Abstract
Nature contains a tremendous diversity of forms both at the organismal and genomic levels. This diversity motivates the twin central questions of molecular evolution: what are the molecular mechanisms of adaptation, and what are the functional consequences of genomic diversity. We report a 22-species comparative analysis of tropomyosin (PPM) genes, which exist in a variety of forms and are implicated in the emergence of a wealth of cellular functions, including the novel muscle functions integral to the functional diversification of bilateral animals. TPM genes encode either or both of long-form [284 amino acid (aa)] and short-form (approximately 248 aa) proteins. Consistent with a role of TPM diversification in the origins and radiation of bilaterians, we find evidence that the muscle-specific long-form protein arose in proximal bilaterian ancestors (the bilaterian ‘stem’). Duplication of the 5′ end of the gene led to alternative promoters encoding long- and short-form transcripts with distinct functions. This dual-function gene then underwent strikingly parallel evolution in different bilaterian lineages. In each case, recurrent tandem exon duplication and mutually exclusive alternative splicing of the duplicates, with further association between these alternatively spliced exons along the gene, led to long- and short-form–specific exons, allowing for gradual emergence of alternative “internal paralogs” within the same gene. We term these Mutually exclusively Alternatively spliced Tandemly duplicated Exon sets “MATEs”. This emergence of internal paralogs in various bilaterians has employed every single TPM exon in at least one lineage and reaches striking levels of divergence with up to 77% of long- and short-form transcripts being transcribed from different genomic regions. Interestingly, in some lineages, these internal alternatively spliced paralogs have subsequently been “externalized” by full gene duplication and reciprocal retention/loss of the two transcript isoforms, a particularly clear case of evolution by subfunctionalization. This parallel evolution of TPM genes in diverse metazoans attests to common selective forces driving divergence of different gene transcripts and represents a striking case of emergence of evolutionary novelty by alternative splicing.
Keywords: genome innovation, alternative splicing, intron sliding, exon duplication, tropomyosin
Introduction
The genetic mechanisms of evolutionary adaptation constitute arguably the largest question in molecular evolution. Under most conditions, a gene must continue to encode the ancestral function, imposing constraints on the emergence of new functions by simple changes in the gene sequence. Much of the debate about the emergence of new functions has focused on two possibilities: changes in gene expression patterns across environmental conditions, developmental stages, tissues, or subcellular locations; or emergence of new genic products, primarily by genomic duplication and/or alternative splicing (AS). These last two mechanisms are to some extent interchangeable in evolution and there is a clear inverse correlation between the occurrence of the two phenomena (Kopelman et al. 2005; Irimia, Rukov et al. 2008). In this regard, alternatively spliced forms can be considered “internal paralogs” (Modrek and Lee 2003), in analogy to true paralogs (gene duplicates), because alternative forms can also diverge while maintaining the ancestral gene function.
Tropomyosin (TPM) genes provide a striking case study of the intersection of these processes. TPM genes are present in all characterized fungi and animals (Vrhovski et al. 2008) and have been extensively studied at the functional level (Gunning et al. 2005; Gooding and Smith 2008). Tropomyosin (Tm) proteins bind actin filaments and play important roles in the different functions and specializations of the actin cytoskeleton (Gunning et al. 2005), the formation of sarcomeres in striated muscles in bilaterians perhaps being the most prominent example.
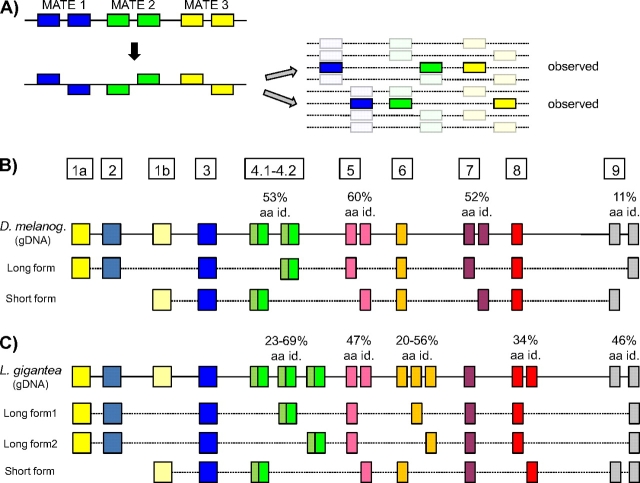
Tm proteins in mammals and studied model invertebrates can be divided into two classes, the so-called long [approximately 284 amino acid (aa)] and short (approximately 248 aa) forms. Cytoskeletal Tms are found in the cytoskeleton of all cell types examined and are usually ‘short’ (although they can also be ‘long’), whereas the Tms found in the contractile apparatus of striated and smooth muscle are only of the long form (Gunning et al. 2008). A TPM gene may encode either one or both forms (fig. 1a provides a general scheme). Genes often encode both long and short forms, deriving from different promoters with associated 5′ exons: the genomic region encoding the 5′ end of the long form (including a promoter plus two long-form–specific exons) lies upstream of the region encoding the 5′ end of the short-form (promoter plus one short-form–specific exon). These alternative 5′ regions are then typically spliced to downstream regions (beginning with the exon traditionally called exon 3; fig. 1a).
FIG. 1.
TPM gene structures in metazoans. (A) General gene structures for bilaterian TPM genes. Bilaterian genes contain two promoters. Short-form transcripts are transcribed from the downstream promoter. Long-form transcripts are transcribed from an upstream promoter, contain two additional exons (1a and 2), and do not include the first exon from short-form transcripts (exon 1b). Nonbilaterian genes encode only short-form transcripts. (B) TPM gene structures from nine bilaterians and two nonbilaterians. TPM genes in most bilaterians contain copies of various exons (boxes with the same color), in contrast to the simpler genes of nonbilaterians; these tandemly duplicated exon sets (MATEs) are alternatively spliced in a mutually exclusive manner. The bilaterians Ciona intestinalis and Hellobdella robusta represent exceptions to this pattern: TPM genes in these species lack duplicated exons, and encode either long- or short-form transcripts, but not both. Boxes/lines indicate exons/introns. Homologous exons (either orthologous or paralogous) are indicated by the same color. Number after the species name, when present, correspond to the paralog represented in the figure (i.e., Homo sapiens 1 represents human TPM1). Full species names are given in the Methods.
Differences between long- and short-form transcripts extend to downstream sequences. For instance, human TPM genes undergo extensive AS and alternative termination of transcription. TPM gene repertoires also differ significantly in number, with four copies in mammals, three of which encode multiple transcripts (Gunning et al. 2005), but only one gene in Caenorhabditis elegans (Vrhovski et al. 2008). The diversity of isoforms resulting from this web of interacting processes is subtly regulated in embryonic development. Gene expression often shows tissue, cellular, or even subcellular specificity (e.g., Karlik and Fyrberg 1986; Lin et al. 1988; Weinberger et al. 1996; Lin and Storti 1997; Schevzov et al. 1997; Hannan et al. 1998; Dalby-Payne et al. 2003; Li and Gao 2003; Vrhovski et al. 2008), and different transcripts within the same species encode a vast array of specialized functions (Gunning et al. 2005).
To probe the evolutionary history of this genetic and functional diversity, we studied gene and transcript structures for 69 TPM genes from 22 sequenced genomes. We find strikingly parallel evolution in a wide variety of bilaterians. In almost all studied bilaterians, TPM genes have evolved by tandem duplication of different exons and mutually exclusive AS of these duplicates (i.e., each transcript contains only one copy of a given duplicated exon). Splicing of each pair (or set) of exons is closely associated both with the AS of other exon pairs/sets and with the alternative promoter usage leading to the long/short distinct gene products. Thus, many bilaterian TPM genes provide striking cases of internal paralogs: long and short isoforms transcribed from the same gene have up to 77% of homologous protein sequence deriving from different exons. We also report cases in which long- and short-form transcripts have been resolved into separate genes, apparently by gene duplication and loss of one promoter (and associated AS regions) in each of the gene duplicates. We designate such cases of conversion of internal paralogs into actual or external paralogs “externalization.” These cases represent a particularly clear example of “subfunctionalization,” partitioning of multiple ancestral gene functions into gene duplicates.
Methods
Genomic and Expressed Sequence Tag sources
We used the following genome sequence assemblies and expression data from the following sources: Monosiga brevicollis v1.0, Trichoplax adhaerens Grell-BS-1999 v1.0, Nematostella vectensis v1.0, Branchiostoma floridae v1.0, Ciona intestinalis v2.0 and v1.0, Takifugu rubripes v4.0, Xenopus tropicalis v4.1, Daphnia pulex v1.0, Hellobdella robusta v1.0, Lottia gigantea v1.0 and Capitella capitata v1.0 at DOE Joint Genome Institute (JGI) webpage (http://genome.jgi-psf.org/euk_home.html); and of Strongylocentrotus purpuratus Build 2.1, Apis mellifera Amel_4.0, Tribolium castaneum Build 2.1, Anopheles gambiae AgamP3.3, Drosophila melanogaster Build Fb5.3, Homo sapiens Build GRCh37, Mus musculus Build 37.1 at the NCBI webpage (http://www.ncbi.nlm.nih.gov/blast/Blast.cgi), and/or Ensembl webpage (http://www.ensembl.org), Hydra magnipapillata v1.0 at Metazome webpage (http://hydrazome.metazome.net/cgi-bin/gbrowse/hydra/), Brugia malayi BMA1 at TIGR webpage (http://blast.jcvi.org/er-blast/index.cgi?project=bma1), C. elegans WS197 and Caenorhabditis briggsae WS197 at WormBase (www.wormbase.org); for Saccoglossus kowalevskii we performed a BLASTN search against the traces at NCBI and then manually assembly the genomic locus through walking in silico.
Search for TPM Genes and Tandemly Duplicated Exons
We used TBlastN against the genome sequences using different TPM genes as queries and e-values. We inspected several hits to detect fast evolving true TPM gene copies. We also performed BlastP against annotated protein databases because the small length of most tropomyosin exons precluded the identification of divergent members of the family when using only TBlastN.
To identify tandemly duplicated exons, we took two complementary approaches. First, for each species, we aligned each identified exon against the upstream and downstream intronic sequences using ClustalW. As homologous exons always had the same nucleotide lengths, reliability of the splice sites for putative exons could be easily assessed. Then, for each newly identified tandemly duplicated exon, we again aligned the exon sequence against the new upstream and downstream intronic sequence, until we could not identify any putative duplicated exon in the intronic sequence. Second, we searched for expressed sequence tags (ESTs) supporting the expression of the different duplicated exons. In some cases, new exons with lower sequence similarity, and thus not confidently detected by ClustalW alignments were identified. All exons for all studied species are provided in supplementary figure S1 (Supplementary Material online).
Phylogenetic Analyses
To assess the global phylogenetic relationships among the TPM genes, phylogenetic trees were generated using exons common to all TPM genes (exons 3–9). Due to the extensive tandem duplication of these exons, there is a high risk of comparing paralogous exons instead of true orthologous sequences across metazoans. In order to randomize this potential effect, for each species with mutually exclusively alternatively spliced tandemly duplicated exon sets (MATEs, see below) at a given exon locus, we randomly selected which of the duplicated exons were included in the alignment. Two independent randomly selected sets of exons 3–9 were generated and trees were inferred for these two replicates. For this approach, accurate alignment was fundamentally important and therefore, some of the highly divergent nonbilaterian intronless genes with varying lengths were not included in the analysis (Podocoryne carinensis TPM2, H. magnipapillata TPMA and C and N. vectensis TPM13, 54a, 54b, and 115).
Individual exon trees were also inferred for each of the ancestral 10 TPM exons (1, 2, 3, 4.1, 4.2, 5, 6, 7, 8, and 9). Exons 1A and 1B were analyzed together. In addition to their respective individual trees, exons 4.1 and 4.2 were also analyzed as a joint exon (exon 4) as they are a single exon in most of the species due to specific intron losses in protostomes and some chordates (fig. 1). All internal exons always had the exact same nucleotide length and therefore alignments were ungapped and reliable. In the case of exons 1 and 9, sequences were aligned using ClustalW and manually curated. For simplicity, we only used exons from two nonbilaterian genes, T. adhaerens TPMI and N. vectensis TPMI. The rest of nonbilaterian genes were very divergent in sequence and length and in many cases intronless and thus could not be aligned to the rest of TPM exons with full confidence.
To study the externalization of short and long TPM isoforms in Lophotrochozoans, three different kinds of trees were constructed. One was inferred using only two exons that are not duplicated (3 and 7) and are therefore constitutive for all the isoforms. Another tree was generated using only the two alternative initial exons, 1A and 1B. The third tree was built for MATE exons (4, 5, 6, 8, and 9). Isoform merges were built according to EST information from the mutually exclusive MATE patterns.
Similarly, to study the externalization of the long isoform in insects, a tree was build using the MATE exons from insect TPM1 and D. pulex TPM that, in the case of insect TPM2, were constitutive (exons 4, 5, and 7).
All trees in the present study were performed using both Bayesian inference (BI) and maximum likelihood (ML). BI trees were inferred using MrBayes 3.1.2 (Huelsenbeck and Ronquist 2001; Ronquist and Huelsenbeck 2003), with the model recommended by ProtTest 1.4 (Drummond and Strimmer 2001; Guindon and Gascuel 2003; Abascal et al. 2005) under the Akaike information and the Bayesian information criterions. Two independent runs were performed, each with four chains. For convention, convergence was reached when the value for the standard deviation of split frequencies stayed below 0.01. Burn-in was determined by plotting parameters across all runs for a given analysis: All trees prior to stationarity and convergence were discarded, and consensus trees were calculated for the remaining trees (from at least 1,000,000 generations).
ML analyses were performed using RAxML version 7.0.3 (Stamatakis 2006) with the model recommended by ProtTest, 10,000 bootstrap replicates and the rapid Bootstrapping algorithm. Alignments in Nexus format are provided in supplementary figure S2 (Supplementary Material online).
Syntenic analysis for TPM4 in rodents was done using Ensembl genome browsers and Genomicus v55.1 (http://www.dyogen.ens.fr/genomicus/cgi-bin/search.pl).
Results
Extensive Alternative Splicing of TPM is Common and Restricted to Bilaterians
We studied TPM genes and transcripts across 22 species, including 18 bilaterians, 3 nonbilaterian metazoans, and 1 choanoflagelate (fig. 1b). There were two sharp distinctions between bilaterian and nonbilaterian species. First, all 18 surveyed bilaterian genomes encode both the short and long forms of the TPM genes, whereas all nonbilaterian genomes encode only the short form, consistent with the long form having arisen in proximal ancestors of bilaterians. Second, nearly all studied bilaterian genomes but no nonbilaterian species exhibited AS. AS was observed in at least one TPM gene in 16 of 18 bilaterian species and was typically extensive, with 16 of 26 alternatively spliced genes having at least three alternative regions (alternatively spliced groups of exons or alternative promoters; see below). In some species, AS was ubiquitous, leading to cases in which the genomic regions giving rise to two transcripts from the same locus had little overlap (for instance in L. gigantea, only 23% of protein-coding regions are shared between the long- and short-form transcripts, see below). The clear relationship between AS and alternative promoters (encoding long or short forms) also held across bilaterian genes: All bilaterian genes encoding both long and short forms also exhibited AS, whereas nearly no genes encoding only short or long forms were alternatively spliced (with the exceptions of a few genes in vertebrates and insects; supplementary tables S1 and S2, Supplementary Material online). Representative examples are shown in figure 1b.
MATEs in Bilaterians
The specific regions undergoing AS varied considerably across bilaterians. For instance, exons 2 and 6 are alternatively spliced in human TPM1, whereas exons 4, 5, and 7 are alternatively spliced in D. melanogaster (note that throughout, exons are numbered according to convention in vertebrate and Drosophila genes rather than actual exon number within a given gene, thus exon 4 refers to homologous coding regions in all species). However, the mode (or mechanism) of AS was the same across all observed cases of AS (with the exception of terminal exons; see below). The nearly universal mode of AS in bilaterian TPM genes is exon creation by tandem duplication of an existing exon, and AS of the resulting exons, such that only one of them is contained in all the transcripts (mutually exclusive AS); in nearly every case, the splicing pattern is remarkably precise, with all available ESTs containing exactly one exon from the same pair/set of duplicated exons (in the rare exceptions, two copies of the exon present in very few ESTs, as with TPM1 and TPM3 of mouse). This pattern is shown in figure 2a, in which exons of the same color show clear homology (including identical length in all cases), consistent with tandem duplication. The mutual exclusion extends to cases with more than two exon copies. For instance, the single TPM gene of L. gigantea contains three copies of exon 6 (figs. 1 and 2c). The three exons are present in 10.2%, 50.2%, and 39.6% of 285 EST sequences, respectively, and no transcript shows zero or more than two copies of the exon (supplementary table S3, Supplementary Material online). We refer to such Mutually exclusively Alternatively spliced Tandemly duplicated Exon sets as MATEs.
FIG. 2.
Production of long and short forms by coordinated processing of alternative transcript regions. (A) General scenario. For three two-exon MATEs, there are eight possible combinations (at right). However, typically only two reciprocal combinations dominate available transcript sequences, with other forms never or almost never observed. (B) The TPM1 gene of D. melanogaster contains five alternative regions: alternative promoters, three two-exon MATEs, and a pair of alternative terminal exons (top line: gDNA). Among the 32 (= 25) possible combinations, only two are observed, with each alternative region found in just one transcript. Exon duplicates range in amino acid identity from 11% to 60%. (C) The more complex structure of the TPM gene of L. gigantea. The gene contains alternative promoters, two two-exon MATE, two three-exon mates, and two terminal exons. Of the 144 possible structures, only three are observed.
The ubiquity and diversity of these MATEs within bilaterians was striking. First, each of the ancestral TPM exons has duplicated to form a MATE at least in one species during evolution (fig. 1 and supplementary table S1, Supplementary Material online). Second, some of the MATEs are species or clade specific (e.g., amphioxus exons 4.1 and 8), whereas others are much more ancient and shared by higher taxonomic groups (e.g., exon 4, 5, and 7 in arthropods) (supplementary fig. S3, Supplementary Material online). Among 70 MATEs comprising 160 exons in 16 species, most consisted of 2 (78.6%) or 3 exons (17.1%), with 4 (2.9%) or more exons (1.4%) being rare.
Internal Paralogy of Bilaterian TPM Genes: Association of Alternative Regions
We also found that AS patterns of MATEs are highly nonindependent, with clear associations between splicing of exons from different MATEs (fig. 2a). For instance, D. melanogaster has two-exon MATEs at exons 4 (exons 4a and 4b) and 5 (exons 5a and 5b). Of the four possible combinations (4a/5a, 4a/5b, 4b/5a, and 4b/5b), two combinations (4a/5b and 4b/5a) account for all 79 available EST sequences, whereas the others (4a/5a and 4b/5b) are not observed. This extends to the two copies of exon 7 as well: exon 7a is associated with 4b/5a, whereas exon 7b is associated with 4a/5b (fig. 2b). In total, there were eight pairs of internal two-exon MATEs with significant EST coverage, from five species; all eight exhibited complete or nearly complete association (supplementary table S3, Supplementary Material online). In additional, there were eight pairs of internal MATEs in which one MATE contained more than two exons. In these cases, each exon within the more than two-exon MATE associates with one exon within the other MATE (table S3). The example of L. gigantea is shown in figure 2c.
AS of MATEs is also closely associated with alternative promoter usage (i.e., long-/short-form transcripts). As mentioned above, long- and short-form transcripts from the same gene are transcribed from different promoters: long forms are transcribed from a promoter upstream of the gene, whereas short forms are transcribed from a proximal promoter located downstream of the long form–specific exon 2 (fig. 1a). In addition, long-form transcripts include a different first exon as well as an additional exon (exon 2 in fig. 1). We found a global association between alternative long/short promoter usage and AS of downstream MATEs (fig. 2 and supplementary table S3, Supplementary Material online). In all 13 cases in 11 species with significant EST coverage, there was a significant association between exons in the first internal MATE and specific promoters. The strength of this association varied among taxa. Most cases in protostomes showed a complete (100%) association, whereas in the case of the TPM1 in mouse, this association was more quantitative (80%, P = 0.0026; supplementary table S3, Supplementary Material online). Moreover, in all (5/5) independent cases the long-form (upstream) promoter was associated with the most downstream exon in the first MATE, whereas the short-form promoter (proximal) was associated with the upstream first exon. For instance, the C. elegans gene has its first MATE at exon 3: among 60 ESTs, the upstream exon copy (exon 3a) is always short-form specific (always spliced to exon 1b; 49 ESTs), and the downstream copy (exon 3b) is always long-form specific (always spliced to exon 2; 11 ESTs).
Thus, association of alternative promoter usage and AS of MATEs and of AS of different MATEs leads to different transcripts from the same genomic locus being largely encoded by different regions within the locus. In some species, the extent of this “internal paralogy” is truly striking. For instance, in D. melanogaster, 67% of homologous regions between short- and long-form transcripts are transcribed from different genomic regions (compared with 100% in the case of true paralogs); in the most extreme case, in the species L. gigantea, this percentage rises up to 77%, and the sequences of the proteins encoded by the two main transcript types are 50% different (fig. 2).
Alternative Splicing of Terminal Exons
The terminal protein-encoding exons of TPM genes show a similar but slightly more complex scenario. Terminal exons typically include the terminal 28 codons of the coding sequence and the stop codon, as well as a variable amount of downstream untranslated sequence (untranslated region[UTR]). As with the internal MATEs, this terminal exon has been extensively duplicated in most bilaterians, with two to six copies per gene (fig. 1 and supplementary table S1, Supplementary Material online). Some duplication events appear to be ancestral to major metazoan groups, whereas others were lineage specific; in the most extreme case of B. floridae, six duplicates of the last exon were identified, and alternative inclusion of the various copies demonstrated by ESTs and RT–PCR (data not shown).
As with the internal MATEs, the terminal exons exhibit extensive AS; however, the splicing patterns are somewhat different. As opposed to the mutually exclusive case for internal exons, in some species some gene transcripts contain multiple copies of the 3′ exon (fig. 3a). However, this pattern still leads to mutually exclusive exon usage at the protein level: in such cases, the first exon in a transcript copy encodes the protein terminus and stop site, and any additional downstream copies are UTR (light gray exons in fig. 3a). Thus, in these cases, UTR length varies with the number of included terminal exons. In other species, this mutually exclusive pattern is achieved directly by alternative polyadenylation processing (fig. 3b). In these genes, each copy of the 3′ exon may contain the polyadenylation site, thus resulting in mutually exclusive polyadenylation choice coupled to AS. Thus, in these cases, length of UTRs is more constant.
FIG. 3.
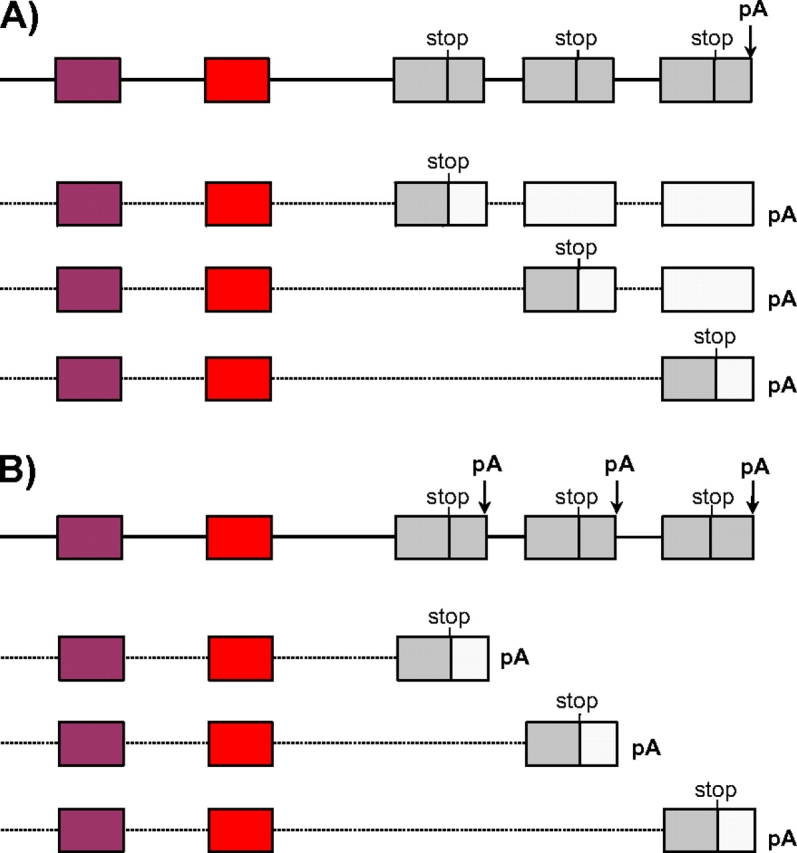
AS and polyadenylation of terminal exons. (A) In some species such as C. elegans, AS leads to different numbers of copies of the terminal exon, with different exons encoding the protein terminus (gray) with downstream copies being entirely untranslated regions (white). (B) On other species such as D. melanogaster, alternative polyadenylation coupled to AS leads to differential usage of 3′ exons and polyadenylation, but less variation in UTR length. In each case, the top line represents genomic DNA, and subsequent lines represent observed transcripts.
Notably, in both cases, splicing/polyadenylation patterns of the terminal exons show clear correspondence with upstream MATEs—there is a close correspondence between exon usage at upstream MATEs and which of the terminal exons encodes the protein terminus in all 13 genes from nine species with extensive EST coverage (supplementary table S3, Supplementary Material online). There is also a close correspondence between MATE exon usage and polyadenylation site and/or UTR length (supplementary table S3, Supplementary Material online). Thus, although AS patterns of 3′ exons are somewhat different than internal exons, there is still a strong correspondence between 3′coding exon choice to AS of different regions. In total, mutually exclusive alternative regions of transcripts in bilaterians extend across the entire transcript, from the promoters to AS of MATEs, to protein termination, and to UTR length and polyadenylation sites.
Gene Duplication in Bilaterians
The above suggests that significant selective pressure has driven the production of increasingly divergent TPM transcripts. As shown by a wealth of previous studies, such transcript diversity is often generated by whole gene duplication and sequence divergence. Accordingly, we also found extensive TPM gene duplication across metazoans (supplementary table S1, Supplementary Material online). Whereas some species harbored a single gene, others had multiple copies, ranging from two in insects and sea urchin to eight in the leech Hellobdela robusta and seven in the ascidian Ciona intestinalis (including a partial duplicate and an atypical shorter TPM gene).
There was a clear inverse association between AS and gene duplication across species. In all bilaterians with a single TPM gene, this gene was highly alternatively spliced and encoded both long and short forms by alternative promoters. Conversely, in the most extreme cases of gene duplication, the ascidian Ciona intestinalis and the leech Hellobdella robusta, each gene contained a single promoter, encoded either long- or short-form transcripts, and did not contain MATEs. The rest of the bilaterian species were intermediate between these extremes, typically containing a single highly alternatively spliced gene with both promoters, as well as additional gene copies with no promoter/splicing variation (supplementary table S1, Supplementary Material online). The only clear exception to this pattern was in vertebrates, which harbor several copies of alternatively spliced genes apparently dating to whole genome duplications in vertebrate ancestors (Putnam et al. 2008).
Subfunctionalization of Long- and Short-TPM Forms After Gene Duplication
We next examined the relationship between gene duplicates in species with multiple gene copies. In a simple case of gene duplication, a single gene duplicates and the duplicates then diverge, thus the gene duplicates are more closely related (and thus similar) to each other than either is to the single unduplicated copy in a distantly related species (fig. 4b, top box). In the case of TPM, the extensive divergence between different transcripts of the same gene greatly complicates the matter. In particular, one possible outcome of a duplication of a dual long-/short-encoding gene could be partitioning of the two forms into the two duplicates: one copy would encode the long form, but lose the short-form sequences, whereas the other would keep the short-form sequences and lose the long form–specific exons. The phylogenetic signal of such a case would be that each gene duplicate would consistently more closely resemble the alternative sequence of one of the two transcripts from the ancestral dual-encoding gene, both for the first exon and for exons from MATEs (fig. 4b, bottom right panels). In particular, duplicates encoding only long-/short-form transcripts would more closely resemble the corresponding transcript from dual-encoding genes. On the other hand, for gene regions that are shared between the two transcripts in dual-encoding genes (i.e., constitutively spliced exons), the gene duplicates would be most closely related to each other (fig. 4b, bottom left panel).
FIG. 4.
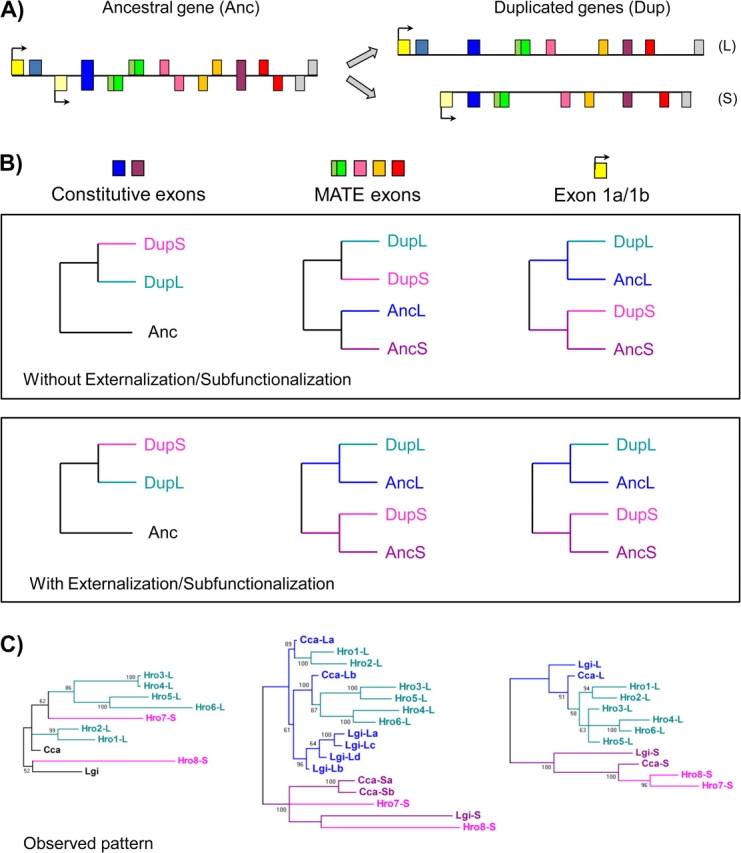
Externalization of internal paralogs by subfunctionalization. (A) General scenario. A single ancestral gene encodes both long (exons above the line) and short (below) forms of the gene. Gene duplication and reciprocal loss of the two forms leads to two descendent genes each encoding just one form. (B) Expected phylogenetic signals under “simple” gene duplication and subfunctionalization. Under both scenarios, ancestrally constitutive exons in the duplicate genes should form a clade. In the absence of externalization/subfunctionalization, the case should be the same for exons that are MATEs in the ancestral gene. However, in the presence of subfunctionalization, for ancestral MATE exons, the gene duplicates should group with the MATE exons of like type: the descendent long-/short-form gene should more closely resemble the ancestral long-/short-form transcript. (C) Phylogenetic trees for gene duplicates in the Hellobdella robusta genome and alternative promoter/AS genes from other Lophotrochozoans. Long-form genes from Hellobdella robusta group with long-form transcripts from other species, whereas short-form genes group with short-form transcripts from other species. Numbers at the nodes correspond to posterior probabilities.
The leech Hellobdella robusta represents a clear case of complete subfunctionalization of the single AS ancestral TPM gene. Related lophotrochozoan species (Lottia and Capitella) have a single gene encoding both long and short forms with several MATEs, whereas Hellobdella robusta contains separate genes encoding the two forms (six long-form genes and two short-form genes). We performed phylogenetic analyses of the various lophotrochozoan genes. We found distinctly different patterns for regions that are and are not shared between the two transcript forms in ‘typical’ (dual transcript encoding) lophotrochozoan genes (from L. gigantea and Capitella capitata). For regions in which both long and short forms are transcribed from the same genomic locus for typical genes (constitutive exons 3 and 7), there is no clear phylogenetic signal (fig. 4c, left). The case was clearly different for regions in which the homologous long- and short-form transcripts in typical lophotrochozoan genes are transcribed from different genomic regions (MATE exons 4, 5, 6, and 9). For these regions, short-form Hellobdella robusta genes grouped with other lophotrochozoan short-form transcripts, whereas long-form Hellobdella robusta genes grouped with long-form transcripts (fig. 4c, middle). The same pattern was found for exons 1a/1b (fig. 4c, right).
This pattern is exactly as expected from gene duplication and reciprocal retention of short and long forms in the two resulting duplicates but is not expected from loss of AS and divergence of transcript forms following gene duplication (fig. 4b, top). When extending the phylogenetic analysis to the orthologous alternative regions of other protostomes (restricted to MATE exons 4 and 9), we obtain the same pattern, consistent with these alternative exons having been present at the origin of protostomes (supplementary fig. S4, Supplementary Material online). Thus, the internal paralogs encoded in the single ancestral lophotrochozoan gene by a pattern of MATEs associated with alternative promoters and persisting for at least half a billion years has become “externalized” to multiple single transcript–encoding genes in Hellobdella robusta.
The case in the tunicate Ciona intestinalis appears to be similar, with five canonical TPM genes, three coding only for long-form proteins and two for short ones. However, in this case, the lack of unambiguously conserved MATEs in the related species (amphioxus and vertebrates) does not allow for a similar phylogenetic analysis as described for Hellobdella robusta.
In other species, including vertebrates or insects, some gene duplicates contain either the short or long form, although both isoforms are also maintained in additional alternatively spliced copies (supplementary table S1, Supplementary Material online). For example, in insects, TPM2 shows a clear case of specific retention of the long muscle-specific isoform. TPM2 is expressed only from the long-form promoter, and phylogenetic analysis of MATE exons 4, 5, and 7 from arthropods shows that single exons present in TPM2 group specifically with the corresponding long form–specific exons of the AS paralog TPM1 (Supplementary fig. S5, Supplementary Material online). Consistent with its genome structure, TPM2 is known to form heterodimers with the long forms of TPM1 to build the contractile units in striated muscle (Molloy et al. 1993; Mateos et al. 2006).
Finally, we also found an apparent case of very recent or ongoing subfunctionalization in the TPM4 gene of rodents. TPM4 in rodents including mouse, rat, and guinea pig, are only expressed from a short form–specific promoter, in contrast to the AS found in all other related mammalian groups. In mouse and rat, sequence searches for exons 1a and 2 give unquestionable hits upstream of exon 1b, but with single indels in either exon 1a (in mouse) or exon 2 (in rat), which would introduce a frameshift in the long form (supplementary fig. S6, Supplementary Material online). This suggests that the first step in subfunctionalization was the loss of expression from the long form–specific promoter, followed by ongoing pseudogenization of the long form–specific exons. In the case of guinea pig, only exon 2 is found in the genomic sequence near the TPM4 (also containing several indels), whereas exon 1a has likely been lost or translocated, as suggested by the lack of conserved synteny for the upstream region.
TPM Genes in Nonbilaterians and the Origin of the Muscular/Long Form
In stark contrast to the complex transcriptional outputs of TPM genes in bilaterians, all surveyed nonbilaterians TPMs encoded only the short-form Tm protein. Gene number across nonbilaterians was highly variable: We found many gene duplicates in some non-bilaterian animals, particularly in cnidarians. Nematostella vectensis and H. vulgaris show six and five TPM genes, respectively, and Podocoryne carinensis was reported to have at least two gene copies with different known functions (Gröger et al. 1999); the placozoan T. adhaerens has two TPM genes. In general, there are two broad types of TPM genes in nonbilaterians. Some TPM genes are similar to the canonical TPM genes (i.e., having the same protein length as most bilaterian short forms, 248 aa), and typically have an intron–exon structure largely conserved with bilaterian genes (supplementary fig. S7, Supplementary Material online, green branches). Other genes have a variety of protein lengths (217–281 aa), typically lack introns (supplementary table S1, Supplementary Material online) and show lower sequence similarity to bilaterian TPMs (supplementary fig. S7, Supplementary Material online, red branches).
The clearest difference between bilaterian and nonbilaterian TPM repertoires is the lack of long forms in nonbilaterian genomes, suggesting that the long form is a bilaterian innovation. We sought to determine how this new gene structure arose. The first exon in the long-form transcript (exon 1a) shows a clear similarity to the first exon of the short form (1b), especially in slow-evolving species (supplementary fig. S8, Supplementary Material online). In addition, we found that the second exon of the long-form transcript also shows significant sequence similarity, and similar length (126 vs. 134 nucleotides), to the second exon of the short form (exon 3; fig. 5a and b). For instance, a Blast search of different bilaterian exon 2 against the Nematostella genome consistently retrieved the Nematostella equivalent of exon 3 (supplementary fig. S8, Supplementary Material online). Thus, the structure of AS bilaterian genes is much as expected if the single ancestral promoter and the first two exons were duplicated in tandem in early bilaterian evolution, leading to the emergence of a new transcript including the upstream copy of the duplicated two exons, the downstream copy of the second exon, and the remainder of the gene (fig. 5b). This scenario is consistent with previous models for the structural evolution of the TPM gene structure (Wieczorek et al. 1988).
FIG. 5.
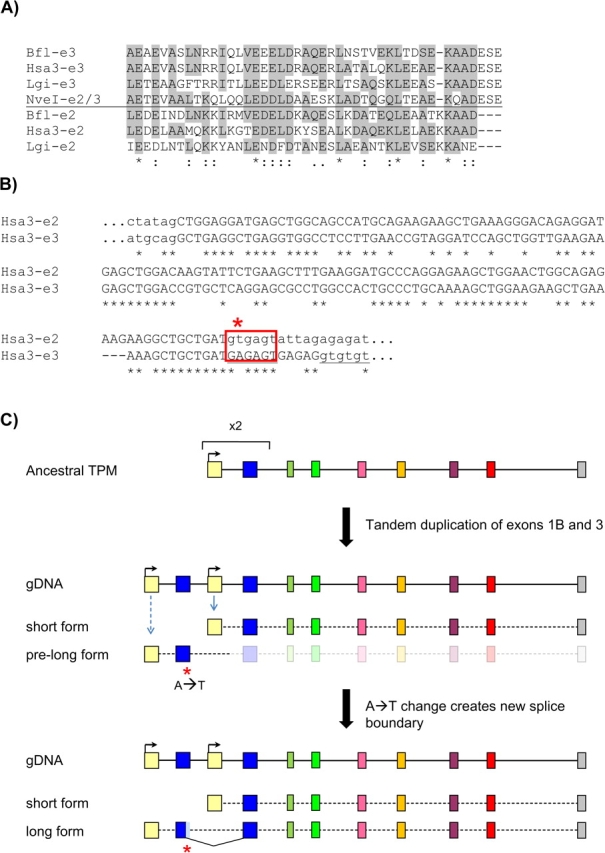
Model for the origin of the long TPM form in bilaterians. (A) Alignment of translated sequences for exons 2 and 3 from different bilaterian species and exon 2/3 from the nonbilaterian N. vectensis. Grey boxes indicate residues that are shared between the ancestral exon/exon 3 and the upstream exon 2. (B) Nucleotide sequence of the human TPM3 exons 2 and 3 and adjoining intronic sequences. A single nucleotide change at the exon 2 could have created the new splice site boundary. (C) Proposed two-step scenario for the origin of the long TPM isoform by tandem exon duplication of exon 1B and 3 (top) and frameshifting sliding in the newly formed exon 2 (bottom). Nve, Nematostella vectensis; Hsa3, Homo sapiens TPM3; Bfl, B. floridae; Lgi, Lottia gigantea.
However, there is one complication to this scenario. Although exons 2 and 3 show clear sequence similarity, they are not only of different lengths (134 and 126) but they have a different relationship to coding frame. Both exons begin with a full codon (i.e., the upstream exon falls between the last codon of exon 1a (1b) and the first of exon 2 (3)). However, although exon 2 also ends with a full codon, exon 3 ends with two bases out of a codon, with the first base of exon 4 completing that codon (i.e., the intron between exons 3 and 4 is in phase 2). Alignment of the two exons shows that this difference represents a truncation in exon 2, with 11 base pairs from exon 3 falling beyond the end of the alignment with exon 3 (fig. 5a and b).
This scenario raises two questions: What was the functional significance of this change, and how did the change occur at a molecular level. The answer to the first question is straightforward: Inclusion of the entirety of both duplicated exons would have induced a frameshift, thus truncation of exon 2 was necessary to put it in frame with exon 3 (because exon 3 begins with a full codon, for exon 2 to be joined to exon 3, exon 2 must end in a full codon). Interestingly, the sequence of exon 3 also provides a probable answer for the second question, of how the new boundary arose. Relative to exon 3, the 3′ terminus of exon 2 lacks three codons. The first two codons encode glutamic acid (E) followed by serine (S) (fig. 5a), which are conserved across bilaterians and with N. vectensis, suggesting that the protein sequence was the same at the time of the genomic duplication. These amino acids are encoded by GAR AGY, thus a single A→T base pair substitution at the second position would lead to the consensus splice site boundary, GTRAGY. In particular, if the sequence at the time of duplication happened to be GAA AGT, a single change would yield the most common splice boundary in most eukaryotes, GTAAGT (Irimia et al. 2007). These considerations suggest that the bilaterian genome structure arose by (1) genomic duplication of the single ancestral promoter and first two exons; followed by (2) creation of a new splice boundary by an A→T base pair change (fig. 5c). Such “sliding” of the intron–exon boundary along the gene is apparently a rare event in evolution, particular for sliding distances that are not a multiple of three bases (Rogozin et al. 2000; Roy 2009). Notably, this scenario resembles the patterns of altered splicing recently reported by Gao and Lynch (2009), in which internally duplicated gene regions were found to utilize splicing boundaries different from those utilized in the ancestral gene.
Conservation of Intron–Exon Structures
We found striking conservation of intron–exon structures across most of the length of TPM genes across a wide range of metazoans (fig. 1). Protein sequences typically show more than 50% amino acid similarity, with no gaps, allowing for confident alignments. Equivalent exons in the different transcripts show clear sequence similarity and are typically of exactly conserved lengths (i.e., exon 2 is 128 nt, exon 3 is 134 nt, exon 4 is 71 nt, etc.; fig. 1); intron positions show exact conservation, including phase. This overall conservation holds across AS variants within a given gene as well as between genes of different species.
Among the few cases in which intron–exon structures are not conserved, the differences reflect a combination of intron losses and intron gains. For instance, genic regions corresponding (i.e., homologous) to human exons 6 and 7 (fig. 1) are each independent exons with conserved lengths across a wide variety of bilaterians. However, in Caenorhabditis nematodes, the intervening intron has been lost, producing a single longer exon (fig. 1). On the other hand, the first exon of the long form (exon 1a), which is conserved as a single exon across almost all bilaterians, is interrupted by an intron in Caenorhabditis nematodes, likely representing a nematode-specific intron gain. Interestingly, the intron separating exon 1a and exon 2 has also been lost in nematodes.
The high degree of conservation of intron positions across species is unexpected. For most genes, species such as flies or nematodes have experienced large amounts of intron loss and gain since their split from the common ancestors, typically leading to only small minorities of intron positions being conserved with vertebrates (Rogozin et al. 2003). The reason for the high conservation in exon–intron structure for the TPM genes could be the widespread AS: For a given MATE, none of the intervening introns can be lost without affecting the AS pattern, which would likely be strongly disfavored. Consistent with this idea, in species with genes displaying no MATEs, a significant number of introns have been lost (supplementary table S1, Supplementary Material online). In other words, the ubiquity of MATEs in most bilaterians would seem to imply that intron–exon structure is highly constrained.
Discussion
The Evolutionary History of TPM
Our results support a detailed picture of the evolutionary history of TPM genes. Beginning from a single-transcript TPM gene that encoded the cytoplasmic form of Tm proteins, as found in all studied nonbilaterians, tandem genomic duplication of the promoter and first two exons of the ancestral gene in a bilaterian ancestor led to alternative transcripts derived from different promoters—the universal ancestral short form and the bilaterian-specific long form, using the upstream copy of the promoter and exon 1, both copies of duplicated exon 2/3, and all downstream exons (fig. 5). Then, whereas the short form maintained the ancestral function of the cytoplasmic Tm proteins and widespread expression, the new long isoform evolved a new function and expression pattern likely related to the origin of the sarcomeres present in bilaterian striated muscular cells. Due to their different functions and expression patterns, these two transcripts experienced different selective pressures; however, because the majority of the two transcripts were initially transcribed from the same genomic sequence, the transcripts could only differentiate at the (duplicated) 5′ end of the gene. Throughout the history of bilaterians, tandem duplication of individual exons and mutually exclusive AS associated with the two promoters then allowed the transcripts to diverge according to differential selective forces, leading to repeated parallel evolution of internal paralogy between the two transcripts. Finally, in some lineages, the transcripts achieved more complete independence, with the entire gene undergoing gene duplication followed by loss of one form from each gene, leading to long- and short-form transcripts encoded by different gene copies. This is a particularly clear case of subfunctionalization, whereby multiple functions of an ancestral gene are partitioned between gene duplicates.
Origin of a New TPM Gene Function and the Evolution of Organismal Complexity
The origin of the long-form Tm protein in bilaterians may be at least in part related to the evolution of a new cellular structure, the sarcomere, which constitutes the basis of striated muscles in bilaterians. Despite the fact that both long- and short-Tm proteins bind actin, they have radically different properties (Gunning et al. 2005). Muscle cells primarily express long muscle-specific Tms that form a highly specialized complex with several other proteins to build the core of the contractile apparatus of muscle fibers. The cytoplasmic forms usually have a complementary expression pattern (e.g., Stark et al. 2005), although small amounts of cytoplasmic forms in skeletal muscle form a cytoskeleton independent of the contractile apparatus in muscle (Gunning et al. 2008). High-level expression of a cytoplasmic Tm protein not normally expressed in skeletal muscle, however, has been shown to compromise the structural integrity of skeletal muscle (Kee et al. 2009).
Some nonbilaterian animals also have striated-like muscles, but these are built on different proteins and have different ultrastructure. For example, in the medusa Podocoryne one of the two isolated TPM genes is specifically expressed in muscle, whereas the other is not expressed in this tissue type, despite the fact that both are short-form TPMs (Gröger et al. 1999). Therefore, one short form TPM was specifically and independently recruited for muscular function (yet without sarcomeres).
The evolution of this novel isoform occurred by tandem exon duplication increasing the length of the TPM, resulting in a protein with seven binding sites to actin instead of six (McLachlan and Stewart 1976; Phillips 1986). Interestingly, metazoan short forms are already longer than homologs in fungi, which are usually 161–199 and have four to five binding sites to actin (Maytum et al. 2008), suggesting that a similar evolutionary process may have account for the origin of the cytoskeletal Tm proteins form itself before the origin of metazoans (Wieczorek, Smith, and Nadal-Ginard 1988).
Remarkably Parallel Evolution in Bilaterian TPM Genes
Deeply diverged bilaterian lineages evolved strikingly similar yet apparently independent patterns for TPM diversification to resolve the functional tension imposed by the long/short functional duality. Throughout the metazoan tree, we repeatedly found evolution of species- and lineage-specific MATEs along the whole TPM gene structure; in total, every TPM exon was duplicated at least in one lineage. In the most parsimonious scenario, 13 independent origins of MATEs would be needed to account for the patterns observed in the studied species. Moreover, new exons were added to preexisting MATEs in a convergent manner in several lineages, requiring up to 13 extra tandem exon duplications of already duplicated exons. Gain of alternative polyadenylation sites, resulting in the mutually exclusive inclusion of different last exons, also occurred in diverse lineages such as vertebrates and arthropods.
Finally, we found extensive whole gene duplication in all major metazoan groups, in many cases leading to convergent subfunctionalization through loss of ancestral exons and splicing patterns, and resulting in the complete externalization of the internal paralogy in lineages as divergent as tunicates and leeches.
Extensive convergent evolution of (tandem) gene duplication across metazoans has been previously reported for several gene families (e.g., D'Aniello et al. 2008; Irimia, Maeso, et al. 2008; Negre and Simpson 2009); however, we are not aware of a similarly extensive case of parallel exon duplication and AS throughout the full gene and full protein length, being perhaps only comparable with the tandem exon duplications of three Dscam exons in Ecdysozoans (Brites et al. 2008).
Alternative Splicing and Gene Duplication
The evolutionary emergence of novel protein functions is presumably greatly constrained by the need to retain old functions. Forty years ago, Ohno (1970) pointed out that gene duplication could resolve this tension: After duplication, one gene copy would continue to produce the original product while the other could evolve new functions. The discovery of AS offered another path for protein innovation: the single gene produces the initial product, but new splicing isoforms could evolve new functions by mutations within isoform-specific regions (Modrek and Lee 2003).
These two paths to genetic novelty seem to have different levels of importance in different lineages (Rukov et al. 2007; Irimia, Rukov et al. 2008; Irimia et al. 2009). More surprisingly, previous evidence suggests that different gene families within a given lineage may have different propensities to evolve via these two pathways, and negative correspondences between gene duplication and AS across gene families have been reported in nematodes and mammals (Kopelman et al. 2005; Hughes and Friedman 2008; Irimia, Rukov et al. 2008).
The TPM gene exemplifies the relation between AS and gene duplication. Both mechanisms seem to be used in different lineages to achieve superficially similar outputs, that is, the optimization and specific divergence of the short- and long-Tm proteins. The origin of the muscle-specific form in bilaterians likely imposed a need for the single TPM gene to optimize the new function as well as constrained the evolution of the short form because both proteins initially shared most of the sequence. Our results suggest that this tension was resolved in most lineages by the evolution of internal exon duplications that are mutually exclusively spliced and subsequently associated with specific promoters. In this way, each specific MATE exon could evolve freely, allowing the optimization of both the specific long- and short-form functions independently. The relation between the origin of MATEs and the existence of alternative promoters is attested to by their close association in modern bilaterian genomes: all genes with both promoters have MATEs and nearly no genes with a single promoter have MATEs, with the exception of a few vertebrate genes.
The initial redundancy produced by AS thus facilitated the evolution of a new gene function (performed by the long form) while keeping the old one (that of the short form). As mentioned above, this was achieved in some lineages by whole gene duplication and subfunctionalization, as it seems to be the extreme case in Hellobdella robusta and Ciona intestinalis in which not a single MATE is found in the full duplicative TPM repertoire, exemplifying the inverse correspondence between AS and gene duplication.
We term this process of subfunctionalization of the different isoforms into independent genes externalization of the internal paralogy. Interestingly, it may mechanistically resemble the duplication-degeneration-complementation model for the evolution of paralogs after gene duplication (Force et al. 1999). In this case, if one of the gene duplicates losses the ability to generate one of the isoforms and the other duplicate the other, both genes must be conserved to account for the full function of the ancestral gene, as described by Force et al. (1999) for the case of mutations in cis regulatory elements of gene duplicates.
Distinct Evolution of TPM Gene Structures and Functions in Vertebrates
Vertebrate TPM genes have been by far the most studied TPMs—nearly all we know about TPM function and regulation comes from the study of the dozens of mammalian Tm protein isoforms. However, the behavior of vertebrate TPMs seems atypical among bilaterians. First, the vertebrate TPM2 encodes only long isoforms but contains both an internal MATE and alternative terminal exons (Table S1). Second, the association between different alternative regions, though generally strong, is less complete than in any other lineages (supplementary table S3, Supplementary Material online). For example, the TPM3 gene encodes 10 isoforms consisting of exon 1a or 1b, exon 6a or 6b, and one of the different copies exon 9. Northern blot analysis shows that they are all expressed in the brain at roughly similar levels, although 6a containing isoforms go down with development while 6b containing isoforms come up (Dufour et al. 1998). A similar situation is reported for TPM1 gene in mammals and TPM2 gene in birds.
These differences between vertebrates and other bilaterians could be related to both genomic and organismal architecture. Vertebrates contain four or more ancient duplicates produced in the rounds of whole genome duplication, and most exhibit at least moderately high levels of AS. In addition, long Tms in vertebrates have been extensively recruited for new cytoskeletal functions, a type of innovation generally associated with short-form Tms in other studied species (Stark et al. 2005). Thus, in vertebrates, there may be more functional divergence between genes (i.e., apart from the long/short distinction), as supported by knockout experiments showing little redundancy among different TPM genes, but significant overlapping between some products of the same genes (Blanchard et al. 1997; Rethinasamy et al. 1998; Robbins 1998; Hook et al. 2004).
Functional Implications of Tm proteins Diversification
The drive to create isoform diversity in the TPM gene family is remarkably consistent and has utilized both gene duplication and AS extensively. Different Tms differ in their ability to interact with myosin motors (Fanning et al. 1994; Bryce et al. 2003), actin severing proteins (Ishikawa et al. 1989; Bryce et al. 2003), capping proteins (Watakabe et al. 1996; Kostyukova and Hitchcock-DeGregori 2004) and cross-linking proteins. There is increasing evidence that the entire actin/tropomyosin filament is the unit of function (Holmes and Lehman 2008) and that the binding of a single type of Tm to a filament provides both fidelity of function and functional characteristics to that filament (Gunning et al. 2005, 2008). Therefore, the functional consequences of Tm proteins diversification are mainly 2-fold. First, spatial segregation of isoforms has provided a mechanism to independently regulate different actin filament populations; second, the Tm proteins isoforms can directly regulate the functional properties of the actin filament.
Single exon changes are sufficient to account for altered spatial segregation (Percival et al. 2004; Schevzov et al. 2005) and functional characteristics such as ability to restore stress fibers to transformed cells (Gimona et al. 1996). This means that a single change in the primary exon sequence of a Tm proteins isoform could be manifested as a change in the functional characteristics of a specific actin filament, and thus the change in the use of an exon will be directly manifested as the generation of a new type of actin filament with novel functional characteristics. This suggests that a single MATE could greatly alleviate the functional tensions produced by the long/short duality.
Interestingly, the presence of extensive convergent evolution of AS at different exons seems to indicate that AS has been of central importance in the evolution of these genes but that the specific exon(s) subject to AS matters considerably less. This suggests that a specific change in one region of a protein can impact the function across the whole protein. Because the Tm proteins polymer is a repeating structure and the whole actin/tropomyosin filament seems to be the unit of function (Holmes and Lehman 2008), any exon change could produce a repeated alteration along the entire length of the filament, giving an explanation for the remarkable convergent evolution of AS at different exons.
Concluding Remarks
Whether morphological innovation arises mainly through the recruitment of nearly unchanged proteins and functional gene networks or through changes in protein functions is the subject of a hot debate (Wagner and Lynch 2008), and only few examples of protein neofunctionalization were reported in recent literature. Our overall analysis of TPM genes in metazoans illustrates a striking case of formation of new genic products, which then underwent significant changes in expression patterns. Through tandem duplication of two exons and AS of a single gene, two distinct proteins evolved independently, and one of them acquired a new function likely associated with a bilaterian-specific innovation, the sarcomere. Evolution of MATEs has allowed the gradual divergence of two functionally distinct gene products encoded in a single gene. Furthermore, across bilaterian evolution, short and long Tm proteins became encoded by distinct genes, after gene duplication and subfunctionalization, a process that we denominate externalization of internal paralogs. Globally, we show that MATEs and gene duplication are two distinct mechanisms for the same purpose: the generation of novel proteins and the acquisition of new, evolutionary relevant functions. On the other hand, these results focus attention on the importance of mutually exclusive splicing of tandemly duplicated exons in the emergence of new functions. The evolutionary forces underlying this mechanism and its uses in the evolution of organismal complexity are priorities in understanding gene evolution.
Supplementary Material
Supplementary figures S1–S8 and tables S1–S3 are available at Molecular Biology and Evolution online (http://www.mbe.oxfordjournals.org/).
Supplementary Material
Acknowledgments
We would like to thank Eugene V. Koonin, Barbara Pernaute, Jakob L. Rukov, Senda Jiménez-Delgado and members of Jordi Garcia-Fernàndez Lab for helpful comments and discussions. M.I., I.M. and J.G.F. were funded by grants BFU2005-00252 and BMC2008-03776 from the Spanish Ministerio de Educación y Ciencia (MEC), M.I. and I.M. hold FPI and FPU grants, respectively; P.G. is supported by grants from the NHMRC and is a Principal Research fellow of the NHMRC; S.W.R. was funded by the Intramural Research Program of the National Library of Medicine at National Institutes of Health/DHHS.
References
- Abascal F, Zardoya R, Posada D. ProtTest: selection of best-fit models of protein evolution. Bioinformatics. 2005;21:2104–2105. doi: 10.1093/bioinformatics/bti263. [DOI] [PubMed] [Google Scholar]
- Blanchard EM, Iizuka K, Christe M, Conner DA, Geisterfer-Lowrance A, Schoen FJ, Maughan DW, Seidman CE, Seidman JG. Targeted ablation of the murine alpha-tropomyosin gene. Circ Res. 1997;81:1005–1010. doi: 10.1161/01.res.81.6.1005. [DOI] [PubMed] [Google Scholar]
- Brites D, McTaggart S, Morris K, Anderson J, Thomas K, Colson I, Fabbro T, Little T, Ebert D, Du Pasquier L. The Dscam homologue of the Crustacean Daphnia is diversified by alternative splicing like in insects. Mol Biol Evol. 2008;25:1429–1439. doi: 10.1093/molbev/msn087. [DOI] [PubMed] [Google Scholar]
- Bryce NS, Schevzov G, Ferguson V, et al. 11 co-authors. Specification of actin filament function and molecular composition by tropomyosin isoforms. Mol Biol Cell. 2003;14:1002–1016. doi: 10.1091/mbc.E02-04-0244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- D'Aniello S, Irimia M, Maeso I, Pascual-Anaya J, Jiménez-Delgado S, Bertrand S, Garcia-Fernàndez J. Gene expansion and retention leads to a diverse tyrosine kinase superfamily in amphioxus. Mol Biol Evol. 2008;25:1841–1854. doi: 10.1093/molbev/msn132. [DOI] [PubMed] [Google Scholar]
- Dalby-Payne JR, O'Loughlin EV, Gunning P. Polarization of specific tropomyosin isoforms in gastrointestinal epithelial cells and their impact on CFTR at the apical surface. Mol Biol Cell. 2003;14:4365–4375. doi: 10.1091/mbc.E03-03-0169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drummond A, Strimmer K. PAL: an object-oriented programming library for molecular evolution and phylogenetics. Bioinformatics. 2001;17:662–663. doi: 10.1093/bioinformatics/17.7.662. [DOI] [PubMed] [Google Scholar]
- Dufour C, Weinberger RP, Schevzov G, Jeffrey PL, Gunning P. Splicing of two internal and four carboxyl-terminal alternative exons in nonmuscle tropomyosin 5 pre-mRNA is independently regulated during development. J Biol Chem. 1998;273:18547–18555. doi: 10.1074/jbc.273.29.18547. [DOI] [PubMed] [Google Scholar]
- Fanning AS, Wolenski JS, Mooseker MS, Izant JG. Differential regulation of skeletal muscle myosin-II and brush border myosin-I enzymology and mechanochemistry by bacterially produced tropomyosin isoforms. Cell Motil Cytoskeleton. 1994;29:29–45. doi: 10.1002/cm.970290104. [DOI] [PubMed] [Google Scholar]
- Force A, Lynch M, Pickett FB, Amores A, Yan YL, Postlethwait J. Preservation of duplicate genes by complementary, degenerative mutations. Genetics. 1999;151:1531–1545. doi: 10.1093/genetics/151.4.1531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao X, Lynch M. Ubiquitous internal gene duplication and intron creation in eukaryotes. Proc Natl Acad Sci U S A. 2009 doi: 10.1073/pnas.0911093106. Advance Access published November 19, 2009, doi:10.1073/pnas.0911093106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gimona M, Kazzaz JA, Helfman DM. Forced expression of tropomyosin 2 or 3 in v-Ki-ras-transformed fibroblasts results in distinct phenotypic effects. Proc Natl Acad Sci U S A. 1996;93:9618–9623. doi: 10.1073/pnas.93.18.9618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gooding C, Smith CW. Tropomyosin exons as models for alternative splicing. Adv Exp Med Biol. 2008;644:27–42. doi: 10.1007/978-0-387-85766-4_3. [DOI] [PubMed] [Google Scholar]
- Gröger H, Callaerts P, Gehring WJ, Schmid V. Gene duplication and recruitment of a specific tropomyosin into striated muscle cells in the jellyfish Podocoryne carnea. J Exp Zool. 1999;285:378–386. [PubMed] [Google Scholar]
- Guindon S, Gascuel O. A simple, fast, and accurate algorithm to estimate large phylogenies by maximum likelihood. Syst Biol. 2003;52:696–704. doi: 10.1080/10635150390235520. [DOI] [PubMed] [Google Scholar]
- Gunning P, O'Neill G, Hardeman E. Tropomyosin-based regulation of the actin cytoskeleton in time and space. Physiol Rev. 2008;88:1–35. doi: 10.1152/physrev.00001.2007. [DOI] [PubMed] [Google Scholar]
- Gunning PW, Schevzov G, Kee AJ, Hardeman EC. Tropomyosin isoforms: divining rods for actin cytoskeleton function. Trends Cell Biol. 2005;15:333–341. doi: 10.1016/j.tcb.2005.04.007. [DOI] [PubMed] [Google Scholar]
- Hannan AJ, Gunning P, Jeffrey PL, Weinberger RP. Structural compartments within neurons: developmentally regulated organization of microfilament isoform mRNA and protein. Mol Cell Neurosci. 1998;11:289–304. doi: 10.1006/mcne.1998.0693. [DOI] [PubMed] [Google Scholar]
- Holmes KC, Lehman W. Gestalt-binding of tropomyosin to actin filaments. J Muscle Res Cell Motil. 2008;29:213–219. doi: 10.1007/s10974-008-9157-6. [DOI] [PubMed] [Google Scholar]
- Hook J, Lemckert F, Qin H, Schevzov G, Gunning P. Gamma tropomyosin gene products are required for embryonic development. Mol Cell Biol. 2004;24:2318–2323. doi: 10.1128/MCB.24.6.2318-2323.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huelsenbeck JP, Ronquist F. MRBAYES: Bayesian inference of phylogenetic trees. Bioinformatics. 2001;17:754–755. doi: 10.1093/bioinformatics/17.8.754. [DOI] [PubMed] [Google Scholar]
- Hughes AL, Friedman R. Alternative splicing, gene duplication and connectivity in the genetic interaction network of the nematode worm Caenorhabditis elegans. Genetica. 2008;134:181–186. doi: 10.1007/s10709-007-9223-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Irimia M, Maeso I, Garcia-Fernàndez J. Convergent evolution of clustering of Iroquois homeobox genes across metazoans. Mol Biol Evol. 2008;25:1521–1525. doi: 10.1093/molbev/msn109. [DOI] [PubMed] [Google Scholar]
- Irimia M, Penny D, Roy SW. Coevolution of genomic intron number and splice sites. Trends Genet. 2007;23:321–325. doi: 10.1016/j.tig.2007.04.001. [DOI] [PubMed] [Google Scholar]
- Irimia M, Rukov JL, Penny D, Garcia-Fernandez J, Vinther J, Roy SW. Widespread evolutionary conservation of alternatively spliced exons in Caenorhabditis. Mol Biol Evol. 2008;5:375–382. doi: 10.1093/molbev/msm262. [DOI] [PubMed] [Google Scholar]
- Irimia M, Rukov JL, Roy SW, Vinther JL, Garcia-Fernàndez J. Quantitative regulation of alternative splicing in evolution and development. BioEssays. 2009;31:40–50. doi: 10.1002/bies.080092. [DOI] [PubMed] [Google Scholar]
- Ishikawa R, Yamashiro S, Matsumura F. Differential modulation of actin-severing activity of gelsolin by multiple isoforms of cultured rat cell tropomyosin. Potentiation of protective ability of tropomyosins by 83-kDa nonmuscle caldesmon. J Biol Chem. 1989;264:7490–7497. [PubMed] [Google Scholar]
- Karlik CC, Fyrberg EA. Two Drosophila melanogaster tropomyosin genes: structural and functional aspects. Mol Cell Biol. 1986;6:1965–1973. doi: 10.1128/mcb.6.6.1965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kee AJ, Gunning PW, Hardeman EC. A cytoskeletal tropomyosin can compromise the structural integrity of skeletal muscle. Cell Motil Cytoskeleton. 2009;66:710–720. doi: 10.1002/cm.20400. [DOI] [PubMed] [Google Scholar]
- Kopelman NM, Lancet D, Yanai I. Alternative splicing and gene duplication are inversely correlated evolutionary mechanisms. Nat Genet. 2005;37:588–589. doi: 10.1038/ng1575. [DOI] [PubMed] [Google Scholar]
- Kostyukova AS, Hitchcock-DeGregori SE. Effect of the structure of the N terminus of tropomyosin on tropomodulin function. J Biol Chem. 2004;279:5066–5071. doi: 10.1074/jbc.M311186200. [DOI] [PubMed] [Google Scholar]
- Li W, Gao FB. Actin filament-stabilizing protein tropomyosin regulates the size of dendritic fields. J Neurosci. 2003;23:6171–6175. doi: 10.1523/JNEUROSCI.23-15-06171.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin JJ, Hegmann TE, Lin JL. Differential localization of tropomyosin isoforms in cultured nonmuscle cells. J Cell Biol. 1988;107:563–572. doi: 10.1083/jcb.107.2.563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin SC, Storti RV. Developmental regulation of the Drosophila tropomyosin I (TmI) gene is controlled by a muscle activator enhancer region that contains multiple cis-elements and binding sites for multiple proteins. Dev Genet. 1997;20:297–306. doi: 10.1002/(SICI)1520-6408(1997)20:4<297::AID-DVG1>3.0.CO;2-2. [DOI] [PubMed] [Google Scholar]
- Mateos J, Herranz R, Domingo A, Sparrow J, Marco R. The structural role of high molecular weight tropomyosins in dipteran indirect flight muscle and the effect of phosphorylation. J Muscle Res Cell Motil. 2006;27:189–201. doi: 10.1007/s10974-005-9044-3. [DOI] [PubMed] [Google Scholar]
- Maytum R, Hatch V, Konrad M, Lehman W, Geeves MA. Ultra short yeast tropomyosins show novel myosin regulation. J Biol Chem. 2008;283:1902–1910. doi: 10.1074/jbc.M708593200. [DOI] [PubMed] [Google Scholar]
- McLachlan AD, Stewart M. The 14-fold periodicity in alpha-tropomyosin and the interaction with actin. J Mol Biol. 1976;103:271–298. doi: 10.1016/0022-2836(76)90313-2. [DOI] [PubMed] [Google Scholar]
- Modrek B, Lee CJ. Alternative splicing in the human, mouse and rat genomes is associated with an increased frequency of exon creation and/or loss. Nat Genet. 2003;34:177–180. doi: 10.1038/ng1159. [DOI] [PubMed] [Google Scholar]
- Molloy J, Kreuz A, Miller R, Tansey T, Maughan D. Effects of tropomyosin deficiency in flight muscle of Drosophila melanogaster. Adv Exp Med Biol. 1993;332:165–171. doi: 10.1007/978-1-4615-2872-2_15. [DOI] [PubMed] [Google Scholar]
- Negre B, Simpson P. Evolution of the achaete-scute complex in insects: convergent duplication of proneural genes. Trends Genet. 2009;25:147–152. doi: 10.1016/j.tig.2009.02.001. [DOI] [PubMed] [Google Scholar]
- Ohno S. Evolution by gene duplication. New York: Springer-Verlag; 1970. [Google Scholar]
- Percival JM, Hughes JA, Brown DL, Schevzov G, Heimann K, Vrhovski B, Bryce N, Stow JL, Gunning PW. Targeting of a tropomyosin isoform to short microfilaments associated with the Golgi complex. Mol Biol Cell. 2004;15:268–280. doi: 10.1091/mbc.E03-03-0176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phillips GNJ. Construction of an atomic model for tropomyosin and implications for interactions with actin. J Mol Biol. 1986;192:128–131. doi: 10.1016/0022-2836(86)90469-9. [DOI] [PubMed] [Google Scholar]
- Putnam N, Butts T, Ferrier DEK, et al. 37 co-authors. The amphioxus genome and the evolution of the chordate karyotype. Nature. 2008;453:1064–1071. doi: 10.1038/nature06967. [DOI] [PubMed] [Google Scholar]
- Rethinasamy P, Muthuchamy M, Hewett T, Boivin G, Wolska BM, Evans C, Solaro RJ, Wieczorek DF. Molecular and physiological effects of alpha-tropomyosin ablation in the mouse. Circ Res. 1998;82:116–123. doi: 10.1161/01.res.82.1.116. [DOI] [PubMed] [Google Scholar]
- Robbins J. Alpha-tropomyosin knockouts: a blow against transcriptional chauvinism. Circ Res. 1998;82:134–136. doi: 10.1161/01.res.82.1.134. [DOI] [PubMed] [Google Scholar]
- Rogozin IB, Lyons-Weiler J, Koonin EV. Intron sliding in conserved gene families. Trends Genet. 2000;16:430–432. doi: 10.1016/s0168-9525(00)02096-5. [DOI] [PubMed] [Google Scholar]
- Rogozin IB, Wolf YI, Sorokin AV, Mirkin BG, Koonin EV. Remarkable interkingdom conservation of intron positions and massive, lineage-specific intron loss and gain in eukaryotic evolution. Curr Biol. 2003;13:1512–1517. doi: 10.1016/s0960-9822(03)00558-x. [DOI] [PubMed] [Google Scholar]
- Ronquist F, Huelsenbeck JP. MrBayes 3: Bayesian phylogenetic inference under mixed models. Bioinformatics. 2003;19:1572–1574. doi: 10.1093/bioinformatics/btg180. [DOI] [PubMed] [Google Scholar]
- Roy SW. Intronization, de-intronization and intron sliding are rare in Cryptococcus. BMC Evol Biol. 2009;9:192. doi: 10.1186/1471-2148-9-192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rukov JL, Irimia M, Mork S, Lund VK, Vinther J, Arctander P. High qualitative and quantitative conservation of alternative splicing in Caenorhabditis elegans and Caenorhabditis briggsae. Mol Biol Evol. 2007;24:909–917. doi: 10.1093/molbev/msm023. [DOI] [PubMed] [Google Scholar]
- Schevzov G, Gunning P, Jeffrey PL, Temm-Grove C, Helfman DM, Lin JJ, Weinberger RP. Tropomyosin localization reveals distinct populations of microfilaments in neurites and growth cones. Mol Cell Neurosci. 1997;8:439–454. doi: 10.1006/mcne.1997.0599. [DOI] [PubMed] [Google Scholar]
- Schevzov G, Vrhovski B, Bryce NS, Elmir S, Qiu MR, O'neill GM, Yang N, Verrills NM, Kavallaris M, Gunning PW. Tissue-specific tropomyosin isoform composition. J Histochem Cytochem. 2005;53:557–570. doi: 10.1369/jhc.4A6505.2005. [DOI] [PubMed] [Google Scholar]
- Stamatakis A. RAxML-VI-HPC: maximum likelihood-based phylogenetic analyses with thousands of taxa and mixed models. Bioinformatics. 2006;22:2688–2690. doi: 10.1093/bioinformatics/btl446. [DOI] [PubMed] [Google Scholar]
- Stark A, Brennecke J, Bushati N, Russell RB, Cohen SM. Animal microRNAs confer robustness to gene expression and have a significant impact on 3′UTR evolution. Cell. 2005;123:1133–1146. doi: 10.1016/j.cell.2005.11.023. [DOI] [PubMed] [Google Scholar]
- Vrhovski B, Thézé N, Thiébaud P. Structure and evolution of tropomyosin genes. Adv Exp Med Biol. 2008;644:6–26. doi: 10.1007/978-0-387-85766-4_2. [DOI] [PubMed] [Google Scholar]
- Wagner GP, Lynch VJ. The gene regulatory logic of transcription factor evolution. Trends Ecol Evol. 2008;23:377–385. doi: 10.1016/j.tree.2008.03.006. [DOI] [PubMed] [Google Scholar]
- Watakabe A, Kobayashi R, Helfman DM. N-tropomodulin: a novel isoform of tropomodulin identified as the major binding protein to brain tropomyosin. J Cell Sci. 1996;109:2299–2310. doi: 10.1242/jcs.109.9.2299. [DOI] [PubMed] [Google Scholar]
- Weinberger R, Schevzov G, Jeffrey P, Gordon K, Hill M, Gunning P. The molecular composition of neuronal microfilaments is spatially and temporally regulated. J Neurosci. 1996;16:238–252. doi: 10.1523/JNEUROSCI.16-01-00238.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wieczorek DF, Smith CW, Nadal-Ginard B. The rat alpha-tropomyosin gene generates a minimum of six different mRNAs coding for striated, smooth and nonmuscle isoforms by alternative splicing. Cell Biol. 1988;8:679–694. doi: 10.1128/mcb.8.2.679. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.