Abstract
Flexibility of the HIV-specific T-cell receptor repertoire is a hallmark of HIV-1 infection. Altered differentiation of HIV-specific CD45RO+/CCR7− (TemRO) CD8+ effector-memory T cells into CD45RA+/CCR7− (TemRA) CD8+ effector-memory T cells as well as increased expression of the senescence marker CD57 has been frequently observed HIV-1 infection, but the structural relationship between clonal expansion and T-cell differentiation has not been defined. In this study, we demonstrate that HIV-specific clonotypes have differing degrees of TemRA differentiation but always maintain a significant proportion of TemRO-phenotype cells. These data indicate that structural constraints of the TCR/peptide major histocompatibility complex interaction play a central role in the TemRA differentiation of HIV-specific CD8+ T cells in chronic HIV-1 infection. Clonotypes with a predominantly TemRA phenotype had a substantial fraction of cells without expression of CD57; and in contrast to the high clonotypic variability of TemRA differentiation, expression of CD57 was highly correlated among T-cell clonotypes within epitope-specific responses, indicating TCR-independent expression of CD57 in vivo. Our data highlight the importance of the structural composition of the TCR repertoire for the effector-memory differentiation of the immune response in chronic viral infections and suggest that TCR-dependent and -independent homeostasis shapes the pathogen-specific effector-memory repertoire in vivo.
Introduction
Virus-specific CD8+ T cells are an important effector arm of the immune defense against chronic infections, such as HIV-1/SIV-infection1,2 and hepatitis C virus infection,3 Accumulating data highlight the importance of qualitative aspects of the CD8+ T-cell response in these infections, such as the clonal composition of the pathogen-specific T-cell receptor (TCR) repertoire.4–10
To acquire antiviral function, naive CD8+ T cells must undergo a maturation process after TCR recognition of their cognate epitope. The CD8+ T-cell maturation process has been delineated in several mouse and human experimental systems, and various differentiation models have been suggested to describe the progression from a naive T cell (Tn) to an effector-memory (Tem) state on encounter with cognate epitope.11,12 Within the different models, antigen-specific and nonspecific stimuli have been described which shape the memory repertoire in vivo. These include the strength and duration of TCR signals, TCR-independent signals such as cytokines, or environmental signals provided by antigen-specific cells or T helper cells.13,14 Differing degrees of expression of a particular set of surface markers on different T-cell clones within a defined epitope-specific response suggest a TCR-driven process, whereas expression of highly similar T-cell differentiation markers on all T-cell clones within an epitope-specific response suggests a TCR-independent process. Here, we determined whether surface markers associated with CD8+ T-cell effector-memory differentiation show evidence of clonal distribution within epitope-specific responses.
CD8+ memory T cells can be distinguished by a wide array of surface markers, which are expressed during the different phases of memory differentiation.15–17 The earliest markers of memory differentiation have been the different high- and low-molecular weight isoforms of the surface-expressed tyrosine phosphatase CD45.18–20 CD45 regulates Src kinases required for T-cell antigen receptor signal transduction, and mutations within this protein are associated with severe immunodeficiencies,21 increased risk of HIV infection,22 and autoimmune diseases.23 During T-cell memory maturation, the reexpression of CD45RA has been defined to represent a “terminally differentiated” effector state, where effector-memory cells display lytic properties and have limited proliferative potential11,24; however, several groups have described a “block” toward terminal differentiation of HIV-specific CD8+ T cells in chronic HIV-1 infection.25,26 In addition, terminal differentiation of total CD8+ T cells27 and HIV-specific CD8+ T cells28 has been associated with slowly progressive HIV-1 disease.
CD57 is a marker for replicative senescence and indicates a history of numerous cell divisions as shown by shorter telomere lengths.29 In addition, CD57 expression is associated with terminal differentiation and altered functional capacities in CD8+ T cells as well as in NK cells.30,31 Expansion of CD57+/CD8+ T cells has been observed in HIV-1 infection,32 and expression of CD57 on CD8+ T cells has been associated with reduced viral load in HIV-1 infection.27 Thus, both CD57 and CD45RA on CCR7-negative effector-memory CD8+ T cells have been associated with terminal differentiation, raising the question as to how these 2 differentiation markers are related within an epitope-specific T-cell response.
In this study, we investigate the effector-memory differentiation of HIV-specific CD8+ T cells at the clonotype level. We have previously shown that HIV-specific clonotypes can differentially respond to circulating virus as measured by their ability to secrete IFN-γ in response to peptides representing HIV-1 epitopes.5 These cells also undergo differing degrees of expansion in vivo in response to fluctuating levels of viremia. In contrast, the TCR repertoire of pathogen-specific recall responses has a very similar structural composition in an animal model of hepatitis C infection.4 In this study, we confirm that HIV-1-infected subjects have lower frequencies of HIV-specific TemRA T cells compared with total CD8+ TemRA T cells, but TemRA phenotype cells still make up a substantial portion of HIV-specific CD8+ T cells in a cohort of subjects with slow disease progression. In a detailed TCR clonotype analysis, we found that clonotypes within an epitope-specific response have differing degrees of TemRA differentiation, indicating that the differentiation of HIV-1 epitope-specific CD8+ T cells is TCR-dependent. In contrast, the degree of CD57 expression within an epitope-specific response was equally distributed among different clonotypes. Not only was CD57 expression similar among different clonotypes targeting the same epitope, but also among epitope-specific clonotypes with different degrees of differentiation as defined by the percentage of TemRO and TemRA cells. This indicates a TCR-independent maturation process for the expression of the senescence marker CD57.
Methods
Subjects studied
All subjects were enrolled in Vanderbilt University and Medizinische Hochschule Hannover. The study was approved by the Institutional Review Board of Vanderbilt University and Medizinische Hochschule Hannover, and each subject gave informed consent for participation in the study in accordance with the Declaration of Helsinki.
Human leukocyte antigen class I tissue typing
The human leukocyte antigen type of the persons was determined by sequence-specific primer polymerase chain reaction (SSP-PCR) performed at the DCI Laboratory, Nashville, TN.
Flow cytometric evaluation of surface antigens and tetramer staining
Antibodies specific for human CD3, CD4, CD8, CD45RA, CD45RO, and CD57 were purchased from BD Biosciences, and antibodies specific for CCR7 were purchased from R&D Systems. For staining of TCR variable regions, the following antibodies have been used: BL37.2 (TRBV9), MPB2D5 (TRBV20-1), CH92 (TRBV28), WJF24 (TRBV29-1), IMMU157 (TRBV5-1), 36 213 (TRBV5-8), 3D11 (TRBV5-5), ZOE (TRBV4-1), ZIZOU4 (TRBV4-3), 56C5.2 (TRBV12), FIN9 (TRBV3), C21 (TRBV25), VER2.32 (TRBV10), IMMU222 (TRBV6-5), H132 (TRBV6-2), JU74.3 (TRBV6-6), CAS1.1.3 (TRBV27), TAMAYA1.2 (TRBV14), E17.5F3 (TRBV19), BA62.6 (TRBV18), ELL1.4 (TRBV30), IG125 (TRBV11-2), IMMU546 (TRBV2), and AF23 (TRBV13) (all derived from Beckman Coulter). HIV-specific tetramers were purchased from Beckman Coulter or obtained from the National Institutes of Health tetramer facility (Atlanta, GA). Antibodies and tetramers were added to freshly isolated peripheral blood mononuclear cells at room temperature for 30 minutes. After washing, stained samples were acquired on a FACSAria flow cytometer (BD Biosciences) and were analyzed with FACS Diva (BD Biosciences) and FlowJo Version 7.5 (TreeStar) software.
cDNA synthesis and TCR sequencing
RNA was extracted from purified T cells using STAT-60 (Tel-Test B). A modified anchored reverse-transcribed PCR was performed with Powerscript Reverse transcriptase (Clontech) from total RNA as previously described4,33 using a gene-specific primer for the β-constant region with a modified cDNA anchor primer (Clontech). Negative controls were included at all amplification steps. Amplification of the cDNA by PCR was performed using TCR constant region-based primers and an anchor-specific primer 5′-AAT CCT TTC TCT TGA CCA TG-3′. PCR products of 500 to 600 bp were gel purified and cloned using the TOPO TA cloning kit (Invitrogen). Selected colonies were sequenced using the Taq DyeDeoxy Terminator cycle sequencing Kit (PE Applied Biosystems) and capillary electrophoresis on an ABI 3700 PRISM automated sequencer (PE Applied Biosystems). After editing and alignment using Sequencher (Gene Codes Corp), sequences were compared with sequences in the human TCR gene database (http://imgt.cines.fr/). The TCR variable and joining region classification system of the international ImMunoGeneTics database34 was used throughout the paper.
Statistical analysis
Values are expressed as mean plus or minus SD. Comparisons between groups were performed using Mann-Whitney test or 2-tailed Student t test, where appropriate. Comparison analyses were performed with SPSS, Version 14.0. All reported P values are 2-tailed, and a P value of less than .05 was considered significant.
Results
Overrepresentation of TemRO phenotype cells on HIV-specific CD8+ T cells in HIV-1-seropositive subjects
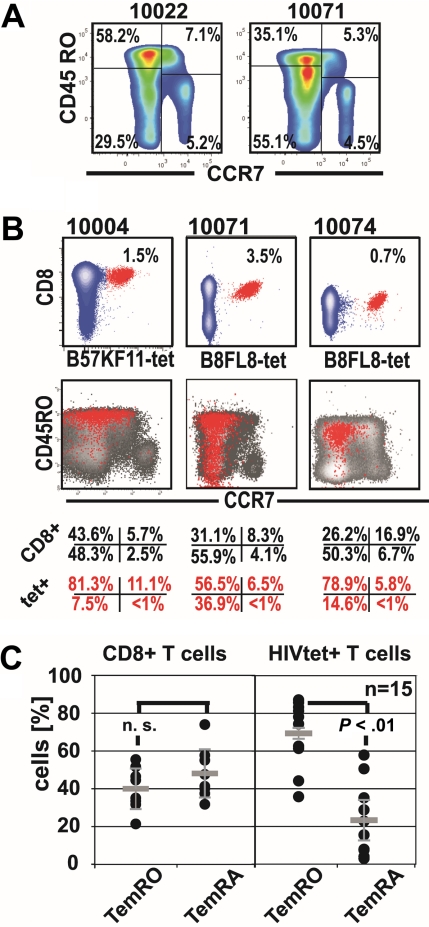
We first analyzed peripheral blood CD8+ T-cell memory phenotypes (Tmem) of CD8+ T cells and HIV-specific CD8+ T cells by CCR7+ and CD45RO staining in combination with tetramer staining in HIV-1-seropositive subjects (Figure 1; Table 1). This analysis revealed an under-representation of HIV-specific TemRA phenotype cells (mean ± SD, 22.71% ± 17.00%) compared with HIV-specific CD8+ TemRO phenotype cells (69.41% ± 15.54%, P < .01 paired t test). However, in 8 of 15 HIV-specific CD8+ T-cell responses, we found the TemRA phenotype comprised more than or equal to 20% of the Tmem repertoire; and in 2 of these subjects, we observed TemRA populations made up more than or equal to 50% of the total Tmem repertoire (Figure 1C). These findings indicate that CD8+ Tmem differentiation in HIV-1 infection is biased toward effector-memory phenotypes, with an over-representation of cells with TemRO phenotype on HIV-specific CD8+ T cells. Some HIV-1-seropositive subjects have substantial proportions of HIV-specific TemRA phenotype CD8+ T cells, indicating that impaired TemRA differentiation does not apply to all HIV-specific CD8+ T-cell responses.
Figure 1.
Expansions of HIV-specific TemRA phenotype cells are present to various degrees in chronic infection. (A) Representative staining of CCR7 and CD45RO on CD8+ T cells in 2 representative HIV-seropositive persons. (B) Identification of the HIV-1-tetramer+ cells (red) and their corresponding Tmem phenotype (overlay: red tet+, black CD8+ T cells). (C) Comparison of effector-memory populations (paired t test) on CD8+ T cells and on HIV-1-tetramer+ T cells.
Table 1.
Demographic data of the cohort
| Patient no. | Year of diagnosis | CD4 count | Log viral load | HLA | Tetramer response studied | Antiretroviral therapy |
|---|---|---|---|---|---|---|
| 10001 | 2002 | 598 | 1.7 | A*02,*02, B*14, *15 | B15-TY11, B15-GY10 | AZT, 3TC, LPV/r |
| 10002 | 1984 | 1026 | 1.8 | A*03,*31, B*27, *57 | B27-KK10, B57-QW9, A3-RK9 | None |
| 10004 | 1984 | 360 | 2.4 | A*03,*30, B*07, *57 | B57-KF11 | None |
| 10015 | 2003 | 1080 | 4.4 | A*01, *03, B*08, *52 | B8-FL8 | None |
| 10022 | 1993 | 573 | 2.5 | A*01, *31, B*08, *27 | B27-KK10, B8-FL8, B8-EI8 | None |
| 10027 | 1993 | 775 | 3.3 | A*01, *02, B*08, *57 | B57-KF11, B8-FL8 | None |
| 10031 | 1997 | 612 | 1.7 | A*03, *03, B*14, *57 | A3-RK9, B15-QW9 | None |
| 10040 | 1991 | 1161 | 1.7 | A*01, *31, B*44, *57 | B57-KF11, B57-QW9 | None |
| 10060 | 2002 | 1085 | 2.9 | A*01, *33, B*42, *57 | B57-KF11 | None |
| 10067 | 1991 | 661 | 2.3 | A*30. *33, B*13, *57 | B57-KF11 | None |
| 10070 | 1996 | 950 | 3.8 | A*23, *74, B*57, *58 | B57-KF11, B57-QW9 | None |
| 10071 | 1992 | 658 | 1.7 | A*01, *66, B*08, *57 | B8-FL8, B57-KF11 | None |
| 10074 | 1999 | 210 | 1.7 | A*01, *26, B*08, *57 | B8-FL8, B57-KF11 | None |
| 10076 | 1999 | 700 | 4.3 | A*02, *30, B*35, *57 | B57-KF11 | None |
| 20011 | 2004 | 570 | 3.51 | A*01, *03, B *08, *08 | B8-EI8 | None |
| 20018 | 2004 | 1353 | 3.52 | A*02, *26, B*40, *57 | B57-KF11 | None |
CD4 count data are reported as cells/mm3.
Analysis of the TCR repertoire and quantification of the clonotype distribution between different effector-memory populations
In chronic viral infection models, effector-memory differentiation of CD8+ T cells could involve simultaneous differentiation of all T-cell clones within an epitope-specific cytotoxic T lymphocyte response. Alternatively, differentiation could be asynchronous and restricted to distinct cytotoxic T lymphocyte clones, which evolve separately into the different effector-memory compartments. It is currently not known whether effector-memory T cells develop from precursors, such as naive and/or Tcm, directly into either the TemRO or TemRA population or whether individual T-cell clones transition between the TemRA and TemRO phenotypes. Each model would predict substantial differences in the composition of the TCR repertoires within the different effector-memory populations; for instance, a highly similar TCR repertoire in each effector-memory compartment would imply simultaneous differentiation of T cells, whereas distinct TCR repertoires in the TemRO and TemRA compartments would imply asynchronous T-cell differentiation.
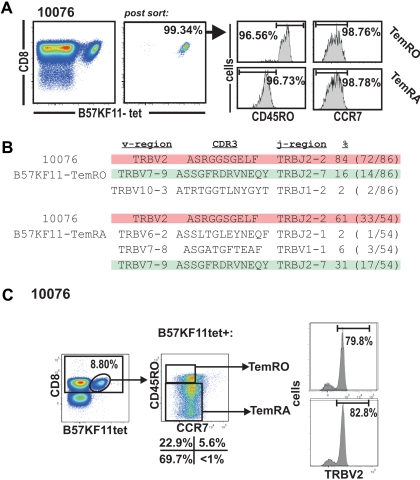
To investigate the structural relationship between effector-memory compartments, we compared the TCR repertoire of sorted HIV-specific Tem populations (> 95% purity) by direct TCR sequencing (Figure 2A-B) as well as by analysis of T-cell receptor-β variable region (TRBV) expression by costaining with TRBV-specific monoclonal antibodies (Figure 2C). Purification of HIV-specific TemRO and TemRA subpopulations and subsequent sequencing of their TCR repertoire revealed identical clonotypes within the distinct Tem populations (Figure 3). The B57KF11 tetramer+ population of subject 10076 is dominated by 2 distinct clonotypes. The TRBV2 clonotype composes 85% of the TemRO tetramer+ population and 61% of the TemRA tetramer+ population, whereas the TRBV7 clonotype composes 16% of the tetramer+ TemRO population and 31% of the tetramer+ TemRA population (Figure 2B). In this case, we did not consider the frequency of clonotypes to be substantially different between memory populations, but the presence of these dominant TCR clonotypes within each memory phenotype population suggests that T-cell clonotypes do not differentiate into discrete memory compartments. This study of the structural composition of the TCR repertoire allowed us to identify TRBV usage of discrete clonotypes within a tetramer+ population. We next stained samples with monoclonal antibodies specific for distinct TRBV regions, which allowed us to quantify the clonal distribution between distinct effector-memory populations, thus avoiding potential PCR bias by sequencing (Figure 2C). The most prevalent clone within the B57KF11 tetramer+ CD8+ T-cell population could be identified by TRBV2 expression (Figures 2B, 3). This population was evenly distributed between TemRO and TemRA phenotype cells. Thus, a combination of sequencing of purified populations and TRBV staining allows a quantitative analysis of the clonal distribution within distinct HIV-specific Tem subsets.
Figure 2.
Analysis of effector-memory clonotype composition on HIV-specific effector-memory subsets. (A) Purification of B57-KF11 tetramer+ CD8+ T cells based on the expression of CCR7 and CD45RO in subject 10076. (B) Sequence analysis of the purified Tmem populations. (C) Analysis of the CCR7/CD45RO expression on B57-KF11 tetramer+ CD8+ T-cell expression TRBV2.
Figure 3.
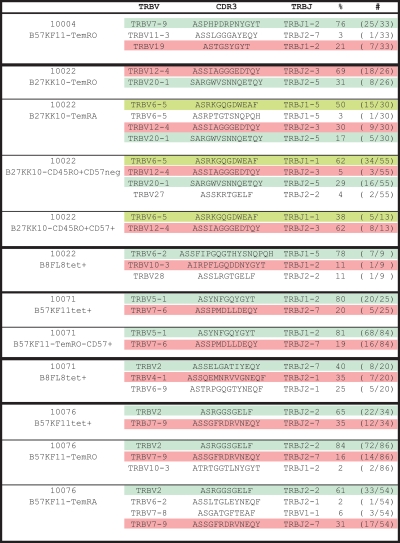
TCR sequences of directly sorted total tetramer+ CD8+ T cells, or tetramer+ cells sorted by effector-memory phenotype and/or CD57 expression.
Clonotype-specific expression of TemRA-phenotype cells on tetramer+ cells indicates TCR-driven effector-memory differentiation of HIV-specific CD8+ T cells
Differentiation of HIV-specific CD8+ T cells to a TemRA phenotype could involve a fraction of each clonotype within an HIV-1-epitope–specific CD8+ T-cell response or could be restricted to distinct clonotypes. Similar expression of CD45RA on all clonotypes would imply a synchronous expression of Tem phenotype marker, which is independent of the TCR/peptide major histocompatibility complex (pMHC) interaction. In contrast, expression of CD45RA+ on distinct clonotypes within an HIV-1–specific Tem population, where other clonotypes fail to express CD45RA, indicates a TCR-driven process of Tem differentiation, which may not apply to all clonotypes within an HIV-epitope–specific CD8+ T-cell response.
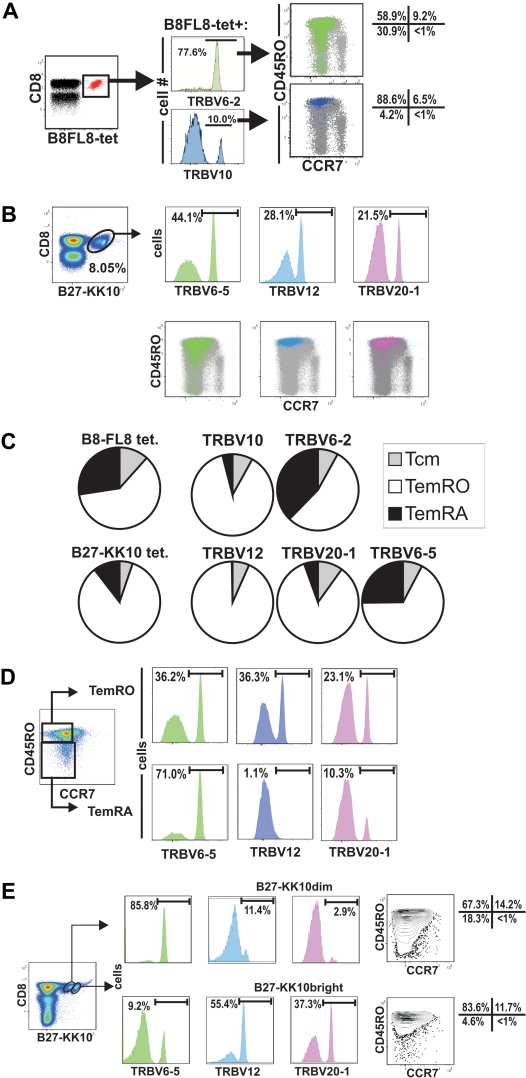
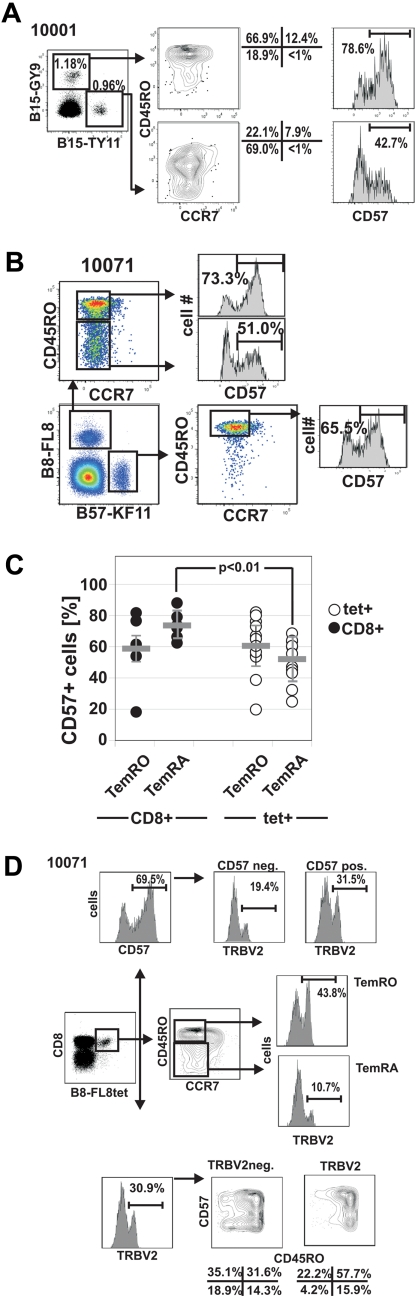
In subject 10022, the B8FL8-tet+ population is made up of 3 clonotypes, each expressing a different TRBV (Figures 3, 4A). The majority (78%) of tetramer+ CD8+ T cells express TRBV6-2, whereas a minority (10%) express TRBV10 (Figure 4A). Simultaneous tetramer, TRBV, and memory phenotype staining showed that 31% of the TRBV6-2 population had a TemRA phenotype, compared with 4% of the TRBV10 population.
Figure 4.
Selective expansions of distinct HIV-epitope–specific clonotypes into terminally differentiated effector-memory phenotypes (CCR7−/CD45RA+) and association of B27-KK10 tetramer binding with expression of distinct clonotypes and effector-memory differentiation. (A) Analysis of the TRBV populations representing single clonotypes of B8-FL8 tetramer+ cells by flow cytometry. (B) TRBV usage of distinct clonotypes and CCR7/CD45RO expression on B27-KK10 tetramer cells in subject 10022. (C) Pie charts comparing the relative T-cell memory distribution of B8-FL8+ and B27-KK10+ clonotypes identified by TRBV expression in subject 10022. (D) Frequency of clonotypes within the B27-KK10 tetramer+ population in subject 10022. (E) TRBV usage and CCR7/CD45RO expression on B27-KK10 tetramer dim and bright cells in subject 10022.
As we analyzed the clonal composition on another epitope (B27-KK10) in subject 10022, we identified 3 clonotypes, which composed the majority of this HIV-epitope–specific response. Namely, TRBV6-5, TRBV12, and TRBV 20-1 represented 44.1%, 28.1%, and 21.5%, respectively, of the B27-KK10 specific T cells (Figure 4B). Again, there were major differences between the different clonotypes regarding the differentiation into the TemRA phenotype ranging from less than 1% (TRBV12) up to 25% (TRBV6-5; Figure 4C). Although “only” 25% of TRBV6-5 displays a TemRA phenotype, it is important to note that this clonotype composes 71.0% of the TemRA population (Figure 4D).
Tetramer staining of the B27-KK10 epitope of subject 10022 revealed 2 cell populations with different magnitudes of tetramer binding (Figure 4B,D-E). Costaining with TRBV antibodies revealed a different distribution of TCR clonotypes within each tetramer+ population (Figure 4B). Namely, we found the TRBV6-5 clonotype made up the majority of the B27-KK10dim population (85.8%), whereas TRBV12 and TRBV20-1 dominated the B27-KK10bright population (Figure 4E). Approximately 15% of the overall B27-KKtet+ population displayed the TemRA phenotype, and a combination of staining with TRBV, CCR7, and CD45RO antibodies revealed that differentiation to the TemRA phenotype was mainly restricted to the TRBV6-5 population (Figure 4D).
Subject 10071 also recognizes the B8FL8 epitope; likewise, 3 clonotypes, each expressing a different TRBV gene, make up this response (Figure 3; supplemental Figure 1, available on the Blood Web site; see the Supplemental Materials link at the top of the online article). A total of 46% of tetramer+ CD8+ T cells express TRBV2 and 7% express TRBV4-1. As in subject 10022, there was differential maturation of clonotypes within this response. A total of 11% of the TRBV2+ cells had a TemRA phenotype, compared with 44% of the TRBV4-1+ cells. We costained tetramer and TRBV clonotypes simultaneously with anti-CD45RO and anti-CD45RA antibodies and confirmed that there was no substantial coexpression of the 2 isoforms at the clonotype level (supplemental Figure 1). To further confirm that clonotype differentiation into TemRA is defined by CD45RO down-regulation, we measured and compared the frequencies of TRBV populations in CD45ROintermediate populations with those in CD45ROhigh or CD45RO− populations (supplemental Figure 1). Clonotypes that differentiated into the TemRA phenotype were more prominent in the CD45RO− population compared with the CD45ROintermediate population, indicating that TemRA differentiation includes loss of CD45RO expression.
Thus, differentiation into the TemRA phenotype was restricted to distinct clonotypes and, in one subject, to different levels of tetramer binding ex vivo, which indicates that effector-memory differentiation can have high variability at the clonal level. In summary, these data indicate that the composition of the CD8+ Tmem phenotype is driven by selective expansion of distinct TCR clonotypes within individual HIV-specific CD8+ T memory populations. These differences could be explained either by a different pace of Tem differentiation or by failure to differentiate into TemRA at the clonotype level.
TemRO phenotype cells are most frequent population at the clonotype level within HIV-epitope specific CD8+ T-cell responses
We extended the analysis of the relative clonal distribution within the HIV-specific effector-memory repertoire to a total of 11 different HIV-specific CD8+ T-cell responses in 9 subjects in whom we analyzed 24 clonotypes (supplemental Figure 2). The frequency of TemRA phenotype within a distinct HIV-specific clonotype ranged between less than 1% (eg, 10004, B57-QW57, TRBV28, and TRBV3-2) to more than 70% (eg, 10076, B57-KF11, and TRBV2). Importantly, the frequency of the TemRO phenotype, which rarely decreases below 50% (5 of 24 clonotypes) and was never below 20%, contrasts with the finding for the TemRA phenotype where we observed a TemRA frequency of less than 10% in 15 of 24 clonotypes (supplemental Figure 2), indicating that HIV-specific effector-memory without a substantial TemRO phenotype is not generated in vivo. Within intraindividual T-cell clones recognizing the same HIV epitope, there was a wide variation of the frequency of TemRO and TemRA phenotypes. We could not detect a pattern for the TemRA phenotype differentiation of HIV-specific T-cell clones. For example, we did not find that the dominant T-cell clonotype within an HIV-epitope–specific response was more or less prone to differentiate into a TemRA phenotype, nor did we find that HIV-epitope–specific responses with a high frequency in the peripheral blood were more prone to contain TemRA phenotype cells.
Differential expression of CD57 on HIV-specific CD8+ effector-memory populations
The data in Figures 3 and 4 indicate selective TemRA differentiation of distinct HIV-specific CD8+ T-cell clones. CD57 is another differentiation marker that has been associated with the proliferative history of individual clonotypes and with replicative senescence.29,35 To understand whether effector-memory differentiation is associated with altered expression of CD57, we compared the expression of CD57 on HIV-1 tetramer+ CD8+ T cells with different CD8+ Tmem phenotypes (Figure 5). We observed variable levels of CD57 expression between different HIV-specific CD8+ T-cell responses, even within the same subject as shown for B15-TY11 and B15-GY9 in subject 10001 (Figure 5A), and there was considerable variation of CD57 expression on HIV-specific CD8+ T-cell responses ranging from 22.3% for B57-KF11 in subject 20018 up to 85.1% for A3-RK9 in subject 10031 (supplemental Figure 3). We then analyzed CD57 expression on TemRO and TemRA phenotype cells, recognizing the same HIV-1-epitope as shown for subject 10071 who recognizes B8-FL8 (HIV-1 nef 90-97, FLKEKGGL; Figure 5B). Overall, the expression of CD57 was very similar on HIV-specific TemRO and TemRA populations (52.0% ± 13.0% vs 60.5% ± 17.2%; P = .204), indicating a similar fraction of senescent cells in both effector-memory subsets (Figure 5C). However, as we compared CD57 expression of different effector-memory subsets between HIV-specific tetramer+ T cells and total CD8+ T cells, we observed that HIV-specific TemRA phenotype cells had significantly lower CD57 expression compared with total TemRA CD8+ T cells (52.04% ± 13.01% vs 73.52% ± 7.82%; P < .01; Figure 5C). This difference in the frequency of CD57 expression offers an insight into the distribution of memory phenotypes and suggests that, although HIV-specific TemRA cells are relatively under-represented in HIV-1 infection, when present these cells exhibit a phenotype suggesting continued potential for expansion.
Figure 5.
Differential CD57 expression on HIV-1-tetramer+ CD8+ T-cell memory subsets. (A) CD57 expression on 2 HIV-epitope specific CD8+ T-cell responses in subject 10001. (B) Lower expression of CD57 on human leukocyte antigen B8-FL8+ TemRA phenotype cells compared with TemRO phenotype cells in subject 10071. (C) Summary of CD57 expression on different memory populations on total CD8+ T cells and HIV-specific CD8+ T cells. (D) Distribution of CD57 on different memory subsets on TRBV+ and TRBV− HIV-specific CD8+ T cells in subject 10071.
We next wished to determine whether there was evidence that CD57 expression was related to the level of Tem differentiation as determined by CCR7/CD45RO expression. To address this question, we performed TCR sequencing of HIV tetramer+ CD57− and CD57+ subpopulations (Figure 3) as well as TRBV staining in combination with CD57 and CCR7/CD45RO on tetramer+ CD8+ T-cell populations (Figure 5D; supplemental Figure 4).
In contrast to differentiation into TemRA cells, where we frequently observed skewed maturation of distinct clonotypes, expression of CD57 was more often equally distributed among clonotypes within an HIV-1-epitope–specific CD8+ T-cell response as shown for the representative subjects 10071 (Figure 5D), 10022 (supplemental Figure 4A-B), and 10004 (supplemental Figure 4C).
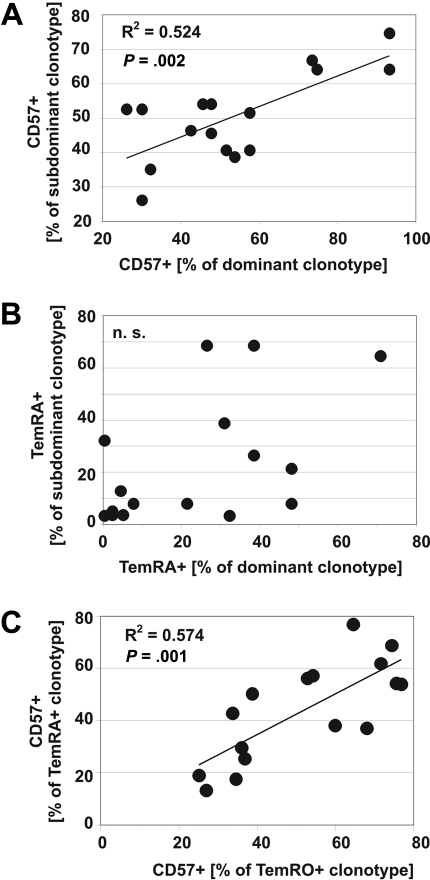
To determine the relationship between CD57 expression and effector-memory differentiation at the clonotype level in more detail, we compared the frequency of CD57 expression between 16 dominant (most frequent) and 16 subdominant (less frequent) T-cell clonotypes recognizing 12 HIV epitopes in 9 HIV-seropositive patients. If one assumes a similar distribution of CD57 expression on each clonotype within each HIV epitope response, a positive correlation would be expected, which was the case for CD57 expression (Figure 6A). In contrast, as we analyzed the degree of TemRA differentiation between T-cell clonotypes, we found no correlation (Figure 6B). This analysis confirmed our observations that, although individual clonotypes are commonly skewed toward different degrees of memory differentiation, CD57 expression on different T-cell clonotypes within a response are highly correlated (Figure 6). In addition, there was a strong positive correlation between the frequency of CD57 expression on the same clonotypes expressing either TemRO or TemRA phenotype (Figure 6C), indicating that TemRA differentiation does not result in substantial alterations of CD57 expression within the same clonotype. These data indicate that TemRA differentiation, as determined by CCR7/CD45RO expression, is not associated with distinct patterns of CD57 expression at the clonal level and therefore argue against replicative senescence29 as an explanation for the relative absence of specific clonotypes within the terminally differentiated memory population. Indeed, the relatively low expression of CD57 on “terminally differentiated” clonotypes indicates the proliferative potential of these cells and provides a mechanism by which these cells are maintained in vivo. In addition, these data suggest that the expression of CD57 is independent of the TemRO/TemRA differentiation pathway.
Figure 6.
Different expression patterns of CD57 and effector-memory marker on HIV-specific clonotypes. Relative distribution of CD57+ (A) and TemRA-phenotype cells (B) between dominant (most frequent) clonotypes and subdominant (less frequent) clonotypes within HIV-epitope–specific CD8+ T-cell responses. (C) Relationship of CD57+ expression on the same clonotype expressing either TemRO or TemRA phenotype marker.
Discussion
In this study, we demonstrate that, in subjects with chronic HIV-1 infection, most of them with slow disease progression, individual T-cell clonotypes within an epitope-specific CD8+ T-cell response can exhibit different degrees of effector-memory differentiation at the clonotype level. We defined effector-memory differentiation by the expression of either CD45RO or CD45RA on CCR7− CD8+ T cells according to the original characterization of T-cell differentiation described by Sallusto et al.24,36 Different models of CD8+ T-cell memory differentiation in the context of viral infections have been previously described.12,37 Chronic viral infections with persistent viruses are characterized by severely impaired T-cell function and highly skewed patterns of effector-memory differentiation that differ between different viral infections. HIV-specific T cells preferentially accumulate as preterminally differentiated cells (CD45RO+/CCR7−; TemRO) or CD27+/CD28− (intermediate) phenotype cells.25,26,38 The interpretation of these data has typically described the accumulation of TemRO cells in HIV-1 infection as a “block” toward terminal differentiation. Subsequent studies have demonstrated that other markers, such as CD57, are more predictive of the ability of cells to proliferate in response to cognate peptide.29 Nevertheless, studies of chronic HIV-1 infection consistently demonstrate a relative absence of HIV-specific TemRA+ CD8+ T cells.
Previous studies have analyzed TCR expression of virus-specific CD8+ T-cell clones and their contribution to the memory repertoire in humans.39–41 However, some of the studies sequenced in vitro expanded virus-specific CD8+ T-cell clones and are thus restricted to clones with proliferative capacity. Vargas et al investigated the relationship between CD45 isoform expression and TRBV expression by antibody staining and found that different TRBV populations might differ in their CD45 isoform expression patterns.42 However, no sequence analysis of the TRBV populations was performed, so it is difficult to draw any conclusion regarding the clonal composition of the TCR repertoire.
Through detailed T-cell sorting and TRBV antibody staining, we have been able to characterize the clonotypic makeup of epitope-specific T cells at several stages of memory T-cell maturation. Our data confirm a dominant HIV-specific TemRO phenotype in CD8+ T cells in the majority of our study subjects. However, we observed a significant proportion of HIV-specific CD8+ T cells with a terminally differentiated effector-memory phenotype in several subjects (Figure 1). It has been hypothesized that high antigen load might serve as an explanation for lack of terminal differentiation on HIV-1-infected subjects.43 Our subjects were generally healthy with CD4 counts more than 350, with low levels of viremia, and, with the exception of one subject, were off antiretroviral therapy. The frequent occurrence of HIV-specific TemRA phenotype cells is in accordance with studies demonstrating an over-representation of these cells in subjects with slow disease progression.28 We observed that CD8+ T-cell responses targeting different epitopes within the same subjects had differing frequencies of cells with TemRA and TemRO phenotype, indicating substantial intraindividual heterogeneity of differentiation status in HIV-1-seropositive subjects.
As we analyzed the structural composition of HIV-specific CD8+ T-cell responses, it became evident that, in the majority of persons, distinct clonotypes preferentially displayed the TemRA+ phenotype (Figures 5, 6). This indicates that maturation to the TemRA phenotype may be influenced by the TCR/pMHC interaction and may therefore apply only to a fraction of the antigen-specific TCR repertoire. This may be the result of differential fine specificity of each TCR for variant epitopes10 or may reflect different TCR/pMHC affinities of distinct clonotypes to the same HIV epitope as we have demonstrated recently.5
It should be emphasized that HIV-specific CD8+ T-cell responses in which a substantial fraction of TemRA phenotypes cells were present still had a significant proportion of CD8+ TemRO phenotype cells (Figures 2, 5). Sequence analysis of TemRA and TemRO cells revealed that identical clonotypes were present in both effector-memory populations. These data support a strong structural relationship between TemRO and TemRA cells and indicate that the TemRA/TemRO differentiation pathways are intertwined such that a fraction of an expanded T-cell clone might differentiate into a TemRA phenotype, whereas a significant proportion might still maintain a TemRO phenotype, which has been previously described in HCMV infection.40 If maturation proceeded directly from the naive T cells or Tcm population into TemRA, one would expect that a proportion of clonotypes should be present in TemRA but not in TemRO. However, our data do not exclude the possibility that TemRA cells could revert to a TemRO state.40,44,45 Indeed, it has suggested that CD8+ TemRA cells might represent a subset of resting memory cells, which have not encountered antigen for some time but might reexpress CD45RO on antigenic stimulation.45
The presence of distinct clonotypes within persons, each with differing degrees of maturation, and the finding that terminal differentiation is not restricted to distinct epitopes support a model where individual TCR/pMHC interactions may be important for effector-memory differentiation as defined by the expression of CD45RO/RA on CCR7− CD8+ T cells. This is also supported by our more detailed analysis of the TCR composition, which demonstrates that preferential maturation takes place at the clonotype level within an epitope-specific response and confirms a central role of the TCR/pMHC interaction for the differentiation of epitope-specific T cells. Maintenance of functional heterogeneity of T cells expressing the same TCR has been hypothesized to complement the benefits of TCR structural diversity to fight pathogens46; however, so far evidence for the generation of functional diversity of individual T-cell clones generated in vivo during chronic viral infections in humans has been lacking.
The presence of “terminally differentiated” HIV-specific CD8+ T cells, as defined by the CCR7−/CD45RA+, in subjects with slow HIV-1 disease progression28 could be considered contradictory to the observed proliferative capacity in these persons.47 The current model of T-cell maturation would predict that “terminal differentiation” as defined by these markers is inevitably associated with lack of proliferation.48 CD57 is a marker for senescent T cells that has recently been proposed to be a superior marker for the identification of terminally differentiated effector-memory cells that lack proliferative capacity.29 We observed CD57 expression on HIV-specific CD8+ T cells with both the TemRO and TemRA phenotype (Figures 3, 5, and 6). The level of CD57 expression was similar on both populations with slightly, but not significantly, decreased expression on HIV-specific CD8+ TemRA+ T cells compared with TemRO phenotype cells (Figure 3D). Interestingly, compared with total CD8+ TemRA cells, CD57 expression on HIV-specific CD8+ T cells with the TemRA phenotype was significantly reduced in HIV-1-seropositive subjects, indicating increased proliferative capacity of HIV-specific CD8+ T cells compared with total CD8+ T cells with the TemRA phenotype. The presence of substantial populations of CD57− cells among terminally differentiated T-cell clonotypes provides a mechanism by which these cells are able to expand and persist in vivo, and our finding indicates that terminal differentiation, which has been observed in subjects with slow HIV-1-disease progression, is not in contradiction with the presence of strong proliferative responses in these persons.47 Our data are most consistent with the hypothesis that alteration of the maturation status of HIV-specific T cells in HIV-1-infection might not necessarily be caused by a “block” in maturation but by an altered redistribution caused by the high turnover of HIV-specific CD8+ T cells in response to persistent infection.49,50 This would explain the heterogeneous expression of CD57, with CD57low and CD57bright populations on HIV-specific TemRA phenotype cells, and the observation that TemRA differentiation was restricted to distinct clonotypes. CD57 expression has been shown to be associated with telomere lengths and reflects the “replicative history” at the clonal level.35 Our findings probably reflect the selective clonotype expansions and contractions within the HIV-specific CD8+ TCR repertoire, which we described previously.5
Despite our observation that some HIV-specific immune responses preferentially demonstrated terminal differentiation as defined by the expression of CD45RA on CCR7 negative CD8+ T cells at the clonotype level, CD57 expression was evenly distributed among individual clonotypes within the epitope-specific responses. In contrast to the TemRA differentiation where we observed frequent clonal expansions, there was a strong correlation of the CD57 expression between different clonotypes within the same epitope-specific responses. CD57 expression could differ between epitope-specific responses in a person, but not between the clonotypes within each of these responses. These data indicate distinct differentiation pathways between the TCR-independent regulation of CD57 and the TCR-driven differentiation of expression of CD45RA within antigen-specific responses. Our data suggest that TCR-dependent and TCR-independent differentiation pathways at the clonotype level shape the structural and functional diversity of the pathogen-specific TCR repertoire in chronic viral infections. Future studies of epitope-specific T cells at the phenotypic and clonotypic level will help determine the relationship between T-cell maturation, the clonotypic makeup of the TCR repertoire, and the ability of T cells to control viremia over the course of chronic infection.
Supplementary Material
Acknowledgments
The authors thank Michael Vetter and Lorraine Sutton for their help with TCR sequencing.
This work was supported by the Bundesministerium für Bildung und Forschung/Kompetenznetz HIV (DMO/01Kl0501), Helmholtz-Zentrum für Infektionsforschung (DMO/IG-SCID-TwinPro-02), the European AIDS Treatment Network NEAT (DMO/WP3), the National Institutes of Health (SAK R01AI39966), and the Vanderbilt-Meharry Center for AIDS Research Immunopathogenesis Core Facility (2P30AI054999). S.A.K. is an Elizabeth Glaser Scientist of the Elizabeth Glaser Pediatric AIDS Foundation.
Footnotes
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Authorship
Contribution: D.M.-O. designed and performed experiments, analyzed the data, and wrote the manuscript; B.C.S., J.A.C., R.M.S., L.B., S.L.L., C.B.D., and R.R. performed experiments; and S.A.K. designed experiments in the study, analyzed the data, and wrote the manuscript.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Spyros A. Kalams, Vanderbilt University Medical Center, 1211 Medical Center Dr, MCN A2207, Nashville, TN 37232; e-mail: spyros.a.kalams@vanderbilt.edu; or Dirk Meyer-Olson, Medizinische Hochschule Hannover, Carl-Neuberg-Str 1, 30625 Hanover, Germany; e-mail: meyer.dirk@mh-hannover.de.
References
- 1.Koup RA, Safrit JT, Cao Y, et al. Temporal association of cellular immune responses with the initial control of viremia in primary human immunodeficiency virus type 1 syndrome. J Virol. 1994;68(7):4650–4655. doi: 10.1128/jvi.68.7.4650-4655.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kalams SA. Cellular immunity for prevention and clearance of HIV infection. Curr Mol Med. 2003;3(3):195–208. doi: 10.2174/1566524033479807. [DOI] [PubMed] [Google Scholar]
- 3.Shoukry NH, Cawthon AG, Walker CM. Cell-mediated immunity and the outcome of hepatitis C virus infection. Annu Rev Microbiol. 2004;58:391–424. doi: 10.1146/annurev.micro.58.030603.123836. [DOI] [PubMed] [Google Scholar]
- 4.Meyer-Olson D, Shoukry NH, Brady KW, et al. Limited T-cell receptor diversity of HCV-specific T-cell responses is associated with CTL escape. J Exp Med. 2004;200(3):307–319. doi: 10.1084/jem.20040638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Meyer-Olson D, Brady KW, Bartman MT, et al. Fluctuations of functionally distinct CD8+ T-cell clonotypes demonstrate flexibility of the HIV-specific TCR repertoire. Blood. 2006;107(6):2373–2383. doi: 10.1182/blood-2005-04-1636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Price DA, Brenchley JM, Ruff LE, et al. Avidity for antigen shapes clonal dominance in CD8+ T cell populations specific for persistent DNA viruses. J Exp Med. 2005;202(10):1349–1361. doi: 10.1084/jem.20051357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Price DA, West SM, Betts MR, et al. T Cell receptor recognition motifs govern immune escape patterns in acute SIV Infection. Immunity. 2004;21(6):793–803. doi: 10.1016/j.immuni.2004.10.010. [DOI] [PubMed] [Google Scholar]
- 8.Yu XG, Lichterfeld M, Chetty S, et al. Mutually exclusive T-cell receptor induction and differential susceptibility to human immunodeficiency virus type 1 mutational escape associated with a two-amino-acid difference between HLA class I subtypes. J Virol. 2007;81(4):1619–1631. doi: 10.1128/JVI.01580-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lichterfeld M, Yu XG, Mui SK, et al. Selective depletion of high-avidity human immunodeficiency virus type 1 (HIV-1)-specific CD8+ T cells after early HIV-1 infection. J Virol. 2007;81(8):4199–4214. doi: 10.1128/JVI.01388-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kalams SA, Johnson RP, Dynan MJ, et al. T-cell receptor usage and fine specificity of human immunodeficiency virus 1-specific cytotoxic T lymphocyte clones: analysis of quasispecies recognition reveals a dominant response directed against a minor in vivo variant. J Exp Med. 1996;183(4):1669–1679. doi: 10.1084/jem.183.4.1669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lanzavecchia A, Sallusto F. Understanding the generation and function of memory T cell subsets. Curr Opin Immunol. 2005;17(3):326–332. doi: 10.1016/j.coi.2005.04.010. [DOI] [PubMed] [Google Scholar]
- 12.Kaech SM, Wherry EJ. Heterogeneity and cell-fate decisions in effector and memory CD8+ T cell differentiation during viral infection. Immunity. 2007;27(3):393–405. doi: 10.1016/j.immuni.2007.08.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Masopust D, Kaech SM, Wherry EJ, Ahmed R. The role of programming in memory T-cell development. Curr Opin Immunol. 2004;16(2):217–225. doi: 10.1016/j.coi.2004.02.005. [DOI] [PubMed] [Google Scholar]
- 14.Williams MA, Tyznik AJ, Bevan MJ. Interleukin-2 signals during priming are required for secondary expansion of CD8+ memory T cells. Nature. 2006;441(7095):890–893. doi: 10.1038/nature04790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Selin LK, Cornberg M, Brehm MA, et al. CD8 memory T cells: cross-reactivity and heterologous immunity. Semin Immunol. 2004;16(5):335–347. doi: 10.1016/j.smim.2004.08.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Williams MA, Holmes BJ, Sun JC, Bevan MJ. Developing and maintaining protective CD8+ memory T cells. Immunol Rev. 2006;211:146–153. doi: 10.1111/j.0105-2896.2006.00389.x. [DOI] [PubMed] [Google Scholar]
- 17.Joshi NS, Kaech SM. Effector CD8 T cell development: a balancing act between memory cell potential and terminal differentiation. J Immunol. 2008;180(3):1309–1315. doi: 10.4049/jimmunol.180.3.1309. [DOI] [PubMed] [Google Scholar]
- 18.Thomas ML. The leukocyte common antigen family. Annu Rev Immunol. 1989;7:339–369. doi: 10.1146/annurev.iy.07.040189.002011. [DOI] [PubMed] [Google Scholar]
- 19.Bell EB. Function of CD4 T cell subsets in vivo: expression of CD45R isoforms. Semin Immunol. 1992;4(1):43–50. [PubMed] [Google Scholar]
- 20.Clement LT. Isoforms of the CD45 common leukocyte antigen family: markers for human T-cell differentiation. J Clin Immunol. 1992;12(1):1–10. doi: 10.1007/BF00918266. [DOI] [PubMed] [Google Scholar]
- 21.Tchilian EZ, Wallace DL, Wells RS, Flower DR, Morgan G, Beverley PC. A deletion in the gene encoding the CD45 antigen in a patient with SCID. J Immunol. 2001;166(2):1308–1313. doi: 10.4049/jimmunol.166.2.1308. [DOI] [PubMed] [Google Scholar]
- 22.Tchilian EZ, Wallace DL, Dawes R, et al. A point mutation in CD45 may be associated with an increased risk of HIV-1 infection. AIDS. 2001;15(14):1892–1894. doi: 10.1097/00002030-200109280-00024. [DOI] [PubMed] [Google Scholar]
- 23.Tchilian EZ, Beverley PC. Altered CD45 expression and disease. Trends Immunol. 2006;27(3):146–153. doi: 10.1016/j.it.2006.01.001. [DOI] [PubMed] [Google Scholar]
- 24.Sallusto F, Lenig D, Forster R, Lipp M, Lanzavecchia A. Two subsets of memory T lymphocytes with distinct homing potentials and effector functions. Nature. 1999;401(6754):708–712. doi: 10.1038/44385. [DOI] [PubMed] [Google Scholar]
- 25.Appay V, Dunbar PR, Callan M, et al. Memory CD8+ T cells vary in differentiation phenotype in different persistent virus infections. Nat Med. 2002;8(4):379–385. doi: 10.1038/nm0402-379. [DOI] [PubMed] [Google Scholar]
- 26.Champagne P, Ogg GS, King AS, et al. Skewed maturation of memory HIV-specific CD8 T lymphocytes. Nature. 2001;410(6824):106–111. doi: 10.1038/35065118. [DOI] [PubMed] [Google Scholar]
- 27.Lieberman J, Trimble LA, Friedman RS, et al. Expansion of CD57 and CD62L-CD45RA+ CD8 T lymphocytes correlates with reduced viral plasma RNA after primary HIV infection. AIDS. 1999;13(8):891–899. doi: 10.1097/00002030-199905280-00004. [DOI] [PubMed] [Google Scholar]
- 28.Addo MM, Draenert R, Rathod A, et al. Fully differentiated HIV-1 specific CD8+ T effector cells are more frequently detectable in controlled than in progressive HIV-1 infection. PLoS ONE. 2007;2:e321. doi: 10.1371/journal.pone.0000321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Brenchley JM, Karandikar NJ, Betts MR, et al. Expression of CD57 defines replicative senescence and antigen-induced apoptotic death of CD8+ T cells. Blood. 2003;101(7):2711–2720. doi: 10.1182/blood-2002-07-2103. [DOI] [PubMed] [Google Scholar]
- 30.Chattopadhyay PK, Betts MR, Price DA, et al. The cytolytic enzymes granyzme A, granzyme B, and perforin: expression patterns, cell distribution, and their relationship to cell maturity and bright CD57 expression. J Leukoc Biol. 2009;85(1):88–97. doi: 10.1189/jlb.0208107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hong HS, Eberhard JM, Keudel P, et al. HIV infection is associated with a preferential decline in less-differentiated CD56dim CD16+ NK cells. J Virol. 2010;84(2):1183–1188. doi: 10.1128/JVI.01675-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Mollet L, Sadat-Sowti B, Duntze J, et al. CD8hi+CD57+ T lymphocytes are enriched in antigen-specific T cells capable of down-modulating cytotoxic activity. Int Immunol. 1998;10(3):311–323. doi: 10.1093/intimm/10.3.311. [DOI] [PubMed] [Google Scholar]
- 33.Meyer-Olson D, Brady KW, Blackard JT, et al. Analysis of the TCR beta variable gene repertoire in chimpanzees: identification of functional homologs to human pseudogenes. J Immunol. 2003;170(8):4161–4169. doi: 10.4049/jimmunol.170.8.4161. [DOI] [PubMed] [Google Scholar]
- 34.Giudicelli V, Duroux P, Ginestoux C, et al. IMGT/LIGM-DB, the IMGT comprehensive database of immunoglobulin and T-cell receptor nucleotide sequences. Nucleic Acids Res. 2006;34:D781–D784. doi: 10.1093/nar/gkj088. (Database issue) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Monteiro J, Batliwalla F, Ostrer H, Gregersen PK. Shortened telomeres in clonally expanded CD28-CD8+ T cells imply a replicative history that is distinct from their CD28+CD8+ counterparts. J Immunol. 1996;156(10):3587–3590. [PubMed] [Google Scholar]
- 36.Sallusto F, Geginat J, Lanzavecchia A. Central memory and effector-memory T cell subsets: function, generation, and maintenance. Annu Rev Immunol. 2004;22:745–763. doi: 10.1146/annurev.immunol.22.012703.104702. [DOI] [PubMed] [Google Scholar]
- 37.Wherry EJ, Ahmed R. Memory CD8 T-cell differentiation during viral infection. J. Virol. 2004;78(11):5535–5545. doi: 10.1128/JVI.78.11.5535-5545.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ellefsen K, Harari A, Champagne P, Bart PA, Sekaly RP, Pantaleo G. Distribution and functional analysis of memory antiviral CD8 T-cell responses in HIV-1 and cytomegalovirus infections. Eur J Immunol. 2002;32(12):3756–3764. doi: 10.1002/1521-4141(200212)32:12<3756::AID-IMMU3756>3.0.CO;2-E. [DOI] [PubMed] [Google Scholar]
- 39.Wills MR, Okecha G, Weekes MP, Gandhi MK, Sissons PJ, Carmichael AJ. Identification of naive or antigen-experienced human CD8(+) T cells by expression of costimulation and chemokine receptors: analysis of the human cytomegalovirus-specific CD8(+) T-cell response. J Immunol. 2002;168(11):5455–5464. doi: 10.4049/jimmunol.168.11.5455. [DOI] [PubMed] [Google Scholar]
- 40.Wills MR, Carmichael AJ, Weekes MP, et al. Human virus-specific CD8+ CTL clones revert from CD45ROhigh to CD45RAhigh in vivo: CD45RAhighCD8+ T cells comprise both naive and memory cells. J Immunol. 1999;162(12):7080–7087. [PubMed] [Google Scholar]
- 41.Weekes MP, Carmichael AJ, Wills MR, Mynard K, Sissons JG. Human CD28-CD8+ T cells contain greatly expanded functional virus-specific memory CTL clones. J Immunol. 1999;162(12):7569–7577. [PubMed] [Google Scholar]
- 42.Vargas AL, Lechner F, Kantzanou M, Phillips RE, Klenerman P. Ex vivo analysis of phenotype and TCR usage in relation to CD45 isoform expression on cytomegalovirus-specific CD8+ T lymphocytes. Clin Exp Immunol. 2001;125(3):432–439. doi: 10.1046/j.1365-2249.2001.01614.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Harari A, Dutoit V, Cellerai C, Bart PA, Du Pasquier RA, Pantaleo G. Functional signatures of protective antiviral T-cell immunity in human virus infections. Immunol Rev. 2006;211:236–254. doi: 10.1111/j.0105-2896.2006.00395.x. [DOI] [PubMed] [Google Scholar]
- 44.Weekes MP, Wills MR, Sissons JG, Carmichael AJ. Long-term stable expanded human CD4+ T-cell clones specific for human cytomegalovirus are distributed in both CD45RAhigh and CD45ROhigh populations. J Immunol. 2004;173(9):5843–5851. doi: 10.4049/jimmunol.173.9.5843. [DOI] [PubMed] [Google Scholar]
- 45.Carrasco J, Godelaine D, Van Pel A, Boon T, van der Bruggen P. CD45RA on human CD8 T cells is sensitive to the time elapsed since the last antigenic stimulation. Blood. 2006;108(9):2897–2905. doi: 10.1182/blood-2005-11-007237. [DOI] [PubMed] [Google Scholar]
- 46.Nikolich-Zugich J, Slifka MK, Messaoudi I. The many important facets of T-cell repertoire diversity. Nat Rev Immunol. 2004;4(2):123–132. doi: 10.1038/nri1292. [DOI] [PubMed] [Google Scholar]
- 47.Migueles SA, Laborico AC, Shupert WL, et al. HIV-specific CD8+ T cell proliferation is coupled to perforin expression and is maintained in nonprogressors. Nat Immunol. 2002;3(11):1061–1068. doi: 10.1038/ni845. [DOI] [PubMed] [Google Scholar]
- 48.Appay V, Rowland-Jones SL. Lessons from the study of T-cell differentiation in persistent human virus infection. Semin Immunol. 2004;16(3):205–212. doi: 10.1016/j.smim.2004.02.007. [DOI] [PubMed] [Google Scholar]
- 49.Gamadia LE, ten Berge IJ, Picker LJ, van Lier RA. Skewed maturation of virus-specific CTLs? Nat Immunol. 2002;3(3):203. doi: 10.1038/ni0302-203. [DOI] [PubMed] [Google Scholar]
- 50.Douek DC, Picker LJ, Koup RA. T cell dynamics in HIV-1 infection. Annu Rev Immunol. 2003;21:265–304. doi: 10.1146/annurev.immunol.21.120601.141053. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.