Abstract
An important challenge in microbial ecology is developing methods that simultaneously examine the physiology of organisms at the molecular level and their ecosystem level interactions in complex natural systems. We integrated extensive proteomic, geochemical, and biological information from 28 microbial communities collected from an acid mine drainage environment and representing a range of biofilm development stages and geochemical conditions to evaluate how the physiologies of the dominant and less abundant organisms change along environmental gradients. The initial colonist dominates across all environments, but its proteome changes between two stable states as communities diversify, implying that interspecies interactions affect this organism's metabolism. Its overall physiology is robust to abiotic environmental factors, but strong correlations exist between these factors and certain subsets of proteins, possibly accounting for its wide environmental distribution. Lower abundance populations are patchier in their distribution, and proteomic data indicate that their environmental niches may be constrained by specific sets of abiotic environmental factors. This research establishes an effective strategy to investigate ecological relationships between microbial physiology and the environment for whole communities in situ.
Keywords: community structure, metaproteomics, microbial ecology, model community, succession
Introduction
Advances in cultivation-independent ‘-omics’ techniques are beginning to allow for the study of microbial populations in natural environments at both a functional biomolecular level and a whole-community level (proposed in Prosser et al, 2007; Little et al, 2008). The combination of environmental metagenomic and proteomic approaches opens the possibility for exploration of basic ecological rules underlying the functioning of microorganisms and the communities they form in nature (proposed by Warnecke and Hugenholtz, 2007; Raes and Bork, 2008). Recent metagenomic studies have revealed the phylogenetic diversity and functional capacity of microbial systems (Tyson et al, 2004; Tringe et al, 2005; DeLong et al, 2006; Rusch et al, 2007; Kunin et al, 2008), and community proteomics can enable the analysis of physiological characteristics in situ (Lacerda et al, 2007; Delmotte et al, 2009; VerBerkmoes et al, 2009). Although this work has greatly advanced our knowledge of natural microbial communities, comprehensive analyses of these systems are often limited by their inherent complexity. To circumvent this issue, one option is to study model communities of reduced complexity, such as biofilm communities found growing in acid mine drainage (AMD) environments (Denef et al, 2010b). Here, we integrated community proteomic and environmental data (physical, chemical, and biological) from 28 natural AMD biofilm communities to reveal relationships of the environment with the whole community, and with the molecular physiology of the individual populations.
Chemoautotrophic biofilm communities sustained by oxidation of iron and sulfur grow underground at the interface between air and sulfuric acid solutions within the Richmond Mine at Iron Mountain, California. In general, initial colonists form few microns thick films that mature into much thicker, differentiated communities with higher species richness (Wilmes et al, 2009). Although the thickness of the biofilm is correlated with maturity, other abiotic factors, such as local flow rates, seem to influence the thickness of individual films. Biofilms develop over the weeks to months and either sink and degrade or are flushed from the system by high flow rates following seasonal rainfall events. Their high biomass production rates and low species richness have enabled extensive metagenomic and proteomic characterization (Tyson et al, 2004; Lo et al, 2007; Denef et al, 2009) and make them an ideal system for exploring principles of microbial ecology at the molecular level (Denef et al, 2010b).
Results
Sample collection and characterization
Twenty-eight biofilm communities were collected from the air–solution interface at seven sites (Supplementary Figure S1) between January 2004 and August 2007. Temperature and solution chemical parameters (including pH, sulfate, metal, nitrate, and nitrite concentrations) were measured at each sample site (Supplementary Table S1). The average temperature and pH values were 40.9±2.8°C and 0.93±0.18 (±s.d.), respectively, and mine discharge was 196±176 l/min. ATP synthesis and all carbon fixation are driven by the high H+ and Fe2+ concentrations (182±0.07 mM) that result from the dissolution of pyrite (FeS2) (Druschel et al, 2004). The community structure (CS) (i.e. composition and population abundance) of biofilms was defined using fluorescent in situ hybridization (FISH; Supplementary Table S2) (Amann et al, 1990).
Community membership patterns
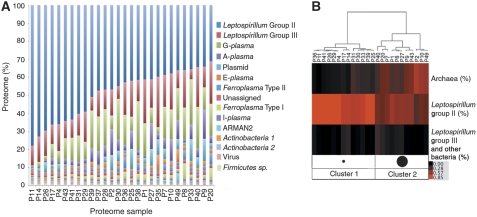
Proteomic and FISH data were used to determine community membership across the 28 biofilm communities. 2D-LC MS/MS-based proteomics performed on each biofilm sample identified an average of 2182±411 proteins from each community (Supplementary Table S3), and 6296 proteins across all communities (http://compbio.ornl.gov/biofilm_amd_ecological_succession). Proteins from iron-oxidizing Leptospirillum Group II bacteria dominated all proteomic data sets (49.6±11.5% of proteins; Figure 1A). Mature biofilms with proportionally lower representation of Leptospirillum Group II had higher abundances of proteins from later biofilm colonizers. The subdominant groups included another iron-oxidizing species, Leptospirillum Group III (13.7±5.5%), and two potentially mixotrophic archaea, G-plasma (9.0±4.9%), and A-plasma (4.6±3.8%) (Figure 1A). A small number of proteins also derived from Actinobacterium 1, Actinobacterium 2, and Firmicutes sp. and other archaea (E-plasma, I-plasma, Ferroplasma Types I and II, and ARMAN-2). Proteins without an organismal affiliation were grouped as unassigned. This category is associated with many different low abundance organisms, and represented a relatively constant fraction of each proteome (Figure 1A; CV=0.11).
Figure 1.
Community structure of biofilm samples. (A) Percent composition of each community proteome based on the organismal assignments for each protein identified. X-axis labels represent biofilm community names. (B) Clustering of biofilm communities (column labels) using community structure data collected by FISH. Color scale is based on the percent composition of each community.
The relative abundances of community members determined by FISH were in good agreement with proteomic relative abundance data (R2=0.80; Supplementary Figure S2). Perfect concordance is not observed, though, as proteins from high abundance organisms are slightly underrepresented in proteome samples and proteins from low abundance organisms are overrepresented. The reason for this deviation from a 1:1 relationship is partly due to the dynamic exclusion filters set during the mass spectrometer run, which will bypass highly abundant peptides after they have been selected once for MS/MS analyses.
Using only FISH data (Supplementary Table S2), biofilms clustered into two distinct patterns of CS that represent different maturation stages, and are distinguished primarily by changes in the relative abundance of archaea to Leptospirillum Group II (Figure 1B). Biofilms from cluster 1 were typically thin (∼10–30 μm in thickness) (Wilmes et al, 2009), and mostly composed of Leptospirillum Group II (78±9.0% of cells). Cluster 2 biofilms were more mature, thicker (∼30–200 μm) (Wilmes et al, 2009), and had a more complex composition, with significantly higher concentrations of archaea (43±11%, t-test—P<2e−6).
Leptospirillum Group II's wide distribution, dominance across all samples, and high proteomic activity shows its wide niche breadth within the AMD system. In contrast, Leptospirillum Group III organisms, like many other low abundance community members, seemed to have a patchier distribution as determined by FISH (Figure 1B), and their proteomes were highly variable across all communities (Figure 1A; CV=0.40). The lower abundance and more restricted distribution of these subdominant populations relative to Leptospirillum Group II indicates their comparatively smaller ecological niches within the AMD system.
Changes in organismal physiology and distribution with environmental conditions
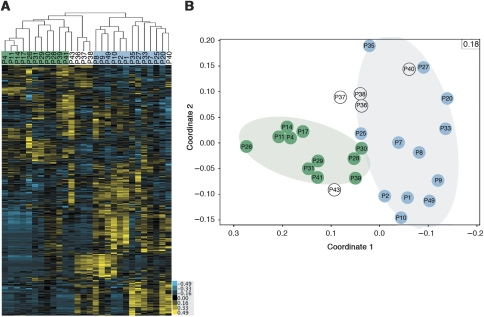
To explore possible relationships between environmental factors and species distribution trends, we tested for significant correlations between proteomic data and biotic and abiotic environmental data. Semiquantitative measurements of protein abundance (normalized spectral abundance factor (NSAF) values) were derived from normalized spectral count values for peptides (Florens et al, 2006). Hierarchical clustering of these values allows for comparison of relative abundances of proteins across samples, enabling efficient detection of patterns within these large multivariate data sets. Communities (columns in Figure 2A) were clustered based on the abundances of all proteins within each proteome and individual proteins (rows in Figure 2A) were clustered based on their relative abundance patterns across all communities. To avoid biases due to the presence or absence of proteins in individual communities and to ensure the fidelity of the results, clustering was performed multiple times with specific subsets of the data (Supplementary Figure S3A). Twenty-three of the communities reproducibly clustered into two groups based on their protein abundance levels (green and blue highlights in Figure 2A). Five communities could not be consistently grouped (P36, P37, P38, P40, and P43) due in part to unusual abundance patterns for some A-plasma and plasmid proteins (Supplementary Figure S3B). A separate form of analysis, non-metric multidimensional scaling (MDS) (Torgerson, 1952), supported the division of protein expression patterns among communities into two groups (Figure 2B).
Figure 2.
Hierarchical and MDS clustering of whole-community proteomes. (A) Clustering of samples using protein abundance data for proteins detected in at least 30% of samples. Column labels represent sample names and are highlighted in green and blue to represent the two groups of samples reproducibly clustered together. Unhighlighted samples showed variability in their clustering patterns. (B) MDS of samples using all protein abundance data. Sample names are given for each point and stress value, which represents the goodness-of-fit, is reported in top-right corner of the graph (values range from 0.00 to 1.00 and lower values indicate better fit). Green and blue groups from (A) (pale oval regions) are discriminated along coordinate 1. Notably, the blue cluster has a wider distribution along the second coordinate (Y-axis) than the green cluster, suggesting a higher degree of variability in the expression pattern of communities in this group. Source data is available for this figure at www.nature.com/msb.
Changes in measured environmental parameters (temperature, flow, sample collection site, pH, and CS; see Supplementary Table S1) were correlated with variations in the protein abundance patterns using the BIOENV statistical procedure (Clarke and Ainsworth, 1993; Oksanen et al, 2007). Significant correlations emerged despite the use of highly complex proteomic data sets comprised of hundreds of variables. ‘Flow’ was the only factor to show consistent correlation with the protein abundance patterns for all organisms (Table I).
Table 1. BIOENV results displaying the correlations of combinations of environmental factors with organismal proteomes.
| Organism proteome | Environmental factorsa | r¯ (All) | r¯ (Single) |
|---|---|---|---|
| aFactors in bold represent the single, strongest correlating factor. | |||
| bA numerical measure of community structure for each biofilm was determined using FISH and included as a factor for the Leptospirillum Group II proteome. This factor was not used in correlations with proteomes of low abundance organisms (A-plasma, G-plasma, Ferroplasma Type I, Ferroplasma Type II, and Leptospirillum Group III) as their presence only in mature biofilms is inherently linked to community structure shifts. | |||
| cPhysical and geochemical factors include pH, temperature, solution discharge rates (flow) on the day of sample collection, and a numerical measure representing the site of collection for each sample (site). | |||
| † P-value <0.10; *<0.05; **<0.01; ***<0.001. | |||
| (a) All factors consideredb | |||
| Leptospirillum Group II | Community structure, flow | 0.48*** | 0.39*** |
| (b) Physical and geochemical factors consideredc | |||
| Leptospirillum Group II | Flow, temperature | 0.34** | 0.28* |
| A-plasma | Site, temperature, flow | 0.40** | 0.26† |
| G-plasma | Temperature, flow | 0.31* | 0.25† |
| Ferroplasma Type I | Temperature, flow | 0.40** | 0.25† |
| Ferroplasma Type II | Temperature, flow | 0.42*** | 0.30* |
| Leptospirillum Group III | pH, flow | 0.24* | 0.15 |
The founding and dominant species
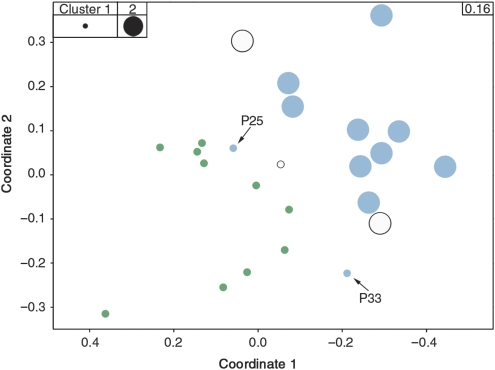
For Leptospirillum Group II, CS bore the strongest correlation to protein expression patterns (Table Ia). This relationship is illustrated by an MDS created using proteomic data for this organism (Figure 3), with circles superimposed over each sample point representing the two CS clusters from Figure 1B.
Figure 3.
Correlation of the community structure factor with Leptospirillum Group II proteomes. An MDS separating samples using only Leptospirillum Group II protein abundance data is shown. Symbols for each point represent community structure clusters from Figure 1B and blue and green highlights represent expression groups from Figure 2. All biofilms of the protein expression group labeled green in Figure 2 represent low developmental stage biofilms and correspond to cluster 1 of Figure 1B (i.e. all green samples are small circles). All but two communities (P25 and P33, noted by arrows) from the high developmental stage protein expression group labeled blue in Figure 2 are consistent with biofilms from cluster 2 of Figure 1B (i.e. all but two blue samples are large circles). P25 and P33 are classified as the high developmental stage samples because their whole-community proteomes included many proteins from low abundance organisms and their Leptospirillum Group II proteomes fall at cluster edges. Stress value is reported in top-right corner of the graph.
Clear metabolic differences in Leptospirillum Group II that coincided with changes in the composition of the surrounding community were detected for the 23 whole-proteome data sets classified as either high or low developmental stage. Use of the significance analysis of microarrays (SAM) technique (Tusher et al, 2001) revealed 474 proteins of Leptospirillum Group II with significant abundance level differences between stages (<4.7% false-discovery rate (FDR); Table II). These Leptospirillum Group II proteins were usually widely distributed across all proteomes, with a majority being detected in >95% of samples (median=96.4%). In addition, these proteins generally comprised a large proportion of each individual proteome (38±2%), suggesting that the presence of surrounding community members influences the overall metabolism of the Leptospirillum Group II population.
Table 2. Number of Leptospirillum Group II proteins determined to be significantly associated with different environmental factors.
| Factor | Total proteins | (+) Correlation/ high dev. stage | (−) Correlation/ low dev. stage | Unique correlationsa | FDRb |
|---|---|---|---|---|---|
| aNumbers in parentheses represent the percent of total correlations that are unique to each respective factor. | |||||
| bFalse-discovery rate (FDR) estimates the percentage of proteins falsely identified to exhibit protein expression changes at an assigned level of significance (i.e. false positives, Tusher et al, 2001). | |||||
| Dev. stage | 474 | 370 | 104 | 250 (53%) | 0.05 |
| Calcium | 360 | 40 | 198 | 58 (16%) | 0.04 |
| Sulfate | 247 | 241 | 6 | 24 (10%) | 0.08 |
| Nitrate | 150 | 101 | 49 | 40 (27%) | 0.09 |
| Temperature | 129 | 122 | 7 | 48 (37%) | 0.08 |
| Arsenic | 109 | 87 | 22 | 1 (1%) | 0.10 |
| pH | 13 | 13 | 0 | 0 (0%) | 0.08 |
| Copper | 0 | 0 | 0 | 0 | — |
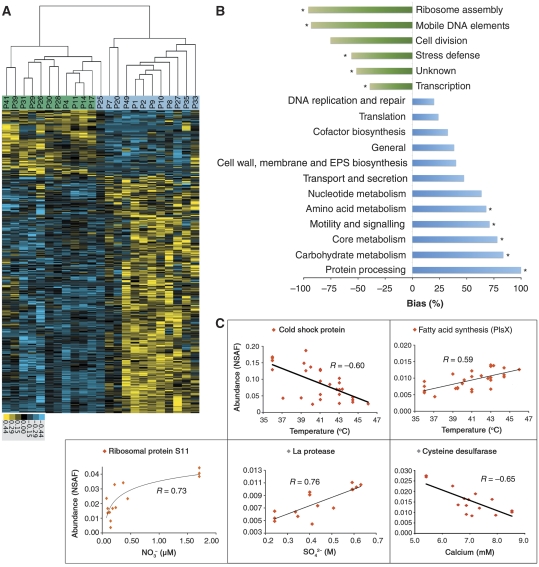
Of the 474 Leptospirillum Group II proteins that showed a significant relationship with CS, 370 are overrepresented in high and 104 in low developmental stage biofilms. Clustering of samples using these proteins revealed two metabolic states when growing within low versus high developmental stage biofilms with high within-cluster correlations (r¯=0.89±0.05; Pearson correlation coefficient; Figure 4A). Leptospirillum Group II proteins associated with this metabolic reorganization were grouped into functional categories and significant biases were defined. Ribosome biosynthesis, which includes ribosome structural proteins, and transcription, which includes RNA polymerase proteins and proteins involved in transcriptional regulation, were significantly elevated in low developmental stage biofilms, as well as proteins involved in physical and chemical stress defense, unknown functions, and associated with mobile genetic elements (Figure 4B). In the high developmental stage biofilms, we detected increased investment in proteins involved in chaperone and protein turnover functions, and environmental signaling, chemotaxis, and motility (Figure 4B). There was also a shift in metabolism away from ribosome biosynthesis towards proteins involved in biosynthesis of extracellular components, carbohydrates, and amino acids.
Figure 4.
Correlations of proteins of Leptospirillum Group II with environmental factors. (A) Clustering of samples using abundance values of differentially detected proteins (significance analysis of microarrays with a false-discovery rate <0.05) of Leptospirillum Group II (474 proteins, see Supplementary Table S4 for the complete list of proteins). Column labels signify sample names and green and blue highlights represent expression group designation from Figure 2. (B) Functional differences of Leptospirillum Group II between developmental stages. Values represent the bias in total proteins overrepresented in either high or low developmental stages. Positive values (blue bars) signify categories overrepresented in high developmental stage biofilms and negative values (green bars) signify categories overrepresented in low developmental stage biofilms. Asterisks note categories significantly overrepresented (98% confidence interval). (C) Pairwise scatter plots of measurements of selected environmental factors strongly correlated to the abundances of a given protein. Source data is available for this figure at www.nature.com/msb.
When the effects of only physical and geochemical parameters on the Leptospirillum Group II proteome were considered, it was noted that the overall physiology of Leptospirillum Group II was largely uncorrelated with individual abiotic variables (Table II). Although some significant correlations between protein abundances and specific abiotic factors were uncovered, the total numbers of proteins were smaller than for those correlated with changes in CS, and very few proteins were uniquely correlated to a given abiotic factor (Table II). For example, only 59 of the 360 proteins with significant correlations to calcium levels were uniquely correlated with this parameter.
Of the proteins that were significantly correlated with specific physical and geochemical parameters, many perform related roles. For example, abundances of the cold shock protein from Leptospirillum Group II decreased as temperatures increased (Supplementary Table S5a) and abundances of a greater than expected number of proteins involved in fatty acid biosynthesis positively correlated with increased temperatures. Also, ∼40% of all detected ribosomal proteins showed positive correlations with nitrate (Figure 4C; Supplementary Table S5b). Additional significant and strong correlations were observed between sulfate levels and protein processing components and calcium levels and proteins involved in various biosynthesis pathways (Figure 4C; Supplementary Table S5c and S5d). Many of these proteins may ameliorate physical stresses associated with the range of abiotic conditions encountered.
Subdominant, later-colonizing taxa
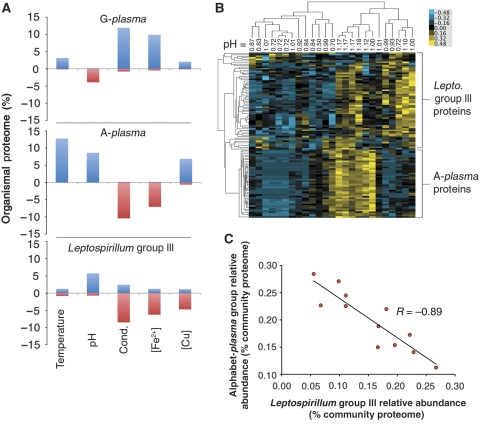
The variable distribution of community members other than Leptospirillum Group II across samples (Figure 1) may result from constraints on their environmental niches. We found that different sets of physical and geochemical parameters were best correlated with each organism's proteome. Temperature showed the strongest correlation with protein abundance patterns of most archaea (A-plasma, G-plasma, and Ferroplasma Types I and II), whereas pH showed a correlation with the proteome of Leptospirillum Group III (Table Ib). We further examined these relationships by analyzing the skew in the total numbers of proteins of A-plasma, G-plasma, and Leptospirillum Group III that correlated positively or negatively with temperature, pH, conductivity, Fe2+ and Cu concentrations. Biases reflect differences in how individual factors may influence the abundance, activity level, or both of each organism (Figure 5A).
Figure 5.
Correlations of the proteins of low abundance organisms with geochemical factors. (A) Percent of A-plasma (n=509 proteins), G-plasma (n=639 proteins), and Leptospirillum Group III (n=936 proteins) identified proteomes either positively (positive values) or negatively correlated (negative values) to a various environmental factors (temperature, pH, conductivity, [Fe2+], and [Cu]). (B) Hierarchical clustering of proteins and samples from A-plasma and Leptospirillum Group III with high correlation to pH using protein abundance data. Values reported at the top of each column represent pH measurements recorded for each sample. Unsupervised clustering of proteins (Y-axis tree) resulted in well-defined groups of A-plasma and Leptospirillum Group III proteins. (C) Scatter plot of relative abundances of Leptospirillum Group III proteins versus those of the Alphabet-plasma Group Proteins (i.e. A-, E-, G-, I-plasma) for high developmental stage whole-community proteomes. Source data is available for this figure at www.nature.com/msb.
The associations of A-plasma and G-plasma proteins to physical factors were generally opposite, despite their close phylogenetic relationship. Temperature and copper were the exceptions, with more proteins from both organisms being positively than negatively correlated with increases in these factors. This is consistent with the BIOENV results that showed a high correlation of temperature with both organisms’ proteomes (Table Ib). In contrast to both these archaea, correlations of Leptospirillum Group III proteins with abiotic factors suggest that this organism favors lower stress environments (i.e. higher pH, lower ionic strength, lower temperature, lower copper; Figure 5A).
Although more Leptospirillum Group III and A-plasma proteins correlated positively than negatively with increasing pH (Figure 5A), their pH optima seemed to be different. Clustering of proteins with strong correlations to pH revealed that Leptospirillum Group III proteins were most abundant at pH ∼0.95 and A-plasma proteins at pH ∼1.12 (Figure 5B).
Further support for the conclusion that Leptospirillum Group III's environmental niche is distinct from other subdominant populations was revealed when the relative abundances of this organism's proteome was considered against those of the Alphabet-plasma group of archaea (i.e. A-, E-, G-, and I-plasma). Here, a strong negative relationship was observed between these groups, demonstrating that as one group increases in activity the other concomitantly decreases (r¯=−0.89; Pearson correlation coefficient; Figure 5C).
Discussion
Value of environmental proteomic approaches for the study of microbial communities
We have used environmental proteomic techniques to examine ecological and physiological processes in a natural model microbial community. Extensive sampling of communities across environmental and temporal gradients provided insight into relationships between environmental parameters and physiological state. Importantly, this work has examined these processes in situ, capturing processes ongoing in the natural environment. An advantage of this approach is that it is cultivation independent, enabling analysis of many coexisting difficult to culture populations. Insights were attained at a community ecology level (i.e. how population abundances within a community change as the environment changes) and at a functional molecular level (i.e. how an individual population changes its physiology as the environment changes). The approach differs from traditional CS profiling (e.g. 16S rRNA gene surveys) used in many microbial ecology studies. Although CS profiling could have shown that Leptospirillum Group II remains the dominant population throughout succession, demonstration that its metabolism changes significantly as biofilms mature relied on the application of proteomic analysis.
Community assembly patterns in AMD biofilms
The finding that Leptospirillum Group II is dominant in all biofilm developmental stages is atypical relative to most patterns of ecological succession for plant and animal communities (Connell and Slatyer, 1977). Similar to patterns of forest establishment in clearings (Glenn-Lewin et al, 1992) and colonization of lava fields (Del Moral and Bliss, 1993), the founding colonist in the AMD system conditions the environment—in this case by fixing carbon and initiating biofilm construction (Goltsman et al, 2009)—enabling the propagation of secondary colonizers. However, in marked contrast to these macroscale examples, Leptospirillum Group II is not ultimately outcompeted and replaced by these later arrivals qas more diverse communities develop (Figure 1). Another exception to this model from macroecology are the ‘fertile islands’ of the African savanna established by Acacia trees (Dean et al, 1999). As in the ‘fertile islands,’ secondary colonists in AMD biofilms most likely depend on persistence of the initial colonist to maintain their niche. This strong and continuing dependence in these systems may be linked to environmental constraints. In both AMD communities and the ‘fertile islands,’ the combination of many physical challenges and the low variety of energy resources probably acts to restrict the diversity of competitors able to perform similar community-essential roles, aiding in the initial colonist's persistence.
Another feature of plant and animal communities is that the most widely distributed species are also commonly the most abundant locally (Hanski et al, 1993). Similarly, Leptospirillum Group II was found in all communities and was also the most abundant population member (Figure 1). The ecological specialization hypothesis proposes that this is due to an ability to ‘tolerate conditions and acquire sufficient resources’ (Brown, 1984), an explanation that also may apply to this organism. Adaptation of this type could be accomplished either by evolution of a stable metabolism that ensures functionality over a wide range of conditions or by continuous modulation of the core metabolism as geochemical parameters change. The finding that the Leptospirillum Group II proteome is not strongly correlated with abiotic environmental perturbations indicates adaptation through a relatively stable, broadly adapted metabolism (Tables I and II). Notably, we identified two stable proteome states that are strongly correlated to microbial community composition. This finding strongly indicates that interspecies interactions directly impact the physiology of this organism.
In summary, we propose that the physiological state of Leptospirillum Group II is largely predicated on interactions with other organisms, which may include resource competition, but that its core metabolism is relatively unchanged by abiotic conditions, such as pH, temperature, and measured geochemical factors. Thus, by augmenting biotic surveys that document the spatial and temporal distribution of a bacterium with direct information about its physiological responses over a range of growth conditions, we were able to evaluate the ecological specialization hypothesis at both a physiological and species level.
Leptospirillum Group II physiological changes: ecological and evolutionary implications
The nature of the Leptospirillum Group II population proteome shift as communities diversify (Table I; Figure 4) provides insight into how biologically imposed constraints may affect physiology during ecological succession. The shift may be related to density-dependent or complexity-dependent effects that typify the transition from an r- to K-environment (MacArthur and Wilson, 1967). In an r-like environment where resources are abundant and predation is low (e.g. early succession where community diversity is low), it is proposed that traits such as rapid growth and dispersal are favored. Conversely, in K-environments (e.g. late succession stages), higher biological diversity, competition for resources, and predation should favor slower growth and the ability to take up and use diminishing resources.
We propose that the functional biases comprising the metabolic shift observed for Leptospirillum Group II (Figure 4B) reflect growth in r- versus K-environments. Ribosome biosynthesis, cell division, and transcription are favored in early succession stages, as expected for rapidly growing cells in thin biofilms that are not limited by resource diffusion constraints, biological competition, and/or predation. In later succession stages, processes such as the metabolism of cellular components (e.g. carbohydrates, nucleotides, amino acids) and environmental sensing are favored. Increases in the abundance of carbohydrate biosynthesis proteins may be a result of increased biomass accumulation in exopolymeric biofilm matrix. Also, deficits in the intracellular amino acid pool may be driven by the demands of general protein synthesis in earlier developmental stages. This is supported by the result that tRNA synthetases are often found in greater abundances in late developmental stage biofilms. This diversification of metabolism and enhancement of the ability to detect chemical gradients in thickening biofilms may be advantageous in a K-like environment where density-dependent effects are higher.
It was also observed that proteins involved in iron oxidation and the electron transport chain are overabundant in late developmental stages. These include nine subunits from the NADH dehydrogenase complex and various cytochrome-containing enzymes (e.g. Cytochrome oxidase, Cytochrome b/b6, and Cytochrome 572, which may oxidize Fe(II) in Leptospirillum Group II; Jeans et al, 2008). Differential abundances of these enzymes may reflect increased iron oxidation by this organism in high developmental stages. In addition, some enzymes involved in the reductive TCA cycle, likely the major carbon fixation pathway in Leptospirillum Group II (Goltsman et al, 2009), are more abundant in late compared with early developmental stages. These include subunits of pyruvate synthase, aconitate hydratase, and succinyl-CoA synthetase. However, as these constitute only a small portion of the overall pathway and given that some may have multiple roles in different metabolic pathways, it is difficult to hypothesize whether carbon fixation rates change with developmental stage.
Despite the relative stability of the Leptospirillum Group II proteome with respect to changes in abiotic factors, specific subsets of proteins correlated with abiotic perturbations and may be involved in specific environmental adaptation mechanisms (Supplementary Table S5). For example, the abundances of lipid biosynthesis proteins correlate with increasing temperature, consistent with observations of lipid composition changes in membranes of organisms under temperature stress (Allen and Bartlett, 2002). In addition, it was found that ribosomal proteins increase with nitrate levels, which are normally at nanomolar levels within the ecosystem and may be growth limiting for Leptospirillum Group II. We also observed significant and strong correlations of protein processing enzymes with sulfate concentrations and of many different metabolic pathways with calcium concentrations (Supplementary Table S5). It is not known what physiochemical interactions may be underlying these relationships, and it may be that other unmeasured abiotic factors that correlate strongly with calcium and sulfate levels may be true causal participants.
It is interesting to note that fine-scale genetic variation occurs within Leptospirillum Group II, some of which may have functional significance in responding to changes in environmental conditions during succession (Tyson et al, 2004; Lo et al, 2007; Simmons et al, 2008). Two genotypic groups have been identified in AMD biofilms, but they do not always co-occur (Denef et al, 2009). In most early succession stage biofilms, only the UBA genotypic group occurs. The five-way genotypic group co-occurs with the UBA type in most late succession stage biofilms. On the basis of strain-resolved proteomic analysis, it was proposed that niche adaptation is associated with both differential regulation of shared genes and differences in gene content (Denef et al, 2010a). As only 4 of the 12 high developmental stage samples from our current study show greater numbers of five-way cells than UBA cells (determined by FISH counts, Denef et al, 2010a and see Supplementary Table S2), we conclude that the proteome changes observed in here can be attributed partially to strain composition changes, but mainly to an overall shift in the metabolism of both strains found in each community. We propose that the synthesis of our results with those of Denef et al (2010a) suggests that the effects of community interactions that motivate proteomic shifts are driving genotypic divergence that will ultimately lead to speciation.
Environmental niche selection of low abundance community members
The abundances and activities of subdominant bacterial and archaeal populations that grow in late succession stages are more variable than those of Leptospirillum Group II (Figure 1). We used changes in proteomic patterns of these organisms to explore their relationships with various abiotic factors and to develop hypotheses about their possible environmental niches. The BIOENV analysis (Table Ib) revealed that the factor showing the most consistent correlation pattern for all organisms was ‘flow,’ which measures AMD solution discharge on the day of sampling and mirrors precipitation cycles (Supplementary Figure S4). Flow likely impacts many solution geochemical and physical parameters (Edwards et al, 1999; Druschel et al, 2004), accounting for the wide correlation with all proteomes. Temperature generally correlated strongly with the proteomes of the archaeal populations, whereas pH correlated with the proteins abundance patterns of the Leptospirillum Group III population.
To further evaluate the inferences that high temperature selects for archaea and high pH for Leptospirillum Group III in more mature biofilms, we tested for significant correlations between the abundances of individual proteins and specific abiotic factors (Figure 5A). Results indicated that the closely related archaea, A-plasma and G-plasma, have different environmental niches. For example, A-plasma growth may be favored in hot, higher pH, lower ionic strength solutions, whereas G-plasma may prefer hot, lower pH, higher ionic strength solutions (Figure 5A). In addition, the analyses demonstrated that Leptospirillum Group III activity/abundance was inversely related to the activities/abundances of various archaeal populations (Figure 5C) and that the former may prefer lower stress environments (Figure 5A). Preference for less stressful environments is consistent with this Leptospirillum Group III's patchy distribution in biofilms and its growth as small microcolonies within the biofilm interior (Wilmes et al, 2009). Other support for these niche hypotheses derives from experimentally manipulated laboratory bioreactors where biofilms were grown under low and high pH conditions (pH=0.8 and 1.4). The abundances of archaeal proteins were greater in lower pH bioreactors, whereas Leptospirillum Group III proteins were more abundant at high pH (Belnap, 2009). Thus, we conclude that specific combinations of geochemical factors restrict the environmental niches of subdominant populations within mature biofilms.
Summary
Community proteomics applied to a suite of biofilm communities enabled an unprecedented level of insight into the activities of multiple coexisting microbial members across the range of physiochemical conditions. A striking finding is that the activities of lower abundance organisms are predicated on restrictive environmental conditions. Perhaps more importantly, the dominant community member exhibits a shift from one physiological state to another that correlates with increased activity of less abundant bacteria and archaea. The strong correlation between physiological state and community membership outweighs any correlation with a single or combination of measured physical or geochemical factors. These findings support a long held, but rarely quantified, axiom in microbial ecology stating that interspecies interactions strongly shape physiological responses in microbial communities.
Materials and methods
Sample collection and metadata measurements
Collection sites for the 28 samples used in this study are shown on the map of the Richmond Mine, Iron Mountain, CA (40°40′38.42″N, 122°31′19.90″W, elevation of ∼900 m) in Supplementary Figure S1. Sample collection was performed as described earlier (Denef et al, 2009). The pH, conductivity, temperature, and Eh of all samples were determined in situ. The standards used for pH measurement were pH 1.00 and pH 1.68 (Ricca Chemical Company) together with standards prepared from standardized sulfuric acid (Fisher Scientific) for the pH range 0–2 as described (Nordstrom et al, 2000). Each in situ sample measured was bracketed by the standard with closest pH and repeated until the pH reading of the bracketing standard agreed within 0.05 pH units. The platinum electrode for the Eh measurements was checked against Zobell's solution (Ricca Chemical Company). Conductivity standards (12.9 and 111 mS/cm, Thermo Scientific Orion), corrected to in situ temperature, and used to calibrate the conductivity probe on-site. Values of ‘flow’ were measured as the total outflow of AMD (l/min) from the mine on the day of sampling.
All water samples were collected in HDPE bottles and filtered as soon as possible. Samples for nitrate, nitrite and iron (II)/total iron were filtered within the mine with 0.1 μm syringe filters (Supor membrane, Pall Corporation). Samples for all other analyses were 0.1 μm filtered (Supor membrane, Pall Corporation) with either a 47 or 142 mm filtering manifold. Filtering was competed within 6 h of sample collection. Water samples were preserved for each analysis according to U.S.G.S. protocols (McCleskey et al, 2004), except for nitrate, which was stored at 4°C in a completely filled 4 ml amber glass bottle, and nitrite, which was frozen on dry ice immediately after filtering and stored at −80°C until analyzed.
Ferrous and total iron concentrations were measured spectrometrically following the ferrozine method (To et al, 1999). Nitrate and nitrite were determined at U.S.G.S., Boulder with a chemiluminescent nitric oxide detector (Sievers Analytical, Model NOA 280), using a method modified from Baeseman et al (2006). The detection limit was 10 nM for nitrate and 5 nM for nitrite. Metals were determined by inductively coupled plasma-optical emission spectroscopy (ICP-OES), run at either UC Berkeley with a Perkin Elmer 5300 DV or at U.S.G.S., Boulder with a Leeman Labs Direct Reading Echelle. Replicate samples run in both laboratories agreed within 5%. Sulfur was determined by ICP-OES with the UC Berkeley instrument. Given the geochemical relationships within the AMD system, all the sulfur measured in solution is assumed to be sulfate (Druschel et al, 2004). U.S.G.S. acidic reference waters AMW 4, SCREE, and PPREE were run several times during each analytical run to monitor analytical accuracy for metals and sulfate.
Numerical representations of sampling site were calculated by using the AB drift site as the seed and assigning whole numbers to the remaining sites based on approximate distances relative to AB drift. For example, the UBA site where P35 was collected was given a value of −3 due to its outlying location relative to other sites. Conversely, samples from the C75 site (P8, P9, P10, P36, P37, P38, and P49) were given a value of +3 as this site lies on the opposite end of the mine from UBA. Values for all parameters are listed in Supplementary Table S1.
FISH of AMD biofilm samples
Characterization of the CS of each biofilm was performed using FISH (Amann et al, 1990). Protocols for biofilm disruption and oligonucleotide probe design, hybridization, microscopy, and CS estimation were followed as described earlier (Bond et al, 2000). Oligonucleotide probes and fluorescent labels used in this study for identification of individual species and groups were as follows: (1) Cy5-LF655 (all Leptospirillum Group bacteria), (2) Cy3-L2UBA353 (Leptospirillum Group II—UBA type bacteria), (3) FITC-L2CG353 (Leptospirillum Group II—five-way type bacteria), (4) Cy5-ARC915 (all Archaea), (5) Cy3-EUBMIX (all Eubacteria), and (6) FITC-LF1252 (Leptospirillum Group III bacteria).
Protein extraction, mass spectrometry, and peptide identification
Whole-cell protein fractions from each sample were extracted, prepared, and approximately equal amounts of total protein were analyzed through 24 h nano-2D-LC (strong cation exchange-reversed phase)—MS/MS on a hybrid LTQ-Orbitrap mass spectrometer (Thermo Fisher Scientific, San Jose, CA) for 23 samples using a previously described protocol (Denef et al, 2009). The remaining proteome samples (P1, P2, P26, P27, and P35) were the first proteomes collected for this project and, as such, were originally run on a stand-alone LTQ mass spectrometer (Thermo Fisher Scientific) using similar amounts of total protein (Ram et al, 2005; Denef et al, 2009). At least three technical replicates were analyzed for each sample.
Each spectrum obtained from tandem MS scans was searched against the Biofilm_AMD_CoreDB_04232008 database containing peptide sequence information derived from the previously published UBA and five-way community genomic data sets (Tyson et al, 2004; Lo et al, 2007) using the Sequest program (Eng et al, 1994) as outlined earlier (Tyson et al, 2004; Denef et al, 2009). Proteins contained within the search database belong to 16 genomic classifications, including: Leptospirillum Group II five-way (2760 proteins) and UBA strains (2629 proteins), Leptospirillum Group III (2730 proteins), A-plasma (2379 proteins), E-plasma (1846 proteins), G-plasma (1893 proteins), I-plasma (1855 proteins), Ferroplasma Type I (2140 proteins) and Type II (2409 proteins), ARMAN-2 (1006 proteins), Actinobacterium 1 (2590 proteins), Actinobacterium 2 (1770 proteins), Firmicutes sp. (1409 proteins), and plasmid (471 proteins), viral (663 proteins), and unassigned (22 045 proteins) categories. The unassigned category is highly redundant and includes many sequences for variants of proteins already included in the main assemblies, many of which occur at low abundance. Also included in this category are unresolved fragments from low abundance organisms and mobile elements. The database also included common contaminants such as trypsin, human keratins, and so on, to ensure that these would not mistakenly be classified as biofilm components.
The DTASelect program (Tabb et al, 2002) was used to parse the results using the filters described earlier (Lo et al, 2007). Proteins from Leptospirillum Group II five-way and UBA strains were condensed into one Leptospirillum Group II genome containing unique proteins and orthologous proteins from each strain. For orthologous proteins, single spectral counts of shared peptides were summed along with unique peptides from each strain to obtain the number of total spectral counts for a given protein. Protein identification was based on the following criteria: (1) at least two peptides identified within the same run, and (2) each matched spectra must have Xcorr values >1.8 (+1), 2.5 (+2), and 3.5 (+3) and ΔCN values >0.08. Similar numbers of total spectra (∼150 000–160 000) were obtained for all runs. We have established false-positive rates with this system at ∼1–5% with these filters in earlier studies (Ram et al, 2005; Denef et al, 2009). All databases, peptide and protein results, MS/MS spectra and Supplementary Tables are archived and made available as open access through the following link: http://compbio.ornl.gov/biofilm_amd_ecological_succession.
Proteome data preparation for statistical analyses
Spectral counts for filtered peptides with scores exceeding the above cutoffs were summed for a given protein across all three technical replicates to obtain the total number of spectra corresponding to that protein. An NSAF was calculated for each protein of the 28 different proteomes according to the method proposed by Florens et al (2006). Two values of the NSAF were calculated for each protein. The first is the result of the normalization with respect to all of the proteins in a community proteome (wcNSAF) and determines each protein's relative abundance compared with all proteins in the sample. The second normalizes individual proteins to the total proteins from the specific organism that particular protein is derived from (orgNSAF), allowing for a relative abundance within each organism's proteome to be determined. As both orgNSAF and wcNSAF values essentially represent percentages, all data were arcsine transformed (ASIN) to approximate a normal distribution. In addition, zeros were substituted for missing values across all 28 proteomes, as proteins not detected in the mass spectrometry analysis are assumed to be below the detection limit and in low abundance. Clustering of data without zeros gave similar results (data not shown). For each possible combination of paired technical replicates in each sample, a test of concordance was conducted using R2 values, which represent the proportion of variability in that is accounted for by a linear regression model for each pairwise comparison. Replicates demonstrated high reproducibility with an average R2 value of 0.974±0.010 for all pairwise comparisons across all samples (Supplementary Table S6).
Hierarchical clustering and non-metric MDS of proteomic and FISH data
ASIN-transformed, mean-centered and scaled wcNSAF and orgNSAF values for each protein were clustered with the Cluster v 3.0 program (de Hoon et al, 2004). Hierarchical clustering was performed on proteins and samples using an uncentered Pearson correlation distance matrix and distances between groups were calculated using average or centroid linkage clustering methods. Cluster files were visualized using Java TreeView (Saldanha, 2004).
As biases can be introduced into clustering results based on the presence or absence of proteins across samples, multiple clustering analyses were performed on subsets of the whole-proteome data. These subsets included proteins detected in >1% of all samples up to 100% of samples using a step size of 10% (i.e. sets containing only proteins present in >10% of samples, >20%…100%). Consistent clustering of a sample within 10 of the 11 trials allowed for confident assignment of that sample to its designated cluster (Supplementary Figure S3).
Using the vegan (Oksanen et al, 2007) and MASS packages of the R software distribution, non-metric MDS was performed on ASIN-transformed wcNSAF and orgNSAF values for all samples. Distance matrices were created using the Bray–Curtis dissimilarity index. This allows for a graphical representation of the relatedness between different samples based on their protein expression patterns within a two-dimensional plane. Hierarchical clustering and MDS using ASIN-transformed orgNSAF values gave similar results.
Hierarchical clustering of untransformed FISH data was performed similar to the protocol for proteomic data. Clustering was performed on samples using an uncentered, absolute Pearson correlation distance matrix and distances between branches were calculated using the complete linkage method. A numerical representation of the relatedness between samples was calculated by measuring the branch length between each sample and sample P4. Values of branch length are reported in Supplementary Table S1 and are used in the BIOENV analysis as the measure of CS.
Correlation of measured variables with protein expression profiles using the BIOENV analysis
Using a method developed by Clarke and Ainsworth (1993), various measured environmental (both abiotic and biotic) variables were correlated to the wcNSAF and orgNSAF values for all samples. Similar results were obtained for both and the results of the orgNSAF analyses are reported. Environmental variables included: sampling site (i.e. site), CS, total mine drainage outflow on the day of sample collection (i.e. flow), pH, and temperature (°C). Raw values of flow and temperature were log-transformed. Values for site and CS were obtained as described above.
These correlation analyses were performed using the ‘BIOENV’ function within the vegan package of the R software distribution (Oksanen et al, 2007). This method involves obtaining Spearman's rank correlations between dissimilarity matrices for samples based on all environmental variables in all combinations or by themselves and a dissimilarity matrix for samples based on protein abundance data. Distance matrices using NSAF values of all samples were made using the Bray–Curtis dissimilarity index, whereas dissimilarity matrices were constructed for measured environmental variables using a Euclidean distance metric. Permutation tests were performed using an in-house script to determine the significance of each correlation. Briefly, this script creates 1000 random permutations of the matrix of environmental variables and compares these with the real protein expression dissimilarity matrix. Top scoring random correlations are recorded and the distribution of these is compared with the real correlations to obtain an approximate P-value for each real value.
Use of SAM for detecting significant differences in protein abundance
Although the SAM software was originally developed for detection of significant expression differences between microarray experiments (Tusher et al, 2001), it has previously been applied to proteomics data (Roxas and Li, 2008). This analysis was performed on ASIN-transformed wcNSAF and orgNSAF values of all samples, and orgNSAF results for Leptospirillum Group II are reported. The two groups in this analysis were defined as the low and high developmental stage samples as defined in Figure 2 and Supplementary Figure S3 (blue and green highlight). The SAM software was run using the ‘sam’ function of the siggenes package of the R programming environment. Tests were performed using a paired experimental design and the standard F-statistic, and at least 100 permutations were run. Differentially detected proteins identified with a <4.7% FDR were considered statistically significant.
Significant proteins from the SAM analysis were grouped into functional categories based largely on their cluster of orthologous groups classifications (Tatusov et al, 2003), and biases were evaluated. In this analysis, the total number of proteins from each functional category overrepresented in either low or high developmental stage biofilms was summed separately and divided by the total proteins for each developmental stage to give a percentage. Biases in each category were calculated by separately normalizing each percentage to the total and then subtracting the low developmental stage normalized percentage from the high developmental stage normalized percentage. Significant biases for each category were determined using a bootstrap re-sampling technique, implemented by the perl script ‘All_scrambler.pl’ (Lauro et al, 2009). Re-sampling the pool of total proteins identified in SAM created 1000 replicate subsamples (n=250 proteins). Significant differences between the numbers of proteins identified to be overrepresented in either high or low developmental stages were assessed at a 98% confidence level.
Correlations of individual protein abundances with geochemical and physical parameters
Proteins of Leptospirillum Group II with significant correlations to various environmental factors were also determined using the significance of microarrays technique (samr package of R). This analysis was performed using all ASIN-transformed orgNSAF values from samples with corresponding metadata measurements (e.g. temperature, pH, [NO3−], [SO42−], [Cu], [As], and [Ca]). Correlations with other factors were not considered due to strong intercorrelations between them (e.g. total iron concentrations correlate with [Fe2+] levels). Tests were generally performed using a quantitative experimental design and the standard test. The exception was for tests with nitrate, where a rank test was performed due to the exponential nature of the measured nitrate values. One thousand permutations were run for tests with each parameter. Groups of proteins identified with <0.10 FDR and with ∣r¯AB∣>0.4 were considered to have a strong and significant correlations to each individual factor. Proteins meeting these thresholds were grouped by functional categories and differences between observed and expected numbers of proteins were detected based on the distribution of the number of proteins in each functional category for the entire detected proteome of Leptospirillum Group II.
Spearman rank correlations of the abundances of proteins from subdominant populations (Leptospirillum Group III, A-plasma, and G-plasma) with environmental factors were determined using the ‘cor’ function of R. Samples were not included in this analysis if a given protein was not detected (i.e. no zero NSAF values were included in correlations). Also, proteins that were present in less than half of the samples with corresponding measures of environmental variables were not included in this analysis to prevent false correlations due to under-sampling. Similar results were seen when wcNSAF and orgNSAF values were used (data not shown).
Strong correlations between environmental factors and abundances of proteins from low abundance organisms were defined as ∣r¯AB∣>0.4 (Ideker et al, 2001). Biases in the number of proteins of each proteome either strongly positively or negatively correlated to individual parameters were determined. The total number of proteins with strong negative and strong positive correlations with each environmental factor was summed separately, and each sum was divided by the total number of proteins from each respective proteome (‘Total’ column, Supplementary Table S3). Strong biases in the total number of proteins from an organism either positively or negatively correlated with a specific factor were inferred to represent a response of that organism to that factor.
Supplementary Material
Figures S1–S4 and Tables S1–S6
Acknowledgments
We thank Mr TW Arman, President, Iron Mountain Mines and Dr R Sugarek, EPA, for site access and Mr R Carver for on-site assistance, P Abraham, M Lefsrud (Oak Ridge National Laboratory) for their assistance with proteomic measurements and analysis, and F Lauro for providing computational assistance. R Barnes, C Miller, and M Power are thanked for helpful reviews. This project was funded by Grant No. DE-FG02-05ER64134 from the US Department of Energy Genomics: GTL project (Office of Science).
Footnotes
The authors declare that they have no conflict of interest.
References
- Allen EE, Bartlett DH (2002) Structure and regulation of the omega-3 polyunsaturated fatty acid synthase genes from the deep-sea bacterium Photobacterium profundum strain SS9. Microbiology 148: 1903–1913 [DOI] [PubMed] [Google Scholar]
- Amann RI, Krumholz L, Stahl DA (1990) Fluorescent-oligonucleotide probing of whole cells for determinative, phylogenetic, and environmental-studies in microbiology. J Bacteriol 172: 762–770 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baeseman J, Smith R, Silverstein J (2006) Denitrification potential in stream sediments impacted by acid mine drainage: effects of pH, various electron donors, and iron. Microb Ecol 51: 232–241 [DOI] [PubMed] [Google Scholar]
- Belnap C (2009) Quantitative Proteomic Comparison of Biofilm versus Planktonic Communities and Biofilm Response to pH Perturbation. Berkeley: University of California [Google Scholar]
- Bond PL, Smriga SP, Banfield JF (2000) Phylogeny of microorganisms populating a thick, subaerial, predominantly lithotrophic biofilm at an extreme acid mine drainage site. Appl Environ Microbiol 66: 3842–3849 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown JH (1984) On the relationship between abundance and distribution of species. Am Nat 124: 255–279 [Google Scholar]
- Clarke KR, Ainsworth M (1993) A method of linking multivariate community structure to environmental variables. Mar Ecol Prog Ser 92: 205–219 [Google Scholar]
- Connell JH, Slatyer RO (1977) Mechanisms of succession in natural communities and their role in community stability and organization. Am Nat 111: 1119–1144 [Google Scholar]
- de Hoon MJL, Imoto S, Nolan J, Miyano S (2004) Open source clustering software. Bioinformatics 20: 1453–1454 [DOI] [PubMed] [Google Scholar]
- Dean WRJ, Milton SJ, Jeltsch F (1999) Large trees, fertile islands, and birds in arid savanna. J Arid Environ 41: 61–78 [Google Scholar]
- Del Moral R, Bliss LC (1993) Mechanisms of primary succession—insights resulting from the eruption of Mount St. Helens. In Advances in Ecological Research, Begon M (ed), Vol. 24, pp 1–66. London: Academic Press Ltd [Google Scholar]
- Delmotte Nl, Knief C, Chaffron S, Innerebner G, Roschitzki B, Schlapbach R, von Mering C, Vorholt JA (2009) Community proteogenomics reveals insights into the physiology of phyllosphere bacteria. Proc Natl Acad Sci 106: 16428–16433 [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeLong EF, Preston CM, Mincer T, Rich V, Hallam SJ, Frigaard N-U, Martinez A, Sullivan MB, Edwards R, Brito BR, Chisholm SW, Karl DM (2006) Community genomics among stratified microbial assemblages in the ocean′s interior. Science 311: 496–503 [DOI] [PubMed] [Google Scholar]
- Denef VJ, Kalnejais LH, Mueller RS, Wilmes P, Baker BJ, Thomas BC, Hettich RL, VerBerkmoes NC, Banfield JF (2010a) Proteogenomic basis for ecological divergence of closely related bacteria in natural acidophilic microbial communities. Proc Natl Acad Sci USA 107: 2383–2390 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Denef VJ, Mueller RS, Banfield JF (2010b) AMD biofilms: using model communities to study microbial evolution and ecological complexity in nature. ISME J 4: 599–610 [DOI] [PubMed] [Google Scholar]
- Denef VJ, VerBerkmoes NC, Shah MB, Abraham P, Lefsrud M, Hettich RL, Banfield JF (2009) Proteomics-inferred genome typing (PIGT) demonstrates inter-population recombination as a strategy for environmental adaptation. Environ Microbiol 11: 313–325 [DOI] [PubMed] [Google Scholar]
- Druschel GK, Baker BJ, Gihring TM, Banfield JF (2004) Acid mine drainage biogeochemistry at Iron Mountain, California. Geochem Trans 5: 13–32 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edwards KJ, Gihring TM, Banfield JF (1999) Seasonal variations in microbial populations and environmental conditions in an extreme acid mine drainage environment. Appl Environ Microbiol 65: 3627–3632 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eng JK, McCormack AL, Yates JR (1994) An approach to correlate tandem mass spectral data of peptides with amino acid sequences in a protein database. J Am Soc Mass Spectrom 5: 976–989 [DOI] [PubMed] [Google Scholar]
- Florens L, Carozza MJ, Swanson SK, Fournier M, Coleman MK, Workman JL, Washburn MP (2006) Analyzing chromatin remodeling complexes using shotgun proteomics and normalized spectral abundance factors. Methods 40: 303–311 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glenn-Lewin DC, Peet RK, Veblen TT (1992) Plant Succession: Theory and Prediction, Vol. 11. London, UK: Chapman and Hall [Google Scholar]
- Goltsman DSA, Denef VJ, Singer SW, VerBerkmoes NC, Lefsrud M, Mueller RS, Dick GJ, Sun CL, Wheeler KE, Zemla A, Baker BJ, Hauser L, Land M, Shah MB, Thelen MP, Hettich RL, Banfield JF (2009) Community genomic and proteomic analysis of chemoautotrophic, iron-oxidizing ‘Leptospirillum rubarum’ (Group II) and Leptospirillum ferrodiazotrophum (Group III) in acid mine drainage biofilms. Appl Environ Microbiol 75: 4599–4615 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanski I, Kouki J, Halkka A (1993) Three explanations of the positive relationship between distribution and abundance of species. In Species Diversity in Ecological Communities: Historical and Geographical Perspectives, Ricklefs RE, Schluter D (eds), pp 108–116. Chicago, IL: University of Chicago Press [Google Scholar]
- Ideker T, Thorsson V, Ranish JA, Christmas R, Buhler J, Eng JK, Bumgarner R, Goodlett DR, Aebersold R, Hood L (2001) Integrated genomic and proteomic analyses of a systematically perturbed metabolic network. Science 292: 929–934 [DOI] [PubMed] [Google Scholar]
- Jeans C, Singer SW, Chan CS, VerBerkmoes NC, Shah M, Hettich RL, Banfield JF, Thelen MP (2008) Cytochrome 572 is a conspicuous membrane protein with iron oxidation activity purified directly from a natural acidophilic microbial community. ISME J 2: 542–550 [DOI] [PubMed] [Google Scholar]
- Kunin V, Raes J, Harris JK, Spear JR, Walker JJ, Ivanova N, von Mering C, Bebout BM, Pace NR, Bork P, Hugenholtz P (2008) Millimeter-scale genetic gradients and community-level molecular convergence in a hypersaline microbial mat. Mol Syst Biol 4: 198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lacerda CMR, Choe LH, Reardon KF (2007) Metaproteomic analysis of a bacterial community response to cadmium exposure. J Proteome Res 6: 1145–1152 [DOI] [PubMed] [Google Scholar]
- Lauro FM, McDougald D, Thomas T, Williams TJ, Egan S, Rice S, DeMaere MZ, Ting L, Ertan H, Johnson J, Ferriera S, Lapidus A, Anderson I, Kyrpides N, Munk AC, Detter C, Han CS, Brown MV, Robb FT, Kjelleberg S et al. (2009) The genomic basis of trophic strategy in marine bacteria. Proc Natl Acad Sci USA 106: 15527–15533 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Little AEF, Robinson CJ, Peterson SB, Raffa KF, Handelsman J (2008) Rules of engagement: interspecies interactions that regulate microbial communities. Annu Rev Microbiol 62: 375–401 [DOI] [PubMed] [Google Scholar]
- Lo I, Denef VJ, VerBerkmoes NC, Shah MB, Goltsman D, DiBartolo G, Tyson GW, Allen EE, Ram RJ, Detter JC, Richardson P, Thelen MP, Hettich RL, Banfield JF (2007) Strain-resolved community proteomics reveals recombining genomes of acidophilic bacteria. Nature 446: 537–541 [DOI] [PubMed] [Google Scholar]
- MacArthur RH, Wilson EO (1967) The Theory of Island Biogeography. Princeton, New Jersey: Princeton University Press [Google Scholar]
- McCleskey RB, Nordstrom DK, Naus CA (2004) Questa baseline and pre-mining ground-water-quality investigation. 16. Quality assurance and quality control for water analyses. In U.S. Geological Survey Open File Report 2004, p 119 [Google Scholar]
- Nordstrom DK, Alpers CN, Ptacek CJ, Blowes DW (2000) Negative pH and extremely acidic mine waters from Iron Mountain, California. Environ Sci Technol 34: 254–258 [Google Scholar]
- Oksanen J, Kindt R, Legendre P, O’Hara B, Stevens MHH (2007) Vegan: Community Ecology Package. R package version 1.8–8
- Prosser JI, Bohannan BJM, Curtis TP, Ellis RJ, Firestone MK, Freckleton RP, Green JL, Green LE, Killham K, Lennon JJ, Osborn AM, Solan M, van der Gast CJ, Young JPW (2007) The role of ecological theory in microbial ecology. Nat Rev Microbiol 5: 384–392 [DOI] [PubMed] [Google Scholar]
- Raes J, Bork P (2008) Molecular eco-systems biology: towards an understanding of community function. Nat Rev Microbiol 6: 693–699 [DOI] [PubMed] [Google Scholar]
- Ram RJ, VerBerkmoes NC, Thelen MP, Tyson GW, Baker BJ, Blake RC II, Shah M, Hettich RL, Banfield JF (2005) Community proteomics of a natural microbial biofilm. Science 308: 1915–1920 [PubMed] [Google Scholar]
- Roxas B, Li Q (2008) Significance analysis of microarray for relative quantitation of LC/MS data in proteomics. BMC Bioinformatics 9: 187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rusch DB, Halpern AL, Sutton G, Heidelberg KB, Williamson S, Yooseph S, Wu D, Eisen JA, Hoffman JM, Remington K, Beeson K, Tran B, Smith H, Baden-Tillson H, Stewart C, Thorpe J, Freeman J, Andrews-Pfannkoch C, Venter JE, Li K et al. (2007) The Sorcerer II global ocean sampling expedition: Northwest Atlantic through Eastern Tropical Pacific. PLoS Biol 5: e77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saldanha AJ (2004) Java Treeview—extensible visualization of microarray data. Bioinformatics 20: 3246–3248 [DOI] [PubMed] [Google Scholar]
- Simmons SL, DiBartolo G, Denef VJ, Goltsman DSA, Thelen MP, Banfield JF (2008) Population genomic analysis of strain variation in Leptospirillum Group II bacteria involved in acid mine drainage formation. PLoS Biol 6: e177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tabb DL, McDonald WH, Yates JR (2002) DTASelect and Contrast: tools for assembling and comparing protein identifications from shotgun proteomics. J Proteome Res 1: 21–26 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tatusov R, Fedorova N, Jackson J, Jacobs A, Kiryutin B, Koonin E, Krylov D, Mazumder R, Mekhedov S, Nikolskaya A, Rao BS, Smirnov S, Sverdlov A, Vasudevan S, Wolf Y, Yin J, Natale D (2003) The COG database: an updated version includes eukaryotes. BMC Bioinformatics 4: 41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- To TB, Nordstrom DK, Cunningham KM, Ball JW, McCleskey RB (1999) New method for the direct determination of dissolved Fe(III) concentration in acid mine waters. Environ Sci Technol 33: 807–813 [Google Scholar]
- Torgerson W (1952) Multidimensional scaling: I. Theory and method. Psychometrika 17: 401–419 [DOI] [PubMed] [Google Scholar]
- Tringe SG, von Mering C, Kobayashi A, Salamov AA, Chen K, Chang HW, Podar M, Short JM, Mathur EJ, Detter JC, Bork P, Hugenholtz P, Rubin EM (2005) Comparative metagenomics of microbial communities. Science 308: 554–557 [DOI] [PubMed] [Google Scholar]
- Tusher VG, Tibshirani R, Chu G (2001) Significance analysis of microarrays applied to the ionizing radiation response. Proc Natl Acad Sci USA 98: 5116–5121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tyson GW, Chapman J, Hugenholtz P, Allen EE, Ram RJ, Richardson PM, Solovyev VV, Rubin EM, Rokhsar DS, Banfield JF (2004) Community structure and metabolism through reconstruction of microbial genomes from the environment. Nature 428: 37–43 [DOI] [PubMed] [Google Scholar]
- VerBerkmoes NC, Denef VJ, Hettich RL, Banfield JF (2009) Systems biology: functional analysis of natural microbial consortia using community proteomics. Nat Rev Microbiol 7: 196–205 [DOI] [PubMed] [Google Scholar]
- Warnecke F, Hugenholtz P (2007) Building on basic metagenomics with complementary technologies. Genome Biol 8: 231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilmes P, Remis JP, Hwang M, Auer M, Thelen MP, Banfield JF (2009) Natural acidophilic biofilm communities reflect distinct organismal and functional organization. ISME J 3: 266–270 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figures S1–S4 and Tables S1–S6