Abstract
Mutations in the human gene ALMS1 cause Alström syndrome, a disorder characterised by neurosensory degeneration, metabolic defects and cardiomyopathy. ALMS1 encodes a centrosomal protein implicated in the assembly and maintenance of primary cilia. Expression of ALMS1 varies between tissues and recent data suggest that its transcription is modulated during adipogenesis and growth arrest. However the ALMS1 promoter has not been defined. This study focused on identifying and characterising the ALMS1 proximal promoter, initially by using 5' RACE to map transcription start sites. Luciferase reporter assay and EMSA data strongly suggest that ALMS1 transcription is regulated by the ubiquitous factor Sp1. In addition, reporter assay, EMSA, chromatin immunoprecipitation and RNA interference data indicate that ALMS1 transcription is regulated by regulatory factor X (RFX) proteins. These transcription factors are cell-type restricted in their expression profile and known to regulate genes of the ciliogenic pathway. We show binding of RFX proteins to an evolutionarily conserved X-box in the ALMS1 proximal promoter and present evidence that these proteins are responsible for ALMS1 transcription during growth arrest induced by low serum conditions. In summary, this work provides the first data on transcription factors regulating general and context-specific transcription of the disease-associated gene ALMS1.
Abbreviations: CAGE, cap-analysis of gene expression; cDNA, DNA complementary to RNA; ChIP, chromatin immunoprecipitation; EMSA, electrophoretic mobility shift assay; FCS, fetal calf serum; LMC, low-mobility complex; NE, nuclear extract; PCR, polymerase chain reaction; RACE, rapid amplification of cDNA ends; RFX, regulatory factor X; RNAi, RNA interference; RT-PCR, reverse transcription-PCR; s.d., standard deviation; SDS-PAGE, sodium dodecyl sulfate polyacrylamide gel electrophoresis; s.e.m., standard error of the mean; siRNA, small interfering RNA; TBP, TATA-binding protein; TSS, transcription start site
Keywords: transcription, promoter, X-box, GC-box, cilia
1. Introduction
Alström syndrome is a rare genetic disorder characterised by retinal dystrophy, childhood obesity, type 2 diabetes and sensorineural hearing loss, with other common features including dilated cardiomyopathy and hepatic and renal failure (Alstrom et al., 1959; Marshall et al., 2005). Alström syndrome is caused by mutations in a novel gene, ALMS1 (Collin et al., 2002; Hearn et al., 2002), which encodes a centrosomal protein (Andersen et al., 2003; Hearn et al., 2005). Recent data indicate that ALMS1 is required for the proper formation and/or maintenance of primary cilia (Graser et al., 2007; Li et al., 2007), sensory cell-surface organelles whose dysfunction is implicated in the pathogenesis of a number of human genetic disorders (Ansley et al., 2003; Badano et al., 2006).
Although ALMS1 is widely expressed (Collin et al., 2005; Hearn et al., 2005), northern blot analysis shows evidence of variation in expression level between tissues, with particularly high detection in testis (Collin et al., 2002; Hearn et al., 2002). Spatially restricted expression has been reported in mouse olfactory epithelium (Genter et al., 2003) and recent studies using cellular models indicate that ALMS1 mRNA levels are upregulated in response to serum starvation (Yabuta et al., 2006) and downregulated during adipogenesis (Romano et al., 2008). Alterations in transcription are likely to contribute to these changes in mRNA abundance, however the promoter of ALMS1 has not been characterised and therefore the mechanisms controlling ALMS1 transcription are unknown.
We previously defined the exon/intron structure of ALMS1 by RT-PCR using primers in predicted exons, and RACE (Hearn et al., 2002). This approach indicated a larger gene than previously predicted, with 23 exons and a 12.9-kb mRNA, and broadly placed the putative start of transcription within a CpG island. Here, we have discovered and precisely mapped variable transcriptional start sites (TSSs) for ALMS1. We have defined a minimal promoter region and present evidence that Sp1 and RFX proteins regulate ALMS1 transcription under generalised and specific conditions respectively.
2. Materials and methods
2.1. Rapid amplification of cDNA ends (RACE)
PCRs were performed on Firstchoice Race-Ready cDNA (Ambion) derived from adult human pancreas, liver and testes. We used 5' RACE Outer and Inner forward primers corresponding to the 5' RACE adapter sequence (Ambion), in combination with an ALMS1-specific reverse primer spanning the exon 4/5 junction (5'-aggaattcccctcagaggtgcaaaacttag-3') and a nested reverse primer in exon 4 (5'-caaattcttggtcttgtgtcaaac-3') respectively. Nested PCR products were gel extracted and cloned using a BamHI site in the 5' RACE Inner primer and a BglII site in exon 3 of ALMS1. Ten to thirteen clones were sequenced for each tissue.
2.2. DNA sequence alignment
Genomic sequences upstream of ALMS1 orthologues were aligned with human genomic sequence using ClustalW within Lasergene software (DNASTAR, Inc.), with default parameters.
2.3. ALMS1 promoter constructs
Base pairs −412 to + 36 (relative to the liver transcription start site defined here by RACE) were amplified by PCR from genomic DNA using restriction site-tagged primers and cloned in pGL3-Basic (Promega) using KpnI and BglII. Deletion constructs were cloned by PCR using this construct as template. Point mutations (Fig. 3A) were introduced into construct −232/+36 using a PCR based method. All constructs were verified by sequencing.
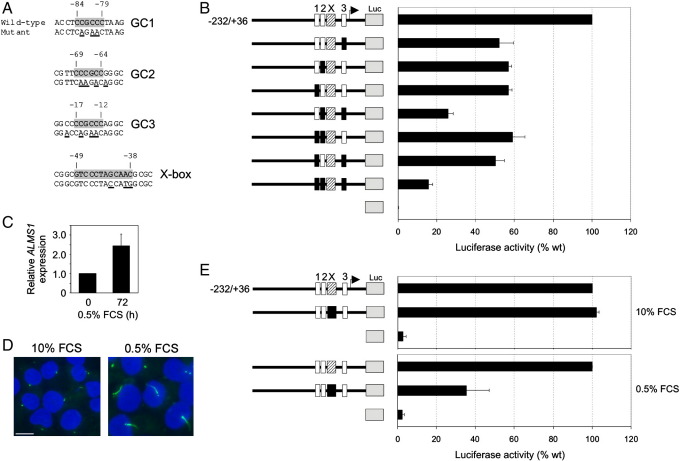
Fig. 3.
Reporter assay analysis of targeted mutations in GC-boxes 1–3 and X-box. (A) Summary of point mutations made in luciferase reporter construct −232/+36. Wild-type nucleotide sequences encompassing predicted X-box and GC-box transcription factor binding sites (shaded) are shown above corresponding mutant sequences. All sequences are 5' to 3'; mutations are underlined. (B) Luciferase assays using PANC-1 cells transfected with wild-type or GC-box-mutant promoter constructs. GC-boxes are labelled 1 to 3; mutant GC-boxes are shaded black. Luciferase activity is expressed relative to wild-type. (C) Quantitative RT-PCR analysis of endogenous ALMS1 expression in PANC-1 cells before and after serum-starvation. (D) Primary cilium assembly in serum-deprived PANC-1 cells. Cells were incubated in growth medium containing either 10% or 0.5% FCS for 72 h and primary cilia visualized by immunostaining for acetylated tubulin (green), which labels ciliary axonemes and centrioles. Nuclear DNA was stained with DAPI (blue). Size bar, 10 µm. (E) Luciferase assays using PANC-1 cells transfected with wild-type or X-box-mutant promoter constructs. The mutant X-box is shaded black. Cells were incubated in either normal (10% FCS) or serum-starved (0.5% FCS) conditions for 48 h prior to lysis. Luciferase activity is expressed relative to wild-type. All data are from three independent experiments; error bars denote s.e.m.
2.4. Cell culture, transfection and reporter assays
Cells were maintained in Dulbecco's modified Eagle medium supplemented with 10% fetal calf serum (FCS) and antibiotics at 37 °C and 5% CO2 (reagents from PAA Laboratories). PANC-l and HepG2 cells were seeded in 12-well culture plates (1 × 105 cells/well) and transiently transfected using TransFast reagent (Promega). Transfections were performed in triplicate. One hour post-transfection, 1 ml DMEM 10% FCS was added and cells incubated for 4 h. Growth medium was then replaced with medium containing either 10% or 0.5% FCS. Cells were lysed 48 h later in 1× Passive Lysis Buffer (Promega) and luciferase expression quantified using the Luciferase Reporter Assay System (Promega) with a TD-20/20 luminometer (Turner Biosystems). Each experiment was performed at least three times, in triplicate. Significant reductions in luciferase activity were verified by using at least two independent preparations of the relevant plasmids.
2.5. Immunofluorescence microscopy
PANC-1 cells were seeded in chamber slides (VWR International) and incubated in medium containing 10% or 0.5% FCS for 72 h. To allow clear visualization of primary cilia, prior to fixation cells were incubated on ice for 50 min to depolymerise cytoplasmic microtubules. Cells were fixed in chilled methanol for 5 min. Immunofluorescence microscopy was performed as described (Hearn et al., 2005), using mouse anti-acetylated tubulin antibody (1:2000; Sigma-Aldrich). Cells were mounted in Vectashield containing DAPI (Vector Laboratories).
2.6. In vitro transcription/translation
pCDNA3-Flag-RFX1 was kindly provided by Zijie Sun, Stanford University (Iwama et al., 1999). In vitro transcription/translation was performed using the T7 TNT Quick Coupled Translation System (Promega). Protein expression was verified by SDS-PAGE and immunoblotting as described (Hearn et al., 2005), using goat anti-RFX1 (Santa Cruz Biotechnology) and peroxidase-conjugated anti-goat (Sigma-Aldrich) antibodies.
2.7. Electrophoretic mobility shift assay (EMSA)
Nuclear extracts were prepared as described (Schreiber et al., 1989). Synthetic oligonucleotides (Qiagen) were annealed and end-labelled with [γ-32P]dCTP. Point mutations designed to disrupt transcription factor binding sites are shown in Fig. 3A. Nuclear extract (4 μg) was incubated in reaction mixture (10 mM Tris–HCl pH 7.5, 1 mM MgCl2, 50 mM NaCl, 0.5 mM EDTA, 0.5 mM DTT, 4% glycerol, and 0.05 mg/ml poly dI–dC) for 15 min at room temperature. For supershift assays, nuclear extracts were pre-incubated on ice for 1 h with 2 µg anti-RFX1 or anti-RFX2 antibody (Santa Cruz Biotechnology). Following addition of radiolabelled probe, reaction mixtures were incubated at 37 °C for 30 min. The resulting protein/DNA complexes were resolved by electrophoresis on non-denaturing 4% polyacrylamide gels in 0.5× TBE buffer. Gels were then dried and exposed to X-ray film.
2.8. Chromatin immunoprecipitation (ChIP)
PANC-1 cells were incubated in DMEM containing 0.5% FCS for 48 h. ChIP was performed with goat anti-RFX1, anti-RFX2, IgG (Santa Cruz Biotechnology) or no antibody. The method was essentially as described (Sanchez-Elsner et al., 2004), except that Protein G-agarose beads (GE Healthcare) were used, and Protein G agarose/antibody/protein complexes were washed as follows: four washes with lysis buffer (25 mM Tris–HCl pH 7.5, 140 mM NaCl, 1 mM EDTA, 1% (v/v) Triton-X-100, 0.1% (w/v) SDS); four washes with lysis buffer containing 500 mM NaCl; four washes with IP wash solution (10 mM Tris–HCl pH 8.0, 250 mM LiCl, 0.5% (v/v) NP-40, 0.5% (w/v) sodium deoxycholate, 1 mM EDTA); and four washes with TE (pH 8.0). Recovered DNA was amplified with custom Taqman Assays (Applied Biosystems) spanning a 90-bp region encompassing the X-box of the ALMS1 promoter (forward: 5'-GCGGCGTCCCTAGCAA-3', reverse: 5'-CCCTGACTGGCGGTTGT-3', probe: 5'-CACTGCGCCTAAGCTG-3') and an 87 bp region upstream of GAPDH (forward: 5'-CCTAATTATCAGGTCCAGGCTACAG-3', reverse: 5'-CGGGAGGCGGCTTGA-3', probe: 5'-CTGCAGGACATCGTG-3'). Triplicate qPCRs were performed using an ABI 7900HT Fast Real-Time System (Applied Biosystems). Percentage of input was calculated as 100×2[Ct(input) – Ct(IP)], after adjusting the mean input Ct value for 1/5 starting material (fraction of input chromatin reserved).
2.9. RNA interference
PANC-1 cells were transfected with pools of three siRNA duplexes targeting RFX1, 2 or 3 (Ambion) or siRNA targeting Lamin (Dharmacon) at 5 nM using HiPerFect transfection reagent (Qiagen). Four hours later growth medium was replaced with medium containing either 10% or 0.5% FCS. Cells were harvested 72 h post-transfection. For quantitative reverse transcription-PCR analysis, total RNA was extracted using TRI reagent (Sigma-Aldrich). RNA was treated with RNAse-free DNAseI (Promega) prior to reverse transcription with oligo d(T)15 primer and Superscript III (Invitrogen) according to the manufacturer's instructions. The resulting cDNAs were amplified using pre-designed TaqMan Gene Expression Assays (Applied Biosystems) and an ABI 7900HT System. Assays were performed in triplicate and TBP was used as endogenous control for relative quantification. For immunoblot analysis, cells were lysed in RIPA buffer (Sigma-Aldrich) supplemented with protease inhibitors (Complete Mini; Roche) for 30 min on ice, and cell lysates then centrifuged at 13,000 rpm for 10 min at 4 °C. Proteins were separated by SDS-PAGE (40 µg/lane) and analysed by immunoblotting, as described (Hearn et al., 2005), using goat anti-RFX1 (0.4 µg/ml; Santa Cruz Biotechnology), peroxidase-conjugated anti-goat (1:50,000) and peroxidase-conjugated anti-β-actin (1:50,000; Sigma-Aldrich) antibodies.
3. Results
3.1. Transcription start site mapping
Our previous TSS prediction for ALMS1 was based on the presence of a potential initiator element close to the most extreme 5' cDNA sequence obtained by RT-PCR (Hearn et al., 2002; our unpublished observations). This did not exclude the possibility that transcription is initiated further upstream, either in contiguous sequence or at undiscovered exon(s). Defining the TSS of ALMS1 is therefore important both to help identify the promoter and to resolve whether additional exons lie upstream of the previously defined exon 1. To achieve this we performed 5'-RACE on cDNA generated from full-length, capped mRNAs. We used cDNAs derived from human liver, testes and pancreas, all of which express the ALMS1 gene (Collin et al., 2002; Hearn et al., 2002). We cloned 10–13 RACE products for each tissue and compared their sequences to the publically available human genome sequence to identify putative TSSs. All of the sequences obtained began within a 48-bp region overlapping the previously predicted start of exon 1 (Fig. 1). All sequences were continuous with the genome from their 5'-ends to the splice donor site of exon 1, i.e. they provided no evidence of novel exons. Notably, all ten liver-derived sequences started at the same nucleotide (Fig. 1), hereinafter used as a reference and termed position + 1. Six out of thirteen testis-derived sequences also started at position + 1, while others started at positions −19, + 10, + 19 and + 20 (Fig. 1). In contrast to both the liver and testis clones, all ten pancreas-derived sequences started at position + 29. Notably, all transcripts are predicted to utilise the previously identified methionine start codon at position + 101. In summary, these data suggest multiple and possibly tissue-specific TSSs for exon 1.
Fig. 1.
Transcription start site mapping data. Summary of ALMS1 transcription start sites suggested by cloning 5'-RACE products from pancreas, testes and liver, and by the cap-analysis of gene expression (CAGE) database (Kawaji et al., 2006). Only the most extreme 5' CAGE tag cluster (> 2 tags) associated with ALMS1 is represented. x-axis, start position relative to the liver TSS predicted by RACE; y-axis, cumulative total of RACE clones/CAGE tags supporting each start site; asterisk, position of previously predicted TSS (Hearn et al., 2002). The predicted translation start codon of ALMS1 is at position + 101 (not shown).
We compared our results with data from the cap-analysis of gene expression (CAGE) database, a transcriptome-wide analysis of sequenced capped 5' ends of transcripts, termed CAGE tags (Shiraki et al., 2003; Kawaji et al., 2006). Of the 143 CAGE tags mapped to ALMS1, 77 were dispersed at 63 different positions throughout the gene. These scattered CAGE tags may represent post-transcriptional processing events (exonic tags; Fejes-Toth et al., 2009) or artefacts rather than TSSs, and therefore we focused our attention on larger clusters of tags. Notably, the most extreme 5' cluster of more than two tags (bp −43 to + 20; 24 tags) overlapped the TSS region identified by RACE (bp −19 to + 29) and suggested a broad TSS distribution (Fig. 1). The most common start site in the cluster, position −21, was supported by six tags, five of which were from a skin cell line; other tags in the cluster were derived from intestine, cerebrum, cecum and adipose. There were two nearby clusters of CAGE tags, at bp + 71 to + 95 (32 tags) and bp + 259 to + 305 (10 tags), suggesting alternative promoters in exon 1. However, we did not find evidence of TSSs in either region by RACE.
3.2. Genomic context of putative transcription start sites
In silico (MatInspector) analyses of genomic sequence upstream of the TSSs defined by 5'-RACE failed to identify a TATA box. To aid discovery of cis-acting regulatory elements we aligned bp −1000 to + 100 of the human genomic sequence with corresponding sequences from dog, mouse and rat. This revealed several evolutionarily conserved regions between bp −200 and + 100, but less conservation further upstream (Fig. 2A and data not shown). Notably, the region −1000 to −394 is largely composed of interspersed repeats, while the region −393 to + 100 is devoid of these elements (data not shown), although it does contain a C-rich stretch (bp + 37 to + 84) that is partially conserved between species (Fig. 2A). MatInspector analysis identified potential transcription factor binding sites in upstream conserved or partially conserved regions, including an X-box (bp −49/−38) and three GC-box-like elements (bp −84/−79, −69/−64 and −17/−12; hereinafter termed GC-box 1, 2 and 3, respectively) (Fig. 2A). Notably, X-boxes are target sites for RFX transcription factors, key regulators of genes important for the assembly and maintenance of primary cilia (Swoboda et al., 2000; Dubruille et al., 2002). GC-boxes are target sites for Sp/KLF transcription factors including Sp1, a ubiquitously expressed factor implicated in the activation of a very large number of genes (Philipsen and Suske, 1999; Wierstra, 2008).
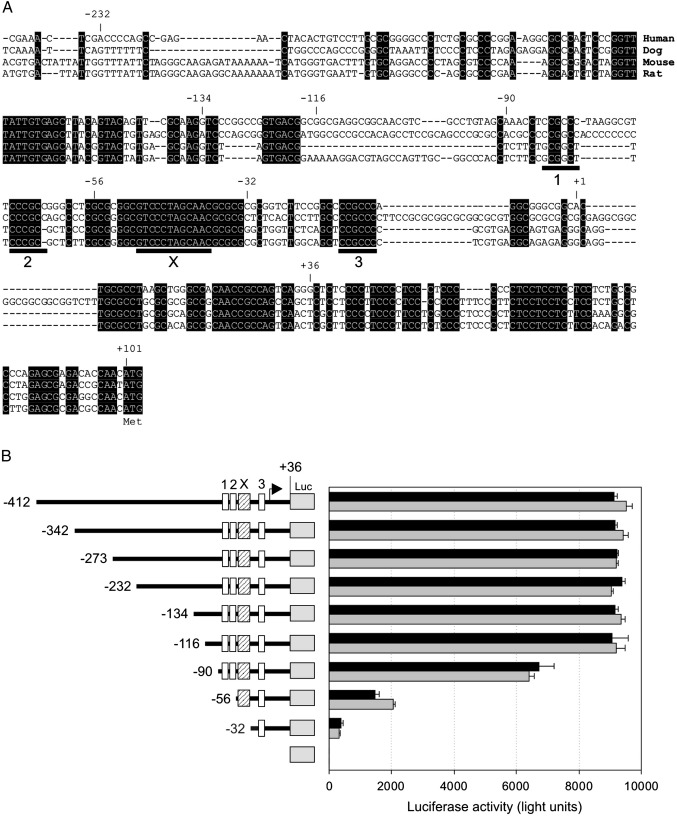
Fig. 2.
Evolutionary conservation and reporter assay analysis of the ALMS1 promoter. (A) Alignment of human genomic sequence (bp −241 to + 103) with equivalent sequences from dog (C. familiaris), mouse (M. musculus) and rat (R. norvegicus). Shading represents conservation in all species. The positions of the predicted X-box and three GC-box transcription factor binding sites (labelled 1–3) are indicated. (B) Luciferase reporter assays using constructs representing 5' deletions of the ALMS1 promoter. Cells were transiently transfected and luciferase activity quantified 48 h post-transfection (black bars, HepG2; grey bars, PANC-1). Positions of the consensus X-box (striped shading, X) and GC-boxes 1, 2 and 3 are shown. Data represent four independent experiments; error bars denote s.d.
3.3. Reporter assay analysis of the proximal promoter
To identify functional regions of the promoter we made a panel of 5' deletion constructs within the context of the pGL3-Basic luciferase reporter vector. The longest construct (−412/+36) included the evolutionarily conserved regions identified upstream of putative TSSs (Fig. 2). We assayed the transcriptional activity of promoter constructs in PANC-1 and HepG2 cells, both of which express ALMS1 (Fig. 6; and Hearn et al., 2005) and represent tissues (pancreas and liver) affected in Alström syndrome. As shown in Fig. 2B, construct −412/+36 exhibited strong promoter activity in these cell lines. Deletion of bp −412 to −117 had little effect on luciferase activity. Additional deletion of bp −116 to −91, a poorly conserved region, reduced activity by approximately 30%. Deletion of bp −90 to −57, including GC-boxes 1 and 2, further reduced promoter activity to ∼ 20% of the activity of construct −412/+36. Finally, deletion of bp −56 to −33, including the conserved X-box, further reduced activity to ∼ 5% of the activity of construct −412/+36 (Fig. 2B). These data suggest that the region −116 to + 36, which includes evolutionarily conserved predicted transcription factor binding sites, is important for basal transcription of ALMS1.
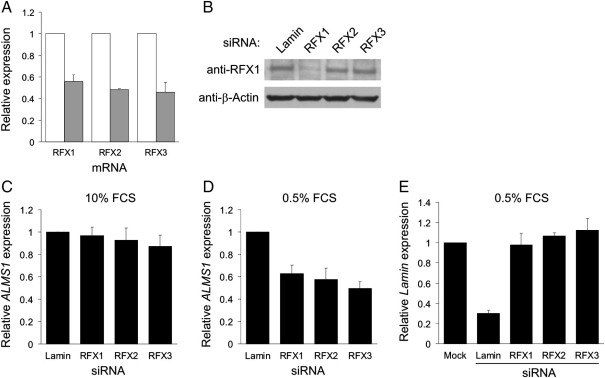
Fig. 6.
Effect of siRNA-mediated knockdown of RFX transcripts on ALMS1 expression. (A) qRT-PCR analysis of RFX gene transcript levels in PANC-1 cells treated with siRNAs targeting RFX1, RFX2 or RFX3. Each gray bar represents the level of the indicated RFX transcript in cells treated with the corresponding RFX siRNA, normalised to the level in cells treated with a negative control siRNA (targeting Lamin; white bars). Data represent mean ± s.e.m. from three independent experiments. (B) Immunoblot analysis of RFX1 protein expression in siRNA-treated PANC-1 cells. β-actin was used as a loading control. (C, D) qRT-PCR analysis of ALMS1 expression in cells treated with RFX siRNAs under either normal (C) or serum-starved (D) growth conditions; results were normalised to the expression level in cells treated with Lamin-directed siRNA. Data are from three independent experiments; error bars denote s.e.m. (E). qRT-PCR analysis of Lamin mRNA levels in cells treated with RFX or Lamin siRNAs. Results (mean ± s.d. from two independent experiments) were normalized to the level in mock-transfected cells.
3.4. Reporter assay analysis of X-box and GC-box mutations
To further investigate the functions of the X-box and GC-boxes 1–3 we generated reporter constructs with point mutations in these elements. Mutations were made to specifically disrupt binding of RFX proteins or Sp1 respectively (Fig. 3A) within the context of the −232/+36 construct (Fig. 2B). We compared the activities of mutant and wild-type constructs in PANC-1 cells. As shown in Fig. 3B, mutating any of the individual GC-boxes in isolation reduced transcriptional activity by approximately 50%. No additional effect was observed on mutating site 1 in combination with site 2 or 3, however, mutating sites 2 and 3 together decreased activity by ∼ 70%. Mutating all three sites within the same construct decreased reporter activity by ∼ 80% (Fig. 3B). In summary, these data indicate that: each GC-box has a positive regulatory role; site 1 seems slightly less important compared to sites 2 and 3; however, together they account for most of the transcriptional activity of construct −232/+36.
Under the 10% FCS cell growth conditions used for the GC-box analysis, mutations in the X-box did not have a significant effect on promoter activity (Fig. 3E). It has been reported that levels of ALMS1 mRNA increase in response to serum starvation (13). Since RFX transcription factors including mammalian RFX3 regulate genes involved in cilium assembly (Bonnafe et al., 2004), a process which is itself promoted on entry of cells into a quiescent state following serum-starvation, we asked if the X-box mediates upregulation of ALMS1 following serum deprivation. We used PANC-1 cells, which show marked serum-dependence of cell growth (Giehl et al., 2000). We first confirmed that serum deprivation for 48–72 h increased levels of ALMS1 mRNA and induced assembly of primary cilia in these cells (Fig. 3C, D); the latter finding has also been independently reported (Nielsen et al., 2008). Under these conditions, reporter assays showed that mutations in the X-box reduced transcriptional activity of the reporter gene by ∼ 60% (Fig. 3E). This strongly implies that the X-box is required for transcription of ALMS1 in serum-starved conditions.
3.5. EMSA analysis of X-box and GC-box-binding proteins
To investigate the protein-binding properties of the X-box and GC-boxes 1–3, we performed EMSAs with duplex oligonucleotide probes spanning each site. As shown in Fig. 4A, probe −23/−8 (containing GC-box 3) formed several complexes with PANC-1 nuclear proteins. The formation of three of these complexes was competed by including an excess of unlabelled probe (Fig. 4A). Preincubation with Sp1 antibody lessened the intensity of the uppermost of these three bands and gave rise to a further retarded (“supershifted”) band (Fig. 4A), supporting the presence of Sp1 in this upper complex. The other bands were unaltered by Sp1 antibody, suggesting that they represent binding of other factors. Probes encompassing GC-boxes 1 and 2 also formed complexes with PANC-1 nuclear proteins, however the formation or mobility of these complexes was not altered by Sp1 antibody (data not shown), suggesting that they also represent the binding of other factors, which remain uncharacterised.
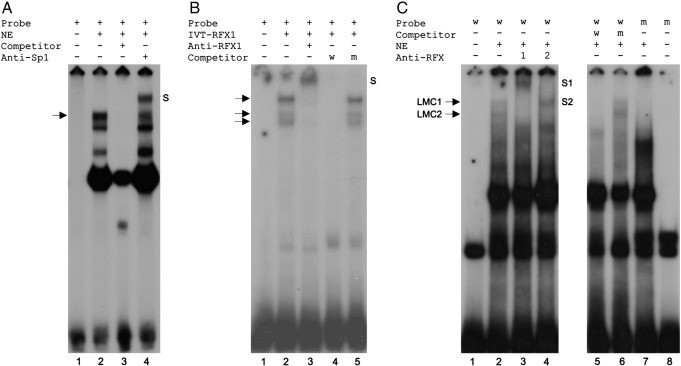
Fig. 4.
EMSA analysis of GC-box 3 and X-box. (A) Electrophoretic mobility shift assay with PANC-1 nuclear extract and probe −23/−8, containing GC-box 3. Positions of the protein-DNA complex with the lowest mobility (lane 2, arrow) and the complex supershifted by Sp1 antibody (lane 4, S) are indicated. Fifty-fold excess unlabelled probe was used as competitor (lane 3). NE, nuclear extract. (B) EMSA with in vitro translated (IVT) RFX1 and probe −56/−33, containing the conserved X-box at bp −49 to −38. Positions of protein-DNA complexes (lane 2, arrows) and complexes supershifted by RFX1 antibody (lane 3, S) are indicated. 100-fold excess unlabelled wild-type (w) or mutant (m) probe was used as competitor, as indicated. (C) EMSA with HeLa nuclear extract and either wild-type or mutant probe −56/−33. Positions of two low-mobility complexes termed LMC1 and LMC2 (lane 2), and complexes supershifted by RFX1 antibody (lane 3, S1) and RFX2 antibody (lane 4, S2), are indicated. Fifty-fold excess unlabelled wild-type or mutant probe was used as competitor, as indicated.
To investigate if RFX proteins bind to the X-box at bp −49 to −38, we first performed an EMSA with in vitro transcribed and translated (IVT) RFX1, a prototypical member of the RFX family. As shown in Fig. 4B, a probe containing the X-box (probe −56/−33) formed three complexes with IVT-RFX1, all of which were retarded by incubation with RFX1 antibody (Fig. 4B, lane 3). Formation of complexes was also competed by excess unlabelled wild-type, but not mutant, probe (Fig. 4B, lanes 4 and 5 respectively).
To test for interaction of endogenous RFX proteins with probe −56/−33 we performed EMSAs with nuclear extract from HeLa cells, which are known to express RFX1, 2 and 3 (Iwama et al., 1999). Of the proteins encoded by the seven known mammalian RFX genes (RFX1–7), RFX1–3 are the most closely related to Caenorhabditis elegans DAF-19, a RFX protein that regulates genes of the ciliogenic pathway (Emery et al., 1996; Swoboda et al., 2000; Aftab et al., 2008). As shown in Fig. 4C (lane 2), probe −56/−33 formed several complexes with HeLa nuclear proteins. RFX1 antibody supershifted the two uppermost complexes (Fig. 4C, lane 3), indicating the presence of RFX1 in these complexes. We termed these two low-mobility complexes LMC1 (upper band) and LMC2 (lower band). LMC2 was abolished by inclusion of anti-RFX2 antibody, possibly yielding a retarded complex co-migrating with LMC1 (Fig. 4C, compare lanes 2 and 4). It is unclear whether anti-RFX2 antibody disrupted the formation of LMC1, although clearly it did not supershift this complex (compared to the effect of RFX1 antibody). These results are consistent with LMC1 representing RFX1 homodimers, and with LMC2 containing RFX2 and possibly representing RFX1/RFX2 heterodimers, similar to previous reports (Reith et al., 1994; Iwama et al., 1999). We did not investigate binding of RFX3 by this approach since we were unable to demonstrate immunoreactivity of an RFX3 antibody. Formation of LMC1 and LMC2 was competed by unlabelled wild-type, but not mutant, probe (Fig. 4C, lanes 5 and 6 respectively), and neither complex formed with labelled mutant probe (Fig. 4C, lane 7), supporting the specificity of binding. In summary, these data indicate that RFX1 and RFX2 can both bind to the X-box in probe −56/−33.
3.6. Chromatin immunoprecipitation analysis of RFX1 and RFX2 binding to the ALMS1 promoter
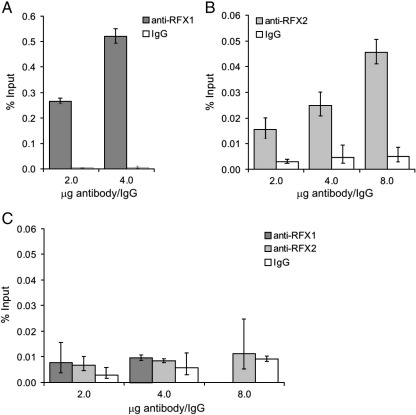
To determine if RFX1 and RFX2 bind to the ALMS1 promoter in vivo, we employed ChIP. We chose to analyse serum-deprived PANC-1 cells, since our reporter assays suggested that RFX proteins activate ALMS1 transcription in these cells/conditions (Fig. 3E). qPCR analysis showed that genomic DNA fragments containing the ALMS1 proximal promoter were highly enriched in anti-RFX1 ChIP fractions, compared to IgG negative control fractions (Fig. 5A). Levels of enrichment obtained with anti-RFX2 were much lower than those obtained with anti-RFX1, though still appeared significant (Fig. 5B). Neither antibody yielded significant enrichment of a negative control region (Fig. 5C). These results indicate that RFX1 and RFX2 bind to the ALMS1 promoter in vivo. The lower level of enrichment obtained with anti-RFX2 may be due, at least in part, to antibody-related limitations. We have been unable to detect endogenous RFX2 on immunoblots, although the antibody does detect exogenous, overexpressed RFX2 (data not shown).
Fig. 5.
Chromatin immunoprecipitation analysis of RFX1 and RFX2 binding to the ALMS1 proximal promoter. PANC-1 cells were incubated in growth medium containing 0.5% FCS for 48 h prior to analysis. (A,B) qPCR analysis of anti-RFX1 (A), anti-RFX2 (B) and IgG ChIP fractions with a Taqman assay encompassing the evolutionarily conserved X-box within the ALMS1 promoter. Note the differing scales of the y-axes. (C) qPCR analysis of anti-RFX1, anti-RFX2 and IgG ChIP fractions with a Taqman assay targeting a negative control region upstream of GAPDH. ChIP assays were performed with 2 or 4 µg of anti-RFX1, and with 2, 4 or 8 µg of anti-RFX2 and IgG, as indicated. DNA recovery is expressed as percentage of input (mean ± s.d. from triplicate qPCR assays).
3.7. RNAi-mediated knockdown of RFX transcription factors downregulates ALMS1 expression in serum-starved cells
To further examine the role of RFX transcription factors in regulating ALMS1 we abrogated their expression by RNA interference. We targeted RFX1, 2 or 3 in PANC-1 cells and confirmed RFX transcript depletion by qRT-PCR (Fig. 6A). We also confirmed depletion of RFX1 protein by immunoblot analysis (Fig. 6B). Due to antibody-related limitations (see above) we were unable to monitor RFX2 or RFX3 protein depletion. Under normal growth conditions (10% serum), knockdown of RFX transcripts did not have a significant effect on ALMS1 mRNA levels, relative to cells transfected with a negative control siRNA (Fig. 6C). However, under serum-starved conditions, knockdown of RFX1, 2 or 3 reduced ALMS1 mRNA levels by approximately 40–50% (Fig. 6D). As a control, we also quantified levels of Lamin mRNA in these cells (Fig. 6E). Lamin expression was not altered by RFX siRNAs, suggesting that the observed decrease in ALMS1 mRNA was not the result of a general effect. These data are consistent with our promoter reporter assays, in which X-box mutations decreased ALMS1 promoter activity only in serum-starved cells (Fig. 3E), and further imply that RFX proteins, acting via the X-box, activate ALMS1 transcription following serum starvation.
4. Discussion
Understanding why ALMS1 expression varies in different contexts requires identification and investigation of the promoter region regulating its transcription. Cloning 5'-RACE products from three different tissues, combined with data from the CAGE project (Kawaji et al., 2006), suggests that transcription of ALMS1 is initiated at numerous sites within a ∼ 70 base pair region. This region contains the previously predicted start of exon 1 and is therefore consistent with the reported gene structure (Collin et al., 2002; Hearn et al., 2002). The genomic context of this region indicates that the ALMS1 promoter belongs to the TATA-less, CpG island-associated class of mammalian promoters. This type of promoter typically contains multiple GC-boxes and has a broad TSS distribution (∼ 100 bp), in contrast to the tightly defined TSSs of most TATA-containing promoters (Carninci et al., 2006). Taken together, the CAGE and RACE data support a broad TSS distribution for ALMS1, yet the RACE data for liver and pancreas suggest tightly defined TSSs in these particular tissues. CAGE data also suggest two downstream TSS clusters in exon 1. Recent data suggest that many CAGE tags mapped to annotated first exons are derived from promoter-associated short RNAs (PASRs; Kapranov et al., 2007; Fejes-Toth et al., 2009). These < 200 nt RNAs are unlikely to be represented in the cDNAs we used for RACE due to selection for > 200 nt synthesis products during cDNA preparation. PASRs may be capped post-transcriptional processing products of longer mRNAs, or independent transcription products from promoters that also generate long mRNAs (Fejes-Toth et al., 2009). Therefore, the question of whether downstream CAGE tag clusters represent processing products or usage of alternative ALMS1 promoters requires further investigation.
Reporter assays using a series of 5' deletion constructs suggest that elements important for basal ALMS1 transcription lie between bp −116 and + 36, relative to the liver TSS defined by RACE. We did not analyse the role of sequences downstream of position + 36 and therefore have not excluded the possibility that the proximal promoter extends downstream of this position. Nevertheless, we have identified several elements between bp −116 and + 36 that significantly influence transcriptional activity in reporter assays. Of three GC-box-like elements analysed in this region, EMSAs indicate that Sp1 binds to the most proximal, GC-box 3. Mutation of this conserved site significantly reduced promoter activity, consistent with an activatory role for Sp1. Notably, mutation of either of the two upstream GC-box-like elements also significantly reduced promoter activity, however EMSAs suggest that other, unidentified factors bind to these elements. Candidates include other members of the Sp/KLF family, e.g. Sp3, Sp4, KLF9 and KLF11 (Philipsen and Suske, 1999).
The promoter also contains an evolutionarily conserved X-box, suggesting regulation by RFX transcription factors. These factors are characterised by a conserved winged-helix DNA-binding domain (Emery et al., 1996; Gajiwala et al., 2000). The sole RFX gene in C. elegans, daf-19, and one of the two known Drosophila RFX genes, dRfx, regulate genes involved in the assembly, maintenance and/or function of primary cilia (Swoboda et al., 2000; Haycraft et al., 2001; Dubruille et al., 2002; Efimenko et al., 2005; Laurencon et al., 2007). C. elegans genes whose human orthologs are mutated in the ciliopathy Bardet–Biedl syndrome are thought to be regulated by DAF-19 (Ansley et al., 2003). Of the products of the seven known mammalian RFX genes (RFX1–7), RFX1, 2 and 3 are the most similar to DAF-19 at the amino acid sequence level (Emery et al., 1996; Aftab et al., 2008) and notably RFX3-deficient mice have defective cilia, indicating conserved ciliogenic functions for this protein in mammals (Bonnafe et al., 2004; Baas et al., 2006; Ait-Lounis et al., 2007). With the exception of RFX5, which regulates MHC II genes (Reith and Mach, 2001), the functions of the remaining mammalian RFX proteins are less clear. It is notable however that RFX1 and RFX2 have similar functional domains and DNA-binding specificities to RFX3 (Reith et al., 1994; Aftab et al., 2008), suggesting that they also activate genes of the ciliogenic pathway (Bonnafe et al., 2004).
Binding of RFX proteins to the X-box in the ALMS1 promoter was supported by EMSAs, both with nuclear extracts and in vitro synthesized RFX1, and by ChIP. Furthermore, our reporter assay and RNAi data implicate these factors, acting through the X-box, in the up-regulation of ALMS1 following serum-starvation (Yabuta et al., 2006). Since serum-starvation promotes primary cilium formation, and RFX family members activate genes of the ciliogenic pathway, these findings are consistent with a specific role for ALMS1 in cilium formation, maintenance and/or function (Hearn et al., 2005; Graser et al., 2007; Li et al., 2007). Interestingly, a transient ciliated phase in differentiating human preadipocytes has recently been described (Marion et al., 2009). We speculate that a similar transition in 3 T3-L1 preadipocytes (Marion et al., 2009) may explain the reduction in Alms1 expression observed during differentiation of these cells (Romano et al., 2008), although the ciliated status of 3 T3-L1 cells throughout differentiation remains to be fully characterised. We also note that ALMS1 expression is not restricted to ciliated cells, suggesting that the protein has additional functions.
Our ChIP data indicate that RFX1 and RFX2 bind to the ALMS1 promoter in vivo. This is consistent with our EMSA results, which suggest that both RFX1 homodimers and RFX1/RFX2 heterodimers are able to interact with the X-box in vitro. Together with the RNAi data, these findings support the notion that RFX1 and RFX2 are directly involved in regulating ALMS1 transcription. It remains to be determined if RFX3 (and/or RFX4–7, whose DNA-binding domains share less sequence similarity with DAF-19; Emery et al., 1996; Aftab et al., 2008) also interact with the ALMS1 X-box. Interestingly, regulation by RFX family members may provide an explanation for the strong expression of ALMS1 in testis (Collin et al., 2002; Hearn et al., 2002) since these factors (in particular RFX2) show elevated expression in testis and have been implicated in activating the transcription of several testis-expressed genes including Spag6, which encodes a component of the axoneme of the sperm flagellum (Reith et al., 1994; Wolfe et al., 2004; Horvath et al., 2009; Kistler et al., 2009). Also of note, both RFX1 and RFX2 are thought to contain auto-regulatory domains that inhibit transcriptional activation, implying that context-dependent transcriptional activation by these factors is mediated by relief of the inhibitory function, possibly via interactions with other factors (Katan et al., 1997; Horvath et al., 2009). Understanding this mechanism could therefore provide further insight into the context-dependent expression of ALMS1.
In summary, our data provide the first analysis of the ALMS1 proximal promoter. The results predict variable TSSs for exon 1, suggest that Sp1 is important for basal ALMS1 transcription, and that RFX proteins activate ALMS1 transcription during serum starvation-induced growth arrest, most likely as part of the ciliogenic program.
Acknowledgments
This work was funded by the British Heart Foundation (project grant PG/04/020). We are also grateful for support from the Leverhulme Trust and Diabetes UK. NH acknowledges support from the NIHR Manchester Biomedical Research Centre. We thank Zijie Sun for RFX expression constructs.
Received by Lynn Jorde
References
- Aftab S., Semenec L., Chu J.S., Chen N. Identification and characterization of novel human tissue-specific RFX transcription factors. BMC Evol Biol. 2008;8:226. doi: 10.1186/1471-2148-8-226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ait-Lounis A., Baas D., Barras E., Benadiba C., Charollais A., Nlend Nlend R., Liegeois D., Meda P., Durand B., Reith W. Novel function of the ciliogenic transcription factor RFX3 in development of the endocrine pancreas. Diabetes. 2007;56:950–959. doi: 10.2337/db06-1187. [DOI] [PubMed] [Google Scholar]
- Alstrom C.H., Hallgren B., Nilsson L.B., Asander H. Retinal degeneration combined with obesity, diabetes mellitus and neurogenous deafness: a specific syndrome (not hitherto described) distinct from the Laurence–Moon–Bardet–Biedl syndrome: a clinical, endocrinological and genetic examination based on a large pedigree. Acta Psychiatr Neurol Scand. 1959;34:1–35. [PubMed] [Google Scholar]
- Andersen J.S., Wilkinson C.J., Mayor T., Mortensen P., Nigg E.A., Mann M. Proteomic characterization of the human centrosome by protein correlation profiling. Nature. 2003;426:570–574. doi: 10.1038/nature02166. [DOI] [PubMed] [Google Scholar]
- Ansley S.J., Badano J.L., Blacque O.E., Hill J., Hoskins B.E., Leitch C.C., Kim J.C., Ross A.J., Eichers E.R., Teslovich T.M., Mah A.K., Johnsen R.C., Cavender J.C., Lewis R.A., Leroux M.R., Beales P.L., Katsanis N. Basal body dysfunction is a likely cause of pleiotropic Bardet–Biedl syndrome. Nature. 2003;425:628–633. doi: 10.1038/nature02030. [DOI] [PubMed] [Google Scholar]
- Baas D., Meiniel A., Benadiba C., Bonnafe E., Meiniel O., Reith W., Durand B. A deficiency in RFX3 causes hydrocephalus associated with abnormal differentiation of ependymal cells. Eur J Neurosci. 2006;24:1020–1030. doi: 10.1111/j.1460-9568.2006.05002.x. [DOI] [PubMed] [Google Scholar]
- Badano J.L., Mitsuma N., Beales P.L., Katsanis N. The ciliopathies: an emerging class of human genetic disorders. Annu Rev Genomics Hum Genet. 2006;7:125–148. doi: 10.1146/annurev.genom.7.080505.115610. [DOI] [PubMed] [Google Scholar]
- Bonnafe E., Touka M., AitLounis A., Baas D., Barras E., Ucla C., Moreau A., Flamant F., Dubruille R., Couble P., Collignon J., Durand B., Reith W. The transcription factor RFX3 directs nodal cilium development and left–right asymmetry specification. Mol Cell Biol. 2004;24:4417–4427. doi: 10.1128/MCB.24.10.4417-4427.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carninci P., Sandelin A., Lenhard B., Katayama S., Shimokawa K., Ponjavic J., Semple C.A., Taylor M.S., Engstrom P.G., Frith M.C., Forrest A.R., Alkema W.B., Tan S.L., Plessy C., Kodzius R., Ravasi T., Kasukawa T., Fukuda S., Kanamori-Katayama M., Kitazume Y., Kawaji H., Kai C., Nakamura M., Konno H., Nakano K., Mottagui-Tabar S., Arner P., Chesi A., Gustincich S., Persichetti F., Suzuki H., Grimmond S.M., Wells C.A., Orlando V., Wahlestedt C., Liu E.T., Harbers M., Kawai J., Bajic V.B., Hume D.A., Hayashizaki Y. Genome-wide analysis of mammalian promoter architecture and evolution. Nat Genet. 2006;38:626–635. doi: 10.1038/ng1789. [DOI] [PubMed] [Google Scholar]
- Collin G.B., Cyr E., Bronson R., Marshall J.D., Gifford E.J., Hicks W., Murray S.A., Zheng Q.Y., Smith R.S., Nishina P.M., Naggert J.K. Alms1-disrupted mice recapitulate human Alstrom syndrome. Hum Mol Genet. 2005;14:2323–2333. doi: 10.1093/hmg/ddi235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collin G.B., Marshall J.D., Ikeda A., So W.V., Russell-Eggitt I., Maffei P., Beck S., Boerkoel C.F., Sicolo N., Martin M., Nishina P.M., Naggert J.K. Mutations in ALMS1 cause obesity, type 2 diabetes and neurosensory degeneration in Alstrom syndrome. Nat Genet. 2002;31:74–78. doi: 10.1038/ng867. [DOI] [PubMed] [Google Scholar]
- Dubruille R., Laurencon A., Vandaele C., Shishido E., Coulon-Bublex M., Swoboda P., Couble P., Kernan M., Durand B. Drosophila regulatory factor X is necessary for ciliated sensory neuron differentiation. Development. 2002;129:5487–5498. doi: 10.1242/dev.00148. [DOI] [PubMed] [Google Scholar]
- Efimenko E., Bubb K., Mak H.Y., Holzman T., Leroux M.R., Ruvkun G., Thomas J.H., Swoboda P. Analysis of xbx genes in C. elegans. Development. 2005;132:1923–1934. doi: 10.1242/dev.01775. [DOI] [PubMed] [Google Scholar]
- Emery P., Durand B., Mach B., Reith W. RFX proteins, a novel family of DNA binding proteins conserved in the eukaryotic kingdom. Nucleic Acids Res. 1996;24:803–807. doi: 10.1093/nar/24.5.803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fejes-Toth K., Sotirova V., Sachidanandam R., Assaf G., Hannon G.J., Kapranov P., Foissac S., Willingham A.T., Duttagupta R., Dumais E., Gingeras T.R. Post-transcriptional processing generates a diversity of 5'-modified long and short RNAs. Nature. 2009;457:1028–1032. doi: 10.1038/nature07759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gajiwala K.S., Chen H., Cornille F., Roques B.P., Reith W., Mach B., Burley S.K. Structure of the winged-helix protein hRFX1 reveals a new mode of DNA binding. Nature. 2000;403:916–921. doi: 10.1038/35002634. [DOI] [PubMed] [Google Scholar]
- Genter M.B., Van Veldhoven P.P., Jegga A.G., Sakthivel B., Kong S., Stanley K., Witte D.P., Ebert C.L., Aronow B.J. Microarray-based discovery of highly expressed olfactory mucosal genes: potential roles in the various functions of the olfactory system. Physiol Genomics. 2003;16:67–81. doi: 10.1152/physiolgenomics.00117.2003. [DOI] [PubMed] [Google Scholar]
- Giehl K., Skripczynski B., Mansard A., Menke A., Gierschik P. Growth factor-dependent activation of the Ras-Raf-MEK-MAPK pathway in the human pancreatic carcinoma cell line PANC-1 carrying activated K-ras: implications for cell proliferation and cell migration. Oncogene. 2000;19:2930–2942. doi: 10.1038/sj.onc.1203612. [DOI] [PubMed] [Google Scholar]
- Graser S., Stierhof Y.D., Lavoie S.B., Gassner O.S., Lamla S., Le Clech M., Nigg E.A. Cep164, a novel centriole appendage protein required for primary cilium formation. J Cell Biol. 2007;179:321–330. doi: 10.1083/jcb.200707181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haycraft C.J., Swoboda P., Taulman P.D., Thomas J.H., Yoder B.K. The C. elegans homolog of the murine cystic kidney disease gene Tg737 functions in a ciliogenic pathway and is disrupted in osm-5 mutant worms. Development. 2001;128:1493–1505. doi: 10.1242/dev.128.9.1493. [DOI] [PubMed] [Google Scholar]
- Hearn T., Renforth G.L., Spalluto C., Hanley N.A., Piper K., Brickwood S., White C., Connolly V., Taylor J.F., Russell-Eggitt I., Bonneau D., Walker M., Wilson D.I. Mutation of ALMS1, a large gene with a tandem repeat encoding 47 amino acids, causes Alstrom syndrome. Nat Genet. 2002;31:79–83. doi: 10.1038/ng874. [DOI] [PubMed] [Google Scholar]
- Hearn T., Spalluto C., Phillips V.J., Renforth G.L., Copin N., Hanley N.A., Wilson D.I. Subcellular localization of ALMS1 supports involvement of centrosome and basal body dysfunction in the pathogenesis of obesity, insulin resistance, and type 2 diabetes. Diabetes. 2005;54:1581–1587. doi: 10.2337/diabetes.54.5.1581. [DOI] [PubMed] [Google Scholar]
- Horvath G.C., Kistler M.K., Kistler W.S. RFX2 is a candidate downstream amplifier of A-MYB regulation in mouse spermatogenesis. BMC Dev Biol. 2009;9:63. doi: 10.1186/1471-213X-9-63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iwama A., Pan J., Zhang P., Reith W., Mach B., Tenen D.G., Sun Z. Dimeric RFX proteins contribute to the activity and lineage specificity of the interleukin-5 receptor alpha promoter through activation and repression domains. Mol Cell Biol. 1999;19:3940–3950. doi: 10.1128/mcb.19.6.3940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kapranov P., Cheng J., Dike S., Nix D.A., Duttagupta R., Willingham A.T., Stadler P.F., Hertel J., Hackermuller J., Hofacker I.L., Bell I., Cheung E., Drenkow J., Dumais E., Patel S., Helt G., Ganesh M., Ghosh S., Piccolboni A., Sementchenko V., Tammana H., Gingeras T.R. RNA maps reveal new RNA classes and a possible function for pervasive transcription. Science. 2007;316:1484–1488. doi: 10.1126/science.1138341. [DOI] [PubMed] [Google Scholar]
- Katan Y., Agami R., Shaul Y. The transcriptional activation and repression domains of RFX1, a context-dependent regulator, can mutually neutralize their activities. Nucleic Acids Res. 1997;25:3621–3628. doi: 10.1093/nar/25.18.3621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawaji H., Kasukawa T., Fukuda S., Katayama S., Kai C., Kawai J., Carninci P., Hayashizaki Y. CAGE Basic/Analysis Databases: the CAGE resource for comprehensive promoter analysis. Nucleic Acids Res. 2006;34:D632–636. doi: 10.1093/nar/gkj034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kistler W.S., Horvath G.C., Dasgupta A., Kistler M.K. Differential expression of Rfx1-4 during mouse spermatogenesis. Gene Expr Patterns. 2009;9:515–519. doi: 10.1016/j.gep.2009.07.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laurencon A., Dubruille R., Efimenko E., Grenier G., Bissett R., Cortier E., Rolland V., Swoboda P., Durand B. Identification of novel regulatory factor X (RFX) target genes by comparative genomics in Drosophila species. Genome Biol. 2007;8:R195. doi: 10.1186/gb-2007-8-9-r195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li G., Vega R., Nelms K., Gekakis N., Goodnow C., McNamara P., Wu H., Hong N.A., Glynne R. A role for Alstrom syndrome protein, alms1, in kidney ciliogenesis and cellular quiescence. PLoS Genet. 2007;3:e8. doi: 10.1371/journal.pgen.0030008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marion V., Stoetzel C., Schlicht D., Messaddeq N., Koch M., Flori E., Danse J.M., Mandel J.L., Dollfus H. Transient ciliogenesis involving Bardet–Biedl syndrome proteins is a fundamental characteristic of adipogenic differentiation. Proc Natl Acad Sci U S A. 2009;106:1820–1825. doi: 10.1073/pnas.0812518106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marshall J.D., Bronson R.T., Collin G.B., Nordstrom A.D., Maffei P., Paisey R.B., Carey C., Macdermott S., Russell-Eggitt I., Shea S.E., Davis J., Beck S., Shatirishvili G., Mihai C.M., Hoeltzenbein M., Pozzan G.B., Hopkinson I., Sicolo N., Naggert J.K., Nishina P.M. New Alstrom syndrome phenotypes based on the evaluation of 182 cases. Arch Intern Med. 2005;165:675–683. doi: 10.1001/archinte.165.6.675. [DOI] [PubMed] [Google Scholar]
- Nielsen S.K., Mollgard K., Clement C.A., Veland I.R., Awan A., Yoder B.K., Novak I., Christensen S.T. Characterization of primary cilia and Hedgehog signaling during development of the human pancreas and in human pancreatic duct cancer cell lines. Dev Dyn. 2008;237:2039–2052. doi: 10.1002/dvdy.21610. [DOI] [PubMed] [Google Scholar]
- Philipsen S., Suske G. A tale of three fingers: the family of mammalian Sp/XKLF transcription factors. Nucleic Acids Res. 1999;27:2991–3000. doi: 10.1093/nar/27.15.2991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reith W., Mach B. The bare lymphocyte syndrome and the regulation of MHC expression. Annu Rev Immunol. 2001;19:331–373. doi: 10.1146/annurev.immunol.19.1.331. [DOI] [PubMed] [Google Scholar]
- Reith W., Ucla C., Barras E., Gaud A., Durand B., Herrero-Sanchez C., Kobr M., Mach B. RFX1, a transactivator of hepatitis B virus enhancer I, belongs to a novel family of homodimeric and heterodimeric DNA-binding proteins. Mol Cell Biol. 1994;14:1230–1244. doi: 10.1128/mcb.14.2.1230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Romano S., Milan G., Veronese C., Collin G.B., Marshall J.D., Centobene C., Favaretto F., Dal Pra C., Scarda A., Leandri S., Naggert J.K., Maffei P., Vettor R. Regulation of Alstrom syndrome gene expression during adipogenesis and its relationship with fat cell insulin sensitivity. Int J Mol Med. 2008;21:731–736. [PubMed] [Google Scholar]
- Sanchez-Elsner T., Ramirez J.R., Sanz-Rodriguez F., Varela E., Bernabeu C., Botella L.M. A cross-talk between hypoxia and TGF-beta orchestrates erythropoietin gene regulation through SP1 and Smads. J Mol Biol. 2004;336:9–24. doi: 10.1016/j.jmb.2003.12.023. [DOI] [PubMed] [Google Scholar]
- Schreiber E., Matthias P., Muller M.M., Schaffner W. Rapid detection of octamer binding proteins with “mini-extracts,” prepared from a small number of cells. Nucleic Acids Res. 1989;17:6419. doi: 10.1093/nar/17.15.6419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shiraki T., Kondo S., Katayama S., Waki K., Kasukawa T., Kawaji H., Kodzius R., Watahiki A., Nakamura M., Arakawa T., Fukuda S., Sasaki D., Podhajska A., Harbers M., Kawai J., Carninci P., Hayashizaki Y. Cap analysis gene expression for high-throughput analysis of transcriptional starting point and identification of promoter usage. Proc Natl Acad Sci U S A. 2003;100:15776–15781. doi: 10.1073/pnas.2136655100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swoboda P., Adler H.T., Thomas J.H. The RFX-type transcription factor DAF-19 regulates sensory neuron cilium formation in C. elegans. Mol Cell. 2000;5:411–421. doi: 10.1016/s1097-2765(00)80436-0. [DOI] [PubMed] [Google Scholar]
- Wierstra I. Sp1: emerging roles—beyond constitutive activation of TATA-less housekeeping genes. Biochem Biophys Res Commun. 2008;372:1–13. doi: 10.1016/j.bbrc.2008.03.074. [DOI] [PubMed] [Google Scholar]
- Wolfe S.A., Wilkerson D.C., Prado S., Grimes S.R. Regulatory factor X2 (RFX2) binds to the H1t/TE1 promoter element and activates transcription of the testis-specific histone H1t gene. J Cell Biochem. 2004;91:375–383. doi: 10.1002/jcb.10748. [DOI] [PubMed] [Google Scholar]
- Yabuta N., Onda H., Watanabe M., Yoshioka N., Nagamori I., Funatsu T., Toji S., Tamai K., Nojima H. Isolation and characterization of the TIGA genes, whose transcripts are induced by growth arrest. Nucleic Acids Res. 2006;34:4878–4892. doi: 10.1093/nar/gkl651. [DOI] [PMC free article] [PubMed] [Google Scholar]