Abstract
The epitype of a single gene or entire genome is determined by cis-linked differences in chromatin structure. I explore the hypothesis that “epitype and associated phenotypes evolve by gene duplication, divergence, and subfunctionalization” parallel to models for the evolution of genotype. This hypothesis is dissected by considering the relationship between epigenetic control and phenotype, the phylogenetic evidence that epitype evolves from ancestral genes following gene duplication, and the possible evolutionary rates of change for different epitypes. Initial supporting arguments for this hypothesis are discussed based on conserved patterns of nucleosome phasing, DNA methylation, and histone variant H2AZ deposition that appear to contribute to the inheritance of epitype in plants and animals. However, patterns of histone modification in recent segmental chromosome duplications are not well conserved. A continued experimental examination of the link between gene phylogeny and epitype and the evolution of epigenetically determined phenotypes is needed to further explore this hypothesis.
INTRODUCTION
Theodosius Dobzhansky stated that “Nothing in biology makes sense except in the light of evolution” (Dobzhansky, 1973). The concepts surrounding epigenetic control would be greatly enhanced if epigenetics could be placed in a full evolutionary context. Epigenetics is concerned with mitotically and/or meiotically heritable differences between cells that are not due to changes in DNA sequence (Haig, 2004). In the 50 years since the inception of epigenetics (Waddington, 1957; Nanney, 1958a), it has become evident that transient changes to chromatin structure are the primary mechanism behind epigenetic controls. David Nanney introduced the term epigenetic controls to describe the various inherited differences between genetically identical daughter cells (Nanney, 1958a). In genetics, genotype and phenotype describe the relationship between DNA sequence and the elaboration of that code as differences in gene expression as well as cell, tissue, organ, and organismal development. In the field of epigenetics, epitype and phenotype describe the parallel relationships between information in chromatin structure and its manifestation during development. Herein, the epitype of a gene or genome is defined as the sum of all cis-linked chromatin structures that distinguish it from naked DNA, including but not limited to nucleosome phasing, base methylation (e.g., 5-methylcytosine [5MeC]), various histone side chain modifications, and deposition of exceptional histone variants within nucleosomes. Cis-linked refers to those changes to chromatin structure in the chromosomal vicinity of a gene that might alter its expression, as distinct from trans-acting diffusible products produced from other genes acting on the gene in question. As outlined, Nanney's epigenetic controls would include cis-linked changes to epitype, trans-acting epigenetic effects, and maternally inherited differences between daughter cells that may give them distinct phenotypes (Nanney, 1958a, 1958b; Haig, 2004) and thus it is a broader concept than epitype. A gene's cis-linked chromatin structure, its epitype, is expected to potentiate, restrict, or otherwise regulate gene expression, thus playing a major role in directing phenotype. It is accepted that genotype and phenotype evolve together, with evolutionary forces acting principally on phenotype, but to some extent on DNA (e.g., GC composition and codon bias). It is essential to find evidence that epitype and associated phenotypes are also acted on by defined evolutionary mechanisms if we are to make evolutionary sense of epigenetic controls.
The hypothesis explored in this article is that epitype and associated phenotypes evolve by gene duplication, divergence, and subfunctionalization. This hypothesis stems from several questions. (1) What is the relationship between epigenetic control and developmental phenotype? (2) What is the relationship between epigenetic control and the evolution of organismal phenotype? (3) What is the phylogenetic evidence that epitype evolves from ancestral genes following gene duplication? (4) What are the evolutionary rates of change for different epitypes? Although all of these questions will remain only partially answered, each serves as a means of dissecting this hypothesis.
WHAT IS THE RELATIONSHIP BETWEEN EPIGENETIC CONTROL AND DEVELOPMENTAL PHENOTYPE?
Epigenetic controls are essential to metabolic, cell, tissue, organ, and organismal development. In yeast, cellular phenotypes, such as gene expression and mating type, are under strong epigenetic control (Thon and Friis, 1997). Early examples of multicellular phenotypes under epigenetic control include chromosome rearrangements in Drosophila melanogaster leading to eversporting displacements of eye color, later called the position variegation effect (Henikoff, 1979), and DNA rearrangement leading to variegation in seed coat color in maize (Zea mays; McClintock, 1956). X chromosome inactivation was viewed early on as an epigenetic phenomenon in mammals (Lyon, 1993). Following this pioneering work, it became clear that mammalian phenotypes as diverse as changes in gene expression, stem cell fate, and heart and limb development are at least partially determined by epigenetics (Grzeschik, 2002; Abbosh et al., 2006; Haaf, 2006; Haston et al., 2009; Mackem and Lewandoski, 2009). In plants, global gene expression, nutrient metabolism, disease resistance, as well as root, shoot, and floral development are all under epigenetic control (Yi and Richards, 2008; Zhang et al., 2008; Kandasamy et al., 2009; Meagher et al., 2009; Smith et al., 2010). The epigenetic control of cell and organ development is now the focus of hundreds of research laboratories. Moreover, it is believed that many disease- and cancer-related phenomena are due to epigenetic changes to chromatin structure that are transiently inherited by abnormal cells (Wallrath et al., 2008; Iacobuzio-Donahue, 2009). Hence, epigenetic control of normal and aberrant developmental phenotypes is widely accepted and has far-reaching scientific, agricultural, and medical significance. A long-term goal in both medicine and agriculture is the intervention and repair of damaged or suboptimal epigenetic controls, but first a better understanding is needed for how epitype is inherited, how it evolves, and the cause and effect relationships between epigenetic control and phenotype.
WHAT IS THE RELATIONSHIP BETWEEN EPIGENETIC CONTROL AND THE EVOLUTION OF ORGANISMAL PHENOTYPE?
Alan Wilson, a pioneer of research on the molecular clock, observed that the rates of DNA mutation and the corresponding rates of change in protein sequences are not rapid enough to account for the rapid anatomical evolution observed among mammals or birds (Wilson et al., 1974; Wyles et al., 1983). The quandary is particularly obvious in comparisons between chimpanzees and humans, where there are very few differences in protein sequence (King and Wilson, 1975). Wilson used protein sequence and quantitative immunological cross reactivity data on a large number of proteins from many species to support this view. He and his colleagues suggested that the relatively high rate of genome rearrangement and associated changes in gene regulation in mammals might account for the rapid changes to gene regulation necessary to affect rapid morphological evolution, whereas some amphibians, such as frogs, show little chromosomal rearrangement and slow morphological evolution (Wilson et al., 1974; King and Wilson, 1975; Wyles et al., 1983).
The problem of explaining rapid morphological evolution relative to sequence evolution becomes even more acute with the recent awareness that complete genome sequence data from diverse mammals reveal a common set of ∼35,000 genes. There appear to be only small differences in gene number due to gene duplication and loss and relatively few bona fide novel gene sequences (Pennacchio, 2003; Siepel et al., 2007). Similarly, in spite of the extreme anatomical differences between monocots such as rice (Oryza sativa) and eudicots such as Arabidopsis thaliana and their common ancestry more than 140 million years (MY) ago, comparisons of these distant angiosperm genomes suggest they too contain similar overall gene composition, intron-exon structures, and ∼35,000 genes (Bennetzen et al., 2004; Tripathi and Sowdhamini, 2006; Schnable et al., 2009).
Interesting mechanistic arguments addressing Alan Wilson's dilemma of explaining rapid anatomical evolution among some groups of higher organisms have been presented recently. First, it has become accepted that rapid rates of change to cis-regulatory elements are essential to rapid morphological evolution (Hoekstra and Coyne, 2007; Carroll, 2008). Second, West et al. (2007) demonstrate that the levels of half of all easily detected Arabidopsis transcripts exhibit heritable transcript level variation that is controlled by quantitative genetic trait loci. Thus, it may be proposed that small differences in the sequences of a large number of genes may combine to produce many divergent gene expression patterns and phenotypes. Third, analysis of recombinant inbred Arabidopsis lines defective in different genes essential to cytosine methylation suggest that complex traits, such as flowering time, plant height, biomass, and bacterial pathogen resistance, behave as quantitative epigenetic trait loci (Johannes et al., 2009; Reinders et al., 2009; Richards, 2009). Hence, small differences in epitype of a large number of loci may combine to produce rapid rates of morphological change. It should therefore be noted that the degree to which variation in any one gene sequence and its epitype contribute to phenotypic variation would differ among loci. Whether rapid changes in cis-elements and recombination of quantitative genetic and epigenetic loci fully account for rapid anatomical evolution remains to be seen.
WHAT IS THE PHYLOGENETIC EVIDENCE THAT EPITYPE EVOLVES FROM ANCESTRAL GENES FOLLOWING GENE DUPLICATION?
Given that epigenetic controls are important to organ and organismal development and species evolution, then epitype must be inherited, not just between two dividing cells in a tissue or an organ, but from one generation of organisms to the next. The meiotic inheritance of epitype between generations has been called variously “epigenetic memory” or “epigenetic inheritance” (Rakyan et al., 2001; Bronner et al., 2007). However, the evolution of epitype remains essentially unexplored and inadequately documented. The hypothesis put forward to focus this discussion states that epitype evolves by gene duplication, divergence, and subfunctionalization and is analogous to the widely accepted view for the evolution of gene, RNA, and protein sequences (Force et al., 1999; Ohno, 1999). This hypothesis does not suggest any mechanisms for the inheritance of epigenetic information following gene duplication nor how such patterns might be dynamic in particular tissues or organs during ontogeny. It rather suggests that patterns of epigenetic changes to chromatin, such as nucleosome position and the locations of 5MeC residues, particular histone side chain modifications, and uncommon histone variants may be conserved among closely related genes in a gene family. In other words, recently duplicated genes should have a conserved epitype and epitype should correlate with gene phylogeny. The evidence for the conservation of different categories of chromatin modification following gene duplication is discussed separately.
Nucleosome Phasing
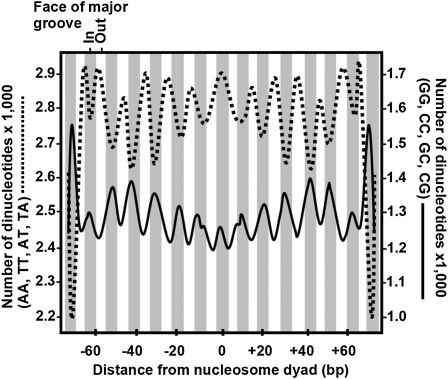
Nucleosome phasing and density refers to the likelihood of finding a nucleosome bound to invariant DNA sequence and at a fixed distance from two flanking nucleosomes. Because phasing determines the accessibility of DNA to the binding of diverse regulatory proteins that silence or activate gene expression, phasing is fundamental to epitype and the control of gene expression. Phasing is precisely controlled, with differences of a few nucleotides having profound effects on gene expression, particularly in the promoter of a gene (Angermayr et al., 2003; Sekinger et al., 2005; Whitehouse et al., 2007; Parnell et al., 2008; Ay and Arnosti, 2010; Bai et al., 2010). Indirect and direct evidence that nucleosome phasing is often conserved after gene duplication comes from a number of sources. One of the most impressive results is shown in Figure 1. Nucleosome position for histone variant H2AZ-enriched nucleosomes is strongly determined by DNA sequence, with G+C-rich dinucleotides inside contacting the nucleosomes as the DNA helix wraps around, and with A+T-rich dinucleotides on the average outside as shown for fungal and vertebrate nucleosomes (Segal et al., 2006; Albert et al., 2007). The G+C- and A+T-rich dinucleotides are each phased in repeats 10 bp apart in the 10-bp helical twist of the DNA. Similarly, a dinucleotide code determines nucleosome position and exclusion, access to transcription factor binding sites, and the transcriptional activity of early neural enhancer genes in the urocordate Cionia (Khoueiry et al., 2010). This code, its impact of nucleosome exclusion, and transcription of early neural enhancer genes are conserved in Drosophila, a very distant metazoan.
Figure 1.
Nucleotide Sequence Composition Directs the Positioning of H2AZ-Enriched Nucleosomes in Saccharomyces.
The frequency distribution of the combined number of AA, TT, AT, and TA dinucleotides (dashed line) or GG, CC, GC, and CG dinucleotides (solid line) at each base pair along the 147 bp of nucleosomal DNA. Repeats of G+C-rich dinucleotides are more commonly in the major groove (gray shaded areas) facing inward against the nucleosome. A+T-rich dinucleotides are generally in the major grooves facing outward from the nucleosome. The bottom axis shows the distance in base pairs from zero at the center of dyad symmetry. (Adapted by permission from Macmillan Publishers Limited: Nature, Albert et al. [2007], copyright 2007.)
These data suggest that patterns in DNA sequence alone may determine a significant percentage of intrinsic nucleosome positioning in a genome and that this in turn could strongly influence other aspects of epitype. It is reasonable to expect that DNA with distinct nucleotide sequences and repeats might wrap around nucleosomes with different defined histone variant compositions (Henikoff et al., 2001). Consistent with these results on phasing, repeats of the 10-bp motif TATAAACGCC control the phasing of some nucleosomes (Roychoudhury et al., 2000). The DNA between nucleosomes is in a distinctly different conformation from nucleosomal DNA, enabling it to recognize specific binding and modifying proteins. In addition, nucleosome phasing directly correlates with the site of RNA polymerase II (Pol II) binding in human CD4+ T cells (Schones et al., 2008). Considering that Pol II binding relative to the transcription start sites is strongly affected by the TATA sequence located approximately at minus 35 bp prior to the transcriptional start site of most genes, the binding of TATA binding factor and Pol II also may influence nucleosome phasing. Hence, it is reasonable to propose that nucleosome phasing is partially dependent upon DNA sequence and evolves by gene duplication.
Cytosine Methylation
The evolutionary conservation of the cytosine methylation epitype among duplicated genes seems reasonable to consider because the inheritance of cytosine methylation is partially understood. Hemimethylated 5′-5MeCpG/3′-GpC sequences are transmitted by cytosine methylation of homologous daughter strands during DNA replication. Families of cytosine methyltransferases and methylcytosine glycosylases and extensive epigenetic machinery are known that maintain dynamic cytosine methylation in plants and animals (Ooi and Bestor, 2008; Kim et al., 2009). In addition, RNA-directed sequence-specific de novo methylation and demethylation of DNA has been reported in angiosperms and mammals (Imamura et al., 2004; Matzke et al., 2009).
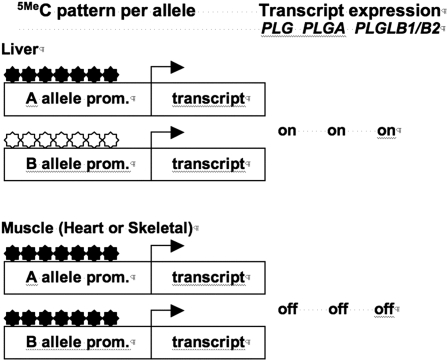
Cortese et al. (2008) compared promoter CpG methylation patterns among members of the ∼35-MY-old human plasminogen precursor gene family and the ∼700- to 900-MY-old human T-Box (TBX) gene family. Plasminogens are blood-clotting factors found in hominids. The four recently evolved human plasminogen precursor genes, PGL, PGLA, and PGLB1/B2, are all on chromosome two and differ from each other by 4 to 5% in DNA sequence, except PGLB1 and PGLB2 that are nearly indistinguishable in DNA sequence. Cytosine DNA methylation patterns are well conserved among seven CpG sites located −171 to −378 nucleotides from the start of transcription within all four promoters. In liver, where transcripts for all four genes are detected, 50% of the three easily distinguished gene sequences are essentially unmethylated at all seven sites. Segregation of an allele-specific marker linked to the PGL gene indicated that methylation in liver was not allele specific; thus, either PGL allele might be fully methylated or unmethylated in any cell as shown in Figure 2. In heart muscle and in skeletal muscle, where the four genes are turned off, nearly 100% of the seven sites are fully cytosine methylated for all four plasminogen genes. In other words, promoter cytosine methylation inversely correlates with the levels of transcript expression in the three tissues examined, and the family members share common promoter methylation and expression patterns.
Figure 2.
Cytosine Methylation Patterns Are Conserved among the Members of the Human Plasminogen Precursor Gene Family PGL, PGLA, and PGLB1/B2.
In human liver, where all four genes are expressed, one of the two alleles for each gene are cytosine methylated 5MeC (black stars) at nearly all seven CpG sites. The other allele remains unmethylated (white stars). In skeletal and heart muscle, where the genes are not expressed, both alleles for each gene are nearly all cytosine methylated at all seven sites. (Adapted from Cortese et al. [2008], with permission from Elsevier.)
Cytosine methylation in promoter regions correlates with gene silencing, and yet most active and inactive genes have moderately strong cytosine methylation throughout the gene body and a sharp dip in cytosine methylation levels just prior to the start of transcription (Zemach et al., 2010). This valley with low cytosine methylation inversely correlates with the location of a commonly found spike in H2AZ deposition. Zemach et al. (2010) found this pattern in most genes in diverse plant and animal species and in a protist green alga, but not in the fungal species examined.
These data provide strong initial support for the above hypothesis in that the potential to display defined 5MeC patterns appears to be well conserved and inherited among some duplicate gene copies. The differential methylation of plasminogen alleles in liver fits well with the concept that epiallelic divergence might precede and promote gene duplication similar to the concept that “allelic divergence precedes and promotes gene duplication” put forward by Proulx and Phillips (2006). Thus, strong selection acting on segregating alleles or epialleles of a single locus may favor the selection of duplicate genes, once gene duplication occurs.
Three sets of cytosine methylation data neither support nor deny the above evolutionary hypothesis but are relevant to understanding the complexity of 5MeC epitypes. The TBX genes encode an ancient family of transcription factors with DNA binding properties that are found among vertebrates and invertebrates. In vertebrates, TBX proteins are crucial in regulating numerous particular pathways of tissue and organ development, including those affecting the development of brain, limb, heart, retina, and T cells. The 15 human TBX genes examined in Cortese et al. (2008) comprise five ancient subfamilies that are expressed principally in the subset of tissues and organs, where many of their developmental activities are characterized (Naiche et al., 2005). The promoters of most of these 15 TBX family members are essentially unmethylated in the eight tissues and organs examined (Cortese et al., 2008). For the seven genes where some tissue-specific cytosine methylation was detected, there was no conservation of methylation patterns that correlated with proximal phylogenic relationships of those genes. Considering that even the most closely related TBX genes, such as TBX1 and TBX10, are 450 MY divergent from a common ancestral gene, it might be concluded that cytosine methylation patterns, if inherited, are not necessarily conserved over the evolutionary timescale represented by this ancient family. Alternatively, cytosine methylation patterns may simply not be important to TBX gene expression and, hence, are not conserved. Cortese et al. (2008) found a similar lack of conservation of methylation patterns for processed pseudogenes, which represent much more recent gene duplications. However, processed pseudogenes are cDNAs of transcripts inserted back into genomes and lack flanking DNA and intron sequences that may be necessary for the inheritance of epitype. Finally, transgene silencing and reactivation by cytosine methylation and demethylation, respectively, is known to occur rapidly, often in the first organismal generation after the transgene is introduced (Chawla et al., 2007; Goll et al., 2009; Mehta et al., 2009). Transgene silencing is a stochastic process that is dependent upon the sequence of the transgene and its context of surrounding chromatin (Dorer and Henikoff, 1997; Talbert and Henikoff, 2000).
Global analysis of sequence-specific cytosine methylation in Arabidopsis revealed that ∼6% of all C residues are methylated (Lister et al., 2008). Widman et al. (2009) show that 20% of CpG methylations are conserved among the numerous gene duplications remaining from a recent polyploidization event that occurred 40 to 80 million years ago (McDowell et al., 1996; Blanc and Wolfe, 2004). In addition, these 5MeC residues are more likely to be conserved between duplicated genes than nonmethylated Cs or the other nucleotides, G, A, or T. It should be noted that the sequence context of the CpG dinucleotides (Cokus et al., 2008) and the expression status of each pair of duplicated genes as both active, both silent, or differentially expressed are likely to further refine interpretations of 5MeC conservation in the near future.
Histone Modification
Rodin and Riggs (2003) propose that epigenetic controls aid “evolution by gene duplication” by silencing recent gene duplicates until beneficial mutations and subfunctionalization occur. Circumstantial evidence supporting this view comes from Zheng's (2008) recent examination of data from Barski et al.'s (2007) determination of the epitype of human T cells. Barski et al. (2007) performed a genome-wide analysis of numerous histone modifications on chromatin immune-precipitated nucleosomal DNA. Zheng (2008) used these data to scan the histone methylation patterns of 1646 random segmental duplications (SDs) with >90% sequence identity that have occurred in the human genome in the last ∼25 MY. They found a strong statistical bias toward histone methylation of one copy of each SD relative to the other copy, particularly for the presence of H2bK5Me1, H3K4Me2, H3K9Me1, H3K36Me3, and H3K79Me1. These data argue against my hypothesis at least as it relates to histone modification epitypes. However, evidence that gene duplicates initially may be silenced by one epigenetic mechanism(s) does not imply that these genes will not be activated later making use of other epigenetic information inherited from their ancestral parent gene. Indeed, Rodin and Riggs (2003) suggest that such a mechanism of epigenetic silencing of SDs exists to preserve duplicate alleles for future activation and use. The inheritance of site-specific histone side chain modification is not understood and may be context dependent, reliant upon information from nucleosome phasing and the location of RNA polymerases, histone variants, and cytosine methylation. Bronner et al. (2007) propose that this kind of “epigenetic memory” is propagated via special macromolecular complexes (epigenetic code replication machinery) containing many of the known histone modifying enzymes, such as DNA methyltransferases, histone acetyltransferases, and histone deacetylases. The somatic inheritance of histone modifications requires their propagation on daughter DNA strands after passage of the replication fork. Margueron and Reinberg (2010) recently discussed an elegant model for this activity and its relevance to epigenetic inheritance. However, confusing cause-and-effect relationships between histone modifications and gene expression further complicates any examination of the evolutionary inheritance of this class of chromatin structures following gene duplication.
Histone Variants
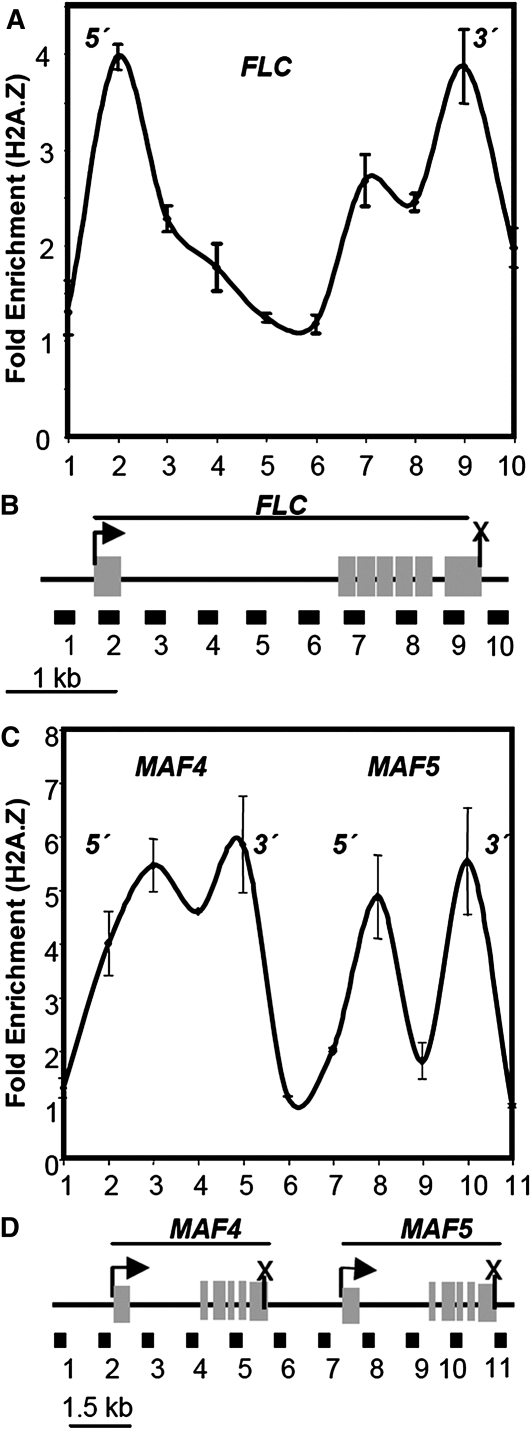
Most classes of histones are comprised of several ancient histone varients. Histone H2AZ variants may substitute for the more common H2A variant(s) within a nucleosome. In yeast, vertebrates, and plants, H2AZ is found at very high levels in a few to several nucleosomes at the 5′ ends of nearly half of all active genes (Meneghini et al., 2003; Li et al., 2005; Albert et al., 2007). My laboratory previously examined H2AZ histone variant deposition among the closely related members of a subfamily of MADS box transcription factors, FLC, MAF4, and MAF5 (Deal et al., 2007). These three genes are all expressed in the apical meristem of Arabidopsis shoots and act as repressors of flowering. They may be estimated to have diverged from common ancestry in the eudicot lineage in the last 140 MY. The expression of all three genes requires normal H2AZ deposition, being significantly downregulated in plants lacking the essential ACTIN-RELATED PROTEIN6 subunit of the SWR1 chromatin remodeling complex or lacking the defining DNA dependent ATPase subunit of SWR1, PIE1/Swr1 (Deal et al., 2007). In the wild type, all three MADS box genes show a striking bimodal distribution of H2AZ deposition, with peaks of H2AZ histones at their 5′ and 3′ ends as shown in Figure 3 (Deal et al., 2007). This is quite distinct from the single 5′ spike of H2AZ observed for most genes. Furthermore, none of the four MADS box genes examined in yeast show bimodal distribution of H2AZ distribution, having either a 5′ peak of H2AZ or no enrichment of H2AZ at either end of the gene (Albert et al., 2007). The Arabidopsis data are consistent with the bimodal distribution of H2AZ among the FLC, MAF4, and MAF5 genes being inherited following gene duplication from a common ancestral MADS box gene. We do not know if the DNA sequences in these peaks of H2AZ-enriched nucleosomes follow the nucleotide base composition rules discussed in the above section on nucleosome phasing.
Figure 3.
MADS Box Repressors of Flowering FLC, MAF4, and MAF5 Share an Uncommon Bimodal H2AZ Distribution Pattern in Arabidopsis.
(A) and (C) All three MADS box transcription factor genes share in bimodal pattern of H2AZ deposition with peaks at their 5′ and 3′ ends in seedlings.
(B) and (D) Maps of FLC, MAF4, and MAF5 genes and their exon structures (black boxes), and regions PCR amplified to quantify H2AZ-enriched sequences (1 to 10 for FLC and 1 to 11 for MAF4 and MAF5). (Redrawn from Deal et al. [2007].)
WHAT ARE THE EVOLUTIONARY RATES OF CHANGE FOR DIFFERENT EPITYPES?
To approach this question, it is useful to recall the wide variation in the rates of accumulation of DNA base substitutions for protein encoding sequences. Within codons, synonymous nucleotide substitution (SNS) and nonsynonymous or replacement nucleotide substitution (RNS) generally evolve at very different rates (Meagher et al., 1989). SNS does not alter the encoded protein sequence. Accumulated changes in SNS proceed rapidly, and although the rate may vary, it is generally approximately half of the rate of DNA base mutation because some SNS is selected against. SNS has been variously estimated to occur in plants and animals at ∼1% per MY per lineage (Sakoyama et al., 1987; Meagher et al., 1989). The rate of RNS varies among individual genes, being dependent upon the degree of conservation of the particular encoded protein sequence. Well-conserved proteins like actin have slow RNS rates (e.g., 0.01% per MY per lineage) (McDowell et al., 1996; Kusakabe et al., 1997), while RNS rates for highly conserved core histone sequences are significantly slower still. By contrast, RNS rates for some poorly conserved proteins like the mammalian serum albumins are much faster, approaching the rate of change for pseudogenes or SNS (Minghetti et al., 1985) and exceeding these rates for genes under directional selection.
In a similar manner, different epigenetic marks (i.e., different changes to chromatin structure) might be expected to accumulate at different rates. Cytosine methylation patterns appear to be conserved among human plasminogen sequences that are <35 MY diverged from a common ancestor. Since the four plasminogen sequences are all expressed and functional, their pattern of 5MeC may all be under strong negative selection. Indeed, the divergence of the CpG dinucleotide sequences and the 5MeCpG epitype must evolve together, when there is selective pressure for cytosine methylation (Widman et al., 2009). Gene families containing both very young and slightly older members need to be examined to further explore these relationships.
Because the position of H2AZ-enriched nucleosomes is largely determined by the location of G+C- and A+T-rich motifs in DNA, nucleosome phasing has the potential to evolve at some rate slightly slower than the DNA base mutation rate. Finally, MADS box genes that regulate flowering appear to have a bimodal H2AZ distribution pattern conserved over ∼140 MY. However, a large percentage of all expressed genes have a single spike of H2AZ at their 5′ end; hence, this 5′ pattern may evolve too slowly or vary too little to be used in estimating an evolutionary rate. Clearly, focused efforts are needed to determine the rates of evolution for different epitypes. However, an important conclusion is that the evolutionary divergence of epitype should be a complex property, comprised of numerous and differentially conserved epigenetic marks evolving at different rates.
THE INFLUENCE OF ENVIRONMENT
Stress and other environmental factors are known to produce a variety of meiotically and mitotically inherited epitypes in plants and animals (Boyko and Kovalchuk, 2008; Chinnusamy et al., 2008; Murgatroyd et al., 2009). Quantitative epigenetic loci, mentioned above, might be particularly suited to respond to environment. In one of the best known examples of environmental influences on one gene's epitype, diet and nutritional supplements produce DNA hypomethylated active and hypermethylated inactive epialleles of the mouse agouti yellow allele that are inherited through meiosis (Morgan et al., 1999, 2008; Martin et al., 2008). The agouti gene and various stress genes must have the right set of CpG sequences to be silenced or activated by changes in cytosine methylation. Hence, DNA sequence plays an essential role in potentiating environmental influences on a DNA methylation epitype. It seems reasonable to expect parallel relationships between DNA sequence and the potential for environmental influences on nucleosome positioning and other epitypes.
GENETICS, EPIGENETICS, AND SEMANTICS
The obvious interpretation of the data discussed herein is that DNA sequence is a major determinant of chromatin structure and that inherited genotype determines the range of epitypes. Accordingly, do we really need to consider epigenetics as a separate level of control for these cases or can genetics alone be used to describe all such inherited states? Consider the example where a G-to-A mutation in a critical G/C-rich repeat that formally bound to an H2AZ nucleosome (Figure 1) shifts the phasing of that nucleosome. This shift in nucleosome position might allow the stochastic activation and resilencing of gene expression by exposing the binding sites for transcriptional activators and/or the TATA binding factor by placing them between nucleosomes. Prior to mutation, these binding sites were covered by nucleosomes causing the gene to be always silent. If the base change itself did not alter the binding site for a specific transcription factor, it would be difficult to describe this mutation as a change in genetic control, without redefining nucleosomes as transcription factors. It is perhaps most logical to view this base change as generating a new genotype and potential epitypes that may be inherited together. This base change would potentiate new epigenetic controls.
CONCLUSIONS AND FUTURE CONSIDERATIONS
The rapid evolution of epigenetic controls may be an important factor in accelerating morphological evolution in higher plants and animals. The hypothesis that epitype and associated phenotypes evolve by gene duplication, divergence, and subfunctionalization was explored. Initial supporting arguments for this hypothesis based on conserved patterns of phasing for H2AZ nucleosomes, cytosine methylation, and H2AZ deposition were discussed. Contrary to this hypothesis, novel histone methylation patterns in most human segmental chromosome duplications appear to be rapidly and uniquely generated and are not conserved.
A deeper understanding of this hypothesis will come from future examinations of epitype in a phylogenetic context, making comparisons of chromatin structures among variously related gene family members. A great deal more could be done with existing genome-wide chromatin structure data sets. Conserved patterns of chromatin modification need to be examined gene by gene, within well-characterized small gene families with widely differently aged members, such as those encoding the plant actins, cofilin/ADFs, and profilins in plants. Search algorithms could be used to find patterns of cis-linked epitype within subclasses of genes in large gene families like those encoding the G protein–coupled receptors, nuclear receptors, ABC transporters, P450 hydroxylases, F-box proteins, and MADS box transcription factors. Enough is now known about the divergence times of various angiosperm and mammalian lineages that the age of gene duplication events may be reasonably estimated; thus, the rates of evolution for various epitypes may be approximated. Finally, examining the evolution of epitype appears to be a valid and important field of study if scientists are to make biological sense of epigenetic controls.
Acknowledgments
This work was supported by a grant from the National Institutes of Health (GM36397). Jonathan Arnold, Kristofer Mussar, Wyatt Anderson, Eileen Roy, Benjamin Nelson, and anonymous reviewers contributed useful editorial comments during the preparation of the manuscript.
References
- Abbosh P.H., Montgomery J.S., Starkey J.A., Novotny M., Zuhowski E.G., Egorin M.J., Moseman A.P., Golas A., Brannon K.M., Balch C., Huang T.H., Nephew K.P. (2006). Dominant-negative histone H3 lysine 27 mutant derepresses silenced tumor suppressor genes and reverses the drug-resistant phenotype in cancer cells. Cancer Res. 66: 5582–5591 [DOI] [PubMed] [Google Scholar]
- Albert I., Mavrich T.N., Tomsho L.P., Qi J., Zanton S.J., Schuster S.C., Pugh B.F. (2007). Translational and rotational settings of H2A.Z nucleosomes across the Saccharomyces cerevisiae genome. Nature 446: 572–576 [DOI] [PubMed] [Google Scholar]
- Angermayr M., Oechsner U., Bandlow W. (2003). Reb1p-dependent DNA bending effects nucleosome positioning and constitutive transcription at the yeast profilin promoter. J. Biol. Chem. 278: 17918–17926 [DOI] [PubMed] [Google Scholar]
- Ay A., Arnosti D.N. (2010). Nucleosome positioning: An essential component of the enhancer regulatory code? Curr. Biol. 20: R404–R406 [DOI] [PubMed] [Google Scholar]
- Bai L., Charvin G., Siggia E.D., Cross F.R. (2010). Nucleosome-depleted regions in cell-cycle-regulated promoters ensure reliable gene expression in every cell cycle. Dev. Cell 18: 544–555 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barski A., Cuddapah S., Cui K., Roh T.Y., Schones D.E., Wang Z., Wei G., Chepelev I., Zhao K. (2007). High-resolution profiling of histone methylations in the human genome. Cell 129: 823–837 [DOI] [PubMed] [Google Scholar]
- Bennetzen J.L., Coleman C., Liu R., Ma J., Ramakrishna W. (2004). Consistent over-estimation of gene number in complex plant genomes. Curr. Opin. Plant Biol. 7: 732–736 [DOI] [PubMed] [Google Scholar]
- Blanc G., Wolfe K.H. (2004). Functional divergence of duplicated genes formed by polyploidy during Arabidopsis evolution. Plant Cell 16: 1679–1691 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boyko A., Kovalchuk I. (2008). Epigenetic control of plant stress response. Environ. Mol. Mutagen. 49: 61–72 [DOI] [PubMed] [Google Scholar]
- Bronner C., Chataigneau T., Schini-Kerth V.B., Landry Y. (2007). The “Epigenetic Code Replication Machinery”, ECREM: A promising drugable target of the epigenetic cell memory. Curr. Med. Chem. 14: 2629–2641 [DOI] [PubMed] [Google Scholar]
- Carroll S.B. (2008). Evo-devo and an expanding evolutionary synthesis: A genetic theory of morphological evolution. Cell 134: 25–36 [DOI] [PubMed] [Google Scholar]
- Chawla R., Nicholson S.J., Folta K.M., Srivastava V. (2007). Transgene-induced silencing of Arabidopsis phytochrome A gene via exonic methylation. Plant J. 52: 1105–1118 [DOI] [PubMed] [Google Scholar]
- Chinnusamy V., Gong Z., Zhu J.K. (2008). Abscisic acid-mediated epigenetic processes in plant development and stress responses. J. Integr. Plant Biol. 50: 1187–1195 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cokus S.J., Feng S., Zhang X., Chen Z., Merriman B., Haudenschild C.D., Pradhan S., Nelson S.F., Pellegrini M., Jacobsen S.E. (2008). Shotgun bisulphite sequencing of the Arabidopsis genome reveals DNA methylation patterning. Nature 452: 215–219 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cortese R., Krispin M., Weiss G., Berlin K., Eckhardt F. (2008). DNA methylation profiling of pseudogene-parental gene pairs and two gene families. Genomics 91: 492–502 [DOI] [PubMed] [Google Scholar]
- Deal R.B., Topp C.N., McKinney E.C., Meagher R.B. (2007). Repression of flowering in Arabidopsis requires activation of FLOWERING LOCUS C expression by the histone variant H2A.Z. Plant Cell 19: 74–83 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dobzhansky T. (1973). Nothing in biology makes sense except in light of evolution. Am. Biol. Teach. 35: 125–129 [Google Scholar]
- Dorer D.R., Henikoff S. (1997). Transgene repeat arrays interact with distant heterochromatin and cause silencing in cis and trans. Genetics 147: 1181–1190 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Force A., Lynch M., Pickett F.B., Amores A., Yan Y.L., Postlethwait J. (1999). Preservation of duplicate genes by complementary, degenerative mutations. Genetics 151: 1531–1545 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goll M.G., Anderson R., Stainier D.Y., Spradling A.C., Halpern M.E. (2009). Transcriptional silencing and reactivation in transgenic zebrafish. Genetics 182: 747–755 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grzeschik K.H. (2002). Human limb malformations; an approach to the molecular basis of development. Int. J. Dev. Biol. 46: 983–991 [PubMed] [Google Scholar]
- Haaf T. (2006). Methylation dynamics in the early mammalian embryo: Implications of genome reprogramming defects for development. Curr. Top. Microbiol. Immunol. 310: 13–22 [DOI] [PubMed] [Google Scholar]
- Haig D. (2004). The (dual) origin of epigenetics. Cold Spring Harb. Symp. Quant. Biol. 69: 67–70 [DOI] [PubMed] [Google Scholar]
- Haston K.M., Tung J.Y., Reijo Pera R.A. (2009). Dazl functions in maintenance of pluripotency and genetic and epigenetic programs of differentiation in mouse primordial germ cells in vivo and in vitro. PLoS One 4: e5654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henikoff S. (1979). Position effects and variegation enhancers in an autosomal region of Drosophila melanogaster. Genetics 93: 105–115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henikoff S., Ahmad K., Malik H.S. (2001). The centromere paradox: Stable inheritance with rapidly evolving DNA. Science 293: 1098–1102 [DOI] [PubMed] [Google Scholar]
- Hoekstra H.E., Coyne J.A. (2007). The locus of evolution: Evo devo and the genetics of adaptation. Evolution 61: 995–1016 [DOI] [PubMed] [Google Scholar]
- Iacobuzio-Donahue C.A. (2009). Epigenetic changes in cancer. Annu. Rev. Pathol. 4: 229–249 [DOI] [PubMed] [Google Scholar]
- Imamura T., Yamamoto S., Ohgane J., Hattori N., Tanaka S., Shiota K. (2004). Non-coding RNA directed DNA demethylation of Sphk1 CpG island. Biochem. Biophys. Res. Commun. 322: 593–600 [DOI] [PubMed] [Google Scholar]
- Johannes F., et al. (2009). Assessing the impact of transgenerational epigenetic variation on complex traits. PLoS Genet. 5: e1000530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kandasamy M.K., McKinney E.C., Deal R.B., Smith A.P., Meagher R.B. (2009). Arabidopsis actin-related protein ARP5 in multicellular development and DNA repair. Dev. Biol. 335: 22–32 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khoueiry P., Rothbacher U., Ohtsuka Y., Daian F., Frangulian E., Roure A., Dubchak I., Lemaire P. (2010). A cis-regulatory signature in ascidians and flies, independent of transcription factor binding sites. Curr. Biol. 20: 792–802 [DOI] [PubMed] [Google Scholar]
- Kim J.K., Samaranayake M., Pradhan S. (2009). Epigenetic mechanisms in mammals. Cell. Mol. Life Sci. 66: 596–612 [DOI] [PMC free article] [PubMed] [Google Scholar]
- King M.C., Wilson A.C. (1975). Evolution at two levels in humans and chimpanzees. Science 188: 107–116 [DOI] [PubMed] [Google Scholar]
- Kusakabe T., Araki I., Satoh N., Jeffery W.R. (1997). Evolution of chordate actin genes: Evidence from genomic organization and amino acid sequences. J. Mol. Evol. 44: 289–298 [DOI] [PubMed] [Google Scholar]
- Li B., Pattenden S.G., Lee D., Gutierrez J., Chen J., Seidel C., Gerton J., Workman J.L. (2005). Preferential occupancy of histone variant H2AZ at inactive promoters influences local histone modifications and chromatin remodeling. Proc. Natl. Acad. Sci. USA 102: 18385–18390 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lister R., O'Malley R.C., Tonti-Filippini J., Gregory B.D., Berry C.C., Millar A.H., Ecker J.R. (2008). Highly integrated single-base resolution maps of the epigenome in Arabidopsis. Cell 133: 523–536 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lyon M.F. (1993). Epigenetic inheritance in mammals. Trends Genet. 9: 123–128 [DOI] [PubMed] [Google Scholar]
- Mackem S., Lewandoski M. (2009). Limb development takes a measured step toward systems analysis. Sci. Signal. 2: pe33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Margueron R., Reinberg D. (2010). Chromatin structure and the inheritance of epigenetic information. Nat. Rev. Genet. 11: 285–296 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin D.I., Cropley J.E., Suter C.M. (2008). Environmental influence on epigenetic inheritance at the Avy allele. Nutr. Rev. 66(Suppl 1): S12–S14 [DOI] [PubMed] [Google Scholar]
- Matzke M., Kanno T., Daxinger L., Huettel B., Matzke A.J. (2009). RNA-mediated chromatin-based silencing in plants. Curr. Opin. Cell Biol. 21: 367–376 [DOI] [PubMed] [Google Scholar]
- McClintock B. (1956). Intranuclear systems controlling gene action and mutation. Brookhaven Symp. Biol. 8: 58–74 [PubMed] [Google Scholar]
- McDowell J.M., Huang S., McKinney E.C., An Y.Q., Meagher R.B. (1996). Structure and evolution of the actin gene family in Arabidopsis thaliana. Genetics 142: 587–602 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meagher R.B., Berry-Lowe S., Rice K. (1989). Molecular evolution of the small subunit of ribulose bisphosphate carboxylase: Nucleotide substitution and gene conversion. Genetics 123: 845–863 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meagher R.B., Kandasamy M.K., McKinney E.C., Roy E. (2009). Chapter 5. Nuclear actin-related proteins in epigenetic control. Int. Rev. Cell Mol. Biol. 277: 157–215 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mehta A.K., Majumdar S.S., Alam P., Gulati N., Brahmachari V. (2009). Epigenetic regulation of cytomegalovirus major immediate-early promoter activity in transgenic mice. Gene 428: 20–24 [DOI] [PubMed] [Google Scholar]
- Meneghini M.D., Wu M., Madhani H.D. (2003). Conserved histone variant H2A.Z protects euchromatin from the ectopic spread of silent heterochromatin. Cell 112: 725–736 [DOI] [PubMed] [Google Scholar]
- Minghetti P.P., Law S.W., Dugaiczyk A. (1985). The rate of molecular evolution of alpha-fetoprotein approaches that of pseudogenes. Mol. Biol. Evol. 2: 347–358 [DOI] [PubMed] [Google Scholar]
- Morgan H.D., Jin X.L., Li A., Whitelaw E., O'Neill C. (2008). The culture of zygotes to the blastocyst stage changes the postnatal expression of an epigentically labile allele, agouti viable yellow, in mice. Biol. Reprod. 79: 618–623 [DOI] [PubMed] [Google Scholar]
- Morgan H.D., Sutherland H.G., Martin D.I., Whitelaw E. (1999). Epigenetic inheritance at the agouti locus in the mouse. Nat. Genet. 23: 314–318 [DOI] [PubMed] [Google Scholar]
- Murgatroyd C., Patchev A.V., Wu Y., Micale V., Bockmuhl Y., Fischer D., Holsboer F., Wotjak C.T., Almeida O.F., Spengler D. (2009). Dynamic DNA methylation programs persistent adverse effects of early-life stress. Nat. Neurosci. 12: 1559–1566 [DOI] [PubMed] [Google Scholar]
- Naiche L.A., Harrelson Z., Kelly R.G., Papaioannou V.E. (2005). T-box genes in vertebrate development. Annu. Rev. Genet. 39: 219–239 [DOI] [PubMed] [Google Scholar]
- Nanney D.L. (1958a). Epigenetic control systems. Proc. Natl. Acad. Sci. USA 44: 712–717 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nanney D.L. (1958b). Epigenetic factors affecting mating type expression in certain ciliates. Cold Spring Harb. Symp. Quant. Biol. 23: 327–335 [DOI] [PubMed] [Google Scholar]
- Ohno S. (1999). Gene duplication and the uniqueness of vertebrate genomes circa 1970–1999. Semin. Cell Dev. Biol. 10: 517–522 [DOI] [PubMed] [Google Scholar]
- Ooi S.K., Bestor T.H. (2008). The colorful history of active DNA demethylation. Cell 133: 1145–1148 [DOI] [PubMed] [Google Scholar]
- Parnell T.J., Huff J.T., Cairns B.R. (2008). RSC regulates nucleosome positioning at Pol II genes and density at Pol III genes. EMBO J. 27: 100–110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pennacchio L.A. (2003). Insights from human/mouse genome comparisons. Mamm. Genome 14: 429–436 [DOI] [PubMed] [Google Scholar]
- Proulx S.R., Phillips P.C. (2006). Allelic divergence precedes and promotes gene duplication. Evolution 60: 881–892 [PubMed] [Google Scholar]
- Rakyan V.K., Preis J., Morgan H.D., Whitelaw E. (2001). The marks, mechanisms and memory of epigenetic states in mammals. Biochem. J. 356: 1–10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reinders J., Wulff B.B., Mirouze M., Mari-Ordonez A., Dapp M., Rozhon W., Bucher E., Theiler G., Paszkowski J. (2009). Compromised stability of DNA methylation and transposon immobilization in mosaic Arabidopsis epigenomes. Genes Dev. 23: 939–950 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richards E.J. (2009). Quantitative epigenetics: DNA sequence variation need not apply. Genes Dev. 23: 1601–1605 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodin S.N., Riggs A.D. (2003). Epigenetic silencing may aid evolution by gene duplication. J. Mol. Evol. 56: 718–729 [DOI] [PubMed] [Google Scholar]
- Roychoudhury M., Sitlani A., Lapham J., Crothers D.M. (2000). Global structure and mechanical properties of a 10-bp nucleosome positioning motif. Proc. Natl. Acad. Sci. USA 97: 13608–13613 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakoyama Y., Hong K.J., Byun S.M., Hisajima H., Ueda S., Yaoita Y., Hayashida H., Miyata T., Honjo T. (1987). Nucleotide sequences of immunoglobulin epsilon genes of chimpanzee and orangutan: DNA molecular clock and hominoid evolution. Proc. Natl. Acad. Sci. USA 84: 1080–1084 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schnable P.S., et al. (2009). The B73 maize genome: Complexity, diversity, and dynamics. Science 326: 1112–1115 [DOI] [PubMed] [Google Scholar]
- Schones D.E., Cui K., Cuddapah S., Roh T.Y., Barski A., Wang Z., Wei G., Zhao K. (2008). Dynamic regulation of nucleosome positioning in the human genome. Cell 132: 887–898 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Segal E., Fondufe-Mittendorf Y., Chen L., Thastrom A., Field Y., Moore I.K., Wang J.P., Widom J. (2006). A genomic code for nucleosome positioning. Nature 442: 772–778 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sekinger E.A., Moqtaderi Z., Struhl K. (2005). Intrinsic histone-DNA interactions and low nucleosome density are important for preferential accessibility of promoter regions in yeast. Mol. Cell 18: 735–748 [DOI] [PubMed] [Google Scholar]
- Siepel A., et al. (2007). Targeted discovery of novel human exons by comparative genomics. Genome Res. 17: 1763–1773 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith A.P., Jain A., Deal R.B., Nagarajan V.K., Poling M.D., Raghothama K.G., Meagher R.B. (2010). Histone H2A.Z regulates the expression of several classes of phosphate starvation response genes but not as a transcriptional activator. Plant Physiol. 152: 217–225 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Talbert P.B., Henikoff S. (2000). A reexamination of spreading of position-effect variegation in the white-roughest region of Drosophila melanogaster. Genetics 154: 259–272 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thon G., Friis T. (1997). Epigenetic inheritance of transcriptional silencing and switching competence in fission yeast. Genetics 145: 685–696 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tripathi L.P., Sowdhamini R. (2006). Cross genome comparisons of serine proteases in Arabidopsis and rice. BMC Genomics 7: 200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waddington C.H. (1957). The Strategy of the Genes. (London: Ruskin House, George Allen and Unwin Ltd; ), pp. 11–59 [Google Scholar]
- Wallrath L.L., Nagy P.L., Geyer P.K. (2008). Editorial. Epigenetics of development and human disease. Mutat. Res. 647: 1–2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- West M.A., Kim K., Kliebenstein D.J., van Leeuwen H., Michelmore R.W., Doerge R.W., St Clair D.A. (2007). Global eQTL mapping reveals the complex genetic architecture of transcript-level variation in Arabidopsis. Genetics 175: 1441–1450 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitehouse I., Rando O.J., Delrow J., Tsukiyama T. (2007). Chromatin remodelling at promoters suppresses antisense transcription. Nature 450: 1031–1035 [DOI] [PubMed] [Google Scholar]
- Widman N., Jacobsen S.E., Pellegrini M. (2009). Determining the conservation of DNA methylation in Arabidopsis. Epigenetics 4: 119–124 [DOI] [PubMed] [Google Scholar]
- Wilson A.C., Sarich V.M., Maxson L.R. (1974). The importance of gene rearrangement in evolution: Evidence from studies on rates of chromosomal, protein, and anatomical evolution. Proc. Natl. Acad. Sci. USA 71: 3028–3030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wyles J.S., Kunkel J.G., Wilson A.C. (1983). Birds, behavior, and anatomical evolution. Proc. Natl. Acad. Sci. USA 80: 4394–4397 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yi H., Richards E.J. (2008). Phenotypic instability of Arabidopsis alleles affecting a disease Resistance gene cluster. BMC Plant Biol. 8: 36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zemach A., McDaniel I.E., Silva P., Zilberman D. (2010). Genome-wide evolutionary analysis of eukaryotic DNA methylation. Science 328: 916–919 [DOI] [PubMed] [Google Scholar]
- Zhang X., Shiu S., Cal A., Borevitz J.O. (2008). Global analysis of genetic, epigenetic and transcriptional polymorphisms in Arabidopsis thaliana using whole genome tiling arrays. PLoS Genet. 4: 1–12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng D. (2008). Asymmetric histone modifications between the original and derived loci of human segmental duplications. Genome Biol. 9: R105. [DOI] [PMC free article] [PubMed] [Google Scholar]