Abstract
Plant fatty acids can be completely degraded within the peroxisomes. Fatty acid degradation plays a role in several plant processes including plant hormone synthesis and seed germination. Two multifunctional peroxisomal isozymes, MFP2 and AIM1, both with 2-trans-enoyl-CoA hydratase and l-3-hydroxyacyl-CoA dehydrogenase activities, function in mouse ear cress (Arabidopsis thaliana) peroxisomal β-oxidation, where fatty acids are degraded by the sequential removal of two carbon units. A deficiency in either of the two isozymes gives rise to a different phenotype; the biochemical and molecular background for these differences is not known. Structure determination of Arabidopsis MFP2 revealed that plant peroxisomal MFPs can be grouped into two families, as defined by a specific pattern of amino acid residues in the flexible loop of the acyl-binding pocket of the 2-trans-enoyl-CoA hydratase domain. This could explain the differences in substrate preferences and specific biological functions of the two isozymes. The in vitro substrate preference profiles illustrate that the Arabidopsis AIM1 hydratase has a preference for short chain acyl-CoAs compared with the Arabidopsis MFP2 hydratase. Remarkably, neither of the two was able to catabolize enoyl-CoA substrates longer than 14 carbon atoms efficiently, suggesting the existence of an uncharacterized long chain enoyl-CoA hydratase in Arabidopsis peroxisomes.
Keywords: β-Oxidation, Crystal Structure, Fatty Acid Metabolism, Fatty Acid Oxidation, Peroxisomes, Multifunctional Protein, Plant Hormones, Plant Lipids, Redox Regulation
Introduction
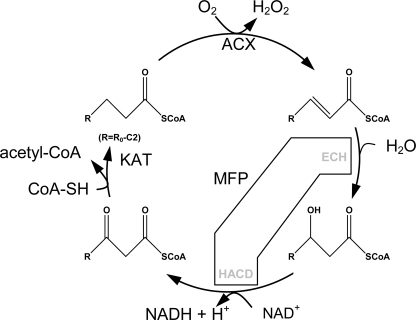
Fatty acids are degraded by the sequential removal of two carbon units in a process known as β-oxidation (Fig. 1). This process is ubiquitous and takes its name from the oxidation taking place at the carbon atom β to the carboxyl group. The discovery of a peroxisomal β-oxidation system was made in plants (1), and whereas peroxisomal β-oxidation in animals appears to be merely a fatty acid chain-shortening machine feeding mitochondrial β-oxidation, plant and fungal β-oxidation takes place almost entirely in the peroxisomes. The products of the reaction are H2O2, acetyl-CoA, reducing equivalents, and a variety of chain length-shortened acyl-containing molecules like the plant hormones jasmonic acid (2) and indole-3-acetic acid (3, 4).
FIGURE 1.
β-Oxidation reaction cycle. ACX oxidizes acyl-CoA to 2-enoyl-CoA generating H2O2 via reduction of FAD. MFP adds H2O over the newly formed double bond (ECH activity) and oxidizes the hydroxyl-acyl group to a ketoacyl (HACD activity). Finally, ketoacyl-CoA thiolase cleaves of a two-carbon unit in a reverse Claisen condensation reaction producing acetyl-CoA and leaving a shortened acyl-CoA ready for another chain-shortening reaction.
The conversion of storage triacylglycerols by β-oxidation provides metabolic energy and carbon skeletons for germination and early post-germinative seedling growth in oil seed plants (5) and metabolic energy and carbon for the production of hydrolytic enzymes in cereals (6). It is a salvage pathway for fatty acids during foliar senescence (7), supplies respiratory substrates to carbohydrate-deprived tissue (8), and at a lower level, is a constitutive property of all plant tissues most likely involved with membrane lipid turnover (7).
Three proteins (each present as several isozymes) that host a total of four enzyme activities constitute the core of peroxisomal β-oxidation. Acyl-CoA oxidase (ACX)6 oxidizes acyl-CoA to 2-trans-enoyl-CoA using FAD as co-enzyme. A multifunctional protein (MFP) adds water over the 2-trans-enoyl-CoA double bond and oxidizes the resultant l-3-hydroxyacyl-CoA using NAD+ to 3-keto-acyl-CoA. Finally, 3-keto-acyl-CoA thiolase (KAT) cleaves off acetyl-CoA thereby shortening the original acyl-CoA by two carbon atoms. Cucumis sativus (cucumber) MFPa (CsMFPa) possesses 2-trans-enoyl-CoA hydratase (ECH) and l-3-hydroxyacyl-CoA dehydrogenase (HACD) activities and, in addition, the d-3-hydroxyacyl-CoA epimerase and Δ3,Δ2-enoyl-CoA isomerase activities needed for oxidation of 4-cis-unsaturated fatty acids (9). Arabidopsis produces two peroxisomal MFPs, AIM1 (3) and MFP2 (10), which share 56 and 75% sequence identity, respectively, with CsMFPa over the entire length of the proteins. Both Arabidopsis thaliana AIM1 (AtAIM1) and AtMFP2 have 2-trans-enoyl-CoA hydratase and 3-hydroxyacyl-CoA dehydrogenase activity (3). Δ3,Δ2-Enoyl-CoA isomerase and d-3-hydroxyacyl-CoA epimerase activities have been inferred on the basis of high sequence identity to CsMFPa. Both AtAIM1 and AtMFP2 are expressed at approximately equal levels in roots, rosette leaves, flowers, and siliques (3). A complete block in β-oxidation by the introduction of an AtAIM1/AtMFP2 double mutation results in nonviable embryos aborting at an early stage of embryo development (11).
AtMFP2 is induced during germination and expressed primarily during post-germinative growth (10), whereas AtAIM1 is expressed predominantly in siliques, flowers, and seedlings older than 8 days (3). Mutations in the AtMFP2 gene give rise to a sucrose-dependent seedling phenotype (11), whereas AtAIM1 disruption causes abnormal inflorescence development resulting in low fertility (3). The different phenotypes reveal the diverse physiological roles of the two genes. In vivo and in vitro characterization of the chain length specificity of AtMFP2 suggests a long chain hydratase coupled to a short, medium, and long chain-specific dehydrogenase (11). AtAIM1 seems to have higher affinity for short chain acyl-CoA, although aim1 plants have elevated levels of C18:1 and C18:2 unsaturated fatty acids (3). The aim1 plants cannot convert 4-(2,4-dichlorophenoxy)butyric acid efficiently to the toxic indole-3-acetic acid analogue, 2,4-dichlorophenoxyacetic acid (3), and AIM1 activity is essential for wound-induced jasmonate production (2).
The molecular organization of the β-oxidation system differs between organisms and organelles. In the yeast lipolytica, the five ACX isoforms present form a heteropentameric complex and are imported into the peroxisomes as such (12). Bacterial and mammalian mitochondrial multifunctional complexes include ECH, HACD, and KAT activities (13, 14). Monofunctional enoyl-CoA isomerases/hydratases, HACD, and KAT, with a preference for shorter chain length substrates, are present in mammalian mitochondria in addition to the MFPs. In mammalian and plant peroxisomes, ACX and KAT activities reside on soluble monofunctional isozymes with different substrate specificities, whereas the ECH and HACD activities are catalyzed by MFPs (15, 16). In plants, MFPs are targeted for the peroxisomes only; no MFPs have been observed in mitochondria or as having mitochondrial targeting signals.
Channeling has been shown to occur between the active sites in bacterial and mammalian MFPs (17–19), but no tight binding protein-protein complexes of β-oxidation enzymes have been isolated from peroxisomes to date. By channeling, the intermediates need not equilibrate with the bulk solvent but are immediately directed from one active site to the next. In plants, an indication of specific isozyme functions by reduced jasmonate formation in the AtACX1/AtACX5 double mutant (20) and other evidence for specific involvement of AtACX1 and AtKAT2 in jasmonate synthesis (21–24) suggest that weak or transient protein-protein interactions and channeling could be mechanisms by which plants control the flow of metabolites through β-oxidation.
The structure of the full-length AtMFP2 reported here is the first structure to be determined for a full-length peroxisomal MFP. AtMFP2 is a 79-kDa protein composed of three domains and a linker helix. It forms a stable monomeric structure and resembles the mammalian peroxisomal MFE-1 in domain organization and size (25). The substrate specificity profiles published here elucidate significant differences between the hydratase domains. In AtAIM1 the hydratase substrate specificity peaks at crotonoyl-CoA extending up to 2-trans-tetradecenoyl-CoA, whereas AtMFP2 hosts a short to medium chain-length hydratase peaking around 2-trans-octenoyl-CoA and 2-trans-dodecenoyl-CoA. Surprisingly, the substrate specificity profiles of AtMFP2 and AtAIM1 reveal that neither of the two is capable of efficiently catabolizing straight chain substrates longer than 2-trans-tetradecenoyl-CoA. The structure of AtMFP2 is compared with mono- and multifunctional enzymes, providing insight into the catalytic mechanism and substrate-binding sites as well as the molecular basis for isozyme specificity.
EXPERIMENTAL PROCEDURES
Expression and Protein Purification
The AtMFP2 was recloned from the Riken database clone pda05150 (26) into a pET24b expression vector (NdeI and NotI restriction sites, Novagen) after PCR amplification using forward primer 5′-GCGGCGATTAATGGATTCACGAACCAAGGGGAAGACG-3′, reverse primer 5′-ATAGTTTAGCGGCCGCTTAGTGATGGTGATGGTGATGCAACCGTGAGCTGGC-3′ (AseI and NotI restriction sites are underlined; coding regions are in bold), and blunt-end ligation in pSTblue-1 (Novagen). Escherichia coli BL21(DE3) cells were transformed with the resultant vector, which codes for the full-length protein and a C-terminal His6 tag. The integrity of the recloned AtMFP2 was confirmed by DNA sequencing (Agowa). Gene expression was achieved by incubating the cells in shaking flask in 2×YT medium, 50 mg/ml kanamycin at 35 °C until a cell density (A600) of 0.6 was reached. The induction of gene expression was achieved by adding isopropyl 1-thio-β-d-galactopyranoside to a final concentration of 0.4 mm and persisted overnight. Cells (∼10 g of cell paste/liter of medium) were harvested by centrifugation. The pET24b-AtMFP2 plasmid was additionally transformed into the methionine auxotroph E. coli B834(DE3) strain (Novagen) for production of selenomethionine-substituted protein for phase determination. In this case the starter cultures were grown in Luria-Bertani-Broth at 35 °C over night. Cells from the starter cultures were harvested by centrifugation and resuspended in Milli-Q water prior to transfer to prewarmed SelenoMet Medium Base with SeleonoMet Nutrient Mix and 40 mg/liter selonomethionine added (Athena Enzyme Systems, Baltimore) plus 100 mg/liter kanamycin. Gene expression was induced by adding isopropyl 1-thio-β-d-galactopyranoside to a final concentration of 0.4 mm after the cell density had reached an A600 of 0.5. The cultures were allowed to produce protein overnight at 35 °C after which they were harvested by centrifugation. This procedure gave a yield of ∼5 g of cell paste/liter of medium. Native and selenomethionine-substituted protein batches were purified following the same purification scheme. Approximately 10 g of wet cell paste was resuspended in 30 ml of buffer A1 (20 mm imidazole, 500 mm NaCl, 30 mm phosphate buffer, pH 7.4), and cell membranes and DNA were disrupted by the addition of 180 mg of lysozyme (Sigma-Aldrich), 3 ml of 10× BugBuster (Novagen), and 1500 units of Benzonase (Sigma-Aldrich). The cell extract was cleared by centrifugation for 40 min at 18,000 × g and 4 °C, filtered through a 0.45-μm syringe filter (Millipore), and applied to a 5-ml HisTrap column (GE Healthcare) pre-equilibrated with buffer A1. The column was washed with a step gradient going from 20 mm to 35 mm and finally to 70 mm imidazole in 500 mm NaCl, 30 mm phosphate buffer, pH 7.4. Each gradient step lasted for 5 column volumes with a flow rate of 1 ml/min. Finally, the bound AtMFP2 was eluted with 500 mm imidazole, 500 mm NaCl, and 30 mm phosphate buffer, pH 7.4. The AtMFP2-containing fractions were pooled and concentrated in a Vivaspin20 30,000 MWCO filter device (Sartorius). The concentrated sample (2 ml) was filtered through a 0.22-μm syringe filter (Millipore) and applied to a Superdex 200 prep grade (26/60) gel filtration column (GE Healthcare) equilibrated and run with 20 mm Hepes, 150 mm NaCl, pH 7.5. The sample was collected at an elution volume of ∼235 ml. The fractionated AtMFP2 was pooled and concentrated to 10 mg/ml in a Vivaspin20 filter device before dialysis against 0.5 liters of 20 mm EDTA, 20 mm Hepes, pH 7.5, in a 10,000 MWCO dialysis tube (SpectraPor) followed by exhaustive dialysis against 20 mm Hepes, pH 7.5. The sample was mixed with 100% glycerol in a 1:1 volume ratio before storage at −20 °C.
Arabidopsis peroxisomal acyl-CoA oxidases, AtACX, were produced and purified for substrate preparation. AtACX1 and AtACX4 were prepared as described by Pedersen and Henriksen (27). N-terminally His6-tagged AtACX3 was expressed from a pET24b vector construct prepared from the pda08625 clone (Riken) in RosettaGami2 (DE3) cells (Novagen). The cells were cultured, and gene expression was induced as described for AtMFP2, except induction was done for 18 h at 14 °C. The cells were harvested by centrifugation and resuspended in buffer A2 (300 mm NaCl, 10 μm FAD, 30 mm phosphate buffer, pH 7.8), to which was added 2% buffer B2 (buffer A2 + 500 mm imidazole), 5% (w/w) lysozyme (Sigma-Aldrich), 100 units/g Benzonase (Sigma-Aldrich), and BugBuster (Novagen) (30 ml of buffer A2/10 g of wet cell paste). The cells were lyzed at room temperature for 20 min on a shaking platform. Cell debris and the insoluble fraction were removed by centrifugation, and the supernatant was fractionated on a 5-ml HisTrap column (GE Healthcare) equilibrated with 2% buffer B2 at 1 ml/min. The column was washed with 15% buffer B2 prior to elution of the bound AtACX3 with 50% buffer B2. The λ280 peak fractions were assayed by SDS-PAGE analysis; the AtACX3-containing fractions were pooled, and the buffer was exchanged for the buffer used for size exclusion chromatography (30 mm MOPS, 300 mm NaCl, 10 μm FAD, pH 7.0) in a Vivaspin20 filter device. The concentrated sample was fractionated on a Superdex 75 16/60 prep grade column (GE Healthcare) run at a flow rate of 0.2 ml/min. The peak fractions were pooled and glycerol added to a final concentration of 5% (v/v) to stabilize the protein and sampled by SDS-PAGE. Pure AtACX3 fractions were pooled and concentrated to 2 mg/ml in a Vivaspin20 filter device.
AtAIM1 was expressed in RosettaGami2 cells harboring a pET46 Ek/LIC (Novagen) construct encoding full-length N-terminally His6-tagged AtAIM1 prepared from clone pda02497 (Riken). The cells were treated as described for AtACX3, except that the induction temperature was 18 °C. The supernatant was fractionated like AtACX3, except buffer A2 was supplemented with 500 mm NaCl, pH 7.4, and it contained no FAD. The column was washed with 10% and eluted with 25% buffer B2. The buffer was exchanged for MonoS buffer (30 mm MES, 200 mm NaCl, pH 6.0) in a Vivaspin20 filter device, and the sample was applied to a MonoS 10/100 GL column (GE Healthcare) running at 5 ml/min. After the column was washed with MonoS buffer, fractionation was achieved using a 0–50% isocratic gradient over 30 column volumes. The buffer for the gradient was 30 mm MES, 1.8 m NaCl, pH 6.0. AtAIM1 eluted at 350 mm NaCl; it was concentrated to 10 mg/ml over the Vivaspin20 filter device and stored at −20 °C in MonoS buffer supplemented with 5% (v/v) glycerol.
Crystallization and Data Collection
AtMFP2 was dialyzed thoroughly against 20 mm Hepes, pH 7.5 (10,000 MWCO), to remove any glycerol prior to crystallization. NAD+ and acetoacetyl-CoA were added to a final concentration of 1 mm. Large, well diffracting crystals were obtained when the protein sample was mixed with mother liquor (4.2 m NaCHO2, 2% (v/v) glycerol) and 40% (v/v) polypropylene glycol in 2:2:0.4 μl micro-seeded drops at 20 °C. Crystals grew to a maximal size of 0.3 × 0.02 × 0.02 mm3 within 3 days after which they were transferred to a cryoprotective solution consisting of 4 m NaCHO2, 10% (v/v) glycerol and stored in N2(l). In the case of the selenomethionine-substituted protein, the mother liquor consisted of 3.9 m NaCHO2, 4% (v/v) glycerol, and the drop was made at a 2:2:0.5 ratio. Data were integrated with Mosflm (28) and scaled with Scala (29) (Table 1). Truncate (29) was used to generate amplitudes. Both data sets suffered from anisotropy; a diffraction anisotropy server (30) was employed to correct for the anisotropy at the model building stage but did not improve the map quality, so anisotropy correction was not incorporated in the refinement.
TABLE 1.
AtMFP2 data collection and refinement statistics
| Statistics | Data set |
|
|---|---|---|
| Native | SeMet peak | |
| Data collection | ||
| Collection site | 14.1 BESSY, Germany | 14.1 BESSY, Germany |
| Wavelength (Å) | 0.95373 | 0.9797 |
| Space group | P3221 | P3221 |
| Cell dimensions | a = b = 110.5 Å | a = b = 112.2 Å |
| c = 125.5 Å | c = 125.0 Å | |
| Resolution (Å) | 25.4-2.50 (2.61-2.50)a | 26.9-2.70 (2.85-2.70 Å)a |
| Rmerge (%) | 7.1 (48.7)a | 8.3 (41.5)a |
| Rpimb (%) | 5.7 (40.5)a | 5.6 (25.4)a |
| Mean (I)/S.D. (I) | 12.2 (1.8)a | 14.6 (1.8)a |
| Completeness (%) | 94.9 (67.5)a | 99.4 (100)a |
| No. of unique reflections | 30,461 (4860)a | 25,324 (3674)a |
| Multiplicity | 3.5 (3.1)a | 4.2 (4.3)a |
| Anomalous completeness (%) | 83.3 (83.9)a | |
| Wilson Bc (Å2) | 51 | 77 |
| Refinement and quality | ||
| No. of non-hydrogen protein atoms | 4,999 | |
| No. of water molecules | 25 | |
| Rworkd (%) | 22 (33)e | |
| Rfreed (%) | 27 (40)e | |
| S.D.d | ||
| Bond angles (°) | 0.581 | |
| Bond lengths (Å) | 0.002 | |
| Mean Bd | ||
| Main chain (Å2) | 77.1 | |
| Side chains (Å2) | 79.8 | |
| Solvent (Å2) | 56.6 | |
| Ramachandran plotf | ||
| Most favored (%) | 89.0 | |
| Additionally allowed (%) | 10.6 | |
| Generously allowed (%) | 0.2 | |
| Disallowed (%) | 0.2 | |
Phase Determination and Refinement
Nineteen selenium sites of the possible 22 were identified with the AutoSol procedure from PHENIX (31) with a partial model of the Pseudomonas fragi (Pf) MFP (individual domains from the poly(Ala) model, Protein Data Bank code 1WDK) (13) from a previous molecular replacement run in PHASER (32) as input for AutoSol SAD selenium-site identification. Phase extension using Resolve (31) produced a partially interpretable electron density map, which was subjected to automated model building with the phenix.autobuild module (33) using keyworded scripts. This resulted in a 74% complete structural model (Rfac/Rfree = 34/37%), which was further refined and optimized using phenix.refine and Coot (34). Structure validation was done using Coot and MolProbity (35). Side chain atoms with poor real space correlation have been omitted from the model. As a consequence of the high overall B-factor, only a very limited number (25 solvent molecules) of solvent molecules could be identified. Neither co-factor nor substrate-like molecules were identified in the electron density despite the presence of NAD+ and acetoacetyl-CoA in the crystallization conditions. The final AtMFP2 model covers amino acid residues 7–719, but three loops had poor or very disordered density and were omitted from the model. Those loops are 71–82 (ECH domain), 365–368 (HACDN domain), and 576–596 (HACDC domain). In addition, amino acid residues 597–623 have poorly defined side chain electron density, and most of the side chain atoms in this region were omitted from the model. Main chain regions 68–87, 236–242, 342–385, 537–621, and 692–695 have B-factors larger than 100 Å2. Pro26-Pro27 and Ser452-Pro453 are cis-peptides (supplemental Fig. S4 online). Final refinement statistics are shown in Table 1.
Enzyme Activity
AtMFP2 and AtAIM1 2-trans-enoyl-CoA hydratase activity was determined using in situ synthesized substrates from C4- to C18-CoA. Aliquots of 50 μm saturated acyl-CoA esters were oxidized with a mixture consisting of AtACX1, AtACX3, and AtACX4 at 50 nm each in 175 mm Tris-HCl, pH 8.5, 2.5% (w/v) polyethylene glycol 400, and 40 pm catalase from bovine liver (Sigma-Aldrich). AtACX2 was not included because the AtACX1 substrate profile covers the C16- and C18-CoAs (Ref. 36 and this study). The reactions were incubated in a FluorSTAR Optima plate reader (BMG Labtech GmbH, Offenburg, Germany) at 27 °C, and the oxidation was monitored by recording the increase in absorbance at 260 nm due to the resonance between the introduced β-double bond and the CoA-thioester bond (37). Upon maximum conversion the absorbance had increased ∼40% as expected from the ratio between the ϵ260 of saturated acyl-CoA and crotonyl-CoA (the ϵ260 of saturated and 3-hydroxy-acyl-CoA used was 16,400 and 22,600/m cm, respectively, for the 2-trans-enoyl-CoA substrates (38)). The hydratase assay was started by dispensing equal volumes of buffer, AtAIM1, or AtMFP2 with a multichannel pipette diluting the acyl-CoA substrates present by 10%. The final concentration of AtAIM1 or AtMFP2 in the reactions was 0.5 nm, and hydration was monitored by recording the decline in absorbance at 260 nm in the FluoSTAR in kinetics mode. To ensure maximal conversion for the dehydrogenase assay, the MFP concentration was adjusted to 5.5 nm. After full conversion, the dehydrogenase reaction was started by the addition of saturating amounts of NAD+ to a final concentration of 1 mm. The plate was read at 340 nm in kinetics mode in a SpetraMAX 340PC 384 plate reader (Molecular Devices) at 27 °C. Experiments with 25 μm substrate were conducted under conditions similar to those described above. To examine the effect of Tween 20 and delipidated bovine serum albumin (BSA; fraction V, 96% purity, Sigma), dehydrogenase assays were set up with C16-CoA and AtMFP2 as described above but titrated with 0–4.9% (w/v) Tween 20 or 0–22 μm delipidated BSA. The concentration of MFP in the Tween 20 assay was 1 nm MFP, and it was 0.5 nm MFP in the BSA assay adjusted to 75 nm MFP in the C16- and C18-CoA dehydrogenase reactions. BSA was delipidated by incubation in 90% methanol at 4 °C overnight. The protein was sedimented by centrifugation at 20,000 × g and washed twice before residual methanol was removed under vacuum.
Structure Analysis
The Coot program (34) was used for structure evaluation. PyMOL was used to prepare Figs. 2, 3, and 7. Domain interfaces and packing was evaluated using the PISA (39), Protorp (40), Prism (41), and Hotsprint (Protein Data Bank code 1WDK) (42) servers. Secondary structure matching (SSM) superpositions were carried out in Coot. Sequence alignment including structural information was done in STRAP (43). Sequence identities were calculated by the FFAS (fold and function assignment system) server (44), consensus motifs were analyzed using WebLogo (45), and alignments and phylogenetic tree analysis were calculated by ClustalW (46) and depicted by TreeView (47). Protein surface topography was calculated using MOE (Chemical Computing Group).
FIGURE 2.
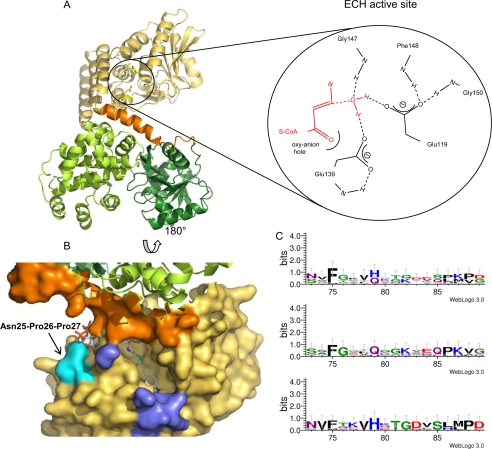
The AtMFP2 ECH-domain. A, the ECH domain is yellow, the linker to the HACD domains orange, the N-terminal HACD domain green, and the C-terminal HACD domain light green. The position of the ECH active site is indicated by a sketch of the hydrogen bonds to the active site Glu amino acid residues (see inset). The red molecules in the sketch are hypothetical and based on the active site of monofunctional RnECH (59). B, solvent-accessible surface of the AtMFP2-ECH acyl-binding pocket rotated 180° around a horizontal axis. The 4-(N,N-dimethylamino)cinnamoyl-CoA substrate from a superposition of RnECH and the AtMFP-ECH domain is included. The N- and C-terminal parts of the flexible loop (Ser71–Tyr88) are colored blue, and the cis-Pro27 is cyan. C, mapping of consensus sequence to the ECH flexible loop region including all of the plant MFP sequences (top) from supplemental Fig. S2 online, the AtMFP2-like sequences (middle), and the AtAIM1-like sequences (bottom).
FIGURE 3.
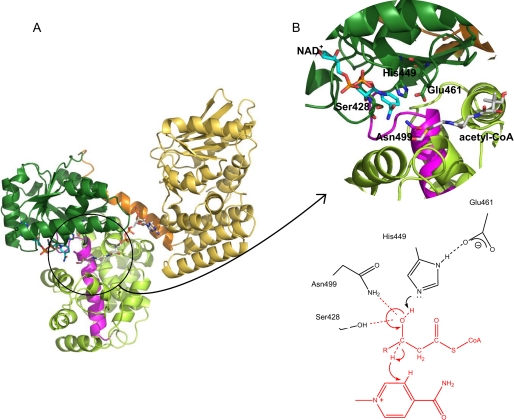
The AtMFP2-HACD domain. A, overall structure of AtMFP2. The NAD+ co-factor (cyan) and an acetyl-CoA molecule (white) from the superimposed structure of PfMFP (Protein Data Bank code 1WDM) are included. B, close-up view of the HACD active site and the conserved active site residues (Ser428, His449, Glu461, and Asn499). The 3-hydroxyacyl-CoA dehydrogenase signature is indicated by the magenta color of the backbone trace. The proposed reaction mechanism of AtMFP2 is based on the present crystal structure and the reaction mechanism and structures of human HACD NAD+ and its substrate analogue complexes (56, 70).
FIGURE 7.
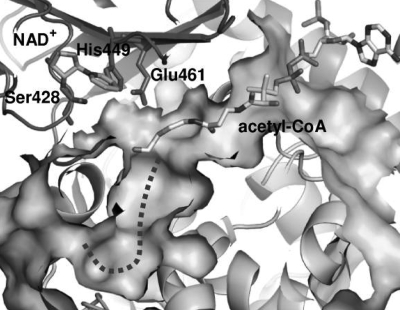
A slice of the solvent-exposed surface of the AtMFP2-HACD active site. The acetoacetyl-CoA and NAD+ included are superimposed molecules from the structure of PfMFP (13). An unoccupied pocket marked with a dotted line is observed extending from the 3-hydroxyl end of the substrate.
RESULTS
Overall Structure
AtMFP2 is a 725-amino acid residue enzyme organized in two compact structural entities separated by an α-helical linker. The two AtMFP2 entities correspond to the two regions in the sequence to which ECH and HACD activities have been mapped (9) (Figs. 1 and 2 and supplemental Fig. S1 online). The interaction area between the N-terminal ECH domain (not including the linker region) and C-terminal HACD entity is only 152 Å2, whereas the helical linker has a 420 Å2 interface with the ECH domain and an 817 Å2 interface with the HACD entity, showing that the helical linker is absolutely essential for the spatial arrangement of the multifunctional protein complex (Fig. 2, orange). The HACD entity can be divided further into two domains (HACDN and HACDC) with substantial interdomain interaction (730 Å2). The HACD domains interface is dominated by hydrophobic contacts (55% nonpolar residues in the interface) but has a relatively loose interface fit (gap volume index, 3.9) (48).
ECH Domain
The ECH domain is a crotonase-like fold with a crotonase fold core of four turns of β-β-α structure extended by a large, mostly α-helical C terminus. Ten β-strands form two mixed β-sheets lying almost perpendicular to one another. On one side, the larger sheet is packed against a single α-helix, whereas it is stacked with a three-layered arrangement of α-helices on the other side of the sheet. The crotonase-like family also includes the ECH domain of the PfMFP complex (13), the monofunctional hexameric or trimeric enoyl-CoA hydratases (49) and enoyl-CoA isomerases (50, 51), the 6-oxo camphor hydrolases (52), the monofunctional hexameric 4-chlorobenzoyl-CoA dehalogenases (53), and the monofunctional monomeric methylmalonyl CoA decarboxylase from E. coli (54). They all share the four turns of β-β-α structure plus the following α-β-α-β structural motif called the T1 domain. It was originally characterized as a motif involved in trimerization of monofunctional proteins. A final common α-helical extension, HE, connects the T1 motif with a trimerization module in monofunctional proteins and represents the α-helical domain linker in AtMFP2 and PfMFP.
Crotonase-like enzymes catalyze diverse reactions but share the use of a CoA-thioester substrate and the formation of a carbanion species as part of their reaction scheme. The enzymes have a low overall sequence identity but have been shown to contain a common sequence pattern, the enoyl-CoA hydratase/isomerase signature, 106(LIVM)-(STAG)-X-(LIVM)-(DENQRHSTA)-G-X3-(AG)3-X4-(LIVMST)-X-(CSTA)-(DQHP)-(LIVMFYA)126 (ProSite entry PS00166). (The AtMFP2 sequence is indicated by bold letters.) The signature makes up the third β-β-α section and is part of the interface binding the pantetheine, thioester, and acyl groups of the substrate but not the nucleotide part of CoA. Along with the helical linker, the small β-sheet embodies the CoA-binding site in MFP-ECHs (Fig. 2), whereas in monofunctional enzymes the last α-helix making up the CoA-binding site originates from a neighboring subunit.
The SSM superposition (55) of AtMFP2-ECH/monofunctional mitochondrial rat, Rattus norvegicus, RnECH (Protein Data Bank code 1MJ3), and AtMFP2-ECH/PfMFP-ECH gives core r.m.s.d. values of 1.5 and 1.7 Å, respectively, including AtMFP2 amino acid residues 8–183 in the former case and 8–244 in the latter (amino acid residues numbers will refer to AtMFP2 in the following discussion unless otherwise specified).
HACD Domains
The two HACD domains form a globular entity with the supposed active site residing at the bottom of a cleft (Fig. 3 and supplemental Fig. S1 online). The N-terminal domain (residues 308–496) is 6-phosphogluconate dehydrogenase-like with eight strands in the central β-sheet flanked by α-helices. It has a Rossmann fold NAD binding motif, but no co-factor is bound in the crystal structure. The structure of the N-terminal AtMFP2-HACD domain is similar to the structures of the HACD domains found in PfMFP, peroxisomal RnMFE-1 (15), and human mitochondrial HACD (56) with SSM superposition r.m.s.d. values of 1.1, 1.6, and 1.1 Å, respectively, by superposition of residues 308–496. The C-terminal HACD domain (residues 497–719) is a dimer of 5-helix motifs belonging to the hydroxyacyl-CoA dehydrogenase-like protein family. The first helical motif (residues 497–575) is connected to the other (residues 624–719) by a long α-helix. Also the C-terminal HACD domains of MFPs are very similar (e.g. the SSM r.m.s.d. to PfMFP is 1.6 Å). Monofunctional mitochondrial dehydrogenases contain only one 5-helix motif, but the functional dimer of these enzymes creates an entity very similar to the C-terminal HACD domain (e.g. the SSM r.m.s.d. to human heart HACD (56) is 2.2 Å when including 177 of 222 amino acid residues). In AtMFP2, the first α-helices of each 5-helix motif run antiparallel and create an interface corresponding to the dimerization interface in monofunctional HACDs. Salt bridges between Arg500 Nϵ and Glu638 Oϵ2 and between Arg500 Nη1/η2 and Glu645 Oϵ1/ϵ2 are conserved in the otherwise hydrophobic interface. The HACD signature, 492(DNES)-X2-(GA)-F-(LIVMFYA)-X-(NT)-R-X 3-(PA)-(LIVMFY)-(LIVMFYST)-X5,6-(LIVMFYCT)-(LIVMFYEAH)-X2-(GVE)516 (ProSite entry PS00067 (57)), is located at the interface between the N- and C-terminal HACD domains and in between the two halves of the C-terminal domain (Fig. 3A). It carries the conserved active site residue Asn499. One highly conserved amino acid residue, Asp523, is engaged in hydrogen bonds important for the structural integrity of the C-terminal domain. The Asp523 carboxylate accepts hydrogen bonds tying the second and third α-helix together. The side chain of another highly conserved residue, Asp540, is disordered in the current structure.
Plant MFPs
There is still not a vast amount of GenBankTM data available for plant MFPs, but two subfamilies of peroxisomal MFPs are emerging (supplemental Fig. S2 online). Although monocots seem to have more MFP isozymes than dicots, the isozymes appear to fall within either an AtAIM1- or AtMFP2-like subgroup. A single barley isozyme (GenBankTM GI: 151418576) identified in expressed sequence tags from inflorescence, developing caryopsis, apex, and germinating seeds could represent a third subfamily of plant MFPs, but the physiological significance of this gene product is unknown. The MFP2-like subfamily is characterized by a large proportion of Gly residues (∼15%) in the ECH flexible loop and only few conserved residues (Fig. 2C). The AtAIM1-like subfamily is characterized by an ECH-flexible loop having only one Gly residue and high sequence conservation (Fig. 2C).
Substrate Specificity
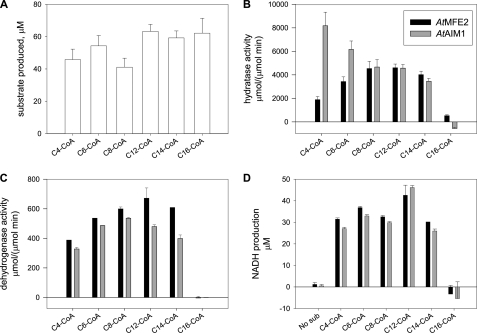
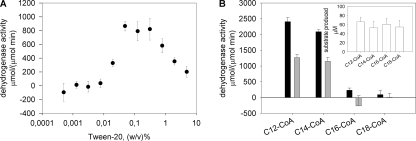
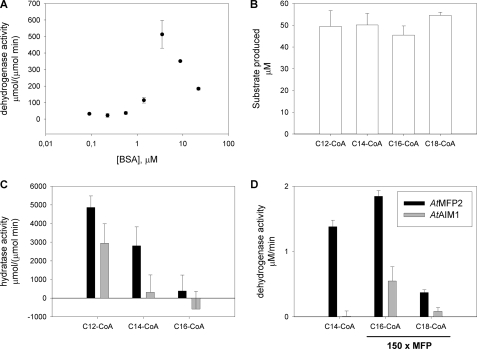
The substrate specificities of AtMFP2 and AtAIM1 were determined in vitro using recombinant acyl-CoA oxidases to synthesize the 2-trans-enoyl-CoA substrates immediately before sampling with AtMFP2 and AtAIM1. The oxidase reaction was monitored by UV absorbance spectroscopy at 260 nm confirming equal amounts of freshly prepared enoyl-CoA substrates to be available in the assays (Fig. 4A). Both AtMFP2 and AtAIM1 have short to long chain (C4–C14) acyl-CoA ECH activity, but the AtAIM1 substrate profile is shifted toward the shorter substrates peaking at C4-CoA compared with AtMFP2 peaking around C8- and C12-CoA (Fig. 4B). The dehydrogenase substrate profile of AtMFP2 is slightly skewed toward the longer chain lengths compared with its hydratase profile and is about 10 times slower (Fig. 4C). The AtAIM1 dehydrogenase substrate profile lacks its C4 hydratase peak and peaks at C8-CoA instead of the C12-CoA of AtMFP2. The total production of NADH with C16-CoA after 1 h of incubation at 27 °C and 10-fold more MFP remains indistinguishable from zero (Fig. 4D). Incubation with 25 instead of 50 μm C16- or C18-CoA did not change this (data not shown). In the presence of Tween 20, some dehydrogenase activity is detected with C16-CoA peaking at around 0.1% (w/v) Tween 20 (Fig. 5A). However, the activity of the MFPs with C14-CoA is equally affected, maintaining the picture of negligible activity with substrates longer than C14-CoA of either AtAIM1 or AtMFP2. Delipidated BSA also affects the activity of AtMFP2 with C16-CoA up to around 6 μm above which the positive effect declines (Fig. 6A). With the preparation of substrates unaffected (Fig. 6B), the presence of 6 μm BSA in the reactions adversely affected the hydratase reaction of AtAIM1 (compared with Fig. 4B), whereas the activity with C16-CoA was below the increased noise level of the assay due to the presence of BSA absorbing in the UV range. With 150 times more MFP present in the C16- and C18-CoA wells than in the C14-CoA wells, AtMFP2 dehydrogenase activity with C18-CoA substrate is detected (Fig. 6D). Under these conditions, the activity of 0.5 nm AtMFP2 is 1.4 ± 0.1 μm/min with C14-CoA, whereas with C16- and C18-CoA it is 1.8 ± 0.1 and 0.4 ± 0.04 μm/min, respectively, for 75 nm AtMFP2. For 75 nm AtAIM1 the activity with C16-CoA is 0.5 ± 0.2 μm/min. Thus, in the presence of BSA the activity of AtMFP2 with C16-CoA is 116 times lower than with C14-CoA.
FIGURE 4.
Hydratase and dehydrogenase activity of recombinant AtMFP2 and AtAIM1. Neither AtMFP2 nor AtAIM1 efficiently degrade enoyl chains longer than C14-CoA. A, in situ 2-trans-enoyl-CoA substrate synthesis with a mixture of acyl-CoA oxidases were monitored at A260. B, upon full conversion 0.5 nm MFP2 (black), AIM1 (gray), or buffer were added and the hydratase activity determined by recording A260 decrease at 27 °C. After the initial determination, additional 5 nm MFP was added to the reactions to secure full conversion. C, after the reactions had run to completion, 1 mm NAD+ was added to each reaction, and the dehydrogenase activity was determined by recording the A340 increase at 27 °C. D, after 1 h at 27 °C total production of NADH was determined by recording the A340. No C16-CoA substrate was fed to this reaction from the hydratase reaction in B. Each reaction consisted of about 50 μm acyl-CoA substrate, AtACX1, AtACX3 and AtACX4 each at 150 nm, 175 mm Tris-HCl, pH 8.5, 2.5% (w/v) polyethylene glycol 400, and 40 pm catalase.
FIGURE 5.
Effect of Tween 20 on MFP activity and substrate profile. A, AtMFP2 dehydrogenase activity was assayed with increasing concentrations of Tween 20. B, the dehydrogenase activities of AtMFP2 (black) and AtAIM1 (gray) with C14-CoA and C16-CoA in the presence of 0.1% (w/v) Tween 20 were determined. The insert illustrates 2-trans-enoyl-CoA production of each substrate. Conditions in both assays were the same as in Fig. 4.
FIGURE 6.
BSA effect on MFP activity and substrate profile. A, AtMFP2 was titrated with BSA, and dehydrogenase activity was determined. B, substrate was prepared in the presence of 6 μm delipidated BSA, and hydratase activity was subsequently determined, C. D, dehydrogenase activity was determined with 150 times more MFP in the C16- and C18-CoA wells compared with 0.5 nm in the C14-CoA well. The data are not normalized to μm enzyme to show residual long chain dehydrogenase activity at high enzyme concentrations. Conditions otherwise are as described for Fig. 4.
DISCUSSION
Hydratase Activity
AtMFP-ECH catalyzes the syn addition of water across the double bond of an α,β-unsaturated thioester converting 2-trans-enoyl-CoA to l-3-hydroxyacyl-CoA (58). Whether the addition of water proceeds via a concerted reaction or a stepwise mechanism is not clear (59, 60). AtMFP-ECH can also catalyze the isomerization of β,γ-unsaturated CoA thioesters to form the α,β-unsaturated thioesters for further β-oxidation (9). The homologous RnECH also catalyzes both hydration and isomerization reactions (61).
Two active site glutamic acids have been proposed to act in catalysis (62, 63), and an oxyanion hole has been proposed to polarize the thioester carbonyl group. The N-terminal Glu activates a water molecule for nucleophilic attack at the C3 position (hereafter called GluN (Glu119 in AtMFP2)). The C-terminal Glu acts as a proton or, alternatively, as a hydrogen bond acceptor (hereafter called GluC (Glu139 in AtMFP2); Fig. 2 and supplemental Fig. S3 online). An active site water molecule hydrogen bonded to GluC and Gly147 is found at a position from where it could execute a nucleophilic attack on the enoyl-CoA substrate. The hydrogen bonding network used to deduce a reaction mechanism for RnECH is only partly conserved in AtMFP-ECH (Fig. 2A). The Gln162(rat) residue observed to donate hydrogen bonds to the active site GluC in the rat enzyme is replaced by a hydrophobic residue in many crotonase-like enzymes including both AtMFP2 and PfMFP. The hydrogen bonds from Gln to GluC provide an argument for a negatively charged GluC in the reaction mechanism (59). The occurrence of a more hydrophobic GluC environment would be expected to raise the pKa of the active site GluC and, consequently, favor the concerted reaction mechanism over a stepwise mechanism, but it could also indicate an adaption to plant peroxisomal pH.
The development of a partial positive charge on the thioester carbonyl during the reaction could be stabilized by the oxyanion hole created by hydrogen bonds donated by the amide nitrogen atoms of Phe68 and Gly116. Gly116 is positioned like the backbone nitrogen atom with a similar function in monofunctional RnECH, methylmelanyl-CoA decarboxylase (54), and 4-chlorobenzoyl-CoA dehalogenase (53), whereas Phe68 is somewhat misplaced due to the disorder of the 71–82 loop and high temperature factors of the 68–87 region.
The acyl-binding pocket appears wide open in AtMFP2 because of the disorder of the 71–82 loop (Fig. 2B and supplemental Fig. S3A online). The corresponding loop is observed to be disordered or flexible in all other crystal structures of crotonase-fold enzymes as well. The flexibility could reflect an ability to adapt the acyl-binding pocket structure to the substrate in accordance with the relatively broad substrate specificity profiles of the enzymes with regards to acyl chain length and the presence of conjugated systems. The part of the acyl-binding pocket that is included in the structure of AtMFP2-ECH is dominated by hydrophobic residues (Phe33, Phe68, Ile89, Val144, Ile145, Gly147, Phe148, Ala274, and Val278) with a few polar atoms/charged residues facing the expected ω-end of the pocket (Glu83, Ala86 O, Ser90 N, Gly147 O, Ala271 O, and Ser295 OH).
An MFP channeling mechanism, with the nucleotide being the pivot shaft for transfer of the acyl chain from ECH to the HACD active site, has been suggested in bacteria (13). If channeling is indeed a common phenomenon among the MFPs, the proposed nucleotide environment of the MFPs is expected to be conserved and would likely include an extended conformation of the acyl-CoA in order to bridge the 40 Å span from the ECH to the HACD active site. The linker and additionally the AtMFP2-ECH motif, -Asn25-Pro26-Pro27- (Fig. 2B), conserved among peroxisomal MFPs and some bacterial MFPs, would interfere with the binding of acyl-CoA in the bent conformation observed in monofunctional ECHs (Fig. 2B). Pro26-Pro27 carries one of the two cis-prolines found in the AtMFP2 structure. There is no crystal structure of an acyl-CoA·MFP-ECH complex. PfMFP was crystallized in the presence of acetoacetyl-CoA and pentaethylene glycol n-octyl ether (C8E5) (13), and the C8E5 molecules were bound in the ECH/HACD domains. The adenine-binding pocket as defined by the PfMFP-HACD·acyl-CoA complex (Protein Data Bank code 1WDM; acetoacetyl-CoA B factor, 160 Å2) (64) consists of residues that are not strictly conserved between AtMFP2 and PfMFP. This pocket resides at the ECH/HACD interface. The three-dimensional shape of this interface is highly conserved however, and the hydrophobicity patches and potential for hydrogen bonds to the adenine moiety persists. A highly conserved MFP pattern of Arg/Lys amino acid residues in the helical linker (290FFSQRGTAKVP300; bold residues are highly conserved) and the α-helix from the HACD domain facing the domain interface (481KKIKK485) could support the existence of a common ECH/HACD adenine-binding site, with positively charged amino acid residues supporting the relocation of the pyrophosphate in the region proposed by Ishikawa et al. (13) as the center of rotation, but supportive mutagenesis, structural, or biophysical evidence is not available yet.
Dehydrogenase Activity
AtMFP2-HACD catalyzes the oxidation of the l-3-hydroxyacyl-CoA hydroxyl group to a keto group while reducing NAD+ to NADH. For mitochondrial short chain HACDs, the reaction takes place by hydride transfer at the si face of the nicotinamide ring (65). The NAD+-binding site, defined by the presence of a 319Gly-X-Gly321-X-X-G324 phosphate binding motif (66), is located in the first β-α transition in the N-terminal AtMFP2-HACD domain adjacent to the active site Ser428, His449, Glu461, and Asn499 logo (Fig. 3A). Amino acid residues observed to hydrogen bond to the co-factor in monofunctional HACDs (67, 68) and PfMFP are conserved and have similar conformations in AtMFP2-HACD (Glu342, Glu401, Lys406, and Asn624), but no significant electron density is observed in the AtMFP2 co-factor-binding site. A similar absence of positive electron density for the co-factor is observed in the crystal structure of the truncated recombinant RnMFE-1-HACD (15), whereas NAD+ is present in the active site of the PfMFP β-oxidation complex but with an average B-factor of 98 Å2 (13). As evidenced by the enzyme activity characterization included in this study, the AtMFP2 used for crystallization experiments is an active enzyme transferring electrons to NAD+. Interestingly, at position 347 in AtMFP2 and AtAIM1, a relatively well conserved Gly is replaced by a bulky Phe colliding with the expected position of a ribose hydroxyl group of NAD+ and therefore likely impeding NAD+ binding and lowering the affinity of AtAIM1 and AtMFP2 for NAD+. The NAD+-binding site, as defined by the structure of PfMFP, is included in Fig. 3 by superposition with AtMFP2. Also included is the superimposed acetyl-CoA from the same PfMFP structure. The conserved residues His449 and Glu461 have been identified as being important for dehydrogenase activity by site-directed mutagenesis of E. coli MFP, and a reaction mechanism has been proposed and later modified (56, 69, 70). The reaction mechanism implies that the hydrogen bond between Glu461 and His449 would orientate His449 for optimal proton subtraction from the substrates hydroxyl group. Ser428 and Asn499 side chains could stabilize the developing negative charge on O3 of the substrate (Fig. 3B). A conserved cis-peptide bond (Ser452–Pro453) adjacent to the active site His449 appears essential for orienting the substrate with backbone hydrogen bonds to O1 and the mercaptoethylamino group.
The relative orientation of the N- and C-terminal domains of AtMFP2-HACD results in the formation of a relatively open Y-shaped NAD+ and acyl binding cleft between the three domains, thereby exhibiting a relatively solvent-exposed binding site not designed for substrate discrimination based on acyl chain length or conjugation of double bonds. The N- and C-terminal domains of human monofunctional HACD have been shown to undergo a small rearrangement upon the binding of substrate and a larger rearrangement upon the formation of the ternary complex with NAD+. The large rearrangement sequesters the active site from solvent and brings the active site residues into optimal position for catalysis to occur (56). The conserved active site residues of AtMFP2-HACD seem too far apart to facilitate the electron/hydride transfer depicted in Fig. 3B. The His449 Nϵ to Asn499 Nδ2 distance is 4.3 versus 3.7 Å in the ternary human HACD complex. The structures of the bacterial PfMFP represent an ensemble of slightly different domain packing (64), suggesting that the HACD subdomains are likely to move upon substrate binding. The domain movements inferred to occur upon substrate/co-factor binding in PfMFP affects only the ECH/HACDN interface and not the HACDN/HACDC interface. This is not consistent with the HACDN/HACDC domain closure upon NAD+/substrate binding observed in monofunctional human HACD and suggests that either the monofunctional enzymes and MFP have different mechanisms of action or that not all functional states of PfMFP are covered by the current crystal structures.
Substrate Specificity
Our data clearly characterize AtAIM1 as a short to medium chain length-specific hydratase coupled to a broad chain length-specific dehydrogenase, whereas AtMFP2 is characterized as a medium chain length-specific hydratase coupled to a broad chain length-specific dehydrogenase (Fig. 4, B–D). Because of the difficulties in obtaining 2-trans-enoyl-CoA substrates, few studies have addressed the actual substrate specificities of AtMFP2 and AtAIM1. However, our data showing the in vitro substrate profiles of AtMFP2 and AtAIM1 for short chain substrates are in accord with the comments on substrate preference made by Richmond and Bleecker (3) and the observation that MFP2 mutants have unaltered crotonyl-CoA hydratase activity levels (11). However, at the other end of the profile, neither AtMFP2 nor AtAIM1 has significant activity with enoyl-CoA substrates longer than C14 (tested with 2-trans-enoyl-C16- and -C18-CoA). Considering the abundance of C18–C22 fatty acids in Arabidopsis seeds, with eicosenoic acid (C20:1) being the predominant seed-specific fatty acid (73), this is remarkable.
To address potential solubility or micelle formation issues as the reason for the lack of activity with longer chain length acyl-CoA substrates, dehydrogenase activity was assayed at 25 μm substrate or in the presence of either Tween 20 or delipidated BSA. Changing the substrate concentration alone did not result in detectable activity, whereas both Tween 20 and delipidated BSA affected the activities of AtMFP2 in a complex manner with a concentration threshold for a positive effect to occur followed by a peak and a gradual decline of effect (Figs. 5A and 6A). The activity enhancing effect may be due to solubilization of the substrates by the detergent or lipid-binding BSA. It is, however, noticeable that the effect was the same for long and medium chain substrates. It is also possible that it represents a direct effect on the dynamics of the enzymes. The decline of the positive effect in the case of the detergent is likely because of denaturation of the enzymes, whereas the declining effect of BSA is possibly because of competitive binding of the acyl-CoA substrates. In the presence of either 0.1% (w/v) Tween 20 or 6 μm BSA, corresponding to the concentrations at maximum activity enhancement with C16-CoA, the activity with C16-CoA and C18-CoA remains negligible relative to the activity with C14-CoA (Figs. 5B and 6C). The addition of excess MFP to the C16- and C18-CoA reactions clearly illustrates that AtMFP2 activity with these substrates is significantly (about 116 and 560 times) lower than with C14-CoA (Fig. 6D). It thus seems unlikely that the lack of activity with substrates longer than C14-CoA is due to solubility issues.
MFPs limited to short to medium chain hydratase activities are known to exist in cucumber peroxisomes and rat liver mitochondria, but they are usually supplemented with isozymes harboring long chain activity as well (14, 71). The existence of additional hydratases has been identified and inferred in Arabidopsis peroxisomes (11, 72), but none of these has been shown or suggested to be specific for long chain substrates. Our data suggest that an additional long chain hydratase is active in Arabidopsis peroxisomes.
Relying on the endogenous ACXs present in the seedlings, Rylott et al. (11) addressed the substrate specificity of AtMFP2 in vivo by feeding crude seedling homogenates with substrates of various chain lengths and detecting the resultant acyl-CoA products by mass spectrometry. Interestingly, they conclude that AtMFP2 is a long chain-specific hydratase coupled to a broad range dehydrogenase. However, when feeding with C18-CoA, 3-hydroxy-octadecanoyl-CoA accumulates at wild-type concentrations in the MFP2 mutant, suggesting that wild-type 2-trans-octadecenoyl-CoA hydratase activity remains in the MFP2 mutant. In a complementary experiment, the pools of long chain fatty acids in Arabidopsis seedlings were quantified. The data show an initial increase and consecutive decrease in virtually all fatty acid pools identified from C16 to C20. The wild-type and mutant seedlings exhibited the same tendencies, but the tendencies were stronger in the wild type, increasing more and ending with lower concentrations of eicosanoic acid (C20:0) and eicosenoic acid (C20:1) after 5 days, relative to the mutants. That the decrease also appears in the mutant insinuates the presence of long chain hydratase activity in the MFP2 mutant seedlings. The weaker tendencies observed in the MFP2 mutants could be attributed to the observed 35% reduction in overall fatty acid levels in the mutants, indicating that the state of the seedlings is affected in a complicated manner. The data illustrate the inherent difficulties encountered when interpreting in vivo data in a complex background such as β-oxidation with its numerous isozymes and involvement in hormone metabolism.
In vitro experiments employing recombinant enzymes may correspond to an incomplete biological system. However, because all experiments were done at concentrations lower than the critical micelle concentration of C16-CoA (202 ± 5 μm (74)), all chain lengths of 2-trans-enoyl-CoA were synthesized equally well by the ACX mixtures employed (Fig. 4A), and the lower substrate concentration or added Tween 20 or BSA did not alter the ratio between conversion of C14-CoA and the longer chain lengths, it is difficult to envisage how the observed substrate profiles could be artificial. Complexes between the MFPs and thiolases could potentially influence the tertiary structure and substrate binding of the MFPs similar to the situation in PfMFP, but it has not been possible to replicate this situation in vitro.
The dehydrogenase substrate profiles of AtMFP2 and AtAIM1 are skewed toward the longer chain lengths compared with their hydratase profiles and are about 10 times slower (Fig. 4C). The AtAIM1 dehydrogenase substrate profile lacks its C4 hydratase peak and peaks at C8-CoA instead. The hydratase reaction being an order of magnitude faster than the dehydrogenase activity is common in multifunctional enzymes (18, 71). The dehydrogenase assay was of course impeded by the fact that hydroxyacyl-CoA substrates longer than C14-CoA were inefficiently synthesized, but even upon incubation with 75 nm MFP for 1 h at 27 °C, when full hydration had occurred (not shown), AtMFP2 dehydrogenation of C16-CoA proceeded about 120 times slower than with C14-CoA (Fig. 6D). It is thus safe to conclude that both AtAIM1 and AtMFP2 are broad range dehydrogenases, with AtMFP2 skewed slightly to the longer substrates, and that the two isozymes have less diverging substrate preference profiles with respect to the dehydrogenase reaction than they have with respect to the hydratase reaction.
Untouched by this study, because of the difficulty of acquiring substrates, is the activity of AtMFP2 and AtAIM1 toward branched and polyunsaturated fatty acid CoA esters. It is very likely that AtMFP2 and AtAIM1 differ in this respect considering the proposed role of AtAIM1 in the metabolism of the bulky substrates jasmonate and indole-3-acetic acid (2, 3).
ECH Acyl Binding
Because AtAIM1-ECH has increased activity with short chain substrates compared with AtMFP2, we would expect to find corresponding changes in the proposed substrate binding. The flexible loop region (Ser71–Tyr88) is shorter by one residue in AtAIM1, but what appears more striking is that the flexible loop in the substrate-binding site is far more conserved between the AIM1-like sequences than between the MFP2-like sequences. The consensus sequence of AIM1-like MFPs is 71NVFxxVHcTGDxSxxPD87 (lowercase signifies a less conserved residue). This could indicate a narrower substrate preference for AIM1-like MFPs and possibly a stronger interaction with the substrates. The larger proportion of Gly residues in this part of the structure in MFP2-like sequences implies a more flexible acyl-binding pocket, which agrees well with the less conserved sequence. The flexible loop is not dominated by hydrophobic residues in either MFP2-like or AIM1-like sequences. In the ECH domain of PfMFP, the ω-end of the binding pocket is lined with the hydrophobic residues Ile86, Leu90, Gly148, Ala275, and Phe278 representing a possible extension of the binding pocket, whereas the side chains of Glu272, Gln276, and Ser295 occupy this space in AtMFP2. This is likely to limit the length of the substrates that fit in this pocket.
HACD Acyl Binding
The HACD acyl-binding site is not explored by the short acetoacetyl-CoA analogues used in most crystallization studies of HACDs. Even in the 3-hydroxybutyryl-CoA complex of human HACD, only the shortest acyl chain possible is included, and it gives no clues to an obvious acyl-binding pocket in this enzyme (56). In PfMFE-HCAD a hydrophobic corridor delineated by Phe505, Phe560, Val555, Met556, and Pro496 extends toward the back of the dehydrogenase domain. In AtMFP2, this hydrophobic corridor is obstructed by Gln553.
A search of the solvent-exposed surface area of AtMFP2 revealed a rather large interior pocket in the HACD active site (Fig. 7). Knowing the location of this pocket, a similar pocket but with a much narrower access channel can be identified in PfMFP. No pocket but only a shallow depression was observed in the structures of human HACDs including both apoenzyme and complexes. Because the AtMFP2 structure is in an open conformation, it is possible that the apparent pocket is not related to acyl binding and is a consequence of the absence of NAD+ and substrate. Future mutagenesis studies will hopefully provide greater understanding of the interaction involved in HACD substrate binding. The putative acyl-binding pocket is ∼15 Å long and 7 Å wide. It is lined primarily by hydrophobic residues: Phe503, Tyr505, Thr506, Gln507, Met510, Cys539, Ala547, Ile548, Thr550, Ala551, Phe554, Ile555, Tyr563, Lys564 (not Nϵ), Ser565, and Ile568. The size and polarity of the residues are conserved among the plant MFPs and are similar between the two MFP groups. The observed difference in substrate specificity of MFPs is therefore likely to be the result of differences in ECH substrate specificity and channeling.
In conclusion, the crystal structure of AtMFP2 in its ground state with no co-factors or substrates bound shows an ECH domain with a very flexible acyl chain-binding pocket and an ECH-HACDN interface that draws on interactions to the adenine moiety of the substrate to generate a pivotal point for substrate transfer between the ECH and the HACD active sites. AtMFP2 and AtAIM1 substrate preference profiles show AtAIM1-ECH to have a preference for short chain acyl-CoAs compared with AtMFP2-ECH. The inability of either AtAIM1 or AtMFP2 to efficiently catabolize substrates longer than C14-enoyl-CoA indicates that an uncharacterized long chain 2-trans-enoyl-CoA hydratase exists in Arabidopsis peroxisomes or that conditions unaccounted for in in vitro experiments are required to create the right substrate binding environment for long acyl chains.
Supplementary Material
Acknowledgments
We acknowledge beamline scientists at the BESSY 14.1 beamline for technical assistance during data collection and Annette Kure Andreassen for protein purification.
This work was supported by the Danish Natural Science Research Council, which provided a postdoctoral stipend through DANSYNC (to S. A.) and synchrotron beam time.

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. S1–S4.
The atomic coordinates and structure factors (code 2WTB) have been deposited in the Protein Data Bank, Research Collaboratory for Structural Bioinformatics, Rutgers University, New Brunswick, NJ (http://www.rcsb.org/).
- ACX
- acyl-CoA oxidase
- At
- Arabidopsis thaliana
- BSA
- bovine serum albumin
- Cs
- Cucumis sativus (cucumber)
- ECH
- 2-trans-enoyl-CoA hydratase
- HACD
- l-3-hydroxyacyl-CoA dehydrogenase
- KAT
- 3-keto-acyl-CoA thiolase
- MES
- 4-morpholinoethanesulfonic acid
- MFP
- multifunctional protein
- MOPS
- 4-morpholinopropanesulfonic acid
- MWCO
- molecular weight cut-off
- Pf
- Pseudomonas fragi
- r.m.s.d.
- root mean square deviation
- Rn
- Rattus norvegicus
- SSM
- secondary structure matching
- YT medium
- 16 g of Bacto Tryptone, 10 g of Bacto Yeast Extract, 5 g of NaCl per liter of distilled H2O, pH 7.0 adjusted with 5 m NaOH.
REFERENCES
- 1.Cooper T. G., Beevers H. (1969) J. Biol. Chem. 244, 3514–3520 [PubMed] [Google Scholar]
- 2.Delker C., Zolman B. K., Miersch O., Wasternack C. (2007) Phytochemistry 68, 1642–1650 [DOI] [PubMed] [Google Scholar]
- 3.Richmond T. A., Bleecker A. B. (1999) Plant Cell 11, 1911–1924 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Zolman B. K., Martinez N., Millius A., Adham A. R., Bartel B. (2008) Genetics 180, 237–251 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kindl H. (1987) in Lipids: Structure and Function (Stumpf P. K., Conn E. E. eds) Vol. 9, pp. 31–52, Academic Press, London [Google Scholar]
- 6.Fath A., Bethke P., Lonsdale J., Meza-Romero R., Jones R. (2000) Plant Mol. Biol. 44, 255–266 [DOI] [PubMed] [Google Scholar]
- 7.Graham I. A., Eastmond P. J. (2002) Prog. Lipid Res. 41, 156–181 [DOI] [PubMed] [Google Scholar]
- 8.Hooks M. A., Bode K., Couee I. (1995) Phytochemistry 40, 657–660 [Google Scholar]
- 9.Preisig-Müller R., Gühnemann-Schäfer K., Kindl H. (1994) J. Biol. Chem. 269, 20475–20481 [PubMed] [Google Scholar]
- 10.Eastmond P. J., Graham I. A. (2000) Biochem. Soc. Trans. 28, 95–99 [DOI] [PubMed] [Google Scholar]
- 11.Rylott E. L., Eastmond P. J., Gilday A. D., Slocombe S. P., Larson T. R., Baker A., Graham I. A. (2006) Plant J. 45, 930–941 [DOI] [PubMed] [Google Scholar]
- 12.Titorenko V. I., Nicaud J. M., Wang H., Chan H., Rachubinski R. A. (2002) J. Cell Biol. 156, 481–494 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ishikawa M., Tsuchiya D., Oyama T., Tsunaka Y., Morikawa K. (2004) EMBO J. 23, 2745–2754 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Uchida Y., Izai K., Orii T., Hashimoto T. (1992) J. Biol. Chem. 267, 1034–1041 [PubMed] [Google Scholar]
- 15.Taskinen J. P., Kiema T. R., Hiltunen J. K., Wierenga R. K. (2006) J. Mol. Biol. 355, 734–746 [DOI] [PubMed] [Google Scholar]
- 16.Ylianttila M. S., Pursiainen N. V., Haapalainen A. M., Juffer A. H., Poirier Y., Hiltunen J. K., Glumoff T. (2006) J. Mol. Biol. 358, 1286–1295 [DOI] [PubMed] [Google Scholar]
- 17.Yang S. Y., Bittman R., Schulz H. (1985) J. Biol. Chem. 260, 2862–2868 [PubMed] [Google Scholar]
- 18.Yang S. Y., Cuebas D., Schulz H. (1986) J. Biol. Chem. 261, 15390–15395 [PubMed] [Google Scholar]
- 19.Yao K. W., Schulz H. (1996) J. Biol. Chem. 271, 17816–17820 [DOI] [PubMed] [Google Scholar]
- 20.Schilmiller A. L., Koo A. J., Howe G. A. (2007) Plant Physiol. 143, 812–824 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Cruz Castillo M., Martínez C., Buchala A., Métraux J. P., León J. (2004) Plant Physiol. 135, 85–94 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Afitlhile M. M., Fukushige H., Nishimura M., Hildebrand D. F. (2005) Plant Physiol. Biochem. 43, 603–609 [DOI] [PubMed] [Google Scholar]
- 23.Pinfield-Wells H., Rylott E. L., Gilday A. D., Graham S., Job K., Larson T. R., Graham I. A. (2005) Plant J. 43, 861–872 [DOI] [PubMed] [Google Scholar]
- 24.Li C., Schilmiller A. L., Liu G., Lee G. I., Jayanty S., Sageman C., Vrebalov J., Giovannoni J. J., Yagi K., Kobayashi Y., Howe G. A. (2005) Plant Cell 17, 971–986 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Palosaari P. M., Hiltunen J. K. (1990) J. Biol. Chem. 265, 2446–2449 [PubMed] [Google Scholar]
- 26.Sakurai T., Satou M., Akiyama K., Iida K., Seki M., Kuromori T., Ito T., Konagaya A., Toyoda T., Shinozaki K. (2005) Nucleic Acids Res. 33, D647–D650 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Pedersen L., Henriksen A. (2004) Acta Crystallogr. D Biol. Crystallogr. 60, 1125–1128 [DOI] [PubMed] [Google Scholar]
- 28.Leslie A. G. (1992) Joint CCP4/ESF-EAMCB Newsletter on Protein Crystallography 26 [Google Scholar]
- 29.Collaborative Computing Project, No. 4 (1994) Acta Crystallogr. D Biol. Crystallogr. 50, 760–763 [DOI] [PubMed] [Google Scholar]
- 30.Strong M., Sawaya M. R., Wang S., Phillips M., Cascio D., Eisenberg D. (2006) Proc. Natl. Acad. Sci. U.S.A. 103, 8060–8065 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Adams P. D., Grosse-Kunstleve R. W., Hung L. W., Ioerger T. R., McCoy A. J., Moriarty N. W., Read R. J., Sacchettini J. C., Sauter N. K., Terwilliger T. C. (2002) Acta Crystallogr. D Biol. Crystallogr. 58, 1948–1954 [DOI] [PubMed] [Google Scholar]
- 32.McCoy A. J., Grosse-Kunstleve R. W., Adams P. D., Winn M. D., Storoni L. C., Read R. J. (2007) J. Appl. Crystallogr. 40, 658–674 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Terwilliger T. C., Grosse-Kunstleve R. W., Afonine P. V., Moriarty N. W., Zwart P. H., Hung L. W., Read R. J., Adams P. D. (2008) Acta Crystallogr. D Biol. Crystallogr. 64, 61–69 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Emsley P., Cowtan K. (2004) Acta Crystallogr. D Biol. Crystallogr. 60, 2126–2132 [DOI] [PubMed] [Google Scholar]
- 35.Davis I. W., Leaver-Fay A., Chen V. B., Block J. N., Kapral G. J., Wang X., Murray L. W., Arendall W. B., 3rd, Snoeyink J., Richardson J. S., Richardson D. C. (2007) Nucleic Acids Res. 35, W375–W383 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hooks M. A., Kellas F., Graham I. A. (1999) Plant J. 20, 1–13 [DOI] [PubMed] [Google Scholar]
- 37.Lazarow P. B. (1978) J. Biol. Chem. 253, 1522–1528 [PubMed] [Google Scholar]
- 38.Dawson R. M. C., Elliot D. C., Elliot W. H., Jones K. M. (1978) Data for Biochemical Research, pp. 191–215, Oxford University Press, Oxford, United Kingdom [Google Scholar]
- 39.Krissinel E., Henrick K. (2007) J. Mol. Biol. 372, 774–797 [DOI] [PubMed] [Google Scholar]
- 40.Reynolds C., Damerell D., Jones S. (2009) Bioinformatics 25, 413–414 [DOI] [PubMed] [Google Scholar]
- 41.Ogmen U., Keskin O., Aytuna A. S., Nussinov R., Gursoy A. (2005) Nucleic Acids Res. 33, W331–W336 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Guney E., Tuncbag N., Keskin O., Gursoy A. (2008) Nucleic Acids Res. 36, D662–D666 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Gille C., Lorenzen S., Michalsky E., Frömmel C. (2003) Bioinformatics 19, 2489–2491 [DOI] [PubMed] [Google Scholar]
- 44.Jaroszewski L., Rychlewski L., Li Z., Li W., Godzik A. (2005) Nucleic Acids Res. 33, W284–W288 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Crooks G. E., Hon G., Chandonia J. M., Brenner S. E. (2004) Genome Res. 14, 1188–1190 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Larkin M. A., Blackshields G., Brown N. P., Chenna R., McGettigan P. A., McWilliam H., Valentin F., Wallace I. M., Wilm A., Lopez R., Thompson J. D., Gibson T. J., Higgins D. G. (2007) Bioinformatics 23, 2947–2948 [DOI] [PubMed] [Google Scholar]
- 47.Page R. D. (1996) Comput. Appl. Biosci. 12, 357–358 [DOI] [PubMed] [Google Scholar]
- 48.Jones S., Thornton J. M. (1996) Proc. Natl. Acad. Sci. U.S.A. 93, 13–208552589 [Google Scholar]
- 49.Engel C. K., Kiema T. R., Hiltunen J. K., Wierenga R. K. (1998) J. Mol. Biol. 275, 847–859 [DOI] [PubMed] [Google Scholar]
- 50.Partanen S. T., Novikov D. K., Popov A. N., Mursula A. M., Hiltunen J. K., Wierenga R. K. (2004) J. Mol. Biol. 342, 1197–1208 [DOI] [PubMed] [Google Scholar]
- 51.Mursula A. M., Hiltunen J. K., Wierenga R. K. (2004) FEBS Lett. 557, 81–87 [DOI] [PubMed] [Google Scholar]
- 52.Leonard P. M., Grogan G. (2004) J. Biol. Chem. 279, 31312–31317 [DOI] [PubMed] [Google Scholar]
- 53.Benning M. M., Taylor K. L., Liu R.-Q., Yang G., Xiang H., Wesenberg G., Dunaway-Mariano D., Holden H. M. (1996) Biochemistry 35, 8103–8109 [DOI] [PubMed] [Google Scholar]
- 54.Benning M. M., Haller T., Gerlt J. A., Holden H. M. (2000) Biochemistry 39, 4630–4639 [DOI] [PubMed] [Google Scholar]
- 55.Krissinel E., Henrick K. (2004) Acta Crystallogr. D Biol. Crystallogr. 60, 2256–2268 [DOI] [PubMed] [Google Scholar]
- 56.Barycki J. J., O'Brien L. K., Strauss A. W., Banaszak L. J. (2000) J. Biol. Chem. 275, 27186–27196 [DOI] [PubMed] [Google Scholar]
- 57.Hulo N., Bairoch A., Bulliard V., Cerutti L., de Castro E., Langendijk-Genevaux P. S., Pagni M., Sigrist C. J. (2006) Nucleic Acids Res. 34, D227–D230 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Willadsen P., Eggerer H. (1975) Eur. J. Biochem. 54, 247–252 [DOI] [PubMed] [Google Scholar]
- 59.Bahnson B. J., Anderson V. E., Petsko G. A. (2002) Biochemistry 41, 2621–2629 [DOI] [PubMed] [Google Scholar]
- 60.Bahnson B. J., Anderson V. E. (1991) Biochemistry 30, 5894–5906 [DOI] [PubMed] [Google Scholar]
- 61.Kiema T. R., Engel C. K., Schmitz W., Filppula S. A., Wierenga R. K., Hiltunen J. K. (1999) Biochemistry 38, 2991–2999 [DOI] [PubMed] [Google Scholar]
- 62.Müller-Newen G., Janssen U., Stoffel W. (1995) Eur. J. Biochem. 228, 68–73 [DOI] [PubMed] [Google Scholar]
- 63.Feng Y., Hofstein H. A., Zwahlen J., Tonge P. J. (2002) Biochemistry 41, 12883–12890 [DOI] [PubMed] [Google Scholar]
- 64.Tsuchiya D., Shimizu N., Ishikawa M., Suzuki Y., Morikawa K. (2006) Structure 14, 237–246 [DOI] [PubMed] [Google Scholar]
- 65.Noyes B. E., Glatthaar B. E., Garavelli J. S., Bradshaw R. A. (1974) Proc. Natl. Acad. Sci. U.S.A. 71, 1334–1338 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Möller W., Amons R. (1985) FEBS Lett. 186, 1–7 [DOI] [PubMed] [Google Scholar]
- 67.Barycki J. J., O'Brien L. K., Bratt J. M., Zhang R., Sanishvili R., Strauss A. W., Banaszak L. J. (1999) Biochemistry 38, 5786–5798 [DOI] [PubMed] [Google Scholar]
- 68.Adams M. J., Ellis G. H., Gover S., Naylor C. E., Phillips C. (1994) Structure 2, 651–668 [DOI] [PubMed] [Google Scholar]
- 69.He X. Y., Deng H., Yang S. Y. (1997) Biochemistry 36, 261–268 [DOI] [PubMed] [Google Scholar]
- 70.Barycki J. J., O'Brien L. K., Strauss A. W., Banaszak L. J. (2001) J. Biol. Chem. 276, 36718–36726 [DOI] [PubMed] [Google Scholar]
- 71.Gühnemann-Schäfer K., Kindl H. (1995) Planta 196, 642–646 [Google Scholar]
- 72.Goepfert S., Hiltunen J. K., Poirier Y. (2006) J. Biol. Chem. 281, 35894–35903 [DOI] [PubMed] [Google Scholar]
- 73.Lemieux B., Miquel M., Somerville C., Browse J. (1990) Theor. Appl. Genet. 80, 234–240 [DOI] [PubMed] [Google Scholar]
- 74.Constantinides P. P., Steim J. M. (1985) J. Biol. Chem. 260, 7573–7580 [PubMed] [Google Scholar]
- 75.Deleted in proof
- 76.Deleted in proof
- 77.Evans P. (2006) Acta Crystallogr. D Biol. Crystallogr. 62, 72–82 [DOI] [PubMed] [Google Scholar]
- 78.French S., Wilson K. (1978) Acta Crystallogr. A Found. Crystallogr. 34, 517–525 [Google Scholar]
- 79.Laskowski R. A., Macarthur M. W., Moss D. S., Thornton J. M. (1993) J. Appl. Crystallogr. 26, 283–291 [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.