Abstract
Inteins mediate protein splicing, which has found many applications in biotechnology and protein engineering. A single valine-to-leucine mutation (V67L) can globally enhance splicing and related cleavage reactions in minimized Mtu RecA inteins. However, the V67L mutation causes little change in crystal structures. To test if protein dynamics contribute to activity enhancement in the V67L mutation, the conformations and dynamics of the minimized and engineered intein ΔΔIhh-V67CM and a single V67L mutant, ΔΔIhh-L67CM, have been studied by solution NMR. Chemical shift perturbations established that the V67L mutation causes global changes, including changes at the N- and C-termini of the intein, which are active sites for protein splicing. The single V67L mutation significantly slows down hydrogen exchange rates globally, indicating a shift to more stable conformations and reduction in the ensemble distribution. Whereas the V67L mutation causes little change for motions on the ps-ns timescale, the motions on the μs-ms timescale affect a region involving the conserved F-block histidine and C-terminal asparagine, which are important residues for C-terminal cleavage. The V67L mutation is proposed to activate splicing by reducing ensemble distribution of the intein structure and by modifying the active sites.
Keywords: Intein, protein dynamics, hydrogen exchange, chemical shift perturbation, chemical exchange
Introduction
Protein splicing is an auto-catalytic posttranslational process in which an in-frame protein fusion, called an intein, is excised from the precursor protein with the concomitant ligation of the two flanking exteins, the N- and C-exteins.1 Inteins have been widely used in protein engineering, labeling, purification, and control of protein functions. 2–6 Inteins are also attractive drug targets as they are only found in unicellular organisms. In principle, intein inhibitors, as a novel class of antibiotics, would have minimal toxic effects to human cells.7 In Mycobacterium tuberculosis (Mtu), two important enzymes, RecA recombinase and DnaB helicase are interrupted by intein sequences. Active RecA and DnaB enzymes are only generated after intein-mediated protein splicing, making protein splicing a desirable target for new anti-tuberculosis drugs.
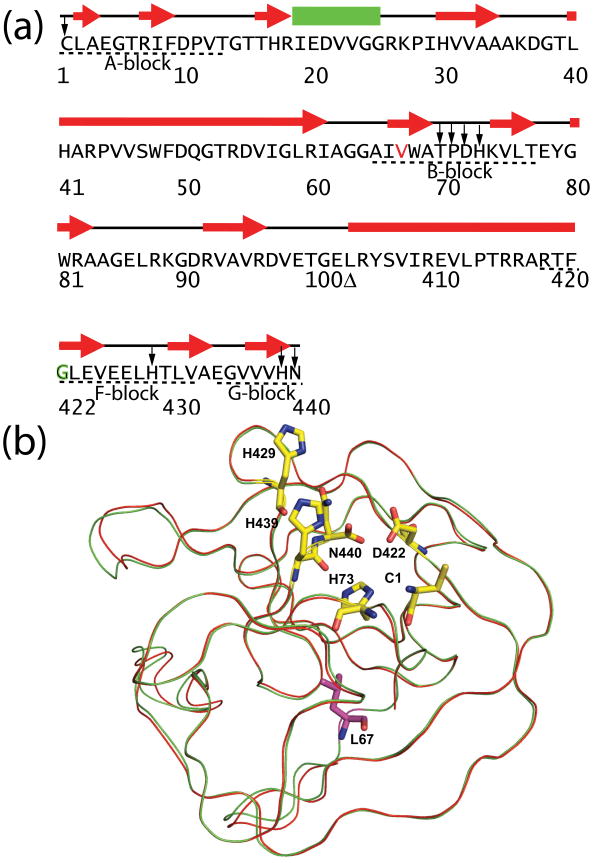
Intein sequences are characterized by conserved blocks (Fig. 1a and 1b).1 The A-block contains the first residue of the intein, usually a cysteine or serine which acts as a nucleophile for N-X acyl shift, the first step of protein splicing. The B-block is characterized by the highly conserved TXXH motif. In the Mtu RecA intein, there are conserved residues, histidine (H429) and aspartate (D422), in the F-block. When D422 is mutated, e.g. to a glycine, C-terminal cleavage occurs instead of protein splicing.8 The penultimate histidine (H439) and the C-terminal residue N440 in the G-block take part in asparagine cyclization, which is coupled to C-terminal cleavage.
Figure 1.
Sequence and structure of minimized Mtu RecA inteins. (a) Sequences, secondary structures, and conserved blocks of engineered and minimized intein ΔΔIhh-V67CM. The residues in the minimized inteins are numbered according to the full length Mtu RecA intein, which results in a discontinuity (Δ) in residue numbers due to the deletion of the endonuclease domain and the replacement of a disordered loop. ΔΔIhh-L67CM is different from ΔΔIhh- V67CM only by the single V67L mutation (V67 colored in red). Conserved blocks are denoted by dashed lines underneath the sequence. In the CM mutant, the conserved D422 is replaced by G422 which is marked in green. Highly conserved residues are marked by arrows. (b) V67L mutation does not cause major changes in X-ray crystal structure. 3D structure of ΔΔIhh-L67SM50 is overlaid with that of ΔΔIhh-V67SM (red). Shown in stick representation (yellow) are the active site residues, C1, H73 (B-block histidine), D422 (F-block aspartate), H429 (F-block histidine), H439 (penultimate histidine) and the C-terminal residue N440. The L67 side-chain is colored purple.
The RecA intein, composed of 440 residues, has been extensively engineered and characterized.9–14 An active 168 residue mini-intein (ΔI; 110Δ383, containing residues 1–110 and 383–440 of the RecA intein) was created by the removal of the homing endonuclease domain, which, however, resulted in decreased splicing activity.9 To restore intein activity, random mutagenesis was carried out with the mini-intein (ΔI) and two interesting mutations, ΔI-SM (Splicing Mutant) and ΔI-CM (Cleavage Mutant), were discovered.8 A single V67L mutation in ΔI-SM restores the protein splicing activity to almost the same level as that of the full-length intein. In contrast, ΔI-CM promotes C-terminal cleavage with no obvious splicing activity, primarily due to D422G and V67L mutations.8 Guided by crystallographic structures of ΔI-SM, a long disordered loop in the mini-intein was replaced with a corresponding short β-turn in the hedgehog proteins to generate the 139 residue “mini-mini intein” or the ΔΔIhh derivatives (Fig. 1), which maintained protein splicing activity.10
The activity enhancement of the V67L mutation was universal for a variety of minimized Mtu RecA inteins with more than 137 residues.10 This effect was also independently observed by Buskirk et al. using directed evolution.15 Wood et al. proposed that the removal of the endonuclease domain destabilized the intein and the V67L mutation restored intein stability.8 It was shown by cavity analysis that V67L improved packing interactions in the hydrophobic core.16 Hiraga et al. demonstrated that the V67L mutant is more resistant to thermolysin digestion in the presence of denaturing reagent, and hence appears more stable.10 Although increased stability may contribute to the activating effect of V67L, enzyme stability and activity are often uncoupled.17 For example, highly stable thermophilic enzymes usually have lower activity at room temperature compared with their mesophilic counterparts.
V67 was located in the conserved B-block and sequence alignment showed that this position was almost always a hydrophobic amino acid.18 Crystal structure of ΔΔIhh revealed that V67 was located in a hydrophobic core and adjacent to the highly conserved TXXH motif (Fig. 1a).12 Structures of Δ ΔIhh (V67) and Δ ΔIhh-SM (V67L), crystallized in the same space group, were found to have an RMSD of 0.2 Å and no detectable differences were observed for the N-terminal and C-terminal splicing junctions, or active site residues (Fig. 1b).12 Thus, the precise mechanism of V67L activation and stabilization remained unclear.
In this paper, we applied solution NMR to two ΔΔIhh inteins with a single mutation at 67 position, V67 vs L67 (Fig. 1), to explore the mechanism of the V67L stabilization and activation of protein splicing. We chose to study ΔΔIhh-V67CM and ΔΔIhh-L67CM because ΔΔIhh-L67SM was found to be more prone to aggregation, which would complicate the NMR relaxation analysis. In contrast, ΔΔIhh-V67CM and ΔΔIhh-L67CM were predominantly monomers in solution, as shown by analytical ultracentrifugation (AUC) (Supplementary Data Fig. S1). Although a few NMR studies have been carried out for inteins, 13–14; 19–22 no in-depth characterization of intein dynamics have been performed at atomic resolution and at multiple timescales. We used chemical shift perturbation, hydrogen deuterium exchange, 15N spin relaxation and residual dipolar coupling (RDC) to show that V67L causes global changes in solution and inteins have unique dynamic properties which contribute to their splicing activity.
Results
Chemical shift perturbation shows global changes affecting active sites
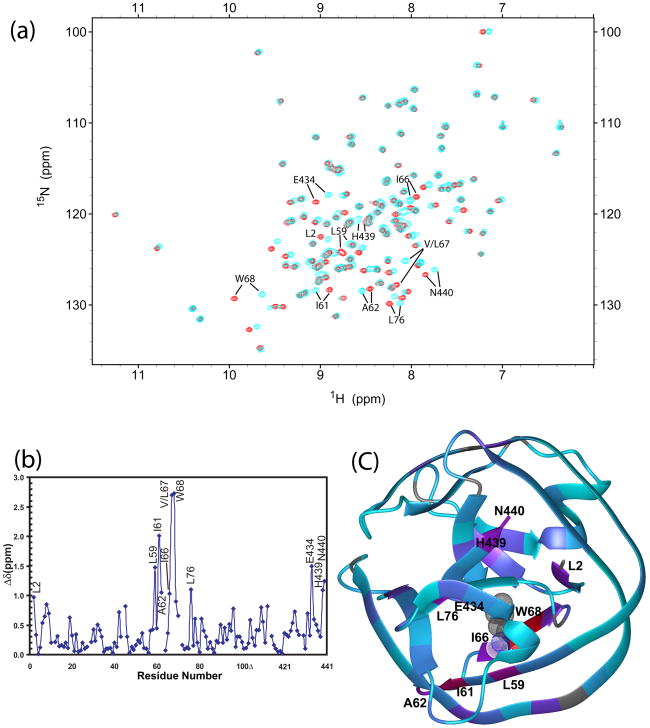
V67 mutation causes extensive chemical shift perturbation in ΔΔIhh-L67CM (Fig. 2a-2b). Excluding residue 67 at the mutation site, ten residues with the strongest chemical shift changes (Δδ), with Δδ defined as , were W68, I61, E434, L59, N440, L76, H439, A62, I66, and L2, listed in decreasing order of Δδ. Large Δδ values were not only observed for residues close to V67 (L59, 4.1 Å, amide N to N distance from V67; I66, 3.4 Å; W68, 3.5 Å; I61, 5.8 Å; E434, 7.0 Å), but also for residues located quite far away from the mutation site (A62, 9.1 Å; L2, 10.9 Å; H439, 12.4 Å; N440, 14.7 Å; L76, 11.9 Å). Δδ values were mapped onto the ribbon diagrams of ΔΔIhh-L67CM crystal structure in Fig. 2c, which demonstrated that chemical shift perturbations radiated from the mutation site continuously to remote sites of the protein through β-strands.
Figure 2.
The V67L mutation causes global changes, affecting the N- and C-terminus of the intein. Peaks with large chemical shift perturbations are labeled. (a) Overlay of 15N-1H HSQC spectra of ΔΔIhh-V67CM (cyan) and ΔΔIhh-L67CM (red) mutants. (b) Chemical shift difference between ΔΔIhh-V67CM and ΔΔIhh-L67CM mutants plotted against residue number. (c) Projection of V67L chemical shift perturbation onto the crystal structure ofΔΔIhh-L67CM (pdb code: 2IN8). The ribbon color is linearly interpolated between red and purple for Δδ between 3.0 and 1.0 ppm; and between purple and cyan for Δδ between 1.0 and 0 ppm. The ribbon is colored grey when chemical shift differences are not available, for prolines, N-terminal residue and unassigned residues. Residue L67 is displayed in meshed CPK mode.
Interestingly, residues from both the N- and C-terminal splice junction (L2, H439 and N440) experienced strong chemical shifts perturbations, indicating transmission of structural and/or dynamic changes to the intein active sites at the N- and C-terminal splicing junctions. Such perturbations at the intein active site may account for the global activity enhancement effect of the V67L mutation. C1 amide was not observed due to fast exchange with solvent. No significant chemical shift changes were observed for residues at the most conserved TXXH motif, even though the motifs are close to V67 based on the crystal structure of ΔΔIhh.12
Hydrogen exchange demonstrates large changes in protein motion and stability
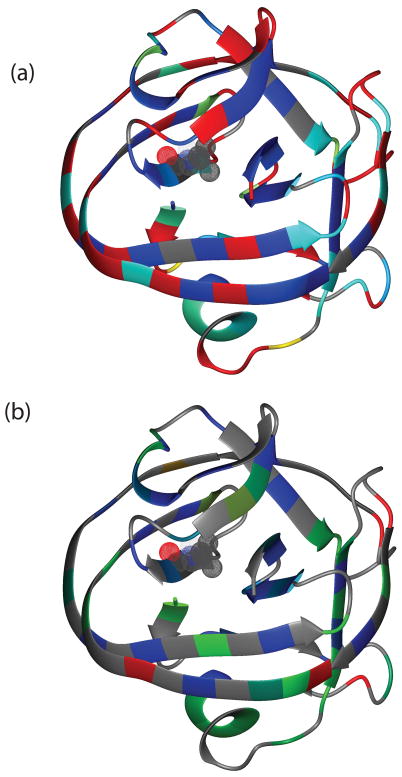
Labile protons on proteins are constantly exchanging with protons from solvent at different rates depending on various aspects of their environment, such as their exposure to the solvent or their involvement in H-bonds, secondary and global structures. Amide hydrogen exchange experiments allow us to determine the dynamics of a protein on a timescale of minutes to days. 15N-1H HSQC spectra were acquired at various times after lyophilized samples were reconstituted in D2O for both ΔΔIhh-V67CM and ΔΔIhh-L67CM. For ΔΔIhh-V67CM, 20 min following the dissolution in D2O, the amides of 45 residues (37.5% of 120 observable backbone amides) were already completely exchanged and not visible in the HSQC spectrum (Supplementary Data Table S2). These fast-exchanging residues are mainly located on the surface of the protein (Fig. 3a). Thereafter, 31 residues (26%) were exchanged between 20 min and 32 hours and 44 (37%) backbone amides have an exchange time longer than 32 hours. These three classes of amide protons were characterized as fast, intermediate and slow in solvent exchange. Similarly for ΔΔIhh-L67CM, 20 min following the dissolution in D2O, the amides of 45 residues (38%) were already exchanged and were therefore not visible in the HSQC spectrum, 21 residues (18%) exchanged between 20 min and 32 hours and 54 (45%) took longer than 32 hours. Therefore the V67L mutation substantially increases the number of slowly exchanging amides (Table 1).
Figure 3.
V67L causes significant changes in hydrogen-deuterium exchange rates kex. (a) Projection of kex for ΔΔIhh-V67CM onto the 3D structure of the intein. The ribbon color is linearly interpolated between yellow and cyan for kex between 0.025 and 0.0005 min−1; and between cyan and blue for kex between 0.0005 and 0.000005 min−1. Residues with fast hydrogen exchange rates, kex> 0.025 min−1, are colored in red. The ribbon is colored grey when kex is not available. Residue V67 is displayed in meshed CPK mode. (b) Projection of the change in the protection factor (ΔP=PΔΔIhh-L67CM–PΔΔIhh-V67CM) caused by the V67L mutation onto the intein 3D structure. The ribbon color is linearly interpolated between blue and green for protection difference between 1 and 0; and between green and red for protection factor difference between 0 and −1. The ribbon is colored grey when protection factor enhancement data were not available.
Table 1.
Number of residues with fast, intermediate and slow solvent exchange rates.
| Classification | Exchange Time | ΔΔIhh-V67CM | ΔΔIhh-L67CM) |
|---|---|---|---|
| fast | 0 to 20 minutes | 45 | 45 |
| intermediate | 20 minutes to 32 hours | 31 | 21 |
| slow | Greater than 32 hours | 44 | 54 |
The protection factor(s) P for ΔΔIhh-V67CM and ΔΔIhh-L67CM are summarized in Table S1 and Fig. S2 in Supplementary Data. Although the absolute values of the protection factors may have uncertainties due to compounding factors in the calculation in kint (see materials and methods), the comparison of the P values between the two intein variants is accurate because of the cancellation of kint in calculatingΔP. The average protection factor for ΔΔIhh-V67CM was about 0.4 smaller than that for ΔΔIhh-L67CM (5.9 vs 6.3), corresponding to an energy difference of 0.5 kcal/mol (ΔGop = 8.1 kcal/mol and 8.6 kcal/mol for ΔΔIhh-V67CM and ΔΔIhh-L67CM, respectively). The differences in protection factor, i.e. the protection enhancement caused by the V67L mutation, were color-coded onto the ribbon diagram of the crystal structure of ΔΔIhh-L67CM (Fig. 3b), showing ΔΔIhh-L67CM has generally enhanced protection factors overΔΔIhh-V67CM throughout the protein. This observation indicates that V67L mutation reduces the dynamic events responsible for solvent exchange, such as global and/or local folding of the protein.
The inteins have unusually rigid termini on the ps-ns timescale
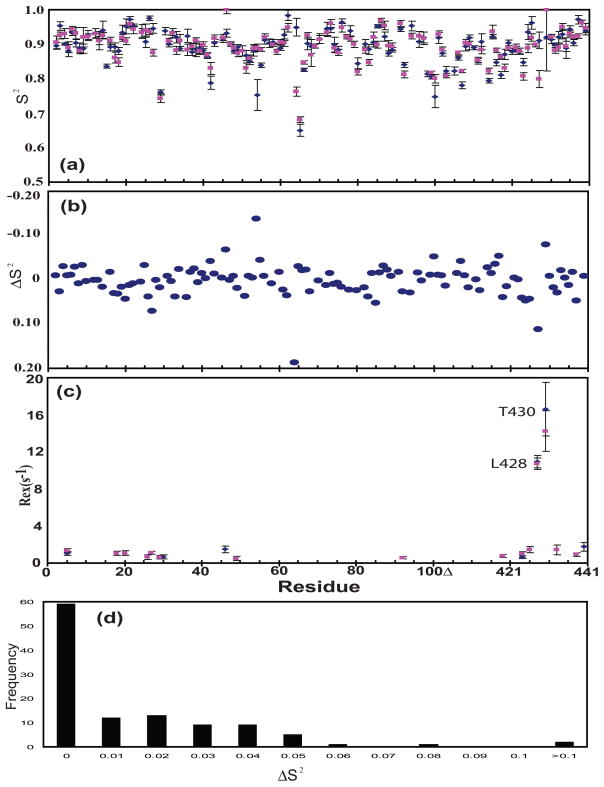
NMR spin relaxation was carried out in order to compare the dynamics of V67 and L67 on the ps-ns and μs-ms timescales. To ensure an appropriate comparison, the aggregation properties of the two proteins in solution were studied by sedimentation equilibrium experiments (Supplementary Data Fig. S1). The molecular weights obtained for ΔΔIhh-V67CM and ΔΔIhh-L67CM from the global fit were 16.2±0.1 kDa and 16.0±0.1 kDa, respectively, in good agreement with the theoretical molecular weight for the monomeric proteins (15.5 kDa, expected for uniformly 15N-labeled proteins). These results indicated that both proteins remained predominantly monomeric in solution, therefore allowing a fair comparison through 15N spin relaxation. Little difference was observed between the relaxation rates of these two proteins (Supplementary Data Fig. S3). The average generalized order parameters, S2, obtained for both proteins are 0.89 (Fig. 4a). As expected from the relaxation rates, the site-specific S2 values were similar over almost the whole intein sequence (Fig. 4a). ΔS2 values caused by the V67L mutation were distributed around zero with small isolated fluctuations (Fig. 4b,4d). Thus the V67L mutation did not perturb protein dynamics on the ps-ns timescale.
Figure 4.
V67L does not change protein dynamics on the ps-ns timescale but inteins have rigid termini. (a) Generalized order parameter, S2. (b) Difference of generalized order parameter, ΔS2, between ΔΔIhh-V67CM and Δ ΔIhh-V67CM, ΔS2 = S2(Δ ΔIhh-L67CM) –S2(Δ ΔIhh-V67CM). (c) Rex, chemical exchange contribution to 15N R2. (d) Histogram plot of frequency against the difference of the difference of generalized order parameter, ΔS2, between ΔΔIhh-V67CM and Δ ΔIhh-V67CM, ΔS2 = S2(Δ ΔIhh-L67CM) –S2(Δ ΔIhh-V67CM).
Although 15N spin relaxation did not uncover dynamic effects of the V67L mutation, it did reveal unique features of intein dynamics on the ps-ns timescale. The average S2 of 0.89 was exceptionally high. The values of backbone amide bond length (rNH) and chemical shift anisotropy (CSA) used in Modelfree analysis (1.02 Å and 172 ppm, respectively) typically resulted in a generalized order parameter of 0.85 for the structured regions (α-helices and β-strands), and lower values for exposed loops and termini regions.23 Both N- and C-termini had NOE values similar to the rest of the protein (Supplementary Data Fig. S3d), indicating that the inteins were rigid at both N- and C-termini on the ps–ns timescale. Modelfree analysis further demonstrated that N- and C-terminal residues of both proteins exhibited high S2 (Fig. 4a), showing no increase in flexibility as commonly observed at the termini of proteins.24 The termini in proteins usually have ~20–30% lower order parameters compared with the rest of the protein.24
Motions on the μs-ms timescale affect F-block histidine and surrounding residues
For both proteins, the amide nitrogen of L428 and T430 had significantly elevated R2 values with R2 > Rave + 5σ, where σ is the standard deviation of R2 (Supplementary Data Fig. S3b), indicating that these residues experience chemical exchange caused by motions on the μs-ms timescale. As expected, Modelfree analysis resulted in large Rex values for these two residues (Rex> 10 s−1; Fig. 4c).
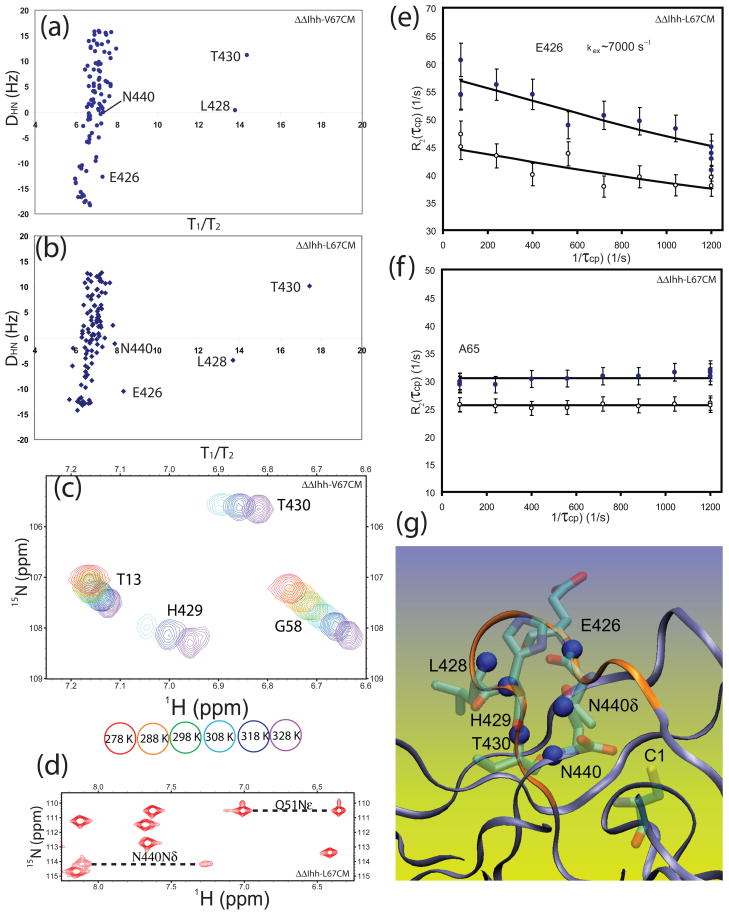
Residual dipolar couplings (RDC) of the NH groups were measured in both ΔΔIhh-V67CM andΔΔIhh-L67CM at 600 MHz. The RDC values agreed well with predictions based on the crystal structure of ΔΔIhh-L67CM (Supplementary Data Fig. S4), suggesting that there are no major structural changes between the solution and crystal structures, and between the V67 and L67 variants. For amide groups without chemical exchange RDC values have a similar orientation dependence on the rotational diffusion tensor as T1/T2. Indeed, a significant correlation between amide RDCs and T1/T2 has been observed for proteins and deviation from this correlation was proposed as a signature for chemical exchange.25 L428 and T430 were clear outliers in the plot of residual dipolar couplings (RDCs) DNH vs T1/T2 for both ΔΔIhh-V67CM (Fig. 5a) and ΔΔIhh-L67CM (Fig. 5b). Thus RDC data confirmed that the two neighboring residues of the F-block H429, L428 and T430, experience chemical exchange broadening caused by motions on the μs- ms timescale. In addition, RDC vs T1/T2 plots (Fig. 5a and 5b) suggested E426 and N440 experienced chemical exchange. This is especially a clear-cut for E426 in ΔΔIhh-L67CM (Fig. 5b).
Figure 5.
Motions at μs - ms timescale in inteins. 15N R2/R1values are plotted versus amide proton-nitrogen residual dipolar coupling, DHN, for ΔΔIhh-V67CM (a) and Δ ΔIhh-L67CM (b). L428 and T430 clearly have contributions from chemical exchange due to motions at μs to ms timescale. (c) Temperature dependence of 15N-1H HSQC. The H429 peak becomes stronger at higher temperature, demonstrating the effect of chemical exchange and the transition from intermediate to fast timescale for chemical exchange with increasing temperature. (d) NH2 groups in 15N-1H HSQC of inteins. The cross peaks for the NH2 group of N440 are much weaker than other peaks and those of Q51, suggesting the effect of chemical exchange. (e) CPMG dispersion curve for E426 obtained at 293 K and at both 800 MHz (solid circles ‘●’) and 600 MHz (open circles ‘○ ’) and their associated fitting curves. (e) CPMG curve for A65 obtained at 293 K and at both 800 MHz (solid circles ‘● ’) and 600 MHz (open circles ‘○ ’) shows no dispersions. (g) Locations of nitrogen nuclei experiencing μs - ms timescale motion in the intein structure. Amide nitrogen nuclei with Rex (blue spheres) are clustered near the turn formed by the hydrogen bond (red dashed line) between E426 CO and H429 NH. Residues with Rex, including amides of conserved H429 and N440 are located at the C-terminal splicing junction. It is noteworthy that the N-terminal splicing junction (C1) is nearby as well.
The backbone amide of H429 was not observed in 15N-1H HSQC spectra between 278–298 K, most likely due to chemical exchange broadening. As temperature increases, the cross peak for H429 started to appear in the 15N-1H HSQC spectra (Fig. 5c). The assignment of H429 at 328 K was confirmed by 3D 15N HSQC-NOESY and can be traced at other high temperatures through the natural temperature dependence of the amide cross peak. The dramatic increase in peak intensity with temperature from 278 K to 328 K (Fig. 5c) clearly demonstrates the existence of strong chemical exchange effects, and the transition of chemical exchange timescale from intermediate towards fast for H429. Chemical exchange also affects the C-terminal residue N440. The sidechain NH2 group of the C-terminal residue N440 was weak when compared with a regular NH2 group, e.g. that of Q51 (Fig. 5d). The backbone amide bond of N440 also experienced small chemical exchange, as shown in Fig. 5b.
To further confirm the presence of chemical exchange and to obtain more details about the chemical exchange process, Carr-Purcell-Meiboom-Gill (CPMG) relaxation dispersion 26; 27 was carried out (Fig. 5e-f). We obtained similar R2 values measured from a Hahn echo experiment and CPMG experiment with τCPMG of 1 ms at 298 K, suggesting the exchange time scale is fast and kex is significantly faster than 1000 Hz at 298 K (data not shown). In order to obtain a CPMG dispersion curve with the smallest τCPMG of ~ 1 ms, we had to slow down the exchange process by lowering the temperature. The minimized inteins aggregate at higher concentrations, limiting the available temperature range. At 293 K, a dispersion curve was obtained for E426, with a kex of ~7000 sec−1 (Fig. 5e). Meanwhile, residue A65, shows no dispersion and can serve as a control residue. Signals from L428, H429, T430 and N440 were too weak for dispersion measurement.
All these amide groups with chemical exchange, E426, L428, H429, T430 and N440, are clustered near the C-terminal splicing junction in the 3D structure of the intein (Fig. 5g). These residues are located near a tight turn maintained by a hydrogen bond between H429 HN and E426 CO. Because the F-block histidine H429 and the C-terminal residue N440 play key roles in the C-terminal cleavage step of protein splicing, motions on the μs-ms timescale may modulate protein splicing activity.
Discussion
X-ray and NMR provide complementary views of the V67L mutation
Although X-ray crystal structures revealed essentially no structural change due to the V67L mutation, NMR chemical shift perturbation uncovered global changes caused by this mutation, in particular, changes at the intein active sites. The absence of macroscopic rearrangement due to the V67 mutation in the X-ray structure was confirmed by the good agreement between residual dipolar coupling (RDC) values determined by solution NMR and those predicted by the X-ray structure (Supplementary Data Fig. S4). However, chemical shifts of amide protons and nitrogen are sensitive to minute structural changes, such as variations in hydrogen bond length on the order 0.1 Å, 28 which can not be detected by X-ray crystallography, and which maybe important to enhanced intein catalysis.
The vast majority of X-ray structures report a single structure for a protein. Although protein dynamics can manifest itself as reduced electron density in crystallography, conventional X-ray crystallography is not a preferred method for characterizing protein dynamics. In contrast, solution NMR chemical shifts of a protein are ensemble averaged quantities and are therefore sensitive to protein dynamics and the redistribution of the ensemble of protein conformations in solution, upon mutation or ligand binding. Structures in a crystal lattice are selected by the crystallization process and crystal contacts may introduce a bias toward a certain conformation that is common to the ensemble of both V67 and L67 variants.
These effects likely explain why extensive chemical shift perturbation is observed with the V67L mutation in the absence of significant change in crystal structures. Such perturbation caused by V67L may reflect underlying conformational and dynamic changes important for enhanced intein catalysis. There could be subtle alterations in the active site geometry, promoting catalysis. V67L may shift the ensemble population towards a state more conducive to catalysis, consistent with the redistribution of the conformational ensemble to more stable structures as shown by our HD exchange results and previous studies of protease digestion with thermolysin. 10
Interestingly, many residues that showed strong chemical shift perturbations, L2, L59, I61, W68, and E434, also exhibited strong chemical shift changes induced by a F421Y mutation, which was isolated by a phage display screening for inteins with high activity.20 Thus, there is a network of residues with highly correlated structural and dynamic properties in the intein,29–30 including residues near the active site. This network of residues could be responsible for allosteric communication, also termed the “ripple effect”,20 between remote sites in inteins.
Protein dynamics and intein catalysis
Motions probed by hydrogen exchange
Amide protons with fast exchange rates may exchange with solvent directly if they are on the surface of the protein, or through a localized unfolding process. In contrast, amide protons with intermediate to slow exchange rates would require cooperative unfolding of one or more secondary structures, or global unfolding. In the EX2 mechanism 31 for amide exchange, the greater the protection of the amide proton from hydrogen exchange, the higher the energy for protein unfolding, the more stable the structure of the protein. The fact that ΔΔIhh-L67CM is thermodynamically more stable than ΔΔIhh-V67CM, as deduced from a slower amide proton exchange rates, demonstrates that the V67L mutation stabilizes the protein. This is consistent with the previous finding that ΔΔIhh-L67CM has greater resistance to thermolysin proteolysis than ΔΔIhh-V67, as unstable or denatured proteins are more prone to proteolytic digestion.10 Interestingly, a few residues with important roles in intein catalysis have slow hydrogen exchange rates. For example, residues in the TXXH motif and G422 (Fig. 1a) have protection factors as high as 7 (Table S1). Even more striking is that residues next to the C-terminus, residues 437–439, have high protection factors. This is consistent with the idea that a rigid C-terminus may be important for intein catalysis.
Motions at the ps-ns timescale
Motions at the ps-ns timescale have been probed by 15N spin relaxation. The enhanced rigidity of the N- and C-termini of the intein is most likely related to their function as both the active sites and substrates for the protein splicing reaction. The N-terminal residue of the intein, usually a cysteine or serine, contributes a nucleophile for N-S acyl shift, the first step of protein splicing, whereas the C-terminal residue, almost always an asparagine, plays a key role in the C-terminal cleavage step of protein splicing. Thus the precise orientation of both termini relative to each other and other active site residues, such as the most conserved B-block histidine, is likely crucial for the activity of the enzyme. Such high S2 values at the termini are extremely rare, but have been observed in ferricytochrome c55132 and TEM-1 β-lactamase33.
Motions at the μs-ms timescale
Motions at the μs-ms timescale manifest themselves in the NMR spectra as the chemical exchange effect, enhancing the transverse relaxation rate (R2). A cluster of amides near the F-block histidine has been found to experience motions on the μs to ms timescale (Fig. 5g). The dynamics of this region will likely affect intein catalysis because the F-block contains the conserved histidine H429 and the C-terminal asparagine N440. H429 has been shown to modulate C-terminal cleavage, while the side chain NH2 group of the C-terminal asparagine directly participates in C-terminal cleavage.11; 34 It was suggested that H429 can deprotonate the NH2 side chain of N440 through a structurally conserved water molecule to activate the Nδ of N440, to initiate the nucleophilic attack of the scissile bond and asparagine cyclization.11;13–14 The observed motions may reflect a conformational exchange between a low energy ground state and a catalysis-competent excited state, e.g. the on and off interaction between H429 with the catalytic water or the equilibrium between the cyclization of N440 and the hydrolysis of succinamide.
Summary
We report the first in-depth analysis of protein dynamics in inteins with atomic resolution and at multiple timescales. We used chemical shift perturbation, hydrogen exchange, 15N spin relaxation and CPMG to study the effects of an activity enhancing mutation V67L. Although previous X-ray studies did not reveal any structural effect of this single mutation, our chemical shift perturbation mapping indicated that there were changes in the N-terminal and C-terminal splicing junction of the intein, which are the active sites for protein splicing. Hydrogen exchange experiments showed that the V67L mutation significantly slowed down amide proton exchange rates and stabilized the intein. The two inteins were rigid in backbone with averaged order parameters of 0.89 across the protein sequence. Surprisingly, the N- and C-terminal amide bonds in the inteins displayed unusual rigidity, which may be a prerequisite for intein catalysis by maintaining optimal active site geometry. Residues exhibiting chemical exchange clustered near the F-block histidine and the C-terminal asparagine and such μs-ms time scale motion likely contributes to the mechanism of intein activity.
Material and Methods
Protein overexpression, purification and NMR sample preparation
NMR samples of ΔΔIhh-L67CM and ΔΔIhh-V67CM were prepared as described previously.13 The E. coli strain JM101 transformed with the overexpressing plasmid was grown at 37 °C to an OD600 of 0.3–0.4 and induced with 1 mM isopropyl β-D-thiogalactoside (IPTG) at 20 °C, then grown for additional 20–24 hours at 20 °C. Isotope labeling was accomplished by growing cultures in M9 minimal medium containing either 1 g/L 15NH4Cl for a uniformly 15N-labeled sample, or 1 g/L 15NH4Cl and 1 g/L 13C6-D-glucose (Cambridge Isotope Laboratories) for a uniformly 15N- and 13C-labeled sample. The pure protein was concentrated and exchanged by ultrafiltration with Amicon Ultra centrifugal filter devices (Millipore Cooperation) (10-kDa molecular weight cutoff) into 50 mM sodium phosphate buffer, pH 7.0, with 100 mM NaCl and 1 mM NaN3. The final protein concentration in the 500 μl NMR samples was 0.15-0.3 mM.
NMR resonance assignment
We have previously carried out the backbone sequential assignment for ΔΔIhh-L67CM (BMRB accession number 15560)13 and the backbone assignment was accomplished forΔΔIhh-V67CM in a similar manner. For both ΔΔIhh-V67CM and ΔΔIhh-L67CM, we were able to carry out the backbone amide assignments for 131 of the 134 observable amides, while C1, G63, and H429 remain unassigned at 298 K. There are five proline residues, P11, P28, P44, P71, and P414 in the 139 residues of minimized intein sequence (Fig. 1a). All amide chemical shifts are listed in Supplementary Data Table S2. For ΔΔIhh-V67CM, T15, V23, A35, I57, W81, G90, V93, V95, Y406, E411, and F421 show spectral overlap. For ΔΔIhh-L67CM, A35, D50, L59, L69, W81, G90, V93, V95, R405, E427, and H439 show spectral overlap. Because of the spectral overlap, all residues aforementioned were only used in chemical shift perturbation analysis, but not in deuterium exchange or detailed relaxation analysis.
Hydrogen Exchange Measurement and Analysis
Aqueous samples containing 0.15–0.30 mM 15N-labeled proteins were lyophilized and then dissolved in 99.98 % D2O prior to 15N-1H HSQC NMR measurements for amide hydrogen exchange rate determination. Spectra were taken every 60 minutes for a total duration of ~32 hours. Peak intensities were fit to a single exponential decay function for hydrogen exchange rate constant (kex) according to I (t) = I0e−Rt.
The exchange data can be used to compare the relative thermodynamic stabilities between ΔΔIhh-V67CMand ΔΔIhh-L67CM. Hydrogen exchange usually occurs through the following mechanism:
where kop and kcl are the rates of a global or local unfolding event that exposes the amide proton to exchange with solvent with a rate of kint. The values of kint are considered as the sum of the contributions by acid catalyzed, base catalyzed, and water catalyzed hydrogen deuterium exchange rate and can be readily obtained based on protein primary sequence, pH, and salt concentration, as shown in equation 1.35
| (1) |
where kA,ref, kB,ref, and kW,ref stand for specific acid, specific base, and water catalyzed exchange rate constants. AL, AR, BL, and BR are the effects of neighboring amino acid side chains on acid (A) and base (B) catalysis with L and R refer to peptide groups to the left and right. To predict exchange rates at different temperatures, activation energy of 14, 17, and 19 kcal/mol are used to as acid catalyzed, base catalyzed, and water catalyzed activation energy, respectively.35 Some uncertainties in kint may be expected because of the uniform Ea used without considering primary sequence effects. However, the comparison of the P values between the two intein variants is accurate because of the cancellation of kint in calculating ΔP. The opening event can be global unfolding, local unfolding, or the isolated breakage of a hydrogen bond. Assuming the EX2 mechanism where kcl ≫ kint,31; 35–36, which is commonly observed at neutral pH conditions for folded proteins, the protection factor and ΔGop, can be calculated through:
| (2) |
| (3) |
where kex stands for observed exchange rates.
Residual dipolar coupling (RDC) measurement
RDCs were extracted from the change in J splitting as a function of the minute changes in molecular alignment in magnetic field. The observed splitting between the amide nitrogen doublets corresponds to the sum of the scalar and dipolar interactions, JHN and DHN. JHN is independent of alignment, to a very good approximation whereas DHN is dependent on the direction of the N-H bond in the alignment frame. DHN can be obtained from the difference between the N–H splitting measured with and without alignment medium. Both ΔΔIhh-V67CM and ΔΔIhh-L67CM were aligned in 7.5% polyacrylamide gels with a stretch ratio (dO/dN) of 1.29 using apparatus described by Chou et al. 37 to measure splitting using the IPAP method described by Ottiger et al. 38 To compare the global fold of solution structure with the crystal structure of ΔΔIhh-L67CM,12 backbone amide RDCs calculated from the crystal structure were compared with experimentally determined RDCs using PALES.39 Only the ΔΔIhh-L67CM crystal structure (pdb code: 2IN8) is used because the crystal structure of ΔΔIhh-V67CMis not available. Either singular value decomposition or non-linear minimization for best alignment orientation with fixed Da or Dr was carried out.
15N relaxation rates and Analysis
All NMR experiments were carried out at 298 K on either a Bruker 600 or a Bruker 800 MHz spectrometer, each equipped with a triple-resonance cryogenic probe. Relaxation properties of ΔΔIhh-V67CM and ΔΔIhh-L67CM were characterized by 15N R1, R2 and heteronuclear steady-state NOE experiments (Supplementary Data Fig. S3). These relaxation parameters were sensitive to motions occurring at the timescale of protein tumbling or faster, on the order of ps to ns. T1, T2 and NOE experiments were performed using the pulse sequence described by Farrow et al.40 At 600 MHz, NMR spectra were acquired with 2048 (t2) × 256 (t1) complex data points, spectral widths of 9615 Hz in 1H and 2432 Hz in 15N, and 16 scans. The recycle delay was 3.0 s. T1 relaxation times of 10 (×2), 100(×2), 200, 300, 400(×2), 500, 600, 700, 800, and 900 ms were used. T2 relaxation times of 6(×2), 20, 34, 48, 62, 76, 90(×2), and 104 ms were used. {1H} 15N steady-state heteronuclear NOEs were obtained by interleaving the proton saturation experiment and no proton saturation experiment at each t1 point. Recycle delay was 8 s and proton saturation was achieved by applying 120 degree proton pulse at 5 ms delay. All NMR data were processed with NMRPipe41 and analyzed with Sparky (T. D. Goddard and D. G. Kneller, SPARKY 3, University of California, San Francisco). For each T1 and T2 experiment, the spectrum with the shortest relaxation time (highest intensities) was peak picked with Sparky. Fitting for relaxation rates was accomplished using the program CURVEFIT (A. G. Palmer, Columbia University, New York, NY). Uncertainties for R1 and R2 were calculated by the Jackknife procedure using CURVEFIT (A. G. Palmer, Columbia University, New York, NY). The heteronuclear NOE values were obtained from the ratio of the peak heights for 1H-saturated and unsaturated spectra. The mean and standard deviation for the heteronuclear-NOE data was calculated from three pairs of experiments. The uncertainties on the NOEs were set to 2 times the standard deviation between intensities of duplicate experiments, as previously used by Savard et al.33 For both ΔΔIhh-L67CM and ΔΔIhh-V67CM, we found 120 residues with well-resolved peaks in the 15N-1H HSQC spectrum to warrant quantitative relaxation analysis on a per-residue basis. In addition to three unassigned amides, 11 more residues were not adequately resolved to permit relaxation analysis. The residues not included for relaxation analysis were: T15, V23, A35, I57, W81, G90, V93, V95, Y406, E411, and F421 in ΔΔIhh-V67CM; A35, D50, L59, L69, W81, G90, V93, V95, R405, E427, and H439 in ΔΔIhh-L67CM.15N NMR relaxation was analyzed using rNH of 1.02 Å as the mean amide nitrogen-hydrogen bond length, and Δσ = σ||−σ⊥ is the CSA of 172 ppm. The amplitudes and timescales of the internal motions of the protein were determined from the relaxation data according to the model-free formalism pioneered by Lipari and Szabo42; 43 and extended by Clore et al.,44 by using the program Modelfree (version 4.15, A. G. Palmer, Columbia University) in combination with FastModelfree.45 The generalized order parameters (S2) obtained by Modelfree described the amplitude of internal motion for individual amide bonds.
CPMG relaxation dispersion data acquisition and analysis
CPMG relaxation dispersion data were acquired at both 800 MHz and 600 MHz magnetic fields using the relaxation-compensated pulse sequences described by Loria et al. 46 and Tollinger et al.47 CPMG frequencies of 80, 240, 400, 560, 720, 880, 1040, and 1200 Hz were used along with total relaxation period of 50 ms. The carrier frequency was set to 126 ppm to accommodate the chemical shift of E426 and minimize off-resonance effects due to the high-power 180 CPMG pulses. Effective transverse relaxation rates were extracted by a two-point estimate and data were analyzed using the full Carver-Richards equation 48 and program CPMGFIT 49 assuming a two-site exchange.
Supplementary Material
Acknowledgments
We are thankful for financial support from the National Institutes of Health (R01GM81408 to C.W. and R01GM44844 to M.B.). Y.L. acknowledges funding from National Science Foundation of China NSFC (No. 20873135).
Footnotes
We have supplied supplementary methods and materials describing the detailed procedure for AUC results and Modelfree analysis. Figures for AUC (Fig. S1), protection factors and their differences (Fig. S2), 15N relaxation rates (Fig. S3) and comparisons of RDC values obtained experimentally and calculated using a crystal structure (Fig. S4) were supplied. We have also supplied tables for chemical shifts of backbone and sidechain amide group (Table S2), hydrogen exchange rates (Table S1), statistics for relaxation parameters (Table S3), 15N relaxation data and residual dipolar coupling (RDC) data (Table S4), statistics for Modelfree analysis (Table S5), and Modelfree analysis results (Table S6). 1H and 15N chemical shifts of backbone and sidechain amides in ΔΔIhh-V67CM were deposited to Biological Magnetic Resonance Bank (access code 16634).
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Paulus H. Protein splicing and related forms of protein autoprocessing. Annu Rev Biochem. 2000;69:447–496. doi: 10.1146/annurev.biochem.69.1.447. [DOI] [PubMed] [Google Scholar]
- 2.Youngeun Kwon MACJAC. Selective Immobilization of Proteins onto Solid Supports through Split-Intein-Mediated Protein Trans-Splicing13. Angew Chem Int Edit. 2006;118:1758–1761. doi: 10.1002/anie.200503475. [DOI] [PubMed] [Google Scholar]
- 3.Seyedsayamdost MR, Yee CS, Reece SY, Nocera DG, Stubbe J. pH Rate Profiles of FnY356-R2s (n = 2, 3, 4) in Escherichia coli Ribonucleotide Reductase: Evidence that Y356 Is a Redox-Active Amino Acid along the Radical Propagation Pathway. J Am Chem Soc. 2006;128:1562–1568. doi: 10.1021/ja055927j. [DOI] [PubMed] [Google Scholar]
- 4.Evans TC, Jr, Benner J, Xu MQ. The Cyclization and Polymerization of Bacterially Expressed Proteins Using Modified Self-splicing Inteins. J Biol Chem. 1999;274:18359–18363. doi: 10.1074/jbc.274.26.18359. [DOI] [PubMed] [Google Scholar]
- 5.Zueger S, Iwai H. Intein-based biosynthetic incorporation of unlabeled protein tags into isotopically labeled proteins for NMR studies. Nat Biotechnol. 2005;23:736–740. doi: 10.1038/nbt1097. [DOI] [PubMed] [Google Scholar]
- 6.Romanelli A, Shekhtman A, Cowburn D, Muir TW. Semisynthesis of a segmental isotopically labeled protein splicing precursor: NMR evidence for an unusual peptide bond at the N-extein-intein junction. Proc Natl Acad Sci USA. 2004;101:6397–6402. doi: 10.1073/pnas.0306616101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Paulus H. Protein splicing inhibitors as a new class of antimycobacterial agents. Drugs of the Future. 2007;32:973–984. [Google Scholar]
- 8.Wood DW, Wu W, Belfort G, Derbyshire V, Belfort M. A genetic system yields self-cleaving inteins for bioseparations. Nat Biotechnol. 1999;17:889–92. doi: 10.1038/12879. [DOI] [PubMed] [Google Scholar]
- 9.Derbyshire V, Wood DW, Wu W, Dansereau JT, Dalgaard JZ, Belfort M. Genetic definition of a protein-splicing domain: functional mini-inteins support structure predictions and a model for intein evolution. Proc Natl Acad Sci USA. 1997;94:11466–11471. doi: 10.1073/pnas.94.21.11466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hiraga K, Derbyshire V, Dansereau JT, Van Roey P, Belfort M. Minimization and stabilization of the Mycobacterium tuberculosis recA intein. J Mol Biol. 2005;354:916–926. doi: 10.1016/j.jmb.2005.09.088. [DOI] [PubMed] [Google Scholar]
- 11.Shemella P, Pereira B, Zhang Y, Van Roey P, Belfort G, Garde S, Nayak SK. Mechanism for intein C-terminal cleavage: a proposal from quantum mechanical calculations. Biophys J. 2007;92:847–853. doi: 10.1529/biophysj.106.092049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Van Roey P, Pereira B, Li Z, Hiraga K, Belfort M, Derbyshire V. Crystallographic and mutational studies of mycobacterium tuberculosis recA mini-inteins suggest a pivotal role for a highly conserved aspartate residue. J Mol Biol. 2007;367:162–173. doi: 10.1016/j.jmb.2006.12.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Du Z, Liu Y, Zheng Y, McCallum S, Dansereau J, Derbyshire V, Belfort M, Belfort G, Roey P, Wang C. 1H, 13C, and 15N NMR assignments of an engineered intein based on Mycobacterium tuberculosis RecA. Biomol NMR Assign. 2008;2:111–113. doi: 10.1007/s12104-008-9098-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Du Z, Shemella PT, Liu Y, McCallum SA, Pereira B, Nayak SK, Belfort G, Belfort M, Wang C. Highly conserved histidine plays a dual catalytic role in protein splicing: a pKa shift mechanism. J Am Chem Soc. 2009;131:11581–11589. doi: 10.1021/ja904318w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Buskirk AR, Ong YC, Gartner ZJ, Liu DR. Directed evolution of ligand dependence: Small-molecule-activated protein splicing. Proc Natl Acad Sci USA. 2004;101:10505–10510. doi: 10.1073/pnas.0402762101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Pereira B, Jain S, Garde S. Quantifying the protein core flexibility through analysis of cavity formation. J Chem Phys. 2006;124:74704. doi: 10.1063/1.2149848. [DOI] [PubMed] [Google Scholar]
- 17.Shu Q, Frieden C. Relation of enzyme activity to local/global stability of murine adenosine deaminase: 19F NMR studies. J Mol Biol. 2005;345:599–610. doi: 10.1016/j.jmb.2004.10.057. [DOI] [PubMed] [Google Scholar]
- 18.Perler FB. InBase: the Intein Database. Nucleic Acids Res. 2002;30:383–384. doi: 10.1093/nar/30.1.383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Heinamaki K, Oeemig JS, Djupsjobacka J, Iwai H. NMR resonance assignment of DnaE intein from Nostoc punctiforme. Biomol NMR Assign. 2009;3:41–3. doi: 10.1007/s12104-008-9137-1. [DOI] [PubMed] [Google Scholar]
- 20.Hiraga K, Soga I, Dansereau JT, Pereira B, Derbyshire V, Du Z, Wang C, Van Roey P, Belfort G, Belfort M. Selection and Structure of Hyperactive Inteins: Peripheral Changes Relayed to the Catalytic Center. J Mol Biol. 2009;393:1106–1117. doi: 10.1016/j.jmb.2009.08.074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Johnson MA, Southworth MW, Herrmann T, Brace L, Perler FB, Wuthrich K. NMR structure of a KlbA intein precursor from Methanococcus jannaschii. Protein Sci. 2007;16:1316–1328. doi: 10.1110/ps.072816707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Oeemig JS, Aranko AS, Djupsjobacka J, Heinamaki K, Iwai H. Solution structure of DnaE intein from Nostoc punctiforme: structural basis for the design of a new split intein suitable for site-specific chemical modification. FEBS Lett. 2009;583:1451–6. doi: 10.1016/j.febslet.2009.03.058. [DOI] [PubMed] [Google Scholar]
- 23.Palmer AG., III NMR probes of molecular dynamics: overview and comparison with other techniques. Annu Rev Bioph Biom. 2001;30:129–155. doi: 10.1146/annurev.biophys.30.1.129. [DOI] [PubMed] [Google Scholar]
- 24.Goodman JL, Pagel MD, Stone MJ. Relationships between protein structure and dynamics from a database of NMR-derived backbone order parameters. J Mol Biol. 2000;295:963–78. doi: 10.1006/jmbi.1999.3419. [DOI] [PubMed] [Google Scholar]
- 25.De Alba E, Baber JL, Tjandra N. The Use of Residual Dipolar Coupling in Concert with Backbone Relaxation Rates to Identify Conformational Exchange by NMR. J Am Chem Soc. 1999;121:4282–4283. [Google Scholar]
- 26.Carr HY, Purcell EM. Effects of diffusion on free precession in nuclear magnetic resonance experiments. Phys Rev. 1954;94:630–8. [Google Scholar]
- 27.Meiboom S, Gill D. Modified spin-echo method for measuring nuclear relaxation times. Rev Sci Instrum. 1958;29:688–91. [Google Scholar]
- 28.Wagner G, Pardi A, Wuethrich K. Hydrogen bond length and proton NMR chemical shifts in proteins. J Am Chem Soc. 1983;105:5948–9. [Google Scholar]
- 29.Goodey NM, Benkovic SJ. Allosteric regulation and catalysis emerge via a common route. Nat Chem Biol. 2008;4:474–482. doi: 10.1038/nchembio.98. [DOI] [PubMed] [Google Scholar]
- 30.Lockless SW, Muir TW. Traceless protein splicing utilizing evolved split inteins. Proc Natl Acad Sci USA. 2009;106:10999–11004. doi: 10.1073/pnas.0902964106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hvidt A, Nielsen SO. Hydrogen exchange in proteins. Adv Protein Chem. 1966;21:287–386. doi: 10.1016/s0065-3233(08)60129-1. [DOI] [PubMed] [Google Scholar]
- 32.Russell BS, Zhong L, Bigotti MG, Cutruzzola F, Bren KL. Backbone dynamics and hydrogen exchange of Pseudomonas aeruginosa ferricytochrome c551. J Biol Inorg Chem. 2003;8:156–166. doi: 10.1007/s00775-002-0401-z. [DOI] [PubMed] [Google Scholar]
- 33.Savard PY, Gagne SM. Backbone Dynamics of TEM-1 Determined by NMR: Evidence for a Highly Ordered Protein. Biochemistry-US. 2006;45:11414–11424. doi: 10.1021/bi060414q. [DOI] [PubMed] [Google Scholar]
- 34.Sun P, Ye S, Ferrandon S, Evans TC, Xu MQ, Rao Z. Crystal Structures of an Intein from the Split dnaE Gene of Synechocystis sp PCC6803 Reveal the Catalytic Model Without the Penultimate Histidine and the Mechanism of Zinc Ion Inhibition of Protein Splicing. J Mol Biol. 2005;353:1093–1105. doi: 10.1016/j.jmb.2005.09.039. [DOI] [PubMed] [Google Scholar]
- 35.Bai Y, Milne JS, Mayne L, Englander SW. Primary structure effects on peptide group hydrogen exchange. Proteins. 1993;17:75–86. doi: 10.1002/prot.340170110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Englander SW, Kallenbach NR. Hydrogen exchange and structural dynamics of proteins and nucleic acids. Q Rev Biophys. 1983;16:521–655. doi: 10.1017/s0033583500005217. [DOI] [PubMed] [Google Scholar]
- 37.Chou JJ, Gaemers S, Howder B, Louis JM, Bax A. A simple apparatus for generating stretched polyacrylamide gels, yielding uniform alignment of proteins and detergent micelles. J Biomol NMR. 2001;21:377–382. doi: 10.1023/a:1013336502594. [DOI] [PubMed] [Google Scholar]
- 38.Ottiger M, Delaglio F, Bax A. Measurement of J and dipolar couplings from simplified two-dimensional NMR spectra. J Magn Reson. 1998;131:373–378. doi: 10.1006/jmre.1998.1361. [DOI] [PubMed] [Google Scholar]
- 39.Zweckstetter M, Bax A. Prediction of Sterically Induced Alignment in a Dilute Liquid Crystalline Phase: Aid to Protein Structure Determination by NMR. J Am Chem Soc. 2000;122:3791–3792. [Google Scholar]
- 40.Farrow NA, Muhandiram R, Singer AU, Pascal SM, Kay CM, Gish G, Shoelson SE, Pawson T, Forman-Kay JD, Kay LE. Backbone dynamics of a free and phosphopeptide-complexed Src homology 2 domain studied by 15N NMR relaxation. Biochemistry-US. 1994;33:5984–6003. doi: 10.1021/bi00185a040. [DOI] [PubMed] [Google Scholar]
- 41.Delaglio F, Grzesiek S, Vuister GW, Zhu G, Pfeifer J, Bax A. NMRPipe: a multidimensional spectral processing system based on UNIX pipes. J Biomol NMR. 1995;6:277–93. doi: 10.1007/BF00197809. [DOI] [PubMed] [Google Scholar]
- 42.Lipari G, Szabo A. Model-free approach to the interpretation of nuclear magnetic resonance relaxation in macromolecules. 1 Theory and range of validity. J Am Chem Soc. 1982;104:4546–4559. [Google Scholar]
- 43.Lipari G, Szabo A. Model-free approach to the interpretation of nuclear magnetic resonance relaxation in macromolecules. 2 Analysis of experimental results. J Am Chem Soc. 1982;104:4559–4570. [Google Scholar]
- 44.Clore GM, Szabo A, Bax A, Kay LE, Driscoll PC, Gronenborn AM. Deviations from the simple two-parameter model-free approach to the interpretation of nitrogen-15 nuclear magnetic relaxation of proteins. J Am Chem Soc. 1990;112:4989–4991. [Google Scholar]
- 45.Cole R, Loria JP. FAST-Modelfree: A program for rapid automated analysis of solution NMR spin-relaxation data. J Biomol NMR. 2003;26:203–213. doi: 10.1023/a:1023808801134. [DOI] [PubMed] [Google Scholar]
- 46.Loria JP, Rance M, Palmer AG. A relaxation-compensated Carr-Purcell-Meiboom-Gill sequence for characterizing chemical exchange by NMR spectroscopy. J Am Chem Soc. 1999;121:2331–2332. [Google Scholar]
- 47.Tollinger M, Skrynnikov NR, Mulder FA, Forman-Kay JD, Kay LE. Slow dynamics in folded and unfolded states of an SH3 domain. J Am Chem Soc. 2001;123:11341–52. doi: 10.1021/ja011300z. [DOI] [PubMed] [Google Scholar]
- 48.Carver JP, Richards RE. General two-site solution for the chemical exchange produced dependence of T2 upon the Carr-Purcell pulse separation. J Magn Reson. 1972;6:89–105. [Google Scholar]
- 49.Korzhnev DM, Salvatella X, Vendruscolo M, Di Nardo AA, Davidson AR, Dobson CM, Kay LE. Low-populated folding intermediates of Fyn SH3 characterized by relaxation dispersion NMR. Nature. 2004;430:586–90. doi: 10.1038/nature02655. [DOI] [PubMed] [Google Scholar]
- 50.Rao NS, Legault P, Muhandiram DR, Greenblatt J, Battiste JL, Williamson JR, Kay LE. NMR Pulse Schemes for the Sequential Assignment of Arginine Side-Chain H[epsilon]Protons. J Magn Reson Ser B. 1996;113:272. doi: 10.1006/jmrb.1996.0188. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.