Abstract
Polycomb proteins are known to be of great importance in human cancer pathogenesis. SUZ12 is a component of the Polycomb PRC2 complex that, along with EZH2, is involved in embryonic stem cell differentiation. EZH2 plays an essential role in many cancer types, but an equivalent involvement of SUZ12 has not been as thoroughly demonstrated. Here we show that SUZ12 is anomalously expressed in human primary tumors, especially in mantle cell lymphoma (MCL), pulmonary carcinomas and melanoma, and is associated with gene locus amplification in some cases. Using MCL as a model, functional and genomic studies demonstrate that SUZ12 loss compromises cell viability, increases apoptosis, and targets genes involved in central oncogenic pathways associated with MCL pathogenesis. Our results support the hypothesis that the abnormal expression of SUZ12 accounts for some of the unexplained features of MCL, such as abnormal DNA repair and increased resistance to apoptosis.
The Polycomb group of proteins (PcG) are transcriptional repressors essential for regulation of embryogenesis, tissue development, stem cell self-renewal, and preservation of cell identity (reviewed in1). PcG proteins modify histone tails to repress gene expression. Two major PcG complexes have been described in humans: the polycomb repressive complex 1 (PRC1), which contains BMI1, MEL18, RING1, RNF2, HPC1, and others, and the polycomb repressive complex 2 (PRC2), which typically contains EZH2, SUZ12 and various isoforms of EED.2 PRC2 has histone methyltransferase (HMTase) activity that allows the complex to trimethylate chromatin specifically at lysine 27 of histone H3. PRC1 recognizes this mark and recruits the machinery necessary to remodel chromatin structure.3,4,5,6
There is mounting evidence of the pathogenic role of PcG in human cancer.7,8,9,10 This is the case for murine Bmi1, which collaborates with c-Myc in transforming lymphoid cells.11,12 Human BMI1 has been found to be deregulated in mantle cell lymphoma (MCL) and in Hodgkin’s and diffuse large B-cell lymphomas.10,13,14,15,16 EZH2 is involved in progression in prostate cancer and in neoplastic transformation of breast epithelial cells.17,18 This member of the PRC2 complex has HMTase activity and is therefore essential for gene transcription regulation. SUZ12, another important member of this complex, in conjunction with EED and RBAP48, is up-regulated in colon and breast tumors,19 but its specific function in human cancer is unknown. SUZ12 is a zinc finger protein that has been found at the breakpoints of a recurrent chromosomal translocation in endometrial stromal sarcoma.20 SUZ12 is essential in mouse development and is required for the proliferation of cultured cells.21 Within the PRC2 complex, SUZ12 is required for the HMTase activity of the complex.21,22
MCL is a lymphoid malignancy with an aggressive clinical behavior, whose study has critically improved our understanding of the pathogenic role of multiple oncogenes and survival pathways.23,24,25 It accounts for around 5% to 8% of non-Hodgkin’s lymphomas, and is associated with a chromosomal translocation t(11;14)(q13;q32) that puts the CCND1 gene under the control of the immunoglobulin heavy chain locus regulatory elements.23 However, this characteristic molecular event does not explain fully the clinical and biological features of the tumor and is not sufficient for tumoral transformation, as has been demonstrated in experimental models.26 Several studies suggest that other molecular events play a pathogenic role in MCL pathogenesis, such as ATM loss or nuclear factor κB pathway activation.24,27 Nevertheless, there are still various MCL oncogenic features that are not explained by the alterations so far identified.
In this study we have investigated the expression pattern of SUZ12 and EZH2 in a large cohort of human normal tissues and tumors in search of patterns associated with transformation events. We demonstrate that SUZ12 is anomalously expressed in several human primary tumors, and that it is especially relevant in specific tumors such as MCL, melanoma and pulmonary carcinomas, where it is associated with gene amplification in some cases. The use of an integrated approach combining genome-wide location assays, functional studies, and gene expression profiling, leads us to conclude that SUZ12 may be involved in MCL pathogenesis.
Materials and Methods
Production of SUZ12 Monoclonal Antibody
A cDNA encoding the full-length human SUZ12 protein was obtained from the laboratory of Dr Yi Zhang (pGEX-KG-SUZ12). The human SUZ12 gene was amplified by polymerase chain reaction (PCR) and introduced into the pDEST-TH1 expression vector (Invitrogen, Carlsbad, CA) by means of Gateway technology. The MBP-SUZ12 fusion protein was then expressed in Escherichia coli strain BL21 (DE3) with 0.4 mmol/L IPTG overnight at 30°C. The bacteria were lysed with BugBuster reagent (Novagen, Madison, WI). The soluble fraction was purified with amylase resin (New England Biolabs, Ipswich, MA), and the joined protein was eluted with 10 mmol/L maltose. The protein-containing fractions were concentrated by Vivaspin ultrafiltration (Sartorius Stedim Biotech, Aubagne, France) and used as an immunogen.
Three BALB/c mice were injected intraperitoneally (three times at 14-day intervals) with 100 μg 6 × MBP-SUZ12 fusion protein and Freund’s adjuvant. A 150-μg booster of the recombinant SUZ12 protein was injected intraperitoneally, and fused three days later, as described previously.28,29 Hybridoma supernatants were screened by enzyme-linked immunosorbent assay. The mouse mAb raised against SUZ12 (220A/A3) was cloned by the limiting dilution technique. Animal experiments were performed under the experimental protocol approved by the Institutional Committee for Care and Use of Animals, CEUCA no. 001/02.
To confirm that 220A/A3 mAb recognized the human SUZ12 protein, immunohistochemistry on frozen cytospin preparations of V5-tagged human SUZ12 expressed in HEK-293T cells was performed. Labeling with the anti-V5 mAb confirmed the efficiency of transfection. A cytospin preparation of V5-tagged human SOX4 protein was used as a negative control (Supplemental Figure 1 at http://ajp.amjpathol.org).
Tissue Microarrays and Immunostaining Techniques
Immunohistochemical expression of SUZ12 and EZH2 were assessed using tissue microarray (TMA) technology for 150 normal and 569 tumoral samples. To this end, we used a tissue array device (Beecher Instruments, Sun Prairie, WI), as previously described.13,30 An additional TMA including 76 MCL cases was also used.31 Immunohistochemical staining was performed on these TMA sections using the following antibodies: McAb SUZ12 (220A/A3) and EZH2 polyclonal antibody (Zymed, San Francisco, CA).32,33 Proliferation indices in MCL cases were evaluated by means of Ki-67 expression using MIB1 monoclonal antibody by DAKO (DAKO, Glostrup, Denmark). M.S.-B. and E.S. evaluated the staining of TMA sections for SUZ12, EZH2, and Ki-67 proteins using uniform criteria. Discrepancies in the scoring of cases were resolved after joint examination on a multiheaded microscope. To ensure the reproducibility of this method, we used straightforward, clear-cut criteria, and cases were scored as positive (1) or negative (0) for SUZ12 and EZH2 antibodies. The threshold was 5% of positive cells for both antibodies. For Ki-67, the values were scored as negative (0) for fewer than 5% of proliferating cells, positive (1) for 5% to 25%, and strongly positive (2) if more than 25% of cells were positive for Ki-67.13 Whole-tissue sections from reactive lymph node, thymus, spleen, and tonsillectomy specimens were used for the examination of benign lymphocyte subpopulations.
FISH
Fluorescence in situ hybridization (FISH) was used to detect SUZ12 copy number changes, as previously reported.34 To study SUZ12 amplification we used the bacterial artificial chromosome clones RP11-290N17 and RP11-640N20 from the BACPAC resources center (Children’s Hospital Oakland Research Institute, Oakland, CA), which spans the entire 17q11.2 genomic region, and a commercial centromeric probe for chromosome 17 (Vysis Inc., Downers Grove, IL), which was used as a control for the ploidy level of chromosome 17.
FISH evaluation was performed by J.S. with no previous knowledge of other genetic, clinical, or immunohistochemical results. Fluorescence signals were scored in each sample by counting the number of single-copy gene and centromeric signals in an average of 130 (60–210) well-defined nuclei. SUZ12 amplification was recognized if the SUZ12/chromosome 17 ratio was greater than 2 in at least 20% of tumor cells. A sample was considered to feature a SUZ12 gene gain if the SUZ12/chromosome 17 ratio was greater than 1.5 but less than 2.35,36
Cell Lines
Human cell lines derived from MCL patients Jeko-1 and Z138 were kindly provided by Dr. Martínez-Climent and cultured in RPMI 1640 medium supplemented with 1% l-glutamine, 10% fetal bovine serum (Invitrogen), 0.5% penicillin/streptomycin, and 0.1% Fungizone. Cells were maintained at 37°C in a humidified 5% CO2 incubator.
The HEK293T cell line was obtained from the American Type Culture Collection and was cultured in Dulbecco’s modified Eagle’s medium supplemented with 1% l-glutamine, 10% fetal bovine serum (Invitrogen), 0.5% penicillin/streptomycin and 0.1% Fungizone. Cells were maintained at 37°C in a humidified 5% CO2 incubator.
Virus Production and Cell Line Infection
Viruses were produced by transient transfection in the HEK293T cell line. Plasmids were produced in the TOP10 E. coli strain (Invitrogen) and grown in low-salt Luria Bertani medium. Plasmids were isolated using Qiagen EndoFree Plasmid Maxi Kit (Qiagen, Venlo, The Netherlands).
For co-transfection, plasmids pCMVdeltaR8.91 (derived from pCMVR8.937), pMD.G and the lentiviral vector pA179.Helix38 containing the shRNAi sequences were used. Plasmids were co-transfected using FuGene 6 (Roche, Basel, Switzerland) following the manufacturer’s recommendations. Target cells (5 × 105) were transduced by spinoculation using the viral supernatant.
Vectors and shRNA Design
Different shRNAs were designed using the SIDE program (http://side.bioinfo.cipf.es, last accessed December 4, 2008). The shRNAs were designed within the ORF sequence of SUZ12 to avoid off-target effects associated with imperfect matching in the 3′ UTR of the target gene. shRNAs were cloned as previously described.39 shRNA sequences used for control and SUZ12 knockdown were as follows: Scramble: (5′-GAGGAACCAAACCATAACA-3′); shSUZ12.783: (5′-GGATGTAAGTTGTCCAATA-3′); shSUZ12.2076: (5′-GCTGACAATCAAATGAATCAT-3′).
Cell Competition Assays
GFP expression was analyzed by FACS 72 hours after infection of the cell lines with lentivirus carrying either the empty vector, or a scrambled control or either of the two shRNAs against SUZ12. This was done every 2 to 4 days. The evolution of GFP expression was compared with the third day using the following formula:
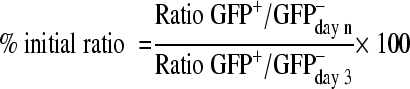 |
Cell Growth and Apoptosis
Cell growth was assessed by counting cells in a Neubauer chamber and using trypan blue dye to exclude dead cells. For cell cycle analysis, 106 cells were washed with PBS and fixed with chilled 70% ethanol added drop by drop and incubated in the cold for at least 1 hour. The cells were then washed again with PBS and resuspended in 500 μl of PBS. RNase A was added at a final concentration of 200 ng/μl and incubated for 30 minutes. Cells were stained with 10 μl propidium iodide (1 mg/ml) before acquisition in the cytometer.
Cell death was quantitated by annexin V–APC (BD PharMingen, Franklin Lakes, NJ) staining according to the manufacturer’s protocol. Briefly, cells were washed in PBS, resuspended in 500 μl of binding buffer (BD PharMingen) containing 0.5 μg/ml annexin V-APC and 25 μg/ml propidium iodide, and then analyzed by flow cytometry. Cell cycle and apoptosis assays were analyzed with a FACSCalibur flow cytometer (BD PharMingen).
Histone Extraction
Histones were isolated by acidic extraction in 0.25 mol/L HCl and precipitation with acetone. Briefly, 106 cells were harvested and incubated overnight in 200 μl of 0.25 mol/L HCl at 4°C with shaking. For histone precipitation, 8 volumes of acetone were added to the supernatant, and histones were pelleted by centrifugation and washed with acetone. Histones were air-dried and resuspended in 0.25 mol/L HCl for subsequent immunoblot analysis.
Western Blot Analysis
Total protein extracts were prepared using radioimmunoprecipitation assay lysis buffer supplemented with protease inhibitors. Antibody detection was performed using fluorescent-labeled secondary antibodies (Alexa 680 and Alexa 800, Rockland, Gilbertsville, PA) and an Odyssey infrared system scanner (LI-COR Biosciences, Lincoln, NE).
Antibodies for immunoblot analysis included mouse anti-SUZ12 mAb (clone 220A/A3), rabbit anti-histone H3 trimethylated at lysine 27 (07-499, Upstate Biotechnology, Lake Placid, NY), mouse anti-H3 mAb (clone 6.6.2, Upstate Biotechnology), mouse anti-PARP (P248, Sigma-Aldrich Inc., St. Louis, MO), and mouse anti-α-tubulin (clone DM1A, Sigma-Aldrich). Band intensities were quantified using ImageJ 1.34S software (National Institutes of Health, Bethesda, MD).
ChIP-on-Chip
Chromatin Immunoprecipitation
Chromatin immunoprecipitation (ChIP) was assayed using the ChIP assay kit (Upstate Biotechnology, Billerica, MA) following the manufacturer’s recommendations. The antibodies used here were specific for SUZ12 (220A/A3 and Upstate, 07-379), histone H3 trimethylated at lysine 27, and IgG (Upstate Biotechnology).
For ChIP-on-chip experiments, three biological replicates were amplified using the GenomePlex whole genome amplification kit (Sigma-Aldrich) following the protocol provided by the manufacturer. The same quantity of each replicate was mixed in a single tube. Input material and samples were labeled with Cy3 and Cy5, respectively, and hybridized onto the human promoter ChIP-on-chip microarray set (Agilent Technologies Inc., Santa Clara, CA).
DNA Microarray Analysis ChIP-on-Chip
The human promoter ChIP-on-chip microarray set covers −5.5 kb upstream to +2.5 kb downstream of the transcriptional start sites with a total of 487,008 probes. Agilent’s ChIP analytics program (v. 1.3.1) was used for the analyses.
A whole-chip error model was used to calculate confidence values from the enrichment ratio and the signal intensity of each probe (probe P value) and of each set of three neighboring probes (probe set P value). Probe sets with significant probe set P values (P < 0.001) and significant individual probe P values (P < 0.01) were judged to be bound. Bound regions were assigned to genes if they were within 1 kb of the transcription start site registered in at least one of five genomic databases.
PCR of Immunoprecipitated Material
Original sequences for primer design were extracted from the March 2006 human reference sequence (NCBI Build 36.1). Immunoprecipitated DNA was subjected to semiquantitative PCR using the following primers: ATM: sense strand 5′-GTTGTGCAAAGGGGTCAACT-3′, antisense strand 5′-TTGGCGGAACTGAAAGAAG-3′; BCOR: sense strand 5′-GCAAAAGACAGGCGAGCAAG-3′, antisense strand 5′-ACCCCCAGAAAGACCAGGAA-3′; BIRC2: sense strand 5′-CCCAGGTGCATTTTTGGAAG-3′, antisense strand 5′-TGCCTGCCAGTCAGTCACAG-3′; CBX2: sense strand 5′-TTCTCCCCGCTGTAACCTGA-3′, antisense strand 5′-GCCCGAGATCCAGAACAATG-3′; E2F5: sense strand 5′-TGGATTGCAGTGGCAGGA-3′, antisense strand 5′-GGCGTGGTAGTGCACACTTG-3′; GADD45G: sense strand 5′-GTGCCAGCGTGTATGGTCAA-3′, antisense strand 5′-CGAGTAAGGGCTGCAAAACG-3′; H2AFZ: sense strand 5′-AGGGCCTGGGAGTTTTCTTG-3′, antisense strand 5′-CTGTGTACAGCGCAGCCATC-3′; HDAC2: sense strand 5′-CTGGAGAAGGAGGCCGTTTC-3′, antisense strand 5′-GCAGACCTGAGGGGGAGAAC-3′; JMJD2D: sense strand 5′-AAATATGTACGGGGCAACCA-3′, antisense strand 5′- TGACATCTCCCCTCCCACTA-3′; VAV3: sense strand 5′-GCTCAGCGCACCTAGACGTT-3′, antisense strand 5′- GGCTCAGGTGTTCGACCTTG-3′.
Functional Gene Classification with Ingenuity Pathways Analysis
We identified functions/pathways classification terms enriched for SUZ12-bound genes using Ingenuity Pathways analysis (Redwood City, CA). The probability associated with a biological process is a measure of its statistical significance with respect to the functions/pathways/lists eligible molecules for the dataset and a reference set of molecules that defines the molecules that could possibly have been functions/pathways/lists eligible. The probability is that associated with a right-tailed Fisher’s exact test.
Gene Expression Profile
For gene expression profiling, total RNA was extracted from cell lines and MCL frozen tumoral samples using the Qiagen RNeasy kit (Qiagen). 500 ng of RNA were labeled with cyanine 5-conjugated dUTP (Cy5) and hybridized onto the Agilent 44K whole genome microarray chip (Agilent Technologies) against a universal human reference RNA (Stratagene, La Jolla, CA) previously labeled with cyanine 3-conjugated dUTP (Cy3). Slides were scanned in an Agilent G2565AA microarray scanner system and data were extracted with feature extraction software (Agilent Technologies).39
Statistical Analysis
To validate ChIP-on-chip results functionally in MCL tumoral samples, Pearson correlations between identified SUZ12 target genes and SUZ12 expression were calculated using the T-Rex program included in the Gene Expression Pattern Analysis Suite (http://www.gepas.org/).40
Gene Set Enrichment Analysis
The gene set enrichment analysis (GSEA) tool (http://www.broad.mit.edu/gsea, last accessed April 7, 2008)41,42 was used to explore functional gene sets, allowing the interpretation of complete gene expression data in relation to SUZ12 expression. The gene sets co-regulated with SUZ12 expression were identified using Pearson correlation, with a minimum of 10 and a maximum of 500 genes in a gene set being required to qualify them for further analysis. The selected gene sets corresponded to Biocarta pathways (http://www.biocarta.com, last accessed April 7, 2008), excluding those that were not relevant to either lymphoid cell biology or cancer. Gene sets with an false discovery rate of less than 0.25 were considered significant.
Results
SUZ12 Protein Expression Is Restricted to Proliferating Cells in Normal Human Tissues
To screen for potential abnormalities in the expression of SUZ12, we compared expression patterns in tumoral samples with those in normal tissues. To this end, we first generated a monoclonal antibody (mAb) against SUZ12 protein (clone 220A/A3; described in Materials and Methods). Demonstration of 220A/A3 mAb specificity against SUZ12 in cytospin preparations can be found in the supplementary information (Supplemental Figure 1 at http://ajp.amjpathol.org). These results were confirmed by Western blot using cell lysate of SUZ12 and SOX4-transfected cells (data not shown). The specificity of the mAb generated is also demonstrated in the shRNA experiments described below. The antibody was found to be suitable for immunohistochemistry, immunofluorescence (data not shown), Western blot detection, and chromatin immunoprecipitation.
With this mAb, we characterized the expression of SUZ12 in a large cohort of non-tumoral human tissues using a TMA containing a panel of around 50 different normal tissues.13 As SUZ12 binds to EZH2 in PRC2 complex, we also characterized EZH2 protein expression in these same tissues. Surprisingly, we found that whereas EZH2 protein is widely detected in a large proportion of tissues, with few exceptions, SUZ12 expression is restricted to proliferating cells in reactive lymphoid tissue, germinal cells in the testis, and the epithelium of (Figure 1,A–D, and Supplemental Table 1 and Supplemental Figure 2 at http://ajp.amjpathol.org).
Figure 1.
Expression of SUZ12 in non-tumoral human tissues: SUZ12 protein is preferentially detected in proliferating cells in various non-tumoral human tissues such as proliferating cells in tonsil germinal center (M, mantle zone; GC, germinal center) (A); proliferating cells in tonsil epithelium (B); germinal cells of the testis (C); and the hair follicle (D). Objective, ×100, immersion).
SUZ12 Is Overexpressed in a Subset of Human Tumors
We also studied SUZ12 and EZH2 expression in TMAs containing representative paraffin sections from 569 cases of multiple (up to 69) human tumor types.13 EZH2 and SUZ12 expression patterns were also different in tumoral samples and we found no significant association between these two proteins. While EZH2 protein was almost ubiquitously expressed in most tumors (489 out of 521, 94%, were positive) although with different intensities, SUZ12 was more restricted to lymphoid, lung, vascular, germinal ,and skin tumors (250 out of 497, 50.3%, had detectable expression). Therefore, those cases positive for SUZ12 are usually EZH2-positive, but not vice versa. We also observed a small fraction of tumors of different types (16/497, 3.2%) in which we could detect SUZ12 but not EZH2. Results are summarized in Supplemental Table 2 and Supplemental Figure 2 at http://ajp.amjpathol.org.
Essentially, SUZ12 expression was more frequent in lymphomas (91%) (notably in MCL); germinal cell-derived tumors (70%) (seminomas, teratocarcinomas, and embryonal carcinomas); skin tumors (88%) (melanomas and skin carcinomas); vascular tumors (83%); and pulmonary neuroendocrine small-cell carcinomas (70%) (Supplemental Table 2, A and B, and Supplemental Figure 2 at http://ajp.amjpathol.org). Therefore, SUZ12 was expressed in tumors characterized by a high growth fraction.
SUZ12 Overexpression Is Associated with Gene Locus Amplification
SUZ12 gene is located at the 17q11.2 locus, which is frequently found to be translocated in endometrial stromal tumors.20 Other cytogenetic alterations have been described at this locus in various types of tumor, such as additions, deletions and translocations,43,44,45,46,47 meaning that these cytogenetic alterations could be related to tumorigenesis.
To determine whether SUZ12 is amplified in primary human tumors, we used FISH analysis in TMA paraffin sections. 17q11.2 amplification or gain was analyzed in the same TMAs as were used for IHQ analysis. Five cases showed gene amplification as determined by the standard criterion35,36 (SUZ12/centromeric 17 ratio >2.0) and two cases showed increased gene copy number (ratio > 1.5) distributed in several tumoral types such as mesothelioma (amplification in 1/10), melanoma (amplification in 1/10), skin basal cell carcinoma (gain in 1/10), thyroid follicular carcinoma (amplification in 1/8), leiomyosarcoma (gain in 1/6), MCL (amplification in 1/10), and ovary serous cystoadenocarcinoma (amplification in 1/6). The results are summarized in Supplemental Table 2, A and B, at http://ajp.amjpathol.org.
Although not exclusive to MCL, the findings of the anomalous expression of SUZ12 in most MCL cases (9/10) compared with its absence in non-tumoral mantle zone cells, and the detection of SUZ12 locus amplification associated with strong SUZ12 expression prompted us to extend the study to a larger cohort of cases for further validation.
Additionally, comparing SUZ12 expression in MCL with that in other lymphoma types, it was detected in those lymphomas with a high growth fraction and that are derived from germinal center B cells (diffuse large B-cell and Burkitt lymphomas), or was restricted to proliferating cells in chronic lymphocytic leukemia, follicular lymphoma, and splenic marginal zone lymphoma cases (Figure 2A). However, in MCL, a tumor characterized by lower or intermediate proliferation, we observed a high level of expression of SUZ12 in most (9/10) cases, in contrast with the absence of SUZ12 from the mantle zone cells in reactive lymphoid tissue, which is the normal counterpart of this tumor type (Figure 2B and Supplemental Figure 2 at http://ajp.amjpathol.org).
Figure 2.
SUZ12 in B-cell lymphomas. A: SUZ12 and Ki-67 expression in different types of B-cell lymphoma, showing the correlation between the strength of SUZ12 expression and proliferation index. B: However, in samples of MCL, a tumor with a relatively low proliferation index, some cases showed increased SUZ12 expression, in contrast to the absence of its expression from normal mantle zone cells (Figure 1A), which are the benign counterpart of MCL. C: SUZ12 gene amplification detected by FISH in an SUZ12-positive MCL case. Several copies of SUZ12 (in red) are detected, compared with only two centromeric copies for chromosome 17 (green). (Objective, ×100, immersion).
SUZ12 in Mantle Cell Lymphomas
Therefore, we checked SUZ12 expression and gene locus alterations in an additional group of 76 MCL cases (Supplemental Table 2C at http://ajp.amjpathol.org). In total, 46 out of 81 (56%) MCL cases (10 in the general multitumor TMA and 71 new evaluable cases in MCL-specific TMA) showed SUZ12 protein expression.
Sixty-seven new MCL samples gave valuable results in the FISH study. In total, three MCL samples (the previous one and two cases in the new TMA) showed gene amplification (Figure 2C) and four cases had a high gene copy number (Supplemental Table 2, B and C at http://ajp.amjpathol.org). The three cases with 17q11.2 locus amplification showed SUZ12 expression in more than 80% of tumoral cells and, remarkably, two of these cases were diagnosed as aggressive (blastoid) MCL cases. These data indicate that around 9% of MCL cases had cytogenetic alterations at the SUZ12 locus. Additionally, polysomy of chromosome 17 was found in 12 MCL samples.
SUZ12 Silencing by shRNA Compromises Cellular Viability
To further assess the significance of SUZ12 overexpression in MCL we performed an RNAi analysis using a lentiviral (HIV)-based RNA interference vector in two MCL-derived cell lines: Jeko-1 and Z138. The vector contains EGFP as a selection marker.
Two sequences (shSUZ12.783 and shSUZ12.2076) correctly induced SUZ12 silencing with a residual expression of less than 20% (Figure 3A). Specificity of the shRNAs was also assessed by measuring trimethylation levels at lysine 27 of histone H3 (H3K27me3), a hallmark of PRC2 activity. Levels of H3K27me3 were significantly lower 72 hours after infection with the lentivirus carrying shRNAs against SUZ12 compared with controls (Figure 3B). To determine whether SUZ12 inhibition had any effect on cell survival or cell growth we designed a strategy based on competitive proliferation similar to that previously described (see Materials and Methods).48 Thus, we infected Jeko-1 and Z138 cells and a slight but constant decrease in GFP+ cell number was observed, specifically in those cells transduced with lentivirus carrying any of the shRNAs against SUZ12 (Figure 3C).
Figure 3.
Effects of SUZ12 depletion in MCL cell lines. A: Left, anti-SUZ12 immunoblot of Jeko-1 and Z138 cell lines transduced either with the empty vector, a scrambled sequence or with two different hairpins against SUZ12. Band signals were normalized with tubulin as a loading control (right). B: Depletion of SUZ12 affects the function of the PRC2 complex as assessed by immunoblot using an antibody against histone H3 trimethylation at lysine 27 (left). Total histone H3 was used to normalize band signals (right). Cropped blots are shown. C–F: SUZ12 knockdown compromises cell viability: C: Expression of EGFP that marks transduced cells was tracked over time to observe differences in viability between cells transduced either with the empty vector or with a scramble control and cells transduced with two different hairpins against SUZ12. D: GFP+-sorted Jeko1 and Z138 proliferation was assessed by counting viable cells using trypan blue exclusion along the time. Mean and SD are shown. E: Immunoblot showing specific cleavage of poly (ADP-ribose) polymerase (cPARP) in SUZ12-deficient cells after sorting. F: Flow cytometric analysis of apoptosis using double staining of Annexin V and propidium iodide after cell sorting.
Additionally, to assess the effect of SUZ12 silencing on cell growth directly, we sorted the GFP+ fraction in each case and measured cell number by trypan blue exclusion counting. Results showed that cells expressing shRNAs against SUZ12 grew less than cells either expressing a scramble control or infected with the empty vector (Figure 3D). Cell cycle analysis by FACS in these GFP+-sorted cells only showed subtle differences: either a slight decrease in G2/M or an increase in SubG1 phase in cells deficient in SUZ12 (data not shown). Levels of apoptosis were augmented on silencing of SUZ12 as demonstrated by specific annexin V staining and cleavage of PARP (Figure 3, E and F). One of the hairpins was more efficient in killing the cells, and this was associated with the levels of H3K27me3 demethylation. In fact, we were not able to detect PARP cleavage in Jeko-1 with one of the hairpins, probably because of the sensitivity of the antibody, since we could still detect a mild increment in apoptosis with annexin V staining.
Identification and Functional Classification of SUZ12 Genomic Target Genes in MCL
To look for potential targets that could explain the role of SUZ12 in MCL we performed ChIP and subsequent hybridization on Agilent’s human promoter microarray (ChIP-on-chip) with the Z138 MCL-derived cell line. We found 17,605 (3.6%) bound probes (peak P < 0.01) corresponding to 1806 genes. Those probes with a normalized log2 ratio >1 were considered to be potential SUZ12 targets (1424 genes including known SUZ12 target genes such as CDKN2A, GADD45G, BMP2, and WNT49,50,51).
To determine the accuracy of SUZ12 target gene discovery, single-locus semiquantitative PCR was performed on the immunoprecipitated material with SUZ12 and H3K27me3 antibodies on 18 candidate SUZ12 target genes, including genes known to be relevant in MCL or lymphoma pathogenesis (ATM, BCOR, VAV), in the control of significant processes such as apoptosis and DNA repair (BIRC2, GADD45), and transcriptional regulation (JMJD2). GADD45G (a previously described target)49 was used as the positive control (Figure 4). Eleven of 18 were validated for SUZ12 and H3K27me3 antibodies, including ATM, CBX2, VAV3, JMJD2, and BIRC2.
Figure 4.
Validation of SUZ12 target genes by semiquantitative ChIP. Single-locus semiquantitative PCR on ChIP samples was performed on several SUZ12 candidate target genes, with SUZ12 and H3K27me3 antibodies. Mouse IgG was used as a negative control.
To determine which relevant pathways could be altered by SUZ12 overexpression, we examined the functions of SUZ12 target genes using the Ingenuity Pathway Analysis program. Of 1424 genes identified by ChIP-on-chip, 658 have annotated functions in the Ingenuity Pathway Analysis database and were functionally classified. The results showed significant enrichment of genes controlling gene expression, cell cycle and proliferation, DNA replication and repair, and development (Figure 5), consistent with the findings of other studies.49,50
Figure 5.
Functional classification of SUZ12-targeted genes. Functions of SUZ12 target genes were analyzed using the Ingenuity Pathway Analysis program. Of 1424 genes identified by ChIP-on-chip, 658 have annotated functions in the Ingenuity Pathway Analysis database. The number of genes identified as belonging to each category is included. The probability is that associated with a right-tailed Fisher’s exact test.
The group of SUZ12 target genes controlling gene expression includes transcription factors and regulators, such as E2F5, POU domain proteins, and SUV39H1, and enzymes regulating transcription and translation, such as DNA and RNA polymerases, jumonji domain (JMJD) proteins, and several eukaryotic translation initiation factors. Development regulators have also been found among SUZ12 targets in MCL, some of which have been previously published as SUZ12 targets in embryonic cells, such as SOX and FOX family genes, POU domain transcription factors, and BMP2. Another relevant finding is the detection of SUZ12 in the promoter region of several miRs genes, two of which have been identified as SUZ12 targets (hsa-mir-124a and hsa-mir-18349,52) (identified genes listed in Supplemental Table 3 at http://ajp.amjpathol.org).
However, the most noteworthy finding was that there were significant genes among the top SUZ12 targets that are known to be involved in MCL pathogenesis, some of which were not previously known to be SUZ12 targets. These included those regulating cell cycle (CDKN2A and other INK4 family genes, cyclins, CDKs, CHEK1, MAD2L1, and BUB3); DNA damage and repair genes (ATM, GADD45, several DNA polymerases and topoisomerases, MLH1, XRCC family genes, and ERCC family genes); apoptosis regulators (BCL2 and BCL2 regulator proteins, BID, several BIRC family members, and others); and we also found members of nuclear factor κB pathway (BCL10, NFKB2, and IKBKG) to be regulated by SUZ12.
SUZ12 Target Gene Expression in Silenced SUZ12 MCL-Derived Cell Lines and MCL Tumoral Samples
To validate functionally and elucidate the relevance of these ChIP-on-chip findings, we looked at the changes in expression of the targets associated with SUZ12 expression in both Z138 cell line after SUZ12 silencing and in tumoral samples from MCL cases using whole genome expression microarrays.
SUZ12 Targets in SUZ12-Silenced Z138 Cell Line
We first analyzed the expression of SUZ12 targets in SUZ12-silenced cells, comparing the expression profile of SUZ12-depleted cells with those infected with the control vector. After SUZ12 silencing in the Z138 cell line, some SUZ12 targets were actually unrepressed. 140 transcripts showed an up-regulation or down-regulation of at least 0.6 (log2 scale) and were considered to be significantly deregulated after SUZ12 silencing. These included, among others, CDKN2A, GADD45G, genes involved in development, such as BMP2, several GATA binding proteins or differentiation factors like MLLT3 and CBX2 (Figure 6A).
Figure 6.
SUZ12 target gene expression was affected in SUZ12-silenced cell lines and MCL cases. A: Gene expression profiling was performed in duplicate 3 and 5 days after SUZ12 depletion in Z138, with two different shRNAs (2076 and 783) versus empty vector. The heat map represents expression levels for each sample. A total of 140 significant genes [log2 (Cy5/Cy3) cut-off = ±0.6 in at least three samples] were ranked by Euclidean squared distance. B: Relative expression levels of 126 inversely correlated SUZ12 genes in MCL tumoral samples (false discovery rate < 0.15). C: Relative expression levels of 62 directly correlated SUZ12 genes in MCL tumoral samples (false discovery rate < 0.15). Pearson correlation coefficients between identified SUZ12 target genes and SUZ12 expression were calculated using the T-Rex program available from the GEPAS site (v. 3.1).
SUZ12 Targets in MCL Samples
We also examined, in MCL tumoral samples, the relation between SUZ12 expression and that of genes identified by ChIP-on-chip. This analysis revealed that 188 of 642 known genes suitable for the analysis (30%) were significantly correlated with SUZ12 expression (Pearson R > ±0.4, false discovery rate <0.15). Many of the SUZ12 targets were actually down-regulated in SUZ12-positive MCL samples (126 inversely correlated with SUZ12 expression versus 62 with a direct correlation) (Figure 6, B and C).
Pathways Co-Regulated with SUZ12 in MCL
Finally, we wanted to determine which characteristics of tumors were associated with changes in SUZ12 levels, identifying functional pathways co-regulated with the expression of SUZ12. To this end we performed a gene set enrichment analysis of SUZ12 expression with all of the genes in MCL tumoral samples, not restricted to SUZ12 targets identified by ChIP, using GSEA software (http://www.broad.mit.edu/gsea/). This analysis revealed a direct correlation with pathways associated with proliferation, such as cell cycle and caspase-apoptosis pathways and an inverse correlation with the proteasome pathway (Table 1). We also found an inverse correlation with the MAPK pathway, but some specific genes included in this pathway such as JUN, FOS, MAPK4, MAPK7, and BRAF showed stronger expression in SUZ12-expressing tumors.
Table 1.
Pathways Co-Regulated with SUZ12 Expression in MCL
| Name | Size | Enrichment score | Normalized enrichment score | P | False discovery rate |
|---|---|---|---|---|---|
| Gene sets positively correlated with SUZ12 expression | |||||
| Mitochondria pathway | 21 | 0.5952 | 2.4673 | 0.0000 | 0.0053 |
| Cell cycle pathway | 21 | 0.5622 | 2.3809 | 0.0000 | 0.0075 |
| ARAP pathway | 19 | 0.5528 | 2.1412 | 0.0020 | 0.0283 |
| Caspase pathway | 21 | 0.5233 | 2.1399 | 0.0040 | 0.0213 |
| CA2+ CAM pathway | 12 | 0.6362 | 1.9820 | 0.0043 | 0.0513 |
| D4GDI pathway | 11 | 0.6060 | 1.8114 | 0.0169 | 0.1118 |
| Gene sets negatively correlated with SUZ12 expression | |||||
| MAPK pathway | 83 | −0.4378 | −3.4409 | 0.0000 | 0.0000 |
| Proteasome pathway | 19 | −0.7375 | −2.8678 | 0.0000 | 0.0000 |
| GH pathway | 26 | −0.4999 | −2.2277 | 0.0000 | 0.0395 |
| IL1R pathway | 29 | −0.4244 | −2.0417 | 0.0080 | 0.0779 |
| CHREBP pathway | 15 | −0.5951 | −2.0912 | 0.0038 | 0.0832 |
| CK1 pathway | 12 | −0.6534 | −2.0431 | 0.0056 | 0.0916 |
| ARF pathway | 16 | −0.5428 | −1.9669 | 0.0056 | 0.1074 |
| RARRXR pathway | 14 | −0.5157 | −1.7385 | 0.0177 | 0.1367 |
| RAB pathway | 10 | −0.5956 | −1.7550 | 0.0220 | 0.1392 |
| ERK pathway | 30 | −0.3620 | −1.7652 | 0.0203 | 0.1402 |
| AKAP-centrosome pathway | 10 | −0.6027 | −1.7413 | 0.0198 | 0.1413 |
| TPO pathway | 22 | −0.4373 | −1.8955 | 0.0096 | 0.1444 |
| PPARA pathway | 51 | −0.2753 | −1.7689 | 0.0249 | 0.1465 |
| Cytokine pathway | 15 | −0.5077 | −1.7904 | 0.0183 | 0.1473 |
| PDGF pathway | 26 | −0.3973 | −1.7772 | 0.0149 | 0.1479 |
| VEGF pathway | 26 | −0.3992 | −1.7929 | 0.0135 | 0.1557 |
| TOLL pathway | 32 | −0.3657 | −1.8513 | 0.0134 | 0.1642 |
| GATA3 pathway | 13 | −0.5419 | −1.7935 | 0.0080 | 0.1678 |
| TOB1 pathway | 16 | −0.4963 | −1.7958 | 0.0189 | 0.1797 |
| P38MAPK pathway | 37 | −0.3407 | −1.8065 | 0.0132 | 0.1876 |
GSEA reporting on statistically significant functionally relevant pathways (P < 0.05, false discovery rate < 0.25) and on positive and negative Pearson correlation coefficients with a high level of SUZ12 expression. P, nominal probability.
Discussion
SUZ12 is a core component of the Polycomb PRC2-HMTase complex that has been shown to be involved in stem cell maintenance and development. Although some studies have demonstrated overexpression of SUZ12 in colon and breast tumors,19,53 its real relevance in human cancer is yet to be established.
In this study, we first explored EZH2 and SUZ12 protein expression in non-tumoral samples. While EZH2 was widely detected in almost every tissue analyzed, SUZ12 was restricted mainly to those tissue compartments with proliferating cells, such as germinal centers in reactive lymphoid tissue, thymic cortex, epithelial basal cells and germinal cells in the testis. All these tissues are characterized by their regenerative capacity, suggesting a role for SUZ12 in tissue homeostasis and in cell cycle and proliferation.
Analysis of tumoral human samples revealed that EZH2 and SUZ12 are not always expressed simultaneously. Actually, those cases positive for SUZ12 are usually EZH2-positive, but not vice versa. We also observed a small fraction of cases in which we could detect SUZ12 but not EZH2. In these cases we cannot rule out the possibility that SUZ12 might have additional EZH2-independent functions.
There was a high level of expression of SUZ12 in a subset of tumoral samples including germinal cell-derived tumors, melanomas, skin basal cell carcinomas, lung neuroendocrine small-cell carcinoma, pituitary and parathyroid adenomas, and lymphomas, most remarkably in MCL, where the high expression of SUZ12 contrasts with its absence in the non-tumoral mantle zone cells in reactive lymph node. Therefore, our results extend previous observations of the strong expression of SUZ12 in human tumors.19,53
SUZ12 locus (17q11.2) has been found amplified, associated with protein overexpression in a small subset of tumors. This finding is especially relevant in MCL, where it seems to be more frequent in blastoid MCL, the aggressive variant of this type of lymphoma, since two of four blastoid-MCL cases showed this amplification. Therefore, our finding of SUZ12 amplification in MCL or melanoma among others types of tumors, along with the presence of SUZ12 translocations in endometrial sarcomas of the cervix,20,54 supports the hypothesis that SUZ12 has an oncogenic function and contributes to tumor formation and maintenance. The findings described here for SUZ12, and the previous results for EZH2,7,17,18 suggest that alteration of the PRC2 complex is a frequent event in human carcinogenesis.
Given the anomalous expression of SUZ12 in MCL tumoral cells compared with the lack of expression in their normal counterparts and the amplification associated with high levels of expression, we decided to perform functional analysis in MCL cell lines as a model to depict SUZ12 role in tumorigenesis. To this end, we silenced SUZ12 expression by RNAi in MCL-derived cell lines, and evaluated its effect on levels of H3K27me3, cell proliferation, apoptosis, and cell survival. A clear decrease in H3K27me3 was detected after SUZ12 silencing, demonstrating the interference with PRC2 activity due to the lower levels of SUZ12 in accordance with previous reports.21 SUZ12 knockdown resulted in an increased apoptosis, as demonstrated by annexin V and PARP cleavage analysis. When we studied the SUZ12 silencing effect over time, we observed that loss of SUZ12 compromised cell viability, as demonstrated by cell counting and competition assays. These results suggest that SUZ12 expression contributes to cell survival in MCL cell lines, avoiding apoptosis and increasing cell proliferation.
SUZ12 is known to exert its function through the direct repression of many target genes. Other studies have shown that SUZ12 targets vary among developmental states, tissues, and cell types.55 Therefore, we decided to identify SUZ12 targets that could mediate the effect on MCL-derived cell line viability and explain their role in MCL pathogenesis. We identified a group of SUZ12 targets with functions in apoptosis (BIRC family genes, BCL2, BID, and cFLIP); cell proliferation (several INK4 family genes, cyclins, and CDKs) and checkpoint regulators (CHK1, BUB3, MADL2, GADD45, and CDK6); DNA damage and repair genes (ATM, GMNN, and MLHL1); gene expression; and cell development. Several of these targets are genes of relevance in MCL (Figure 7). For instance, CDKN2A, ATM, BCL10, and RBL1, identified here as being SUZ12 targets, are frequently deleted or lost in MCL.23 SUZ12 could collaborate to inactivate these genes by epigenetic means. Other molecules, members of pathways of relevance in MCL, are also targets of SUZ12. For example, nuclear factor κB pathway components (BLIMP1, IKBKG, and NFKB2) and proteasomal pathway members have been found to be targets of SUZ12. Some of these targets have been previously found in other cell lines (such as BMP2, CDKN2A, SOX3, and GADD45G) but others have not been described before and could be MCL-specific. This is the case for several of the top identified targets such as ATM, BIRC family genes, BTK, BUB3, MAD2L1, and RBL1, among others, some of which have also been demonstrated with classic ChIP (Figure 4), suggesting that they are bona fide SUZ12 targets, and possibly specific to MCL cells. We also found that SUZ12 targets several microRNAs, some of which, for example, hsa-miR-148a56 and hsa-miR-223,57 were already known to be involved in cancer and differentiation.
Figure 7.
SUZ12 regulates the expression or function of multiple important pathways controlling MCL pathogenesis. Genes identified as SUZ12 targets by ChIP-on-chip in red. Genes marked with an asterisk suffer translocation or overexpression in MCL cases. Genes underlined are frequently deleted in MCL.
Many of these target genes were inversely correlated with SUZ12 expression levels in tumoral samples from MCL patients (Figure 6B), such as BIRC2, TMBIM4, XRCC6, JMJD2D, MAD2L1 and BUB3, among others. Additionally, validation in SUZ12-depleted cell lines revealed that several target genes were re-expressed after SUZ12 silencing (Figure 6A). The fact that some of the SUZ12 targets remained unaltered after SUZ12 depletion has several possible explanations. For instance, many SUZ12 targets may require not only the depletion of SUZ12 but also additional events like DNA demethylation or the presence of an activator to be expressed again.1,50
All these findings indicate that SUZ12 could collaborate in deregulating the expression of many important pathways controlling MCL pathogenesis (Figure 7). We propose that the abnormal expression of SUZ12 may account for some of the still unexplained features of MCL, including abnormal DNA repair and increased resistance to apoptosis.
Interestingly, recent publications have described the capacity of several drugs to block the HMTase activity of PRC2 complexes.58,59 In fact, LBH589 has proved to be effective in acute myelogenous leukemia cells. Patients suffering from other tumors, like MCL and pulmonary neuroendocrine small-cell carcinoma, in which PRC2 alterations are detected, might also benefit from this therapy.
Acknowledgments
We thank Dr. Yi Zhang and Dr. Ru Cao for providing us with the SUZ12 cDNA for antibody production. We express our gratitude to all members of the Monoclonal Antibodies and Histology and Immunohistochemistry Units (CNIO) for their excellent technical contribution and assistance. Special thanks are extended to Esteban Ballestar, Filipe Jacinto, and Amaia Lujambio from the former Cancer Epigenetics Group (CNIO) for their help with the ChIP experiments; to Jorge Luis Martinez from the Protein Technology Unit (CNIO) for SUZ12 protein production; to Orlando Dominguez from the Genomics Unit (CNIO) for ChIP-on-chip hybridization and help in analysis; and to all of the personnel of the Lymphoma Group (CNIO) for their help and discussions.
Footnotes
Address reprint requests to Dr. Margarita Sanchez-Beato, Ph.D., Lymphoma Group, Molecular Pathology Programme, Centro Nacional de Investigaciones Oncológicas (CNIO), C/ Melchor Fernández Almagro 3, E-28029 Madrid, Spain. E-mail: msbeato@cnio.es.
Supported by grants from the Ministerio de Educación y Ciencia, Spain (SAF2004-06952-C02-02, SAF2005-00221, and SAF2007-65957-C02-02) and the Ministerio de Sanidad y Consumo, Spain (G03/179, RD06/0020/0083 and RTICC RD06/0020/0107). D.M.-P. is supported by a fellowship from the Ministerio de Educación y Ciencia (AP2005-3972); L.D.L. is supported by a fellowship from Ministerio de Sanidad y Consumo (FI08/00038); N.M. is supported by a contract from Ministerio de Sanidad y Consumo (CP06/00002).
Supplemental material for this article can be found on http://ajp.amjpathol.org.
References
- Sparmann A, van Lohuizen M. Polycomb silencers control cell fate, development and cancer. Nat Rev Cancer. 2006;6:846–856. doi: 10.1038/nrc1991. [DOI] [PubMed] [Google Scholar]
- Cao R, Wang L, Wang H, Xia L, Erdjument-Bromage H, Tempst P, Jones RS, Zhang Y. Role of histone H3 lysine 27 methylation in Polycomb-group silencing. Science. 2002;298:1039–1043. doi: 10.1126/science.1076997. [DOI] [PubMed] [Google Scholar]
- Czermin B, Melfi R, McCabe D, Seitz V, Imhof A, Pirrotta V. Drosophila enhancer of Zeste/ESC complexes have a histone H3 methyltransferase activity that marks chromosomal Polycomb sites. Cell. 2002;111:185–196. doi: 10.1016/s0092-8674(02)00975-3. [DOI] [PubMed] [Google Scholar]
- Shao Z, Raible F, Mollaaghababa R, Guyon JR, Wu CT, Bender W, Kingston RE. Stabilization of chromatin structure by PRC1, a Polycomb complex. Cell. 1999;98:37–46. doi: 10.1016/S0092-8674(00)80604-2. [DOI] [PubMed] [Google Scholar]
- Muller J, Hart CM, Francis NJ, Vargas ML, Sengupta A, Wild B, Miller EL, O'Connor MB, Kingston RE, Simon JA. Histone methyltransferase activity of a Drosophila Polycomb group repressor complex. Cell. 2002;111:197–208. doi: 10.1016/s0092-8674(02)00976-5. [DOI] [PubMed] [Google Scholar]
- Kuzmichev A, Nishioka K, Erdjument-Bromage H, Tempst P, Reinberg D. Histone methyltransferase activity associated with a human multiprotein complex containing the Enhancer of Zeste protein. Genes Dev. 2002;16:2893–2905. doi: 10.1101/gad.1035902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bracken AP, Pasini D, Capra M, Prosperini E, Colli E, Helin K. EZH2 is downstream of the pRB-E2F pathway, essential for proliferation and amplified in cancer. EMBO J. 2003;22:5323–5335. doi: 10.1093/emboj/cdg542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vonlanthen S, Heighway J, Altermatt HJ, Gugger M, Kappeler A, Borner MM, van Lohuizen M, Betticher DC. The bmi-1 oncoprotein is differentially expressed in non-small cell lung cancer and correlates with INK4A-ARF locus expression. Br J Cancer. 2001;84:1372–1376. doi: 10.1054/bjoc.2001.1791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Kemenade FJ, Raaphorst FM, Blokzijl T, Fieret E, Hamer KM, Satijn DP, Otte AP, Meijer CJ. Coexpression of BMI-1 and EZH2 polycomb-group proteins is associated with cycling cells and degree of malignancy in B-cell non-Hodgkin lymphoma. Blood. 2001;97:3896–3901. doi: 10.1182/blood.v97.12.3896. [DOI] [PubMed] [Google Scholar]
- Bea S, Tort F, Pinyol M, Puig X, Hernandez L, Hernandez S, Fernandez PL, van Lohuizen M, Colomer D, Campo E. BMI-1 gene amplification and overexpression in hematological malignancies occur mainly in mantle cell lymphomas. Cancer Res. 2001;61:2409–2412. [PubMed] [Google Scholar]
- Haupt Y, Alexander WS, Barri G, Klinken SP, Adams JM. Novel zinc finger gene implicated as myc collaborator by retrovirally accelerated lymphomagenesis in E mu-myc transgenic mice. Cell. 1991;65:753–763. doi: 10.1016/0092-8674(91)90383-a. [DOI] [PubMed] [Google Scholar]
- van Lohuizen M, Verbeek S, Scheijen B, Wientjens E, van der Gulden H, Berns A. Identification of cooperating oncogenes in E mu-myc transgenic mice by provirus tagging. Cell. 1991;65:737–752. doi: 10.1016/0092-8674(91)90382-9. [DOI] [PubMed] [Google Scholar]
- Sanchez-Beato M, Sanchez E, Gonzalez-Carrero J, Morente M, Diez A, Sanchez-Verde L, Martin MC, Cigudosa JC, Vidal M, Piris MA. Variability in the expression of polycomb proteins in different normal and tumoral tissues. A pilot study using tissue microarrays. Mod Pathol. 2006;19:684–694. doi: 10.1038/modpathol.3800577. [DOI] [PubMed] [Google Scholar]
- Dukers DF, van Galen JC, Giroth C, Jansen P, Sewalt RG, Otte AP, Kluin-Nelemans HC, Meijer CJ, Raaphorst FM. Unique polycomb gene expression pattern in Hodgkin’s lymphoma and Hodgkin’s lymphoma-derived cell lines. Am J Pathol. 2004;164:873–881. doi: 10.1016/S0002-9440(10)63175-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raaphorst FM, Vermeer M, Fieret E, Blokzijl T, Dukers D, Sewalt RG, Otte AP, Willemze R, Meijer CJ. Site-specific expression of polycomb-group genes encoding the HPC-HPH/PRC1 complex in clinically defined primary nodal and cutaneous large B-cell lymphomas. Am J Pathol. 2004;164:533–542. doi: 10.1016/S0002-9440(10)63143-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanchez-Beato M, Sanchez E, Garcia JF, Perez-Rosado A, Montoya MC, Fraga M, Artiga MJ, Navarrete M, Abraira V, Morente M, Esteller M, Koseki H, Vidal M, Piris MA. Abnormal PcG protein expression in Hodgkin’s lymphoma. Relation with E2F6 and NFkappaB transcription factors. J Pathol. 2004;204:528–537. doi: 10.1002/path.1661. [DOI] [PubMed] [Google Scholar]
- Kleer CG, Cao Q, Varambally S, Shen R, Ota I, Tomlins SA, Ghosh D, Sewalt RG, Otte AP, Hayes DF, Sabel MS, Livant D, Weiss SJ, Rubin MA, Chinnaiyan AM. EZH2 is a marker of aggressive breast cancer and promotes neoplastic transformation of breast epithelial cells. Proc Natl Acad Sci USA. 2003;100:11606–11611. doi: 10.1073/pnas.1933744100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varambally S, Dhanasekaran SM, Zhou M, Barrette TR, Kumar-Sinha C, Sanda MG, Ghosh D, Pienta KJ, Sewalt RG, Otte AP, Rubin MA, Chinnaiyan AM. The polycomb group protein EZH2 is involved in progression of prostate cancer. Nature. 2002;419:624–629. doi: 10.1038/nature01075. [DOI] [PubMed] [Google Scholar]
- Kirmizis A, Bartley SM, Farnham PJ. Identification of the polycomb group protein SU(Z)12 as a potential molecular target for human cancer therapy. Mol Cancer Ther. 2003;2:113–121. [PubMed] [Google Scholar]
- Koontz JI, Soreng AL, Nucci M, Kuo FC, Pauwels P, van Den Berghe H, Cin PD, Fletcher JA, Sklar J. Frequent fusion of the JAZF1 and JJAZ1 genes in endometrial stromal tumors. Proc Natl Acad Sci USA. 2001;98:6348–6353. doi: 10.1073/pnas.101132598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pasini D, Bracken AP, Jensen MR, Lazzerini Denchi E, Helin K. Suz12 is essential for mouse development and for EZH2 histone methyltransferase activity. EMBO J. 2004;23:4061–4071. doi: 10.1038/sj.emboj.7600402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao R, Zhang Y. SUZ12 is required for both the histone methyltransferase activity and the silencing function of the EED-EZH2 complex. Mol Cell. 2004;15:57–67. doi: 10.1016/j.molcel.2004.06.020. [DOI] [PubMed] [Google Scholar]
- Jares P, Colomer D, Campo E. Genetic and molecular pathogenesis of mantle cell lymphoma: perspectives for new targeted therapeutics. Nat Rev Cancer. 2007;7:750–762. doi: 10.1038/nrc2230. [DOI] [PubMed] [Google Scholar]
- Martinez N, Camacho FI, Algara P, Rodriguez A, Dopazo A, Ruiz-Ballesteros E, Martin P, Martinez-Climent JA, Garcia-Conde J, Menarguez J, Solano F, Mollejo M, Piris MA. The molecular signature of mantle cell lymphoma reveals multiple signals favoring cell survival. Cancer Res. 2003;63:8226–8232. [PubMed] [Google Scholar]
- Rosenwald A, Wright G, Wiestner A, Chan WC, Connors JM, Campo E, Gascoyne RD, Grogan TM, Muller-Hermelink HK, Smeland EB, Chiorazzi M, Giltnane JM, Hurt EM, Zhao H, Averett L, Henrickson S, Yang L, Powell J, Wilson WH, Jaffe ES, Simon R, Klausner RD, Montserrat E, Bosch F, Greiner TC, Weisenburger DD, Sanger WG, Dave BJ, Lynch JC, Vose J, Armitage JO, Fisher RI, Miller TP, LeBlanc M, Ott G, Kvaloy S, Holte H, Delabie J, Staudt LM. The proliferation gene expression signature is a quantitative integrator of oncogenic events that predicts survival in mantle cell lymphoma. Cancer Cell. 2003;3:185–197. doi: 10.1016/s1535-6108(03)00028-x. [DOI] [PubMed] [Google Scholar]
- Bodrug SE, Warner BJ, Bath ML, Lindeman GJ, Harris AW, Adams JM. Cyclin D1 transgene impedes lymphocyte maturation and collaborates in lymphomagenesis with the myc gene. EMBO J. 1994;13:2124–2130. doi: 10.1002/j.1460-2075.1994.tb06488.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stilgenbauer S, Winkler D, Ott G, Schaffner C, Leupolt E, Bentz M, Moller P, Muller-Hermelink HK, James MR, Lichter P, Dohner H. Molecular characterization of 11q deletions points to a pathogenic role of the ATM gene in mantle cell lymphoma. Blood. 1999;94:3262–3264. [PubMed] [Google Scholar]
- Garcia JF, Roncador G, Garcia JF, Sanz AI, Maestre L, Lucas E, Montes-Moreno S, Fernandez Victoria R, Martinez-Torrecuadrara JL, Marafioti T, Mason DY, Piris MA. PRDM1/BLIMP-1 expression in multiple B and T-cell lymphoma. Haematologica. 2006;91:467–474. [PubMed] [Google Scholar]
- Montes-Moreno S, Roncador G, Maestre L, Martinez N, Sanchez-Verde L, Camacho FI, Cannata J, Martinez-Torrecuadrada JL, Shen Y, Chan WC, Piris MA. Gcet1 (centerin), a highly restricted marker for a subset of germinal center-derived lymphomas. Blood. 2008;111:351–358. doi: 10.1182/blood-2007-06-094151. [DOI] [PubMed] [Google Scholar]
- Kallioniemi OP, Wagner U, Kononen J, Sauter G. Tissue microarray technology for high-throughput molecular profiling of cancer. Hum Mol Genet. 2001;10:657–662. doi: 10.1093/hmg/10.7.657. [DOI] [PubMed] [Google Scholar]
- Tracey L, Aggarwal M, Garcia-Cosio M, Villuendas R, Algara P, Sanchez-Beato M, Sanchez-Aguilera A, Garcia JF, Rodriguez A, Camacho FI, Martinez N, Ruiz-Ballesteros E, Mollejo M, Piris MA. Somatic hypermutation signature in B-cell low-grade lymphomas. Haematologica. 2008;93:1186–1194. doi: 10.3324/haematol.12999. [DOI] [PubMed] [Google Scholar]
- Caretti G, Di Padova M, Micales B, Lyons GE, Sartorelli V. The Polycomb Ezh2 methyltransferase regulates muscle gene expression and skeletal muscle differentiation. Genes Dev. 2004;18:2627–2638. doi: 10.1101/gad.1241904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raman JD, Mongan NP, Tickoo SK, Boorjian SA, Scherr DS, Gudas LJ. Increased expression of the polycomb group gene. EZH2, in transitional cell carcinoma of the bladder. Clin Cancer Res. 2005;11:8570–8576. doi: 10.1158/1078-0432.CCR-05-1047. [DOI] [PubMed] [Google Scholar]
- Palacios J, Honrado E, Osorio A, Cazorla A, Sarrio D, Barroso A, Rodriguez S, Cigudosa JC, Diez O, Alonso C, Lerma E, Dopazo J, Rivas C, Benitez J. Phenotypic characterization of BRCA1 and BRCA2 tumors based in a tissue microarray study with 37 immunohistochemical markers. Breast Cancer Res Treat. 2005;90:5–14. doi: 10.1007/s10549-004-1536-0. [DOI] [PubMed] [Google Scholar]
- Singh B, Stoffel A, Gogineni S, Poluri A, Pfister DG, Shaha AR, Pathak A, Bosl G, Cordon-Cardo C, Shah JP, Rao PH. Amplification of the 3q26.3 locus is associated with progression to invasive cancer and is a negative prognostic factor in head and neck squamous cell carcinomas. Am J Pathol. 2002;161:365–371. doi: 10.1016/S0002-9440(10)64191-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sholl LM, John Iafrate A, Chou YP, Wu MT, Goan YG, Su L, Huang YT, Christiani DC, Chirieac LR. Validation of chromogenic in situ hybridization for detection of EGFR copy number amplification in nonsmall cell lung carcinoma. Mod Pathol. 2007;20:1028–1035. doi: 10.1038/modpathol.3800946. [DOI] [PubMed] [Google Scholar]
- Lois C, Hong EJ, Pease S, Brown EJ, Baltimore D. Germline transmission and tissue-specific expression of transgenes delivered by lentiviral vectors. Science. 2002;295:868–872. doi: 10.1126/science.1067081. [DOI] [PubMed] [Google Scholar]
- Ruiz-Vela A, Aggarwal M, de la Cueva P. Lentiviral (HIV)-based RNA interference screen in human B-cell receptor regulatory networks reveals MCL1-induced oncogenic pathways. Blood. 2008;111:1665–1676. doi: 10.1182/blood-2007-09-110601. [DOI] [PubMed] [Google Scholar]
- Ruiz-Vela A, Aggarwal M, de la Cueva P, Treda C, Herreros B, Martin-Perez D, Dominguez O, Piris MA. Lentiviral (HIV)-based RNA interference screen in human B-cell receptor regulatory networks reveals MCL1-induced oncogenic pathways. Blood. 2008;111:1665–1676. doi: 10.1182/blood-2007-09-110601. [DOI] [PubMed] [Google Scholar]
- Montaner D, Tarraga J, Huerta-Cepas J, Burguet J, Vaquerizas JM, Conde L, Minguez P, Vera J, Mukherjee S, Valls J, Pujana MA, Alloza E, Herrero J, Al-Shahrour F, Dopazo J. Next station in microarray data analysis: gEPAS. Nucleic Acids Res. 2006;34:W486–491. doi: 10.1093/nar/gkl197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL, Gillette MA, Paulovich A, Pomeroy SL, Golub TR, Lander ES, Mesirov JP. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci USA. 2005;102:15545–15550. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mootha VK, Lindgren CM, Eriksson KF, Subramanian A, Sihag S, Lehar J, Puigserver P, Carlsson E, Ridderstrale M, Laurila E, Houstis N, Daly MJ, Patterson N, Mesirov JP, Golub TR, Tamayo P, Spiegelman B, Lander ES, Hirschhorn JN, Altshuler D, Groop LC. PGC-1alpha-responsive genes involved in oxidative phosphorylation are coordinately downregulated in human diabetes. Nat Genet. 2003;34:267–273. doi: 10.1038/ng1180. [DOI] [PubMed] [Google Scholar]
- Onciu M, Schlette E, Medeiros LJ, Abruzzo LV, Keating M, Lai R. Cytogenetic findings in mantle cell lymphoma cases with a high level of peripheral blood involvement have a distinct pattern of abnormalities. Am J Clin Pathol. 2001;116:886–892. doi: 10.1309/JQMR-323G-71Y9-M7MB. [DOI] [PubMed] [Google Scholar]
- Espinet B, Sole F, Woessner S, Bosch F, Florensa L, Campo E, Costa D, Lloveras E, Vila RM, Besses C, Montserrat E, Sans-Sabrafen J. Translocation (11;14)(q13;q32) and preferential involvement of chromosomes 1, 2, 9, 13, and 17 in mantle cell lymphoma. Cancer Genet Cytogenet. 1999;111:92–98. doi: 10.1016/s0165-4608(98)00228-3. [DOI] [PubMed] [Google Scholar]
- Starczynowski DT, Vercauteren S, Telenius A, Sung S, Tohyama K, Brooks-Wilson A, Spinelli JJ, Eaves CJ, Eaves AC, Horsman DE, Lam WL, Karsan A. High-resolution whole genome tiling path array CGH analysis of CD34+ cells from patients with low-risk myelodysplastic syndromes reveals cryptic copy number alterations and predicts overall and leukemia-free survival. Blood. 2008;112:3412–3424. doi: 10.1182/blood-2007-11-122028. [DOI] [PubMed] [Google Scholar]
- Douglas EJ, Fiegler H, Rowan A, Halford S, Bicknell DC, Bodmer W, Tomlinson IP, Carter NP. Array comparative genomic hybridization analysis of colorectal cancer cell lines and primary carcinomas. Cancer Res. 2004;64:4817–4825. doi: 10.1158/0008-5472.CAN-04-0328. [DOI] [PubMed] [Google Scholar]
- Kraggerud SM, Skotheim RI, Szymanska J, Eknaes M, Fossa SD, Stenwig AE, Peltomaki P, Lothe RA. Genome profiles of familial/bilateral and sporadic testicular germ cell tumors. Genes Chromosomes Cancer. 2002;34:168–174. doi: 10.1002/gcc.10058. [DOI] [PubMed] [Google Scholar]
- Ivanova N, Dobrin R, Lu R, Kotenko I, Levorse J, DeCoste C, Schafer X, Lun Y, Lemischka IR. Dissecting self-renewal in stem cells with RNA interference. Nature. 2006;442:533–538. doi: 10.1038/nature04915. [DOI] [PubMed] [Google Scholar]
- Lee TI, Jenner RG, Boyer LA, Guenther MG, Levine SS, Kumar RM, Chevalier B, Johnstone SE, Cole MF, Isono K, Koseki H, Fuchikami T, Abe K, Murray HL, Zucker JP, Yuan B, Bell GW, Herbolsheimer E, Hannett NM, Sun K, Odom DT, Otte AP, Volkert TL, Bartel DP, Melton DA, Gifford DK, Jaenisch R, Young RA. Control of developmental regulators by Polycomb in human embryonic stem cells. Cell. 2006;125:301–313. doi: 10.1016/j.cell.2006.02.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bracken AP, Dietrich N, Pasini D, Hansen KH, Helin K. Genome-wide mapping of Polycomb target genes unravels their roles in cell fate transitions. Genes Dev. 2006;20:1123–1136. doi: 10.1101/gad.381706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kotake Y, Cao R, Viatour P, Sage J, Zhang Y, Xiong Y. pRB family proteins are required for H3K27 trimethylation and Polycomb repression complexes binding to and silencing p16INK4alpha tumor suppressor gene. Genes Dev. 2007;21:49–54. doi: 10.1101/gad.1499407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marson A, Levine SS, Cole MF, Frampton GM, Brambrink T, Johnstone S, Guenther MG, Johnston WK, Wernig M, Newman J, Calabrese JM, Dennis LM, Volkert TL, Gupta S, Love J, Hannett N, Sharp PA, Bartel DP, Jaenisch R, Young RA. Connecting microRNA genes to the core transcriptional regulatory circuitry of embryonic stem cells. Cell. 2008;134:521–533. doi: 10.1016/j.cell.2008.07.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirmizis A, Bartley SM, Kuzmichev A, Margueron R, Reinberg D, Green R, Farnham PJ. Silencing of human polycomb target genes is associated with methylation of histone H3 Lys 27. Genes Dev. 2004;18:1592–1605. doi: 10.1101/gad.1200204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oliva E, de Leval L, Soslow RA, Herens C. High frequency of JAZF1-JJAZ1 gene fusion in endometrial stromal tumors with smooth muscle differentiation by interphase FISH detection. Am J Surg Pathol. 2007;31:1277–1284. doi: 10.1097/PAS.0b013e318031f012. [DOI] [PubMed] [Google Scholar]
- Squazzo SL, O'Geen H, Komashko VM, Krig SR, Jin VX, Jang SW, Margueron R, Reinberg D, Green R, Farnham PJ. Suz12 binds to silenced regions of the genome in a cell-type-specific manner. Genome Res. 2006;16:890–900. doi: 10.1101/gr.5306606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lujambio A, Calin GA, Villanueva A, Ropero S, Sanchez-Cespedes M, Blanco D, Montuenga LM, Rossi S, Nicoloso MS, Faller WJ, Gallagher WM, Eccles SA, Croce CM, Esteller M. A microRNA DNA methylation signature for human cancer metastasis. Proc Natl Acad Sci USA. 2008;105:13556–13561. doi: 10.1073/pnas.0803055105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stamatopoulos B, Meuleman N, Haibe-Kains B, Saussoy P, Van den Neste E, Michaux L, Heimann P, Martiat P, Bron D, Lagneaux L. MicroRNA-29c and microRNA-223 downregulation has in vivo significance in chronic lymphocytic leukemia and improves disease risk stratification. Blood. 2009;113:5237–5245. doi: 10.1182/blood-2008-11-189407. [DOI] [PubMed] [Google Scholar]
- Fiskus W, Pranpat M, Balasis M, Herger B, Rao R, Chinnaiyan A, Atadja P, Bhalla K. Histone deacetylase inhibitors deplete enhancer of zeste 2 and associated polycomb repressive complex 2 proteins in human acute leukemia cells. Mol Cancer Ther. 2006;5:3096–3104. doi: 10.1158/1535-7163.MCT-06-0418. [DOI] [PubMed] [Google Scholar]
- Tan J, Yang X, Zhuang L, Jiang X, Chen W, Lee PL, Karuturi RK, Tan PB, Liu ET, Yu Q. Pharmacologic disruption of Polycomb-repressive complex 2-mediated gene repression selectively induces apoptosis in cancer cells. Genes Dev. 2007;21:1050–1063. doi: 10.1101/gad.1524107. [DOI] [PMC free article] [PubMed] [Google Scholar]









