Summary
We discuss a Bayesian discovery procedure for multiple comparison problems. We show that under a coherent decision theoretic framework, a loss function combining true positive and false positive counts leads to a decision rule based on a threshold of the posterior probability of the alternative. Under a semi-parametric model for the data, we show that the Bayes rule can be approximated by the optimal discovery procedure (ODP), recently introduced by Storey (2007a). Improving the approximation leads us to a Bayesian discovery procedure (BDP), which exploits the multiple shrinkage in clusters implied by the assumed nonparametric model. We compare the BDP and the ODP estimates in a simple simulation study and in an assessment of differential gene expression based on microarray data from tumor samples. We extend the setting of the ODP by discussing modifications of the loss function that lead to different single thresholding statistics. Finally, we provide an application of the previous arguments to dependent (spatial) data.
Keywords: Multiple Comparison, False Discovery Rate, Loss Function, Bayes Optimal Rule
1. Introduction
A number of different approaches have been introduced in the recent literature to address the multiple comparison problem. Most focus on controlling some error rate. For example, the control of the familywise error rate (FWER) guarantees a bound on the probability of a false rejection among all tests. Benjamini and Hochberg (1995) developed a simple procedure, based on the ordered p-values that controls the false discovery rate (FDR), defined as the expected proportion of rejected null hypotheses which are erroneously rejected. A decision-theoretic approach to the multiple comparison problem requires the explicit statement of a loss function, which weights the relative importance of the different outcomes according to the preferences and inferential focus of the investigators. Cohen and Sackrowitz (2007) prove the inadmissibility of the Benjamini and Hochberg procedure under any loss that is a linear combination of false discoveries and false acceptances and under several sampling models, including the general one-parameter exponential family. Müller et al. (2004, 2007) undertake a decision theoretic approach to multiple testing and discuss several loss functions that lead to the use of FDR-based rules. More recently, Bogdan et al. (2008) compared the Benjamini-Hochberg procedure with several Bayesian rules for multiple testing. They show that whenever the proportion of true nulls is small, the misclassification error of the Benjamini-Hochberg procedure is close to optimal, in the sense of matching a Bayesian oracle. This property is shown to be shared by some of the Bayesian procedures they consider. In addition, through simulations, they show that Bayes rules generally perform better for large or moderate proportions.
The general multiple comparison problem is stated as follows. Assume we observe data sets x1,…,xm, where xi = {x1i,…,xnii}, and for each xi we consider a test of a null hypothesis H0i. Often the data is reduced to a statistic zi with zi ~ f (zi; μi), for some distribution f, indexed by an unknown parameter μi, i = 1,…,m. Assume we wish to test H0i : μi ∈ A vs. H1i : μi ∉ A for i = 1,…,m. We discuss the multiple comparison problem in a Bayesian decision theoretic framework. A Bayesian decision problem is characterized by a sampling model, f (zi; μi) in this case, a prior and a loss function which represents the utility preferences associated with possible outcomes.
Specifically, we use a prior model that includes a random effects distribution G for the μi. Instead of a parametric model for G, we consider G as an unknown random probability measure (RPM). A prior probability model for an RPM is known as a nonparametric Bayesian model. We assume a Dirichlet process (DP) prior, one of the most popular nonparametric Bayesian models.
For sufficiently large m and a small total mass parameter of the DP prior, the posterior RPM can be approximated by the empirical distribution of the maximum likelihood estimates μ̂i. The result is an approximate Bayes rule that is closely related to Storey’s optimal discovery procedure (ODP, Storey, 2007a).
The ODP is based on a thresholding statistic,
| (1) |
The null hypothesis H0i is rejected if SODP(zi) ≥ λ, for some 0 ≤ λ < ∞. Let diODP = I (SODP (zi) ≥ λ). Storey proves that dODP maximizes the expected number of true positives (ETP) among all procedures with equal or smaller expected number of false positives (EFP). For a point null, A = {0}, the test reduces to thresholding SODP (zi) = ∑μj ∉ A f (zi; μj)/f (zi; 0). As stated, the threshold function SODP involves the unknown parameters μj, j = 1,…,m. In practice, SODP (·) has to be estimated. For a point null, i.e., A = {0}, the ODP is evaluated as
| (2) |
where μ̂j is a point estimate for μj (e.g., the maximum likelihood estimate). It is shown that the performance of ŜODP is comparable to the theoretically optimal discovery procedure based on SODP.
For general A the ODP proceeds with the thresholding function
| (3) |
where wj are suitable weights, chosen to estimate the true status of each hypothesis. For example, wj = 1 for all comparisons that are estimated (by some preliminary test) to be null, and wj = 0 otherwise. Storey et al. (2007b) show that (3) outperforms many procedures commonly used in testing a large number of genes for differential expression.
We show that the ODP statistic can be recognized as an approximate Bayes rule under the proposed nonparametric prior model for μi. This result is in accordance with Ferguson’s (1973) observation that nonparametric Bayesian inference often yields results that are comparable to corresponding classical inference. The expectation in Storey’s optimality statement for dODP is under the (frequentist) repeated sampling model. The expectation in the Bayes rule is under the posterior distribution for a given outcome z. Maximization of the conditional expectation for each z implies maximization of the marginal expectation across repeated sampling. A similar argument can be made about the constraint on the expected number of false positives. A bound on the conditional expectation, conditional on each outcome z implies the same bound on the marginal expectation. We conclude that the Bayes rule under the nonparametric prior approximately satisfies the optimality property of the ODP procedure. By the same arguments we show that thresholding the marginal posterior probability amounts to controlling the positive FDR (Storey, 2002; Storey and Tibshirani, 2003).
Once we have established the utility function and the probability model leading to the ODP we can proceed with generalizations in two directions. First, we will consider variations of the ODP to improve the approximation. We show that the resulting rules lead to small improvement in inference. More importantly, once we recognize the ODP as a Bayes rule we can modify the procedure to adapt to variations in the loss function. We provide specific examples, including a study of exceedance regions of a spatial process and the detection of neurodegenerative patterns in MRI scans.
DP priors in the context of multiple hypotheses testing have been considered before by Gopalan and Berry (1993). More recently, Dahl and Newton (2007) have proposed a DP mixture model (BEMMA) for testing correlated hypotheses and showed that the induced clustering information leads to an improved testing power. Unrelated to a discussion of Storey’s ODP, Bogdan et al. (2008) proposed the same semiparametric model we introduce in section 3 and used it for the comparison of misclassification rates among several competing models. The distinction between the previous approaches and ours is that here the DP prior is part of a decision theoretic setup. Besides, both Gopalan and Berry (1993) and Dahl and Newton (2007) restrict inference to hypotheses concerning the configuration of ties among the parameters of interest. A Bayesian implementation of the ODP procedure has been recently considered also by Cao et al. (2009). They develop a parametric Bayes hierarchical mixture model that achieves shrinkage of the gene-specific sampling variances. The resulting posterior means are then plugged in the ODP statistic and this is shown to outperform the original ODP and many widely used competing procedures. The discussion in Cao et al. (2009) is purely empirical, without reference to a decision theoretic setup.
The format of the paper is as follows. In section 2, we introduce the decision problem and the resulting Bayes rule. In section 3 we introduce the nonparametric (NP) probability model and we detail the algorithm for posterior inference. In section 4 we finally discuss the interpretation of the ODP as an approximate Bayes rule and introduce the corresponding BDP statistics. In section 5 we compare the behavior of the ODP and the BDP with simulated data and a microarray dataset. We show that the BDP provides at least some improvement over the frequentist optimal procedure. In section 6 we discuss some extensions of the previous settings for multigroup experiments, different loss functions and different inferential purposes. In particular, we consider a spatially dependent NP model and apply the Bayesian multicomparison decision rule to a simplified MRI dataset.
2. The Decision Problem
For a formal definition of the multiple comparison problem we need some notation and minimal assumptions on the sampling model. Assume that the data are zi | μi ~ f (zi; μi), independently across i, i = 1,…,m. The competing hypotheses are formalized as
using, for example, A = (−∊+ ∊) or A = {0}. Let G denote the distribution of μi, obtained by marginalizing over the two competing hypotheses and let π0 denote the prior probability of the null hypothesis, i.e., π0 ≡ G(A) = p(H0i). Let G(μ | A) ∝ G(μ) I(μ ∈ A) denote G conditional on A. The model can be written as a mixture prior,
where Ac denote the complement set of A. Alternatively, the model can be defined as a hierarchical model by means of a latent indicator parameter ri ∈ {0, 1}, which is interpreted as the (unknown) truth of the i-th comparison.
| (4) |
We will use z = (z1,…,zm) and θ = (G, ri, μi, i = 1,…,m) to refer generically to the data and parameters in model (4).
In addition to a probability model, a Bayesian decision theoretic approach is characterized by a set of actions (decisions) and a loss function corresponding to all possible outcomes of the experiment. Let di ∈ {0, 1} denote the decision for the i-th hypothesis, with di = 1 indicating a decision against H0i, and let d = (d1,…,dm). To define an optimal rule for di we introduce a loss function L(d, θ, z). The optimal rule is defined by minimizing L in expectation with respect to the posterior model p(θ | z). Formally,
We use a loss function that combines the true positive count, TP = ∑ di ri, and false positive count, FP = ∑ di(1 − ri),
| (5) |
The loss (5) can be interpreted as the Lagrangian for maximizing TP subject to a given bound on FP. It is a multiple comparison equivalent of the loss function underlying the Neyman-Pearson paradigm for a simple test.
Let vi = E(ri | z) denote the marginal posterior probability for the i-th alternative hypothesis. It is straightforward to show that the optimal rule under (5) is a threshold on vi,
| (6) |
Alternatively, the threshold on vi can be written as a threshold on the posterior odds vi/(1 − vi). The statement is true for any probability model (subject only to the stated quantities having meaningful interpretations). Moreover rules based on thresholding the marginal posterior probability imply control of the frequentist positive FDR (Storey, 2002; Storey and Tibshirani, 2003):
PROPOSITION 1
Consider m hypothesis tests H0i : μi ∈ A vs H1i : μi ∈ Ac, data z1,…,zm, where zi | μi ~ f (zi; μi), independently across i, i = 1,…,m, and a prior probability model p(μi | G) = π0 G(μi | A) + (1 − π0) G(μi | Ac) for some distribution G and probabilty π0 = p(H0i) = p(ri = 0). Let the rejection region be determined by (6), i.e. . Let . Then,
PROOF
See appendix.
The only substantial assumption in Proposition 1 is that is a threshold on the gene-specific posterior probabilities of differential expression. Bogdan et al. (2008) proved a similar result for loss functions that are a linear combination of FN and FP counts.
Note that rule (6) is different from rules based on local FDR. Local FDR is defined as the posterior probability of the null given that we have observed a certain value zi, and given assumed known sampling models under the null and alternative hypotheses (Efron et al., 2001). In contrast, vi is defined conditionally on the observed values of z across all tests. Hence, the local FDR provides a measure of significance local to zi, while vi is a global measure of significance.
3. A Nonparametric Bayes Decision Rule
3.1. A Semiparametric Bayesian Model
We complete the sampling model (4) with a prior model for G. Prior probability models for unknown distributions, G in this case, are traditionally known as non-parametric (NP) Bayesian models. One of the most commonly used NP Bayesian priors is the DP model. We write G ~ DP (G*,α) to indicate a DP for a random probability measure G. See Ferguson (1973, 1974) for a definition and important properties of the DP model. The model requires the specification of two parameters, the base measure G* and the total mass parameter α. The base measure G* is the prior mean, E(G) = G*. The total mass parameter determines, among other important properties, the variation of the random measure around the prior mean. Small values of α imply high uncertainty. In the following discussion we exploit two key properties of the DP. A random measure G with DP prior is a.s. discrete. This allows us to write G as a mixture of point masses, G = ∑wh δmh. Another important property is the conjugate nature of the DP prior under random sampling. Assume μi ~ G, i = 1,…,m, are an i.i.d. sample from a random measure G with DP prior, p(G) = DP(G*, α). Then, the posterior probability model is . Here, Fm = 1/m ∑ δμi (·) is the empirical distribution of the realized μi’s.
We use a DP prior on G to complete model (4)
| (7) |
Model (7) implies that the prior for the null hypothesis p0 = G(A) is Beta, p0 ~ Be (αG* (A), α[1 − G* (A)]).
3.2. A semiparametric Bayes Rule for Multiple Testing
Many approaches have been proposed in the literature to implement posterior Monte Carlo simulation for DP mixture models. See, for example, Neal (2000) for a review. We outline how these methods can be adapted to compute the posterior probabilities vi.
Let zi, i = 1,⋯,m denote the observed data (or a summary statistic) for test i. We assume
| (8) |
For example, f (zi; μi) ≡ N (zi; μi, σ), a normal distribution with mean μi and and variance σ2. We complete the model with a non-parametric prior
| (9) |
where hA(·) and hAc (·) are distributions with support, respectively, on A and Ac. Equations (8) and (9) define a DP mixture model where G* itself is a mixture of two terms. Unrelated to a discussion of the ODP, Bogdan et al. (2008) proposed the same model for multiple comparison problems. They compare misclassification rates using inference based on pFDR and FDR.
Algorithms for posterior Monte Carlo simulation in DP process mixture models can easily be modified to adapt to the mixture in G*. We will focus on the case A = {0} and outline the necessary changes for general A. We set hA(·) = δ0(·), i.e. a point mass at 0. Also, we choose hAc(·) to be continuous, e.g. N(0, σ2) and will denote it simply by h(·). The a.s. discrete nature of a DP random probability measure implies a positive probability for ties in a sample from G. The ties naturally define a partition of observations into clusters with common values μj. We introduce latent cluster membership indicators si to describe this partition by si = sk if μi = μk. We reserve the label si = 1 for the null distribution, i.e. we set si = 1 if and only if μi = 0. Let z−i and s−i denote the set of observations and the indicators excluding the i-th one. Also, let L be the number of clusters defined by ties (an unmatched single observation counts as a singleton cluster), ms be the size of cluster s, and m− i,s be the size of cluster s without observation i. Finally, let Γs = {i : si = s} denote the s-th cluster, and let denote the common value of μi for cluster s. Then, the i-th observation falls in one of the existing clusters or forms a new cluster according to the following modified Pólya urn scheme,
Here, denotes the posterior distribution of based on the prior h(·) and all observations zh, h ∈ Γs/{i}. Note that the posterior of given all the observations in cluster 1 is still a point mass δ0(·). We described the algorithm for a particular choice of A and G*. But it can be easily extended to more general A and G*. In the general case, clusters are formed either by samples from hA(·) or hAc (·). The algorithm is greatly simplified when A is an interval, hA(μ) ∝ h(μ) IA(μ), hAc (μ) ∝ h(μ) IAc (μ), for some continuous distribution h(μ) and π0 = G*(A). Then, equations (8) and (9) describe a traditional DP mixture of normals model with Gaussian base measure G*. The usual Pólya urn scheme for DP mixtures may be used.
Once we have a posterior Monte Carlo sample of the model parameters it is easy to evaluate the decision rule. Assume we have B posterior Monte Carlo samples of random partitions, , b = 1,…,B. We evaluate vi = E(ri | z) numerically as We evaluate by substituting v̄i in (6)
| (10) |
In the next section we show that SNP can be approximated by a single threshold statistic similar to (1).
4. The Bayesian Discovery Procedure (BDP)
4.1. The ODP as approximate Bayes Rule
We show that, under a DP prior model, d* ≈ dODP, that is the ODP is an approximate Bayes Rule. We start by writing the marginal posterior probability as expectation of conditional posterior probabilities,
and proceed with an approximation of the conditional posterior distribution p(G | z). The conjugate nature of the DP prior under random sampling implies . Recall that Fm ∝ ∑ δμ̂i is the empirical distribution of the maximum likelihood estimates μ̂i. For large m, small α, and an informative sampling model f(zi; μ), E(G | z) ≈ Fm. Further, for large m, the uncertainty of the posterior DP, is negligible, allowing us to approximate the posterior on the random probability measure G with a degenerate distribution at . Therefore,
| (11) |
The connection with the ODP rule is apparent by computing the posterior odds, . Finally, thresholding vi/(1− vi) is equivalent to thresholding
This is (3) with wj = I(μ̂j ∈ A).
Recognizing the ODP as an approximate Bayes rule opens two important directions of generalization. First, we can sharpen the approximation to define a slightly improved procedure, at no cost beyond moderate computational effort. We will do this in sections 4.2 and 4.3. Second, we can improve the ODP by making it more relevant to the decision problem at hand by modifying features of the underlying loss function; we will do this in Section 6.
4.2. The BDP statistic
We can improve the approximation in (11) by conditioning on a cluster configuration s and using cluster specific point estimates . Given model (8)–(9), we can approximate the posterior probability vi by
| (12) |
The μ̂j are point estimates based on (9). For example, one could use the posterior means μ̂j = E(μj | z). Short of posterior means, we propose to use a partition s(b) to evaluate cluster-specific point estimates , using maximum likelihood estimation within each cluster. The choice of the specific partition s(b) is not critical. Finally, we report test i significant if SBDP(zi) > t, for some threshold t. Substituting in (12) the SBDP can be interpreted as a multiple shrinkage version of the SODP statistic. For later reference we note that thresholding SBDP is equivalent to thresholding
| (13) |
By the earlier argument v̂i ≈ vi.
The nature of the approximation (12) is further clarified and formalized by the following result, which justifies the replacement of μi by the cluster specific mle’s . Proving asymptotic results of this kind for the DP is generally not easy. One has to establish that the posterior mass for the random G is concentrated on a set of RPM’s with a small number of discontinuities relative to m. See, for example, Arratia et al. (2003) and Coram and Lalley (2006) for recent discussions. In particular, Coram and Lalley (2006) address the problem from the perspective of “overfitting” for NP Bayes procedures. They conjecture that in a variety of problems, the critical determinant of the consistency of Bayes procedures are the rate functions in associated large deviations problems. We avoid the technical difficulties by proving the result for a finite dimensional Dirichlet prior, that is a random probability measure Gk such that
| (14) |
with (p1,…,pk) ~ D(α/k,…,α/k), and as in (9). Inference under the DP prior and (14) is comparable, since any integrable functional of the Dirichlet Process can be approximated by corresponding functionals of Gk for sufficiently large k (see Iswharan and Zarepour, 2002). Also Rodriguez et al. (2009) discuss the approximation of inference under a DP prior by results under Gk.
THEOREM 2
Assume , i = 1,…,m and a random effects distribution p(μi | Gk) = Gk as in (9), with Gk defined in (14). Assume f, hA(·), and hAc (·) satisfy the conditions for the Laplace approximation for an open set A (see Schervish, 1995, chapter 7.4.3). Then
The expectation is with respect to the posterior distribution over all possible partitions of {1,…,m} with at most k clusters and is the cluster-specific m.l.e.
PROOF
See appendix.
4.3. Multigroup comparisons
The definition of the BDP can easily be extended to the general k samples comparison problem. We assume that data for experimental units i, i = 1,…,m, are arranged by k distinct groups, and we wish to test if the k groups share a common sampling model. Let xi = {xi1,…,xin} be a vector of measurements across k experimental conditions, i = 1,…,m. We denote the subset of data from each condition by . Alternatively, data in each group may be reduced to statistics , and we can write z = {z1,…,zm}, , with similar notation for μ and μi. For notational simplicity, we proceed with the case k = 2. But the arguments hold for general k. The competing hypothesis are against . Typically . Under the loss (5) and the NP model
we can proceed as in section 2.3 and approximate the posterior odds for the i-th comparison by
| (15) |
where and μ̂i are appropriate estimates of the relevant parameters within and across conditions. Expression (15) is an estimated ODP statistics for the multicomparison problem, as discussed in Storey et al. (2007b). As before, substituting cluster-specific estimates for a selected partition s defines the corresponding BDP rule.
5. Comparison of SODP versus SBDP
5.1. A Simulation Study
We conduct a simulation study to compare the ODP with the NP Bayesian approximation outlined in the previous sections. We assume m = 2000 tests of H0 : μi = 0 versus H1 : μ ≠ 0 based on a single observation zi ~ N(μi, 1) for each test. The simulation truth is such that half of the observations are drawn from the null, while the other half are sampled from the following probability distribution:
| μi | −4 | −3 | −2 | −1.5 | 1.2 | 2 | 3 | 4.5 | 5.8 |
|---|---|---|---|---|---|---|---|---|---|
| pi | 0.02 | 0.08 | 0.01 | 0.01 | 0.27 | 0.02 | 0.08 | 0.005 | 0.005 |
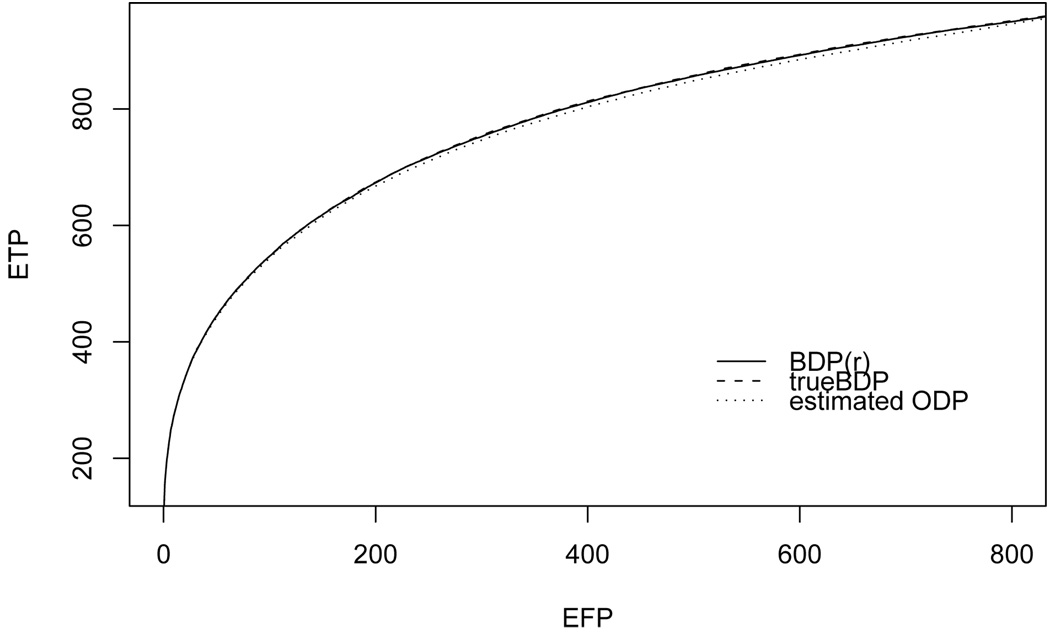
The distribution mimicks the observed distribution of the t-scores in the microarray data example considered in section 5.2, as shown in Figure 1. We use A = {0}, and hAc (·) = N (0, 1). We simulated 1000 datasets. For each simulated data set we ran 2000 iterations of a posterior MCMC sampler (with 1000 iterations burn in). The results confirm the obervation in Storey et al. (2007b) that the SODP outperforms the UMP unbiased procedure in all cases where the alternative means are not arranged in a perfectly symmetric fashion around zero. The SBDP further improves on the SODP by borrowing strength across comparisons with the multiple shrinkage induced by the DP clustering. Figure 2 shows that for any threshold of expected FP, the expected TP is comparable and slightly better under the SBDP than under the SODP. Expectations are over repeated simulations, i.e., the comparison is by the criterion that is being optimized by the oracle version (1) of the ODP. The curves for the SBDP are computed with a random (last) configuration; the true BDP curve refers to the BDP statistics computed based on the (known) true configuration. The differences in Figure 2 are small. However, for many applications with massive multiple comparisons the number of comparisons is much larger, leading to correspondingly larger differences in true positives. Most importantly, the improvements come at no additional experimental cost.
Fig. 1.
The scores zi for the simulation example in 5.1 (left panel) and the the data zi for the BRCA mutation microarray data in section 5.2 (right panel).
Fig. 2.
ETP = E(TP) versus EFP = E(TP) for ŜODP and SBDP. BDP(r) refers to SBDP computed for a random configuration (we use the last configuration in the MCMC simulation). TrueBDP refers to the BDP statistics computed on the basis of the simulation truth. The three curves are almost undistinguishable. See 5.1 for details.
We also considered a similar comparison using (12) with posterior means substituted for μ̂i, and using A = (−ε, ε), for several (small) values of ε, instead of the point null. The plot (not shown) of expected TP versus FP showed no substantial differences to Figure 2.
5.2. A Microarray Data Example
We first compare the ŜODP versus the SBDP test by analyzing a microarray dataset obtained from breast cancer tumor tissues. The data have been analyzed, among others, in Hedenfalk et al (2001), Storey and Tibshirani (2003) and Storey et al. (2007b) and can be downloaded from http://research.nhgri.nih.gov/microarray/NEJM_Supplement/. The data consist of 3,226 gene expression measurements on n1 = 7 BRCA1 arrays and n2 = 8 BRCA2 arrays (a third “sporadic” group was not used for this analysis). Following Storey and Tibshirani (2003), genes with one or more measurement exceeding 20 were eliminated from the data set, leaving m = 3, 169 genes.
Let xij be the log2 expression measurement for gene i on array j, i = 1,…,m, j = 1,…,n. For illustration purposes, we test differential expression between BRCA1 and BRCA2 mutation genes using the two samples t statistics , where x̄i,k and are respectively the sample mean and sample variance for gene i with respect to the arrays in group k, k = 1, 2. We assume model (8) and (9) with f(zi; μi) = N(μi, σ) and test H0i : μi = 0. For simplicity, we fix σ = 1. A model extension with a prior on unknown σ is straightforward. The results remain almost unchanged (not shown).
We assess the relative performance of the estimated ŜODP versus approximation (12) of the NP Bayes rule. For a fair comparison, we evaluate the SBDP as a frequentist rule with rejection region {SBDP ≥ c}. The power of the test is evaluated in terms of the FDR and the q-value. See Storey (2002) and Storey et al. (2007b) for more discussion of the q-value and its evaluation.
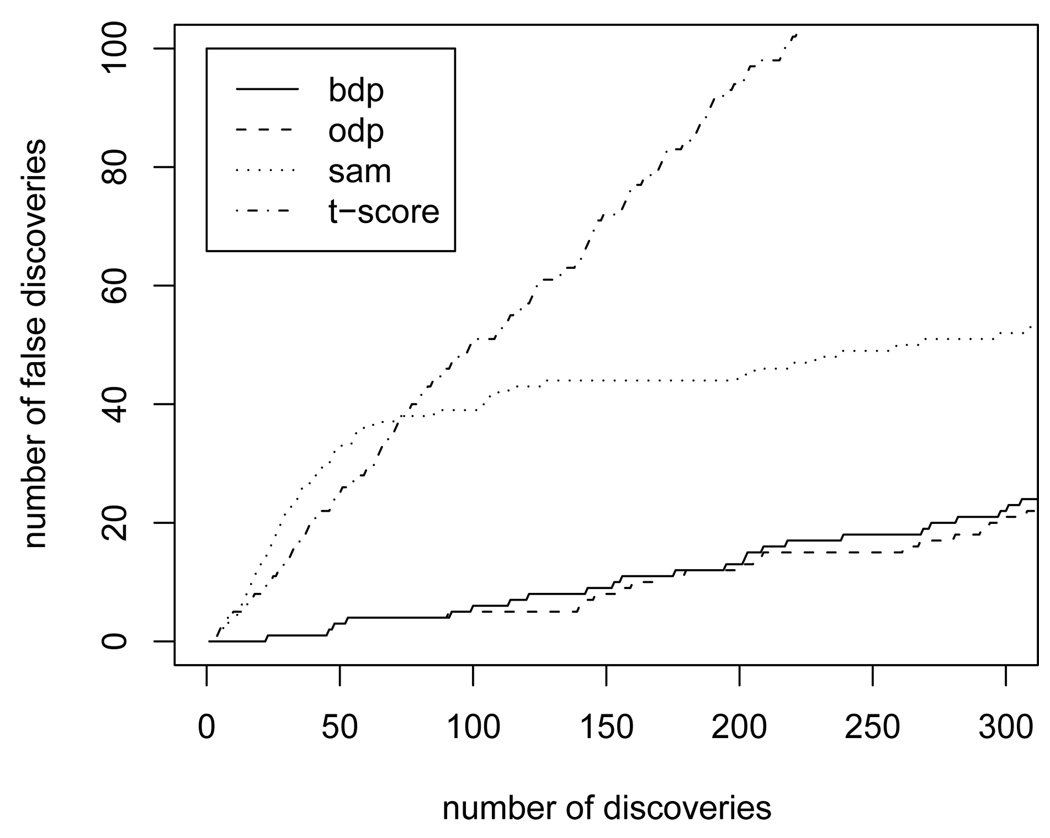
The evaluation of SBDP is based on 2000 iterations of a posterior MCMC sampler (1000 burn in). The number of significant genes detected at each q-value is shown in Figure 3. We report the SBDP as computed on the basis of the cluster configuration of a single iteration of the MCMC sampling scheme. We use the partition from a random iteration and from MAP (maximum a posteriori) partition. Other choices are possible. However, our experience does not suggest significantly different conclusions using alternative choices. In Figure 3 we see that in both cases the SBDP achieves larger numbers of significant genes at the same q-value than the SODP. The result leads us to recommend the SBDP as a useful tool in multicomparison problems where a natural clustering of the tests is expected. In Table 1, we report the percentage of genes that are flagged by both tests for some choices of q-values. For most q-values of practical interest the SBDP procedure identifies all genes that were flagged by the SODP, plus additional discoveries. For example, at q = 0.05, the BDP reveals 98 significant genes, against 87 revealed by the ODP and 47 by the standard FDR procedure devised by Benjamini and Hochberg (1995). Out of the 11 additional genes, 7 had been previously reported in the literature as significant in distinguishing BRCA1 and BRCA2 mutations (see Hedenfalk et al. 2001). The additionally identified genes come at no extra cost beyond moderate computational effort. No additional experimental effort is required, and no trade off in error rates is involved.
Fig. 3.
A comparison of SBDP and SODP for identifying differentially expressed genes (see section 5.2). The BDP(h) curve is is based on the MAP configuration; BDP(r) refers to SBDP computed under the (random) last configuration imputed in the MCMC simulation.
Table 1.
The intersection between the decisions with the ODP and the BDP procedures.
| q-value | ODP | BDP | BDP⋂ODP |
|---|---|---|---|
| 0.05 | 87 | 98 | 100% |
| 0.06 | 124 | 148 | 100% |
| 0.07 | 163 | 176 | 100% |
| 0.08 | 202 | 217 | 100% |
| 0.09 | 234 | 241 | 99.5% |
| 0.10 | 272 | 270 | 98.14% |
| 0.11 | 293 | 298 | 98.63% |
| 0.12 | 318 | 341 | 98.11% |
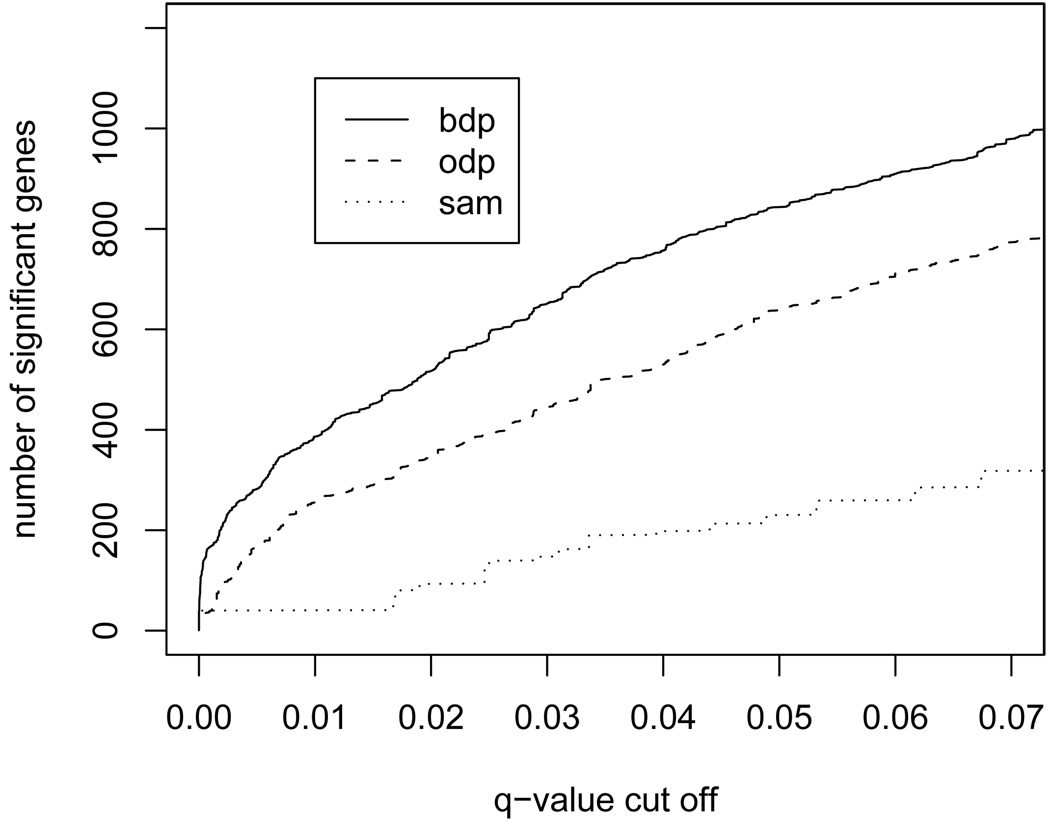
For an alternative comparison we consider the “Golden Spike” data of Choe et al. (2005). They describe an Affymetrix GeneChip experiment, where 1309 individual cRNAs have been “spiked in” at known relative concentrations between the two samples (spike-in and control). We implemented the BDP and ODP as described above. For comparison, we also include alternative comparisons using the SAM procedure (Tusher et al., 2001; Storey, 2002) and independent two-sample t-tests. We used the reduced Golden Spike dataset provided in the R package st. The dataset reports 11,475 genes with 3 replicates per group, including 1,331 genes with known differential expression. Figure 4 shows the results of a comparison of BDP, ODP, SAM and independent t-tests. To compute the SAM statistic we used the samr package in R and alternatively the SAM software from http://www-stat.stanford.edu/~tibs/SAM/. Both implementations give practically identical results.
Fig. 4.
Golden Spike data. A comparison of SBDP, SODP, SAM and independent two-sample t-tests.
Both comparisons, in Figures 3 and 5, are for the ODP and BDP test based on data with a single gene-specific summary statistic zi per gene. The restriction to a single summary statistic in the definition of ODP and BDP was purely for presentation, and can easily be relaxed. Let xijk denote the expression measurement for gene i, sample j and condition k. For simplicity, we still assume only two conditions. Group k = 0 is assumed to be a control/reference group. Let be the gene specific mean of the observations in group 0. We define zijk = xijk − μ̂i0, k = 0, 1, and assume , with μi0 ≡ 0. The hypothesis testing problem becomes:
Fig. 5.
Hedenfalk data: Comparison of SBDP, SODP and SAM under model (16).
Similar to before we define a random effects distribution G for and assume a NP Bayesian prior on G,
| (16) |
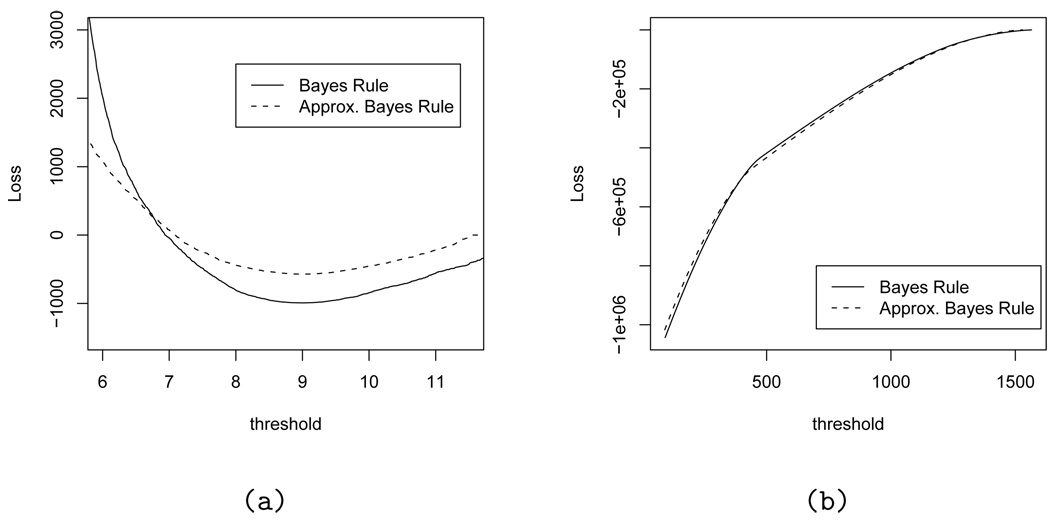
For the DP base measure we use a conjugate normal-inverse χ2 distribution . We can then proceed as in section 4.3 to define the ODP and BDP statistic, respectively. Figure 6 summarizes the results.
Fig. 6.
Expected loss under the exact Bayes rules dm* (left panel) and dp* (right panel) plotted against the cutoff t (solid lines). The dashed lines show the expected loss under the ODP rules based on (left panel) and (right panel).
6. Extensions of the ODP and BDP
6.1. Weighted Loss Functions
Once the optimality criterion and the probability model that lead to the ODP are identified, it is easy to modify the procedure to better accomodate preferences and losses other than (5). Often some discoveries might be considered more important than others. For example if A = {μi = 0} one might be more interested in large deviations from 0. In this case the loss function should include an explicit reward for detecting true signals as a function of some (monotone) function of μi. Scott and Berger (2003) describe a decision theoretic approach based on separate loss functions for false positives and false negatives. Similarly, we consider the following loss function,
| (17) |
where t(μi) is a known function of the mean, e.g. t(μi) = ||μi||, ||·|| being some norm of μi. Let . The posterior expected loss is . The Bayes rule is easily shown to be
| (18) |
for some threshold t. Although the nature of the rule has changed, we can still proceed as before and define a modified SODP statistics that approximates the Bayes rule. Let be an empirical Bayes estimate of , justified similarly to the approximation in (11). As before the point estimates μ̂i are cluster-specific m.l.e.’s for some partition s. By an argument similar to before we can justify the following single thresholding statistic as an approximation of (18):
| (19) |
We use as a single thresholding function for the multiple comparison test in lieu of the SODP.
Any loss function that is written as a sum of comparison-specific terms leads to similar approximations and modifications of the ODP. For example, consider a loss function that involves a stylized description of a follow-up experiment. The loss function is motivated by the following scenario. Many microarray group comparison experiments are carried out as a screening experiment. Genes that are flagged in the microarray experiment are chosen for a follow-up experiment to verify the possible discovery with an alternative experimental platform. For example, Abruzzo et al. (2005) describe a setup where RT-PCR (reverse transcription polymerase chain reaction) experiments are carried out to confirm discoveries proposed by an initial microarray group comparison. Abruzzo et al. (2005) report specific values for correlations across the platforms, error variances etc. On the basis of this setup Müller et al. (2007) consider a loss function that formalizes the consequences of this follow-up experiment. The loss function includes a sampling cost for the RT-PCR experiment and a reward that is realized if the RT-PCR experiment concludes with a significant outcome. The sample size is determined by a traditional power argument for a two-sample comparison, assuming a simple z-test for the difference of two population means. The probability of a significant outcome is the posterior predictive probability of the test statistic in the follow-up experiment falling in the rejection region. Let (μ̄i, si) denote the posterior mean and standard deviation of the difference in mean expression for gene i between the two experimental groups. Let ρ̄, ρ*, pρ denote known parameters of the joint distribution of the microarray gene expression and the outcome of the RT-PCR experiment for the same gene. Details of the sampling model (see Müller et al., 2007) are not required for the following argument. Finally, let qα define the (1 − α) quantile of a standard normal distribution. For a given signficance level α and a desired power (1 − β) at the alternative , we find a minimum sample size for the follow up experiment
Let Φ(z) denote the standard normal c.d.f. The posterior predictive probability for a significant outcome of the follow-up experiment is approximately
We formalize the goal of maximizing the probability of success for the follow-up experiment while controlling the sampling cost by the loss function
Here c2 is a fixed cost per gene for setting up a follow-up experiment, c1 is the (large) reward for a significant outcome in the follow-up experiment, and c3 ≡ 1 is the sampling cost per gene and experiment. The Bayes rule is . As before we can use and approximate the Bayes rule by a modified ODP style statistic. Let π̂i and n̂i denote πi and ni evaluated with the approximations for μ̄i and . We consider the modified ODP threshold statistic
Figure 6 compares the exact Bayes rules (18) and with the tests based on the approximate ODP statistic and , respectively.
6.2. Spatial Dependence
The nature of the ODP as an approximate Bayes rule was based on the semi-parametric model (7). However, the Bayes rules (6) or (18) remain meaningful under any probability model, as long as vi and have meaningful interpretations. For example, in geostatistical applications, we may be interested in isolating the exceedance regions of a spatial process, i.e. where the process has values above a given threshold (Zhang et al., 2008). Similarly, in the analysis of fMRI data, we aim to detect region specific activations of the brain. See Pacifico et al. (2004) and Flandin and Penny (2007) for two recent Bayesian solutions to the problem. In particular, Friston and Penny (2003) propose an empirical Bayes approach to build posterior probability maps of site specific signals. These approaches do not make use of an explicitely defined optimality criterion to support the proposed decision rules.
We consider a variation of the ODP that is suitable for spatial inference problems, using a specific spatial probability model as an example. We use the spatial model proposed by Gelfand et al. (2005). Let {Y (s), s ∈ D} be a random field, where D ⊂ Rd, d ≥ 2. Let s(n) = (s1,…,sn) be the specific distinct locations in D where observations are collected. Assume that we have replicate observations at each location so that the full data set consists of the collection of vectors Yi = {Yi(s1),…,Yi(sn)}T, i = 1,…,m. We assume
| (20) |
where f is some multivariate distribution, μ is a (not necessarily constant across s) regressive term and θi = {θi(s1),…,θi(sn)}T is a spatial random effect, such that
| (21) |
for some base measure G0. See Gelfand et al. (2005) for details. The assumption of the DP prior in the model is unrelated to the DP that we used to justify the nature of the ODP as approximate Bayes rule. In this setting, the inferential problem might be quite general, as it may involve subsets of sites and replicates.
For simplicity, we consider a null hypothesis specific to each location s and replicate i: H0si: θi ∈ Asi, Asi = {θi(s) > b}. For a fixed replicate i, let dj = d(sj) be the decision at site sj, j = 1,…,n. Analogously, let rj = r(sj) denote the unknown truth at sj. We could now proceed as before and consider the loss (5) and the rule, , where vj is the posterior probability of the event A under the chosen probability model. For m sufficiently large and under model (20)–(21), it is possible to use the asymptotic arguments detailed in section 4 and define a BDP statistics for the spatial testing problem.
The loss function (5) is usually not an adequate representation of the investigator’s preferences in a spatial setting. Posterior probability maps may show very irregular patterns that could lead to, for example, flagging very irregular sets of pixels for exceedance θi(s) > b. We may explicitly include into the loss function a penalty for such irregularities, i.e.
| (22) |
where PI is a penalization for irregularities. For example, PI could be the number of connected regions. See the example below. The decision rule corresponding to (22) is
Finding d* requires numerical optimization.
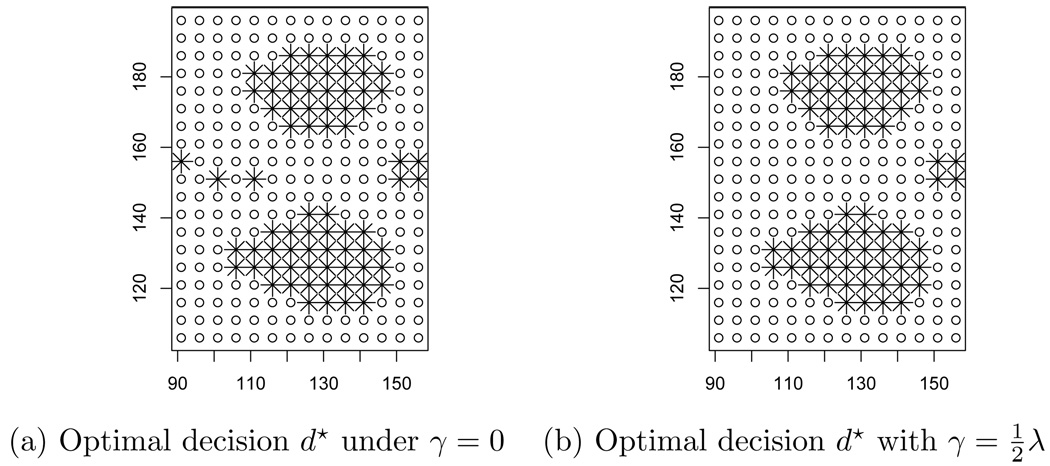
We illustrate the approach with a dataset of 18 individuals who underwent an MRI scan to detect signals of neurodegenerative patterns typical of the Alzheimer’s disease (Ashburner et al., 2003). The data have been provided by the Laboratory of Epidemiology and Neuroimaging, IRCSS, Centro San Giovanni di Dio, Brescia, Italy and have been previously normalized with the freely available SPM5 sowftare (http://www.fil.ion.ucl.ac.uk/spm/, see Friston et al. (1995) and Worsley and Friston (1995)). For simplicity, the dataset is restricted to gray density matter intensity values collected on a regular two-dimensional grid of 14 × 19 pixels encompassing the hippocampus and are treated as continuous. The data have been analyzed in Petrone et al. (2009) before, although with a different inferential aim.
We assume the random effect model (20)–(21), where f is a multivariate gaussian, with mean μ + θ and covariance matrix τ2In. The base measure G0 is a zero-mean stationary Gaussian process with variance σ2 and correlation ρ(s, s′) = exp(−φ||s − s′||), for some range parameter φ. Vague inverse gamma prior distributions on τ2 and σ2, and a vague gamma prior for φ complete the model. Hence, (20)–(21) defines a DP mixture of spatial processes (Gelfand et al., 2005). The model is sufficiently flexible to account for most of the spatial dependence observed in each individual. However, it is known that one of the marks of the Alzheimer’s disease is local hippocampal atrophy. Low grey matter intensity observed in normal neuroanatomical structures of the brain should not be reported as a signal. This consideration may lead to introduce several kinds of penalties into (22) to penalize for detections in non-interesting regions. Local atrophy is a condition that typically affects clusters of sites at the same time. This leads us to consider a penalty PI for the number of isolated signals on D. Specifically, PI is the number of interconnected regions and isolated points for which di(s) = 1. We use a numerical procedures to explore the action space and minimize (22). We find the optimal decision d* by a random search, initialized with the optimal rule under γ = 0.
Figure 7 shows the resulting optimal rule for one individual in the MRI dataset. We are interested in detecting regions of low gray matter intensity in the MRI scans. Hence we consider A = {θ(s) < b}, where b is a fixed constant, corresponding to the first decile of the dataset. The activation threshold for the posterior probability is t = 0.8. Figure 7 shows the activation map for an individual with recognizable signs of hypoccampal atrophy for γ = 0 (panel (a)) and for (panel (b)). The additional penalty term provided a principled and coherent means of removing the singleton clusters that would otherwise be reported.
Fig. 7.
The effect of a loss function disfavoring isolated signals on the decisions taken according to loss (5). See 6.2 for details.
7. Conclusions and Summary
Starting from a decision theoretic framework, we provided an interpretation of the ODP as an approximate Bayes rule and introduced two directions of generalizations. First we improved the rule by sharpening the approximation. In a simulation example and a data analysis example we showed improved performance of the resulting BDP rule, even by the frequentist operating characteristics that are optimized by the oracle version of the ODP. Second, we considered generalizations of the ODP by replacing the original generic loss function by loss functions that better reflect the goals of the specific analysis. For loss functions with similar additive structure as the original loss function the resulting rule can still be approximated by a single thresholding statistic similar to the ODP.
The use of a decision theoretic framework provides a convenient assurance of coherent inference for the proposed approach. However, it also inherits the limitations of any decision theoretic procedure. The optimality is always with respect to an assumed probability model and loss function. The stated loss function is usually a stylized version of the actual inference goals. Often that is sufficient to obtain a reasonable rule. But we still caution to critically validate and if necessary revise the inference in the light of evaluations such as frequentist operating characteristics. Also, the proposed methods are more computation intensive than the original ODP procedure. In the simplest case we require some additional simulation to find a clustering of comparisons to compute cluster-specific m.l.e.’s.
The main strengths of the proposed approach are the generality and the assurance of coherent inference. The approach is general in the sense that the proposed methods are meaningful for any underlying probability model, and in principle for arbitrary loss functions. The approach is coherent in the sense that it derives from minimizing expected loss under a well defined probability model. From a data analysis perspective, an important strength of the proposed approach is the need and the opportunity to explicitely think about the inference goals and formalize them in the loss function. A practical strength is the opportunity for improved inference at no additional experimental cost, and only moderate additional computational cost.
Aknowledgments
The work of the third author is partly supported by the NIH CTSA Grant UL1 RR024982.
Appendix
Proof of Preposition 1
The proof follows closely the proof of Theorem 1 in Storey and Tibshirani (2003). First, rewrite
| (23) |
The expectation is with respect to the distribution of (z1,…,zm), conditionally on the event that some of the comparisons are significant. Hence,
Since di is a function of the sample z1,…,zm, the inner expectation is just
and since ,
Proof of Theorem 2
Because of the exchangeability of samples from a Pólya Urn, without loss of generality, we may consider vm = p(rm = 1|z1,…,zm). First, note that
Both numerator and denominator take the form of
for a Borel set B. Let so(m) be a vector of cluster indicators, that is , i = 1,…,m, j = 1,…,k.
For any fixed m, let {so(m)} denote the set of all partitions of {1,…,m} into at most k clusters. From the discussions in Ferguson (1983), Lo (1984), Ishwaran and James (2003), it follows that
| (24) |
where is the number of elements in cluster j. If . Expression (24) highlights that any partition of {1,…,m} can be obtained from a corresponding partition of {1,…,m−1} by adding the m-th observation to any of the previous cluster or by forming a new one.
Now, note that for each j = 1,…,k, either . If , we can use Laplace approximation arguments (see Schervish, 1995, chapter 7.4.3 or Ghosh et al., 2006, pag. 115) to obtain
where is the MLE estimate computed in cluster j, j = 1,…k, obtained by solving and Φ(·) denotes a standard gaussian probability distribution.
Next we relabel the non-empty clusters by identifying the set as the set of unique values .
The proof is completed after noting that, since , and because of the asymptotic consistency of posterior distributions, as m → ∞, Φ(μ̂j ∈ B) → IB(μ̂j).
Contributor Information
Michele Guindani, University of New Mexico, Alberquerque, NM 87111, U.S.A..
Peter Müller, University of Texas M.D. Anderson Cancer Center, Houston, TX 77030, U.S.A..
Song Zhang, University of Texas Southwestern Medical Center, Dallas, TX 75390, U.S.A..
References
- Arratia R, Tavaré S, Barbour A. Logarithmic Combinatorial Structures: A Probabilistic Approach. EMS Monographs in Mathematics; 2003. [Google Scholar]
- Ashburner J, Csernansky J, Davatzikos C, Fox N, Frisoni G, Thompson P. Computer-assisted imaging to assess brain structure in healthy and diseased brains. The Lancet Neurology. 2003;2:79–88. doi: 10.1016/s1474-4422(03)00304-1. [DOI] [PubMed] [Google Scholar]
- Benjamini Y, Hochberg Y. Controlling the false discover rate – a practical and powerful approach to multiple testing. J. R. Statist. Soc. B. 1995;75:289–300. [Google Scholar]
- Bogdan M, Gosh J, Tokdar S. A comparison of the benjamini-hochberg procedure with some Bayesian rules for multile testing. In: Balakrishnan N, Peña E, Silvapulle M, editors. Beyond Parametrics in Interdisciplinary Research: Festshcrift in Honor of Professor Pranab K. Sen. Beachwood, Ohio, USA: Institute of Mathematical Statistics; 2008. pp. 211–230. IMS Collections. [Google Scholar]
- Cao J, Jie X, Zhang S, Whitehurst A, White M. Bayesian optimal discovery procedure for simultaneous significance testing. BMC, Bioinformatics. 2009;10:5. doi: 10.1186/1471-2105-10-5. to appear. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choe S, Boutros M, Michelson A, Church G, Halfon M. Preferred analysis methods for affymetrix genechips revealed by a wholly defined control dataset. Genome Biology. 2005;6:R16. doi: 10.1186/gb-2005-6-2-r16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohen A, Sackrowitz H. More on the inadmissibility of step-up. Journal of Multivariate Analysis. 2007;98:481–492. [Google Scholar]
- Coram M, Lalley S. Consistency of bayes estimators of a binary regression function. Ann. Statist. 2006;34:1233–1269. [Google Scholar]
- Dahl D, Newton M. Multiple hypothesis testing by clustering treatment effects. J. Am. Statistic. Assoc. 2007;102:517–526. [Google Scholar]
- Efron B, Tibshirani R, Storey J, Tusher V. Empirical bayes analysis of a microarray experiment. J. Am. Statist. Assoc. 2001;96:1151–1160. [Google Scholar]
- Ferguson T. A Bayesian analysis of some nonparametric problems. Ann. Statist. 1973;1:209–230. [Google Scholar]
- Ferguson T. Prior distributions on spaces of probability measures. Ann. Statist. 1974;2:615–629. [Google Scholar]
- Ferguson TS. Bayesian density estimation by mixtures of normal distributions. In: Rizvi H, Rustagi J, editors. Recent Advances in Statistics. New York: Academic Press; 1983. pp. 287–302. [Google Scholar]
- Flandin G, Penny W. Bayesian fmri data analysis with sparse spatial basis function priors. Neuroimage. 2007;34:1108–1125. doi: 10.1016/j.neuroimage.2006.10.005. [DOI] [PubMed] [Google Scholar]
- Friston KJ, Ashburner L, Poline J, Frith C, Frackowiak R. Spatial registration and normalization of images. Humain Brain Mapping. 1995;2:165–189. [Google Scholar]
- Friston KJ, Penny W. Posterior probability maps and spms. NeuroImage. 2003;19:1240–1249. doi: 10.1016/s1053-8119(03)00144-7. [DOI] [PubMed] [Google Scholar]
- Gelfand A, Kottas A, MacEachern S. Bayesian nonparametric spatial modeling with Dirichlet processes mixing. J. Am. Statist. Ass. 2005;100:1021–1035. [Google Scholar]
- Ghosh J, Delampady M, Tapas S. An Introduction to Bayesian Analysis: Theory and Methods. Springer; 2006. [Google Scholar]
- Gopalan R, Berry D. Bayesian multiple comparisons using dirichlet process priors. J. Am. Statist. Assoc. 1993;93:1130–1139. [Google Scholar]
- Hedenfalk I, Duggan D, Chen Y, Radmacher M, Bittner M, Simon R, Meltzer P, Gusterson B, M E, Raffeld M, Yakhini Z, Ben-Dor A, Dougherty E, Kononen J, Bubendorf L, Fehrle W, Pittaluga S, Gruvberger S, Loman N, Johannsson O, Olsson H, Wilfond B, Sauter G, Kallioniemi Olli-P, Borg A, Trent J. Gene-expression profiles in hereditary breast cancer. New England Journal of Medicine. 2001;344:539–549. doi: 10.1056/NEJM200102223440801. [DOI] [PubMed] [Google Scholar]
- Ishwaran H, James L. Some further developments for stick-breaking priors: finite and infinite clustering and classification. Sankhya Series A. 2003;65:577–592. [Google Scholar]
- Lo A. On a class of bayesian nonparametric estimates: I density estimates. Ann. Statist. 1984;12(1):351–357. [Google Scholar]
- Müller P, Parmigiani G, Rice K. Fdr and bayesian multiple comparisons rules. In: Bernardo J, Bayarri M, Berger J, Dawid A, Heckerman, Smith AD, West M, editors. Bayesian Statistics 8. Oxford, UK: Oxford University Press; 2007. [Google Scholar]
- Müller P, Parmigiani G, Robert CP, Rousseau J. Optimal sample size for multiple testing: the case of gene expression microarrays. J. Am. Statist. Assoc. 2004;99:990–1001. [Google Scholar]
- Neal RM. Markov chain sampling methods for dirichlet process mixture models. Journal of Computational and Graphical Statistics. 2000;9:249–265. [Google Scholar]
- Pacifico MP, Genovese C, Verdinelli I, Wasserman L. False discovery control for random fields. J. Am. Statistic. Assoc. 2004;99:1002–1014. [Google Scholar]
- Petrone S, Guindani M, Gelfand A. Hybrid Dirichlet processes for functional data, to appear. J. R. Statist. Soc. B. 2009 to appear. [Google Scholar]
- Rodriguez A, Dunson D, Gelfand A. The nested Dirichlet process. J. Am. Statist. Assoc. 2009 to appear. [Google Scholar]
- Schervish MJ. Theory of Statistics. Springer; 1995. [Google Scholar]
- Scott J, Berger J. An exploration of aspects of bayesian multiple testing. Journal of Statistical Planning and Inferece. 2003;136:2144–2162. [Google Scholar]
- Storey J. A direct approach to false discovery rates. J. R. Statist. Soc. B. 2002;64:479–498. [Google Scholar]
- Storey J. The optimal discovery procedure: a new approach to simultaneous significance testing. J. R. Statist. Soc. B. 2007a;69(3):347–368. [Google Scholar]
- Storey J, Dai J, Leek J. The optimal discovery procedure for large-scale significance testing, with applications to comparative microarray experiments. Biostatistics. 2007b;8:414–432. doi: 10.1093/biostatistics/kxl019. [DOI] [PubMed] [Google Scholar]
- Storey J, Tibshirani R. Statistical significance for genomewide studies. Proc. Natn. Acad. Sci. USA. 2003;100(16):9440–9445. doi: 10.1073/pnas.1530509100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tusher V, Tibshirani R, Chu G. Significance analysis of microarrays applied to the ionizing radiation response. PNAS. 2001;98:5116–5121. doi: 10.1073/pnas.091062498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Worsley KJ, Friston KJ. Analysis of fmri time-series revisited-again. NeuroImage. 1995;2:173–181. doi: 10.1006/nimg.1995.1023. [DOI] [PubMed] [Google Scholar]
- Zhang J, Craigmile P, Cressie N. Loss function approaches to predict a spatial quantile and its exceedance region. Technometrics. 2008;50:216–227. [Google Scholar]