 2002; Verma et al., 2002). Four additional MPN+ domain proteins, namely, AMSH (associated molecule with the SH3 domain of STAM) and the related AMSH-LP (AMSH-LIKE PROTEIN) as well as Brcc36/c6.1 and 2A-DUB/KIAA1915, have so far been characterized only in mammalian cells (Tanaka et al., 1999; Cope et al., 2002; Maytal-Kivity et al., 2002; Dong et al., 2003; Kikuchi et al., 2003; McCullough et al., 2004; Cooper et al., 2009).
Human (Homo sapiens) AMSH and AMSH-LP hydrolyze K63-linked ubiquitin chains, suggesting that these proteins function in processes, such as endocytosis, that require K63-linked ubiquitin chains (Tanaka et al., 1999; Ishii et al., 2001; McCullough et al., 2004, 2006; Sato et al., 2008). The proposed role of Hs-AMSH in endocytosis is supported by the finding that Hs-AMSH interacts with clathrin, with ESCRT-0, and with ESCRT-III subunits (Agromayor and Martin-Serrano, 2006; Kyuuma et al., 2007; Ma et al., 2007; Row et al., 2007). The nature of these interactions together with a number of functional studies has given rise to a model whereby Hs-AMSH has two mutually nonexclusive functions at distinct steps of endocytosis (Clague and Urbe, 2006): First, the Hs-AMSH–dependent deubiquitination of endocytosed cargo at ESCRT-0 may promote cargo recycling to the plasma membrane; second, Hs-AMSH may recycle ubiquitin by deubiquitinating endocytosed cargo prior to its sorting to the multivesicular bodies at ESCRT-III. The homolog of Hs-AMSH in mouse (Mus musculus) is essential for postnatal growth; however, its role in development remains to be elucidated (Ishii et al., 2001).
In Arabidopsis thaliana, three proteins, AMSH1, AMSH2, and AMSH3, have significant sequence similarity to Hs-AMSH and Hs-AMSH-LP. Through the analysis of plants expressing inactive variants of these proteins, we identified AMSH3 as a major Arabidopsis DUB that hydrolyzes K48- and K63-linked ubiquitin chains in vitro and in vivo. Furthermore, we found that AMSH3 is essential for proper vacuole biogenesis, trafficking from the Golgi to the vacuole and the vacuolar trafficking of endocytosed cargo.
RESULTS
AMSH3 Is a Major Deubiquitinating Enzyme in Arabidopsis
The Arabidopsis genome encodes three hitherto uncharacterized proteins with homology to the MPN+ domain of Hs-AMSH and Hs-AMSH-LP, designated AMSH1, AMSH2, and AMSH3 (see Supplemental Figure 1 online) (Maytal-Kivity et al., 2002; Tsang et al., 2006; Hurley and Yang, 2008). AMSH1 and AMSH3, but not AMSH2, share homology with their human counterparts also in the N-terminal MIT (microtubule interacting and trafficking molecule) domain, but all three plant AMSH proteins lack domains of functional importance for Hs-AMSH, namely, a bipartite nuclear localization signal (NLS), a clathrin binding site, and a STAM binding motif (SBM) (see Supplemental Figure 1 online) (Kikuchi et al., 2003; Nakamura et al., 2006).
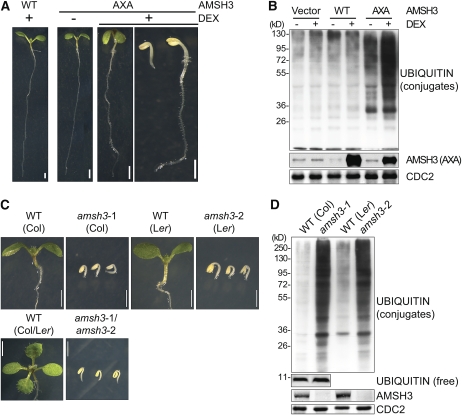
To examine the biological function of the Arabidopsis AMSH proteins, we expressed wild-type or enzymatically inactive variants of AMSH1, AMSH2, and AMSH3 with an active site AXA mutation in Arabidopsis using a dexamethasone (DEX)-inducible system (Aoyama and Chua, 1997; Verma et al., 2002; Gusmaroli et al., 2004). We subsequently found that specifically the expression of AMSH3-AXA induced a seedling growth arrest that was also accompanied by a strong accumulation of ubiquitin conjugates (Figures 1A and 1B). We then also found that two independent amsh3 null alleles, amsh3-1 and amsh3-2, as well as an amsh3-1/amsh3-2 trans-heterozygote displayed the same morphological and molecular phenotype as did the most severe DEX-induced AMSH3-AXA seedlings (Figures 1C and 1D). We thus concluded that AMSH3 is a major DUB in Arabidopsis and that its function is necessary for seedling development. Since the inducible expression of the enzymatically inactive AMSH3-AXA was sufficient to generate the amsh3 null mutant phenotype, we further concluded that the loss of DUB activity is the molecular cause for the phenotype observed in the amsh3 mutants. Since it had been previously reported that the phenotype of a specific DUB mutant from Saccharomyces cerevisiae may be caused by the depletion of free ubiquitin (Springael et al., 1999; Swaminathan et al., 1999), we examined free ubiquitin levels in the amsh3-1 mutant allele. Since we found comparable levels of free ubiquitin in the wild type and amsh3, we excluded the possibility that free ubiquitin levels are limiting in the mutant (Figure 1D).
Figure 1.
amsh3 Mutants Are Seedling Lethal and Accumulate Ubiquitin Conjugates.
(A) and (B) Phenotypes (A) and immunoblots (B) with an anti-ubiquitin P4D1 antibody from protein extracts of 7-d-old Arabidopsis seedlings overexpressing DEX-inducible AMSH3 (wild type [WT]) or AMSH3-AXA (AXA) grown in the absence (−) or presence (+) of 30 μM DEX. Seedlings with severe and weak phenotypes are shown for the DEX-treated AMSH3-AXA seedlings, which in strongly expressing transgenic lines represent up to 35 and 65% of the seedlings, respectively. CDC2 is a loading control. Bars = 1 mm.
(C) and (D) Phenotypes (C) and immunoblots (D) with an anti-ubiquitin P4D1 antibody from protein extracts of 7-d-old seedlings of amsh3-1, amsh3-2, and their trans-heterozygotes in comparison to wild-type seedlings of the same age. Bars = 1 mm.
[See online article for color version of this figure.]
AMSH3 Hydrolyzes K48- and K63-Linked Ubiquitin Chains
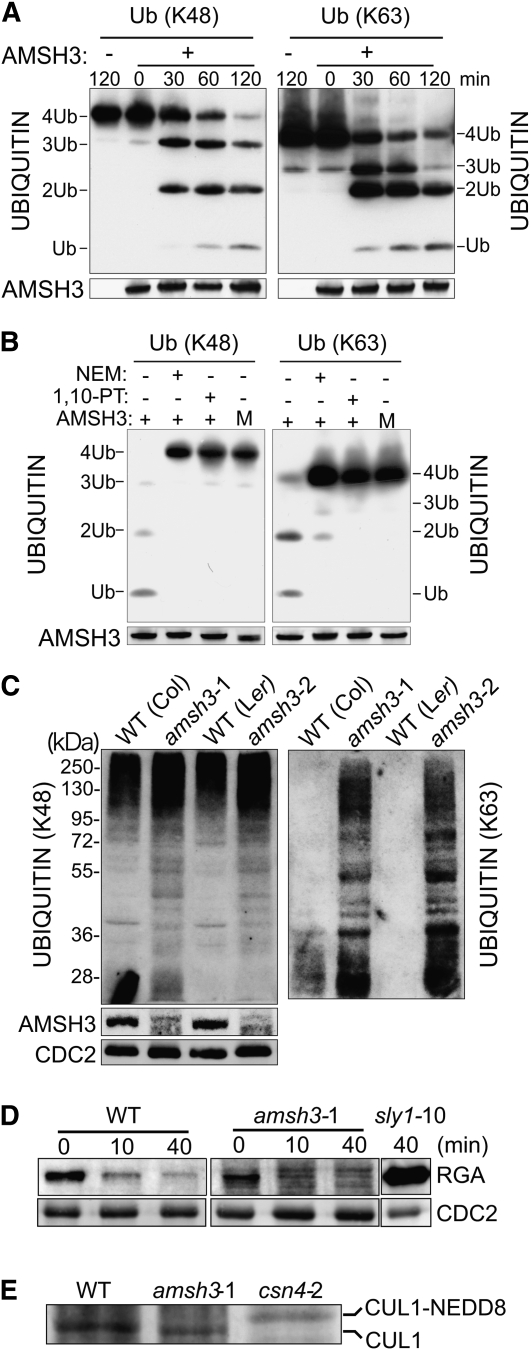
To determine the substrate specificity of AMSH3, we tested the DUB activity of recombinant AMSH3 and AMSH3-AXA toward K48- and K63-linked ubiquitin chains. In these experiments, we found AMSH3 but not AMSH3-AXA to efficiently hydrolyze both chain types in vitro (Figures 2A and 2B). Furthermore, we found AMSH3 to be able to remove ubiquitin from an in vitro polyubiquitinated T7-Sic1PYp substrate (see Supplemental Figure 2A online) (Saeki et al., 2005). Using K48- and K63-ubiquitin chain-specific antibodies (Newton et al., 2008), we then showed that both chain types accumulate in amsh3 mutants in vivo (Figure 2C). At the same time, we noticed that the two antibodies detect high and low/medium molecular weight ubiquitin conjugates, respectively.
Figure 2.
AMSH3 Hydrolyzes K48- and K63-Linked Ubiquitin Chains.
(A) In vitro deubiquitination assay with tetrameric K48- (left panel) or K63-linked (right panel) ubiquitin chains. WT, wild type.
(B) In vitro deubiquitination assay in the absence and presence of 5 mM NEM and 1 mM 1,10-PT. M, AMSH3-AXA mutant variant.
(C) Immunoblots of total protein extracts from 7-d-old amsh3-1 and amsh3-2 mutants in comparison to their respective wild types using antibodies specific for K48- and K63-linked ubiquitin chains (left and right panels, respectively).
(D) Immunoblots of total protein extracts (30 μg) from 7-d-old wild-type (Col) and amsh3-1 mutant seedlings that had been treated with 100 μM GA3 for the times indicated. The sly1-10 mutant extract (6 μg) serves as a positive control.
(E) Immunoblots of total protein extracts from 7-d-old wild type (Col), amsh3-1, and csn4-1 mutant seedlings. Neddylation results in a size shift of CUL1 (CUL1-NEDD8).
Since the accumulation of ubiquitinated proteins is also a phenotype of 26S proteasome mutants (Kurepa et al., 2008; Book et al., 2009), we were interested in testing proteasomal degradation in amsh3. We found, however, that the gibberellic acid–induced degradation of REPRESSOR-OF-ga1-3 (RGA), an established proteasomal substrate (McGinnis et al., 2003), is not altered in amsh3 mutants (Figure 2D). We also tested whether amsh3 mutants are defective in the deconjugation of the ubiquitin-related protein NEDD8 by comparing the abundance of unneddylated and neddylated CULLIN1 (CUL1) between the wild type and amsh3. In contrast with mutants of CSN, such as csn4, in which neddylated CUL1 accumulates due to a destabilization of CSN (Dohmann et al., 2008), we detected no change in the abundance of neddylated and unneddylated CUL1 in amsh3 (Figure 2E). Finally, we found that AMSH3 is unable to hydrolyze chains of SUMO protein, another ubiquitin-related protein (see Supplemental Figure 2B online). In summary, we conclude that AMSH3 is a K48- and K63-ubiquitin chain DUB that is not implicated in protein degradation by the 26S proteasome, deneddylation, or desumoylation.
Loss of AMSH3 Impairs Vacuole Formation and Vacuolar Transport
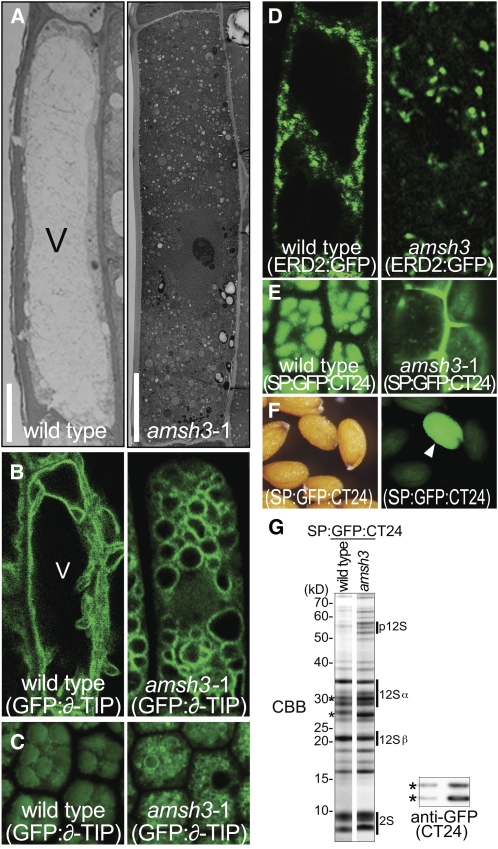
The seedling growth arrest phenotype of amsh3 mutants suggested that these mutants are defective in a major cellular mechanism. By transmission electron microscopy (TEM) of ultrathin sections of amsh3 mutant seedlings, we observed that the central lytic vacuole, which in differentiated epidermis cells of wild-type roots occupies most of the intracellular space, is absent in amsh3 mutants (Figure 3A). Instead, the cytosol of amsh3 mutants is filled with numerous small round structures. The lack of a central vacuole is the only apparent defect in amsh3 mutants since our further analyses of cellular organelles by TEM or using green fluorescent protein (GFP) marker lines (Geldner et al., 2009) did not reveal significant changes in their morphology, number, size, or shape (see Supplemental Figures 3A and 3B online).
Figure 3.
amsh3 Mutants Lack Large Central Vacuoles and Are Defective in Protein Transport.
(A) Transmission electron micrographs of root epidermis cells of wild-type and amsh3 mutant seedlings after high-pressure freezing, freeze-substitution, and epon embedding. Wild-type and amsh3 mutant seedlings were grown under continuous light for 2 and 5 d, respectively, to compare seedlings of the same developmental stage. V, vacuole. Bars = 5 μm (wild type) and 10 μM (amsh3).
(B) Confocal microscopy images of the vacuolar membrane marker GFP:y-TIP in wild-type (2-d-old) and amsh3 (5-d-old) mutant cells.
(C) Confocal microscopy images of the vacuolar membrane marker GFP:y-TIP and protein storage vacuole autofluorescence in dissected walking stick stage embryos of the wild type and amsh3. Note that the amsh3 mutant has smaller protein storage vacuoles than the wild type.
(D) Confocal microscopy images of ERD2:GFP in wild-type (2-d-old) and amsh3 (5-d-old) mutant cells.
(E) SP:GFP:CT24 in walking stick stage embryos of the wild type and amsh mutants.
(F) Light (left) and fluorescence (right) microscopy images of an amsh3-1/AMSH3 segregating population carrying the vacuolar cargo marker SP:GFP:CT24. Arrowhead indicates the amsh3 mutant seed.
(G) Coomassie blue–stained (CBB) SDS-PAGE gel of total protein extracts of wild type and amsh3 mutants carrying the vacuolar cargo marker SP:GFP:CT24 (left panel). p12S, 12S globulin precursors; 12Sα, 12S globulin α-subunits; 12Sβ, 12S globulin β-subunits; 2S, 2S albumin. Immunoblot using anti-GFP of the same extracts to detect GFP:CT24 (right panel, asterisks).
To gain insight into the fate of the central vacuole in amsh3 mutants, we examined amsh3 cells with regard to the distribution of the vacuolar membrane marker GFP:y-TONOPLAST INTRINSIC PROTEIN (GFP:y-TIP) (Cutler et al., 2000). In contrast with the wild type in which GFP:y-TIP outlines the dynamic vacuolar membrane structure, we found GFP:y-TIP to reside in the membranes of numerous small round structures in amsh3 mutant cells (Figure 3B). We thus concluded that they are small vacuoles that form as a consequence of impaired vacuole biogenesis.
A prominent mutant in plant vacuole biogenesis is vacuoleless1 (vcl1). vcl1 mutants fail to form a vacuole in early embryos and also have defects in cytokinesis (Rojo et al., 2001). Since vcl1 is an embryo-lethal mutant but amsh3 is seedling lethal, we were interested in examining the cellular phenotypes of amsh3 during embryogenesis. Interestingly, none of our microscopy analyses provided evidence for incomplete or abnormal cell divisions, and we also found no evidence for vacuole biogenesis defects in early stages of embryo development (see Supplemental Figure 3C online). We noticed, however, that 28% (n = 32) of late stage embryos (walking stick stage) segregating from an amsh3/AMSH3 parent line have small fragmented protein storage vacuoles (Figure 3C). This observation suggests that AMSH3 function becomes critical for vacuole formation at late stages of embryo development.
The vacuole formation defect of amsh3 mutants may be the consequence of impaired transport from the endoplasmic reticulum (ER) to the vacuole. The first step of this transport is from the ER to the Golgi apparatus via COPII vesicles. To examine the functionality of the COPII pathway, we made use of a GFP reporter of ENDOPLASMIC RETICULUM RETENTION DEFECTIVE2 (ERD2:GFP), which accumulates in the ER when COPII is dysfunctional (Takeuchi et al., 2000). Since we did not observe this accumulation of ERD2:GFP in the ER of the amsh3 mutant, we conclude that the ER-to-Golgi transport via the COPII vesicle transport pathway is not defective in the amsh3 mutant (Figure 3D).
We then examined protein transport from the Golgi to the vacuole. Several studies had shown that the loss of vacuole formation or defects in vacuolar transport cause the mis-sorting of vacuolar cargo from the trans-Golgi network to the intercellular space via a default secretion pathway (Rojo et al., 2001; Fuji et al., 2007; Ebine et al., 2008). We therefore monitored the transport of the protein storage vacuole cargo SP:GFP:CT24 in amsh3 mutant cells (Nishizawa et al., 2003). Consistent with the notion that vacuolar transport is impaired in amsh3 mutants, we found the GFP marker to accumulate in the intercellular space and to some extent also in smaller cytoplasmic structures of amsh3 mutant embryos (Figure 3E).
The secretion of the SP:GFP:CT24 marker in amsh3 allowed us to identify homozygous amsh3 mutants based on their fluorescence at the seed stage (Figure 3F). We used this phenotype to isolate amsh3 mutant seeds and to then compare their storage protein profile to that of the wild type. Here, we detected additional bands in amsh3, which according to their pattern and molecular weight correspond to the 12S globulin propeptide (p12S), indicating that amsh3 mutants are partially impaired in 12S globulin processing (Figure 3G, top panel). We also observed that the GFP marker accumulates in the amsh3 mutant (Figure 3G, bottom panel), a feature that was also observed in the vacuolar sorting receptor1 (vsr1) mutant (Fuji et al., 2007). However, in contrast with vsr1, which had been shown to accumulate exclusively p2S albumin and p12S globulin propeptides, propeptide accumulation in amsh3 mutants was comparatively moderate, possibly reflecting the fact that AMSH3 function becomes critical when protein storage vacuole filling is largely completed (Fuji et al., 2007; Craddock et al., 2008).
amsh3 Mutants Accumulate Autophagosomes
In addition to the loss of the central lytic vacuole, our TEM analysis also revealed that amsh3 mutants accumulate large numbers of vesicles, many of which could be identified as autophagosomes based on their double- or multimembrane structure (Figures 4A and 4B). Further support for the accumulation of autophagosomes in the amsh3 mutants came from the observation that the mutants have increased ATG8e levels (Figure 4C). ATG8 proteins are components of autophagosomes, and their accumulation correlates with the number of autophagosomes present in the cell (Kirisako et al., 1999; Thompson et al., 2005). Since autophagosomes are known to fuse with the vacuole in order to degrade and recycle their cytoplasmic contents, their accumulation could be indicative for defects in membrane fusion or, alternatively, represent a starvation response as a consequence of vacuolar malfunction (Kim and Klionsky, 2000).
Figure 4.
amsh3 Mutants Accumulate Autophagosomes.
(A) and (B) TEM image of amsh3 root epidermis cells after high-pressure freezing, freeze-substitution, and epon bedding. Arrowheads indicate autophagosomes. G, Golgi. Bars = 0.5 μm.
(C) Immunoblots of total protein extracts from wild-type and amsh3 seedlings using anti-ATG8e and anti-CDC2 (loading control) antibodies.
AMSH3 Is Required for Efficient Endocytosis
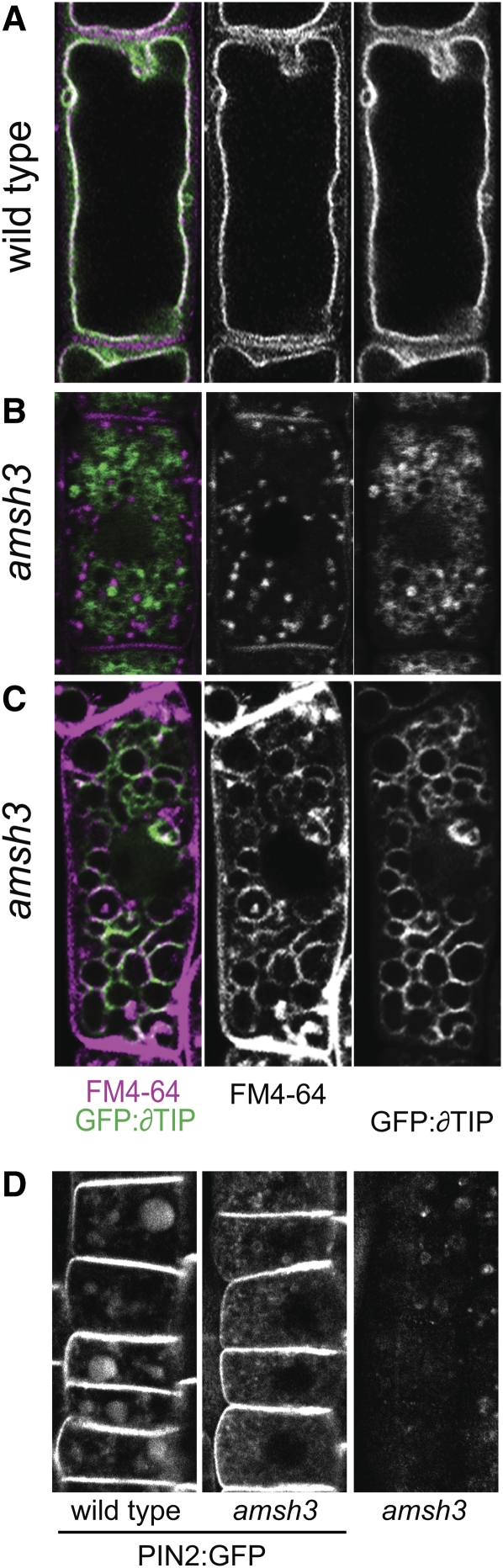
Since the vacuole is also the target organelle of endocytosis, we wanted to examine whether this transport route is functional in amsh3 mutants. To this end, we examined the endocytosis of the lipophilic dye FM4-64 and its transport to the vacuolar membrane. In wild-type cells, FM4-64 was typically endocytosed to detectable levels within 20 min and accumulated in the vacuolar membrane after 5 h (Figure 5A). Whereas we found the rate of FM4-64 uptake to be indistinguishable between wild-type and amsh3 mutant cells, we noticed differences in the intracellular transport between the two genotypes. The majority of amsh3 cells (83%) accumulated FM4-64 predominantly in smaller vesicles and, even after prolonged incubation, FM4-64 was not delivered to the vacuolar structures of the mutant (Figure 5B). In a minor fraction of amsh3 cells (17%), however, FM4-64 was transported within 5 h to the ϖ-TIP:GFP-positive vacuolar structures (Figure 5C). We thus concluded that AMSH3 is required for efficient transport from the plasma membrane to the vacuole or for membrane fusion events during endocytosis.
Figure 5.
amsh3 Mutants Have Defects in the Endocytosis Pathway.
(A) to (C) Confocal microscopy images of GFP:y-TIP (right panels, green in the left panels) and FM4-64 staining (middle panels, purple in the left panels) after 5 h in the wild type (A) and in the amsh3 mutant ([B] and [C]). Representative images of the two types of targeting observed in amsh3 are shown in (B) and (C), respectively. In most amsh3 mutants, FM4-64 remains in vesicles (85%; n = 18) and is not transported to the GFP:y-TIP stained structures.
(D) Confocal images of PIN2:GFP in wild-type (left) and amsh3 mutant (middle) cells. Vacuole-localized PIN2:GFP signals are visible in the wild type but not in the amsh3 mutant. Because amsh3 seedlings exhibit background fluorescence, a control image of an amsh3 seedling without PIN2:GFP is shown on the right.
We also examined the endocytosis of the integral membrane protein and auxin efflux facilitator PIN-FORMED2 (PIN2). The polarly localized PIN2 is constitutively endocytosed, and vacuolar PIN2:GFP can be visualized in the vacuoles of seedlings that have been grown in the dark for several hours (Figure 5D) (Laxmi et al., 2008). When we examined PIN2:GFP endocytosis and vacuolar transport in amsh3 seedlings, we observed vacuolar GFP signals in the wild type but no accumulation of PIN2 in vacuolar structures of the amsh3 mutant (Figure 5D). We thus concluded that PIN2:GFP cannot be efficiently targeted to the vacuolar structures in amsh3 mutants, indicating that the intracellular transport of PIN2:GFP from the membrane to the vacuole is disturbed in amsh3. At the same time, polar PIN2:GFP targeting was not affected in amsh3 mutant cells, suggesting that the transport of PIN2 to the plasma membrane and possibly also its recycling are not defective in the mutant (Figure 5D).
A Role for AMSH3 in Intracellular Transport
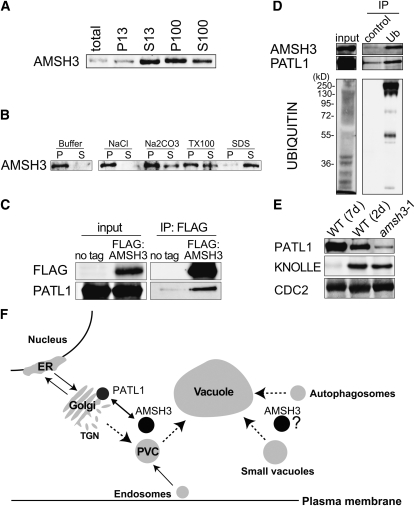
The MPN+ domain proteins RPN11 and CSN5 are part of the 26S proteasome and CSN protein complex, respectively. To examine whether AMSH3 associates with a stable protein complex in vivo, we performed size exclusion chromatography of AMSH3 and found that the vast majority of AMSH3 (61 kD) elutes with a molecular mass of <158 kD, suggesting that AMSH3 is not stably associated with a high molecular mass protein complex (see Supplemental Figure 4A online). Cellular fractionation using ultracentrifugation revealed that AMSH3 is found in the soluble (S100) and the membrane (P100) fraction (Figure 6A). To examine the biochemical nature of the interaction of AMSH3 with membranes, we treated the membrane fraction (P100) with different buffers and found that AMSH3 is partially solubilized by Na2CO3 or Triton X-100, suggesting that it is not an integral membrane protein (Figure 6B).
Figure 6.
AMSH3 Is a Membrane-Associated Protein That Interacts with PATL1.
(A) Immunoblots of total protein extracts after differential centrifugation. The low-speed pellet (P13) and supernatant (S13) and the high-speed pellet (P100) and supernatant (S100) were subjected to immunoblotting with anti-AMSH3 antibody.
(B) The P100 fraction obtained in (A) was incubated with the buffers containing one of 1 M NaCl, 0.1 M Na2CO3, 1% Triton X-100, or 1% SDS, and solubilization of AMSH3 was analyzed using anti-AMSH3. P, pellet; S, supernatant.
(C) Anti-FLAG immunoprecipitation from 7-d-old Arabidopsis wild-type seedlings (no tag) or seedlings expressing FLAG:AMSH3. Total extracts (input) and immunoprecipitates (IP) were subjected to immunoblot using anti-FLAG and anti-PATL1 antibodies.
(D) Ubiquitin immunoprecipitation using an anti-ubiquitin P4D1-conjugated agarose (Ub) or a control ProteinA/G-agarose immunoprecipitation (control) from 7-d-old seedlings expressing AMSH3-AXA. Immunoprecipitates were subjected to immunoblot using anti-AMSH3, anti-PATL1, and anti-ubiquitin antibodies.
(E) Immunoblot with an anti-PATL1 antibody on total extracts of amsh3-1 in comparison with 7- and 2-d-old wild-type seedlings. Note that the amount of PATL1 is reduced in the amsh3 mutant, whereas the amount of the cell plate protein KNOLLE is unaltered.
(F) Model of AMSH3 interactions. Protein–protein interaction data suggest a role for AMSH3 in intracellular transport involving the SEC14-homolog PATL1. AMSH3 dysfunction results in the accumulation of ubiquitinated proteins and defects in intracellular vesicle transport. Dotted lines mark the pathways for which AMSH3 function may be required. PVC, prevacuolar compartment; TGN, trans-Golgi network.
To understand the cellular function of AMSH3, we next sought to find AMSH3 interaction partners. To this end, we immunoprecipitated FLAG:AMSH3 from transgenic plants and analyzed the coimmunoprecipitated proteins by liquid chromatography–tandem mass spectrometry (LC-MS/MS). Consistent with our observation that AMSH3 is not part of a protein complex, the FLAG:AMSH3 immunoprecipitation yielded a limited number of interaction partners, which appeared to be bound with comparatively low affinity (see Supplemental Table 1 and Supplemental Figure 4B online). In view of the apparent trafficking defect of the amsh3 mutant, we found it particularly noteworthy that several AMSH3 interactors are proteins with a reported or expected function in intracellular transport processes: a predicted clathrin binding protein (Rothman and Schmid, 1986); two HSC70 proteins, which are involved in clathrin uncoating (Xing et al., 2010); and PATELLIN1 (PATL1) and PATL2, two Arabidopsis proteins related to yeast SEC14 (Peterman et al., 2004) (see Supplemental Table 1 online). PATL1 has been implicated in transport processes in Arabidopsis, notably, cell plate formation during cytokinesis, and recently has also been identified independently in two plant ubiquitinome studies (Peterman et al., 2004; Igawa et al., 2009; Saracco et al., 2009). With a PATL1-specific antibody, we could subsequently confirm the AMSH3-PATL1 coimmunoprecipitation (Figure 6C). PATL1 and AMSH3 could also be immunoprecipitated with ubiquitin, although we could not find evidence for a direct ubiquitination of PATL1 (Figure 6D). Interestingly, we found PATL1 to be less abundant in amsh3 than in wild-type seedlings of the same developmental stage, suggesting an interrelationship between the two proteins (Figure 6E).
DISCUSSION
In this study, we analyzed the Arabidopsis DUB AMSH3, which hydrolyzes K48- and K63-linked ubiquitin chains in vitro. Using ubiquitin chain-type specific antibodies, we further show that amsh3 mutants accumulate protein conjugates with both ubiquitin chain topologies in vivo. Hs-AMSH and Hs-AMSH-LP had previously been reported to hydrolyze specifically K63-linked ubiquitin chains, and in the case of Hs-AMSH-LP, this finding is supported by a structural analysis, which had identified several amino acids in Hs-AMSH-LP that participate in the binding of K63-linked diubiquitin (Sato et al., 2008). Interestingly, we found that most relevant residues are also conserved in Arabidopsis AMSH3, thus suggesting that AMSH3 and Hs-AMSH employ a similar mechanism for the hydrolysis of K63-linked ubiquitin chains (see Supplemental Figure 1 online). However, the identity of the AMSH3 residues that may participate in the binding of K48-linked chains cannot be predicted based on the present data; thus, the apparent difference between the substrate specificities of human AMSH and AMSH3 remains to be elucidated.
AMSH3 lacks three functional domains that have been reported for Hs-AMSH, namely, an NLS, an SBM, and a clathrin binding site. In agreement with the absence of an NLS in AMSH3, and in contrast with Hs-AMSH, we found AMSH3 to localize exclusively to the cytoplasm. In the context of the absence of the SBM, it is also noteworthy that there are no apparent homologs of the ESCRT-0 subunits STAM and HRS in plants. Whereas plant proteins have been identified as predicted functional homologs of all ESCRT-I, -II, and -III subunits, plant ESCRT-0 subunits share only very limited homology to their yeast and human counterparts (Winter and Hauser, 2006). In contrast with the human AMSH, which had been reported to interact with ESCRT subunits, we did not find any ESCRT subunit interactions following AMSH3 immunoprecipitation, suggesting that AMSH3 is not stably associated with ESCRT complexes in planta.
Vacuoles are organelles that occupy most of the cellular volume in plant and yeast cells, and they are indispensable for growth in plants but not in yeast (Rojo et al., 2001). In contrast with yeast, in which vacuolar biogenesis is comparatively well understood, little is known about vacuolar biogenesis in plants. Many of the >40 vacuolar protein sorting proteins required for vacuolar protein sorting in yeast participate in the transport to the vacuole, either through the Golgi or by endocytosis (Bankaitis et al., 1986; Rothman and Stevens, 1986; Raymond et al., 1992). However, as yet, no DUB has been shown to be required for vacuolar biogenesis in this organism. In turn, apparent VPS protein homologs, such as VPS16 (VCL1), VPS29, VPS35, and VPS45, have been implicated in vacuolar biogenesis in plants, particularly in Arabidopsis, suggesting that there are significant parallels between yeast and plant vacuolar biogenesis (Rojo et al., 2001; Shimada et al., 2006; Sanmartin et al., 2007; Ebine et al., 2008; Yamazaki et al., 2008; Zouhar et al., 2009). Interestingly, AMSH3 itself does not have a recognizable homolog in S. cerevisiae. The yeast mutant of the DUB Doa4p, which has a role in the deubiquitination of endocytosed cargo and is homologous to human UBPY, has normal vacuolar morphology (Reggiori and Pelham, 2001). Based on our data, we propose that vacuole biogenesis and possibly membrane fusion events required for vacuole biogenesis require deubiquitination by AMSH3 in planta. Future experiments are needed to reveal the identity of the ubiquitin conjugates whose deubiquitination is essential for proper cellular trafficking and vacuole biogenesis.
METHODS
Biological Material
All experiments were performed with Arabidopsis thaliana. The T-DNA insertion lines of AMSH3, designated amsh3-1 (WiscDsLox412D09; Columbia ecotype, Col-0) and amsh3-2 (CSHL_GT9764; Landsberg erecta ecotype) were obtained from the ABRC at Ohio State University and from Cold Spring Harbor Laboratory, respectively. The mutants were backcrossed two times to the respective wild types. The amsh3-1 allele was complemented by the transgenic construct 35S:HA:AMSH3.
sly1-10 and csn4-2 were previously described (McGinnis et al., 2003; Dohmann et al., 2008). Plant transformations were performed using the floral dip method (Clough and Bent, 1998). Seedlings were grown in continuous light on standard Murashige and Skoog growth medium (Duchefa Biochemie) supplemented with 1% sucrose; adult plants were grown on soil. When indicated, 30 μM DEX (Sigma-Aldrich) was added to the medium. The following previously published GFP marker lines were crossed into the amsh3-1 mutant: PIN2:GFP (Laxmi et al., 2008), SP:GFP:CT24 (Fuji et al., 2007), GFP:ϖ-TIP (Cutler et al., 2000), and Wave lines (Geldner et al., 2009).
Cloning Procedures
For GST:AMSH3, the AMSH3 open reading frame (ORF) was PCR amplified using primers EI13 and EI14, and the resulting fragment was cloned into the BamHI and SalI sites of pGEX-6-P1 (GE Healthcare). To obtain GST:AMSH3-AXA, AXA mutations were introduced with primers EI19 and EI20 using DpnI-based site-directed mutagenesis.
To yield pTA:AMSH and pTA:AMSH3-AXA, the wild-type and mutant AMSH3 ORFs were amplified with primers EI25 and EI26 using GST:AMSH3 and GST:AMSH-AXA as a template, respectively, and the resulting fragments were cloned into the XhoI and SpeI sites of pTA7002 (Aoyama and Chua, 1997). 35S:FLAG:AMSH3 and 35S:HA:AMSH3 were generated in pEarley202 (Earley et al., 2006) and pJawohl2B-3xHA-GW (I. Somssich, Cologne, Germany) using Gateway cloning (Invitrogen) using fragments amplified with EI7 and EI8 (for FLAG and HA) or EI7 and EI52 (for GFP) and Gateway adapter primers. To generate 35S:ERD2:GFP, the ERD2 coding sequence was inserted in frame using Gateway technology into 35S-GFP (Kan) (J. Parker, Cologne, Germany), with the ERD2 ORF amplified with primers EI165 and EI167. All primer sequences can be found in Supplemental Table 2 online.
DUB Assays
For deubiquitination assays, GST:AMSH3 and GST:AMSH3-AXA were expressed and purified from Escherichia coli Rosetta (DE3) (Merck Chemicals) using standard procedures. The purification was performed using Glutathione Sepharose 4B, and the GST moiety was cleaved with PreScission protease (GE Healthcare). Deubiquitination experiments were performed as described previously with slight modifications as listed below (McCullough et al., 2006). Either tetrameric K48- or K63-ubiquitin chains (125 ng; Boston Biochem) and 50 ng of recombinant AMSH3 or AMSH3-AXA were used in each reaction. The T7-Sic1PYp DUB assay was performed as previously described (Saeki et al., 2005). For inhibitor experiments, 5 mM NEM and 1 mM 1,10-PT were added. For desumoylation assays, 1.25 μg of either SUMO2 or SUMO3 chains (Boston Biochem) were mixed with 0.5 μg of recombinant AMSH3 protein and incubated for 150 min at 37°C.
Cell Biology
Seedlings were analyzed and photographed using a Leica MZ16 stereomicroscope with a PLAN-APO ×1 objective (Leica). Fluorescence microscopy for identifying SP:GFP:CT24-containing seeds was performed using an Olympus SZX12 fluorescence microscope with a digital color camera XC10 and a DFPLAPO 1.2xPF2 objective (Olympus) or a FV1000/IX81 laser scanning confocal microscope with a UPlanSApo ×60/1.20 objective (Olympus). Images were obtained and processed using the Cell-F or FluoView software. For FM4-64 staining, seedlings were incubated for 10 min in 1 μM FM4-64 (Invitrogen) and then transferred to liquid Murashige and Skoog medium and incubated for 5 h at 26°C in the dark. For ultrastructural analysis, high-pressure frozen, freeze-substituted, and epon-embedded samples were used (Reichardt et al., 2007).
Differential Centrifugation and Solubilization
Total protein extracts were prepared in extraction buffer from 2.5 g (fresh weight) of 7-d-old seedlings. Differential centrifugation was performed in a Sorvall WX 80 ultracentrifuge (Thermo) as previously described (Tamura et al., 2005). For the solubilization experiment, the P100 pellet was incubated in buffers containing 0.1 M Na2CO3, 1M NaCl, or 1% Triton X-100 (30 min on ice) or 1% SDS (30 min at 20°C).
Protein Blots and Antibodies
SDS-PAGE and immunoblots were performed following standard methods using SuperSignal West Femto Maximum Sensitivity Substrate (Thermo Fisher Scientific) and ECL Western Blotting Detection Reagents (GE Healthcare). An anti-AMSH3 antibody was raised in rabbit (Eurogentec) against the purified full-length protein expressed and purified from a clone obtained by insertion of an AMSH3 cDNA amplified with the primers EI7 and EI8 and cloned into pDEST17 using Gateway technology (Invitrogen). The serum was purified with a Hi-Trap IgG column (GE Healthcare) and an AMSH3-loaded NHS-activated HP column (GE Healthcare). The anti-AMSH3 antibody was used in a dilution of 1:1500. In addition, the following antibodies and dilutions were used in this study: anti-FLAG M2 (1:2000; Sigma-Aldrich), anti-GST (1:2000; GE Healthcare), anti-HA-HRP conjugated (1:2500; Roche), anti-GFP (1:5000; Roche), anti-Ub (K48) (1:2000; Millipore), anti-Ub (K63) (1:2000; Millipore), anti-Ub P4D1 (1:2500; Cell Signaling Technology), anti-CDC2 (1:5000; Santa Cruz Biotechnology), anti-T7-HRP (1:2000; Merck), anti-PATL1 (1:2500) (Peterman et al., 2004), anti-ATG8e (1:2000) (Thompson et al., 2005), anti-KNOLLE (1:3000) (Lauber et al., 1997), anti-CUL1 (1:2000) (Schwechheimer et al., 2002), and anti-RGA (1:2000) (Willige et al., 2007).
Immunoprecipitations
For immunoprecipitation, protein extracts were prepared from 500 μg of plant material (fresh weight) using a HomGen homogenizer (Schuett) and buffer A (50 mM Tris-HCl, pH 7.5, 100 mM NaCl, and 10% glycerol) supplemented with Complete protease inhibitor (Roche), 5 mM NEM, and 1 mM 1,10-PT. Subsequently, the total extract was centrifuged for 10 min at 4°C, and the supernatant was incubated with the 20 μL of either anti-Ubiquitin- (Santa Cruz Biotechnology) or anti-FLAG (Sigma-Aldrich) antibody-conjugated beads for 1 h. FLAG:AMSH3 was immunoprecipitated for LC-MS/MS analysis (Biqualys) from 8 g of 10-d-old seedlings. After centrifugation for 10 min at 4°C, the supernatant was mixed and incubated for preclearing with 120 μL Protein A/G Agarose (Santa Cruz Biotechnology). The supernatant was then incubated for 1 h at 4°C with 120 μL of anti-FLAG M2 Agarose (Sigma-Aldrich). After three washes in ice-cold buffer A, the FLAG immunoprecipitate was eluted with FLAG peptide (Sigma-Aldrich). The eluate was subjected to SDS-PAGE, and samples for LC-MS/MS were prepared as described elsewhere (http://biochemistry.wur.nl/Biochem). Samples were separated using an nLC-LTQ-Orbitrap MS-MS at Biqualys, and the data were analyzed using the Bioworks software (Thermo Fisher).
Gel Filtration
Total plant protein extract (2.5 mg) was loaded onto a Superose6 gel filtration column (GE Healthcare) attached to an ÄKTA FPLC system (GE Healthcare). Gel filtration chromatography was performed as previously described (Isono et al., 2004), and protein standards from a High Molecular Weight Calibration Kit (GE Healthcare) were used.
Accession Numbers
Sequence data from this article can be found in the Arabidopsis Genome Initiative database under the following accession numbers: AMSH1 (AT1G48790), AMSH2 (AT1G10600), AMSH3 (AT4G16144), ATG8e (AT2G45170), CDC2 (AT3G48750), CULLIN1 (AT4G02570), ERD2 (AT1G29330), KNOLLE (AT1G08560), PATL1 (AT1G72150), PIN2 (AT5G57090), RGA (AT2G01570), and y-TIP (AT3G16240).
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure 1. Alignment of AMSH1, AMSH2, and AMSH3 with Their Counterparts from Human and Drosophila melanogaster.
Supplemental Figure 2. AMSH3 Deconjugates Ubiquitin but Not SUMO Chains.
Supplemental Figure 3. The amsh3 Mutant Is Specifically Defective in Vacuole Formation.
Supplemental Figure 4. AMSH3 Is Not Incorporated into a Stable Complex.
Supplemental Table 1. Proteins Identified by Mass Spectrometry following the Immunoprecipitation of FLAG:AMSH3.
Supplemental Table 2. List of Primers Used in This Study.
Acknowledgments
We thank Björn C. Willige and Marie-Theres Hauser (Universität für Bodelkultur, Vienna, Austria) for critical comments on the manuscript and Natasha Raikhel (University of California, Riverside), Takashi Ueda (University of Tokyo), and Miyo Morita (Nara Institute of Science and Technology) for discussions. We acknowledge the receipt of antibodies and GFP marker lines from Kaye Peterman (Wellesley College, MA), Richard Vierstra (University of Wisconsin-Madison), and Shigeru Utsumi (Kyoto University, Japan). E.I. was supported by a fellowship of the Japan Society for the Promotion of Science. C.S. and Y.-D.S. are supported by grants from the Deutsche Forschungsgemeinschaft (SFB446 and SPP1365).
References
- Acconcia F., Sigismund S., Polo S. (2009). Ubiquitin in trafficking: The network at work. Exp. Cell Res. 315: 1610–1618 [DOI] [PubMed] [Google Scholar]
- Agromayor M., Martin-Serrano J. (2006). Interaction of AMSH with ESCRT-III and deubiquitination of endosomal cargo. J. Biol. Chem. 281: 23083–23091 [DOI] [PubMed] [Google Scholar]
- Ambroggio X.I., Rees D.C., Deshaies R.J. (2004). JAMM: A metalloprotease-like zinc site in the proteasome and signalosome. PLoS Biol. 2: E2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aoyama T., Chua N.H. (1997). A glucocorticoid-mediated transcriptional induction system in transgenic plants. Plant J. 11: 605–612 [DOI] [PubMed] [Google Scholar]
- Bankaitis V.A., Johnson L.M., Emr S.D. (1986). Isolation of yeast mutants defective in protein targeting to the vacuole. Proc. Natl. Acad. Sci. USA 83: 9075–9079 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Book A.J., Smalle J., Lee K.H., Yang P., Walker J.M., Casper S., Holmes J.H., Russo L.A., Buzzinotti Z.W., Jenik P.D., Vierstra R.D. (2009). The RPN5 subunit of the 26s proteasome is essential for gametogenesis, sporophyte development, and complex assembly in Arabidopsis. Plant Cell 21: 460–478 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Z.J., Sun L.J. (2009). Nonproteolytic functions of ubiquitin in cell signaling. Mol. Cell 33: 275–286 [DOI] [PubMed] [Google Scholar]
- Clague M.J., Urbe S. (2006). Endocytosis: The DUB version. Trends Cell Biol. 16: 551–559 [DOI] [PubMed] [Google Scholar]
- Clough S.J., Bent A.F. (1998). Floral dip: A simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 16: 735–743 [DOI] [PubMed] [Google Scholar]
- Cooper E.M., Cutcliffe C., Kristiansen T.Z., Pandey A., Pickart C.M., Cohen R.E. (2009). K63-specific deubiquitination by two JAMM/MPN+ complexes: BRISC-associated Brcc36 and proteasomal Poh1. EMBO J. 28: 621–631 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cope G.A., Suh G.S., Aravind L., Schwarz S.E., Zipursky S.L., Koonin E.V., Deshaies R.J. (2002). Role of predicted metalloprotease motif of Jab1/Csn5 in cleavage of Nedd8 from Cul1. Science 298: 608–611 [DOI] [PubMed] [Google Scholar]
- Craddock C.P., Hunter P.R., Szakacs E., Hinz G., Robinson D.G., Frigerio L. (2008). Lack of a vacuolar sorting receptor leads to non-specific missorting of soluble vacuolar proteins in Arabidopsis seeds. Traffic 9: 408–416 [DOI] [PubMed] [Google Scholar]
- Cutler S.R., Ehrhardt D.W., Griffitts J.S., Somerville C.R. (2000). Random GFP:cDNA fusions enable visualization of subcellular structures in cells of Arabidopsis at a high frequency. Proc. Natl. Acad. Sci. USA 97: 3718–3723 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dohmann E.M., Levesque M.P., Isono E., Schmid M., Schwechheimer C. (2008). Auxin responses in mutants of the Arabidopsis CONSTITUTIVE PHOTOMORPHOGENIC9 signalosome. Plant Physiol. 147: 1369–1379 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong Y., Hakimi M.A., Chen X., Kumaraswamy E., Cooch N.S., Godwin A.K., Shiekhattar R. (2003). Regulation of BRCC, a holoenzyme complex containing BRCA1 and BRCA2, by a signalosome-like subunit and its role in DNA repair. Mol. Cell 12: 1087–1099 [DOI] [PubMed] [Google Scholar]
- Earley K.W., Haag J.R., Pontes O., Opper K., Juehne T., Song K., Pikaard C.S. (2006). Gateway-compatible vectors for plant functional genomics and proteomics. Plant J. 45: 616–629 [DOI] [PubMed] [Google Scholar]
- Ebine K., Okatani Y., Uemura T., Goh T., Shoda K., Niihama M., Morita M.T., Spitzer C., Otegui M.S., Nakano A., Ueda T. (2008). A SNARE complex unique to seed plants is required for protein storage vacuole biogenesis and seed development of Arabidopsis thaliana. Plant Cell 20: 3006–3021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuji K., Shimada T., Takahashi H., Tamura K., Koumoto Y., Utsumi S., Nishizawa K., Maruyama N., Hara-Nishimura I. (2007). Arabidopsis vacuolar sorting mutants (green fluorescent seed) can be identified efficiently by secretion of vacuole-targeted green fluorescent protein in their seeds. Plant Cell 19: 597–609 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geldner N., Denervaud-Tendon V., Hyman D.L., Mayer U., Stierhof Y.-D., Chory J. (2009). Rapid, combinatorial analysis of membrane compartments in intact plants with a multicolor marker set. Plant J. 59: 169–178 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gusmaroli G., Feng S., Deng X.W. (2004). The Arabidopsis CSN5A and CSN5B subunits are present in distinct COP9 signalosome complexes, and mutations in their JAMM domains exhibit differential dominant negative effects on development. Plant Cell 16: 2984–3001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hershko A., Ciechanover A. (1998). The ubiquitin system. Annu. Rev. Biochem. 67: 425–479 [DOI] [PubMed] [Google Scholar]
- Hurley J.H., Yang D. (2008). MIT domainia. Dev. Cell 14: 6–8 [DOI] [PubMed] [Google Scholar]
- Igawa T., Fujiwara M., Takahashi H., Sawasaki T., Endo Y., Seki M., Shinozaki K., Fukao Y., Yanagawa Y. (2009). Isolation and identification of ubiquitin-related proteins from Arabidopsis seedlings. J. Exp. Bot. 60: 3067–3073 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishii N., Owada Y., Yamada M., Miura S., Murata K., Asao H., Kondo H., Sugamura K. (2001). Loss of neurons in the hippocampus and cerebral cortex of AMSH-deficient mice. Mol. Cell. Biol. 21: 8626–8637 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Isono E., Saeki Y., Yokosawa H., Toh-e A. (2004). Rpn7 Is required for the structural integrity of the 26 S proteasome of Saccharomyces cerevisiae. J. Biol. Chem. 279: 27168–27176 [DOI] [PubMed] [Google Scholar]
- Kerscher O., Felberbaum R., Hochstrasser M. (2006). Modification of proteins by ubiquitin and ubiquitin-like proteins. Annu. Rev. Cell Dev. Biol. 22: 159–180 [DOI] [PubMed] [Google Scholar]
- Kikuchi K., Ishii N., Asao H., Sugamura K. (2003). Identification of AMSH-LP containing a Jab1/MPN domain metalloenzyme motif. Biochem. Biophys. Res. Commun. 306: 637–643 [DOI] [PubMed] [Google Scholar]
- Kim J., Klionsky D.J. (2000). Autophagy, cytoplasm-to-vacuole targeting pathway, and pexophagy in yeast and mammalian cells. Annu. Rev. Biochem. 69: 303–342 [DOI] [PubMed] [Google Scholar]
- Kirisako T., Baba M., Ishihara N., Miyazawa K., Ohsumi M., Yoshimori T., Noda T., Ohsumi Y. (1999). Formation process of autophagosome is traced with Apg8/Aut7p in yeast. J. Cell Biol. 147: 435–446 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Komander D., Clague M.J., Urbe S. (2009). Breaking the chains: Structure and function of the deubiquitinases. Nat. Rev. Mol. Cell Biol. 10: 550–563 [DOI] [PubMed] [Google Scholar]
- Kurepa J., Toh E.A., Smalle J.A. (2008). 26S proteasome regulatory particle mutants have increased oxidative stress tolerance. Plant J. 53: 102–114 [DOI] [PubMed] [Google Scholar]
- Kyuuma M., Kikuchi K., Kojima K., Sugawara Y., Sato M., Mano N., Goto J., Takeshita T., Yamamoto A., Sugamura K., Tanaka N. (2007). AMSH, an ESCRT-III associated enzyme, deubiquitinates cargo on MVB/late endosomes. Cell Struct. Funct. 31: 159–172 [DOI] [PubMed] [Google Scholar]
- Lauber M.H., Waizenegger I., Steinmann T., Schwarz H., Mayer U., Hwang I., Lukowitz W., Jurgens G. (1997). The Arabidopsis KNOLLE protein is a cytokinesis-specific syntaxin. J. Cell Biol. 139: 1485–1493 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laxmi A., Pan J., Morsy M., Chen R. (2008). Light plays an essential role in intracellular distribution of auxin efflux carrier PIN2 in Arabidopsis thaliana. PLoS One 3: e1510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma Y.M., Boucrot E., Villén J., el Affar B., Gygi S.P., Göttlinger H.G., Kirchhausen T. (2007). Targeting of AMSH to endosomes is required for epidermal growth factor receptor degradation. J. Biol. Chem. 282: 9805–9812 [DOI] [PubMed] [Google Scholar]
- Maytal-Kivity V., Reis N., Hofmann K., Glickman M.H. (2002). MPN+, a putative catalytic motif found in a subset of MPN domain proteins from eukaryotes and prokaryotes, is critical for Rpn11 function. BMC Biochem. 3: 28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCullough J., Clague M.J., Urbe S. (2004). AMSH is an endosome-associated ubiquitin isopeptidase. J. Cell Biol. 166: 487–492 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCullough J., Row P.E., Lorenzo O., Doherty M., Beynon R., Clague M.J., Urbe S. (2006). Activation of the endosome-associated ubiquitin isopeptidase AMSH by STAM, a component of the multivesicular body-sorting machinery. Curr. Biol. 16: 160–165 [DOI] [PubMed] [Google Scholar]
- McGinnis K.M., Thomas S.G., Soule J.D., Strader L.C., Zale J.M., Sun T.P., Steber C.M. (2003). The Arabidopsis SLEEPY1 gene encodes a putative F-box subunit of an SCF E3 ubiquitin ligase. Plant Cell 15: 1120–1130 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mukhopadhyay D., Riezman H. (2007). Proteasome-independent functions of ubiquitin in endocytosis and signaling. Science 315: 201–205 [DOI] [PubMed] [Google Scholar]
- Nakamura M., Tanaka N., Kitamura N., Komada M. (2006). Clathrin anchors deubiquitinating enzymes, AMSH and AMSH-like protein, on early endosomes. Genes Cells 11: 593–606 [DOI] [PubMed] [Google Scholar]
- Newton K., et al. (2008). Ubiquitin chain editing revealed by polyubiquitin linkage-specific antibodies. Cell 134: 668–678 [DOI] [PubMed] [Google Scholar]
- Nishizawa K., Maruyama N., Satoh R., Fuchikami Y., Higasa T., Utsumi S. (2003). A C-terminal sequence of soybean beta-conglycinin alpha' subunit acts as a vacuolar sorting determinant in seed cells. Plant J. 34: 647–659 [DOI] [PubMed] [Google Scholar]
- Peterman T.K., Ohol Y.M., McReynolds L.J., Luna E.J. (2004). Patellin1, a novel Sec14-like protein, localizes to the cell plate and binds phosphoinositides. Plant Physiol. 136: 3080–3094 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raymond C.K., Howald-Stevenson I., Vater C.A., Stevens T.H. (1992). Morphological classification of the yeast vacuolar protein sorting mutants: Evidence for a prevacuolar compartment in class E vps mutants. Mol. Biol. Cell 3: 1389–1402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reggiori F., Pelham H.R. (2001). Sorting of proteins into multivesicular bodies: Ubiquitin-dependent and -independent targeting. EMBO J. 20: 5176–5186 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reichardt I., Stierhof Y.-D., Mayer U., Richter S., Schwarz H., Schumacher K., Jurgens G. (2007). Plant cytokinesis requires de novo secretory trafficking but not endocytosis. Curr. Biol. 17: 2047–2053 [DOI] [PubMed] [Google Scholar]
- Reyes-Turcu F.E., Ventii K.H., Wilkinson K.D. (2009). Regulation and cellular roles of ubiquitin-specific deubiquitinating enzymes. Annu. Rev. Biochem. 78: 363–397 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rojo E., Gillmor C.S., Kovaleva V., Somerville C.R., Raikhel N.V. (2001). VACUOLELESS1 is an essential gene required for vacuole formation and morphogenesis in Arabidopsis. Dev. Cell 1: 303–310 [DOI] [PubMed] [Google Scholar]
- Rothman J.E., Schmid S.L. (1986). Enzymatic recycling of clathrin from coated vesicles. Cell 46: 5–9 [DOI] [PubMed] [Google Scholar]
- Rothman J.H., Stevens T.H. (1986). Protein sorting in yeast: Mutants defective in vacuole biogenesis mislocalize vacuolar proteins into the late secretory pathway. Cell 47: 1041–1051 [DOI] [PubMed] [Google Scholar]
- Row P.E., Liu H., Hayes S., Welchman R., Charalabous P., Hofmann K., Clague M.J., Sanderson C.M., Urbe S. (2007). The MIT domain of UBPY constitutes a CHMP binding and endosomal localization signal required for efficient epidermal growth factor receptor degradation. J. Biol. Chem. 282: 30929–30937 [DOI] [PubMed] [Google Scholar]
- Saeki Y., Isono E., Toh E.A. (2005). Preparation of ubiquitinated substrates by the PY motif-insertion method for monitoring 26S proteasome activity. Methods Enzymol. 399: 215–227 [DOI] [PubMed] [Google Scholar]
- Sanmartin M., Ordonez A., Sohn E.J., Robert S., Sanchez-Serrano J.J., Surpin M.A., Raikhel N.V., Rojo E. (2007). Divergent functions of VTI12 and VTI11 in trafficking to storage and lytic vacuoles in Arabidopsis. Proc. Natl. Acad. Sci. USA 104: 3645–3650 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saracco S.A., Hansson M., Scalf M., Walker J.M., Smith L.M., Vierstra R.D. (2009). Tandem affinity purification and mass spectrometric analysis of ubiquitylated proteins in Arabidopsis. Plant J. 59: 344–358 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sato Y., Yoshikawa A., Yamagata A., Mimura H., Yamashita M., Ookata K., Nureki O., Iwai K., Komada M., Fukai S. (2008). Structural basis for specific cleavage of Lys 63-linked polyubiquitin chains. Nature 455: 358–362 [DOI] [PubMed] [Google Scholar]
- Schwechheimer C., Serino G., Deng X.W. (2002). Multiple ubiquitin ligase-mediated processes require COP9 signalosome and AXR1 function. Plant Cell 14: 2553–2563 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimada T., Koumoto Y., Li L., Yamazaki M., Kondo M., Nishimura M., Hara-Nishimura I. (2006). AtVPS29, a putative component of a retromer complex, is required for the efficient sorting of seed storage proteins. Plant Cell Physiol. 47: 1187–1194 [DOI] [PubMed] [Google Scholar]
- Springael J.Y., Galan J.M., Haguenauer-Tsapis R., Andre B. (1999). NH4+-induced down-regulation of the Saccharomyces cerevisiae Gap1p permease involves its ubiquitination with lysine-63-linked chains. J. Cell Sci. 112: 1375–1383 [DOI] [PubMed] [Google Scholar]
- Swaminathan S., Amerik A.Y., Hochstrasser M. (1999). The Doa4 deubiquitinating enzyme is required for ubiquitin homeostasis in yeast. Mol. Biol. Cell 10: 2583–2594 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takeuchi M., Ueda T., Sato K., Abe H., Nagata T., Nakano A. (2000). A dominant negative mutant of sar1 GTPase inhibits protein transport from the endoplasmic reticulum to the Golgi apparatus in tobacco and Arabidopsis cultured cells. Plant J. 23: 517–525 [DOI] [PubMed] [Google Scholar]
- Tamura K., Shimada T., Kondo M., Nishimura M., Hara-Nishimura I. (2005). KATAMARI1/MURUS3 is a novel golgi membrane protein that is required for endomembrane organization in Arabidopsis. Plant Cell 17: 1764–1776 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka N., Kaneko K., Asao H., Kasai H., Endo Y., Fujita T., Takeshita T., Sugamura K. (1999). Possible involvement of a novel STAM-associated molecule “AMSH” in intracellular signal transduction mediated by cytokines. J. Biol. Chem. 274: 19129–19135 [DOI] [PubMed] [Google Scholar]
- Thompson A.R., Doelling J.H., Suttangkakul A., Vierstra R.D. (2005). Autophagic nutrient recycling in Arabidopsis directed by the ATG8 and ATG12 conjugation pathways. Plant Physiol. 138: 2097–2110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsang H.T., Connell J.W., Brown S.E., Thompson A., Reid E., Sanderson C.M. (2006). A systematic analysis of human CHMP protein interactions: Additional MIT domain-containing proteins bind to multiple components of the human ESCRT III complex. Genomics 88: 333–346 [DOI] [PubMed] [Google Scholar]
- Verma R., Aravind L., Oania R., McDonald W.H., Yates J.R., III, Koonin E.V., Deshaies R.J. (2002). Role of Rpn11 metalloprotease in deubiquitination and degradation by the 26S proteasome. Science 298: 611–615 [DOI] [PubMed] [Google Scholar]
- Willige B.C., Ghosh S., Nill C., Zourelidou M., Dohmann E.M., Maier A., Schwechheimer C. (2007). The DELLA domain of GA INSENSITIVE mediates the interaction with the GA INSENSITIVE DWARF1A gibberellin receptor of Arabidopsis. Plant Cell 19: 1209–1220 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winter V., Hauser M.T. (2006). Exploring the ESCRTing machinery in eukaryotes. Trends Plant Sci. 11: 115–123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xing Y., Bocking T., Wolf M., Grigorieff N., Kirchhausen T., Harrison S.C. (2010). Structure of clathrin coat with bound Hsc70 and auxilin: Mechanism of Hsc70-facilitated disassembly. EMBO J. 29: 655–665 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamazaki M., Shimada T., Takahashi H., Tamura K., Kondo M., Nishimura M., Hara-Nishimura I. (2008). Arabidopsis VPS35, a retromer component, is required for vacuolar protein sorting and involved in plant growth and leaf senescence. Plant Cell Physiol. 49: 142–156 [DOI] [PubMed] [Google Scholar]
- Zouhar J., Rojo E., Bassham D.C. (2009). AtVPS45 is a positive regulator of the SYP41/SYP61/VTI12 SNARE complex involved in trafficking of vacuolar cargo. Plant Physiol. 149: 1668–1678 [DOI] [PMC free article] [PubMed] [Google Scholar]