Abstract
We report a new assay of human δ-aminolevulinic acid dehydratase (ALAD), an enzyme converting δ-aminolevulinic acid (ALA) into porphobilinogen. The assay is developed for use in the clinical diagnosis of δ-aminolevulinic acid dehydratase-deficient porphyria, a rare enzymatic deficiency of the heme biosynthetic pathway. The assay involves the incubation of erythrocyte lysate with the natural substrate, ALA, followed by quantitative in situ conversion of porphobilinogen to its butyramide, and liquid-liquid extraction into a mass spectrometer-friendly solvent. Quantitation of the butyrylated porphobilinogen is done by electrospray ionization tandem mass spectrometry, using a deuterium labeled internal standard. The assay stays well within the range wherein ALAD activity is linear with time. The Km of ALAD for ALA was measured as 333 μM, and the Vmax was 19.3 μM/hr. Average enzyme activity among a random sample of 36 anonymous individuals was 277 μmol/L erythrocyte lysate/hour with a standard deviation of 90 μmol/L erythrocyte lysate/hour. The tandem mass spectrometric assay should easily detect the enzyme deficiency, which causes a reduction of activity by 95–99%. The assay shows good reproducibility, low background, requires a simple workup, and uses a commercially available substrate.
INTRODUCTION
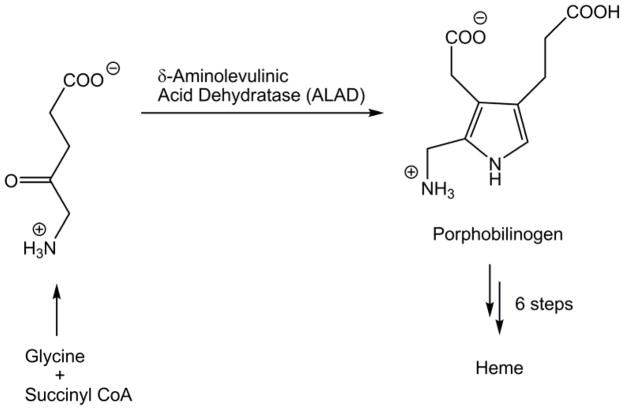
Heme-based proteins, such as the cytochrome enzymes and hemoglobin, play a vital role in human life. The central molecule of these proteins, heme, is made by the human body in an eight-step, enzyme-assisted pathway. Genetic deficiency in any one of the last seven of these enzymes defines any one of seven rare genetic disorders known collectively as the porphyrias.1 Deficiency in the second enzyme, δ-aminolevulinic acid dehydratase (ALAD), is the cause of ALAD-deficient porphyria (ADP), sometimes referred to as Doss porphyria.2 ALAD catalyzes the formation of porphobilinogen (PBG), a pyrrole intermediate, from two molecules of δ-aminolevulinic acid (ALA) (Scheme 1). ALAD is the first enzyme in the pathway to be found in the cytosol of the cell, rather than the mitochondrion.
Scheme 1.
ADP is passed on in an autosomal recessive pattern. Homozygous patients experience a drop in enzyme activity of 95–99%. A heterozygous deficiency results in an enzyme activity level roughly 50% of non-affected people but does not cause ADP. However, the heterozygotes, thought to be as prevalent as 1–2% of certain populations,1 are potentially at risk for greater negative effects of lead poisoning. Lead poisoning results in a lack of ALAD activity as lead reversibly replaces zinc at the enzyme’s active sites.3 Clinically, ADP typically presents with acute abdominal pain and peripheral neuropathy, similar to other acute porphyrias.4
Given its similarities to other porphyrias, ADP is difficult to diagnose. It is differentiated from acute intermittent porphyria (AIP), deficiency in the subsequent enzyme of the pathway, only by increased urinary excretion of PBG. Current assays for ALAD deficiency are, for the most part, based on a method developed by Mauzerall and Granick in 1956.5 This technique involves reacting the enzymatic product with p-dimethylaminobenzaldehyde, or Erlich’s reagent, to create a compound detectable by colorimetry or fluorimetry.6,7,8,9,10 A common alternative is a coupled-enzyme assay,11,12 wherein porphobilinogen is carried further down the biosynthetic pathway to produce hydroxymethylbilane, which spontaneously cyclizes to uroporphyrinogen I, which is then oxidized and detected as uroporphyrin I by fluorimetry. None of the current methods detect porphobilinogen directly, putting them at a clear disadvantage compared to a method that does. Furthermore, it is exceedingly difficult by these methods to distinguish this specific enzyme deficiency from those upstream or downstream.
Previous work in our laboratory has focused on developing procedures for testing enzyme activity levels in human samples using tandem mass spectrometry as a common analytical platform for both clinical diagnostics and newborn screening.13,14,15 The methods involve selecting an appropriate biological source of enzyme, typically either isolated and lysed blood cells for diagnostics or dried blood spots for screening, and incubating this sample with the enzyme’s substrate, which is often synthetically designed in-lab. The enzymatic product is then quantified by selected-reaction monitoring tandem mass spectrometry, using a mass-differentiated internal standard such as a deuterium-labeled isotopologue or a homologue. This method takes advantage of the speed and sensitivity offered by mass spectrometry over spectroscopic methods, as well as the specificity and selectivity offered by the use of a tandem instrument. The unique masses used for each assay’s product and standard allow for easy multiplexing, as the products of many different assays can be combined into one injection without any need for chromatographic separation. In particular, we have developed tandem mass spectrometry assays for the detection of acute intermittent porphyria,14 porphyria cutanea tarda, hepatoerythropoietic porphyria, and hereditary coproporphyria,15 which are caused by deficient enzymes in the later stages of the heme biosynthetic pathway. In an effort to provide clinical laboratories with a complete cassette of porphyria assays based on a single analytical platform, we developed a new procedure for the direct assay of ALAD which is reported here.
EXPERIMENTAL
Materials
All water used was purified by a Millipore Milli-Q 18MΩ filtering system. Porphobilinogen was purchased from Frontier Scientific (Provo, UT). Butyryl-d7 chloride and sodium butyrate-d7 were supplied by C/D/N Isotopes (Pointe-Claire, Quebec). δ-Aminolevulinic acid, dithiothreitol, and all other chemicals were obtained from Sigma-Aldrich (St. Louis, MO).
Isolation of Erythrocytes
Red blood cells were isolated and lysed according to a procedure previously used in this laboratory.14 All procedures regarding human blood followed Institutional Review Board (IRB) protocols and received an approval (IRB 25200). Blood was drawn into a vacuum sealed tube with heparin. Whole blood (3 mL) was then transferred to a 15-mL polypropylene centrifuge tube, 9 mL of 0.9% w/v saline solution was added, the sample was gently shaken, and the tube was centrifuged at 600 × g for 10 minutes. The non-erythrocyte-containing supernatant was discarded, followed by another addition of 9 mL saline. The tube was inverted a few times to gently re-mix the sample, followed by a second centrifugation at 600 × g for 10 minutes. Again the supernatant was discarded, and the cells were washed and centrifuged a third time. Following this third washing, 2.5 mL of red blood cells were drawn from the bottom of the tube and placed in a new 15 mL centrifuge tube. The tube was frozen in a dry ice/acetone bath for 10 minutes and then brought back to room temperature in a water bath. This was repeated two additional times in order to ensure complete cell lysis. The resulting lysate was centrifuged at 12,000 × g for 10 minutes, prior to being split into 50 μL aliquots, stored in 600 μL polypropylene microfuge tubes at −80°C. Prior to use in assays, the sample tubes were brought back to room temperature and diluted with 450 μL of 18MΩ deionized water. A 50 μL aliquot of this solution was used for assays corresponding to 5 μL of red blood cell lysate per assay.
Internal Standard Synthesis
d7-Butyrylated porphobilinogen (d7-But-PBG) was synthesized using d14-butyric anhydride, which was made from butyryl d7-chloride and sodium butyrate-d7 (both C/D/N Isotopes, 98% D) as follows. Sodium butyrate-d7 (1 g, 8.54 mmol) was added under stirring to anhydrous tetrahydrofuran (50mL) under an argon atmosphere. Butyryl d7-chloride (1 g, 8.80 mmol) was added drop-wise, and the system was allowed to react for 72 hours at room temperature. Sodium chloride was filtered off, the solvent was evaporated in vacuo, and the residue was purified by short-path vacuum distillation. GC-MS analysis showed that the synthesized d14-butyric anhydride was >98% pure and its D content was 98%.
The d7-butyrylated porphobilinogen internal standard was synthesized from 25.5 mg (8.4 × 10−5 mol) of porphobilinogen using three molar equivalents of d14-butyric anhydride in 5 mL of 0.25 M sodium phosphate buffer (pH 6.8) for 30 minutes at room temperature. HCl (1 M) was added to reduce the pH to ~2.0, followed by ammonium sulfate (50% w/w of aqueous phase). The product was extracted with ten 2-mL portions of ethyl acetate. The solvent was removed by evaporation under a stream of N2 and the resulting solid was reconstituted in water for purification by HPLC. A 10 mL/min gradient of 100:0 to 0:100 H2O:ACN over 30 minutes using a 100 × 20 mm reverse-phase C18 column achieved adequate separation to isolate d7-But-PBG, which eluted at 10.4 minutes. No acid was necessary in the HPLC solvents to separate the compound of interest from the impurities. The chemical stability of the internal standard was checked by HPLC-MS and 1H-NMR after 16 months of storage at −80 ºC and the sample was found to be unchanged.
Assay Protocol
An amalgamation of previously reported assays was adapted to be compatible with electrospray ionization and tandem mass spectrometry.5,11 Three hundred microliters of 0.25 M sodium phosphate buffer (pH 6.80), 50 μL of 20 mM dithiothreitol in buffer, and 50 μL of 10-fold diluted red blood cell lysate were combined in a 2.0-mL polypropylene microfuge tube and pre-incubated at 37°C for 15 minutes. Then 100 μL of 5 mM ALA in buffer was added and the mixture was incubated for 60 minutes. After incubation, 5 μL of n-butyric anhydride was added and allowed to react for 30 minutes. Following this in situ derivatization, 5 μL of a 275 μM solution of d7-But-PBG in buffer was added, as well as 100 μL 1 M HCl to reduce the pH to ~2. Four hundred and fifty microliters of water-saturated n-butanol and ca. 300 mg ammonium sulfate were then added, the mixture was vortexed for 30 seconds until the ammonium sulfate dissolved, followed by centrifugation for 3 minutes at 13,200 × g to separate the layers. Two hundred microliters of the n-butanol supernatant was removed with a syringe to a new 600-μL polypropylene tube, and stored at −80°C. The solution was mixed with 200 μL methanol containing 2% (v/v) formic acid immediately before analysis.
Mass Spectrometry
Analysis was conducted by flow injection with 10 μL injections using a Waters Quattro Micro triple quadrupole mass spectrometer in positive ion mode. Solvent flow to the spectrometer’s ESI ion source came from a Waters 1525u HPLC pump, flowing methanol containing 1% (v/v) formic acid at 100 μL/min The instrument was operated using MassLynx software with the following settings: electrospray ionization capillary voltage, 4.00 kV; cone voltage, 38 V; extractor, 2 V; RF lens voltage, 200 mV; source temperature, 80 °C; desolvation temperature, 300 °C; desolvation gas flow, 500 L/hr; dwell time, 250 μs; collision energy, 20 eV; collision gas was argon at 1.97 mTorr.
RESULTS AND DISCUSSION
Assay and Sample Work-Up Conditions
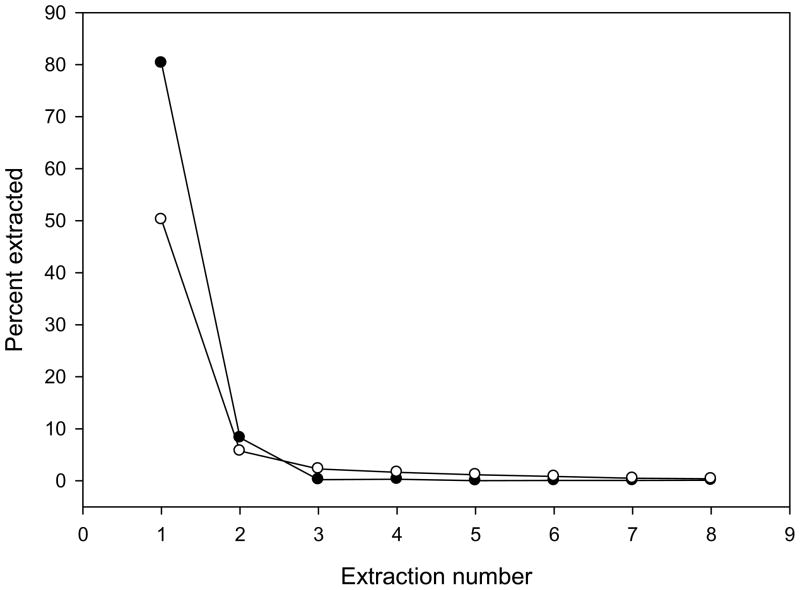
Analysis by electrospray ionization mass spectrometry mandates that the sample be in a compatible solvent free of involatile salts and buffers. Therefore, extraction of the enzymatic product into a suitable solvent was necessary. However, porphobilinogen is ionic or zwitterionic at all relevant pH, so derivatization was necessary to allow extraction. We found that porphobilinogen butyramide was effectively extracted into n-butanol. In situ conversion of porphobilinogen to a butyramide was quantitative by treating the assay mixture with 5 μL of butyric anhydride for 30 min. Of the extraction solvents used, n-butanol was superior to ethyl acetate. The cross-solubility of water and n-butanol (roughly 10% soluble) was overcome by using n-butanol that had previously been saturated with water. Under these conditions, 89% of porphobilinogen butyramide was extracted in two steps, including 80% in the first extraction (Figure 1). One extraction was therefore deemed to be sufficient for the assay. There was no significant difference in extractability between the enzymatic product and the deuterated internal standard. Using anhydrous n-butanol decreased the extraction yield to 52% in the first step (Figure 1).
Figure 1.
Fraction of porphobilinogen butyramide extracted in consecutive extractions by anhydrous n-butanol (open circles) and water-saturated n-butanol (full circles).
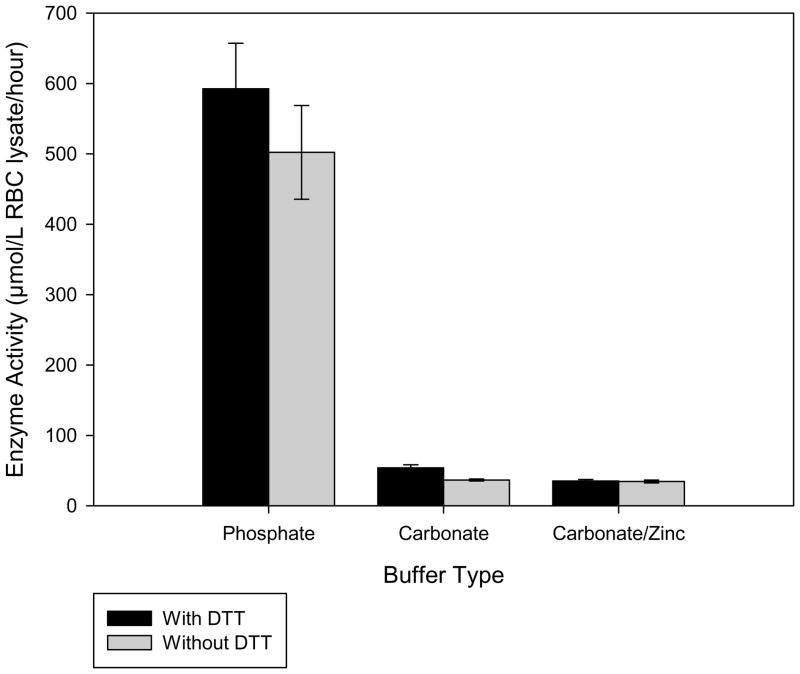
Previous fluorimetric assays for ALAD have shown that dithiothreitol (DTT) increased enzymatic activity16, by preventing possible inhibition caused by the level of lead in the analyzed blood. However, DTT may suppress the ESI-MS analyte signal, and as such could negatively affect the assay results. Experimental runs with and without DTT showed that the inclusion of 2 mM DTT resulted in a 16% increase of enzyme activity; thus, DTT was included in the final assay mixture. Zinc was also considered as an assay additive, since zinc is used by the enzyme as an active site cofactor. Due to the insolubility of zinc phosphate, this additive required the use of a carbonate buffer. However, using a carbonate buffer rather than phosphate caused nearly a 90% drop in activity and addition of zinc resulted in a further decrease by up to 35% (Figure 2). Thus, the assays were run in the phosphate buffer system, without any added zinc.
Figure 2.
Comparison of enzyme activity levels using different buffer systems with or without the inclusion of dithiothreitol.
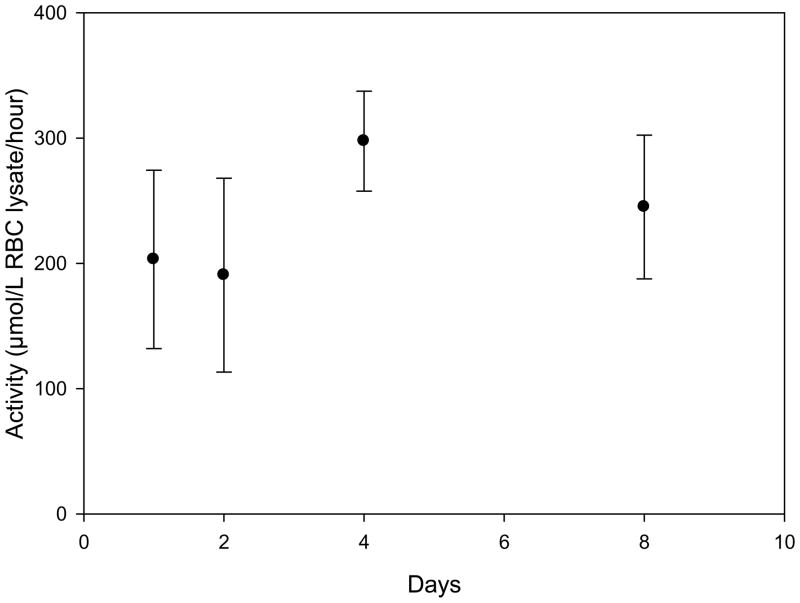
Since a tandem quadrupole mass spectrometer may not be available for immediate analysis in some clinical laboratories, a study was carried out to determine the viability of transporting blood sample to a laboratory with the necessary instrumentation. The relevant shipping conditions were duration and temperature; as such, freshly-drawn whole blood samples were stored at either room temperature or at 4 °C for a set of time intervals ranging up to 8 days (Figure 3). Temperatures below 0 °C were not considered, as any such storage conditions would prematurely lyse the desired erythrocytes, making it impossible to isolate them. This experiment was conducted using two separate sets of blood samples, one for each of the two most common blood preservatives (heparin and EDTA). Results show that while some activity is lost within a week at room temperature (regardless of preservative), storing with heparin at 4 °C had no effect on the measured enzyme activity level (Figure 3). Furthermore, both preservatives allowed the retention of enzyme activity, although heparinized samples gave slightly higher levels. Thus, samples should be preserved in heparin and kept at 4 °C for any required shipping, to ensure the most reliable determination of enzyme activity.
Figure 3.
Measured activity levels of heparinized blood stored at 4°C for a number of days before erythrocyte isolation and enzyme assay measurement.
Mass Spectrometry
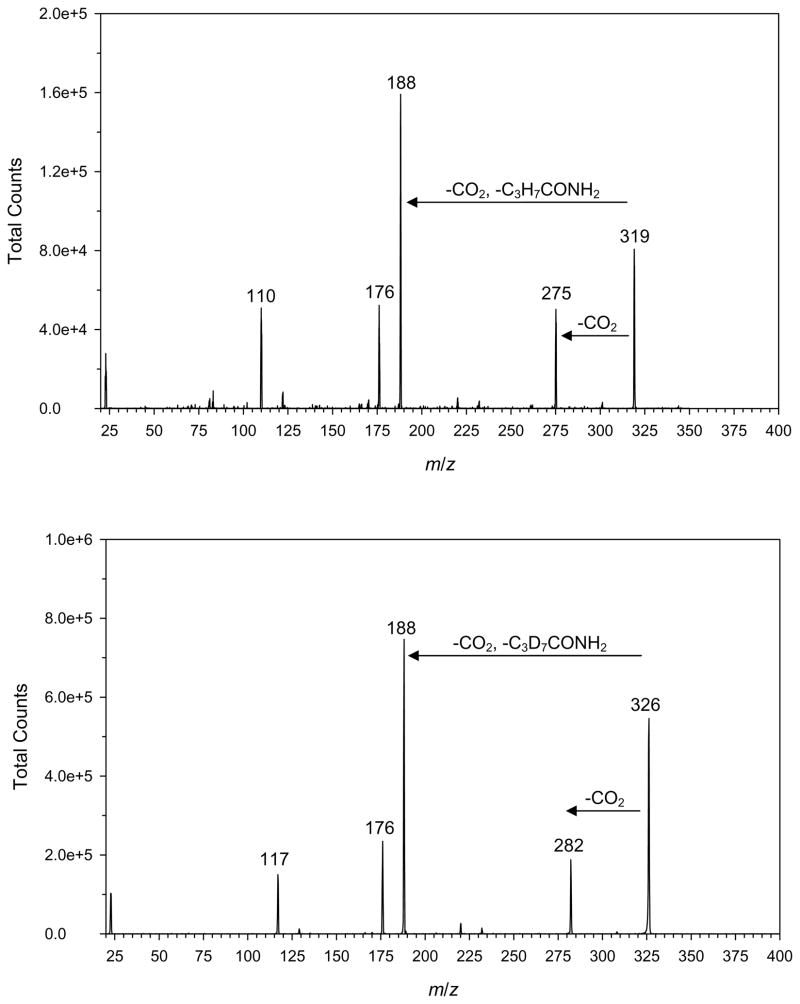
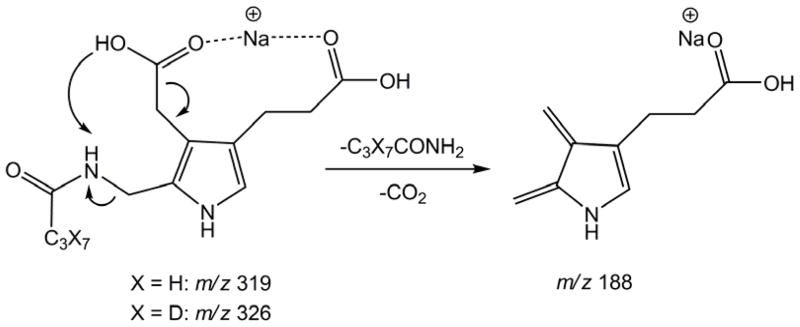
Porphobilinogen was found to be readily protonated, sodiated, or potassiated by electrospray ionization. Protonated porphobilinogen readily eliminated ammonia in the electrospray source and on collisional activation. For example, at 5 eV laboratory collision energy, the (M+H)+ ion completely dissociated. However, due to the above-described solubility issues, underivatized porphobilinogen was not a suitable analyte for this assay. Electrospray of porphobilinogen butyramide mainly produced (M + Na)+ and (M + K)+ ions which were less prone to dissociation than the (M + H)+ ion. The (M + Na)+ ion showed the highest intensity for both the derivatized product and internal standard and was therefore used for MS/MS analysis. The (M + K)+ ions, albeit also abundant, showed intensity variations in different assay runs. Collision-induced dissociation of the (M + Na)+ ions gave fragments at m/z 275 and 282 for the butyrylated enzymatic product and internal standard, respectively, due to loss of 45 Da (COOH) (Figure 4). The most abundant fragment ion at m/z 188 by loss of 131 Da from the butyrylated product was due to a combined elimination of butyramide and CO2. The transitions monitored by MS/MS were m/z 319 → 188 for the enzymatic product and 326 → 188 for the internal standard (Figure 4). Scheme 2 shows the tentative scheme for the elimination involving a proton transfer from the proximate carboxyl group onto the butyramide moiety. The fact that both the deuterated and non-deuterated (M + Na)+ precursor ions fragmented to form the same product ion was beneficial because it increased the duty cycle of the measurements, thereby increasing the assay efficiency.
Figure 4.
Collision induced dissociation mass spectra at 20 eV of (M + Na)+ ions from top: porphobilinogen butyramide (m/z 319) and bottom: porphobilinogen d7-butyramide (m/z 326).
Scheme 2.
Blank measurements were carried out with samples that lacked erythrocytes or the δ-aminolevulinic acid substrate. In each of these blank measurements, no detectable signal for the m/z 319 → 188 transition was found by MS/MS. Blank correction in the ALAD assays was therefore unnecessary.
ALAD Enzyme Kinetics
The assay conditions were optimized by varying the assay time, amount of sample, and concentration of substrate to determine the Km and Vmax. The time course of the assay was found to be linear up to 4 h (Figure S1, Supporting Information). A 15 min pre-incubation period was necessary to warm up the blood sample before the addition of the substrate. Based on the time dependence we chose a 60 minute incubation for standard assays which produced 800–1000 pmol of porphobilinogen to be readily quantified by tandem mass spectrometry. Experiments were done to find the optimal amount of red blood cell lysate to use. The assay showed a linear product formation with erythrocyte lysate volume (Figure S2, Supporting Information). The most convenient amount was found to be 50 μL of diluted lysate corresponding to 5 μL of the erythrocyte fraction.
Taking into account Km values previous reported17,18 for this enzyme from slightly different biological sources, which ranged from 160 μM in bovine liver to 600 μM in human fetal erythrocytes, he Michaelis-Menten enzyme kinetics parameters were determined by varying the concentration of substrate in the range of 100 μM to 3 mM, with triplicate assays at each concentration level, and the enzyme activity at each concentration was fitted to a non-linear least-squares model (Figure S3, Supporting Information). The Km was found to be 340 μM, and the Vmax was found to be 19 μM/hr. Give these values, the concentration used for the assay was set at 1 mM, roughly three times the Km, to ensure enzyme saturation and reduce any possibility of inter-sample variance in enzyme velocity.
Clinical Sample Analysis
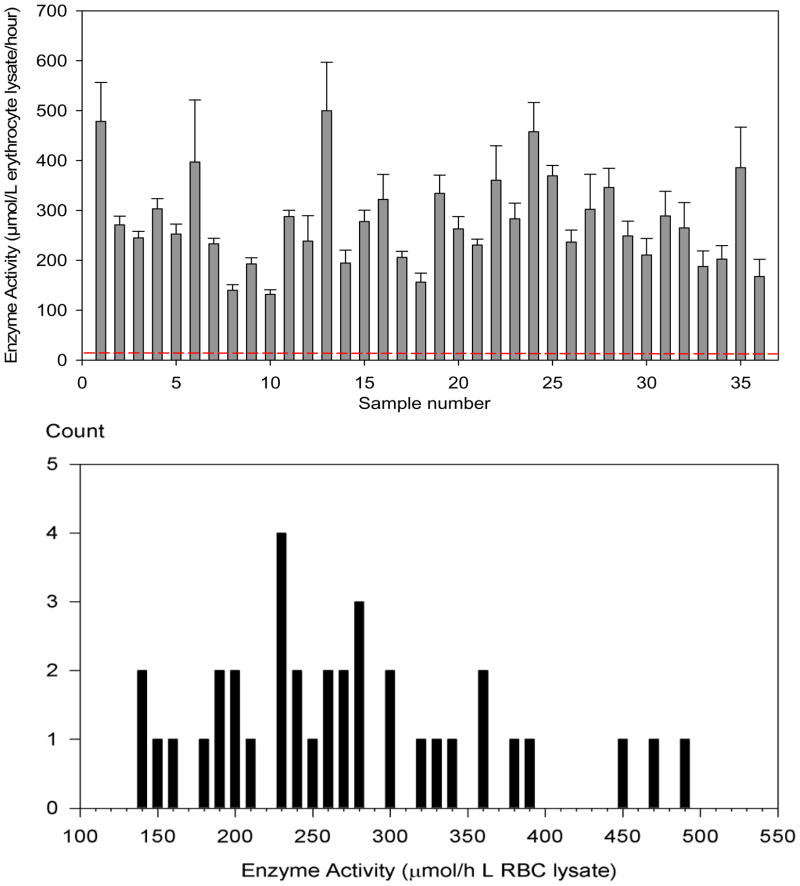
Following the determination of the relevant parameters, the assay was applied to 35 clinical blood samples, of unknown but varied ages and storage conditions, along with the original blood sample used to elucidate the assay parameters. An equal volume of red blood cells, and subsequently of red blood cell lysate, was used for all samples. The 36 samples showed ALAD activities ranging from 140 to 500 μmol/L erythrocyte lysate/hour. The distribution of activities is shown in Figure 5. The mean enzyme activity was 277 μmol/L erythrocyte lysate/hour, with a standard deviation of 90 μmol/L erythrocyte lysate/hour. The relative standard deviation between injections of the same assay was 3%, while assay-to-assay (using the same enzyme source sample) RSD was 13%. Figure 5 further shows that a 5% enzyme activity due to an ADP-affected patient would appear at 2.9 standard deviations of the sample mean and would be readily distinguished from those of healthy individuals.
Figure 5.
Top panel: Chart of enzyme activity of 36 random erythrocyte samples assayed by tandem mass spectrometry. The data error bars are one standard deviation of triplicate measurements. The red dashed line indicates the presumed ALAD activity in a sample from a homozygotic affected patient. Bottom panel: Distribution of activities among the 36 samples.
CONCLUSIONS
Tandem mass spectrometry has been shown to be an effective and efficient means for the quantitation of ALAD in red blood cells. The method requires only a one-step liquid-liquid extraction out of aqueous reaction buffer into n-butanol and dilution with acidified methanol. Blood samples need not be processed or assayed immediately if stored with heparin at 4 °C, and thus can be shipped to a laboratory capable of tandem mass spectrometric analysis without difficulty. Due to the high specificity of tandem mass spectrometry, resulting from the analytes differing in mass from other compounds in the heme biosynthetic pathway, the assay extract could easily be combined with extracts from other porphyria assays for multiplexed detections in a single injection into the mass spectrometer, improving efficiency. This method offers advantages over earlier more cumbersome, time-consuming, and less specific methods. In spite of some efforts, we have been unable to locate patients who had been previously diagnosed with Doss porphyria.19 It is hoped that the specific assays developed in this laboratory, together with the increasing availability of tandem mass spectrometers in clinical laboratories, will contribute to improved diagnostics of porphyrias in clinical practice.
Supplementary Material
Acknowledgments
Support of this work by the NIH-NIDDK (Grant R01 DK067859) is gratefully acknowledged. We thank Dr. Martin Sadilek for technical assistance with mass spectrometric measurements.
References
- 1.Anderson KE, Sassa S, Bishop DF, Desnick RJ. Disorders of Heme Biosynthesis: X-Linked Sideroblastic Anemia and the Porphyrias. In: Scriver CR, Beaudet AL, Sly WS, Valle D, editors. The Metabolic and Molecular Basis of Inherited Disease. 8. Chapter 124. McGraw-Hill; New York: 2000. pp. 2993–2999.pp. 3005–3008. [Google Scholar]
- 2.Doss M, von Tiepermann R, Schneider J, Schmid H. Klin Wochenschr. 1979;57:1123–1127. doi: 10.1007/BF01481493. [DOI] [PubMed] [Google Scholar]
- 3.Bird TD, Hamernyik P, Nutter JY, Labbe RF. Am J Hum Genet. 1979;31:662–668. [PMC free article] [PubMed] [Google Scholar]
- 4.Doss M, Sassa S. The Porphyrias. In: Noe DA, Rock RC, editors. Laboratory Medicine. The Selection and Interpretation of Clinical Laboratory Studies. Chapter 26. Williams and Wilkins; Baltimore, MD: 1994. pp. 535–553. [Google Scholar]
- 5.Mauzerall D, Granick S. J Biol Chem. 1955;219:435–446. [PubMed] [Google Scholar]
- 6.Lüönd RM, Walker J, Neier RW. J Org Chem. 1992;57:5005–5013. [Google Scholar]
- 7.Sassa S, Fujita H, Kappas A. Pediatrics. 1990;86:84–86. [PubMed] [Google Scholar]
- 8.Anderson PM, Desnick RJ. J Biol Chem. 1979;254:6924–6930. [PubMed] [Google Scholar]
- 9.Wigfield DC, Farant JP. Clin Chem. 1981;27:100–103. [PubMed] [Google Scholar]
- 10.Wigfield DC, Farant JP, Goldberg C, MacKeen JE. J Anal Tox. 1981;5:57–61. doi: 10.1093/jat/5.2.57. [DOI] [PubMed] [Google Scholar]
- 11.Giampetro PF, Desnick RJ. Anal Biochem. 1983;131:83–92. doi: 10.1016/0003-2697(83)90138-0. [DOI] [PubMed] [Google Scholar]
- 12.Bishop DF, Desnick RJ. Method Enzymol. 1986;123:339–345. doi: 10.1016/s0076-6879(86)23040-2. [DOI] [PubMed] [Google Scholar]
- 13.Gelb MH, Tureček F, Scott CR, Chamoles NA. J Inherited Metab Dis. 2006;29:397–404. doi: 10.1007/s10545-006-0265-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wang Y, Gatti P, Sadilek M, Scott CR, Tureček F, Gelb MH. Anal Chem. 2008;80:2599–2605. doi: 10.1021/ac702130n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wang Y, Scott CR, Gelb MH, Tureček F. Anal Chem. 2008;80:2606–2611. doi: 10.1021/ac702244x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sassa S, Granick S, Kappas A. Ann NY Acad Sci. 1975;244:419–439. doi: 10.1111/j.1749-6632.1975.tb41546.x. [DOI] [PubMed] [Google Scholar]
- 17.Sassa S. Enzyme. 1982;28:133–145. doi: 10.1159/000459097. [DOI] [PubMed] [Google Scholar]
- 18.Chang CS, Sassa S. Blood. 1985;65:939–944. [PubMed] [Google Scholar]
- 19.Akagi R, Kato N, Inoue R, Anderson KE, Jaffe EK, Sassa S. Mol Gen Metabol. 2006;87:329–336. doi: 10.1016/j.ymgme.2005.10.011. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.