In yeast and vertebrates, the essential telomere binding protein POT1 protects chromosome ends, but in Arabidopsis, POT1 proteins have evolved to bind telomerase instead. This study examines the function of POT1 in the moss Physcomitrella patens. The findings show that moss POT1 functions in a manner similar to yeast and vertebrate POT1. Thus, POT1 proteins are evolving very rapidly in plants.
Abstract
In vertebrates, the single-stranded telomeric DNA binding protein Protection of Telomeres 1 (POT1) shields chromosome ends and prevents them from eliciting a DNA damage response. By contrast, Arabidopsis thaliana encodes two divergent full-length POT1 paralogs that do not exhibit telomeric DNA binding in vitro and have evolved to mediate telomerase regulation instead of chromosome end protection. To further investigate the role of POT1 in plants, we established the moss Physcomitrella patens as a new model for telomere biology and a counterpoint to Arabidopsis. The sequence and architecture of the telomere tract is similar in P. patens and Arabidopsis, but P. patens harbors only a single-copy POT1 gene. Unlike At POT1 proteins, Pp POT1 efficiently bound single-stranded telomeric DNA in vitro. Deletion of the P. patens POT1 gene resulted in the rapid onset of severe developmental defects and sterility. Although telomerase activity levels were unperturbed, telomeres were substantially shortened, harbored extended G-overhangs, and engaged in end-to-end fusions. We conclude that the telomere capping function of POT1 is conserved in early diverging land plants but is subsequently lost in Arabidopsis.
INTRODUCTION
Telomeres are nucleoprotein complexes that physically cap the ends of eukaryotic chromosomes. Telomeric DNA promotes genome stability through elaborate interactions with a plethora of telomere-associated proteins. The evolutionarily conserved Protection of Telomeres 1 (POT1) protein is a key component of shelterin, the vertebrate telomere capping complex (de Lange, 2005). POT1 binds to the single-stranded G-rich region of the chromosome terminus via structurally conserved N-terminal oligonucleotide/oligosaccharide binding folds (OB-folds) (Lei et al., 2003, 2004). Through interactions with its binding partner TPP1 (Houghtaling et al., 2004; Liu et al., 2004; Ye et al., 2004), POT1 forms a bridge to the double-stranded region of the telomere tract. Deletion of POT1 in Schizosaccharomyces pombe causes rapid telomere loss and cell death, suggesting that its main function is to protect telomeres from degradation (Baumann and Cech, 2001). In vertebrates, the most critical POT1 function appears to be its ability to block ATR (Ataxia Telangiectasia and Rad3 related)-dependent recognition of telomeric DNA as sites of DNA damage (Denchi and de Lange, 2007; Churikov and Price, 2008). POT1 also plays a role in both telomere length regulation by modulating telomerase activity and in chromosome end protection by preventing nucleolytic attack of the C-rich telomeric DNA strand (reviewed in de Lange, 2009).
Arabidopsis thaliana is the reference species for plant biology; consequently, telomere research has focused on this model. Although orthologs of several yeast and vertebrate telomere-related genes have been described (Riha et al., 2001; Song et al., 2008), a technical obstacle for the functional characterization of other candidates is that T-DNA insertion mutants are currently unavailable for a substantial fraction of Arabidopsis genes (O'Malley and Ecker, 2010). Genetic analysis is also complicated by the multiple whole-genome duplications associated with Brassicaceae, the family to which Arabidopsis belongs (Schranz and Mitchell-Olds, 2006). For example, sequence homologs of the mammalian shelterin subunits TRF1 and TRF2 are encoded by at least six different TRFL (TRF-like) genes in Arabidopsis, which appear to be partially redundant (Chen et al., 2001; Karamysheva et al., 2004; Hwang and Cho, 2007). Furthermore, Arabidopsis encodes three divergent POT1 paralogues, including two full-length proteins, At POT1a and At POT1b, and a truncated isoform termed At POT1c (Shakirov et al., 2005; Rossignol et al., 2007).
This pattern of extensive gene duplication and divergence in Brassicaceae suggests that lessons learned from Arabidopsis may not always be applicable to telomere biology in other plants. Indeed, while POT1 is critical for telomere integrity in yeast and vertebrates, this is not true for At POT1a and At POT1b. Like the other Brassicaceae POT1 proteins examined to date, At POT1a and At POT1b exhibit no detectable binding to single-stranded telomeric DNA in vitro (Surovtseva et al., 2007; Shakirov et al., 2009a). Notably, At POT1a physically associates with the telomerase RNP and is critical for telomere length maintenance in vivo (Surovtseva et al., 2007). Although initial experiments with overexpression of dominant-negative mutants suggested that At POT1b is involved in chromosome end protection (Shakirov et al., 2005), recent analysis of a null allele revealed that At POT1b is a negative regulator of telomerase activity and plays no significant role in telomere protection (E. Shakirov, A. Nelson, and D. Shippen, unpublished data). Thus, At POT1a and At POT1b have evolved to function in the telomerase pathway. To fully elucidate the spectrum of POT1 functions in the plant kingdom, it will be necessary to examine the contribution of POT1 in other plant species.
Mosses (bryophytes) belong to the oldest diverging clade of extant land plants, whose ancestors made the first move from aquatic to terrestrial habitats some 450 million years ago (Zimmer et al., 2007). Unlike its younger cousins angiosperms (flowering plants), the moss Physcomitrella patens possesses a remarkably efficient system of homologous recombination, which accounts for an unprecedented success rate in targeted gene replacement among higher eukaryotes (Quatrano et al., 2007). Gene targeting in P. patens is not only five orders of magnitude more efficient than in angiosperms, but also two orders of magnitude more efficient than in the embryonic stem cells of mice and comparable with that observed in Saccharomyces cerevisiae (Schaefer, 2002). Virtually any nonessential P. patens gene can be deleted, and the resulting moss phenotype can be screened within several weeks following transformation.
With its recently sequenced genome (Rensing et al., 2008) and excellent molecular and genetic tools (Quatrano et al., 2007), P. patens is poised to become the new green yeast of plant biology (Schaefer, 2001). Due to its phylogenetic position within the plant kingdom, P. patens also provides a unique opportunity to trace the evolution of telomere-related genes from the first primitive land plants to the most developmentally advanced angiosperms. Here, we describe the initial characterization of telomere biology in P. patens and show that this species possesses canonical plant TTTAGGG telomere repeats. However, unlike Arabidopsis, P. patens harbors only a single POT1 gene. Deletion of Pp POT1 results in catastrophic loss of bulk telomeric DNA, increased G-overhangs, and end-to-end chromosome fusions. These findings point to an essential role for Pp POT1 in chromosome end protection similar to its yeast and vertebrate orthologs but distinct from Arabidopsis.
RESULTS
Telomere Length and Composition in P. patens
The length of the telomere tract varies considerably among different species of land plants and green algae, ranging from 0.5 kb in Chlorella vulgaris to over 150 kb in tobacco (Nicotiana tabacum; Fajkus et al., 1995; Higashiyama et al., 1995). In most Arabidopsis ecotypes, telomeres span 2 to 5 kb (Shakirov and Shippen, 2004). To evaluate telomere length in P. patens, terminal restriction fragment (TRF) analysis was performed using a radioactive probe consisting of four canonical plant TTTAGGG repeats. The most commonly used isolate of P. patens, Gransden (Gd), harbored telomere tracts in the range of 0.6 to 3 kb (Figure 1A, lane 2). Telomeres in the second isolate, Villersexel (Vx-1), were slightly longer, in the range of 1 to 3.5 kb (Figure 1A, lane 3). Thus, P. patens telomeres are approximately twofold shorter than in Arabidopsis (Figure 1A, lane 1) but similar in length to the telomere tracts in another moss species, Barbula unguiculata (Suzuki, 2004). We verified that TTTAGGG-hybridizing repeats are located at chromosomal ends using Bal31 exonuclease. A true telomeric signal should disappear over time, while interstitial cross-hybridizing regions are resistant to enzyme treatment. With the exception of a single interstitial band (indicated by an asterisk), the bulk hybridization signal was lost by 30 min of Bal31 treatment (Figure 1B), confirming that these Bal31-sensitive TTTAGGG-hybridizing repeats are located at chromosome ends.
Figure 1.
Moss Telomere Length Analysis.
(A) TRF analysis of telomeric DNA in P. patens isolates Gd (lane 2) and Vx-1 (lane 3). Molecular weight markers are shown on the left. Genomic DNA was digested with TruII and hybridized with a plant telomere-specific (TTTAGGG)4 probe. Arabidopsis DNA was used as a control (lane 1).
(B) Bal31 analysis of genomic DNA from P. patens isolate Gd. TruII digestion of genomic DNA was performed without prior Bal31 treatment (0 min) or after various incubation periods with Bal31 exonuclease. Asterisk indicates cross-hybridizing interstitial telomeric DNA band that is not sensitive to Bal31 digestion for up to 60 min.
Identification of P. patens POT1
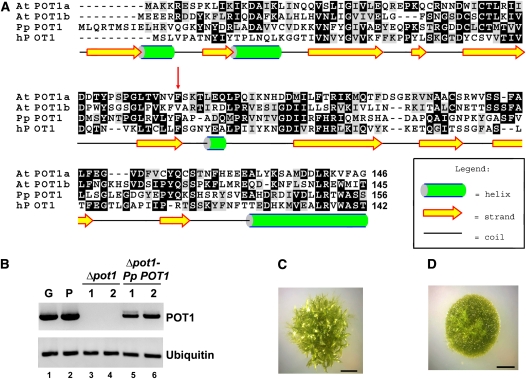
The publicly available P. patens genome database (http://moss.nibb.ac.jp/) was searched with the Arabidopsis POT1a and POT1b sequences as the queries using BLASTp and tBLASTn programs. Similar to most other plants analyzed to date (Shakirov et al., 2009b), P. patens harbors a single POT1 gene, which encodes a 497–amino acid protein with 50 and 45% sequence similarity to Arabidopsis POT1a and POT1b, respectively. The Pp POT1 protein is predicted to contain two N-terminal DNA binding OB-folds with secondary structures similar to the human POT1 protein (Figure 2A). Like At POT1a and At POT1b, Pp POT1 consists of 10 exons with conserved exon-intron junctions, indicating that P. patens and Arabidopsis POT1 genes are indeed orthologous. As noted for other P. patens genes (Rensing et al., 2005), Pp POT1 introns are on average twice as long as in Arabidopsis. RT-PCR revealed that Pp POT1 is continuously expressed during gametophytic development (protonemata and gametophore tissues) (Figure 2B, lanes 1 and 2).
Figure 2.
Identification and Analysis of P. patens POT1.
(A) Amino acid alignment of the predicted Pp POT1 OB1 region with Arabidopsis (At POT1a and At POT1b) and human POT1 proteins. The secondary structure of Pp POT1 OB1 was predicted with PsiPred software (McGuffin et al., 2000). Numbers indicate amino acid positions relative to the start codon. Red arrow indicates the position of a biochemically important F62 residue in human POT1 and the corresponding amino acid F74 in Pp POT1. Alignment was generated with MEGA 3 software (Kumar et al., 2004) and visualized in the BOXSHADE format.
(B) RT-PCR results of Pp POT1 gene expression (top panel) in the wild type (lanes 1 and 2), Δpot1 (lanes 3 and 4), and Δpot1-Pp-POT1 complemented line (lanes 5 and 6). Ubiquitin (bottom panel) was used to normalize for RNA loading.
(C) and (D) General morphology of wild-type (C) and Δpot1 (D) colonies at the 4-week-old stage. The wild-type colony harbors multiple leafy-like gametophores, but only filamentous protonemata tissue is visible in Δpot1.
Characterization of Pp POT1 DNA Binding Activity
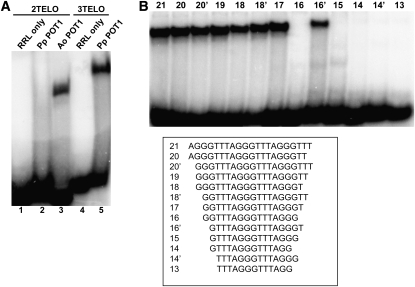
Although POT1 proteins from Brassicaceae have thus far failed to bind single-stranded telomeric DNA in vitro (Surovtseva et al., 2007; Shakirov et al., 2009a), a recent survey revealed that recombinant POT1 proteins from asparagus (Asparagus officinalis), maize (Zea mays), and the green alga Ostreococcus lucimarinus are capable of efficient telomeric DNA binding (Shakirov et al., 2009b). To examine the nucleic acid binding properties of Pp POT1, we expressed Pp POT1 in rabbit reticulocyte lysate (RRL) and performed electrophoretic mobility shift assays (EMSA) with radioactively labeled cocktails of telomeric oligonucleotides containing seven different permutations of either two (2TELO) or three (3TELO) plant TTTAGGG repeats (Figure 3A). A. officinalis POT1 (Ao POT1) served as a positive control (Figure 3A, lane 3) (Shakirov et al., 2009b). While no binding of Pp POT1 to the 2TELO probe was observed (Figure 3A, lane 2), Pp POT1 efficiently bound the 3TELO cocktail (Figure 3A, lane 5). As expected (Surovtseva et al., 2007; Shakirov et al., 2009a), POT1 proteins from Arabidopsis and several other vascular plants showed no detectable telomeric DNA binding (see Supplemental Figure 1 online). Of the seven individual permutations in the 3TELO cocktail probe, Pp POT1 bound (AGGGTTT)3 most efficiently (see Supplemental Figure 2 online).
Figure 3.
Characterization of Pp POT1 Interaction with Telomeric DNA in Vitro.
(A) EMSA was performed with a cocktail of seven 32P-labeled oligonucleotides corresponding to two (2TELO) (lanes 1 to 3) or three (3TELO) (lanes 4 and 5) TTTAGGG repeats. Unprogrammed RRL reactions lacking external DNA template were used as negative controls (lanes 1 and 4), and a reaction with asparagus POT1 protein (Ao POT1) was performed as a positive control (lane 3). Pp POT1 binds 3TELO probe (lane 5) but not 2TELO probe (lane 2).
(B) Identification of the Pp POT1 minimum DNA binding site. Equal amounts of RRL-expressed Pp POT1 were incubated with the indicated radioactively labeled oligonucleotides (bottom panel), and protein-DNA complexes were separated by native PAGE.
The minimum DNA binding site (MBS) in vertebrate POT1 proteins is highly conserved and consists of 10 to 12 nucleotides (Lei et al., 2004; Loayza et al., 2004; Wei and Price, 2004; He et al., 2006). To define the Pp POT1 MBS, we performed EMSA with (AGGGTTT)3 and a series of single nucleotide truncations from either the 5′ or 3′ end of this substrate (Figure 3B). Removal of the first two nucleotides from both ends of the substrate (oligonucleotide 17) did not decrease binding. However, deletion of one additional nucleotide from the 3′ end, but not from the 5′ end of this substrate, completely abolished binding (Figure 3B, compare oligonucleotides 16 and 16′). Since Pp POT1 does not bind to two different oligonucleotides containing 15 nucleotides each (see Supplemental Figure 3A online), we conclude that its minimum tight binding substrate is the 16-nucleotide sequence, GTTTAGGGTTTAGGGT.
We next asked which domain(s) in the P. patens POT1 are required for MBS recognition. Like POT1 proteins from vertebrates and several plants (Lei et al., 2004; He et al., 2006; Wu et al., 2006; Shakirov et al., 2009b), both N-terminal OB-folds were necessary for telomeric DNA binding (see Supplemental Figure 3B online, lanes 4 to 7). Interestingly, the EMSA profile changed from a single band to a smear in the absence of Pp POT1 C terminus (see Supplemental Figure 3B online, compare lanes 2 and 3), suggesting that this region (amino acids 323 to 497) modulates Pp POT1 interaction with DNA in vitro.
In mammalian and fission yeast POT1 proteins, an invariant Phe in the first OB-fold (F62 in human POT1; Figure 2A) is required for efficient DNA binding (Lei et al., 2004; He et al., 2006; Wu et al., 2006). To test if the corresponding residue (F74) is required for telomeric DNA interaction by Pp POT1, we performed EMSA with an F74A Pp POT1 mutant. The mutant protein failed to bind telomeric DNA in vitro (see Supplemental Figure 3B online, lane 1). Altogether, these data indicate that P. patens POT1 is a bona fide single-stranded telomeric DNA binding protein with structural and biochemical properties similar to POT1 proteins from yeast and vertebrates but distinct from Arabidopsis.
Morphological and Developmental Defects in the Δpot1 Strain
We generated a deletion of P. patens POT1 (Δpot1) by replacing its complete open reading frame (ORF) with the hygromycin resistance gene HptII (see Supplemental Figure 4A online). The absence of the POT1 ORF in hygromycin-resistant transformants was verified by PCR genotyping (see Supplemental Figure 4B online), DNA gel blot analysis (see Supplemental Figure 4C online), and by sequencing across the entire Δpot1 locus. RT-PCR confirmed the absence of POT1 mRNA in Δpot1 (Figure 2B, lanes 3 and 4). Hereafter, we refer to Δpot1 as a null mutant.
P. patens was able to grow vegetatively in the absence of POT1, suggesting that this gene is not essential for long-term moss survival. Nevertheless, the Δpot1 strain displayed a striking loss of developmental program. Although the juvenile protonemata filaments initially grew efficiently, aberrant morphological defects emerged and worsened progressively as the Δpot1 advanced to more mature stages of development, ultimately culminating in complete sterility. The growth of gametophores (leafy-like tissue on top of the colony) was significantly delayed in the Δpot1 strain (Figures 2C and 2D), but the overall number of emerging gametophores was reduced only slightly (see Supplemental Figure 5 online). Furthermore, upon cold treatment, which in wild-type P. patens induces the formation of sex organs, the female archegonium and the male antheridium never formed in Δpot1, explaining the sterility phenotype. These data indicate that POT1 is necessary for the normal development of P. patens.
Telomere Length Maintenance Defects in Δpot1
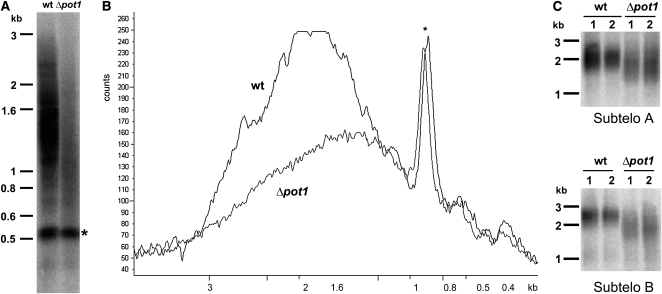
We next asked if the developmental and morphological defects in Δpot1 are accompanied by deficiencies in telomere maintenance. TRF analysis revealed a dramatic decrease (∼72%) in the overall intensity of telomeric hybridization signal in Δpot1 relative to the wild type, indicating that the majority of chromosome ends experienced a massive loss of telomeric DNA (Figure 4A). The average length of the remaining telomere tracts was reduced by ∼200 bp (Figure 4B). Bal31 digestion of Δpot1 DNA confirmed that the residual TTTAGGG hybridization signal was terminally located (see Supplemental Figure 6 online). Complementation with a wild-type Pp POT1 gene (hereafter termed Δpot1-Pp-POT1) restored telomere length to wild-type, demonstrating that telomere loss was the result of POT1 deletion (see Supplemental Figures 7A and 8A online). As expected, RT-PCR confirmed that POT1 mRNA expression was restored in the Δpot1-Pp-POT1 line (Figure 2B, lanes 5 and 6). We conclude that POT1 is necessary for proper telomere length maintenance in P. patens.
Figure 4.
Defects in Telomere Length Regulation in Δpot1.
(A) TRF analysis of DNA from the wild type (wt) and Δpot1. The asterisk indicates an interstitial cross-hybridizing band used as a loading control.
(B) Quantification of results in (A) with ImageQuant software.
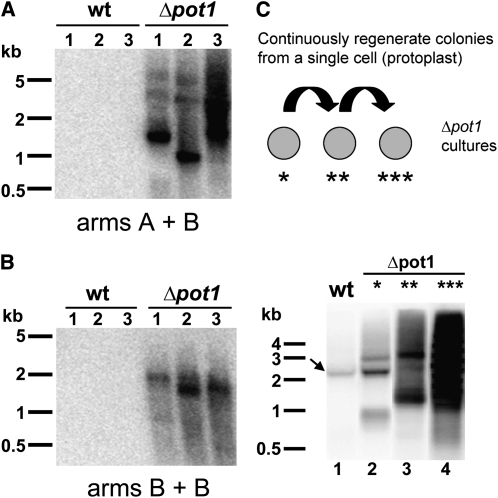
(C) PETRA results of telomere lengths on two individual chromosome arms, designated A and B, in the wild-type and Δpot1 moss. A telomeric probe (TTTAGGG)4 was used to detect TRF and PETRA signals. Molecular weight markers are shown on the left of each panel.
Notably, the Δpot1-Pp-POT1 construct did not complement Δpot1 morphological and developmental defects. Although PCR genotyping of Δpot1-Pp-POT1 strain confirmed the presence of the POT1 gene in its original locus (see Supplemental Figure 4B online), DNA gel blot analysis indicated that additional copies of the complementation construct are present in the Δpot1-Pp-POT1 strain (see Supplemental Figure 7B online). Thus, increased gene dosage may explain the persistence of developmental defects. Alternatively, the severe genome instability that arises from telomere uncapping (see below) may have resulted in irreversible aberrations in cellular pathways controlling moss morphology and development, as previously reported for Arabidopsis mutants with dysfunctional telomeres (Riha et al., 2001; Song et al., 2008; Surovtseva et al., 2009).
The TRF assay measures bulk telomere length changes. To explore the dynamics of individual telomere tracts in Δpot1, we employed the primer extension telomere repeat amplification (PETRA) assay originally developed to study Arabidopsis telomeres (Heacock et al., 2004). PETRA amplifies individual telomeric DNA tracts using PCR primers directed at the G-overhang and a unique subtelomeric sequence. Although the sequenced P. patens genome is not yet assembled into its 27 chromosomes, two unique subtelomeric regions (arbitrarily designated A and B; see Methods) were identified. PETRA reactions conducted with wild-type P. patens generated a smear spanning up to 0.5 kb (Figure 4C), but in Δpot1, both A and B telomeres were >200 bp shorter (Figure 4C). Importantly, the wild-type PETRA profile was restored in Δpot1-Pp-POT1 (see Supplemental Figure 8B online).
Telomerase activity is reduced by ∼10-fold in Arabidopsis plants null for At POT1a (Surovtseva et al., 2007). To determine if in vitro telomerase activity is altered in Δpot1, we performed the quantitative telomere repeat amplification protocol (Kannan et al., 2008) with protein extracts from wild-type and Δpot1 strains. No statistically significant difference in enzyme activity was observed (see Supplemental Figure 9 online). We conclude that POT1 proteins make fundamentally different contributions to telomere maintenance in Arabidopsis and P. patens.
Defects in Telomere Architecture and Genome Stability in Δpot1
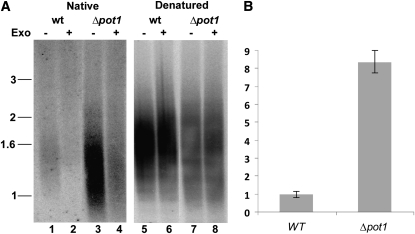
Mutations in telomere capping proteins can lead to increased G-overhang length, resulting from increased nucleolytic attack on the C-rich telomeric strand or uncoupled replication of the G-rich and C-rich strands (Linger and Price, 2009). We analyzed G-overhang status in Δpot1 mutants using the in-gel hybridization assay. DNA gel blotting with a telomeric DNA probe was performed under native conditions to measure single-stranded telomeric DNA. The gel was rehybridized with the same probe under denaturing conditions to normalize for the total amount of double- and single-stranded telomeric DNA loaded. Relative to the wild type, the G-overhang signal in Δpot1 increased by approximately eightfold (Figures 5A, lanes 1 and 3, and 5B). Exonuclease treatment confirmed that hybridization was associated with the chromosome terminus (Figure 5A, lanes 2 and 4). We conclude that P. patens POT1 is critical for proper chromosome end structure.
Figure 5.
Increased G-Overhang Signals in Δpot1 Mutants.
(A) In-gel hybridization results using a G-strand specific probe (C3TA3)4 to detect telomeric DNA under native (lanes 1 to 4) and denaturing (lanes 5 to 8) conditions in the wild type (wt) and Δpot1. T4 DNA Polymerase (a 3′→5′ exonuclease [Exo]) was added to reactions shown in lanes designated by (+).
(B) Quantification of the G-overhang signal in (A). Δpot1 signal intensity relative to the wild type is shown (error bars measured for n = 4, sd is ± 0.835).
S. pombe and mammalian POT1 proteins are required to protect chromosomes from end-to-end fusions (Baumann and Cech, 2001; Veldman et al., 2004; He et al., 2006; Hockemeyer et al., 2006). By contrast, Arabidopsis POT1a and POT1b null mutants do not exhibit telomere fusions (Surovtseva et al., 2007; E.V. Shakirov, A.D. Nelson, and D.E. Shippen, unpublished data). To evaluate chromosome stability in Δpot1, we employed the telomere fusion PCR assay (TF-PCR) (Heacock et al., 2004). As with PETRA, TF-PCR uses unique subtelomere-specific PCR primers directed toward the chromosome terminus. If the target chromosome ends are covalently fused, the fusion junction will be amplified and detected by DNA gel blot analysis. As expected, no TF-PCR products were observed with DNA samples from wild-type P. patens or in the complemented Δpot1-Pp-POT1 strain (see Supplemental Figure 8C online). However TF-PCR products were abundant in reactions with Δpot1 DNA (Figure 6A). Interestingly, chromosome fusions were also detected in Δpot1 PCR reactions containing only one subtelomere-specific primer (Figure 6B). Since P. patens spends most of its lifecycle in the haploid form, these data suggest that sister chromatids fuse in the absence of POT1.
Figure 6.
Accumulation of Chromosome Fusions in Δpot1.
(A) and (B) TF-PCR detects multiple instances of end-to-end chromosome fusions in Δpot1 moss but not in the wild type (wt). TF-PCR results with subtelomeric primers specific for chromosome arms A and B and DNA from three individual colonies of the wild type or Δpot1 (A). TF-PCR results with one subtelomeric primer specific for chromosome arm B (B).
(C) Increased incidence of chromosome fusions in Δpot1 cultures continuously regrown from a single protoplast. Top panel: Diagram of the experimental design. Bottom panel: TF-PCR results with DNA from wild-type (lane 1) and Δpot1 colonies (lanes 2-4). Δpot1 colonies were regenerated from a single cell through protoplasting once (lane 2), twice (lane 3), or three times (lane 4) (see asterisks in top panel). Arrow indicates a nonspecific PCR product detected in some wild-type samples. Molecular weight markers are shown on the left of each panel.
Chromosome end maintenance defects worsen over time in mouse models deficient for either mPOT1a or mPOT1b (Wu et al., 2006; Hockemeyer et al., 2008). By contrast, we found no evidence of progressive telomere shortening or increased incidence of telomere fusions in long-term vegetatively growing Δpot1 cultures. However, since relatively few cultured P. patens cells are proliferating (Cove et al., 2006), it is possible that the telomere deprotection phenotype would be exacerbated if the majority of Δpot1 cells were forced to divide. To investigate this possibility, we subjected Δpot1 cells to several consecutive rounds of colony regrowth from a single progenitor cell (protoplast) (Figure 6C). As predicted, the abundance of TF-PCR products was elevated in Δpot1 cultures as the number of protoplasting and new colony formation events increased (Figure 6C). Taken together, our data indicate that POT1 is essential for telomere integrity and chromosome end protection in P. patens.
DISCUSSION
P. patens, a New Model System for Plant Telomere Research
The ability of eukaryotic cells to distinguish between double-stranded DNA breaks and natural chromosome termini is essential to avoid aberrant genome rearrangements and to ensure continued cell proliferation. The factors responsible for telomere stability are evolving rapidly (Linger and Price, 2009). In vertebrates, shelterin represses ATR- and ATM-dependent DNA damage pathways, preventing telomere-specific homologous recombination and nonhomologous end-joining reactions (de Lange, 2009). While only a subset of the shelterin components can be discerned in plant genomes, single-stranded telomeric DNA binding proteins are an essential component of the protective telomere cap in plants (Surovtseva et al., 2009) and many other model organisms (Gao et al., 2007; Raices et al., 2008; Linger and Price, 2009; Miyake et al., 2009). POT1 orthologs have been described in a wide variety of eukaryotic lineages, from ciliated protozoa and yeast to humans (Baumann and Cech, 2001; Baumann et al., 2002; Jacob et al., 2007). Despite substantial amino acid sequence conservation, POT1a and POT1b from Arabidopsis appear to have lost this critical chromosome capping function, raising the possibility that POT1 plays a fundamentally different role in the plant kingdom. The analysis of POT1 in the moss P. patens reported here rejects this hypothesis and indicates that in early land plant lineages, such as bryophytes, POT1 functions in a similar fashion to its fungal and vertebrate counterparts.
P. patens has the potential to emerge as a powerful new model for telomere biology. Like Arabidopsis, P. patens has short telomere tracts abutted by unique sequences on at least some chromosome ends. These characteristics allow detection of subtle defects in telomere maintenance and chromosome end protection using the highly sensitive PCR assays of PETRA and TF-PCR (Heacock et al., 2004). Like Arabidopsis, P. patens is genetically tractable, but it has the added advantage of a streamlined genome with much less redundancy. Besides a single-copy POT1 gene, P. patens harbors only two TRFL genes (as opposed to six in Arabidopsis). Finally, P. patens has an exceptionally high rate of homologous recombination, and we exploited this property for targeted deletion of POT1. Our results not only provide direct genetic evidence of an evolutionarily conserved role for POT1 in chromosome end protection in plants, but they also demonstrate that P. patens can serve as a critical counterpoint to Arabidopsis, facilitating a deeper understanding of plant telomere composition and evolution.
Conservation of POT1 Functions in Eukaryotes
While the functions of POT1 differ between Arabidopsis and P. patens, together they encompass several of the roles previously ascribed to POT1 orthologs in mammals and fission yeast. Several studies indicated that mammalian POT1 proteins modulate telomerase action in vitro (Wang et al., 2007; Latrick and Cech, 2010; Zaug et al., 2010) and in vivo (Colgin et al., 2003; Loayza and de Lange, 2003). Although the details of how POT1 influences telomerase are likely to be organism specific, this function is conserved in plants, as illustrated by the phenotypes of Arabidopsis POT1a-deficient plants (Surovtseva et al., 2007). Similar to the budding yeast ever-shorter-telomere mutants (Lundblad and Szostak, 1989), Arabidopsis POT1a null mutants display progressive telomere shortening at a rate of ∼200 to 500 bp per plant generation (Surovtseva et al., 2007). As previously described for plants lacking the telomerase catalytic subunit TERT (Riha et al., 2001), this gradual telomere erosion leads to a delayed onset of developmental and cell proliferation defects apparent only after multiple plant generations. By contrast, P. patens Δpot1 mutants exhibit a rapid onset of telomere uncapping characterized by extensive telomere shortening, increased G-overhang length and chromosome fusions. These phenotypes are reminiscent of the situation in S. pombe Pot1-deficient cells, where chromosome ends rapidly lose all of their telomeric DNA and undergo chromosome circularization (Baumann and Cech, 2001). Thus, the function of P. patens POT1 is consistent with an essential role in chromosome end protection.
Like the surviving POT1-deficient S. pombe cells, which cannot undergo meiosis (Baumann and Cech, 2001), P. patens Δpot1 cultures fail to undergo sexual reproduction and display a partial loss of developmental program. However, unlike S. pombe, P. patens Δpot1 mutants do not lose all telomeric DNA and instead telomeres stabilize at a new, shorter length. This outcome may reflect the establishment of a new equilibrium between telomere-shortening activities and extension by telomerase. It is also possible that homologous recombination contributes to the maintenance of telomere tracts in Δpot1 P. patens, as proposed for telomerase-deficient Arabidopsis mutants (Watson and Shippen, 2007). Mouse POT1 proteins prevent recombination of telomeric DNA (He et al., 2006; Wu et al., 2006; Palm et al., 2009), and considering the naturally high rate of homologous recombination in P. patens, this alternative mode of telomere maintenance is certainly feasible.
Although we could detect chromosome fusions in long-term dividing P. patens Δpot1 cultures, accumulation of these aberrant structures was slow, required multiple cell divisions, and did not result in complete senescence. A similar phenotype is associated with POT1 deletion in chicken cells (Churikov et al., 2006), implying that some aspects of Pp POT1 function are more similar to vertebrate POT1 than to the S. pombe POT1. It is also possible that P. patens POT1 functions redundantly with another telomere protection factor, such as a homolog of the putative Arabidopsis double-stranded telomere binding protein TBP1 (Hong et al., 2007) or the newly identified plant CST complex (Surovtseva et al., 2009).
Evolution of POT1 Function in Plants
Our data underscore remarkable divergence of POT1 functions in lower versus higher plants and provide a framework for elucidating evolutionary changes responsible for the switch from telomere protection to telomerase regulation. We speculate that much of the functional divergence is reflected by changes in the nucleic acid binding properties of plant POT1 proteins. Only a subset of the plant POT1 proteins examined to date display single-stranded telomeric DNA binding activity in vitro (Surovtseva et al., 2007; Shakirov et al., 2009a, 2009b). Among those that do bind telomeric DNA, significant variation exists within their respective minimal DNA binding site. For example, while Pp POT1 and Ol POT1 (from green alga O. lucimarinus) prefer telomeric DNA substrates terminating with a T on the 3′ end, POT1 proteins from the flowering plants prefer oligonucleotides terminating in a G (Shakirov et al., 2009b). The length of the POT1 MBS also varies dramatically among different plant species, ranging from seven to 16 nucleotides (Shakirov et al., 2009b; this study). Previously described MBS sequences were reported in the size range 7 to 12 (Croy and Wuttke, 2006; Shakirov et al., 2009b), but here we show that the P. patens POT1 MBS is 16 nucleotides in length. The C terminus of Pp POT1 enhances telomeric DNA binding. It is possible that Pp POT1 oligomerizes through its C-terminal domain, accounting for its unusually long MBS. Notably, the A. officinalis POT1, whose MBS is only nine nucleotides, does not require the C-terminal domain for efficient DNA binding in vitro (Shakirov et al., 2009b). Alternatively, the extended minimum DNA binding site in Pp POT1 might reflect the contribution of an additional OB-fold in the C terminus, as has been postulated in yeast Cdc13 and vertebrate POT1 (Theobald and Wuttke, 2004; Wei and Price, 2004).
The most compelling evidence of the rapid evolution of POT1 is illustrated by the Arabidopsis POT1 proteins. Recent data indicate that At POT1a and At POT1b physically interact with telomerase ribonucleoprotein complexes instead of telomeric DNA (Surovtseva et al., 2007; C. Cifuentes-Rojas, K. Kannan, L. Tseng, and D. Shippen, unpublished data). Thus, we speculate that the switch in plant POT1 proteins from binding telomeric DNA to telomerase fueled their evolution from telomere capping proteins into telomerase regulatory factors. Further analysis of POT1 in Arabidopsis and P. patens may more clearly elucidate the dynamic and evolving interactions between the chromosome terminus and the telomerase enzyme.
METHODS
Moss Techniques and Generation of POT1 Deletion and Complementation Constructs
Growth of Physcomitrella patens isolates Gd and Vx-1 and protoplast generation and transformation were performed as described previously (Perroud and Quatrano, 2008). The gene deletion construct was designed to remove the full ORF of Pp POT1 from start codon to stop codon. Genomic DNA (1.2 kb) immediately upstream of the start codon was PCR amplified with primers 5′-GCATCCTAGGCGTGTGATCCCGCAAT-3′ and 5′-GCATCTCGAGGAGAACAGACGATTATGTAAG-3′ and inserted 5′ to the hygromycin resistance HptII gene in the pBHRF vector (Schaefer et al., 2010), using AvrII and XhoI restriction enzymes. Similarly, 1.2 kb of genomic DNA immediately downstream of the Pp POT1 stop codon was PCR amplified with primers 5′-GCATGCGGCCGCGAGCCAATTTTTTTTTTGGCTTTG-3′ and 5′-GCATACTAGTATGAAGATAATAGTGC-3′ and inserted 3′ to the hygromycin resistance cassette using NotI and SpeI restriction enzymes. The final vector containing the hygromycin resistance cassette, flanked by the targeting P. patens genomic DNA sequence on both sides, was cut with AvrII and SpeI and introduced into wild-type P. patens isolate Gd by protoplast transformation (Schaefer and Zryd, 1997). Double crossing-over with this construct results in the complete deletion of POT1 ORF, and the entire Δpot1 locus was subsequently PCR amplified with primers 5′-GCTCATACAACAAGCACATTGAC-3′ and 5′-CAACTTCATCCAACCATGCAG-3′, flanking the POT1 locus, and sequenced to verify its absence.
To generate the complementation construct Δpot1-Pp-POT1, the entire Pp POT1 ORF was PCR amplified with 5′-GAATTCTCTTGGAAAGATGCGAGCGGCTGGTCTATGTTGCAGAGGACCATGTCG-3′ and 5′-GATCCTCGAGTCATCCCGGAAATCGTGTAC-3′ and cloned into the pBNRF vector (Schaefer et al., 2010) upstream of the G418 (kanamycin) resistance gene NptII using the same restriction sites. The targeting P. patens genomic DNA sequence on both sides of the complementation construct was the same as in pBHRF and contained the native Pp POT1 promoter. Δpot1 strain was transformed as described above. Stable transformants were selected on G418-containing medium and genotyped for the presence of the rescuing construct in the correct location. DNA gel blot analysis was performed with DIG-labeled PCR probes spanning the targeting sequences as described by Perroud and Quatrano (2008).
TRF, PETRA, and TF-PCR
P. patens DNA was extracted as described (Cocciolone and Cone, 1993). TRF analysis was performed using TruII (Fermentas) restriction enzyme and a 32P 5′ end-labeled (T3AG3)4 oligonucleotide probe (Fitzgerald et al., 1999). For the Bal31 exonuclease assay, 100 μg of P. patens genomic DNA was incubated with 30 units of Bal31 (New England Biolabs) or with water (0 min time point) in 1× Bal31 reaction buffer at 30°C. Equal amounts of sample were removed at specified intervals for 60 min. Reactions were stopped by the addition of 20 mM EGTA and heating to 65°C for 15 min. DNA in each sample was precipitated with isopropanol and ammonium acetate, followed by TruII digestion and DNA gel blotting as described above. TF-PCR was performed essentially as described (Heacock et al., 2004) using primers 5′-CACTACATCGCTGGTCAGAAACGA-3′ (specific for chromosome arm A, P. patens scaffold_2162) and 5′-GAAGGTATGTCATGGCCTCAAAGCT-3′ (specific for chromosome arm B, P. patens scaffold_229). Primer B can also be substituted with primer B1 (5′-GATTGCACCATCATTGCCATCGCA-3′), which is located on the same scaffold 32 nucleotides closer to the start of the telomeric tract. Two modifications to the TF-PCR protocol were made: primer annealing was performed at 58°C, and PCR was run for 30 cycles. PETRA was performed for 19 cycles with primers A or B as described (Heacock et al., 2004). Radioactive signals were scanned by a Pharos FX Plus Molecular Imager (Bio-Rad Laboratories), and the data were analyzed by either IMAGEQUANT software (Molecular Dynamics) or by Quantity One v.4.6.5 software (Bio-Rad). G-overhang analysis and site-directed mutagenesis were performed as described (Song et al., 2008). The quantitative telomere repeat amplification protocol procedure can be found in the Supplemental Methods online.
EMSA Assays
Expression of plant POT1 proteins in RRL and EMSA assays were conducted as described (Shakirov et al., 2009a). Briefly, each reaction (15 μL total volume) contained equal amounts (4 μL) of RRL-translated plant POT1 protein, 0.5 pmol of 32P-labeled telomeric oligonucleotide, 3 μL of 5× DNA binding buffer (100 mM Tris-HCl, pH 7.8, 250 mM NaCl, 50 mM MgCl, 5 mM EDTA, 5 mM DTT, and 25% glycerol) and 1 μL each of Herring sperm DNA (50 μg/mL), DNA oligonucleotide 5′-TTAATTAACCCGGGGATCCGGCTTGATCAACGAATGATCC-3′ and RNA oligonucleotide (UUUAGGG)5, which were added as nonspecific competitors (2.5 μM each) as described by Baumann et al. (2002). Reactions were incubated at room temperature for 15 min, and the complexes were separated on 5% polyacrylamide gel (acrylamide:bisacrylamide, 29:1) for 2 h at 150 V in 0.8× TBE at room temperature, dried, and exposed to phosphor imager screens. Screens were scanned by a Pharos FX Plus molecular imager, and signal intensity was quantified by Quantity One v.4.6.5 software.
RACE and RT-PCR Conditions
Total RNA was extracted from plant tissue using Tri Reagent solution (Sigma-Aldrich). Reverse transcription was performed using Superscript III reverse transcriptase (Invitrogen) as described (Shakirov et al., 2005). To deduce the full-length Pp POT1 cDNA, 5′ and 3′ rapid amplification of cDNA ends (FirstChoice RLM-RACE kit; Ambion) was performed according to the manufacturer's instructions. To analyze Pp POT1 expression, RT-PCR was initially performed with control reactions for Ubiquitin gene expression as described (Harries et al., 2005) to normalize for RNA loading, followed by POT1 RT-PCR with primers 5′-TCGACCCGGGATGTTGCAGAGGACCATGTC-3′ and 5′-GATCCTCGAGTCATCCCGGAAATCGTGTAC-3′, which span the entire ORF.
Accession Numbers
Sequence data from this article can be found in the Arabidopsis Genome Initiative or GenBank/EMBL databases under the following accession numbers: AY884593 (At POT1a), AY884594 (At POT1b), Q9NUX5 (h POT1), and XP_001766873 (Pp Ubiquitin). Pp POT1 cDNA was deposited into GenBank under accession number EU880302.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure 1. Analysis of DNA Binding Capacity of Recombinant Plant POT1 Proteins.
Supplemental Figure 2. Pp POT1 Efficiently Binds the AGGGTTT Permutation of the Plant Telomeric Repeat in Vitro.
Supplemental Figure 3. DNA and Protein Domain Requirements for Efficient DNA Binding of Pp POT1 Protein.
Supplemental Figure 4. Generation of the Δpot1 Strain.
Supplemental Figure 5. Colony Morphology and Growth Defects in Δpot1 P. patens.
Supplemental Figure 6. Bal31 Analysis of Telomeres in the Δpot1 Strain of P. patens.
Supplemental Figure 7. Generation of Δpot1-Pp POT1 Complementation Strain.
Supplemental Figure 8. Complementation of Δpot1-Specific Telomere Phenotypes in the Δpot1-Pp POT1 Complementation Strain.
Supplemental Figure 9. In Vitro Telomerase Activity Levels Are Not Affected in Δpot1 Mutants.
Supplemental Methods. Quantitative Telomerase Activity Assay (Q-TRAP).
Acknowledgments
We thank Johnny Croy and Debbie Wuttke for help with modeling P. patens POT1, Jessica Joseph and Lauren Gunther for help with maintenance of moss cultures, and other members of the Shippen and Quatrano laboratories for insightful discussions. This work was supported by National Institutes of Health Grant GM065383 to D.E.S. and a Washington University in St. Louis grant to R.S.Q.
References
- Baumann P., Cech T.R. (2001). Pot1, the putative telomere end-binding protein in fission yeast and humans. Science 292: 1171–1175 [DOI] [PubMed] [Google Scholar]
- Baumann P., Podell E., Cech T.R. (2002). Human Pot1 (protection of telomeres) protein: Cytolocalization, gene structure, and alternative splicing. Mol. Cell. Biol. 22: 8079–8087 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen C.M., Wang C.T., Ho C.H. (2001). A plant gene encoding a Myb-like protein that binds telomeric GGTTAG repeats in vitro. J. Biol. Chem. 276: 16511–16519 [DOI] [PubMed] [Google Scholar]
- Churikov D., Price C.M. (2008). Pot1 and cell cycle progression cooperate in telomere length regulation. Nat. Struct. Mol. Biol. 15: 79–84 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Churikov D., Wei C., Price C.M. (2006). Vertebrate POT1 restricts G-overhang length and prevents activation of a telomeric DNA damage checkpoint but is dispensable for overhang protection. Mol. Cell. Biol. 26: 6971–6982 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cocciolone S.M., Cone K.C. (1993). Pl-Bh, an anthocyanin regulatory gene of maize that leads to variegated pigmentation. Genetics 135: 575–588 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colgin L.M., Baran K., Baumann P., Cech T.R., Reddel R.R. (2003). Human POT1 facilitates telomere elongation by telomerase. Curr. Biol. 13: 942–946 [DOI] [PubMed] [Google Scholar]
- Cove D., Bezanilla M., Harries P., Quatrano R. (2006). Mosses as model systems for the study of metabolism and development. Annu. Rev. Plant Biol. 57: 497–520 [DOI] [PubMed] [Google Scholar]
- Croy J.E., Wuttke D.S. (2006). Themes in ssDNA recognition by telomere-end protection proteins. Trends Biochem. Sci. 31: 516–525 [DOI] [PubMed] [Google Scholar]
- de Lange T. (2005). Shelterin: The protein complex that shapes and safeguards human telomeres. Genes Dev. 19: 2100–2110 [DOI] [PubMed] [Google Scholar]
- de Lange T. (2009). How telomeres solve the end-protection problem. Science 326: 948–952 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Denchi E.L., de Lange T. (2007). Protection of telomeres through independent control of ATM and ATR by TRF2 and POT1. Nature 448: 1068–1071 [DOI] [PubMed] [Google Scholar]
- Fajkus J., Kovarik A., Kralovics R., Bezdek M. (1995). Organization of telomeric and subtelomeric chromatin in the higher plant Nicotiana tabacum. Mol. Gen. Genet. 247: 633–638 [DOI] [PubMed] [Google Scholar]
- Fitzgerald M.S., Riha K., Gao F., Ren S., McKnight T.D., Shippen D.E. (1999). Disruption of the telomerase catalytic subunit gene from Arabidopsis inactivates telomerase and leads to a slow loss of telomeric DNA. Proc. Natl. Acad. Sci. USA 96: 14813–14818 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao H., Cervantes R.B., Mandell E.K., Otero J.H., Lundblad V. (2007). RPA-like proteins mediate yeast telomere function. Nat. Struct. Mol. Biol. 14: 208–214 [DOI] [PubMed] [Google Scholar]
- Harries P.A., Pan A., Quatrano R.S. (2005). Actin-related protein2/3 complex component ARPC1 is required for proper cell morphogenesis and polarized cell growth in Physcomitrella patens. Plant Cell 17: 2327–2339 [DOI] [PMC free article] [PubMed] [Google Scholar]
- He H., Multani A.S., Cosme-Blanco W., Tahara H., Ma J., Pathak S., Deng Y., Chang S. (2006). POT1b protects telomeres from end-to-end chromosomal fusions and aberrant homologous recombination. EMBO J. 25: 5180–5190 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heacock M., Spangler E., Riha K., Puizina J., Shippen D.E. (2004). Molecular analysis of telomere fusions in Arabidopsis: Multiple pathways for chromosome end-joining. EMBO J. 23: 2304–2313 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Higashiyama T., Maki S., Yamada T. (1995). Molecular organization of Chlorella vulgaris chromosome I: Presence of telomeric repeats that are conserved in higher plants. Mol. Gen. Genet. 246: 29–36 [DOI] [PubMed] [Google Scholar]
- Hockemeyer D., Daniels J.P., Takai H., de Lange T. (2006). Recent expansion of the telomeric complex in rodents: Two distinct POT1 proteins protect mouse telomeres. Cell 126: 63–77 [DOI] [PubMed] [Google Scholar]
- Hockemeyer D., Palm W., Wang R.C., Couto S.S., de Lange T. (2008). Engineered telomere degradation models dyskeratosis congenita. Genes Dev. 22: 1773–1785 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hong J.P., Byun M.Y., Koo D.H., An K., Bang J.W., Chung I.K., An G., Kim W.T. (2007). Suppression of RICE TELOMERE BINDING PROTEIN 1 results in severe and gradual developmental defects accompanied by genome instability in rice. Plant Cell 19: 1770–1781 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Houghtaling B.R., Cuttonaro L., Chang W., Smith S. (2004). A dynamic molecular link between the telomere length regulator TRF1 and the chromosome end protector TRF2. Curr. Biol. 14: 1621–1631 [DOI] [PubMed] [Google Scholar]
- Hwang M.G., Cho M.H. (2007). Arabidopsis thaliana telomeric DNA-binding protein 1 is required for telomere length homeostasis and its Myb-extension domain stabilizes plant telomeric DNA binding. Nucleic Acids Res. 35: 1333–1342 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacob N.K., Lescasse R., Linger B.R., Price C.M. (2007). Tetrahymena POT1a regulates telomere length and prevents activation of a cell cycle checkpoint. Mol. Cell. Biol. 27: 1592–1601 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kannan K., Nelson A.D., Shippen D.E. (2008). Dyskerin is a component of the Arabidopsis telomerase RNP required for telomere maintenance. Mol. Cell. Biol. 28: 2332–2341 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karamysheva Z.N., Surovtseva Y.V., Vespa L., Shakirov E.V., Shippen D.E. (2004). A C-terminal Myb extension domain defines a novel family of double-strand telomeric DNA-binding proteins in Arabidopsis. J. Biol. Chem. 279: 47799–47807 [DOI] [PubMed] [Google Scholar]
- Kumar S., Tamura K., Nei M. (2004). MEGA3: Integrated software for Molecular Evolutionary Genetics Analysis and sequence alignment. Brief. Bioinform. 5: 150–163 [DOI] [PubMed] [Google Scholar]
- Latrick C.M., Cech T.R. (2010). POT1-TPP1 enhances telomerase processivity by slowing primer dissociation and aiding translocation. EMBO J. 29: 924–933 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lei M., Podell E.R., Baumann P., Cech T.R. (2003). DNA self-recognition in the structure of Pot1 bound to telomeric single-stranded DNA. Nature 426: 198–203 [DOI] [PubMed] [Google Scholar]
- Lei M., Podell E.R., Cech T.R. (2004). Structure of human POT1 bound to telomeric single-stranded DNA provides a model for chromosome end-protection. Nat. Struct. Mol. Biol. 11: 1223–1229 [DOI] [PubMed] [Google Scholar]
- Linger B.R., Price C.M. (2009). Conservation of telomere protein complexes: Shuffling through evolution. Crit. Rev. Biochem. Mol. Biol. 44: 434–446 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu D., Safari A., O'Connor M.S., Chan D.W., Laegeler A., Qin J., Songyang Z. (2004). PTOP interacts with POT1 and regulates its localization to telomeres. Nat. Cell Biol. 6: 673–680 [DOI] [PubMed] [Google Scholar]
- Loayza D., de Lange T. (2003). POT1 as a terminal transducer of TRF1 telomere length control. Nature 424: 1013–1018 [DOI] [PubMed] [Google Scholar]
- Loayza D., Parsons H., Donigian J., Hoke K., de Lange T. (2004). DNA binding features of human POT1: A nonamer 5′-TAGGGTTAG-3′ minimal binding site, sequence specificity, and internal binding to multimeric sites. J. Biol. Chem. 279: 13241–13248 [DOI] [PubMed] [Google Scholar]
- Lundblad V., Szostak J.W. (1989). A mutant with a defect in telomere elongation leads to senescence in yeast. Cell 57: 633–643 [DOI] [PubMed] [Google Scholar]
- McGuffin L.J., Bryson K., Jones D.T. (2000). The PSIPRED protein structure prediction server. Bioinformatics 16: 404–405 [DOI] [PubMed] [Google Scholar]
- Miyake Y., Nakamura M., Nabetani A., Shimamura S., Tamura M., Yonehara S., Saito M., Ishikawa F. (2009). RPA-like mammalian Ctc1-Stn1-Ten1 complex binds to single-stranded DNA and protects telomeres independently of the Pot1 pathway. Mol. Cell 36: 193–206 [DOI] [PubMed] [Google Scholar]
- O'Malley R.C., Ecker J.R. (2010). Linking genotype to phenotype using the Arabidopsis unimutant collection. Plant J. 61: 928–940 [DOI] [PubMed] [Google Scholar]
- Palm W., Hockemeyer D., Kibe T., de Lange T. (2009). Functional dissection of human and mouse POT1 proteins. Mol. Cell. Biol. 29: 471–482 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perroud P.F., Quatrano R.S. (2008). BRICK1 is required for apical cell growth in filaments of the moss Physcomitrella patens but not for gametophore morphology. Plant Cell 20: 411–422 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quatrano R.S., McDaniel S.F., Khandelwal A., Perroud P.F., Cove D.J. (2007). Physcomitrella patens: Mosses enter the genomic age. Curr. Opin. Plant Biol. 10: 182–189 [DOI] [PubMed] [Google Scholar]
- Raices M., Verdun R.E., Compton S.A., Haggblom C.I., Griffith J.D., Dillin A., Karlseder J. (2008). C. elegans telomeres contain G-strand and C-strand overhangs that are bound by distinct proteins. Cell 132: 745–757 [DOI] [PubMed] [Google Scholar]
- Rensing S.A., Fritzowsky D., Lang D., Reski R. (2005). Protein encoding genes in an ancient plant: Analysis of codon usage, retained genes and splice sites in a moss, Physcomitrella patens. BMC Genomics 6: 43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rensing S.A., et al. (2008). The Physcomitrella genome reveals evolutionary insights into the conquest of land by plants. Science 319: 64–69 [DOI] [PubMed] [Google Scholar]
- Riha K., McKnight T.D., Griffing L.R., Shippen D.E. (2001). Living with genome instability: Plant responses to telomere dysfunction. Science 291: 1797–1800 [DOI] [PubMed] [Google Scholar]
- Rossignol P., Collier S., Bush M., Shaw P., Doonan J.H. (2007). Arabidopsis POT1A interacts with TERT-V(I8), an N-terminal splicing variant of telomerase. J. Cell Sci. 120: 3678–3687 [DOI] [PubMed] [Google Scholar]
- Schaefer D.G. (2001). Gene targeting in Physcomitrella patens. Curr. Opin. Plant Biol. 4: 143–150 [DOI] [PubMed] [Google Scholar]
- Schaefer D.G. (2002). A new moss genetics: targeted mutagenesis in Physcomitrella patens. Annu. Rev. Plant Biol. 53: 477–501 [DOI] [PubMed] [Google Scholar]
- Schaefer D.G., Delacote F., Charlot F., Vrielynck N., Guyon-Debast A., Le Guin S., Neuhaus J.M., Doutriaux M.P., Nogue F. (2010). RAD51 loss of function abolishes gene targeting and de-represses illegitimate integration in the moss Physcomitrella patens. DNA Repair (Amst.) 9: 526–533 [DOI] [PubMed] [Google Scholar]
- Schaefer D.G., Zryd J.P. (1997). Efficient gene targeting in the moss Physcomitrella patens. Plant J. 11: 1195–1206 [DOI] [PubMed] [Google Scholar]
- Schranz M.E., Mitchell-Olds T. (2006). Independent ancient polyploidy events in the sister families Brassicaceae and Cleomaceae. Plant Cell 18: 1152–1165 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shakirov E.V., McKnight T.D., Shippen D.E. (2009a). POT1-independent single-strand telomeric DNA binding activities in Brassicaceae. Plant J. 58: 1004–1015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shakirov E.V., Shippen D.E. (2004). Length regulation and dynamics of individual telomere tracts in wild-type Arabidopsis. Plant Cell 16: 1959–1967 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shakirov E.V., Song X., Joseph J.A., Shippen D.E. (2009b). POT1 proteins in green algae and land plants: DNA-binding properties and evidence of co-evolution with telomeric DNA. Nucleic Acids Res. 37: 7455–7467 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shakirov E.V., Surovtseva Y.V., Osbun N., Shippen D.E. (2005). The Arabidopsis Pot1 and Pot2 proteins function in telomere length homeostasis and chromosome end protection. Mol. Cell. Biol. 25: 7725–7733 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song X., Leehy K., Warrington R.T., Lamb J.C., Surovtseva Y.V., Shippen D.E. (2008). STN1 protects chromosome ends in Arabidopsis thaliana. Proc. Natl. Acad. Sci. USA 105: 19815–19820 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Surovtseva Y.V., Churikov D., Boltz K.A., Song X., Lamb J.C., Warrington R., Leehy K., Heacock M., Price C.M., Shippen D.E. (2009). Conserved telomere maintenance component 1 interacts with STN1 and maintains chromosome ends in higher eukaryotes. Mol. Cell 36: 207–218 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Surovtseva Y.V., Shakirov E.V., Vespa L., Osbun N., Song X., Shippen D.E. (2007). Arabidopsis POT1 associates with the telomerase RNP and is required for telomere maintenance. EMBO J. 26: 3653–3661 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki K. (2004). Characterization of telomere DNA among five species of pteridophytes and bryophytes. J. Bryol. 26: 175–180 [Google Scholar]
- Theobald D.L., Wuttke D.S. (2004). Prediction of multiple tandem OB-fold domains in telomere end-binding proteins Pot1 and Cdc13. Structure 12: 1877–1879 [DOI] [PubMed] [Google Scholar]
- Veldman T., Etheridge K.T., Counter C.M. (2004). Loss of hPot1 function leads to telomere instability and a cut-like phenotype. Curr. Biol. 14: 2264–2270 [DOI] [PubMed] [Google Scholar]
- Wang F., Podell E.R., Zaug A.J., Yang Y., Baciu P., Cech T.R., Lei M. (2007). The POT1-TPP1 telomere complex is a telomerase processivity factor. Nature 445: 506–510 [DOI] [PubMed] [Google Scholar]
- Watson J.M., Shippen D.E. (2007). Telomere rapid deletion regulates telomere length in Arabidopsis thaliana. Mol. Cell. Biol. 27: 1706–1715 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei C., Price C.M. (2004). Cell cycle localization, dimerization, and binding domain architecture of the telomere protein cPot1. Mol. Cell. Biol. 24: 2091–2102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu L., et al. (2006). Pot1 deficiency initiates DNA damage checkpoint activation and aberrant homologous recombination at telomeres. Cell 126: 49–62 [DOI] [PubMed] [Google Scholar]
- Ye J.Z., Hockemeyer D., Krutchinsky A.N., Loayza D., Hooper S.M., Chait B.T., de Lange T. (2004). POT1-interacting protein PIP1: A telomere length regulator that recruits POT1 to the TIN2/TRF1 complex. Genes Dev. 18: 1649–1654 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zaug A.J., Podell E.R., Nandakumar J., Cech T.R. (2010). Functional interaction between telomere protein TPP1 and telomerase. Genes Dev. 24: 613–622 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zimmer A., Lang D., Richardt S., Frank W., Reski R., Rensing S.A. (2007). Dating the early evolution of plants: detection and molecular clock analyses of orthologs. Mol. Genet. Genomics 278: 393–402 [DOI] [PubMed] [Google Scholar]