Abstract
Nodulin 26 (nod26) is a major intrinsic protein that constitutes the major protein component on the symbiosome membrane (SM) of N2-fixing soybean nodules. Functionally, nod26 forms a low energy transport pathway for water, osmolytes, and NH3 across the SM. Besides their transport functions, emerging evidence suggests that high concentrations of major intrinsic proteins on membranes provide interaction and docking targets for various cytosolic proteins. Here it is shown that the C-terminal domain peptide of nod26 interacts with a 40-kDa protein from soybean nodule extracts, which was identified as soybean cytosolic glutamine synthetase GS1β1 by mass spectrometry. Fluorescence spectroscopy assays show that recombinant soybean GS1β1 binds the nod26 C-terminal domain with a 1:1 stoichiometry (Kd = 266 nm). GS1β1 also binds to isolated SMs, and this binding can be blocked by preincubation with the C-terminal peptide of nod26. In vivo experiments using either a split ubiquitin yeast two-hybrid system or bimolecular fluorescence complementation show that the four cytosolic GS isoforms expressed in soybean nodules interact with full-length nod26. The binding of GS, the principal ammonia assimilatory enzyme, to the conserved C-terminal domain of nod26, a transporter of NH3, is proposed to promote efficient assimilation of fixed nitrogen, as well as prevent potential ammonia toxicity, by localizing the enzyme to the cytosolic side of the symbiosome membrane.
Keywords: Ammonia, Glutamine, Membrane Proteins, Nitrogen Metabolism, Protein-Protein Interactions, Aquaporin, Major Intrinsic Proteins, Nitrogen Fixation, Symbiosis, Transporters
Introduction
The formation of legume-rhizobia N2-fixing symbioses in root nodules leads to the development of a novel organelle known as the “symbiosome,” which houses the endosymbiotic rhizobia bacteroids. In mature nodules, the host infected cells are occupied by thousands of symbiosomes, which constitute the major organelle within this specialized cell type. The symbiosome is delimited by the symbiosome membrane (SM),2 which controls the transport of all metabolites between the symbiont and the plant host. These transport activities include the efflux of the primary metabolic product of nitrogen fixation (NH3/NH4+) and the uptake of dicarboxylates as an energy source to support bacterial N2 fixation (1, 2). Because of the unique role of the SM, a number of host nodulin (“nodule-enhanced”) proteins are synthesized and localized to this membrane (3, 4), where they serve transport and regulatory functions in the symbiosis.
Early studies revealed that nodulin 26 (nod26), a member of the major intrinsic protein (MIP)/aquaporin superfamily, is among the ensemble of soybean SM nodulins produced during symbiosome biogenesis (4–7). Since the original identification of nod26 over 20 years ago (4), the role of this channel in the symbiosis has been debated. Functional analyses show that nod26 is a multifunctional “aquaglyceroporin” that transports multiple substrates including water, formamide, and glycerol (6, 8). As a symbiosome-specific aquaglyceroporin, nod26 has been proposed to serve as a low energy transport pathway for water as well as osmolytes within the infected cell, potentially to aid in cell volume regulation and to facilitate infected cell adaptation to osmotic stresses (6, 8, 9). nod26 has also been proposed to transport ammonia gas (10) and may serve a metabolic function as a facilitated transport pathway for fixed NH3 efflux from the symbiosome to the plant cytosol for assimilation.
Similar to a number of MIPs, nod26 is the most abundant protein on its resident membrane (4, 5) and constitutes more than 10% of the total SM protein (6). Due to the high concentration of MIPs on their resident membrane, an alternative function for these proteins as interaction and membrane-docking targets for other cytosolic proteins has emerged. For example, the cytosolic C-terminal region of mammalian AQP2 interacts with a multicomponent motor protein complex (11–13) that is involved in trafficking this protein from intracellular vesicles to the plasma membrane in response to the antidiuretic hormone vasopressin (14, 15). The homologous C-terminal region of AQP0, the most abundant protein in lens fiber membranes, interacts with multiple lens proteins including regulatory and cytoskeletal proteins (16–18), gap junction proteins (19, 20), and lens crystallins (21, 22). In the present study, nod26 was investigated as a potential SM-docking target for proteins in the nodule cytosol. It is shown that the C-terminal domain of nod26 serves as a site for the association of cytosolic glutamine synthetase (GS) with the SM, suggesting novel roles for nod26 in ammonia transport and assimilation.
EXPERIMENTAL PROCEDURES
Purification of Nodule Glutamine Synthetase
Soybean (Glycine max cv Bragg) nodulated with Bradyrhizobium japonicum USDA110 were grown as described in Ref. 5. Symbiosome membranes were isolated as described in Refs. 5 and 6. For the preparation of soybean nodule GS, nodules were homogenized in 100 mm Tris-HCl (pH 8.4), 10 mm MgOAc, 10% v/v glycerol, 0.05% v/v Triton X-100 (3 ml/g of nodules) on ice. The extract was centrifuged at 35,000 × g at 4 °C for 30 min, and the supernatant proteins were precipitated with an equal volume of chilled acetone. The precipitate was collected and resuspended in 10 mm Tris-HCl (pH 7.5), 10 mm MgOAc, 10% v/v glycerol (Sephacryl Buffer) and subjected to differential (NH4)2SO4 precipitation. The pellet obtained from the 30–60% saturation cut was resuspended in Sephacryl Buffer and chromatographed at 4 °C on Sephacryl S300 (50 × 2-cm column). Fractions with maximal GS activity were pooled and stored at −80 °C.
Molecular Cloning of GS1 Isoforms from Nitrogen-fixing Soybean Root Nodules
Total RNA was extracted from 35-day-old soybean nodules by using the PLANT RNA extraction reagent (Invitrogen). Total cDNA was prepared using the SuperScript III reverse transcription kit (Invitrogen). PCR amplification of cDNAs encoding GS1 isoforms GS1β1, GS1β2, GS1γ1, and GS1γ2 (23) was done from soybean nodule cDNA using primers against the 5′- and 3′-untranslated regions of each isoform (supplemental Table 1) based on sequences available from the genomic data base Phytozome. PCR products were cloned into pCR2.1-TOPO (Invitrogen) and transformed into Escherichia coli DH5α. Isoform identity (supplemental Fig. 1) was verified by automated DNA sequence analysis.
Expression and Purification of Recombinant GS1 β1
A cDNA containing the full-length open reading frame (ORF) of soybean cytosolic GS1β1 was cloned into pDEST17 in-frame with an amino-terminal His tag linker by using the Gateway Cloning Technology (Invitrogen). Recombinant GS1β1 expression in E. coli BL21* was induced in mid-log cultures with 1 mm isopropyl β-d-1-thiogalactopyranoside. Cultures were grown with shaking for 16 h at room temperature. Cells were collected and resuspended in 20 mm Tris-HCl (pH 7.9), 300 mm NaCl, 25 mm imidazole, 10% (v/v) glycerol, 1 mm phenylmethylsulfonyl fluoride, 1 μg/ml leupeptin, 0.1% (w/v) Triton X-100 and lysed in a French press pressure cell (SLM-AMINCO Spectronic Instruments). The lysate was centrifuged at 150,000 × g for 30 min at 4 °C, and the supernatant fraction was chromatographed on Ni2+-nitrilotriacetic acid (Ni-NTA)-agarose pre-equilibrated in NTA Buffer (20 mm Tris-HCl, pH 7.5, 300 mm NaCl, 5% (v/v) glycerol) containing 20 mm imidazole. The column was washed with NTA Buffer containing 60 mm imidazole and eluted with NTA Buffer containing 500 mm imidazole. Fractions containing GS activity were pooled, dialyzed against 50 mm Tris-HCl, pH 7.5, 150 mm NaCl, 5 mm MgCl2, and stored at −80 °C.
Affinity Chromatography on Peptide Resins
A synthetic peptide (CK-25), which contains the entire C-terminal region of soybean nod26 (see Fig. 1) with an additional N-terminal cysteine, was obtained from GenScript (Piscataway, NJ). The CI-14 peptide, corresponding to the C terminus of Lotus japonicus nod26, was prepared as described in Ref. 9. For the purpose of this study, identical results were obtained regardless of which nod26 peptide was used. Immobilized peptide resins were prepared by covalent coupling to ω-aminohexyl-agarose by the protocol in Ref. 9. Soybean nodule extracts were prepared as described above, and 5 ml (1 mg/ml protein) were applied to resin (0.2 ml) equilibrated in 50 mm Tris-HCl, 150 mm NaCl, pH 7.5 (Binding Buffer). After extensive washing, the resin was eluted with 50 mm Tris-HCl, pH 7.5, 6 m urea, and the eluent was analyzed by SDS-PAGE on 12.5% (w/v) polyacrylamide gels using the buffer system of Laemmli (24).
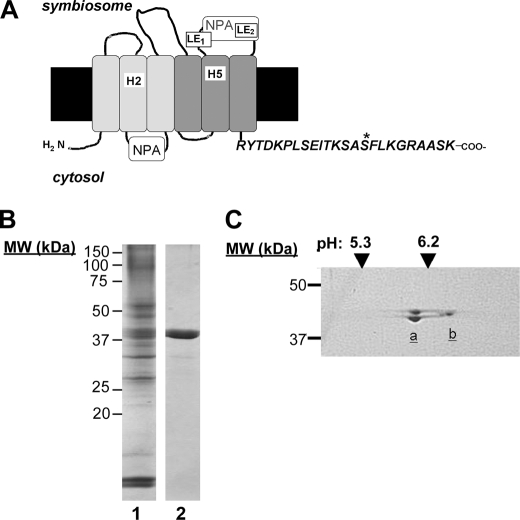
FIGURE 1.
Isolation of soybean nodule proteins interacting with the C-terminal domain of nod26. A, topology of soybean nod26 in the SM. The conserved NPA loops and the amino acid positions of the ar/R selectivity filter (H2, H5, LE1, and LE2) are indicated based on Wallace et al. (7). The sequence of the C-terminal cytosolic domain is shown with the site of phosphorylation (55) indicated with an asterisk. B, results of affinity chromatography of a soluble extract of 28 day old nodules on the nod26 C-terminal peptide affinity resin. Lane 1, soybean nodule extract (10 μg of protein). Lane 2, affinity resin-bound protein eluted with 6 m urea. MW, molecular mass markers. C, results of two-dimensional gel electrophoresis of 3 μg of the affinity resin-purified protein from panel B. The positions of pH markers in the first dimension and molecular mass standards in the second dimension are shown. Lowercase a and b designations show the positions of migration of soybean GS1 β and γ isoforms, respectively, based on Ref. (23).
For resin association assays, 50 μl of CK-25 peptide resin or underivatized ω-aminohexyl-agarose (negative control) were incubated with 50 units (1 unit = 1 nmol of γ-hydroxyglutamate/min at 37 °C) of purified soybean GS in Binding Buffer for 30 min at 25 °C with intermittent mixing. The resin was separated from the soluble fraction by centrifugation and washed with 10 ml of Binding Buffer. The fraction of the GS activity bound to the resin or present in the unadsorbed supernatant fractions was determined.
Two-dimensional Electrophoresis
For two-dimensional electrophoresis, protein samples (5 μg) were dissolved in a final volume of 150 μl of 8 m urea, 2% (w/v) octylglucoside, 4% (w/v) CHAPS, 1% (w/v) dithiothreitol, 0.16% (v/v) BioLytes 5–7, 0.04% (v/v) BioLytes 3–10. Separation of samples in the first dimension was done by isoelectric focusing on 7-cm ReadyStrip IPG strips (immobilized linear pH 5–8 gradient from Bio-Rad) in a PROTEAN isoelectric focusing cell apparatus (Bio-Rad) at 20 °C. Isoelectric focusing was done using the following five steps: 100 V for 200 V-h, 500 V for 500 V-h, 1000 V for 1000 V-h, 1000–8000 V for 1 h, and maintained at 8000 V for 8000 V-h. The IPG strips were equilibrated twice for 15 min in 5 ml of 50 mm Tris-HCl, pH 6.8, 6 m urea, 20% (v/v) glycerol, and 2% (w/v) SDS. Two percent (w/v) dithiothreitol was added during the first equilibration step followed by the addition of 2.5% (w/v) iodoacetamide during the second equilibration step. The IPG strips were placed on top of the 8.5% (w/v) polyacrylamide gels for second-dimension separation by SDS-PAGE.
Mass Spectrometry
Proteins resolved by SDS-PAGE were identified by in-gel tryptic digestion followed by peptide mass fingerprinting as described in Ref. 25. Digested peptides were extracted in 0.1% (v/v) trifluoroacetic acid and 60% (v/v) acetonitrile, and the extracted samples were desalted and concentrated with a 10-μl ZipTipC18 (Millipore, Bedford, MA) following the instructions provided by the manufacturer. The final sample was mixed with α-cyano-4-hydroxy-cinnamic acid dissolved in 60% (v/v) acetonitrile, 0.1% (v/v) trifluoroacetic acid. Peptide mass spectra were acquired on a Bruker microflex time-of-flight mass spectrometer (Bruker Daltonics) with a nitrogen laser operating at 337 nm on a positive ion mode. The masses of tryptic peptides were analyzed by searching the National Center for Biotechnology Information (NCBI) nonredundant protein data base with ProFound tool from The Rockefeller University Laboratory of Mass Spectrometry and Gaseous Ion Chemistry web site. Peptide masses were assumed to be monoisotopic, and methionine residues were assumed to be partially oxidized. A maximum of two missed tryptic cleavages were allowed. Tandem mass spectrometry sequence analysis of in-gel tryptic digests was done at the University of Georgia Proteomics Facility.
Peptide and SM Binding Assays
CK-25 peptide (0.42 μmol) was dissolved in 75 μl of 50 mm Tris-HCl, pH 7.5, and labeled at 4 °C overnight with a 10-fold molar excess of N,N′-dimethyl-N-(iodoacetyl)-N′-(7-nitrobenz-2-oxa-1,3-diazol-4-yl)ethylenediamine (Molecular Probes). The nitrobenzoxadiazole (NBD)-labeled peptide was isolated from excess N,N′-dimethyl-N-(iodoacetyl)-N′-(7-nitrobenz-2-oxa-1,3-diazol-4-yl)ethylenediamine reagent by chromatography on a Sephadex G-25 in 50 mm Tris-HCl (pH 7.5). The labeled peptide concentration was calculated by A497 (NBD ϵ = 26,000 m−1cm−1), and residual unlabeled peptide was quantitated by using Ellman's assay (26). Under these conditions, labeling of the N-terminal cysteine was stoichiometric.
Fluorescence measurements were performed using a QuantaMaster UV VIS (Photon Technology International) spectrofluorometer at 22 °C. Peptide and GS binding assays were done in 50 mm Tris-HCl, pH 7.5, 150 mm NaCl, 5 mm MgCl2 with the fluorescent peptide kept constant at 0.67 μm, and the concentration of GS was varied. Binding reactions were incubated at room temperature for 5 min prior to fluorescence measurements (excitation λ = 480 nm, emission λ = 545 nm). The increase in fluorescence intensity as a function of added GS was fit to the following binding expression
 |
where ΔF is the change in fluorescence intensity, ΔFmax is the maximal change in fluorescent intensity at saturation, [GS] is the concentration of GS, and Kd is the dissociation constant.
For SM binding experiments, 100 μg of SM were incubated with 50 units of purified soybean nodule GS in Binding Buffer with 5 mm MgCl2 for 1 h at 4 °C. Membranes were collected by centrifugation at 200,000 × g for 30 min and washed with 2 ml of Binding Buffer with 5 mm MgCl2. The centrifugation/washing cycle was repeated three additional times, and the membranes were resuspended in 100 μl of Binding Buffer and assayed for GS activity. For peptide inhibition experiments, 10 μm CI-14 peptide was preincubated with GS for 30 min at 4 °C prior to incubation with SM samples.
Yeast Split Ubiquitin Analyses
Split ubiquitin yeast two-hybrid experiments were performed as described in Ref. 27. THY.AP4 and THY.AP5 Saccharomyces cerevisiae strains as well as the pMetYCgate, pNXgate32, pNubWT, KAT1/pMetYCgate, and KAT1/pNXgate32 vectors were obtained from the Arabidopsis Biological Resource Center (Ohio State University). The nod26 bait construct containing the C terminus of ubiquitin (Cub) was generated by cloning the soybean nod26 ORF into the pMetYCgate vector in the THY.AP4 strain by in vivo recombination (Basic protocol 1 in Ref. 28). Prey constructs (GS1 and nod26) containing the N-terminal ubiquitin fragment (Nub) were generated by cloning the respective ORFs in the pNXgate32 vector in the THY.AP5 strain. The primers used in the preparation of all the constructs are listed in supplemental Table 1. Interaction assays were done by mating various bait and prey pairs (Basic protocol 2 in Ref. 28). Diploids were selected by replica plating on synthetic medium plates lacking tryptophan, leucine, and uracil. For β-galactosidase selection, diploids were grown on selective medium for 2 days at 30 °C and overlaid with 60 mm sodium phosphate, pH 7.2, 0.2% sodium dodecyl sulfate, 2 mg/ml 5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside (X-gal), 0.5% (w/v) agarose. Plates were stored at 37 °C overnight to allow color development. For HIS3 selection, diploid cells were replica-plated and grown on a synthetic dextrose medium lacking histidine, leucine, tryptophan, and uracil for 2 days at 30 °C. Growth was scored by comparison of test yeast colonies to control yeast colonies containing empty pMetYCgate and pNXgate vectors.
Bimolecular Fluorescence Complementation (BiFC) Assay
Constructs containing soybean nodule GS1β1 or nod26 coding sequences as translational fusions with either the N-terminal 154 residue fragment (YFP-N) or the C-terminal 84 residue fragment (YFP-C) of the yellow fluorescent protein were prepared in pd35S-YFP-N or pd35S-YFP-C vectors (a kind gift from Dr. Andreas Nebenführ, the University of Tennessee, Knoxville, TN). Prior to cloning into BiFC plasmids, the BamHI restriction site in GS1β1 was removed by PCR-based site-directed mutagenesis (29). The N-terminal fusions of GS1β1 and nod26 (YFP-C-GS1β1 and YFP-N-nod26) were prepared by cloning ORFs of GS1β1 and nod26 into BamHI-NotI-digested pd35S-YFP-C and pd35S-YFP-N, respectively. The C-terminal fusion of nod26 (nod26-YN) was prepared by cloning the nod26 ORF into XbaI-BamHI-digested pd35S-YFP-N. In all constructs, a linker region of 10 residues (GGHHHHHHGG) was introduced between the YFP fragment and the ORF of interest. All primers used for molecular cloning are listed in supplemental Table 1.
Transient expression of fusion proteins and visualization of BiFC interactions were done by tungsten particle bombardment of onion bulb epidermal cells as described previously (30, 31). Bombarded onion cells were incubated for 24 h at 28 °C prior to microscopic examination with an Axiovert 200 M microscope (Zeiss) equipped with filters for YFP fluorescence (Chroma, filter set 52017). Images were captured with a digital camera (Hamamatsu Orca-ER) controlled by the Openlab software (Improvision).
Other Analytical Methods
GS activity was assayed by the hydroxylamine colorimetric method (32) or by the determination of inorganic phosphate release (33). GS kinetic studies were performed using the microtiter assay described in Ref. 33. Analysis of DNA sequences was done by automated sequencing using a PerkinElmer Life Sciences Applied Biosystems 373 DNA sequencer and the Prism dye terminator reaction at the University of Tennessee, Molecular Biology Resource Facility, Knoxville, TN. Protein analyses were done using the BCA (Pierce Biochemicals) or the Bradford (Bio-Rad) methods and the manufacturer's protocol.
RESULTS
Isolation of a 40-kDa Soybean Nodule Protein Interacting with the C-terminal Domain of Nodulin 26
The nod26 C terminus is composed of a hydrophilic 24-amino acid extension (Fig. 1), which is exposed to the cytosolic side of the symbiosome (5), and has a sequence that is conserved among group I of nod26-like intrinsic proteins (7). Previous studies (11–13, 16–22) have shown that the C-terminal domain of MIPs is a site for protein-protein interaction. To investigate the possibility that the nod26 cytosolic C-terminal extension serves as a protein interaction site, resins with immobilized synthetic peptides with the nod26 C-terminal sequence were used in affinity chromatography experiments with soybean nodule extracts. Chromatography of a nodule soluble extract on nod26 C-terminal peptide-agarose resulted in the adsorption of a major 40-kDa protein band, which was bound tightly to the resin, requiring 6 m urea for elution (Fig. 1B). This was the only detectable protein in the urea eluent. Resolution by two-dimensional electrophoresis revealed that the 40-kDa protein band actually consisted of a collection of bands with distinct pIs and slightly different molecular masses (Fig. 1C). The significance of this two-dimensional electrophoretic profile is discussed below.
Identification of the 40-kDa Nodulin 26-interacting Protein as Cytosolic Glutamine Synthetase
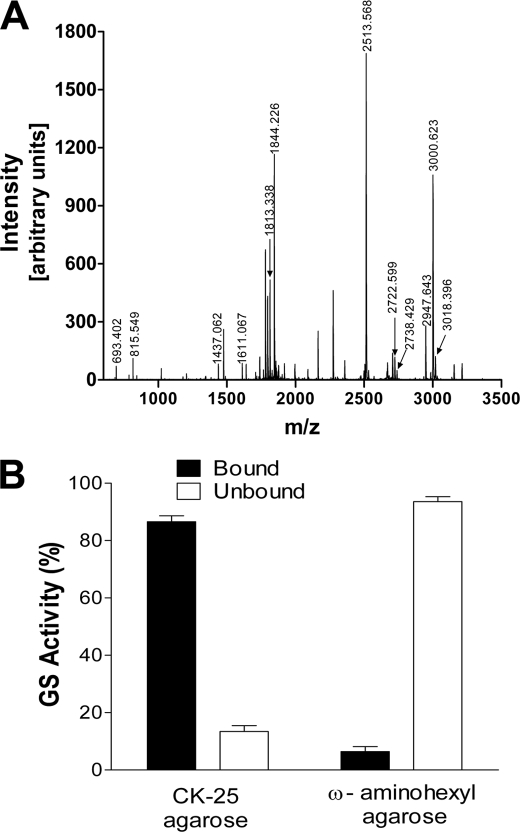
The 40-kDa protein band was excised from an SDS-PAGE gel, digested with trypsin, and subjected to MALDI-TOF mass spectrometric analysis (Fig. 2A). Analysis of the peptide mass fingerprint (Table 1) identified soybean cytosolic glutamine synthetase GS1 β1 as the most likely candidate protein (E value = 6.1 × 10−5, 56% sequence coverage). Confirmation of this assignment was obtained by tandem mass spectrometry analysis of a 1610.022-Da tryptic peptide, which yielded a sequence (277HKEHIAAYGEGNER290) characteristic of soybean GS1β1. In addition, the proposed molecular weight of soybean GS1β1 (Mr = 38,759) is in agreement with the observed 40-kDa molecular mass of the protein on an SDS-PAGE gel (Fig. 1B). To further confirm the identity of the 40-kDa protein, GS was purified from soybean nodule extracts (Fig. 3A) and used in interaction assays with the nod26 C-terminal peptide affinity resin (Fig. 2B). Nodule GS was bound quantitatively to nod26 peptide resins but not to a negative control resin (Fig. 2B). Overall, these experiments show that the major 40-kDa interacting protein from soybean nodule extracts is cytosolic GS.
FIGURE 2.
Identification of the 40-kDa protein as soybean cytosolic glutamine synthetase. A, MALDI-TOF spectrum of protonated tryptic peptides (MH+) of the 40-kDa protein isolated by affinity chromatography on nod26 peptide-agarose. The 40-kDa protein was resolved by electrophoresis as in Fig. 1B and subjected to in-gel tryptic digestion and mass spectroscopic analysis. The y axis shows the intensity as arbitrary units. The mass-to-charge ratio is plotted on the x axis. The results of mass fingerprinting analysis are in Table 1. B, interaction of GS with the C-terminal domain of nod26. Purified native soybean nodule GS (49 units) was incubated with nod26 peptide-agarose (CK-25-agarose) or with underivatized ω-aminohexyl-agarose. The fraction of GS activity bound to the resins (solid bars) as well as in the unadsorbed fraction (open bars) was measured (n = 6, error bars show S.E.).
TABLE 1.
Mass fingerprint of 40 kDa protein tryptic peptides from MALDI-TOF spectroscopy
| Measured massa | Predicted mass | Residue indicesb | Sequencec |
|---|---|---|---|
| 692.392 | 692.385 (0.007)d | 219–223 | YILER |
| 785.382 | 785.334 (0.048) | 327–332 | GYFEDR |
| 814.532 | 814.491 (0.041) | 268–275 | AAIDKLGK |
| 1436.042 | 1435.755 (0.287) | 39–52 | TLPGPVSDPSELPK |
| 1610.022 | 1609.759 (0.263) | 277–290 | HKEHIAAYGEGNERe |
| 1737.602 | 1737.854 (−0.252) | 276–290 | KHKEHIAAYGEGNER |
| 1779.212 | 1778.902 (0.310) | 19–34 | VIAEYIWIGGSGMMMDLR |
| 1812.332 | 1812.039 (0.293) | 224–240 | ITEIAGGGVVVSFDPKIPK |
| 1843.212 | 1842.901 (0.312) | 296–311 | HETADINTFLWGVANR |
| 2356.362 | 2356.172 (0.190) | 85–106 | GNNILVICDAYTPAGEPIPTNK |
| 2512.562 | 2512.273 (0.289) | 85–107 | GNNILVICDAYTPAGEPIPTNKR |
| 2512.562 | 2512.273 (0.289) | 84–106 | RGNNILVICDAYTPAGEPIPTNK |
| 2668.442 | 2668.374 (0.068) | 84–107 | RGNNILVICDAYTPAGEPIPTNKR |
| 2946.632 | 2946.479 (0.153) | 113–137 | VFSHPDVVAEVPWYGIEQEEEYTLLQK |
| 2999.612 | 2999.392 (0.220) | 53–79 | WNYDGSSTGQAPGEDSEVILYPQAIFR |
| 3017.392 | 3017.416 (−0.023) | 138–165 | DIQWPLGWPVGGFPGPQGPYYCGVGADK |
a The experimental mass of each peptide from the MALDI-TOF spectrum (Fig. 2) is shown along with the theoretical mass of the corresponding tryptic digest peptide from soybean GS1β1. Mass is reported in daltons.
b The amino acid sequence index of soybean GS1β1 corresponding to each peptide is shown.
c The derived primary sequence of each soybean GS1β1 peptide is shown.
d The error between the experimental and theoretical masses of each peptide is shown parenthetically.
e The sequence of this peptide was confirmed by tandem mass spectrometry analysis.
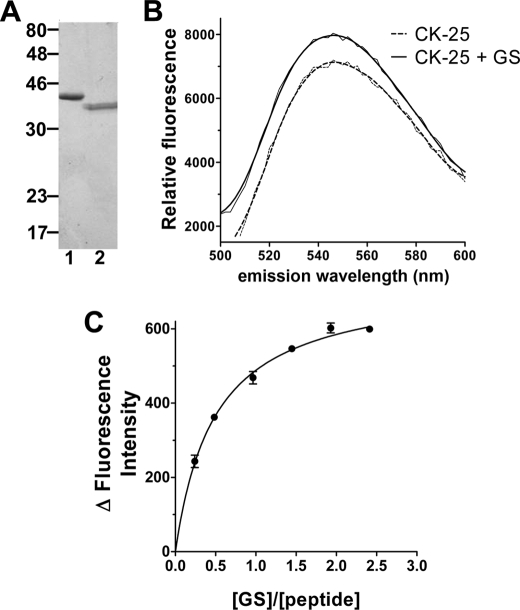
FIGURE 3.
Quantitation of the interaction between GS1 and the C-terminal peptide of soybean nod26 (CK-25). A, SDS-PAGE profile of purified GS. Lane 1, purified recombinant soybean GS1 β1; lane 2, purified native soybean nodule GS. Each lane contains 0.5 μg of purified protein. B, fluorescence emission spectra of 0.64 μm NBD-labeled CK-25 in the presence (solid line) or absence (dotted line) of 1.36 μm recombinant soybean GS1 β1. λex = 480 nm. C, binding curve of NBD-labeled CK-25 and recombinant soybean GS1 β1. The peptide was kept constant at 0.67 μm, and the change in the intensity of fluorescence emission at 545 nm (excitation wavelength = 480 nm) was monitored in response to an increase in the concentration of GS1 β1. Error bars show S.E. (n = 4).
To quantify the interaction of the C-terminal nod26 domain with cytosolic GS1β1, a fluorescence spectroscopy approach was used (Fig. 3). Recombinant GS1β1 was expressed with an amino-terminal His tag in E. coli and purified by Ni2+ chelate chromatography (Fig. 3A). The CK-25 peptide containing the full-length soybean nod26 C-terminal domain was labeled on its amino-terminal cysteine with the fluorescent label NBD. In the presence of an equal molar concentration of purified GS1β1, the labeled CK-25 peptide showed an increase in fluorescence intensity at its emission maximum of 545 nm (Fig. 3B), which was used as an index of peptide-enzyme interaction. The peptide showed saturable binding with half-saturation occurring at a [GS]/[NBD-CK-25] ratio of 0.51, suggesting a binding stoichiometry of one peptide:one GS monomer (Fig. 3C). Assuming 1:1 binding stoichiometry, a fit of the binding data yielded a Kd of 266 nm (S.E. = 18 nm) for peptide binding to recombinant GS1 β1.
To assay the ability of native nod26 to interact with GS, SMs were isolated from soybean nodules by the Percoll step gradient method, which produces vesicles with the hydrophilic nod26 C terminus exposed on the outer surface of the vesicle (5). Control SMs contained a small but significant amount of GS activity (Fig. 4C). This observation is consistent with previous proteomic analyses that showed that SMs possess peripherally associated GS (34). Incubation of SMs with purified native soybean nodule GS resulted in additional membrane adsorption of GS. Binding of the enzyme to SMs was competitively inhibited by preincubating the purified native soybean nodule GS with 10 μm nod26 peptide (Fig. 4C), suggesting that the nod26 C terminus is responsible for SM binding of GS.
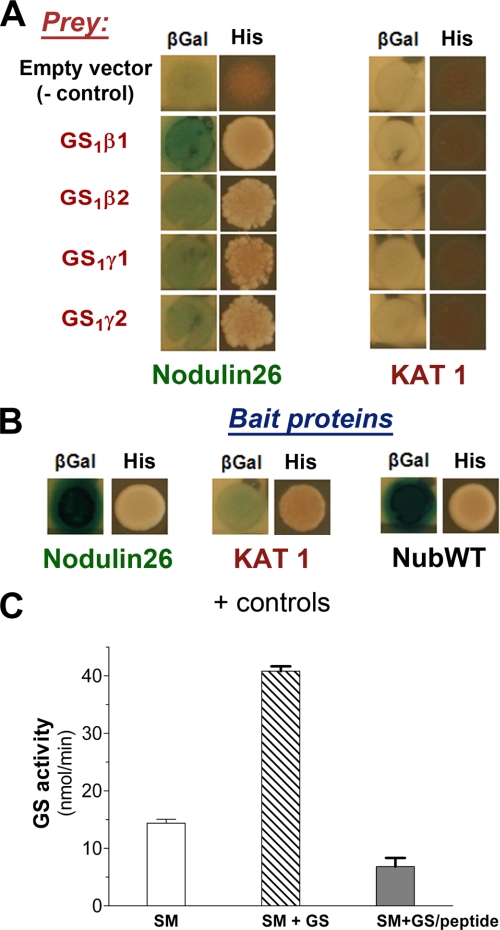
FIGURE 4.
Analysis of interactions between glutamine synthetase and nod26 on SMs and in vivo using the yeast split ubiquitin system. A, S. cerevisiae strains (THY.AP4) containing bait constructs consisting of the nod26 cDNA or the Arabidopsis KAT1 potassium channel cDNA cloned as translational fusions to the C-terminal fragment of ubiquitin (Cub) fused to a synthetic LexA-VP16 transcription factor were mated with THY.AP5 strains containing the GS prey constructs shown. Interaction of bait and prey proteins was tested by activation of two reporter genes; βGal (left panel), shows the results of a β-galactosidase overlay assay, and His (right panel) shows growth on selection medium lacking histidine. The empty vector control shows the results of mating of the indicated bait vectors to the empty pNXgate32 vector. B, the results of positive control matings are shown. In the case of nod26, the mating of homologous bait and prey constructs (resulting in homooligomerization) was performed. A similar mating with homologous KAT1 bait and prey constructs yield positive results because of dimerization (27). NubWT shows the results of using wild-type ubiquitin, which constitutively interacts with Cub fragments and has been used previously as positive control in this system (27). C, isolated purified soybean SMs were incubated with purified native soybean nodule GS (SM + GS) as well as soybean nodule GS preincubated with 10 μm nod26 peptide (SM+GS/peptide). As a control, SMs were incubated with an equivalent volume of binding buffer without added GS (SM). Membranes were washed, and the GS activity bound was assayed with the error bars showing S.E. (n = 6).
Cytosolic Glutamine Synthetase Interacts with Full-length Nodulin 26 in Vivo
Soybean nodules contain four cytosolic GS isoforms (GS1β1, GS1β2, GS1γ1, and GS1γ2) that are distinguished by molecular weight and isoelectric point (23). Analysis of the two-dimensional electrophoretic profile in this study (Fig. 1C) suggests that both β and γ isoforms are represented in the GS fraction that binds to the C-terminal nod26 peptide. To investigate the isoform specificity and to test the interaction of full-length nod26 and GS in vivo, a split ubiquitin yeast two-hybrid screen (27) was employed. The full-length cDNAs corresponding to GS1β1, GS1β2, GS1γ1, and GS1γ2 “preys” were cloned as translational fusions to a modified N-terminal fragment of ubiquitin (NubG). nod26 (bait) was translationally fused to the C-terminal fragment of ubiquitin (Cub) followed by a synthetic LexA-VP16 transcription factor. Due to a reduced affinity of NubG for Cub, interaction of these fragments and the reconstitution of ubiquitin is only possible when these fragments are placed in close proximity due to an interaction between the bait and prey proteins. Cleavage of the reconstituted ubiquitin releases the LexA-VP16 transcription factor, which diffuses into the nucleus and induces the transcription of reporter genes under control of the LexA promoter (β-galactosidase and HIS3).
nod26 forms homotetramers like other MIPs (35–38), and the Arabidopsis potassium channel AtKAT1 has also been demonstrated to form oligomers (27). Therefore, the subunit-subunit interactions between monomers of these proteins served as excellent positive controls and were among the most robust interactions reported in this screen (Fig. 4B). The homooligomerization results also suggest that both of these proteins are properly expressed and folded in yeast. Additionally, the wild-type N-terminal fragment of ubiquitin (NubWT) served as a system control because it constitutively interacts with Cub and activates both reporter genes without prey protein attached (Fig. 4B).
Mating of yeast strains containing the four soybean GS isoform prey constructs with strains containing the nod26 bait construct resulted in a positive interaction as indicated by β-galactosidase expression and growth on histidine selection medium (Fig. 4A). As a negative control, an AtKAT1 potassium channel bait construct (27) was used in mating experiments with the GS1 prey constructs. These matings showed no apparent expression of β-galactosidase or growth on histidine selection medium (Fig. 4B). Overall, the data suggest that all four cytosolic soybean nodule GS isoforms form a complex with soybean nod26. This conclusion is supported by the observation that recombinant forms of both β and γ isoforms were bound to isolated SM (supplemental Fig. 2).
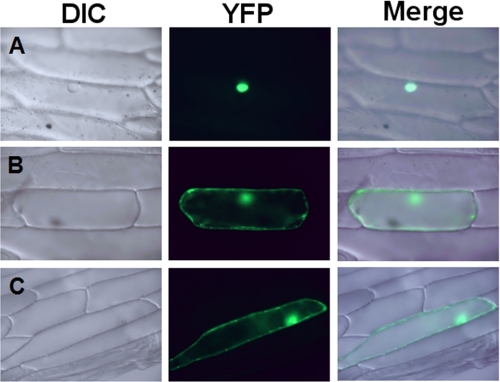
To determine whether nod26 interacts with GS in planta, BiFC experiments were performed (Fig. 5). BiFC is a protein interaction technique in which two putative interacting proteins are fused to either an N-terminal or a C-terminal fragment of yellow fluorescent protein (YFP-N and YFP-C) and are expressed separately. If the two proteins interact, the N- and C-terminal fragments will be brought into close proximity, resulting in the reconstitution of YFP, which can be measured by fluorescence microscopy. nod26 was translationally fused to the YFP-N fragment at either its amino-terminal (YFP-N·nod26) or its carboxyl-terminal (nod26·YFP-N) end. The YFP-C-terminal fragment was translationally fused to the amino-terminal end of GS1β1 (YFP-C·GS1 β1). The Arabidopsis transcription factor HY5 has been previously demonstrated to dimerize in BiFC experiments and localize to the nucleus (30) and was used as a positive control for this assay. Transient expression of the various constructs in onion epidermal cells was done by particle bombardment. Individual transformation of the each nod26 and GS construct yielded negative results. However, co-transformation of YFP-C·GS1β1 with either the nod26·YFP-N or the YFP-N·nod26 constructs reconstituted the YFP signal (Fig. 5), suggesting an interaction between both the nod26 construct and GS1β1. Overall, the findings show that cytosolic nodule GS interacts with soybean nod26 with the site of interaction likely to include the C-terminal cytosolic domain.
FIGURE 5.
Visualization of the interaction of nod26 with soybean GS1 β1 by BiFC in onion cells. Onion epidermal cells were transformed with the following BiFC pairs: A, positive control construct consisting of the YFP-N·HY5/YFP-C·HY5 pair; B, an amino-terminal fusion of YFP-N·nod26 with an amino-terminal fusion of YFP-C·GS1β1; C, a carboxyl-terminal fusion of nod26·YFP-N with YFP-C·GS1β1. DIC, differential interference contrast optics; YFP, fluorescent images using the Yellow Fluorescent Protein filter set; Merge, the superimposition of both images.
DISCUSSION
The SM is a unique symbiotic interface between legume hosts and endosymbiotic rhizobia bacteria in N2-fixing symbioses. Biogenesis of the SM occurs early in the rhizobia infection process and is accompanied by the biosynthesis of a variety of nodulin proteins. Several of these proteins become integral components of the mature SM and mediate transport and regulatory processes associated with metabolite exchange between the symbiotic partners (1, 2). Among these proteins is nod26, which is a major component of the mature symbiosome (4–6). nod26 confers a high intrinsic water permeability (6, 8) to the SM and fulfills a secondary function as a transporter of neutral metabolites such as glycerol and NH3 (6, 8, 10). In the present study, evidence is provided for an additional function of nod26 as a site for the interaction of cytosolic nodule GS.
GS (EC 6.3.1.2) is the critical enzyme for assimilation of environmental ammonia as well as endogenous ammonia produced metabolically. Plant GSs are divided into two isoform classes that are distinguished by their subcellular location, with GS1 found in the cytosol, whereas GS2 resides in plastids (39–41). Plant cytosolic GS1 is encoded by a small, highly conserved gene family in angiosperms (41), with three GS1 isoform classes (designated α, β, and γ) typically present in legumes (23, 42). Gmgln-α, Gmgln-β, and Gmgln-γ show differential expression during development and in response to environmental and metabolic cues. In mature N2-fixing soybean nodules, four GS1 isoforms (β1, β2, γ1, γ2) exhibit high expression (23). The β isoforms are characterized as the “constitutive” GS1 subclass that exhibit a broad expression pattern in soybean tissues but show particularly high expression in nodules and are inducible by high levels of ammonia (23, 43). The γ isoforms are selectively expressed as nodulin proteins in a developmentally regulated fashion in soybean nodules (23, 44) and other legumes (39, 43, 45). The expression of the four GS1 isoforms during soybean nodule development coincides with the onset of nitrogen fixation (23), consistent with a role as the major enzymes responsible for the ATP-dependent assimilation of fixed ammonia transported from the symbiosome to the cytosolic compartment. The expression of GS1 (occurring at approximately day 10 in nodule development (23)) also parallels the appearance of nod26 protein in developing nodules (9).
From a structural perspective, questions remain regarding the composition and binding determinants for the association of the nod26·GS complex. The four soybean GS1 isoforms expressed in nodules share more than 85% amino acid sequence identity (supplemental Fig. 1), and the observation that all interact with nod26 suggests that these proteins contain a conserved interaction site for the nod26 C-terminal domain. Thus, any isoform could conceivably form a complex with nod26 in vivo. X-ray crystallography of plant cytosolic GS (46) shows a homodecameric structure of two stacked pentameric subunit rings with catalytic sites shared between adjacent monomeric subunits. The finding of a one-to-one binding stoichiometry suggests that each GS monomer possesses a binding site for the nod26 C-terminal domain. The high apparent affinity (Kd = 266 nm) for the interaction of C-terminal nod26 domain for GS1β1, combined with the observation that both nod26 (as high as 15% of the symbiosome membrane protein (6)) and GS (2% of the nodule cytosolic protein (47)) are major components of the soybean infected cell, strongly suggest that cytosolic GS binds to the nod26 C terminus at biologically relevant concentrations. This is supported by the yeast split ubiquitin and BiFC experiments.
Analysis of the effects of the nod26 CK-25 peptide on the activity and kinetics of GS (supplemental Fig. 3) suggests that the interaction exhibits a modest effect on enzyme activity with a 25% increase in Vmax and no significant effect on the Km for NH4+. Although the actual site of interaction on the GS decamer remains to be elucidated, the findings suggest that interaction with nod26 could serve principally to localize GS to the surface of the SM rather than exerting an effect on enzyme activity.
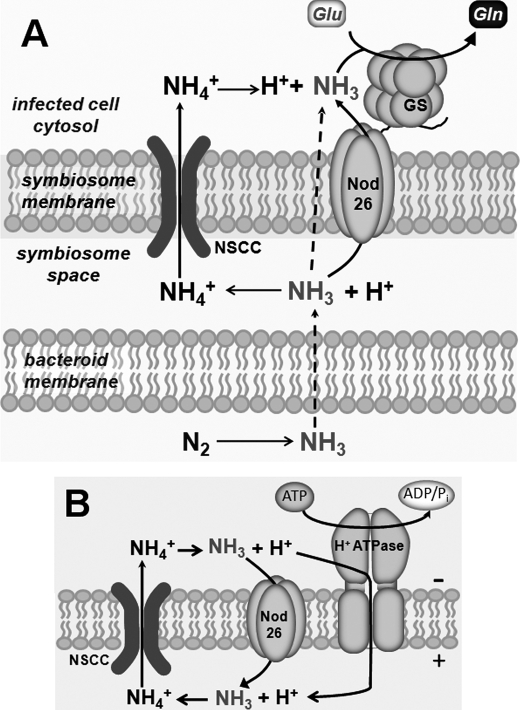
The potential symbiotic significance of nod26 interaction with GS can be understood from the perspective of the known transporters and pumps on the SM (1, 2, 10, 48–52) and the inherent toxicity of ammonia/ammonium transport across energized membranes (53), and it is summarized in Fig. 6. N2 fixation by rhizobium bacteroids results in the production of NH3, which diffuses across the bacteroid membrane into the symbiosome space. Efflux of NH3/NH4+ from the symbiosome space to the cytosol can occur by 1) directional transport of NH4+ cation to the cytosol by an inwardly rectified, voltage-activated cation channel (48–50) or 2) passive diffusion of uncharged NH3 through the SM (52) with facilitated diffusion of NH3 through nod26 potentially providing a low energy efflux pathway (10). The relative contributions of these pathways remain a subject of debate and depend upon the pH of the symbiosome space and the resting potential of the SM, both of which are primarily controlled by an energizing H+-pumping ATPase on the SM (51). An interaction between nod26 and GS would localize this critical assimilatory enzyme to the surface of the symbiosome, the site of fixed NH3/NH4+ release into the infected cell cytosol. Direct interaction of GS with nod26 could facilitate rapid assimilation of reduced nitrogen in the form of unprotonated NH3 transported through the nod26 channel, potentially as a “metabolic funnel” (Fig. 6). Additionally, because nod26 is the most abundant SM protein, interaction with GS would increase the local concentration of the enzyme at the symbiosome surface, which would enhance the rate of assimilation of NH3/NH4+ that leaves the symbiosome through other efflux pathways.
FIGURE 6.
Metabolic model for interaction of nodulin 26 and glutamine synthetase and its effect on nitrogen assimilation in nitrogen-fixing nodules. A, a model for efflux and assimilation of fixed nitrogen in symbiosomes is shown. Ammonia produced by the action of nitrogenase in the bacteroid moves into the symbiosome space by simple diffusion (52). Efflux of fixed nitrogen from the symbiosome space can occur as either NH4+ or NH3. NH4+ is directionally transported to the cytosolic side of the SM by a nonselective cation channel (NSCC), which is voltage-activated and is inwardly rectified (48–50). A diffusive pathway for NH3 efflux also exists with nod26 representing a facilitated pathway for this gas (10). Binding of GS to the C-terminal domain increases the concentration of this assimilatory enzyme at the symbiosome surface and also serves as a potential site for rapid assimilation of ammonia traversing nodulin 26. B, a potential mechanism for ammonia futile cycling through the SM. The SM is energized by an H+-ATPase, which generates a proton gradient by pumping H+ into the symbiosome space (51). When the SM is hyperpolarized, the nonselective cation channel is activated, which directionally transports NH4+ into the cytosolic compartment (48). Because the cytosol is more alkaline than the symbiosome space, NH4+ can release a H+ with NH3 potentially reentering the symbiosome space through nodulin 26. Maintenance of cytosolic NH4+ levels at low concentrations by rapid assimilation via GS would be one approach to prevent this potential metabolite cycling.
An additional advantage of the nod26-GS association may stem from the observation that high levels of ammonium are toxic to plants, which is potentially the result of wasteful “ammonia futile cycling” (53). In the case of the symbiosome, such a process could operate due to the acidic pH of the symbiosome space and the high concentrations of ammonium that accumulate during active nitrogen fixation (54) (Fig. 6B). Entry of NH4+ into the more alkaline plant cytosol would result in a loss of a proton generating NH3, which could reenter the symbiosome space, possibly through nod26, which could represent the lowest energy pathway for NH3 in the SM. The result would be a net transport of a proton from the symbiosome space to the cytosol, which would dissipate the proton motive force generated by the SM H+-ATPase (51) and lead to hydrolysis of ATP and futile cycling. As stated above, the interaction of nod26 with GS could facilitate rapid NH4+ assimilation, preventing its accumulation in the cytosol. The maintenance of low cytosolic concentrations of NH4+, which are estimated to be 50-fold lower than the NH4+ concentration in nitrogen-fixing symbiosomes (54), would prevent potential futile cycling.
Another potential level of complexity in the nod26-GS interaction comes from the observation that both binding partners are subject to post-translational phosphorylation (5, 55–57). In the case of nod26, the unique site of phosphorylation is Ser262 (55), which resides in the C-terminal domain, which is the site of GS interaction. Ser262 phosphorylation is catalyzed by a calcium-dependent protein kinase that is localized to the SM (5). nod26 phosphorylation is developmentally regulated, becoming apparent at the onset of nitrogen fixation, and is maintained at steady-state levels throughout the N2-fixing portion of the nodule lifespan (9). In addition, phosphorylation is regulated by osmotic stress signals (9), which may reflect the regulation of nod26 transport as part of an osmoregulatory response. Preliminary analyses suggest that nod26 peptides that are phosphorylated at Ser262 retain the ability to bind to GS in vitro, but the influence of phosphorylation on the assembly and activity of the complex in vivo remains to be addressed. Cytosolic GS1 is also a target for posttranslational phosphorylation by various protein kinases in plant tissues (56, 57), with phosphorylation potentially leading to interaction with 14-3-3 proteins (56) and other unidentified phosphoproteins (57). The interplay between phosphorylation, GS regulation, and interaction with nod26 and other potential regulatory targets and the effects of these on N2 fixation and assimilation in response to environmental cues remain a topic for future investigation.
This work was supported by National Science Foundation Grant MCB-0618075 (to D. M. R.) and by a National Science Foundation Graduate Fellowship (to I. S. W.).
This article was selected as a Paper of the Week.

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. 1–3 and Table 1.
- SM
- symbiosome membrane
- nod26
- nodulin 26
- GS
- glutamine synthetase
- MIP
- major intrinsic protein
- ORF
- open reading frame
- Ni-NTA
- nickel-nitrilotriacetic acid
- BiFC
- bimolecular fluorescence complementation
- NBD
- nitrobenzoxadiazole
- YFP
- yellow fluorescent protein
- MALDI-TOF
- matrix-assisted laser desorption/ionization-time of flight
- CHAPS
- 3-[(3-cholamidopropyl)dimethylammonio]-1-propanesulfonic acid.
REFERENCES
- 1.Udvardi M. K., Day D. A. (1997) Annu. Rev. Plant Physiol. Plant Mol. Biol. 48, 493–523 [DOI] [PubMed] [Google Scholar]
- 2.Day D. A., Poole P. S., Tyerman S. D., Rosendahl L. (2001) Cell. Mol. Life Sci. 58, 61–71 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Fortin M. G., Zelechowska M., Verma D. P. S. (1985) EMBO J. 4, 3041–3046 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Fortin M. G., Morrison N. A., Verma D. P. (1987) Nucleic Acids Res. 15, 813–824 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Weaver C. D., Crombie B., Stacey G., Roberts D. M. (1991) Plant Physiol. 95, 222–227 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Rivers R. L., Dean R. M., Chandy G., Hall J. E., Roberts D. M., Zeidel M. L. (1997) J. Biol. Chem. 272, 16256–16261 [DOI] [PubMed] [Google Scholar]
- 7.Wallace I. S., Choi W. G., Roberts D. M. (2006) Biochim. Biophys. Acta 1758, 1165–1175 [DOI] [PubMed] [Google Scholar]
- 8.Dean R. M., Rivers R. L., Zeidel M. L., Roberts D. M. (1999) Biochemistry 38, 347–353 [DOI] [PubMed] [Google Scholar]
- 9.Guenther J. F., Chanmanivone N., Galetovic M. P., Wallace I. S., Cobb J. A., Roberts D. M. (2003) Plant Cell 15, 981–991 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Niemietz C. M., Tyerman S. D. (2000) FEBS Lett. 465, 110–114 [DOI] [PubMed] [Google Scholar]
- 11.Noda Y., Horikawa S., Furukawa T., Hirai K., Katayama Y., Asai T., Kuwahara M., Katagiri K., Kinashi T., Hattori M., Minato N., Sasaki S. (2004) FEBS Lett. 568, 139–145 [DOI] [PubMed] [Google Scholar]
- 12.Noda Y., Horikawa S., Katayama Y., Sasaki S. (2004) Biochem. Biophys. Res. Com. 322, 740–745 [DOI] [PubMed] [Google Scholar]
- 13.Noda Y., Sasaki S. (2005) Biol. Cell 97, 885–892 [DOI] [PubMed] [Google Scholar]
- 14.Brown D., Hasler U., Nunes P., Bouley R., Lu H. A. (2008) Curr. Opin. Nephrol. Hypertens. 17, 491–498 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Takata K. (2006) Cell. Mol. Biol. 52, 34–39 [PubMed] [Google Scholar]
- 16.Girsch S. J., Peracchia C. (1991) Curr. Eye Res. 10, 839–849 [DOI] [PubMed] [Google Scholar]
- 17.Lindsey Rose K. M., Gourdie R. G., Prescott A. R., Quinlan R. A., Crouch R. K., Schey K. L. (2006) Invest Ophthalmol. Vis. Sci. 47, 1562–1570 [DOI] [PubMed] [Google Scholar]
- 18.Rose K. M., Wang Z., Magrath G. N., Hazard E. S., Hildebrandt J. D., Schey K. L. (2008) Biochemistry 47, 339–347 [DOI] [PubMed] [Google Scholar]
- 19.Yu X. S., Jiang J. X. (2004) J. Cell Sci. 117, 871–880 [DOI] [PubMed] [Google Scholar]
- 20.Yu X. S., Yin X., Lafer E. M., Jiang J. X. (2005) J. Biol. Chem. 280, 22081–22090 [DOI] [PubMed] [Google Scholar]
- 21.Liu B. F., Liang J. J. (2008) J. Cell. Biochem. 104, 51–58 [DOI] [PubMed] [Google Scholar]
- 22.Fan J., Fariss R. N., Purkiss A. G., Slingsby C., Sandilands A., Quinlan R., Wistow G., Chepelinsky A. B. (2005) Mol. Vis. 11, 76–87 [PubMed] [Google Scholar]
- 23.Morey K. J., Ortega J. L., Sengupta-Gopalan C. (2002) Plant Physiol. 128, 182–193 [PMC free article] [PubMed] [Google Scholar]
- 24.Laemmli U. K. (1970) Nature 227, 680–685 [DOI] [PubMed] [Google Scholar]
- 25.Jensen O. N., Wilm M., Shevchenko A., Mann M. (1999) Methods Mol. Biol. 112, 513–530 [DOI] [PubMed] [Google Scholar]
- 26.Sedlak J., Lindsay R. H. (1968) Anal. Biochem. 25, 192–205 [DOI] [PubMed] [Google Scholar]
- 27.Obrdlik P., El-Bakkoury M., Hamacher T., Cappellaro C., Vilarino C., Fleischer C., Ellerbrok H., Kamuzinzi R., Ledent V., Blaudez D., Sanders D., Revuelta J. L., Boles E., André B., Frommer W. B. (2004) Proc. Natl. Acad. Sci. U.S.A. 101, 12242–12247 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Grefen C., Lalonde S., Obrdlik P. (2007) Curr. Protoc. Neurosci. 41, 5.27.1–5.27.41 [DOI] [PubMed] [Google Scholar]
- 29.Sambrook J., Russell D. W. (2001) in Molecular Cloning: A Laboratory Manual, pp. 13.19–13.25, Cold Spring Harbor Laboratory, Cold Spring Harbor, NY [Google Scholar]
- 30.Li J. F., Park E., von Arnim A. G., Nebenführ A. (2009) Plant Methods 5, 6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Li J. F., Nebenführ A. (2007) J. Biol. Chem. 282, 20593–20602 [DOI] [PubMed] [Google Scholar]
- 32.Minet R., Villie F., Marcollet M., Meynial-Denis D., Cynober L. (1997) Clinica. Chimica. Acta 268, 121–132 [DOI] [PubMed] [Google Scholar]
- 33.Gawronski J. D., Benson D. R. (2004) Anal. Biochem. 327, 114–118 [DOI] [PubMed] [Google Scholar]
- 34.Catalano C. M., Lane W. S., Sherrier D. J. (2004) Electrophoresis 25, 519–531 [DOI] [PubMed] [Google Scholar]
- 35.Fu D., Libson A., Miercke L. J., Weitzman C., Nollert P., Krucinski J., Stroud R. M. (2000) Science 290, 481–486 [DOI] [PubMed] [Google Scholar]
- 36.Sui H., Han B. G., Lee J. K., Walian P., Jap B. K. (2001) Nature 414, 872–878 [DOI] [PubMed] [Google Scholar]
- 37.Harries W. E., Akhavan D., Miercke L. J., Khademi S., Stroud R. M. (2004) Proc. Natl. Acad. Sci. U.S.A. 101, 14045–14050 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Törnroth-Horsefield S., Wang Y., Hedfalk K., Johanson U., Karlsson M., Tajkhorshid E., Neutze R., Kjellbom P. (2006) Nature 439, 688–694 [DOI] [PubMed] [Google Scholar]
- 39.Forde B. G., Day H. M., Turton J. F., Shen W. J., Cullimore J. V., Oliver J. E. (1989) Plant Cell 1, 391–401 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Miflin B. J., Habash D. Z. (2002) J. Exp. Bot. 53, 979–987 [DOI] [PubMed] [Google Scholar]
- 41.Bernard S. M., Habash D. Z. (2009) New Phytol. 182, 608–620 [DOI] [PubMed] [Google Scholar]
- 42.Gebhardt C., Oliver J. E., Forde B. G., Saarelainen R., Miflin B. J. (1986) EMBO J. 5, 1429–1435 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Temple S. J., Heard J., Ganter G., Dunn K., Sengupta-Gopalan C. (1995) Mol. Plant Microbe Interact 8, 218–227 [DOI] [PubMed] [Google Scholar]
- 44.Temple S. J., Kunjibettu S., Roche D., Sengupta-Gopalan C. (1996) Plant Physiol. 112, 1723–1733 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Stanford A. C., Larsen K., Barker D. G., Cullimore J. V. (1993) Plant Physiol. 103, 73–81 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Unno H., Uchida T., Sugawara H., Kurisu G., Sugiyama T., Yamaya T., Sakakibara H., Hase T., Kusunoki M. (2006) J. Biol. Chem. 281, 29287–29296 [DOI] [PubMed] [Google Scholar]
- 47.McParland R. H., Guevara J. G., Becker R. R., Evans H. J. (1976) Biochem. J. 153, 597–606 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Tyerman S. D., Whitehead L. F., Day D. A. (1995) Nature 378, 629–632 [Google Scholar]
- 49.Roberts D. M., Tyerman S. D. (2002) Plant Physiol. 128, 370–378 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Obermeyer G., Tyerman S. D. (2005) Plant Physiol. 139, 1015–1029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Udvardi M. K., Day D. A. (1989) Plant Physiol. 90, 982–987 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Udvardi M. K., Day D. A. (1990) Plant Physiol. 94, 71–76 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Britto D. T., Siddiqi M. Y., Glass A. D., Kronzucker H. J. (2001) Proc. Natl. Acad. Sci. U.S.A. 98, 4255–4258 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Streeter J. G. (1989) Plant Physiol. 90, 779–782 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Weaver C. D., Roberts D. M. (1992) Biochemistry 31, 8954–8959 [DOI] [PubMed] [Google Scholar]
- 56.Finnemann J., Schjoerring J. K. (2000) Plant J. 24, 171–181 [DOI] [PubMed] [Google Scholar]
- 57.Lima L., Seabra A., Melo P., Cullimore J., Carvalho H. (2006) J. Exp. Bot. 57, 2751–2761 [DOI] [PubMed] [Google Scholar]