Inherited transpositions of the endogenous Ds create stable insertion lines, a resource for targeting gene knockouts and examining mechanisms of transposition. Ds preferentially inserts into genes, at target sites within 16-bpair segments of DNA with specific structural properties. These results suggest approaches to predict insertion sites in transposon mutagenesis experiments.
Abstract
The maize (Zea mays) transposable element Dissociation (Ds) was mobilized for large-scale genome mutagenesis and to study its endogenous biology. Starting from a single donor locus on chromosome 10, over 1500 elements were distributed throughout the genome and positioned on the maize physical map. Genetic strategies to enrich for both local and unlinked insertions were used to distribute Ds insertions. Global, regional, and local insertion site trends were examined. We show that Ds transposed to both linked and unlinked sites and displayed a nonuniform distribution on the genetic map around the donor r1-sc:m3 locus. Comparison of Ds and Mutator insertions reveals distinct target preferences, which provide functional complementarity of the two elements for gene tagging in maize. In particular, Ds displays a stronger preference for insertions within exons and introns, whereas Mutator insertions are more enriched in promoters and 5′-untranslated regions. Ds has no strong target site consensus sequence, but we identified properties of the DNA molecule inherent to its local structure that may influence Ds target site selection. We discuss the utility of Ds for forward and reverse genetics in maize and provide evidence that genes within a 2- to 3-centimorgan region flanking Ds insertions will serve as optimal targets for regional mutagenesis.
INTRODUCTION
The maize (Zea mays) transposable elements Activator (Ac) and Dissociation (Ds) were the first transposable genetic elements identified, and studies of these elements led to a model of cut-and-paste transposition that is still widely accepted today (McClintock, 1949; Greenblatt and Brink, 1962; Greenblatt, 1966). The autonomous 4.5-kb Ac encodes for an 807–amino acid transposase protein that is capable of mobilizing Ac and nonautonomous Ds elements (Coupland et al., 1988; Houba-Herin et al., 1990; Becker and Kunze, 1997). Most Ds elements are simple deletion derivatives of Ac, although a variety of Ds structures have been cataloged (Kunze and Weil, 2002). Studies of Ac/Ds have contributed to our understanding of epigenetic regulation of gene expression (Schwartz and Dennis, 1986; Chen et al., 1987; Chomet et al., 1987), to the mechanisms that restructure genomes (McClintock, 1947; Zhang et al., 2009), and, to our understanding of gene function when used as insertional mutagens, molecular tags, and selectable marker shuttles in two-component gene tagging (Sundaresan et al., 1995; Kolesnik et al., 2004; Qu et al., 2008). Thus, the Ac/Ds system has become one of the best-characterized transposable element families in plants (Kunze and Weil, 2002).
Ac is a member of the hAT superfamily of transposable elements, named after the founding members hobo, Activator, and Tam3 (Calvi et al., 1991). Studies of the hAT element Hermes indicate that this family of elements moves via a cut-and-paste mechanism involving staggered cuts one nucleotide 5′ of each end of the duplex DNA transposon molecule and the formation of a hairpin intermediate (Zhou et al., 2004). Studies of Ac and Ds excision in maize suggest a similar mechanism of transposon excision (Scott et al., 1996; Bai et al., 2007). A recent three-dimensional structure of the Hermes transposase suggests a model of transposition in which a hexameric ring of transposase subunits forms the active hAT transposon complex (Hickman et al., 2005). Relative position of the active binding sites of adjacent monomers likely dictates the 8-bp size of the target site duplication generated upon element insertion in the genome. A higher-order protein complex is also likely necessary for Ac excision (Feldmar and Kunze, 1991; Kunze et al., 1993). Studies in vitro have defined a consensus AAACGG binding site for the Ac transposase in the subterminal repeats of the element that may help nucleate the formation of such a higher-order transposase complex (Kunze and Starlinger, 1989). In contrast with this strong binding site preference for the element ends, no strong target site consensus sequences have been identified for Ds or Ac elements in maize (Grotewold et al., 1991; Dellaporta and Moreno, 1994).
Despite this detailed understanding of Ac/Ds biology and regulation, these elements have seen limited use in maize as a basis for large-scale insertional mutagenesis (Brutnell and Conrad, 2003). This is largely due to relatively low copy number and poor distribution of the elements throughout the maize genome. Both Ac and Ds display a strong reinsertion bias to regions of the genome that are in close genetic linkage to the donor site. In studies of Ac transposition from the p1 and bz1 loci, ∼60% of transpositions were to linked sites, and the majority of transpositions were within 10 centimorgans (cM) of the donor locus (Greenblatt, 1984; Dooner and Belachew, 1989). Notably, this propensity for short-range transpositions can be exploited to create multiple alleles when a target locus is close to an Ac or Ds donor (Hake et al., 1989; Kermicle et al., 1989; Athma et al., 1992; Dellaporta and Calderon-Urrea, 1993; Colasanti et al., 1998; Shen et al., 2000; Vollbrecht et al., 2000; Singh et al., 2003; Bai et al., 2007). In an elegant regional mutagenesis study, >250 Ac-induced alleles of the p1 locus were recovered from Ac insertions flanking the gene (Moreno et al., 1992). These alleles defined exon-intron boundaries, 5′- and 3′-noncoding regions, and a proximal enhancer element nearly 4 kb upstream of the transcription start site.
To exploit Ac/Ds in gene tagging experiments, we developed a genetic scheme to distribute Ds insertions throughout the genome by enriching for transpositions that are unlinked to a donor Ds. An advantage of Ds insertions is that, once mobilized, they may be maintained as stable insertions by removing the Ac transposase source. We analyzed >18,000 Ds excision events and calculated relative frequencies of pre- and postmeiotic transposition. Sequences flanking just over 2000 Ds elements were recovered, and 1616 insertions were placed on maize BAC sequences. Insertions mapped to each of the 10 maize chromosomes. However, regional insertion site preferences were detected using both physical and genetic maps. These insertion site biases were compared and contrasted with those of the highly active Mutator transposon. An analysis of Ds insertion sites revealed that DNA at and surrounding the 8-bp target site duplication displayed specific structural features relative to flanking DNA. These features include a symmetrical arrangement of a particular combination of flexible and rigid conformational domains in a 16- to 18-bp window centered on the 8-bp target site.
RESULTS
Genetic Screen for Linked and Unlinked Transpositions
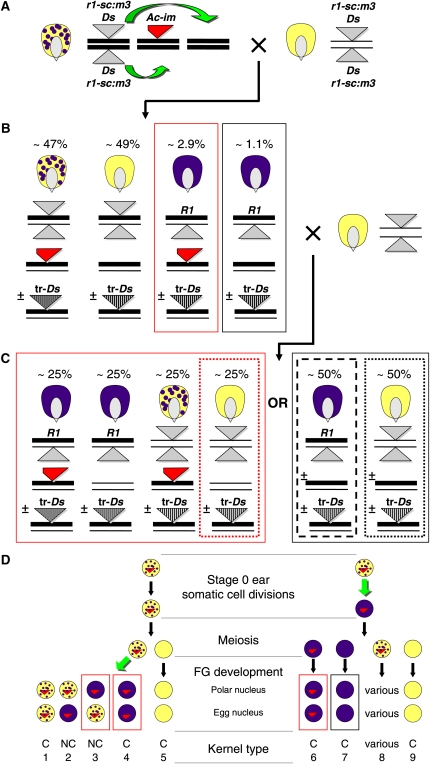
To distribute Ds insertions throughout the maize genome, an immobilized source of Activator, (Ac-im) was used (Conrad and Brutnell, 2005). Ac-im provides transposase to mobilize Ds elements but is incapable of excision due to a terminal 10-bp deletion. In the genetic scheme, described in detail in Figure 1, Supplemental Figure 1, and Supplemental Methods, Ds excisions were first selected from a donor locus that regulates anthocyanin biosynthesis in the kernel, then propagated for one generation prior to screening for Ds reinsertions. This two-step strategy was adopted to enrich for germinal transposition events and reduce the frequency of cloning somatic insertions. Lines containing Ac-im (chromosome 7S) and a Ds insertion at the r1 locus (chromosome 10L; the r1-sc:m3 allele) were the donor (stage 0) lines for transposition events. These lines were test crossed by an r1-sc:m3 line (Figure 1A). Among the resulting stage 1 progeny were germinal Ds excisions that restored a functional R1 allele (Figure 1B) and potentially contained a Ds reinsertion. To recover germinally inherited events and to segregate the source of transposase away from the transposed Ds (tr-Ds, Figure 1B), each fully colored (R1) stage 1 kernel was sown and test crossed to create corresponding stage 2 progeny with (Figure 1C, left group) or without (Figure 1C, right group) Ac-im. Thus, the vast majority of Ds insertions recovered resulted from pre- or postmeiotic transpositions of Ds from the r1-sc:m3 locus. Overall, 49% of Ac-im mediated excisions of Ds from r1-sc:m3 occurred during female gametophyte development, similar to the frequency reported previously from a much smaller sample size (Conrad and Brutnell, 2005). However, among R1 excisions that were concordant (R1 inherited in both embryo and endosperm lineages), there was an ∼2:1 ratio of sporophyte to gametophyte excisions (see Supplemental Methods online).
Figure 1.
Pedigree Representation of Genetic Scheme to Enrich for Unlinked Ds insertions.
(A) Ds transposition, signified by the green arrows, occurs in stage 0 (zero) of the pedigree. Spotted kernels hemizygous for Ac-im (red triangle with a broken end) are sown and then test crossed to provide opportunity for the Ds element (gray triangle) in the r1-sc:m3 allele to transpose to new locations in the genome. Chromosomes from stage 0 plants are represented as thick lines to track them through subsequent generations.
(B) Putative stage 0 Ds transposition events are selected as fully colored stage 1 progeny kernels and test crossed to validate excision and detect segregating Ac-im elements. Each F1 (stage 1) ear segregates nearly 1:1 for coarsely spotted (inherited Ac-im) and colorless (without Ac-im) kernels. These ears also segregate a low frequency of fully colored kernels that arise from Ds excision in stage 0 to generate and transmit a functional R1 allele. If the excised Ds reinserted, then a transposed Ds (tr-Ds, barred gray triangle inserted into a stage 0–derived chromosome) may segregate into any stage 1 kernel class, as dictated by when the excision occurred and where the element reinserted (see text). Relative frequencies of each kernel class were calculated from our populations. All fully colored, stage 1 kernels are selected (red and black boxes) and test crossed again by r1-sc:m3 pollen, creating one stage 2 ear per putative Ds excision event.
(C) Stage 2 progeny are the source for cloning the stage 0 transpositions and for seed distribution. Each stage 2 ear either contains Ac-im (left, derived from red boxed kernel class in stage 1) or lacks it (right, derived from black boxed class in stage 1). Progeny kernels are selected from each stage 2 ear and sown for DNA isolation from the seedlings, to identify and clone inherited, transposed Ds elements. To select for unlinked transpositions, kernels without Ac-im (light dotted red and light dotted black boxes) are selected for DNA isolation. To enrich for transpositions linked to the revertant R1 allele from stage 0, fully colored progeny are selected only from ears that lacked Ac-im entirely (dashed black box). This selection program identifies stage 2 ears that segregate germinal Ds transposition(s) from stage 0, while excluding somatic reinsertions. Remaining kernels from stage 2 ears are retained as a source for the identified, new Ds insertions. Kernel class frequencies for stage 2 ears are estimated.
(D) Transposition in stage 0 may occur at various times in development, with different genetic consequences. Cells or cell lineages (large circles) are shown in the indicated tissues; a lineage is spotted and/or contains a red triangle when it carries Ac-im. At the r1 locus, a spotted or yellow lineage contains only the r1-sc:m3 allele, while a purple lineage contains an R1 excision allele. Arrows denote cell divisions during developmental stage. A green arrow indicates Ds excision from r1-sc:m3 occurring during cell divisions. Bottom rows of female gametophyte (FG) cells are fertilized to produce stage 1 kernels as in (B). When Ds excision occurs premeiotically (right side) Ac-im and the R1 excision allele segregate independently during meiosis, and after fertilization, all fully colored kernels are concordant (C), with pigmented endosperm and an R1 excision allele in the embryo (boxed, 6 and 7). If Ds excises after meiosis (left side) in FG tissue, then in the soma of the haploid gametophyte the nontransposing Ac-im neither is lost nor segregates away from R1 so the corresponding stage 1 kernels will always contain Ac-im. However, excision may occur during female gametophyte development (see Supplemental Figure 3 online), such that fertilized nuclei may or may not contain the R1 excision allele and progeny may be concordant (C; e.g., box 4) or nonconcordant (NC; e.g., box 3), respectively. Boxed FG genotypes, numbered 1 to 9, are colored to correspond with the progeny type they produce in (B).
To enrich for stable Ds insertions to sites unlinked from the r1 locus, colorless kernels lacking Ac-im (i.e., -Ac-im kernels, Figure 1C, fine-dotted boxes) were selected from each stage 2 ear and processed to recover new Ds insertion(s). By selecting these r1 kernels, transpositions to the same chromatid as the donor locus were selected against and populations were biased toward insertions at unlinked sites (see Supplemental Figure 1 online). And because r1 kernels lack a transposase source, somatic transpositions did not confound the cloning of sequences flanking heritable Ds insertions. Processing new insertion lines (see Methods and Supplemental Figure 2 online) consisted of bar-coding seed packets, extracting highly purified DNA by a high-throughput method for small pools of freshly germinated seedlings (Ahern et al., 2009), and then performing DNA gel blot analysis with methylation-sensitive restriction enzymes to identify lines containing transposed Ds elements. Inverse PCR with a primer specific to the Ds under study and with a methylation-sensitive restriction enzyme was used to recover DNA flanking the Ds, and PCR fragments were sequenced. Sequences flanking Ds were called fDs (for flanking-Ds) tags.
Reinsertion: Genome-Wide Distribution of Transposed Ds Elements
hAT family transposable elements transpose by a cut-and-paste mechanism comprising the coupled but distinct processes of element excision from the donor and reinsertion into the target locus (Zhou et al., 2004). From 18,510 R1 kernel selections that produced high quality and concordant stage 2 ears, 12,317 lines were processed to produce 2636 inverse PCR fragments, each a candidate Ds reinsertion. In many lines, no reinserted Ds was detected. From the inverse PCR fragments, we obtained 2439 fDs sequence tags derived from 2028 new, sequence-indexed Ds insertions (Table 1). The insertions were recovered from 1900 different lines; most lines contained just one new Ds insertion. Some insertions were recovered more than once. In 78 instances, an insertion was recovered twice, in 34 instances three times, in 18 instances four times, in three instances five times, in two instances six times, and in one instance each seven, eight, and nine times, respectively. Because redundant insertions had intact Ds terminal sequences and were detected experimentally at low frequency in the pedigrees, most appear to be independent new transpositions rather than multiple recoveries of cryptic elements (Leu et al., 1992) or other preexisting Ds or Ds-related sequences in the parental lines. Thus, redundancies either represent hotspots of preferential insertion or transpositions that arose as somatic sectors and were falsely captured as independent events in the selection scheme (see Methods). After eliminating redundancy, there were 1785 new Ds insertions. The integrity of the molecular and genetic procedures was tested by genotyping 260 field-grown lines with chromosomally placed transpositions using a PCR assay (see Methods). The PCR product of the expected size was identified in 216 of 260 lines (83.1%). Sequencing of the PCR products from seven lines showed that all contained the expected junction with Ds. Failure to confirm the presence of the Ds in the PCR assay may indicate poor primer design or that the Ds insertion was not recovered in the stage 2 population. In negative controls where primer-template combinations were purposely mismatched, spurious products were detected in zero of 24 lines. These results suggest a minimum of 83% of the 1785 unique, new transpositions are accurately characterized and pedigreed.
Table 1.
fDs Sequence Tag Placement and Repeat Analysis
| fDs Tag Length (bp) |
|||||||||
| BLASTN (Locations)a | Placed?b | Insertions | Fraction | n, Tagsc | Average | Min | Max | Repeat Content | Repetitive Sequences |
| One or two | Yes | 1616 | 79.7% | 1980 | 418 | 15 | 1676 | 7.6% | 1.7% |
| Multiple | No | 139 | 6.9% | 169 | 533 | 21 | 1313 | 19.1% | 7.7% |
| None; e-value < 10−10 | No | 114 | 5.6% | 115 | 117 | 19 | 568 | 3.9% | 0.9% |
| None; e-value > 10−10 | No | 159 | 7.8% | 175 | 244 | 23 | 860 | 11.9% | 5.1% |
| Totals | 2028 | 100.0% | 2439 | 399 | 15 | 1676 | 8.8% | 2.3% | |
Sequence tags grouped into categories based on number of locations they are placed to by our BLAST pipeline.
Genome placement status in public displays at http://plantgdb.org/prj/AcDsTagging/records.php.
Number of sequence tags in this category.
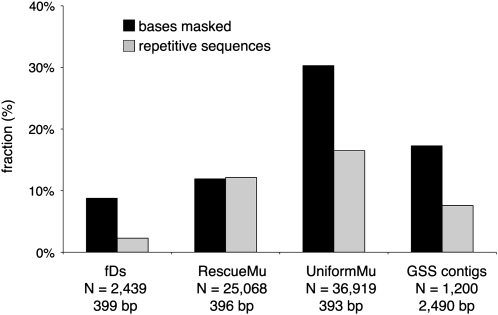
The maize genome consists of a gene-poor repetitive component, in which clusters of Class I and Class II transposable elements constitute ∼84% of the genome and a small fraction of interspersed gene-rich, low-copy regions (Schnable et al., 2009). To provide an estimate of transposed Ds placement in the maize genome, repeat content of the 2439 fDs sequence tags was measured and found to be exceptionally low (8.8% of bases could be repeat masked and 2.3% of sequences classified as repetitive; Figure 2; see Methods). For similarly sized tags that flank engineered (RescueMu) or endogenous (UniformMu) Mutator family transposon insertion sites (Fernandes et al., 2004; McCarty et al., 2005) or for random GSS sequences enriched by methyl-filter and high-C0t methods (Rabinowicz et al., 1999; Yuan et al., 2003), repeat content was substantially higher (Figure 2). These results suggest that large-scale cloning of hypomethylated Ds reinsertions samples a specific component of the maize gene space that differs from that sampled by large scale Mutator cloning. This result may reflect an inherent property of Ds that differs from Mutator. Alternatively, this result may indicate a bias in the distribution of Ds insertions due to the use of methylation-sensitive restriction enzymes (REs) to clone sequences flanking Ds insertions. When a methylation-insensitive RE was used to clone sequences flanking Ac insertions, a higher percentage mapped to repetitive regions in the genome than when methylation-sensitive REs were used (Kolkman et al., 2005).
Figure 2.
Repeat Content of Sequence Tags That Flank Transposon Insertions.
N, number of tags; bp, average tag size in base pairs. Closed bars, percentage of bases masked in each dataset; open bars, percentage of the sequences in a given collection scored as repeat derived. The fDs, RescueMu, and GSS contig data sets contain assembled contigs, while the UniformMu data set consists of raw sequence reads.
New Ds insertions were assigned to BAC locations in the B73 reference genome using BLASTN with criteria that accommodated genome differences between the W22 and B73 inbred lines (see Methods). Of 2028 insertions, 1501 (74%) placed to unique locations and 115 (5.7%) mapped with similar probability to exactly two distinct locations (Table 1), possibly reflecting insertions into homoeologous regions (Gaut and Doebley, 1997). Among the 412 insertions (20.3%) that were not placed by these criteria, 139 (6.9%) mapped to three or more locations in the genome and had a higher repeat content (Table 1). These were tentatively classified as insertions into repetitive regions. One hundred and fourteen insertions (5.6%) could not be placed by our criteria despite having a relatively low BLASTN e-value (<10−10); these appear to represent sequences with more extensive indel polymorphisms between W22 and the reference B73 genome. The 159 unplaced insertions (7.8%) with no good match (e-value > 10−10) may derive from regions that are not yet sequenced in B73 or that are unique to W22. To examine the properties of Ds transposition throughout the genome from a known donor locus, a detailed characterization was performed on the 1482 unique, nonredundant placements generated in this study.
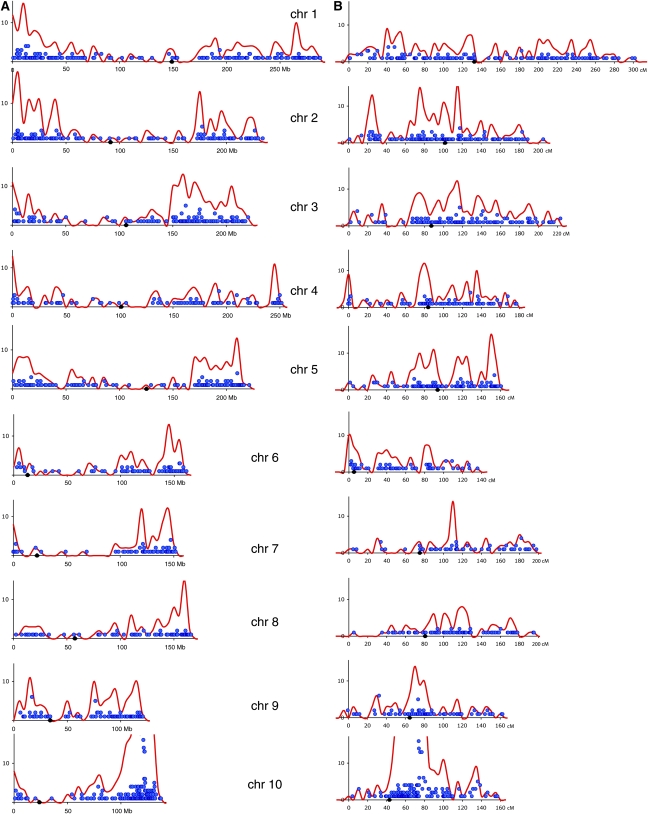
Ds showed a clear preference for reinsertion toward the telomeres and away from the centromeres (Figure 3A). This preference is apparent on every chromosome and likely reflects the higher gene density near the telomeres (Fengler et al., 2007) and a bias for transpositions into the hypomethylated, gene-rich regions of the genome (Chen et al., 1987; Bennetzen et al., 1994; Rabinowicz et al., 1999). As expected, many insertions were recovered near the donor r1 locus on 10L. Therefore, chromosome 10 was excluded when performing statistical analyses of the genome-wide distributions (Table 2). On the basis of their estimated physical length, the other nine chromosomes contained significantly nonrandom Ds reinsertion content, and Ds distribution was also significantly nonrandom when the 18 chromosome arms were considered as bins (P values < 0.0001). Chromosomes 3 and 9 contained more insertions per base pair than expected by chance, and chromosome 4 had fewer (P value < 0.01). At the level of chromosome arms, 2S and 3L contained more insertions per base pair than expected by chance and 4L and 8S contained fewer. On the genetic map, the genome-wide distribution of Ds insertions was smoother, but regional hot and cold spots were apparent (Figure 3B). Among the nine nondonor chromosomes, the overall distribution was nonrandom when either the whole chromosomes or the chromosome arms were considered as bins (P value < 0.0001). In particular, four chromosome arms had more insertions than expected by chance (2S, 3L, 5L, and 6S, range of 0.86 to 1.87 insertions/cM), and three arms had fewer than expected (1L, 7S, and 8S, range of 0.15 to 0.47 insertions/cM). By either metric, physical or genetic, insertions were enriched on 2S and 3L. Interestingly, no marked bias against reinsertion around the site of Ac-im at ∼89 cM on 7L was detected, even though we selected against insertions that were closely linked to Ac-im (i.e., spotted kernels). In summary, Ds transposes to all parts of the maize genome with a likely bias for gene-rich regions, but there may be regional hot and cold spots.
Figure 3.
Distribution of Ds Insertions on the Maize Chromosomes.
(A) Ds insertions plotted on the physical map. The x axis indicates base pair position in megabases (Mb) along each chromosome with short arms on the left and long arms on the right. Centromere positions are indicated by black diamonds. Number of Ds insertions is indicated on the y axis. Each blue dot represents a BAC in the assembly hit by at least one Ds element, with the dot's vertical position corresponding to the number of Ds placed to that BAC. The red curve is a smoothed plot of insertions per adjacent 5-Mb bin. Insertions with redundancy >2 are omitted, and only one BAC match per insertion location is depicted.
(B) Genetic map positions. Axes and symbols are as in (A), but the coordinate basis is centimorgan units, and the red curve shows insertions per adjacent 5-cM bin.
Table 2.
Genome-Wide Distribution of Ds Insertions
| Location | No. of Ds | Genetic Map P Valuea | Physical Map P Valuea |
| Chromosome | |||
| 1 | 188 | 0.617 | 0.399 |
| 2 | 171 | <0.0001b | 0.015 |
| 3 | 172 | 0.006b | 0.002b |
| 4 | 110 | 0.517 | <0.0001c |
| 5 | 139 | 0.0003b | 0.764 |
| 6 | 85 | 0.888 | 0.140 |
| 7 | 74 | <0.0001c | 0.039c |
| 8 | 84 | 0.001c | 0.094 |
| 9 | 100 | 0.791 | 0.004b |
| 10 | 349 | N/A | N/A |
| Overall | 1472 | <0.0001 | <0.0001 |
| Chromosome Arm | |||
| 1S | 102 | 0.019 | 0.237 |
| 1L | 86 | 0.008c | 1.000 |
| 2S | 97 | <0.0001b | <0.0001b |
| 2L | 74 | 0.251 | 0.131 |
| 3S | 48 | 0.431 | 0.028 |
| 3L | 124 | <0.0001b | <0.0001b |
| 4S | 46 | 0.413 | 0.033 |
| 4L | 64 | 0.920 | 0.001c |
| 5S | 75 | 0.021 | 0.842 |
| 5L | 64 | 0.006 | 0.842 |
| 6S | 10 | 0.0002b | 0.475 |
| 6L | 75 | 0.527 | 0.080 |
| 7S | 11 | <0.0001c | 0.484 |
| 7L | 63 | 0.116 | 0.055 |
| 8S | 17 | <0.0001c | 0.002c |
| 8L | 67 | 0.532 | 1.000 |
| 9S | 29 | 0.081 | 0.071 |
| 9L | 71 | 0.284 | 0.025 |
| 10S | 16 | 0.040 | 0.699 |
| 10L | 333 | N/A | N/A |
χ2 test of independence.
Significant and more Ds than expected by chance.
Significant and fewer Ds than expected by chance.
N/A, not applicable.
Quantifying Interchromosomal and Intrachromosomal Ds Transposition: Linked versus Unlinked Transpositions and Regional Asymmetry among Transpositions to Linked Sites
When distributing Ds insertions throughout the genome, stage 1 individuals containing Ac-im were used and tissue was harvested only from colorless stage 2 progeny lacking Ac-im (Figure 1C, dotted and hyphenated boxes). We hypothesized that by restricting selections to colorless kernels, Ds insertions genetically linked to the revertant R1 allele at the donor locus would be poorly represented in the collection, as discussed earlier (see Supplemental Figure 1 online). Thus, to examine an overall, unfiltered distribution of Ds transpositions from the r1 locus, the subpopulation of all stage 2 ears that lacked Ac-im was examined (Figure 1C, right side, two black boxes). In this subpopulation, we first selected and analyzed the fully colored stage 2 kernels (Figure 1C, black hyphened box, purple kernel) that should enrich for Ds insertions in the proximity of the donor r1 locus and then selected and analyzed the colorless stage 2 kernels (Figure 1C, black dotted box, yellow kernel) that should enrich for unlinked Ds insertions. Because this sampling was inclusive for almost all events, to linked and unlinked sites, we refer to these selections as inclusive screening (by contrast, standard screening was biased to unlinked sites). Those transpositions not sampled using inclusive screening were Type II events linked to the donor locus (see Supplemental Figure 1 online); these were systematically excluded by both screening approaches.
Using the inclusive screening method (∼8000 stage 1 ears were screened to identify 2000 that did not contain Ac but segregated R1 kernels), Ds insertions were recovered on chromosome 10 at a rate of 42% (123 of 293 genome placements; Table 3), which was significantly higher than the rate using the standard approach (17.2% or 93 of 541 placements; P value < 10−11). Moreover, in the inclusive and standard populations, the distributions of transposition among the nine, nondonor chromosomes were similar (P value = 0.26), but distributions differed either when defined as the complete, 10 chromosomes (P value < 10−14) or as two fractions, “on” and “not on” chromosome 10 (P value < 10−18). These data confirm the efficacy of the genetic strategies to select for and against linked transpositions.
Table 3.
Stage 2 Selection Enriches for Unlinked Transpositions
| Screen Approach, Ds Recovered |
||
| Chromosome | Inclusive | Standarda |
| 1 | 38 | 75 |
| 2 | 31 | 64 |
| 3 | 22 | 65 |
| 4 | 14 | 45 |
| 5 | 13 | 65 |
| 6 | 16 | 33 |
| 7 | 11 | 28 |
| 8 | 12 | 29 |
| 9 | 13 | 44 |
| 10 | 123 | 93 |
| Total | 293 | 541 |
Only includes data from seasons where both standard and inclusive screens were performed.
Data from inclusive screening are ideally suited to investigate relationships between Ds transposition frequencies and genetic linkage. Any recovered Ds insertions (Figure 1B, black box) were the result of premeiotic transpositions on stage 0 plants, followed by segregation of Ac-im away from the donor locus on stage 1 ears. Thus, of all unlinked, premeiotic Ds transpositions generated in stage 0 plants, approximately half cosegregated with Ac-im and were not recovered, and half of the remainder cosegregated with the donor locus and were not recovered. Thus, inclusive screening resulted in the recovery of ∼25% of all premeiotic transpositions to unlinked sites (Figure 1D, right panel). In total, 170 unlinked Ds events were recovered from the 2000 ears. As only 25% of the total population was recovered, we estimate the number of premeiotic, unlinked transpositions from r1-sc:m3 at 680/2000 excision events. On the other hand, of the closely linked Ds insertions, nearly all are expected to be recovered among colored stage 1 kernels (see Supplemental Figure 1 online). Thus, selection for R1 kernels likely resulted in the recovery of half of the very tightly linked transpositions (i.e., those that did not cosegregate with Ac-im), and 0.5*((1 − p)/100) of less tightly linked reinsertions at a recombination distance p between the reinsertion and the donor. We applied these frequencies to the 117 linked transpositions (placed to locations within ±50 cM of the donor) that were recovered from the 2000 lines by binning the insertions in progressively larger centimorgan intervals around the r1 locus and then calculating for each bin the number of corresponding reinsertions that segregated into other kernel classes (see Supplemental Table 1 online). Thus, the number of premeiotic, linked transpositions was estimated as 377/2000 excision events. These calculated frequencies are likely an underestimate of the total number of Ds transpositions as they assume all transposed Ds elements were detected in the molecular assay that is clearly overoptimistic; moreover, single R1 kernels were selected when they occurred in sectors of three or more. However, each class, unlinked and linked, should be equally affected by those detection limits, and the data may be used to compare frequencies to linked and unlinked sites.
To estimate the relative frequency of premeiotic, linked, and unlinked Ds transpositions in plants hemizygous for Ac-im, we compared the population of unlinked transpositions (680 at sites within ±50 cM of the donor) to the detected (377) and undetected linked transpositions. As discussed above (see Supplemental Figure 1 online), we did not capture Type II events that are tightly linked to the R locus. If we assume those type II Ds transpositions occur at the same frequency as Type I, as seen for Ac (Greenblatt and Moreno), then within a ±10 cM window around r1 we missed ∼169 Type II events (see Supplemental Table 3 online). We therefore calculate the frequency of unlinked reinsertions as 680/(680 + 376 + 169) = 56%. This frequency calculation is limited to transpositions prior to meiosis and excludes intragenic transpositions that fail to restore R function but does suggest that transposition to unlinked sites may occur more frequently for Ds than Ac (Greenblatt, 1984; Dooner and Belachew, 1989).
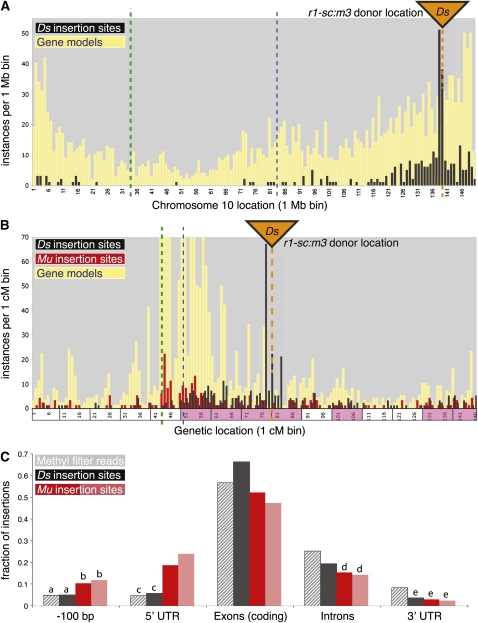
To analyze regional patterns of Ds transposition on chromosome 10, fDs tags were positioned onto the March 2009 pseudomolecule release (4a.53 from maizesequence.org), which consists of 149.7 Mb in a genetic map of 146 cM (see Methods), with the r1-sc:m3 donor locus located at position 138.1 Mb or 78.9 cM. A total of 362 unique insertions on chromosome 10 were examined, from both standard and inclusive screens. Insertions clustered around the donor locus as expected, but surprisingly, significantly more insertions were located on the proximal side of r1 (P values < 0.05; Figure 4A, black bars; see Supplemental Table 2 online). On the genetic map, an even more pronounced asymmetry favoring proximal transpositions was evident (Figure 4B, black bars) and statistically significant (P values < 0.001; see Supplemental Table 2 online). Overall, 323 of the 362 Ds insertions were within 50 cM of the donor locus. To examine the extent of the skew, these 323 insertions were grouped into adjacent, 10-cM bins across the chromosome using the r1 locus as an endpoint for phasing the bins (Figure 4B, boxes on the x axis). Within a continuous 30 cM proximal to r1 and 10 cM distal, the density of reinsertions per bin was much higher than expected by chance (magenta bins in Figure 4B; see Supplemental Table 3 online). While an affinity of Ds for genetically linked regions during intrachromosomal transposition is perhaps anticipated, these data demonstrate that this affinity may extend over vast physical distances, on the order of 50 Mb. As map positions were not derived from direct genetic mapping experiments, the observed asymmetry around the donor locus could be an artifact of inferring B73 placements from W22 sequence. For example, chromosome 10 may be structurally and/or recombinationally dissimilar in W22 and B73. Nevertheless, from these data a distinct polarity to regional Ds transposition from the r1 locus was observed, with a directional bias toward the centromere.
Figure 4.
Distribution of Ds Reinsertions Relative to Chromosome Annotations.
(A) and (B) Detail analysis of intrachromosomal reinsertions on chromosome 10.
(A) Vertical bars indicate the number (y axis) of gene models (yellow, from maizesequence.org release 4a.53) or Ds insertions (black) relative to their binned positions in the pseudomolecule assembly (1-Mb bin size, x axis); bin 1 refers to coordinates 0 to 1 Mb, bin 2 refers to 1 to 2 Mb, etc. Orange triangle and dotted line both indicate the position of the donor locus, r1-sc:m3; heavy, green dotted line is the centromere region; thin, blue dashed line is a landmark location for referencing between (A) and (B).
(B) A genetic map plot is constructed similarly to (A), except the x axis is converted to 1-cM units. Frequency distribution of a subset of Mutator insertions is also plotted (red bars). Adjacent, 10-cM super-bins are shaded magenta on the x axis if their density of Ds insertions is greater than expected by chance (P value < 0.001). The y axis is truncated at 100 instances per 1-cM bin, so the gene model is not shown for nine of the 146 bins (maximum value = 349 genes/cM).
(C) Genome-wide analysis of insertion site positions relative to gene annotation on maize pseudochromosomes. Gray bars are random hypomethylated (control) sequences; other bars indicate Ds (black), UniformMu (dark red), and RescueMu (light red) insertion sites. Each subcategory is mutually exclusive; for example, exons with UTRs are excluded from “coding exons.” Statistically indistinguishable values are marked with the same letter.
Ds Insertion Patterns Relative to Genes and Other Transposons
Because Ds targets hypomethylated regions of the genome (Chen et al., 1987) that are enriched for genes (Bennetzen et al., 1994; Rabinowicz et al., 1999), we compared Ds placement to a recent public release of gene models (version 4a.53 filtered gene set, June 1, 2009) from maizesequence.org. The redundancy in these preliminary gene models was condensed to a core set including all ab initio (FGENESH) genes and only the longest evidence-based gene model per locus. On chromosome 10, gene density (Figure 4A, yellow bars) was highest toward the telomeres, as expected (Fengler et al., 2007). Interestingly, when these data were superimposed onto the genetic map, gene density per centimorgan (Figure 4B, yellow bars) was highest between the centromere (Figure 4, green dotted line) and r1, especially within a region extending proximal to r1 by 35 cM (boundary marked by blue dashed line, Figures 4A and 4B) that coincides with the region enriched for Ds reinsertions. The same patterns were observed when genes were called on the basis of alignment of sorghum (Sorghum bicolor) or rice (Oryza sativa) peptides (data not shown), indicating the asymmetry in gene density is not due to an abundance of transposon sequences in the gene model set. The higher density of genes per centimorgan proximal to r1 relative to distal may be a primary driver of asymmetric Ds distribution. If that were the case, other transposons with genic insertion preferences, such as Mutator (Fernandes et al., 2004; McCarty et al., 2005), should show a similar regional insertion bias. To investigate this hypothesis, we mapped public RescueMu and UniformMu insertion flanking sequences (Fernandes et al., 2004; McCarty et al., 2005) on the pseudomolecules and compared their distribution with Ds. The 260 unique Mu insertions on chromosome 10 (Figure 4B, red bars) also showed a bias for the gene-rich region between r1 and the centromere. Mu insertion density is higher near the centromere, but Ds coverage is much higher in bins closer to the donor position (Figure 4B). For distal, unlinked bins on 10L that showed high frequency of Ds insertions, Mu insertion frequency is also high, suggesting that increased frequency of Ds on distal 10L may be unrelated to the physical connection of this region to the donor. These results support the hypothesis that local gene density acts as a sink to help drive the directional polarity of intrachromosomal Ds reinsertions and highlight potentially complementary, regional differences between Ds and Mu insertion preferences.
To better characterize the potential of Ds for genome-wide mutagenesis in maize, Ds insertion sites were also examined relative to annotated gene features, such as exons, introns, and untranslated regions (UTRs) in the full set of chromosome pseudomolecules. Random hypomethylated clone sequences were used as a control to represent a similar fraction of the genome as the hypomethylated Ds insertion sites recovered in this study. A total of 893 genes contained a Ds insertion, with 66% of the insertions predicted to reside in coding exons (Figure 4C). A similar frequency (75%) was reported for analysis of a much smaller set of Ac insertions (Cowperthwaite et al., 2002). Ds insertions in our collection map within genes (defined as segments from 100 bp upstream to the end of each transcribed region) at a significantly higher rate than do Mutator insertions (P values < 10−10) or the hypomethylated DNA controls (P value < 10−4). This stronger insertion bias for Ds compared with Mu correlates with a relatively increased insertion frequency of Ds into both exons (P values < 10−9) and introns (P values < 0.02). A significantly greater proportion of Ds than of hypomethylated DNA controls in exons implies the enrichment is transposon based and not simply a result of selective recovery of hypomethylated target sites. Conversely, a significantly higher proportion of Mu insertions were in 5′-UTRs and the near promoter region (Figure 4C, −100 bp), consistent with a comprehensive analysis of Mu transposition patterns that per base pair rates of Mu insertion are highest in this segment (Liu et al., 2009). Ds inserted into 3′-UTRs significantly less frequently than expected by chance compared with the hypomethylated controls (P value < 10−4). To investigate whether or not frequency differences derived from each transposon targeting a distinct class of gene, we compared mean lengths of genic compartments for Mu and Ds target genes (see Supplemental Methods online). These analyses indicate a bias of Mu for genes with long UTRs and of Ds for more compact genes with short introns. Thus, both Mu (Liu et al., 2009) and Ds favor genes as insertion sites, but Ds and Mu have preferences for different genic regions, with a significantly larger proportion of Ds insertions in the collection found in genes and in exons.
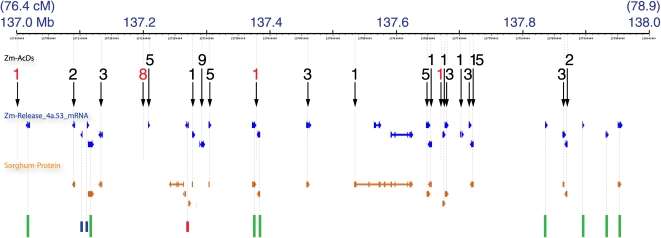
Given the tendency of Ds to transpose to linked sites and the potential to exploit that property for mutagenesis, the dynamics of transposition in the immediate vicinity of a Ds donor was examined. Bin 138 was chosen as 74 nonredundant Ds insertions resided in the 1-Mb and ∼2.5-cM interval, the highest density of insertions and among the highest densities of raw gene models (41 genes/Mb; Figure 4A). There were 28 nonredundant and nonretrotransposon gene models, most of which were supported by sequence similarity to sorghum peptides. Within the bin, Ds insertions distribute as 21 clusters over the gene models, with one to 15 Ds insertions per cluster (Figure 5, one black arrow per cluster). Seventeen of the clusters place at least one Ds insertion in or within a few hundred base pairs of a gene model (Figure 5, black numerals above black arrows). Four additional locations in the region contain Ds insertions but are not closely associated with a gene model, three as singletons, and one as a cluster of eight insertions (Figure 5, red numerals above black arrows). Of the remaining 11 gene models that do not contain at least one Ds insertion, there are eight with cDNA support and/or a sorghum match (Figure 5, green bars, bottom). The other three of these gene models have poor evidence to support them (Figure 5 bottom, short blue and red lines; status left to right: one EST and no sorghum match, two ESTs and no sorghum match, several ESTs but all have a better sequence match to a paralog on chromosome 7), and we tentatively classify these three as possible misannotations. Some of the eight missed genes have Ds insertions in close proximity (e.g., two gene models near 137.38 Mb are separated by 5 kb) with a Ds almost midpoint between them. Thus, in the 1-Mb region, 17/25 well-supported gene models contain insertions (Ds/gene: average = 3.7; range = 1 to 15), while 8/25 do not, and there are four Ds insertion clusters outside gene models. To compare Ds distribution relative to genes within this 1-Mb interval, we modeled the region as 25 equivalent genes, used the Poisson distribution to calculate the expected frequency of insertions, and evaluated our data for goodness of fit using the χ2 test with Yates' correction. The observed insertion distribution differed significantly from expectations under a Poisson model (P value < 0.002). These results demonstrate that within the context of a nearby, 2.5-cM interval, Ds is a potent mutagen with local, gene-specific hot and cold spots. Interestingly, we placed only two Mu elements to this interval, one coincident with a Ds cluster but the other within one of the eight gene models that was not hit by Ds.
Figure 5.
Fine-Scale Relationship of 74 Ds Insertions to Genes in a 1-Mb Interval of Chromosome 10: Ds Hits Most Genes in the Interval.
View of the chromosome 10 pseudomolecule (build 1) based on the genome browser at PlantGDB (http://www.plantgdb.org/ZmGDB/cgi-bin/getRegion.pl). Top, scale for interval. First track, positions of 74 nonredundant Ds insertions in 21 insertion clusters (black arrows) with the number of Ds per cluster. Black numeral, cluster aligns with a gene model; red numeral, cluster is not within a gene model. Second track (blue), nonredundant gene models from maizesequence.org release 4a.53. Third track (orange), sorghum peptides mapped to maize. Bottom track (colored lines), annotated genes that are not hit by Ds. Green lines, well-supported gene models; short blue lines, gene models with only one or two ESTs as support and no sorghum peptide; red line, gene model whose supporting ESTs have a better match on a different chromosome. Faint, dotted lines connect coincident gene and transposon features and indicate their approximate genome coordinates.
Local Reinsertion Preferences: Structural Features of Ds Insertion Sites
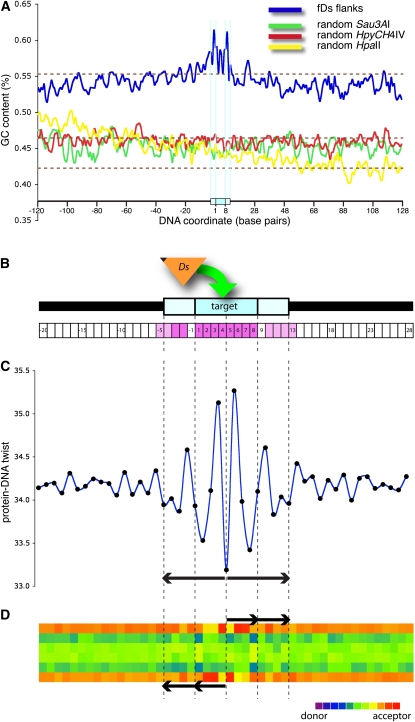
Collections of Ac/Ds transposon flanking sequences from maize and heterologous systems have consistently reported that there is no strong consensus sequence for target site insertion (Moreno et al., 1992; Kuromori et al., 2004; Ito et al., 2005). To investigate potential insertion site preferences for endogenous Ds elements in maize, we analyzed a multiple sequence alignment (MSA) of 1741 unique flanking sequence tags that contained an intact Ds-genomic DNA junction. Each tag derives from either the 5′ or 3′ side of Ds and contains a putative target site duplication (TSD), created by Ds when it inserted (Fedoroff et al., 1983). We analyzed in detail a 248-base window of the MSA, containing the eight bases of the TSD and ±120 bases on each side (see Methods). The segment had high (54.6%) average guanine-cytosine (GC) content, remarkably similar to the average for exon space (55.4%; Figure 6A) (Haberer et al., 2005). By contrast, the hypomethylated, control clones contained lower GC content, more similar to intergenic or intron space. GC content diverged at base positions in and near the TSD (Figure 6A, blue curve). In fact, total sequence content was nonrandom at only 18 of the 248 positions in the alignment, 16 of which were arrayed symmetrically around the TSD: all eight positions of the TSD (base positions 1 to 8) were highly nonrandom (P values < 10−36, Figure 6B); the adjacent bases at positions −1 and 9 were random; and the next adjacent, paired blocks of four bases (positions 10 to 13 and −2 to −5) were nonrandom (P values < 0.01; Figure 6B). Despite this symmetrical pattern of nonrandom bases, the information content (Schneider and Stephens, 1990; Crooks et al., 2004) of the segment was extremely low (i.e., there was no meaningful consensus sequence in the data; see Supplemental Figure 4 online).
Figure 6.
DNA Structural Features and Patterns of Hydrogen Bond Presentation at Insertions Sites.
(A) GC content (blue) of 1741 aligned fDs sequences across a 248-bp window including the 8-bp Ds target site (darker blue box on the x axis, coordinates 1 to 8), 120 bases adjacent to the 5′ side of Ds (coordinates −1 to −120) and 120 bases to the other side (coordinates 9 to 128). GC content for alignments of random hypomethylated sequences are plotted as controls in green (Sau3AI), red (HpyCHIV), and yellow (HpaII). Dotted, brown lines indicate average GC content for maize regions, including exons (55.4%), the overall genome (46.5%), and introns (42.3%) (Haberer et al., 2005).
(B) Schematic of a hypothetical segment of genomic DNA (black line) with an 8-bp Ds insertion site target (base pairs 1 to 8, darker blue box) and flanking 4-bp segments (light-blue boxes). A Ds element (orange triangle) is marked at its 5′ end to indicate polarity of the fDs alignments in all panels. Numerals in the x axis boxes are base pair coordinates that extend across all panels (vertical, dotted lines). Base positions with nonrandom composition are colored dark (P value < 10−36) or light (P value < 10−2) magenta.
(C) Average values of protein-DNA twist (black dots) for each position (x axis) in the multiple sequence alignment are plotted according to their values (y axis) and connected by a smoothed line to depict changes between adjacent positions. The structural pattern has twofold symmetry about the center of the target site extending across at least 16 base positions (paired, black arrows below plot). Supplemental Figure 5 online includes additional structural plots, including for the full 248-bp region.
(D) Heat map display of hydrogen bond (h-bond) presentation in the major groove of DNA, displaying average values for each position in the multiple sequence alignment. Each base pair in a double helix may contribute to h-bonding at any of six locations, depending on the base pair, so there are six rows for each base pair position. Strongest h-bonding donor locations are purple, while strongest acceptor positions are red. A symmetrical and palindromic pattern is evident across at least 16 base positions, consisting of two subrepeats (black arrows above and below graphic) on each side of the center of the target site.
The combination of no consensus base sequence and highly nonrandom bases at most positions suggested that some parameter other than primary DNA sequence may guide Ds insertion. For example, features of DNA three-dimensional structure guide nucleosome positioning in eukarytotes (Kaplan et al., 2009). Local interactions between adjacent bases and among base triplets are known to induce deformations from the regular double helix structure, especially in the context of protein–DNA interactions (Olson et al., 1998), such as that occurring between a transposase enzyme and target DNA during transposition. Thus, a variety of structural features and biophysical properties, originally measured or derived from analysis of protein-DNA crystal structures and other experimental data, may be computed for a sequence or sequence alignment by analyzing di- and trinucleotide pairs. The various parameters are computationally independent (Liao et al., 2000; Hackett et al., 2007). Such structural computations predict elements of DNA three-dimensional structure, such as local degrees of double helix flexibility versus rigidity (Ornstein et al., 1978; Brukner et al., 1995; Gorin et al., 1995; Olson et al., 1998), and have been used to predict in genomic DNA sequence biologically functional regions, including promoters and transcription start sites (Ohler et al., 2002; Wang and Benham, 2006; Abeel et al., 2008), as well as to profile insertion sites of P element transposons in Drosophila melanogaster and of retroviral and transposon vectors in transgenic animals (Liao et al., 2000; Vigdal et al., 2002; Hackett et al., 2007).
For more than a dozen structural features of DNA that we computed from the MSA of fDs tags, outlying values consistently arrayed symmetrically in and near the 8-bp target site. For example, protein DNA-twist predicts twist angle torsion based on data from protein-DNA complexes (Olson et al., 1998). A region with a high value of protein-DNA twist is more likely to be deformed by protein–DNA interaction than a region with a lower value. In the MSA of Ds insertion sites, both the highest and lowest values for protein-DNA twist were located among the nonrandom bases in and adjacent to the TSD (Figure 6C), and the structural profile was symmetrical about the center of the TSD (Figure 6C, black arrows). By contrast, for similar MSAs of control, hypomethylated DNA sequence reads, the computed structural plots showed similar averages but minimal variation (see Supplemental Figure 4 online). Similar analysis of >3000 perfect TSDs and flanking sequences from Ac transposition in Arabidopsis revealed highly similar structural profiles to those for Ds in maize, although overall flanking sequence content in the MSAs differed between species (data not shown). These results suggest that during reinsertion in Ac/Ds transposition, at least 16 bp of contiguous DNA sequence form a structural feature that is recognized by the Ac-encoded transposase and that the preferred structural conformation includes a pattern of alternating sites with more and less than average DNA deformability.
Hydrogen-bonding contact between protein chains and DNA bases in the major groove of the double helix is a critical component of the specificity and stability of protein-DNA complexes. As for structural features, similar hydrogen bonding presentations in the major groove may result from different DNA sequences (Seeman et al., 1976). We therefore also examined the collection of aligned Ds target sites for patterns of hydrogen bonding presentation by calculating potential hydrogen-bonding capacity of positions in the major groove. The analyses may be displayed in a heat map that shows each position's propensity to serve as a hydrogen bond donor or acceptor (Liao et al., 2000). The heat map (Figure 6D) contains six rows for each base pair, corresponding to the six positions in the major groove that may serve as hydrogen bond donors (coolest color or purple) or acceptors (warmest color or red). For the MSA of 1741 insertion sequences, at each position the heat map shows the average hydrogen bonding presentation for all aligned bases at that position (Figure 6D). Across the 248-bp window, the profile is uniform except for at 16 positions centered on the TSD, in which there is a palindromic pattern of four 4-base repeating units (Figure 6D, arrows). The regular, symmetric arrangement of paired hydrogen bond donor and acceptor motifs suggests a specific, preferred interaction during transposon insertion that includes a homodimeric or homotetrameric protein complex.
DISCUSSION
Ds as a Resource for Regional Mutagenesis
The maize transposable elements Ac and Ds have been extensively characterized but not widely used in maize as insertional mutagens (Brutnell and Conrad, 2003). This is largely attributable to two inherent characteristics of this transposable element family: (1) both Ac and Ds are maintained at relatively low copy numbers in the genome (Chen et al., 1987), and (2) a tendency to insert at sites genetically linked to the donor locus has been demonstrated for both Ac (Van Schaik and Brink, 1959; Greenblatt, 1984; Dooner and Belachew, 1989) and Ds (this study). The low copy number and germinal insertion frequency of Ac/Ds (Brutnell and Dellaporta, 1994) limits the utility of the system in random mutagenesis (Brutnell and Conrad, 2003) but the preference of these elements for closely linked transpositions can be exploited in targeted, regional mutagenesis (Hake et al., 1989; Athma et al., 1992; Moreno et al., 1992; Colasanti et al., 1998; Vollbrecht et al., 2000; Ahern et al., 2009).
Here, we describe the placement of 1501 Ds insertions to unique locations on the maize physical map (http://www.plantgdb.org/prj/AcDsTagging/records.php). Importantly, the collection is maintained as a series of W22 inbreds, with one or two new Ds insertions in each line, ensuring a low mutational load for any given line. Lines segregate Ac-im, so that stable, Ds insertion phenotypes may be easily examined. Crosses to reference alleles maintained in W22 will also permit near-isogenic comparisons of any mutants recovered with wild-type siblings. Even though Ds distribution is nonrandom, 85% of the maize gene space now resides within 4 cM of at least one Ds insertion described in this collection. Due to the cloning strategy used to recover Ds flanking sequences, the collection is enriched for elements residing in the hypomethylated gene-rich regions of the genome (Bennetzen et al., 1994; Rabinowicz et al., 1999). Thus, we anticipate that the majority of these Ds elements will be capable of remobilization by Ac, where linked (regional) sites would be excellent targets. Indeed, we have remobilized several elements in regional mutagenesis experiments to generate Ds insertion alleles of genes encoding anthocyanin and starch biosynthetic enzymes (data not shown). Tandem Ac/Ds elements can undergo alternative transposition, which leads to a variety of genome rearrangements (Zhang et al., 2009). Thus, a collection of localized Ds launch pads could also be used to generate local deletions (Page et al., 2004). In summary, we generated an extremely comprehensive collection of sequence-indexed Ds insertions to serve as genome-wide platforms for regional mutagenesis.
We anticipate that this collection will have great value as a tool for forward- and reverse-genetics approaches (Ahern et al., 2009). As such, we extensively compared and contrasted insertion site preferences of Ds and Mutator elements. Mutator is a prolific maize transposable element, generating up to 10 germinal insertions in each gametophyte (May et al., 2003), has been the foundation for several mutagenesis programs in maize (Brutnell, 2002), and has complementary strengths to Ac/Ds approaches for functional genomics (Cowperthwaite et al., 2002). Our comparisons of Ds and Mutator insertion sites also reveal complementary insertion site biases that may be exploited in mutagenesis programs. A striking difference between Mutator and Ac/Ds transposition is the proximity of the donor locus to the preferred target sites. Whereas Mutator prefers targets that are unlinked to the donor element (Lisch et al., 1995), >60% of all Ac transpositions are to sites genetically linked to the donor locus (Van Schaik and Brink, 1959; Greenblatt, 1984; Dooner and Belachew, 1989) and of those, nearly half are to sites within 10 cM of the donor. Surprisingly, a majority (56%) of premeiotic Ds transpositions from r1-sc:m3 mapped to unlinked sites in the genome. This may reflect a difference between Ac and Ds insertion site selection or the fact that the Ds estimate derives from a subpopulation of transpositions, ignoring an expected large number of intragenic reinsertions at r1 and the approximately one-third of all events that are postmeiotic. Similar to Ac, among linked transpositions, Ds showed a strong preference (61/117 elements) for sites within 10 cM of the donor locus r1-sc:m3.
At a regional level on chromosome 10, Mutator and Ds also displayed different insertion site biases. In a 1-Mb region, 74 Ds insertions were mapped to 21 unique clusters. Seventeen cluster locations coincided with evidence-based gene models, strongly supporting the view that Ds specifically targets gene-rich regions. However, Ds insertions were not detected in eight evidence-supported genes that also reside in this interval, a larger number of missed genes than expected (P value < 0.002) that suggests Ds has local target site preferences. Alternatively, the missed genes may represent sequences present on chromosome 10 in the reference B73 genome, but absent from the corresponding region of the W22 line in which Ds mutagenesis was performed. Furthermore, several target sites were populated by multiple independent Ds insertions, a bias that could be inherent to Ds transposition or reflect a relationship to the Ds donor locus. In the latter case, using multiple Ds insertions in regional mutagenesis may help to ensure a broader range of target site insertions. Our examination of publicly available Mu flanking sequences revealed two insertions within this interval. One Mu insertion mapped to a gene model without a Ds insertion, the other to a location that was also identified by Ds and is not supported by a gene model. The former demonstrates complementarity of the two systems. The latter insertion is of particular interest, given the tendency of both Mutator and Ds to insert into or near genes. It is possible that this target site and the other hot spots identified by Ds reveal additional features of the transcriptional landscape that are not represented in ab initio or EST gene models, such as enhancer and promoter elements and small noncoding RNAs. A more detailed analysis of these Ds insertions could prove fruitful in probing gene function and regulation in maize.
An ideal outcome of a mutagenesis experiment is the recovery of multiple strong and weak mutant alleles. A survey of Mutator insertions (Figure 4; also see Liu et al., 2009) revealed that many fall upstream of coding sequences in promoter or 5′-UTR sequences. This preference for upstream insertion may explain the relatively high frequency of Mu-suppressible alleles (May et al., 2003; McCarty et al., 2005), where interaction of Mutator transposase with the element ends likely affects flanking gene expression (Martienssen et al., 1989; Barkan and Martienssen, 1991; May et al., 2003). By contrast, very few suppressible Ac or Ds insertion alleles have been described (Vollbrecht et al., 2000). Similarly, while regulatory mutations have been described in which 5′ Ac or Ds insertion influences gene expression at a particular locus (Klein et al., 1988; Schiefelbein et al., 1988; Sullivan et al., 1989; Moreno et al., 1992, 1997; Bai et al., 2007), they are vastly outnumbered by those conditioning a complete loss-of-function phenotype (Kunze et al., 1993).
Another important consideration for gene tagging experiments is the stability of the insertion. Remobilizing transposable element insertions to generate additional stable excision alleles is a powerful approach to linking phenotypic with genotypic variation (Schultes et al., 1996; Bai et al., 2007). In the absence of Ac, insertions of Ds are stable, but they can be remobilized in the presence of Ac. Excisions of Ac or Ds usually result in the insertion of six, seven, or eight nucleotides originating from the target site duplication, which may generate additional mutant alleles, including in-frame insertions (Scott et al., 1996; Bai et al., 2007). This feature of Ac/Ds has been exploited to alter the activity of a starch biosynthetic enzyme (Giroux et al., 1996) as well as the cellular localization of a transcription factor (Liu et al., 1996) and has great potential for defining functional domains of proteins in planta (Brutnell and Conrad, 2003). The relatively uniform distribution of Ds elements observed throughout the gene space and their high frequency of exon targeting suggest that most genes can be targeted for Ds insertional and excision mutagenesis. By contrast, Mutator insertions show no propensity for intragenic transposition and are generally stable, though it is possible to identify abortive transposition events through a PCR-based screen to generate deletions flanking the site of Mutator insertion (Das and Martienssen, 1995).
Finally, the ability to predict local insertion sites greatly enhances the utility of a gene tagging program. Ds shows a preference for distinct structural features of the DNA double helix at the point of insertion. Mu insertion sites also display a nonrandom DNA composition (Dietrich et al., 2002; Liu et al., 2009) and contain a clear DNA structural signature (E. Vollbrecht, unpublished data). However, structural features at the Mu TSD are less pronounced and have different composition and symmetry properties than at the Ds TSD. The latter differences may reflect the likely heteromeric composition of the transposase complex that mediates Mu insertion (Lisch et al., 1995; Raizada and Walbot, 2000), while Ac/Ds requires only the Ac-encoded transposase. These differences also reinforce the notion that a given transposase's preference is for a distinct and specific DNA structural signature, rather than just for regions of increased DNA helix flexibility or deformability. Finally, different DNA structural preferences for these transposons may also correlate with their different affinities for particular genic regions. For example, promoter regions contain unique DNA structural signatures (Wang and Benham, 2006; Abeel et al., 2008) and are differentially targeted by Mu and Ac/Ds. Overall, it will be useful to exploit a combination of knowledge about local, structural, and gene-related preferences with the known regional preference of Ds for hypomethylated regions of the genome to more efficiently design transposon targeting experiments for creating specific gene knockouts.
Patterns of structural features of DNA at aligned Ds insertion sites suggest an Ac transposase–DNA interaction region spanning at least 16 nucleotides. Most notably, periodicity of hydrogen-bonding presentation suggests four repeats of a single motif, palindromic about the center of the TSD. These data are consistent with an interaction between dimeric or tetrameric Ac transposase units and up to 16 bp of DNA at the target site. hAT transposases, including Ac and Hermes, can form homomeric complexes through a highly conserved C-terminal domain (Essers et al., 2000; Michel et al., 2003), and while no protein crystal structure for Ac has been solved, data suggest that Ac–Ac interaction is critical for transposition in planta (Kunze et al., 1993). Thus, by analogy to the crystal structure for Hermes, the functional Ac transposase complex is likely a hexameric ring, with threefold symmetry from assembly of three homodimeric units (Hickman et al., 2005). Only hexameric Hermes is active, but in that structure only two of six active sites, one each from two adjacent dimers, are proposed to interact with DNA and catalyze the two cuts staggered by 8 bp that produce the target site duplication upon insertion. If Ac interacts similarly, the structural features and hydrogen-bonding patterns we observed may represent the palindromic arrangement of just two, extended (8 bp) DNA recognition motifs, for example, to position two, dimeric Ac subunits over two DNA cleavage sites. Alternatively, one core interaction surface may be in the central 8 bp of the TSD with adjacent nucleotides providing structural information that is propagated to the site of direct transposase binding.
Dooner et al. (1994) observed that among unlinked transpositions from an Ac donor on chromosome 9S, unlinked regions of 9L were preferred over regions in the other chromosomes. In this study, a Ds donor on 10L showed no bias for 9L, but did show a similar preference for some intrachromosomal, genetically unlinked bins (on 10L). Ds also showed a preference for transposition to linked sites but with asymmetry: within equivalent linkage (recombinational) space on either side of the r1-sc:m3 donor locus, Ds showed a striking preference (81 proximal versus 36 distal, by inclusive screening) for the proximal recombination (centimorgan) space, which was also much higher in gene density per centimorgan. Thus, Ds prefers linked, gene-rich regions over linked, gene-poor regions. The lower frequency of reinsertions in linked, distal segments with fewer suitable (genic) targets per centimorgan could also drive the increased insertion frequency in unlinked, distal locations. If there are relatively few linked, genic targets, then Ds may instead distribute more distantly and potentially even to other chromosomes. In the Ac study (Dooner et al., 1994), linked transpositions distributed symmetrically around the donor (14 proximal versus 13 distal insertions), but the sample size was much smaller and those Ac and our Ds distributions cannot be statistically distinguished (χ2 homogeneity test, P value = 0.086). Thus, if a similar mechanism acted on chromosome 9 in that study, one would expect gene density per centimorgan to be higher on 9S distal to bz1 than in the other flanking region toward and across the centromere onto 9L. On the other hand, the distal 10L region could be a general hot spot for Ds, as may be determined by monitoring reinsertions from other donor loci. Based on the available data, we speculate a hierarchical nature of components that govern Ac/Ds reinsertion preferences. We propose that a physically proximate, genetic linkage space, presumably correlating with sites of recombination, initially defines a constrained and roughly symmetrical regional window within which Ds is likely to transpose and then within that window a density of euchromatic gene space acts as an additional and perhaps independent driver of Ds insertion. If a transposing element escapes local reinsertion, then presumably other chromosomes become viable targets, in which case architecture of the nucleus becomes the relevant component of physical proximity. Finally, at the most fine-scale level, within the preferred chromatin space there is a preference for sites of integration with a particular DNA structure.
METHODS
Field Genetics
All stocks are maintained in an inbred W22 background. A small set of founder lines, homozygous for Ac-im (Conrad and Brutnell, 2005) and for the mutable r1-sc:m3 allele, which contains a Ds in an intron (Alleman and Kermicle, 1993; this study), were generated and characterized by DNA gel blot for Ds copy number. The backcross parent for all crosses (Figure 1, right side parent) was homozygous for the r1-sc:m3 allele. After an initial round of backcrossing in nurseries to produce the first stage 1 progeny (similar to Figure 1B, left side, but without –Ac-im segregants), nonconcordant (colored aleurone, mutable embryo type) progeny among the stage 1, colored kernels (Figure 1B, red box) in our backcrossing scheme recapitulated the stage 0 cross. These nonconcordant stage 1, hemizygous for Ac-im, progeny were the source of all subsequent stage 0 lines (Figure 1A). Field locations included Ames, IA; Ithaca, NY; Molokai, HI; and Puerto Vallarta, Mexico.
From stage 1 ears, fully colored kernels were selected as the source of Ds transpositions (Figure 1B). When fully colored kernels were arranged on the ear in contiguous groups three or more, we only selected one of the kernels to avoid potentially selecting multiple kernels from a sector of sporophyte tissue derived from a single, somatic event.
Processing of Transpositions to Isolate fDs DNA Sequence
Steps in processing from field genetics, through wet laboratory and informatics steps, are as previously described (Ahern et al., 2009) and/or depicted in Supplemental Figure 2 online. These steps produced fDs sequence tags. Throughout development of the resource, we confirmed the presence of insertions and tracked insertions by spot testing original seed packets with a PCR assay using a Ds and a flanking sequence primer and/or DNA gel blots. Approximately 80% of the events were confirmed in this way. In later stages of the project, we improved on this figure by implementing an earlier QC check. Two independent seedling pools were assayed in initial DNA gel blots, and only when a transposed Ds fragment was detected in both did we process tissue for cloning. For large-scale confirmation of the presence of insertions, we developed a standardized PCR assay including a public web interface (http://plantgdb.org/cgi-bin/prj/AcDsTagging/getPrimers.pl) as described in detail in Supplemental Methods online.
Placing Ds to Chromosomes and Genetic Maps
To position Ds locations across the whole genome, BLASTN was used to place repeat masked fDs sequences to maize (Zea mays) BAC and BAC-sized (i.e., >10,000 bp) sequences in the htgs and nr sections of GenBank, as well as to the initial pseudomolecule build of B73 made available by the Arizona Genomics Institute (http://www2.genome.arizona.edu/genomes/maize). To account for sequence differences between W22 and the reference B73 genome sequence, we empirically developed a set of quality criteria that formed the basis of placement scores. BLAST hits with an e-value above a cutoff of 10−10 were discarded. Quality classification, based on the BLASTN percentage identity and coverage scores, was applied hierarchically to retained BLAST hits; once a sequence satisfied a higher-level criterion, lower level criteria were not applied. The highest level classification was HQ (high quality); the classifications were defined as follows: HQ (high quality): ≥95% coverage and ≥95% identity with no gaps; MQ1 (medium quality 1): between 90 and 95% coverage and ≥95% identity with no gaps; MQ2 (medium quality 2): >90% coverage and ≥95% cumulative identity, with one or more gap openings; MQ3 (medium quality 3): ≥95% identity over at least 200 bp of sequence, only assigned when there is only one MQ hit; MQ4 (medium quality 4): >90% identity over at least 200 bp of sequence, only assigned when there is only one MQ hit; MP (multiple placement): the first satisfied HQ or MQ criterion matched three or more locations; NGH (no good hits): none of the HQ or MQ criteria were satisfied. After quality criteria were applied, a placement score was assigned to each HQ and MQ sequence tag as follows: Single Placement implied the assigned quality score matched a BAC at a single location in the sequence assembly; for BAC placements, Single Duplicated Placement implied matches to more than one BAC, but in a region where those BACs likely overlapped in the assembly and hence likely a single genome location. Dual Placement implied matches to BACs or chromosomes at two locations in the genome. The same procedures were used to place Mutator and Ds elements and with similar results for each transposon; ∼40% of the placements were by each of HQ and MQ2 quality criteria, with the remaining 20% of the placements divided among MQ1, MQ3, and MQ4 criteria classes.
To estimate genetic map locations of Ds placements on the genome sequence assembly, we assigned a genetic (centimorgan) coordinate to each BAC or base pair in the assembly as follows. We first selected 618 unique, somewhat evenly spaced, skeleton markers from the ISU-IBM7 genetic map, a multimeiotic map that is corrected for its IRIL origins (Fu et al., 2006) and whose length closely matches that of traditional maize genetic maps (http://www.maizegdb.org/map.php). Mean separation between selected markers was 3.02 cM (minimum 0.8 cM; maximum 10.4 cM). Each genetic marker was placed to BAC(s) by BLASTN, and its centimorgan coordinate was assigned to the base coordinate of the left end of the right-most BAC that it hit. We then similarly assigned a genetic coordinate to the base coordinate of the left end of every BAC in the assembly, by linear extrapolation using the two closest flanking, anchored BACs. To accommodate base pairs distal to the most telomeric genetic markers, we expanded the genetic map. Each Ds element was then assigned the centimorgan value for the right-most BAC it was placed to. Genetic map inferences used release 3b.50 of the maize genome. The maize tiling path update of March 20, 2009 (http://www.genome.arizona.edu/cgi-bin/jzhang/mtp.cgi) contained a major correction for the region of chromosome 10 that contains the donor r1 locus, so we used the pseudomolecule release (build 1) for all detailed analysis of placements to chromosome 10. A total of 253 of the chromosome 10 markers from ISU-IBM7 were placed to the pseudomolecule by BLASTN, the set was trimmed to 226 that were colinear, and base positions were assigned a centimorgan value either directly when resident between two markers with the same centimorgan coordinate or by linear interpolation when between two markers with different centimorgan coordinates.
DNA Sequence Analysis
For analysis of average mono-, di-, and trinucleotide features of Ds insertion sites, we aligned 1741 unique, unpaired (i.e., one sequence tag per insertion) fDs sequences. A total of 1122 sequence tags derived from the 5′-side of Ds and 619 tags were from the 3′-side. The multiple sequence alignment retained phase and polarity relative to the TSD, so that 5′-side sequences were oriented to terminate in the eight bases adjacent to the 5′ end of Ds, and 3′-side sequences were oriented to begin with the eight bases adjacent to the Ds. In the multiple sequence alignment, the two groups overlapped in the eight bases of the TSD, and up to ±120 bases outside of the TSD were retained for a total length of 248 bases. Positions in the first 120 bases of the alignment contained from 883 to 1122 members, the central eight bases were present in all 1741 sequences, and the last 120 base positions contained between 619 and 491 members. This multiple sequence alignment was used for various analyses of the target site and flanking regions. Average content for each position(s) (i.e., or for each mono-, di-, or trinucleotide) was calculated as the membership at that position(s) in the alignment. As controls, random, hypomethylated clones were retrieved from GenBank, trimmed of contaminating vector and enzyme recognition site sequences, and their first 248 bases aligned and analyzed as three independent, 248-bp MSAs: 912 Sau3AI sequences, 1360 HpyCH4IV sequences from hypomethylated partial restriction clones, and 1195 HpaII sequences from hypomethylated partial restriction clones (Emberton et al., 2005).
To compare Ds and Mu insertion sites on chromosome 10, we sought a Mu flanking sequence data set as similar as possible to the fDs data. Thus, among all RescueMu sequencing reads at GenBank (n = 191,715), we identified all paired reads containing perfect target site duplications, took the reads for only one side of Mu, collapsed them to nonredundant sequences, repeat masked them, then used our BLAST pipeline to make 2936 unique placements to the pseudochromosomes, of which 160 were on chromosome 10. From publicly available UniformMu sequences, a set of 2667 curated, 454 paired-end read assemblies treated the same way identified 1846 unique placements, 100 on chromosome 10.
Repeat masking used RepeatMasker Open-3.0 version 3.2.7 (Smit et al., 2004) and the TIGR 4.0 repeat database (http://maize.jcvi.org/repeat_db.shtml) with the –nolow option. In Figure 2, a sequence was classified as repetitive if more than 70% of its bases were masked.
Online Resources
All pedigree and molecular information about each insertion line is publicly available from the project website at PlantGDB (http://plantgdb.org/prj/AcDsTagging/index.php). All seed stocks are also available through this website until deposited at the Maize Genetics Co-op stock center.
Accession Numbers
Flanking Ds sequences are classified with the library name “Maize transposon insertions from ISU and BTI” and deposited to the GSS division of the National Center for Biotechnology Information.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure 1. Proposed Mechanism of Ds Transpositions Selected in Stage 1 Ears and Classes of Recovered Events Based on Kernel Selections in Stage 2 Ears.
Supplemental Figure 2. Processing Workflow for Integrated Field Genetics, Wet Lab, and Informatics Pipeline.
Supplemental Figure 3. Transposition during Development of Female Gametophytes That Contain Ac-im and r1-sc:m3 May Lead to Nonconcordant Embryo and Endosperm Genotypes.
Supplemental Figure 4. Information Content at Aligned Ds Target Site Duplications.
Supplemental Figure 5. DNA Structural Features at Insertion Sites Are Divergent.
Supplemental Table 1. Intrachromosomal Transpositions Recovered by Inclusive Screening That Locate to Intervals Linked to the r1-sc:m3 Donor.
Supplemental Table 2. Intrachromosomal Ds Transpositions from r1-sc:m3 Are Distributed Asymmetrically.
Supplemental Table 3. The 10-cM Bins on Chromosome 10 That Are Ds Insertion Hot Spots.
Supplemental Methods. Genetics, Calculations, and Genotyping Methods for Working with Ds Lines.
Acknowledgments
This research was supported by the National Science Foundation (NSF DBI-0501713 to T.P.B., V.P.B., and E.V.) and startup funds from Iowa State University (to E.V.).
References
- Abeel T., Saeys Y., Bonnet E., Rouzé P., Van de Peer Y. (2008). Generic eukaryotic core promoter prediction using structural features of DNA. Genome Res. 18: 310–323 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ahern K.R., Deewatthanawong P., Schares J., Muszynski M., Weeks R., Vollbrecht E., Duvick J., Brendel V.P., Brutnell T.P. (2009). Regional mutagenesis using Dissociation in maize. Methods 49: 248–254 [DOI] [PubMed] [Google Scholar]
- Alleman M., Kermicle J.L. (1993). Somatic variegation and germinal mutability reflect the position of transposable element Dissociation within the maize R gene. Genetics 135: 189–203 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Athma P., Grotewold E., Peterson T. (1992). Insertional mutagenesis of the maize P gene by intragenic transposition of Ac. Genetics 131: 199–209 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bai L., Singh M., Pitt L., Sweeney M., Brutnell T.P. (2007). Generating novel allelic variation through Activator insertional mutagenesis in maize. Genetics 175: 981–992 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barkan A., Martienssen R.A. (1991). Inactivation of maize transposon Mu suppresses a mutant phenotype by activating an outward-reading promoter near the end of Mu1. Proc. Natl. Acad. Sci. USA 88: 3502–3506 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Becker H.A., Kunze R. (1997). Maize Activator transposase has a bipartite DNA binding domain that recognizes subterminal sequences and the terminal inverted repeats. Mol. Gen. Genet. 254: 219–230 [DOI] [PubMed] [Google Scholar]
- Bennetzen J.L., Schrick K., Springer P.S., Brown W.E., SanMiguel P. (1994). Active maize genes are unmodified and flanked by diverse classes of modified, highly repetitive DNA. Genome 37: 565–576 [DOI] [PubMed] [Google Scholar]
- Brukner I., Sanchez R., Suck D., Pongor S. (1995). Trinucleotide models for DNA bending propensity: Comparison of models based on DNaseI digestion and nucleosome packaging data. J. Biomol. Struct. Dyn. 13: 309–317 [DOI] [PubMed] [Google Scholar]
- Brutnell T.P. (2002). Transposon tagging in maize. Funct. Integr. Genomics 2: 4–12 [DOI] [PubMed] [Google Scholar]
- Brutnell T.P., Conrad L.J. (2003). Transposon tagging using Activator (Ac) in maize. Methods Mol. Biol. 236: 157–176 [DOI] [PubMed] [Google Scholar]
- Brutnell T.P., Dellaporta S.L. (1994). Somatic inactivation and reactivation of Ac associated with changes in cytosine methylation and transposase expression. Genetics 138: 213–225 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calvi B.R., Hong T.J., Findley S.D., Gelbart W.M. (1991). Evidence for a common evolutionary origin of inverted repeat transposons in Drosophila and plants: Hobo, Activator, and Tam3. Cell 66: 465–471 [DOI] [PubMed] [Google Scholar]
- Chen J., Greenblatt I.M., Dellaporta S.L. (1987). Transposition of Ac from the P locus of maize into unreplicated chromosomal sites. Genetics 117: 109–116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chomet P.S., Wessler S., Dellaporta S.L. (1987). Inactivation of the maize transposable element Activator (Ac) is associated with its DNA modification. EMBO J. 6: 295–302 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colasanti J., Yuan Z., Sundaresan V. (1998). The Indeterminate gene encodes a zinc finger protein and regulates a leaf-generated signal required for the transition to flowering in maize. Cell 93: 593–603 [DOI] [PubMed] [Google Scholar]
- Conrad L.J., Brutnell T.P. (2005). Ac-immobilized, a stable source of Activator transposase that mediates sporophytic and gametophytic excision of Dissociation elements in maize. Genetics 171: 1999–2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coupland G., Baker B., Schell J., Starlinger P. (1988). Characterization of the maize transposable element Ac by internal deletions. EMBO J. 7: 3653–3659 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cowperthwaite M., Park W., Xu Z., Yan X., Maurais S.C., Dooner H.K. (2002). Use of the transposon Ac as a gene-searching engine in the maize genome. Plant Cell 14: 713–726 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crooks G.E., Hon G., Chandonia J.M., Brenner S.E. (2004). WebLogo: A Sequence Logo Generator. (Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; ). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Das L., Martienssen R. (1995). Site-selected transposon mutagenesis at the Hcf106 locus in maize. Plant Cell 7: 287–294 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dellaporta S.L., Calderon-Urrea A. (1993). Sex determination in flowering plants. Plant Cell 5: 1241–1251 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dellaporta S.L., Moreno M.A. (1994). Gene tagging with Ac/Ds elements in maize. The Maize Handbook, Freeling M., Walbot V., (New York: Springer-Verlag; ), 219–233 [Google Scholar]
- Dietrich C.R., Cui F., Packila M.L., Li J., Ashlock D.A., Nikolau B.J., Schnable P.S. (2002). Maize Mu transposons are targeted to the 5′ untranslated region of the gl8 gene and sequences flanking Mu target-site duplications exhibit nonrandom nucleotide composition throughout the genome. Genetics 160: 697–716 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dooner H.K., Belachew A. (1989). Transposition pattern of the maize element Ac from the Bz-m2(Ac) Allele. Genetics 122: 447–457 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dooner H.K., Belachew A., Burgess D., Harding S., Ralston M., Ralston E. (1994). Distribution of unlinked receptor sites for transposed Ac elements from the bz-m2(Ac) allele in maize. Genetics 136: 261–279 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emberton J., Ma J., Yuan Y., SanMiguel P., Bennetzen J.L. (2005). Gene enrichment in maize with hypomethylated partial restriction (HMPR) libraries. Genome Res. 15: 1441–1446 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Essers L., Adolphs R.H., Kunze R. (2000). A highly conserved domain of the maize Activator transposase is involved in dimerization. Plant Cell 12: 211–224 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fedoroff N., Wessler S., Shure M. (1983). Isolation of the transposable maize controlling elements Ac and Ds. Cell 35: 235–242 [DOI] [PubMed] [Google Scholar]
- Feldmar S., Kunze R. (1991). The ORFa protein, the putative transposase of maize transposable element Ac, has a basic DNA binding domain. EMBO J. 10: 4003–4010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fengler K., Allen S.M., Li B., Rafalski A. (2007). Distribution of genes, recombination, and repetitive elements in the maize genome. Crop Sci. 47: S-83–S-95 [Google Scholar]
- Fernandes J., Dong Q., Schneider B., Morrow D.J., Nan G.L., Brendel V., Walbot V. (2004). Genome-wide mutagenesis of Zea mays L. using RescueMu transposons. Genome Biol. 5: R82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu Y., Wen T.-J., Ronin Y.I., Chen H.D., Guo L., Mester D.I., Yang Y., Lee M., Korol A.B., Ashlock D.A., Schnable P.S. (2006). Genetic dissection of intermated recombinant inbred lines using a new genetic map of maize. Genetics 174: 1671–1683 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaut B.S., Doebley J.F. (1997). DNA sequence evidence for the segmental allotetraploid origin of maize. Proc. Natl. Acad. Sci. USA 94: 6809–6814 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giroux M.J., Shaw J., Barry G., Cobb B.G., Greene T., Okita T., Hannah L.C. (1996). A single mutation that increases maize seed weight. Proc. Natl. Acad. Sci. USA 93: 5824–5829 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gorin A.A., Zhurkin V.B., Wilma K. (1995). B-DNA twisting correlates with base-pair morphology. J. Mol. Biol. 247: 34–48 [DOI] [PubMed] [Google Scholar]
- Greenblatt I. (1966). Transposition and replication of Modulator in maize. Genetics 53: 361–369 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greenblatt I.M. (1984). A chromosome replication pattern deduced from pericarp phenotypes resulting from movements of the transposable element, Modulator, in maize. Genetics 108: 471–485 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greenblatt I.M., Brink R.A. (1962). Twin mutations in medium variegated pericarp maize. Genetics 47: 489–501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grotewold E., Athma P., Peterson T. (1991). A possible hot-spot for Ac insertion in the maize P gene. Mol. Gen. Genet. 230: 329–331 [DOI] [PubMed] [Google Scholar]
- Haberer G., Young S., Bharti A.K., Gundlach H., Raymond C., Fuks G. (2005). Structure and architecture of the maize genome. Plant Physiol. 139: 1612–1624 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hackett C., Geurts A., Hackett P. (2007). Predicting preferential DNA vector insertion sites: Implications for functional genomics and gene therapy. Genome Biol. 8: S12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hake S., Vollbrecht E., Freeling M. (1989). Cloning Knotted, the dominant morphological mutant in maize using Ds2 as a transposon tag. EMBO J. 8: 15–22 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hickman A.B., Perez Z.N., Zhou L., Musingarimi P., Ghirlando R., Hinshaw J.E., Craig N.L., Dyda F. (2005). Molecular architecture of a eukaryotic DNA transposase. Nat. Struct. Mol. Biol. 12: 715–721 [DOI] [PubMed] [Google Scholar]
- Houba-Herin N., Becker D., Post A., Larondelle Y., Starlinger P. (1990). Excision of a Ds-like maize transposable element (Ac delta) in a transient assay in Petunia is enhanced by a truncated coding region of the transposable element Ac. Mol. Gen. Genet. 224: 17–23 [DOI] [PubMed] [Google Scholar]
- Ito T., Motohashi R., Kuromori T., Noutoshi Y., Seki M., Kamiya A., Mizukado S., Sakurai T., Shinozaki K. (2005). A resource of 5,814 dissociation transposon-tagged and sequence-indexed lines of Arabidopsis transposed from start loci on chromosome 5. Plant Cell Physiol. 46: 1149–1153 [DOI] [PubMed] [Google Scholar]
- Kaplan N., Moore I.K., Fondufe-Mittendorf Y., Gossett A.J., Tillo D., Field Y., LeProust E.M., Hughes T.R., Lieb J.D., Widom J., Segal E. (2009). The DNA-encoded nucleosome organization of a eukaryotic genome. Nature 458: 362–366 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kermicle J.L., Alleman M., Dellaporta S.L. (1989). Sequential mutagenesis of a maize gene, using the transposable element Dissociation. Genome 31: 712–716 [Google Scholar]
- Klein A.S., Clancy M., Paje-Manalo L., Furtek D.B., Hannah L.C., Nelson O.E., Jr (1988). The mutation bronze-mutable4 derivative 6856 in maize is caused by the insertion of a novel 6.7-kilobase pair transposon in the untranslated leader region of the bronze-1gene. Genetics 120: 779–790 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kolesnik T., Szeverenyi I., Bachmann D., Kumar C.S., Jiang S., Ramamoorthy R., Cai M., Ma Z.G., Sundaresan V., Ramachandran S. (2004). Establishing an efficient Ac/Ds tagging system in rice: Large-scale analysis of Ds flanking sequences. Plant J. 37: 301–314 [DOI] [PubMed] [Google Scholar]
- Kolkman J.M., Conrad L.J., Farmer P.R., Hardeman K., Ahern K.R., Lewis P.E., Sawers R.J., Lebejko S., Chomet P., Brutnell T.P. (2005). Distribution of Activator (Ac) throughout the maize genome for use in regional mutagenesis. Genetics 169: 981–995 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kunze R., Behrens U., Courage-Franzkowiak U., Feldmar S., Kuhn S., Lutticke R. (1993). Dominant transposition-deficient mutants of maize Activator (Ac) transposase. Proc. Natl. Acad. Sci. USA 90: 7094–7098 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kunze R., Starlinger P. (1989). The putative transposase of transposable element Ac from Zea mays L. interacts with subterminal sequences of Ac. EMBO J. 8: 3177–3185 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kunze R., Weil C.F. (2002). The hAT and CACTA superfamily of plant transposons. Mobile DNA, Craig N.L., (Washington, D.C.: ASM Press; ), 565–610 [Google Scholar]
- Kuromori T., Hirayama T., Kiyosue Y., Takabe H., Mizukado S., Sakurai T., Akiyama K., Kamiya A., Ito T., Shinozaki K. (2004). A collection of 11 800 single-copy Ds transposon insertion lines in Arabidopsis. Plant J. 37: 897–905 [DOI] [PubMed] [Google Scholar]
- Leu J.Y., Sun Y.H., Lai Y.K., Chen J. (1992). A maize cryptic Ac-homologous sequence derived from an Activator transposable element does not transpose. Mol. Gen. Genet. 233: 411–418 [DOI] [PubMed] [Google Scholar]
- Liao G.C., Rehm E.J., Rubin G.M. (2000). Insertion site preferences of the P transposable element in Drosophila melanogaster. Proc. Natl. Acad. Sci. USA 97: 3347–3351 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lisch D., Chomet P., Freeling M. (1995). Genetic characterization of the Mutator system in maize: Behavior and regulation of Mu transposons in a minimal line. Genetics 139: 1777–1796 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu S., Yeh C.-T., Ji T., Ying K., Wu H., Tang H.M., Fu Y., Nettleton D., Schnable P.S. (2009). Mu transposon insertion sites and meiotic recombination events co-localize with epigenetic marks for open chromatin across the maize genome. PLoS Genet. 5: e1000733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y.H., Alleman M., Wessler S.R. (1996). A Ds insertion alters the nuclear localization of the maize transcriptional activator R. Proc. Natl. Acad. Sci. USA 93: 7816–7820 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martienssen R.A., Barkan A., Freeling M., Taylor W.C. (1989). Molecular cloning of a maize gene involved in photosynthetic membrane organization that is regulated by Robertson's Mutator. EMBO J. 8: 1633–1639 [DOI] [PMC free article] [PubMed] [Google Scholar]
- May B.P., Liu H., Vollbrecht E., Senior L., Rabinowicz P.D., Roh D., Pan X., Stein L., Freeling M., Alexander D., Martienssen R. (2003). Maize-targeted mutagenesis: A knockout resource for maize. Proc. Natl. Acad. Sci. USA 100: 11541–11546 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCarty D.R., et al. (2005). Steady-state transposon mutagenesis in inbred maize. Plant J. 44: 52–61 [DOI] [PubMed] [Google Scholar]
- McClintock B. (1947). Cytogenetic studies of maize and Neurospora. Year B. Carnegie Inst. Wash. 46: 146–152 [Google Scholar]
- McClintock B. (1949). Mutable loci in maize. Year B. Carnegie Inst. Wash. 48: 142–154 [PubMed] [Google Scholar]
- Michel K., O'Brochta D.A., Atkinson P.W. (2003). The C-terminus of the Hermes transposase contains a protein multimerization domain. Insect Biochem. Mol. Biol. 33: 959–970 [DOI] [PubMed] [Google Scholar]
- Moreno M., Harper L., Kreuger R., Dellaporta S., Freeling M. (1997). liguleless1 encodes a nuclear-localized protein required for induction of ligules and auricles during maize leaf organogenesis. Genes Dev. 11: 616–628 [DOI] [PubMed] [Google Scholar]
- Moreno M.A., Chen J., Greenblatt I., Dellaporta S.L. (1992). Reconstitutional mutagenesis of the maize P gene by short-range Ac transpositions. Genetics 131: 939–956 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohler U., Liao G.C., Niemann H., Rubin G.M. (2002). Computational analysis of core promoters in the Drosophila genome. Genome Biol. 3: RESEARCH0087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olson W.K., Gorin A.A., Lu X.J., Hock L.M., Zhurkin V.B. (1998). DNA sequence-dependent deformability deduced from protein-DNA crystal complexes. Proc. Natl. Acad. Sci. USA 95: 11163–11168 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ornstein R.L., Rein R., Breen D.L., MacElroy R.D. (1978). An optimized potential function for the calculation of nucleic acid interaction energies. I- Base stacking. Biopolymers 17: 2341–2360 [DOI] [PubMed] [Google Scholar]
- Page D.R., Kohler C., Da Costa-Nunes J.A., Baroux C., Moore J.M., Grossniklaus U. (2004). Intrachromosomal excision of a hybrid Ds element induces large genomic deletions in Arabidopsis. Proc. Natl. Acad. Sci. USA 101: 2969–2974 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qu S., Desai A., Wing R., Sundaresan V. (2008). A versatile transposon-based activation tag vector system for functional genomics in cereals and other monocot plants. Plant Physiol. 146: 189–199 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rabinowicz P., Schutz K., Dedhia N., Yordan C., Parnell L., Stein L., McCombie W., Martienssen R. (1999). Differential methylation of genes and retrotransposons allows shotgun sequencing of the maize genome. Nat. Genet. 23: 305–308 [DOI] [PubMed] [Google Scholar]
- Raizada M.N., Walbot V. (2000). The late developmental pattern of Mu transposon excision is conferred by a cauliflower mosaic virus 35S-driven MURA cDNA in transgenic maize. Plant Cell 12: 5–21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schiefelbein J.W., Furtek D.B., Dooner H.K., Nelson O.E., Jr (1988). Two mutations in a maize bronze1 allele caused by transposable elements of the Ac-Ds family alter the quantity and quality of the gene product. Genetics 120: 767–777 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schnable P.S., et al. (2009). The B73 maize genome: Complexity, diversity, and dynamics. Science 326: 1112–1115 [DOI] [PubMed] [Google Scholar]
- Schneider T.D., Stephens R.M. (1990). Sequence logos: A new way to display consensus sequences. Nucleic Acids Res. 18: 6097–6100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schultes N.P., Brutnell T.P., Allen A., Dellaporta S.L., Nelson T., Chen J. (1996). Leaf permease1 gene of maize is required for chloroplast development. Plant Cell 8: 463–475 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwartz D., Dennis E. (1986). Transposase activity of the Ac controlling element in maize is regulated by its degree of methylation. Mol. Gen. Genet. 205: 476–482 [Google Scholar]
- Scott L., LaFoe D., Weil C.F. (1996). Adjacent sequences influence DNA repair accompanying transposon excision in maize. Genetics 142: 237–246 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seeman N.C., Rosenberg J.M., Rich A. (1976). Sequence-specific recognition of double helical nucleic acids by proteins. Proc. Natl. Acad. Sci. USA 73: 804–808 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen B., Zheng Z., Dooner H.K. (2000). A maize sesquiterpene cyclase gene induced by insect herbivory and volicitin: Characterization of wild-type and mutant alleles. Proc. Natl. Acad. Sci. USA 97: 14807–14812 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh M., Lewis P.E., Hardeman K., Bai L., Rose J.K., Mazourek M., Chomet P., Brutnell T.P. (2003). Activator mutagenesis of the pink scutellum1/viviparous7 locus of maize. Plant Cell 15: 874–884 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smit A.F.A., Hubley R., Green P. (2004). RepeatMasker Open-3.0. Institute for Systems Biology. http://www.repeatmasker.org. (January 10, 2007)
- Sullivan T.D., Schiefelbein J.W., Jr., Nelson O.E., Jr (1989). Tissue-specific effects of maize bronze gene promoter mutations induced by Ds1 insertion and excision. Dev. Genet. 10: 412–424 [DOI] [PubMed] [Google Scholar]
- Sundaresan V., Springer P., Volpe T., Haward S., Jones J.D., Dean C., Ma H., Martienssen R. (1995). Patterns of gene action in plant development revealed by enhancer trap and gene trap transposable elements. Genes Dev. 9: 1797–1810 [DOI] [PubMed] [Google Scholar]
- Van Schaik N.W., Brink R.A. (1959). Transposition of Modulator, a component of the variegated pericarp allele in maize. Genetics 44: 725–738 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vigdal T.J., Kaufman C.D., Izsvák Z., Voytas D.F., Ivics Z. (2002). Common physical properties of DNA affecting target site selection of Sleeping Beauty and other Tc1/mariner transposable elements. J. Mol. Biol. 323: 441–452 [DOI] [PubMed] [Google Scholar]
- Vollbrecht E., Reiser L., Hake S. (2000). Shoot meristem size is dependent on inbred background and presence of the maize homeobox gene, knotted1. Development 127: 3161–3172 [DOI] [PubMed] [Google Scholar]
- Wang H., Benham C.J. (2006). Promoter prediction and annotation of microbial genomes based on DNA sequence and structural responses to superhelical stress. BMC Bioinformatics 7: 248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan Y., SanMiguel P.J., Bennetzen J.L. (2003). High-Cot sequence analysis of the maize genome. Plant J. 34: 249–255 [DOI] [PubMed] [Google Scholar]
- Zhang J., Yu C., Pulletikurti V., Lamb J., Danilova T., Weber D.F., Birchler J., Peterson T. (2009). Alternative Ac/Ds transposition induces major chromosomal rearrangements in maize. Genes Dev. 23: 755–765 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou L., Mitra R., Atkinson P.W., Hickman A.B., Dyda F., Craig N.L. (2004). Transposition of hAT elements links transposable elements and V(D)J recombination. Nature 432: 995–1001 [DOI] [PubMed] [Google Scholar]