Abstract
The dependence of the fluorescence of catalase upon the concentration of added superoxide dismutase (SOD) indicates that SOD binds to saturable sites on catalase. The affinity of SOD for these sites varies with temperature, and with the concentration of each of three nominally inert polymeric additives—dextran 70, Ficoll 70, and polyethylene glycol 2000. At room temperature (25.0°C) and higher, the addition of high concentrations of polymer is found to significantly enhance the affinity of SOD for catalase, but with decreasing temperature the enhancing effect of polymer addition diminishes, and at 8.0°C, addition of polymer has little or no effect on the affinity of SOD for catalase. The results presented here provide the first experimental evidence for the existence of competition between a repulsive excluded volume interaction between protein and polymer, which tends to enhance association of dilute protein, and an attractive interaction between protein and polymer, which tends to inhibit protein association. The net effect of high concentrations of polymer upon protein associations depends upon the relative strength of these two types of interactions at the temperature of measurement, and may vary significantly between different proteins and/or polymers.
Introduction
Protein-protein interactions play important roles in many essential biological processes, such as the regulation of enzymatic activities and signal transduction (1,2). Most physiological fluid media contain between 5 and 40% by volume of macromolecules, and are often referred to as macromolecularly crowded rather than concentrated, inasmuch as no single species of macromolecule is necessarily concentrated (3). Thus, a crowded physiological environment often serves as the field of action for protein-protein interactions in vivo (3,4). Crowding can be achieved in vitro by adding high concentrations of nominally inert macromolecules or macromolecular crowding agents to a solution containing the reactants of interest (5–9). Reduction of configurational space due to steric exclusion and other repulsive intermolecular interactions (e.g., electrostatic) in crowded media can have a significant effect upon the conformation and state of association of each macromolecular species, dilute as well as concentrated, in the medium (10,11). In recent years, it has gradually become recognized that there may exist fundamental differences between protein-protein interactions in crowded and dilute solutions (9,12–16).
Until now, the focus of attention in experimental and theoretical studies of macromolecular crowding has been on the role of repulsive steric interactions, due to their undeniable ubiquity (5,10,17). Although the likely presence of additional weak and nonspecific attractive interactions between macromolecules—in contrast to strong attractive interactions leading to the formation of specific complexes—has been acknowledged (7,18–23), their significance and influence on specific interactions has, to our knowledge, received only cursory attention. As steric interactions are athermal, their influence is expected to be essentially independent of temperature. The influence of attractive interactions, on the other hand, would be expected to be manifested in a temperature dependence of any observed crowding effect (24). This study was undertaken to detect and characterize any significant modulating effect of temperature with respect to the influence of high concentrations of inert polymeric crowding agents upon the association of dilute proteins, and to quantitate the respective contributions of weak attractive interactions and volume exclusion to the total crowding effect.
As a model system, we have studied the effect of several polymers on the interaction between catalase (EC 1.11.1.6) and copper, zinc superoxide dismutase, or SOD (EC 1.15.1.1). These enzymes normally act in concert in vivo, whereby SOD protects catalase against inhibition by the superoxide anion radical while catalase provides protection for SOD against inactivation by its own reaction product, hydrogen peroxide (H2O2) (25,26). A number of studies (27–31) have indirectly suggested that the two enzymes may form a transient physical complex, although a direct study of complex formation has not heretofore been undertaken. Our initial fluorescence titration experiments established that the interaction between catalase and SOD in dilute solution may be described quantitatively as the equilibrium association between SOD and an undetermined number of equivalent and independent binding sites on catalase.
The effect of temperature and varying concentrations of three different polymeric crowding agents (dextran 70, Ficoll 70, and polyethylene glycol (PEG) 2000) upon the affinity of SOD for the catalase binding sites was then measured. It was found that variations in temperature between 8.0°C and 37.0°C substantially modulate the effect of all three polymers on the affinity of SOD for catalase binding sites. We present a simplified statistical-thermodynamic model, according to which polymer interacts with each protein species via a nonspecific repulsive interaction (steric volume exclusion) and an attractive semispecific interaction that scales with the solvent-accessible surface area of that species. The experimental data are then interpreted in the context of this model.
Materials and Methods
Materials
The following reagents were used in this study: bovine liver catalase (Sigma-Aldrich, St. Louis, MO) was used without further purification. The A1%1cm value of 13.5 at 405 nm (32) was used for protein concentration measurements. Bovine erythrocyte SOD was prepared and purified as described previously (33,34). Purified SOD was homogenous on SDS-PAGE. The concentration of SOD was determined at 258 nm using a molar extinction coefficient of 10,300 L·mol−1·cm−1 (34). The specific activity of SOD was determined by using a pyrogallol autooxidation inhibition assay (35) to be 4680 ± 90 U mg−1 (n = 3). Catalase activity was measured at 405 nm by the absorbance of a stable complex of ammonium molybdate with H2O2 that remained after the decomposition of H2O2 catalyzed by catalase (36) as 2240 ± 60 U mg−1 (n = 3). The crowding agents, dextran 70, Ficoll 70, PEG 2000, and glycerol, were obtained from Sigma. All other chemicals used were made in China and were of analytical grade. All reagent solutions used were prepared in 50 mM NaH2PO4-Na2HPO4 buffer containing 15 mM NaCl (pH 7.4). It was verified that the pH of a buffer did not change significantly upon addition of any of the nonionic additives (polymers, glycerol) at concentrations of up to 300 g/L or change in temperature over the range 8.0–37.0°C.
Tryptophan intrinsic fluorescence
Intrinsic fluorescence spectroscopic experiments on the interaction of catalase with SOD in the absence and in the presence of glycerol, dextran 70, Ficoll 70, or PEG 2000, were carried out at 8.0, 15.0, 25.0, 30.0, and 37.0°C using a model No. LS-55 luminescence spectrometer (PerkinElmer Life Sciences, Shelton, CT). Because SOD contains a single tyrosine residue (Tyr-108), which is completely conserved, and no tryptophan per subunit (37), and catalase contains 21 tyrosine and 6 tryptophan residues per subunit (38,39), the excitation wavelength at 295 nm was used for the intrinsic fluorescence measurements, and the emission spectra were recorded between 300 and 400 nm. At 295 nm, no tyrosine fluorescence is elicited, nor its energy transferred to tryptophan (40). The excitation and emission slits were both 10 nm, and the scan speed was 250 nm/min. A quantity of 2.70 mL of catalase was placed in a 1-cm thermostated quartz fluorescence cuvette and titrated with 10-μL aliquots of SOD with continuous stirring. Both enzyme solutions were prepared in phosphate buffer containing different concentrations of crowding agents (100–300 g/L). After each sample addition, the solution was mixed thoroughly and allowed to equilibrate thermally for 5 min before the fluorescence measurements. The fluorescence measurements of catalase and SOD were carried out with optical density below 0.1 at 295 and 258 nm, respectively, to avoid the inner filter effects (9,41).
The fluorescence data for the control system, the buffer titrated with 10 μL aliquots of SOD, was also measured under the same condition and used to correct the observed fluorescence and the dilution effects. Each spectrum was scanned three times to acquire the final fluorescence emission spectra. Although the absolute intensity and wavelength of maximum emission may vary with temperature or concentration of a particular additive, the wavelength of maximum emission in a given medium at fixed temperature is independent of SOD concentration under all conditions.
Analysis of the dependence of fluorescence intensity on SOD concentration
We postulate equilibrium association of SOD with one or more equivalent and independent binding sites on catalase (a single binding site is hereafter referred to as “site”), the affinity of which is characterized by an apparent equilibrium association constant defined as
| (1) |
where cSOD, csite, and cSOD⋅site denote the molar concentrations of free SOD, free site, and bound SOD, respectively. In this study, the concentration of catalase (i.e., sites) was much smaller than that of added SOD, so that cSOD ≈ cSOD,tot is the total molar concentration of S. We denote the fluorescence intensity of catalase in the absence of SOD by F0 and in the presence of saturating amounts of SOD by Fsat. When all sites are equivalent and independent, the dependence of catalase fluorescence intensity at the emission maximum upon SOD concentration will be given by (42)
| (2) |
where fsat is the fractional saturation of binding sites, given by
| (3) |
Equations 2 and 3 were fit to the experimentally measured dependence of F upon Stot via nonlinear least-squares to obtain best-fit values of F0, Fsat, and log10 Ka. Least-squares curve fitting was performed using either Origin (MicroCal, Northampton, MA) or MATLAB (The MathWorks, Natick, MA). The reported uncertainty of the derived best-fit value of log10 Ka corresponds to 95% confidence limits or ±2 SE of estimate. Depending upon the measured affinity, which varies substantially with temperature and the concentration of crowding agent, the fractional saturation obtained at the highest feasible SOD concentrations varied between ∼40% and >95%. It is shown in Table S1 in the Supporting Material that the data are sufficiently precise that the value of log10 Ka may be estimated to within acceptable limits of uncertainty even under the lowest affinity conditions encountered in this study.
Thermodynamic analysis of the dependence of equilibrium binding constant upon additive concentration
The thermodynamic equilibrium site-binding constant in the absence of additive, Koa, is given by
| (4) |
where aX and γx, respectively, denote the thermodynamic activity and activity coefficient of species X. We thus define the crowding factor (10,20) as
| (5) |
where the various activity coefficients are functions of additive concentration (10,20).
We note that Eqs. 4 and 5 are strictly applicable only to the case of a protein bearing a single binding site. In the Supporting Material, we present an extended treatment applicable to multiple identical binding sites and justification for the approximation employed here.
The crowding factor so-defined is a measure of the effect of additive upon the standard Gibbs free energy changes accompanying site binding,
| (6) |
where R denotes the molar gas constant and T the absolute temperature. This expression may be further decomposed into its enthalpic and entropic contributions,
| (7) |
where ΔΔH and ΔΔS, respectively, denote the effect of additive upon the standard enthalpy and entropy changes accompanying site binding. A molecular interpretation of these quantities will be introduced after presentation of the experimental results.
Results
The interaction between catalase and SOD is saturable
The dependence of fluorescence intensity at the emission maximum upon the concentration of SOD at 25.0°C in the absence and in the presence of 200 g/L Ficoll 70 is plotted in Fig. 1, together with the best fit of Eqs. 2 and 3 to each data set. These plots typify the results of many titrations of catalase with SOD carried out under conditions of varying polymer concentration and temperature. The best-fit values of F0, Fsat, and log10 Ka vary with temperature and with the concentration and type of each polymer added, but under all conditions the raw data are fit to within experimental precision by Eqs. 2 and 3. We may thus interpret the results as indicating that SOD binds to one or more specific sites on a catalase molecule, and that these binding sites are equivalent and independent.
Figure 1.
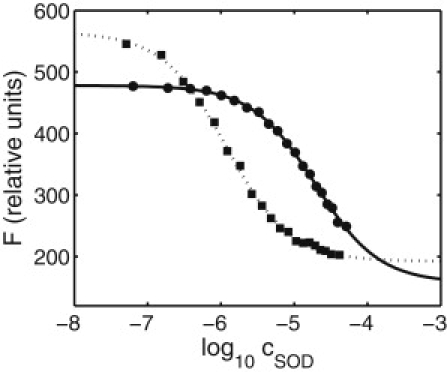
Dependence of catalase fluorescence intensity at 25.0°C on the concentration of added SOD. (Circles) No additive. (Solid curve) Best-fit of Eqs. 2 and 3, with F0 = 478, Fsat = 157, and log10Ka = 4.71. (Squares) 200 g/L Ficoll 70 added. (Dotted curve) Best-fit of Eqs. 2 and 3, with F0 = 565, Fsat = 192, and log10Ka = 5.93. The concentration of catalase in the cuvette was 0.09 μM.
The quenching of fluorescence intensity of the catalase tryptophan upon the addition of SOD suggests that reporter groups are either directly involved in SOD binding or are in the vicinity of a binding site. Because catalase is a homotetramer, symmetry considerations suggest the presence of either two or four binding sites, but the fluorescence titrations contain no information regarding the stoichiometry. Subsequent calculations relating to the effect of crowding will be formulated in the context of a model for binding of SOD to an individual site, and crowding effects are assumed to be equal for all sites.
Titration curves measured under all conditions of varying temperature and additive concentration could be fit to within experimental precision by Eqs. 2 and 3, indicating that while changes in temperature and additive concentration do affect the affinity of SOD for catalase, they do not alter the basic mode of association. The precision with which the value of Ka can be determined depends upon the extent of change in saturation over the experimentally accessible range of SOD concentrations, which in turn depends upon the affinity. Best-fit values of log10 Ka obtained under each set of experimental conditions are reported in Table S1, together with uncertainty limits corresponding to ±2 SE of estimate (95% confidence limits).
Effect of polymer concentration upon site-binding at constant temperature
The influence of various concentrations of three nonspecific macromolecular crowding agents, dextran 70, Ficoll 70, and PEG 2000, and one low molecular mass compound, glycerol, on the interaction of catalase with SOD, was determined by fluorescence titration at 25.0°C. The dependence of ln Γ upon the concentration of each of these additives is plotted in Fig. 2. It is evident that at 25.0°C, the three polymeric additives have a significant and qualitatively similar enhancing effect upon the association equilibrium, whereas glycerol has a modestly inhibiting effect. Within the uncertainty of measurement, the magnitude of each of these effects increases approximately linearly with additive concentration.
Figure 2.

Dependence of ln Γ on additive concentration. Error bars correspond to ±2 SE of estimate (95% confidence limits). Lines plotted in each panel corresponding to a polymeric additive represent the best-fits of special case 1 (dashed) and special case 2 (dotted) of the molecular model introduced below to the full set of data representing both concentration and temperature dependence. (The two lines calculated for PEG superimpose.) The lines are calculated using parameter values specified in the section describing the molecular model and in Table 2.
Combined effect of temperature and high polymer concentration on site-binding
In Fig. 3, the value of ln Γ obtained in the presence of 200 g/L of each of the polymeric additives is plotted as a function of 1/T, in accordance with the functional form suggested by Eq. 7. In all three cases, the observed dependence is linear to within experimental uncertainty, indicating that the corresponding values of ΔΔH and ΔΔS are approximately independent of temperature over this range of temperatures. The values of these parameters, obtained for each polymer by linear least-squares fit to the data, are given in the figure caption. Both ΔΔH and ΔΔS are positive, indicating that the overall effect of enthalpic interaction between each polymer and protein is to inhibit heteroassociation of catalase and SOD, whereas the overall effect of entropic interaction between polymer and protein is to enhance heteroassociation.
Figure 3.
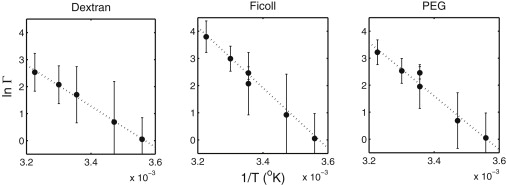
Dependence of ln Γ on inverse temperature (K−1) at an additive concentration of 200 g/L. The dotted lines in each panel represent the best fit of Eq. 7, with the following parameter values. Dextran 70: ΔΔH/R = 7500 ± 1500 K; ΔΔS/R = 25.6 ± 5.0. Ficoll 70: ΔΔH/R = 11,000 ± 1500 K; ΔΔS/R = 39.3 ± 5.0. PEG 2000: ΔΔH/R = 10,100 ± 1400 K; ΔΔS/R = 35.3 ± 4.6. Indicated uncertainties represent ±1 SE of estimate.
Molecular model for the combined effect of enthalpic and entropic interactions between protein and polymer upon protein association
Equation 5 may be rewritten as
| (8) |
When the total concentration of protein is sufficiently dilute, as in the present set of experiments, the activity coefficient of each protein species depends only upon its interaction with polymer (17),
| (9) |
where δGi,polymer represents the increment of Gibbs free energy characterizing the interaction between protein and polymer under standard state conditions, which depend upon polymer concentration.
Early studies of the thermodynamic interaction of protein and polymer (43–46) focused almost exclusively on excluded volume interactions, although the possible contribution of additional attractive interactions was acknowledged (18). Here we introduce explicitly an additional contribution to the activity coefficient or chemical potential of the ith protein species arising from attractive interactions between protein and polymer, which is assumed to be independent of excluded volume interactions:
| (10) |
For simplicity, we shall represent each protein species as an equivalent sphere. The properties of these equivalent spherical particle representations are presented in Table 1. For purposes of estimating the activity coefficient, the dimensions of (site) are taken to be those of catalase tetramer in the absence of SOD. Extension to arbitrary convex hard-particle representations is straightforward (17), but complicates calculations and adds undetermined parameters without additional insight into underlying principles. The magnitude of the excluded volume contribution to the chemical potential of the ith protein species due to interaction between that species and polymer is estimated using Ogston available volume theory (20,47,48),
| (11) |
where ri denotes the radius of the effective hard sphere representing protein species i, rp the radius of the effective hard rod representing polymer, vex,p the specific volume excluded by polymer to protein, and wp the w/v concentration of polymer. The free energy of attractive interaction between polymer and protein is assumed to be proportional to the surface area of the protein and the w/v concentration of polymer,
| (12) |
where δGi(attract) denotes the free energy of attractive interaction per unit surface area of the ith protein species and per unit w/v concentration of polymer. It follows that
| (13) |
Table 1.
Geometric properties of the spherical equivalent particles representing each protein species, calculated assuming that the partial specific volume of each species is 0.73 cm3/g
| MW | V (cm3) | r (cm) | s (cm2) | |
|---|---|---|---|---|
| SOD | 30,000 | 2.64 × 10−20 | 2.06 × 10−7 | 5.3 × 10−13 |
| Site (catalase) | 240,000 | 2.91 × 10−19 | 4.11 × 10−7 | 2.12 × 10−12 |
| SOD⋅site | 270,000 | 3.27 × 10−19 | 4.28 × 10−7 | 2.30 × 10−12 |
From Eqs. 8 and 10, we obtain
| (14) |
where
| (15) |
and
| (16) |
In the Appendix, general expressions for ln Γ(exvol) and ln Γ(attract) are presented that are functions of δHi(attract) and δSi(attract) for each of the three species indicated. Also presented are two special cases that resulted in simplified expressions for ln Γ(exvol) and ln Γ(attract).
To examine the ability of each of these special cases to account for the experimental data presented here, the values of ri and si for each of the protein species were set equal to the corresponding values in Table 1. The values of rp and vex,p were set equal to 7 × 10−8 cm and 7.5 × 10−4 L/g, as obtained from previous analyses of dextran-induced excluded volume effects using Ogston available volume theory (49–51).
For special case 1, Eqs. 14, 22, and 23 were globally fit to all of the concentration-dependent and temperature-dependent results presented in Table S1 to obtain the best-fit values for and for each polymeric additive given in Table 2.
Table 2.
Best-fit parameters obtained by global fitting of special cases 1 and 2 of the molecular model described in the text to the experimentally measured dependence of ln Γ on temperature and the concentration of each polymeric additive
| Special case 1 | Special case 2 | |||
|---|---|---|---|---|
| (K L g−1 cm−2) | (L g−1 cm−2) | (K L g−1 cm−2) | (L g−1 cm−2) | |
| Dextran 70 | −6.8 × 1013 | −2.2 × 1011 | −10.1 × 1013 | −3.4 × 1011 |
| Ficoll 70 | −10.4 × 1013 | −3.5 × 1011 | −15.5 × 1013 | −5.2 × 1011 |
| PEG 2000 | −9.6 × 1013 | −3.2 × 1011 | −14.2 × 1013 | −4.8 × 1011 |
For special case 2, Eqs. 14, 15, and 22 were globally fit to all of the concentration-dependent and temperature-dependent results presented in Table S1 to obtain the best-fit values for δH(attract) and δS(attract) for each polymeric additive given in Table 2.
In Fig. 2, the dependence of ln Γ on polymer concentration at 25.0°C, calculated for each special case using the corresponding global best-fit parameter values, is plotted together with the data. In Fig. 4, the dependence of ln Γ upon temperature at a polymer concentration of 200 g/L, calculated for each special case using the corresponding global best-fit parameter values, is plotted together with the data.
Figure 4.
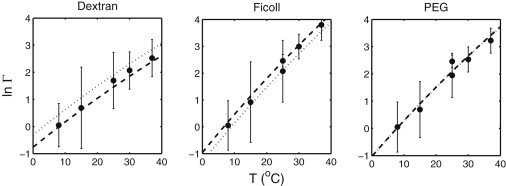
Dependence of ln Γ on temperature (°C) at an additive concentration of 200 g/L. Lines plotted in each panel corresponding to a polymeric additive represent the best-fits of special case 1 (dashed) and special case 2 (dotted) of the molecular model introduced below to the full set of data representing both concentration and temperature dependence. (The two lines calculated for PEG superimpose.) The lines are calculated using parameter values specified in the section describing the molecular model and in Table 2.
Discussion
The extensive fluorescence titrations on which this study was based were carried out over an extremely broad range of SOD concentration—up to three orders of magnitude. All of these data, obtained under widely varying conditions of additive concentration and temperature, are precisely accounted for by the simple hypothesis that under each set of conditions SOD binds reversibly to independent and equivalent binding sites on catalase, although the number of such sites cannot be determined from these or any other spectroscopic data alone. Examination of the homotetrameric structure of the catalase molecule (39) reveals three twofold axes of rotational symmetry. Therefore there may exist two or four identical binding sites situated at an interface between protomers or four binding sites situated entirely within an individual protomer. Mao et al. (31) have estimated that four molecules of SOD binds to one molecule of catalase to form SOD-catalase conjugate. Our additional isothermal titration calorimetric experiments (data not shown) also suggest that SOD can bind to four independent and equivalent binding sites on catalase with a moderate affinity (105 M−1).
The affinity of SOD for catalase was found to increase significantly with increasing concentration of each of three nonionic polymeric additives at 25.0°C, whereas it decreased slightly upon addition of the same w/v concentrations of glycerol, which has an elemental composition comparable to that of the polymers. This indicates that the affinity enhancement observed in the presence of polymeric additive is due to the polymeric nature of the additive rather than to modification of bulk solvent properties (e.g., alteration of dielectric constant, ionic strength) per se.
Perhaps the most important and novel finding of our study is that the enhancement of catalase-SOD binding by added polymer decreases significantly with decreasing temperature. According to Eq. 7, the effect is predicted to vanish at an absolute temperature
| (17) |
that we call the θ-temperature by analogy to a comparable situation encountered in the theory of polymer solutions (52), when attractive and repulsive interactions between polymer segments cancel, and the polymer chain behaves like an ideal random coil. The existence of a θ-temperature for the interaction of protein and polymer was predicted recently by Douglas et al. (24) on the basis of a Flory-Huggins type lattice model incorporating attractive interactions as well as excluded volume repulsive interactions between protein and polymer. The values of Tθ for each of the three polymers, calculated using the above equation with values for ΔΔH and ΔΔS given in the caption to Fig. 3, fall in the range 6–8°C.
The molecular model proposed here requires a number of simplifying assumptions. The qualitative or semiquantitative validity of the equivalent hard-particle representation of proteins has been demonstrated previously in numerous publications cited in several reviews (3,7,11). The Ogston model, which we used to estimate the excluded volume contribution of polymers to thermodynamic activity of proteins (Eq. 11), has been used before to successfully model the experimentally observed dependence of the activity of proteins upon polymer concentration (49–51). However, one may ask whether the values of the model parameters rp and vex,p used here are appropriate, given that the earlier analyses did not take into account the possible concurrent existence of enthalpic interactions.
We believe that the values used here are sufficiently realistic for our purpose, because the value of rp used here (7 × 10−8 cm) was obtained by Laurent and Killander (50) using the Ogston model to successfully model the dependence of the partition coefficient (thermodynamically equivalent to the activity coefficient) of a series of proteins in dextran upon their molecular weight. It is extremely unlikely that the strength of enthalpic interactions between dextran and all of these proteins was identical, unless it was negligible in all cases. Moreover, the dependence of the activity coefficient of bovine serum albumin upon the concentrations of Ficoll 70 and dextran 70 was measured via tracer sedimentation equilibrium (51) in the laboratory of one of the coauthors very recently, and the activity coefficient of bovine serum albumin in either polymer was found to be independent of temperature between 5 and 37°C (A. Fodeke and A. P. Minton, unpublished). It is clear that the temperature-dependent crowding effect observed in this study is not a universal property of all protein-polymer systems, but a property of this particular system (catalase and SOD) deriving from a sequence-dependent enthalpic interaction between one or both of these proteins and the polymeric crowders. Thus we feel that consensus values of the Ogston model parameters derived from analysis of the interaction between polymers and a broad variety of highly soluble proteins should provide an acceptable semiquantitative estimate of the magnitude of the excluded volume contribution to total protein-polymer interaction in those systems (such as the one studied here) exhibiting a nonnegligible enthalpic contribution to this interaction.
The validity of additional simplifying assumptions introduced here remains to be examined. The first of these is embodied in Eq. 12, which states that the attractive interaction of polymer with each protein species is proportional to the solvent-accessible surface area of that species. A more atomistic analysis might assume that the attractive interaction of polymer with protein reflects binding of polymer segments to specific sites on the surface of each protein species, and that when the proteins associate, some of the sites become inaccessible to polymer. Because we have no way of estimating the number of sites on each protein and the extent to which these sites become inaccessible upon association, it is reasonable to assume that the number of accessible sites is proportional to solvent-accessible surface area. Because the interaction of polymer and protein is assumed to be weak, as it is manifested only at very high polymer concentrations, we elect to treat the interaction as equivalent to physical adsorption (53), and the parameter δGi(attract) thus represents an affinity of polymer for protein species i that is averaged over the solvent-accessible surface area.
The second simplifying assumption, employed in special case 1 of the molecular model, is that the activity coefficients of site and SOD⋅site are approximately identical, i.e., equally affected by temperature changes and the addition of polymer. We rationalize this assumption on the grounds that SOD is only ∼1/8 the mass of catalase, so that the size of catalase and a catalase-SOD complex would be of similar size (Table 1).
The third simplifying assumption, employed in special case 2 of the molecular model, is that values of δHi(attract) and δSi(attract) are approximately identical for all species. We do not claim any physical justification for this assumption. Special case 2 is presented as an alternative to special case 1 that is employed to test the sensitivity of the model to alternative simplifying assumptions.
With only two adjustable parameters, the simplified molecular model presented here successfully accounts for both the observed dependence of SOD⋅site binding affinity on polymer concentration at fixed temperature, and the observed dependence of binding affinity on temperature at fixed polymer concentration, to within the uncertainty of measurement. Although the numerical values of molecular parameters derived from modeling the data (in contrast to the thermodynamic parameters ΔΔH and ΔΔS) depend upon approximations employed in order to render the molecular model computationally tractable, and hence should be regarded as having order-of-magnitude significance only, the essential qualitative validity of the model does not appear to depend upon the particulars of these approximations.
The data and model presented in this article provide a possible explanation for the diversity of results reported in the literature regarding the effect of high concentrations of added polymers upon the association of dilute macromolecules at constant temperature. Although many observations of a large effect of inert polymer on protein association have been reported (reviewed in (3,7,11,20)), other studies have found little or no effect of polymer (22,54,55), even under conditions when comparable concentrations of inert proteins were observed to have a substantial effect (54,55).
The results presented here indicate that three different polymeric crowding agents have a qualitatively similar crowding effect. However, the molecular model proposed suggests that the values of ΔΔH and ΔΔS may depend upon the size and nature of the solvent-accessible surface of the particular macromolecules undergoing association and of their putative complexes. Hence, ΔΔH and/or ΔΔS, and consequently Tθ, may be subject to significant variation between different associating systems and polymeric crowding agents. (A modest difference of only 5% in ΔΔH or ΔΔS between two different experimental systems would result in an ∼15° difference in Tθ.)
The effect of polymer on association at a given temperature T thus depends upon the relationship between T and Tθ. When T is >>Tθ, excluded volume effects will predominate, and polymer will have an enhancing effect upon protein association. When T is <<Tθ, attractive interactions between polymer and protein will predominate, and polymer would be expected to have an inhibiting effect upon protein association. In addition, when T is approximately equal to Tθ, the two effects would be expected to approximately cancel and, as a result, the addition of polymer would have little or no effect on protein association.
The great majority of experiments carried out with the intention of investigating crowding effects on protein association or folding employ one of the polymeric crowding agents used in this study, because these materials are relatively inexpensive and transparent in the visible and near-ultraviolet. An often-asked question is whether such additives provide a credible simulation of the local environment of proteins in physiological media that contain high concentrations of other proteins, as opposed to flexible chain polymers (R. J. Ellis, Emeritus Professor of Biochemistry, Warwick University, personal communication). It has been reported that high concentrations of several inert proteins appear to induce self-association of a dilute protein under experimental conditions where addition of high concentrations of added PEG has no effect (54,55). At the time of this writing, little is known about the temperature dependence of protein-induced crowding effects. The only relevant data available indicate that self-crowding of hemoglobin by other hemoglobin molecules is not significantly affected by variation in temperature over the temperature range 5–37°C (56). More-thorough investigation of the temperature dependence of crowding by high concentrations of protein is clearly called for.
The influence of temperature and temperature-dependent interactions between macromolecules on the magnitude of crowding effects has been largely neglected until now. Our results reveal the possible importance of an additional factor that must be taken into account when assessing the physiological consequences of crowding (11,57). The results of a very recent Brownian dynamics simulation of a complex, highly volume-occupied mixture of proteins and nucleic acids designed to mimic eukaryotic cytoplasm, incorporating model hydrophobic and electrostatic attraction between macromolecules (58), suggest that the overall effect of crowding upon protein associations and stability in such an environment may depend upon sequence-specific properties of individual proteins in addition to factors such as size and shape affecting excluded volume. This conclusion is supported by the results of our study. However, it would be premature to draw broad conclusions from either the simulation or the results of our highly restricted study. We hope that these results will help to establish the importance of studying the temperature dependence of crowding effects on the association state and stability of proteins and other macromolecules, and the importance of employing inert protein crowding agents (see, for example, (51,59,60)) in addition to, or instead of, polymeric crowding agents in future experimental studies.
Appendix: A Molecular Model for the Combined Effect of Enthalpic and Entropic Interactions on Binary Hetero-Association
It follows from text Eqs. 11, 13, 15, and 16 that in the general case
| (18) |
and
| (19) |
where
| (20) |
and
| (21) |
Equations 18–21 may be rearranged into the form of Eq. 7, but the resulting expressions for ΔΔH and ΔΔS contain too many undetermined parameters to be useful. However, there are two special cases that are accessible to further analysis, albeit highly approximate.
Special case 1
In the limit that SOD is much smaller than site, it is reasonable to assume that the properties of site and SOD⋅site are very similar. According to Table 1, the radii of the equivalent spherical particles representing site and SOD⋅site differ by <5% and their surface areas differ by <10%. We shall therefore make the additional approximation that attractive interactions between polymer and these two species are comparably similar. It follows that the contributions of the activity coefficients of site and SOD⋅site to Eqs. 15 and 16 approximately cancel, leading to the simplified expressions
| (22) |
and
| (23) |
Equations 14, 22, and 23 thus specify the concentration and temperature dependence of ln Γ as a function of and and the structural parameters rSOD, sSOD, rP, and vex,p.
Special case 2
If we assume that δH(attract) and δS(attract) are approximately the same for all species, then Eqs. 19–21 simplify to
| (24) |
Equations 14, 18, and 24 thus specify the concentration and temperature dependence of ln Γ as a function of δH(attract), δS(attract), the radii, and the surface areas of each protein, rp, and vex,p.
Acknowledgments
A.P.M. thanks Y.L. and the College of Life Sciences, Wuhan University, for hospitality extended during a visit during which this collaborative study was initiated.
Research of Y.L., M.J., H-T.L., and J.C. is supported by National Key Basic Research Foundation of China (Grant 2006CB910301 to Y.L.), National Natural Science Foundation of China (Grants 30770421 and 30970599 to Y.L.), and Chinese 111 Project Grant B06018. The research of A.P.M. is supported by the Intramural Research Program of the National Institute of Diabetes and Digestive and Kidney Diseases.
Footnotes
Ming Jiao's present address is School of Pharmacy, Xianning University, Xianning, China.
Contributor Information
Allen P. Minton, Email: minton@helix.nih.gov.
Yi Liang, Email: liangyi@whu.edu.cn.
Supporting Material
References
- 1.Schreiber G. Kinetic studies of protein-protein interactions. Curr. Opin. Struct. Biol. 2002;12:41–47. doi: 10.1016/s0959-440x(02)00287-7. [DOI] [PubMed] [Google Scholar]
- 2.Stites W.E. Protein-protein interactions: interface structure, binding thermodynamics, and mutational analysis. Chem. Rev. 1997;97:1233–1250. doi: 10.1021/cr960387h. [DOI] [PubMed] [Google Scholar]
- 3.Zimmerman S.B., Minton A.P. Macromolecular crowding: biochemical, biophysical, and physiological consequences. Annu. Rev. Biophys. Biomol. Struct. 1993;22:27–65. doi: 10.1146/annurev.bb.22.060193.000331. [DOI] [PubMed] [Google Scholar]
- 4.Fulton A.B. How crowded is the cytoplasm? Cell. 1982;30:345–347. doi: 10.1016/0092-8674(82)90231-8. [DOI] [PubMed] [Google Scholar]
- 5.Ellis R.J. Macromolecular crowding: obvious but underappreciated. Trends Biochem. Sci. 2001;26:597–604. doi: 10.1016/s0968-0004(01)01938-7. [DOI] [PubMed] [Google Scholar]
- 6.Ellis R.J. Macromolecular crowding: an important but neglected aspect of the intracellular environment. Curr. Opin. Struct. Biol. 2001;11:114–119. doi: 10.1016/s0959-440x(00)00172-x. [DOI] [PubMed] [Google Scholar]
- 7.Hall D., Minton A.P. Macromolecular crowding: qualitative and semiquantitative successes, quantitative challenges. Biochim. Biophys. Acta. 2003;1649:127–139. doi: 10.1016/s1570-9639(03)00167-5. [DOI] [PubMed] [Google Scholar]
- 8.Minton A.P. Implications of macromolecular crowding for protein assembly. Curr. Opin. Struct. Biol. 2000;10:34–39. doi: 10.1016/s0959-440x(99)00045-7. [DOI] [PubMed] [Google Scholar]
- 9.Zhou Y.L., Liao J.M., Liang Y. Macromolecular crowding enhances the binding of superoxide dismutase to xanthine oxidase: implications for protein-protein interactions in intracellular environments. Int. J. Biochem. Cell Biol. 2006;38:1986–1994. doi: 10.1016/j.biocel.2006.05.012. [DOI] [PubMed] [Google Scholar]
- 10.Minton A.P. Excluded volume as a determinant of macromolecular structure and reactivity. Biopolymers. 1981;20:2093–2120. [Google Scholar]
- 11.Zhou H.X., Rivas G., Minton A.P. Macromolecular crowding and confinement: biochemical, biophysical, and potential physiological consequences. Annu. Rev. Biophys. 2008;37:375–397. doi: 10.1146/annurev.biophys.37.032807.125817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kozer N., Schreiber G. Effect of crowding on protein-protein association rates: fundamental differences between low and high mass crowding agents. J. Mol. Biol. 2004;336:763–774. doi: 10.1016/j.jmb.2003.12.008. [DOI] [PubMed] [Google Scholar]
- 13.Minton A.P. How can biochemical reactions within cells differ from those in test tubes? J. Cell Sci. 2006;119:2863–2869. doi: 10.1242/jcs.03063. [DOI] [PubMed] [Google Scholar]
- 14.Rivas G., Ferrone F., Herzfeld J. Life in a crowded world. EMBO Rep. 2004;5:23–27. doi: 10.1038/sj.embor.7400056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Stevens A., Wang S.X., Schleich T. 13C-NMR off-resonance rotating frame spin-lattice relaxation studies of bovine lens γ-crystalline self-association: effect of macromolecular crowding. Biochim. Biophys. Acta. 1995;1246:82–90. doi: 10.1016/0167-4838(94)00172-d. [DOI] [PubMed] [Google Scholar]
- 16.Zorrilla S., Rivas G., Lillo P. Protein self-association in crowded protein solutions: a time-resolved fluorescence polarization study. Protein Sci. 2004;13:2960–2969. doi: 10.1110/ps.04809404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Minton A.P. Molecular crowding: analysis of effects of high concentrations of inert cosolutes on biochemical equilibria and rates in terms of volume exclusion. Methods Enzymol. 1998;295:127–149. doi: 10.1016/s0076-6879(98)95038-8. [DOI] [PubMed] [Google Scholar]
- 18.Comper W.D., Laurent T.C. An estimate of the enthalpic contribution to the interaction between dextran and albumin. Biochem. J. 1978;175:703–708. doi: 10.1042/bj1750703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Du F., Zhou Z., Liang Y. Mixed macromolecular crowding accelerates the refolding of rabbit muscle creatine kinase: implications for protein folding in physiological environments. J. Mol. Biol. 2006;364:469–482. doi: 10.1016/j.jmb.2006.09.018. [DOI] [PubMed] [Google Scholar]
- 20.Minton A.P. The effect of volume occupancy upon the thermodynamic activity of proteins: some biochemical consequences. Mol. Cell. Biochem. 1983;55:119–140. doi: 10.1007/BF00673707. [DOI] [PubMed] [Google Scholar]
- 21.Minton A.P. A molecular model for the dependence of the osmotic pressure of bovine serum albumin upon concentration and pH. Biophys. Chem. 1995;57:65–70. doi: 10.1016/0301-4622(95)00046-z. [DOI] [PubMed] [Google Scholar]
- 22.Phillip Y., Sherman E., Schreiber G. Common crowding agents have only a small effect on protein-protein interactions. Biophys. J. 2009;97:875–885. doi: 10.1016/j.bpj.2009.05.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Li C., Pielak G.J. Using NMR to distinguish viscosity effects from nonspecific protein binding under crowded conditions. J. Am. Chem. Soc. 2009;131:1368–1369. doi: 10.1021/ja808428d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Douglas J.F., Dudowicz J., Freed K.F. Crowding induced self-assembly and enthalpy-entropy compensation. Phys. Rev. Lett. 2009;103:135701. doi: 10.1103/PhysRevLett.103.135701. [DOI] [PubMed] [Google Scholar]
- 25.Kwon H.Y., Choi S.Y., Kang J.H. Oxidative modification and inactivation of Cu,Zn-superoxide dismutase by 2,2′-azobis(2-amidinopropane) dihydrochloride. Biochim. Biophys. Acta. 2000;1543:69–76. doi: 10.1016/s0167-4838(00)00197-7. [DOI] [PubMed] [Google Scholar]
- 26.Liang Y., Qu S.S., Liang Y. An on-line calorimetric study of the dismutation of superoxide anion catalyzed by SOD in batch reactors. Chem. Eng. Sci. 2000;55:6071–6078. [Google Scholar]
- 27.Amstad P., Peskin A., Cerutti P. The balance between Cu,Zn-superoxide dismutase and catalase affects the sensitivity of mouse epidermal cells to oxidative stress. Biochemistry. 1991;30:9305–9313. doi: 10.1021/bi00102a024. [DOI] [PubMed] [Google Scholar]
- 28.Beckman J.S., Minor R.L., Jr., Freeman B.A. Superoxide dismutase and catalase conjugated to polyethylene glycol increases endothelial enzyme activity and oxidant resistance. J. Biol. Chem. 1988;263:6884–6892. [PubMed] [Google Scholar]
- 29.D'Agnillo F., Thomas M.S.C. Polyhemoglobin-superoxide dismutase-catalase as a blood substitute with antioxidant properties. Nat. Biotechnol. 1998;16:667–671. doi: 10.1038/nbt0798-667. [DOI] [PubMed] [Google Scholar]
- 30.Keller G.A., Warner T.G., Hallewell R.A. Cu,Zn superoxide dismutase is a peroxisomal enzyme in human fibroblasts and hepatoma cells. Proc. Natl. Acad. Sci. USA. 1991;88:7381–7385. doi: 10.1073/pnas.88.16.7381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mao G.D., Thomas P.D., Poznansky M.J. Superoxide dismutase (SOD)-catalase conjugates. Role of hydrogen peroxide and the Fenton reaction in SOD toxicity. J. Biol. Chem. 1993;268:416–420. [PubMed] [Google Scholar]
- 32.Prajapati S., Bhakuni V., Jain S.K. Alkaline unfolding and salt-induced folding of bovine liver catalase at high pH. Eur. J. Biochem. 1998;255:178–184. doi: 10.1046/j.1432-1327.1998.2550178.x. [DOI] [PubMed] [Google Scholar]
- 33.Bannister J., Bannister W., Wood E. Bovine erythrocyte cupro-zinc protein. 1. Isolation and general characterization. Eur. J. Biochem. 1971;18:178–186. doi: 10.1111/j.1432-1033.1971.tb01228.x. [DOI] [PubMed] [Google Scholar]
- 34.McCord J.M., Fridovich I. Superoxide dismutase. An enzymic function for erythrocuprein (hemocuprein) J. Biol. Chem. 1969;244:6049–6055. [PubMed] [Google Scholar]
- 35.Marklund S., Marklund G. Involvement of the superoxide anion radical in the autooxidation of pyrogallol and a convenient assay for superoxide dismutase. Eur. J. Biochem. 1974;47:469–474. doi: 10.1111/j.1432-1033.1974.tb03714.x. [DOI] [PubMed] [Google Scholar]
- 36.Góth L. A simple method for determination of serum catalase activity and revision of reference range. Clin. Chim. Acta. 1991;196:143–151. doi: 10.1016/0009-8981(91)90067-m. [DOI] [PubMed] [Google Scholar]
- 37.Ferreira S.T., Stella L., Gratton E. Conformational dynamics of bovine Cu, Zn superoxide dismutase revealed by time-resolved fluorescence spectroscopy of the single tyrosine residue. Biophys. J. 1994;66:1185–1196. doi: 10.1016/S0006-3495(94)80901-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Fielden E.M., Roberts P.B., Rotilio G. Mechanism and inactivation of superoxide dismutase activity. Biochem. Soc. Trans. 1973;1:52–53. [Google Scholar]
- 39.Reid T.J., 3rd, Murthy M.R., Rossmann M.G. Structure and heme environment of beef liver catalase at 2.5 Å resolution. Proc. Natl. Acad. Sci. USA. 1981;78:4767–4771. doi: 10.1073/pnas.78.8.4767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lakowicz J.R. Springer Science and Business Media; New York: 2006. Principles of Fluorescence Spectroscopy. [Google Scholar]
- 41.Eftink M.R., Ghiron C.A. Fluorescence quenching studies with proteins. Anal. Biochem. 1981;114:199–227. doi: 10.1016/0003-2697(81)90474-7. [DOI] [PubMed] [Google Scholar]
- 42.Tanford C. Wiley & Sons; New York: 1961. Physical Chemistry of Macromolecules. [Google Scholar]
- 43.Laurent T.C., G. Ogston A. The interaction between polysaccharides and other macromolecules. 4. The osmotic pressure of mixtures of serum albumin and hyaluronic acid. Biochem. J. 1963;89:249–253. doi: 10.1042/bj0890249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Laurent T.C. The interaction between polysaccharides and other macromolecules. 5. The solubility of proteins in the presence of dextran. Biochem. J. 1963;89:253–257. doi: 10.1042/bj0890253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ogston A.G., Phelps C. The partition of solutes between buffer solutions and solutions containing hyaluronic acid. Biochem. J. 1961;78:827–833. doi: 10.1042/bj0780827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Atha D.H., Ingham K.C. Mechanism of precipitation of proteins by polyethylene glycols. Analysis in terms of excluded volume. J. Biol. Chem. 1981;256:12108–12117. [PubMed] [Google Scholar]
- 47.Ogston A.G. The spaces in a uniform random suspension of fibers. Trans. Faraday Soc. 1958;54:1754–1757. [Google Scholar]
- 48.Ogston A.G. On the interaction of solute molecules with porous networks. J. Phys. Chem. 1970;74:668–669. [Google Scholar]
- 49.Hatters D.M., Minton A.P., J. Howlett G. Macromolecular crowding accelerates amyloid formation by human apolipoprotein C-II. J. Biol. Chem. 2002;277:7824–7830. doi: 10.1074/jbc.M110429200. [DOI] [PubMed] [Google Scholar]
- 50.Laurent T.C., Killander J. A theory of gel filtration and its experimental verification. J. Chromatogr. A. 1964;14:317–330. [Google Scholar]
- 51.Rivas G., Fernández J.A., Minton A.P. Direct observation of the self-association of dilute proteins in the presence of inert macromolecules at high concentration via tracer sedimentation equilibrium: theory, experiment, and biological significance. Biochemistry. 1999;38:9379–9388. doi: 10.1021/bi990355z. [DOI] [PubMed] [Google Scholar]
- 52.Flory P.J. Cornell University Press; Ithaca, NY: 1953. Principles of Polymer Chemistry. [Google Scholar]
- 53.Chatelier R.C., Minton A.P. Adsorption of globular proteins on locally planar surfaces: models for the effect of excluded surface area and aggregation of adsorbed protein on adsorption equilibria. Biophys. J. 1996;71:2367–2374. doi: 10.1016/S0006-3495(96)79430-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Minton A.P., Wilf J. Effect of macromolecular crowding upon the structure and function of an enzyme: glyceraldehyde-3-phosphate dehydrogenase. Biochemistry. 1981;20:4821–4826. doi: 10.1021/bi00520a003. [DOI] [PubMed] [Google Scholar]
- 55.Wilf J., Minton A.P. Evidence for protein self-association induced by excluded volume. Myoglobin in the presence of globular proteins. Biochim. Biophys. Acta. 1981;670:316–322. doi: 10.1016/0005-2795(81)90103-3. [DOI] [PubMed] [Google Scholar]
- 56.Ross P.D., Briehl R.W., Minton A.P. Temperature dependence of nonideality in concentrated solutions of hemoglobin. Biopolymers. 1978;17:2285–2288. doi: 10.1002/bip.1978.360170920. [DOI] [PubMed] [Google Scholar]
- 57.Zimmerman S.B., Trach S.O. Estimation of macromolecule concentrations and excluded volume effects for the cytoplasm of Escherichia coli. J. Mol. Biol. 1991;222:599–620. doi: 10.1016/0022-2836(91)90499-v. [DOI] [PubMed] [Google Scholar]
- 58.McGuffee S.R., Elcock A.H. Diffusion, crowding and protein stability in a dynamic molecular model of the bacterial cytoplasm. PLoS Comput. Biol. 2010;6:e1000694. doi: 10.1371/journal.pcbi.1000694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Rivas G., Fernández J.A., Minton A.P. Direct observation of the enhancement of noncooperative protein self-assembly by macromolecular crowding: indefinite linear self-association of bacterial cell division protein FtsZ. Proc. Natl. Acad. Sci. USA. 2001;98:3150–3155. doi: 10.1073/pnas.051634398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Zhou B.R., Liang Y., Chen J. Mixed macromolecular crowding accelerates the oxidative refolding of reduced, denatured lysozyme: implications for protein folding in intracellular environments. J. Biol. Chem. 2004;279:55109–55116. doi: 10.1074/jbc.M409086200. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


