Abstract
Cell wall peptidoglycan assembly is a tightly regulated process requiring the combined action of multienzyme complexes. In this study we provide direct evidence showing that substrate transformations occurring at the different stages of this process play a crucial role in the spatial and temporal coordination of the cell wall synthesis machinery. Peptidoglycan substrate alteration was investigated in the Gram-positive bacterium Lactococcus lactis by substituting the peptidoglycan precursor biosynthesis genes of this bacterium for those of the vancomycin-resistant bacterium Lactobacillus plantarum. A set of L. lactis mutant strains in which the normal d-Ala-ended precursors were partially or totally replaced by d-Lac-ended precursors was generated. Incorporation of the altered precursor into the cell wall induced morphological changes arising from a defect in cell elongation and cell separation. Structural analysis of the muropeptides confirmed that the activity of multiple enzymes involved in peptidoglycan synthesis was altered. Optimization of this altered pathway was necessary to increase the level of vancomycin resistance conferred by the utilization of d-Lac-ended peptidoglycan precursors in the mutant strains. The implications of these findings on the control of bacterial cell morphogenesis and the mechanisms of vancomycin resistance are discussed.
Keywords: Antibiotics, Bacterial Metabolism, Carboxypeptidase, Cell Wall, Lactic Acid, Bacterial Morphogenesis, Ovococcus, Penicillin-binding Protein, Peptidoglycan, Vancomycin Resistance
Introduction
Peptidoglycan (or murein) is a major stress-bearing component of the bacterial cell wall. This polymer is made of glycan chains interconnected by covalent cross-links between short peptides. It acts as an exoskeleton and plays a key role in determining and maintaining cell shape throughout the cell cycle. The first steps of peptidoglycan biosynthesis occur in the cytoplasm where specific enzymes catalyze the formation of the uridinediphospho-n-acetylmuramyl-pentapeptide (UDP-MurNAc-pentapeptide)4 precursor (1). After the addition of n-acetyl-glucosamine (GlcNAc), this precursor is transported through the plasma membrane and incorporated into the peptidoglycan by penicillin-binding proteins (PBPs) (2). High molecular weight PBPs act in transglycosylation and transpeptidation reactions required for peptidoglycan polymers formation and cross-linking, whereas low molecular weight PBPs are suggested to be carboxypeptidases and endopeptidases involved in peptide chains processing during peptidoglycan synthesis and maturation. PBPs collaborate with peptidoglycan hydrolases, possibly through the assembly of multienzymatic complexes, which together constitute the peptidoglycan biosynthesis machinery (3–7).
Bacterial cell morphology is the result of a tightly regulated process coupling peptidoglycan synthesis with cell growth and cell division. Proper cell shape requires proper localization and activity of the peptidoglycan biosynthesis machinery. PBP localization studies performed in bacilli and cocci suggest that recruitment of these proteins is mediated by structural determinants of the cytoskeleton (e.g. the FtsZ ring at the septum and actin-like filaments along the longitudinal axis of the cell) (3, 8, 9). Substrate availability also appears to be important for proper localization and activity of PBPs, as was reported for Escherichia coli (10) and more recently for Streptococcus pneumoniae (11) and Staphylococcus aureus (12). Although the exact mechanism of this control is still unclear, it has been suggested that PBP localization depends on the activity of the carboxypeptidases that trim the peptidoglycan pentapeptide chains, providing the appropriate acceptor substrates for high molecular weight transpeptidases (10, 11) or eliminate these side chains, thereby preventing new peptidoglycan synthesis in specific regions of the cell wall (11, 12). Likewise, proper recruitment of carboxypeptidases and other peptidoglycan hydrolases may also depend on the specific location of their substrate during cell growth (11, 13).
In this study the flexibility of the cell wall biosynthesis machinery toward a new substrate was examined in the Gram-positive bacterium Lactococcus lactis. In terms of shape and PBP content, L. lactis is closely related to other ovoid cocci, such as streptococci and enterococci, including a number of pathogenic species. The primary structure of L. lactis peptidoglycan monomer is GlcNAc-MurNAc-l-Ala-γ-d-Glu (or isoglutamine)-l-Lys-d-Ala-d-Ala, with a d-Asn (or a d-Asp) cross-bridge residue attached to l-Lys (14–17). The d-Ala C-terminal residue of L. lactis peptidoglycan peptide chains was changed to d-Lac. To this end, specific genes involved in peptidoglycan precursor synthesis from Lactobacillus plantarum, another lactic acid bacterium, were transferred and expressed in L. lactis (Fig. 1). In L. plantarum, the peptide chain ended with d-Ala-d-Lac, which makes this bacterium naturally resistant to vancomycin due to the fact that d-Ala-d-Lac termini are not targets for the antibiotic (18, 19). The resulting L. lactis mutants produced different levels of precursors terminated by d-Ala-d-Lac instead of d-Ala-d-Ala and showed different levels of resistance to vancomycin. Utilization of the new substrate forced L. lactis peptidoglycan biosynthetic pathway to adapt, leading to significant changes in peptidoglycan structure and cell morphology.
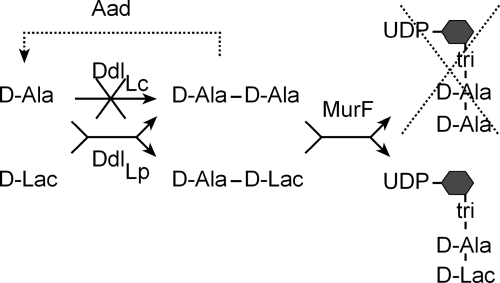
FIGURE 1.
The strategy used for the production of d-Lac-ended peptidoglycan precursors in L. lactis. d-Ala-d-Lac was produced in L. lactis by expressing the ddlLp ligase from L. plantarum. In a second step, the endogenous ddlLc ligase of L. lactis was inactivated to reduce the level of precursors ended by d-Ala-d-Ala. The complete substitution of d-Ala-ended precursors by d-Lac-ended precursors was achieved by expressing the Aad dipeptidase that eliminates d-Ala-d-Ala dipeptides produced by ddlLp (dashed lines). MurF, uridinediphospho-n-acetylmuramyl-tripeptide:d-Ala-d-Lac ligase; tri, l-Ala-d-Glu-m-DAP; hexagon, n-acetylmuramic acid.
EXPERIMENTAL PROCEDURES
Bacterial Strains, Plasmids, and Growth Conditions
The bacterial strains and plasmids used in this study are listed in supplemental Table S1. The L. lactis subsp. cremoris strain NZ3900 and its derivatives were grown at 28 °C in M17 broth (BD Biosciences) containing 0.5% glucose (M17-glucose). Strains carrying the expression vectors pGIM020, pGIM022, pGIM023, pGIM024, pGIM025, and pGIM026 were grown in the presence of 10 μg/ml chloramphenicol, whereas their ddlLc d-Ala-d-Ala ligase mutant derivatives MD001, MD002, MD003, MD004, and MD006 were selected and cultured on 5 μg/ml chloramphenicol and 5 μg/ml erythromycin (Sigma). For induction of genes under the control of the nisA expression signals, nisin A (Sigma) was used at a concentration of 0.5 ng/ml. d-Lactate (15 mm; Fluka) was added in the cultures, except for strains expressing the ldhDLh d-Lac dehydrogenase gene from Lactobacillus helveticus.
Analysis of growth dependence of the ddlLc mutants toward nisin and d-Lac was performed as followed; overnight cultures were grown in the presence of nisin (0.5 ng/ml), d-Lac (15 mm), erythromycin (5 μg/ml), and chloramphenicol (5 μg/ml), washed once, and diluted at an A600 nm of 0.05 in M17-glucose broth containing erythromycin and chloramphenicol supplemented with nisin and d-Lac or just nisin.
Construction of Expression Plasmids
For pGIM020, a DNA fragment containing the ddlLp d-Ala-d-Lac ligase open reading frame (ORF) and its ribosome binding site was amplified from L. plantarum strain NCIMB8826 with primers DDLPLXP3 and DDLPLXP4 (supplemental Table S1). The 1187-bp PCR fragment was digested with BamHI and ScaI and inserted into the pNZ2650 plasmid (20) digested with BamHI and PvuII. The resulting plasmid (pGIM020) contains ddlLp under the control of the PnisA promoter followed by the L. plantarum ldhL transcriptional terminator. For co-expressing ddlLp with the ldhDLh gene of L. helveticus, a 1434-bp fragment encompassing the ldhDLh ORF and promoter sequences was PCR-amplified from L. helveticus CNRZ32 with primers PNZLDHD1-PNZLDHD2 (supplemental Table S1), digested with XbaI and ScaI, and inserted into the XbaI/NspI-digested pGIM020 plasmid to yield pGIM022. Construction of the pGIM023 vector for co-expressing ddlLp and the aad d-Ala-d-Ala dipeptidase gene from L. plantarum as a bicistronic unit under the control of the PnisA promoter was previously described (18). The same procedure was used for the construction of pGIM024 to co-express ddlLp with the VanXA d-Ala-d-Ala dipeptidase gene of the vancomycin-resistant Enterococcus faecium strain BM4147. A 671-bp fragment encompassing the ribosome binding site and ORF of VanXA was amplified from pAT83 plasmid (21) using primers VANX1 and VANX2 (supplemental Table S1). The resulting PCR product was digested with BsrGI and BamHI and cloned into similarly digested pGIM020.
For the complementation control plasmid pGIM026 expressing the ddlASt d-Ala-d-Ala ligase gene from Streptococcus thermophilus, primers SthddlABam1 and SthddlAKpnI (supplemental Table S1) were used to amplify the ddlASt ribosome binding site and ORF directly from S. thermophilus LMG18311 colonies. The resulting 1104-bp PCR fragment was digested with BamHI and KpnI and inserted into pGIM020 cut with the same enzymes, replacing the ddlLp insert and placing ddlASt under the transcriptional control of the PnisA promoter.
Construction of L. lactis Knock-out Mutants
Disruption of the ddlLc ligase gene was carried out by homologous recombination-mediated integration of the disruptive plasmid pGIL001. For pGIL001 construction, an internal 574-bp fragment of ddlLc was generated by PCR using the LCDDLDISR1 and LCDDLDISR2 primers (supplemental Table S1). The fragment was digested with PstI and XbaI and cloned into the suicide plasmid pGIZ907 (19) digested with NsiI and XbaI. Chromosomal integration of pGIL001 generated 5′- and 3′-terminal-truncated copies of ddlLc separated by vector sequences. These include an erythromycin resistance gene as well as an outward-facing promoter PLdh (19) to ensure expression of the murF gene located downstream of ddlLcin L. lactis. Inactivation of the dacA gene was carried out using the disruptive plasmid pGIBD006. A PCR fragment containing the dacA ORF was generated using the BLD_DacAUpNcoI and BLD_DacADownSacI primers (supplemental Table S1) and cloned into the pGMTEasy vector (Invitrogen). The resulting plasmid was digested with SphI and KasI to provide an internal fragment of the dacA gene that was then cloned into pUC18Ery (22). Chromosomal integration of pGIBD006 produced two truncated copies of the dacA gene separated by vector sequences.
Determination of Vancomycin Resistance Level and Population Analysis
Minimal inhibitory concentrations (MICs) of vancomycin displayed by the wild type and mutant strains of L. lactis were determined by the E-test method (AB-Biodisk). Cells were plated on solid medium and incubated for 24 h at 28 °C in the presence of an E-test strip containing a gradient of vancomycin (from 0.016 to 256 μg/ml). MIC values are indicated by the position where the edge of the inhibition ellipse intersects the side of the strip. For population heterogeneity analysis, serial cultures of the primary mutant MD003 and the control strains MD006 and NZ3900 (pNZ8048) were performed by 100-fold diluting overnight-grown cultures into 10 ml of fresh medium every day (∼10 generations intervals). At each passage diluted samples were plated on increasing concentrations of vancomycin (from 0 to 1024 μg/ml), and subpopulations displaying different levels of vancomycin resistance were estimated by expressing the number of colony forming units obtained for each antibiotic concentration per ml of culture and A600 nm unit.
Preparation and Analysis of UDP-linked Peptidoglycan Precursors
Cells were grown in M17-glucose broth supplemented with antibiotics, nisin, and d-Lac (when appropriate) to an A600 nm of 0.7 and treated with bacitracin (200 μg/ml; Sigma) for 90 min before harvesting by centrifugation. Peptidoglycan precursors were then extracted with 20% trichloroacetic acid and analyzed by reverse-phase high pressure liquid chromatography (HPLC) as previously described (19).
Determination of Peptidoglycan Composition
The reference strain used, PH8076, is an erythromycin-resistant L. lactis strain isogenic to NZ3900.5 The erythromycin resistance marker carried by this strain was inserted together with the nisR and nisK regulatory genes in the pepN locus using the integration vector pNZ9573 (23). This strain also contains a mutation in the α-acetolactate decarboxylase gene aldB that has no effect on cell wall peptidoglycan composition. PH8076 was transformed with the pNZ8048 empty vector and used as a wild type reference for muropeptide analysis.
Peptidoglycan from L. lactis strains PH8076 (pNZ8048), MD003, and MD003-128 was prepared as previously described (14, 16). Purified peptidoglycan (4 mg dry weight in 500 μl) was then digested with mutanolysin (2500 units/ml; Sigma) in 25 mm sodium phosphate buffer (pH 5.5) for 19 h at 37 °C under rotational shaking. Muropeptides were reduced with sodium borohydride and separated by reverse phase HPLC using a Hypersil ODS column (C18, 250 × 4.6 mm; 5 μm; ThermoHypersil-Keystone) at 50 °C as described by Courtin et al. (14). Fractions were collected, and 1 μl of the fractions containing muropeptides of interest was analyzed by matrix-assisted laser desorption ionization time-of-flight (MALDI-TOF) mass spectrometry with a Voyager DE STR mass spectrometer (Applied Biosystems) with α-cyano-4-hydroxycinnamic acid matrix. The structures of the muropeptides I and II were determined by mass spectrometry as described in Arbeloa et al. (24) using an electrospray time-of-flight mass spectrometer operating in positive mode (Qstar Pulsar I, Applied Biosystems).
Microscopic Imaging and Van-FL Staining
Van-FL staining was essentially performed as previously described by Daniel and Errington (25). Cells from an overnight culture were diluted into fresh M17-glucose medium and grown to early exponential phase (A600 nm = 0.2–0.3) at 30 °C. Culture samples (200 μl) were incubated for 30 min with a mixture of equal amount of vancomycin BODIPY-FL conjugate (Molecular Probes) and unlabeled vancomycin (Sigma) at a final concentration of 3 μg/ml. Cells were fixed in 1.6% formaldehyde (in phosphate-buffered saline) and mounted on poly-lysine-coated slides (25). 4′,6-Diamidino-2-phenylindole (0.5 μg/ml; Sigma) was added for nucleoid staining. Images were taken using a SonyCoolSnap HQ cooled CCD camera (Roper Scientific Ltd) attached to a Zeiss Axiovert microscope. For filamentation induction by methicillin treatment, the antibiotic was added to exponentially growing cultures (A600 nm = 0.1) at concentrations of 0.1 and 0.5 μg/ml, and cells were incubated at 28 °C for 4.5 h before microscope observation.
Complete Genome Sequencing of MD003-128 L. lactis Mutant
For SOLiD sequencing, approximately 5 μg of chromosomal DNA was sheared and size-selected to an average size of 100 bp. P1 and P2 adaptors were ligated and amplified for 15 cycles; 0.2 pg/μl double-stranded DNA library was added to the emulsion with one billion beads according to manufacturer instructions. From 11 million positive beads, corresponding to the MD003-128 strain DNA, 4.6 millions (42%) matched to the L. lactis MG1363 sequence (26) used as reference for the analysis by Corona-Lite (Applied Biosystems) or MAQ version 0.7.1 software (27). It produced 231 million nucleotides corresponding to a total coverage of 91.6 of the 2,529,478-bp genome. 127 kb (5%) of 2.5 Mb were not covered, and 30.9 kb (1.2%) were determined with ambiguity. Usually the latter were the regions of genome poorly covered with the sequencing reads. 29 kb of these had coverage of less than 5, most of the others less than 10. It should also be mentioned that with the data produced and the analysis tools that were applied, we were not able to detect small indel-like mutations.
RESULTS
Production of d-Lac-ended Peptidoglycan Precursors in L. lactis
A key enzyme for determining the C-terminal composition of the peptidoglycan peptide chain is the Ddl ligase that links together the two last residues before their incorporation into the precursor by the MurF enzyme (Fig. 1). In L. lactis, the ddlLc ligase is specific for d-Ala-d-Ala dipeptides formation, whereas the ddlLp ligase of L. plantarum synthesizes d-Ala-d-Lac depsipeptides (Fig. 1). To produce d-Lac-ended peptidoglycan precursors in L. lactis, the L. plantarum ddlLp gene was cloned as a transcriptional fusion with the nisin-inducible promoter PnisA and introduced in NZ3900, a L. lactis strain containing the nisR and nisK regulatory genes required for nisin induction (Fig. 1a, see also “Experimental Procedures”). Because L. lactis exclusively produces the l-isomer of lactate, d-Lac (15 mm) was added to the culture medium to serve as a substrate for the ddlLp ligase. Alternatively, ddlLp was co-expressed with the ldhDLh gene encoding the d-Lac dehydrogenase of L. helveticus to produce d-Lac directly within the cell by pyruvate conversion.
Precursors terminated by d-Ala-d-Lac were not detected in the cytoplasm of the strain overexpressing ddlLp alone (NZ3900 (pGIM020), Table 1). However, they represented 25% of the soluble precursors purified from the strain expressing both ddlLp and ldhDLh (NZ3900 (pGIM022), Table 1). This shows that the L. plantarum ddlLp ligase is active in L. lactis and that the MurF enzyme of L. lactis is capable of incorporating d-Ala-d-Lac depsipeptides in the precursors (Fig. 1).
TABLE 1.
Production of peptidoglycan precursors ended by d-Ala-d-Lac in L. lactis subsp. cremoris and impact on vancomycin resistance
All the strains were cultured in M17-glucose broth containing nisin (0.5 ng/ml) and d -Lac (15 mm) except NZ3900 (pGIM022) and MD002, cultured without adding d -Lac in the medium. Peptidoglycan precursors were prepared and analyzed by reverse-phase HPLC as previously described (19). MICs of vancomycin were determined on solid medium by the E-test method (AB-Biodisk). The presence of highly resistant colonies in the inhibition zone of the E-test plate is indicated.
| Strain (plasmid) | Genotype |
d-Ala-d-Lac ended precursora | Vanco MIC | Highly resistant colonies | ||||
|---|---|---|---|---|---|---|---|---|
|
ddl ligase gene |
d-Lac dehydrogenase ldhDLh | Dipeptidase gene | ||||||
| ddlLc | ddlLp | ddlASt | ||||||
| % | μg/ml | |||||||
| NZ3900 (pNZ8048) | + | − | − | − | − | 0b | 1.5 | − |
| NZ3900 (pGIM020) | + | + | − | − | − | 0b | 1.5 | − |
| MD001 | − | + | − | − | − | 64 | 1.5 | + |
| NZ3900 (pGIM022) | + | + | − | + | − | 25 | 1.5 | − |
| MD002 | − | + | − | + | − | 42 | 1.5 | + |
| NZ3900 (pGIM023) | + | + | − | − | aad | 100 | 3–4 | + |
| MD003 | − | + | − | − | aad | 100 | 3–24 | + |
| NZ3900 (pGIM024) | + | + | − | − | vanXA | 91 | 2–3 | + |
| MD004 | − | + | − | − | vanXA | 100 | 6 | + |
| NZ3900 (pGIM006) | + | − | + | − | − | NDc | 1.5 | − |
| MD006 | − | − | + | − | − | NDc | 1.5 | − |
a Percentage of penta-substituted UDP-MurNAc-pent▵depsi)peptide precursors terminated by d-Ala-d -Lac.
b No UDP-MurNAc-tri-d -Ala-d -Lac could be detected.
c ND, not determined.
To increase the ratio of d-Lac- over d-Ala-ended precursors in NZ3900 (pGIM020) and NZ3900 (pGIM022), the endogenous d-Ala-d-Ala ligase gene of L. lactis was disrupted in both strains, yielding the mutated MD001 and MD002 derivatives, respectively (Table 1, see also Fig. 1). The ddlLc gene was also disrupted in a strain (MD006) expressing an heterologous d-Ala-d-Ala ligase from S. thermophilus (ddlASt) to ascertain that the phenotypes observed after changing the peptidoglycan precursor are not simply the consequence of nonspecific polar effects affecting the activity of other genes in the ddlLc locus (see below).
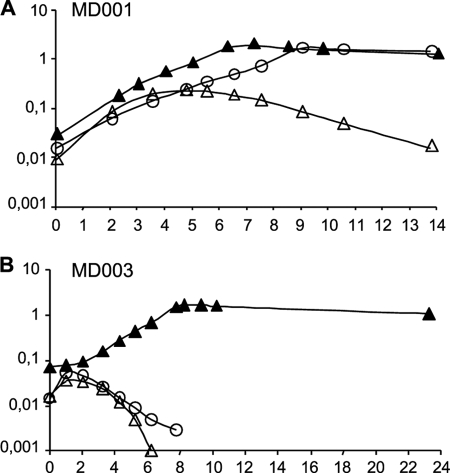
Growth of the MD001 and MD002 strains required the presence of nisin in the culture, showing that peptidoglycan synthesis in these two mutants absolutely relies on the heterologous expression of the L. plantarum ligase gene ddlLp (Fig. 2A, data not shown) Prolonged incubation in nisin-deprived medium resulted in a decrease in the optical density of the culture, which is indicative of cell lysis caused by a defect in peptidoglycan synthesis (Fig. 2A). Growth of the control strain MD006 was not dependent on nisin, presumably because of sufficient residual expression of the ddlASt gene in the absence of inducer (data not shown). As expected, the MD002 mutant co-expressing ddlLp with the ldhDLh gene from L. helveticus grew equally well whether d-Lac was added in the culture or not (data not show). However, growth of MD001, which does not produces d-Lac, was also insensitive to the absence of d-Lac in the medium (Fig. 2A).
FIGURE 2.
Growth dependence of the MD001 and MD003 mutants toward nisin and d-lactate. The L. lactis MD001 (A) and MD003 (B) mutants were cultured in M17-glucose in the presence and absence of d-Lac (15 mm) and nisin (0.5 ng/ml), shown as closed and open triangles, respectively, and in the presence of nisin (0.5 ng/ml) without d-Lac (open circles). Growth of the cultures was followed by measuring the A600 nm.
Peptidoglycan precursor analysis showed that MD001 and MD002 still produced a certain amount of d-Ala-ended UDP-MurNac-pentapeptides, although the proportion of d-Lac-ended precursors expressed by the two mutants was significantly increased when compared with the non-mutated strains (Table 1). These results are consistent with our previous data showing that the ddlLp ligase is bispecific, being capable of synthesizing both d-Ala-d-Lac and d-Ala-d-Ala in L. plantarum (18). It is, thus, likely that MD001 utilizes the d-Ala-d-Ala dipeptides produced by ddlLp to sustain cell wall synthesis in a d-Lac-deprived medium.
In vancomycin-resistant enterococci, exclusive replacement of d-Ala-d-Ala termini by d-Ala-d-Lac termini requires the production of a d-Ala-d-Ala dipeptidase (VanX) in addition to the d-Ala-d-Lac ligase (VanA/B) and the d-Lac dehydrogenase (VanH) (28). A VanX-like d-Ala-d-Ala dipeptidase (Aad) was identified in L. plantarum and was shown to contribute to peptidoglycan precursor selectivity by eliminating the d-Ala-d-Ala dipeptides that are synthesized by the ddlLp ligase (18). To see whether this dipeptidase could play a VanX-like role by reprogramming peptidoglycan precursor synthesis in L. lactis (Fig. 1), Aad and ddlLp were co-produced in the wild type strain (NZ3900 (pGIM023)) as well as in a ddlLc mutant background (MD003). Equivalent strains were constructed for the co-expression of ddlLp and the VanXA gene from E. faecium BM4147 (N23900 (pGIM024) and MD004, respectively).
Low amounts of d-Ala-ended UDP-MurNac-pentapeptides (9%) were still detected in the cytoplasm of the NZ3900-derivative expressing ddlLp and VanXA (NZ3900 (pGIM024), Table 1), but they were totally undetectable in the strain co-expressing ddlLp and aad (NZ3900 (pGIM023)) and in the ddlLc mutant derivatives MD003 and MD004 (Table 1). This shows that the Aad dipeptidase of L. plantarum is at least as efficient as the enterococcal VanX dipeptidase in reorienting the biosynthesis of peptidoglycan precursors when expressed in a heterologous host. Growth of MD003 and MD004 was dependent on the presence of both nisin and d-Lac in the culture medium (Fig. 2B, data not shown), indicating that d-Lac-ended peptidoglycan precursors have become essential for cell wall synthesis in these two mutants.
Changing the Peptidoglycan Precursor Is Not Sufficient to Confer a High Level of Resistance to Vancomycin
Processing of UDP-MurNAc-pentadepsipeptides by the cell wall biosynthesis machinery was first examined by looking at the effect of modulating the pool of d-Ala/d-Lac-ended precursors on the level of resistance to vancomycin expressed by the different strains of L. lactis (Table 1). All the strains producing a mixture of d-Lac- and d-Ala-ended precursors remained sensitive to vancomycin as determined by the E-test method (Table 1, see “Experimental Procedures”). Intriguingly, the complete replacement of d-Ala-ended UDP-MurNAc-pentapeptides by d-Lac-ended UDP-MurNac-pentadepsipeptides only slightly increased the resistance level, giving minimal vancomycin inhibitory concentrations ranging from 3 to 24 μg/ml (Table 1). However, inspection of the plates after 24 h of further incubation revealed that all the primary mutant strains that produced UDP-MurNAc-pentadepsipeptides (except NZ3900 (pGIM022), which accumulated the lowest amount of these modified precursors), gave rise to isolated colonies in the inhibition zone of the E-test (Table 1, Fig. 3A). Population analysis performed by plating serial dilutions of a MD003 culture on increasing concentrations of vancomycin revealed that derivatives expressing higher levels of resistance accumulated during growth and up to completely dominated the cell population after ∼40 generations (Fig. 3B). No accumulation of vancomycin-resistant derivatives was observed for the wild type strain or for the MD006 reference strain exclusively producing d-Ala-ended peptidoglycan precursors (Fig. 3B, data not shown).
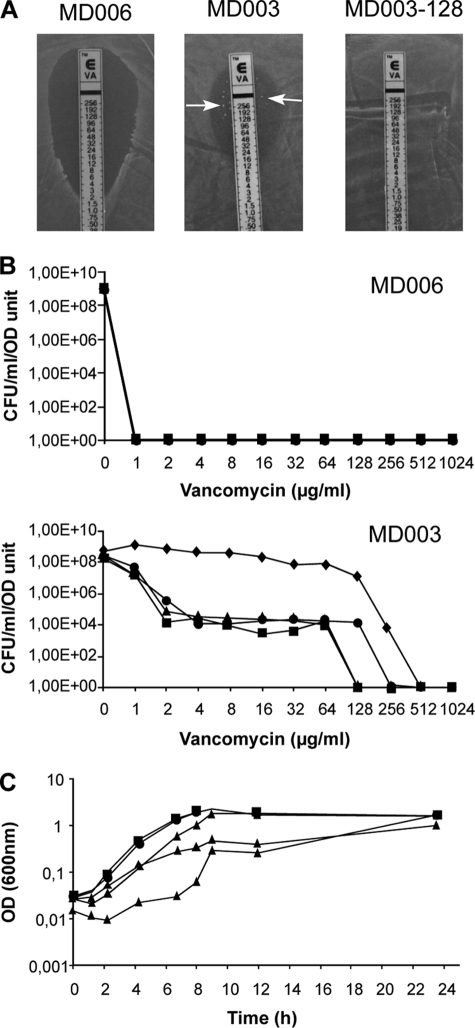
FIGURE 3.
Phenotypic adaptation to d-Lac-ended peptidoglycan precursors in L. lactis. A, shown are representative vancomycin antibiograms obtained for the control strain MD006, the primary mutant MD003, and its vancomycin-resistant derivative MD003-128 by the E-test method (see “Experimental Procedures”). Arrows indicate the presence of highly resistant colonies in the E-test inhibitory zone obtained for MD003. B, evolution of vancomycin resistance within cell populations of the MD006 (top) and MD003 (bottom) primary mutants is shown. Dilutions of serial cultures of both mutant strains were plated on increasing concentrations of vancomycin. Graphs express the number of colony forming units (CFU) per ml and A600 nm units of the cultures obtained at each vancomycin concentration after 10 (squares), 20 (circles), 30 (triangles), and 40 (diamonds) generations. C, shown is growth rate comparison between the MD003 primary mutant and its vancomycin-resistant derivative MD003-128. Three independent cultures of the control strain MD006, the vancomycin-resistant clone MD003-128, and the MD003 primary isolate were performed in M17-glucose supplemented with erythromycin (5 μg/ml), chloramphenicol (5 μg/ml), d-Lac (20 mm), and nisin (0.5 ng/ml). Growth of the cultures was followed by measuring the A600 nm as a function of time. For MD006 (squares) and MD003-128 (circles), mean values obtained for the three cultures are plotted. For MD003 (triangles), the growth curves obtained for the three independent cultures are shown.
MD003-derived clones that were recovered from the inhibition zone of the E-tests or from prolonged cultures showed high levels of vancomycin resistance, with MIC values exceeding 256 μg/ml (Fig. 3A). Their growth was dependent on the presence nisin and d-Lac in the culture medium, showing that peptidoglycan synthesis in these variants still relied on the activity of the exogenous ddlLp ligase (data not shown). To get further insight on the origin of the observed change in resistance level, plasmid DNA extracted from several independent sub-clones was back-introduced into NZ3900 cells, and the endogenous ddlLc ligase gene was then inactivated as described above for the initial MD003 mutant. This new generation of mutants reproduced the phenotype of the parental strain, giving rise to highly resistant cells emerging from a relatively low level of resistance background. This demonstrates that the increased vancomycin resistance displayed by the MD003 sub-clones did not result from a mutation affecting the ddlLp/aad expression vector but, rather, from some genetic and/or physiological adaptation that occurred during cultivation of the primary mutant so as to improve the fitness of the bacterial cells. Supporting this conclusion, the growth rate of “adapted” vancomycin-resistant sub-clones was indistinguishable from that of the control strain MD006, whereas independent cultures performed with the primary MD003 mutant showed delayed and sometimes multiphasic growth curves indicative of bacterial population heterogeneity (Fig. 3C).
To get further insight on the mechanism that led to this physiological adaptation of L. lactis cells, the complete genome of a selected vancomycin-resistant MD003 derivative (MD003-128) was sequenced by high throughput SOLiD sequencing (see “Experimental Procedures”. The result revealed six changes compared with the wild type strain MG1363, including two silent mutations (supplemental Table S2). Two of the sense mutations affected the amino acid sequence of the catabolite control protein A, a global metabolic regulator of L. lactis (29). The remaining two mutations were in a phage protein and in a protein of unknown function, respectively (supplemental Table S2). Thus, none of the specific changes found in the MD003-128 genome affected a gene known to be directly involved in cell wall biosynthesis.
d-Ala/d-Lac Substitution of the C Terminus of Peptidoglycan Peptide Precursors Leads to Major Modifications of Peptidoglycan Structure
To determine the effect of changing the precursors on peptidoglycan structure, muropeptides extracted from the cell wall of the MD003 primary mutant and the highly vancomycin-resistant MD003-128 derivative were analyzed by reverse-phase HPLC and MALDI-TOF mass spectrometry (14) (see “Experimental Procedures”). Intriguingly, peptidoglycan composition established for MD003-128 was essentially the same as that of MD003 (data not shown). This indicates that the adaptive process that led to increased vancomycin resistance in MD003-128 was not accompanied with further alterations of the peptidoglycan synthetic pathway than those already taking place in the parental strain MD003. In contrast, the muropeptide elution profile obtained for both mutants was strikingly different from that of the reference strain cultured under the same conditions, the latter being virtually identical to the profile reported for the parental L. lactis strain MG1363 (14) (Fig. 4, supplemental Table S3).
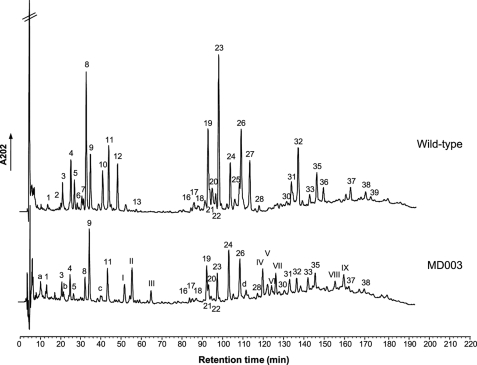
FIGURE 4.
Effect of changing the precursor on peptidoglycan structure. Peptidoglycan extracted from the reference strain PH8076 (pNZ8048) (top) and from the d-Lac-ended precursor-producing MD003 mutant (bottom) were digested with the mutanolysin muramidase and analyzed by HPLC as described under “Experimental Procedures.” The chemical structures assigned to the identified picks of muropeptides (numbered) are given in supplemental Table S3 and Table 2.
Consistent with the absence of d-Ala-containing precursors in the cytoplasm, the peptidoglycan of MD003 lacked the wild type muropeptide species containing d-Ala-ended pentapeptide side chains. They were replaced by six novel peaks showing the expected mass increase (1 mass unit) for the replacement of the C-terminal d-Ala residue by a d-Lac residue (Fig. 4, Table 2). The presence of d-Lac termini in the muropeptide assigned to the peak II was confirmed by tandem mass spectrometry (MS/MS) (data not shown). Three other novel species differed by 57 units from the mass of the disaccharide tripeptide substituted with a d-Asp cross-bridge residue and that of the two dimers generated from this monomer, respectively (Fig. 4; Table 2). MS/MS analysis assigned this mass difference to a glycine residue at the fourth position of the tetrapeptide side chain (Table 2, data not shown).
TABLE 2.
Molecular masses and proposed structures for muropeptides present (roman numbers) and absent (arabic numbers) from the peptidoglycan of the MD003 mutant strain
| Peaka | Mass (M+Na)+ |
Identificationc | |
|---|---|---|---|
| Observed | Calculatedb | ||
| 6 | NDd | 990.46 | Penta |
| 10 | NDd | 1105.49 | Penta-D |
| 12 | NDd | 1104.50 | Penta-N |
| I | 1020.52 | NAe | Tetra/Gly-D |
| II | 1106.48 | 1106.49 | Penta/d -Lac-D |
| III | 1105.55 | 1105.51 | Penta/d -Lac-N |
| 25 | NDd | 2097.95 | Penta-N-Tetra-Df |
| 27 | NDd | 2096.96 | Penta-N-Tetra-N |
| IV | 2098.95 | 2098.95 | Penta/d -Lac-N-Tetra-Df |
| V | 2012.89 | NAe | Tetra/Gly-N-Tetra-Df |
| VI | 2011.99 | NAe | Tetra/Gly-N-Tetra-N |
| VII | 2097.86 | 2097.97 | Penta/d -Lac-N-Tetra-N |
| 35b | NDd | 3090.42 | Penta-N-Tetra-N-Tetra-Df |
| 36 | NDd | 3089.44 | Penta-N-Tetra-N-Tetra-N |
| VIII | 3091.31 | 3091.42 | Penta/d -Lac-N-Tetra-N-Tetra-Df |
| IX | 3090.41 | 3090.44 | Penta/d -Lac-N-Tetra-N-Tetra-N |
| 39 | NDd | 4081.90 | Penta-N-Tetra-N-Tetra-N-Tetra-N |
a Peak numbers refer to Fig. 4.
b Sodiated molecular ions were the most abundant on MALDI-TOF mass spectra for all muropeptides. m/z values correspond to monoisotopic masses.
c Penta, disaccharide pentapeptide (l-Ala-d-iGln-l-Lys-d-Ala-d-Ala; iGln is iso-glutamine); Tetra, disaccharide tetrapeptide (l-Ala-d-iGln-l-Lys-d-Ala); Tetra/Gly, disaccharide tetrapeptide (l-Ala-d-iGln-l-Lys-Gly); Penta/d-Lac, disaccharide pentadepsipeptide (l-Ala-d-iGln-l-Lys-d-Ala-d-Lac); disaccharide, GlcNAc-MurNAc; D, d-Asp; N, d-Asn; iGln, α-amidated isoGlu (or γGlu).
d ND, not determined. MALDI-TOF masses were determined only for MD003; structures from the control peptidoglycan were assigned by comparing peaks with retention times observed for the corresponding muropeptides previously identified in the parental strain MG1363 (14).
e NA, not applicable.
f Positions of N and D in peptide chains of oligomeric forms are arbitrarily given.
Intriguingly, in addition to the formation of novel structures, the peptidoglycan of MD003 was characterized by a dramatic decrease in disaccharides with tripeptide side chains, mainly in favor of disaccharides tetrapeptide, whereas the ratio of disaccharides pentadepsipeptide was identical to the ratio of disaccharides pentapeptide measured in the control (supplemental Table S4). Muropeptide containing a disaccharide tripeptide at the acceptor position for the transpeptidation reaction were also under-represented among the dimers, trimmers, and tetramers, without modification of the degree of cross-linking (supplemental Table S4). Another important structural modification observed for MD003 was the lack of amidation of the d-Asp cross-bridge residue bound to the side chains (supplemental Table S5). Together, these data show that a d-Ala to d-Lac substitution at the C-terminal position of the peptide chain affects the activity of multiple enzymes involved in peptidoglycan assembly and processing, either directly or indirectly.
Utilization of Altered Peptidoglycan Precursors for Cell Wall Synthesis Induces Morphological Changes in L. lactis
Incorporation of peptidoglycan precursors in the cell wall of L. lactis strains expressing different ratios of UDP-MurNAc-pentadepsipeptides and different levels of vancomycin resistance was examined by fluorescence microscopy after staining with Van-FL, a fluorescent derivative of vancomycin. In bacteria that produce d-Ala-d-Ala-ended precursors, this approach is used to visualize biosynthetically active regions of the cell wall that are usually richer in d-Ala-d-Ala termini than mature peptidoglycan (25).
When wild type NZ3900 cells were stained with the Van-FL probe, fluorescence was mainly distributed at mid-cell and near the poles, indicating that these regions contain the highest concentrations in d-Ala-d-Ala termini. Compilation of cells according to cell cycle progression suggests that bright poles resulted from newly separated division septa, with fluorescence remaining apparent after cell constriction (Fig. 5A). Consistent with this persistence of fluorescence, ∼15% of L. lactis muropeptides contain intact, d-Ala-d-Ala-ended pentapeptide side chains that are not engaged in cross-linking reactions (14). This pattern is, thus, slightly different from that previously reported for S. pneumoniae where 100% of the pentapeptide side chains are processed during cell wall synthesis (25, 30).
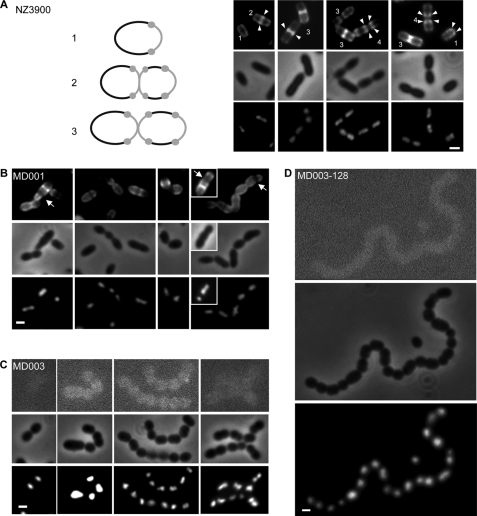
FIGURE 5.
Van-FL staining and morphology of L. lactis mutant strains expressing different proportions of d-Lac-ended versusd-Ala-ended peptidoglycan precursors. For each strain micrographs of representative cells visualized by Van-FL staining (upper panels) and 4′,6-diamidino-2-phenylindole staining (lower panels) are presented. Scale bars: 1 μm. A, wild type NZ3900 cells transformed with the empty expression vector pNZ8048 are shown. Cells are numbered according to cell cycle progression. Arrowheads show brighter fluorescence spots that appear at the division site and then separate at a later stage of cell division to occupy a polar position in the newborn cells. A schematic representation of the different stages of the cell cycle is shown on the left, with the Van-FL-stained zones and spots colored gray. B, cells of the MD001 mutant producing a mixture of d-Ala-ended and d-Lac-ended peptidoglycan precursors are shown. Arrows indicate morphological defects associated with dark areas not stained by the Van-FL probe. C, cells of the MD003 mutant exclusively producing d-Lac-ended peptidoglycan precursors are shown. Contrast of the fluorescence images was increased to visualize the position of the cell in the field. D, shown is a representative cell chain formed by the highly vancomycin resistant MD003 derivative MD003-128.
Van-FL staining was still observed for the MD001 strain producing both d-Ala- and d-Lac-ended precursors (Fig. 5B). However, the staining pattern appeared to be much more heterogeneous than that seen for the wild type strain, suggesting differential utilization and/or processing of the peptidoglycan precursors in different cells. A specific class of elongated cells (representing 22% of the cell population, n = 119) showed an asymmetrically positioned dark band dividing the cell into two unequal compartments (Fig. 5B). This morphological anomaly was sometimes associated with mis-positioned nucleoids as revealed by 4′,6-diamidino-2-phenylindole staining. On average, MD001 cells were slightly shorter and thicker than the wild type cells, giving them a swollen appearance (compare Fig. 5, A and B, Table 3). In addition, daughter cells often remained attached to each other after cell division, generating short chains that were not or rarely observed with the wild type strain (Fig. 5, supplemental Fig. S1).
TABLE 3.
Effect of peptidoglycan precursors modification on cell shape (length/width ratio)
| Strain | Length average ± ICa | Width average ± ICa | Length/Widthb |
|---|---|---|---|
| μm | μm | ||
| NZ3900 | 1.7 ± 0.05 | 0.7 ± 0.01 | 2.4 |
| MD006 | 1.9 ± 0.05 | 0.8 ± 0.02 | 2.4 |
| MD001 | 1.6 ± 0.05 | 0.8 ± 0.01 | 2.0 |
| MD003 | 1.5 ± 0.06 | 0.9 ± 0.02 | 1.6 |
| MD003–128 | 1.3 ± 0.04 | 1.0 ± 001 | 1.3 |
| BLD006 | 1.3 ± 0.04 | 1.0 ± 0.02 | 1.3 |
| VES2065 | 1.7 ± 0.06 | 0.97 ± 0.02 | 1.8 |
a n ≥ 200 cells. IC, interval of confidence, α = 0.01.
b Ratio of average cell length to average cell width.
These morphological alterations were found strongly accentuated in the MD003 mutant (Fig. 5C) and even further pronounced in the vancomycin-resistant derivative MD003-128 (Fig. 5D). These two strains were not stained by the Van-FL probe, consistent with the fact that d-Ala-d-Lac-ended precursors are the only substrates available for cell wall synthesis. However, MD003 and MD003-128 cells showed an increased tendency to adopt a spherical or almost spherical shape (Fig. 5, C and D) characterized by a reduced length/width ratio compared with the wild type cells (Table 3). The defect in cell separation was also more pronounced for MD003 and MD003-128, resulting in the formation of long and curly chains (Fig. 5 and supplemental Fig. S1). In contrast, the MD006 mutant expressing the d-Ala-d-Ala ligase of S. thermophilus produced normal and well individualized ovoid cells comparable with the wild type cells (data not shown). This demonstrates that the progressive alteration of cell shape and cell separation observed with the MD001 and MD003 (MD003-128) mutants is the direct consequence of an increased utilization of d-Lac-ended precursors for cell wall synthesis.
Implication of Penicillin-binding Proteins
Muropeptides analysis showed that a distinctive alteration of MD003 and MD003-128 peptidoglycan is the decrease in disaccharides with tripeptide side chains and the concomitant increase of disaccharides with tetrapeptide side chains. A similar enrichment of tetrapeptides at the expense of tripeptides was previously reported for a L. lactis mutant deficient for the l,d-carboxypeptidase DacB (VES2065) (14). This suggests that changing the nature of the peptidoglycan precursors may have altered the activity of L. lactis carboxypeptidases. Analysis of the L. lactis genome revealed the presence of two carboxypeptidases. In addition to DacB, L. lactis encodes DacA, a protein homologous to PBP3 from S. pneumoniae (31). A dacA mutant strain (BLD006) was constructed, and the morphology of both the dacA and dacB mutants was analyzed. The dacA mutant produced cells that were spherical or almost spherical as observed for MD003 (Table 3; Fig. 6), whereas dacB cells were thicker than the wild type cells and tended to remain associated into chains (Table 3; Fig. 6). Together these phenotypes are reminiscent of those observed for the MD003 and MD003-128 precursor mutants (Fig. 6; see also Fig. 5).
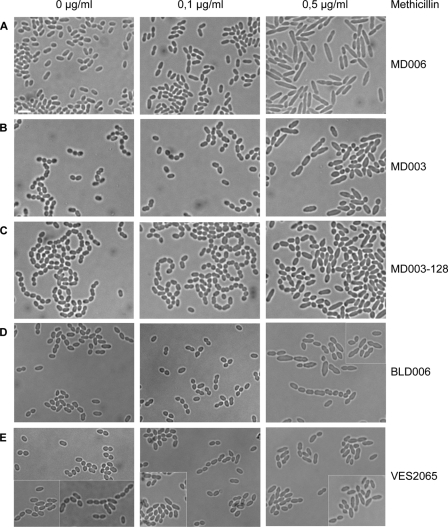
FIGURE 6.
Effect of methicillin on cell morphology. Exponentially growing cells of the control strain MD006 (A), the MD003 primary mutant (B), the highly vancomycin-resistant MD003-derived clone MD003-128, and the carboxypeptidase dacA (BLD006; D) and dacB (VES2065; E) mutants were incubated for 4h30 with the indicated concentrations of methicillin before be examined by light microscopy. Scale bar = 3μm.
To get further insight on the mechanism that led to the observed morphological defects of the mutant strains, bacterial cells were treated with methicillin, a β-lactam antibiotic known to specifically block PBPs involved in cell division (32). When incubated with increasing concentrations of methicillin, the wild type and control strain MD006 stopped dividing, generating elongated cells reminiscent of bacterial filaments (Fig. 6A). In contrast, cells of the MD003 and MD003-128 mutants remained more compact and started to swell to adopt a puffy phenotype (Fig. 6, B and C). A similar phenotype was observed with the dacA mutant and to a lower extent with the dacB mutant (Fig. 6, D and E). These results demonstrate that the cell elongation and division processes can be separated in L. lactis and that changing the peptidoglycan precursors affects cell elongation, presumably by perturbing the activity of specific PBPs, including carboxypeptidases.
DISCUSSION
This study reports the production of peptidoglycan precursors ending by d-Ala-d-Lac instead of d-Ala-d-Ala in L. lactis through the heterologous expression of genes involved in peptidoglycan synthesis from L. plantarum. Modification of the last residue of the stem peptide strongly affected cell wall peptidoglycan composition and cell morphology. Cells became spherical instead of ovoid and remained assembled into chains. Our data support the view that peptidoglycan composition plays a central role in the coordination of cell wall biosynthesis and processing enzymes.
Cell Wall Biosynthesis Reprogramming and Morphogenesis; Regulation by the Substrate
L. lactis mutants exclusively producing d-Lac-ended precursors are viable, showing that the cell wall synthesis machinery can accept the surrogate precursors. Analysis of the muropeptide composition of the MD003 mutant showed that all the d-Ala-ended pentapeptide side chains were replaced by an equivalent amount of pentadepsipeptide side chains ending by d-Ala-d-Lac. In addition, the level of cross-linking was not affected by the precursor alteration. These results show that modified stem peptides can efficiently be used as a donor but also as an acceptor for transpeptidation in L. lactis. By comparison, d-Ala-d-Lac termini were not detected in the peptidoglycan extracted from vancomycin-resistant strains of enterococci and S. aureus expressing the vanA/B gene cluster, although the level of cross-linking was unchanged (33–37). It was proposed that removing d-Lac from the peptide chains is necessary in these bacterial species to supply transpeptidases with an appropriate acceptor substrate, which seems not to be the case in L. lactis (33, 37).
Although the level of transpeptidation was not impaired in the mutant strains, changing the last residue of the peptidoglycan precursor peptide chain significantly modified the muropeptide composition of the bacterium. Several activities involved in the processing and/or exportation of the peptidoglycan precursors may be altered in the mutants, whereas structural changes in the muropeptides may have affected the activity of other enzymes acting at a later stage of peptidoglycan synthesis and maturation. We propose that altering one of these specific activities or the coordination of multiple components of the cell wall machinery is responsible for the morphological changes observed in the L. lactis precursor mutants.
A marked difference that was revealed by the muropeptide analysis is the decrease in amidation of the d-Asp cross-bridge in the MD003 mutant compared with wild type L. lactis. Although little is known on the physiological significance of peptidoglycan amidation during cell wall synthesis, the asparagine synthase (AsnH) responsible for d-Asp amidation was recently identified in L. lactis (17). Amidation is thought to occur after d-Asp incorporation into the peptidoglycan precursor by a d-Asp-specific ligase (16, 17). A d-Ala to d-Lac substitution at the fifth position of the stem peptide may, therefore, interfere with this reaction by affecting the structure of its substrate. However, the lack of amidation alone cannot explain the morphological defects that characterize the mutant strains expressing d-Lac-ended precursors, as an asnH null mutant that is completely deficient for d-Asp amidation exhibits normal cell shape.6
In contrast, disruption of the carboxypeptidase genes dacA and dacB markedly affected cell morphology producing phenotypes that phenocopied those of MD003 and MD003-128. Mutation in the dacB gene was shown to alter the ratio of tripeptide versus tetrapeptide disaccharides as we observed in the precursor mutants (14). The DacA protein of L. lactis is homologous to the PBP3 carboxypeptidase of S. pneumoniae. PBP3 was proposed to trim the last residue of pentapeptides all over the cell except in the equatorial zone. This activity is required to properly position high molecular weight PBPs in this region by providing them with the appropriate donor substrates for the transpeptidation reaction (11). As a consequence, disruption of PBP3 was found to alter the morphology of S. pneumoniae as we observed here for the dacA mutant of L. lactis (31). It is, thus, likely that changing the structure of the precursor altered the activity of peptidoglycan processing enzymes such as DacB and/or DacA carboxypeptidases. This may in turn have affected the recruitment of other PBPs. Heterogeneous distribution of carboxypeptidases and/or transpeptidases in the cell wall of L. lactis is supported by the Van-FL staining pattern obtained for the wild type strain, with some regions of the wall appearing more fluorescent than others at different stages of the cell cycle.
The morphological defects observed for the MD003 mutant of L. lactis are also reminiscent of those reported for S. thermophilus mutants deficient in the transpeptidase PBP2b or the transmembrane RodA protein, two enzymes thought to be specifically involved in cell elongation (38). We show here that cell elongation and cell division can be uncoupled in L. lactis by treating the cells with methicillin. Wild type bacteria in which cell division was blocked by the antibiotic sustained active longitudinal growth, giving rise to long filaments. This process was severely compromised in the MD003 and MD003-128 mutants, indicating that changing the nature of the peptidoglycan precursor specifically perturbed the cell elongation machinery. This may occur directly by affecting the activity of key enzymes of the elongation process (e.g. PBP2b) or indirectly by altering their recruitment within the cell wall (e.g. through DacA and/or DacB activity). Changing the substrate may also affected the activity and/or location of other peptidoglycan-processing enzymes, such as peptidoglycan hydrolases that are required for cell separation, as was reported in E. coli (13). Elucidation of the exact mechanism that is responsible for the observed morphological changes in the precursor mutants of L. lactis will undoubtedly provide new insight into how the different components of the cell wall biosynthesis machinery cooperate during cell morphogenesis in ovococci and how their activity is controlled by substrate processing during peptidoglycan synthesis.
Resistance to Vancomycin; Further Adaptation to Peptidoglycan Reprogramming Is Required
Key enzymes for specific d-Lac incorporation into the cell wall of L. plantarum include the d-lactate dehydrogenase LdhDLp, the d-Ala-d-Lac ligase ddlLp, and the d-Ala-d-Ala dipeptidase Aad (18, 19, 39). Unlike their enterococcal van homologues, the genes encoding these different activities are not clustered within an operon, and they appear to have a more fundamental role in cell physiology than being simply required for vancomycin resistance (18, 19, 39). We, therefore, proposed that genes functionally equivalent to those of L. plantarum were initially recruited by glycopeptide producers to protect themselves against the antibiotic and were subsequently transmitted to enterococci and other bacterial species to confer resistance to vancomycin (18). In support of this view, we show here that the co-expression of these metabolic genes in L. lactis allows peptidoglycan precursor synthesis to be remodeled, just as the expression of the vancomycin resistance vanA or vanB operons in enterococci.
However, an intriguing finding of this study is the fact that vancomycin resistance is not an immediate consequence of changing the precursor but the result of an adaptive process after its utilization for cell wall synthesis. Similar observations were made when the vanHAX resistance operon from E. faecium BM4147 was transferred in L. lactis (data not shown), indicating that the ability to develop vancomycin resistance depends on properties of the bacterial cells and not on the genes that were used to alter precursor biosynthesis. Consistent with this, the growth rate of vancomycin resistant MD003 derivatives was significantly improved when compared with that of the primary mutant, suggesting that cells were adapted to better utilize the new peptidoglycan substrate, thereby increasing their fitness. Sequencing of the MD003-128 complete genome revealed that this adaptation process did not directly affect enzymes specifically involved in peptidoglycan synthesis or cell wall processing (supplemental Table S2). However, two mutations were detected in catabolite control protein A, a pleiotropic regulator known to regulate multiple cellular processes including virulence and the level of antibiotics resistance in S. aureus (40). Thus, these data support the conclusion that phenotypic adaptation leading to high levels of vancomycin resistance resulted from global optimization of L. lactis cell wall biosynthetic machinery rather than from specific alterations of the peptidoglycan synthetic pathway. Also supporting this view is the finding that the muropeptide composition of the adapted MD003-128 strain is unchanged when compared with that of the mother strain MD003.
The impact that the nature of the precursor may have on peptidoglycan synthesis and cell morphology together with the requirement for further optimization of the biosynthesis machinery has an important and more general implication on vancomycin resistance. Not all bacteria may exhibit the same flexibility to cope with altered peptidoglycan precursors, and one may, therefore, anticipate that some species would be more prone to become resistant to vancomycin than others.
Supplementary Material
Acknowledgments
We warmly thank J. Delcour for helpful discussions and scientific advice. We are grateful to M. Arthur for the MS/MS determination of muropeptides I and II and to A. Marnef for the construction of the MD006 strain. We thank Dr. S. Kulakauskas for help in L. lactis MD003-128 sequencing and Dr. S. Kennedy and N. Galleron (MetaQuant sequencing platform) for preparing the amplicon libraries and for preliminary data analysis. Sequencing of MD003-128 was supported in part by the program AIP BioRessources 2009 of INRA (project SOLIMUT).
This work was supported by Fonds Spéciaux de la Recherche (Université Catholique de Louvain), Fonds National de la Recherche Scientifique, and Actions de Recherche Concertées (Communauté Française de Belgique). This work was also supported by a grant from the Biotechnology and Biological Sciences Research Council, Swindon, United Kingdom, and a short-term European Molecular Biology Organization fellowship (to M. D.).

The on-line version of this article (available at http://www.jbc.org) contains supplemental Tables S1–S5 and Fig. 1.
P. Hols, unpublished data.
M. Deghorain, unpublished data.
- MurNAc
- N-acetylmuramic acid
- GlcNAc
- N-acetylglucosamine
- PBP
- penicillin-binding protein
- MIC
- minimal inhibitory concentration
- Van-FL
- vancomycin BODIPY-FL conjugate
- ddlLc
- d-Ala-d-Ala ligase gene from L. lactis subsp. cremoris
- ddlLp
- d-Ala-d-Lac ligase gene from L. plantarum
- ddlASt
- d-Ala-d-Lac ligase gene from S. thermophilus
- ldhDLh
- d-Lac dehydrogenase gene from L. helveticus
- aad
- d-Ala-d-Ala dipeptidase gene from L. plantarum
- VanXA
- d-Ala-d-Ala dipeptidase gene from E. faecium
- HPLC
- high pressure liquid chromatography
- MALDI-TOF
- matrix-assisted laser desorption ionization time-of-flight
- ORF
- open reading frame
- MS
- mass spectroscopy
- d-Lac
- d-lactate.
REFERENCES
- 1.Barreteau H., Kovac A., Boniface A., Sova M., Gobec S., Blanot D. (2008) FEMS Microbiol. Rev. 32, 168–207 [DOI] [PubMed] [Google Scholar]
- 2.Sauvage E., Kerff F., Terrak M., Ayala J. A., Charlier P. (2008) FEMS Microbiol. Rev. 32, 234–258 [DOI] [PubMed] [Google Scholar]
- 3.Carballido-López R., Formstone A. (2007) Curr. Opin. Microbiol. 10, 611–616 [DOI] [PubMed] [Google Scholar]
- 4.den Blaauwen T., de Pedro M. A., Nguyen-Distèche M., Ayala J. A. (2008) FEMS Microbiol. Rev. 32, 321–344 [DOI] [PubMed] [Google Scholar]
- 5.Höltje J. V. (1998) Microbiol. Mol. Biol. Rev. 62, 181–203 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Vollmer W., Joris B., Charlier P., Foster S. (2008) FEMS Microbiol. Rev. 32, 259–286 [DOI] [PubMed] [Google Scholar]
- 7.Zapun A., Vernet T., Pinho M. G. (2008) FEMS Microbiol. Rev. 32, 345–360 [DOI] [PubMed] [Google Scholar]
- 8.Cabeen M. T., Jacobs-Wagner C. (2007) J. Cell Biol. 179, 381–387 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Scheffers D. J., Pinho M. G. (2005) Microbiol. Mol. Biol. Rev. 69, 585–607 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Begg K. J., Takasuga A., Edwards D. H., Dewar S. J., Spratt B. G., Adachi H., Ohta T., Matsuzawa H., Donachie W. D. (1990) J. Bacteriol. 172, 6697–6703 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Morlot C., Noirclerc-Savoye M., Zapun A., Dideberg O., Vernet T. (2004) Mol. Microbiol. 51, 1641–1648 [DOI] [PubMed] [Google Scholar]
- 12.Pinho M. G., Errington J. (2005) Mol. Microbiol. 55, 799–807 [DOI] [PubMed] [Google Scholar]
- 13.Priyadarshini R., Popham D. L., Young K. D. (2006) J. Bacteriol. 188, 5345–5355 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Courtin P., Miranda G., Guillot A., Wessner F., Mézange C., Domakova E., Kulakauskas S., Chapot-Chartier M. P. (2006) J. Bacteriol. 188, 5293–5298 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Schleifer K. H., Kandler O. (1972) Bacteriol. Rev. 36, 407–477 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Veiga P., Piquet S., Maisons A., Furlan S., Courtin P., Chapot-Chartier M. P., Kulakauskas S. (2006) Mol. Microbiol. 62, 1713–1724 [DOI] [PubMed] [Google Scholar]
- 17.Veiga P., Erkelenz M., Bernard E., Courtin P., Kulakauskas S., Chapot-Chartier M. P. (2009) J. Bacteriol. 191, 3752–3757 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Deghorain M., Goffin P., Fontaine L., Mainardi J. L., Daniel R., Errington J., Hallet B., Hols P. (2007) J. Bacteriol. 189, 4332–4337 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Goffin P., Deghorain M., Mainardi J. L., Tytgat I., Champomier-Vergès M. C., Kleerebezem M., Hols P. (2005) J. Bacteriol. 187, 6750–6761 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hols P., Kleerebezem M., Schanck A. N., Ferain T., Hugenholtz J., Delcour J., de Vos W. M. (1999) Nat. Biotechnol. 17, 588–592 [DOI] [PubMed] [Google Scholar]
- 21.Arthur M., Molinas C., Courvalin P. (1992) J. Bacteriol. 174, 2582–2591 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.van Kranenburg R., Marugg J. D., van Swam I. I., Willem N. J., de Vos W. M. (1997) Mol. Microbiol. 24, 387–397 [DOI] [PubMed] [Google Scholar]
- 23.de Ruyter P. G., Kuipers O. P., de Vos W. M. (1996) Appl. Environ. Microbiol. 62, 3662–3667 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Arbeloa A., Hugonnet J. E., Sentilhes A. C., Josseaume N., Dubost L., Monsempes C., Blanot D., Brouard J. P., Arthur M. (2004) J. Biol. Chem. 279, 41546–41556 [DOI] [PubMed] [Google Scholar]
- 25.Daniel R. A., Errington J. (2003) Cell 113, 767–776 [DOI] [PubMed] [Google Scholar]
- 26.Wegmann U., O'Connell-Motherway M., Zomer A., Buist G., Shearman C., Canchaya C., Ventura M., Goesmann A., Gasson M. J., Kuipers O. P., van Sinderen D., Kok J. (2007) J. Bacteriol. 189, 3256–3270 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Li H., Ruan J., Durbin R. (2008) Genome Res. 18, 1851–1858 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Arthur M., Depardieu F., Reynolds P., Courvalin P. (1996) Mol. Microbiol. 21, 33–44 [DOI] [PubMed] [Google Scholar]
- 29.Zomer A. L., Buist G., Larsen R., Kok J., Kuipers O. P. (2007) J. Bacteriol. 189, 1366–1381 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ng W. L., Kazmierczak K. M., Winkler M. E. (2004) Mol. Microbiol. 53, 1161–1175 [DOI] [PubMed] [Google Scholar]
- 31.Schuster C., Dobrinski B., Hakenbeck R. (1990) J. Bacteriol. 172, 6499–6505 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lleo M. M., Canepari P., Satta G. (1990) J. Bacteriol. 172, 3758–3771 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.de Jonge B. L., Handwerger S., Gage D. (1996) Antimicrob. Agents Chemother. 40, 863–869 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bouhss A., Josseaume N., Severin A., Tabei K., Hugonnet J. E., Shlaes D., Mengin-Lecreulx D., Van Heijenoort J., Arthur M. (2002) J. Biol. Chem. 277, 45935–45941 [DOI] [PubMed] [Google Scholar]
- 35.de Jonge B. L., Gage D., Handwerger S. (1996) Microb. Drug Resist. 2, 225–229 [DOI] [PubMed] [Google Scholar]
- 36.Billot-Klein D., Shlaes D., Bryant D., Bell D., van Heijenoort J., Gutmann L. (1996) Biochem. J. 313, 711–715 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Severin A., Tabei K., Tenover F., Chung M., Clarke N., Tomasz A. (2004) J. Biol. Chem. 279, 3398–3407 [DOI] [PubMed] [Google Scholar]
- 38.Thibessard A., Fernandez A., Gintz B., Leblond-Bourget N., Decaris B. (2002) J. Bacteriol. 184, 2821–2826 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ferain T., Hobbs J. N., Jr., Richardson J., Bernard N., Garmyn D., Hols P., Allen N. E., Delcour J. (1996) J. Bacteriol. 178, 5431–5437 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Seidl K., Stucki M., Ruegg M., Goerke C., Wolz C., Harris L., Berger-Bächi B., Bischoff M. (2006) Antimicrob. Agents Chemother. 50, 1183–1194 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.