Summary
Directed evolution studies often make use of water-in-oil compartments, which conventionally are prepared by bulk emulsification, a crude process that generates non-uniform droplets and can damage biochemical reagents. A microfluidic emulsification circuit was devised that generates uniform water-in-oil droplets (21.9 ± 0.8 μm radius) with high throughput (107–108 droplets per hour). The circuit contains a radial array of aqueous flow nozzles that intersect a surrounding oil flow channel. This device was used to evolve RNA enzymes with RNA ligase activity, selecting enzymes that could resist inhibition by neomycin. Each molecule in the population had the opportunity to undergo 108-fold selective amplification within its respective compartment. Then the progeny RNAs were harvested and used to seed new compartments. During five rounds of this procedure, the enzymes acquired mutations that conferred resistance to neomycin and caused some enzymes to become dependent on neomycin for optimal activity.
Introduction
Darwinian evolution involves the amplification and mutation of genetic information, coupled with the selection of corresponding phenotypic traits. In biological organisms this coupling is achieved through processes of transcription and translation, and by the co-localization of genes and gene products within a common cellular environment. In multicellular organisms phenotypic traits may be expressed at higher levels of organization, but still must be coupled to their corresponding genes through physical co-localization. The in vitro Darwinian evolution of molecules does not employ cells or organisms, but nonetheless must provide a way to couple genes and their corresponding traits. For certain functional nucleic acids the genotype and phenotype are embodied within the same molecule and compartmentalization is unnecessary. For other nucleic acids, however, the phenotype is expressed through the action of separate molecules, and it is necessary to devise a means for co-localizing the gene and its functional consequences.
The two most prominent approaches in directed evolution for coupling gene and trait are phage display (Scott and Smith, 1990; Smith and Petrenko, 1997) and in vitro compartmentalization (IVC) (Tawfik and Griffiths, 1998; Miller et al., 2006). In phage display, phage particles are assembled within a host cell, where a modified phage gene is packaged together with a corresponding modified phage coat protein. Phage particles are harvested based on the function of the modified coat protein, and the genes responsible for that function are recovered and amplified. This method is limited by the requirement to transform host cells, but nonetheless has proven very powerful in evolving peptides (Cwirla et al., 1990; Devlin et al., 1990), antibodies (McCafferty et al., 1990; Barbas et al., 1991), and protein enzymes (McCafferty et al., 1991; Soumillon et al., 1994) that can be displayed on the surface of phage. Related display methods have been devised for bacterial and eukaryotic cells (Francisco et al., 1993; Boder and Wittrup, 1997), ribosome particles (Hanes and Pluckthun, 1997), and mRNAs that are directly linked to their translation product (Roberts and Szostak, 1997).
IVC involves the production of artificial cell-like compartments, most commonly by forming a water-in-oil emulsion, where each water droplet contains one or a few genetic molecules together with their corresponding gene products. The droplets can be subjected to a high-throughput screen (e.g., by fluorescence-activated cell sorting) (Bernath et al., 2004; Aharoni et al., 2005; Mastrobattista et al., 2005) or selection based on modification of a co-encapsulated genetic element (Ghadessy et al., 2001; Lee et al., 2002). IVC has been applied primarily to the directed evolution of proteins, in which case the droplets must contain a gene and the entire transcription-translation machinery. IVC also has been used to evolve RNA enzymes, for example, that catalyze a Diels-Alder cycloaddition reaction with multiple turnover (Agresti et al., 2005), that ligate two separate RNA substrates (Levy et al., 2005), or that catalyze RNA polymerization on a separate template (Zaher and Unrau, 2007). In all these studies the water-in-oil emulsions were prepared in mayonnaise-like fashion, employing a chilled oil-surfactant mixture to which the aqueous components were slowly added while stirring continuously. This procedure has considerable variability, but can be mastered by a skilled practitioner. Even with best practices, however, the biochemical constituents of the mixture may become damaged due to the vigorous mixing that occurs during homogenization.
The yield of droplets from IVC is typically 107 μL−1, with a mean droplet diameter of 2 μm (30 fL volume). However, there is typically 5- to 10-fold variation in droplet diameter, with the degree of polydispersity depending on the oil-surfactant formulation and the stirring procedure (Miller et al., 2006). For certain applications, such as multiple-turnover reactions or compartmentalized replication, functional molecules that happen to reside in a larger droplet will enjoy a selective advantage because of the greater carrying capacity of their local environment.
Efforts have been made to employ microfluidic technology to produce water-in-oil emulsions that are highly uniform (Umbanhowar et al., 2000; Thorsen et al., 2001). The general approach is to cause a narrow aqueous stream (10–100 μm diameter) to intersect with an oil stream, relying on interfacial surface tension to break up the laminar flow and form individual droplets. However, even at flow rates of 0.1 μL min−1 (requiring 10–20 psi), the rate of production of 20 μm droplets is only ~105 droplets per hour (Umbanhowar et al., 2000). This would not be sufficient to maintain a sizeable population of evolving molecules that are contained within separate fluidic compartments. Recently, a variety of more sophisticated droplet generators have been described (for review, see Christopher and Anna, 2007), including devices that can operate with substantially higher throughput (Mazutis et al. 2009; Agresti et al. 2010; Zeng et al., 2010). However, the complicated procedures required to fabricate these devices may limit their widespread use in chemical biology applications. In addition, back pressure issues and topological constraints on the oil inputs make these circuits technically challenging to implement in arrayed formats.
An alternative method for fabricating circuits that generate microfluidic droplets employs soft lithography and does not require access to a cleanroom facility (Duffy et al., 1998). Devices of this kind have been used to conduct protein crystallization screens (Zheng et al., 2003), to synthesize semiconductor nanocrystals (Chan et al., 2005), and to culture cells and even multicellular organisms within microfluidic droplets (Clausell-Tormos et al., 2008). However, low throughput continues to be a challenge in applying this technology to IVC for the purposes of in vitro evolution experiments.
Here a microfluidic device is described that is easily fabricated using soft lithography and generates uniform water-in-oil droplets with high throughput and scalability. This device was used to carry out the compartmentalized directed evolution of an RNA enzyme, with multiple rounds of selective amplification occurring within individual droplets. As a selective pressure, neomycin was introduced at a concentration that inhibited the catalytic activity of the parental RNA enzymes. The evolving population developed resistance to the inhibitor through the acquisition of phenotypic traits that could be understood in terms of the structural properties of the RNA.
Results
A multichannel droplet generator was fabricated in polydimethylsiloxane (PDMS), which can be used to produce highly uniform water-in-oil emulsions for compartmentalized directed evolution. As shown in the circuit schematic (Figure 1A), the device consists of a circular nozzle array with 110 fluidic channels (20 μm wide, 20 μm step height) that fan out from a central aqueous input reservoir. The tips of the nozzles empty into an oil flow channel (500 μm wide, 150 μm step height) that is supplied by an oil-surfactant input reservoir. Droplets form at the nozzle tips and are swept into the oil phase for collection at the output reservoir. The oil phase, which is driven by a syringe pump, bifurcates at the entrance to the oil flow channel. No nozzles are present at the poles of the channel because there is lower shear at these positions, which would would yield non-uniform droplets.
Figure 1.
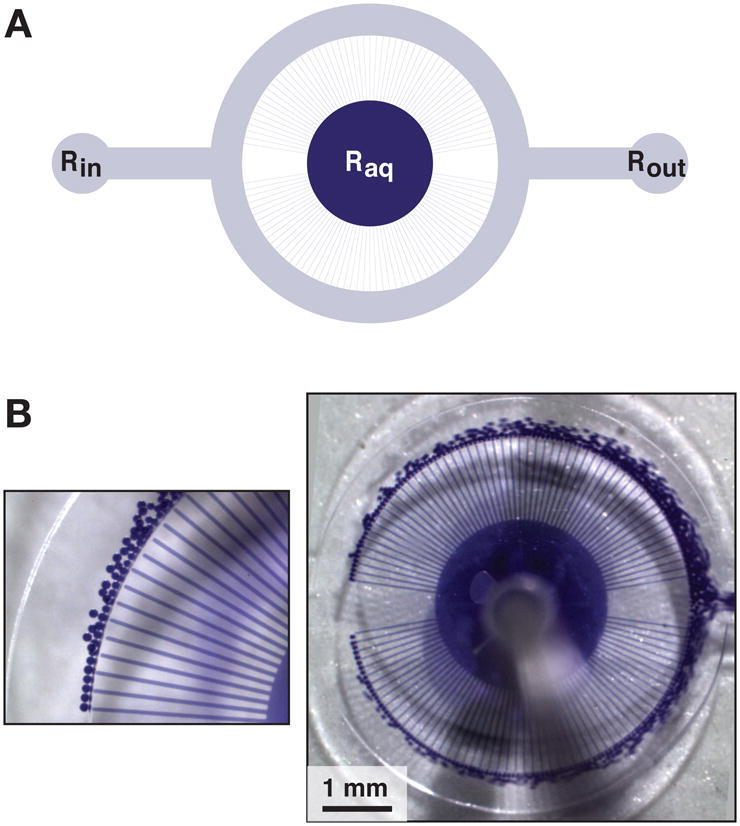
Microfluidic device for generating uniform water-in-oil emulsions. (A) Schematic of the device, showing central aqueous input reservoir (Raq) with 110 radial nozzles that empty into an annular oil flow channel. An oil-surfactant mixture is supplied at the oil input reservoir (Rin), and the emulsion is collected at the output reservoir (Rout). The reservoirs are contacted by microbore tubing, which delivers material to and from the device. (B) Photograph of the device during operation, with magnified view on the left. For visualization, the aqueous phase contained bromophenol blue in a solution of 15 mM MgCl2, 50 mM KCl, and 50 mM EPPS (pH 7.5).
Droplet generation in the device was assessed by dispersing a buffered aqueous solution that contained bromophenol blue dye to aid in visualizing individual droplets. The oil phase contained 70% Ar20 silicone oil (polyphenyl-methylsiloxane), 26% mineral oil, and 4% Abil EM90 (a non-ionic silicone-based emulsifier). The flow rates of the aqueous and oil phases were 5 and 70 μL min−1, respectively. Micrographs depicting the formation of the dye-containing droplets (Figure 1B) illustrate the uniformity of droplets produced by this method. The images were processed to assess the droplet size in situ. The droplets have a radius of 21.9 ± 0.8 μm (4% standard deviation), each containing a volume of 44 ± 2 pL.
Using the silicone oil/mineral oil/silicone-based emulsifier combination, the device can generate droplets continuously at a rate of 1.9 kHz (~107 droplets per hour). It can emulsify aqueous solutions at even higher flow rates, exceeding 100 μL min−1, which generates droplets at a frequency of nearly 40 kHz (108 droplets per hour). At these very high flow rates, however, droplet shearing in the output channel starts to become a problem. The device is amenable to various aqueous and oil formulations, and it is likely that higher flow rates could be achieved with customized formulations and optimized circuit designs. The formulation used here differs from commonly used oil/detergent combinations (Miller at al., 2006) because the latter were found to generate unstable droplets when emulsifying samples containing polymerase proteins. The combination of mineral oil and silicone-based emulsifier has been used previously to form stable emulsions that support eukaryotic cell-free translation systems (Ghadessy and Holliger, 2004). However, that combination was unsatisfactory in the present system, and inclusion of 70% silicon oil was found to be necessary to obtain highly stable droplets.
The microfluidic nozzle array circuit was used to generate a highly uniform water-in-oil emulsion in which the individual droplets were seeded with a starting population of RNA enzymes derived from the class I RNA ligase (Ekland et al., 1995). The droplets also contained an oligonucleotide substrate for RNA-catalyzed ligation and a combination of polymerase proteins and nucleotide building blocks necessary to bring about the selective amplification of reactive RNA enzymes. These are the components of the previously described continuous in vitro evolution system (Wright and Joyce, 1997), which has been applied to the class I ligase (Schmitt and Lehman, 1999; Ordoukhanian and Joyce, 1999; Paegel and Joyce, 2008), as well as the DSL ligase (Voytek and Joyce, 2007), but only in bulk solution. The droplets were seeded with a low concentration of RNA enzymes so that each droplet contained none or a single parent molecule. The droplet volume and the concentration of reactants allowed each parent molecule to give rise to ~108 progeny through repeated rounds of selective amplification within the droplet. Afterwards, the droplets were harvested and the RNAs (or their corresponding cDNAs) were collected and processed.
The microfluidic compartmentalized evolution system was used to develop variants of the class I ligase that are resistant to inhibition by the antibiotic neomycin. Neomycin has a strong propensity to bind to RNA (Hendrix et al., 1997) and has been shown to inhibit the activity of several RNA enzymes (von Ahsen et al., 1991; Stage et al., 1995; Rogers et al., 1996; Mikkelsen et al., 1999). The starting ligase enzyme exhibited a catalytic rate, kcat, of 21 min−1 in the absence of neomycin, but this rate decreased to 6.5 min−1 in the presence of 100 μM neomycin and to 0.3 min−1 in the presence of 300 μM neomycin (measured in the presence of 10 mM MgCl2 and 50 mM KCl at pH 7.5 and 37 °C). In the continuous evolution system the starting ligase enzyme amplified 10-fold in 15 min in the absence of neomycin, but exhibited no measurable amplification in the presence of 100 μM neomycin. Accordingly, the experiment to evolve neomycin-resistant enzymes was initiated in the presence of 10 μM neomycin, and this concentration was increased progressively to 100 μM over the course of several rounds of compartmentalized selective amplification.
A starting population of variant RNA enzymes was constructed by introducing random mutations at a frequency of 12% per nucleotide position throughout 124 positions of a previously isolated variant of the class I ligase (Paegel and Joyce, 2008). The starting population of 1 × 1011 molecules therefore included only ~104 copies of the wild-type enzyme, together with a very sparse sampling of mutants containing an average of 15 mutations per molecule. It was not possible to initiate continuous evolution with this population when only a single founder molecule was seeded into each compartment because nearly all compartments contained non-functional molecules. Instead, 105 molecules were seeded into each compartment so that a significant fraction of compartments would contain at least one functional molecule that could react and undergo selective amplification in the presence of 10 μM neomycin. The total volume of 500 μL contained 107 such compartments, which were incubated at 37 °C for 6 h.
Following this incubation the emulsion was centrifuged, extracted with hexane to remove the oil components, and heated at 90 °C for 10 min to inactivate the polymerase proteins and hydrolyze the RNA. The remaining cDNAs then were used as the input for error-prone PCR (Cadwell and Joyce, 1992), followed by in vitro transcription to generate a daughter population of RNA enzymes. These molecules in turn were used to produce a second microfluidic water-in-oil emulsion, but this time seeding only a single RNA enzyme into each compartment, still in the presence of 10 μM neomycin. In the third and fourth rounds the concentration of neomycin was increased to 30 μM, and in the fifth round it was increased to 100 μM, again seeding a single molecule into each compartment.
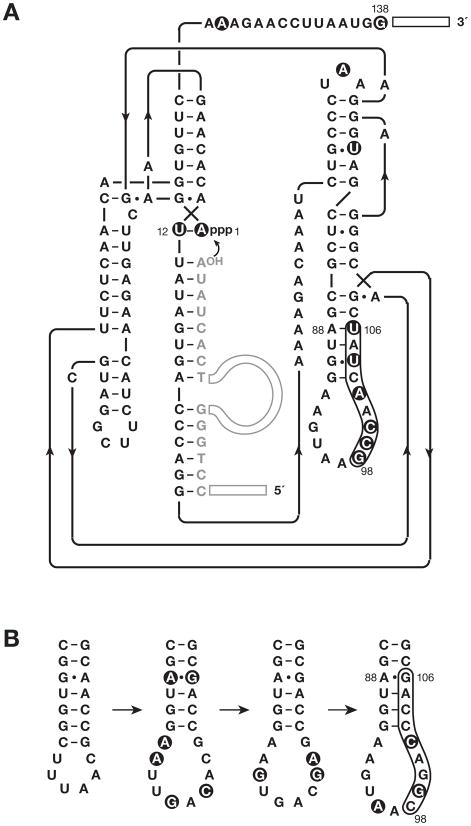
After the fifth round of compartmentalized selective amplification the cDNAs were cloned and sequenced, and the corresponding RNAs were prepared and analyzed. Among 20 clones that were characterized, three distinct sequence classes were observed. All clones contained three common mutations: a G→A change at position 1, a compensatory C→U change at position 12, and a C→G change at position 138 (Figure 2A; see also Table S1). Also notable were changes within a portion of the P7 stem-loop region (positions 98–106), which had mutated from the wild-type sequence 5′-CGGACCCAG-3′ (still present in six clones) to either 5′-GCCAGCCAG-3′ (in seven clones) or 5′-GCCAACUAU-3′ (in six clones; changes underlined). All three major classes of variants, together with the starting wild-type enzyme, were evaluated in a selective amplification assay performed in bulk solution in either the absence or presence of 100 μM neomycin.
Figure 2.
Sequence and secondary structure of the neomycin-resistant form of the class I ligase. (A) Representative individual from the class of variants that are dependent on neomycin for optimal activity. The secondary structure depiction follows that of the recently reported crystal structure (Shechner et al., 2009). The oligonucleotide substrate is shown in gray. Primer binding sites at the 5′ end of the substrate and 3′ end of the enzyme are depicted as open rectangles. Curved arrow indicates the site of ligation. Filled circles highlight mutations relative to the starting wild-type enzyme. (B) Evolutionary maturation of the P7 stem-loop region, beginning (at left) with the b1-207 form of the enzyme (Ekland et al., 1995), then the evolved variant used to initiate continuous in vitro evolution (Wright and Joyce, 1997), then the E100-3 variant that emerged following 100 successive transfers of continuous evolution (Wright and Joyce, 1997), and finally (at right) the evolved variant that emerged following 500 successive rounds of chip-based continuous evolution (Paegel and Joyce, 2008). The latter variant was used to initiate the present study. Mutations relative to the previous variant in the series are highlighted by filled circles.
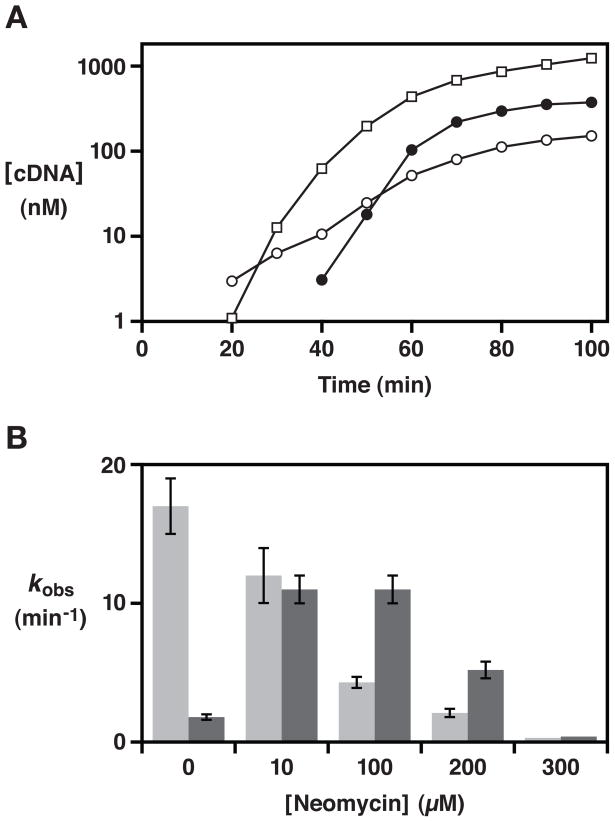
As stated above, in the absence of neomycin the wild-type enzyme amplified ~10-fold in 15 min. The enzymes containing either no or four mutations in the P7 region amplified somewhat more efficiently, while the enzyme containing six mutations in this region amplified less efficiently, requiring 37 min to achieve 10-fold amplification. In the presence of 100 μM neomycin the wild-type enzyme showed no detectable amplification, while all three major variants amplified efficiently. The enzyme containing six mutations in the P7 region amplified 10-fold in 16 min, which is considerably faster than its rate of amplification in the absence of neomycin (Figure 3A; see also Figure S1).
Figure 3.
Biochemical properties of the wild-type and neomycin-resistant forms of the class I ligase. (A) Amplification profiles comparing the wild-type enzyme in the absence of neomycin (open squares) and the evolved enzyme in either the absence (open circles) or presence (filled circles) of 100 μM neomycin. Amplification was initiated with 1 nM RNA enzyme and 2.5 μM substrate, and the yield of full-length cDNA was determined at various times. No amplification of the wild-type enzyme was observed in the presence of 100 μM neomycin. (B) Catalytic activity of the wild-type (light gray) and evolved (dark gray) enzymes in the presence of various concentrations of neomycin. Reaction conditions: 30 nM enzyme, 2.5 μM substrate, 10 mM MgCl2, and 50 mM KCl at pH 7.5 and 37 °C.
The rate of RNA-catalyzed RNA ligation was measured for both the wild-type enzyme and the enzyme containing six mutations in the P7 region, determined as a function of neomycin concentration. The wild-type enzyme exhibited progressive loss of activity with increasing neomycin concentration, whereas the evolved enzyme exhibited optimal activity in the presence of 10–100 μM neomycin, declining in activity at both higher and lower concentrations of neomycin (Figure 3B). Formal kinetic analyses were carried out for both the wild-type and evolved enzyme, in either the absence or presence of 100 μM neomycin (Table 1). In the absence of neomycin, the wild-type enzyme exhibited a kcat of 21 min−1, while the evolved enzyme had a kcat of 4.7 min−1. In the presence of 100 μM neomycin these rates were approximately reversed, with a kcat of 6.5 min−1 for the wild-type and 25 min−1 for the evolved enzyme.
Table 1.
Kinetic properties of the wild-type and evolved enzymes.
| [Neomycin] | Wild-type | Evolved | ||
|---|---|---|---|---|
| 0 | 100 μM | 0 | 100 μM | |
| kcat (min−1) | 21 ± 0.8 | 6.5 ± 0.3 | 4.7 ± 0.1 | 25 ± 1 |
| KM (μM) | 0.4 ± 0.05 | 0.7 ± 0.1 | 4.2 ± 0.4 | 3.1 ± 0.7 |
Reaction conditions: 10 mM MgCl2, 50 mM KCl, pH 7.5, 37 °C.
In both the presence and absence of neomycin, the evolved enzyme has a significantly higher Km for the oligonucleotide substrate compared to that of the wild-type enzyme. The wild-type enzyme is itself the result of a long heritage of continuous in vitro evolution, in which the population was provided with 5 μM substrate during 10298-fold selective amplification (Wright and Joyce, 1997), then 2.5 μM substrate during 1060-fold selective amplification (Schmitt and Lehman, 1999), then progressively decreasing concentrations of substrate, ultimately reaching 0.05 μM, during 10500-fold selective amplification (Paegel and Joyce, 2008). It was during the latter era of decreasing substrate concentrations when the enzyme evolved a submicromolar Km. In the present study the substrate concentration was 2.5 μM throughout, thereby relieving selective pressure on Km. Accordingly, the Km of the evolved enzyme, although 5–10-fold elevated compared to that of the wild-type, does not substantially exceed the concentration of substrate that was available during the evolution process.
The wild-type and evolved enzymes were tested for catalytic activity in the presence of 100 μM concentration of three other aminoglycosides: kanamycin, paromomycin, and tobramycin. The wild-type is inhibited by all three of these compounds, but to a lesser extent than it is inhibited by neomycin (Figure 3B; see also Figure S2). The evolved enzyme, which is stimulated by neomycin, is also stimulated by the other three aminoglycosides, but to a lesser extent. Thus the neomycin dependence of the evolved enzyme is somewhat specific for that ligand. The wild-type and evolved enzymes also were tested in the presence of various concentrations of Mg2+, measured in either the absence or presence of 100 μM neomycin (Figure 3B; see also Figure S2). In the absence of neomycin, the wild-type enzyme showed no dependence on Mg2+ concentration, whereas the neomycin-activated enzyme showed a slight increase in activity with increasing Mg2+ over the range of 10–50 mM. In the presence of neomycin, both enzymes showed no significant dependence on Mg2+ concentration. Thus it is unlikely that neomycin and Mg2+ occupy the same binding site in the evolved enzyme. Rather, the enzyme appears to have evolved a specific binding site for neomycin that enhances catalytic activity.
Discussion
In vitro compartmentalization provides a way to link genotype and phenotype during the course of a directed evolution experiment (Tawfik and Griffiths, 1998). Compartmentalization is achieved by emulsifying the aqueous phase, which contains the biochemical components, in an oil phase that includes emulsion stabilizers. This procedure results in large numbers of water-in-oil droplets, each harboring one or a few individuals from the population. In past practice, however, the droplets were highly variable in size, which can bias the effect of selection because individuals that happen to occupy a larger droplet will benefit from the greater carrying capacity of their local environment. This bias is especially a concern when carrying out multiple-turnover reactions or iterative selective amplification within the compartments. In some screening applications it is possible to apply a correction factor to the observed phenotype to compensate for differences in compartment volume, or to consider only those droplets that fall within a particular size range (Aharoni et al., 2005). It would be far preferable, however, to generate droplets that are highly uniform in size, provided that large numbers of such droplets can be obtained.
Microfluidic emulsification has proven to be a viable approach for preparing highly uniform droplet trains, but the yield has been much lower than what can be obtained by bulk emulsification (Thorsen et al., 2001). The nozzle array circuit described here achieves substantial throughput, generating 107–108 droplets per hour with the initial design. It does so by significantly reducing backpressure, which is the primary cause of device failure and would otherwise limit the flow rate and therefore the population size. Vigorous mixing during homogenization is eliminated, preventing damage to the biochemical reagents. The nozzle array is topologically equivalent to single-channel circuits for generating slugs of fluid (Song et al., 2003), but the depth and width of the oil flow channel are greater to minimize fluidic resistance at the sites of droplet formation, which prevents oil from intruding into the nozzles. The circuit is easily fabricated in PDMS using a simple dual-height mold. Multiple such circuits could be made to operate in parallel to provide even greater throughput.
The directed evolution of RNA enzymes was carried out within microfluidic water-in-oil droplets to demonstrate the capabilities of the new system. The system allows large numbers of functional molecules to operate within separate compartments, although the demonstration case reported here did not critically depend on compartmentalization and could have been carried out in bulk solution. As a selection pressure, the evolving RNA enzymes were challenged to escape inhibition by neomycin. This compound is a member of the family of aminoglycoside antibiotics, many of which exhibit potent antimicrobial activity through their ability to bind 16S ribosomal RNA and thereby inhibit protein synthesis (Moazed and Noller, 1987). Aminoglycosides have a strong propensity to bind to structured RNAs and have been shown to inhibit the function of various biological RNAs and ribonucleoproteins (Walter et al., 1999; Schroeder et al., 2000). The catalytic activity of the starting class I RNA ligase is substantially inhibited in the presence of 10–100 μM neomycin, which prevents it from undergoing selective amplification in the continuous in vitro evolution system. Following five rounds of compartmentalized evolution, however, novel variants arose that are resistant to neomycin. Compared to the wild-type, the evolved enzymes have a four-fold faster catalytic rate in the presence of 100 μM neomycin, which enables them to undergo efficient selective amplification.
Genetic changes within the evolved molecules were clustered mainly in the P7 stem-loop region of the enzyme (Figure 2A). Over the course of several previous in vitro evolution experiments involving the class I ligase (Wright and Joyce, 1997; Schmitt and Lehman, 1999; Paegel and Joyce, 2008), this region underwent significant changes (Figure 2B). The changes were confined largely to the loop portion of the region, the only exception being inversion of a non-canonical G88•A106 pair in the stem (to an A•G pair), which occurred several hundred rounds of evolution ago. In the present study, this same base pair became mutated to a Watson-Crick pair for the first time, accompanied by mutation of the G90•C104 pair to a G•U wobble pair. Elimination of the A•G pair within the stem is especially notable in light of a recent study demonstrating that such motifs have a special propensity to bind neomycin (Disney et al., 2008).
It is intriguing that the evolved RNA enzyme depicted in Figure 2A has a five-fold faster catalytic rate (kcat) and eight-fold greater catalytic efficiency (kcat Km−1) in the presence compared to the absence of 100 μM neomycin (Table 1). Other evolved individuals that contain fewer mutations within the P7 stem-loop also are resistant to neomycin, but are not dependent on neomycin for optimal activity. This result echoes a theme that has been observed in the evolution of antibiotic resistance in bacteria, where dependence can accompany resistance. Microbial resistance to the inhibitory effect of aminoglycosides can be attained through one or two point mutations within the 16S rRNA sequence (Melancon, 1988; De Stasio, 1989), and additional mutations can result in dependence on the aminoglycoside for optimal growth rates (Hummel and Böck, 1983; Honoré et al., 1995). The two strategies of either resistance alone or resistance combined with dependence both confer selective advantage within the microfluidic compartmentalized evolution system.
Significance
The preparation of water-in-oil emulsions using a microfluidic device, in contrast to emulsions prepared by bulk homogenization, results in highly uniform water droplets. The size and production rate of these droplets is precisely determined by the geometry and programmed flow rate of the device. High throughput is achieved using a radial nozzle array, which is straightforward to fabricate and operate, and could be scaled to achieve even higher rates of droplet production. An important application of microfluidic compartmentalization is in directed evolution studies, especially where uniformity of droplet size is critical for avoiding selection bias. The nozzle array circuit could be coupled with other computer-controlled fluidic handling steps to provide an integrated system for molecular evolution and analytical biochemistry.
Experimental Procedures
Library construction
Template DNAs for transcribing the starting pool of RNAs were prepared by PCR cross-extension of two synthetic oligodeoxynucleotides having the sequence 5′-TAATACGACTCACTATAGGA-AGAACACACTATAGTGACCCAGGAAAAGACAAATCTGCCCTTAGAGCTTGAGAACATCTT-CGGATGCACGGGA-3′ and 5′-AGTCCCTCCATGCTCGTGCATTAAGGTTCTGTGAACACTG-TTGAGAACGCTGGGTCCGTTACTTCCATCGCGAGCTGCCTCCCGTGCATCC-3′ (T7 RNA polymerase promoter sequence underlined; nucleotides randomized at 12% degeneracy italicized). The cross-extension products, consisting of ~1013 distinct sequences, were used as input for in vitro transcription, and the resulting RNAs were purified by denaturing polyacrylamide gel electrophoresis (PAGE) and desalted on a Sephadex G-25 column prior to use.
Microfluidic fabrication
Master molds of SU-8 photoresist fabricated on silicon wafers were obtained from the Stanford Microfluidics Foundry. The aqueous and oil fluid flow layers were fabricated with 20- and 150-μm step height, respectively. PDMS replicates were cast from the silicon master mold (Duffy et al., 1998), diced into individual circuits, and reservoirs were carved using a 0.75-mm biopsy punch (Harris Uni-Core, Ted Pella, Redding, CA). The device and a glass microscope slide were cleaned, chemically hydrolyzed for 30 min (Sui et al., 2006), bonded, and baked at 70 °C for 1 h. Tygon microbore tubing (0.01″ i.d. × 0.03″ o.d; Saint-Gobain, Valley Forge, PA) was inserted directly into the reservoirs. Syringes (1 mL TB and 5 mL Luer-lock; BD, Franklin Lakes, NJ) were fitted with 30 gauge blunt syringe needles and inserted into the microbore tubing. The syringes were mounted on digital syringe pumps (SP101i, World Precision Instruments, Sarasota, FL), which were used to drive fluids through the circuit.
Microfluidic compartmentalization
The oil phase syringe was loaded with a mixture of 4% (w/w) Abil EM90 (Evonik Degussa, Essen, Germany) (Ghadessy and Holliger, 2004), 26% Ar20 silicone oil (Sigma-Aldrich, St. Louis, MO), and 70% mineral oil (Sigma-Aldrich). The aqueous phase syringe was loaded with a mixture of 15 mM MgCl2, 50 mM KCl, and 50 mM EPPS (pH 7.5). The aqueous phase was pumped into the circuit at the central reservoir (Raq) at a rate of 5 μL min−1 and dispersed into the oil phase, which was pumped into the circuit at the input reservoir (Rin) at a rate of 70 μL min−1. Prior to use, each circuit was rinsed for 30 min under these conditions. In order to measure droplet size, bromophenol blue (0.1% w/v) was added to the aqueous mixture. Droplet generation was observed using a stereozoom microscope (S8, Leica Microsystems, Bannockburn, IL), equipped with a CCD for image acquisition (DFC290, Leica). Droplets were imaged in situ, and droplet diameters were determined for 200 droplets using ImageJ (National Institutes of Health, Bethesda, MD), rejecting objects with circularity <0.95.
Compartmentalized selective amplification
Each round of compartmentalized selective amplification was carried out by first mixing on ice the population of RNA enzymes, 2.5 μM oligonucleotide substrate having the sequence 5′-CCGAAG-CCTGGGATCAATAATACGACTCACUAUA-3′ (T7 RNA polymerase promoter sequence underlined, RNA residues bolded), 10–100 μM neomycin B (Sigma-Aldrich), 2 U μL−1 Superscript II reverse transcriptase (Invitrogen, Carlsbad, CA), 5 U μL−1 T7 RNA polymerase, 2.5 μM 5′-fluorescein-labeled cDNA primer having the sequence 5′-AGTCCCTCCATGCTCG-3′, 2 mM each NTP, 0.2 mM each dNTP, 15 mM MgCl2, 50 mM KCl, 4 mM DTT, and 50 mM EPPS (pH 7.5). The reaction mixture then was loaded into a syringe, which was kept chilled using freezer packs, and dispersed continuously into the oil phase. 500 μL of aqueous emulsion product was collected over 100 min, then incubated at 37 °C for 6 h. Following incubation, 50 μL of 1 N NaOH was added to the emulsion and the sample was vortexed to stop the amplification process. The emulsion then was centrifuged, extracted with hexane, transferred to a vacuum concentrator to remove residual hexane, and incubated at 90 °C for 10 min. The cDNAs that remained following alkaline hydrolysis were used as input for error-prone PCR (Cadwell and Joyce, 1992). The PCR products were transcribed, and the resulting RNAs were purified by PAGE and desalted to provide the population used to initiate the next round of compartmentalized selective amplification. No mutagenesis was performed prior to the final round.
Analysis of individual clones
Following the fifth round of compartmentalized selective amplification, the PCR products were cloned and sequenced. Plasmid DNA from individual clones was amplified by PCR, transcribed, and the resulting RNAs were purified by PAGE and desalted. Amplification assays were performed starting with 1 nM RNA enzyme, in either the presence or absence of 100 μM neomycin, with all of the other components of the amplification mixture as described above. Aliquots were removed from the mixture at various times and the reaction was quenched by adding EDTA and urea. The reaction products were separated by PAGE, and the yield of cDNA was determined based on extension of the 5′-fluorescein-labeled cDNA primer, as quantitated using a PharosFX imager (Bio-Rad, Hercules, CA).
Values for kcat and Km were obtained by reacting 10 nM [α-32P]-labeled RNA enzyme with various concentrations of substrate in either the presence or absence of 100 μM neomycin, and in the presence of 10 mM MgCl2, 50 mM KCl, and 50 mM EPPS (pH 7.5) at 37 °C. Reaction rates were determined for each concentration of substrate based on at least six data points, which were fit to a biphasic equation: y = a − be−k1t − ce−k2t, where b and k1 are the amplitude and rate of the initial fast phase, and c and k2 are the amplitude and rate of the slow phase. The amplitude of the fast phase was typically ~50% of the overall maximum extent. Values for kcat and KM were obtained from a Michaelis-Menten plot of k1 versus substrate concentration. Neomycin dose-dependence studies were performed similarly, but using 30 nM RNA enzyme, 2.5 μM oligonucleotide substrate, and 0, 10, 100, 200, 300, or 500 μM neomycin. Analysis of other aminoglycosides was performed using 100 μM kanamycin, tobramycin (both from Sigma-Aldrich), or paromomycin (Research Products International, Mount Prospect, IL).
Supplementary Material
Acknowledgments
This work was supported by National Science Foundation grant no. MCB-0614614 (G.F.J.), and NIH Pathway to Independence Career Development Award no. GM083155 (B.M.P.). The authors declare that there is no conflict of interest.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Agresti JJ, Kelly BT, Jäschke A, Griffiths AD. Selection of ribozymes that catalyse multiple-turnover Diels-Alder cycloadditions by using in vitro compartmentalization. Proc Natl Acad Sci USA. 2005;102:16170–16175. doi: 10.1073/pnas.0503733102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Agresti JJ, Antipov E, Abate AR, Ahn K, Rowat AC, Baret JC, Marquez M, Klibanov AM, Griffiths AD, Weitz DA. Ultrahigh-throughput screening in drop-based microfluidics for directed evolution. Proc Natl Acad Sci USA. 2010;107:4004–4009. doi: 10.1073/pnas.0910781107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aharoni A, Amitai G, Bernath K, Magdassi S, Tawfik DS. High-throughput screening of enzyme libraries: thiolactonases evolved by fluorescence-activated sorting of single cells in emulsion compartments. Chem Biol. 2005;12:1281–1289. doi: 10.1016/j.chembiol.2005.09.012. [DOI] [PubMed] [Google Scholar]
- Barbas CF, Kang AS, Lerner RA, Benkovic SJ. Assembly of combinatorial antibody libraries on phage surfaces: the gene III site. Proc Natl Acad Sci USA. 1991;88:7978–7982. doi: 10.1073/pnas.88.18.7978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernath K, Hai MT, Mastrobattista E, Griffiths AD, Magdassi S, Tawfik DS. In vitro compartmentalization by double emulsions: sorting and gene enrichment by fluorescence activated cell sorting. Anal Biochem. 2004;325:151–157. doi: 10.1016/j.ab.2003.10.005. [DOI] [PubMed] [Google Scholar]
- Boder ET, Wittrup KD. Yeast surface display for screening combinatorial polypeptide libraries. Nat Biotechnol. 1997;15:553–557. doi: 10.1038/nbt0697-553. [DOI] [PubMed] [Google Scholar]
- Cadwell RC, Joyce GF. Randomization of genes by PCR mutagenesis. PCR Methods Applic. 1992;2:28–33. doi: 10.1101/gr.2.1.28. [DOI] [PubMed] [Google Scholar]
- Chan EM, Alivisatos AP, Mathies RA. High-temperature micro uidic synthesis of CdSe nanocrystals in nanoliter droplets. J Am Chem Soc. 2005;127:13854–13861. doi: 10.1021/ja051381p. [DOI] [PubMed] [Google Scholar]
- Christopher GF, Anna SL. Microfluidic methods for generating continuous droplet streams. J Phys D: Appl Phys. 2007;40:R319–R336. [Google Scholar]
- Clausell-Tormos J, et al. Droplet-based micro uidic platforms for the encapsulation and screening of mammalian cells and multicellular organisms. Chem Biol. 2008;15:427–437. doi: 10.1016/j.chembiol.2008.04.004. [DOI] [PubMed] [Google Scholar]
- Cwirla SE, Peters EA, Barrett RW, Dower WJ. Peptides on phage: a vast library of peptides for identifying ligands. Proc Natl Acad Sci USA. 1990;87:6378–6382. doi: 10.1073/pnas.87.16.6378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Stasio EA, Moazed D, Noller HF, Dahlberg AE. Mutations in 16S ribosomal RNA disrupt antibiotic-RNA interactions. EMBO J. 1989;8:1213–1216. doi: 10.1002/j.1460-2075.1989.tb03494.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Devlin JJ, Panganiban LC, Devlin PE. Random peptide libraries: a source of specific protein binding molecules. Science. 1990;249:404–406. doi: 10.1126/science.2143033. [DOI] [PubMed] [Google Scholar]
- Disney MD, Labuda LP, Paul DJ, Poplawski SG, Pushechnikov A, Tran T, Velagapudi SP, Wu M, Childs-Disney JL. Two-dimensional combinatorial screening identifies specific aminoglycoside-RNA internal loop partners. J Am Chem Soc. 2008;130:11185–11194. doi: 10.1021/ja803234t. [DOI] [PubMed] [Google Scholar]
- Duffy DC, McDonald JC, Schueller OJA, Whitesides GM. Rapid prototyping of micro uidic systems in poly(dimethylsiloxane) Anal Chem. 1998;70:4974–4984. doi: 10.1021/ac980656z. [DOI] [PubMed] [Google Scholar]
- Ekland EH, Szostak JW, Bartel DP. Structurally complex and highly active RNA ligases derived from random RNA sequences. Science. 1995;269:364–370. doi: 10.1126/science.7618102. [DOI] [PubMed] [Google Scholar]
- Francisco JA, Campbell R, Iverson BL, Georgiou G. Production and fluorescence-activated cell sorting of Escherichia coli expressing a functional antibody fragment on the external surface. Proc Natl Acad Sci USA. 1993;90:10444–10448. doi: 10.1073/pnas.90.22.10444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghadessy FJ, Ong JL, Holliger P. Directed evolution of polymerase function by compartmentalized self-replication. Proc Natl Acad Sci USA. 2001;98:4552–4557. doi: 10.1073/pnas.071052198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghadessy FJ, Holliger P. A novel emulsion mixture for in vitro compartmentalization of transcription and translation in the rabbit reticulocyte system. Protein Eng Des Sel. 2004;17:201–204. doi: 10.1093/protein/gzh025. [DOI] [PubMed] [Google Scholar]
- Hanes J, Pluckthun A. In vitro selection and evolution of functional proteins by using ribosome display. Proc Natl Acad Sci USA. 1997;94:4937–4942. doi: 10.1073/pnas.94.10.4937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hendrix M, Alper PB, Priestley ES, Wong CH. Hydroxyamines as a new motif for the molecular recognition of phosphodiesters: implications for aminogloycoside-RNA interactions. Angew Chemie. 1997;36:95–98. [Google Scholar]
- Honoré N, Marchal G, Cole ST. Novel mutation in 16S rRNA associated with streptomycin dependence in Mycobacterium tuberculosis. Antimicrob Agents Chemother. 1995;39:769–770. doi: 10.1128/AAC.39.3.769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hummel H, Böck A. On the basis of aminoglycoside-dependent growth of mutants from E. coli: physiological studies. Mol Gen Genet. 1983;191:167–175. doi: 10.1007/BF00334809. [DOI] [PubMed] [Google Scholar]
- Lee YF, Tawfik DS, Griffiths AD. Investigating the target recognition of DNA cytosine-5 methyltransferase HhaI by library selection using in vitro compartmentalisation. Nucleic Acids Res. 2002;30:4937–4944. doi: 10.1093/nar/gkf617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levy M, Griswold KE, Ellington AD. Direct selection of trans-acting ligase ribozymes by in vitro compartmentalization. RNA. 2005;11:1555–1562. doi: 10.1261/rna.2121705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mastrobattista E, Taly V, Chanudet E, Treacy P, Kelly BT, Griffiths AD. High-throughput screening of enzyme libraries: in vitro evolution of a β-galactosidase by fluorescence-activated sorting of double emulsions. Chem Biol. 2005;12:1291–1300. doi: 10.1016/j.chembiol.2005.09.016. [DOI] [PubMed] [Google Scholar]
- Mazutis L, et al. Droplet-based micro uidic systems for high-throughput single DNA molecule isothermal ampli cation and analysis. Anal Chem. 2009;81:4813–4821. doi: 10.1021/ac900403z. [DOI] [PubMed] [Google Scholar]
- McCafferty J, Griffiths AD, Winter G, Chiswell DJ. Phage antibodies: filamentous phage displaying antibody variable domains. Nature. 1990;348:552–554. doi: 10.1038/348552a0. [DOI] [PubMed] [Google Scholar]
- McCafferty J, Jackson RH, Chiswell DJ. Phage-enzymes: expression and affinity chromatography of functional alkaline phosphatase on the surface of bacteriophage. Protein Eng. 1991;4:955–961. doi: 10.1093/protein/4.8.955. [DOI] [PubMed] [Google Scholar]
- Melancon P, Lemieux C, Brakier-Gingras L. A mutation in the 530 loop of Escherichia coli 16S ribosomal RNA causes resistance to streptomycin. Nucleic Acids Res. 1988;16:9631–9639. doi: 10.1093/nar/16.20.9631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mikkelsen NE, Brännvall M, Virtanen A, Kirsebom LA. Inhibition of RNase P RNA cleavage by aminoglycosides. Proc Natl Acad Sci USA. 1999;96:6155–6160. doi: 10.1073/pnas.96.11.6155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller OJ, Bernath K, Agresti JJ, Amitai G, Kelly BT, Mastrobattista E, Taly V, Magdassi S, Tawfik DS, Griffiths AD. Directed evolution by in vitro compartmentalization. Nat Methods. 2006;3:561–570. doi: 10.1038/nmeth897. [DOI] [PubMed] [Google Scholar]
- Moazed D, Noller HF. Interaction of antibiotics with functional sites in 16S ribosomal RNA. Nature. 1987;327:389–394. doi: 10.1038/327389a0. [DOI] [PubMed] [Google Scholar]
- Ordoukhanian P, Joyce GF. A molecular description of the evolution of resistance. Chem Biol. 1999;6:881–889. doi: 10.1016/s1074-5521(00)80007-1. [DOI] [PubMed] [Google Scholar]
- Paegel BM, Joyce GF. Darwinian evolution on a chip. PLoS Biol. 2008;6:900–906. doi: 10.1371/journal.pbio.0060085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberts RW, Szostak JW. RNA-peptide fusions for the in vitro selection of peptides and proteins. Proc Natl Acad Sci USA. 1997;94:12297–12302. doi: 10.1073/pnas.94.23.12297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rogers J, Chang AH, von Ahsen U, Schroeder R, Davies J. Inhibition of the self-cleavage reaction of the human hepatitis delta virus ribozyme by antibiotics. J Mol Biol. 1996;259:916–925. doi: 10.1006/jmbi.1996.0369. [DOI] [PubMed] [Google Scholar]
- Schmitt T, Lehman N. Non-unity molecular heritability demonstrated by continuous evolution in vitro. Chem Biol. 1999;6:857–869. doi: 10.1016/s1074-5521(00)80005-8. [DOI] [PubMed] [Google Scholar]
- Schroeder R, Waldisch C, Wank H. Modulation of RNA function by aminoglycoside antibiotics. EMBO J. 2000;19:1–9. doi: 10.1093/emboj/19.1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scott JK, Smith GP. Searching for peptide ligands with an epitope library. Science. 1990;249:386–390. doi: 10.1126/science.1696028. [DOI] [PubMed] [Google Scholar]
- Shechner DM, Grant RA, Bagby SC, Koldobskaya Y, Piccirilli JA, Bartel DP. Crystal structure of the catalytic core of an RNA-polymerase ribozyme. Science. 2009;326:1271–1275. doi: 10.1126/science.1174676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith GP, Petrenko VA. Phage display. Chem Rev. 1997;97:391–410. doi: 10.1021/cr960065d. [DOI] [PubMed] [Google Scholar]
- Song H, Tice JD, Ismagilov RF. A micro uidic system for controlling reaction networks in time. Angew Chemie. 2003;42:768–772. doi: 10.1002/anie.200390203. [DOI] [PubMed] [Google Scholar]
- Soumillion P, Jespers L, Bouchet M, Marchand-Brynaert J, Winter G, Fastrez J. Selection of β-lactamase on filamentous bacteriophage by catalytic activity. J Mol Biol. 1994;237:415–422. doi: 10.1006/jmbi.1994.1244. [DOI] [PubMed] [Google Scholar]
- Stage TK, Hertel KJ, Uhlenbeck OC. Inhibition of the hammerhead ribozyme by neomycin. RNA. 1995;1:95–101. [PMC free article] [PubMed] [Google Scholar]
- Sui GD, et al. Solution-phase surface modi cation in intact poly(dimethylsiloxane) micro uidic channels. Anal Chem. 2006;78:5543–5551. doi: 10.1021/ac060605z. [DOI] [PubMed] [Google Scholar]
- Tawfik DS, Griffiths AD. Man-made cell-like compartments for molecular evolution. Nat Biotechnol. 1998;16:652–656. doi: 10.1038/nbt0798-652. [DOI] [PubMed] [Google Scholar]
- Thorsen T, Roberts RW, Arnold FH, Quake SR. Dynamic pattern formation in a vesicle-generating micro uidic device. Phys Rev Lett. 2001;86:4163–4166. doi: 10.1103/PhysRevLett.86.4163. [DOI] [PubMed] [Google Scholar]
- Umbanhowar PB, Prasad V, Weitz DA. Monodisperse emulsion generation via drop break off in a co owing stream. Langmuir. 2000;16:347–351. [Google Scholar]
- Von Ahsen U, Davies J, Schroeder R. Antibiotic inhibition of group I ribozyme function. Nature. 1991;353:368–370. doi: 10.1038/353368a0. [DOI] [PubMed] [Google Scholar]
- Voytek SB, Joyce GF. Emergence of a fast-reacting ribozyme that is capable of undergoing continuous evolution. Proc Natl Acad Sci USA. 2007;104:15288–15293. doi: 10.1073/pnas.0707490104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walter F, Vicens Q, Westhof E. Aminoglycoside RNA interactions. Curr Opin Chem Biol. 1999;3:694–704. doi: 10.1016/s1367-5931(99)00028-9. [DOI] [PubMed] [Google Scholar]
- Wright MC, Joyce GF. Continuous in vitro evolution of catalytic function. Science. 1997;276:614–617. doi: 10.1126/science.276.5312.614. [DOI] [PubMed] [Google Scholar]
- Zaher HS, Unrau PJ. Selection of an improved RNA polymerase ribozyme with superior extension and fidelity. RNA. 2007;13:1017–1026. doi: 10.1261/rna.548807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeng Y, Novak R, Shuga J, Smith MT, Mathies RA. High-performance single cell genetic analysis using microfluidic emulsion generator arrays. Anal Chem. 2010;82 doi: 10.1021/ac902683t. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng B, Roach LS, Ismagilov RF. Screening of protein crystallization conditions on a micro uidic chip using nanoliter-size droplets. J Am Chem Soc. 2003;125:11170–11171. doi: 10.1021/ja037166v. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.