Abstract
Smooth muscle (SM) is a spontaneously contractile tissue that provides physical support and function to organs such as the uterus. Uterine smooth muscle–related neoplasia comprise common well-differentiated benign lesions called leiomyomas (ULM), and rare, highly aggressive and pleomorphic tumors named leiomyosarcomas (ULMS). MicroRNAs (miRNAs) are small non-coding RNAs that play essential roles in normal cellular development and tissue homeostasis that can be used to accurately subclassify different tumor types. Here, we demonstrate that miRNAs are required for full smooth muscle cell (SMC) differentiation of bone marrow–derived human mesenchymal stem cells (hMSCs). We also report a miRNA signature associated with this process. Moreover, we show that this signature, along with miRNA profiles for ULMS and ULM, are able to subclassify tumors of smooth muscle origin along SM differentiation. Using multiple computational analyses, we determined that ULMS are more similar to hMSCs as opposed to ULM, which are linked with more mature SMCs and myometrium. Furthermore, a comparison of the SM differentiation and ULMS miRNA signatures identified miRNAs strictly associated with SM maturation or transformation, as well as those modulated in both processes indicating a possible dual role. These results support separate origins and/or divergent transformation pathways for ULM and ULMS, resulting in drastically different states of differentiation. In summary, this work expands on our knowledge of the regulation of SM differentiation and sarcoma pathogenesis.
Leiomyosarcoma, a rare, highly aggressive malignancy of smooth muscle (SM) tissues, makes up approximately 10% of soft tissue sarcomas, tumors of mesenchymal origin. As a consequence of diverse cytogenetic and molecular profiles, the etiology and molecular basis of leiomyosarcoma remain poorly understood. Fibroids or leiomyomas (ULM), more common SM neoplasia, are benign malignancies of the uterine smooth muscle or myometrium (MM).1 Although generally accepted as tumors of SM origin, it has been hypothesized that ULM and uterine leiomyosarcomas (ULMS) are separate entities of independent cellular and/or molecular origin.2
Smooth muscle is an involuntary nonstriated muscle that can be found throughout the body, within the vasculature and organs such as the uterus. The broad function of smooth muscle cells (SMCs) in these organs is to maintain organ structure and elicit necessary contractions, but specific actions, such as unique responses to environmental cues or rate of contractility, depend on the particular anatomical location.3 The multitude of SMC types and the inherent differences in their molecular regulation have hampered the study of the mechanisms responsible for SMC development. In addition, the lack of standardized differentiation systems, both in vitro and in vivo, has also hindered the progress of such studies.
Mesenchymal stem cells (MSCs) are an adult stem cell population originally identified as a minor population of bone marrow cells that have the ability to adhere to tissue culture plastic.4 MSC-like cells have since been isolated from various tissues and are loosely defined by adherence to tissue culture plastic, the combinatorial expression of numerous surface markers (most commonly CD29, CD73, CD105, CD54, Stro-1, and CD44),5 and their capacity to differentiate into multiple lineages, such as osteoblasts, chondrocytes, adipocytes, and myoblasts.6 Recent publications have validated the use of human MSCs (hMSCs) isolated from adipose tissues as a source for generating mature SMCs in vitro,7,8 but the molecular regulation of this process is only partly understood.
MicroRNAs (miRNAs), small, noncoding RNAs, continue to display an increasingly prominent role in tissue development and homeostasis, as evidenced by the lethality of mice lacking Dicer, an enzyme essential for production of mature miRNAs.9 Additionally, miRNA expression profiles, or ‘signatures,’ have been used to successfully classify many tumor types, including sarcomas,10,11 because they faithfully recapitulate the cell or lineage of origin. Moreover, aberrant miRNA expression has also been implicated in the development of cancer by altering the levels of oncogenes and tumor suppressors.12,13 More significantly, recent studies have revealed a functional link between a miRNA’s dual role in regulation of differentiation and oncogenesis.14,15,16
In this study, we establish two efficient models of SM differentiation from bone marrow–derived hMSCs, demonstrate that Dicer or Drosha down-regulation impair SM maturation, supporting a critical role for miRNAs in the control of SM differentiation, and identify a miRNA signature associated with this process. In addition, we define miRNA signatures of ULMS and ULM compared with normal MM. These signatures are then used to assess the miRNA’s ability to subclassify SM lesions according to their differentiation status. Using a number of independent computational tools we find that ULMS more closely resemble hMSCs than ULM, normal MM, or in vitro SMCs. Importantly, juxtaposing the two miRNA signatures (SM differentiation and ULMS) also allow us to identify miRNAs modulated in either, or both, processes of differentiation and transformation, rendering a list of possible molecular regulators of those events, as well as revealing a novel link between the two. Our data demonstrate that miRNAs are required for smooth muscle differentiation of hMSCs and that they can serve as markers of SM differentiation that robustly subclassify ULMS and ULM in relation to their differentiation status.
Materials and Methods
Cell Culture
hMSCs were obtained from Dr. Darwin Prockop (Texas A&M University, College Station, TX) and maintained in subconfluent conditions as described.17 An adaptation of a previously published protocol8 was used to achieve SMC differentiation. Briefly, hMSCs were seeded at 75–100 cells/mm2 and, on near confluency, smooth muscle differentiation medium (SMDM) [MCDB131 (Sigma, St. Louis, MO) supplemented with 1% FBS and 100U/ml Heparin] was added. The thromboxane A2 (TxA2)-induced method of differentiation was adapted from Kim et al.7 In brief, hMSCs were plated at 0.2 × 106 cells per well in a 6-well plate, re-fed with fresh α minimal essential media (αMEM) with 0.25% FBS 24 hours before addition of 1.0 μmol/L of the TxA2 chemical analog U46619 (Enzo Life Sciences, Farmingdale, NY). Primary uterine smooth muscle cells (UtSMC) (Lonza, Basel, Switzerland) were maintained as per the manufacturer’s protocol. Isolation of human myometrial smooth muscle cells was achieved by incubating manually dissected fresh myometrium with collagenase V at 37°C while shaking for 4 to 6 hours. Large debris was discarded after centrifugation, and the remaining cells (SMCs) were plated in smooth muscle media (Lonza).
RNA Extraction, RT-PCR, and Real-Time PCR Analyses
Total RNA was extracted using the mirVana microRNA extraction kit (Ambion, Austin, TX) or the miRNeasy Mini Kit (Qiagen, Valencia, CA). Semiquantitative reverse transcription PCR (RT-PCR) was performed by reverse transcribing 250 ng of total RNA with Superscript III Kit (Invitrogen, Carlsbad, CA), followed by standard PCR using RNA specific primers. Quantitative real-time PCR (qRT-PCR) was performed with same RT reaction using FastStart SYBR Green MasterMix (Roche, Basel, Switzerland) and BioRad iCycler equipment. RT-PCR primers: CD73 F 5′-GCACTATCTGGTTCACCGTGT-3′, CD73 R 5′-CCATTTCAAAACCCGAATGT-3′; Calponin F 5′-ATGTCCTCTGCTCACTTCA-3′, Calponin R 5′-TTTCCGCTCCTGCTTCTCT-3′; SM-MHC F 5′-GGACGACCTGGTTGTTGATT-3′, SM-MHC R 5′-GTAGCTGCTTGATGGCTTCC-3′; GAPDH F 5′-ACCACAGTCCATGCCATCAC-3′, GAPDH R 5′-TCCACCACCCTGTTGCTGTA-3′. qRT-PCR primers: SM-MHC F 5′-GACAAGTTTGTGGCCGACCT-3′, SM-MHC R 5′-CTCCGTCATCTTGGCCATCT-3′; GAPDH F 5′-CGCTCTCTGCTGGTGGTGTT-3′, GAPDH R 5′-CCATGGTGTCTGAGCGATGT-3′. qRT-PCR for miRNA expression levels was performed with the TaqMan miRNA Assay kit (Applied Biosystems, Austin, TX) according to manufacturer’s protocol using 12.5 ng of total RNA for reverse transcription. The small RNA RNU44 was used for normalization.
Western Blot Analyses
Cell pellets were lysed using RIPA buffer (Sigma) with protease inhibitors (Roche). Proteins were resolved on 3 to 8% NuPAGE gels (Invitrogen) or 4 to 20% Tris-glycine SDS PAGE gels (Invitrogen) for large and average molecular weight proteins, respectively. Proteins were transferred to PVDF or nitrocellulose membranes, blocked with 5% milk in PBS for 1 hour, and incubated overnight at 4°C with primary antibody (CD105 #611314, BD Biosciences, San Jose, CA; Calponin #M3556, Dako, Denmark; SM22α #ab10135, Abcam, Cambridge, MA; ASMA #ab5694, Abcam; Dicer #3363, Cell Signaling, Danvers, MA; Drosha #ab12286, Abcam; α-Actin #A2066, Sigma; β-Tubulin #T9026, Sigma). Membranes were incubated with HRP-conjugated secondary antibodies for 1 hour before being developed using enhanced chemiluminiscence (ECL) plus Western blotting detection kit (GE Health care, Piscataway, NJ).
Immunofluorescence and Fluorescence-Activated Cell Sorting Analyses
Cells grown onto coverslips were fixed with 4% formaldehyde for 20 minutes at room temperature. Fixed cells were incubated with primary antibody (CD105-APC #17-1057, eBioscience, San Diego, CA; ASMA #ab5694, Abcam) for 2 hours at room temperature, washed with PBS, incubated with secondary antibody when applicable (Alexa Fluor 610, Invitrogen, Carlsbad, CA) for 30 minutes at room temperature, washed with PBS, and counterstained with 0.5 μg/ml Hoechst 3358 (Invitrogen). Zeiss Imager M1 microscope with attached AxioCam MRm camera was used to acquire images. Fluorescence-activated cell sorting (FACS) for membrane staining of CD105 (CD105-APC #17-1057, eBiosciences) was conducted by trypsinizing cells and incubating with antibody for 30 minutes. Intracellular staining for ASMA (ASMA-FITC #F377, Sigma) was performed as previously described.18 In each instance, a BD LSRII flow cytometer and FlowJo software were used.
Contraction Assay
Assays were performed as previously described.7 Briefly, cells were trypsinized and resuspended in αMEM with 1% FBS at a concentration of 0.4 × 106 cells/ml. Cell suspension was mixed in a 1:1 ratio with 3 mg/ml collagen type I (BD Biosciences, San Jose, CA) as per manufacturer’s protocol. Cell/collagen mix (500 μl) was pipetted into a 24-well plate in triplicate. Gel was allowed to solidify at 37°C for 1 hour before carefully being dislodged from the well and placed in a 6-well plate with 3 ml of αMEM with 1% FBS. A picture was taken every hour for 12 hours. Image J was used to measure the area of the gel and that of the well, which was used for normalization. Extent of contraction was calculated by dividing the area of the gel by the area of the well at each time point and then indicating the percentage of area lost referred to the initial gel size.
Oligonucleotide Transfection
hMSCs were transfected with 50 nmol/L of mimic or inhibitor molecules (Dharmacon, Lafayette, CO) to overexpress or knockdown miRNA expression respectively, or Stealth siRNA (Invitrogen, Carlsbad, CA) designed for efficient knockdown of Dicer1 or Drosha, using Lipofectamine 2000 (Invitrogen, Carlsbad, CA) according to manufacturer’s recommendation.
miRNA Arrays
RNA labeling and array hybridization were conducted using protocols optimized at the MSKCC Genomics Core facility using 100 ng of total RNA and Agilent (Santa Clara, CA) ‘early access’ miRNA microarrays comprising mature miRNA sequences according to miRBase version 9.2. Raw intensity values were obtained from the arrays with Agilent’s Feature Extractor software and translated to ‘total gene signal’ values by subtracting the background. Total gene signals found to be below background were flagged as undetectable or absent. Equivalent time points for four independent time courses were used as biological replicates. The data were quantile normalized and log transformed using the generalized logarithm (to deal with negative values from the background subtraction). miRNAs found to be below background in 50% or more of the samples across the time course were eliminated from further analysis. For ULMS, ULM, and MM array analysis, miRNAs determined by Feature Extractor to be below background signal in more than 25% of the samples were eliminated from further analysis.19
Bioinformatic and Statistical Analyses
Analysis of ULMS, ULM, and MM Samples
The Bioconductor LIMMA package was used to identify miRNAs differentially expressed between normal and tumor samples (ULMS and ULM signatures). An unsupervised hierarchical clustering with Pearson correlation, distance function, and complete linkage was used. To assess the robustness, a parametric bootstrap resampling technique was used in which 1000 bootstrap replicas for the original dataset were generated and clustered. A consensus tree was built from these 1000 bootstrap trees; the number at each node in the consensus tree indicates how many of the bootstrap trees contain that subcluster.
Identifying miRNAs Monotonically Trending in SM Differentiation
To identify miRNAs that were significantly and monotonically changing over time, we used the nonparametric Jonckheere-Terpstra (J.T.) test. We grouped adjacent time points to create a number of equally sized time groups. The set of miRNAs which scored significant by the J.T.-test was then used to compute a time course progression value that assigned to any sample its effective position along the differentiation time course.
Clusters of miRNAs Differentially Expressed during SM Differentiation
miRNAs (from two time courses) who had a fold change of 2.5 or greater between maximum and minimum expression values were identified (129 miRNAs). Time points were split into 10 groups, and the expression was averaged and standardized to have mean 0 and SD 1. Expression profile clusters were calculated using the mfuzz algorithm,20 a fuzzy c-means R package commonly used for clustering profiles of timeseries. The number of clusters was set to 10 with a fuzzification parameter of 1.1.
Phylogenetic Tree Reconstruction Methods and Heatmap Generation
To identify differentially expressed miRNAs among samples of ULMS, ULM, and MM (defined as the ‘classifier list’) we used the analysis of variance algorithm as implemented in the R package20 with Benjamini-Hochberg correction for false positives and a P value cutoff of 0.01 or 0.05. Phylogenetic trees were then generated by applying the Weighted Least Squares (WLS)21 algorithm as implemented in the Phylip package version 3.6722 to the Pearson correlation distance of subsets of differentially expressed miRNAs (the ‘classifier list’). To assess the statistical significance of the phylogeny, the reconstruction was repeated 1000 times with random subsets of miRNAs. We drew the bootstrap sample size n from the discrete uniform distribution on the interval [50, N], where N is the total number of differentially expressed miRNAs. Further, we bootstrapped the ULMS, ULM, and MM samples to incorporate the uncertainty of tumor classification. Therefore, we sampled for each tumor subtype ni experiments with replacement from the set of the ni experiments of this subtype. Once a consensus tree was determined, it was rooted and visualized with Dendroscope version 2.2.2.23 Heatmap of gene expression was generated with the heatmap plus function of the R statistical package using the ‘classifier list.’
Clinical Specimens
Human tissue (ULMS, ULM, and MM) specimens were collected at the time of surgery and snap frozen in liquid nitrogen then transferred to −80°C for storage. Approval to collect specimens was granted by MSKCC IRB protocol number # 97-134. Normal human myometrium was obtained under NYU IRB protocol H10457-01A.
Results
Development and Characterization of an in Vitro SMC Differentiation Model
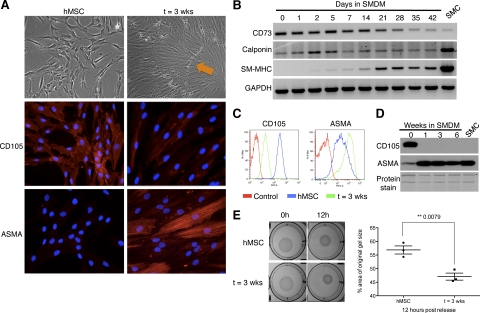
We developed an in vitro SMC differentiation system by inducing differentiation of bone marrow–derived hMSCs to SMCs, adapting a previously published protocol.8 To confirm the generation of functional SMCs from hMSCs, various samples were collected for expression or functional assays throughout the six-week time course of differentiation. By phase contrast microscopy we observed the appearance of “hills” (Figure 1A, orange arrow) and “valleys” at approximately 21 days postaddition of SMDM. Reverse transcriptase-polymerase chain reaction (RT-PCR) analysis showed induction of SM markers calponin and SM-myosin heavy chain (SM-MHC), paralleling a down-regulation of hMSC marker CD73 (Figure 1B). Immunofluorescence, FACS, and Western blot analyses demonstrated that expression of CD105, another marker of hMSCs, is lost shortly after the addition of SMDM, followed by induction of alpha-smooth muscle actin (ASMA), an early marker of SM (Figure 1, A, C, and D). To assess whether hMSC-derived SMCs were functional, we performed a contraction assay using published protocols7 (Figure 1E). Overall, our data prove that this method of differentiation renders mature contractile SMCs from bone marrow–derived hMSCs.
Figure 1.
Cells derived from SM differentiation of hMSCs reproduce the phenotypic and molecular characteristics of mature SMCs. A: Brightfield images (×100 magnification) and immunofluorescence for CD105 and α-smooth muscle actin (ASMA) in human mesenchymal stem cells (hMSC) and after SM differentiation (t = 3 weeks) (×200 magnification). Characteristic ‘hills (orange arrow) and valleys’ are observed by day 21. B: RT-PCR analysis of CD73, calponin, and smooth muscle myosin heavy chain (SM-MHC) during SM differentiation. GAPDH is used as RNA loading control. SMC: RNA from smooth muscle tissue used as a control. FACS (C) and Western blot (D) analyses of CD105 and ASMA during SM differentiation. For FACS, an isotypic control for each respective antibody is used to detect nonspecific staining (Control). SMC: protein lysate of smooth muscle cells isolated from human myometrial tissue. Amido black (protein staining) serves as loading control. E: Gel-contraction assay on hMSCs maintained in MSC medium (hMSC) or smooth muscle differentiation medium (t = 3 weeks), visualized at 0 hours and 12 hours post release (pictures, left); differences in contraction quantified at 12 hours (right). Image J software was used to measure the area of the gel and of the well, which was used for normalization.
miRNA Processing Is Necessary for Efficient Smooth Muscle Differentiation
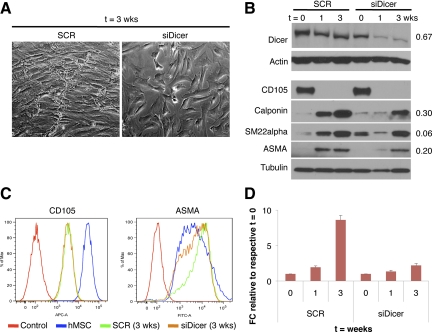
Recent findings have revealed that Dicer and Drosha, critical components of the cellular miRNA processing machinery, are essential for hMSC differentiation into the adipo- and osteocyte lineages.24 As a proof-of-principle of the functional requirement of miRNAs in SMC differentiation, we investigated the effect of knocking down Dicer in our in vitro model, using siRNA oligonucleotides. Dicer down-regulation impaired full SMC differentiation, as attested by morphological changes, reduced SM markers such as calponin, smooth muscle-22-alpha (SM22α), and ASMA protein levels, and more importantly, lack of SM-MHC mRNA up-regulation, considered the hallmark of mature SMCs (Figure 2, A–D). This result strongly supports a requisite of miRNAs for efficient SMC differentiation.
Figure 2.
miRNAs are required for full SM differentiation. A: Brightfield images of hMSCs transfected with scramble (SCR) (left) or siDicer (right) oligonucleotides (oligo) at t = 3 weeks (×100 magnification) B: Western blot analysis of Dicer, CD105, calponin, SM22α, and ASMA at different time points of SM differentiation, upon Dicer silencing. Actin or tubulin is used as loading control. Numbers on the right indicate the level of reduction of protein levels at t = 3 weeks compared with the corresponding scramble control, after normalization to actin or tubulin. C: FACS analysis of CD105 and ASMA expression in SCR- and siDicer-transfected cells at t = 3 weeks, and hMSC (t = 0) used as control. Control: isotype control. D: qRT-PCR of SM-MHC in hMSCs transfected with scramble oligo or siDicer. Data were normalized to GAPDH expression. Error bars represent standard deviation (SD) between two experimental replicates. FC indicates fold change.
A miRNA Signature Associated with Smooth Muscle Differentiation
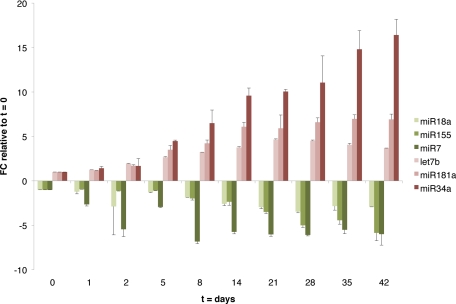
To define the miRNA ‘signature’ of SMC differentiation, miRNA microarray analysis was performed at multiple time points of differentiation, and a J.T. trend test was used to determine which miRNAs were modulated during this process. This analysis identified 30 miRNAs either significantly up-regulated or down-regulated during SMC differentiation (Table 1). Validation of a subset of these miRNAs was performed in two independent time courses one of which is represented on Figure 3. Because of the nature of this analysis, only miRNAs whose expression increased (or decreased) between consecutive time points were considered, limiting the outcome to miRNAs that display a monotonic trend. To identify those miRNAs that display a more dynamic expression pattern (ie, up-regulated miRNAs that reach a plateau, miRNAs initially induced and then repressed, etc), we defined expression profile clusters (see Materials and Methods and Supplemental Figure 1 at http://ajp.amjpathol.org). miRNAs in cluster 3 (with an overall ascending pattern) and clusters 6, 7, and 9 (demonstrating a descending pattern) showed a strong correlation with SM maturation over time and were considered the core of the SM differentiation signature (see Supplemental Table 1 at http://ajp.amjpathol.org). Importantly, 11 (of 12) miRNAs found by J.T. trend analysis to increase during differentiation are found in cluster 3 (P value = 9.296e-13, Fisher’s exact test); similarly, of 18 descending miRNAs determined by J.T. trend analysis, 16 overlap with clusters 6, 7, and 9 (P value <2.2e-16). These results validate the two methods used to determine the SM miRNA signature.
Table 1.
miRNAs that Trend Monotonically during SM Differentiation
| miRNA | FDR | FC | Chr. location | Overlapping transcripts |
|---|---|---|---|---|
| hsa-miR-181c | 0.0000000 | 8.50 | 19p13 | Intergenic |
| hsa-miR-204 | 0.0000007 | 8.30 | 9q21 | TRPM3 |
| hsa-miR-181a | 0.0000000 | 8.07 | 1q31 (a-1), 9q33 (a-2) | RP11-31E23.1 (a-1), RP11-348K2.1 (a-2, sense), NR6A1 (a-2, antisense) |
| hsa-miR-34a | 0.0000001 | 6.73 | 1p36 | Intergenic |
| hsa-miR-373* | 0.0000001 | 6.61 | 19q13 | Intergenic |
| hsa-miR-34b | 0.0000004 | 5.57 | 11q23 | Intergenic |
| hsa-miR-498 | 0.0000152 | 3.61 | 19q13 | Intergenic |
| hsa-miR-663 | 0.0000275 | 3.59 | 20p11 | RP3-410C9.1 |
| hsa-miR-542-5p | 0.0000068 | 3.04 | Xq26 | Intergenic |
| hsa-let-7b | 0.0001959 | 2.72 | 22q13 | RP4-695O20__B.10 |
| hsa-miR-152 | 0.0007280 | 2.37 | 17q21 | COPZ2 |
| hsa-miR-324-5p | 0.0001593 | 2.03 | 17p13 | ACADVL |
| hsa-miR-93 | 0.0000616 | −2.30 | 7q22 | MCM7 |
| hsa-miR-155 | 0.0007701 | −2.51 | 21q21 | MIRHG2 |
| hsa-miR-202 | 0.0000001 | −2.57 | 10q26 | RP13-49I15.3 |
| hsa-miR-431 | 0.0000275 | −2.59 | 14q32 | RTL1 |
| hsa-miR-376b | 0.0000603 | −2.82 | 14q32 | Intergenic |
| hsa-miR-323 | 0.0007280 | −2.89 | 14q32 | Intergenic |
| hsa-miR-758 | 0.0000000 | −2.94 | 14q32 | Intergenic |
| hsa-miR-130b | 0.0000023 | −3.09 | 22q11 | PPIL2 (sense), TOP3B (antisense) |
| hsa-miR-625 | 0.0000025 | −3.31 | 14q23 | FUT8 |
| hsa-miR-106a | 0.0000001 | −3.37 | Xq26 | Intergenic |
| hsa-miR-92b | 0.0001959 | −3.43 | 1q22 | Intergenic |
| hsa-miR-20a | 0.0006004 | −3.50 | 13q31 | MIRHG1 |
| hsa-miR-409-3p | 0.0000053 | −3.75 | 14q32 | Intergenic |
| hsa-miR-17-5p | 0.0000018 | −4.24 | 13q31 | MIRHG1 |
| hsa-miR-138 | 0.0000002 | −5.78 | 3p21 (−1), 16q12 (−2) | Intergenic (−1, −2) |
| hsa-miR-18a | 0.0000616 | −5.90 | 13q31 | MIRHG1 |
| hsa-miR-224 | 0.0000715 | −6.90 | Xq28 | GABRE |
| hsa-miR-7 | 0.0005677 | −10.38 | 9q21 (−1), 15q25 (−2), 19p13 (−3) | HNRNPK (−1), Intergenic (−2), C19orf30 (−3) |
A trend test identified 30 miRNAs that monotonically trend up or down during SM differentiation. False discovery rate (FDR) cutoff was set at 0.001. FC indicates fold change between t = 0 and the end time point. Chromosomal and intra/intergenic locations are also indicated.
denotes a miRNA derived from the antisense strand.
Figure 3.
Validation of miRNA array data in SM time course. Fold change (FC) of miRNAs monotonically increasing (red color palette) or decreasing (green palette) during SM differentiation of hMSCs, relative to t = 0, as measured by qRT-PCR. RNU44 was used for normalization. Error bars represent SD between two experimental replicates.
The miRNA SM ‘Signature’ Is Confirmed in an Independent Differentiation Model
To demonstrate that the miRNA signature found was not restricted to the heparin-based differentiation method outlined above, an additional method of SMC differentiation was characterized. It was recently reported that thromboxane A2 (TxA2) is able to efficiently induce SMC differentiation of hMSCs isolated from adipose tissue.7 We successfully adapted that protocol to induce SMC differentiation of bone marrow–derived hMSCs (see Supplemental Figure 2 at http://ajp.amjpathol.org). Again, miRNA processing proved to be essential for efficient SM differentiation in this independent model, as demonstrated by the effect of down-regulation of Dicer (see Supplemental Figure 3 at http://ajp.amjpathol.org) or Drosha (see Supplemental Figure 4 at http://ajp.amjpathol.org). Importantly, miRNAs identified and validated as associated with SM differentiation of hMSC using the heparin-based method (Figure 3) were also found to trend similarly in the TxA2-induced differentiation system (see Supplemental Figure 5 at http://ajp.amjpathol.org). The reproducibility of the results between two independent systems strongly reinforces the value of our miRNA signature of SM differentiation. Moreover, we showed that ectopic expression of a signature component, miR-155, normally down-regulated during SM differentiation, inhibits SM maturation, as assessed by lack of SM-MHC induction (Supplemental Figure 4D at http://ajp.amjpathol.org). These results further confirm the ability of our methodology to identify miRNAs functionally involved in SM differentiation.
An miRNA Signature of ULMS and ULM, Compared with Normal Myometrium
In addition to using miRNA profiles to identify potential regulators of SM differentiation, we sought to determine whether miRNAs are dysregulated in ULMS and ULM, because some of those may play a role in the biology of these tumors. To this end, miRNA expression profiles were elucidated by hybridizing 10 frozen samples of each tissue type (ULMS, ULM, and MM) to Agilent arrays. miRNAs significantly altered between ULMS and MM (miRNAs defining the ‘ULMS signature’) were limited to an FDR of less than 1%. The resulting signature was composed of 32 up-regulated and 40 down-regulated miRNAs (see Supplemental Table 2 at http://ajp.amjpathol.org). Similarly, miRNAs differentially expressed in ULM versus normal MM were determined (see Supplemental Table 3 at http://ajp.amjpathol.org).
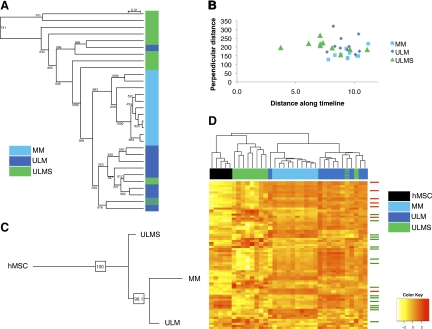
Unsupervised hierarchical clustering was also performed to investigate the ability of miRNA signatures of normal and tumor samples to faithfully classify the tissue type. The miRNA profile was able to accurately cluster ULMS, ULM, and MM (Figure 4A). We found one misclassified ULMS case that had been erroneously diagnosed by histopathological criteria. This analysis strongly supports that miRNAs are able to subclassify smooth muscle tumors and segregate leiomyomas and leiomyosarcomas into two distinct identifiable categories.
Figure 4.
Phylogenetic and clustering analyses of SM tumor subtypes using miRNA profiles. A: Cluster analysis of ULMS, ULM, and MM samples (n = 10 each) based on their miRNA expression profiles. Note that miRNAs are able to accurately segregate benign and malignant uterine mesenchymal lesions. B: Distance analysis of ULMS, ULM, and MM along a timeline based on SM miRNA signature. C: Consensus tree derived from phylogenetic analysis of ULMS, ULM, and MM tissues, rooted by hMSC samples. D: Unsupervised clustering analysis of five hMSC samples along with ULMS, ULM, and MM tissue samples based on the expression of the ‘classifier list’ of miRNAs differentially expressed between the three tissue subtypes. Green and blue hash marks (4A and D) represent a ULMS case that was misclassified and later found to be ULM-like. Arrows indicate SM differentiation signature miRNAs (red: up-regulated miRNAs, green: down-regulated miRNAs).
miRNAs Classify ULMS and ULM Based on the Distance Along SM Differentiation
We hypothesized that the above-defined miRNA profile associated with SM differentiation could be used to identify the differentiation state of the various SM tumors. To this end, we used monotonically trending miRNAs from the time course analysis (Table 1) to define a timeline from 0 to the final time point (N). Two distances were computed for every sample as well as for the mean of the time course samples: the distance along the timeline and the distance perpendicular to it. We then depicted the tumor samples along the same timeline and found that MM and ULM were localized in one grouping near the later time points of differentiation while ULMS appeared more scattered and several of them associated with earlier time points (Figure 4B).
Phylogenetic Analyses Based on miRNA Profiles Cluster ULMS with hMSCs
We recently developed a computational algorithm to categorize tumor subtypes according to the dissimilarity of their gene expression from that of stem cells and fully differentiated tissue, and thereby construct a phylogenetic tree of cancer subtypes.25 We used this tool to analyze the differentiation status of ULMS and ULM using miRNA expression profiles. miRNAs differentially expressed among ULMS, ULM, and MM samples ‘classifier list’ were found using analysis of variance with a P value cutoff of 0.01. The weighted least squares tree reconstruction algorithm was then used to reconstruct 1000 trees with bootstrapping of the subgroup average expression of the differentially expressed miRNAs. Fconsensus from the Phylip package was used to find the consensus tree, which was rooted with hMSC expression. In the consensus tree, ULMS were placed closer to hMSCs than ULM and MM. The consensus tree was very robust, being consistent in more than 98% of the reconstructed trees in the process of bootstrapping (Figure 4C). A separate unsupervised hierarchical clustering (based on the ‘classifier list’ used for phylogenetic tree construction) showed that hMSCs cluster with ULMS, whereas ULM group together with MM (Figure 4D), independently confirming the results shown above using the time progression and phylogenetic analyses (Figure 4, B and C). Interestingly, we noted that miRNAs differentially expressed between hMSCs, MM, ULM, and ULMS were enriched in components of the SM differentiation signature, and that their expression often correlates with the level of maturation of the tissue (red arrows, miRNAs up-regulated during differentiation; green arrows, those that are down-regulated). Overall, these analyses support the conclusion that miRNA expression links ULMS to a poorly differentiated status and suggests that ULMS and ULM are separate entities, distinguishable by a miRNA profile associated with their stage of differentiation.
Identification of miRNAs Involved in SM Differentiation and/or Neoplastic Transformation
The generation of miRNA profiles for SM differentiation and ULMS allowed us to discriminate miRNAs that are exclusively associated with either SM maturation or neoplastic transformation, from miRNAs that appear to be modulated in both processes, suggesting they may play a dual role. We found that 20 out of 72 miRNAs altered in ULMS (Supplemental Table 2 at http://ajp.amjpathol.org) are also modulated during SM differentiation (clusters 3, 6, 7, and 9, Supplemental Figure 1 at http://ajp.amjpathol.org), suggesting that these miRNAs may play a role in both differentiation and transformation (Table 2). The remaining 52 miRNAs altered in ULMS but not modulated during differentiation may be strictly associated to transformation. This information cannot be achieved by traditional profiling of tumor and normal tissue and provides insights into both processes and their intersection at LMS pathogenesis.
Table 2.
Comparison of miRNA Signatures Identifies miRNAs Altered in SM Differentiation and ULMS
| miRNA | FC | Cluster |
|---|---|---|
| hsa-miR-490 | 17.98 | 6 |
| hsa-miR-630 | 5.97 | 7 |
| hsa-miR-130b | 5.47 | 6 |
| hsa-miR-15b | 5.03 | 9 |
| hsa-miR-663 | 3.92 | 3 |
| hsa-miR-19a | 3.55 | 9 |
| hsa-miR-18a* | 2.34 | 7 |
| hsa-miR-18b | 2.27 | 6 |
| hsa-miR-18a | 2.15 | 6 |
| hsa-let-7b | −1.82 | 3 |
| hsa-miR-23b | −1.91 | 3 |
| hsa-miR-26a | −1.98 | 3 |
| hsa-miR-193a | −2.23 | 3 |
| hsa-miR-509 | −3.68 | 6 |
| hsa-miR-29c | −4.29 | 3 |
| hsa-miR-299-3p | −4.31 | 6 |
| hsa-miR-202 | −4.44 | 6 |
| hsa-miR-495 | −12.69 | 9 |
| hsa-miR-508 | −16.88 | 6 |
| hsa-miR-126* | −52.67 | 3 |
List of the 20 miRNAs found in both SM differentiation and ULMS signatures. FC indicates fold change between ULMS and MM tissues. The number in “Cluster” column refers to the cluster (Supplemental Figure 1 at http://ajp.amjpathol.org) that best represents an miRNA’s pattern during SM differentiation.
Discussion
Our data reveal a pivotal role for miRNAs in the process of SM differentiation and demonstrate a remarkable ability of miRNA patterns to subclassify different tumors of the smooth muscle lineage. It has previously been shown that miRNA expression and processing are required for other differentiation processes of the mesenchymal lineage, including osteocyte, adipocyte,24 skeletal muscle (miR-181a),26 and cardiac muscle (miR-1 and miR-133).27 This is the first report, however, of miRNA requirement for smooth muscle differentiation of mesenchymal stem cells.
Not surprisingly, miRNAs modulated during SM differentiation include some that have already been associated with other developmental processes, including mesenchymal and hematopoietic differentiation. For instance, miR-181a has a known role in skeletal muscle26 and hematopoietic maturation,28 which points to a general role for this miRNA in the exit from a multipotent state and/or in the commitment to a differentiated stage. MiR-181a controls HOXA11, a repressor of myogenin during skeletal muscle differentiation26; whether this miRNA acts on the same or specific targets to mediate each of these differentiation processes remains to be determined. Similarly, several let-7 members, including 7b, are also modulated during osteocytic and adipocytic differentiation.24 Other miRNAs in the SM differentiation signature include miR-155, with a known role in B-cell differentiation,29 and miR-18a, a member of the miR-17–92 family, which regulates stem cell renewal and displays pro-proliferative and oncogenic potential.13 Our experiments show that at least some of these miRNAs (ie, miR-155) play an active role in SM differentiation and are not mere byproducts of the actual ‘driver’ alterations. Ongoing experiments should clarify whether key miRNAs have common functions in the control of multidifferentiation capacity, cell cycle regulation, or proliferation, and whether they suppress the same or exclusive targets in different maturation processes.
The lack of a suitable standardized in vitro model of differentiation, similar to the C2C12 cell system commonly used to investigate the molecular mechanisms governing skeletal myogenesis, has limited the study of SM differentiation to date. In the absence of a generally accepted model of in vitro SM differentiation, the usefulness and applicability of a miRNA signature inferred from bone marrow–derived hMSC remains disputable. Nonetheless, hMSCs isolated from adipose tissue or bone marrow, as well as stem cells residing in the hair follicle,30,31 have been generally recognized as reliable sources of vascular smooth muscle. Another caveat of our in vitro differentiation systems is that while they recapitulate a general conversion of hMSCs into cells with SM properties, specific requirements for vascular, genitourinary, or intestinal SM function may not be fully mimicked by any of them. This may explain the absence of miR-145, a phenotypic modulator of VSMCs32,33 or miR-133a, a repressor of the SM phenotype in the heart,34 from our SM miRNA signature.
miRNAs have shown a surprising capacity to recapitulate the cell, lineage, or tissue of origin,11,35 and even to discriminate the source of metastases of unknown primary origin.36 Our data reveal the capacity of miRNAs to subclassify tumors of smooth muscle lineage with distinct histological, clinical, and biological features (ULM and ULMS), confirming the hypothesis that the latter are two separate entities. It has been discussed whether ULMS evolve from ULM into malignant tumors or whether these have independent cellular or molecular etiology.1 A leiomyoma origin for ULMS lesions has been postulated to occur in only 5 to 10% of cases in which a tumor presents areas histologically remnant of both types of lesions.37 However, the coexistence of both tumor types does not suffice as proof of tumor evolution. The recent identification of cells with stem cell properties in the myometrium38 has raised the possibility that ULMS and ULM derive from stem cells and mature smooth muscle cells of the uterus, respectively. However, the characteristic plasticity of smooth muscle cells would also support a model based on dedifferentiation. In our hands, two separate analyses based on miRNA expression confirmed that ULM and ULMS are indeed distinct entities that can be segregated based on their differentiation status. First, a miRNA-based time course progression value showed that ULM cluster next to normal MM around later stages of the time course, whereas ULMS are distributed along the timeline of progression. Second, hierarchical clustering based on the subset of miRNAs that most significantly segregate ULMS from ULM and MM, placed ULMS next to hMSC, whereas ULM and normal myometria grouped separately and apart from ULMS.
Note that this methodology cannot directly inform of the cell of origin of a tumor type. Both the position of a subtype in a differentiation-based phylogeny and the similarity of a subtype to an in vitro differentiation time course provide information about the bulk of tumor cells only. It can therefore be concluded from this study that the bulk of ULMS cells resides in a more immature stage than the bulk of ULM cells or normal SMCs. Whether those cells derive from a differentiation blockade of a SM progenitor or from the ‘de-differentiation’ of mature SM cells warrants in vivo lineage tracing experimentation. The importance of this cross talk between developmental and transformation mechanisms is exemplified by the differential effect produced by the incorporation of the SYT-SSX fusion gene, pathognomonic of synovial sarcomas, at specific stages of skeletal myogenesis using conditional alleles.39 Similar lineage tracing-tumor models in which oncogenic hits are introduced at specific stages of SM differentiation—possibly having a different outcome in maturation and neoplastic transformation—would shed light onto the cell-of-origin of ULMS and ULM, respectively.
Moreover, our analyses are able to identify miRNA alterations related to the transformation process while segregating them from those strictly linked to the differentiation stage. However, these alterations may not be necessarily mutually exclusive; thus, not coincidentally, almost a third of the miRNAs abnormally expressed in ULMS compared with MM (20 of 72) are miRNAs associated with SM differentiation (Table 2). Of these, 6 miRNAs that increase during SM differentiation (from cluster 3 in Supplemental Figure 1 and Supplemental Table 1 at http://ajp.amjpathol.org) were found to be down-regulated in ULMS, which this study has identified as being mostly undifferentiated, further confirming that low expression level of these miRNAs is strongly associated with an immature phenotype. Conversely, 8 miRNAs down-regulated during SM differentiation (from clusters 6, 7, and 9 in Supplemental Figure 1 and Supplemental Table 1 at http://ajp.amjpathol.org), including several components of the miR-17-92 cluster (18a, 18a*, 19a), are overexpressed in ULMS compared with MM. These correlations may only be consequential, or they may identify candidate miRNAs that play a causative role in transformation by impacting the differentiation stage of a progenitor or mature SMC. Such a phenomenon was recently described for miR-29 and miR-206, two miRNAs that contribute to the oncogenic properties of rhabdomyosarcoma by altering the differentiation and proliferative state of tumor cells.14,15,16 Interestingly, miR-29c was identified as one of the 6 miRNAs from cluster 3 up-regulated in SM differentiation and concomitantly altered in ULMS compared with MM (Table 2), suggesting a potential common involvement in these two sarcoma subtypes.
Our study has several biomedical implications. A better understanding of miRNAs modulated during smooth muscle generation may facilitate current attempts to artificially engineer or regenerate damaged smooth muscle tissues. Further, it expands our comprehension of the molecular alterations underlying leiomyosarcoma pathogenesis, identifying new differences with leiomyomas, and revealing an intimate connection between developmental, differentiation, and transformation processes. Finally, the ability of miRNAs to modulate differentiation renders them attractive therapeutic agents against poorly differentiated sarcomas.
Acknowledgments
We thank Dr. Agnes Viale and members of the MSKCC Genomics Facility for array hybridization, and Edna Foundation’s members for unconditional support.
Footnotes
Address reprint requests to Eva Hernando, Ph.D., New York University School of Medicine, Department of Pathology, New York, NY 10016. E-mail: eva.hernando@med.nyu.edu.
See related Commentary on page 541
Supported by grants of the NYSTEM (C023058), American Cancer Society (RSG-08-161-01-DDC), the Liddy Shriver Sarcoma Initiative, and the Edna’s Foundation of Hope. M.V.G. has been supported by a Ministerio de Educacion y Ciencia (MEC, Spain) postdoctoral fellowship; F.M. is a Leon Levy Foundation Young Investigator and is funded by the National Cancer Institute (R01CA138234).
CME disclosure: None of the authors disclosed any relevant financial relationships.
Supplemental material for this article can be found on http://ajp.amjpathol.org.
References
- Hornick JL, Fletcher CD. Criteria for malignancy in nonvisceral smooth muscle tumors. Ann Diagn Pathol. 2003;7:60–66. doi: 10.1053/adpa.2003.50010. [DOI] [PubMed] [Google Scholar]
- Skubitz KM, Skubitz AP. Differential gene expression in leiomyosarcoma. Cancer. 2003;98:1029–1038. doi: 10.1002/cncr.11586. [DOI] [PubMed] [Google Scholar]
- Halayko AJ, Solway J. Molecular mechanisms of phenotypic plasticity in smooth muscle cells. J Appl Physiol. 2001;90:358–368. doi: 10.1152/jappl.2001.90.1.358. [DOI] [PubMed] [Google Scholar]
- Colter DC, Class R, DiGirolamo CM, Prockop DJ. Rapid expansion of recycling stem cells in cultures of plastic-adherent cells from human bone marrow. Proc Natl Acad Sci USA. 2000;97:3213–3218. doi: 10.1073/pnas.070034097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dominici M, Le Blanc K, Mueller I, Slaper-Cortenbach I, Marini F, Krause D, Deans R, Keating A, Prockop D, Horwitz E. Minimal criteria for defining multipotent mesenchymal stromal cells. The International Society for Cellular Therapy position statement, Cytotherapy. 2006;8:315–317. doi: 10.1080/14653240600855905. [DOI] [PubMed] [Google Scholar]
- Kolf CM, Cho E, Tuan RS. Mesenchymal stromal cells. Biology of adult mesenchymal stem cells: regulation of niche, self-renewal and differentiation. Arthritis Res Ther. 2007;9:204. doi: 10.1186/ar2116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim MR, Jeon ES, Kim YM, Lee JS, Kim JH. Thromboxane A2 induces differentiation of human mesenchymal stem cells to smooth muscle-like cells. Stem Cells. 2009;27:191–199. doi: 10.1634/stemcells.2008-0363. [DOI] [PubMed] [Google Scholar]
- Rodriguez LV, Alfonso Z, Zhang R, Leung J, Wu B, Ignarro LJ. Clonogenic multipotent stem cells in human adipose tissue differentiate into functional smooth muscle cells. Proc Natl Acad Sci USA. 2006;103:12167–12172. doi: 10.1073/pnas.0604850103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernstein E, Kim SY, Carmell MA, Murchison EP, Alcorn H, Li MZ, Mills AA, Elledge SJ, Anderson KV, Hannon GJ. Dicer is essential for mouse development. Nat Genet. 2003;35:215–217. doi: 10.1038/ng1253. [DOI] [PubMed] [Google Scholar]
- Subramanian S, Lui WO, Lee CH, Espinosa I, Nielsen TO, Heinrich MC, Corless CL, Fire AZ, van de Rijn M. MicroRNA expression signature of human sarcomas. Oncogene. 2008;27:2015–2026. doi: 10.1038/sj.onc.1210836. [DOI] [PubMed] [Google Scholar]
- Volinia S, Calin GA, Liu CG, Ambs S, Cimmino A, Petrocca F, Visone R, Iorio M, Roldo C, Ferracin M, Prueitt RL, Yanaihara N, Lanza G, Scarpa A, Vecchione A, Negrini M, Harris CC, Croce CM. A microRNA expression signature of human solid tumors defines cancer gene targets. Proc Natl Acad Sci USA. 2006;103:2257–2261. doi: 10.1073/pnas.0510565103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He L, He X, Lim LP, de Stanchina E, Xuan Z, Liang Y, Xue W, Zender L, Magnus J, Ridzon D, Jackson AL, Linsley PS, Chen C, Lowe SW, Cleary MA, Hannon GJ. A microRNA component of the p53 tumour suppressor network. Nature. 2007;447:1130–1134. doi: 10.1038/nature05939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He L, Thomson JM, Hemann MT, Hernando-Monge E, Mu D, Goodson S, Powers S, Cordon-Cardo C, Lowe SW, Hannon GJ, Hammond SM. A microRNA polycistron as a potential human oncogene. Nature. 2005;435:828–833. doi: 10.1038/nature03552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taulli R, Bersani F, Foglizzo V, Linari A, Vigna E, Ladanyi M, Tuschl T, Ponzetto C. The muscle-specific microRNA miR-206 blocks human rhabdomyosarcoma growth in xenotransplanted mice by promoting myogenic differentiation. J Clin Invest. 2009;119:2366–2378. doi: 10.1172/JCI38075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H, Garzon R, Sun H, Ladner KJ, Singh R, Dahlman J, Cheng A, Hall BM, Qualman SJ, Chandler DS, Croce CM, Guttridge DC. NF-kappaB-YY1-miR-29 regulatory circuitry in skeletal myogenesis and rhabdomyosarcoma. Cancer Cell. 2008;14:369–381. doi: 10.1016/j.ccr.2008.10.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan D, Dong XD, Chen X, Wang L, Lu C, Wang J, Qu J, Tu L. MicroRNA-1/206 targets c-Met and inhibits rhabdomyosarcoma development. J Biol Chem. 2009;284:29596–29604. doi: 10.1074/jbc.M109.020511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sekiya I, Larson BL, Smith JR, Pochampally R, Cui JG, Prockop DJ. Expansion of human adult stem cells from bone marrow stroma: conditions that maximize the yields of early progenitors and evaluate their quality. Stem Cells. 2002;20:530–541. doi: 10.1634/stemcells.20-6-530. [DOI] [PubMed] [Google Scholar]
- Sinha S, Hoofnagle MH, Owens GK. Derivation of contractile smooth muscle cells from embryonic stem cells. Methods Mol Biol. 2009;482:345–367. doi: 10.1007/978-1-59745-060-7_22. [DOI] [PubMed] [Google Scholar]
- Ach RA, Wang H, Curry B. Measuring microRNAs: comparisons of microarray and quantitative PCR measurements, and of different total RNA prep methods. BMC Biotechnol. 2008;8:69. doi: 10.1186/1472-6750-8-69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Futschik ME, Carlisle B. Noise-robust soft clustering of gene expression time-course data. J Bioinform Comput Biol. 2005;3:965–988. doi: 10.1142/s0219720005001375. [DOI] [PubMed] [Google Scholar]
- Fitch WM, Margoliash E. Construction of phylogenetic trees. Science. 1967;155:279–284. doi: 10.1126/science.155.3760.279. [DOI] [PubMed] [Google Scholar]
- Felsenstein J. Mathematics vs. evolution: mathematical evolutionary theory. Science. 1989;246:941–942. doi: 10.1126/science.246.4932.941. [DOI] [PubMed] [Google Scholar]
- Huson DH, Richter DC, Rausch C, Dezulian T, Franz M, Rupp R. Dendroscope: an interactive viewer for large phylogenetic trees. BMC Bioinformatics. 2007;8:460. doi: 10.1186/1471-2105-8-460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oskowitz AZ, Lu J, Penfornis P, Ylostalo J, McBride J, Flemington EK, Prockop DJ, Pochampally R. Human multipotent stromal cells from bone marrow and microRNA: regulation of differentiation and leukemia inhibitory factor expression. Proc Natl Acad Sci USA. 2008;105:18372–18377. doi: 10.1073/pnas.0809807105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riester M, Stephan-Otto Attolini C, Downey RJ, Singer S, Michor F. A differentiation-based phylogeny of cancer. PLoS Comput Biol. 2010;6:e100777. doi: 10.1371/journal.pcbi.1000777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naguibneva I, Ameyar-Zazoua M, Polesskaya A, Ait-Si-Ali S, Groisman R, Souidi M, Cuvellier S, Harel-Bellan A. The microRNA miR-181 targets the homeobox protein Hox-A11 during mammalian myoblast differentiation. Nat Cell Biol. 2006;8:278–284. doi: 10.1038/ncb1373. [DOI] [PubMed] [Google Scholar]
- Chen JF, Mandel EM, Thomson JM, Wu Q, Callis TE, Hammond SM, Conlon FL, Wang DZ. The role of microRNA-1 and microRNA-133 in skeletal muscle proliferation and differentiation. Nat Genet. 2006;38:228–233. doi: 10.1038/ng1725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen CZ, Li L, Lodish HF, Bartel DP. MicroRNAs modulate hematopoietic lineage differentiation. Science. 2004;303:83–86. doi: 10.1126/science.1091903. [DOI] [PubMed] [Google Scholar]
- Thai TH, Calado DP, Casola S, Ansel KM, Xiao C, Xue Y, Murphy A, Frendewey D, Valenzuela D, Kutok JL, Schmidt-Supprian M, Rajewsky N, Yancopoulos G, Rao A, Rajewsky K. Regulation of the germinal center response by microRNA-155. Science. 2007;316:604–608. doi: 10.1126/science.1141229. [DOI] [PubMed] [Google Scholar]
- Liu JY, Swartz DD, Peng HF, Gugino SF, Russell JA, Andreadis ST. Functional tissue-engineered blood vessels from bone marrow progenitor cells. Cardiovasc Res. 2007;75:618–628. doi: 10.1016/j.cardiores.2007.04.018. [DOI] [PubMed] [Google Scholar]
- Liu JY, Peng HF, Andreadis ST. Contractile smooth muscle cells derived from hair-follicle stem cells. Cardiovasc Res. 2008;79:24–33. doi: 10.1093/cvr/cvn059. [DOI] [PubMed] [Google Scholar]
- Cheng Y, Liu X, Yang J, Lin Y, Xu DZ, Lu Q, Deitch EA, Huo Y, Delphin ES, Zhang C. MicroRNA-145, a novel smooth muscle cell phenotypic marker and modulator. controls vascular neointimal lesion formation. Circ Res. 2009;105:158–166. doi: 10.1161/CIRCRESAHA.109.197517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cordes KR, Sheehy NT, White MP, Berry EC, Morton SU, Muth AN, Lee TH, Miano JM, Ivey KN, Srivastava D. miR-145 and miR-143 regulate smooth muscle cell fate and plasticity. Nature. 2009;460:704–710. doi: 10.1038/nature08195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu N, Bezprozvannaya S, Williams AH, Qi X, Richardson JA, Bassel-Duby R, Olson EN. microRNA-133a regulates cardiomyocyte proliferation and suppresses smooth muscle gene expression in the heart. Genes Dev. 2008;22:3242–3254. doi: 10.1101/gad.1738708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu J, Getz G, Miska EA, Alvarez-Saavedra E, Lamb J, Peck D, Sweet-Cordero A, Ebert BL, Mak RH, Ferrando AA, Downing JR, Jacks T, Horvitz HR, Golub TR. MicroRNA expression profiles classify human cancers. Nature. 2005;435:834–838. doi: 10.1038/nature03702. [DOI] [PubMed] [Google Scholar]
- Rosenfeld N, Aharonov R, Meiri E, Rosenwald S, Spector Y, Zepeniuk M, Benjamin H, Shabes N, Tabak S, Levy A, Lebanony D, Goren Y, Silberschein E, Targan N, Ben-Ari A, Gilad S, Sion-Vardy N, Tobar A, Feinmesser M, Kharenko O, Nativ O, Nass D, Perelman M, Yosepovich A, Shalmon B, Polak-Charcon S, Fridman E, Avniel A, Bentwich I, Bentwich Z, Cohen D, Chajut A, Barshack I. MicroRNAs accurately identify cancer tissue origin. Nature Biotechnol. 2008;26:462–469. doi: 10.1038/nbt1392. [DOI] [PubMed] [Google Scholar]
- Mittal KR, Chen F, Wei JJ, Rijhvani K, Kurvathi R, Streck D, Dermody J, Toruner GA. Molecular and immunohistochemical evidence for the origin of uterine leiomyosarcomas from associated leiomyoma and symplastic leiomyoma-like areas. Mod Pathol. 2009;22:1303–1314. doi: 10.1038/modpathol.2009.96. [DOI] [PubMed] [Google Scholar]
- Ono M, Maruyama T, Masuda H, Kajitani T, Nagashima T, Arase T, Ito M, Ohta K, Uchida H, Asada H, Yoshimura Y, Okano H, Matsuzaki Y. Side population in human uterine myometrium displays phenotypic and functional characteristics of myometrial stem cells. Proc Natl Acad Sci USA. 2007;104:18700–18705. doi: 10.1073/pnas.0704472104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haldar M, Hancock JD, Coffin CM, Lessnick SL, Capecchi MR. A conditional mouse model of synovial sarcoma: insights into a myogenic origin. Cancer Cell. 2007;11:375–388. doi: 10.1016/j.ccr.2007.01.016. [DOI] [PubMed] [Google Scholar]