ABSTRACT
It is not understood why only some infections with Entamoeba histolytica result in disease. The calcium-regulated transcription factor upstream regulatory element 3-binding protein (URE3-BP) was initially identified by virtue of its role in regulating the expression of two amebic virulence genes, the Gal/GalNac lectin and ferredoxin. Here we tested whether this transcription factor has a broader role in regulating virulence. A comparison of in vivo to in vitro parasite gene expression demonstrated that 39% of in vivo regulated transcripts contained the URE3 motif recognized by URE3-BP, compared to 23% of all promoters (P < 0.0001). Amebae induced to express a dominant positive mutant form of URE3-BP had an increase in an elongated morphology (30% ± 6% versus 14% ± 5%; P = 0.001), a 2-fold competitive advantage at invading the intestinal epithelium (P = 0.017), and a 3-fold increase in liver abscess size (0.1 ± 0.1 g versus 0.036 ± 0.1 g; P = 0.03). These results support a role for URE3-BP in virulence regulation.
IMPORTANCE
Amebic dysentery and liver abscess are caused by Entamoeba histolytica. Amebae colonize the colon and cause disease by invading the intestinal epithelium. However, only one in five E. histolytica infections leads to disease. The factors that govern the transition from colonization to invasion are not understood. The transcription factor upstream regulatory element 3-binding protein (URE3-BP) is a calcium-responding regulator of the E. histolytica Gal/GalNAc lectin and ferredoxin genes, both implicated in virulence. Here we discovered that inducible expression of URE3-BP changed trophozoite morphology and promoted parasite invasion in the colon and liver. These results indicate that one determinant of virulence is transcriptional regulation by URE3-BP.
INTRODUCTION
Entamoeba histolytica, the causative agent of amebiasis, is prevalent in areas of poor sanitation (1). For example, a prospective study of children in an urban slum in Dhaka, Bangladesh, detected E. histolytica at least once in 80% of the children (1). Disease only occurs in ~20% of infections, but an asymptomatic infection can develop into invasive disease (2, 3). Symptomatic infection can manifest itself as amebic dysentery, where trophozoites invade the intestinal wall, or as a liver abscess, which occurs, on average, 3 months after travel to an area of endemicity (2). The variable outcome of infection and the latency period between colonization and disease suggest that adaption of the parasite to the host via altered gene expression could contribute to the virulent phenotype (2).
The DNA transcription element upstream regulatory element 3 (URE3) was originally identified as a positive regulatory sequence by linker-scanner mutagenesis of the promoter of the E. histolytica Gal/GalNAc lectin gene hgl5 (4). The URE motif was also found in the ferredoxin 1 gene promoter; however, mutation of the URE3 motif within the ferredoxin 1 gene promoter produced a 2-fold drop in gene expression, indicating that URE3 could be either a positive or a negative regulator of gene expression (5). The URE3 consensus motif was defined using an electrophoretic mobility shift assay to measure the binding affinity of a mutated URE3 motif for proteins isolated from the E. histolytica nucleus. The consensus motif was, in turn, validated by the use of gene reporter assays (6).
The transcription factor specific for the URE3 motif, URE3-BP, was identified in a yeast one-hybrid screen using URE3 as bait (7). URE3-BP contains two calcium-binding motifs (EF hands), and in the presence of calcium, URE3-BP dissociates from DNA containing the URE3 motif (8). Increased levels of calcium inhibited URE3-BP binding to DNA and promoted its interaction with the E. histolytica C2A (EhC2A) phospholipid binding protein and the sequestration of URE3-BP to the amebic plasma membrane (9). Mutation in one of the URE3-BP EF hand motifs resulted in a dominant positive mutant protein that remained bound to the URE3 motif in the presence of an intracellular [Ca2+] signal. To empirically define additional genes regulated by URE3-BP, this URE3-BP EF hand mutant was expressed in E. histolytica trophozoites using an inducible promoter system. This altered the transcript level of 50 genes and trophozoite motility (6, 10).
To investigate the impact of URE3-BP regulation on virulence, the occurrence of a URE3 consensus motif in the promoters of genes specifically induced during in vivo infection was measured by microarrays. In addition, the effect of inducible expression of a dominant positive mutant form of URE3-BP on parasite morphology and virulence was tested.
RESULTS
Enrichment of the URE3 cis-acting sequence in promoters expressed in vivo.
A genome-wide transcriptional analysis of E. histolytica was previously performed on trophozoites isolated from the ceca of three mice 1 day after infection and from an in vitro culture (11). These data were reanalyzed using updated annotation of the E. histolytica genome. The promoters of transcripts modulated in amebae from the cecum 1 day after infection were analyzed for the presence or absence of the URE3 consensus motif (6) as described in Materials and Methods. Consistent with URE3-BP being a regulator of virulence, the URE3 motif was observed in the promoters of 39% of in vivo modulated genes. This was a statistically significant increase over the incidence of the URE motif (23%) in all E. histolytica promoters (Table 1) (two-sided P value, <0.0001). For details of transcripts changed over 4-fold, see Table S1 in the supplemental material. Promoters of the URE3-associated transcripts modulated both in vivo and by overexpression of dominant positive URE3-BP contained not only the URE3 consensus (T[ATG]T[TC][CG]T[AT][TGC][TG]) but also an additional novel motif [CA]GATG[TA]T[TC][AG][AG] (see Table S1 in the supplemental material).
TABLE 1 .
Enrichment of the URE3 consensus motif in transcripts changed 2-fold in vivoa
| Parameter | No. of promoters containing a URE3 matrix 375 to 25 bases 5′ of ATG start | No. of promoters without a URE3 matrix | Total no. (%) of promoters analyzed | P value |
|---|---|---|---|---|
| No. of promoters of transcripts modulated in luminal amebae | 54 | 85 | 139 (39) | <0.0001 |
| No. of promoters of unchanged E. histolytica transcripts | 1,931 | 6,452 | 8,383 (23) | |
| Total no. (%) of promoters analyzed | 1,985 (2.7) | 6,537 (1.3) | 8,522 (100) |
A contingency table χ2 test was used to compare the occurrences of the URE3 motif in transcripts significantly modulated by at least 2-fold at day 1 in vivo. These were identified by the use of the Linear Model for Microarray Data, and the false-discovery rate was corrected by the use of the Benjamini and Hochberg equation as described in Materials and Methods. Promoters were defined as the region from −375 bp to −25 bp 5′ of the initiating ATG of the structural gene. Background motif levels in E. histolytica promoters were determined as previously described (6).
Inducible expression of constitutively active URE3-BP altered trophozoite morphology.
Microarray analysis of amebae expressing the dominant positive mutant form of URE3-BP identified 50 modulated transcripts. Fifteen of these transcripts encoded potential membrane proteins, which suggested that URE3-BP had a role in modulating the surface composition of the parasite (6). Host-parasite interactions are key during invasive disease and involve such virulence activities as contact-dependent cytolysis, phagocytosis of cells, and motility (12, 13).
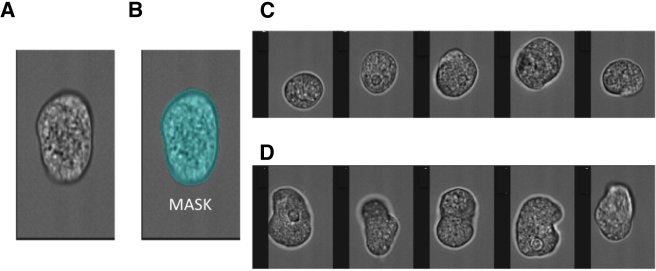
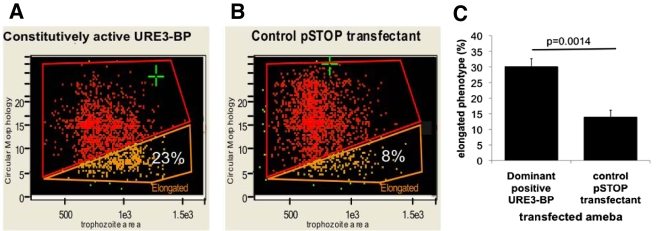
One measure of alterations in surface composition is changes in cell morphology. The morphological impact of URE3-BP was measured in trophozoites expressing dominant positive URE3-BP. The Amnis Imagestream cytometer was used to compare trophozoites expressing dominant positive URE3-BP [pEF(2)URE3-BP] with those of the equivalent control strain (induced pSTOP) (Fig. 1 and 2.) A two-dimensional bright-field image of the three-dimensional ameba was analyzed (Fig. 1A). To minimize the effects of the orientation of the amebae in flow, extended-depth-of-field imaging was used. The IDEAS program was used to identify single in-focus trophozoites and to derive the aspect ratio or circularity of the image (Fig. 1B to D). To achieve the latter goal, the IDEAS software repeatedly measured cell radius; a high “circularity score” resulted from an internally consistent measurement. The automated process allowed detailed information on cell morphology to be gathered at the population level. To define the amebic shape, two different functions were used: trophozoite area and circular morphology (Fig. 2A and B). An arbitrary gate was drawn to capture the most elongated amebae. Transfectants that expressed dominant positive URE3-BP had a statistically significant 2-fold increase in the number of trophozoites with an elongated phenotype [30% ± 6% elongated in pEF(2)URE3-BP versus 14% ± 5% in pSTOP; n = 5, P = 0.0014] (Fig. 2C). The elongated phenotype could reflect surface changes related to motility and chemotaxis. Induction of URE3-BP had previously been shown to increase transwell migration, suggesting a possible role in the regulation of cellular motility (6).
FIG 1 .
Measurement of amebic morphology. The Amnis Imagestream imaging cytometer was used to measure the morphology of fixed amebic trophozoites. (A) Bright-field image of an elongated trophozoite. (B) The pixels which constituted the bright-field image of the trophozoites (MASK). From the mask, the computational features of area and circular morphology were derived. A high circularity score resulted from an internally consistent measurement of cell radius. Representative images show trophozoites characterized and “tagged” as either circular (C) or elongated (D).
FIG 2 .
Amebae expressing constitutively active URE3-BP exhibited an elongated trophozoite morphology. A representative graph is shown with the output of the circular morphology feature (y axis) and trophozoite area (x axis). (A) Trophozoites expressing dominant positive URE3-BP. (B) Induced control pSTOP transfectants. There was a statistically significant increase in the number of elongated amebae detected in trophozoites expressing dominant positive URE3-BP (χ2 P value, <0.0001). (C) Graph of average data derived from experiments on 2 separate days. The y axis indicates the percentage of elongated amebae, and the x axis shows the plasmid carried (n = 5; P = 0.0014; standard errors are shown).
Expression of dominant positive URE3-BP conferred a competitive advantage at the host tissue interface in the murine model of amebic colitis.
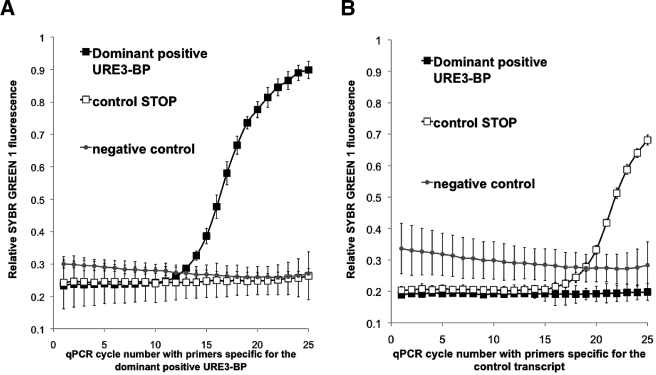
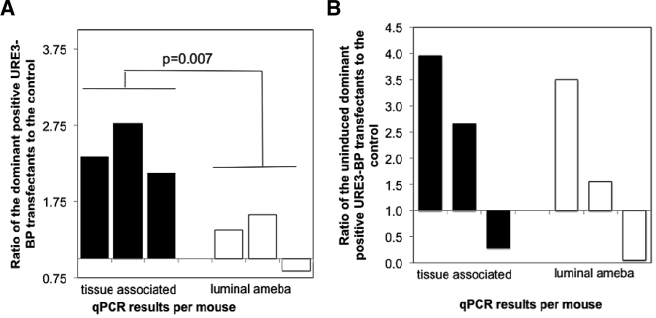
To test whether the expression of dominant positive URE3-BP conferred a competitive advantage in establishing an invasive infection, CBA/J mice were infected intracecally with a 1:1 ratio of amebae induced to express dominant positive URE3-BP to the control pSTOP strain. We first established that we were able to distinguish dominant positive URE3-BP transcripts from the induced control pSTOP plasmid in mRNA isolated from the initial mixed inocula (Fig. 3). Lower levels of control mRNA transcribed from pSTOP were consistently observed in vitro, as shown in Fig. 3B, presumably due to the reduced stability of the untranslated transcripts, and verified in vivo the inducible expression of the URE3-BP transcript 7 days after infection (data not shown). We compared the ratios of amebic transfectants isolated from the intestinal lumen and the epithelium. Amebae were considered luminal if they were isolated from the cecal contents and tissue associated if they were isolated from the mucosa removed from the luminal surface of the cecum by scraping. Mice were sacrificed 3 weeks after infection. Primer specificity allowed quantitative PCR (qPCR) to be used as a surrogate marker for the proportion of the amebic population carrying the dominant URE3-BP expression construct. The ratio of infecting amebae determined by qPCR was used to correct the qPCR ratio of amebae isolated from three different infected mice (Fig. 3). Comparisons of uninduced amebae showed no statistically significant difference. In the induced amebae, the ratio of dominant positive URE3-BP to the induced pSTOP transfectant control doubled in the tissue-associated amebae but not in the amebae isolated from the cecal lumen (2.4 ± 0.3 versus 1.3 ± 0.4; unpaired t test P = 0.017, paired t test P = 0.0071) (Fig. 4). Amebae expressing the dominant positive URE3-BP protein were therefore more effective at associating with the host epithelium, indicating a greater potential for establishing an invasive infection within the gut.
FIG 3 .
qRT-PCR measurement of in vitro expression levels and verification of primer specificity. qRT-PCR was conducted on the URE3-BP mRNAs. The y axis indicates double-stranded DNA-dependent SYBR green 1 fluorescence at 530 nm. The x axis represents the PCR cycle number. (A) Primers specific for the dominant positive URE3-BP expression plasmid. (B) Primers specific for the control transcript of the induced pSTOP transfectant. ■, RNA isolated from dominant positive URE3-BP; ☐, induced pSTOP transfectant control; ●, background. Lower control mRNA levels were consistently observed in vitro, as shown in panel B, presumably due to the reduced stability of the untranslated transcript.
FIG 4 .
Competitive advantage in intestinal invasion conferred by the expression of dominant positive URE3-BP in the murine model of amebic colitis. CBA mice were infected intracecally with a 1:1 ratio of amebae induced to express dominant positive URE3-BP to the control induced pSTOP strain (A) or with uninduced amebae (B). Mice were sacrificed 3 weeks after infection, and the ratios of dominant positive URE3-BP to the control pSTOP strain were determined by qPCR for both the tissue-associated and luminal amebae (n = 3). Values were corrected by the input data, and statistical significance was determined using a paired t test (P = 0.007).
Induction of URE3-BP-dependent gene expression increased liver abscess size in the gerbil liver abscess model of amebiasis.
To examine the effects of overexpression of URE3-BP on the ability to form amebic liver abscesses, experimental or control pSTOP transfectants induced as described in Materials and Methods were alternately injected into either the right or the left anterior lobe of a gerbil liver. To verify the experimental design, the identity of the trophozoite strain initiating an abscess was confirmed by isolation of amebae from the affected site and subsequently by qPCR. Seven days postinfection, the gerbils were sacrificed and liver abscess weights were measured.
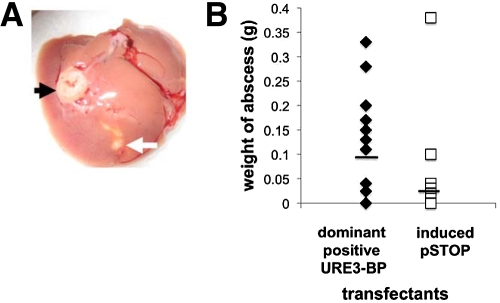
Infection with URE3-BP-expressing amebae produced liver abscesses in 69% of the gerbils versus 31% with the induced pSTOP transfectant control strain, which was not a statistically significant difference (P = 0.07) (Fig. 5A). The average abscess produced by infection with the URE3-BP transfectants was three times larger than that produced by infection with the induced pSTOP transfectant controls (0.1 ± 0.1 g versus 0.036 ± 0.1 g; n = 16). Because the distribution of liver abscess size was not Gaussian, statistical significance was determined by the Wilcoxon matched-pair signed-rank test (P = 0.0342) and the unpaired Mann-Whitney test (P = 0.02) (Fig. 5B). The capacity of URE3-BP-activated amebae to promote an increase in amebic liver abscess size supported a role for URE3-BP in regulating the virulence potential of the parasite.
FIG 5 .
Increase in liver abscess size following infection with amebae expressing dominant positive URE3-BP. (A) Infected liver abscess from amebae expressing the dominant positive URE3-BP protein injected into the right lobe of the liver (black arrow) and abscess from the control induced pSTOP transfectants injected in the left liver lobe (white arrow). (B) Effect of induction of dominant positive URE3-BP on abscess size (n = 16; statistical significance [P = 0.0342] was determined using the Wilcoxon matched-pair signed-rank test). The use of the unpaired Mann-Whitney test also gave statistical significance (P = 0.019).
DISCUSSION
The important conclusion of this work is that URE3-BP has a role in the regulation of virulence. Amebae expressing the dominant positive mutant form of URE3-BP were more virulent in two different animal models of amebiasis. An increase in liver abscess size resulted when trophozoites expressing dominant positive URE3-BP were injected into the gerbil liver (Fig. 5). In the mouse model of amebic colitis, a competitive advantage at the host tissue interface was conferred upon amebae expressing the dominant positive URE3-BP protein (Fig. 4). URE3-BP therefore promoted the expression of the E. histolytica virulence phenotype.
Distinct patterns of E. histolytica gene expression have been observed under a variety of experimental conditions (14–17). A comparison of the transcripts expressed during in vitro culture trophozoite mRNA levels one day after infection in the mouse model of amebiasis identified changes in parasite gene expression (11). Consistent with a role for URE3-BP in virulence, a statistically significant enrichment of the URE3 motif was observed in the promoters of in vivo (Table 1) (17).
URE3-BP activity is controlled not only by direct calcium binding but also by membrane sequestration due to the formation of a calcium-dependent complex with the phospholipid binding protein EhC2A (9). It is interesting to speculate whether a signaling defect is present in the avirulent E. histolytica “A” strain, where EhC2A was overexpressed 1,000-fold (18). A rise in either EhC2A levels or intracellular Ca2+ should decrease motility, as the dominant positive URE3-BP protein increased trophozoite chemotaxis (6, 19). Our data support the hypothesis that URE3-BP and EhC2A are key participants in a calcium-signaling pathway that regulates virulence.
In the switch between asymptomatic colonization and invasive disease, the regulation of trophozoite motility may be crucial. During invasive amebiasis, amebae are in close contact with both the fibronectin-rich basal membrane and the epithelial cells of the gastrointestinal tract. Changes at the level of transcription or translation often occur in response to environmental cues, such as contact with epithelial cells. Only motile amebae are able to cause disease in the liver abscess animal model (20). In the ex vivo human model of Bansal et al., motile E. histolytica invaded through the gut and penetrated the lamina propria (12).
In response to chemoattractants, neutrophils adopt a highly polarized morphology, the inhibition of which impairs motility and hence their capacity to invade injured host tissue (21). Stimulation with chemoattractants causes polymerization and reorganization of actin; this has been correlated with the extension of new pseudopods during chemotaxis. Formation of a leading front and acquisition of polarity in Dictyostelium are coordinated by secondary messengers, such as Ca2+ (22). Ameboid chemotaxis obviously requires polarization; however, even in the absence of chemoattractants, neutrophils remain highly polarized and it is hypothesized that the preexisting polarity potentiates the neutrophil chemotactic response (22). The URE3-BP-mediated change in the equilibrium of elongated versus round trophozoite shapes may reflect an increase in the number of trophozoites adopting a similar polarized morphology and be correlated with the proportion of amebae actively motile and extending lamellipodia.
The refashioning of cell shape in response to an upstream signal in cells typically involves many proteins with functions in actin binding, adhesion (such as integrins), and cell signaling (23). Several potential glycosylphosphatidylinositol-anchored cell surface proteins are repressed by dominant positive URE3-BP. It is possible that they could act in a manner similar to that of the glycosylphosphatidylinositol-anchored cell surface glycoprotein CEA. This expression of this protein leads to an increase in cell adhesion mediated via the α5β1 integrin interaction with fibronectin (24). Therefore, changes in protein expression initiated by the URE3-BP transcription factor could lead to remodeling of the cell surface, repression of cell adhesion, and hence possibly an increase in motility (Fig. 2) (14).
Further studies are required to understand the particular contribution of URE3-BP-regulated proteins to this complex process and their contribution to trophozoite virulence in different host milieux. From this work, however, it is clear that URE3-BP control plays an important role in these processes.
MATERIALS AND METHODS
Strain and culture.
E. histolytica strain HM1:IMSS trophozoites were grown at 37°C in TYI-S-33 medium containing penicillin (100 U/ml) and streptomycin (100 µg/ml) (Gibco/BRL) (25).
Stable transfection of E. histolytica trophozoites.
Stable transfection of E. histolytica trophozoites was achieved by the previously described lipofection technique (26, 27). Briefly, amebae (2.2 × 105/ml) were washed in medium 199 (Invitrogen, CA) supplemented with 5.7 mM cysteine, 1 mM ascorbic acid, 25 mM HEPES (pH 6.8), 10 µg of DNA, and 30 µl of Superfect (Qiagen). Treated amebae were incubated for 3 h at 37°C, followed by overnight culture in TYI-S-33 medium at 37°C.
The two-plasmid inducible system and the vectors used in this study are described in references 6 and 10. Briefly, DNA encoding a myc-tagged dominant positive mutant form of URE3-BP was ligated into a tetracycline-inducible vector to create pEF(2)URE3-BP. The control plasmid, pSTOP, derived from pEF(2)mutURE3-BP, has two stop codons added after the myc tag, which results in a truncated protein (6). Transfected amebae were selected for either hygromycin alone (Tet-Repressor expression plasmid; 15 µg/ml) or G418 (6 µg/ml) and hygromycin (Tet-Repressor expression plasmid and the inducible expression construct).
qRT-PCR.
One milliliter of Trizol (Invitrogen) was added to a pellet of 2 × 106 amebae, and an initial RNA isolation was performed according to the manufacturer’s directions. RNA greater than 200 nucleotides in length was separated from the total RNA by the use of the RNeasy protocol (Qiagen). RNA was isolated from at least two independent cultures. Reverse transcription using the Superscript II enzyme (Invitrogen) was followed by quantitative real-time PCR (qRT-PCR) to confirm the induction of the recombinant mRNA using a forward primer specific to the myc tag (ATTTCAGAAGAAGATTAATGC) and a construct-specific reverse primer (expression plasmid, CTGATGGTGGCATTTGA; induced pSTOP transfectant control, TAACTGATGTTAGCTATTGT). The cDNA was subjected to 40 amplification cycles of 94°C for 30 s, annealing at 61.8°C for 30 s, and extension at 72°C for 30 s. Triton X-100 (0.15%) was added to the PCR mixture to improve the amplification efficiency of the HotStar Taq reaction (Qiagen).
Murine model of amebic colitis.
Male CBA/J mice (The Jackson Laboratory) were infected by luminal injection of 2 × 106 trophozoites into the cecum, which was exposed by laparoscopic surgery as described by Houpt et al. (28). After recovery from surgery, mice were given drinking water containing 0.2 mg/ml doxycycline and 5% sucrose. mRNA isolated from the cecum of a selected mouse was subjected to qRT-PCR to confirm tetO derepression. At 30 days postinjection, the mice were sacrificed and amebae were collected either from the cecal contents (luminal) or by scraping the mucosal surface three times with a razor blade (tissue proximal). Grubbs’ test was used to identify significant outlier values (GraphPad Software QuickCalcs).
Gerbil model of amebic liver abscess formation.
Amebic liver abscesses were induced by the method of Chadee and Meerovitch (29). Amebae (1 × 106) transfected with either the control plasmid pSTOP or the plasmid expressing dominant positive URE3-BP were pretreated with 5 µg/ml tetracycline for 14 to 20 h. Induced transfectants containing the control plasmid pSTOP and the plasmid containing dominant positive URE3-BP [pEF(2)mutURE3-BP] were inoculated into the same 50- to 60-day-old Mongolian male gerbils (Meriones unguiculatus). One transfectant was injected into the right anterior lobe of the liver and the other into the left lobe. The order in which the transfectants were injected was reversed in half of the animals. After recovery from surgery, the gerbils received drinking water containing 2 mg/ml doxycycline. The higher concentration of doxycycline was required to maintain derepression of the tetracycline repressor in these desert animals (30). Gerbils were sacrificed 7 days after challenge, livers were extracted, and abscess weights were measured.
qPCR.
Closed circular DNA was isolated from cecal contents by a modification of the methods of Pontes et al. and Sikorski et al. (31, 32). Amebae were collected by a low-speed spin (900 × g for 5 min), washed with phosphate-buffered saline (PBS), and lysed by the addition of 700 μl Rapid One-Step Extraction buffer (10 mM Tris [pH 8.0], 300 mM EDTA [pH 8.0], 1% sodium lauryl sulfate, 1% polyvinylpolypyrrolidone). The lysate was heated to 95°C for 10 min, vortexed briefly, and returned to 95°C for 10 min. The cell lysate was then centrifuged at 14,000 × g for 10 min at room temperature. Five hundred microliters of the supernatant was then collected in a separate tube, mixed with 50 µl of cold 3 M sodium acetate (pH 5.3, 4°C) and 1 ml of cold ethanol (−20°C), and immediately centrifuged at 14,000 × g for 15 min. The supernatant was then discarded, and the pellet was washed once with 500 µl of 70% ethanol (−20°C), dried under vacuum, and resuspended in 50 mM MOPS buffer (50 mM MOPS [morpholinepropanesulfonic acid], 750 mM NaCl [pH 7.5]) to give a final volume of 0.75 ml. The sample was then purified using the Zymo plasmid purification kit (Zymo, Inc.). The eluted DNA was then twice passed over a Zymo-Spin IV-HRC filtration column, and PCR was performed as described above.
Analysis of amebic morphology.
Late-log-phase E. histolytica trophozoites were induced to express dominant positive URE3-BP with tetracycline for 16 h in normal TYI medium, followed by a 4-h incubation in serum-free medium. The induced control pSTOP transfectants were treated identically. Cells were rinsed in situ with 20 mM HEPES sodium salt (pH 7.2)–140 mM NaCl–5 mM EDTA–5 mM MgCl2 prewarmed at 37°C and fixed with 4% paraformaldehyde in PBS for 30 min at room temperature. Following fixation, samples were washed and resuspended in PBS and filtered through a 70-µm nylon cell strainer (BD Falcon). Growth arrest was confirmed by nuclear staining with a 1-min incubation with 4′,6-diamidino-2-phenylindole (33). The Amnis Imagestream imaging cytometer was used to collect at least 2,000 images of each sample (Amnis Corporation). The ImageStream data exploration and analysis software was used for data analysis. Spectral compensation was performed using the dark-field image. Automated quantitative image analysis of each sample in an experiment used a common template to quantify the “elongated” amebae using identical parameters for both control (induced pSTOP transfectant) and experimental [induced pEF(2)mutURE3-BP, transfectants which express dominant positive URE3-BP] samples. In each template, gating was performed to generate a population of single, in-focus images (usually yielding at least 1,000 images per sample), and masking was used to identify trophozoite morphology. Within the template, the surface area and circularity measurements were used to plot the sample, and tagging of the most morphologically changed amebae identified the noncircular population.
Reanalysis of microarray data and URE3-BP matrix detection.
Previously published E. histolytica microarray data sets from in vitro-cultured amebae and amebae isolated from the cecal lumen 1 day after infection were downloaded from the National Institutes of Health Gene Expression Omnibus (accession numbers: platform, GPL9693; data set, GSE8484) (11, 34). The data were reanalyzed using the reannotated E. histolytica genome (accession no. NZ_AAFB00000000; http://amoebadb.org/), the new Array Data Analysis and Management System (Virginia Bioinformatics Institute; http://pathport.vbi.vt.edu/main/microarray-tool.php) (35), and the Linear Model for Microarray Data, and the method of Benjamini and Hochberg was used to determine the false discovery rate; q values of less than 0.05 were considered significant as previously described by Gilchrist et al. (6, 36–39). The DNA Pattern Find program (http://bioinformatics.org/sms/) was used to locate the URE3 consensus motif. The strategy employed allowed a one-base divergence from the canonical TATTCTATT motif to the URE3 consensus of T[ATG]T[TC][CG]T[AT][TGC][TG] in the putative promoters of URE3-BP responsive genes (−375 to −25 from the ATG start), as described by Gilchrist et al. (6).
Supplemental material
ACKNOWLEDGMENT
This work was supported by NIH grant AI-37941 to W. A. Petri, Jr.
Footnotes
Citation Gilchrist, C. A., E. S. Moore, Y. Zhang, C. B. Bousquet, J. A. Lannigan, et al. 2010. Regulation of virulence of Entamoeba histolytica by the URE3-BP transcription factor. mBio 1(1):e00057-10. doi:10.1128/mBio.00057-10.
Editor John Boothroyd, Stanford University
REFERENCES
- 1. Haque R., Mondal D., Duggal P., Kabir M., Roy S., Farr B. M., Sack R. B., Petri W. A. 2006. Entamoeba histolytica infection in children and protection from subsequent amebiasis. Infect. Immun. 74:904–909 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Haque R., Huston C. D., Hughes M., Houpt E., Petri W. A. 2003. Amebiasis. N. Engl. J. Med. 348:1565–1573 [DOI] [PubMed] [Google Scholar]
- 3. Mandell G. L., Douglas R. G., Bennett J. E., Dolin R. 2005. Mandell, Douglas, and Bennett’s principles and practice of infectious diseases. Churchill Livingstone/Elsevier, Philadelphia, PA [Google Scholar]
- 4. Purdy J. E., Pho L. T., Mann B. J., Petri W. A. 1996. Upstream regulatory elements controlling expression of the Entamoeba histolytica lectin. Mol. Biochem. Parasitol. 78:91–103 [DOI] [PubMed] [Google Scholar]
- 5. Gilchrist C. A., Mann B. J., Petri W. A. 1998. Control of ferredoxin and Gal/GalNAc lectin gene expression in Entamoeba histolytica by a cis-acting DNA sequence. Infect. Immun. 66:2383–2386 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Gilchrist C. A., Baba D. J., Zhang Y., Crasta O., Evans C., Caler E., Sobral B. W. S., Bousquet C. B., Leo M., Hochreiter A., Connell S. K., Mann B. J., Petri W. A. 2008. Targets of the Entamoeba histolytica transcription factor URE3-BP. PLoS Negl. Trop. Dis. 2:e282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Gilchrist C. A., Holm C. F., Hughes M. A., Schaenman J. M., Mann B. J., Petri W. A. 2001. Identification and characterization of an Entamoeba histolytica upstream regulatory element 3 sequence-specific DNA-binding protein containing EF-hand motifs. J. Biol. Chem. 276:11838–11843 [DOI] [PubMed] [Google Scholar]
- 8. Gilchrist C. A., Leo M., Line C. G., Mann B. J., Petri W. A. 2003. Calcium modulates promoter occupancy by the Entamoeba histolytica Ca2+-binding transcription factor URE3-BP. J. Biol. Chem. 278:4646–4653 [DOI] [PubMed] [Google Scholar]
- 9. Moreno H., Linford A. S., Gilchrist C. A., Petri W. A. 18 December 2009, posting date. Phospholipid-binding protein EhC2A mediates calcium-dependent translocation of transcription factor URE3-BP to the plasma membrane of Entamoeba histolytica. Eukaryot. Cell. doi: 10.1128/EC.00346-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Ramakrishnan G., Vines R. R., Mann B. J., Petri W. A. 1997. A tetracycline-inducible gene expression system in Entamoeba histolytica. Mol. Biochem. Parasitol. 84:93–100 [DOI] [PubMed] [Google Scholar]
- 11. Gilchrist C. A., Houpt E., Trapaidze N., Fei Z., Crasta O., Asgharpour A., Evans C., Martino-Catt S., Baba D. J., Stroup S., Hamano S., Ehrenkaufer G., Okada M., Singh U., Nozaki T., Mann B. J., Petri W. A. 2006. Impact of intestinal colonization and invasion on the Entamoeba histolytica transcriptome. Mol. Biochem. Parasitol. 147:163–176 [DOI] [PubMed] [Google Scholar]
- 12. Bansal D., Ave P., Kerneis S., Frileux P., Boché O., Baglin A. C., Dubost G., Leguern A., Prevost M., Bracha R., Mirelman D., Guillén N., Labruyère E. 2009. An ex-vivo human intestinal model to study Entamoeba histolytica pathogenesis. PLoS Negl. Trop. Dis. 3:e551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. McCoy J. J., Mann B. J., Petri W. A. 1994. Adherence and cytotoxicity of Entamoeba histolytica or how lectins let parasites stick around. Infect. Immun. 62:3045–3050 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Gilchrist C. A., Petri W. A. 2009. Using differential gene expression to study Entamoeba histolytica pathogenesis. Trends Parasitol. 25:124–131 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Ehrenkaufer G. M., Singh U. 2008. Transcriptional regulatory networks in Entamoeba histolytica. Curr. Drug Targets 9:931–937 [DOI] [PubMed] [Google Scholar]
- 16. Vicente J. B., Ehrenkaufer G. M., Saraiva L. M., Teixeira M., Singh U. 2009. Entamoeba histolytica modulates a complex repertoire of novel genes in response to oxidative and nitrosative stresses: implications for amebic pathogenesis. Cell. Microbiol. 11:51–69 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Singh U., Ehrenkaufer G. M. 2009. Recent insights into Entamoeba development: identification of transcriptional networks associated with stage conversion. Int. J. Parasitol. 39:41–47 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Biller L., Davis P., Tillack M., Matthiesen J., Lotter H., Stanley S., Tannich E., Bruchhaus I. 2010. Differences in the transcriptome signatures of two genetically related Entamoeba histolytica cell lines derived from the same isolate with different pathogenic properties. BMC Genomics 11:63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Jain R., Kumar S., Gourinath S., Bhattacharya S., Bhattacharya A. 2009. N- and C-terminal domains of the calcium binding protein EhCaBP1 of the parasite Entamoeba histolytica display distinct functions. PLoS ONE 4:e5269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Coudrier E., Amblard F., Zimmer C., Roux P., Olivo-Marin J., Rigothier M., Guillén N. 2005. Myosin II and the Gal-GalNAc lectin play a crucial role in tissue invasion by Entamoeba histolytica. Cell. Microbiol. 7:19–27 [DOI] [PubMed] [Google Scholar]
- 21. Anderson S. I., Behrendt B., Machesky L. M., Insall R. H., Nash G. B. 2003. Linked regulation of motility and integrin function in activated migrating neutrophils revealed by interference in remodelling of the cytoskeleton. Cell Motil. Cytoskeleton 54:135–146 [DOI] [PubMed] [Google Scholar]
- 22. Franca-Koh J., Devreotes P. N. 2004. Moving forward: mechanisms of chemoattractant gradient sensing. Physiology (Bethesda) 19:300–308 [DOI] [PubMed] [Google Scholar]
- 23. Small J. V., Stradal T., Vignal E., Rottner K. 2002. The lamellipodium: where motility begins. Trends Cell Biol. 12:112–120 [DOI] [PubMed] [Google Scholar]
- 24. Camacho-Leal P., Zhai A. B., Stanners C. P. 2007. A co-clustering model involving alpha5beta1 integrin for the biological effects of GPI-anchored human carcinoembryonic antigen (CEA). J. Cell. Physiol. 211:791–802 [DOI] [PubMed] [Google Scholar]
- 25. Diamond L. S. 1961. Axenic cultivation of Entamoeba histolytica. Science 134:336–337 [DOI] [PubMed] [Google Scholar]
- 26. Olvera A., Olvera F., Vines R. R., Recillas-Targa F., Lizardi P. M., Dhar S., Bhattacharya S., Petri W., Alagón A. 1997. Stable transfection of Entamoeba histolytica trophozoites by lipofection. Arch. Med. Res. 28:49–51 [PubMed] [Google Scholar]
- 27. Asgharpour A., Gilchrist C., Baba D., Hamano S., Houpt E. 2005. Resistance to intestinal Entamoeba histolytica infection is conferred by innate immunity and Gr-1+ cells. Infect. Immun. 73:4522–4529 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Houpt E. R., Glembocki D. J., Obrig T. G., Moskaluk C. A., Lockhart L. A., Wright R. L., Seaner R. M., Keepers T. R., Wilkins T. D., Petri W. A. 2002. The mouse model of amebic colitis reveals mouse strain susceptibility to infection and exacerbation of disease by CD4+ T cells. J. Immunol. 169:4496–4503 [DOI] [PubMed] [Google Scholar]
- 29. Chadee K., Meerovitch E. 1989. Entamoeba histolytica: diffuse liver inflammation in gerbils (Meriones unguiculatus) with experimentally induced amebic liver abscess. J. Protozool. 36:154–158 [DOI] [PubMed] [Google Scholar]
- 30. Vines R. R., Ramakrishnan G., Rogers J. B., Lockhart L. A., Mann B. J., Petri W. A. 1998. Regulation of adherence and virulence by the Entamoeba histolytica lectin cytoplasmic domain, which contains a beta2 integrin motif. Mol. Biol. Cell 9:2069–2079 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Pontes L. A., Dias-Neto E., Rabello A. 2002. Detection by polymerase chain reaction of Schistosoma mansoni DNA in human serum and feces. Am. J. Trop. Med. Hyg. 66:157–162 [DOI] [PubMed] [Google Scholar]
- 32. Sikorski J., Möhle M., Wackernagel W. 2002. Identification of complex composition, strong strain diversity and directional selection in local Pseudomonas stutzeri populations from marine sediment and soils. Environ. Microbiol. 4:465–476 [DOI] [PubMed] [Google Scholar]
- 33. Mukherjee C., Clark C. G., Lohia A. 2008. Entamoeba shows reversible variation in ploidy under different growth conditions and between life cycle phases. PLoS Negl. Trop. Dis. 2:e281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Barrett T., Troup D. B., Wilhite S. E., Ledoux P., Rudnev D., Evangelista C., Kim I. F., Soboleva A., Tomashevsky M., Marshall K. A., Phillippy K. H., Sherman P. M., Muertter R. N., Edgar R. 2009. NCBI GEO: archive for high-throughput functional genomic data. Nucleic Acids Res. 37:D885–D890 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Loftus B., Anderson I., Davies R., Alsmark U. C. M., Samuelson J., Amedeo P., Roncaglia P., Berriman M., Hirt R. P., Mann B. J., Nozaki T., Suh B., Pop M., Duchene M., Ackers J., Tannich E., Leippe M., Hofer M., Bruchhaus I., Willhoeft U., Bhattacharya A., Chillingworth T., Churcher C., Hance Z., Harris B., Harris D., Jagels K., Moule S., Mungall K., Ormond D., Squares R., Whitehead S., Quail M. A., Rabbinowitsch E., Norbertczak H., Price C., Wang Z., Guillén N., Gilchrist C., Stroup S. E., Bhattacharya S., Lohia A., Foster P. G., Sicheritz-Ponten T., Weber C., Singh U., Mukherjee C., El-Sayed N. M., Petri W. A., Clark C. G., Embley T. M., Barrell B., Fraser C. M., Hall N. 2005. The genome of the protist parasite Entamoeba histolytica. Nature 433:865–868 [DOI] [PubMed] [Google Scholar]
- 36. Gentleman R. C., Carey V. J., Bates D. M., Bolstad B., Dettling M., Dudoit S., Ellis B., Gautier L., Ge Y., Gentry J., Hornik K., Hothorn T., Huber W., Iacus S., Irizarry R., Leisch F., Li C., Maechler M., Rossini A. J., Sawitzki G., Smith C., Smyth G., Tierney L., Yang J. Y. H., Zhang J. 2004. Bioconductor: open software development for computational biology and bioinformatics. Genome Biol. 5:R80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Tusher V. G., Tibshirani R., Chu G. 2001. Significance analysis of microarrays applied to the ionizing radiation response. Proc. Natl. Acad. Sci. U. S. A. 98:5116–5121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Smyth G. K., Michaud J., Scott H. S. 2005. Use of within-array replicate spots for assessing differential expression in microarray experiments. Bioinformatics 21:2067–2075 [DOI] [PubMed] [Google Scholar]
- 39. Benjamini Y., Hochberg Y. 1995. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J. R. Stat. Soc. B Stat. Methodol. 57:289–300 [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.