Abstract
Within the circulatory system, blood flow regulates vascular remodeling1, stimulates blood stem cell formation2, and plays a role in the pathology of vascular disease3. During vertebrate embryogenesis, vascular patterning is initially guided by conserved genetic pathways that act prior to circulation4. Subsequently, endothelial cells must incorporate the mechanosensory stimulus of blood flow with these early signals to shape the embryonic vascular system4. However, few details are known about how these signals are integrated during development. To investigate this process, we focused on the aortic arch (AA) blood vessels, which are known to remodel in response to blood flow1. By using 2-photon imaging of live zebrafish embryos, we observe that flow is essential for angiogenesis during AA development. We further find that angiogenic sprouting of AA vessels requires a flow-induced genetic pathway in which the mechano-sensitive zinc finger transcription factor klf2a5-7 induces expression of an endothelial-specific microRNA, mir-126, to activate Vegf signaling. Taken together, our work describes a novel genetic mechanism in which a microRNA facilitates integration of a physiological stimulus with growth factor signaling in endothelial cells to guide angiogenesis.
Within a blood vessel, flow exerts tangential and perpendicular forces upon endothelial cells, leading to cytoskeletal rearrangements and changes in gene expression4. While initial embryonic vascular patterning is largely independent of these hemodynamic forces, the onset of circulation drives subsequent remodeling of the circulatory system4. For example, flow plays an important role in the unilateral regression of the sixth AA during mouse development1. In zebrafish, the fifth and sixth AA arise after flow begins and form a persistent connection to the lateral dorsal aortae (LDA) that provides circulation to the trunk8, 9. These vessels continue to undergo angiogenesis throughout larval stages to comprise the gill vasculature9. To investigate how flow affects angiogenesis, we observed development of AA5 and 6 in zebrafish embryos by 2-photon time-lapse imaging (Supplementary Fig. 1a-c). To co-visualize endothelial cells and flow, we performed microangiography on Tg(kdrl:egfp)la116 embryos, which display fluorescent green endothelial cells, using unconjugated Quantum dots (QDots). At 46 hpf, we observed AA perfusion, but no connection between the fifth and sixth AA and the LDA (data not shown and Supplementary Fig. 1d, 46h). Several hours later, the AA 5/6 connecting vessel (referred to as AA5x according to reference 8) sprouted from the left and right AAs (Supplementary Movie 1). At this point, the sprouts were sufficiently lumenized to allow perfusion with Qdots (Supplementary Fig. 1d, 53.75 magnified; Supplementary Movies 1 and 2). However, blood cells entering from the ventral aorta (VA) became trapped in AA5 and 6 (Supplementary Movie 3). AA5x sprouts then fused with the LDA to form a patent circulatory connection (Supplementary Fig. 1d, 59.75h, Supplementary Movie 1). Subsequently, the AA5x fully lumenized and blood flow through AA5 and 6 commenced (Supplementary Movie 4). These observations indicated that the AA5x develops via concomitant angiogenesis and lumenization in the presence of flow.
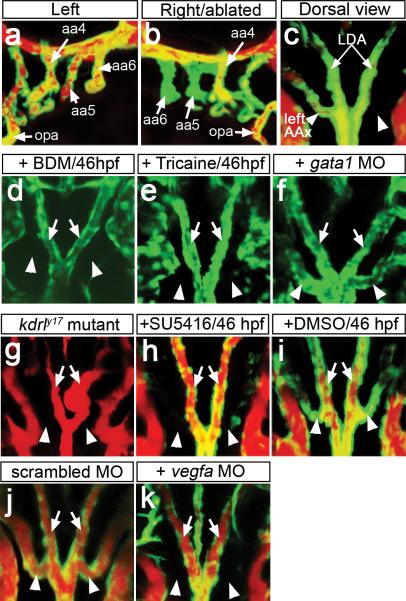
To determine if flow was required for this process, we performed unilateral laser microsurgery on Tg(kdrl:egfp)la116 embryos to sever the connection between the VA and AA5 and 6 prior to AA5x sprouting (Supplementary Figs. 1e, 2a). Following microsurgery at 46 hpf, we observed normal AA perfusion on the unoperated side by microangiography at 72 hpf (Fig. 1a). By contrast, on the operated side (right) AA5 and 6 failed to bear flow (Fig. 1b), although cranial blood vessels and the AAs appeared morphologically normal (Fig. 1b; Supplementary Figure 2b). A dorsal view of the same embryo revealed that the AA5x formed on the left side of the embryo, but not on the right side where flow was blocked (Fig. 1c, Supplementary Table 1). To support these results, we treated Tg(kdrl:egfp)la116 embryos beginning at 46 hpf with the myosin ATPase inhibitor 2,3-butanedione 2-monoxime (BDM) or the anesthetic Tricaine methanesulfonate to arrest the heart and block circulation10. In both treatments, embryos failed to form the AA5x (Fig. 1d, e; Supplementary Table 1), although vascular morphogenesis in other anatomical locations appeared normal (Supplementary Figure 3). 2-photon time-lapse microscopy of embryos without flow suggested that a failure to initiate sprouting, rather than vessel regression, was responsible for loss of AA5x (Supplementary Movies 5 and 6). Time lapse analysis using Tg(fli1a:negfp)y7 embryos, in which endothelial cell nuclei are labeled with Egfp, revealed decreased migratory activity of cells within the aortic arches in the absence of flow when compared to wild type (Supplementary Movies 7 and 8). Interestingly, embryos injected with a gata1 Morpholino displayed normal AA5x development (Fig 1f, Supplementary Table 1), suggesting that shear stress from blood cells was dispensable for AA5x angiogenesis. Together, these results indicate that the AA5x forms via angiogenesis and that this process is dependent on flow.
Figure 1. AA5x angiogenesis requires flow and Vegf signaling.
a-k, Tg(kdrl:egfp)la116 embryos at 72 hpf (a-c), 60 hpf (d-f) or 65 hpf (g-k). a-c, g-k Embryos subjected to microangiography. Endothelial cells are green, flow is red. a, b, Lateral views, anterior to left (a), or right (b), dorsal is up. c-k, dorsal view, anterior is up. a, b, aortic arches (AA, numbered, indicated by arrows) after severing right AA5 and 6 from ventral aorta; opa – opercular artery. c, dorsal view of embryo in a, b. d, e, Stills from Supplementary Movies 5 and 6. Embryos treated beginning at 46 hpf with BDM (d), Tricaine (e), or injected with gata1 MO (f). g, kdrly17 mutant embryo at 65 hpf. h-k, Embryos treated with 2.5 μM SU5416 (h) or 0.1% DMSO (i) beginning at 46 hpf or injected with 3 ng of scrambled MO (j) or 3 ng of Vegfa MO (k). c-k, Arrows: lateral dorsal aortae (LDA), arrowheads: AA5x.
Vascular endothelial growth factor (Vegf) signaling has been implicated in flow-mediated AA remodeling in mouse embryos1. Accordingly, we observed AA expression of the zebrafish Vegf receptor-2 ortholog, kdrl, including expression in the developing AA5x at 48 hpf (Supplementary Fig. 4a). We also observed vegfa expression in the developing glomerulus (Supplementary Fig. 4b, c), which is located near the branch point of the dorsal aorta and towards which the AA5x sprouts (Supplementary Fig. 4d), and in cells surrounding the AA blood vessels (Supplementary Fig. 4e). Consistent with a role for Vegf signaling during AA5x angiogenesis, embryos bearing a kinase-dead mutation in Kdrl (referred to as kdrly17; ref 11) failed to form a patent AA5x (Fig. 1g; Supplementary Table 1). Furthermore, treatment with the Vegf receptor inhibitor SU5416 from 46 to 65 hpf resulted in a block in AA5x formation, while DMSO had no effect (Fig. 1h, i; Supplementary Table 1). Similarly, partial reduction of Vegfa using a low Morpholino dose (3 ng; see reference 12) blocked AA5x development (Fig. 1j, k). Overall vascular morphology and circulatory function, including initial perfusion of the aortic arches, were normal following these manipulations (Supplementary Fig. 3). These observations demonstrate that AA5x formation requires Vegf signaling. In other developmental settings, Notch signaling coordinates Vegf-stimulated angiogenesis13, 14. However, we did not detect expression of Notch signaling molecules or Notch activation in the AAs (Supplementary Fig. 5a-c) and AA5x was not affected by loss of the Notch ligand dll4 (Supplementary Fig 5d). These results suggest that a Notch-independent mechanism is responsible for Vegf-stimulated AA5x angiogenesis.
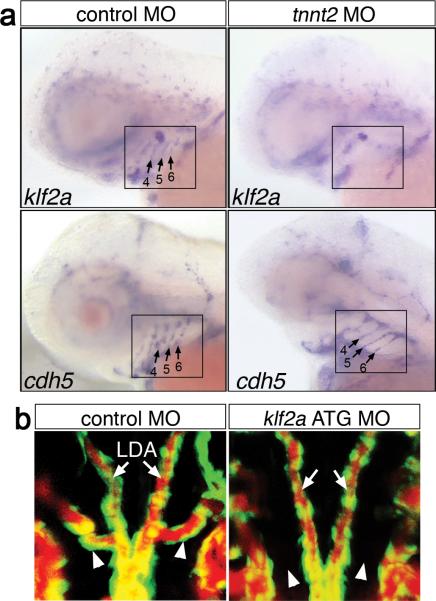
A possible candidate gene responsible for integrating flow and Vegf signaling during AA5x formation was the zinc finger transcription factor, klf2, which is induced by flow in endothelial cells6, 7. We observed that zebrafish klf2a was expressed in the AA in a pattern similar to the endothelial marker, vascular-endothelial cadherin (cdh5; Fig. 2a) and was expressed in the developing AA5x (Supplementary Fig. 4f). Furthermore, AA expression of klf2a, but not cdh5, was reduced in cardiac troponin T2 (tnnt2)-deficient embryos, which lack circulation (Fig. 2a; Supplementary Table 2; Supplementary Fig. 6a, ref 15) and in embryos treated with Tricaine (Supplementary Fig. 6b; Supplementary Table 2). To determine if klf2a was required for AA5x angiogenesis, we utilized Morpholinos targeting either the klf2a exon 3 splice acceptor site (Supplementary Fig. 7a, b) or the klf2a start codon. Embryos injected with either Morpholino displayed normal morphology and grossly normal circulatory patterns, including perfusion of the aortic arches following angiography (Supplementary Fig. 7c, d and data not shown), consistent with recent work demonstrating relatively normal flow patterns and heart rate in klf2a-deficient zebrafish embryos at 48 hpf 16. However, the normal transient AA circulatory block persisted in klf2a-deficient embryos (compare Supplementary Movies 3, 4, and 9), suggesting a defect in AA5x formation. Indeed, while embryos injected with control Morpholino appeared normal, klf2a-deficient siblings failed to develop the AA5x (Fig. 2b; Supplementary Fig. 7e, f; Supplementary Table 3). Thus, despite the presence of flow, loss of klf2a mimics the AA5x defect observed in embryos lacking flow or Vegf signaling.
Figure 2. AA5x angiogenesis requires klf2a.
a, Embryos subjected to whole mount in situ hybridization at 65 hpf using indicated riboprobes (klf2a, cdh5). Lateral view, anterior to the left, dorsal is up. Embryos were injected with 2 ng of scrambled control (left) or tnnt2 (right) MO. Aortic arch region is denoted by black boxes; aortic arches 4-6 are indicated by numbered arrows in cases where staining is present. b, Microangiogram of 72 hpf Tg(kdrl:egfp)la116 embryo injected with 11 ng-embryo scrambled control MO (top) or klf2a ATG MO (bottom); arrowheads indicate position of normal AA5x formation, arrows denote lateral dorsal aortae. Dorsal views, anterior is up.
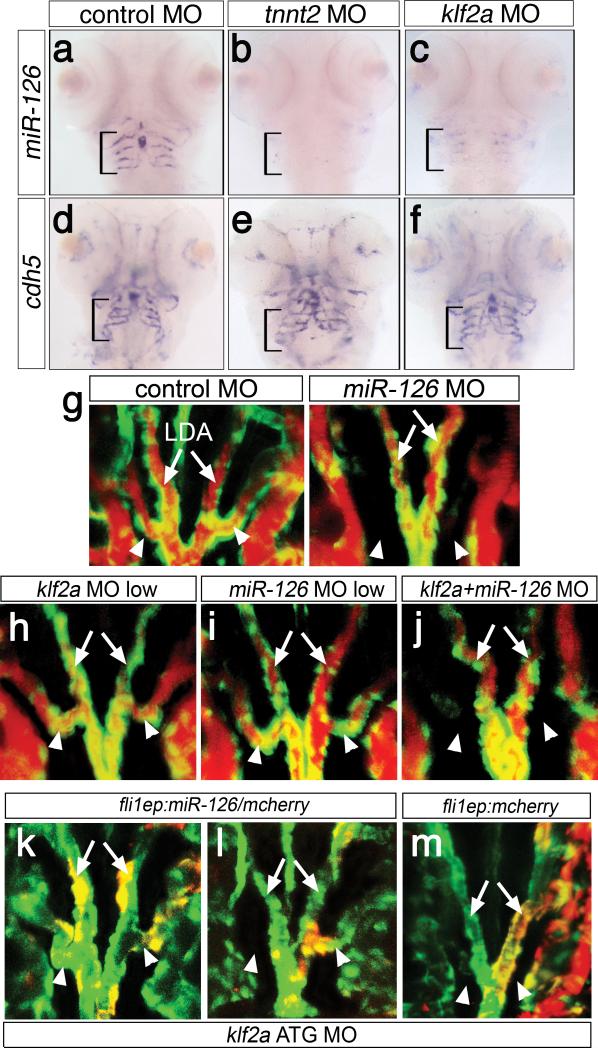
In Xenopus laevis embryos, klf2 is important for Vegf receptor-2 expression 17. However, kdrl expression appeared normal in klf2a-deficient zebrafish embryos (Supplementary Fig 6a, c). Similarly, neither kdrl nor vegfa were altered in embryos lacking circulation (Supplementary Fig. 6a, c) and we did not observe consistent reduction in other known klf2 responsive genes5-7 in the absence of flow or klf2a (Supplementary Fig. 6a). These results raised the possibility that a post-transcriptional mechanism linked flow, klf2a, and Vegf signaling. A candidate for this role was the endothelial-restricted microRNA, miR-12618, which can enhance Vegf signaling19, 20. While miR-126 expression was apparent in the embryonic vasculature prior to circulation (Supplementary Fig. 8a), at later stages its expression appeared much higher in the AAs (Fig 3a, Supplementary Fig. 8a). Strikingly, we found that AA miR-126 expression was dependent on both flow and klf2a expression. While control embryos expressed high levels of miR-126 within the AAs, tnnt2- or klf2a-deficient embryos did not (Fig. 3a-c; Supplementary Figs. 6a and 8b, Supplementary Table 2). By contrast, expression of cdh5 and let-7a was unchanged in the absence of flow or klf2a (Fig. 3d-f, Supplementary Fig. 8b), ruling out a general defect in endothelial gene expression or microRNA processing, respectively. Tricaine treatment to block flow similarly reduced miR-126 AA expression (Supplementary Fig. 6b; Supplementary Table 2). Embryos injected with a Morpholino to prevent miR-126 processing (Supplemental Fig. 8c) displayed blocked AA circulation (Supplementary Movie 10) and hemorrhage in this region by 60 hpf (Supplementary Fig. 8d). Similar to loss of klf2a, the AA5x did not form in miR-126-deficient embryos (Fig. 3g; Supplementary Table 3; Supplementary Movie 11). We also observed ectopic branching of segmental vessels and abnormal patterning of cranial blood vessels in miR-126-deficient embryos (Supplementary Fig. 8e). These results demonstrate that AA expression of miR-126 requires flow and klf2a and that miR-126 itself is required for AA5x angiogenesis.
Figure 3. miR-126 acts downstream of klf2a during AA5x development.
a-f, Ventral view, anterior is up. Bracket: AA3-6. Expression of miR-126 (a-c) or cdh5 (d-f). Embryos injected with 11 ng control MO (a, d), 2 ng tnnt2 MO (b, e), 11 ng klf2a ATG MO (c, f). g-j, Tg(kdrl:egfp)la116 embryos at 65 hpf, dorsal view, anterior is up. Endothelial cells are green, blood flow is red. Embryos injected with 20 ng of control or miR-126 MO (g), 2 ng klf2a ATG MO (h), 7 ng miR-126 MO (i), or co-injected with 2 ng klf2a ATG MO and 7 ng miR-126 MO (j). k-m, Tg(kdrl:egfp)la116 embryos co-injected with 11 ng klf2a ATG MO and pTol-fli1ep:miR-126/mcherry (k, l) or 11 ng klf2a ATG MO and pTol-fli1ep:mcherry (m). k-m, Yellow indicates egfp and mcherry co-expression; dorsal views, anterior is up. g-m, Arrows: lateral dorsal aortae (LDA), arrowheads: AA5x.
Our results suggested that klf2a acted upstream of miR-126 to induce flow-stimulated angiogenesis. Consistent with this possibility, exogenous klf2a in embryos lacking blood flow restored AA miR-126 expression (Supplementary Fig. 9a-c). To further test their genetic interaction, we co-injected klf2a and miR-126 Morpholinos at suboptimal doses that individually caused no, or mild low penetrant aortic arch defects (Fig. 3h, i; Supplementary Fig. 9d,e; Supplementary Table 3). Co-injection of both Morpholinos in this case caused a drastic increase in the penetrance of AA5x defects, suggesting that miR-126 and klf2a act in a common pathway (Fig. 3j, Supplementary Fig. 9e, Supplementary Table 3). Interestingly, other vascular defects observed in miR-126-deficient embryos were not apparent in co-injected embryos (Supplementary Fig. 8e, data not shown), suggesting a specific genetic interaction between miR-126 and klf2a during AA5x development. To further confirm that miR-126 functioned downstream of klf2a, we drove mosaic endothelial expression of a miR-126/monomeric cherry (mcherry) transgene in klf2a-deficient embryos using the fli1ep promoter fragment (Supplementary Fig. 10a; ref 21). This construct drove flow-independent endothelial expression of mature miR-126 (Supplementary Fig. 10b, c and data not shown) and led to an increased proportion of klf2a-deficient embryos with AA5x formation as compared to injection of klf2a Morpholino alone (Supplementary Table 3). Rescued embryos displayed miR-126/mcherry transgene expression in AA5x endothelial cells, including cases of bi- and uni-lateral rescue (Fig. 3k, l), while the control fli1ep:mcherry transgene failed to rescue (Fig. 3m). These results indicate that miR-126 acts downstream of klf2a to drive flow-stimulated angiogenesis.
miR-126 promotes angiogenesis by repressing spred1 and pik3r2, which normally inhibit Vegf signaling19, 20. Our observations suggested that in the absence of flow and klf2a, reduced miR-126 expression allows upregulation of these molecules thereby preventing Vegf-induced AA5x angiogenesis. While miR-126 can repress the zebrafish spred1 3’UTR, it had no effect on pik3r2 in whole embryo miRNA sensor assays (Supplementary Fig. 11a). Using an endothelial autonomous miRNA sensor assay (Supplementary Fig. 11b), we further found that the spred1 3’ UTR prevented expression of a mcherry transcript in blood vessels, while egfp fused to a control 3’UTR was expressed (Fig. 4a, b; Supplementary Fig. 11c). By contrast, the mcherry-spred1-3’UTR transgene was robustly expressed in embryos lacking miR-126, blood flow, or klf2a (Fig. 4c-h, Supplementary Fig. 11c). These results support a genetic pathway in which spred1 repression is mediated by klf2a and miR-126 in response to flow. Accordingly, over-expression of mRNA encoding Spred1 blocked AA5x formation (Fig. 4i, j, Supplementary Table 3), while reducing Spred1 in miR-126-deficient embryos rescued AA5x development (Fig. 4k, l, Supplementary Table 3). Taken together, our findings support the existence of a genetic pathway in which flow induces klf2a and miR-126 (Fig. 4m). While our data suggest that the interaction between these genes occurs in AA endothelial cells, we cannot rule out the possibility of an indirect role for klf2a upstream of miR-126. Nevertheless, flow-stimulated miR-126 subsequently inhibits spred1 in endothelial cells to allow angiogenesis to proceed in response to Vegf (Fig 4m). In the absence of flow, klf2a and miR-126 are reduced allowing spred1 to repress Vegf-stimulated angiogenesis. Thus, miR-126 provides a crucial link between flow and Vegf signaling to promote angiogenesis. Importantly, flow, klf2a, and miR-126 were similarly required for angiogenesis in the zebrafish-xenograft model22 (Supplementary Fig 12), suggesting that this pathway may represent a general mechanism for flow-stimulated angiogenesis in the zebrafish.
Figure 4. Flow-mediated repression of Spred1 is required for AA5x angiogenesis.
a-h, Cranial vessel expression of a miR-126 sensor at 65 hpf; lateral views, anterior to the left, dorsal is up. a, c, e, g, Expression of Egfp fused to control 3’UTR (green) and mCherry-spred1-3’UTR (red), coexpression is yellow. b, d, f, h, Expression of mCherry-spred1-3’UTR (red). Embryos co-injected with miR-126 sensor construct and 20 ng control MO (a, b), 20 ng miR-126 MO (c, d), 2 ng tnnt2 MO (e, f) , or 11 ng klf2a ATG MO (g, h). i-l, Tg(kdrl:egfp)la116 embryos at 65 hpf, dorsal view, anterior is up. Endothelial cells in green, circulation in red. Arrow - Lateral dorsal aortae; AA5x - arrowheads. Embryos left uninjected (i), injected with 100 pg spred1 mRNA (j), 20 ng control MO (k), or 1 ng spred1 MO and 20 ng miR-126 MO (l). m, Pathway responsible for flow-stimulated angiogenesis.
The stereotyped pattern of the vertebrate circulatory system is initially established by conserved genetic pathways that act before circulation to drive endothelial differentiation and provide guidance cues. How haemodynamic forces subsequently modulate these pathways in vivo is largely unknown. Our current work provides new insights into how an endothelial cell's response to flow can be integrated with early developmental signals to drive angiogenesis in the presence of flow.
Methods summary
Zebrafish and their embryos were handled according to standard protocols23 and in accordance with University of Massachusetts Medical School IACUC guidelines. For laser-assisted microsurgery, embryos at 46 hpf were anesthetized and immobilized in 0.5% of low-melt agarose (Biorad). The connection between AA5 and AA6 and the ventral aorta was ablated using a Micropoint laser (Photonic Instrument, Inc) mounted on a Zeiss AX10 Imager M1. SU5416 (Calbiochem) was prepared and used as described previously11. Control embryos were treated with 0.1% dimethyl sulfoxide (DMSO). To arrest heartbeat, embryos were treated with 15 mM of 2,3-butanedione 2-monoxime (BDM; Sigma-Aldrich) or with buffered Tricaine methanesulfonate (Sigma-Aldrich) at 0.66 mg/ml in egg water for the indicated times. Two-photon time-lapse imaging, confocal microscopy and microangiography was performed as previously13, 24, with additional modifications as noted in Supplementary Methods. Antisense riboprobes against dll4, vegfa, kdrl, fli1a, and cdh5 were generated and used for whole mount in situ hybridization as described elsewhere25. A klf2a fragment was PCR amplified and cloned by Gateway recombination. The resulting clone was linearized with BglII and a DIG-labeled riboprobe was synthesized using T7 polymerase. Digoxigenin (DIG)-labeled locked nucleic acid (LNA) probes (Exiqon, Copenhagen) were used to detect mature miR-126 and let-7 using in situ hybridization or Northern analysis as described elsewhere18. Morpholinos, mRNA and Tol2-based plasmids were prepared and injected as previously11,21. In cases of co-injection with Morpholinos, Tol2-plasmids and transposase, a DNA/transposase mRNA mixture was initially injected, followed by Morpholino. Plasmid construction details are provided in the full methods section. Morpholinos against vegfa, tnnt2 and gata1 have been described elsewhere15, 26, 25; all other Morpholino and oligonucleotide sequences are provided in the full methods section.
Supplementary Material
Acknowledgements
We would like to thank Beth Roman, Victor Ambros, and Charles Sagerstrom for critical review of the manuscript. We thank Jau-Nian Chen for providing the Tg(kdrl:egfp)la116 zebrafish line. We greatly appreciate the assistance of Tobias Stork and Marc Freeman for help in performing laser assisted microsurgery. We also thank Clemens Grabher for the kind gift of gata1 Morpholino. We are grateful to Monica Beltrame for providing the cdh5 plasmid and Thomas Smith and Sarah Sheppard for technical assistance. We thank Michael Green and Narendra Wajapeyee for the Ras-transformed NIH3T3 cell line. This work was supported in part by grants from the National Heart, Lung, and Blood Institute (N. D. L.), National Cancer Institute (N. D. L.) and the National Institute of Diabetes and Digestive and Kidney Disease (C.S. and K. E. F.). We apologize to researchers whose work we were unable to cite due to space constraints.
Methods
Zebrafish lines
The Tg(kdrl:egfp)la116 and kdrly17 lines have been described elsewhere 8,11. The Notch sensor line, Tg(tp1bglob:egfp)um14, has been described and validated as Notch-responsive in previous studies 27. A description of the Tg(fli1ep:dsRedEx)um13 line can be found in Covassin et al 28 while the Tg(fli1a:negfp)y7 line is described in Roman et al 29.
Two-photon time lapse imaging and confocal microscopy
Embryos were treated with 0.003% mM 1-phenyl-2-thiourea to prevent pigmentation and immobilized using 0.1% Tricaine as elsewhere 13. Microangiography was performed using Qtracker 655 non-targeted quantum dots (Invitrogen) or Rhodamine-labeled Dextran as described elsewhere 13. Embryos were mounted in low-melt agarose containing PTU and Tricaine as previously13. Two-photon imaging was performed as previously 24, except that GFP and unconjugated Qdots (655 nm emission) were simultaneously excited at 830 nm and the emitted fluorescence separated using a two-photon specific filter cube with 510/50 nm and 645/60 nm emission bandpasses. Both the sample and the objective were kept at 28°C during the experiment. Laser scanning confocal microscopy was performed as described elsewhere 11. To generate time-lapse movies and vertical projections, we utilized Imaris (Bitplane). Flash Professional 8 (Macromedia) was used to label movies.
Transgenic miRNA expression construct
To create a vector capable of expressing a zebrafish mature miRNA in a cell type-specific manner, we relied on multisite Gateway cloning. For the miRNA cassette, we constructed a middle entry (pME) clone containing a 753bp fragment of the zebrafish EF1alpha gene (accession number NM_131263) encompassing the TATA box, first non-coding exon 1, intron 1 and partial exon 2, 5' of the ATG. This fragment was PCR-amplified from genomic zebrafish DNA using high fidelity DNA polymerase (forward 5'-CGCTCGGTCCTCCTCTCGAGTATAAATTCTC-3' and reverse 5'-CTTTCCCATGTCGACTAAGTTTCTGCGGACC-3'), cloned into a TOPO TA cloning vector (Invitrogen) and validated by sequencing. A multicloning site was inserted between the StuI and ClaI restriction sites of intron 1 by annealing the primers 5'-CCTCTGTGGTACCATTCTACATGTGTTGATTTTCTGTATTTTAGTGAATTCTGT GAT-3' and 5'-CGATCACAGAATTCACTAAAATACAGAAAATCAACACATGTAGAATGGTACC ACAGAGG-3' to allow insertion of a pri-miRNA with KpnI and EcoRI restriction sites. Finally, the recombinant fragment was shuttled into pDONR 221 (Invitrogen) by first amplifying the insert using the primers 5'-GGGACAAGTTTGTACAAAAAAGCAGGCTCTCGAGTATAAATTCTCCAACCAA AGC-3' (forward) and 5'-GGGGACCACTTTGTACAAGAAAGCTGGGTCGATACCGTCGACTAAGTTTCTG CGGACC-3' (reverse). The resulting PCR fragment was used in a BP reaction with pDONR221 to generate a miRNA middle entry vector (pME-miR) and confirmed by sequencing.
To generate a miR-126 middle entry cassette, 600 bp of genomic sequence flanking the mature miR-126 sequence on chromosome 8 (miRBase accesion number MI0001979,Zv7) was amplified by PCR (see Table below for primer sequences). The resulting fragment was cloned by Zero Blunt TOPO PCR Cloning (Invitrogen) and validated by sequencing. The miR-126 fragment was subcloned using flanking EcoRI sites into pME-miR to give pME-miR126 (see above). To generate an endothelial autonomous miRNA expression construct, we utilized multisite Gateway LR cloning. The following plasmids were included in the multisite reaction: p5E-fli1ep28, pME-miR-126, p3E-mcherry 21, 30, and pDestTol2pA 30. A control vector was constructed by using the empty pME-miR in a parallel LR multisite reaction. The reaction was performed using LR clonase II plus (Invitrogen) as previously 21.
miRNA sensor assays
We amplified the 3’UTR of zebrafish spred1 or pik3r2 by PCR and cloned them via BP Gateway recombination with pDONR P2-P3 (Invitrogen) to generate 3’ entry plasmids (p3E-spred1-3’utr, p3E-pik3r2-3’utr, see Table below for primer sequences). For whole embryo assays, we generated a pCS2-based construct by performing multisite Gateway LR reactions with pCS2Dest2 21, pENTR-egfp 21 and the p3E-spred1-3’utr or p3E-pik3r2-3’utr entry clones. The resulting plasmids were referred to as pCS-egfp-spred1-3’utr or pCS-egfp-pik3r2-3’utr. These plasmids were linearized by digestion with NotI and used as templates to synthesize capped mRNA using SP6 polymerase (mMessage Machine, Ambion). As a control, we synthesized mRNA encoding the monomeric red fluorescent protein, mcherry containing the SV40 late polyadenylation sequence found in pCS2. 60 pg of mcherry mRNA was co-injected with 60 pg of egfp sensor mRNA with or without 60 fmol of miR-126 duplex (see below for sequences) into 1-cell stage zebrafish embryos. Embryos were observed at 24 hpf for MCherry and Egfp expression using a MZFLIII microscope equipped with epifluorescence. Digital images were captured using a Zeiss mRC digital camera and Axiovision software.
To generate an endothelial autonomous miRNA sensor construct, we cloned the basal promoter-EGFP-polyA cassette from plasmid pENTRbasEGFP 21 into the PspOMI site of pDest Tol2pA 30, in an opposite orientation to the attR4-attR3 Gateway cassette in the parent vector (see Supplementary Figure 11b); this plasmid is referred to as pTolbasPegfprev-R4R3. To generate the injection construct, we performed a multisite Gateway reaction using pTol-basPegfprev-R4R3, a 5’ entry clone containing the fli1ep fragment (p5e-fli1ep 28), a middle entry clone encoding mcherry (pME-mcherry), and p3E-spred1-3’utr.
For endothelial autonomous assays, we co-injected 25 pg of Tol2 transposase mRNA, 25 pg of Tol2-reporter plasmid, with or without miR-126, tnnt2, or klf2a Morpholino, as indicated. Embryos were observed for mCherry and Egfp expression using a MZFLIII microscope equipped with epifluorescence. Subsequently, confocal stacks were acquired as previously 28 and voxel intensity of red and green fluorescence was quantified in 3-dimensional reconstructions using Imaris.
klf2a mRNA expression construct
We amplified the open reading frame of zebrafish klf2a (NM_131856) by PCR and cloned it via BP Gateway recombination with pDONR 221 to generate a middle entry (referred to as pME-klf2a; see Table below for primer sequences). In parallel, we constructed a Destination vector in which mCherry was placed upstream of the viral 2A sequence and an attB1/attB2 Gateway cassette (pCSmCherry2ADest). Transfer of an in-frame entry clone into this vector generates a cassette in which two proteins are expressed from a single transcript. To generate an injection construct, we performed a LR Gateway reaction with pCSmCherry2ADest and pME-klf2a. The resulting plasmid is referred to as pCS-mcherry-2A-klf2a. This plasmid was linearized by digestion with NotI and used as template to synthesize capped mRNA using SP6 polymerase (mMessage Machine, Ambion). As a control, we synthesized mRNA encoding the monomeric red fluorescent protein, mcherry. Wild type embryos were injected with 400 pg of mRNA into one cell stage embryos with or without tnnt2 MO (3 ng) or klf2a MO splice site (2.5 ng) and control MO (3ng). We assayed for expression and presence of the injected mRNA at 48 hpf by visualization of mCherry fluorescence and RT-PCR, respectively (see Supplementary Fig. 9a, b).
Xenografts
Implantation of Ras-transformed NIH3T3 cells into zebrafish embryos was performed as previously described22. Cells were labeled with PKH26 red fluorescent linker (Sigma) and subsequently grafted into 48 hpf Tg(kdrl:egfp)la116 embryos that had been injected with control (20 ng), tnnt2 (3 ng), klf2a splice blocking MO (2.5 ng) or miR-126 (20 ng) morpholino. Microangiography was performed on embryos 24 hours following the graft followed by confocal microscopy. Imaris (Biplane) software was use to measure and compare vascular tumor volume in 3-dimensional reconstructions of confocal stacks.
Quantitative RT-PCR
Total RNA was extracted from 20 to 40 embryos per group in Trizol (Invitrogen) according to the manufacturer's instructions. Products were amplified in a real-time PCR reaction with StepOne Plus Real-Time PCR System (Applied Biosystems) using a Power SYBR Green mix (Applied Biosystems) according to the manufacturer's instructions. Mature miR-126 was amplified using the miScript Reverse Transcription Kit and miScript SYBR Green PCR Kit (Qiagen) with primers indicated below.
| Morpholino | sequences |
| klf2a ATG MO | GGACCTGTCCAGTTCATCCTTCCAC |
| klf2a splice blocking MO | CTCGCCTATGAAAGAAGAGAGGATT |
| miR-126 MO | TGCATTATTACTCACGGTACGAGTTTGAGTC |
| spred1 MO | AAACCTGTGGAAGGAGAAGGAAACC |
| control MO | Oligo-31N |
| tnnt2 MO | CATGTTTGCTCTGATCTGACACGCA |
| Primers | |
| Klf2a attb1 | gggg aca agt ttg tacaaaaaagcaggctacaggtcc ATGCATCTCAGC |
| Klf2a attb2 | GGGGACCACTTTGTACAAGAAAGCTGGGTcattttccagagtccgttcC |
| Spred 3′UTR attb2 | GGGG ACA GCT TTC TTG TAC AAA GTG G |
| CGCTTGTGCCACCGCTGC | |
| Spred 3′UTR attb3 | GGGG AC AAC TTTGTATAATAA AGT TG |
| GCAGAGCGTTTGGCGGTGT | |
| PIk3r2 3UTR attb2 | GGGG ACA GCT TTC TTG TAC AAA GTG G |
| CACAACGACTCGCTGAATGT | |
| PIK3r2 3UTR attb3 | GGGG AC AAC TTT GTA TAA TAA AGT TG |
| TCCTCAAGCTGGGATCATGT | |
| Spred ORF attb1 | ggggacaagtttgtacaaaaaagcaggctacatgagcgaa gaa cca aac |
| Spred ORF attb2 | ggggaccactttgtacaagaaagctgggttcatcctgcagccttgt |
| miR126 chm8 F | TGCCATGCCTTGACGAGAAG |
| miR126 chm8 R | GTGATATTTAATGTAACATGCC |
| qpcr Bact F | AGCTTGAAACTCGCCAAGTG |
| qpcr Bact R | CAGCTTTATAGCCGGCACTG |
| qpcr vegf F | GCCAAAGGCAGAAGTCAAAG |
| qpcr vegf R | TGCAGGAGCATTTACAGGTG |
| qpcr Kdrl F | TCTACTGGGCTTTTCCCTCTC |
| qpcr Kdrl R | AGGGTTACTATGGTGACGTTGC |
| qpcr Zelastin F | GCTCGTCTCCATACAAAGCA |
| qpcr Zelastin R | CAGAACTCCTCCTGGTAGCC |
| qpcr Z Tie2 F | GCCGTCAAGAGGATGAAAGA |
| qpcr Z Tie2 R | GCTGCTGTGAGGAGAGTGTG |
| qpcr Znos1 F | TCAAATACGCCACCAACAAA |
| qpcr Znos1 R | GAGAAGAAGGGGCAAAACATC |
| qpcr ZItgb5 F | TGGGAAGGATGGACAAAGAG |
| qpcr ZItgb5 R | CGGGTGAATGAGGAGACACT |
| qpcr ZEdg1 F | CCACCGTATTCAGCGTTATT |
| qpcr ZEdg1 R | GTCAAGGAGGAGCAGGATGA |
| qpcr ZEdn 1 F | ATGAGGCGAAACCACAGAAG |
| qpcr ZEdn 1 R | AGAAACCACTTGAGCGAGG |
| F-qpcr klf2a | CCAACGCCAACCAAAGAGTA |
| R-qpcr klf2a | CTCGCCTGTGTGTGTTCTGT |
| Mir 126 qpcr F for miRscript | CGT ACC GTG AGT AAT AAT GC |
| R-RT-PCR klf2a | ACGTGGTACCCGCACGGCGAACTCACACTTG |
| F-RT-PCR klf2a | ACGTGGTACCacgcacggcgactcacacttg |
| miRNA Duplex | |
| miR-126 sense | ucguaccgugaguaauaaugc |
| miR-126 antisense | gcauuauuacucacgguacga |
| miR-126 scramble antisense | gcaccacaacucaagguucga |
Footnotes
Supplementary Information is linked to the online version of the paper at www.nature.com/nature.
References
- 1.Yashiro K, Shiratori H, Hamada H. Haemodynamics determined by a genetic programme govern asymmetric development of the aortic arch. Nature. 2007;450:285–8. doi: 10.1038/nature06254. [DOI] [PubMed] [Google Scholar]
- 2.Adamo L, et al. Biomechanical forces promote embryonic haematopoiesis. Nature. 2009;459:1131–5. doi: 10.1038/nature08073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Gimbrone MA, Jr., Topper JN, Nagel T, Anderson KR, Garcia-Cardena G. Endothelial dysfunction, hemodynamic forces, and atherogenesis. Ann N Y Acad Sci. 2000;902:230–9. doi: 10.1111/j.1749-6632.2000.tb06318.x. discussion 239-40. [DOI] [PubMed] [Google Scholar]
- 4.le Noble F, Klein C, Tintu A, Pries A, Buschmann I. Neural guidance molecules, tip cells, and mechanical factors in vascular development. Cardiovasc Res. 2008;78:232–41. doi: 10.1093/cvr/cvn058. [DOI] [PubMed] [Google Scholar]
- 5.Dekker RJ, et al. Prolonged fluid shear stress induces a distinct set of endothelial cell genes, most specifically lung Kruppel-like factor (KLF2). Blood. 2002;100:1689–98. doi: 10.1182/blood-2002-01-0046. [DOI] [PubMed] [Google Scholar]
- 6.Parmar KM, et al. Integration of flow-dependent endothelial phenotypes by Kruppel-like factor 2. J Clin Invest. 2006;116:49–58. doi: 10.1172/JCI24787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lee JS, et al. Klf2 is an essential regulator of vascular hemodynamic forces in vivo. Dev Cell. 2006;11:845–57. doi: 10.1016/j.devcel.2006.09.006. [DOI] [PubMed] [Google Scholar]
- 8.Anderson MJ, Pham VN, Vogel AM, Weinstein BM, Roman BL. Loss of unc45a precipitates arteriovenous shunting in the aortic arches. Dev Biol. 2008;318:258–67. doi: 10.1016/j.ydbio.2008.03.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Isogai S, Horiguchi M, Weinstein BM. The vascular anatomy of the developing zebrafish: an atlas of embryonic and early larval development. Dev Biol. 2001;230:278–301. doi: 10.1006/dbio.2000.9995. [DOI] [PubMed] [Google Scholar]
- 10.Serluca FC, Drummond IA, Fishman MC. Endothelial signaling in kidney morphogenesis: a role for hemodynamic forces. Curr Biol. 2002;12:492–7. doi: 10.1016/s0960-9822(02)00694-2. [DOI] [PubMed] [Google Scholar]
- 11.Covassin LD, Villefranc JA, Kacergis MC, Weinstein BM, Lawson ND. Distinct genetic interactions between multiple Vegf receptors are required for development of different blood vessel types in zebrafish. Proc Natl Acad Sci U S A. 2006;103:6554–9. doi: 10.1073/pnas.0506886103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Nasevicius A, Larson J, Ekker SC. Distinct requirements for zebrafish angiogenesis revealed by a VEGF-A morphant. Yeast. 2000;17:294–301. doi: 10.1002/1097-0061(200012)17:4<294::AID-YEA54>3.0.CO;2-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Siekmann AF, Lawson ND. Notch signalling limits angiogenic cell behaviour in developing zebrafish arteries. Nature. 2007;445:781–4. doi: 10.1038/nature05577. [DOI] [PubMed] [Google Scholar]
- 14.Hellstrom M, et al. Dll4 signalling through Notch1 regulates formation of tip cells during angiogenesis. Nature. 2007;445:776–80. doi: 10.1038/nature05571. [DOI] [PubMed] [Google Scholar]
- 15.Sehnert AJ, et al. Cardiac troponin T is essential in sarcomere assembly and cardiac contractility. Nat Genet. 2002;31:106–10. doi: 10.1038/ng875. [DOI] [PubMed] [Google Scholar]
- 16.Vermot J, et al. Reversing blood flows act through klf2a to ensure normal valvulogenesis in the developing heart. PLoS Biol. 2009;7:e1000246. doi: 10.1371/journal.pbio.1000246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Meadows SM, Salanga MC, Krieg PA. Kruppel-like factor 2 cooperates with the ETS family protein ERG to activate Flk1 expression during vascular development. Development. 2009;136:1115–25. doi: 10.1242/dev.029538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wienholds E, et al. MicroRNA expression in zebrafish embryonic development. Science. 2005;309:310–1. doi: 10.1126/science.1114519. [DOI] [PubMed] [Google Scholar]
- 19.Fish JE, et al. miR-126 regulates angiogenic signaling and vascular integrity. Dev Cell. 2008;15:272–84. doi: 10.1016/j.devcel.2008.07.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wang S, et al. The endothelial-specific microRNA miR-126 governs vascular integrity and angiogenesis. Dev Cell. 2008;15:261–71. doi: 10.1016/j.devcel.2008.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Villefranc JA, Amigo J, Lawson ND. Gateway compatible vectors for analysis of gene function in the zebrafish. Dev Dyn. 2007;236:3077–87. doi: 10.1002/dvdy.21354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Nicoli S, Ribatti D, Cotelli F, Presta M. Mammalian tumor xenografts induce neovascularization in zebrafish embryos. Cancer Res. 2007;67:2927–31. doi: 10.1158/0008-5472.CAN-06-4268. [DOI] [PubMed] [Google Scholar]
- 23.Westerfield M. The Zebrafish Book. University of Oregon Press; Eugene, Oregon: 1993. [Google Scholar]
- 24.Siekmann AF, Standley C, Fogarty KE, Wolfe SA, Lawson ND. Chemokine signaling guides regional patterning of the first embryonic artery. Genes Dev. 2009;23:2272–7. doi: 10.1101/gad.1813509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lawson ND, Vogel AM, Weinstein BM. sonic hedgehog and vascular endothelial growth factor act upstream of the Notch pathway during arterial endothelial differentiation. Dev Cell. 2002;3:127–36. doi: 10.1016/s1534-5807(02)00198-3. [DOI] [PubMed] [Google Scholar]
- 26.Rhodes J, et al. Interplay of pu.1 and gata1 determines myelo-erythroid progenitor cell fate in zebrafish. Dev Cell. 2005;8:97–108. doi: 10.1016/j.devcel.2004.11.014. [DOI] [PubMed] [Google Scholar]
- 27.Parsons MJ, et al. Notch-responsive cells initiate the secondary transition in larval zebrafish pancreas. Mech Dev. 2009;126:898–912. doi: 10.1016/j.mod.2009.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Covassin LD, et al. A genetic screen for vascular mutants in zebrafish reveals dynamic roles for Vegf/Plcg1 signaling during artery development. Dev Biol. 2009;329:212–26. doi: 10.1016/j.ydbio.2009.02.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Roman BL, et al. Disruption of acvrl1 increases endothelial cell number in zebrafish cranial vessels. Development. 2002;129:3009–19. doi: 10.1242/dev.129.12.3009. [DOI] [PubMed] [Google Scholar]
- 30.Kwan KM, et al. The Tol2kit: a multisite gateway-based construction kit for Tol2 transposon transgenesis constructs. Dev Dyn. 2007;236:3088–99. doi: 10.1002/dvdy.21343. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.