Abstract
S100A6 has been implicated in a variety of biological functions as well as tumorigenesis. In this study, we investigated the expression status of S100A6 in relation to the clinicopathological features and prognosis of patients with gastric cancer and further explored a possible association of its expression with epigenetic regulation. S100A6 expression was remarkably increased in 67.5% of gastric cancer tissues as compared with matched noncancerous tissues. Statistical analysis demonstrated a clear correlation between high S100A6 expression and various clinicopathological features, such as depth of wall invasion, positive lymph node involvement, liver metastasis, vascular invasion, and tumor-node metastasis stage (P < 0.05 in all cases), as well as revealed that S100A6 is an independent prognostic predictor (P = 0.026) significantly related to poor prognosis (P = 0.0004). Further exploration found an inverse relationship between S100A6 expression and the methylation status of the seventh and eighth CpG sites in the promoter/first exon and the second to fifth sites in the second exon/second intron. In addition, the level of histone H3 acetylation was found to be significantly higher in S100A6-expressing cancer cells. After 5-azacytidine or trichostatin A treatment, S100A6 expression was clearly increased in S100A6 low-expressing cells. In conclusion, our results suggested that S100A6 plays an important role in the progression of gastric cancer, affecting patient prognosis, and is up-regulated by epigenetic regulation.
S100A6, also known as calcyclin, is a low-molecular-weight acidic protein containing 2 EF-hand calcium-binding motifs.1,2 It was discovered on the basis of its cell cycle-dependent expression and is preferentially expressed in G1 phase of the cell cycle after mitogenic stimuli.3,4 S100A6 is a member of the S100 family, which are found localized to the cytoplasm and/or nucleus in a wide range of cell types.5 S100A6 may interact with binding or target proteins, thereby regulating dynamics of cytoskeleton constituents, cell growth and differentiation, and calcium homeostasis.5,6,7,8,9,10
Subsequent studies showed that S100A6 may also be involved in the regulation of cancer progression.11 The deregulation of S100A6 expression during malignant transformation has thus far been described in human pancreatic cancer, malignant thyroid neoplasms, malignant melanoma, breast cancer, hepatocellular carcinoma, lung cancer, prostate cancer, and colorectal carcinoma.12,13,14,15,16,17,18,19,20,21 S100A6 overexpression has been reported to link with metastasis in colon cancer14,15 and is a well-established marker of melanoma in which its level correlates with tumor invasiveness and poor prognosis. Although the high expression of S100A6 was reported in gastric cancer, its correlation with patient prognosis and clinicopathological features has not been fully investigated.22 Like other S100 proteins, S100A6 may promote cancer progression through specific roles in cell survival and apoptotic pathways,6 however the exact mechanism is unclear.
In this study, we performed a detailed analysis of S100A6 expression in primary gastric cancer, matched metastatic lymph nodes, and liver metastatic nodules. Then we analyzed the relationship between S100A6 overexpression and clinicopathological features and patients prognosis. And to gain insight into the mechanism of the regulation of S100A6 expression in gastric cancer, we examined DNA methylation and histone modifications along the S100A6 gene, which may affect S100A6 expression in cancer cell lines by previous reports.23,24
Materials and Methods
Patients and Tissue Specimens
One hundred sixty-six patients with gastric cancer were studied, including 114 males and 52 females (mean age, 58 years; range, 27–82 years) diagnosed and surgically treated in Beijing Cancer Hospital between 1995 and 2001. The depth of tumor invasion, histological grade, lymph node metastasis, liver metastasis, and vascular invasion were obtained from histopathological reports. Stage of gastric cancer was classified according to 2002 tumor-node metastasis (TNM) classification recommended by the American Joint Committee on Cancer.25 None of the patients received chemotherapy or radiation therapy preoperatively. All patients were followed up until January 2007.
After gastrectomy, resected specimens were processed routinely for macroscopic pathological assessment, and tissues were sampled and snap-frozen in liquid nitrogen. In addition, 28 matched liver metastases and 30 matched metastatic lymph nodes were also collected from these patients. Fresh human tissues were stored at −80°C for Western blot analyses and also fixed with 10% formalin in PBS for immunohistochemistry.
This investigation was performed after approval by Ethics Committee of Peking University. Informed consent was obtained from each patient.
Cell Culture
Gastric cancer cell lines KATO3, RF1, RF48, MKN45, and AGS were obtained from ATCC (American Type Culture Collection, Manassas, VA), and cell lines BGC823 and SGC7901 were established in China and obtained from Cell Research Institute, Shanghai, China.26 Cancer cells were routinely grown as a monolayer in RPMI-1640 medium (GIBCO BRL, Carlsbad, CA), supplemented with 10% (v/v) fetal calf serum (FCS, GIBCO) and antibiotics at 37°C in a humidified 5% CO2 atmosphere. Cells seeded on glass slides were fixed by 4% formaldehyde for immunocytochemistry.
Immunohistochemistry Analysis
Four-micrometer sections from formalin-fixed paraffin-embedded tissues were mounted on poly-L-lysine-coated slides and then deparaffinized in xylene and rehydrated through alcohol to distilled water. Endogenous peroxidase activity was blocked with 3% hydrogen peroxide for 15 minutes at room temperature. After pressure cooking the slides in 10 mmol/L EDTA (pH 8.0) for 3 minutes, the sections were incubated overnight at 4°C with mouse anti-S100A6 antibody (1:500, H00006277-M16, Abnova, Taiwan). Primary antibodies were detected using a two-step EnVision System (Dako, Denmark). Positive and negative immunohistochemistry controls were routinely used. For negative controls, the primary antibody was replaced by nonimmune mouse serum to confirm its specificity. Moreover, we used internal positive control in immunohistochemistry for quality assurance.27
Evaluation of S100A6 staining was principally according to the scoring criteria described previously.28 The information recorded included subcellular location of S100A6 staining (nuclear and/or cytoplasmic), intensity of staining (negative, weak, moderate, and strong), and percentage of cells demonstrating positive immunoreactivity. High expression cases were moderately or strongly stained with >20% of cells demonstrating positive immunoreactivity in nucleus and/or cytoplasm, and the rest (low expression) were negatively or weakly stained.
Laser Confocal Scanning
For double staining, mouse anti-human S100A6 antibody (1:500, Abnova) and rabbit anti-human Ki-67 antibody (1:100, RM-9106-S0, LabVision) were used as primary antibodies. The secondary antibodies used for double staining were FITC-conjugated goat anti-mouse antibody and Rhodamine (TRITC)-conjugated goat anti-rabbit antibody (Jackson ImmunoResearch, West Grove, PA). Confocal images were acquired using Leica TCS SP5 confocal microscope (Leica, Mannheim, Germany). Excitation wavelength for TRITC was 586 nm, and that for FITC 488 nm. In addition, the nuclei of specimens were simultaneously stained with DAPI, excitation at 358 nm.
Immunoelectron Microscopy
A postembedding immunogold method was applied for the detection of S100A6 proteins. The anti-S100A6 antibody was obtained from Abnova and used at a dilution of 1:40. Cases immunohistochemically positive for S100A6 were selected. Then small samples (1 mm3 in size) of these positive cases were immersed 6 hours in fresh glutaraldehyde fixative solution, washed in 0.2 mol/L sucrose, and then fixed in osmium tetroxide. These samples were then dehydrated through a graded acetone series (−4°C), embedded in epoxy resin, and polymerized at 37°C for 24 hours and 60°C for 48 hours. Ultrathin sections were obtained on a ultramicrotome and collected on nickel grids. All sections were incubated in 10% H2O2 then rinsed with PBS enriched with 1% bovine serum albumin (BSA) and 0.5% powdered skim milk three times for 5 minutes each and incubated in the primary antibodies at 4°C overnight. After being washed three times in TBS (pH 7.6), the sections were incubated in the goat anti-mouse IgG conjugated to 10 nm gold particles antibody (Sigma-Aldrich, St. Louis, MO) at a dilution of 1:100 in TBS (pH 7.6). Sections were rinsed three times for 10 minutes each in enriched TBS and three times for 10 minutes each in distilled water, and stained with uranyl acetate and lead citrate. The sections were observed and photographed in a Zeiss 900 transmission electron microscope (Oberkochen, Germany). For negative controls, the primary antibody was replaced by nonimmune mouse serum.
Separation of Nuclear and Cytoplasmic Fractions
According to manufacturer’s instructions, total cell lysates from 10 paired fresh human tissues (primary gastric cancer tissues and matched adjacent nonneoplastic mucosa) were separated into cytoplasmic and nuclear fractions using a NE-PER Nuclear and Cytoplasmic Extraction Kit (Pierce, Rockford, IL).
Protein Extraction and Immunoblot Analysis
Total proteins were extracted by Trizol according to manufacturer’s instructions. Protein concentration was determined using a BCA Protein Assay Kit (Pierce), and samples were normalized by dilution with appropriate lysis buffer. Samples were denatured in SDS sample loading buffer by heating to 100°C for 3 minutes and resolved by SDS-PAGE (12% minigels), followed by transfer to Immobilon PVDF transfer membranes (Millipore, Watford, UK), and then subjected to Western blot analysis. Blots were blocked by 5% nonfat milk in PBS for 30 minutes and incubated in mouse anti-S100A6 monoclonal antibody (1:2000) and mouse anti-α-tubulin monoclonal antibody (1:5000, T5168, Sigma-Aldrich) for 2 hours at room temperature. After washing, membranes were incubated with horseradish peroxidase (HRP)-conjugated anti-mouse antibody (Santa Cruz Biotechnology, Santa Cruz, CA) at a 1:1000 dilution. Protein–antibody complexes conjugated with peroxidase were visualized with SuperSignal West Pico Chemiluminscent Substrate (Pierce). S100A6 levels were quantified by a computerized densitometer.
Real-Time PCR
Quantitative assessment of S100A6 mRNA levels by real-time PCR. Total RNA was extracted from BGC823, SGC7901, KATO3, AGS, MKN45, RF1, and RF48 cells by Trizol reagent (Invitrogen, Carlsbad, CA) according to the manufacturer’s instructions. Specific primers for S100A6 and actin were as reported.29 Quantitative RT-PCR was done with a SYBR Green PCR kit (ABI, USA). Each sample was run thrice. Expression of each gene was given as the ratio of expression of each target gene mRNA to that of β-actin mRNA.
DNA Methylation Status Analysis
DNA was isolated according to the standard phenol/chloroform protocol from MKN45, BGC823, SGC7901, KATO3, AGS, RF1, RF48 cell lines, and 53 pairs fresh human tissues (primary gastric cancer tissues and matched nonneoplastic gastric mucosa). Bisulfite modification was performed in accordance with the standard procedures of the EZ DNA methylation kit (Zymo Research, Orange, CA). The methylation profile of S100A6 gene (accession number: NM_014624, and the base number was according to the UCSC genome browser), comprising 1000 bp upstream of the transcription start site and 1646 bp transcribed sequence comprising three exons and two introns (47 CpG sites, 151775549-151775008 and 151774423-151774103), were detected by nested PCR to amplify the bisulfite-modified DNA. The primers were as reported.23 Then the PCR products of cell lines were subcloned into the pMD18-T vector and sequenced, while the tissue PCR products were sequenced directly. The methylated percentages of the CpG sites in primary gastric cancer tissues and adjacent nonneoplastic gastric mucosa were calculated separately. The base number was according to the UCSC Genome Browser.
5-Azacytidine Treatment and Trichostatin A Expression Analysis
MKN45, RF1, and RF48 cells were incubated for 4 days in RPMI 10% (v/v) fetal calf serum containing 5, 10 μmol/L 5-azacytidine (Sigma-Aldrich) or 4 days with 1 μg/ml trichostatin A (Sigma-Aldrich), a histone deacetylase inhibitor. Media were changed every 24 hours. Total RNA was extracted by Trizol reagent, and then the mRNA expression of S100A6 was detected by real-time PCR.
The ChIP Assay
Ten primary gastric cancer tissues and matched nonneoplastic gastric mucosa were chopped into small pieces, the AGS, SGC7901, RF1, RF48 cell lines were washed with PBS, and the samples were crosslinked by 1% formaldehyde and lysated in the lysis buffer. Chromatin of the samples was sonicated to small fragments, then the sonicated chromatin was immunoprecipitated with the antibodies of anti-acetyl-H3 (06-599, Upstate Biotech), anti-acetyl-H3 (lysine 9) (07-352, Upstate Biotech), anti-trimethyl-H3 (lysine 9) (07-442, Upstate Biotech), and anti-trimethyl-H3 (lysine 27) (07-449, Upstate Biotech). Then the protein A Sepharose Fast Flow (Sigma-Aldrich) was added to each sample. The samples were incubated in the 65°C waterbath to reverse formaldehyde crosslink. DNA was extracted and dissolved in 50 μl TE buffer. PCR was applied to amplify the products above. The subsequent PCR was performed with the forward: 5′-CTGGCTCGAGGGTCA-3′ and backward: 5′-GGCTGTTCCGGGTTAGGA-3′ primers.23 Then the average percentage of input signal of the primary gastric cancer and nonneoplastic mucosa were compared.
Statistical Analysis
Clinicopathologic variables were extracted from histopathological reports. To obtain associations between S100A6 expression and clinicopathologic variables, data were cross-tabulated and a χ2 test performed, except for age parameter, which was assessed by Student’s t-test. Cumulative survival was estimated by Kaplan–Meier method, and comparisons between groups were done using a log-rank test. Overall survival was measured from date of initial surgery to date of death, counting death from any cause as the end point, or the last date of information as the end point if no event was documented. A multivariate analysis of Cox proportional hazards regression model (backward, stepwise) was created to assess the influence of each variable on survival. Significance was set at P < 0.05.
The variables of the histone H3 modification was assessed by Student’s t-test between the primary gastric cancer tissues and nonneoplastic gastric mucosa.
Results
S100A6 Overexpression in Primary Gastric Cancer, Metastatic Lymph Node, and Liver Metastases
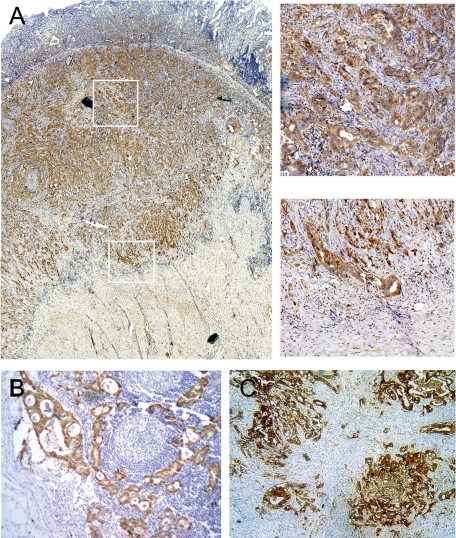
The intensity of S100A6 staining was remarkably higher in primary gastric cancer tissues compared with matched noncancerous mucosa, which had either absent or weak cytoplasmic staining (see supplemental Figure 1 at http://ajp.amjpathol.org). The high expression of S100A6 was detected in 67.5% (112 of 166) of gastric cancer tissues. Immunohistochemical staining of S100A6 in gastric cancer tissues was graded and classified by criteria described in Materials and Methods (see supplemental Figure 2 at http://ajp.amjpathol.org). In primary lesions where tumor invading muscularis propria, S100A6 nuclear staining was frequently found more intense in the invading fronts than in central portions (Figure 1A). Moreover, infiltrating neutrophils and macrophages were also sporadically S100A6-positive.
Figure 1.
Immunohistochemical staining of S100A6 in primary gastric carcinoma, lymph node metastases, and liver metastatic nodules. A: The left large image is the strong S100A6 expression in tumor (×40), and the right two images are high-power images, showing S100A6 staining in the central part (upper) and invasion front (lower) of primary tumor tissues (×400). B and C: Strong S100A6 staining in lymph node metastases (×100) and liver metastatic nodules (×200).
High S100A6 expression was also observed in both lymph node metastases (Figure 1B) and liver metastases (Figure 1C). High S100A6 immunostaining was detected in all lymph node metastases (100%, 30/30), infiltrating into the marginal sinus or diffusing into the lymph nodes. However, only 66.7% (20/30) of matched primary gastric cancer expressed high S100A6. The difference was significant between the two groups (P = 0.001). Twenty-six of 28 (92.9%) liver metastases showed high S100A6 expression, as did 24 of 28 (85.7%) matched primary gastric cancer, without statistical difference (P = 0.388).
S100A6 Overexpression Associated with Clinically Aggressive Phenotype and Short Survival and Served as an Independent Prognostic Indicator
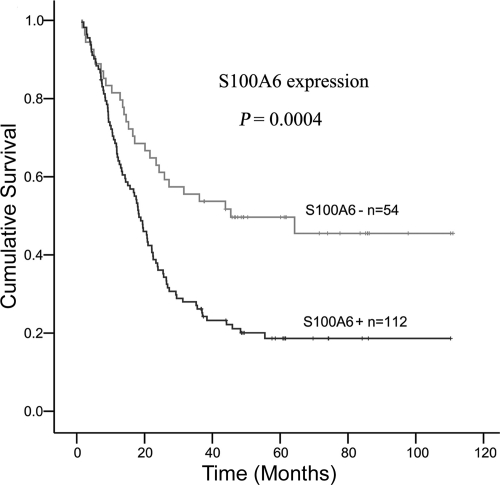
Table 1 shows the correlation between immunohistochemical expression of S100A6 and clinicopathological parameters. Statistical analysis showed that S100A6 expression was strongly associated with depth of wall invasion (P = 0.030), lymph node metastasis (P = 0.037), liver metastasis (P = 0.024), vascular invasion (P = 0.044), and TNM stage (P = 0.048). The analysis of S100A6 expression and survival was depicted in Figure 2. Kaplan–Meier survival curves showed high S100A6 expression group had significantly lower 5-year survival rate (18.62%) than low S100A6 expression group (49.65%; log-rank = 12.52, P = 0.0004; Figure 2). A multivariate Cox proportional hazards model (Table 2) using variables associated with survival in our study revealed that S100A6 expression was a significant independent prognostic indicator (P = 0.026).
Table 1.
Association of S100A6 Expression with Clinicopathological Parameters in Gastric Cancer Patients
| Variables | Cases | S100A6 expression
|
P | |
|---|---|---|---|---|
| Negative − (n = 54) | Positive + (n = 112) | |||
| Sex | ||||
| Male | 114 | 40 | 74 | 0.298 |
| Female | 52 | 14 | 38 | |
| Age (y), mean ± SE | 166 | 59.9 ± 1.4 | 57.2 ± 1.1 | 0.149 |
| Depth of wall invasion | ||||
| T1 + T2 | 28 | 14 | 14 | 0.03 |
| T3 + T4 | 138 | 40 | 98 | |
| Differentiation | ||||
| Well and moderated | 45 | 14 | 31 | 0.812 |
| Poorly | 121 | 40 | 81 | |
| Lymph node metastasis | ||||
| Negative | 31 | 15 | 16 | 0.037 |
| Positive | 135 | 39 | 96 | |
| Liver metastasis | ||||
| M0 | 138 | 50 | 88 | 0.024 |
| M1 | 28 | 4 | 24 | |
| Vascular invasion | ||||
| V (−) | 60 | 25 | 35 | 0.044 |
| V (+) | 99 | 26 | 73 | |
| Not recorded* | 7 | |||
| TNM stages | ||||
| I + II | 37 | 17 | 20 | 0.048 |
| III+ IV | 129 | 37 | 92 | |
Data incomplete.
Figure 2.
Kaplan-Meier survival curves for 166 patients with gastric cancer according to S100A6 expression.
Table 2.
Multivariate Analysis of Prognostic Factors by Cox Proportional Hazard Model
| Variables | P | Relative risk | CI (95%) |
|---|---|---|---|
| Depth of wall invasion | |||
| T3 + T4 vs. T1 + T2 | 0.005 | 2.91 | 1.38–6.14 |
| Differentiation | |||
| Poor versus well/moderate | 0.437 | 0.84 | 0.55–1.30 |
| Lymph node metastasis | |||
| Positive vs. negative | 0.031 | 2.10 | 1.07–4.12 |
| Vascular invasion | |||
| Positive vs. negative | 0.334 | 1.25 | 0.80–1.95 |
| Liver metastasis | |||
| Positive vs. negative | 0.000 | 2.83 | 1.70–4.71 |
| S100A6 expression | |||
| High versus low | 0.026 | 1.67 | 1.06–2.61 |
CI indicates confidence interval.
Localization of S100A6 in Gastric Cancer Tissues
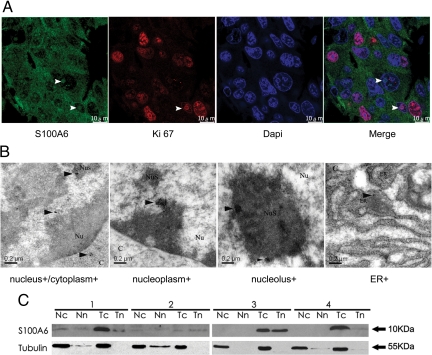
By both immunohistochemistry and immunofluorescence, S100A6 distribution was found throughout the cytoplasm and/or nucleus. In addition, by laser confocal scanning, the result of Ki-67 antigen30 and S100A6 double-staining showed S100A6 nuclear distribution was either evenly throughout the whole nucleus or mainly in the nucleolus in gastric caner tissues (Figure 3A). In nonneoplastic mucosa, Ki-67 antigen was weakly expressed in the nucleus (see supplemental Figure 3 at http://ajp.amjpathol.org). Ultrastructural images captured by immunoelectron microscope demonstrated that S100A6 was deposited throughout the cytoplasm staining endoplasmic reticulum, mitochondria, and lysosome, and within the nucleus where it distributed to both nucleoplasmic and nucleolar structures in gastric cancer tissues (Figure 3B). There was no S100A6 deposition in the negative control staining (data not shown).
Figure 3.
Ultrastructural localization of S100A6 expression in gastric cancer tissues and protein expression in human gastric carcinoma (T) and matched nonneoplastic mucosa (N). A: Immunofluorescence staining captured by laser confocal scanning in gastric cancer tissue, including S100A6, Ki-67, DAPI, and merged image. S100A6 subcellular localization includes cytoplasm, whole nucleus, and nucleolus (indicated by white arrowheads). Scale bars, 10 μm. B: Immunoelectron microscopy: cancer cells display strong immunoreactivity as indicated by gold labeling particles throughout the cytoplasm including the endoplasmic reticulum (ER+), and within the nucleus such as nucleoplasm and nucleolus (indicated by black arrowheads). Scale bars, 0.2 μm. C indicates cytoplasm; Nu, nucleus; Nus, nucleolus. C: S100A6 expression in nuclear extracts (n) and cytoplasmic exactions (c) of matched T and N from four gastric cancer patients.
S100A6 Protein Expression in Primary Gastric Cancer by Immunoblotting
In addition, Western blot was performed to detect the expression of S100A6. It revealed a single band of 10.5 kDa, which was correctly positioned for S100A6. In primary gastric cancer, 96.8% (31/32) was S100A6-positive and 71.9% (23/32) of cancer showed higher expression (average 2.9-fold) than did the nonneoplastic mucosa. Moreover, the localization of S100A6 protein in nuclear and/or cytoplasmic compartments was confirmed by immunoblotting of nuclear and/or cytoplasmic fractions from ten paired gastric cancer samples (Figure 3C).
S100A6 Expression in Gastric Cancer Cell Lines
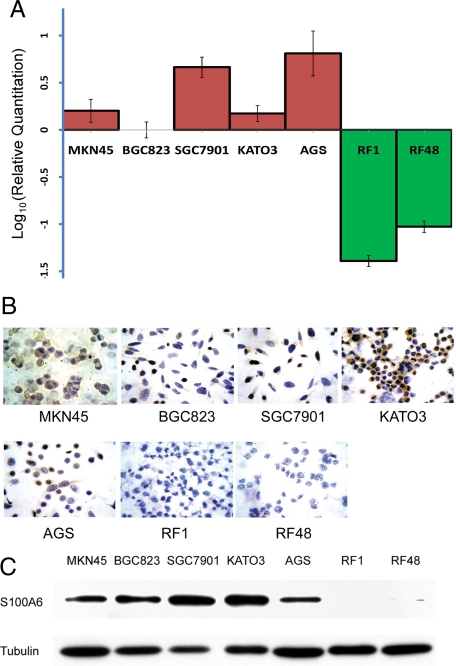
Because previous studies have shown that epigenetic factors may regulate S100A6 gene expression,23 to further explore the mechanism of the up-regulation of S100A6, epigenetic regulation was investigated. First, we chose the S100A6 high- and low-expressing cell lines by measuring S100A6 expression levels in seven gastric cancer cell lines. The real-time PCR results showed that the expression of S100A6 mRNA was significantly higher in the AGS, MKN45, KATO3, BGC823, and SGC7901 cells than RF1 and RF48 cells (Figure 4A), and S100A6 protein levels in the seven cell lines were detected by immunocytochemistry and Western blot. Results showed that protein expression roughly paralleled its mRNA level in these cell lines. RF1 and RF48 cells were found S100A6-negative, whereas AGS, BGC823, KATO3, SGC7901 and MKN45 were positive for S100A6 (Figure 4, B and C). Immunocytochemistry staining showed all of the positive cell lines displayed both cytoplasmic and a varied extent of nuclear staining.
Figure 4.
The expression of S100A6 in the gastric cancer cell lines. A: Quantitative assessment (Log10 value) of S100A6 mRNA levels by real-time PCR in BGC823, SGC7901, KATO3, AGS, MKN45, RF1, and RF48 cell lines, each sample was run thrice, SD are shown. B: Immunocytochemical staining of S100A6 in gastric cancer cell lines (×400). C: The S100A6 protein expression in gastric cancer cell lines by Western blot.
Methylation Profile of S100A6 Gene in Gastric Cancer Cell Lines
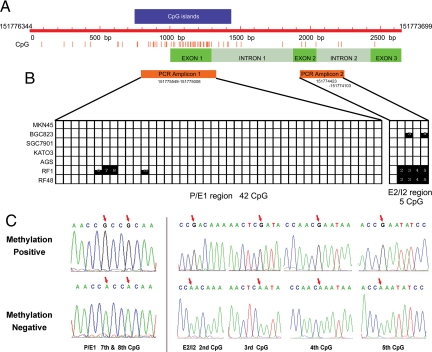
Then we investigated the methylation status of S100A6 in above seven gastric cancer cell lines. Figure 5A shows the distribution of CpG pairs in the region comprising 1000 bp upstream of the transcription start site and in the 1646 bp transcribed sequence comprising three exons and two introns. In total, there are 65 CpG dinucleotides present in this 2646 bp long DNA stretch (151776344-151773699). The CpG island was predicted by CpG Island Searcher (available at http://www.cpgislands.com, last accessed May, 2010), and the proximal S100A6 gene promoter fragment, the first exon and a part of the first intron, are in a CpG-rich region that fulfills all of the criteria for a CpG island as previously reported.23 CpG islands are defined here as sequences longer than 500 bp, with an average GC level higher than 50% and a frequency of CpG greater than 60% of the statistically expected value.31
Figure 5.
The CpG sites methylation status in the MKN45, BGC823, SGC7901, KATO3, AGS, RF1, and RF48 cell lines. A: Schematic representation of CpG dinucleotide distribution in 1646bp S100A6 gene and its 1000 bp upstream. B: The methylation status of the CpG sites in the P/E1 and E2/I2 regions. Each bar represents one CpG pair: black fully filled bar, cytosine residue methylated; white bar, cytosine residue unmethylated; half-filled bar: cytosine residue partially methylated. C: The different methylation status of the seventh and eighth CpG sites in the P/E1 region and the second to fifth CpG sites in the E2/I2 region. The red arrows represent the methylation positive (upper) and methylation negative (lower) CpG sites respectively.
Methylation status of S100A6 in above seven gastric cancer cell lines was investigated by clone sequencing of two regions amplified from bisulfite-modified DNA, the promoter/first exon (P/E1) region, which contains the upstream promoter (11 CpG) and first exon region (31 CpG), and the second exon/second intron (E2/I2) region, which contains the second exon (3 CpG) and second intron (2 CpG). HEK293 cells were used as a positive control,23 with 43 of 47 CpG sites were fully methylated, while the sixth, 13th, and 14th CpG pairs in the P/E1 and first in the E2/I2 incompletely methylated in this study.
The difference of methylation status between S100A6 high-expressing cells (AGS, MKN45, KATO3, BGC823, and SGC7901) and low-expressing cells (RF1 and RF48) was obvious. In the P/E1 region, only RF1 cells was found fully methylated at the seventh and eighth CpG pairs and partially methylated at the sixth and 12th CpG pairs, while the other six cell lines maintained the demethylation status at all CpG sites. And in the E2/I2 region, the second to fifth, a total of four CpG pairs were fully methylated in RF1 and RF48 cell lines, and the third and fifth CpG pairs incompletely methylated in the BGC823 cells, while the CpG pairs of the other five cell lines in this region showed demethylation status (Figure 5B). The positive and negative methylation in these sites are depicted in Figure 5C.
Methylation Profile of S100A6 Gene in Gastric Tissues
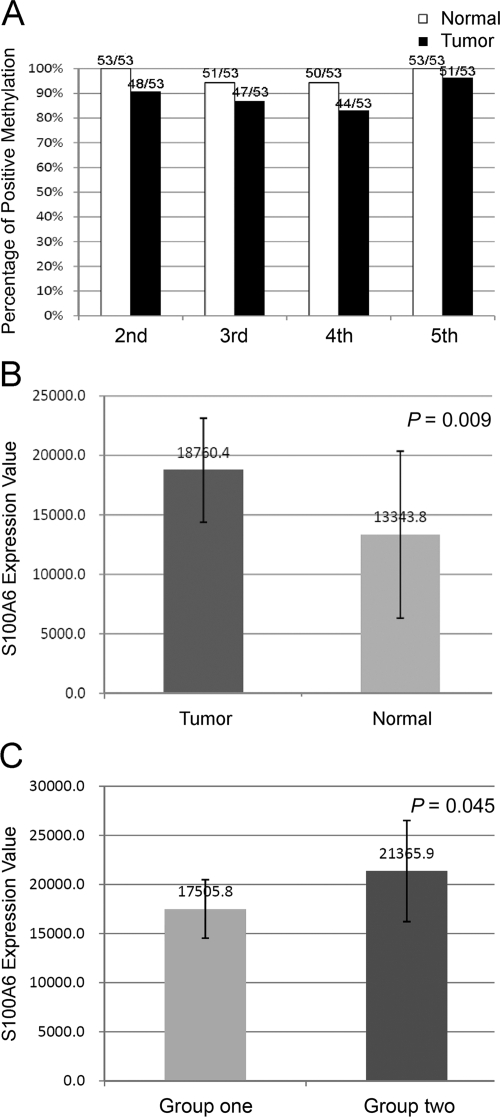
Subsequently, methylation status of the seventh and eighth CpG sites in P/E1 region and the second to fifth CpG sites in the E2/I2 region was investigated in 53 paired primary gastric cancer and nonneoplastic mucosa. The results showed that both the primary gastric cancer and nonneoplastic mucosa were almost entirely methylation-free in the P/E1 region except that only one of 53 gastric cancer was found methylated in the seventh CpG site, and that both were of hypermethylation in the second to fifth CpG sites of the E2/I2 region. In the four sites of the E2/I2 region, cancer tissues were found with relatively lower percentage of positive methylation rate when compared with the adjacent nonneoplastic gastric mucosa (48/53:53/53, 47/53:51/53, 44/53:50/53, 51/53:53/53, respectively), however, the difference did not reach statistical significance (Figure 6A).
Figure 6.
Methylation status of CpG sites in the gastric cancer and nonneoplastic mucosa. A: Percentage of positive methylation in the second to fifth CpG sites of E2/I2 region in paired gastric cancer and nonneoplastic mucosa. B: Average S100A6 expression value of 46 gastric cancer tissues and 20 nonneoplastic mucosa. Error bars indicate SD. C: The expression difference of S100A6 between group one and group two. Group one: cancer tissues with the second to fifth sites of the E2/I2 region entirely methylated; group two: cancer tissues with at least a single CpG site of this region unmethylated. Error bars indicate SD.
Then the correlation between the methylation status of the second to fifth CpG sites in E2/I2 region and the S100A6 expression level in gastric cancer tissues was analyzed. The S100A6 expression value of 46 gastric cancers and 22 nonneoplastic mucosa from above 53 paired tissues were available from our microarray database (by illumina Sentrix BeadChip cDNA microarray, manuscript in preparation). Similar to our previous immunohistochemistry results, the average expression level of S100A6 between tumor and nonneoplastic mucosa was significantly different (P = 0.009, Figure 6B), and then based on methylation status in E2/I2 region, 46 gastric cancer tissues were divided into two groups, group one (n = 36) with all of the four CpG sites methylated and group two (n = 10) with at least a single CpG site unmethylated. Analysis showed that the average S100A6 expression level in group two was significantly higher than that in group one (21365.9 ± 5150.3 versus 17505.8 ± 2985.6; P = 0.045, Figure 6C).
Modified Histone H3 Binding to the S100A6 Gene Promoter in Gastric Cell Lines and Tissues
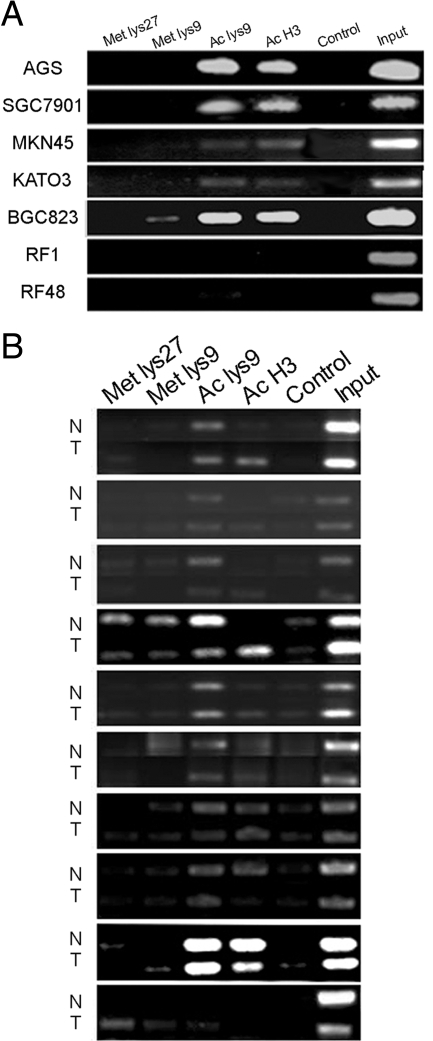
We further examined whether the histone H3 modification plays a role in the expression of the S100A6 in above seven gastric cancer cell lines. The ChIP method and antibodies were indicated in the Materials and Methods. The results showed the levels of acetylated histone H3 and acetylated histone H3 (lysine 9) bound to the S100A6 gene promoter were higher in S100A6 high-expressing cells such as AGS, SGC7901, BGC823, KATO3, and MKN45 than those in low-expressing cells such as RF1 and RF48 (Figure 7A), while trimethylated histone H3 (lysine 27) and trimethylated histone H3 (lysine 9) modification profiles were not significantly different between high- and low-expressing cells.
Figure 7.
Histone H3 modification profile. A: Results of PCR amplification of the ChIP products from seven gastric cancer cell lines, after precipitating the sonicated lysate with antibodies against modified histone H3: Met lys27, anti-trimethyl-H3 (lysine 27); Met lys9, anti-trimethyl-H3 (lysine 9); Ac lys9, anti-acetyl-H3 (lysine 9); and Ac H3, anti-acetyl-H3. B: Results of PCR amplification of the ChIP products from paired gastric cancer and nonneoplastic mucosa.
Moreover, the average percentage of input signal of acetylated histone H3 was significantly different (P = 0.043) between matched cancer and nonneoplastic mucosa. More acetylated H3 bound to S100A6 gene promoter was found in cancer tissues than that in matched nonneoplastic mucosa. The modification of trimethylated histone H3 (lysine 27), trimethylated histone H3 (lysine 9), and acetylated histone H3 (lysine 9) were of no significant difference between the cancer and nonneoplastic mucosa (Figure 7B).
S100A6 Expression Up-Regulated by 5-Azacytidine or Trichostatin A in RF1 and RF48 Cells
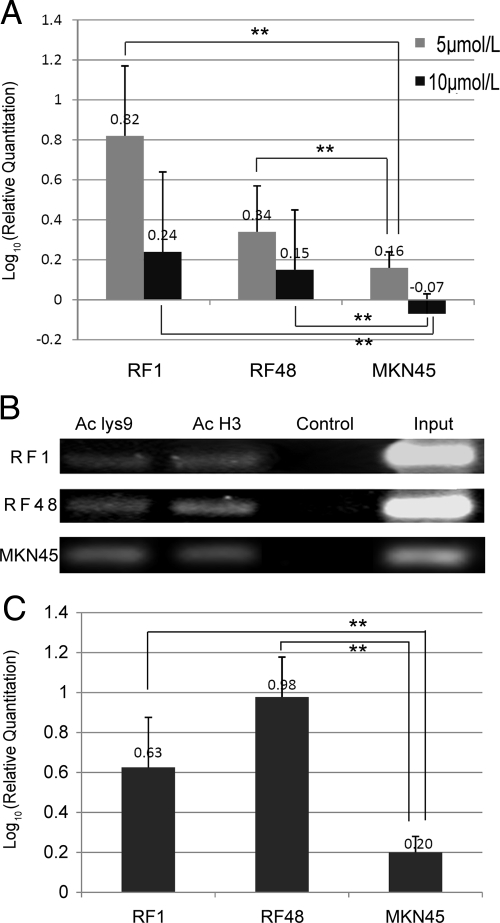
To further make sure that DNA methylation controls the S100A6 expression in gastric cancer, we treated S100A6 methylation-positive cells RF1 and RF48 and methylation-negative cells MKN45, with methyltransferase inhibitor 5-azacytidine. Four days after incubated the RF1 and RF48 cells with 5-azacytidine, the CpG sites in the E2/I2 region was partially demethylated, and the CpG sites in the P/E1 region was also found demethylated in some of the clones of RF1 cells. S100A6 expression was detected at higher level in the treated cells than that in the untreated cells by real-time PCR. The elevation in RF1 cells were more obvious than that in RF48 cells, and the highest expression of S100A6 was in the cells treated by 5 μmol/L 5-azacytidine. However, as the negative control, after MKN45 cells were treated by 5-azacytidine, S100A6 level was slightly increased in the 5 μmol/L 5-azacytidine-treated cells and slightly decreased in the 10 μmol/L 5-azacytidine-treated cells (Figure 8A). The relative quantification of S100A6 mRNA increase in the 5, 10 μmol/L 5-azacytidine-treated RF1, RF48 was higher than that in MKN45 cells (P < 0.01).
Figure 8.
The influence of the 5-azacytidine or TSA on the S100A6 expression in MKN45, RF1, and RF48 cells. A: The relative quantification (Log10 value) of S100A6 mRNA increases in the 5, 10 μmol/L 5-azacytidine-treated RF1, RF48, and MKN45 cells. B: PCR amplification of the ChIP products from TSA-treated RF1, RF48, and MKN45 cells, after precipitating the sonicated lysate with antibodies against modified histone H3: Ac lys9, anti-acetyl-H3 (lysine 9); and Ac H3, anti-acetyl-H3. C: The relative quantification of S100A6 mRNA change in the TSA-treated RF1, RF48, and MKN45 cells. Statistical analysis was performed using the Student’s t-test. Data shown are the mean ± SD of at least three independent experiments. **P < 0.01.
Moreover, we then examined whether inhibition of histone deacetylation by use of trichostatin A (TSA) would also lead to transcriptional activation of the S100A6 gene promoter. Our results showed treatment of RF1 and RF48 cells with TSA was enough to increase the histone acetylation modification (Figure 8B) and to stimulate S100A6 expression (Figure 8C). Whereas in the MKN45 cells (S100A6 high-expressing), the change of histone acetylation modification was not obvious (Figure 8B) and the S100A6 expression was increased slightly (Figure 8C). The relative quantification of S100A6 mRNA increase in the TSA-treated RF1, RF48 was higher than that in MKN45 cells (P < 0.01).
Discussion
In this study, we found that S100A6 expression was remarkably increased in gastric cancer compared with matched noncancerous mucosa. Further analysis showed that S100A6 overexpression was associated with a clinically aggressive phenotype such as depth of wall invasion, lymph node metastasis, liver metastasis, vascular invasion, and TNM stage. Most importantly, S100A6 overexpression correlated with patient prognosis and was an independent prognostic predictor. Previous publications showed that the overexpression of S100A6 was also found in other cancer types. For example, high levels of S100A6 expression related to a significant decrease in survival time for patients with pancreatic cancer,28 and in melanoma, a significant correlation was found between S100A6 mRNA levels in metastases and survival time of patients,32 and in non-small-cell lung cancer, S100A6-positive cases showed a trend of longer survival compared with S100A6-negative cases.21 And a recent report found that S100A6 was significantly up-regulated in gastric cancers and associated with gastric cancer tumorigenesis.22
Our finding supported the involvement of S100A6 in the invasion and metastasis of gastric cancer. High S100A6 in lymph node metastases (100%, 30/30) was significantly higher than matched primary gastric cancer (66.7%, 20/30), and in primary lesions, S100A6 nuclear staining was found more intense in invasion fronts than in central portions. This phenomenon was also found in colorectal cancer.15 Although little is known about the exact mechanism of its expression with aggressive phenotype, it does interact with the actin cytoskeleton and through tropomyosin.7,33 S100A6 also interacts indirectly with chaperone proteins such as Hsp70 in the nucleus.34 In addition, Nedjadi et al suggested S100A6 can interact with the annexin 2 and then promotes pancreatic cancer cell motility.35 Therefore, these interactions may play an important role in cancer metastasis.
It is known that the effect of DNA methylation, which can compress chromatin structure, can affect transcription process.36 Expression of several members of the S100 family, including S100A2, S100A4, S100A6, and S100P, is known to be regulated epigenetically.18,37,38,39 The normal regulation of individual S100 family members can be disrupted during tumorigenesis, which has been demonstrated in diverse cancer types including lung, prostate, pancreas, and colon cancer.18,19,24,37,38,39,40,41 Therefore, to find out the mechanism of S100A6 up-regulation in gastric cancer, we investigated the CpG islands methylation profile and histone H3 modification in gastric cancer cell lines and gastric cancer and adjacent nonneoplastic mucosa.
First, five S100A6-high-expressing gastric cancer cell lines (AGS, SGC7901, MKN45, KATO3, BGC823) and two S100A6-low-expressing gastric cancer cell lines (RF1, RF48) were identified by real-time RCR. The protein level was roughly consistent with the mRNA level. Then we examined the methylation status of 47 of 65 CpG dinucleotides present within a 2646-bp-long region encompassing the proximal promoter, the coding sequence in genomic DNA isolated from cells. The clone sequencing results showed that in the P/E1 region the S100A6 high-expressing cells were entirely methylation-free while the S100A6-low-expressing cells RF1 (with the lowest expression) was methylated in the seventh and eighth CpG sites. In the E2/I2 region, the S100A6-high-expressing cell lines was almost methylation-free except BGC823 was incompletely methylated in the third and fifth CpG pairs, while the S100A6-low-expressing cells were all heavily methylated in the second to fifth CpG sites. Thus, a clear difference exists in cytosine methylation in the S100A6 gene among the examined cells, conforming to the transcriptional status of the S100A6 gene. Previous results by Lesniak et al also showed that the methylation profile of the CpG sites in the P/E1 region may be related to the expression regulation of S100A6 low-expressing cells and that the CpG pairs in the E2/I2 regions were highly methylated in the S100A6 gene inactive cells.23
There is slight dissimilarity of methylation status between cultured cells and tissues. In P/E1 region, only one of 53 gastric cancer was found methylated in the seventh CpG site. And in E2/I2 region, cancer tissues were found with slightly lower percentage of positive methylation rate in the second to fifth sites when compared with the adjacent nonneoplastic mucosa (P = NS), however the average S100A6 expression in cancer tissues with all of the four sites entirely methylated was significantly higher than those with part of the sites methylated (P = 0.045). This dissimilarity may be because of mixture of normal and cancer cells in the cancer tissues. Therefore after combining the results from cells and the patient tissues, our results suggested that an inverse relationship may exist between S100A6 expression and the methylation status of the seventh and eighth sites in P/E1 and the second to fifth CpG sites in E2/I2.
To further make sure that DNA methylation indeed involves the S100A6 expression regulation in gastric cancer, we treated S100A6 low-expressing cells RF1 and RF48 with 5-azacytidine. 5-azacytidine can inhibit DNA methyltransferase, which is an important regulatory mechanism of gene expression.42 Our data showed that RF1 and RF48 cells grown for 4 days in 5-azacytidine started to synthesize S100A6 mRNA. S100A6 was expressed highest in the 5 μmol/L 5-azacytidine-treated cells, and in the 10 μmol/L 5-azacytidine-treated cells the expression was lower, which suggested 5-azacytidine may also affect cell proliferation with the increase in the concentration.43 Thus, a dependence of the induction of S100A6 expression on 5-azacytidine treatment again indicates that DNA methylation is the mechanism involved in the control of activity of this gene. An understanding of these intimately correlated epigenetic changes may be of importance for predicting the outcome of patients with gastric cancer.
In addition to DNA methylation, it has become ever more obvious that histone modification can contribute to gene regulation.44,45 We found that acetylated histone H3, including acetylated histone H3 (lysine 9), was much more often associated with S100A6 transcription in S100A6-high-expressing cell lines than S100A6 low-expressing cell lines, while the trimethylated histone H3 (lysine 27) and trimethylated histone H3 (lysine 9) failed to show any association. We further made sure the effect of histone H3 acetylation on S100A6 expression by treating cells with TSA. After TSA treatment, the S100A6 expression was elevated evidently in RF1 and RF48 cells. Thus, a dependence of the induction of S100A6 expression on TSA treatment reconfirms that histone H3 acetylation involves in the control of activity of this gene.
A little bit different from the results of the cell lines, acetylated histone H3, not acetylated histone H3 (lysine 9), was much more often associated with S100A6 transcription in tumor tissues than that in nonneoplastic mucosa. One possible explanation for this difference between the cell lines and tissues may lie in the genetic heterogeneity of the tissues and the dissimilar environment of cells cultured in vitro.46,47,48 Another possible explanation is that different acetyl-lysine positions of histones H3 other than lysine 9 may also affect the transcription. In most species, acetylation of histone H3 occurs at several different lysine positions including 9, 14, 18, and 23 in the histone tail and is performed by a family of enzymes known as histone acetyltransferases (HATs). Acetylation at lysine 9 appears to have a dominant role in histone deposition and chromatin assembly in some organisms,49,50 and acetylation of lysine 14 is commonly seen in genes that are being actively transcribed into RNA.51 Therefore, our results implicated that the acetylation of histone H3 (lysine 14) may be of importance in regulating S100A6 expression in tissues and deserves further exploration.
In summary, our study showed that high S100A6 expression in gastric cancer is associated with aggressive clinicopathological features, such as depth of wall invasion, positive lymph node involvement, liver metastasis, TNM stage, and poor clinical outcome. The methylation profile of specific CpG sites and the acetylated histone H3 modification may affect its transcription. These observations suggested that S100A6 plays an important role in the progression of gastric cancer and the association of its expression with aggressive phenotype warrants in-depth studies.
Acknowledgments
We thank Professor Da-jun Deng (Department of Cancer Prevention and Treatment Institute, Peking University School of Oncology) for his helpful suggestions and comments on epigenetic regulations.
Footnotes
Address reprint requests to Jia-Fu Ji, M.D., Ph.D., Department of Surgery, Beijing Cancer Hospital & Institute, Peking University School of Oncology, Key Laboratory of Carcinogenesis and Translational Research (Ministry of Education), Beijing 100142, P. R. China. E-mail: jiafuj@gmail.com.
Supported by National Natural Science Foundation of China (No. 30471677) and Foundation of Beijing Municipal Committee of Science & Technology (No. D0905001040631).
X.H.W. and L.H.Z. contributed equally to this work.
Supplemental material for this article can be found on http://ajp.amjpathol.org.
References
- Ferrari S, Calabretta B, deRiel JK, Battini R, Ghezzo F, Lauret E, Griffin C, Emanuel BS, Gurrieri F, Baserga R. Structural and functional analysis of a growth-regulated gene, the human calcyclin. J Biol Chem. 1987;262:8325–8332. [PubMed] [Google Scholar]
- Donato R. S100: a multigenic family of calcium-modulated proteins of the EF-hand type with intracellular and extracellular functional roles. Int J Biochem Cell Biol. 2001;33:637–668. doi: 10.1016/s1357-2725(01)00046-2. [DOI] [PubMed] [Google Scholar]
- Calabretta B, Battini R, Kaczmarek L, de Riel JK, Baserga R. Molecular cloning of the cDNA for a growth factor-inducible gene with strong homology to S-100, a calcium-binding protein. J Biol Chem. 1986;261:12628–12632. [PubMed] [Google Scholar]
- Calabretta B, Kaczmarek L, Mars W, Ochoa D, Gibson CW, Hirschhorn RR, Baserga R. Cell-cycle-specific genes differentially expressed in human leukemias. Proc Natl Acad Sci USA. 1985;82:4463–4467. doi: 10.1073/pnas.82.13.4463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cross SS, Hamdy FC, Deloulme JC, Rehman I. Expression of S100 proteins in normal human tissues and common cancers using tissue microarrays: s100A6. S100A8, S100A9 and S100A11 are all overexpressed in common cancers. Histopathology. 2005;46:256–269. doi: 10.1111/j.1365-2559.2005.02097.x. [DOI] [PubMed] [Google Scholar]
- Emberley ED, Murphy LC, Watson PH. S100 proteins and their influence on pro-survival pathways in cancer. Biochem Cell Biol. 2004;82:508–515. doi: 10.1139/o04-052. [DOI] [PubMed] [Google Scholar]
- Golitsina NL, Kordowska J, Wang CL, Lehrer SS. Ca2+-dependent binding of calcyclin to muscle tropomyosin. Biochem Biophys Res Commun. 1996;220:360–365. doi: 10.1006/bbrc.1996.0410. [DOI] [PubMed] [Google Scholar]
- Mani RS, McCubbin WD, Kay CM. Calcium-dependent regulation of caldesmon by an 11-kDa smooth muscle calcium-binding protein, caltropin. Biochemistry. 1992;31:11896–11901. doi: 10.1021/bi00162a031. [DOI] [PubMed] [Google Scholar]
- Slomnicki LP, Nawrot B, Lesniak W. S100A6 binds p53 and affects its activity. Int J Biochem Cell Biol. 2009;41:784–790. doi: 10.1016/j.biocel.2008.08.007. [DOI] [PubMed] [Google Scholar]
- Tsoporis JN, Izhar S, Parker TG. Expression of S100A6 in cardiac myocytes limits apoptosis induced by tumor necrosis factor-alpha. J Biol Chem. 2008;283:30174–30183. doi: 10.1074/jbc.M805318200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Filipek A, Kuznicki J. Calcyclin–from basic research to clinical implications. Acta Biochim Pol. 1993;40:321–327. [PubMed] [Google Scholar]
- Shekouh AR, Thompson CC, Prime W, Campbell F, Hamlett J, Herrington CS, Lemoine NR, Crnogorac-Jurcevic T, Buechler MW, Friess H, Neoptolemos JP, Pennington SR, Costello E. Application of laser capture microdissection combined with two-dimensional electrophoresis for the discovery of differentially regulated proteins in pancreatic ductal adenocarcinoma. Proteomics. 2003;3:1988–2001. doi: 10.1002/pmic.200300466. [DOI] [PubMed] [Google Scholar]
- Brown LM, Helmke SM, Hunsucker SW, Netea-Maier RT, Chiang SA, Heinz DE, Shroyer KR, Duncan MW, Haugen BR. Quantitative and qualitative differences in protein expression between papillary thyroid carcinoma and normal thyroid tissue. Mol Carcinog. 2006;45:613–626. doi: 10.1002/mc.20193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stulik J, Osterreicher J, Koupilova K, Knizek J, Bures J, Jandik P, Langr F, Dedic K, Schafer BW, Heizmann CW. Differential expression of the Ca2+ binding S100A6 protein in normal, preneoplastic and neoplastic colon mucosa. Eur J Cancer. 2000;36:1050–1059. doi: 10.1016/s0959-8049(00)00043-5. [DOI] [PubMed] [Google Scholar]
- Komatsu K, Kobune-Fujiwara Y, Andoh A, Ishiguro S, Hunai H, Suzuki N, Kameyama M, Murata K, Miyoshi J, Akedo H, Tatsuta M, Nakamura H. Increased expression of S100A6 at the invading fronts of the primary lesion and liver metastasis in patients with colorectal adenocarcinoma. Br J Cancer. 2000;83:769–774. doi: 10.1054/bjoc.2000.1356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weterman MA, Stoopen GM, van Muijen GN, Kuznicki J, Ruiter DJ, Bloemers HP. Expression of calcyclin in human melanoma cell lines correlates with metastatic behavior in nude mice. Cancer Res. 1992;52:1291–1296. [PubMed] [Google Scholar]
- Pedrocchi M, Schafer BW, Mueller H, Eppenberger U, Heizmann CW. Expression of Ca(2+)-binding proteins of the S100 family in malignant human breast-cancer cell lines and biopsy samples. Int J Cancer. 1994;57:684–690. doi: 10.1002/ijc.2910570513. [DOI] [PubMed] [Google Scholar]
- Rehman I, Cross SS, Catto JW, Leiblich A, Mukherjee A, Azzouzi AR, Leung HY, Hamdy FC. Promoter hyper-methylation of calcium binding proteins S100A6 and S100A2 in human prostate cancer. Prostate. 2005;65:322–330. doi: 10.1002/pros.20302. [DOI] [PubMed] [Google Scholar]
- Rehman I, Cross SS, Azzouzi AR, Catto JW, Deloulme JC, Larre S, Champigneuille J, Fromont G, Cussenot O, Hamdy FC. S100A6 (Calcyclin) is a prostate basal cell marker absent in prostate cancer and its precursors. Br J Cancer. 2004;91:739–744. doi: 10.1038/sj.bjc.6602034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J, Kim J, Yoon S, Joo J, Lee Y, Lee K, Chung J, Choe I. S100A6 protein as a marker for differential diagnosis of cholangiocarcinoma from hepatocellular carcinoma. Hepatol Res. 2002;23:274–286. doi: 10.1016/s1386-6346(02)00008-6. [DOI] [PubMed] [Google Scholar]
- De Petris L, Orre LM, Kanter L, Pernemalm M, Koyi H, Lewensohn R, Lehtio J. Tumor expression of S100A6 correlates with survival of patients with stage I non-small-cell lung cancer. Lung Cancer. 2009;63:410–417. doi: 10.1016/j.lungcan.2008.06.003. [DOI] [PubMed] [Google Scholar]
- Yang YQ, Zhang LJ, Dong H, Jiang CL, Zhu ZG, Wu JX, Wu YL, Han JS, Xiao HS, Gao HJ, Zhang QH. Upregulated expression of S100A6 in human gastric cancer. J Dig Dis. 2007;8:186–193. doi: 10.1111/j.1751-2980.2007.00311.x. [DOI] [PubMed] [Google Scholar]
- Lesniak W, Slomnicki LP, Kuznicki J. Epigenetic control of the S100A6 (calcyclin) gene expression. J Invest Dermatol. 2007;127:2307–2314. doi: 10.1038/sj.jid.5700879. [DOI] [PubMed] [Google Scholar]
- Lesniak W, Swart GW, Bloemers HP, Kuznicki J. Regulation of cell specific expression of calcyclin (S100A6) in nerve cells and other tissues. Acta Neurobiol Exp (Wars) 2000;60:569–575. doi: 10.55782/ane-2000-1377. [DOI] [PubMed] [Google Scholar]
- American Joint Committee on Cancer New York: Springer-Verlag,; Gastric Cancer. AJCC Cancer Staging. Manual, (6th ed.) 2002:pp. 99–103. [Google Scholar]
- Ji J, Chen X, Leung SY, Chi JT, Chu KM, Yuen ST, Li R, Chan AS, Li J, Dunphy N, So S. Comprehensive analysis of the gene expression profiles in human gastric cancer cell lines. Oncogene. 2002;21:6549–6556. doi: 10.1038/sj.onc.1205829. [DOI] [PubMed] [Google Scholar]
- Packeisen J, Buerger H, Krech R, Boecker W. Tissue microarrays: a new approach for quality control in immunohistochemistry. J Clin Pathol. 2002;55:613–615. doi: 10.1136/jcp.55.8.613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vimalachandran D, Greenhalf W, Thompson C, Luttges J, Prime W, Campbell F, Dodson A, Watson R, Crnogorac-Jurcevic T, Lemoine N, Neoptolemos J, Costello E. High nuclear S100A6 (Calcyclin) is significantly associated with poor survival in pancreatic cancer patients. Cancer Res. 2005;65:3218–3225. doi: 10.1158/0008-5472.CAN-04-4311. [DOI] [PubMed] [Google Scholar]
- Ohuchida K, Mizumoto K, Ishikawa N, Fujii K, Konomi H, Nagai E, Yamaguchi K, Tsuneyoshi M, Tanaka M. The role of S100A6 in pancreatic cancer development and its clinical implication as a diagnostic marker and therapeutic target. Clin Cancer Res. 2005;11:7785–7793. doi: 10.1158/1078-0432.CCR-05-0714. [DOI] [PubMed] [Google Scholar]
- Kill IR. Localisation of the Ki-67 antigen within the nucleolus. Evidence for a fibrillarin-deficient region of the dense fibrillar component. J Cell Sci. 1996;109 (Pt 6):1253–1263. doi: 10.1242/jcs.109.6.1253. [DOI] [PubMed] [Google Scholar]
- Gardiner-Garden M, Frommer M. CpG islands in vertebrate genomes. J Mol Biol. 1987;196:261–282. doi: 10.1016/0022-2836(87)90689-9. [DOI] [PubMed] [Google Scholar]
- Maelandsmo GM, Florenes VA, Mellingsaeter T, Hovig E, Kerbel RS, Fodstad O. Differential expression patterns of S100A2. S100A4 and S100A6 during progression of human malignant melanoma. Int J Cancer. 1997;74:464–469. doi: 10.1002/(sici)1097-0215(19970822)74:4<464::aid-ijc19>3.0.co;2-9. [DOI] [PubMed] [Google Scholar]
- Breen EC, Tang K. Calcyclin (S100A6) regulates pulmonary fibroblast proliferation, morphology, and cytoskeletal organization in vitro. J Cell Biochem. 2003;88:848–854. doi: 10.1002/jcb.10398. [DOI] [PubMed] [Google Scholar]
- Spiechowicz M, Zylicz A, Bieganowski P, Kuznicki J, Filipek A. Hsp70 is a new target of Sgt1–an interaction modulated by S100A6. Biochem Biophys Res Commun. 2007;357:1148–1153. doi: 10.1016/j.bbrc.2007.04.073. [DOI] [PubMed] [Google Scholar]
- Nedjadi T, Kitteringham N, Campbell F, Jenkins RE, Park BK, Navarro P, Ashcroft F, Tepikin A, Neoptolemos JP, Costello E. S100A6 binds to annexin 2 in pancreatic cancer cells and promotes pancreatic cancer cell motility. Br J Cancer. 2009;101:1145–1154. doi: 10.1038/sj.bjc.6605289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kleinjan DA, van Heyningen V. Long-range control of gene expression: emerging mechanisms and disruption in disease. Am J Hum Genet. 2005;76:8–32. doi: 10.1086/426833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wicki R, Franz C, Scholl FA, Heizmann CW, Schafer BW. Repression of the candidate tumor suppressor gene S100A2 in breast cancer is mediated by site-specific hypermethylation. Cell Calcium. 1997;22:243–254. doi: 10.1016/s0143-4160(97)90063-4. [DOI] [PubMed] [Google Scholar]
- Rosty C, Ueki T, Argani P, Jansen M, Yeo CJ, Cameron JL, Hruban RH, Goggins M. Overexpression of S100A4 in pancreatic ductal adenocarcinomas is associated with poor differentiation and DNA hypomethylation. Am J Pathol. 2002;160:45–50. doi: 10.1016/S0002-9440(10)64347-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sato N, Fukushima N, Matsubayashi H, Goggins M. Identification of maspin and S100P as novel hypomethylation targets in pancreatic cancer using global gene expression profiling. Oncogene. 2004;23:1531–1538. doi: 10.1038/sj.onc.1207269. [DOI] [PubMed] [Google Scholar]
- Feng G, Xu X, Youssef EM, Lotan R. Diminished expression of S100A2, a putative tumor suppressor, at early stage of human lung carcinogenesis. Cancer Res. 2001;61:7999–8004. [PubMed] [Google Scholar]
- Nakamura N, Takenaga K. Hypomethylation of the metastasis-associated S100A4 gene correlates with gene activation in human colon adenocarcinoma cell lines. Clin Exp Metastasis. 1998;16:471–479. doi: 10.1023/a:1006589626307. [DOI] [PubMed] [Google Scholar]
- Rothenburg S, Koch-Nolte F, Thiele HG, Haag F. DNA methylation contributes to tissue- and allele-specific expression of the T-cell differentiation marker RT6. Immunogenetics. 2001;52:231–241. doi: 10.1007/s002510000267. [DOI] [PubMed] [Google Scholar]
- Guo Y, Engelhardt M, Wider D, Abdelkarim M, Lubbert M. Effects of 5-aza-2′-deoxycytidine on proliferation, differentiation and p15/INK4b regulation of human hematopoietic progenitor cells. Leukemia. 2006;20:115–121. doi: 10.1038/sj.leu.2404019. [DOI] [PubMed] [Google Scholar]
- Ito T. Role of histone modification in chromatin dynamics. J Biochem. 2007;141:609–614. doi: 10.1093/jb/mvm091. [DOI] [PubMed] [Google Scholar]
- Bender J. Chromatin-based silencing mechanisms. Curr Opin Plant Biol. 2004;7:521–526. doi: 10.1016/j.pbi.2004.07.003. [DOI] [PubMed] [Google Scholar]
- Sandberg R, Ernberg I. Assessment of tumor characteristic gene expression in cell lines using a tissue similarity index (TSI). Proc Natl Acad Sci USA. 2005;102:2052–2057. doi: 10.1073/pnas.0408105102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perou CM, Jeffrey SS, van de Rijn M, Rees CA, Eisen MB, Ross DT, Pergamenschikov A, Williams CF, Zhu SX, Lee JC, Lashkari D, Shalon D, Brown PO, Botstein D. Distinctive gene expression patterns in human mammary epithelial cells and breast cancers, Proc Natl Acad Sci USA. 1999;96:9212–9217. doi: 10.1073/pnas.96.16.9212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ross DT, Scherf U, Eisen MB, Perou CM, Rees C, Spellman P, Iyer V, Jeffrey SS, Van de Rijn M, Waltham M, Pergamenschikov A, Lee JC, Lashkari D, Shalon D, Myers TG, Weinstein JN, Botstein D, Brown PO. Systematic variation in gene expression patterns in human cancer cell lines. Nat Genet. 2000;24:227–235. doi: 10.1038/73432. [DOI] [PubMed] [Google Scholar]
- Strahl BD, Allis CD. The language of covalent histone modifications. Nature. 2000;403:41–45. doi: 10.1038/47412. [DOI] [PubMed] [Google Scholar]
- Hansen JC, Tse C, Wolffe AP. Structure and function of the core histone N-termini: more than meets the eye. Biochemistry. 1998;37:17637–17641. doi: 10.1021/bi982409v. [DOI] [PubMed] [Google Scholar]
- Kuo MH, Brownell JE, Sobel RE, Ranalli TA, Cook RG, Edmondson DG, Roth SY, Allis CD. Transcription-linked acetylation by Gcn5p of histones H3 and H4 at specific lysines. Nature. 1996;383:269–272. doi: 10.1038/383269a0. [DOI] [PubMed] [Google Scholar]