We show that Kar9p polarity is instructed by a feedback loop that requires astral microtubules, actin cables, and Myo2p-based transport to enforce Kar9p loading to the bud-ward pole. This novel mechanism also provides the basis for a model unifying Kar9p polarity and the stereotyped pattern of spindle pole inheritance known to occur in yeast.
Abstract
In Saccharomyces cerevisiae, Kar9p, one player in spindle alignment, guides the bud-ward spindle pole by linking astral microtubule plus ends to Myo2p-based transport along actin cables generated by the formins Bni1p and Bnr1p and the polarity determinant Bud6p. Initially, Kar9p labels both poles but progressively singles out the bud-ward pole. Here, we show that this polarization requires cell polarity determinants, actin cables, and microtubules. Indeed, in a bud6Δ bni1Δ mutant or upon direct depolymerization of actin cables Kar9p symmetry increased. Furthermore, symmetry was selectively induced by myo2 alleles, preventing Kar9p binding to the Myo2p cargo domain. Kar9p polarity was rebuilt after transient disruption of microtubules, dependent on cell polarity and actin cables. Symmetry breaking also occurred after transient depolymerization of actin cables, with Kar9p increasing at the spindle pole engaging in repeated cycles of Kar9p-mediated transport. Kar9p returning to the spindle pole on shrinking astral microtubules may contribute toward this bias. Thus, Myo2p transport along actin cables may support a feedback loop by which delivery of astral microtubule plus ends sustains Kar9p polarized recruitment to the bud-ward spindle pole. Our findings also explain the link between Kar9p polarity and the choice setting aside the old spindle pole for daughter-bound fate.
INTRODUCTION
The differential fate of intracellular components and organelles coupled to chromosomal segregation along an axis of cell polarity is a recurrent theme observed in asymmetric cell divisions throughout evolution (Segal and Bloom, 2001; Gonczy, 2008). In Saccharomyces cerevisiae, the preanaphase spindle must orient along the mother-bud axis to drive chromosomal segregation across the bud neck. This polarized orientation is controlled by cytoplasmic or astral microtubules (aMTs) generated from each spindle pole by the respective spindle pole bodies (SPBs; Byers, 1981).
An intricate program for temporal and spatial control of aMT–cortex interactions culminates in the establishment of spindle polarity, with one SPB targeted to the bud and the other SPB confined to the mother cell (Smeets and Segal, 2002). Spindle polarity rests on the interplay between events intrinsic to the spindle pathway and the extrinsic asymmetric marking of cortical domains for aMT capture. A built-in asymmetry distinguishes SPBs by their history—the “old” SPB inherited from the preceding cell cycle and the “new” SPB that assembles as cells proceed through the G1/S transition (Pereira et al., 2001; Jaspersen and Winey, 2004). aMTs already present at the old SPB engage in interactions with the bud cortex, cued by the polarity determinant Bud6p (Shaw et al., 1997; Segal et al., 2000a). As the spindle assembles, Bud6p accumulates at the bud neck, thus restricting access to newly formed aMTs from the new SPB to the bud (Amberg et al., 1997; Segal et al., 2002). This program commits the old SPB to become the bud-ward pole or SPBbud (Pereira et al., 2001; Huisman and Segal, 2005).
Another layer of extrinsic control enforcing SPB asymmetric commitment is based on Kar9p, a protein that guides aMTs toward the bud along polarized actin cables. Kar9p is recruited at a SPB and then reaches aMT plus ends by binding the plus-end tracking protein Bim1p, the yeast homologue of EB1 (Vaughan, 2005). From the plus end, Kar9p acts as a cargo of the type V myosin Myo2p, thus linking the aMT to Myo2p transport (Beach et al., 2000; Korinek et al., 2000; Lee et al., 2000; Miller et al., 2000; Yin et al., 2000; Hwang et al., 2003; Pearson and Bloom, 2004). On disengagement from Myo2p, Kar9p may then return to the SPB on an aMT (Liakopoulos et al., 2003; Huisman et al., 2004; Cuschieri et al., 2006). Organization of polarized actin cables requires, among others, the yeast formins Bni1p and Bnr1p, which set up two axes of cell polarity from the bud tip and the bud neck, respectively (Pruyne et al., 2004b). Bud6p also contributes to this process by stimulating actin cable nucleation by formins (Kikyo et al., 1999; Moseley and Goode, 2005; Delgehyr et al., 2008). Conversely, formins are important to control the temporal distribution of Bud6p between the bud tip and the bud neck that is key to the partition of aMT–cortex interactions priming spindle polarity (Delgehyr et al., 2008).
Kar9p is found at both SPBs at onset of spindle assembly but is progressively polarized to mark the SPBbud (Huisman et al., 2004). As a result, Kar9p selectively guides aMT plus ends from one pole to the bud, enforcing spindle polarity (Liakopoulos et al., 2003; Maekawa and Schiebel, 2004). The guidance of aMT plus ends by Myo2p-based transport along actin cables toward the bud mediated by Kar9p is here referred to simply as “delivery of aMT plus ends.”
How polarization of Kar9p is achieved is unknown, although posttranslational modification of Kar9p has been proposed. Kar9p is a substrate of cyclin-dependent kinase (CDK) (Liakopoulos et al., 2003; Maekawa et al., 2003; Moore and Miller, 2007) but phosphorylation does not regulate asymmetric localization. Instead, Clb4p-dependent kinase may control Kar9p deployment to aMT plus ends (Maekawa and Schiebel, 2004). Indeed, a Kar9p mutant lacking CDK phosphorylation sites localized close to the SPB and did not support delivery of aMTs (Moore and Miller, 2007). Moreover, Clb4p-Cdk1p bound to Kar9p translocates along aMTs to modulate dynamic aMT–cortex interactions (Maekawa and Schiebel, 2004). Recently, Kar9p was shown to undergo sumoylation (Leisner et al., 2008; Meednu et al., 2008). Yet, symmetric localization of Kar9p in a smt3 mutant (SMT3 encodes SUMO) was fully reversed by inactivating the spindle assembly checkpoint (Leisner et al., 2008), arguing that symmetry was unrelated to the failure to sumoylate Kar9p. Thus, the significance of this modification for Kar9p polarization remains unclear. Finally, neither posttranslational modification would provide a mechanistic basis for coupling the choice that sets aside the old SPB to become the SPBbud with Kar9p polarization.
A final layer of extrinsic polarity supports coupling of successful chromosomal segregation across the bud neck with mitotic exit. This is based on the asymmetric localization to the committed bud-ward pole of signaling components regulating the mitotic exit network (MEN), such as Bfa1p (Bardin et al., 2000; Pereira et al., 2000; Doxsey et al., 2005). This system operates irrespective of intrinsic spindle polarity or SPB history, as it is always the pole ultimately translocating into the bud that becomes labeled by these components (Pereira et al., 2001).
As indicated above, establishment of spindle polarity entails the partnership between Bud6p and formins to generate spatial cues for outlining two axes of cell polarity and for partitioning aMT interactions during spindle formation (Pruyne et al., 2004a; Delgehyr et al., 2008). We therefore asked whether the direct perturbation of this program might impair Kar9p polarization in the course of spindle assembly. Here, we show that Kar9p polarity was disrupted in a bud6Δ bni1Δ mutant. This ultimately uncovered a novel mechanism involving actin cables and Myo2p-dependent delivery of aMT plus ends to instruct and maintain Kar9p localization at the SPBbud through a potential feedback loop.
MATERIALS AND METHODS
Yeast Strains, Plasmids, and Genetic Procedures
Yeast strains used in this study are listed in Supplemental Table S1 and were isogenic to 15DauA—a ade1 his2 leu2-3112 trp1-1a ura3Dns arg4, unless indicated. myo2-17 and myo2-18 alleles (Schott et al., 1999) were introduced into 15D background by transformation with the plasmid pRS305myo2-17 or pRS305myo2-18 (kindly provided by Felipe Santiago, Cornell University, Ithaca, NY). KAR9-GFP3 was introduced by transformation with pRM3226 (Moore and Miller, 2007; a gift from Rita Miller, Oklahoma State University, Stillwater, OK). Standard yeast genetic procedures and media were used (Guthrie and Fink, 1991). Yeast cultures were grown at 25°C unless stated. Further details on constructs, growth conditions, and cell synchronization are provided in Supplemental Data.
Digital Imaging Microscopy
Still images and time-lapse recordings were carried out using an Eclipse E800 microscope (Nikon, Tokyo, Japan) with a CFI Plan Apochromat 100×, numerical aperture 1.4 objective and a CoolSNAP-HQ charge-coupled device camera as described previously (Huisman et al., 2007). Images of cells expressing cyan fluorescent protein (CFP)-Tub1p or Spc42p-CFP and Kar9p-GFP3 were obtained using a CFP/yellow fluorescent protein filter set (Huisman et al., 2004). Still images were obtained as five-plane Z-stacks at a distance of 0.8 μm between planes using 2 × 2 binning and processed with MetaMorph software (Molecular Devices, Sunnyvale, CA). Digital overlays of images were used for scoring. Kar9p modes of localization were arbitrarily grouped into four categories: 1) one pole: only one SPB or aMTs from one SPB carried label; 2) asymmetric: unequal label between the two SPBs (> 4-fold difference in intensity); 3) partially symmetric: unequal label (<4-fold difference), and 4) symmetric: both poles labeled equally (within a-fold difference). Time lapse analysis was carried out by projecting stacks of three planes at a Z-distance of 1 μm (Huisman et al., 2004). Measurements in digital images were performed using MetaMorph software. Linescan analysis was carried out along the spindle and aMT axes to assess the relative fluorescence label between SPBs to aid in scoring. Spindles were scored as oriented if a line drawn through the long axis of the spindle intersected the bud neck (Theesfeld et al., 1999). Kar9p label intensity was measured as the integrated intensity within the cell limits after subtracting the cell background.
RESULTS
A bud6Δ bni1Δ Mutant Exhibits Impaired Kar9p Polarization to the SPBbud
Given the importance of spatial cues in the program that partitions aMT–cortex interactions such that the old SPB becomes the SPBbud (Segal et al., 2000a; Pereira et al., 2001), we examined the extent of Kar9p polarization when this program is perturbed by mutations disrupting cortical components. This analysis revealed an outstanding interaction between bud6Δ and bni1Δ mutations.
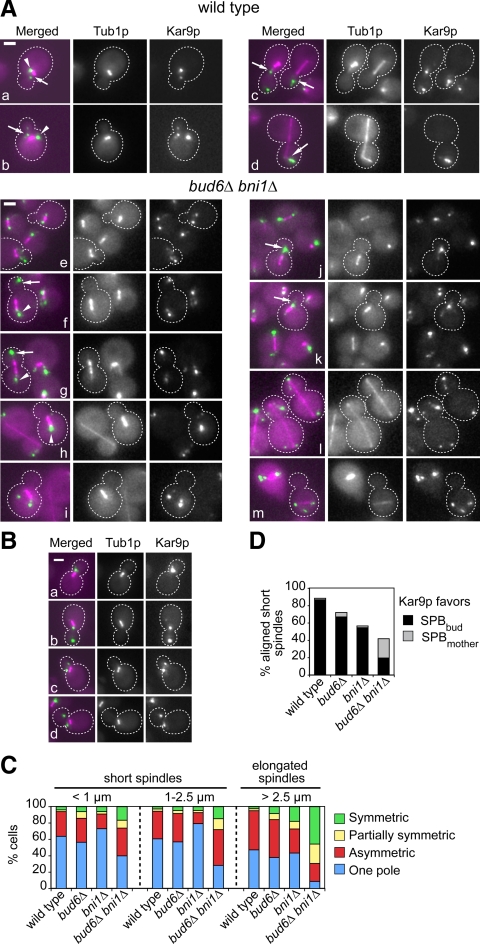
As expected, Kar9p-GFP3 labeled both SPBs of wild-type cells in the early stages of spindle assembly (Figure 1A, a and b, arrow and arrowhead), to then become polarized to the SPBbud (Figure 1Ac, arrows). Polarization favoring the SPBbud was maintained during anaphase (Figure 1Ad, arrow). This trend was also apparent in bni1Δ or bud6Δ single mutants, with a modest effect on <1-μm-long spindles observed in bud6Δ cells. By contrast, bud6Δ bni1Δ cells exhibited a marked loss of Kar9p-GFP3 polarization (Figure 1A, e–m). As the spindle assembled, Kar9p-GFP3 remained associated with both SPBs in nearly 80% of cells containing preanaphase spindles (Figure 1C). Excess symmetry in bud6Δ bni1Δ cells was not accompanied by bulk changes in Kar9p phosphorylation (data not shown). Remarkably, the ability to polarize Kar9p-GFP3 was entirely uncoupled from spindle alignment or SPBbud identity. Indeed, in addition to cells with misaligned spindles labeled at both poles (Figure 1A, e and i), cells also exhibited correctly positioned spindles in which Kar9p-GFP3 was nevertheless symmetric (Figure 1Af, arrow and arrowhead), asymmetric to favor the SPBbud (Figure 1Ag, arrow and arrowhead), asymmetric favoring the mother-bound pole or SPBmother (Figure 1Ah, arrowhead), or strongly polarized to the SPBbud (Figure 1A, j and k, arrows). Lack of correlation between spindle alignment and Kar9p-GFP3 symmetry persisted whether spindle elongation took place across the bud neck (Figure 1Al) or not (Figure 1Am). More than 90% of preanaphase wild-type cells with aligned spindles polarized Kar9p to the SPBbud (i.e., one pole labeled or strongly asymmetric), whereas this correlation was lost in bud6Δ bni1Δ cells (Figure 1D). In the small proportion of preanaphase spindles that were misaligned in wild-type cells (10.7%), most favored one pole, indicating that misalignment per se did not increase symmetric recruitment of Kar9p (Figure 2A). Similarly, misaligned preanaphase spindles in bud6Δ bni1Δ cells (51.3%) were not enriched for symmetry, emphasizing the lack of correlation between Kar9p polarization and spindle alignment in the mutant (Figure 2A). This indicated that the disruption of cell polarity in the bud6Δ bni1Δ mutant (Delgehyr et al., 2008) prevented an instructive mechanism that enforces progressive recruitment of Kar9p onto the SPBbud, in addition to compromising Kar9p function in delivery of aMT plus ends due to perturbation of actin integrity.
Figure 1.
Kar9p polarization to the SPBbud is disrupted in a bud6Δ bni1Δ mutant. (A) Representative images of wild-type (a–d) and bud6Δ bni1Δ cells (e–m) expressing Kar9p-GFP3 (in green) and CFP-Tub1p (in magenta), showing the extent of polarization of Kar9p-GFP3 along the spindle pathway. Wild-type cells exhibited Kar9p-GFP3 label on both poles at onset of spindle assembly (a and b, arrows and arrowheads). Label became polarized to the SPBbud in preanaphase spindles (c, arrows) and continued to favor the SPBbud in anaphase (d, arrow). bud6Δ bni1Δ cells did not coordinate polarization of Kar9p-GFP3 with spindle alignment. (e) Two cells with misaligned spindles marked at both poles by Kar9-GFP3. (f) A cell with a short spindle carrying symmetric Kar9-GFP3 label (arrow and arrowhead) despite spindle alignment. (g) A cell with a correctly aligned spindle and Kar9p-GFP3 label favoring the SPBbud (arrow). (h) Kar9p-GFP3 preferentially marks the SPBmother (arrowhead) despite spindle alignment. (i) Misaligned spindle symmetrically labeled by Kar9-GFP3. (j and k) Kar9p-GFP3 label is associated mainly with the SPBbud (arrows). (l) Two cells with elongated spindle exhibit Kar9p-GFP3 label associated with both poles despite alignment. (m) A cell containing a misaligned anaphase spindle with Kar9p-GFP3 present at both poles. In conclusion, loss of polarity persisted throughout anaphase and seemed unrelated to alignment or SPB identity. (B) Categories of Kar9p localization used in this study: one pole (a), asymmetric (b), partially symmetric (c), and symmetric (d). (C) Distribution of asynchronous cells according to their labeling by Kar9p-GFP3 as depicted in B. Cells with <1-μm-long spindles (n > 282 cells); 1- to 2.5-μm-long spindles (n > 290 cells) or elongated spindles (n > 270 cells) in asynchronous cultures were scored. (D) Distribution of preanaphase cells with correctly aligned spindles according to the relative bias of Kar9p-GFP3 toward the SPBbud or SPBmother, n > 390. The frequency of preanaphase spindle misalignment was 10.7% in wild-type cells, 25% in bud6Δ cells, 40% in bni1Δ cells, and 51.3% in bud6Δ bni1Δ cells. Only in bud6Δ bni1Δ cells Kar9p-GFP3 no longer favored the SPBbud. For a detailed breakdown of all categories scored, see Supplemental Figure S1. Bars, 2 μm.
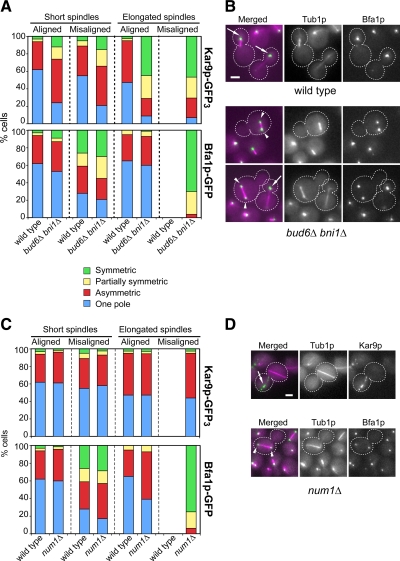
Figure 2.
Kar9p-GFP3 symmetry is specifically caused by disruption of cell polarity irrespective of spindle orientation. (A) Distribution of cells according to spindle alignment and Kar9p-GFP3 (top) or Bfa1p-GFP (bottom) polarization in wild-type versus bud6Δ bni1Δ cells. The frequency of preanaphase spindle misalignment was 10% in wild-type cells (n = 400) and 52% in bud6Δ bni1Δ cells (n = 530). The frequency of misaligned anaphase spindles in bud6Δ bni1Δ cells was 16% (n = 275). No misaligned anaphase spindles were observed in wild-type cells (n = 270). Bfa1p-GFP symmetry strongly correlated with spindle mispositioning. By contrast, Kar9p-GFP3 localized symmetrically to a similar extent among both aligned and misaligned spindles. (B) Representative images for localization of Bfa1p-GFP in wild-type versus bud6Δ bni1Δ cells. Overlays of fluorescence images of Bfa1p-GFP (in green) and CFP-Tub1p (in magenta) are shown. Arrows point to the SPBbud. Arrowheads in images corresponding to bud6Δ bni1Δ cells point to symmetric label in misaligned short (top) and elongated (bottom) spindles. (C) Distribution of num1Δ cells according to spindle alignment and Kar9p-GFP3 (top) or Bfa1p-GFP (bottom) polarization. The frequency of spindle misalignment in the num1Δ mutant was 15% in preanaphase cells (n = 285) and 27% in anaphase cells (n = 151). num1Δ cells were not disrupted for Kar9p polarity irrespective of spindle alignment. By contrast, Bfa1p-GFP was symmetrically localized in misaligned spindles. (D) Representative images for localization of Kar9p-GFP3 or Bfa1p-GFP (shown in green) relative to CFP-Tub1p (shown in magenta) in num1Δ cells with misaligned spindles. An arrow points to Kar9p-GFP3 polarized label. Arrowheads point to Bfa1p-GFP symmetric label. Bar, 2 μm.
Following these results, wild-type and bud6Δ bni1Δ cells were examined for their ability to support correct polarization of Bfa1p, a regulator of the MEN that favors the SPBbud (Caydasi and Pereira, 2009; Monje-Casas and Amon, 2009). In contrast to Kar9p, nearly 90% of preanaphase cells that contained aligned spindles showed a strong bias for Bfa1p-green fluorescent protein (GFP) at the SPBbud (Figure 2A) in both wild-type and bud6Δ bni1Δ cells. Moreover, in 90% of anaphase cells containing elongated spindles across the bud neck, Bfa1p-GFP strongly favored the SPBbud, in sharp contrast to Kar9p-GFP3 symmetric localization in the mutant at the same stage. Yet, consistent with the importance of asymmetric aMT–cortex interactions in enforcing localization of Bfa1p to the SPBbud in response to alignment (Caydasi and Pereira, 2009; Monje-Casas and Amon, 2009), 50% of bud6Δ bni1Δ cells containing misaligned preanaphase spindles, typically away from the bud neck, showed symmetric localization of Bfa1p-GFP (Figure 2, A and B, arrowheads). Misaligned anaphase spindles of bud6Δ bni1Δ cells (16% of all anaphases in the mutant) were also markedly enriched for Bfa1p symmetry. In conclusion, bud6Δ bni1Δ cells were impaired in polarizing Kar9p to the SPBbud (even past anaphase), whereas fully proficient to regulate the recruitment of Bfa1p-GFP upon spindle alignment. Finally, the fact that Kar9p-GFP3 symmetry did not relate to spindle misalignment but specifically to the bud6Δ bni1Δ mutations was further confirmed by the contrasting behavior of Kar9p versus Bfa1p in a num1Δ mutant (in which spindle orientation is impaired by the inactivation of the cortical anchor for dynein). Indeed, a num1Δ mutation did not affect Kar9p polarity irrespective of spindle alignment (Figure 2, C and D, arrow). By contrast, misaligned spindles in the same mutant were symmetrically decorated by Bfa1p-GFP (Figure 2, C and D, arrowhead). In conclusion, Kar9p and Bfa1p polarized localization in response to cell polarity may arise by distinct mechanisms.
Actin Depolymerization Increases Kar9p Symmetric Localization
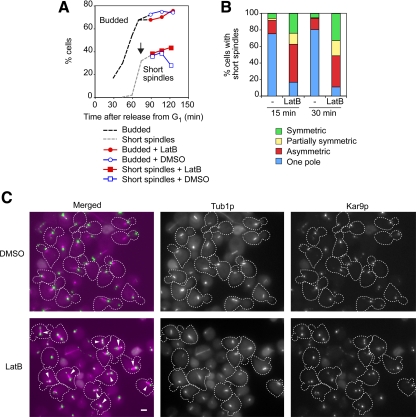
The impact of bud6Δ and bni1Δ mutations on Kar9p distribution suggested the importance of actin organization for polarizing Kar9p. This was directly probed by treating otherwise wild-type cells with the actin-depolymerizing drug latrunculin (Lat). A synchronous cell population was allowed to proceed past bud emergence and was then treated with 100 μM LatB (Figure 3A, arrow) to selectively induce actin cable depolymerization (Irazoqui et al., 2005). More than 90% of budded cells containing a short spindle exhibited Kar9p-GFP3 recruitment to both poles within 30 min (Figure 3, B and C, arrowheads). The effect of LatB was reversible (see below); yet, it randomized the pattern of SPB inheritance (data not shown). Treatment with 200 μM LatA, additionally disrupting actin patches, also increased Kar9p symmetry (Supplemental Figure S2A).
Figure 3.
Effect of actin depolymerization by LatB on Kar9p-GFP3 localization. (A) After arrest in G1 by α factor, wild-type cells were released and allowed to proceed past bud emergence. At the point indicated by the arrow, the culture was divided for treatment with either 100 μM LatB or dimethyl sulfoxide (DMSO) as a control, and aliquots were drawn for scoring Kar9p polarity. Samples also were taken during the time course to monitor budding and progression of the spindle pathway (n = 300 cells). (B) Aliquots from the time course were analyzed for Kar9p distribution in short spindles after treatment for 15 min (nLatB = 268 cells; nDMSO = 204 cells) and 30 min (nLatB = 262 cells; nDMSO = 191 cells). More than 90% of cells exhibited Kar9p label at both poles after 30-min treatment with LatB. (C) Representative fields of cells treated with DMSO or LatB for 30 min from the experiment depicted in A. Treatment with LatB induced both spindle misalignment and symmetric localization of Kar9p (arrowheads). Bar, 2 μm.
Actin depolymerization triggers the cell morphogenesis checkpoint that ultimately inhibits mitotic CDK (Keaton and Lew, 2006). The effect of LatB on Kar9p symmetry, however, was independent of this checkpoint because a swe1Δ mutant maintained a wild-type response to the drug (Supplemental Figure S2B). Symmetry in response to LatB occurred without changes in Kar9p phosphorylation and was unaffected in cyclin mutants (data not shown). Thus, Kar9p symmetric recruitment upon disruption of actin organization did not involve CDKs. Moreover, Kar9p symmetry increased in response to LatB treatment in cells carrying either a tub2C354S allele that reduces microtubule turnover (Gupta et al., 2002) or a mad2Δ mutation (Supplemental Figure S2, C and D), indicating that this increase in Kar9p symmetry did not depend on changes in aMT stability and did not require cross-talk with the spindle assembly checkpoint.
Finally, Kar9p polarization depended on actin integrity even beyond the actin-sensitive period of spindle orientation (Theesfeld et al., 1999). Indeed, LatB treatment after preanaphase spindle orientation was fully accomplished also resulted in increased Kar9p symmetry without affecting spindle alignment (Supplemental Figure S3), indicating that actin integrity is required not only for promoting but also for maintaining polarized Kar9p localization to the SPBbud. Moreover, this observation confirmed that Kar9p symmetry is a direct consequence of actin perturbation and unrelated to spindle alignment.
One Formin-dependent Axis of Cell Polarity Is Sufficient for Kar9p Polarization
The requirement for actin integrity in enforcing Kar9p polarization may underscore the role of cell polarity in promoting the distinctive identity of cortical compartments in mother and daughter cells as evidenced, for example, by the asymmetric control of mating type switching (Cosma, 2004).
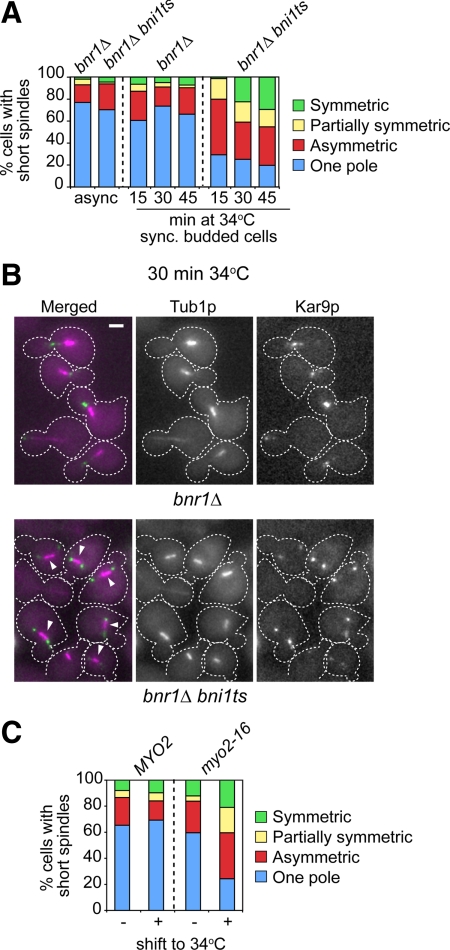
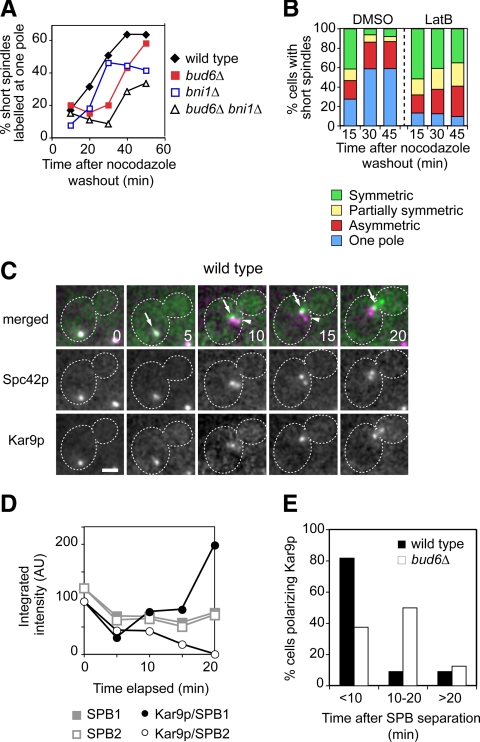
Two formin-dependent axes of cell polarity outline the organization of actin cables in yeast—one set from the bud tip (Bni1p dependent) and the other set from the bud neck (Bnr1p dependent). A bni1Δ mutant supported correct Kar9p polarization to the SPBbud (Figure 1C), even though cell polarity and actin organization within the bud are severely compromised. Similarly, a bnr1Δ mutant exhibited correct Kar9p polarization (Figure 4, A and B) in spite of the reduction in actin cables in the mother cell (Pruyne et al., 2004a). Yet, a bnr1Δ bni1ts mutant exhibited a marked increase in Kar9p symmetry upon shift of synchronized budded cells to 34°C (Figure 4, A and B, arrowheads).
Figure 4.
At least one formin and Myo2p are required for correct Kar9p polarity. (A and B) Effect of inactivation of both yeast formins on Kar9p polarized localization in synchronized budded cells. bnr1Δ or bnr1Δ bni1ts cells were released from a G1 block at 23°C, and samples were taken to monitor budding and progression of the spindle pathway (data not shown). (A) At 45 min after release from G1, budded cells were shifted to 34°C, and still images were acquired for scoring Kar9p localization onto short spindles after a 15-min incubation (nbnr1Δ = 249; nbnr1Δbni1ts = 75), 30 min (nbnr1Δ = 281; nbnr1Δbni1ts = 325), and 45 min (nbnr1Δ = 83; nbnr1Δbni1ts = 305). For reference, Kar9p localization in asynchronous cultures is shown (n > 226 cells). The bni1ts allele bni1-FH2#1 (Sagot et al., 2002) was introduced into 15D background as described in Supplemental Data. Single bnr1Δ or bni1Δ mutants exhibited correct Kar9p polarization, whereas combined inactivation of both formins increased Kar9p symmetry. (B) Representative images of cells from the experiment shown in A after a 30-min incubation at 34°C. Overlays show CFP-Tub1p (in magenta) and Kar9p-GFP3 (in green). Bar, 2 μm. Arrowheads point to cells with disrupted Kar9p polarity. (C) Distribution of Kar9p modes of localization in logarithmic cultures of MYO2 or myo2-16 cells grown at 23°C (nMYO2 = 152; nmyo2-16 = 99) or after 1-h shift to 34°C (nMYO2 = 184; nmyo2-16 = 275). Only budded cells were scored. After a temperature shift, the myo2-16 mutant exhibited a marked disruption of Kar9p polarized distribution.
To further assign elements participating in a mechanism by which Kar9p may single out the SPBbud dependent upon actin cables, the possible roles of the yeast type V myosins Myo2p and the nonessential Myo4p/She1p (Pruyne et al., 2004b) were explored. A shift to 34°C of an asynchronous culture of a myo2-16 mutant, which carries a conditional-lethal allele disrupting the function of the Myo2p cargo domain without directly perturbing actin cable organization (Schott et al., 1999), led to loss of Kar9p polarity (Figure 4C). By contrast, a myo4Δ strain was unaffected (data not shown). Thus, Kar9p polarization required intact actin cables and specifically Myo2p-dependent transport.
Kar9p Repolarization upon Recovery from Nocodazole Treatment Depends on Cell Polarity Determinants and Actin
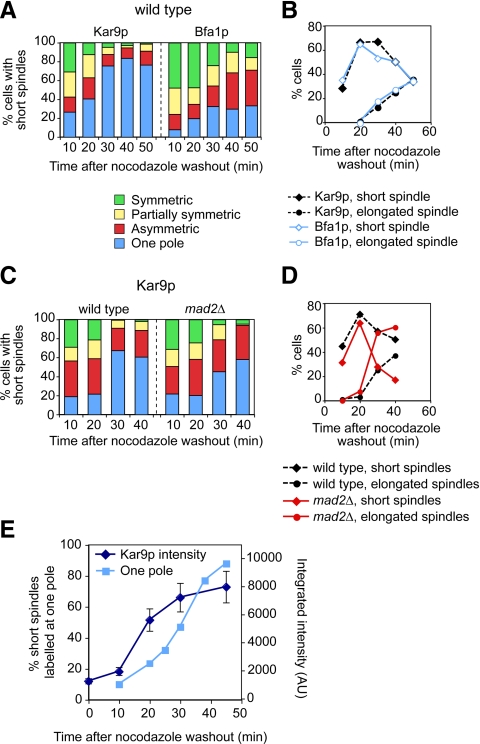
Cortical cues promote spindle polarity by directing the old SPB, already carrying aMTs, to become the SPBbud while confining the new SPB to the mother cell (Segal et al., 2000a; Yeh et al., 2000). This program that helps define SPB identity based on history may instruct Kar9p polarization to the SPBbud. Accordingly, exposure to the MT poison nocodazole erases SPB identity and randomizes SPB fate (Pereira et al., 2001). Moreover, nocodazole induces Kar9p symmetric recruitment onto the now equivalent SPBs (Huisman and Segal, 2005). Yet, a spindle can still reform and align if cells are allowed to recover from the drug treatment, suggesting the ability of the spindle to rebuild polarity even though SPB differential history has been erased.
We therefore asked how symmetry breaking as the spindle reforms from equivalent SPBs relates to Kar9p polarity. To this end, Kar9p localization was monitored (Figure 5A) alongside reformation of the mitotic spindle (Figure 5B), during recovery from nocodazole treatment. Wild-type cells expressing Spc42p-CFP (a marker for the SPB) and either Kar9p-GFP3 or Bfa1p-GFP were released from a treatment with 15 μg/ml nocodazole to follow their recovery. Cells in which SPBs had separated were scored for the extent of polarized localization (Figure 5A). Kar9p initially localized to both SPBs and became polarized within 30 min from the nocodazole washout. By contrast, polarization of Bfa1p, which depends on both correct polarized aMT–cortex interactions and spindle alignment, proceeded more slowly.
Figure 5.
Kar9p repolarization upon recovery from nocodazole treatment highlights the importance of intact MTs for Kar9p localization to the SPBbud. (A and B) Time course for repolarization of Kar9p versus Bfa1p after nocodazole washout in wild-type cells. Asynchronous wild type cells expressing Spc42p-CFP and either Kar9p-GFP3 or Bfa1p-GFP were incubated in 15 μg/ml nocodazole for 45 min. Cells were washed and then resuspended in fresh medium to resume cell cycle progression in the absence of the drug. Localization of Kar9p-GFP3 or Bfa1p-GFP (A) and formation of spindles (B) were scored at the indicated time points. Kar9p polarity was restored by 30 min. By contrast, Bfa1p presence on both spindle poles persisted throughout the time course. (C and D) Intact MTs are required for correct Kar9p polarization to the SPBbud irrespective of the spindle assembly checkpoint. Depolymerization of MTs in a mad2Δ mutant also led to Kar9p-GFP3 symmetry (C). Time course experiment for wild-type or mad2Δ cells expressing Kar9p-GFP3 and Spc42p-CFP as in A except that cells were incubated with nocodazole for only 30 min. Kar9p polarity (C) and formation of spindles in wild-type and mad2Δ cells (D) were scored at the indicated time points. (E) Kar9p-GFP3 recruitment increased after nocodazole washout. Wild-type cells were treated as described in A. Plot depicts Kar9p label intensity per cell along a time course for recovery after nocodazole washout. For reference, the accumulation of cells labeled at a single pole is also shown. Integrated intensity was measured in >50 cells by using digital images, and mean values are plotted. Error bars, SEM.
Nocodazole treatment could evoke the spindle assembly checkpoint (Musacchio and Salmon, 2007). However, a mad2Δ strain also exhibited symmetric Kar9p localization upon nocodazole treatment (Figure 5, C and D), indicating that loss of Kar9p polarity resulted from disrupting MTs without involving this checkpoint.
Kar9p label intensity was relatively low in the presence of nocodazole, ∼15% relative to the maximal label present in asynchronous cells (Figure 5E). After nocodazole washout, the intensity of the label per cell increased, pointing to a contribution of aMTs to Kar9p loading. Kar9p repolarization followed, showing that polarized label stemmed from a net increase in the amount of Kar9p associated with one SPB. The extent of this differential increase could not be measured with statistical significance due to the variability in the label among cells for individual SPBs and associated aMTs in the later part of the time course.
Recovery from nocodazole treatment was then monitored in cell polarity mutants. Relative to wild-type cells, bud6Δ cells exhibited a delay in repolarization of Kar9p but eventually reached a level comparable with that observed in the asynchronous culture (Figure 6A and Supplemental Figure S4A). Failure to break symmetry led to spindle transits into the bud (data not shown). A bud6Δ bni1Δ mutant exhibited a further delay relative to a bud6Δ mutant, whereas a bni1Δ mutant initiated recovery like wild-type cells but leveled off to an intermediate value. Finally, symmetry breaking in wild-type cells after nocodazole washout was prevented by LatB (Figure 6B). The percentage of cells exhibiting label at both poles remained constant, yet absolute symmetry progressively decreased, demonstrating that increased Kar9p accumulation in the presence of aMTs permitted further stochastic variation in Kar9p loading among SPBs over time. This may explain why absolute symmetry may not be achievable following a 30-min LatB treatment that renders >90% of cells exhibiting label on both SPBs (Figure 3).
Figure 6.
Efficient Kar9p repolarization to the SPBbud requires polarity determinants and actin cables. (A) Time course analysis of Kar9p repolarization after treatment with nocodazole in the indicated strains was carried out as described in Figure 5. For simplicity, the “one pole” category is depicted in the plot. For full details on the time course see Supplemental Figure S3A. Cell polarity mutants were delayed in Kar9p repolarization. (B) LatB prevented Kar9p repolarization after nocodazole washout. Time course analysis in wild-type cells was carried out as described in Figure 5 except that after removal of nocodazole, part of the culture was treated with either dimethyl sulfoxide (DMSO) or 100 μM LatB for the indicated times, and aliquots were drawn for scoring (n > 128). In the presence of LatB, symmetry persisted but unequal loading to the SPBs increased over time. (C and D) Time lapse analysis of Kar9p behavior during recovery from nocodazole treatment in a wild-type cell. (C) Unseparated SPBs became repositioned near the bud neck (0–10 min, arrows). At onset of SPB separation (5–10 min) Kar9p preferentially marked one SPB (arrow vs. arrowhead). Bias persisted during alignment (10–20 min). Numbers indicate time elapsed in minutes. Bars, 2 μm. (D) Label intensities for Spc42p-CFP at SPBs (SPB1 is indicated by the arrow in C) and Kar9p-GFP3 associated with each pole during the time lapse depicted in C. When SPB separation began at 5 min, Kar9p labeled SPBs unequally. At 10 min, SPBs are clearly separated and Kar9p label increased at SPB1. Further increase occurred by 20 min. (E) Summary of time lapse analysis for symmetry breaking upon recovery from nocodazole treatment in wild-type versus bud6Δ cells. In wild-type cells, 45.8% cells recorded (n = 24 cells) exhibited Kar9p symmetric label at onset of SPB separation that became polarized within 10 min. In bud6Δ cells, 69.6% (n = 24 cells recorded) exhibited symmetric label that took longer to repolarize. For further representative time lapse sequences see Supplemental Figure S3C.
For further validation, symmetry breaking was also evaluated in single cells by time-lapse recordings at relatively low time resolution. This made it possible to use dual-color imaging to score the approximate time elapsed from the separation of SPBs marked symmetrically until Kar9p polarized distribution became established (Figure 6, C–E, and Supplemental Figure S4C). Although it was not technically possible to fully correlate Kar9p behavior and MT–cortex interactions, the general trend was that, during recovery, SPBs of wild-type cells moved toward the bud neck and often before SPB separation, as Kar9p-mediated delivery of aMT plus ends was restored. This was followed by SPB separation in the presence of polarized Kar9p (Figure 6C, 10 min, arrow and arrowhead; and D) in 54.2% of cells recorded (n = 24 cells). As the spindle became aligned, Kar9p label on the newly designated SPBbud intensified (Figure 6C, 15–20 min, arrows; and D). The observed increase in Kar9p recruitment at the SPBbud validated the results of the population analysis depicted in Figure 5E. In cases in which Kar9p was still symmetric when SPB separation took place, Kar9p polarization seemed to coincide with the reorientation of the spindle (Supplemental Figure S4C). In 82% of cells label was repolarized within 10 min from SPB separation (Figure 6E). By contrast, a bud6Δ mutant exhibited a lower frequency of asymmetric label at the time of SPB separation (30.4% of cells recorded; n = 23 cells). Moreover, 62.5% of cells exhibiting symmetric label took longer than 10 min to break symmetry (Figure 6E) with SPB separation often preceding complete repositioning near the bud neck as cells recovered (Supplemental Figure S4C). In either case, symmetry breaking was never observed without initial SPB movement toward the bud (n > 23 cells), suggesting that the initiation of transports was a key factor in rebuilding Kar9p polarity.
Together, symmetry breaking required aMTs and correlated best with the engagement of SPBs in Kar9p-mediated movement. In wild-type cells, this mechanism led to relatively fast symmetry breaking once aMTs reformed (whether one pole gained access to the bud or not). In bud6Δ cells, initial engagement in Kar9p-mediated delivery of aMT plus ends might be delayed due to the reduction in actin cables within the mother cell. However, once SPBs began to move closer to the bud, Kar9p became repolarized with label intensifying at the SPB eventually directed toward the bud.
Kar9p Behavior during Recovery from LatB Treatment
Given the possible relationship between Myo2p-based transport of aMT plus ends and Kar9p polarity suggested by our results, the behavior of Kar9p during the reestablishment of actin-dependent guidance of aMT plus ends was further examined after recovery from treatment with LatB. Synchronized budded cells were treated with 100 μM LatB, allowed to recover in the presence of fresh medium, and mounted to carry out time lapse recordings.
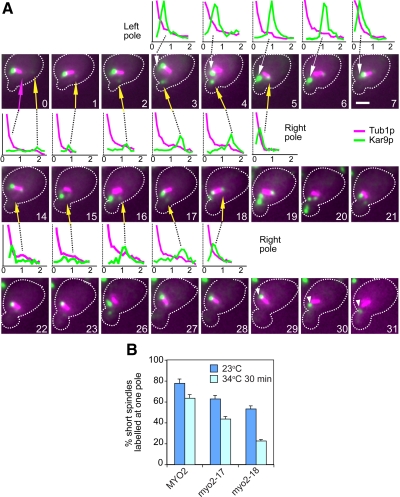
The effect of LatB was indeed reversible, because Kar9p-decorated aMTs emanating from each spindle pole began the characteristic angular displacements relative to the polarity-axis reflecting delivery of aMT plus ends toward the bud (Liakopoulos et al., 2003; Huisman et al., 2004). The concomitant SPB movement led to a seesaw-like behavior of the spindle. During alternating transits, the load of Kar9p onto both SPBs persisted (Figure 7A, arrows). Over time, delivery of plus ends of aMTs emanating from one particular SPB gained momentum and polarity became established (Figure 7A, 28–31 min, arrowheads), suggesting that cycles of aMT delivery enforced the localization of Kar9p to a particular SPB (79%; n = 249 aMTs delivered toward the bud). Enhanced label at the SPB was partly contributed by Kar9p returning to the SPB on a depolymerizing aMT after a transport event (e.g., Figure 7A, 4–5 min, yellow arrow; 6–7 min, white arrow) in agreement with previous observations at higher temporal resolution (Liakopoulos et al., 2003; Huisman et al., 2004; Cuschieri et al., 2006). Even when asymmetric, the residual presence of relatively low amounts of Kar9p at one SPB was sufficient to engage that pole in aMT mediated transports that were followed by an increase in Kar9p loading to the same pole (e.g., Figure 7A, 0–5 min and 14–18 min, yellow arrows). Due to lack of temporal resolution, however, it was not possible to estimate a threshold in Kar9p recruitment below which delivery of aMT plus ends along actin cables would cease. By contrast, Kar9p polarity was quickly restored (<10 min; data not shown) in cells in which symmetrically labeled spindles were already inserted at the bud neck at the start of the time lapse presumably posing a constraint to Kar9p-based movement by the mother-bound pole.
Figure 7.
Kar9p accumulation bias may be enforced by sustained cycles of aMT delivery along actin cables. (A) Kar9p behavior after recovery from treatment with LatB. Wild-type cells were synchronized and released from G1 to allow formation of a bud before treatment with LatB. Overlays of fluorescence images for Kar9p (in green) and Tub1p (in magenta) are shown. Linescans for fluorescence intensity depict Kar9p-GFP3 distribution along an aMT (with the spindle pole close to the origin and the plus end always to the right; distance in micrometers) for the indicated frames. Seesaw-like behavior of the spindle accompanied Kar9p-mediated transport from alternating poles because aMTs decorated by Kar9p displayed the characteristic angular movements. Initially, Kar9p was asymmetric, but a cycle of transport engaging the right pole (yellow arrows; 0–5 min) led to intensified label at that pole as the aMT depolymerized (5 min). Label decreased as the left pole (white arrows) engaged in deliveries (5–7 min). A second cycle from the right pole (14–18 min, yellow arrows) triggered an increase in Kar9p label at this pole. Both poles continued to move in response to alternating transits, although the left pole was favored, restoring the bias. Progressively, label disappeared from the right pole (28–31 min) and persisted on the left pole and further transports began to align the spindle (29–31 min, arrowheads). Numbers indicate time elapsed in minutes. Bars, 2 μm. (B) Inactivation of a myo2ts allele disrupting Kar9p binding to the cargo domain of Myo2p increased Kar9p symmetry. Plot for distribution of cells with short spindles labeled at one pole by Kar9p in asynchronous MYO2, myo2-17 (encoding a cargo domain mutant with defects in polarized secretion, yet able to bind Kar9p) and myo2-18 (encoding a mutant disrupted for Kar9p binding) cultures grown at 23°C or transferred to 34°C for 30 min.
Thus, we asked whether actin cables may promote, at least in part, Kar9p polarization by supporting a bias in Kar9p recruitment to the SPB engaged in Myo2p-dependent delivery of aMT plus ends. Among a collection of temperature-sensitive alleles of MYO2 that disrupt cargo domain functions in vesicular trafficking and polarized growth (Schott et al., 1999), myo2-18 also disrupted the corresponding interaction between Kar9p and the myosin tail, whereas myo2-17 did not (Yin et al., 2000). In support of the importance of Myo2p for a mechanism that promotes asymmetric loading in response to delivery of Kar9p-decorated aMTs along actin cables, myo2-18 led to a marked decrease in cells labeled at a single SPB relative to myo2-17 after a shift to a semipermissive temperature (Figure 7B). By contrast, the same treatment disrupted Sec4p-GFP localization in both mutants (myo2-18, 66.5% ± 4.9; myo2-17 65.5% ± 0.7; MYO2, 13%). Thus, the role of Myo2p in promoting Kar9p polarity was directly linked to the ability of Kar9p to act as cargo in Myo2p-based transport.
DISCUSSION
A Feedback Loop May Instruct Kar9p Polarization to the SPBbud
Kar9p drives spindle orientation by linking aMT plus ends from a designated SPB, the bud-ward pole or SPBbud, to Myo2p-based transport. Here, we show that a bud6Δ bni1Δ mutant exhibited loss of Kar9p polarization throughout the spindle pathway—Kar9p was symmetric or correlated poorly with the SPBbud. Yet, the same mutant supported Bfa1p polarization coupled to spindle alignment. Thus, layers of extrinsic control of Bfa1p polarity still persisted (Smeets and Segal, 2002; Yoshida et al., 2005; Caydasi and Pereira, 2009) consistent with the ability of bud6Δ bni1Δ cells to delay mitotic exit in the presence of misaligned spindles.
The secretory pathway and actin integrity are required to maintain cell polarity (Ayscough et al., 1997; Jin and Amberg, 2000; Pruyne et al., 2004b; Irazoqui et al., 2005). This may contribute to impart distinctive identities to mother and bud compartments. In turn, signals for unequal loading of regulators to the daughter or the mother-bound SPB may arise once each pole establishes interactions with their target cortical domains as recently proposed for Bfa1p (Monje-Casas and Amon, 2009).
The requirement for actin and cell polarity for Kar9p polarization could have suggested that Kar9p itself may be subject to such control. Yet, the phenotype of bud6Δ bni1Δ cells, Kar9p behavior during symmetry breaking and the allele-specific induction of Kar9p symmetry by myo2 mutants are inconsistent with this scenario. Indeed, Kar9p symmetry was not increased in response to misalignment per se. Furthermore, symmetry breaking could take place within the mother cell, even if SPBs were far from the bud neck (Figure 6; data not shown). Thus, Kar9p could become polarized even if both SPBs occupied, and interacted with, the mother compartment, in sharp contrast with the differential dynamic association of Bfa1p to mother and daughter-bound SPBs induced by both asymmetric interactions and/or access of SPBs to their intended compartments (Caydasi and Pereira, 2009; Monje-Casas and Amon, 2009). Indeed, one axis of polarity outlined by actin cables ending at the bud neck in a bni1Δ mutant proved sufficient to sustain Kar9p polarization despite the impairment in spindle alignment arising from lack of actin cables in the bud (Pruyne et al., 2004b; Delgehyr et al., 2008). Finally, polarization best correlated with Kar9p-mediated transports eliciting SPB movement.
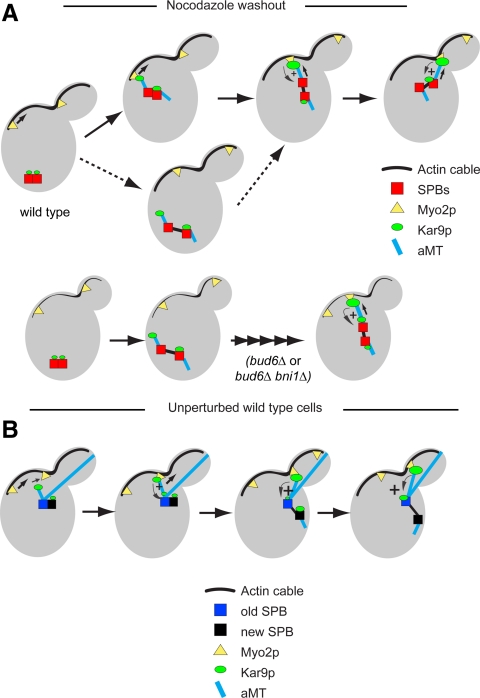
We therefore propose that the impact of actin integrity on Kar9p polarity is a direct consequence of the requirement of Myo2p-dependent transport for inducing cycles of delivery of aMT plus ends that stimulate loading of Kar9p to the pole engaged, thus generating a feedback loop that can sustain further delivery of aMT plus ends from this pole (Figure 8A). Accordingly, bud6Δ or bud6Δ bni1Δ cells would be delayed in symmetry breaking to the extent in which delivery of aMT plus ends is compromised by the perturbation in actin cable organization (Amberg et al., 1997; Pruyne et al., 2004a; Delgehyr et al., 2008). Kar9p can be recruited at the SPB from a cytoplasmic pool, and it may not be readily exchangeable, because recovery after photobleaching at the SPB under otherwise unperturbed conditions may exceed 4 min (Liakopoulos et al., 2003). In addition, once reaching aMT plus ends, Kar9p can return to the SPB along shortening aMTs (Liakopoulos et al., 2003; Huisman et al., 2004; Cuschieri et al., 2006). This recycling on aMTs after disengagement from Myo2p may contribute toward the accumulation of Kar9p upon cycles of delivery of aMT plus ends, as suggested by our time-lapse analysis. Consistent with this notion, the abnormally persistent interaction between Kar9p and Myo2p provoked by a tub4-Δdsyl mutation results in both excessive aMT plus end dwelling at the bud cell cortex and depletion of Kar9p label at the SPB (Cuschieri et al., 2006). Yet, it was not technically possible to assess the respective contributions of both pools of Kar9p (recruitment vs. returning to the SPB) to this recycling by photobleaching experiments and dual-color imaging, in the context of dynamic delivery of aMT plus ends.
Figure 8.
A model for Kar9p polarization driven by actin cables, Myo2p, and aMTs. (A) Nocodazole treatment erases differential SPB history and allows Kar9p symmetric recruitment to a basal level. After removal of the drug, Kar9p recruitment increases after regrowth of aMTs. aMTs from one SPB may stochastically engage in Kar9p-mediated deliveries. These interactions can rapidly evoke a feedback loop that helps symmetry breaking, before (solid arrow) or after (dashed arrows) SPB separation. In a bud6Δ mutant, the reduction in actin cables may lead to a delay in SPB repositioning and symmetry breaking. (B) In an unperturbed wild-type cell, spindle polarity is primed by aMTs from the old SPB tethering the side-by-side SPBs to the incipient bud. The old SPB also engages in Kar9p-mediated transport through its existing aMTs. The new SPB can also recruit Kar9p to a basal level but lacks aMTs. A bias for Kar9p recruitment to the old SPB can become established before aMTs form at the new SPB. This bias prevails after SPB separation leading to the loss of Kar9p from the new SPB.
The fact that this instructive mechanism hinges on the ability of Kar9p to act as a cargo of Myo2p further distinguishes the underlying links between Kar9p versus Bfa1p and cell polarity. Indeed, the presence of asymmetric aMT interactions and spindle alignment were insufficient for Kar9p polarity in the absence of actin cables, which are essential for sustaining Myo2p-based transport. By contrast, Bfa1p polarity senses spindle alignment even in a kar9Δ mutant once aMTs gain access to the bud and spindles align through the functionally redundant dynein pathway (Caydasi and Pereira, 2009; Monje-Casas and Amon, 2009). Thus, Bfa1p polarization cannot arise in direct response to delivery of aMT plus ends powered by Myo2p-dependent transport (absent without Kar9p) but instead responds to yet to be defined signals derived from asymmetric aMT contacts (mother vs. bud cell) as proposed by Monje-Casas and Amon (2009) and by spindle alignment (e.g., num1Δ cells).
Establishment of Spindle Polarity: Committing the Old SPB to Become the SPBbud
We have previously linked S phase CDK with control of intrinsic SPB asymmetry for the establishment of spindle polarity (Segal et al., 2000b). CDK may delay aMT organization at the new SPB until early spindle assembly, thus preventing this SPB from engaging in interactions with the bud (Shaw et al., 1997; Huisman et al., 2007).
Kar9p begins to single out the SPBbud coincident with the establishment of spindle polarity, thus enforcing delivery of aMT plus ends from this pole to the bud after SPB separation (Huisman et al., 2004; Maekawa and Schiebel, 2004). Early in the cell cycle, Kar9p may associate with both side-by-side SPBs already tethered to the growing bud in response to cortical cues. However, stimulation of recruitment through feedback can only engage the old SPB through its existing aMTs (Figure 8B). Cycles of delivery of aMT plus ends would continue over time, encouraged by this feedback. After SPB separation, two mechanisms conspire to confine the new SPB to the mother cell. First, aMTs now formed by the new SPB are restricted from access to the bud (Segal et al., 2000a; Delgehyr et al., 2008). Second, Kar9p recruitment to this pole would fade, because a feedback loop cannot be established, because the history of the two SPBs would have built a pronounced bias in favor of the old SPB, now the SPBbud. Treatment with either nocodazole or LatB renders the SPBs equal to recruit Kar9p. Yet, upon recovery from either drug treatment, cell polarity and actin cables can promote symmetry breaking and repolarization of Kar9p to mark one SPB to become the SPBbud.
The yeast paradigm has effectively predicted the behavior of centrosomes associated with asymmetric divisions during stem cell self renewal in Drosophila male germline and neuroblasts (Rebollo et al., 2007; Yamashita et al., 2007). Moreover, feedback mechanisms have been recently implicated in both spontaneous symmetry breaking to direct random bud emergence in the absence of cortical landmarks for bud site selection (Irazoqui et al., 2003) as well as to ensure the generation of a single mother-bud axis in S. cerevisiae (Howell et al., 2009). As shown here, budding yeast provides an excellent setup to also model spontaneous cell polarization and the controls accounting for asymmetric fate of cellular components in the absence of preestablished asymmetric cues. The ability for spontaneous symmetry breaking is not restricted to yeast (Wedlich-Soldner and Li, 2003; Paluch et al., 2006). Thus, understanding the complex relationship between polarity determinants and the assignment of asymmetric fate to the yeast spindle poles may provide valuable insight into centrosome control in those systems.
Supplementary Material
ACKNOWLEDGMENTS
We thank David Pellman, Felipe Santiago, and Rita Miller for generously providing strains and constructs. We also thank Tanja Herlt and Colin Hockings for contributing constructs and data to this project and Pablo Huertas for fruitful discussions and comments on the manuscript. N. D. was partly supported by a postdoctoral fellowship from the Fondation pour la Recherche Médicale (France), A. Z. by a J.R.S. Fincham Bursary, and M.A.J.O. by a postdoctoral fellowship from the Fundación Ramón Areces (Spain). In addition, M. S. acknowledges the support from The Wellcome Trust and the Biotechnology and Biological Sciences Research Council.
Abbreviations used:
- aMT
astral microtubule
- CDK
cyclin-dependent kinase
- Lat
latrunculin
- MEN
mitotic exit network
- SPB
spindle pole body.
Footnotes
This article was published online ahead of print in MBoC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E10-03-0197) on June 9, 2010.
REFERENCES
- Amberg D. C., Zahner J. E., Mulholland J. W., Pringle J. R., Botstein D. Aip3p/Bud6p, a yeast actin-interacting protein that is involved in morphogenesis and the selection of bipolar budding sites. Mol. Biol. Cell. 1997;8:729–753. doi: 10.1091/mbc.8.4.729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ayscough K. R., Stryker J., Pokala N., Sanders M., Crews P., Drubin D. G. High rates of actin filament turnover in budding yeast and roles for actin in establishment and maintenance of cell polarity revealed using the actin inhibitor latrunculin-A. J. Cell Biol. 1997;137:399–416. doi: 10.1083/jcb.137.2.399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bardin A. J., Visintin R., Amon A. A mechanism for coupling exit from mitosis to partitioning of the nucleus. Cell. 2000;102:21–31. doi: 10.1016/s0092-8674(00)00007-6. [DOI] [PubMed] [Google Scholar]
- Beach D. L., Thibodeaux J., Maddox P., Yeh E., Bloom K. The role of the proteins Kar9 and Myo2 in orienting the mitotic spindle of budding yeast. Curr. Biol. 2000;10:1497–1506. doi: 10.1016/s0960-9822(00)00837-x. [DOI] [PubMed] [Google Scholar]
- Byers B. Cytology of the yeast life cycle. In: Strathern J. N., Jones E. W., Broach J. R., editors. The Molecular Biology of the Yeast Saccharomyces: Life Cycle and Inheritance. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory; 1981. pp. 59–96. [Google Scholar]
- Caydasi A. K., Pereira G. Spindle alignment regulates the dynamic association of checkpoint proteins with yeast spindle pole bodies. Dev. Cell. 2009;16:146–156. doi: 10.1016/j.devcel.2008.10.013. [DOI] [PubMed] [Google Scholar]
- Cosma M. P. Daughter-specific repression of Saccharomyces cerevisiae HO: Ash1 is the commander. EMBO Rep. 2004;5:953–957. doi: 10.1038/sj.embor.7400251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cuschieri L., Miller R., Vogel J. Gamma-tubulin is required for proper recruitment and assembly of Kar9-Bim1 complexes in budding yeast. Mol. Biol. Cell. 2006;17:4420–4434. doi: 10.1091/mbc.E06-03-0245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delgehyr N., Lopes C. S., Moir C. A., Huisman S. M., Segal M. Dissecting the involvement of formins in Bud6p-mediated cortical capture of microtubules in S. cerevisiae. J. Cell Sci. 2008;121:3803–3814. doi: 10.1242/jcs.036269. [DOI] [PubMed] [Google Scholar]
- Doxsey S., McCollum D., Theurkauf W. Centrosomes in cellular regulation. Annu. Rev. Cell Dev. Biol. 2005;21:411–434. doi: 10.1146/annurev.cellbio.21.122303.120418. [DOI] [PubMed] [Google Scholar]
- Gonczy P. Mechanisms of asymmetric cell division: flies and worms pave the way. Nat. Rev. Mol. Cell Biol. 2008;9:355–366. doi: 10.1038/nrm2388. [DOI] [PubMed] [Google Scholar]
- Gupta M. L., Jr, Bode C. J., Thrower D. A., Pearson C. G., Suprenant K. A., Bloom K. S., Himes R. H. beta-Tubulin C354 mutations that severely decrease microtubule dynamics do not prevent nuclear migration in yeast. Mol. Biol. Cell. 2002;13:2919–2932. doi: 10.1091/mbc.E02-01-0003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guthrie C., Fink G. R. Guide to Yeast Genetics and Molecular Biology. San Diego, CA: Academic Press; 1991. [Google Scholar]
- Howell A. S., Savage N. S., Johnson S. A., Bose I., Wagner A. W., Zyla T. R., Nijhout H. F., Reed M. C., Goryachev A. B., Lew D. J. Singularity in polarization: rewiring yeast cells to make two buds. Cell. 2009;139:731–743. doi: 10.1016/j.cell.2009.10.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huisman S. M., Bales O. A., Bertrand M., Smeets M. F., Reed S. I., Segal M. Differential contribution of Bud6p and Kar9p to microtubule capture and spindle orientation in S. cerevisiae. J. Cell Biol. 2004;167:231–244. doi: 10.1083/jcb.200407167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huisman S. M., Segal M. Cortical capture of microtubules and spindle polarity in budding yeast - where's the catch? J. Cell Sci. 2005;118:463–471. doi: 10.1242/jcs.01650. [DOI] [PubMed] [Google Scholar]
- Huisman S. M., Smeets M. F., Segal M. Phosphorylation of Spc110p by Cdc28p-Clb5p kinase contributes to correct spindle morphogenesis in S. cerevisiae. J. Cell Sci. 2007;120:435–446. doi: 10.1242/jcs.03342. [DOI] [PubMed] [Google Scholar]
- Hwang E., Kusch J., Barral Y., Huffaker T. C. Spindle orientation in Saccharomyces cerevisiae depends on the transport of microtubule ends along polarized actin cables. J. Cell Biol. 2003;161:483–488. doi: 10.1083/jcb.200302030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Irazoqui J. E., Gladfelter A. S., Lew D. J. Scaffold-mediated symmetry breaking by Cdc42p. Nat. Cell Biol. 2003;5:1062–1070. doi: 10.1038/ncb1068. [DOI] [PubMed] [Google Scholar]
- Irazoqui J. E., Howell A. S., Theesfeld C. L., Lew D. J. Opposing roles for actin in Cdc42p polarization. Mol. Biol. Cell. 2005;16:1296–1304. doi: 10.1091/mbc.E04-05-0430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaspersen S. L., Winey M. The budding yeast spindle pole body: structure, duplication, and function. Annu. Rev. Cell Dev. Biol. 2004;20:1–28. doi: 10.1146/annurev.cellbio.20.022003.114106. [DOI] [PubMed] [Google Scholar]
- Jin H., Amberg D. C. The secretory pathway mediates localization of the cell polarity regulator Aip3p/Bud6p. Mol. Biol. Cell. 2000;11:647–661. doi: 10.1091/mbc.11.2.647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keaton M. A., Lew D. J. Eavesdropping on the cytoskeleton: progress and controversy in the yeast morphogenesis checkpoint. Curr. Opin. Microbiol. 2006;9:540–546. doi: 10.1016/j.mib.2006.10.004. [DOI] [PubMed] [Google Scholar]
- Kikyo M., Tanaka K., Kamei T., Ozaki K., Fujiwara T., Inoue E., Takita Y., Ohya Y., Takai Y. An FH domain-containing Bnr1p is a multifunctional protein interacting with a variety of cytoskeletal proteins in Saccharomyces cerevisiae. Oncogene. 1999;18:7046–7054. doi: 10.1038/sj.onc.1203184. [DOI] [PubMed] [Google Scholar]
- Korinek W. S., Copeland M. J., Chaudhuri A., Chant J. Molecular linkage underlying microtubule orientation toward cortical sites in yeast. Science. 2000;287:2257–2259. doi: 10.1126/science.287.5461.2257. [DOI] [PubMed] [Google Scholar]
- Lee L., Tirnauer J. S., Li J., Schuyler S. C., Liu J. Y., Pellman D. Positioning of the mitotic spindle by a cortical-microtubule capture mechanism. Science. 2000;287:2260–2262. doi: 10.1126/science.287.5461.2260. [DOI] [PubMed] [Google Scholar]
- Leisner C., Kammerer D., Denoth A., Britschi M., Barral Y., Liakopoulos D. Regulation of mitotic spindle asymmetry by SUMO and the spindle-assembly checkpoint in yeast. Curr. Biol. 2008;18:1249–1255. doi: 10.1016/j.cub.2008.07.091. [DOI] [PubMed] [Google Scholar]
- Liakopoulos D., Kusch J., Grava S., Vogel J., Barral Y. Asymmetric loading of Kar9 onto spindle poles and microtubules ensures proper spindle alignment. Cell. 2003;112:561–574. doi: 10.1016/s0092-8674(03)00119-3. [DOI] [PubMed] [Google Scholar]
- Maekawa H., Schiebel E. Cdk1-Clb4 controls the interaction of astral microtubule plus ends with subdomains of the daughter cell cortex. Genes Dev. 2004;18:1709–1724. doi: 10.1101/gad.298704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maekawa H., Usui T., Knop M., Schiebel E. Yeast Cdk1 translocates to the plus end of cytoplasmic microtubules to regulate bud cortex interactions. EMBO J. 2003;22:438–449. doi: 10.1093/emboj/cdg063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meednu N., Hoops H., D'Silva S., Pogorzala L., Wood S., Farkas D., Sorrentino M., Sia E., Meluh P., Miller R. K. The spindle positioning protein Kar9p interacts with the sumoylation machinery in Saccharomyces cerevisiae. Genetics. 2008;180:2033–2055. doi: 10.1534/genetics.108.095042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller R. K., Cheng S. C., Rose M. D. Bim1p/Yeb1p mediates the Kar9p-dependent cortical attachment of cytoplasmic microtubules. Mol. Biol. Cell. 2000;11:2949–2959. doi: 10.1091/mbc.11.9.2949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monje-Casas F., Amon A. Cell polarity determinants establish asymmetry in MEN signaling. Dev. Cell. 2009;16:132–145. doi: 10.1016/j.devcel.2008.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore J. K., Miller R. K. The CDK, Cdc28p, regulates multiple aspects of Kar9p function in yeast. Mol. Biol. Cell. 2007 doi: 10.1091/mbc.E06-04-0360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moseley J. B., Goode B. L. Differential activities and regulation of Saccharomyces cerevisiae formin proteins Bni1 and Bnr1 by Bud6. J. Biol. Chem. 2005;280:28023–28033. doi: 10.1074/jbc.M503094200. [DOI] [PubMed] [Google Scholar]
- Musacchio A., Salmon E. D. The spindle-assembly checkpoint in space and time. Nat. Rev. Mol. Cell Biol. 2007;8:379–393. doi: 10.1038/nrm2163. [DOI] [PubMed] [Google Scholar]
- Paluch E., van der Gucht J., Sykes C. Cracking up: symmetry breaking in cellular systems. J. Cell Biol. 2006;175:687–692. doi: 10.1083/jcb.200607159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pearson C. G., Bloom K. Dynamic microtubules lead the way for spindle positioning. Nat. Rev. Mol. Cell Biol. 2004;5:481–492. doi: 10.1038/nrm1402. [DOI] [PubMed] [Google Scholar]
- Pereira G., Hofken T., Grindlay J., Manson C., Schiebel E. The Bub2p spindle checkpoint links nuclear migration with mitotic exit. Mol. Cell. 2000;6:1–10. [PubMed] [Google Scholar]
- Pereira G., Tanaka T. U., Nasmyth K., Schiebel E. Modes of spindle pole body inheritance and segregation of the Bfa1p-Bub2p checkpoint protein complex. EMBO J. 2001;20:6359–6370. doi: 10.1093/emboj/20.22.6359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pruyne D., Gao L., Bi E., Bretscher A. Stable and dynamic axes of polarity use distinct formin isoforms in budding yeast. Mol. Biol. Cell. 2004a;15:4971–4989. doi: 10.1091/mbc.E04-04-0296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pruyne D., Legesse-Miller A., Gao L., Dong Y., Bretscher A. Mechanisms of polarized growth and organelle segregation in yeast. Annu. Rev. Cell Dev. Biol. 2004b;20:559–591. doi: 10.1146/annurev.cellbio.20.010403.103108. [DOI] [PubMed] [Google Scholar]
- Rebollo E., Sampaio P., Januschke J., Llamazares S., Varmark H., Gonzalez C. Functionally unequal centrosomes drive spindle orientation in asymmetrically dividing Drosophila neural stem cells. Dev. Cell. 2007;12:467–474. doi: 10.1016/j.devcel.2007.01.021. [DOI] [PubMed] [Google Scholar]
- Sagot I., Rodal A. A., Moseley J., Goode B. L., Pellman D. An actin nucleation mechanism mediated by Bni1 and profilin. Nat. Cell Biol. 2002;4:626–631. doi: 10.1038/ncb834. [DOI] [PubMed] [Google Scholar]
- Schott D., Ho J., Pruyne D., Bretscher A. The COOH-terminal domain of Myo2p, a yeast myosin V, has a direct role in secretory vesicle targeting. J. Cell Biol. 1999;147:791–808. doi: 10.1083/jcb.147.4.791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Segal M., Bloom K. Control of spindle polarity and orientation in Saccharomyces cerevisiae. Trends Cell Biol. 2001;11:160–166. doi: 10.1016/s0962-8924(01)01954-7. [DOI] [PubMed] [Google Scholar]
- Segal M., Bloom K., Reed S. I. Bud6 directs sequential microtubule interactions with the bud tip and bud neck during spindle morphogenesis in Saccharomyces cerevisiae. Mol. Biol. Cell. 2000a;11:3689–3702. doi: 10.1091/mbc.11.11.3689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Segal M., Bloom K., Reed S. I. Kar9p-independent microtubule capture at Bud6p cortical sites primes spindle polarity before bud emergence in Saccharomyces cerevisiae. Mol. Biol. Cell. 2002;13:4141–4155. doi: 10.1091/mbc.02-05-0067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Segal M., Clarke D. J., Maddox P., Salmon E. D., Bloom K., Reed S. I. Coordinated spindle assembly and orientation requires Clb5p-dependent kinase in budding yeast. J. Cell Biol. 2000b;148:441–452. doi: 10.1083/jcb.148.3.441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaw S. L., Yeh E., Maddox P., Salmon E. D., Bloom K. Astral microtubule dynamics in yeast: a microtubule-based searching mechanism for spindle orientation and nuclear migration into the bud. J. Cell Biol. 1997;139:985–994. doi: 10.1083/jcb.139.4.985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smeets M. F., Segal M. Spindle polarity in S. cerevisiae: MEN can tell. Cell Cycle. 2002;1:308–311. doi: 10.4161/cc.1.5.143. [DOI] [PubMed] [Google Scholar]
- Theesfeld C. L., Irazoqui J. E., Bloom K., Lew D. J. The role of actin in spindle orientation changes during the Saccharomyces cerevisiae cell cycle. J. Cell Biol. 1999;146:1019–1032. doi: 10.1083/jcb.146.5.1019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vaughan K. T. TIP maker and TIP marker; EB1 as a master controller of microtubule plus ends. J. Cell Biol. 2005;171:197–200. doi: 10.1083/jcb.200509150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wedlich-Soldner R., Li R. Spontaneous cell polarization: undermining determinism. Nat. Cell Biol. 2003;5:267–270. doi: 10.1038/ncb0403-267. [DOI] [PubMed] [Google Scholar]
- Yamashita Y. M., Mahowald A. P., Perlin J. R., Fuller M. T. Asymmetric inheritance of mother versus daughter centrosome in stem cell division. Science. 2007;315:518–521. doi: 10.1126/science.1134910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yeh E., Yang C., Chin E., Maddox P., Salmon E. D., Lew D. J., Bloom K. Dynamic positioning of mitotic spindles in yeast: role of microtubule motors and cortical determinants. Mol. Biol. Cell. 2000;11:3949–3961. doi: 10.1091/mbc.11.11.3949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin H., Pruyne D., Huffaker T. C., Bretscher A. Myosin V orientates the mitotic spindle in yeast. Nature. 2000;406:1013–1015. doi: 10.1038/35023024. [DOI] [PubMed] [Google Scholar]
- Yoshida S., Guillet M., Pellman D. MEN signaling: daughter bound pole must escape her mother to be fully active. Dev. Cell. 2005;9:168–170. doi: 10.1016/j.devcel.2005.07.006. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.