This manuscript demonstrates for the first time that P-body (PB) formation in response to stress is a regulated process, and that at least two different pathways drive PB assembly. We provide evidence that PB formation and translation attenuation are not strictly linked.
Abstract
mRNA is sequestered and turned over in cytoplasmic processing bodies (PBs), which are induced by various cellular stresses. Unexpectedly, in Saccharomyces cerevisiae, mutants of the small GTPase Arf1 and various secretory pathway mutants induced a significant increase in PB number, compared with PB induction by starvation or oxidative stress. Exposure of wild-type cells to osmotic stress or high extracellular Ca2+ mimicked this increase in PB number. Conversely, intracellular Ca2+-depletion strongly reduced PB formation in the secretory mutants. In contrast to PB induction through starvation or osmotic stress, PB formation in secretory mutants and by Ca2+ required the PB components Pat1 and Scd6, and calmodulin, indicating that different stressors act through distinct pathways. Consistent with this hypothesis, when stresses were combined, PB number did not correlate with the strength of the translational block, but rather with the type of stress encountered. Interestingly, independent of the stressor, PBs appear as spheres of ∼40–100 nm connected to the endoplasmic reticulum (ER), consistent with the idea that translation and silencing/degradation occur in a spatially coordinated manner at the ER. We propose that PB assembly in response to stress occurs at the ER and depends on intracellular signals that regulate PB number.
INTRODUCTION
Cells adapt to stress by varying their proteome. These changes in protein expression can be achieved through transcriptional and translational control or through changes in protein stability. Stress causes attenuation of general translation, whereas the translation of a subset of mRNAs is up-regulated. Many mRNAs are sequestered in processing bodies or P-bodies (PBs) or stress granules (SGs) in response to stress. In SGs, mRNAs are stored until the stress is alleviated and the mRNAs can return to the cytosol (Coller and Parker, 2005). PBs on the other hand, are sites of mRNA storage and turnover. The stress conditions that result in either PB or SG formation are only partially overlapping. Recent evidence suggests that PB formation could precede stress granule formation, and PBs could mature either into stress granules or into mRNA-degrading PBs (Buchan et al., 2008). However, SGs could potentially also form independently from PBs. Although the mechanism of SG assembly still remains elusive, more is known about PB assembly. According to the current model, two separate complexes bind the mRNA: the decapping complex at the 5′ end and the Lsm–Pat1 complex at the 3′ end of the mRNAs, to promote interaction between different mRNPs to allow PB assembly (Decker et al., 2007; Franks and Lykke-Andersen, 2008; Reijns et al., 2008). Thus, loss of one complex may reduce the efficiency with which PBs are formed. In yeast, the decapping complex contains the decapping proteins Dcp1 and Dcp2 and the decapping promoting factor Edc3. The 3′-binding Lsm–Pat1 complex consists of Sm and Sm-like (Lsm) proteins, which form two heptameric rings that encircle the RNA (Salgado-Garrido et al., 1999) and to which the decapping activator Pat1 is recruited. The Lsm–Pat1 complex shows an inherent affinity to deadenylated mRNA sequences (Bouveret et al., 2000; Tharun et al., 2000, 2005; Tharun and Parker, 2001; Chowdhury et al., 2007). PB formation seems to correlate with defects in translation initiation, whereas translation elongation problems do not cause PBs to form (Eulalio et al., 2007; Parker and Sheth, 2007). Despite what is known about PB assembly, it is still debated if PBs are merely aggregations of “unused” mRNA or whether PB assembly is regulated through distinct signals.
Components of the secretory pathway are responsible for the transport of proteins and lipids between cellular compartments as well as to the cell surface. Intracellular trafficking components have been implicated in mRNA transport (Aronov and Gerst, 2004; Trautwein et al., 2004; Bi et al., 2007). Interestingly, Deloche et al. (2004) showed that in yeast at least a subset of secretory transport mutants failed to properly initiate translation. This translation attenuation is likely a consequence of membrane stress caused by blocks along the secretory pathway. Given that blocked translation initiation can lead to PB formation (Eulalio et al., 2007; Parker and Sheth, 2007), we asked whether mutants in the secretory pathway also promote PB formation in S. cerevisiae. We found that many more PBs were formed in secretory transport mutants than after induction of PBs upon starvation in wild-type cells. The PBs observed in secretory mutants and upon starvation were indistinguishable by size or by Dcp2-myc and Dhh1-myc content, as judged by immunoelectron microscopy. Dcp2-myc and Dhh1-myc formed sphere-like structures 40–100 nm in diameter. The multiple PB phenotype was also induced in wild-type cells by the application of hyperosmotic shock or by increasing extracellular Ca2+ levels. The induction of numerous PBs in response to membrane stress required functional calmodulin and the PB components Scd6 and Pat1. Interestingly, mutations in calmodulin or deletion of PAT1, or SCD6, did not interfere with PB induction through starvation or hyperosmotic shock. The effect of inducing PBs under starvation and Ca2+ was additive because the block in translation initiation was much stronger when stresses were combined, but nevertheless multiple PBs were induced. Our results demonstrate that distinct signaling pathways are in place to induce PB production, depending on the stress encountered, and hence PB formation is more than just the mere consequence of a block in translation initiation. In this study we uncover that PB assembly is not the result of aggregation, but is induced through distinct pathways, one of which requires calmodulin.
MATERIALS AND METHODS
Yeast Methods
Standard genetic techniques were used throughout (Sherman, 1991). All modifications were carried out chromosomally, with the exceptions listed below. Chromosomal tagging and deletions were performed as described in Knop et al. (1999) and Gueldener et al. (2002). The temperature-sensitive alleles sec21-1, sec27-1, and cmd1-3 were transferred into the YPH499 or NYY0-1 background according to Erdeniz et al. (1997). The cmd1-3 allele was introduced into the arf1-11 background using plasmid pHS47 (URA3) that was kindly provided by E. Schiebel (University of Heidelberg, Heidelberg, Germany). After plasmid transformation the wild-type allele CMD1 was deleted on the chromosome. The sec6-4 allele was integrated into the SEC6 locus using the pRS406 vector (Sikorski and Hieter, 1989). The sar1-D32G allele was introduced into YPH499 on plasmid pMYY3–1 (TRP1) that was a gift from A. Nakano (RIKEN, Saitama, Japan). For ER costaining, cells were transformed with pSM1959 (LEU2) or pSM1960 (URA3), high-copy plasmids carrying Sec63-RFP, which were kindly provided by S. Michaelis (Johns Hopkins University, Baltimore, MD). Strains used are listed in Table 1.
Table 1.
Yeast strains
| Strain | Designation | Genotype | Reference |
|---|---|---|---|
| YPH499 | WT | MAT a ade2-101 his3-200 leu2-1 lys2-801 trp-63 ura3-52 | Sikorski and Hieter (1989) |
| NYY0-1 | ARF1 | MAT a ade2::ARF1::ADE2 arf1::HIS3 arf2::HIS3 ura3 lys2 trp1 his3 leu2 | Yahara et al. (2001) |
| NYY11-1 | arf1-11 | MAT a ade2::arf1-11::ADE2 arf1::HIS3 arf2::HIS3 ura3 lys2 trp1 his3 leu2 | Yahara et al. (2001) |
| NYY17-1 | arf1-17 | MAT a ade2::arf1-17::ADE2 arf1::HIS3 arf2::HIS3 ura3 lys2 trp1 his3 leu2 | Yahara et al. (2001) |
| NYY18-1 | arf1-18 | MAT a ade2::arf1-18::ADE2 arf1::HIS3 arf2::HIS3 ura3 lys2 trp1 his3 leu2 | Yahara et al. (2001) |
| YAS1031A | ARF1 Dcp2-GFP | MAT a ade2::ARF1::ADE2 arf1::HIS3 arf2::HIS3 ura3 lys2 trp1 his3 leu2 DCP2::DCP2-yEGFP-kanMX4 | This study |
| YAS1032A | arf1-11 Dcp2-GFP | MAT a ade2::arf1-11::ADE2 arf1::HIS3 arf2::HIS3 ura3 lys2 trp1 his3 leu2 DCP2::DCP2-yEGFP-kanMX4 | This study |
| YAS1033A | arf1-17 Dcp2-GFP | MAT a ade2::arf1-17::ADE2 arf1::HIS3 arf2::HIS3 ura3 lys2 trp1 his3 leu2 DCP2::DCP2-yEGFP-kanMX4 | This study |
| YAS1034A | arf1-18 Dcp2-GFP | MAT a ade2::arf1-18::ADE2 arf1::HIS3 arf2::HIS3 ura3 lys2 trp1 his3 leu2 DCP2::DCP2-yEGFP-kanMX4 | This study |
| YAS2428 | ARF1 Edc3-eqFP611 eIFG2-GFP | MAT a ade2::ARF1::ADE2 arf1::HIS3 arf2::HIS3 ura3 lys2 trp1 his3 leu2 EDC3::EDC3-eqFP611-kanMX4 eIFG2::eIFG2-yEGFP-TRP1 (K. lactis) | This study |
| YAS2429 | arf1-11 Edc3-eqFP611 eIFG2-GFP | MAT a ade2::arf1-11::ADE2 arf1::HIS3 arf2::HIS3 ura3 lys2 trp1 his3 leu2 EDC3::EDC3-eqFP611-kanMX4 eIFG2::eIFG2-yEGFP-TRP1 (K. lactis) | This study |
| YAS1681 | sar1-D32G Dcp2-GFP | MAT a ade2-101 his3-200 leu2-1 lys2-801 trp-63 ura3-52 SAR1::LEU2 (K. lactis) pMYY3-1(Ycp[sar1-D32G TRP1]) DCP2::DCP2-yEGFP-kanMX4 | This study |
| YAS1133 | sec21-1 Dcp2-GFP | MAT α ade2-101 his3-200 leu2-1 lys2-801 trp-63 ura3-52 sec21-1 DCP2::DCP2-yEGFP-kanMX4 | This study |
| YAS1134 | sec27-1 Dcp2-GFP | MAT a ade2-101 his3-200 leu2-1 lys2-801 trp-63 ura3-52 sec27-1 DCP2::DCP2-yEGFP-kanMX4 | This study |
| YAS1131 | Δgea2 gea1-19 Dcp2-GFP | MAT a ura3-52 leu2,3-112 his3-200 GEA2::HIS3 gea1-19 DCP2::DCP2-yEGFP-kanMX4 | This study |
| YAS1135 | sec6-4 Dcp2-GFP | MAT a ade2-101 his3-200 leu2-1 lys2-801 trp-63 ura3-52 sec6-4 CHS6::CHS6-9myc-TRP1 (K. lactis) DCP2::DCP2-yEGFP-kanMX4 | This study |
| YAS1877 | sec3-2 Dcp2-GFP | MAT a ura3 leu2 trp1 sec3-2 DCP2::DCP2-yEGFP-kanMX4 | This study |
| YAS1880 | sec4-8 Dcp2-GFP | MAT a ura3 leu2 lys sec4-8 DCP2::DCP2-yEGFP-kanMX4 | This study |
| YAS1878 | sec2-41 Dcp2-GFP | MAT a ura3 leu2 his3 lys2 trp1 ade2 sec2-41 DCP2::DCP2-yEGFP-kanMX4 | This study |
| YAS2235 | ARF1 Pub1-GFP | MAT a ade2::ARF1::ADE2 arf1::HIS3 arf2::HIS3 ura3 lys2 trp1 his3 leu2 PUB1::PUB1-yEGFP-kanMX4 | This study |
| YAS1945 | ARF1 Dcp2-GFP Δhog1 | MAT a ade2::ARF1::ADE2 ARF1::HIS3 ARF2::HIS3 ura3 lys2 trp1 his3 leu2 DCP2::DCP2-yEGFP-kanMX4 HOG::LEU2 (K . lactis) | This study |
| YAS1946 | arf1-11 Dcp2-GFP Δhog1 | MAT a ade2::arf1-11::ADE2 ARF1::HIS3 ARF2::HIS3 ura3 lys2 trp1 his3 leu2 DCP2::DCP2-yEGFP-kanMX4 HOG::LEU2 (K. lactis) | This study |
| YAS1685 | ARF1 Dcp2-GFP Δsho1 | MAT a ade2::ARF1::ADE2 ARF1::HIS3 ARF2::HIS3 ura3 lys2 trp1 his3 leu2 DCP2::DCP2-yEGFP-kanMX4 SHO1::LEU2 (K. lactis) | This study |
| YAS1686 | arf1-11 Dcp2-GFP Δsho1 | MAT a ade2::arf1-11::ADE2 ARF1::HIS3 ARF2::HIS3 ura3 lys2 trp1 his3 leu2 DCP2::DCP2-yEGFP-kanMX4 SHO1::LEU2 (K. lactis) | This study |
| YAS1947 | ARF1 Dcp2-GFP Δsko1 | MAT a ade2::ARF1::ADE2 ARF1::HIS3 ARF2::HIS3 ura3 lys2 trp1 his3 leu2 DCP2::DCP2-yEGFP-kanMX4 SKO1::LEU2 (K. lactis) | This study |
| YAS1948 | arf1-11 Dcp2-GFP Δsko1 | MAT a ade2::arf1-11::ADE2 ARF1::HIS3 ARF2::HIS3 ura3 lys2 trp1 his3 leu2 DCP2::DCP2-yEGFP-kanMX4 SKO1::LEU2 (K. lactis) | This study |
| YAS1967 | ARF1 Dcp2-GFP Δslt2 | MAT a ade2::ARF1::ADE2 ARF1::HIS3 ARF2::HIS3 ura3 lys2 trp1 his3 leu2 DCP2::DCP2-yEGFP-kanMX4 SLT2::LEU2 (K. lactis) | This study |
| YAS1968 | arf1-11 Dcp2-GFP Δslt2 | MAT a ade2::arf1-11::ADE2 ARF1::HIS3 ARF2::HIS3 ura3 lys2 trp1 his3 leu2 DCP2::DCP2-yEGFP-kanMX4 SLT2::LEU2 (K. lactis) | This study |
| YAS1986 | ARF1 Dcp2-GFP Δhog1 Δslt2 | MAT a ade2::ARF11::ADE2 ARF1::HIS3 ARF2::HIS3 ura3 lys2 trp1 his3 leu2 DCP2::DCP2-yEGFP-kanMX4 HOG1::LEU2 (K. lactis) SLT2::URA3 (K. lactis) | This study |
| YAS1987 | arf1-11 Dcp2-GFP Δhog1 Δslt2 | MAT a ade2::arf1-11::ADE2 ARF1::HIS3 ARF2::HIS3 ura3 lys2 trp1 his3 leu2 DCP2::DCP2-yEGFP-kanMX4 HOG1::LEU2 (K. lactis) SLT2::URA3 (K. lactis) | This study |
| YAS2092 | ARF1 Dcp2-GFP Δhog1 Δslt2 Δcnb1 | MAT a ade2::ARF1::ADE2 ARF1::HIS3 ARF2::HIS3 ura3 lys2 trp1 his3 leu2 DCP2::DCP2-yEGFP-kanMX4 HOG1::loxP SLT2::loxP CNB1::LEU2 (K. lactis) | This study |
| YAS2292 | arf1-11 Dcp2-GFP Δhog1 Δslt2 Δcnb1 | MAT a ade2::arf1-11::ADE2 ARF1::HIS3 ARF2::HIS3 ura3 lys2 trp1 his3 leu2 DCP2::DCP2-yEGFP-kanMX4 HOG1::loxP SLT2::loxP CNB1::LEU2 (K. lactis) | This study |
| YAS2575 | ARF1 Dcp2-GFP Δypk1 | MAT a ade2::ARF1::ADE2 ARF1::HIS3 ARF2::HIS3 ura3 lys2 trp1 his3 leu2 DCP2::DCP2-yEGFP-kanMX4 YPK1::LEU2 (K. lactis) | This study |
| YAS2010 | arf1-11 Dcp2-GFP Δypk1 | MAT a ade2::arf1-11::ADE2 ARF1::HIS3 ARF2::HIS3 ura3 lys2 trp1 his3 leu2 DCP2::DCP2-yEGFP-kanMX4 YPK1::LEU2 (K. lactis) | This study |
| YAS2021 | ARF1 Dcp2-GFP Δcnb1 | MAT a ade2::ARF11::ADE2 ARF1::HIS3 ARF2::HIS3 ura3 lys2 trp1 his3 leu2 DCP2::DCP2-yEGFP-kanMX4 CNB1::LEU2 (K. lactis) | This study |
| YAS2051 | arf1-11 Dcp2-GFP Δcnb1 | MAT a ade2::arf1-11::ADE2 ARF1::HIS3 ARF2::HIS3 ura3 lys2 trp1 his3 leu2 DCP2::DCP2-yEGFP-kanMX4 CNB1::LEU2 (K. lactis) | This study |
| YAS2088 | ARF1 Dcp2-GFP Δcmk1 Δcmk2 | MAT a ade2::ARF11::ADE2 ARF1::HIS3 ARF2::HIS3 ura3 lys2 trp1 his3 leu2 DCP2::DCP2-yEGFP-kanMX4 CMK1::LEU2 (K. lactis) CMK2::URA3 (K. lactis) | This study |
| YAS2089 | arf1-11 Dcp2-GFP Δcmk1 Δcmk2 | MAT a ade2::arf1-11::ADE2 ARF1::HIS3 ARF2::HIS3 ura3 lys2 trp1 his3 leu2 DCP2::DCP2-yEGFP-kanMX4 CMK1::LEU2 (K. lactis) CMK2::URA3 (K. lactis) | This study |
| YAS2090 | ARF1 Dcp2-GFP Δcmk1 Δcmk2 Δcnb1 | MAT a ade2::ARF1::ADE2 ARF1::HIS3 ARF2::HIS3 ura3 lys2 trp1 his3 leu2 DCP2::DCP2-yEGFP-kanMX4 CMK1::loxP CMK2::loxP CNB1::LEU2 (K. lactis) | This study |
| YAS2091 | arf1-11 Dcp2-GFP Δcmk1 Δcmk2 Δcnb1 | MAT a ade2::arf1-11::ADE2 ARF1::HIS3 ARF2::HIS3 ura3 lys2 trp1 his3 leu2 DCP2::DCP2-yEGFP-kanMX4 CMK1::loxP CMK2::loxP CNB1::LEU2 (K. lactis) | This study |
| YAS2586 | arf1-11 Dcp2-GFP Δgcn2 | MAT a ade2::arf1-11::ADE2 arf1::HIS3 arf2::HIS3 ura3 lys2 trp1 his3 leu2 DCP2::DCP2-yEGFP-kanMX4 GCN2::URA3 (K. lactic) | This study |
| YAS2588 | arf1-11 Dcp2-GFP Δire1 | MAT a ade2::arf1-11::ADE2 arf1::HIS3 arf2::HIS3 ura3 lys2 trp1 his3 leu2 DCP2::DCP2-yEGFP-kanMX4 IRE1::URA3 (K. lactic) | This study |
| YAS2296 | ARF1 cmd1-3 Dcp2-GFP | MAT a ade2::ARF1::ADE2 ARF1::HIS3 ARF2::HIS3 ura3 lys2 trp1 his3 leu2 DCP2::DCP2-yEGFP-kanMX4 CMD1::cmd1-3 | This study |
| YAS2580 | arf1-11 cmd1-3 Dcp2-GFP | MAT a ade2::arf1-11::ADE2 arf1::HIS3 arf2::HIS3 ura3 lys2 trp1 his3 leu2 DCP2::DCP2-yEGFP-kanMX4 | This study |
| pHS47(cmd1-3 URA3) CMD1::kanMX4 | |||
| YAS2294 | ARF1 Dcp2-GFP Δpat1 | MAT a ade2::ARF1::ADE2 ARF1::HIS3 ARF2::HIS3 ura3 lys2 trp1 his3 leu2 DCP2::DCP2-yEGFP-kanMX4 PAT1::LEU2 (K. lactis) | This study |
| YAS2295 | arf1-11 Dcp2-GFP Δpat1 | MAT a ade2::arf1-11::ADE2 ARF1::HIS3 ARF2::HIS3 ura3 lys2 trp1 his3 leu2 DCP2::DCP2-yEGFP-kanMX4 PAT1::LEU2 (K. lactis) | This study |
| YAS2297 | sec6-4 Dcp2-GFP Δpat1 | MAT a ade2-101 his3-200 leu2-1 lys2-801 trp-63 ura3-52 sec6-4 CHS6::CHS6-9myc-TRP1 (K. lactis) DCP2::DCP2-yEGFP-kanMX4 PAT1::LEU2 (K. lactis) | This study |
| YAS1097 | ARF1 Dcp2-GFP Δscd6 | MAT a ade2::ARF1::ADE2 arf1::HIS3 arf2::HIS3 ura3 lys2 trp1 his3 leu2 DCP2::DCP2-yEGFP-kanMX4 SCD6::LEU2 (K. lactis) | This study |
| YAS1098 | arf1-11 Dcp2-GFP Δscd6 | MAT a ade2::arf1-11::ADE2 arf1::HIS3 arf2::HIS3 ura3 lys2 trp1 his3 leu2 DCP2::DCP2-yEGFP-kanMX4 SCD6::LEU2 (K. lactis) | This study |
| YAS2500 | sec6-4 Dcp2-GFP Δscd6 | MAT a ade2-101 his3-200 leu2-1 lys2-801 trp-63 ura3-52 sec6-4 CHS6::CHS6-9myc-TRP1 (K. lactis) DCP2::DCP2-yEGFP-kanMX4 SCD6::LEU2 (K. lactis) | This study |
| YAS1294 | ARF1 Dcp2-9myc | MAT a ade2::ARF1::ADE2 arf1::HIS3 arf2::HIS3 ura3 lys2 trp1 his3 leu2 DCP2::DCP2-9myc-TRP1 (K. lactis) | This study |
| YAS1295 | arf1-11 Dcp2-9myc | MAT a ade2::arf1-11::ADE2 arf1::HIS3 arf2::HIS3 ura3 lys2 trp1 his3 leu2 DCP2::DCP2-9myc-TRP1 (K. lactis) | This study |
| YAS1693 | ARF1 Dhh1-9myc | MAT a ade2::ARF1::ADE2 ARF1::HIS3 ARF2::HIS3 ura3 lys2 trp1 his3 leu2 DHH1::DHH1-9myc-TRP1 (K. lactis) | This study |
| YAS1694 | arf1-11 Dhh1-9myc | MAT a ade2::arf1-11::ADE2 ARF1::HIS3 ARF2::HIS3 ura3 lys2 trp1 his3 leu2 DHH1::DHH1-9myc-TRP1 (K. lactis) | This study |
| YAS2576 | arf1-11 Pat1-GFP | MAT a ade2::arf1-11::ADE2 arf1::HIS3 arf2::HIS3 ura3 lys2 trp1 his3 leu2 PAT1::PAT1-yEGFP-TRP1 (K. lactis) | This study |
| YAS2578 | arf1-11 Scd6-GFP | MAT a ade2::arf1-11::ADE2 arf1::HIS3 arf2::HIS3 ura3 lys2 trp1 his3 leu2 SCD6::SCD6-yEGFP-kanMX4 | This study |
| YAS1153 | ARF1 Dhh1-GFP | MAT a ade2::ARF1::ADE2 arf1::HIS3 arf2::HIS3 ura3 lys2 trp1 his3 leu2 DHH1::DHH1-yEGFP-kanMX4 | This study |
| YAS1154 | arf1-11 Dhh1-GFP | MAT a ade2::arf1-11::ADE2 arf1::HIS3 arf2::HIS3 ura3 lys2 trp1 his3 leu2 DHH1::DHH1-yEGFP-kanMX4 | This study |
| YAS2153 | ARF1 Cmd1-GFP | MAT a ade2::ARF1::ADE2 arf1::HIS3 arf2::HIS3 ura3 lys2 trp1 his3 leu2 CMD1::CMD1-yEGFP-kanMX4 | This study |
| YAS2154 | arf1-11 Cmd1-GFP | MAT a ade2::arf1-11::ADE2 arf1::HIS3 arf2::HIS3 ura3 lys2 trp1 his3 leu2 CMD1::CMD1-yEGFP-kanMX4 | This study |
Fluorescence Microscopy
Yeast cells were grown in YPD to early log phase and shifted for 1 h to 37°C or subjected to various stresses where indicated; arf1-11 Δslt2 required additional osmotic support for growth and was cultured in medium containing 1 M sorbitol. The cells were taken up in HC-complete medium (without glucose or supplemented with CaCl2 or NaCl, where indicated) and immobilized on concanavalin A–coated slides. Fluorescence was monitored with an Axiocam mounted on an Axioplan 2 fluorescence microscope (Carl Zeiss, Oberkochen, Germany) using Axiovision software. Image processing was performed using Adobe Photoshop CS2 (San Jose, CA). For counting, pictures were exported to Photoshop and inverted, and the tonal range was adjusted using the levels dialog box to facilitate counting; all pictures from the same experiment were treated equally. A minimum of 100 cells from at least two independent experiments was counted for each condition. In the quantification graphs, the size of the box is determined by the 25th and 75th percentiles, the whiskers represent the 5th and 95th percentiles, the horizontal line and the little square mark the median and the mean, respectively.
Denaturing Yeast Extracts and Western Blot
Fifteen milliliters of yeast culture was grown to early log phase (OD600 0.5–0.7) and shifted for 1 h to 37°C where indicated. The cells were harvested and lysed with glass beads in 150 μl of lysis buffer (20 mM Tris/HCl, pH 8.0, 5 mM EDTA) in the presence of 1 mM dithiothreitol (DTT) and protease inhibitors. The lysates were incubated at 65°C for 5 min, and unlysed cells subsequently were removed by centrifugation. The protein concentration was determined using the DC Protein Assay (Bio-Rad, Richmond, CA), and the equivalent of 30 μg of total protein was analyzed by SDS-PAGE and immunoblotting. Total Slt2 was detected using goat anti-Mpk1 antibody (yN-19, Santa Cruz Biotechnology, Santa Cruz, CA) and phospho-Slt2 using rabbit antiphospho-p44/42 MAP kinase (Thr202/Tyr204) antibody (Cell Signaling, Beverly, MA) with horseradish peroxidase-conjugated secondary antibodies (Pierce, Rockford, IL) and enhanced chemiluminescence reagent (GE Healthcare, Freiburg, Germany).
Polysome Profile Analysis
Polysome preparations were performed as described previously (de la Cruz et al., 1997) on 4–47% sucrose gradients prepared with a Gradient Master (Nycomed Pharma, Westbury, NY). Gradient analysis was performed using a gradient fractionator (Labconco, Kansas City, MO) and the Äcta FPLC system (GE Healthcare) and continuously monitored at A254.
Immunoelectron Microscopy
Cells expressing Dcp2–9myc or Dhh1–9myc were grown to early log-phase at 23°C and then shifted to 37°C for 1 h or transferred to a medium lacking a carbon source for 15 min. Cells were fixed and treated for immunoelectron microscopy as described in Prescianotto-Baschong and Riezman (2002). Ten-nanometer gold particles coupled to goat anti-rabbit IgG (BBInternational, Cardiff, United Kingdom) were used to detect the binding of polyclonal rabbit anti-myc antibodies (Abcam, Cambridge, MA).
Flotation of PBs
Flotation of ER membranes was performed according to Schmid et al. (2006). The equivalent of 50 OD600 units was converted into spheroplasts at 37°C and lysed by Dounce homogenization in 3 ml of lysis buffer (20 mM HEPES/KOH, pH 7.6, 100 mM sorbitol, 100 mM KAc, 5 mM Mg(Ac)2, 1 mM EDTA, 100 μg/ml cycloheximide) in the presence of 1 mM DTT and protease inhibitors. After removal of cellular debris (5 min, 300 × g), membranes were pelleted by centrifugation (10 min, 13,000 × g), resuspended in 2 ml of lysis buffer containing 50% sucrose, and layered on top of 2 ml 65% sucrose in lysis buffer. Two additional 5- and 2-ml cushions (40 and 0% sucrose) were layered on top. The step gradient was spun in a TST41.14 rotor for 16 h at 28,000 × g. After centrifugation, 1-ml fractions were collected from each of the cushions and the interphases and were TCA-precipitated. The samples were analyzed by SDS-PAGE and immunoblotting using monoclonal mouse anti-myc antibodies (9E10, Sigma, St. Louis, MO) and polyclonal rabbit anti-Sec61 antibodies (a gift from R. Schekman, University of California at Berkeley, Berkeley, CA).
Northern Blot
Yeast cells were grown in YPD to early log phase and shifted for 1 h to 37°C where indicated. Cells were lysed in lysis buffer by grinding in liquid nitrogen. RNA was extracted from the P13 pellet using TriZOL reagent (Invitrogen, Carlsbad, CA), and 15 μg of RNA was resolved on agarose gels containing formaldehyde. The RNA was transferred onto Hybond N+ (Amersham Biosciences, Piscataway, NJ) and subsequently hybridized to HAC1 probes, which were generated using an AlkPhos direct labeling kit (Amersham Biosciences). The probes were detected using the CDP-Star reagent (GE Healthcare) according to manufacturer's recommendations.
RESULTS
PB Number Is Increased in arf1 Mutants
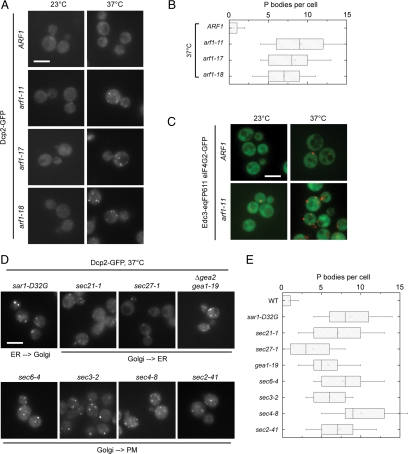
Several mutants in the secretory pathway lead to attenuation of translation (Deloche et al., 2004). In addition, specific mutant alleles of the small GTPase Arf1 are defective in the asymmetric distribution of ASH1 mRNA, and these defects are not caused by disturbances of the actin cytoskeleton (Trautwein et al., 2004). Therefore, we wondered whether arf1 mutants induce PBs, which provide a storage and degradation location for mRNAs in response to translational arrest. As a marker for PBs we used Dcp2 (decapping protein 2), which is required for the decapping of mRNAs and for PB formation (Dunckley and Parker, 1999; Sheth and Parker, 2003; Teixeira and Parker, 2007). We chromosomally appended Dcp2 with green fluorescent protein (GFP) and determined the number of PBs in control and temperature-sensitive arf1 mutant cells (Figure 1A). As expected, few PBs were observed in wild-type cells or in arf1 mutants at the permissive temperature, with Dcp2-GFP largely distributed throughout the cytosol. Strikingly, a large increase in PB number (9–10 on average) was observed in arf1 mutant alleles upon shift to 37°C (Figure 1, A and B). The temperature shift represents considerable stress for the wild type, but does not induce a block in translation, and only 1–2 PBs were present in wild-type cells at 37°C (Figure 1, A and B).
Figure 1.
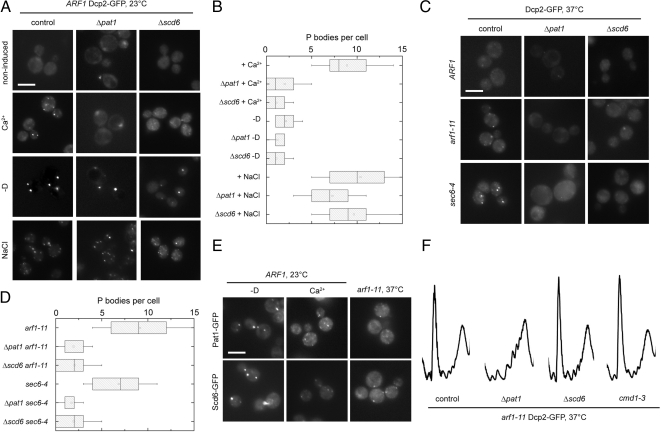
arf1 and secretory pathway mutants have multiple PBs. (A) The PB marker Dcp2 was chromosomally tagged with GFP in the control strain and in several temperature-sensitive arf1 mutants. At the permissive temperature (23°C), no PBs are observed and Dcp2-GFP is dispersed throughout the cytoplasm; upon shift to the nonpermissive temperature (37°C) for 1 h, PB formation is induced in all strains. The increase in PB number is more pronounced in arf1 mutants than in the control. (B) Quantification of the multiple PB phenotype in arf1 mutants at nonpermissive temperature. A minimum of a hundred cells from at least two independent experiments was counted for each condition. The size of the box is determined by the 25th and 75th percentiles; the whiskers represent the 5th and 95th percentiles, the horizontal line and the little square mark the median and the mean, respectively. (C) Wild-type and arf1-11 mutant cells expressing the PB marker Edc3-eqFP611 and the stress granule marker eIF4G2-GFP were shifted to 37°C for 1 h. Although multiple PBs were formed in the arf1-11 mutant, we observed no induction of stress granules in the mutant or the control strain. (D) The number of PBs in different temperature-sensitive mutants in components of the secretory pathway was determined after shift for 1 h to 37°C. All secretory mutants we analyzed displayed a multiple PB phenotype to a varying degree. (E) Quantification of the multiple PB phenotype in secretory mutants after a shift to the nonpermissive temperature. See B for details on the representation. Scale bars, (A, C, and D) 5 μm.
We have previously shown that arf1-11 and arf1-18 but not arf1-17 failed to localize ASH1 mRNA to the bud tip of yeast cells (Trautwein et al., 2004). Strikingly, the arf1-17 mutation also caused a dramatic increase in PB number similar to that detected in arf1-11 and arf1-18, indicating that mislocalization of mRNAs that are dependent on the SHE machinery is not the cause of multiple PB formation (Figure 1, A and B).
The Dcp2 foci we observed in arf1 mutants likely represent P bodies and not stress granules (or EGP-bodies) because, generally, stress granules do not contain Dcp2 (Kedersha et al., 2005; Anderson et al., 2006; Hoyle et al., 2007). To provide corroborating evidence we appended another PB component, the helicase Dhh1, with GFP, which behaved similarly to Dcp2-GFP in the arf1 strains (Supplemental Figure 1). Moreover, deletion of an essential SG component, PUB1, (Buchan et al., 2008; Swisher and Parker, 2010) did not interfere with the formation of Dcp2-GFP-positive structures (Supplemental Figure 2A). Finally, the SG marker eIF4G2 fused to GFP did not accumulate in foci in arf1-11 at 37°C (Figure 1C).
PB Number Is Increased in a Variety of Secretory Transport Mutants
Because the major function of Arf1p is to initiate coat protein I (COPI)- and clathrin-coated vesicle budding events, we asked whether the increase in PBs is a common feature in secretory transport mutants. We monitored Dcp2-GFP in various temperature-sensitive secretory transport mutants covering endoplasmic reticulum (ER)→Golgi, Golgi→ER, and post-Golgi trafficking steps and determined the number of PBs after a shift for 1 h to the nonpermissive temperature (Figure 1D). Interestingly, most of the mutants showed an increase in PB number similar to arf1 mutants (Figure 1E), indicating that this multiple PB phenotype is most likely related to a general defect in secretion.
Hyperosmotic Stress Led to Numerous PBs
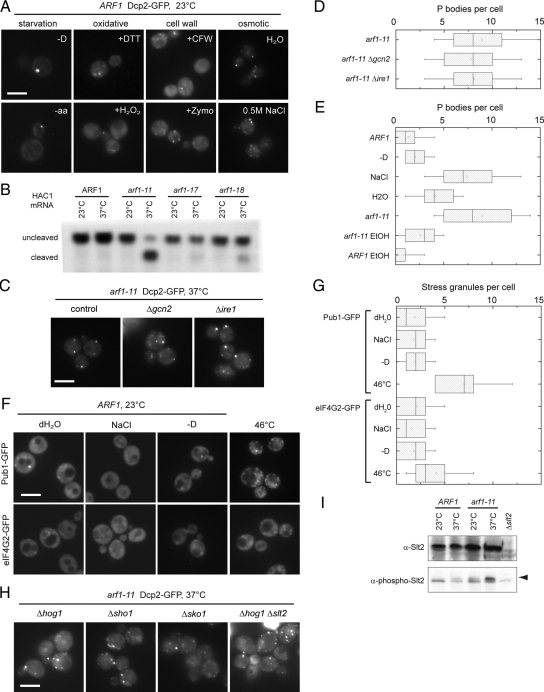
How do secretory transport mutants induce multiple PBs? It is unlikely that this phenotype occurs in response to general cellular stress. As has been reported before (Teixeira et al., 2005), PB formation is induced by starvation and under oxidation/redox conditions (Figure 2A). However, neither starvation nor ox/redox stress led to a similar increase in PB number in wild-type cells as observed for the arf1 or secretory pathway mutants; therefore, these mutants appear to trigger a different type of PB response. Secretory pathway mutants may cause ER stress, which in turn would activate the unfolded protein response (UPR). Alternatively, secretory transport mutants may prevent the correct targeting of proteins to the plasma membrane and hence cause cell wall and osmotic stress. First, we tested whether UPR is coupled to the formation of multiple PBs. The hallmark for the activation of UPR is splicing of the transcriptional activator HAC1 mRNA (Sidrauski and Walter, 1997). Shift of arf1-11 to the restrictive temperature–induced UPR as indicated by the cleavage of the HAC1 mRNA, which was also true for arf1-18, albeit to a lesser extent, but not for wild type and arf1-17 (Figure 2B), indicating that there is no direct correlation between UPR and PB formation. Moreover, deletion of IRE1, the endonuclease that splices HAC1 transcript at the ER and senses unfolded proteins in the ER (Cox and Walter, 1996; Kimata et al., 2006), or GCN2, the single eIF2alpha kinase, which has been implicated in UPR (Patil et al., 2004), did not affect multiple PB formation in arf1-11 (Figure 2, C and D). In contrast, when we challenged control or wild-type cells with cell wall stress, we detected an up-regulation of PB formation. More importantly, after application of osmotic stress by incubating the cells for a short period of time (15 min) in 0.5 M NaCl, a similar increase in the number of PBs was observed as in the arf1 mutants (Figure 2, A and E). We also examined SG formation under the same conditions to determine if these stresses were specific for assembling multiple PBs. Only 1–2 SG were induced under various stresses, except for robust heat stress (46°C), using Pub1-GFP, eIF4G2-GFP, and eIF3B-GFP as markers (Figure 2F, Supplemental Figure 2B), indicating that this stress is PB specific. The foci formed at 46°C represent SG because eIF3 localizes to SGs after robust heat shock (Grousl et al., 2009). These data could indicate that in arf1 mutants the response to osmotic or cell wall stress is activated, and that this specific stress response increases PB number. Our data are consistent with the possibility that components of the cell wall or plasma membrane constituents do not reach the plasma membrane efficiently in arf1 and most secretory pathway mutants at the nonpermissive temperature, and the cells become more susceptible to osmotic stress. If this hypothesis was true, preventing signaling in the osmo-response pathway should rescue the multiple PB phenotype in arf1-11 mutants.
Figure 2.
Different stresses lead to induction of a different number of PBs. (A) Osmotic stress induces an increase in PB number in wild-type cells. Wild-type cells expressing Dcp2-GFP were grown in rich medium to early log phase at 23°C. Starvation was induced by incubating cells in medium either lacking a carbon source (−D) or amino acids (−aa) for 15 min. To induce reductive or oxidative stress, 10 mM DTT or 0.4 mM H2O2 were added to the cultures for 1 h. Spheroplasting by enzymatic digest (+Zymo) or treatment with calcofluor white (+CFW) were used to induce cell wall stress. To induce osmotic stress, cells were harvested and either incubated in H2O or rich medium containing 0.5 M NaCl for 15 min. All stresses were applied at 23°C. Although cell wall stress did cause an increase in PB number, osmotic stress induced a strong multiple PB phenotype similar to arf1 mutants. (B) UPR is not generally activated in arf1 mutant alleles. Northern blot with a probe against HAC1 mRNA on total RNA extracted from ARF1 and arf1 cells grown at 23°C or shifted to 37°C for 1 h. The HAC1 cleavage product indicative of an active UPR is only observed in two of the mutant alleles. (C) The UPR components Gcn2 and Ire1 are not required for assembly of multiple PBs in arf1-11. Cells expressing Dcp2-GFP were deleted for either GCN2 or IRE1 and shifted to 37°C for 1 h. (D) Quantification of PBs in arf1-11 deleted for GCN2 or IRE1 after a shift to the nonpermissive temperature. See Figure 1B for details on the representation. (E) Quantification of the multiple PB phenotype in cells under osmotic stress and in the presence of ethanol. For the ethanol experiment, wild-type or arf1-11 cells expressing Dcp2-GFP were shifted to 37°C for 1 h in the presence or absence of 1.6 M EtOH. No effect was observed for wild-type, whereas the multiple PB phenotype in arf1-11 was rescued in the presence of EtOH. See Figure 1B for details on the representation. (F) Wild-type cells expressing the stress granule markers Pub1-GFP or eIF4G2-GFP were grown in rich medium at 23°C to early log phase and then either incubated in H2O, rich medium containing 0.5 M NaCl, or medium lacking a carbon source (−D) for 15 min, or subjected to high heat shock (10 min at 46°C). We did not observe a marked induction of stress granules under conditions that induce multiple PBs; however, Pub1-GFP containing stress granules were induced by heat shock. (G) Quantification of SGs in the wild-type induced by various stresses. See Figure 1B for details on the representation. (H) Plasma membrane stress signaling pathways are not required for the assembly of multiple PBs. Key components of different signaling pathways were deleted in arf1-11 cells expressing Dcp2-GFP. None of the mutants resulted in a reduction of PBs in arf1-11 at 37°C. See Table 2 for quantification of all mutations tested. (I) Lysates were generated from wild-type or arf1-11 cells after a 1 h shift to 37°C and analyzed by immunoblot to detect total Slt2 and Slt2 phosphorylated in response to cell wall integrity signaling. The band marked with the arrowhead corresponds to phospho-Slt2. A lysate of a strain deleted for SLT2 was loaded as a reference. Scale bars, (A, D, F, and G) 5 μm.
Neither the High-Osmolarity Glycerol Nor the Cell Wall Integrity Pathways Are Required for the Increase in PB Number in arf1 Mutants
The major pathway activated in response to osmotic stress is the high-osmolarity glycerol (HOG) mitogen-activated protein (MAP) kinase signaling pathway (Van Wuytswinkel et al., 2000; Hohmann, 2002). The HOG pathway is at least partially repressed by addition of ethanol to the growth medium (Hayashi and Maeda, 2006). Strikingly, a very strong reduction in PB number was observed when arf1-11 cells were shifted to the restrictive temperature in the presence of ethanol (Figure 2E), suggesting that the HOG pathway may be activated in the arf1-11 mutant. Hog1 is not essential, and therefore we could test the requirement for HOG signaling pathway in PB formation directly by deleting HOG1 in arf1-11. Surprisingly, the number of PBs was unchanged upon loss of Hog1p in arf1-11 at 37°C (Figure 2H, Table 2). Moreover, deletion of the osmo-sensor SHO1, or the HOG-dependent osmo-responsive transcription factor SKO1, did not interfere with PB formation in arf1-11 (Figure 2H, Table 2). The results indicate that Hog1 is not involved in PB assembly in secretory mutants. This is in agreement with recent data, suggesting that Hog1 may play a role in PB disassembly, and that PBs are formed in Δhog1 cells under mild and severe osmotic shock (Romero-Santacreu et al., 2009).
Table 2.
PB phenotype of various mutants
| Supernumerary PBs |
PBs induced by starvation | ||
|---|---|---|---|
| in arf1–11 at 37°C | in ARF1 + Ca2+ | ||
| No deletion | + (9.15 ± 4.16) | + (8.89 ± 3.5) | + (2.16 ± 1.01) |
| Δhog1 | + (8.07 ± 3.5) | + (7.51 ± 3.38) | n.d. |
| Δsho1 | + (7.66 ± 3.62) | + (5.52 ± 2.37) | n.d. |
| Δsko1 | + (7.72 ± 3.32) | + (5.43 ± 2.55) | n.d. |
| Δslt2 | + (5.96 ± 4.47) | + (7.64 ± 3.28) | n.d. |
| Δhog1 Δslt2 | + (6.8 ± 5.18) | + (7.48 ± 2.72) | n.d. |
| Δsli2 | + (7.65 ± 4.05) | + (6.9 ± 3.45) | n.d. |
| Δcnb1 | + (6.41 ± 2.48) | + (6.71 ± 2.74) | n.d. |
| Δcmk1 Δcmk2 | + (7.65 ± 3.45) | + (6.27 ± 2.85) | n.d. |
| Δcmk1 Δcmk2 Δcnb1 | + (7.72 ± 4.05) | + (6.81 ± 2.8) | n.d. |
| Δhog1 Δslt2 Δcnb1 | + (7.97 ± 4.75) | + (8.32 ± 3.35) | n.d. |
| Δpat1 | − (1.93 ± 1.79) | − (2.02 ± 2.42) | + (1.2 ± 0.99) |
| Δscd6 | − (2.04 ± 1.81) | − (1.08 ± 1.27) | + (2.41 ± 1.28) |
| cmd1-3 | n.d. | − (2.02 ± 1.84) | + (2.16 ± 1.07) |
Values in parentheses are average number of PBs counted per cell ± 1 SD. A minimum 100 hundred cells from at least two independent experiments were counted for each strain. n.d., not determined.
Another signaling pathway acting at the plasma membrane is the cell wall integrity pathway, which plays a role in the progression through the cell cycle. The MAP kinase Slt2p, also referred to as Mpk1p, is central to the cell wall integrity pathway (Lee et al., 1993) and is phosphorylated in response to cell wall stress. We observed the phosphorylation of Slt2 in arf1-11 cells after shift to the nonpermissive temperature (Figure 2I). A low level of phosphorylation is already observable in arf1-11 cells at 23°C and in wild-type cells at 37°C, but these levels may be too low to drive numerous PB formation. Similar to Hog1, Slt2 is not essential, and we could determine the number of PBs in an arf1-11 Δslt2 strain at 37°C (Table 2). As in the case of loss of Hog1, deletion of SLT2 did not reduce PB number in arf1-11 cells, showing that under these conditions neither the HOG nor the cell wall integrity pathway is involved in PB formation. Moreover, the two pathways do not cooperate in PB induction, because a deletion of both HOG1 and SLT2 did not reduce PB number in arf1-11 cells (Figure 2H, Table 2). Finally, we deleted the serine/threonine kinase Sli2p (also referred to as Ypk1p), which is involved in the cell integrity and sphingolipid-mediated signaling pathway (Schmelzle et al., 2002). Again, deletion of SLI2 had no effect on PB number in arf1-11 cells (Table 2). Similarly, treating arf1-11 cells with chlorpromazine, a membrane-permeable molecule that binds anionic lipids such as polyphosphoinositides and leads to translation attenuation (De Filippi et al., 2007) did not change the amount of PBs (data not shown). Taken together, these results argue that the increase in PB number seen in arf1 or secretory mutants is independent of the major plasma membrane signaling pathways.
Ca2+ Induces Numerous PBs
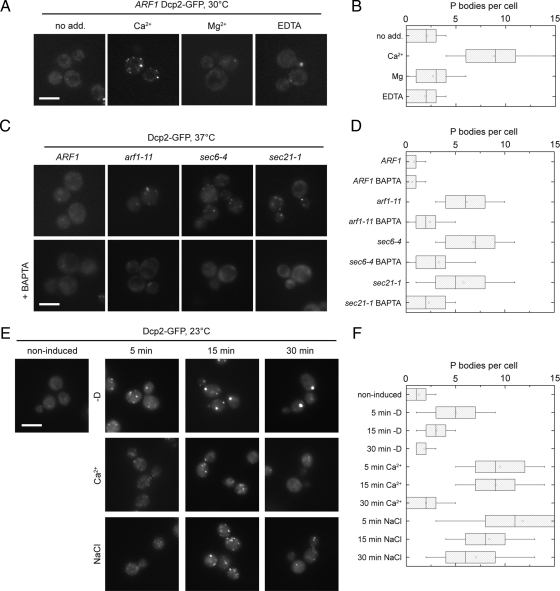
Because hyperosmotic shock leads to a transient increase in intracellular Ca2+ (Batiza et al., 1996; Matsumoto et al., 2002) and a Ca2+/calcineurin-regulated response is required for growth under high osmolarity and cell wall stress (Heath et al., 2004), we asked next whether Ca2+ is required for the formation of multiple PBs. Incubation of wild-type cells with Ca2+-induced rapid PB formation, to the same extent as hyperosmotic shock, within 10 min after CaCl2 addition (Figure 3, A and B). Moreover, the effect was specific to Ca2+ because adding MgCl2 had no effect on PB formation (Figure 3, A and B). If formation of multiple PBs depends on increased Ca2+ levels, arf1-11 cells should not accumulate PBs in the presence of Ca2+-chelating agents. To test this hypothesis, we precultured the strains in the presence of low concentrations of BAPTA before and during the temperature-shift. Under these conditions, only 1–2 PBs were observed in arf1-11 (Figure 3, C and D). Moreover, Ca2+ was less potent in inducing PBs in ARF1. Similar results were also observed in the gamma-COP mutant sec21-1 and the exocyst component mutant sec6-4 upon growth in BAPTA and shift to 37°C (Figure 3, C and D), indicating that the increase in PB number in secretory mutants is dependent on a change in intracellular Ca2+. Interestingly, in contrast to the PBs formed under glucose starvation or NaCl, Ca2+-induced PBs disappear rapidly over time, and after ∼30 min only a few, if any, PBs could be detected (Figure 3, E and F). Most likely transporters are expressed at the plasma membrane, which extrude excess Ca2+ to maintain ion homeostasis. This effect is not observed in secretory pathway mutants, in which PBs persisted, even after a prolonged shift to the nonpermissive temperature. This observation is expected as delivery of transporters to the plasma membrane is blocked in these mutants (Figure 1D).
Figure 3.
Ca2+ induces PB assembly to a similar extent as that of a secretion block. (A) Wild-type cells expressing Dcp2-GFP were treated with 200 mM CaCl2, MgCl2 or 12.5 mM EDTA for 15 min at 30°C, washed and inspected under the microscope. Only Ca2+ treatment induced multiple PBs. (B) Quantification of PBs in wild-type cells treated with Ca2+, Mg2+ or EDTA. See Figure 1B for details on the representation. (C) The Ca2+-chelator BAPTA prevents formation of multiple PBs in secretory transport mutants. Cells expressing Dcp2-GFP were pre-cultured in the presence or absence of 0.6 mM BAPTA and then shifted to 37°C for 1 h before inspection under the microscope. PB induction was strongly reduced. (D) Quantification of PBs in secretory mutants treated with BAPTA after shift to the nonpermissive temperature. See Figure 1B for details on the representation. (E) Time-course analysis of PB induction under various conditions. Wild-type cells expressing Dcp2-GFP were either harvested and resuspended in rich medium without a carbon source (−D), or treated with 200 mM CaCl2 or 0.5 M NaCl. After various time-points, samples were removed, fixed with formaldehyde, and analyzed under the microscope. PB response to Ca2+appears to be transient, whereas PBs induced by osmotic stress or starvation persist longer. (F) Quantification of PBs in the time-course experiment. See Figure 1B for details on the representation. Scale bars, (A, C, and E) 5 μm.
Elevated Ca2+ Levels and Starvation Lead to Translation Attenuation and PB Formation through Different Pathways
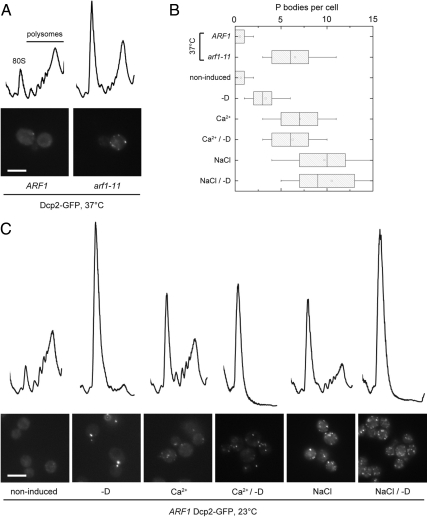
Secretory pathway mutants cause translation attenuation at the nonpermissive temperature (Deloche et al., 2004) and induction of PBs is thought to correlate with translation attenuation (Sheth and Parker, 2003). The hallmark of translation attenuation is a loss of translating polysomes and a concomitant increase in the monosome (80S) fraction in the cell. First, we established that arf1-11 cells indeed attenuate translation by determining their polysome profile after shift to 37°C (Figure 4, A and B). Next we asked whether the number of PBs in the cell could be correlated with the strength of translational block because Parker and Sheth (2007) proposed a model in which the rates of translation and degradation of mRNAs would be governed by a dynamic equilibrium between polysomes and mRNPs in PBs. Therefore, we recorded polysome profiles of wild-type cells that were either starved for glucose or incubated with Ca2+ or a combination of both stresses (Figure 4, B and C). As shown previously (Ashe et al., 2000), glucose starvation causes a pronounced shift of ribosomes from the polysome fraction toward monosomes. A similar change was observed in cells that were treated with Ca2+ or NaCl and in arf1-11, albeit to a lesser extent (Figure 4). According to the dynamic equilibrium model, the weak translational repression induced by high Ca2+, NaCl, or the arf1-11 mutant would yield multiple PBs, whereas the strong translational repression observed upon starvation would result in 1–2 PBs. It is conceivable that the multiple PBs we observe would coalesce to 1–2 PBs that contain more RNA or represent more densely packed, matured PBs, if translational repression was stronger. If this hypothesis was correct, combining starvation and high Ca2+ should lead to the formation of 1–2 PBs. However, combining high Ca2+ or NaCl with starvation led to the almost complete absence of polysomes, and even the monosomes seemed to partially disassemble, but multiple PBs were observed under these conditions (Figure 4C). These results indicate that dynamic equilibrium might not be sufficient to explain different PB numbers observed under starvation and high Ca2+ and suggests the presence of at least two distinct pathways to generate PBs, one of which functions in sensing low glucose levels, whereas the other responds to an increase in Ca2+ levels.
Figure 4.
Translation attenuation and PB induction in response to different stresses. (A) Wild-type or arf1-11 cells expressing Dcp2-GFP were grown in rich medium to early log phase at 23°C and the shifted to 37°C for 1 h. Cells were either inspected under the microscope or subjected to polysome profile analysis. In arf1-11 cells, translation is attenuated. (B) Quantification of the PBs observed in A and C. See Figure 1B for details on the representation. (C) Cells expressing Dcp2-GFP were grown in rich medium to early log phase at 23°C. Starvation was induced by resuspending cells into medium lacking a carbon source (−D). Alternatively, cells were incubated in rich medium containing 0.5 M NaCl or 200 mM CaCl2, or subjected to combinations of these stresses. After 15 min incubation at 23°C cells were either inspected under the microscope or subjected to polysome profile analysis. Although starvation resulted in a marked increase in the monosome/polysome ratio and induction of few PBs, wild-type cells under osmotic stress or in the presence of Ca2+ displayed an intermediate increase in the monosome/polysome ratio and had multiple PBs, as was observed in the arf1-11 mutant shifted to 37°C for 1 h. When starvation was combined with either osmotic stress or Ca2+ treatment, polysomes were almost completely abolished, but cells had multiple PBs. Scale bars, (A and C) 5 μm.
Calmodulin Is Required for the Induction of PBs in Secretory Pathway Mutants
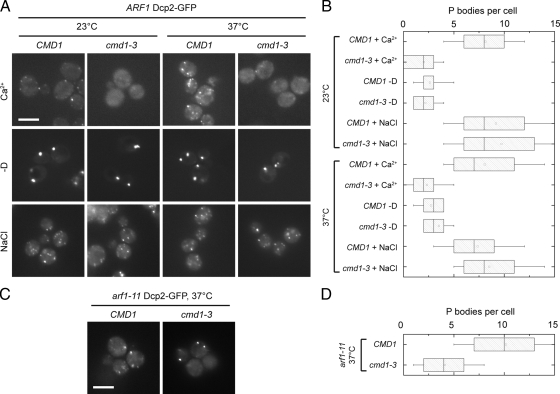
The next question was how the cell would sense the potential intracellular Ca2+ changes. A major player in Ca2+ signaling is calmodulin (Cmd1), which contains four EF hands, three of which can coordinate Ca2+. The temperature-sensitive cmd1-3 mutant does not bind Ca2+ (Geiser et al., 1991). Therefore, to test whether calmodulin is involved in signaling to PBs, we replaced CMD1 by the cmd1-3 mutation in our Dcp2-GFP–expressing wild type and asked if cells would still accumulate PBs in the presence of Ca2+. The cmd1-3 mutant blocked formation of numerous PBs (Figure 5A), demonstrating that PB induction upon elevated Ca2+ levels requires functional calmodulin; however, is calmodulin also required for PB induction by other stresses? To address this question, we analyzed PB assembly after induction by glucose starvation or NaCl addition, but detected no difference between wild-type and cmd1-3 cells (Figure 5, A and B). Similarly, when we introduced the cmd1-3 allele into the arf1-11 mutant, induction of multiple PBs was reduced at the nonpermissive temperature (Figure 5, C and D), indicating that the PBs formed in the secretory mutants do indeed correspond to PBs induced by Ca2+, but differ from those induced by hyperosmotic stress, although they are indistinguishable by light microscopy.
Figure 5.
Calmodulin is required for the assembly of multiple PBs in the presence of Ca2+ and in secretory mutants. (A) PBs were induced in wild-type and cmd1-3 mutant cells expressing Dcp2-GFP by treatment with 200 mM CaCl2, 0.5 M NaCl, or incubation in rich medium lacking a carbon source (−D) for 15 min at 23°C, or after a 1 h shift to 37°C. Although PB induction by osmotic stress or starvation was not affected in the cmd1-3 mutant, a strong reduction of PB number was observed in the mutant after Ca2+ treatment. (B) Quantification of PBs in the ARF1 cmd1-3 mutant induced either at 23°C or after a shift to 37°C for 1 h. See Figure 1B for details on the representation. (C) arf1-11 cells expressing Dcp2-GFP that were deleted for CMD1 but carried a plasmid containing the cmd1-3 allele were shifted to 37°C for 1 h. PB number was reduced if compared with arf1-11 alone. (D) Quantification of PBs in the arf1-11 cmd1-3 mutant induced at 37°C. See Figure 1B for details on the representation. Scale bars, (A and C) 5 μm.
To determine if one of the established Ca2+ signaling pathways is required for PB induction, we screened through mutants in the phosphatase calcineurin– and in calmodulin-dependent kinases (Table 2). Deletion of the catalytic subunit of calcineurin, CNB1, or of the calmodulin-dependent kinases, CMK1 and CMK2, in arf1-11 mutant or wild-type cells had no effect on PB induction by temperature-shift or Ca2+ treatment, respectively. Even concomitant loss of both calmodulin-dependent kinases and the regulatory subunit of calcineurin (Δcmk1 Δcmk2 Δcnb1) or of HOG1, SLT2, and CNB1 (Δhog1 Δslt2 Δcnb1) did not inhibit formation of numerous PBs in arf1-11 cells at 37°C or in ARF1 cells in the presence of Ca2+ (Table 2). Taken together these data indicate that the classical Ca2+ signaling pathways are not involved in inducing PB production, suggesting that calmodulin might act on PBs by an unknown mechanism.
Calmodulin is an abundant protein that performs a plethora of functions in the cell and is localized to sites of polarized growth and the spindle pole body (Ohya and Botstein, 1994; Spang et al., 1996). When we appended Cmd1 with GFP in arf1-11 and wild-type cells and induced PBs by temperature-shift or Ca2+, respectively, we did not observe accumulation of Cmd1-GFP into multiple foci, indicating that Cmd1 is not a bona fide component of PBs. It is important to note, that under these PB inducing conditions Cmd1-GFP was lost from the site of polarized growth, the small bud tip, providing corroborating evidence that the Ca2+ levels might be elevated in secretory mutants (Supplemental Figure 3).
Pat1 and Scd6 Are Required for PB Assembly in Secretory Pathway Mutants
The above data indicate the existence of multiple parallel pathways by which PB formation can occur. The assembly of PBs has been best characterized under starvation conditions (Parker and Sheth, 2007; Teixeira and Parker, 2007; Franks and Lykke-Andersen, 2008). Interestingly, no single known component of PBs was solely responsible for PB formation (Teixeira and Parker, 2007). Thus, if different pathways exist that lead to PB formation, one might be able to identify PB components that are required for PB formation only under specific conditions. To test this hypothesis, we tested two PB components that are not strictly involved in PB formation under starvation, namely Pat1 and Scd6 (Teixeira and Parker, 2007). We chose Pat1 because it contains an EF-hand, which might coordinate a Ca2+ ion. Pat1 binds, together with the Lsm1-7 complex, to the 3′ region of mRNAs, and is involved in PB formation. However, PBs are still formed in Δpat1 cells upon glucose starvation (Teixeira and Parker, 2007). Scd6 is an Sm-like protein (Lsm) most likely involved in the regulation of mRNA translation and/or degradation in PBs (Decker and Parker, 2006) and was first described as a multicopy suppressor of a membrane transport defect (Nelson and Lemmon, 1993). Deletion of PAT1 or SCD6 in wild-type cells strongly suppressed PB assembly induced by Ca2+ (Figure 6, A and B).
Figure 6.
Pat1 and Scd6 are required for PB assembly in secretory transport mutants and upon Ca2+ treatment. (A) PBs were induced in wild type, Δpat1 or Δscd6 mutant cells expressing Dcp2-GFP by treatment with 200 mM CaCl2 or 0.5 M NaCl, or after incubation in rich medium lacking a carbon source (−D) for 15 min at 23°C. Although PB induction by osmotic stress or starvation was not affected in the deletion mutants, a strong reduction of PB number was observed after Ca2+ treatment. (B) Quantification of PBs in the Δpat1 or Δscd6 strains compared with wild-type. See Figure 1B for details on the representation. (C) Pat1 and Scd6 are required for assembly of multiple PBs in secretion mutants. Wild type, arf1-11, and sec6-4 expressing Dcp2-GFP and deleted for either PAT1 or SCD6 were shifted to 37°C for 1 h. Deletion of PAT1 or SCD6 abolished the multiple PB phenotype in secretion mutants. (D) Quantification of PBs in secretory mutants deleted for PAT1 or SCD6 after a shift to the nonpermissive temperature. See Figure 1B for details on the representation. (E) PBs were induced in wild-type or arf1-11 cells expressing either Pat1-GFP or Scd6-GFP by incubation in rich medium lacking a carbon source (−D), treatment with 200 mM CaCl2 for 15 min at 23°C, or a shift to 37°C for 1 h, respectively. Both Pat1-GFP and Scd6-GFP behaved similarly to Dcp2-GFP and were found in multiple PBs after Ca2+ treatment or in the secretory mutant after shift to the nonpermissive temperature. (F) arf1-11 cells deleted for PAT1 or SCD6, or carrying the cmd1-3 allele were shifted to 37°C for 1 h and their polysome profile recorded. Although all three strains do not induce multiple PBs under this condition, translation is derepressed only in the strain deleted for PAT1. Scale bars, (A, C, and E) 5 μm.
In agreement with previously published data, deletion of PAT1 or SCD6 did not influence PB formation upon glucose starvation of hyperosmotic shock, indicating that both proteins are required specifically for the formation of Ca2+-dependent PBs (Figure 6, A and B). Next, we investigated the effect of PAT1 or SCD6 deletion in arf1-11 and sec6-4 after shift to the nonpermissive temperature. As expected, these cells did not form multiple PBs (Figure 6, C and D); however, most cells did contain a single PB, reinforcing the idea that PB assembly is not strictly dependent on Pat1 or Scd6. When we appended Pat1 or Scd6 with GFP in wild-type and arf1-11 mutant cells and subjected them to Ca2+ treatment or shift to 37°C, respectively, the GFP signal remained mostly cytoplasmic. However, multiple GFP foci formed under these conditions, indicating that both proteins are components of the PBs we observe (Figure 6E).
Taken together, our data provide evidence that the multiple PB phenotype in secretory pathway mutants is due to changes in intracellular Ca2+ and that Pat1 and Scd6 are involved in translating these changes into PB assembly.
Rescue of PB Formation in Secretory Pathway Mutants Is Independent of Translation Derepression
Given the relationship between PB formation and block of translation, we next wondered whether translation attenuation in the secretory mutant arf1-11 would also be rescued by deletion of PAT1 or SCD6 or by introducing the cmd1-3 allele. We shifted cells for 1 h to the nonpermissive temperature and recorded the polysome profile. Interestingly, although deletion of PAT1 restored translation even above wild-type levels, neither deletion of SCD6 nor the mutation in calmodulin influenced translation levels (Figure 6F), indicating that PB formation can be prevented independently of translation derepression.
PBs Are in the Vicinity of the ER
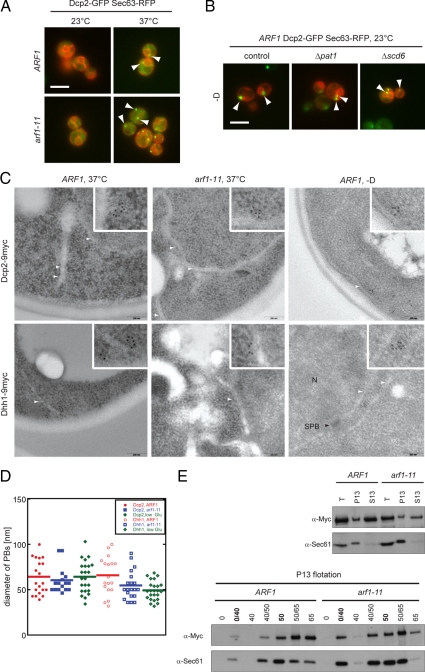
PB formation in secretory pathway mutants requires Scd6. The Lsm Scd6 homologue in Caenorhabditis elegans, CAR-1, is required for ER organization (Squirrell et al., 2006). The Drosophila homologue, trailer hitch, is localized close to ER exit sites, and is required for normal ER-exit site formation (Wilhelm et al., 2005). We therefore hypothesized that PBs induced in secretory mutants localize to the ER. To test this hypothesis, we examined the colocalization of Dcp2-GFP foci with an ER marker, Sec63-RFP, and found that they were in close proximity to the ER, in both wild-type and arf1 mutant cells shifted to 37°C (Figure 7A). Immunoelectron microscopy confirmed this observation (Figure 7C), and gold particles were found next to the ER in both mutant and wild-type cells after shift to 37°C or in wild-type cells under starvation. The labeling was specific, because no gold particle accumulations were found in an untagged strain (Supplemental Figure 4). Neither Scd6 nor Pat1 were required for the juxtaposition of PBs and ER (Figure 7B).
Figure 7.
PBs associate with the ER. (A) Wild-type and arf1-11 mutant cells expressing Dcp2-GFP and the ER marker Sec63p-RFP were shifted to 37°C for 1 h. In both control and arf1-11 cells, PBs were observed next to the ER. White arrowheads point toward a selection of PBs close to the ER. (B) PBs were induced in wild-type cells expressing Dcp2-GFP and the ER marker Sec63p-RFP and deleted for either PAT1 or SCD6 by incubating cells in rich medium without a carbon source for 15 min. In the control as well as in the deletion strains, PBs were observed in proximity to the ER. White arrowheads point toward a selection of PBs close to the ER. White bars (A and B), 5 μm. (C) Dcp2 and Dhh1 are part of spherical structures in proximity to the ER. Wild-type and arf1-11 cells expressing either Dcp2–9myc or Dhh1–9myc were shifted to 37°C for 1 h or incubated in rich medium without a carbon source for 15 min (−D) at 23°C. Cells were fixed and analyzed by immuno-EM for myc. Clusters of gold particles localized next to ER membranes. Scale bar, 200 nm. The inlet is a twofold magnification of the area surrounding PBs. White arrowheads point to ER membranes; N marks a nucleus and SPB with a black arrowhead points to a spindle pole body in the nuclear envelope. (D) Determination of the approximate diameter of PBs. The largest distance between two gold particles in a cluster was determined for Dcp2–9myc and Dhh1–9myc in wild-type and arf1-11 cells. The size of the clusters did not change significantly between different conditions for the two different markers. Each dot represents an individual PB, the horizontal line marks the average. (E) PBs are physically connected to the ER. Yeast lysates of wild-type and arf1-11 cells expressing Dcp2–9myc were prepared after a shift to 37°C for 1 h and separated into a 13,000 × g pellet (P13) and a corresponding supernatant (S13). The pellet was subjected to buoyant density centrifugation. A fraction of Dcp2–9myc cosedimented with the ER marker Sec61p in the P13 fraction and floated with Sec61p to the 0/40% sucrose interphase. Dcp2–9myc behaved similar in wild-type and arf1-11 lysates.
We observed that the Dcp2-myc signal was organized in ring-like structures, which had in all cases a diameter of ∼40 to 100 nm (Figure 7D). Therefore, surprisingly, the PBs induced under various conditions share a strikingly similar organization. To confirm this result, we determined the localization of another PB component, the helicase Dhh1, which is not essential for PB formation (Teixeira and Parker, 2007). Interestingly, Dhh1 accumulated in similar size foci, again close to the ER, suggesting that Dcp2 and Dhh1 are part of the same spherical structure (Figure 7C), independent of the induction condition or the strain background used. A spherical structure for PBs has been previously proposed (Kedersha et al., 2005; Teixeira et al., 2005; Wilczynska et al., 2005); however, the data underlying this hypothesis were obtained by light microscopy. Given the size of 40–100 nm of PBs, the resolution of a standard light microscope does not allow a precise measurement. Interestingly, the size of the PBs was similar in wild type and arf1-11 (Figure 7D). Therefore, PB number could potentially be correlated to the amount of mRNA that has to be silenced and/or to be degraded.
PBs Are Tightly Associated with the ER
The close proximity of PBs to the ER prompted us to test whether PBs are bound to the ER. First, we performed a differential centrifugation of total yeast cell lysate (Figure 7E) and found the PB marker Dcp2-myc segregated into the P13 and the S13 fraction. We would not expect all Dcp2 to be efficiently depleted from the soluble pool, because we always observed cytoplasmic Dcp2-GFP under the microscope. The accumulation of a PB component in the P13 fraction could be explained either by its membrane association or simply by sedimentation of PBs at medium speed because of their large size and mass. To distinguish between these possibilities, we performed a buoyant density centrifugation with the P13 fraction (Figure 7E). Membrane particles would float, whereas large protein complexes should remain in the bottom of the tube. A fraction of Dcp2 floated to the same position in the gradient as the translocon component Sec61p, indicating that PBs do not only localize close to the ER but are physically associated with the membrane (Figure 7E, 0/40 interphase). This localization of PBs to the ER suggests the intriguing possibility that translation of mRNA on one hand and mRNA silencing and degradation on the other hand occur in a spatially coordinated manner at the ER.
DISCUSSION
We have characterized a pathway through which PBs are induced in response to a block in secretion. This pathway is different from PB induction under starvation. Moreover, we found that not all stresses elicit assembly of PBs to the same extent. Although we observed only 1–4 PBs per cell upon starvation or exposure to oxidative stress, numerous PBs were formed in response to secretion defects or treatment with Ca2+ (average 9–10). Although appearance of multiple PBs could also be induced by osmotic stress, this pathway was independent from Ca2+.
Thus far, no specific signaling pathway has been implicated in the formation of PBs. The common view is rather that a number of stresses cause attenuation of translation initiation, and, as a consequence, mRNAs accumulate in the cytoplasm and passively aggregate into PBs (Andrei et al., 2005; Coller and Parker, 2005; Ferraiuolo et al., 2005). However, when we combined stresses, the PB number did not correlate with the strength of the translational block, but rather with the type of stress encountered, indicating that PB number is regulated by an upstream signal dependent on the stressor, but less dependent on the translational response. Interestingly, when other labs studied PB induction under starvation, loss of Dhh1 and Pat1 decreased the number of PBs but corresponded with increased polysome-associated mRNA (Coller and Parker, 2005), implying that an equilibrium between translation initiation an PB formation could exist. When secretory mutants were deleted for PAT1, we observed a similar phenomenon. However, when we deleted SCD6 or introduced cmd1-3 into arf1-11, PB induction was strongly reduced, but the attenuation of translation initiation remained unchanged, indicating an uncoupling of translation initiation and PB formation. Therefore, mRNA release from polysomes might not be the driving force for PB formation in secretory mutants.
The PBs induced in secretory transport mutants were very similar to PBs formed under starvation or heat stress as judged by immunoelectron microscopy. Thus the increase in number of PBs would suggest that more mRNA might have to be silenced and eventually be degraded, if the stress persisted over longer periods. In mammalian cells, PBs appear to be highly dynamic structures in that they can quickly exchange a subset of the constituents with the cytoplasm (Andrei et al., 2005; Kedersha et al., 2005). It is therefore conceivable that the composition of PBs might vary depending on the particular stress that induced their assembly. Individual PB proteins may also have different exchange rates and vary in the extent to which they can be exchanged. In fact, Dcp2 seems to be one of the less exchangeable PB constituents (Aizer et al., 2008). Thus Dcp2 and Dhh1 levels may not change dramatically between different PB subtypes, which may therefore be indistinguishable by immunoelectron microscopy approach. Because the Ca2+-induced PBs are much shorter lived than the starvation-induced PBs, it is plausible that also a maturation pathway for PBs exists. The hallmark of such a pathway would first be mRNA storage and later mRNA degradation.
The data we present here challenge the current prevailing model of PB assembly on various levels. First, different stresses lead to a different number of PBs independent of the level of translation attenuation (Figures 2 and 4). Second, the diameters of Dcp2-myc and Dhh1-myc rings, which we assume represent PBs, do not significantly change under various stresses (Figure 7). If PBs were mere mRNP aggregates, why would they be uniform in size? Third, the formation of PBs after secretory transport block required functional calmodulin, whereas Dcp2-GFP foci were still observed in starved cmd1-3 mutant cells subjected to starvation or osmotic stress (Figure 5). Finally, the Lsm-associated protein Pat1 and the Lsm homologue Scd6 were essential for PB formation in secretory pathway mutants or in Ca2+-treated wild-type cells but not during starvation or NaCl treatment (Figure 6). Therefore our data indicate that parallel pathways for PB induction must exist and that those pathways may modulate the extent to which PBs are formed.
Here, we identified three novel modulators of the PB response: calmodulin, Scd6, and Pat1. The interaction partner for calmodulin in PB assembly remains elusive, but calmodulin does not seem to act through CaM kinases as the concomitant loss of CaM kinase 1 and 2 (Δcmk1 Δcmk2) had no effects on PB formation.
Whether Cmd1, Scd6, and Pat1 act in parallel or whether Cmd1 is upstream of Scd6 and Pat1 remains unclear. The EF-hand contained in Pat1 does not seem to be directly involved in Ca2+ sensing; in a strain in which we mutated the putative Ca2+ coordinating residues, multiple PBs were still induced in response to Ca2+ (unpublished results). We envision a pathway in which secretory pathway mutants would trigger a change in intracellular Ca2+, and this change would be sensed by calmodulin. Calmodulin might then either directly, or through Scd6 and Pat1, promote PB formation. The assembly of PBs induced by starvation or other stresses would follow a different, calmodulin-independent route.
Supplementary Material
ACKNOWLEDGMENTS
We are grateful to E. Schiebel, A. Diepold and members of the Spang lab for discussions, and to E. Hartmann and I. G. Macara for critical comments on the manuscript. S. Michaelis is acknowledged for providing the Sec63-RFP plasmid, and we thank T. Davis (University of Washington, Seattle, WA), M. Hall (University of Basel, Basel, Switzerland), A. Nakano, R. Schekman, and E. Schiebel for reagents. This work was supported by the Boehringer Ingelheim Fonds (C.K.), the Werner Siemens Foundation (J.W.), the Swiss National Science Foundation, and the University of Basel.
Footnotes
This article was published online ahead of print in MBoC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E10-02-0099) on June 2, 2010.
REFERENCES
- Aizer A., Brody Y., Ler L. W., Sonenberg N., Singer R. H., Shav-Tal Y. The dynamics of mammalian P body transport, assembly, and disassembly in vivo. Mol. Biol. Cell. 2008;19:4154–4166. doi: 10.1091/mbc.E08-05-0513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson J. R., Mukherjee D., Muthukumaraswamy K., Moraes K. C., Wilusz C. J., Wilusz J. Sequence-specific RNA binding mediated by the RNase PH domain of components of the exosome. RNA. 2006;12:1810–1816. doi: 10.1261/rna.144606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andrei M. A., Ingelfinger D., Heintzmann R., Achsel T., Rivera-Pomar R., Luhrmann R. A role for eIF4E and eIF4E-transporter in targeting mRNPs to mammalian processing bodies. RNA. 2005;11:717–727. doi: 10.1261/rna.2340405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aronov S., Gerst J. E. Involvement of the late secretory pathway in actin regulation and mRNA transport in yeast. J. Biol. Chem. 2004;279:36962–36971. doi: 10.1074/jbc.M402068200. [DOI] [PubMed] [Google Scholar]
- Ashe M. P., De Long S. K., Sachs A. B. Glucose depletion rapidly inhibits translation initiation in yeast. Mol. Biol. Cell. 2000;11:833–848. doi: 10.1091/mbc.11.3.833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Batiza A. F., Schulz T., Masson P. H. Yeast respond to hypotonic shock with a calcium pulse. J. Biol. Chem. 1996;271:23357–23362. doi: 10.1074/jbc.271.38.23357. [DOI] [PubMed] [Google Scholar]
- Bi J., Tsai N. P., Lu H. Y., Loh H. H., Wei L. N. Copb1-facilitated axonal transport and translation of kappa opioid-receptor mRNA. Proc. Natl. Acad. Sci. USA. 2007;104:13810–13815. doi: 10.1073/pnas.0703805104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bouveret E., Rigaut G., Shevchenko A., Wilm M., Seraphin B. A Sm-like protein complex that participates in mRNA degradation. EMBO J. 2000;19:1661–1671. doi: 10.1093/emboj/19.7.1661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buchan J. R., Muhlrad D., Parker R. P bodies promote stress granule assembly in Saccharomyces cerevisiae. J. Cell Biol. 2008;183:441–455. doi: 10.1083/jcb.200807043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chowdhury A., Mukhopadhyay J., Tharun S. The decapping activator Lsm1p–7p-Pat1p complex has the intrinsic ability to distinguish between oligoadenylated and polyadenylated RNAs. RNA. 2007;13:998–1016. doi: 10.1261/rna.502507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coller J., Parker R. General translational repression by activators of mRNA decapping. Cell. 2005;122:875–886. doi: 10.1016/j.cell.2005.07.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cox J. S., Walter P. A novel mechanism for regulating activity of a transcription factor that controls the unfolded protein response. Cell. 1996;87:391–404. doi: 10.1016/s0092-8674(00)81360-4. [DOI] [PubMed] [Google Scholar]
- De Filippi L., Fournier M., Cameroni E., Linder P., De Virgilio C., Foti M., Deloche O. Membrane stress is coupled to a rapid translational control of gene expression in chlorpromazine-treated cells. Curr. Genet. 2007;52:171–185. doi: 10.1007/s00294-007-0151-0. [DOI] [PubMed] [Google Scholar]
- de la Cruz J., Iost I., Kressler D., Linder P. The p20 and Ded1 proteins have antagonistic roles in eIF4E-dependent translation in Saccharomyces cerevisiae. Proc. Natl. Acad. Sci. USA. 1997;94:5201–5206. doi: 10.1073/pnas.94.10.5201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Decker C. J., Parker R. CAR-1 and trailer hitch: driving mRNP granule function at the ER? J. Cell Biol. 2006;173:159–163. doi: 10.1083/jcb.200601153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Decker C. J., Teixeira D., Parker R. Edc3p and a glutamine/asparagine-rich domain of Lsm4p function in processing body assembly in Saccharomyces cerevisiae. J. Cell Biol. 2007;179:437–449. doi: 10.1083/jcb.200704147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deloche O., de la Cruz J., Kressler D., Doere M., Linder P. A membrane transport defect leads to a rapid attenuation of translation initiation in Saccharomyces cerevisiae. Mol. Cell. 2004;13:357–366. doi: 10.1016/s1097-2765(04)00008-5. [DOI] [PubMed] [Google Scholar]
- Dunckley T., Parker R. The DCP2 protein is required for mRNA decapping in Saccharomyces cerevisiae and contains a functional MutT motif. EMBO J. 1999;18:5411–5422. doi: 10.1093/emboj/18.19.5411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erdeniz N., Mortensen U. H., Rothstein R. Cloning-free PCR-based allele replacement methods. Genome Res. 1997;7:1174–1183. doi: 10.1101/gr.7.12.1174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eulalio A., Behm-Ansmant I., Schweizer D., Izaurralde E. P-body formation is a consequence, not the cause, of RNA-mediated gene silencing. Mol. Cell. Biol. 2007;27:3970–3981. doi: 10.1128/MCB.00128-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferraiuolo M. A., Basak S., Dostie J., Murray E. L., Schoenberg D. R., Sonenberg N. A role for the eIF4E-binding protein 4E-T in P-body formation and mRNA decay. J. Cell Biol. 2005;170:913–924. doi: 10.1083/jcb.200504039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franks T. M., Lykke-Andersen J. The control of mRNA decapping and P-body formation. Mol. Cell. 2008;32:605–615. doi: 10.1016/j.molcel.2008.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geiser J. R., van Tuinen D., Brockerhoff S. E., Neff M. M., Davis T. N. Can calmodulin function without binding calcium? Cell. 1991;65:949–959. doi: 10.1016/0092-8674(91)90547-c. [DOI] [PubMed] [Google Scholar]
- Grousl T., et al. Robust heat shock induces eIF2alpha-phosphorylation-independent assembly of stress granules containing eIF3 and 40S ribosomal subunits in budding yeast, Saccharomyces cerevisiae. J. Cell Sci. 2009;122:2078–2088. doi: 10.1242/jcs.045104. [DOI] [PubMed] [Google Scholar]
- Gueldener U., Heinisch J., Koehler G. J., Voss D., Hegemann J. H. A second set of loxP marker cassettes for Cre-mediated multiple gene knockouts in budding yeast. Nucleic Acids Res. 2002;30:e23. doi: 10.1093/nar/30.6.e23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayashi M., Maeda T. Activation of the HOG pathway upon cold stress in Saccharomyces cerevisiae. J. Biochem. 2006;139:797–803. doi: 10.1093/jb/mvj089. [DOI] [PubMed] [Google Scholar]
- Heath V. L., Shaw S. L., Roy S., Cyert M. S. Hph1p and Hph2p, novel components of calcineurin-mediated stress responses in Saccharomyces cerevisiae. Eukaryot. Cell. 2004;3:695–704. doi: 10.1128/EC.3.3.695-704.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hohmann S. Osmotic stress signaling and osmoadaptation in yeasts. Microbiol. Mol. Biol. Rev. 2002;66:300–372. doi: 10.1128/MMBR.66.2.300-372.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoyle N. P., Castelli L. M., Campbell S. G., Holmes L. E., Ashe M. P. Stress-dependent relocalization of translationally primed mRNPs to cytoplasmic granules that are kinetically and spatially distinct from P-bodies. J. Cell Biol. 2007;179:65–74. doi: 10.1083/jcb.200707010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kedersha N., Stoecklin G., Ayodele M., Yacono P., Lykke-Andersen J., Fritzler M. J., Scheuner D., Kaufman R. J., Golan D. E., Anderson P. Stress granules and processing bodies are dynamically linked sites of mRNP remodeling. J. Cell Biol. 2005;169:871–884. doi: 10.1083/jcb.200502088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimata Y., Ishiwata-Kimata Y., Yamada S., Kohno K. Yeast unfolded protein response pathway regulates expression of genes for anti-oxidative stress and for cell surface proteins. Genes Cells. 2006;11:59–69. doi: 10.1111/j.1365-2443.2005.00921.x. [DOI] [PubMed] [Google Scholar]
- Knop M., Siegers K., Pereira G., Zachariae W., Winsor B., Nasmyth K., Schiebel E. Epitope tagging of yeast genes using a PCR-based strategy: more tags and improved practical routines. Yeast. 1999;15:963–972. doi: 10.1002/(SICI)1097-0061(199907)15:10B<963::AID-YEA399>3.0.CO;2-W. [DOI] [PubMed] [Google Scholar]
- Lee K. S., Irie K., Gotoh Y., Watanabe Y., Araki H., Nishida E., Matsumoto K., Levin D. E. A yeast mitogen-activated protein kinase homolog (Mpk1p) mediates signalling by protein kinase C. Mol. Cell. Biol. 1993;13:3067–3075. doi: 10.1128/mcb.13.5.3067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsumoto T. K., Ellsmore A. J., Cessna S. G., Low P. S., Pardo J. M., Bressan R. A., Hasegawa P. M. An osmotically induced cytosolic Ca2+ transient activates calcineurin signaling to mediate ion homeostasis and salt tolerance of Saccharomyces cerevisiae. J. Biol. Chem. 2002;277:33075–33080. doi: 10.1074/jbc.M205037200. [DOI] [PubMed] [Google Scholar]
- Nelson K. K., Lemmon S. K. Suppressors of clathrin deficiency: overexpression of ubiquitin rescues lethal strains of clathrin-deficient Saccharomyces cerevisiae. Mol. Cell. Biol. 1993;13:521–532. doi: 10.1128/mcb.13.1.521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohya Y., Botstein D. Diverse essential functions revealed by complementing yeast calmodulin mutants. Science. 1994;263:963–966. doi: 10.1126/science.8310294. [DOI] [PubMed] [Google Scholar]
- Parker R., Sheth U. P bodies and the control of mRNA translation and degradation. Mol. Cell. 2007;25:635–646. doi: 10.1016/j.molcel.2007.02.011. [DOI] [PubMed] [Google Scholar]
- Patil C. K., Li H., Walter P. Gcn4p and novel upstream activating sequences regulate targets of the unfolded protein response. PLoS Biol. 2004;2:E246. doi: 10.1371/journal.pbio.0020246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prescianotto-Baschong C., Riezman H. Ordering of compartments in the yeast endocytic pathway. Traffic. 2002;3:37–49. doi: 10.1034/j.1600-0854.2002.30106.x. [DOI] [PubMed] [Google Scholar]
- Reijns M. A., Alexander R. D., Spiller M. P., Beggs J. D. A role for Q/N-rich aggregation-prone regions in P-body localization. J. Cell Sci. 2008;121:2463–2472. doi: 10.1242/jcs.024976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Romero-Santacreu L., Moreno J., Perez-Ortin J. E., Alepuz P. Specific and global regulation of mRNA stability during osmotic stress in Saccharomyces cerevisiae. RNA. 2009;15:1110–1120. doi: 10.1261/rna.1435709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salgado-Garrido J., Bragado-Nilsson E., Kandels-Lewis S., Seraphin B. Sm and Sm-like proteins assemble in two related complexes of deep evolutionary origin. EMBO J. 1999;18:3451–3462. doi: 10.1093/emboj/18.12.3451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmelzle T., Helliwell S. B., Hall M. N. Yeast protein kinases and the RHO1 exchange factor TUS1 are novel components of the cell integrity pathway in yeast. Mol. Cell. Biol. 2002;22:1329–1339. doi: 10.1128/mcb.22.5.1329-1339.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmid M., Jaedicke A., Du T. G., Jansen R. P. Coordination of endoplasmic reticulum and mRNA localization to the yeast bud. Curr. Biol. 2006;16:1538–1543. doi: 10.1016/j.cub.2006.06.025. [DOI] [PubMed] [Google Scholar]
- Sherman F. Getting started with yeast. Methods Enzymol. 1991;194:3–21. doi: 10.1016/0076-6879(91)94004-v. [DOI] [PubMed] [Google Scholar]
- Sheth U., Parker R. Decapping and decay of messenger RNA occur in cytoplasmic processing bodies. Science. 2003;300:805–808. doi: 10.1126/science.1082320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sidrauski C., Walter P. The transmembrane kinase Ire1p is a site-specific endonuclease that initiates mRNA splicing in the unfolded protein response. Cell. 1997;90:1031–1039. doi: 10.1016/s0092-8674(00)80369-4. [DOI] [PubMed] [Google Scholar]
- Sikorski R. S., Hieter P. A system of shuttle vectors and yeast host strains designed for efficient manipulation of DNA in Saccharomyces cerevisiae. Genetics. 1989;122:19–27. doi: 10.1093/genetics/122.1.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spang A., Grein K., Schiebel E. The spacer protein Spc110p targets calmodulin to the central plaque of the yeast spindle pole body. J. Cell Sci. 1996;109:2229–2237. doi: 10.1242/jcs.109.9.2229. [DOI] [PubMed] [Google Scholar]
- Squirrell J. M., Eggers Z. T., Luedke N., Saari B., Grimson A., Lyons G. E., Anderson P., White J. G. CAR-1, a protein that localizes with the mRNA decapping component DCAP-1, is required for cytokinesis and ER organization in Caenorhabditis elegans embryos. Mol. Biol. Cell. 2006;17:336–344. doi: 10.1091/mbc.E05-09-0874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swisher K.D., Parker R. Localization to, and effects of Pbp1, Pbp4, Lsm12, Dhh1, and Pab1 on stress granules in Saccharomyces cerevisiae. PLoS One. 2010;5:e10006. doi: 10.1371/journal.pone.0010006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teixeira D., Parker R. Analysis of P-body assembly in Saccharomyces cerevisiae. Mol. Biol. Cell. 2007;18:2274–2287. doi: 10.1091/mbc.E07-03-0199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teixeira D., Sheth U., Valencia-Sanchez M. A., Brengues M., Parker R. Processing bodies require RNA for assembly and contain nontranslating mRNAs. RNA. 2005;11:371–382. doi: 10.1261/rna.7258505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tharun S., He W., Mayes A. E., Lennertz P., Beggs J. D., Parker R. Yeast Sm-like proteins function in mRNA decapping and decay. Nature. 2000;404:515–518. doi: 10.1038/35006676. [DOI] [PubMed] [Google Scholar]
- Tharun S., Muhlrad D., Chowdhury A., Parker R. Mutations in the Saccharomyces cerevisiae LSM1 gene that affect mRNA decapping and 3′ end protection. Genetics. 2005;170:33–46. doi: 10.1534/genetics.104.034322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tharun S., Parker R. Targeting an mRNA for decapping: displacement of translation factors and association of the Lsm1p–7p complex on deadenylated yeast mRNAs. Mol. Cell. 2001;8:1075–1083. doi: 10.1016/s1097-2765(01)00395-1. [DOI] [PubMed] [Google Scholar]
- Trautwein M., Dengjel J., Schirle M., Spang A. Arf1p provides an unexpected link between COPI vesicles and mRNA in Saccharomyces cerevisiae. Mol. Biol. Cell. 2004;15:5021–5037. doi: 10.1091/mbc.E04-05-0411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Wuytswinkel O., Reiser V., Siderius M., Kelders M. C., Ammerer G., Ruis H., Mager W. H. Response of Saccharomyces cerevisiae to severe osmotic stress: evidence for a novel activation mechanism of the HOG MAP kinase pathway. Mol. Microbiol. 2000;37:382–397. doi: 10.1046/j.1365-2958.2000.02002.x. [DOI] [PubMed] [Google Scholar]
- Wilczynska Z., Happle K., Muller-Taubenberger A., Schlatterer C., Malchow D., Fisher P. R. Release of Ca2+ from the endoplasmic reticulum contributes to Ca2+ signaling in Dictyostelium discoideum. Eukaryot. Cell. 2005;4:1513–1525. doi: 10.1128/EC.4.9.1513-1525.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilhelm J. E., Buszczak M., Sayles S. Efficient protein trafficking requires trailer hitch, a component of a ribonucleoprotein complex localized to the ER in Drosophila. Dev. Cell. 2005;9:675–685. doi: 10.1016/j.devcel.2005.09.015. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.