Abstract
Drosophila telomeres are elongated by transposition of specialized retroelements rather than telomerase activity, and are assembled independently of the terminal DNA sequence. Drosophila telomeres are protected by terminin, a complex that includes the HOAP (Heterochromatin Protein 1/origin recognition complex-associated protein) and Moi (Modigliani) proteins and shares the properties of human shelterin. Here we show that Verrocchio (Ver), an oligonucleotide/oligosaccharide-binding (OB) fold-containing protein related to Rpa2/Stn1, interacts physically with HOAP and Moi, is enriched only at telomeres, and prevents telomere fusion. These results indicate that Ver is a new terminin component; we speculate that, concomitant with telomerase loss, Drosophila evolved terminin to bind chromosome ends independently of the DNA sequence.
Keywords: verrocchio, terminin, telomeres, Rpa2/Stn1, Drosophila
Telomeres are nucleoprotein complexes that protect the ends of linear chromosomes and regulate terminal DNA replication. If telomeres are not properly capped, they are sensed as double-strand breaks, leading to the activation of cell cycle checkpoints and inappropriate DNA repair, which might result in end-to-end fusions (Palm and de Lange 2008). In most organisms, telomeres contain arrays of GC-rich repeats, which are added to chromosome ends by telomerase (Palm and de Lange 2008). Drosophila telomeres are elongated by transposition of three specialized retroelements, rather than telomerase activity; several studies indicate that Drosophila telomeres are epigenetic structures assembled independently of the sequence of terminal DNA (Cenci et al. 2005; Mason et al. 2008; Rong 2008).
In organisms with telomerase, telomeres are protected by two well-characterized protein assemblies: the Cdc13–Stn1–Ten1 (CST) and shelterin complexes. These complexes are evolutionarily conserved, even if they vary in composition and architecture in different phyla. The three subunits of the CST complex all contain oligonucleotide/oligosaccharide-binding (OB) fold domains, and interact to each other to form an RPA-like complex that binds the telomere 3′ overhang. (Mitton-Fry et al. 2002; Gao et al. 2007; Gelinas et al. 2009; Sun et al. 2009). Shelterin has been thoroughly characterized in human cells; it is a six-protein complex that specifically associates with the telomeric TTAGGG repeats. Three of the shelterin subunits interact directly with the TTAGGG repeats: TRF1 and TRF2 bind the TTAGGG duplex, and POT1 binds the 3′ overhang. TRF1, TRF2, and POT1 are interconnected by TIN2 and TPP1, and TRF2 interacts with hRap1, a distant homolog of Saccharomyces cerevisiae Rap1. The shelterin subunits share three properties that distinguish them from the nonshelterin telomere-associated proteins. They are specifically enriched at telomeres, they are present at telomeres throughout the cell cycle, and their functions are limited to telomere maintenance (Palm and de Lange 2008).
The Stn1 and Ten1 subunits of the S. cerevisiae CST complex are conserved in Schizosaccharomyces pombe, plants, and humans, while shelterin-like elements are found in S. pombe and plants but not in S. cerevisiae (Martin et al. 2007; Song et al. 2008; Linger and Price 2009; Lue 2009; Miyake et al. 2009; Surovtseva et al. 2009). S. pombe and plants have both a shelterin-like complex and a CST-like complex, both of which are required for telomere protection. The two complexes are present also in humans, and are thought to collaborate in telomere protection. However, the human CST complex does not share the shelterin properties, and appears to have a relatively minor role in telomere capping. (Miyake et al. 2009; Surovtseva et al. 2009).
In addition to the shelterin and CST components, yeast, plant, and mammalian telomeres contain several conserved polypeptides required for proper telomere function. These polypeptides include many proteins involved in DNA repair, such as the ATM kinase, the Ku70/80 heterodimer, the MRE11/RAD50/NBS1 (MRN) complex, Rad51, the ERCC1/XPF endonuclease, the Apollo exonuclease, and the RecQ family members WRN and BLM. In addition, yeast and mammalian telomeres are enriched in proteins that are homologous to Drosophila HP1 (Heterochromatin Protein 1). All nonshelterin and non-CST proteins function not only at telomeres, but are also involved in several cellular processes that are not related with telomeres (Palm and de Lange 2008; Linger and Price 2009).
The search for Drosophila telomere-capping proteins has relied mainly on the isolation of mutants that display frequent telomeric fusions in larval brain cells. Genetic and molecular analyses thus far have identified nine loci that are required to prevent end-to-end fusion in Drosophila. These are UbcD1, which encodes a highly conserved E2 enzyme that mediates protein ubiquitination; Su(var)205 and caravaggio (cav), which encode HP1 and HOAP (HP1/origin recognition complex [ORC]-associated protein), respectively; the Drosophila homologs of the ATM, RAD50, MRE11, and NBS1 genes; without children (woc), which specifies a transcription factor associated with the initiating form of RNA polymerase II (Pol II); and modigliani (moi, which encodes a novel protein that binds both HOAP and HP1 (for review, see Cenci et al. 2005; Ciapponi and Cenci 2008; Mason et al. 2008; Rong 2008; see also Raffa et al. 2009). HOAP and Moi have properties that distinguish them from the other Drosophila telomere-capping proteins: They localize only at telomeres, and appear to function only in telomere maintenance. These properties are similar to the properties of human shelterin, suggesting that the HOAP–Moi complex, which we called terminin, is a functional analog of shelterin (Raffa et al. 2009).
Here we show that Verrocchio (Ver) is required to prevent telomere fusion. Ver interacts directly with both HOAP and Moi and localizes only at telomeres, suggesting that Ver is a novel terminin component. Bioinformatics analyses showed that Ver contains an OB fold domain that shares structural similarity with the OB fold of Stn1. Mutations in the predicted Ver OB fold cause telomere fusion, suggesting that the Ver OB fold is required for telomere protection.
Results and Discussion
Isolation and characterization of verrocchio
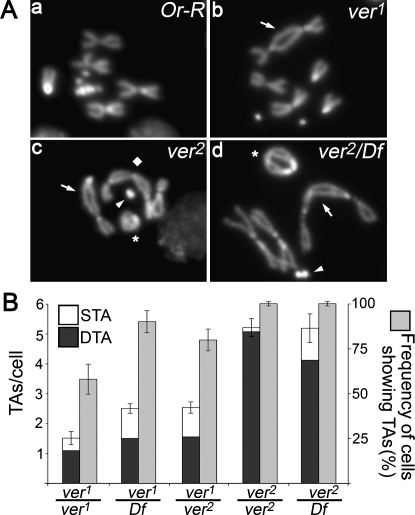
We isolated the verrocchio (ver1) mutation in the course of a cytological screen aimed at the identification of genes required for telomere protection against fusion events (see the Supplemental Material). Flies homozygous for ver1 die at the larval/pupal boundary, allowing cytological analysis of larval brain cells. DAPI-stained brain preparations from ver1 homozygous larvae revealed that mitotic cells display frequent telomeric associations (TAs), some of which generate multicentric chromosomes that resemble little “trains” of chromosomes (Fig. 1A). The verrocchio gene was named after this phenotype, just as caravaggio (Cenci et al. 2003) and modigliani (Raffa et al. 2009), as the names of these artists are names of Italian trains.
Figure 1.
Mutations in ver cause telomeric fusions. (A) Examples of TAs in ver mutant neuroblasts. ver mutants show two types of TAs: single TAs (STAs), in which a single telomere associates with either its sister or a nonsister telomere, and double TAs (DTAs), wherein a pair of sister telomeres joins with another pair. In wild-type cells, the frequency of putative TAs is <0.01 per cell (n = 500). (Panel a) Control (Or-R) metaphase. (Panel b) ver1/ver1 metaphase with a 2-3 dicentric chromosome generated by a DTA (arrow). (Panel c) ver2/ver2 metaphase showing XL-2 (arrow) and 4-4 (arrowhead) dicentric chromosomes, a ring X chromosome (asterisk), and a tricentric 2-3-2 chromosome (diamond), all generated by DTAs. (Panel d) ver2/Df(3L)sex204 metaphase containing 2-2 (arrow) and 4-4 (arrowhead) dicentric chromosomes and a dicentric ring involving both X chromosomes (asterisk), all generated by DTAs. (B) Frequencies of TAs in ver mutants. The TA frequencies in ver1/ver2 and ver1/Df(3L)sex204 mutants are significantly higher than that observed in ver1 homozygotes (P < 0.01, with Students t-test); the TA frequencies observed in ver2/ver2 and ver2/Df brains are not significantly different. At least 200 cells from at least four brains were scored for each genotype.
Recombination analysis with visible markers and deficiency mapping placed ver1 in the 69C4–69F6 interval. Complementation tests with P element-induced insertional mutations mapping to the same interval showed that ver1 fails to complement l(3)S147910, which henceforth will be designated as ver2. ver2 homozygotes and ver2/Df(3L)sex204 hemizygotes exhibit comparable frequencies of TAs (about five per cell) (Fig. 1A,B). These frequencies are significantly higher than those observed in ver1/ver1, ver1/Df(3L)sex204, and ver1/ver2 mutant brains (Fig. 1B), suggesting that ver2 is a genetically null allele, while ver1 is hypomorphic.
The ver2 allele carries a P{lacW} construct inserted into the coding sequence of the CG14121 gene (Carré et al. 2005), which encodes a 214-amino-acid polypeptide. DNA sequencing revealed that, in the ver1 allele, the CG14121 gene contains a T → C transition that converts the natural stop codon into a glutamine codon. This base substitution leads to a conceptual protein 12 amino acids longer than the wild-type gene product. RT–PCR revealed that CG14121 is normally transcribed in ver1 mutants, while no transcript was detectable in ver2 mutants (Supplemental Fig. S1A), consistent with the conclusion that ver2 is a genetically null allele.
Ver specifically localizes at telomeres
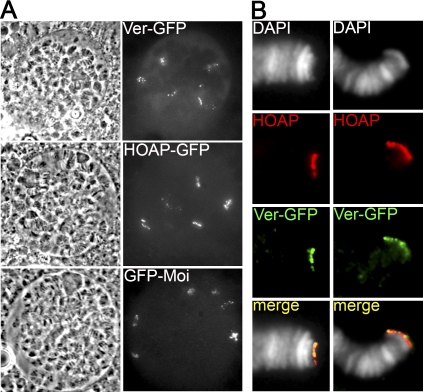
To determine the subcellular localization of Ver, we transformed flies with a construct containing the wild-type ver coding sequence fused in-frame with the GFP sequence. Animals homozygous for ver mutations and bearing this construct did not display telomere fusions in larval brains (we did not observe TAs in 200 cells scored) (Supplemental Fig. S4B), and survived to adulthood. Thus, the Ver-GFP protein rescues both the lethality and the telomere fusion phenotype of ver mutants. Analysis of unfixed polytene chromosome nuclei from Ver-GFP-expressing larvae revealed six discrete fluorescent signals (Fig. 2A). These signals are likely to correspond to telomeres, as the same type and number of signals were observed in unfixed polytene nuclei from larvae that express either HOAP-GFP or GFP-Moi, two telomere-specific proteins required to prevent end-to-end fusions (Cenci et al. 2003; Raffa et al. 2009). Ver localization was also analyzed in formaldehyde/acetic acid-fixed preparations that allow a clear visualization of individual polytene chromosome arms. Immunostaining of these preparations with both anti-GFP and anti-HOAP antibodies revealed that the signals elicited by these antibodies are exclusively telomeric and fully coincident (Fig. 2B). We thus conclude that Ver specifically accumulates at all Drosophila telomeres.
Figure 2.
Ver specifically localizes at polytene chromosome telomeres. (A) Localization of Ver-GFP, HOAP-GFP, and GFP-Moi in live, unsquashed salivary gland nuclei from third instar larvae. Note that these nuclei display six discrete fluorescent signals that are likely to correspond to the euchromatic telomeres (XL, 2L, 2R, 2L, 3R, and 4R). (B) Polytene chromosomes from Ver-GFP-expressing larvae immunostained with anti-GFP (green) and anti-HOAP (red) antibodies. Note the precise colocalization at telomeres of the Ver and HOAP signals.
We also generated flies carrying a ver1-GFP transgene, which was regularly transcribed (Supplemental Fig. S1B). However, the Ver1-GFP mutant protein was not detected by either immunoblotting or immunofluorescence (Supplemental Fig. S1C; data not shown), suggesting that the additional 12 amino acids appended to the C terminus of wild-type Ver render Ver1 unstable in vivo.
Ver interacts with HOAP and Moi
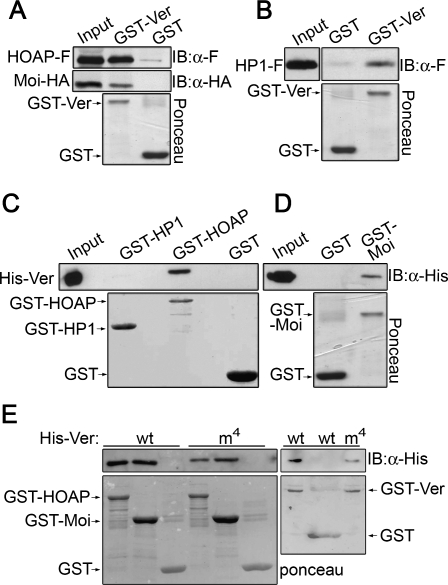
The finding that Ver precisely colocalizes with HOAP, which in turn colocalizes with both Moi and HP1 (Cenci et al. 2003; Raffa et al. 2009), prompted us to investigate whether these proteins interact physically. We thus performed a GST pull-down assay by incubating GST-Ver with extracts from S2 cells expressing both HOAP-Flag and Moi-HA. As shown in Figure 3A, both HOAP and Moi are precipitated by GST-Ver, but not by GST alone. We also found that GST-Ver pulls down HP1-Flag from S2 cell extracts (Fig. 3B). However, pull-down assays using purified bacterially expressed proteins showed that His-Ver is captured efficiently by both GST-HOAP and GST-Moi, but not by GST-HP1 or GST alone (Fig. 3C,D). We thus conclude that Ver directly binds both HOAP and Moi but not HP1. These results are consistent with previous studies showing direct interactions between HOAP and HP1, HOAP and Moi, and Moi and HP1(Shareef et al. 2001; Raffa et al. 2009).
Figure 3.
Ver directly binds HOAP and Moi. (A) GST-Ver precipitates both HOAP-Flag (HOAP-F) and Moi-HA from S2 cell extracts. (B) GST-Ver precipitates HP1-Flag (HP1-F) from S2 cell extracts. (C) GST-HOAP, but not GST-HP1, binds bacterially purified 6His-Ver. (D) GST-Moi binds bacterially purified 6His-Ver. (E) The 6His-Verm4 mutant protein binds GST-HOAP and GST-Moi; both 6His-Ver and 6His-Verm4 bind GST-Ver. Input is 10%. (IB) Immunoblotting.
Ver is an OB fold-containing protein
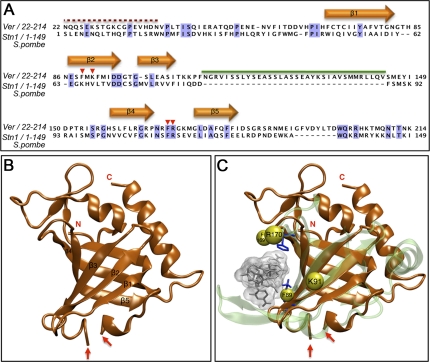
To search for homologs of the Ver protein, we used the procedure outlined in Supplemental Figure S2 and described in detail in the Supplemental Material. Briefly, we first searched the nonredundant (NR) protein sequence databases using CSI-BLAST (Biegert and Soding 2009). We identified 22 proteins sharing significant similarity with Ver, including 11 proteins from different Drosophila species (Clark et al. 2007), and five OBFC1-containing proteins, which share homology with the OB fold domain of Stn1 (Supplemental Fig. S3A). We next searched the Conserved Domain Database (CDD) at the NCBI using a hidden Markov model (HMM) generated on the multiple sequence alignment (MSA) of the 12 Drosophila Ver homologs (Supplemental Fig. S3B). The highest-scoring hit was the hOBFC1-like subfamily of OB fold domains (CDD ID, cd04483; E-value of 1.6 × 10−5; probability score of 97.50%) that belongs to the Rpa2 OB fold family. The HMM was also used to search the Protein Data Bank (PDB) (Supplemental Fig. S2A). The highest scoring hit was the OB fold domain of S. pombe Stn1 (Sun et al. 2009), with an E-value of 8.2 × 10−13 and a probability score of 99.42%, followed by the OB fold domain of human Rpa2. Additional searches using several fold recognition methods (FFAS03, mgenthreader, Phyre, FUGUE, and SAM-T08) in the GeneSilico metaserver yielded the OB fold domain of S. pombe Stn1 as the highest-scoring hit (Fig. 4A; Supplemental Fig. S2).
Figure 4.
Ver contains a Rap2/Stn1-related OB fold domain. (A) Sequence alignment of Ver (residues 22–214) with its structural template, the OB fold domain of S. pombe Stn1 (residues 1–149). Shaded columns indicate identical residues. Red arrowheads indicate residues predicted to be Ver-binding sites to ssDNA. Orange arrows correspond to secondary structure elements (β strands) that are significantly conserved among OB folds. The red dotted line indicates the Ver portion that tends to be intrinsically disordered; the green line corresponds to a protein region for which we failed to find a suitable template. (B) Ribbon representation of the Ver model. The two red arrows indicate the putative boundary of the helix-turn–helix (HTH) motif (residues ∼111–144) inserted between the third (β3) and fourth (β4) β strand. (C) Superimposition of the Ver model (in orange) on the hRPA70 OB fold (in green) bound to ssDNA. The Ver residues F89, K91, F169, and R170 are shown as yellow spheres. The hRPA70 residues K343 and F386, which mediate protein–DNA interactions, are shown as blue sticks.
Based on these results, we used the S. pombe Stn1 structure (Sun et al. 2009) as a template to build a model of the Ver core portion (residues ∼37–205) using the “FRankenstein's monster” approach (Fig. 4B; Supplemental Fig. S2C; Kosinski et al. 2003). We next compared the predicted Ver structure (Fig. 4B) with the protein structures in the PDB database using the DALI program (Holm et al. 2008). We found significant similarities with several OB fold-containing proteins, including human RPA70 (hRPA70), whose structure has been determined in complex with the cognate ssDNA (Bochkarev et al. 1997). Thus, we superimposed the Ver model to the hRPA70 structure to generate a model of the Ver OB fold domain bound to ssDNA (Fig. 4C). Despite the low overall sequence identity, the F89 and F169/R170 (F169 and R170 are next to each other) (Fig. 4C) residues of Ver superimpose reasonably well with the K343 and F386 residues of hRPA70 (Fig. 4C), which are responsible for protein/DNA interactions (Bochkarev et al. 1997). The Ver/DNA model also suggests that K91 might be involved in protein–DNA interactions because it protrudes from the face of the OB fold domain predicted to bind DNA (Fig. 4C).
The Ver OB fold domain is required for telomere protection
To ask whether the Ver OB fold domain plays a role in telomere capping, we generated a mutant version of Ver (Verm4) that carries four amino acid substitutions in the putative DNA-binding sites (F89A/K91E/F169A/R170E) (Fig. 4C). Verm4 interacted normally with HOAP and Moi in GST pull-down experiments. We also found that GST-Ver binds His-Ver and His-Verm4 (Fig. 3E), indicating that wild-type and mutant Ver proteins are all capable of self-interaction.
We next transformed flies with a construct bearing the verm4 sequence fused in-frame with the GFP sequence. Verm4-GFP accumulated regularly at polytene chromosome telomeres (Supplemental Fig. S4A). However, ver2/ver2 mutant brains carrying either one or two copies of the verm4 transgene displayed about five TAs per cell (n = 100) (Supplemental Fig. S4B). These findings suggest that recruitment of Ver to chromosome ends does not require an intact OB fold, but is probably mediated by interactions with HOAP (see below). However, the Ver OB fold appears to be required for telomere capping, as the Verm4 mutant protein does not protect telomeres from fusion events.
Recruitment dependencies of the Ver protein
To define the role of Ver in telomere protection, we first asked whether Ver is required for proper localization of Moi, HP1, and HOAP. Of the latter proteins, only HOAP is clearly detectable at both mitotic and polytene chromosome telomeres; Moi and HP1 can be easily detected at polytene chromosome ends, but not at mitotic telomeres (Fanti et al. 1998; Raffa et al. 2009). Thus, we analyzed HP1 and Moi localization only in polytene chromosomes, and HOAP localization in both mitotic and polytene chromosomes.
Mutations in ver did not substantially affect HOAP localization at mitotic telomeres (Supplemental Fig. S5A), and ver2 mutants showed normal concentrations of HOAP and HP1 at polytene chromosome telomeres (Supplemental Fig. S5B). Thus, the telomere fusion phenotype elicited by ver mutations is not due to the absence of HOAP and HP1.
We next analyzed polytene chromosomes of ver mutants expressing a GFP-moi transgene. In a wild-type background, the product of this transgene accumulates at polytene chromosome ends (Raffa et al. 2009). However, in a ver mutant background, GFP-Moi does not localize at telomeres, suggesting that Moi localization at telomeres requires the ver function (Supplemental Fig. S6).
We also analyzed Ver-GFP localization on polytene chromosomes of cav and moi mutants. Although Ver-GFP was expressed regularly in both of these mutants, it failed to accumulate at the polytene telomeres (Supplemental Figs. S1C, S6). These results indicate that proper Ver localization at telomeres requires HOAP, and that Ver and Moi are mutually dependent for their telomeric localization. These findings are consistent with previous results indicating that HOAP mediates Moi recruitment at telomeres (Raffa et al. 2009).
Ver, Moi, and HOAP are fast-evolving proteins
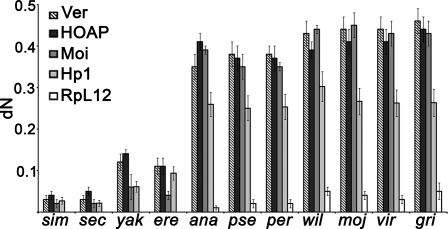
Because HOAP, Moi, and Ver form a telomere-specific capping complex, and because previous studies have shown that HOAP is a rapidly evolving protein (Schmid and Tautz 1997), we asked whether Moi and Ver share this property with HOAP. We calculated the number of nonsynonymous substitutions per nonsynonymous site (dN values) for pairwise comparisons between Drosophila melanogaster genes and orthologous genes of 11 recently sequenced Drosophila species (Clark et al. 2007). This analysis was performed also for cav, Su(var)205 (HP1), and RpL12 (ribosomal protein); cav and RpL12 were used as positive and negative controls, respectively. We estimated the dN values using four different methods: ZRN98 LWL85, PBL93, and KUMAR. These methods gave comparable results; the dN values for cav, moi, and ver increase dramatically with the increase of the evolutionary distance between D. melanogaster and the species used for pairwise comparison. In contrast, the dN values for Su(var)205 exhibit only moderate increases, while those for RpL12 show little or no variation (Fig. 5; Supplemental Fig. S7). Thus, we conclude that HOAP, Moi, and Ver have evolved at faster rates than either RpL12 or HP1.
Figure 5.
The terminin components are encoded by fast-evolving genes. The mean numbers of nonsynonymous substitutions per nonsynonymous site (dN values, represented as bars ±SD) for ver, cav, and moi increase dramatically with the increase of the evolutionary distance between D. melanogaster and each of the 11 Drosophila species used for comparison. The increases in dN values for Su(var)205 are significantly lower than those observed for ver, cav, and moi. dN values have been calculated using the ZRN98 method (Zhang et al. 1998). (sim) Drosophila simulans; (sec) Drosophila sechellia; (yak) Drosophila yakuba; (ere) Drosophila erecta; (ana) Drosophila ananassae; (pse) Drosophila pseudoobscura; (per) Drosophila persimilis; (wil) Drosophila willistoni; (moj) Drosophila mojavensis; (vir) Drosophila virilis; (gri) Drosophila grimshawi.
Ver is a terminin component
We suggested previously that fly telomeres are capped by the HOAP–Moi complex, which we called terminin, and which has the same properties of shelterin: a specific telomeric localization throughout the cell cycle, and a telomere-limited function (Raffa et al. 2009). Here, we showed that ver mutants exhibit a very high frequency of telomeric fusions (about five per cell), comparable with those observed previously in cav (HOAP) and moi mutants (Cenci et al. 2003; Musarò et al. 2008; Raffa et al. 2009). Consistent with these findings, Ver is enriched exclusively at telomeres like HOAP and Moi, and colocalizes precisely and interacts physically with both these proteins. In addition, our current analyses indicate that Ver functions only at telomeres. These findings strongly suggest that Ver is a component of the terminin complex.
Our results indicate that Ver contains an OB fold domain that shares structural similarity with the Rpa2/Stn1 OB fold. Interestingly, the Drosophila genome does not appear to contain homologs of the shelterin subunits and the other CST subunits. However, all of the nonshelterin and non-CST components of human telomeres are conserved in flies. Conversely, with the exception of HOAP and Moi, all of the Drosophila telomere-related proteins identified so far have clear human counterparts (Cenci et al. 2005; Raffa et al. 2009). Thus, we hypothesize that, concomitant with telomerase loss, Drosophila lost the shelterin and the CST homologs that bind DNA in a sequence-specific fashion, and evolved terminin to bind chromosome ends independently of the DNA sequence.
Our hypothesis on terminin evolution generates several expectations. It is logical to assume that telomerase loss resulted in a divergence of terminal DNA sequences, accompanied by a strong selective pressure toward the evolution of sequence-independent telomere-binding factors. It is also conceivable that the evolutionary pressure on these factors was higher than that exerted on telomere proteins not specifically involved in capping. Therefore, one would predict that proteins involved directly and exclusively in telomere capping evolved more rapidly than the other telomere-associated proteins. This prediction is verified by the finding that HOAP, Moi, and Ver are fast-evolving proteins, while the other Drosophila telomere proteins, including HP1, are not (Fig. 5; Supplemental Fig. S7; Supplemental Table S1).
Although the frequencies of telomeric fusions elicited by loss of each terminin component are fully comparable, Ver, Moi, and HOAP do not play identical roles at Drosophila telomeres. HOAP localizes at telomeres independently of Ver and Moi, which are both HOAP-dependent and mutually dependent for telomeric localization. In addition, while loss of HOAP triggers both the DNA damage and the spindle assembly (SAC) response (Musarò et al. 2008), depletion of either Ver (see the Supplemental Material) or Moi (Raffa et al. 2009) does not appear to elicit these checkpoint responses. These results suggest that HOAP is crucial for masking chromosome ends to avoid their recognition as double-strand breaks. Ver and Moi are not required for terminal DNA protection so as to prevent checkpoint responses. However, Ver and Moi are essential to hide chromosome ends from the DNA repair machineries that mediate telomere fusion. A Ver protein with mutations in the OB fold domain is still recruited at telomeres, but is unable to prevent telomere fusion. This suggests that the integrity of the Ver OB fold domain is crucial to prevent inappropriate repair of terminal DNA, and implies that Drosophila telomeres terminate with a single-strand overhang like their yeast, plant, and mammalian counterparts.
Materials and methods
Drosophila strains
The ver1 allele was isolated from a collection of 1680 EMS-induced third chromosome late lethals, generated in Charles Zuker's laboratory. The ver1 allele was characterized by DNA sequence analysis. The l(3)S147910 (ver2) and Df(3L)sex204 are described in FlyBase (http://flybase.org). The moi1 and cav1 mutations have been described previously (Cenci et al. 2003; Raffa et al. 2009).
Chromosome cytology and immunostaining
Preparation and immunostaining of mitotic and polytene chromosomes have been described previously (Cenci et al. 2003; Raffa et al. 2005). In vivo detection and immunostaining of GFP-tagged proteins on polytene chromosomes were carried out according to Raffa et al. (2009). See the Supplemental Material for details.
GFP constructs
To generate the construct for ver-GFP expression, the EGFP CDS fused in-frame with the 3′ end of the ver CDS was cloned into the pJZ4 vector (a derivative of pCASPER4) under the control of a tubulin promoter, as described previously for the GFP-moi construct (Raffa et al. 2009). The UAS-cav-GFP construct was generated using the Gateway strategy (Invitrogen) using a pPWG as destination vector (Drosophila Genomics Resource Center [DGCR], Indiana University). Germline transformation was accomplished by the BestGene Company, using standard methods. actGAL4 was used as a driver for UAS-cav-GFP.
GST pull-down assays
To obtain a GST-Ver fusion protein, full-length ver cDNA was cloned in pGEX-6P vector as described previously for moi, cav (HOAP), and Su(var)205 (HP1) (Raffa et al. 2009). Bacterially expressed GST fusion proteins were purified by incubating crude lysates with glutathione sepharose 4B (Amersham), as recommended by the manufacturer. To obtain HOAP-Flag and Moi-HA-expressing cultures, S2 cells were transfected using Cellfectin (Invitrogen) with pAWF-cav (DGRC, Indiana University) and pJZ4-moi-HA constructs, and were harvested 72 h after transfection. Extracts were prepared by standard methods. 6His proteins were generated and purified from bacteria as described in the Supplemental Material. GST pull-down experiments were performed as described previously (Raffa et al. 2009). HOAP-Flag, Moi-HA, and 6His-Ver were detected with anti-Flag HRP-conjugated (1:1000; Roche), mouse anti-HA 12CA5 (1:1000; Roche), and anti-His HRP-conjugated (1:500; Roche) antibodies, respectively.
Bioinformatic analyses
Detailed methods used for modeling the Ver protein and for calculation of the dN values are given in the Supplemental Material.
Site-directed mutagenesis
The wild-type ver CDS was cloned into the pET200 vector (Invitrogen). Point mutations were introduced using the Quick Change Site-Directed Rapid Mutation kit (Stratagene).
Acknowledgments
We are grateful to R. Kellum and S. Elgin for the anti-HOAP and anti-HP1 antibodies, G. Cestra for the anti-GFP antibody, C. Carrè for fly stocks, T. Ingegnere for technical assistance, M. Savino for helpful discussion, and A. Tramontano for invaluable advice on bioinformatic analysis. G.D.R. was supported by a fellowship from the Fondazione Italiana per la Ricerca sul Cancro (FIRC), and G.C. was supported by an EMBO fellowship (AST 136.00-2007). This work was supported in part by grants from AIRC and Italian Telethon to M.G.
Footnotes
Article is online at http://www.genesdev.org/cgi/doi/10.1101/gad.574810.
Supplemental material is available at http://www.genesdev.org.
References
- Biegert A, Soding J 2009. Sequence context-specific profiles for homology searching. Proc Natl Acad Sci 106: 3770–3775 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bochkarev A, Pfuetzner RA, Edwards AM, Frappier L 1997. Structure of the single-stranded-DNA-binding domain of replication protein A bound to DNA. Nature 385: 176–181 [DOI] [PubMed] [Google Scholar]
- Carré C, Szymczak D, Pidoux J, Antoniewski C 2005. The histone H3 acetylase dGcn5 is a key player in Drosophila melanogaster metamorphosis. Mol Cell Biol 25: 8228–8238 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cenci G, Siriaco G, Raffa GD, Kellum R, Gatti M 2003. The Drosophila HOAP protein is required for telomere capping. Nat Cell Biol 5: 82–84 [DOI] [PubMed] [Google Scholar]
- Cenci G, Ciapponi L, Gatti M 2005. The mechanism of telomere protection: A comparison between Drosophila and humans. Chromosoma 114: 135–145 [DOI] [PubMed] [Google Scholar]
- Ciapponi L, Cenci G 2008. Telomere capping and cellular checkpoints: Clues from fruit flies. Cytogenet Genome Res 122: 365–373 [DOI] [PubMed] [Google Scholar]
- Clark AG, Eisen MB, Smith DR, Bergman CM, Oliver B, Markow TA, Kaufman TC, Kellis M, Gelbart W, Iyer VN, et al. 2007. Evolution of genes and genomes on the Drosophila phylogeny. Nature 450: 203–218 [DOI] [PubMed] [Google Scholar]
- Fanti L, Giovinazzo G, Berloco M, Pimpinelli S 1998. The heterochromatin protein 1 prevents telomere fusions in Drosophila. Mol Cell 2: 527–538 [DOI] [PubMed] [Google Scholar]
- Gao H, Cervantes RB, Mandell EK, Otero JH, Lundblad V 2007. RPA-like proteins mediate yeast telomere function. Nat Struct Mol Biol 14: 208–214 [DOI] [PubMed] [Google Scholar]
- Gelinas AD, Paschini M, Reyes FE, Heroux A, Batey RT, Lundblad V, Wuttke DS 2009. Telomere capping proteins are structurally related to RPA with an additional telomere-specific domain. Proc Natl Acad Sci 106: 19298–19303 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holm L, Kaariainen S, Rosenstrom P, Schenkel A 2008. Searching protein structure databases with DaliLite v.3. Bioinformatics 24: 2780–2781 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kosinski J, Cymerman IA, Feder M, Kurowski MA, Sasin JM, Bujnicki JM 2003. A ‘FRankenstein's monster’ approach to comparative modeling: Merging the finest fragments of fold-recognition models and iterative model refinement aided by 3D structure evaluation. Proteins 53: 369–379 [DOI] [PubMed] [Google Scholar]
- Linger BR, Price CM 2009. Conservation of telomere protein complexes: Shuffling through evolution. Crit Rev Biochem Mol Biol 44: 434–446 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lue NF 2009. Plasticity of telomere maintenance mechanisms in yeast. Trends Biochem Sci 35: 8–17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin V, Du LL, Rozenzhak S, Russell P 2007. Protection of telomeres by a conserved Stn1–Ten1 complex. Proc Natl Acad Sci 104: 14038–14043 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mason JM, Frydrychova RC, Biessmann H 2008. Drosophila telomeres: An exception providing new insights. Bioessays 30: 25–37 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitton-Fry RM, Anderson EM, Hughes TR, Lundblad V, Wuttke DS 2002. Conserved structure for single-stranded telomeric DNA recognition. Science 296: 145–147 [DOI] [PubMed] [Google Scholar]
- Miyake Y, Nakamura M, Nabetani A, Shimamura S, Tamura M, Yonehara S, Saito M, Ishikawa F 2009. RPA-like mammalian Ctc1–Stn1–Ten1 complex binds to single-stranded DNA and protects telomeres independently of the Pot1 pathway. Mol Cell 36: 193–206 [DOI] [PubMed] [Google Scholar]
- Musarò M, Ciapponi L, Fasulo B, Gatti M, Cenci G 2008. Unprotected Drosophila melanogaster telomeres activate the spindle assembly checkpoint. Nat Genet 40: 362–366 [DOI] [PubMed] [Google Scholar]
- Palm W, de Lange T 2008. How shelterin protects mammalian telomeres. Annu Rev Genet 42: 301–334 [DOI] [PubMed] [Google Scholar]
- Raffa GD, Cenci G, Siriaco G, Goldberg ML, Gatti M 2005. The putative Drosophila transcription factor woc is required to prevent telomeric fusions. Mol Cell 20: 821–831 [DOI] [PubMed] [Google Scholar]
- Raffa GD, Siriaco G, Cugusi S, Ciapponi L, Cenci G, Wojcik E, Gatti M 2009. The Drosophila modigliani (moi) gene encodes a HOAP-interacting protein required for telomere protection. Proc Natl Acad Sci 106: 2271–2276 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rong YS 2008. Telomere capping in Drosophila: Dealing with chromosome ends that most resemble DNA breaks. Chromosoma 117: 235–242 [DOI] [PubMed] [Google Scholar]
- Schmid KJ, Tautz D 1997. A screen for fast evolving genes from Drosophila. Proc Natl Acad Sci 94: 9746–9750 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shareef MM, King C, Damaj M, Badagu R, Huang DW, Kellum R 2001. Drosophila heterochromatin protein 1 (HP1)/origin recognition complex (ORC) protein is associated with HP1 and ORC and functions in heterochromatin-induced silencing. Mol Biol Cell 12: 1671–1685 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song X, Leehy K, Warrington RT, Lamb JC, Surovtseva YV, Shippen DE 2008. STN1 protects chromosome ends in Arabidopsis thaliana. Proc Natl Acad Sci 105: 19815–19820 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun J, Yu EY, Yang Y, Confer LA, Sun SH, Wan K, Lue NF, Lei M 2009. Stn1–Ten1 is an Rpa2–Rpa3-like complex at telomeres. Genes Dev 23: 2900–2914 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Surovtseva YV, Churikov D, Boltz KA, Song X, Lamb JC, Warrington R, Leehy K, Heacock M, Price CM, Shippen DE 2009. Conserved telomere maintenance component 1 interacts with STN1 and maintains chromosome ends in higher eukaryotes. Mol Cell 36: 207–218 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J, Rosenberg HF, Nei M 1998. Positive Darwinian selection after gene duplication in primate ribonuclease genes. Proc Natl Acad Sci 95: 3708–3713 [DOI] [PMC free article] [PubMed] [Google Scholar]