Abstract
Peroxisome proliferator-activated receptor-gamma (PPAR-γ) is a member of the nuclear receptor (NR) superfamily of ligand-activated transcriptional factors. Among other functions, PPAR-γ acts as a key regulator of the adipogenesis. Since several cytokines (IL-1, TNF-α, TGF-β) had been known to inhibit adipocyte differentiation in mesenchymal stem cells (MSCs), we examined the effect of these cytokines on the transactivation function of PPAR-γ. We found that the TNF-α/IL-1-activated TAK1/TAB1/NIK (NFκB-inducible kinase) signaling cascade inhibited both the adipogenesis and Tro-induced transactivation by PPAR-γ by blocking the receptor binding to the cognate DNA response elements. Furthermore, it has been shown that the noncanonical Wnts are expressed in MSCs and that Wnt-5a was capable to inhibit transactivation by PPAR-γ. Treatment with Wnt5a-activated NLK (nemo-like kinase) induced physical association of the endogenous NLK and H3K9 histone methyltransferase (SETDB1) protein complexes with PPAR-γ. This resulted in histoneH3K9 tri-methylation at PPAR-γ target gene promoters. Overall, our data show that cytokines and noncanonical Wnts play a crucial role in modulation of PPAR-γ regulatory function in its target cells and tissues.
1. Introduction
Peroxisome proliferator-activated receptor-gamma (PPAR-γ) belongs to the nuclear receptor (NR) superfamily and regulates target gene mRNA expression in the ligand-dependent manner [1]. Similar to most known NRs, PPAR-γ contains distinct domains for binding the DNA (DBD), ligand (LBD), and various cofactor complexes. The structure of PPAR-γ LBD consists of 12 α-helices and 4 β-sheets [2].
For ligand-dependent transcriptional control by PPAR-γ, several distinct classes of transcriptional coregulators/coregulator complexes are indispensable in addition to basic transcription machinery to reorganize chromatin state at the genomic target loci [1, 3]. Transcriptional coregulators for NRs can be divided into two classes in regard to the mechanisms of chromatin reorganization. One class consists of histone modifying enzymes that reversibly modify the N-terminal tails of nucleosomal histone proteins [4, 5]. For example, acetylation and methylation at histone H3K4 and H3K36 are chromatin activating modifications and support transcriptional up-regulation by NRs [6, 7]. In contrast, transcriptional repression by NRs is coupled with inactivating modifications like deacetylation and methylation at histone H3K9 and H3K27 [8]. Accordingly, cognate histone modifying enzymes serve as NR coregulators.
The other class of transcriptional coregulators includes chromatin remodeling factors that directly reorganize nucleosomal arrays using ATP hydrolysis as a source of energy [9, 10]. Chromatin remodelers function as multi-subunit complexes and include ATPase catalytic subunits. Four distinct types of chromatin remodeling complexes (SWI/SNF, ISWF, WINAC, and NURD) have been so far identified as transcriptional coregulators of NRs [11, 12].
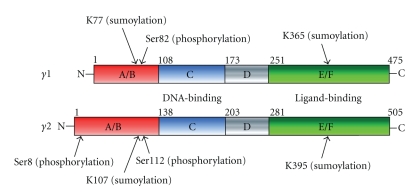
Besides ligand dependency, various signaling pathways modulate the ligand-dependent transactivation function of NRs. For example, phosphorylation in the N-terminal region of estrogen receptor alpha (ER-α) by certain pathway-activated protein kinases enhances the transactivation function of ER-α [13]. The transcriptional activity of PPAR-γ is also modulated through positive and negative crosstalk with other signaling pathways [14]. The molecular mechanisms of the crosstalk include direct and indirect associations of PPAR-γ with intracellular signal transducers or transcriptional factors as well as covalent modifications of PPAR-γ protein, such as phosphorylation by signal-dependent protein kinases [15] or sumoylation by UBC9 [16]. Phosphorylation of PPAR-γ in the N-terminal domain suppresses the transactivation function of PPAR-γ by reducing affinity for PPAR-γ ligands [17], whereas ligand-dependent sumoylation of PPAR-γ represses the NF-κB activation and antagonizes inflammatory responses [16]. These clearly indicate that modifications in the PPAR-γ molecule play a pivotal role in modulation of its physiological action (Figure 1).
Figure 1.
Structure and posttranslational modifications of PPAR-γ1, - γ2 proteins. Although PPAR-γ was ubiquitinated, lysine residues are not determined [18].
2. Signaling Crosstalk between PPAR-γ and Cytokines in MSCs
Mesenchymal stem cells (MSCs) derived from various adult tissues have the potential to differentiate into different lineages, including osteoblasts, chondrocytes, adipocytes, or myocytes [19–21]. Reflecting such pluripotency, a number of regulators involved in the control of MSC differentiation have been identified and characterized [19]. Bone morphogenetic protein (BMP) signaling molecules (particularly BMP-2, -4, -6, and -7) act as major osteogenic inducers and may also influence adipocyte differentiation [22] through induction of PPAR-γ corepressor, TAZ [23]. Recently, the hedgehog signaling has been shown to inhibit adipogenesis and induce osteoblastogenesis [24].
Since several cytokines (IL-1, TNF-α, TGF-β) inhibit adipocyte differentiation in MSC, we examined the effect of their signaling on the transactivation function of PPAR-γ. Treatment with TNF-α or IL-1 inhibited Tro-induced transcriptional activity of PPAR-γ. Interestingly, treatment with both Tro and cytokine (IL-1 or TNF-α) induced osteoblastogenesis in ST2 cells. Thus, cytokines and activated PPAR-γ appeared to stimulate cytodifferentiation of bone marrow progenitor cells into osteoblasts, in addition to cytokine-dependent interference with adipocyte differentiation. Since TNF-α and IL-1 are known to activate the NF-κB in the nucleus, and the nuclear NF-κB is indispensable for osteoclastogenesis from heamatopoetic stem cells, these cytokines appear to be physiologically important for the mesenchymal stem cell fate decision. We therefore studied effects of downstream mediators of the TNFα/IL-1 signaling on the MSC differentiation [14].
In ST2 cells, the TNF-α/IL-1-activated TAK1/TAB1/NIK (NFκB-inducible kinase) signaling cascade inhibited both the adipogenesis and Tro-induced transactivation by PPAR-γ. Though it was previously reported that phosphorylation of PPAR-γ by MAP kinase resulted in repression of the PPAR-γ function [15], we showed that TNF-α/IL-1-induced inhibition of PPAR-γ did not involve its phosphorylation by the NIK.
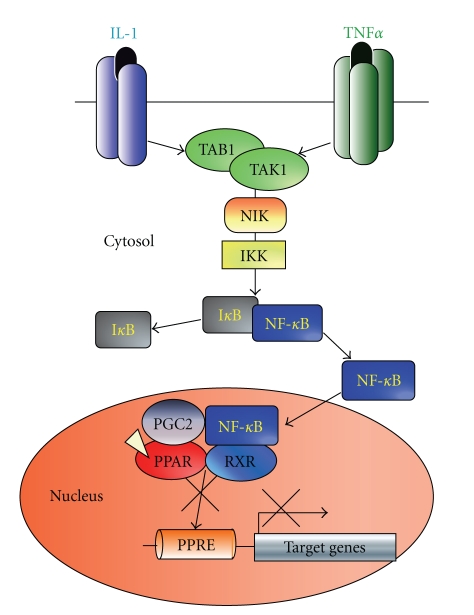
Consistent with suppression of the PPAR-γ-dependent luciferase reporter gene activity, the activated TAK1/TAB1/NIK was found to suppress the Tro-induced expression of endogenous PPAR-γ target genes. We found that treatment with these cytokines or ectopic expression of some of their downstream mediators blocked binding of PPAR-γ to its response element DNA sequences (PPRE) in the target gene promoters (Cbl-associated protein, CAP). CAP is a signaling protein that interacts with both c-Cbl and the insulin receptor that may be involved in the specific insulin-stimulated tyrosine phosphorylation of c-Cbl [25, 26]. Next, we have shown that the TAK1/TAB1/NIK pathway-activated NF-κB blocks the DNA binding of PPAR-γ at the PPRE. Together with the previous reports that agonist-activated PPAR-γ inhibits DNA binding by NF-κB [27], it appears that an association of ligand-activated PPAR-γ with nuclear NF-κB results in a complex incapable to interact with DNA at either corresponding binding sites (Figure 2).
Figure 2.
Schema of the proposed molecular mechanism of adipogenesis inhibition by TNF-α and IL-1 through suppression of PPAR-γ function by NF-κB activated via the NIK-TAK1/TAB1-mediated cascade.
Thus, we presume that TNF-α/IL-1 triggers activation of NF-κB through the TAK1/TAB1/NIK axis, leading to a physical association between PPAR-γ and NF-κB thereby inhibiting the ligand-dependent PPAR-γ transactivation. Since PPAR-γ is a prime regulator of adipogenesis, suppression of the PPAR-γ function may inhibit adipogenesis and consequently, shift the bone marrow cell fate decision towards the osteoblastogenesis [14].
3. Noncanonical Wnt Signaling Induces Osteoblastogenesis through Transrepression of PPAR-γ by Histone Methyltransferase Complex
Our recent studies of the effects of Wnts on the osteoblastogenesis and adipogenesis have shown that Wnt signaling may directly regulate the transactivation function of PPAR-γ in the MSCs [28]. Several frizzled receptors and Wnt ligands have been found expressed at significant levels in the ST2 cells and in mouse bone marrow cell primary culture. Interestingly, noncannonical Wnt ligand (Wnt-5a) and receptors (Frizzled-2 and -5) were found to be expressed in these cells at particular high levels [28]. While Wnt-3a, a canonical Wnt ligand, did not affect transactivation function of Tro-induced PPAR-γ, noncanonical Wnt-5a was capable to repress activation by PPAR-γ recombinant and endogenous PPAR-γ target gene promoters. We then explored an ability of downstream mediators of the Wnt-5a signaling to repress PPAR-γ and determined that CaMKII-TAK1/TAB2-NLK axis members were potent inhibitors of the receptor. This was consistent with reports that NLK-deficient mice exhibited increased adipocyte concentration in the bone marrow [29].
As the NLK acts as a downstream mediator in the Wnt-5a signaling pathway, we explored molecular basis of the transrepressive effects of NLK on the PPAR-γ transcriptional function. Since tricostatine A, an inhibitor of a wide range of HDACs, was unable to reverse the NLK-mediated suppression of PPAR-γ function, this opened a question about possible involvement of other inactivating histone modifying enzymes. NLK-containing protein complexes were biochemically purified from nuclear extracts of KCl-treated HeLa cells expressing FLAG-tagged NLK [9, 30] and a distinct NLK-nuclear protein complex with a molecular weight of around 400–500 kDa was isolated and analysed [28, 31]. In this complex, a 170 kDa component was identified as a SETDB1, a transcription inhibiting histone lysine-methyltransferase (HKMT) that methylates histone H3 at K9 [32, 33]. Importantly, in ST2 cells, treatment with Wnt5a induced a physical association of endogenous NLK-SETDB1 protein complexes with PPAR-γ.
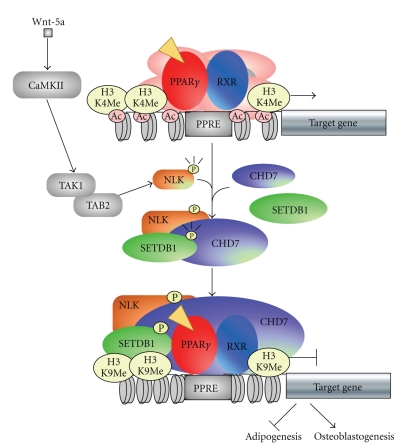
ChIP analysis of endogenous transcriptional factors and histone modifications at the PPAR-γ response element (PPRE) in the aP2 gene promoter [34] has shown that treatment with Tro induced recruitment of known PPAR-γ coactivator SRC-1. However, simultaneous treatment with Wnt-5a and Tro induced recruitment of NLK and SETDB1 at the PPRE region. Consistently, an increase in histone H3 di- and tri-methylation at K9 was observed together with histone hypoacetylation. Such coordinated chromatin silencing histone modifications at the PPAR-γ target genes were more prominent after a 7-day treatment with Wnt-5a that was long enough to induce the osteoblastogenesis. Furthermore, an ectopic expression of either NLK or SETDB1 in the presence of Tro was potent to induce the osteoblastogenesis and inhibit the adipogenesis, whereas a knockdown of either NLK or SETDB1 potentiated the Tro-induced adipogenesis even in the presence of Wnt-5a. Thus, we have shown that Wnt-5a induces the osteoblastogenesis through attenuating the PPAR-γ-induced adipogenesis in the bone marrow MSC (Figure 3).
Figure 3.
Schematic model of crosstalk between PPAR-γ and Wnt-5a signaling in MSC. NLK activated by the Wnt5a signaling pathway phosphorylates SETDB1 and forms a complex with PPAR-γ/RXR and chromodomain containing protein 7 (CHD7).
Upon Wnt-5a-induced activation of the noncanonical Wnt signaling, the SETDB1 HKMT forms a complex with phosphorylated NLK. This NLK/SETDB1 complex associates with PPAR-γ and methylates H3-K9 at the PPAR-γ target gene promoters leading to their transcriptional silencing. Interestingly, the NLK also suppresses the transactivation function of the A-Myb through histone methylation [35], suggesting that the NLK might control gene expression by histone modification through recruitment of SETDB1.
The noncanonical Wnt-5a ligand regulates MSC differentiation through the CaMKII-TAK1/TAB2-NLK signaling cascade that is distinct from the canonical Wnt pathway, which is mediated by the β-catenin/TCF signal transduction. Several recent reports have demonstrated that the canonical Wnt pathway mediated by LRP5/β-catenin is also indispensable for the osteoblastogenesis [36–38]. Hence, both the canonical and noncanonical Wnt pathways are considered to support the osteoblastogenesis in the bone marrow mesenchymal cells. However, only the noncanonical Wnt signaling appears to impair the PPAR-γ-inducible adipogenesis and switch the MSC differentiation into the osteoblastic lineage.
4. Conclusion
In summary, IL-1, TNF-α, and noncanonical Wnt signaling pathways suppress the PPAR-γ function in MSCs and thus, are capable to influence stem cell fate [39]. Interestingly, molecular mechanism of suppression of the PPAR-γ transcriptional activity by the IL-1 and TNF-α is different from that induced by the noncanonical Wnt ligands. IL-1 or TNF-α-activated NF-κB inhibits the DNA binding capacity of the receptor, while Wnt5a-activated NLK promotes PPAR-γ/SETDB1 complex formation leading to silencing epigenetic chromatin modifications at the PPRE. Recent studies show that PPAR-γ also plays pivotal roles in other cells and tissues, such as osteoclasts [40], kidney cells [41], and macrophages [27]. This opens questions about the existence of other mechanisms of modulations of the PPAR-γ physiological activity specific for these types of differentiated cells that may be different from those in stem cells.
Acknowledgments
The first author was supported in part by a Grant-In-Aid for Basic Research on Priority Areas (Dynamics of extracellular environments), The Nakatomi-foundation, The Cell Science Research Foundation. The third author was supported by a Grant-In-Aid for Basic Research Activities for Innovative Biosciences (BRAIN), and Priority Areas from the Ministry of Education, Science, Sports, and Culture of Japan.
References
- 1.Michalik L, Auwerx J, Berger JP, et al. International union of pharmacology. LXI. Peroxisome proliferator-activated receptors. Pharmacological Reviews. 2006;58(4):726–741. doi: 10.1124/pr.58.4.5. [DOI] [PubMed] [Google Scholar]
- 2.Nolte RT, Wisely GB, Westin S, et al. Ligand binding and co-activator assembly of the peroxisome proliferator-activated receptor-γ . Nature. 1998;395(6698):137–143. doi: 10.1038/25931. [DOI] [PubMed] [Google Scholar]
- 3.Rosenfeld MG, Lunyak VV, Glass CK. Sensors and signals: a coactivator/corepressor/epigenetic code for integrating signal-dependent programs of transcriptional response. Genes and Development. 2006;20(11):1405–1428. doi: 10.1101/gad.1424806. [DOI] [PubMed] [Google Scholar]
- 4.Kouzarides T. Chromatin modifications and their function. Cell. 2007;128(4):693–705. doi: 10.1016/j.cell.2007.02.005. [DOI] [PubMed] [Google Scholar]
- 5.Allis CD, Berger SL, Cote J, et al. New nomenclature for chromatin-modifying enzymes. Cell. 2007;131(4):633–636. doi: 10.1016/j.cell.2007.10.039. [DOI] [PubMed] [Google Scholar]
- 6.Fujiki R, Chikanishi T, Hashiba W, et al. GlcNAcylation of a histone methyltransferase in retinoic-acid-induced granulopoiesis. Nature. 2009;459(7245):455–459. doi: 10.1038/nature07954. [DOI] [PubMed] [Google Scholar]
- 7.Huang N, vom Baur E, Garnier J-M, et al. Two distinct nuclear receptor interaction domains in NSD1, a novel SET protein that exhibits characteristics of both corepressors and coactivators. EMBO Journal. 1998;17(12):3398–3412. doi: 10.1093/emboj/17.12.3398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Garcia-Bassets I, Kwon Y-S, Telese F, et al. Histone methylation-dependent mechanisms impose ligand dependency for gene activation by nuclear receptors. Cell. 2007;128(3):505–518. doi: 10.1016/j.cell.2006.12.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kitagawa H, Fujiki R, Yoshimura K, et al. The chromatin-remodeling complex WINAC targets a nuclear receptor to promoters and is impaired in Williams syndrome. Cell. 2003;113(7):905–917. doi: 10.1016/s0092-8674(03)00436-7. [DOI] [PubMed] [Google Scholar]
- 10.Lemon B, Inouye C, King DS, Tjian R. Selectivity of chromatin-remodelling cofactors for ligand-activated transcription. Nature. 2001;414(6866):924–928. doi: 10.1038/414924a. [DOI] [PubMed] [Google Scholar]
- 11.Bao Y, Shen X. SnapShot: chromatin remodeling complexes. Cell. 2007;129(3):p. 632. doi: 10.1016/j.cell.2007.04.018. [DOI] [PubMed] [Google Scholar]
- 12.Kishimoto M, Fujiki R, Takezawa S, et al. Nuclear receptor mediated gene regulation through chromatin remodeling and histone modifications. Endocrine Journal. 2006;53(2):157–172. doi: 10.1507/endocrj.53.157. [DOI] [PubMed] [Google Scholar]
- 13.Kato S, Endoh H, Masuhiro Y, et al. Activation of the estrogen receptor through phosphorylation by mitogen-activated protein kinase. Science. 1995;270(5241):1491–1494. doi: 10.1126/science.270.5241.1491. [DOI] [PubMed] [Google Scholar]
- 14.Suzawa M, Takada I, Yanagisawa J, et al. Cytokines suppress adipogenesis and PPAR-γ function through the TAK1/TAB1/NIK cascade. Nature Cell Biology. 2003;5(3):224–230. doi: 10.1038/ncb942. [DOI] [PubMed] [Google Scholar]
- 15.Hu E, Kim JB, Sarraf P, Spiegelman BM. Inhibition of adipogenesis through MAP kinase-mediated phosphorylation of PPARγ . Science. 1996;274(5295):2100–2103. doi: 10.1126/science.274.5295.2100. [DOI] [PubMed] [Google Scholar]
- 16.Pascual G, Fong AL, Ogawa S, et al. A SUMOylation-dependent pathway mediates transrepression of inflammatory response genes by PPAR-γ . Nature. 2005;437(7059):759–763. doi: 10.1038/nature03988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Shao D, Rangwala SM, Bailey ST, Krakow SL, Reginato MJ, Lazar MA. Interdomain communication regulating ligand binding by PPAR-γ . Nature. 1998;396(6709):377–380. doi: 10.1038/24634. [DOI] [PubMed] [Google Scholar]
- 18.van Beekum O, Fleskens V, Kalkhoven E. Posttranslational modifications of PPAR-γ: fine-tuning the metabolic master regulator. Obesity. 2009;17(2):213–219. doi: 10.1038/oby.2008.473. [DOI] [PubMed] [Google Scholar]
- 19.Takada I, Kouzmenko AP, Kato S. Wnt and PPARgamma signaling in osteoblastogenesis and adipogenesis. Nature Reviews. Rheumatology. 2009;5(8):442–447. doi: 10.1038/nrrheum.2009.137. [DOI] [PubMed] [Google Scholar]
- 20.Arthur A, Zannettino A, Gronthos S. The therapeutic applications of multipotential mesenchymal/stromal stem cells in skeletal tissue repair. Journal of Cellular Physiology. 2009;218(2):237–245. doi: 10.1002/jcp.21592. [DOI] [PubMed] [Google Scholar]
- 21.Hamada H, Kobune M, Nakamura K, et al. Mesenchymal stem cells (MSC) as therapeutic cytoreagents for gene therapy. Cancer Science. 2005;96(3):149–156. doi: 10.1111/j.1349-7006.2005.00032.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Muruganandan S, Roman AA, Sinal CJ. Adipocyte differentiation of bone marrow-derived mesenchymal stem cells: cross talk with the osteoblastogenic program. Cellular and Molecular Life Sciences. 2009;66(2):236–253. doi: 10.1007/s00018-008-8429-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hong J-H, Hwang ES, McManus MT, et al. TAZ, a transcriptional modulator of mesenchymal stem cell differentiation. Science. 2005;309(5737):1074–1078. doi: 10.1126/science.1110955. [DOI] [PubMed] [Google Scholar]
- 24.Spinella-Jaegle S, Rawadi G, Kawai S, et al. Sonic hedgehog increases the commitment of pluripotent mesenchymal cells into the osteoblastic lineage and abolishes adipocytic differentiation. Journal of Cell Science. 2001;114(11):2085–2094. doi: 10.1242/jcs.114.11.2085. [DOI] [PubMed] [Google Scholar]
- 25.Baumann CA, Ribon V, Kanzaki M, et al. CAP defines a second signalling pathway required for insulin-stimulated glucose transport. Nature. 2000;407(6801):202–207. doi: 10.1038/35025089. [DOI] [PubMed] [Google Scholar]
- 26.Ribon V, Johnson JH, Camp HS, Saltiel AR. Thiazolidinediones and insulin resistance: peroxisome proliferator-activated receptor γ activation stimulates expression of the CAP gene. Proceedings of the National Academy of Sciences of the United States of America. 1998;95(25):14751–14756. doi: 10.1073/pnas.95.25.14751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ricote M, Li AC, Willson TM, Kelly CJ, Glass CK. The peroxisome proliferator-activated receptor-γ is a negative regulator of macrophage activation. Nature. 1998;391(6662):79–82. doi: 10.1038/34178. [DOI] [PubMed] [Google Scholar]
- 28.Takada I, Mihara M, Suzawa M, et al. A histone lysine methyltransferase activated by non-canonical Wnt signalling suppresses PPAR-γ transactivation. Nature Cell Biology. 2007;9(11):1273–1285. doi: 10.1038/ncb1647. [DOI] [PubMed] [Google Scholar]
- 29.Kortenjann M, Nehls M, Smith AJH, et al. Abnormal bone marrow stroma in mice deficient for nemo-like kinase, Nlk. European Journal of Immunology. 2001;31(12):3580–3587. doi: 10.1002/1521-4141(200112)31:12<3580::aid-immu3580>3.0.co;2-n. [DOI] [PubMed] [Google Scholar]
- 30.Yanagisawa J, Kitagawa H, Yanagida M, et al. Nuclear receptor function requires a TFTC-type histone acetyl transferase complex. Molecular Cell. 2002;9(3):553–562. doi: 10.1016/s1097-2765(02)00478-1. [DOI] [PubMed] [Google Scholar]
- 31.Vissers LELM, van Ravenswaaij CMA, Admiraal R, et al. Mutations in a new member of the chromodomain gene family cause CHARGE syndrome. Nature Genetics. 2004;36(9):955–957. doi: 10.1038/ng1407. [DOI] [PubMed] [Google Scholar]
- 32.Wang H, An W, Cao R, et al. mAM facilitates conversion by ESET of dimethyl to trimethyl lysine 9 of histone H3 to cause transcriptional repression. Molecular Cell. 2003;12(2):475–487. doi: 10.1016/j.molcel.2003.08.007. [DOI] [PubMed] [Google Scholar]
- 33.Schultz DC, Ayyanathan K, Negorev D, Maul GG, Rauscher FJ., III SETDB1: a novel KAP-1-associated histone H3, lysine 9-specific methyltransferase that contributes to HP1-mediated silencing of euchromatic genes by KRAB zinc-finger proteins. Genes and Development. 2002;16(8):919–932. doi: 10.1101/gad.973302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Tontonoz P, Hu E, Graves RA, Budavari AI, Spiegelman BM. mPPARγ2: tissue-specific regulator of an adipocyte enhancer. Genes and Development. 1994;8(10):1224–1234. doi: 10.1101/gad.8.10.1224. [DOI] [PubMed] [Google Scholar]
- 35.Kurahashi T, Nomura T, Kanei-Ishii C, Shinkai Y, Ishii S. The Wnt-NLK signaling pathway inhibits A-Myb activity by inhibiting the association with coactivator CBP and methylating histone H3. Molecular Biology of the Cell. 2005;16(10):4705–4713. doi: 10.1091/mbc.E05-05-0470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Glass DA, II, Bialek P, Ahn JD, et al. Canonical Wnt signaling in differentiated osteoblasts controls osteoclast differentiation. Developmental Cell. 2005;8(5):751–764. doi: 10.1016/j.devcel.2005.02.017. [DOI] [PubMed] [Google Scholar]
- 37.Gong Y, Slee RB, Fukai N, et al. LDL receptor-related protein 5 (LRP5) affects bone accrual and eye development. Cell. 2001;107(4):513–523. doi: 10.1016/s0092-8674(01)00571-2. [DOI] [PubMed] [Google Scholar]
- 38.Ross SE, Hemati N, Longo KA, et al. Inhibition of adipogenesis by Wnt signaling. Science. 2000;289(5481):950–953. doi: 10.1126/science.289.5481.950. [DOI] [PubMed] [Google Scholar]
- 39.Takada I, Kouzmenko AP, Kato S. Molecular switching of osteoblastogenesis versus adipogenesis: implications for targeted therapies. Expert Opinion on Therapeutic Targets. 2009;13(5):593–603. doi: 10.1517/14728220902915310. [DOI] [PubMed] [Google Scholar]
- 40.Wan Y, Chong L-W, Evans RM. PPAR-γ regulates osteoclastogenesis in mice. Nature Medicine. 2007;13(12):1496–1503. doi: 10.1038/nm1672. [DOI] [PubMed] [Google Scholar]
- 41.Guan Y, Hao C, Cha DR, et al. Thiazolidinediones expand body fluid volume through PPARγ stimulation of ENaC-mediated renal salt absorption. Nature Medicine. 2005;11(8):861–866. doi: 10.1038/nm1278. [DOI] [PubMed] [Google Scholar]