Abstract
Structure determination of membrane proteins by crystallographic means has been facilitated by crystallization in lipidic mesophases. It has been suggested, however, that this so-called in meso method, as originally implemented, would not apply to small protein targets having ≤4 transmembrane crossings. In our study, the hypothesis that the inherent flexibility of the mesophase would enable crystallogenesis of small proteins was tested using a transmembrane pentadecapeptide, linear gramicidin, which produced structure-grade crystals. This result suggests that the in meso method should be considered as a viable means for high-resolution structure determination of integral membrane peptides, many of which are predicted to be coded for in the human genome.
The high-resolution structures of important membrane proteins have been obtained with crystals generated by the so-called in meso method (1). The method involves an initial reconstitution of the target protein into the bilayer of a cubic mesophase followed by the addition of a precipitant that triggers nucleation and crystal growth (2). At its simplest, the cubic phase consists of lipid and water. The lipid exists as a continuous, highly curved bilayer that divides the aqueous component into two interpenetrating but noncontacting channels.
The in meso method has been shown to be quite general in that it has been used to solve crystal structures of prokaryotic and eukaryotic proteins—proteins that are monomeric, homo- and heteromultimeric, chromophore-containing and chromophore-free, and α-helical and β-barrel proteins. Its most recent successes are the human, engineered β2-adrenergic and adenosine A2A G protein-coupled receptors (3).
A proposal has been advanced for how in meso crystallogenesis takes place at a molecular level ((1,4); and see our Fig. 1). Typically, it begins with an isolated biological membrane that is treated with detergent to solubilize the target protein. The protein-detergent complex is purified by standard wet biochemical methods that usually involve chromatography. Homogenizing with a monoacylglycerol effects reconstitution of the purified protein into the bilayer of the cubic phase. The latter is bicontinuous in the sense that both the aqueous and bilayer components are continuous in three-dimensional space (Fig. 1). The protein retains its native conformation and activity and is free to move within the plane of the cubic phase bilayer. A precipitant is added to the mesophase which triggers a phase separation. Under conditions leading to crystallization, one of the separated phases is lamellar and becomes enriched in protein. The locally high concentration of protein (that may or may not include native membrane lipid), in conjunction with an appropriate bathing solution composition and bilayer microstructure, act to facilitate nucleation and crystal growth. Aspects of this model are supported by experiment (1).
Figure 1.
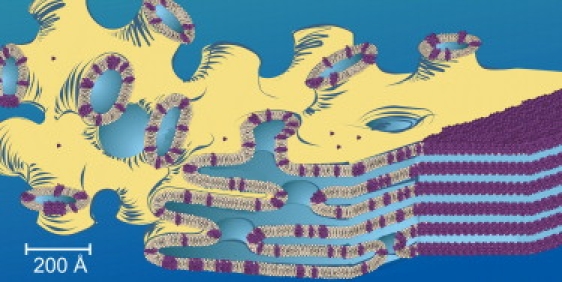
Cartoon representation of the events proposed to take place during the crystallization of linear gramicidin from the lipidic cubic mesophase. Lollipop-shaped objects represent the lipid monoolein, gramicidin dimers are colored purple, and the aqueous medium is shaded blue. See text for details. Adapted from Cherezov et al. (4).
To be a generally applicable method it must work with all sorts of membrane proteins and peptides, both large and small. In a detailed theoretical analysis of the in meso process, Grabe et al. (5) concluded that it will only work with proteins having at least five transmembrane helices and that it was not suitable for “small proteins”. However, given the inherent flexibility of the compartments in a bicontinuous mesophase, we speculated that the in meso method would prove useful for a broad range of membrane protein types and sizes. The in meso structures solved to date (see www.mpdb.tcd.ie (6)) support this view. However, the latter group does not include small proteins of the type referred to by Grabe et al. (5). In this study, we set out to explore the lower size limit of the method and chose to work with linear gramicidin, a transmembrane pentadecapeptide.
Gramicidin is an antibiotic produced nonribosomally by Bacillus brevis (7). It acts, in part, by creating pores in membranes, rendering them incapable of supporting life-sustaining transmembranal gradients. Naturally occurring gramicidin is a mixture of isoforms: gA (80%), gB (6%), and gC (14%). The amino acid sequence of gA is:
In gB and gC, Trp at position 11 is replaced by L-Phe and L-Tyr, respectively (8). The ion-conducting form of gramicidin is generally considered to be a dimer. Controversy exists as to whether this is a head-to-head single-stranded dimer (9) or a left- or right-handed intertwined parallel or antiparallel double helix (10–12). Current structure models (13) are based on macromolecular x-ray crystallography and nuclear magnetic resonance (see Table S2 in the Supporting Material).
To determine whether gramicidin crystallizes in meso, trials were set up by using standard procedures (14,15). The gramicidin was codissolved with monoolein (1 mol gramicidin:20 mol monoolein) in 2,2,2-trifluoroethanol and the solvent was evaporated by flushing with nitrogen gas followed by overnight drying under high vacuum at room temperature (18–23°C). Hydration and cubic phase formation was accomplished by mixing the “dry” gramicidin/lipid with water (lipid/water ratio, 3:2 by weight) at room temperature. In separate spectroscopic and small-angle x-ray scattering studies, gramicidin was shown to reconstitute into the bilayer following this protocol (15).
Crystallization trials were set up by robot using 50 nL mesophase and 800 nL precipitant solution (20%(w/v) polyethylene glycol 6000, 0.1 M Bicine at pH 9.0) on glass sandwich plates, as described (14). Trials were conducted at 20°C, and pyramidal-shaped crystals (Fig. 2 B) measuring 30 × 30 × 30 μm3, which appeared after ∼3–5 days, were harvested directly from the plates. Crystals were cryo-cooled in liquid nitrogen without added cryoprotectant. Diffraction data were collected at The General Medicine and Cancer Institutes Collaborative Access Team (GM/CA-CAT) beamline (23ID-B), the Advanced Photon Source, using a 10-μm collimated beam and a MAR 300 charge-coupled device detector (Rayonix/MAR USA, Evanston, IL). Crystals grew in space group P21 (a = 30.6 Å, b = 62.6 Å, c = 30.6 Å, and β = 100.0°) and diffracted to 1.7 Å. The structure was solved by molecular replacement using the CCP4 program Phaser (16) and the PDB gramicidin model 1AL4 (11). The final R and Rfree values were 0.18 and 0.21, respectively. Further details on crystallization, data collection, and structure determination are described in the Supporting Material.
Figure 2.
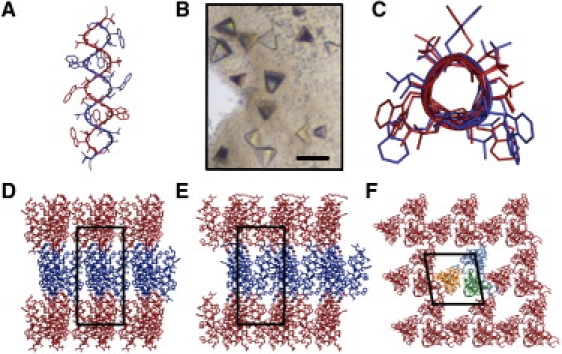
Structure of gramicidin obtained using crystals grown in meso. (A and C) Molecular structures of intertwined gramicidin dimers and (D–F) crystal packing arrangement. Alternate layers in panels D and E are colored red and blue to highlight Type I packing. Individual monomers in panels A and C are colored red and blue for clarity. (B) Crystals of gramicidin growing in meso (scale bar, 80 μm).
As noted, the structure of gramicidin crystallized in meso was solved by molecular replacement with a gramicidin model obtained using crystals grown from n-propanol (11). The corresponding structures are very similar. The peptide exists as an intertwined helical homodimer in an antiparallel arrangement (Fig. 2, A and C). Individual dimers are arranged in layers with their long axis oriented approximately normal to the layer plane (Fig. 2, D and E). This so-called Type I packing is consistent with the proposed mechanism for crystallization in meso; it has been observed in all crystal structures obtained to date by the in meso method (1). By comparison, none of the gramicidin crystal structures obtained by other methods show Type I packing (Table S2).
At first blush, the finding that gramicidin crystallizes in meso might be considered contrary to the conclusion of the analysis by Grabe et al. (5). In that study it was stated
“This poses a problem for crystallizing small proteins and generally limits the broad-based applicability of the in cubo method, although in particular the use of MO (monoolein)-based cubic phases is limited to membrane proteins with five or more transmembrane helices.”
It is important to note, however, that the analysis was performed on the cubic-Pn3m phase. In contrast, the precipitant that facilitated gramicidin crystal growth in meso included polyethylene glycol. The polymer triggers a swelling of the cubic phase and transformation to the sponge phase (17). The latter has enlarged aqueous channels and an irregular, less curved bilayer (Fig. 1) (4). Importantly, the sponge phase retains bicontinuity and thus, can support crystallogenesis in a manner consistent with the mechanistic model outlined above (1). The lessening of curvature in the sponge phase will naturally reduce the energy barrier to translational diffusion within the bilayer which is integral to crystal growth. At the same time, reduced curvature lowers the membrane deformation energy and concomitantly the driving force for protein migration to a flattened bilayer wherein crystallization takes place. The fact that crystals of gramicidin form in meso suggests that the deformation energy does not dominate under conditions of crystallization that involve a sponge phase host.
The most important outcome of this work is the finding that crystals of gramicidin were obtained by the in meso method and that they diffract to high resolution. Thus, a peptide consisting of a β-helix with a diameter of 5.8 Å and that traverses the membrane can be crystallized by the in meso method. This, of course, assumes that the final crystal form of gramicidin is the same as that in the mesophase from which the crystal grew. Nonetheless, the result suggests that membrane proteins with just one or two transmembrane helices are likely to yield to crystallogenesis by the in meso method under appropriate chemical and environmental conditions. The take-home message therefore is that integral membrane peptides should not be ruled out from consideration as targets for crystallization by the in meso method with a view to high-resolution structure determination.
This result opens up the in meso method to a vast array of membrane protein and peptide targets. Indeed, membrane proteins predicted from genome sequence analysis are dominated by those with less than three transmembrane crossings (18). The current structure represents the first time that gramicidin has been crystallized from a lipid bilayer, as opposed to an organic solvent. Thus, the crystallization trials of these new targets can now be carried out in a physiologically more relevant context in meso.
Acknowledgments
The authors thank J. Lyons, V. Pye, and D. Doyle of the Membrane Structural and Functional Biology Group for assistance with data collection and processing, and for helpful discussions.
This work was supported by the Science Foundation Ireland (grant No. 07/IN.1/B1836), FP7 COST Action (grant No. CM0902), and the National Institutes of Health (No. GM75915). Use of the Advanced Photon Source is supported by the US Department of Energy (grant No. DE-AC02-06CH11357). Diffraction data were collected at the GM/CA-CAT beamline, Advanced Photon Source; GM/CA-CAT is funded by the US National Institutes of Cancer (grant No. Y1-CO-1020) and General Medical Sciences (grant No. Y1-GM-1104).
Accession Number
Coordinates and structure factors have been deposited in the Protein DataBank under identification code 2XDC.
Supporting Material
References and Footnotes
- 1.Caffrey M. Crystallizing membrane proteins for structure determination: use of lipidic mesophases. Annu. Rev. Biophys. 2009;38:29–51. doi: 10.1146/annurev.biophys.050708.133655. [DOI] [PubMed] [Google Scholar]
- 2.Landau E.M., Rosenbusch J.P. Lipidic cubic phases: a novel concept for the crystallization of membrane proteins. Proc. Natl. Acad. Sci. USA. 1996;93:14532–14535. doi: 10.1073/pnas.93.25.14532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Blois T.M., Bowie J.U. G-protein-coupled receptor structures were not built in a day. Protein Sci. 2009;18:1335–1342. doi: 10.1002/pro.165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cherezov V., Clogston J., Caffrey M. Room to move: crystallizing membrane proteins in swollen lipidic mesophases. J. Mol. Biol. 2006;357:1605–1618. doi: 10.1016/j.jmb.2006.01.049. [DOI] [PubMed] [Google Scholar]
- 5.Grabe M., Neu J., Nollert P. Protein interactions and membrane geometry. Biophys. J. 2003;84:854–868. doi: 10.1016/S0006-3495(03)74904-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Raman P., Cherezov V., Caffrey M. The membrane Protein DataBank. Cell. Mol. Life Sci. 2006;63:36–51. doi: 10.1007/s00018-005-5350-6. www.mpdb.tcd.ie [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wallace B.A. Recent advances in the high resolution structures of bacterial channels: gramicidin A. J. Struct. Biol. 1998;121:123–141. doi: 10.1006/jsbi.1997.3948. [DOI] [PubMed] [Google Scholar]
- 8.Townsley L.E., Tucker W.A., Hinton J.F. Structures of gramicidins A, B, and C incorporated into sodium dodecyl sulfate micelles. Biochemistry. 2001;40:11676–11686. doi: 10.1021/bi010942w. [DOI] [PubMed] [Google Scholar]
- 9.Urry D.W. The gramicidin A transmembrane channel: a proposed π(L,D) helix. Proc. Natl. Acad. Sci. USA. 1971;68:672–676. doi: 10.1073/pnas.68.3.672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Veatch W., Stryer L. The dimeric nature of the gramicidin A transmembrane channel: conductance and fluorescence energy transfer studies of hybrid channels. J. Mol. Biol. 1977;113:89–102. doi: 10.1016/0022-2836(77)90042-0. [DOI] [PubMed] [Google Scholar]
- 11.Burkhart B.M., Gassman R.M., Duax W.L. Heterodimer formation and crystal nucleation of gramicidin D. Biophys. J. 1998;75:2135–2146. doi: 10.1016/S0006-3495(98)77656-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cotten M., Fu R., Cross T.A. Solid-state NMR and hydrogen-deuterium exchange in a bilayer-solubilized peptide: structural and mechanistic implications. Biophys. J. 1999;76:1179–1189. doi: 10.1016/S0006-3495(99)77282-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.PDB ID codes for published gramicidin structures: MX studies: 1AL4, 1C4D, 1GMK, 1AV2, 1ALX, 1BDW, 1W5U, 1ALZ, 2IZQ; NMR studies: 1GRM, 1JNO, 1JO3, 1JO4, 1KQE, 1MAG, 1NG8, 1NRM, 1NRU, 1NT5, 1NT6, 1TKQ, 1MIC. See Supporting Material for more details.
- 14.Caffrey M., Cherezov V. Crystallizing membrane proteins using lipidic mesophases. Nat. Protoc. 2009;4:706–731. doi: 10.1038/nprot.2009.31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Liu W., Caffrey M. Gramicidin structure and disposition in highly curved membranes. J. Struct. Biol. 2005;150:23–40. doi: 10.1016/j.jsb.2004.12.007. [DOI] [PubMed] [Google Scholar]
- 16.McCoy A.J., Grosse-Kunstleve R.W., Read R.J. Phaser crystallographic software. J. Appl. Cryst. 2007;40:658–674. doi: 10.1107/S0021889807021206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Engström S., Alfons K., Ljusberg-Wahren H. Solvent-induced sponge (L3) phases in the solvent-monoolein-water system. Colloid Polym. Sci. 1998;108:93–98. [Google Scholar]
- 18.Nugent T., Jones D.T. Transmembrane protein topology prediction using support vector machines. BMC Bioinformatics. 2009;10:159. doi: 10.1186/1471-2105-10-159. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


