Abstract
The use of herbal dietary supplements in the United States is rapidly growing, and it is crucial that the quality and safety of these preparations be ensured. To date, it is still a challenge to determine the mechanisms of toxicity induced by mixtures containing many chemical components, such as herbal dietary supplements. We previously proposed that analyses of the gene expression profiles using microarrays in the livers of rodents treated with herbal dietary supplements is a potentially practical approach for understanding the mechanism of toxicity. In this study, we utilized microarrays to analyze gene expression changes in the livers of male B6C3F1 mice administered Ginkgo biloba leaf extract (GBE) by gavage for 2 years, and to determine pathways and mechanisms associated with GBE treatments. Analysis of 31,802 genes revealed that there were 129, 289, and 2,011 genes significantly changed in the 200, 600, and 2,000 mg/kg treatment groups, respectively, when compared with control animals. Drug metabolizing genes were significantly altered in response to GBE treatments. Pathway and network analyses were applied to investigate the gene relationships, functional clustering, and mechanisms involved in GBE exposure. These analyses indicate alteration in the expression of genes coding for drug metabolizing enzymes, the NRF2-mediated oxidative stress response pathway, and the Myc gene-centered network named “cell cycle, cellular movement, and cancer” were found. These results indicate that Ginkgo biloba-related drug metabolizing enzymes may cause herb–drug interactions and contribute to hepatotoxicity. In addition, the outcomes of pathway and network analysis may be used to elucidate the toxic mechanisms of Ginkgo biloba.
Introduction
Since the U.S. Congress passed the Dietary Supplement Health and Education Act (DSHEA) in 1994, herbal products have been the fastest growing segment of the vitamin, mineral supplements, and herbal products industry in the United States. In 2004, the American Herbal Products Association estimated that there were about 3,000 species of plants in as many as 50,000 different products sold as herbal supplements in the United States (Zurer and Hanson, 2004). St. John's wort, Ginkgo biloba, golden seal, panax ginseng, kava, aloe vera, and mild thistle extract are among the most widely used of these products (Chan and Fu, 2007; Chan et al., 2007; Fu et al., 2008; Guo et al., 2009).
Although it has been reported that a number of herbal dietary supplements cause adverse health effects (Chan and Fu, 2007; Chan et al., 2007; Fu, 2007; Fu et al., 2007, 2008; Gurley et al., 2005, 2007; Hu et al., 2005; Singh, 2005), to date, safety issues concerning potential side effects and toxic contamination of herbal products have not been adequately addressed. Thus, assessment of the safety of herbal plants and herbal dietary supplements is timely and important (FDA, 2001, 2004a, 2004b; Fong, 2002; Fu et al., 2002). Recently, a number of herbal dietary supplements and active ingredients have been nominated by the U.S. Food and Drug Administration (FDA) and the U.S. National Institutes of Health (NIH) to the National Toxicology Program (NTP) for determination of their toxicity and tumorigenicity. Ginkgo biloba, panax ginseng, kava, aloe vera, and green tea are among the herbal dietary supplements currently under investigation by the NTP.
In general, approaches for determining the mechanism by which a pure chemical induces toxicities or tumors have been well established. Nevertheless, it is still a challenge to determine the mechanisms of toxicities or tumor induction elicited by a mixture of many chemical components, such as herbal plants and herbal dietary supplements. For chemical mixtures there is a need for new approaches for elucidating mechanisms. Toward this goal, we previously examined the alterations in gene expression of drug metabolizing enzymes in the livers of Fischer 344 rats administered kava extract by gavage for 14 weeks (Guo et al., 2009). Our results indicate that kava extract can significantly modulate drug-metabolizing enzymes, which can potentially cause herb–drug interactions and may be responsible for different types of liver injuries. The gene expression profile correlated well (Guo et al., 2009) with immunohistochemical data collected using the same liver tissues (Clayton et al., 2007). We observed that kava altered the expression of Cyp1a1 (Cytochrome p450 1a1) and many other Cyp enzymes that metabolize various xenobiotics and drugs, and the changes for many of those genes were first reported by us (Guo et al., 2009). These findings illustrate that, without obtaining the whole spectrum of gene expression changes, some important information may be missed. Our study also suggested that analysis of the gene expression profiles using microarrays in the livers of rodents treated with herbal dietary supplements is a potentially practical approach for understanding the mechanism of toxicity (Guo et al., 2009).
Ginkgo biloba has been one of the most widely sold products in health food stores in the United States, with total sales exceeding $100 million (Chan et al., 2007). Several reports have indicated that Ginkgo biloba inhibits Cyp activity and, when taken in combination with prescription and conventional medications, may produce Cyp-mediated herb–drug interactions (Bressler, 2005; Gurley et al., 2005, 2007; Hu et al., 2005; Matthias et al., 2007; Singh, 2005; Williamson, 2005; Wold et al., 2005; Zou et al., 2002). Gingko biloba leaf extract was nominated by the National Cancer Institute to the NTP to conduct toxicological evaluation, mechanistic studies, and a 2-year chronic carcinogenicity bioassay based on the following reasons: (1) Ginkgo biloba is a well-defined product and itself or its active ingredients have clearly demonstrated biological activities, (2) Ginkgo biloba can be consumed in rather large doses for an extended period of time, and (3) some ingredients in Ginkgo biloba are known mutagens (NTP, 1998). As a continuation of our mechanistic studies of herbal dietary supplements, in the present study, gene expression changes were analyzed with the focus on drug metabolizing genes in the livers of male B6C3F1 mice treated orally and chronically with Ginkgo biloba extract. In addition, changes in pathways and networks, which may indicate toxicity, were also explored.
Materials and Methods
Ginkgo biloba leaf extract preparation and animal treatment
Ginkgo biloba leaf extract (GBE) (CAS No. 90045-36-6) was received from the Midwest Research Institute (MRI, Kansas City, MO). Infrared spectroscopy and HPLC/UV analyses confirmed the chemical was GBE by comparing with a reference sample received from MRI. The identity, chemical purity, and formulation analysis evaluation data are available at the archives of the National Institute of Environmental Health Sciences. Male B6C3F1 mice (38–44 days old) were obtained from Taconic Laboratory Animals and Services (Germantown, NY) and were used in the NTP toxicity and carcinogenesis studies of GBE in mice. The animals were quarantined for 14 days before separating at random into four groups. GBE in corn oil was administered to each group by gavage at 0, 200, 600, or 2,000 mg/kg, 5 days a week for 104 weeks. The animals were monitored twice daily during the course of the experiment, and body weights and clinical observations were recorded weekly for the first 13 weeks and every 4 weeks thereafter, and at the study termination. Animal handling and husbandry were conducted in accordance with guidelines of the National Institutes of Health. During necropsy at terminal sacrifice (104 weeks); a piece of the left lateral lobe of the liver was collected from 10 animals from each group, cut into three pieces and frozen in liquid nitrogen until use.
RNA isolation and quality control
Total RNA was isolated from liver tissues of control and GBE-treated mice using an RNeasy kit (Qiagen, Valencia, CA). The RNA was quantified by optical density reading using a NanoDrop ND-1000 spectrophotometer (NanoDrop products, Wilmington, DE). The purity and quality of extracted RNA were evaluated using the RNA 6000 LabChip and Agilent 2100 Bioanalyzer (Agilent Technologies, Palo Alto, CA). RNA samples with RNA integrity numbers (RINs) greater than 9.0 were used for target labeling, microarray experiments, and TaqMan assays.
Preparation of cyanine 5 (Cy5)-labeled aRNA for array hybridization
Five micrograms of RNA from each sample was used for Cy5-labeled aRNA preparation using the Ambion MessageAmp aRNA Amplification Kit (Applied Biosystems Inc, Foster City, CA) with minor modifications to adopt the amino-allyl UTP indirect labeling protocol. In brief, the 4 μL T7 UTP solution (75 mM) was replaced with 2 μL of T7 UTP solution (75 mM) and 3 μL of 50 mM 5-(3-aminoallyl)-UTP in the process of in vitro transcription to synthesize aRNA. The reaction conditions and the purification of unlabeled aRNA remained the same as described in the user's manual. For Cy5 labeling of aRNA, 40 μg of unlabeled aRNA was used in each labeling reaction to generate sufficient Cy5-labeled cRNA for the microarray hybridization in triplicate. The Cy5 dye (GE Healthcare, Piscataway, NJ) was suspended in 32 μL DMSO before use. Eight microliters of Cy5 dye solution and 2.5 μL of 10 μM NaOH were added to the 40 μg aRNA and the volume was adjusted to 25 μL by addition of ddH2O. The mixture was incubated at 25°C in the dark for 2 h. The labeled cRNA was then purified using the Ambion NucAway Spin Column (ABI). The amount and labeling efficiency of the purified Cy5-labeled cRNA were quantified by optical density reading using a NanoDrop ND-1000. The minimal requirement of labeling efficiency was 10 Cy5 dye molecules per 1,000 nucleotides.
Hybridization of labeled cRNA to microarrays and microarray imaging
Microarray experiments were performed using Phalanx Mouse OneArray Version 1.1 (MOA 1.1; Phalanx Biotech Group, Inc., HsinChu, Taiwan). Each microarray contains 31,802 oligonucleotide probes that include 29,922 mouse gene probes for transcription expression profiling and 1,80 experimental control probes. The MOA 1.1 content is based on an abridged version of the Mouse Exonic Evidence Based Oligonucleotide (MEEBO). MEEBO consists of a set of 70-mer probes made available to the public specifically for DNA microarray. For microarray hybridization experiments, 10-μg Cy5-labeled cRNA derived from each RNA sample was applied to each of three MOA 1.1 microarrays. Prior to the microarray hybridization, the Cy5-labeled cRNAs were fragmented using the reagents and protocol provided in Ambion RNA Fragmentation Reagents kit (Ambion Inc., Austin, TX). Each 10-μg fragmented Cy5-labeled cRNA was suspended in OneArray hybridization buffer at a final volume of 180 μL for microarray hybridization. After microarray hybridization, the arrays were scanned using a fluorescence scanner (GenePix 4000B; Molecular Devices, Sunnyvale, CA) and the fluorescent intensities were extracted from the generated images by following the instructions and the conditions described in the MOA 1.1 User's Manual.
Microarray data analysis
Each Cy5-labeled cRNA derived from a liver RNA sample was hybridized to MOA 1.1 microarrays in triplicate. The raw microarray intensity data was imported to into ArrayTrack (www.fda.gov/nctr/science/centers/toxicoinformatics/ArrayTrack/). The intensities of the three technical replicates for each probe were averaged and then normalized by quantile normalization. Differentially expressed genes (DEGs) were selected based on t-test and fold change cutoff criteria. Principal component analysis (PCA) and hierarchical cluster analysis (HCA) were conducted within ArrayTrack. Additional calculations were performed within JMP 7.0 (SAS Institute, Cary, NC). The pathways, networks,and functional analyses were generated through the use of ingenuity pathways analysis (IPA;Ingenuity Systems, www.ingenuity.com). Canonical pathways analysis identified the pathways from the IPA library of canonical pathways that were most significant to the data set. DEGs that associated with a canonical pathway in the IPA Base were considered for the analysis. The significance of the association between the data set and the canonical pathway was measured in two ways: (1) a ratio of the number of genes from the data set that map to the pathway divided by the total number of genes that map to the canonical pathway is displayed, and (2) Fischer's exact test was used to calculate a p-value determining the probability that the association between the genes in the dataset and the canonical pathway is explained by chance alone. DEGs containing gene identifiers and corresponding expression values were uploaded into the application to generate networks. Each gene identifier was mapped to its corresponding gene object in the ingenuity pathways knowledge base. These genes, called focus genes, were overlaid onto a global molecular network developed from information contained in the ingenuity pathways knowledge base. Networks of these focus genes were then algorithmically generated based on their connectivity.
TaqMan gene expression assays
DEGs identified with microarray analysis were selectively confirmed by TaqMan assays (Applied Biosystems, Foster City, CA). Eleven TaqMan probes were used in these assays, including Cyp1a1 (Mm00487218_m1), Cyp1a2 (Mm00487224_m1), Cyp2b10 (Mm00456591_m1), Cp2b13 (Mm00771172_g1), Cyp2c55 (Mm00472168_m1), Cyp2d13 (Mm00775259_gl), Ces2 (Mm00524035_m1), Fmo3 (Mm00514964_m1), Gsta2 (Mm00833353_mH), Actb (Mm00607939_s1), and Polr2a (Mm 00839493_m1). Actb and Polr2a were used as endogenous controls. cDNA was prepared using a High-Capacity cDNA Archive Kit (Applied Biosystems), that is, 2 μg RNA was reverse-transcribed in a final volume of 20 μL with random primers at 25°C for 10 min followed by 37°C for 120 min according to the manufacturer's instructions. Each TaqMan assay was run in triplicate for each RNA sample. Total cDNA (25 ng) in a 25 μL final volume was used for each assay. Assays were run with Universal Master Mix (2 ×) without AmpErase UNG on an Applied Biosystems 7900 HT Real-Time PCR System using universal cycling conditions (10 min at 95°C; 15 s at 95°C, 1 min 60°C, 40 cycles).
TaqMan data normalization and analysis
Two endogenous control genes, β-actin (Actb) and RNA polymerase II A (Polr2a), were used for normalization. Each replicate cycle threshold (CT) was normalized to the average CT of the two endogenous controls on a per sample basis. The comparative CT method (Livak and Schmittgen, 2001) was used to calculate relative quantification of gene expression. The following formula was used to calculate the relative amount of the transcripts in the GBE-treated sample (treated) and the vehicle-treated sample (control), and both were normalized to the endogenous controls: ΔΔCT = ΔCT (treated) − ΔCT (control), where ΔCT is the difference in CT between the target gene and endogenous controls by subtracting the average CT of controls. The fold change for each treated sample relative to the control sample = 2−ΔΔCT. A list of DEGs was identified using a two-tailed t-test. The criteria were P value less than 0.05 and a mean difference equal to or greater than two-fold.
Results
Animals
During the course of the study no clinical findings attributed to GBE administration were observed. After 18–19 months of GBE treatment, the body weights of the 600 and 2,000 mg/kg mice began to decrease gradually. At terminal sacrifice the body weights of these two dose groups of mice were significantly lower (∼17 and ∼25%, respectively) compared with controls.
Microarray data quality assessment
In this study, the global changes of gene expression in mouse liver treated with GBE were determined by high-density oligonucleotide microarray analysis (Mouse OneArray, which contains 31,802 probes covering 29,922 verified mouse genes). Total RNA was isolated from liver tissues of control mice and mice treated with 200, 600, and 2,000 mg/kg GBE. Each group consisted of four animals; therefore, 16 RNA samples were isolated for microarray analysis. Three technical replicate arrays were performed for each RNA sample, and thus a total of 48 microarrays were used in this study.
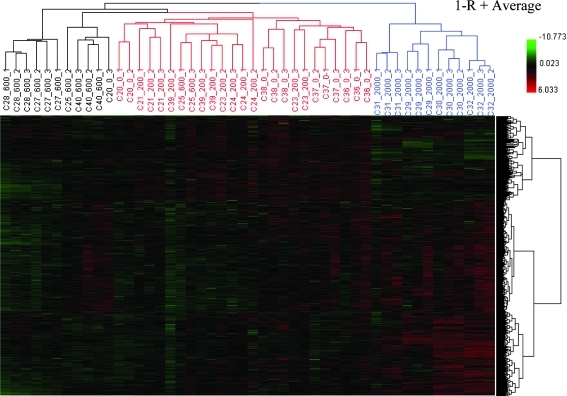
The overall quality of data from the 48 microarrays was first assessed by HCA. The log2 intensity of the entire gene set including 31,802 probes was scaled by Z-score transformation, and then these values were hierarchically clustered using a distance metric of 1−R, where R is the Pearson correlation coefficient between two samples, and the average linkage (Fig. 1). As expected, for most samples, the three technical replicates for the same RNA sample were tightly grouped, indicating close similarity among them. In addition, two apparent clusters were revealed. The first cluster (in black and red) consists of samples from the control, and low- (200 mg/kg) and middle-dose (600 mg/kg) GBE treatments; the second cluster (in blue) includes samples from high-dose (2,000 mg/kg) GBE treatments. Samples in the high-dose group were clearly separated from those in the control, low, and middle groups (Fig. 1). In the first big cluster, the three subgroups (control, low, and middle dose) were also well separated with few exceptions.
FIG. 1.
Hierarchical cluster analysis (HCA) of expression profiles for control and GBE-treated groups. Each column represents the results from an individual hybridization. Each row represents the log2 intensity values of the 48 samples for one particular gene. Samples are labeled according to the convention of Animal ID_Dose (mg/kg)_Technical replicate number.
The reproducibility of the three technical replicates was further assessed by calculating the Pearson correlation coefficient of pairwise log2 intensity. R values of each pairwise comparison were obtained; the median R values of the three technical replicates are shown in Table 1. The median R values across 16 RNA samples range from 0.984–0.997. One array from sample C38 (C38_3) and one from C24 (C24_2) showed dissimilarity in comparison with its other two technical replicates with relatively low correlation coefficients (0.979 and 0.965, respectively). These two hybridizations were considered as outliers and were removed from further calculation and data analysis.
Table 1.
Assessment of Microarray Data Quality (Technical Replicates): The Microarray Data Reproducibility Was Assessed by the Pearson Correlation Coefficient of Pairwise Log2 Intensity Correlation among Three Technical Replicates (1, 2, and 3)
| Sample No. | Dose (mg/kg) | R Values, Median |
|---|---|---|
| C20_1,2,3 | 0 | 0.992, 0.991, 0.998 |
| C36_1,2,3 | 0 | 0.995, 0.991, 0.996 |
| C37_1,2,3 | 0 | 0.992, 0.990, 0.997 |
| C38_1,2,3 | 0 | 0.979, 0.979, 0.997 |
| C21_1,2,3 | 200 | 0.991, 0.987, 0.996 |
| C23_1,2,3 | 200 | 0.992, 0.984, 0.994 |
| C24_1,2,3 | 200 | 0.965, 0.961, 0.997 |
| C39_1,2,3 | 200 | 0.997, 0.996, 0.998 |
| C25_1,2,3 | 600 | 0.990, 0.990, 0.997 |
| C27_1,2,3 | 600 | 0.992, 0.992, 0.997 |
| C28_1,2,3 | 600 | 0.995, 0.993, 0.997 |
| C40_1,2,3 | 600 | 0.993, 0.990, 0.994 |
| C29_1,2,3 | 2,000 | 0.984, 0.982, 0.990 |
| C30_1,2,3 | 2,000 | 0.988, 0.988, 0.997 |
| C31_1,2,3 | 2,000 | 0.990, 0.990, 0.994 |
| C32_1,2,3 | 2,000 | 0.995, 0.995, 0.997 |
The R value of three technical replicates is shown. Pearson correlation matrix of arrays was calculated based on all data points (31,802 probes).
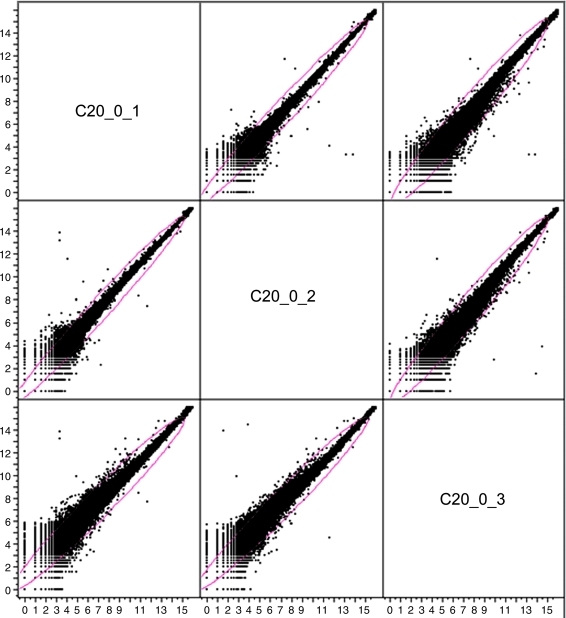
It is worth noting that the intensities of all data points (31,802 probes) were included for the comparison. No specific cutoff was applied for evaluation of microarray quality. For visualization purposes, as an example, the raw log2 intensity data of all 31,802 probes from the three technical replicates (C20_0_1, C20_0_2, and C20_0_3) of animal #C20 were plotted against each other (Fig. 2). As shown in the scatter plot, for most spots, the intensity values from the replicate microarrays accumulated along the diagonal axis with a correlation >0.996, indicating that the data from the technical microarrays were highly reproducible.
FIG. 2.
The log2 raw intensity data of the three technical replicates (C20_0_1, C20_0_2, and C20_0_3) were plotted against each other.
Profiling of gene expression
The intensity values of each probe from the three technical replicates were averaged and log2 transformed. The overall quality of the microarray data from the 16 RNA samples was further assessed by calculating the Pearson correlation coefficients of the averaged log2 intensity data for all pairwise sample comparisons within each control or treatment group (Table 2). The median correlation coefficient was 0.973 across all 16 arrays with a range from 0.926 to 0.989. The median correlation for the control group was 0.987, with a range from 0.983 to 0.988, whereas the median correlation for the 2,000 mg/kg treatment group was 0.970, with a range from 0.961 to 0.977. These results demonstrated the high degree of gene expression similarity for samples within the same group. On the other hand, the median correlation between the control group and the 2,000 mg/kg GBE treated group was 0.957, with a range from 0.933 to 0.964, demonstrating that there were significant differences in the gene expression profiles between the control and high dose GBE treated groups.
Table 2.
Assessment of Microarray Data Quality (Biological Replicates)
| C20-0 | C36-0 | C37-0 | C38-0 | C21-200 | C23-200 | C24-200 | C39-200 | C25-600 | C27-600 | C28-600 | C40-600 | C29-2000 | C30-2000 | C31-2000 | C32-2000 | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| C20-0 |
1.000 |
0.988 |
0.988 |
0.986 |
0.987 |
0.987 |
0.979 |
0.976 |
0.976 |
0.983 |
0.973 |
0.985 |
0.958 |
0.959 |
0.960 |
0.939 |
| C36-0 | 1.000 | 0.988 | 0.985 | 0.988 | 0.987 | 0.978 | 0.977 | 0.973 | 0.978 | 0.971 | 0.985 | 0.957 | 0.963 | 0.964 | 0.944 | |
| C37-0 | 1.000 | 0.983 | 0.985 | 0.986 | 0.976 | 0.976 | 0.973 | 0.974 | 0.965 | 0.985 | 0.956 | 0.958 | 0.960 | 0.940 | ||
| C38-0 | 1.000 | 0.984 | 0.986 | 0.979 | 0.978 | 0.974 | 0.985 | 0.979 | 0.978 | 0.952 | 0.957 | 0.958 | 0.933 | |||
| C21-200 | 1.000 | 0.989 | 0.983 | 0.981 | 0.980 | 0.981 | 0.975 | 0.984 | 0.959 | 0.962 | 0.959 | 0.940 | ||||
| C23-200 | 1.000 | 0.984 | 0.987 | 0.980 | 0.981 | 0.973 | 0.982 | 0.956 | 0.963 | 0.964 | 0.939 | |||||
| C24-200 | 1.000 | 0.980 | 0.977 | 0.978 | 0.972 | 0.973 | 0.961 | 0.966 | 0.957 | 0.940 | ||||||
| C39-200 | 1.000 | 0.986 | 0.973 | 0.967 | 0.969 | 0.945 | 0.954 | 0.952 | 0.926 | |||||||
| C25-600 | 1.000 | 0.975 | 0.969 | 0.972 | 0.951 | 0.953 | 0.949 | 0.930 | ||||||||
| C27-600 | 1.000 | 0.988 | 0.977 | 0.960 | 0.963 | 0.958 | 0.941 | |||||||||
| C28-600 | 1.000 | 0.969 | 0.955 | 0.957 | 0.951 | 0.933 | ||||||||||
| C40-600 | 1.000 | 0.968 | 0.967 | 0.963 | 0.955 | |||||||||||
| C29-2000 | 1.000 | 0.977 | 0.961 | 0.970 | ||||||||||||
| C30-2000 | 1.000 | 0.970 | 0.973 | |||||||||||||
| C31-2000 | 1.000 | 0.962 | ||||||||||||||
| C32-2000 | 1.000 |
The microarray data reproducibility was assessed by the Pearson correlation coefficient of pairwise log2 intensity correlation. Pearson correlation matrix of 16 arrays was calculated based on all data points (31,802 probes).
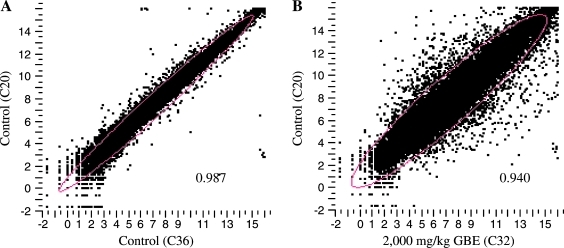
The log2 transformed intensity of any two gene expression profiles was plotted and compared, as shown in Figure 3. When intensities of the entire probe set obtained from a control sample (C20) were plotted against another control sample (C36), most of data points gathered along the diagonal axis of the scatter plot with a correlation coefficient value of 0.987, demonstrating the good repeatability of the two biological replicates (Fig. 3A). However, in comparison between the control sample (C20) and the 2,000 mg/kg GBE-treated sample (C32), many data points were scattered, indicating a great number of genes were altered in response to the treatment of high-dose GBE (Fig. 3B).
FIG. 3.
Overall gene expression profiles. The log2 transformed intensity was used for scatter plotting. (A) Two control samples (C20 and C36); (B) control (C20) and 2,000 mg/kg GBE (C32)-treated sample were plotted against each other.
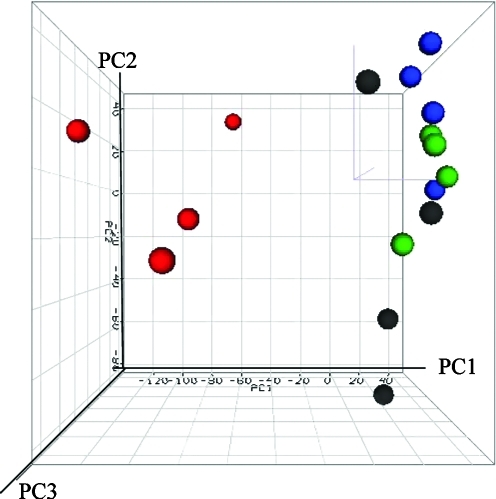
The intensities of the gene expression data were also analyzed by PCA. PCA uses analysis of the principal sources of variance in data and displays this information graphically either in a two-dimensional or three-dimensional space, which is most commonly used to classify gene expression profiles (Wang and Gehan, 2005). Figure 4 displays a PCA 3D view using the first three principal components for gene expression profiles from the samples. The PCA result showed a clear separation between controls and 2,000 mg/kg-treated samples, suggesting that there was a treatment effect on liver gene expression which is consistent with the results from the HCA (Fig. 1). Less separation was observed between controls, low-dose (i.e., 200 mg/kg), and middle-dose (i.e., 600 mg/kg) groups based on PCA.
FIG. 4.
Principal component analysis (PCA) of gene expression profiles for GBE-treated groups and the concurrent controls. The intensity of the entire gene set was used; no specific cutoff was applied for the analysis. The autoscaled method was used for the PCA. The blue, green, black, and red dots indicate control, 200, 600, and 2,000 mg/kg treatment, respectively. PC1, PC2, and PC3 represent first principal component, second principal component, and third principal component.
Differentially expressed genes (DEGs)
The average intensities for each probe from three technical replicates of each sample were normalized by the quantile normalization method, which assumes that a quantile plot of two data vectors with the same distribution will have a straight diagonal line. The normalized data were used for selection of DEGs. We had reported that a straightforward approach of fold change selection plus a nonstringent P cutoff is useful in identifying reproducible gene lists (Guo et al., 2006; Shi et al., 2006). We used a twofold change in gene expression compared to the controls and a P-value less than 0.01 for the difference as minimum requirements for the selection of DEGs. Based on these two criteria a total of 129, 289, and 2,011 genes, respectively, in 200, 600, and 2,000 mg/kg treatment groups satisfied the requirements and were identified as DEGs. Out of 2,011 genes identified in the 2,000 mg/kg treatment group, 817 were upregulated and 1,194 genes were downregulated in response to GBE exposure. Significance analysis of microarray (SAM) (Tusher et al., 2001), the statistical method that calculates difference in gene expression based on permutation analysis of expression data and calculates a false discovery rate, was also applied to generate DEGs. With an FDR of 0.01 and fold change greater than 2, a total of 2,211 genes were identified in the 2,000 mg/kg treatment group compared to control group. Out of 2,011 genes that were indentified as DEGs using the t-test, 1,952 genes (97%) are overlapped with those produced by the SAM, indicating that similar gene lists were produced with two gene selection methods.
Because metabolic activation of chemicals is very important for liver toxicity, and alteration of drug metabolizing genes is suspected to contribute to the toxicity of Gingko biloba, we first focused our investigation on the gene expression changes of drug metabolizing enzymes and found that the expression of a large number of drug-metabolizing genes was altered. Table 3 lists the 68 drug-metabolizing genes whose expression was significantly altered by the 2,000 mg/kg GBE treatment. Among the 68 altered genes, 36 genes were upregulated and 32 genes were downregulated. As tabulated in Table 3, among the 68 drug-metabolizing enzyme associated genes, 33 genes are Phase I metabolizing enzymes; 18 genes are Phase II metabolizing enzymes, and 17 genes are transporters (Phase III). As summarized in Table 4, 21% (33 out of 154) of Phase I genes, 21% (18 out of 87) of Phase II genes, and 24% (17 out of 72) of Phase III genes were altered. Although a relatively small portion of genes (6.7%) were altered at the genome level (2011 DEGs out of 29,922 probes), the percentage of changed drug metabolizing genes (22%, Table 4) was high.
Table 3.
Genes Involved in Three Phases of Liver Drug Metabolism Altered by 2,000 mg/kg GBE Treatment
| Gene symbol | Locus link ID | Gene description | Fold change | p Value |
|---|---|---|---|---|
| Phase I | ||||
| Adh7 | 11529 | alcohol dehydrogenase 7 (class IV), mu or sigma polypeptide | 2.5 | 0.006 |
| Aldh5a1 | 214579 | aldhehyde dehydrogenase family 5, subfamily A1 | −2.9 | 0.002 |
| Aldh8a1 | 237320 | aldehyde dehydrogenase 8 family, member A1 | −2.8 | 0.005 |
| Cyp1a1 | 13076 | cytochrome P450, family 1, subfamily a, polypeptide 1 | 6.8 | 0.001 |
| Cyp1a2 | 13077 | cytochrome P450, family 1, subfamily a, polypeptide 2 | 2.3 | 0.008 |
| Cyp2a12 | 13085 | cytochrome P450, family 2, subfamily a, polypeptide 12 | −2.2 | 0.004 |
| Cyp2a5 | 13087 | cytochrome P450, family 2, subfamily a, polypeptide 5 | 2.9 | 0.005 |
| Cyp2b10 | 13088 | cytochrome P450, family 2, subfamily b, polypeptide 10 | 17.3 | 0.001 |
| Cyp2b13 | 13089 | cytochrome P450, family 2, subfamily b, polypeptide 13 | −38.5 | 0.004 |
| Cyp2b20 | 13088 | cytochrome P450, family 2, subfamily b, polypeptide 10 | 14.6 | 0 |
| Cyp2b9 | 13094 | cytochrome P450, family 2, subfamily b, polypeptide 9 | −18.9 | 0.009 |
| Cyp2c29 | 13095 | cytochrome P450, family 2, subfamily c, polypeptide 29 | 3.6 | 0 |
| Cyp2c55 | 72082 | cytochrome P450, family 2, subfamily c, polypeptide 55 | 50.2 | 0 |
| Cyp2c66 | 69888 | cytochrome P450, family 2, subfamily c, polypeptide 66 | 4.4 | 0.001 |
| Cyp2d13 | 68444 | cytochrome P450, family 2, subfamily d, polypeptide 13 | −29.4 | 0.003 |
| Cyp2f2 | 13107 | cytochrome P450, family 2, subfamily f, polypeptide 2 | −13.2 | 0 |
| Cyp2g1 | 13108 | cytochrome P450, family 2, subfamily g, polypeptide 1 | −2.1 | 0.009 |
| Cyp2j5 | 13109 | cytochrome P450, family 2, subfamily j, polypeptide 5 | −7.1 | 0 |
| Cyp2j9 | 74519 | cytochrome P450, family 2, subfamily j, polypeptide 9 | −14.9 | 0 |
| Cyp2u1 | 71519 | cytochrome P450, family 2, subfamily u, polypeptide 1 | −17.9 | 0.008 |
| Cyp3a16 | 13114 | cytochrome P450, family 3, subfamily a, polypeptide 16 | −8.4 | 0.003 |
| Cyp3a41 | 53973 | cytochrome P450, family 3, subfamily a, polypeptide 41 | −2.3 | 0.009 |
| Cyp46a1 | 13116 | cytochrome P450, family 46, subfamily a, polypeptide 1 | −8.9 | 0.001 |
| Cyp4f14 | 64385 | cytochrome P450, family 4, subfamily f, polypeptide 14 | −4.8 | 0.006 |
| Cyp4f16 | 70101 | cytochrome P450, family 4, subfamily f, polypeptide 16 | 2.9 | 0.001 |
| Cyp51 | 13121 | cytochrome P450, family 51 | 2.0 | 0.002 |
| Dhrs3 | 20148 | dehydrogenase/reductase (SDR family) member 3 | −2.0 | 0.003 |
| Dhrs9 | 241452 | dehydrogenase/reductase (SDR family) member 9 | 3.6 | 0.001 |
| Dpyd | 99586 | dihydropyrimidine dehydrogenase | −3.8 | 0 |
| Fmo3 | 14262 | flavin containing monooxygenase 3 | −76.9 | 0.001 |
| Maoa | 17161 | monoamine oxidase A | 6.3 | 0.008 |
| Maob | 109731 | monoamine oxidase B | −2.4 | 0.001 |
| Ptgs1 | 19224 | prostaglandin-endoperoxide synthase 1 | −2.5 | 0.004 |
| Phase II | ||||
| Acsl1 | 14081 | acyl-CoA synthetase long-chain family member 1 | −3.1 | 0.005 |
| Acsl4 | 50790 | acyl-CoA synthetase long-chain family member 4 | 5.2 | 0 |
| Ces2 | 234671 | carboxylesterase 2 | 24.7 | 0 |
| Ephx1 | 13849 | epoxide hydrolase 1, microsomal | 2.5 | 0.002 |
| Gsta2 | 14858 | glutathione S-transferase, alpha 2 (Yc2) | 34.6 | 0.001 |
| Gsta4 | 14860 | glutathione S-transferase, alpha 4 | 2.6 | 0.006 |
| Gstk1 | 76263 | glutathione S-transferase kappa 1 | −2.2 | 0.004 |
| Gstm1 | 14862 | glutathione S-transferase, mu 1 | 2.8 | 0.002 |
| Gstm2 | 14863 | glutathione S-transferase, mu 2 | 4.4 | 0.004 |
| Gstm3 | 14864 | glutathione S-transferase, mu 3 | 11.5 | 0 |
| Gstm4 | 14865 | glutathione S-transferase, mu 4 | 4.4 | 0.007 |
| Gstm6 | 14867 | glutathione S-transferase, mu 6 | 5.2 | 0.002 |
| Gsto1 | 14873 | glutathione S-transferase omega 1 | −4.7 | 0.003 |
| Gstp1 | 14870 | glutathione S-transferase, pi 1 | 4.4 | 0 |
| Nnmt | 18113 | nicotinamide N-methyltransferase | −10.4 | 0 |
| Nqo3a2 | 72017 | cytochrome b5 reductase 1 | 3.2 | 0 |
| Sult1d1 | 53315 | sulfotransferase family 1D, member 1 | −2.7 | 0.006 |
| Sult3a1 | 57430 | sulfotransferase family 3A, member 1 | −35.7 | 0 |
| Phase III | ||||
| Abcb1a | 18671 | ATP-binding cassette, subfamily B (MDR/TAP), member 1A | 2.3 | 0.004 |
| Abcc12 | 244562 | ATP-binding cassette, subfamily C (CFTR/MRP), member 12 | 5.5 | 0.009 |
| Abcc2 | 12780 | ATP-binding cassette, subfamily C (CFTR/MRP), member 2 | 3.1 | 0.004 |
| Abcc4 | 239273 | ATP-binding cassette, subfamily C (CFTR/MRP), member 4 | 18.9 | 0.001 |
| Abcc5 | 27416 | ATP-binding cassette, subfamily C (CFTR/MRP), member 5 | 7.5 | 0.001 |
| Abcc6 | 27421 | ATP-binding cassette, subfamily C (CFTR/MRP), member 6 | −3.0 | 0.006 |
| Abcd3 | 19299 | ATP-binding cassette, subfamily D (ALD), member 3 | −2.3 | 0.006 |
| Aqp6 | 11831 | aquaporin 6 | 2.2 | 0.003 |
| Aqp7 | 11832 | aquaporin 7 | 17.8 | 0.001 |
| Mvp | 78388 | major vault protein | 2.7 | 0 |
| Slc10a1 | 20493 | solute carrier family 10 (sodium/bile acid cotransporter family), member 1 | −4.4 | 0.002 |
| Slc22a3 | 20519 | solute carrier family 22 (organic cation transporter), member 3 | 10.2 | 0.004 |
| Slc22a7 | 108114 | solute carrier family 22 (organic anion transporter), member 7 | −4.4 | 0.005 |
| Slc29a1 | 63959 | solute carrier family 29 (nucleoside transporters), member 1 | −2.4 | 0.005 |
| Slc2a1 | 20525 | solute carrier family 2 (facilitated glucose transporter), member 1 | 2.0 | 0.002 |
| Slc3a1 | 20532 | solute carrier family 3, member 1 | −4.2 | 0.009 |
| Slc7a5 | 20539 | solute carrier family 7 (cationic amino acid transporter, y+ system), member 5 | 3.3 | 0.002 |
Table 4.
Numbers of Changed Phase I, Phase II, and Phase III Genes after 2,000 mg/kg GBE Treatment
| Genes presented on array | Genes altered by GBE treatment (%) | |
|---|---|---|
| Phase I | 154 | 33 (21%) |
| Phase II | 87 | 18 (21%) |
| Phase III | 72 | 17 (24%) |
| Total drug metabolizing genes | 313 | 68 (22%) |
Real-time polymerase chain reaction (PCR) validation
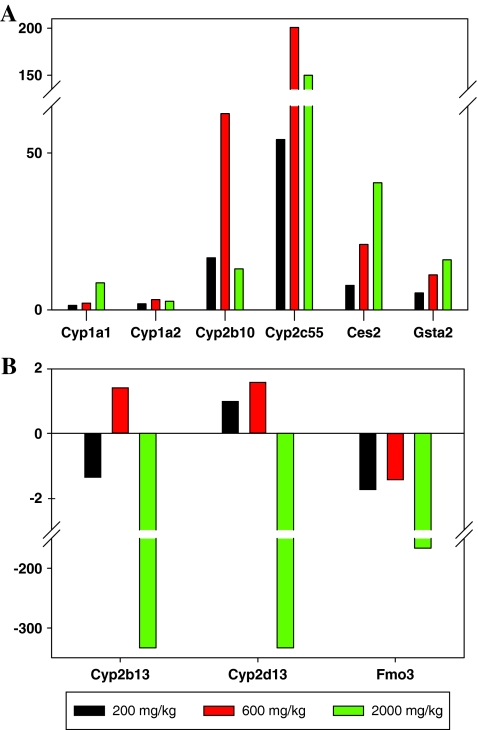
TaqMan assays were used to verify the results for a selected group of genes whose expression changes were seen using microarrays. Nine drug metabolizing genes were selected for the TaqMan validation. The results are shown in Figure 5A. Based on triplicate measurements for each RNA sample, the genes Cyp1a1, Cyp1a2, Cyp2b10, Cyp2c55, Ces2, and Gsta2 were overexpressed, and the changes in Cyp1a1, Ces2, and Gsta2 were dose dependent. The expression of Cyp2b13, Cyp2d13, and Fmo3 genes were decreased, notably for the highest dose treatment (Fig. 5B). Over 100-fold decrease in gene expression was observed for the three downregulated genes in the 2,000 mg/kg treatment group. As expected, TaqMan data were in good agreement with the microarray data (Fig. 5 and Table 3).
FIG. 5.
TaqMan assays of the gene expression changes; (A) Cyp1a1, Cyp1a2, Cyp2b10, Cyp2c55, Ces2, and Gsta2; and (B) Cyp2d13; Cyp2b13, and Fmo3.
Pathway analysis
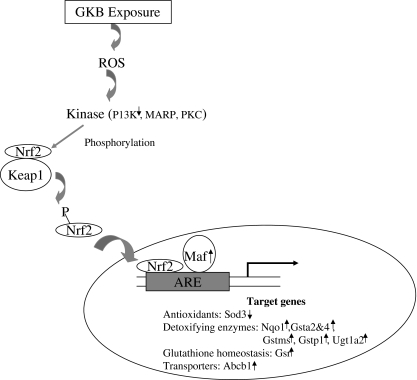
IPA (version 7.0) was used to determine the most relevant biological functions, pathways, and networks of the genes altered by 2,000 mg/kg GBE treatment. Out of 2,011 DEGs, 1,246 were identified by IPA and were overlaid onto a global molecular network developed from information contained in the database. Not to our surprise, the top canonical pathway was “metabolism of xenobiotics by cytochrome P450” with 36 genes involved and a P-value of 1.26E-11 followed by “fatty acid metabolism” (Table 5 and Table 6). Interestingly, NRF2 (nuclear factor erythroid-related factor 2)-mediated oxidative stress response was identified with 23 genes involved and a P-value of 5.37E-03. In the pathway regulated by NRF2, most of the genes were upregulated, whereas two genes were downregulated (Fig. 6). The upregulated genes included those coding for detoxifying enzymes (Nqo1, Gstms, Gstas, Gstp1, and Ugt1a2), glutathione homeostasis (Gsr), and transporter (Abcb1). Maf2, a transcriptional factor forms heterodimers with Nrf2, was also upregulated. The two downregulated genes were P13k, which plays a role in Nrf2 phosphorylation, and Sod3, which is a family member of antioxidants. Nqo1 showed a prominent change in expression, exhibiting 7.5- and 11-fold induction for 600 and 2,000 mg/kg treatments, respectively.
Table 5.
Top 20 Canonical Pathways Altered by GBE Treatment
| Canonical pathway | p-Value | |
|---|---|---|
| 1 | Metabolism of xenobiotics by Cytochrome P450 | 1.26E−11 |
| 2 | Fatty acid metabolism | 4.57E−09 |
| 3 | Tryptophan metabolism | 1.74E−07 |
| 4 | LPS/IL-1 mediated inhibition of RXR function | 2.04E−07 |
| 5 | Arachidonic acid metabolism | 2.57E−07 |
| 6 | Linoleic acid metabolism | 3.09E−06 |
| 7 | Aryl hydrocarbon receptor signaling | 9.77E−06 |
| 8 | Xenobiotic metabolism signaling | 1.91E−04 |
| 9 | Glutathione metabolism | 4.57E−04 |
| 10 | Aminosugars metabolism | 1.38E−03 |
| 11 | PXR/RXR activation | 1.74E−03 |
| 12 | NRF2-mediated oxidative stress response | 5.37E−03 |
| 13 | Hepatic fibrosis/hepatic stellate cell activation | 1.07E−02 |
| 14 | Tyrosine metabolism | 1.35E−02 |
| 15 | Acute phase response signaling | 2.29E−02 |
| 16 | FXR/RXR activation | 2.40E−02 |
| 17 | Sphingolipid metabolism | 3.47E−02 |
| 18 | G-Protein coupled receptor signaling | 4.68E−02 |
| 19 | Coagulation system | 4.79E−02 |
| 20 | Glycosaminoglycan degradation | 5.89E−02 |
Table 6.
Canonical Pathways Altered by GBE Treatment
| Ingenuity canonical pathways | −Log (p-value) | Ratio |
|---|---|---|
| Metabolism of xenobiotics by cytochrome P450 | 1.09E+01 | 1.67E−01 |
| Fatty acid metabolism | 8.34E+00 | 1.64E−01 |
| Tryptophan metabolism | 6.76E+00 | 1.21E−01 |
| LPS/IL-1 mediated inhibition of RXR function | 6.69E+00 | 1.84E−01 |
| Arachidonic acid metabolism | 6.59E+00 | 1.31E−01 |
| Linoleic acid metabolism | 5.51E+00 | 1.68E−01 |
| Aryl hydrocarbon receptor signaling | 5.01E+00 | 1.71E−01 |
| Xenobiotic metabolism signaling | 3.72E+00 | 1.40E−01 |
| Glutathione metabolism | 3.34E+00 | 1.35E−01 |
| Aminosugars metabolism | 2.86E+00 | 1.06E−01 |
| PXR/RXR activation | 2.76E+00 | 1.51E−01 |
| NRF2-mediated oxidative stress response | 2.27E+00 | 1.28E−01 |
| Hepatic fibrosis/hepatic stellate cell activation | 1.97E+00 | 1.30E−01 |
| Tyrosine metabolism | 1.87E+00 | 5.41E−02 |
| Acute phase response signaling | 1.64E+00 | 1.16E−01 |
| FXR/RXR activation | 1.62E+00 | 1.22E−01 |
| Sphingolipid metabolism | 1.46E+00 | 9.78E−02 |
| G-Protein coupled receptor signaling | 1.33E+00 | 1.06E−01 |
| Coagulation system | 1.32E+00 | 1.71E−01 |
| Glycosaminoglycan degradation | 1.23E+00 | 8.20E−02 |
| Axonal guidance signaling | 1.20E+00 | 9.07E−02 |
| Glycerolipid metabolism | 1.17E+00 | 7.64E−02 |
| Glycine, serine, and threonine metabolism | 1.16E+00 | 6.21E−02 |
| One carbon pool by folate | 1.14E+00 | 1.05E−01 |
| Cell cycle: G2/M DNA damage checkpoint regulation | 1.13E+00 | 1.40E−01 |
| C21-steroid hormone metabolism | 1.08E+00 | 5.71E−02 |
| Urea cycle and metabolism of amino groups | 1.07E+00 | 6.25E−02 |
| Fructose and mannose metabolism | 1.03E+00 | 5.04E−02 |
| Neuregulin signaling | 9.93E−01 | 1.10E−01 |
| Phenylalanine metabolism | 9.82E−01 | 4.67E−02 |
| Nitrogen metabolism | 9.67E−01 | 4.51E−02 |
| PPAR signaling | 9.67E−01 | 1.05E−01 |
| PTEN signaling | 9.42E−01 | 1.09E−01 |
| IL-6 signaling | 9.42E−01 | 1.10E−01 |
| LXR/RXR activation | 9.41E−01 | 9.88E−02 |
| Hepatic cholestasis | 9.31E−01 | 8.59E−02 |
| Nucleotide sugars metabolism | 8.31E−01 | 3.08E−02 |
| Glutamate metabolism | 8.24E−01 | 6.41E−02 |
| IL-10 signaling | 8.14E−01 | 1.03E−01 |
| Galactose metabolism | 7.99E−01 | 5.36E−02 |
| Starch and sucrose metabolism | 7.81E−01 | 5.21E−02 |
| Propanoate metabolism | 7.60E−01 | 5.56E−02 |
| Arginine and proline metabolism | 7.57E−01 | 4.49E−02 |
| NF-kB signaling | 7.49E−01 | 9.79E−02 |
| cAMP-mediated signaling | 7.27E−01 | 9.43E−02 |
| Glycolysis/gluconeogenesis | 7.20E−01 | 7.09E−02 |
| Purine metabolism | 7.17E−01 | 6.22E−02 |
| Glyoxylate and dicarboxylate metabolism | 7.11E−01 | 2.63E−02 |
| Bile acid biosynthesis | 6.93E−01 | 5.21E−02 |
| Beta-alanine metabolism | 6.87E−01 | 6.06E−02 |
| Valine, leucine, and isoleucine degradation | 6.86E−01 | 6.54E−02 |
| Histidine metabolism | 6.36E−01 | 4.50E−02 |
| Glutamate receptor signaling | 6.13E−01 | 8.96E−02 |
| Complement system | 6.12E−01 | 1.11E−01 |
| Retinol metabolism | 6.10E−01 | 7.94E−02 |
| Nitric oxide signaling in the cardiovascular system | 5.99E−01 | 8.24E−02 |
| Toll-like receptor signaling | 5.84E−01 | 9.80E−02 |
| PDGF signaling | 5.78E−01 | 9.46E−02 |
| Cell cycle: G1/S checkpoint regulation | 5.69E−01 | 1.00E−01 |
| Methionine metabolism | 5.56E−01 | 3.95E−02 |
| Apoptosis signaling | 5.36E−01 | 9.20E−02 |
| Taurine and hypotaurine metabolism | 5.35E−01 | 4.26E−02 |
| Phenylalanine, tyrosine, and tryptophan biosynthesis | 5.35E−01 | 3.08E−02 |
| Amyotrophic lateral sclerosis signaling | 5.03E−01 | 7.77E−02 |
| B cell receptor signaling | 4.46E−01 | 8.11E−02 |
| O-Glycan biosynthesis | 4.39E−01 | 6.98E−02 |
| VDR/RXR activation | 4.24E−01 | 8.75E−02 |
| Lysine degradation | 4.20E−01 | 4.17E−02 |
| Sonic Hedgehog signaling | 4.14E−01 | 9.68E−02 |
| Selenoamino acid metabolism | 4.14E−01 | 4.35E−02 |
| Biosynthesis of steroids | 4.14E−01 | 2.36E−02 |
| Ascorbate and aldarate netabolism | 3.63E−01 | 2.44E−02 |
| GABA receptor signaling | 3.58E−01 | 7.55E−02 |
| Pyrimidine metabolism | 3.52E−01 | 5.26E−02 |
| TR/RXR activation | 3.43E−01 | 7.61E−02 |
| Serotonin receptor signaling | 3.29E−01 | 6.52E−02 |
| GM-CSF signaling | 3.22E−01 | 8.06E−02 |
| Ceramide signaling | 3.21E−01 | 7.59E−02 |
| Pantothenate and CoA biosynthesis | 3.13E−01 | 3.17E−02 |
| p53 signaling | 2.97E−01 | 8.05E−02 |
| N-Glycan biosynthesis | 2.81E−01 | 4.60E−02 |
| SAPK/JNK signaling | 2.77E−01 | 7.53E−02 |
| Glycosphingolipid biosynthesis—ganglioseries | 2.71E−01 | 3.45E−02 |
| Glycosphingolipid biosynthesis—globoseries | 2.71E−01 | 4.88E−02 |
| Pentose and glucuronate interconversions | 2.67E−01 | 2.68E−02 |
| Nicotinate and nicotinamide metabolism | 2.67E−01 | 5.43E−02 |
| TGF-beta signaling | 2.64E−01 | 7.23E−02 |
| Fatty acid biosynthesis | 2.60E−01 | 2.04E−02 |
| Amyloid processing | 2.55E−01 | 7.69E−02 |
| Pentose phosphate pathway | 2.47E−01 | 3.37E−02 |
FIG. 6.
Alteration of NRF2 (nuclear factor erythroid-related factor 2)-mediated oxidative stress response.
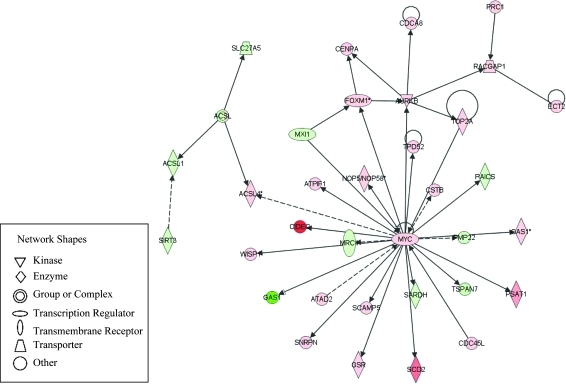
The significantly changed genes after GBE administration were mapped into 87 networks. Each network was associated with specific genes and involved different functions. Twenty-three networks had 20 or more focus molecules. The first network, which contains the largest number of genes (34 genes), with functions related to “cell cycle, cellular movement and cancer,” is illustrated in Figure 7. The central gene in this network is Myc, the pro-oncogene, which was upregulated by about sixfold. Twenty-one genes were found to be associated directly with Myc and these genes function in cell cycle, cell growth/proliferation, and cell death processes. Out of these 21 Myc-associated genes, 15 were upregulated and 6 were downregulated. Notably, Cidec (cell death-inducing DFFA-like effectors c), which promotes apoptosis, exhibited a 47-fold induction. Known molecules of DNA replication checkpoint (Cdca8 and Cdc45l) were also upregulated. Gene Gas1 (growth arrest-specific 1), the tumor suppressor gene that blocks entry to S phase, and prevents cycling of cells, was remarkably downregulated by 20-fold.
FIG. 7.
The first network (cell cycle, cellular movement, and cancer) containing 34 differentially expressed genes. Green indicates downregulation and red indicates upregulation. Solid lines indicate direct interactions and dashed lines indicate indirect interactions between two molecules.
Discussion
Currently there is no established methodology for determining the mechanisms of toxicity (particularly tumorigenicity) induced by a mixture containing many chemical components such as GBE and other herbal plant extracts. Previously, we have proposed that DNA microarray analysis should be a highly practical initial approach for revealing the whole spectrum of gene expression alterations by a chemical mixture (Guo et al., 2009). Microarray technology is a useful tool to rapidly detect the induction/inhibition of drug-metabolizing enzymes after toxicant treatment, and it has greatly contributed to the understanding of drug metabolizing enzyme function and expression (Blomme et al., 2009; Rezen et al., 2007). Although levels of gene expression do not fully represent the levels of enzyme activities, investigations at the gene expression level have revealed that there is a high degree of correlation for Phase I enzymes between the fold inductions of the enzymatic activity and mRNA expression in liver samples (Iyer and Sinz, 1999; Roymans et al., 2004).
In this study, we analyzed gene expression changes in the livers of B6C3F1 mice treated chronically with 200, 600, and 2,000 mg/kg of GBE for 2 years using a genome-wide microarray approach. A total of 31,802 probes covering 29,922 verified mouse genes were analyzed, and three technical replicate arrays were performed for each RNA samples. The results of HCA (Fig. 1), Pearson's correlation coefficient of pairwise log2 intensity correlation (Table 1), and scatter plotting of the overall gene expression profiles (Fig. 2) demonstrated the high quality and reproducibility of DNA microarray data obtained from the three technical replicates.
When the gene expression results of GBE-treated rats were compared to the controls, a total of 2,011 genes in the 2,000 mg/kg treatment group were differentially expressed, of which there were 68 drug-metabolizing enzyme-associated genes, with 33, 18, and 17 genes belonging to Phase I, Phase II, and Phase III, respectively (Table 3). It is worth noting that 23 out of the 33 expressed Phase I metabolism associated genes were the Cyps, in which 2 genes belong to the Cyp1 superfamily (Cyp1a1 and 1a2), 15 genes belong to the Cyp2 superfamily, and 2 genes belong to the Cyp3 superfamily (Table 3). The Cyp1, Cyp2, and Cyp3 superfamilies are the most important metabolizing enzymes in the metabolism of drugs and the metabolic activation of toxic and carcinogenic xenobiotics (Gonzalez and Gelboin, 1994; Gonzalez and Yu, 2006).
One of the most reported adverse effects caused by herbal products is hepatotoxicity. Although the mechanism of hepatotoxicity initiated by herbal products is not clear, modulation of drug-metabolizing enzymes, leading to herb–drug interactions, is probably one of the major causes (Clayton et al., 2007; Clouatre, 2004; Guo et al., 2009). The alteration of a number of drug-metabolizing enzymes by GBE treatment (Table 3) could potentially lead to herb–drug interactions as well as changes in the metabolic activation of carcinogenic chemicals when concomitantly present in the body. Our results are consistent with the report that of Ginkgo biloba modulated Cyp 450 enzyme activity, and consequently may produce Cyp-mediated herb–drug interactions (Dubey et al., 2004; Gurley et al., 2004, 2005; Zou et al., 2002).
It is well established that the Cyp1 superfamily is important in the metabolism of xenobiotics, and the members of Cyp2 and Cyp3 superfamilies catalyze metabolism of drugs and other substances. Cyp1a1, which generally exists in a very low amount in the adult liver (Martignoni et al., 2006), was dramatically induced by GBE treatment, as detected by both the microarray analysis and TaqMan assay (Table 3 and Fig. 3A). Cyp1a1 and Cyp1a2 are regulated by AHR, the aryl hydrocarbon receptor, which plays an important role in cell cycle regulation, apoptosis, and development (Xu et al., 2005). It is evident that some toxins such as tumorigenic polycyclic aromatic hydrocarbons (PAHs) (Martignoni et al., 2006) and carcinogen 2,3,7,8-tetrachlorodibenzo-p-dioxin (TCDD) are able to induce Cyp1a1 through AHR, and the upregulation of Cyp1a1 is considered the indicator of TCDD-induced carcinogenesis (Brunnberg et al., 2006). Thus, induction of Cyp1a1 may contribute to the toxic mechanism of Ginkgo biloba, and/or may enhance metabolic activation of the hazardous tumorigenic xenobiotics and jeopardize health.
Besides Cyp, the flavin-containing monooxygenase (Fmo) is another major group of Phase I drug-metabolizing enzymes catalyzing the oxidative biotransformation of drugs. Fmo enzymes break down compounds that contain nitrogen, sulfur, or phosphorus. Fmo3, a major Fmo isoform, plays an important role in metabolizing drugs such as the anticancer drug tamoxifen, the pain medication codeine, the antifungal drug ketoconazole, and antipsychotic drugs clozapine and olanapine (Ciraulo et al., 2005; Klick and Hines, 2007; Parte and Kupfer, 2005). In our study, GBE inhibited the expression of Fmo3 remarkably with the high dose treatment (77-fold reduction detected by microarray and 166-fold reduction validated by TaqMan assay). These results suggest that downregulated expression of the Fmo3 enzyme by GBE potentially inhibits metabolism of certain drugs resulting in drug–drug or drug–food interactions, thus reducing the therapeutic effects or enhancing the toxicity induced by their metabolites.
It is worth noting that although the microarray studies on the changes of drug-metabolizing enzyme expression in kava-treated rats (Guo et al., 2009) and kava-treated mice (Guo et al., submitted for publication) used a 90-day treatment of kava, the present GBE study was performed over a 2-year period of treatment. The difference in time of herbal treatment suggests that the effect of such a dietary supplement on drug metabolizing enzymes is a persistent phenomenon (up to 2 years). Our previous study with kava and the present study with Ginkgo biloba provide substantial evidence that the two herbal dietary supplements cause remarkable changes in drug metabolizing enzymes, particularly the Cyp isozymes, raising the possibility that these herbal supplements might profoundly affect the pharmacokinetics of many coadministrated drugs or other food supplements, thus leading to hepatotoxicity.
In addition to the investigation of alteration for many drug metabolizing genes, in this study we also explored pathways and networks in response to GBE treatment and found that Ginkgo biloba exposure resulted in the significant stimulation of the Nrf2-mediated oxidative pathway (Fig. 6) and the alteration of the Myc-centered network (Fig. 7).
A previous study found that long-term exposure to kava could result in the perturbation of the Nrf2-mediated pathway and eventually generate reactive oxygen species (ROS). In this study, Ginkgo biloba treatments also stimulated the Nrf2-mediated oxidative stress response pathway (p = 0.005). Keap1–Nrf2–ARE signaling plays a critical role in protecting cells from endogenous and exogenous stresses, and is involved in antioxidative response, detoxification of xenobiotics, and proteome maintenance (Kensler et al., 2007). Under normal physiological conditions the transcription factor Nrf2 localizes in the cytoplasm and interacts with Keap1. Upon oxidative stress, Nrf2 is released from Keap1 and translocates to the nucleus and subsequently activates its various downstream target genes (Kensler et al., 2007). The target genes show a wide spectrum of functions, such as inactivating oxidants, increasing the levels of glutathione, and enhancing toxin export via transporters to enhance cell survival. As illustrated in Figure 6, in response to GBE treatment, Nrf2 regulated genes including various Gst genes which are involved in GSH-conjugate formation were altered. The Gst genes (Gsta2, Gsta4, Gstms, and Gstp1) and Ugt1a2 were all upregulated following GBE treatment. In turn, the resulting enhancement of GST enzymes is used for neutralizing the electrophiles, the process considered generally as an important detoxification mechanism. NADPH-dependent enzyme NQO1 was also upregulated, which has protective roles toward detoxification of xenobiotic carbonyls and quinones. GBE treatment also elevated Gsr (glutathione reductase) gene expression, and GSR regulates cellular GSH homeostasis by catalyzing the reduction of GSSG to GSH using NADPH as a reducing cofactor (Harvey et al., 2009) In addition, GBE treatment reduced the activity of Sod3, which is one of the superoxide dismutases (SODs) that are the most important line of antioxidant enzyme defense systems against ROS and particularly superoxide anion radicals (Zelko et al., 2002). Studies using Nrf2 knock-out mice showed that they are more susceptible to acetaminophen-induced hepatocellular injury (Chan et al., 2001) and benzo[a]pyrene-induced tumor formation with higher levels of DNA adducts (Ramos-Gomez et al., 2001). This susceptibility is partly due to a reduced level in the expression of detoxification enzymes (Aleksunes and Manautou, 2007; Kensler et al., 2007). Activation of detoxification enzymes plays a pivotal role in protecting cells from oxidative insult when cells encounter a toxin challenge.
Based on the network analysis, 34 genes were incorporated into the first network in response to GBE treatment (Fig. 7). The key molecule in this most significantly changed network was Myc pro-oncogene with 5.7-fold increased expression. The protein encoded by this gene is a multifunctional nuclear phosphoprotein that plays a role in cell cycle progression, apoptosis, and cellular transformation. It functions as a transcription factor that regulates transcription of specific target genes. It has been established that stimulation of Myc pathways can cause liver damage, leading to severe lesions up to the tumorigenic level. Overexpression of Myc has been frequently observed in tumors and is the early and critical event in the pathogenesis of hepatocellular carcinoma (Beer et al., 2008; Dang, 1999; Pelengaris et al., 2002). A study using a transgenic model indicated that overexpression of the c-Myc gene sufficiently induced hepatic proliferation and tumorigenesis (Beer et al., 2008; Sandgren et al., 1989). Many studies have shown that Myc gene overexpression during hepatocarcinogenesis induced by carcinogens including choline-devoid diet, arsenite, 5-diethoxycarbonyl-1,4-dihydrocollidine (DDC), and carbon tetrachloride (CCl4) (Beer et al., 2008; Chandar et al., 1989; Chen et al., 2001). In addition, DDC and carbon tetrachloride cooperate with Myc to promote hepatocyte proliferation and rapidly uncover the onset of liver cancer (Beer et al., 2008). Using a toxicogenomic approach to study the molecular mechanisms of nongenotoxic carcinogenicity revealed that major pathways linked to cancer were interconnected via Myc (Nie et al., 2006), and the high percentage of gene expression signature linked to Myc suggested the important role of the Myc gene. Therefore, the alteration of Myc-mediated pathways may serve as potential predictors in carcinogenicity. Among 34 genes incorporated into the Myc-centered network (Fig. 7), over 60% of the genes (21 genes) were reported to be directly associated with Myc and most of these genes function in cell cycle, cell growth/proliferation, and cell death processes. The Myc associated gene, Gas1 (growth arrest-specific 1) was found to be 20-fold downregulated. Gas1, the tumor suppressor gene, which plays role in growth suppression by blocking entry to S phase, is directly inhibited at the transcriptional level by Myc (Lee et al., 1997), and the transcriptional repression of growth arrest genes is one step in the promotion of cell growth. Downregulation was also one of the distinct events in the process of EGF (epidermal growth factor)-induced hepatocarcinogenesis (Borlak et al., 2005). The loss of growth control through Gas1 may be a necessary event in the multi-step neoplastic transformation.
In summary, we analyzed gene expression profiles in the liver of mice treated with Ginkgo biloba, a widely sold product in health food stores in the United States. Using the systematic approaches of HCA, PCA, pathway analysis, and network analysis, we identified 129, 289, and 2,011 genes that were differentially expressed in the livers of 300, 600, and 2,000 mg/kg GBE-treated mice, respectively, and observed that a significant portion of the changed genes were associated with drug metabolism, Nrf2-mediated oxidative stress, and the Myc gene-centered network. Based on our results, we speculate that long-term exposure of Ginkgo biloba may cause hepatic damage, including severe liver lesions and even liver tumors. Further studies are warranted.
Acknowledgments
We thank Drs. Tucker A. Patterson and James C. Fuscoe for their critical review of this manuscript. This article is not an official guidance or policy statement of the National Toxicology Program (NTP) or U.S. Food and Drug Administration (FDA). No official support or endorsement by the FDA or NTP is intended or should be inferred. This research was supported in part by the Intramural Research Program of the NIH, National Institute of Environmental Health Sciences.
Author Disclosure Statement
The authors declare that there is no conflict of interest.
References
- Aleksunes L.M. Manautou J.E. Emerging role of Nrf2 in protecting against hepatic and gastrointestinal disease. Toxicol Pathol. 2007;35:459–473. doi: 10.1080/01926230701311344. [DOI] [PubMed] [Google Scholar]
- Beer S. Komatsubara K. Bellovin D.I. Kurobe M. Sylvester K. Felsher D.W. Hepatotoxin-induced changes in the adult murine liver promote MYC-induced umorigenesis. PLoS ONE. 2008;3:e2493. doi: 10.1371/journal.pone.0002493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blomme E.A. Yang Y. Waring J.F. Use of toxicogenomics to understand mechanisms of drug-induced hepatotoxicity during drug discovery and development. Toxicol Lett. 2009;186:22–31. doi: 10.1016/j.toxlet.2008.09.017. [DOI] [PubMed] [Google Scholar]
- Borlak J. Meier T. Halter R. Spanel R. Spanel-Borowski K. Epidermal growth factor-induced hepatocellular carcinoma: gene expression profiles in precursor lesions, early stage and solitary tumours. Oncogene. 2005;24:1809–1819. doi: 10.1038/sj.onc.1208196. [DOI] [PubMed] [Google Scholar]
- Bressler R. Herb–drug interactions: interactions between kava and prescription medications. Geriatrics. 2005;60:24–25. [PubMed] [Google Scholar]
- Brunnberg S. Andersson P. Lindstam M. Paulson I. Poellinger L. Hanberg A. The constitutively active Ah receptor (CA-Ahr) mouse as a potential model for dioxin exposure—effects in vital organs. Toxicology. 2006;224:191–201. doi: 10.1016/j.tox.2006.04.045. [DOI] [PubMed] [Google Scholar]
- Chan K. Han X.D. Kan Y.W. An important function of Nrf2 in combating oxidative stress: detoxification of acetaminophen. Proc Natl Acad Sci USA. 2001;98:4611–4616. doi: 10.1073/pnas.081082098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan P.C. Fu P.P. Toxicity of Panax ginseng-An herbal medicine and dietary supplements. J Food Drug Anal. 2007;15:416–427. [Google Scholar]
- Chan P.C. Xia Q. Fu P.P. Ginkgo biloba leave extract: biological, medicinal, and toxicological effects. J Environ Sci Health C Environ Carcinog Ecotoxicol Rev. 2007;25:211–244. doi: 10.1080/10590500701569414. [DOI] [PubMed] [Google Scholar]
- Chandar N. Lombardi B. Locker J. c-Myc gene amplification during hepatocarcinogenesis by a choline-devoid diet. Proc Natl Acad Sci USA. 1989;86:2703–2707. doi: 10.1073/pnas.86.8.2703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen H. Liu J. Zhao C.Q. Diwan B.A. Merrick B.A. Waalkes M.P. Association of c-myc overexpression and hyperproliferation with arsenite-induced malignant transformation. Toxicol Appl Pharmacol. 2001;175:260–268. doi: 10.1006/taap.2001.9253. [DOI] [PubMed] [Google Scholar]
- Ciraulo D.A. Shader R.I. Greenblatt D.J. Creelman W. Drug Interaction in Psychiatry. Lippincott Williams & Wilkins; Philaldephia, PA: 2005. [Google Scholar]
- Clayton N.P. Yoshizawa K. Kissling G.E. Burka L.T. Chan P.C. Nyska A. Immunohistochemical analysis of expressions of hepatic cytochrome P450 in F344 rats following oral treatment with kava extract. Exp Toxicol Pathol. 2007;58:223–236. doi: 10.1016/j.etp.2006.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clouatre D.L. Kava kava: examining new reports of toxicity. Toxicol Lett. 2004;150:85–96. doi: 10.1016/j.toxlet.2003.07.005. [DOI] [PubMed] [Google Scholar]
- Dang C.V. c-Myc target genes involved in cell growth, apoptosis, and metabolism. Mol Cell Biol. 1999;19:1–11. doi: 10.1128/mcb.19.1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dubey A.K. Shankar P.R. Upadhyaya D. Deshpande V.Y. Ginkgo biloba—an appraisal. Kathmandu Univ Med J. 2004;2:225–229. [PubMed] [Google Scholar]
- FDA. FDA advises dietary supplement manufacturers to remove comfrey products from the market. 2001. Jul 6, 2001. http://www.cfsan.fda.gov/∼dms/supplmnt.html http://www.cfsan.fda.gov/∼dms/supplmnt.html
- FDA. FDA Public Health News, FDA announced major initiatives for dietary supplements. 2004a. Nov 5, 2004.
- FDA. Final rule declaring dietary supplements containing ephedrine alkaloids adulterated because they present an unreasonable risk federal register. 2004. Feb 11, 2004. [PubMed]
- Fong H.H. Integration of herbal medicine into modern medical practices: issues and prospects. Integr Cancer Ther. 2002;1:287–293. doi: 10.1177/153473540200100313. [DOI] [PubMed] [Google Scholar]
- Fu P.P. Quality assurance and safety of herbal dietary supplements. J Food Drug Anal. 2007;15:333–334. [Google Scholar]
- Fu P.P. Yang Y.C. Xia Q. Chou M.W. Cui Y.Y. Lin G. Pyrrolizidine alkaloids—tumorigenic components in Chinese herbal medicines and dietary supplements. J Food Drug Anal. 2002;10:198–211. [Google Scholar]
- Fu P.P. Xia Q. Chou M.W. Lin G. Detection, hepatotoxicity, and tumorigenicity of pyrrolizidine alkaloids in Chinese herbal plants and herbal dietary supplemnets. J Food Drug Anal. 2007;15:400–415. [Google Scholar]
- Fu P.P. Xia Q. Guo L. Yu H. Chan P.C. Toxicity of kava kava. J Environ Sci Health Part C. 2008;26:89–112. doi: 10.1080/10590500801907407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonzalez F.J. Gelboin H.V. Role of human cytochromes P450 in the metabolic activation of chemical carcinogens and toxins. Drug Metab Rev. 1994;26:165–183. doi: 10.3109/03602539409029789. [DOI] [PubMed] [Google Scholar]
- Gonzalez F.J. Yu A.M. Cytochrome P450 and xenobiotic receptor humanized mice. Annu Rev Pharmacol Toxicol. 2006;46:41–64. doi: 10.1146/annurev.pharmtox.45.120403.100007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo L. Lobenhofer E.K. Wang C. Shippy R. Harris S.C. Zhang L., et al. Rat toxicogenomic study reveals analytical consistency across microarray platforms. Nat Biotechnol. 2006;24:1162–1169. doi: 10.1038/nbt1238. [DOI] [PubMed] [Google Scholar]
- Guo L. Li Q. Xia Q. Dial S. Chan P.C. Fu P. Analysis of gene expression changes of drug metabolizing enzymes in the livers of F344 rats following oral treatment with kava extract. Food Chem Toxicol. 2009;47:433–442. doi: 10.1016/j.fct.2008.11.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gurley B.J. Gardner S.F. Hubbard M.A. Williams D.K. Gentry W.B. Carrier J., et al. In vivo assessment of botanical supplementation on human cytochrome P450 phenotypes: Citrus aurantium, Echinacea purpurea, milk thistle, and saw palmetto. Clin Pharmacol Ther. 2004;76:428–440. doi: 10.1016/j.clpt.2004.07.007. [DOI] [PubMed] [Google Scholar]
- Gurley B.J. Gardner S.F. Hubbard M.A. Williams D.K. Gentry W.B. Cui Y., et al. Clinical assessment of effects of botanical supplementation on cytochrome P450 phenotypes in the elderly: St John's wort, garlic oil, Panax ginseng and Ginkgo biloba. Drugs Aging. 2005;22:525–539. doi: 10.2165/00002512-200522060-00006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gurley B.J. Swain A. Barone G.W. Williams D.K. Breen P. Yates C.R., et al. Effect of goldenseal (Hydrastis canadensis) and kava kava (Piper methysticum) supplementation on digoxin pharmacokinetics in humans. Drug Metab Dispos. 2007;35:240–245. doi: 10.1124/dmd.106.012708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harvey C.J. Thimmulappa R.K. Singh A. Blake D.J. Ling G. Wakabayashi N., et al. Nrf2-regulated glutathione recycling independent of biosynthesis is critical for cell survival during oxidative stress. Free Radic Biol Med. 2009;46:443–453. doi: 10.1016/j.freeradbiomed.2008.10.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu Z. Yang X. Ho P.C. Chan S.Y. Heng P.W. Chan E., et al. Herb–drug interactions: a literature review. Drugs. 2005;65:1239–1282. doi: 10.2165/00003495-200565090-00005. [DOI] [PubMed] [Google Scholar]
- Iyer K.R. Sinz M.W. Characterization of Phase I and Phase II hepatic drug metabolism activities in a panel of human liver preparations. Chem Biol Interact. 1999;118:151–169. doi: 10.1016/s0009-2797(99)00007-1. [DOI] [PubMed] [Google Scholar]
- Kensler T.W. Wakabayashi N. Biswal S. Cell survival responses to environmental stresses via the Keap1–Nrf2–ARE pathway. Annu Rev Pharmacol Toxicol. 2007;47:89–116. doi: 10.1146/annurev.pharmtox.46.120604.141046. [DOI] [PubMed] [Google Scholar]
- Klick D.E. Hines R.N. Mechanisms regulating human FMO3 transcription. Drug Metab Rev. 2007;39:419–442. doi: 10.1080/03602530701498612. [DOI] [PubMed] [Google Scholar]
- Lee T.C. Li L. Philipson L. Ziff E.B. Myc represses transcription of the growth arrest gene gas1. Proc Natl Acad Sci USA. 1997;94:12886–12891. doi: 10.1073/pnas.94.24.12886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Livak K.J. Schmittgen T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2(−Delta Delta C(T)) Method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- Martignoni M. Groothuis G.M. De Kanter R. Species differences between mouse, rat, dog, monkey and human CYP-mediated drug metabolism, inhibition and induction. Expert Opin Drug Metab Toxicol. 2006;2:875–894. doi: 10.1517/17425255.2.6.875. [DOI] [PubMed] [Google Scholar]
- Matthias A. Blanchfield J.T. Penman K.G. Bone K.M. Toth I. Lehmann R.P. Permeability studies of Kavalactones using a Caco-2 cell monolayer model. J Clin Pharm Ther. 2007;32:233–239. doi: 10.1111/j.1365-2710.2007.00810.x. [DOI] [PubMed] [Google Scholar]
- Nie A.Y. McMillian M. Parker J.B. Leone A. Bryant S. Yieh L., et al. Predictive toxicogenomics approaches reveal underlying molecular mechanisms of nongenotoxic carcinogenicity. Mol Carcinog. 2006;45:914–933. doi: 10.1002/mc.20205. [DOI] [PubMed] [Google Scholar]
- NTP. Nomination of Ginkgo biloba extract. 1998. http://ntp.niehs.nih.gov/?objectid=BDCABE80-123F-7908-7BF75015B4AC13D http://ntp.niehs.nih.gov/?objectid=BDCABE80-123F-7908-7BF75015B4AC13D
- Parte P. Kupfer D. Oxidation of tamoxifen by human flavin-containing monooxygenase (FMO) 1 and FMO3 to tamoxifen-N-oxide and its novel reduction back to tamoxifen by human cytochromes P450 and hemoglobin. Drug Metab Dispos. 2005;33:1446–1452. doi: 10.1124/dmd.104.000802. [DOI] [PubMed] [Google Scholar]
- Pelengaris S. Khan M. Evan G. c-MYC: more than just a matter of life and death. Nat Rev Cancer. 2002;2:764–776. doi: 10.1038/nrc904. [DOI] [PubMed] [Google Scholar]
- Ramos-Gomez M. Kwak M.K. Dolan P.M. Itoh K. Yamamoto M. Talalay P., et al. Sensitivity to carcinogenesis is increased and chemoprotective efficacy of enzyme inducers is lost in nrf2 transcription factor-deficient mice. Proc Natl Acad Sci USA. 2001;98:3410–3415. doi: 10.1073/pnas.051618798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rezen T. Contreras J.A. Rozman D. Functional genomics approaches to studies of the cytochrome p450 superfamily. Drug Metab Rev. 2007;39:389–399. doi: 10.1080/03602530701498760. [DOI] [PubMed] [Google Scholar]
- Roymans D. Van Looveren C. Leone A. Parker J.B. McMillian M. Johnson M.D., et al. Determination of cytochrome P450 1A2 and cytochrome P450 3A4 induction in cryopreserved human hepatocytes. Biochem Pharmacol. 2004;67:427–437. doi: 10.1016/j.bcp.2003.09.022. [DOI] [PubMed] [Google Scholar]
- Sandgren E.P. Quaife C.J. Pinkert C.A. Palmiter R.D. Brinster R.L. Oncogene-induced liver neoplasia in transgenic mice. Oncogene. 1989;4:715–724. [PubMed] [Google Scholar]
- Shi L. Reid L.H. Jones W.D. Shippy R. Warrington J.A. Baker S.C., et al. The MicroArray Quality Control (MAQC) project shows inter- and intraplatform reproducibility of gene expression measurements. Nat Biotechnol. 2006;24:1151–1161. doi: 10.1038/nbt1239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh Y.N. Potential for interaction of kava and St. John's wort with drugs. J Ethnopharmacol. 2005;100:108–113. doi: 10.1016/j.jep.2005.05.014. [DOI] [PubMed] [Google Scholar]
- Tusher V.G. Tibshirani R. Chu G. Significance analysis of microarrays applied to the ionizing radiation response. Proc Natl Acad Sci USA. 2001;98:5116–5121. doi: 10.1073/pnas.091062498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang A. Gehan E.A. Gene selection for microarray data analysis using principal component analysis. Stat Med. 2005;24:2069–2087. doi: 10.1002/sim.2082. [DOI] [PubMed] [Google Scholar]
- Williamson E.M. Interactions between herbal and conventional medicines. Expert Opin Drug Saf. 2005;4:355–378. doi: 10.1517/14740338.4.2.355. [DOI] [PubMed] [Google Scholar]
- Wold R.S. Lopez S.T. Yau C.L. Butler L.M. Pareo-Tubbeh S.L. Waters D.L., et al. Increasing trends in elderly persons' use of nonvitamin, nonmineral dietary supplements and concurrent use of medications. J Am Diet Assoc. 2005;105:54–63. doi: 10.1016/j.jada.2004.11.002. [DOI] [PubMed] [Google Scholar]
- Xu C. Li C.Y. Kong A.N. Induction of phase I, II and III drug metabolism/transport by xenobiotics. Arch Pharm Res. 2005;28:249–268. doi: 10.1007/BF02977789. [DOI] [PubMed] [Google Scholar]
- Zelko I.N. Mariani T.J. Folz R.J. Superoxide dismutase multigene family: a comparison of the CuZn–SOD (SOD1), Mn–SOD (SOD2), and EC–SOD (SOD3) gene structures, evolution, and expression. Free Radic Biol Med. 2002;33:337–349. doi: 10.1016/s0891-5849(02)00905-x. [DOI] [PubMed] [Google Scholar]
- Zou L. Harkey M.R. Henderson G.L. Effects of herbal components on cDNA-expressed cytochrome P450 enzyme catalytic activity. Life Sci. 2002;71:1579–1589. doi: 10.1016/s0024-3205(02)01913-6. [DOI] [PubMed] [Google Scholar]
- Zurer P. Hanson D. Chemistry puts herbal supplements to the test. Chemical & Engineering News. 2004;82:16. [Google Scholar]