Abstract
Purpose
This retrospective study was designed to analyze the FSHR gene variants in subjects with primary and secondary amenorrhea with hypergonadotropic hypogonadism.
Materials and methods
Eighty six women with primary or secondary amenorrhea and 100 normally cycling proven fertile women of Indian origin were retrospectively studied. These subjects were systematically screened for entire FSHR gene.
Results
The frequency distribution of polymorphism at −29 position of FSHR gene is altered in women with primary and secondary amenorrhea as compared to controls. AA genotype at −29 position of FSHR gene seems to be associated with increased serum FSH levels in the study subjects. We have identified a novel homozygous mutation C1723T (Ala575Val) in one woman with primary amenorrhea.
Conclusions
Our findings suggest that increased serum FSH levels in subjects with primary amenorrhea correlated to FSHR genotype at position −29. We identified a novel homozygous mutation C1723T (Ala575Val) in a woman with primary amenorrhea.
Keywords: FSHR, Mutations, Polymorphisms, Primary amenorrhea, Secondary amenorrhea
Introduction
Follicle stimulating hormone (FSH) acts through binding to their cognate receptor (FSHR) present on the plasma membrane of granulosa cells in the ovary and Sertoli cells in the testis. This hormone-receptor interaction is crucial for regular gonadal function [1]. The importance of FSHR gene in female reproductive physiology is evident from the studies in FSHR knockout mice, where block in folliculogenesis was observed at preantral stage and these mice showed hypergonadotropic ovarian failure [2]. These findings suggest the importance of FSHR in ovarian physiology.
Importance of FSHR gene in female reproductive physiology is also evident from genetic studies in women with ovarian failure. Diverse inactivating mutations and polymorphisms have been described in FSHR gene in women with ovarian dysfunction. The first FSHR mutation identified was the Ala189Val (C566T) homozygous substitution in Finnish population. The affected women showed poorly developed secondary sexual characteristics, primary amenorrhea and recessively inherited hypergonadotropic ovarian failure [3]. Other naturally occurring inactivating mutations of FSHR gene; Asp224Val (A671T) and Leu601Val (T479C), Pro348Arg (C1043G), Ala419Thr (G1255A), Pro519Thr (A1556C), and Val221Gly (C1801G) have been reported in women with primary amenorrhea [4–8]. A compound mutation Arg573Cys (C1717T) and Ile160Thr (T479C) has been reported in a woman with secondary amenorrhea [9]. These subjects with primary or secondary amenorrhea were hypergonadotropic with hypogonadism. In vitro studies have demonstrated that mutations of FSHR gene have dramatically reduced the receptor expression and impaired proper signal transduction [5, 6, 10]. It has been reported that no inactivating mutations in FSHR gene were identified in women with polycystic ovary syndrome (PCOs), resistant ovary syndrome (ROS) and premature ovarian failure (POF) [11–16].
In addition to mutations, a number of polymorphisms have been reported in the FSHR gene. G−29A, A919G (Thr307Ala) and A2039G (Asn680Ser) polymorphisms have been extensively studied by different investigators. It was reported that the polymorphism A2039G is associated with high basal FSH levels in women undergoing Assisted Reproductive Technology (ART) programme [17–22]. Recently, we have reported association of FSHR gene polymorphism (A919G, G−29A) with altered ovarian response in subjects undergoing gonadotropin treatment during ART programme [23, 24].
In the present study, the known/novel mutations and polymorphisms of FSHR gene were investigated in Indian women with primary and secondary amenorrhea. Further, the prevalence of FSHR gene variations was examined and the association of receptor polymorphisms with gonadotropin levels was analyzed in these subjects.
Materials and method
Subjects
All the subjects included in the present study were from Western-India. The present study was approved by the Ethics Committee for Clinical Research of National Institute for Research in Reproductive Health. All the women included in this study gave informed consent for their medical history to be reviewed, and for the collection of peripheral blood samples for molecular analysis.
A total of eighty-six women visiting the ‘Endocrinology Clinic’ and ‘Genetic Research Centre’ of the Institute were recruited as the study subjects. Out of these, forty-eight women with primary ovarian failure (age: 14–20 years) were diagnosed by the history of primary amenorrhea; poorly developed secondary sex characteristics and streak or small ovaries on ultrasonography. The basal FSH levels (normal follicular phase FSH levels: 2–9 mIU/ml) in these women were found to be elevated >40 mIU/ml (Table 1). Thirty-eight women aged 20–38 years had secondary amenorrhea for 6 months or more. These secondary amenorrhea subjects were found to have FSH concentration of >40 mIU/ml when estimated on two occasions at least a month apart. These subjects showed small or streak ovaries on ultrasonography and poorly developed secondary sex characteristics (Table 1). All the eighty-six female subjects showed no history of mums, surgery. Except three subjects none of the subjects reported family history of primary or secondary amenorrhea. Women with abnormal karyotype, autoimmune disorders, prolactiniemia, infections and exposed to radiation were excluded from the present study.
Table 1.
Characteristics of the subjects with primary amenorrhea, secondary amenorrhea and controls
| Parameters | Primary amenorrhea n = 48 | Secondary amenorrhea n = 38 | Controls n = 100 |
|---|---|---|---|
| Age (years) | 16.0 ± 3.6 (14–20) | 32.0 ± 3.4 (26–38) | 32.2 ± 3.9 (22–40) |
| Age at menarche (years) | NA | 15.10 ± 2.29 (11–22) | 14.67 ± 1.9 (11–17) |
| Normal menstrual cycles (%) | NA | 52.5 | 100 |
| Menstrual cycle length (days) | NA | 26.7 ± 2.5 (23–30) | 27.1 ± 1.8 (23–30) |
| Irregular cycles (%) | NA | 47.5 | 0 |
| Amenorrhoeic since (age, years) | 15.0 ± 2.2 | 25.26 ± 2.42 | NA |
| Normal ovaries (%) | 0 | 62.5 | 100.0 |
| Streak or small ovaries (%) | 74.3 | 37.5 | 0 |
| Basal FSH levels (mIU/ml) | 72.98 ± 12.8a | 80.08 ± 6.11b | 6.16 ± 1.72a,b |
| Basal LH levels (mIU/ml) | 35.65 ± 7.46a | 46.51 ± 4.26b | 5.15 ± 1.00a,b |
| Proven fertile (n) | 0 | 0 | 100 |
Values are means; ±SEM; (range)
NA Not applicable
aValues are significantly different between primary amenorrhea and control subjects (p < 0.05; One-way ANOVA)
bValues are significantly different between secondary amenorrhea and control subjects (p < 0.05; One-way ANOVA)
One hundred normally cycling proven fertile healthy females with a history of regular cyclicity served as the controls. All the control subjects had normal pubarche and menarche and were phenotypically normal, nor did they have a family history of mental retardation, fragile X syndrome or POF.
Serum FSH and luteinizing hormone (LH) levels were measured by radioimmunoassay (RIA) using commercial kit from Diagnostics Systems Laboratories (Webster, Texas, USA).
DNA isolation and polymerase chain reaction
All the enrolled subjects were screened for FSHR gene variations using genomic DNA isolated from whole blood. Exon 1–10 and 5′ untranslated region (UTR) of FSHR gene were amplified using genomic DNA as a template by polymerase chain reaction (PCR) with specific oligonucleotide primers (Table 2). PCR conditions were specifically adjusted for each fragment. These primer sequences have been either adapted from previously published studies [23, 25, 26] or designed for the present study (Table 2). Five microliters of each PCR product was resolved on 10% polyacrylamide gel electrophoresis (PAGE) and visualized by silver staining. Amplified products were used for further analysis.
Table 2.
Primers used to amplify specific region of FSH receptor gene. PCR products were used for restriction fragment length polymorphism, single-strand conformation polymorphism or DNA sequencing
| Region of FSH receptor gene | Primer No. | Primer sequence | Product size (bp) | Primer location | Reference No. |
|---|---|---|---|---|---|
| Promoter | 1F | 5′ TATTCCAGACATGCCTAATGG 3′ | 291 | −263 to +28 | Wunsch et al. 2005 [26] |
| 1R | 5′ CCAGCAAAGAGACCAGGAGC 3′ | ||||
| Exon 1 | 2F | 5′ GGAGCTTCTGAGATCTGTGG C 3′ | 235 | −71 to intronic | Gromoll et al. 2000 [25] |
| 2R | 5′ AAATGCCAGCCATGCAGTTG 3′ | ||||
| Exon 2 | 3F | 5′ TCACTAGTGTAGACAGGATG 3′ | 252 | intronic | Gromoll et al. 2000 [25] |
| 3R | 5′ TTGAGGCATTCACTCACAGC 3′ | ||||
| Exon 3 | 4F | 5′ GCCCTAAAATATTCAAGAG 3′ | 215 | intronic | Gromoll et al. 2000 [25] |
| 4R | 5′ GCCTCCCAGGAATGTAGAA 3′ | ||||
| Exon 4 | 5F | 5′ GCATTCCTTACCATCAAGATC 3′ | 152 | intronic | Gromoll et al. 2000 [25] |
| 5R | 5′ GTGGGGGTACCAAACTACATG 3′ | ||||
| Exon 5 | 6F | 5′ GAAATTCATGCTCTAACAACTG 3′ | 200 | intronic | Gromoll et al. 2000 [25] |
| 6R | 5′ TGGGCAAGACAGATACTGAG 3′ | ||||
| Exon 6 | 7F | 5′ ACATTCAAGATAACATAAAC3′ | 101 | intronic | Present study |
| 7R | 5′ GCGGATCCTTACAGAATCACACTTTC 3′ | ||||
| Exon 7 | 8F | 5′ TTCAGATGGCTGAATAAGA 3′ | 86 | intronic to 655 | Present study |
| 8R | 5′ GCGGATCCGCTCATCTAGTTGGGTTC3′ | ||||
| Exon 8 | 9F | 5′ TACAACAGCAATATTTATGTGGC 3′ | 198 | intronic | Gromoll et al. 2000 [25] |
| 9R | 5′ GAGAGTTGACTTCTAACTTACAC 3′ | ||||
| Exon 9 | 10F | 5′ TGCCTGCTAACCAAGAGCAG 3′ | 289 | intronic | Gromoll et al. 2000 [25] |
| 10R | 5′ TGCCTGAGCAGGGCTTAAAG 3′ | ||||
| Exon 10A | 11F | 5′ GCTATACTGGATCTGAGATG 3′ | 221 | intronic to 1026 | Gromoll et al. 2000 [25] |
| 11R | 5′ ACCACTTCATTGCATAAGTC 3′ | ||||
| Exon 10B | 12F | 5′ TTGACATGACGTACACTGAG 3′ | 289 | 976 to 1264 | Gromoll et al. 2000 [25] |
| 12R | 5′ CAACTGATGCAATGAGCAG 3′ | ||||
| Exon 10C | 13F | 5′ CTGATCTCTGCATTGGAATC 3′ | 265 | 1219 to 1483 | Gromoll et al. 2000 [25] |
| 13R | 5′ ATCCAGCCCATCACCATGAC 3′ | ||||
| Exon 10D | 14F | 5′ AGCTGGACTGCAAGGTGCAG 3′ | 260 | 1426 to 1674 | Gromoll et al. 2000 [25] |
| 14R | 5′ GGTTCCGCACTGTGAGGTAG 3′ | ||||
| Exon 10E | 15F | 5′ CCTTGTGCTCAATGTCCTGG 3′ | 219 | 1601 to 1820 | Gromoll et al. 2000 [25] |
| 15R | 5′ GCTTTGGACACAGTGATGAG 3′ | ||||
| Exon 10F | 16F | 5′ CCATTTCTGCCTCCCTCAAG 3′ | 227 | 1722 to 2000 | Gromoll et al. 2000 [25] |
| 16R | 5′ TGGATGGGTGTTGTGGACAG 3′ | ||||
| Exon 10G | 17F | 5′ GAGCAAGTGTGGCTGCTATG 3′ | 258 | 1922 to −91(3′ region) | Gromoll et al. 2000 [25] |
| 17R | 5′ TGTAGAAGCACTGTCAGCTC 3′ | ||||
| Exon 10 | 18F | 5′ AGCTCTGAGCTTCATCC 3′ | 357 | 856 to 1209 | Present study |
| 18R | 5′ GTTGCACATAAGGAACC 3′ | ||||
| Exon 10 | 19F | 5′ CCCATGTGAAGATATCATG 3′ | 328 | 1061 to 1389 | Present study |
| 19R | 5′ GATAGCTGTCGGAGTG 3′ | ||||
| Exon 10 | 20F | 5′ GGCCTTTGCTGATCCCTG 3′ | 868 | 1211 to 2084 | Present study |
| 20R | 5′ GGGCTAAATGACTTAGAGGG 3′ |
Restriction fragment length polymorphism (RFLP)
RFLP for six known mutations (T479C, C566T, C1043G, G1255A, A1556C, and C1717T), three known polymorphisms (G−29A, A919G and A2039G) and a novel mutation (C1723T) was performed using specific restriction enzymes and optimum conditions described in Table 3. All the enzymes and their respective buffers were purchased from Promega, Madison, WI, USA, except for Msc I which was purchased from New England Biolabs, Ipswich, MA, USA. The digested products were separated on 10% PAGE and visualized by silver staining.
Table 3.
Restriction analysis performed for detection of mutations and polymorphisms of FSH receptor gene giving details of primers and enzymes used along with other experimental conditions
| Mutations/polymorphisms | PCR primer pairs used | Enzyme used | Enzyme units per reaction of 20 μl (U) | Digestion at 37°C (hours) |
|---|---|---|---|---|
| G−29Aa,c | 2F,2R | Mbo II | 0′5 | 20 |
| T479C (Thr160Ile) | 7F,7R | Mun I | 2 | 3 |
| C566T (Ala189Val) | 8F,8R | Bsm I | 7 | 1 |
| A919Ga (Thr307Ala) | 18F,19R | Bsu36 I | 5 | 16 |
| C1043G (Pro348Arg) | 11F,12R | Dde I | 5 | 3 |
| G1255A (Ala419Thr) | 12F,14R | SfaNI | 2′5 | 3 |
| A1556C (Pro519Thr) | 14F,14R | Nco I | 2 | 1 |
| C1717T (Arg573Cys) | 20F,20R | CfoI | 2 | 16 |
| C1723Tb (Ala575Val) | 15F,15R | MscI | 1 | 20 |
| A2039Ga (Asn680Ser) | 17F,17R | BsrS I | 1 | 16 |
aFSH receptor polymorphisms
bNovel FSH receptor mutation detected in the present study
cRFLP protocol designed in the laboratory for detection of polymorphism at position −29 (Reference: Achrekar et al. 2009b)
Single-strand conformation polymorphism (SSCP)
All the subjects recruited in present study were screened for novel mutations and polymorphisms in 5′UTR and entire coding region of FSHR gene by single-strand conformation polymorphism (SSCP). For each exon, seven different electrophoresis conditions were initially employed and of these, two conditions were selected, based on the SSCP migration pattern. Briefly, the PCR products were mixed with formamide and bromophenol blue dye. This mixture was denatured for 8–12 min at 95°C and immediately chilled on ice before loading to SSCP gel [27]. The samples were electrophoresed on 6.5–10% polyacrylamide gels (stock solution of polyacrylamide: 40%) at 250–350 V for 9–10 h at 4°C. The single stranded conformers were visualized by subjecting the gels to silver staining [28]. Different migration patterns observed by SSCP were confirmed by direct DNA sequencing.
DNA sequencing
Nucleotide sequencing of the PCR products was carried out using the ABI Big Dye Terminator protocol on the ABI 3100 Avant system (Applied Biosystems, Foster City, CA, USA) at the DNA sequencing facility of the Institute. Bi-directional sequencing was carried out using specific sets of primers (Table 2).
Statistical analysis
One-way analysis of variance (ANOVA) and post-hoc (LSD) test were employed to analyze the differences in serum gonadotropin levels between the groups. Chi-squared analysis and Fischer’s test were employed to analyze the frequency distribution of polymorphisms between the different groups studied. Statistical analysis was performed using SPSS software. Odds ratio >1.0 was considered as positive association. A value of P < 0.05 was considered statistically significant.
Results
Detection of known mutations/polymorphisms of FSHR gene
Amplicons of the expected size were obtained using genomic DNA as a template from all the subjects and controls. RFLP analysis for T479C, C566T, C1043G, G1255A, A1556C, and C1717T mutations of FSHR gene revealed the absence of these reported mutations in study subjects as well as in controls. As expected, RFLP analysis demonstrated that FSHR gene is polymorphic at −29, 919 and 2039 position in all the subjects recruited in study.
The frequency distribution of the genotypes at −29 position in the 5′UTR of FSHR gene was evaluated in all subjects. The frequency distribution in the control subjects was 5% for homozygous G, 1% for homozygous A and 94% for heterozygous GA. In the case of women with primary amenorrhea it was 31.25% for homozygous G, 16.67% for homozygous A and 52.08% for heterozygous GA; and in case of women with secondary amenorrhea it was 28.94% for homozygous G, 15.78% for homozygous A and 55.28% for heterozygous GA genotypes. In the subjects with primary and secondary amenorrhea the frequency distribution of this polymorphism was observed to be statistically different as compared to that observed in controls (Table 4). However, the frequency distribution for genotypes at position 919 and 2039 did not differ significantly in study subjects as compared to control subjects (Table 4).
Table 4.
Frequency distribution of the FSH receptor genotypes at position −29, 919 and 2039
| Nucleotide position in FSH receptor gene | Genotypes | Controls n = 100 (%) | Primary amenorrhea subjects n = 48 (%) | Secondary amenorrhea subjects n = 48 (%) |
|---|---|---|---|---|
| −29 | GG | 05 (5) | 19 (39.58) | 11 (28.94) |
| GA | 94 (94) | 22 (45.83) | 21 (55.28) | |
| AA | 01 (1)a,b | 07 (14.59)a | 06 (15.78)b | |
| 919 | AA | 24 (24) | 14 (29.16) | 11 (28.94) |
| AG | 53 (53) | 23 (47.93) | 17 (44.75) | |
| GG | 13 (13) | 11 (22.91) | 10 (26.31) | |
| 2039 | AA | 31 (31) | 15 (31.25) | 11 (28.94) |
| AG | 56 (56) | 25 (52.08) | 21 (55.28) | |
| GG | 13 (13) | 08 (16.67) | 06 (15.78) |
aThe frequency distribution of genotype AA at −29 position of FSH receptor gene is significantly different between the controls and primary amenorrhea subjects (p < 0.05; Chi-squared test)
bThe frequency distribution of genotype AA at −29 position of FSH receptor gene is significantly different between the controls and secondary amenorrhea subjects (p < 0.05; Chi-squared test)
Further, we evaluated the frequency distribution of nine haplotypes (variants at amino acid position 307 and 680 of FSHR gene) in all the groups. However, the prevalence of five haplotypes (TT-NS, TT-SS, TA-SS, AA-NN and AA-NS) differed significantly between subjects with primary amenorrhea and controls. The frequency distribution of two haplotypes (TT-NS and TA-SS) differed significantly between subjects with secondary amenorrhea and controls (Table 5). It was further observed that the odds ratio for the above mentioned haplotypes was >1.0 (Table 5).
Table 5.
Frequency distribution of the FSH receptor haplotypes (SNP at nucleotide amino acid position 307 and 680)
| Haplotype | Controls n = 100 (%) | Primary amenorrhea subjects n = 48 (%) | Secondary amenorrhea subjects n = 38 (%) | ||
|---|---|---|---|---|---|
| n (%) | Odds Ratio (95% CI) | n (%) | Odds Ratio (95% CI) | ||
| Thr Thr(919)–Asn Asn (2039) | 13 (13) | 2 (4.16) | 0.29 | 3 (7.89) | 0.56 |
| Thr Thr(919)–Asn Ser (2039) | 12 (12)a,b | 8 (16.66)a | 1.47c | 9 (23.70)b | 2.07c |
| Thr Thr(919)–Ser Ser (2039) | 0 (0)a | 2 (4.16)a | 8.30c | 0 (0) | 0.26 |
| Thr Ala(919)–Asn Asn (2039) | 13 (13) | 2 (4.16) | 0.29 | 3 (7.89) | 0.57 |
| Thr Ala(919)–Asn Ser (2039) | 34 (34) | 15 (31.28) | 0.88 | 12 (31.59) | 0.89 |
| Thr Ala (919)–Ser Ser (2039) | 6 (6)a,b | 7 (14.58)a | 2.67c | 3 (7.89)b | 1.35c |
| Ala Ala(919)–Asn Asn (2039) | 5 (5)a | 6 (12.50)a | 2.71c | 5 (13.15) | 0.29 |
| Ala Ala(919)–Asn Ser (2039) | 10 (10)a | 6 (12.50)a | 1.28c | 02 (5.26) | 0.50 |
| Ala Ala(919)–Ser Ser (2039) | 7 (7) | 0 (0) | 0.13 | 1 (2.63) | 0.28 |
Values are numbers (percentage)
aThe frequency distribution of these haplotypes is significantly different between the controls and primary amenorrhea subjects (p < 0.05; Fischer’s test)
bThe frequency distribution of these haplotypes is significantly different between the controls and secondary amenorrhea subjects (p < 0.05; Fischer’s test)
cOdds ratio>1.0 is statistically significant
Detection of novel mutations and polymorphisms of FSHR gene by SSCP
All the subjects recruited for study were screened for novel variations in FSHR gene by employing SSCP. PCR amplified products of 5′UTR and entire coding region of FSHR gene (primer pairs 1 to 17; Table 2) were subjected to SSCP analysis. SSCP for exon 1 (primer pair 2), exon 10A and exon 10G (primer pair 11 and 17) showed mobility shifts as expected because these fragments harbored the polymorphic sites of FSHR gene; at −29, 919 and 2039 position respectively. Randomly selected samples that showed change in the migration pattern were subjected to direct DNA sequencing. The results of sequencing confirmed the presence of polymorphic sites at positions −29, 919 and 2039 of FSHR gene.
In one of the subjects with primary amenorrhea, SSCP of exon 10E (primer pair 15; Table 2) showed a change in the migration pattern. Direct DNA sequencing of this PCR product identified a homozygous C to T transition at nucleotide position 1723, which predicted a change of Ala 575 to Val in the FSHR protein (Accession No. FJ211856). The affected subject was a 16 year old girl presenting primary amenorrhea. Her serum FSH and LH levels were 76 mIU/ml and 46.6 mIU/ml respectively. Her serum T3, T4, TSH and prolactin levels were in normal range. Ultrasonography revealed a hypoplastic uterus (size 2.7×1.3 × 0.8 cm) with thin endometrium. Both the ovaries were very small in size. This subject did not respond to estrogen and progesterone for induction of menstruation. The reason for this observation is not known. The subject had no family history of amenorrhea.
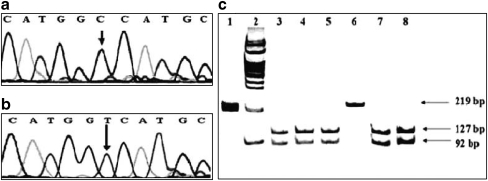
To identify whether C1723T nucleotide change is a mutation or polymorphism, we designed a RFLP (as described below) for detecting this novel (C1723T) change in all the subjects. Further, to confirm the results of RFLP analysis, direct DNA sequencing of the PCR products (exon 10E; primer pair 15) was performed for the controls (n = 100), subjects with primary amenorrhea (n = 15) and subjects with secondary amenorrhea (n = 15). Partial electropherograms of the wild type and mutant genotype for position 1723 of FSHR gene is shown in Fig. 1a and b respectively (Fig. 1).
Fig. 1.
a Partial electropherogram from DNA sequencing of PCR product showing C1723C (Ala at 575 position) indicating wild-type sequence, b Partial electropherogram of PCR product showing T1723T (Val at 575 position) indicating mutant sequence (Accession No FJ211856). c RFLP analysis using Msc I restriction enzyme for detecting C1723T mutation. Lane 1: undigested PCR product of 219 bp; lane 2: 100 bp molecular weight marker; lanes 3–5 and 7–8: two fragments of 127 bp and 92 bp indicating CC genotype (wild-type homozygous); lane 6: 219 bp fragment indicating loss of restriction site (mutant homozygous)
RFLP analysis for the detection of novel mutation C1723T of FSHR gene
For screening C1723T transition in all the subjects enrolled for present study, restriction enzyme Msc I was used in RFLP analysis. The recognition site of Msc I enzyme is TGGCCA and for the wild type sequence this enzyme digests the PCR product (219 bp) into two fragments of 127 bp and 92 bp. The PCR product obtained from one of the primary amenorrhea subject known to harbor this mutation was resistant to digestion by Msc I enzyme (Fig. 1c). Screening of all the controls (n = 100) and subjects with primary amenorrhea (n = 47, excluding the candidate subject) and subjects with secondary amenorrhea (n = 38) by RFLP did not reveal the presence of this mutation.
Hormonal profile
The potential association between FSHR polymorphisms with serum levels of gonadotropins in subjects with primary and secondary amenorrhea was carried out. The subjects were independently segregated based on the FSHR genotypes at −29, 919 and 2039 position. Serum FSH levels were significantly higher in primary amenorrhea subjects with AA genotype as compared to GG and GA genotypes at −29 position (P < 0.05) (Table 6). However, no statistically significant differences were noted in the serum LH levels in these groups. The serum FSH and LH levels showed no statistically significant differences among genotypes at 919 and 2039 position in women with primary or secondary amenorrhea (data not shown).
Table 6.
Serum levels of FSH and LH in the subjects with primary and secondary amenorrhea. The subjects in both the groups were segregated based on the FSH receptor genotypes at position −29
| Hormone | Primary amenorrhea subjects (n = 48) | Secondary amenorrhea subjects (n = 38) | ||||
|---|---|---|---|---|---|---|
| GG (n = 15) | GA (n = 25) | AA (n = 8) | GG (n = 11) | GA (n = 21) | AA (n = 6) | |
| FSH (mIU/ml) | 55.40 ± 10.16a | 65.65 ± 5.96b | 97.90 ± 12.39a,b | 82.81 ± 9.11 | 68.77 ± 7.89 | 88.66 ± 1.33 |
| LH (mIU/ml) | 27.05 ± 4.41 | 34.68 ± 6.10 | 45.23 ± 11.87 | 52.44 ± 1.55 | 44.96 ± 6.18 | 40.13 ± 5.05 |
Values are means (±SEM). Statistical analysis was performed using One-way ANOVA and post-hoc (LSD) test
aValues are significantly different between AA and GG genotypes (P = 0.17)
bValues are significantly different between AA and GA genotypes (P = 0.10)
Discussion
Interaction of FSH with its receptor is crucial for follicular development and maturation, which in turn is indispensable for fertility in females. Any variation in the genotype of FSHR may contribute towards altered ability of the receptor to bind FSH and to induce signal transduction pathway. Interestingly, FSHR knockout mice appear to be a complete phenocopy of the subjects with inactivating FSHR mutations who display blockage of follicular development but intact primordial recruitment [2]. Inactivating mutations in FSHR gene have been linked to the loss of ovarian function in women where these mutations result in impairment of the receptor function [4, 5].
In the present study, 5′UTR and coding region of FSHR gene were screened systematically in total 86 subjects with primary and secondary amenorrhea, presenting hypergonadotropic hypogonadism with normal karyotype and 100 normally cycling proven fertile women served as controls. To the best of our knowledge, ours is one of the few studies [3, 13, 16, 29] comprising the whole coding region of FSHR gene.
The first FSHR gene mutation identified was C566T, in women with ovarian dysgenesis from Finnish population. Other reports from American, German, Brazilian, Mexican and Argentine population [11, 12, 15, 16, 30] showed absence of this FSHR gene mutation in the subjects studied. However, Jiang et al., in 1998 [31], identified only one mutation (C566T) carrier in large scale screening of 1,162 subjects from Switzerland. Thus our results further strengthen the observation that C566T mutation is restricted to Finland and may represent founder effect of this country. The other inactivating mutations reported so far (T479C, C1717T, A671T, C1801G, C1043G, G1255A, A1556C and T662G) are single case reports [4–9]. However, none of the subjects enrolled in present study, harbored any of the reported mutations of FSHR gene.
In the present study, results of RFLP, SSCP and direct DNA sequencing of 5′UTR and exon 1–10 revealed three previously reported polymorphisms, G−29A, A919G and A2039G [1]. We evaluated the frequency distribution of these polymorphic variants in all the subjects recruited. The prevalence of AA genotypes at −29 position was observed to be significantly higher in both the amenorrhic groups as compared to control group (Table 1). It has been reported that the frequency distribution of the polymorphism at −29 position was strikingly different in Indonesian and German women; suggesting putative ethnic variability in the incidence of the polymorphisms [26]. The allelic frequency of ‘A’ allele at the same position was reported to be significantly higher in essential hypertension subjects than in control subjects [32]. We have earlier reported that the frequency distribution of the genotype AA at −29 position was significantly higher in women undergoing ART programme as compared to the control subjects [24].
In the present study, it was observed that the prevalence of genotypes at 919 and 2039 did not differ significantly between study groups (Table 1). Our results are in accordance with the earlier study, where no association of genotypes with gynaecological diseases was observed when the frequency distribution of the genotypes at position 2039 was compared in hypothalamic primary amenorrhea, secondary amenorrhea and POF subjects [18]. Another study has failed to observe differences in the prevalence of FSHR genotype in fertile and infertile women [13]. However, in yet another study, it was reported that FSHR variant Ala/Ala 307 and Ser/Ser 680 were significantly more prevalent among anovulatory women [33].
In accordance with earlier studies [11, 13, 17], two polymorphic variants at 307 and 680 amino acid position were found to occur in linkage disequilibrium in the present study. Frequency distribution of the haplotypes revealed that prevalence of TT-NS, TT-SS, TA-SS, AA-NN and AA-NS was significantly different between controls and primary amenorrhea subjects. However, frequency distribution of TT-NS and TA-SS differed significantly between controls and secondary amenorrhea subjects (Table 5). The haplotype TT-SS was absent in control subjects and showed its presence in two subjects with primary amenorrhea. The odds ratio for haplotypes TT-NS, TT-SS, TA-SS, AA-NN and AA-NS was >1.0 (Table 5) indicating that the subjects with these haplotypes have more tendency to develop amenorrhea.
It has been reported earlier that certain polymorphisms in the FSHR gene are associated with altered responsiveness to FSH in women undergoing ART programme. It has been shown that Ser680 substitution is associated with hypo-response of the ovary to exogenous FSH [17, 18, 20, 22]. However, a few studies have refuted this view [22, 33]. We have recently reported that alteration at 919 position (Thr307Ala) of FSHR gene, is associated with hyper ovarian response [23].
It has been reported earlier that certain polymorphisms in the FSHR gene are associated with altered responsiveness to FSH in women undergoing ART programme. It has been shown that Ser680 substitution is associated with hypo-response of the ovary to exogenous FSH [17, 18, 20, 22]. However, a few studies have refuted this view [22, 33]. We have recently reported that alteration at 919 position (Thr307Ala) of FSHR gene, is associated with hyper ovarian response [23]. We have also reported that the women (undergoing ART programme) with AA genotype at −29 position of FSHR gene are associated with hypo-response to exogenous FSH [24]. These observations prompted us to investigate if any of the polymorphisms (at position −29, 919 and 2039) are associated with the loss of ovarian function resulting in amenorrhea.
In the present study, we have observed that the prevalence of the polymorphisms at −29 position differed significantly in subjects with amenorrhea and controls. The frequency distribution of primary and secondary amenorrhea subjects AA genotype at −29 position was 14.59% and 15.78% respectively. In control subjects the frequency distribution of AA genotype was only 1% (Table 4). Interestingly, the serum FSH levels were significantly higher in primary amenorrhea women with AA genotype as compared to GG and GA genotypes at −29 position, indicative of end organ insensitivity (Table 6). It has also been reported that homozygous A at −29 position of FSHR gene has reduced transcriptional activity [33]. Due to this reason, we hypothesized that in AA genotype the receptor expression may be affected. It is also possible that due to lack of functional FSHR, the negative feedback exhibited by the estradiol on the hypothalamo-pituitary gonadal axis is lost. This could be one of the reasons for increased serum FSH levels in subjects with AA genotypes at −29 position.
To identify novel polymorphisms and mutations in FSHR gene, the 5′UTR and the entire coding region was screened by SSCP in all recruited subjects. It has been proposed earlier that SSCP technique allows identification of a nucleotide change (polymorphism/mutation) with high sensitivity (close to 100%) under two different electrophoresis conditions [34]. In the present study we have used two different conditions for electrophoresis to ensure detection of almost all the changes that would be present. Although, our results indicate absence of alterations in exon 2–9, exon 10 (primer set B-D and F) of FSHR gene; failure to detect changes due to limitations in the technique sensitivity can not be ruled out. Indeed, several other studies on genetic analysis of the subjects with ovarian disorders, have failed to identify any mutations in the subjects studied [12–16, 29, 30, 35]. Similarly, our results suggest that the mutations in FSHR gene are rare events and may not be a cause of loss of ovarian function in subjects with amenorrhea.
In the present study, we identified a novel C1723T transition in the FSHR gene by SSCP followed by direct DNA sequencing. To detect this novel nucleotide change in all the subjects enrolled for present study, a RFLP was designed. RFLP analysis revealed that this novel C1723T transition is present only in one subject with primary amenorrhea. All the samples including controls (n = 100) were further subjected to DNA sequencing to confirm the results of RFLP analysis. We believe that the C1723T transition is a mutation because it was detected in only one case out of the total 236 subjects recruited; controls (n = 100), subjects with amenorrhea (n = 86) and subjects undergoing Assisted Reproductive Technology programme, having normal cyclicity (n = 50; unpublished data), by RFLP analysis and DNA sequencing. This novel mutation has not been reported earlier in any of the studies so far [3, 11, 14, 16, 36].
The C1723T transition results in Ala575Val amino acid change, in the sixth helix of transmembrane domain (TMD) of FSHR. It is also of interest to note that the Ala at position 575 is conserved across species in FSHR gene and also amongst other glycoprotein hormone receptors and may have an important role in receptor function. This variation at 575 amino acid position is potentially a mutation and hence adds up to the spectrum of mutations reported earlier in the FSHR gene. Several mutations in the TMD of FSHR have been reported suggesting the importance of the domain in normal receptor function [37]. Two inactivating mutations reported in the first and the second helix (Pro348Arg and Ala419Thr respectively) affect hormone binding and signal transduction [5, 6]. In the present study, the mutation identified (Ala575Val) resides in the sixth helix of the TMD. To the best of our knowledge this is the third impending inactivating mutation identified in transmembrane helices and the first of its kind in sixth helix.
Most of the FSHR mutations reported in the literature have been studied in depth, with an aim to understand their functional significance. In case of the first mutation (Ala189Val) reported from Finnish population, Ala189 is the amino acid in perfectly conserved stretch of five amino acids (Ala189 Phe Asn Gly Thr) in glycoprotein hormone receptor is substituted with Val. The presence of Val at 189 position in FSHR has been reported to interfere with the efficiency of glycosylation resulting in impaired receptor trafficking and folding [38]. In vitro studies showed that the mutated receptor had normal binding affinity and severely reduced plasma membrane expression and signal transduction [11]. In vitro studies for the two mutations, Ile160Thr (T479C) and Asp224Val (A671T) showed that these mutations almost completely impair FSH binding and marginal response to FSH stimulation [4]. Studies with the other two mutations, Arg573Cys (C1717T) and Leu601Val (C1801G) showed normal hormone binding and decreased signal transduction [9]. The functional significance of the novel mutation identified in the present study (C1723T) is being investigated.
In summary, the results of the present study demonstrate the significantly higher frequency distribution of AA genotype at −29 position of FSHR gene in primary and secondary amenorrhea subjects as compared to controls. The prevalence of haplotypes (polymorphism at 307 and 680 amino acid position) TT-NS, TT-SS, TA-SS, AA-NN and AA-NS was significantly higher in amenorrhea subjects. The odds ratio>1.0 for these above mentioned haplotypes indicated increased probability of developing amenorrhea. The primary amenorrhea subjects with AA genotype at −29 position of FSHR gene showed significantly higher serum FSH levels. Only one single homozygous mutation has been identified in a woman with primary amenorrhea in the present study. These findings indicate that the mutations of FSHR gene are rare in Indian women with primary and secondary amenorrhea and hence may not contribute as a causative factor for these conditions in most cases.
Conclusions
Our findings suggest that increased serum FSH levels in subjects with primary amenorrhea correlated to FSHR genotype at position −29. Screening of subjects recruited in the present study indicated that no known FSHR gene mutations are present in these subjects. Interestingly, we have identified a novel homozygous mutation C1723T (Ala575Val) in a woman with primary amenorrhea.
Acknowledgements
The research work related to this publication was supported by grants from National Institute for Research in Reproductive Health (NIRRH/MS/69/2009) and from the Indian Council of Medical Research, New Delhi, India (63/116/2001-BMS II). We would like to thank Dr. Anurupa Maitra (DNA Sequencing Core Facility, NIRRH) for her help in DNA sequencing. We acknowledge Dr. D. Balaiah and Mr. Prashant Tapse (Division of Biostatistics, NIRRH) for their help in statistical analysis.
Footnotes
Capsule
Novel homozygous FSHR gene mutation C1723T (Ala575Val) is identified in a woman with primary amenorrhea.
References
- 1.Simoni M, Gromoll J, Nieschlag E. The follicle stimulating hormone receptor: biochemistry, molecular biology, physiology, and pathophysiology. Endoc Rev. 1997;18:739–73. doi: 10.1210/er.18.6.739. [DOI] [PubMed] [Google Scholar]
- 2.Dierich A, Sairam MR, Monaco L, Fimia GM, Gansmuller A, LeMeur M, et al. Impairing follicle-stimulating hormone (FSH) signaling in vivo: targeted disruption of the FSHR leads to aberrant gametogenesis and hormonal imbalance. Proc Natl Acad Sci USA. 1998;95:13612–7. doi: 10.1073/pnas.95.23.13612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Aittomäki K, Lucena JL, Pakarinen P, Sistonen P, Tapanainen J, Gromoll J, et al. Mutation in the follicle-stimulating hormone receptor gene causes hereditary hypergonadotropic ovarian failure. Cell. 1995;82:959–68. doi: 10.1016/0092-8674(95)90275-9. [DOI] [PubMed] [Google Scholar]
- 4.Touraine P, Beau I, Gougeon A, Meduri G, Desroches A, Pichard C. New natural inactivating mutations of the follicle-stimulating hormone receptor: correlations between receptor function and phenotype. Mol Endocrinol. 1999;13:1844–54. doi: 10.1210/me.13.11.1844. [DOI] [PubMed] [Google Scholar]
- 5.Allen LA, Achermann JC, Pakarinen P, Kotlar TJ, Huhtaniemi IT, Jameson JL. A novel loss of function mutation in exon 10 of the FSHR gene causing hypergondotrophic hypogonadism: clinical and molecular characteristics. Hum Reprod. 2003;18:251–6. doi: 10.1093/humrep/deg046. [DOI] [PubMed] [Google Scholar]
- 6.Doherty E, Pakarinen P, Tiitinen A, Kiilavuori A, Huhtaniemi I, Forrest S. A novel mutation in the FSHR inhibiting signal transduction and causing primary ovarian failure. J Clin Endocrinol Metab. 2002;87:1151–5. doi: 10.1210/jc.87.3.1151. [DOI] [PubMed] [Google Scholar]
- 7.Meduri G, Touraine P, Beau I, Lahuna O, Desroches A, Vacher-Lavenu MC, et al. Delayed puberty and primary amenorrhea associated with a novel mutation of the human follicle-stimulating hormone receptor: clinical, histological, and molecular studies. J Clin Endocrinol Metab. 2003;88:3491–8. doi: 10.1210/jc.2003-030217. [DOI] [PubMed] [Google Scholar]
- 8.Nakamura Y, Maekawa R, Yamagata Y, Tamura I, Sugino N. A novel mutation in exon8 of the follicle-stimulating hormone receptor in a woman with primary amenorrhea. Gynecol Endocrinol. 2008;24:708–12. doi: 10.1080/09513590802454927. [DOI] [PubMed] [Google Scholar]
- 9.Beau I, Touraine P, Meduri G, Gougeon A, Desroches A, Matuchansky C, et al. A novel phenotype related to partial loss of function mutations of the follicle stimulating hormone receptor. J Clin Invest. 1998;102:1352–9. doi: 10.1172/JCI3795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Rannikko A, Pakarinen P, Manna R, Beau I, Misrahi M, Aittomaki K. Functional characterization of the human FSHR with an inactivating Ala189Val mutation. Mol Hum Reprod. 2002;8:311–7. doi: 10.1093/molehr/8.4.311. [DOI] [PubMed] [Google Scholar]
- 11.Fonte Kohek MB, Batista MC, Russell AJ, Vass K, Giacaglia LR, Mendonca BB, et al. No evidence of the inactivating mutation (C566T) in the follicle-stimulating hormone receptor gene in Brazilian women with premature ovarian failure. Fertil Steril. 1998;70:565–7. doi: 10.1016/S0015-0282(98)00203-9. [DOI] [PubMed] [Google Scholar]
- 12.Liu JY, Gromoll J, Cedars MI, Barbera AR. Identification of allelic variants in the follicle-stimulating hormone receptor genes of females with or without hypergonadotropic amenorrhea. Fertil Steril. 1998;70:326–31. doi: 10.1016/S0015-0282(98)00151-4. [DOI] [PubMed] [Google Scholar]
- 13.Conway GS, Conway E, Walker C, Hoppner W, Gromoll J, Simoni M. Mutation screening and isoform prevalence of the follicle stimulating hormone receptor gene in women with premature ovarian failure, resistant ovary syndrome and polycystic ovary syndrome. Clin Endocrinol (Oxf) 1999;51:97–9. doi: 10.1046/j.1365-2265.1999.00745.x. [DOI] [PubMed] [Google Scholar]
- 14.Takakura K, Takebayashi K, Wang HQ, Kimura F, Kasahara K, Noda Y. Follicle-stimulating hormone receptor gene mutations are rare in Japanese women with premature ovarian failure and polycystic ovary syndrome. Fertil Steril. 2001;75:207–9. doi: 10.1016/S0015-0282(00)01673-3. [DOI] [PubMed] [Google Scholar]
- 15.Chesnaye E, Canto P, Ulloa-Aguirre A, Méndez JP. No evidence of mutations in the follicle-stimulating hormone receptor gene in Mexican women with 46, XX pure gonadal dysgenesis. Am J Med Genet. 2001;98:125–8. doi: 10.1002/1096-8628(20010115)98:2<125::AID-AJMG1020>3.0.CO;2-I. [DOI] [PubMed] [Google Scholar]
- 16.Sundblad V, Chiauzzi VA, Escobar ME, Dain L, Charreau EH. Screening of FSHR gene in Argentine women with premature ovarian failure (POF) Mol Cell Endocrinol. 2004;222:53–9. doi: 10.1016/j.mce.2004.05.002. [DOI] [PubMed] [Google Scholar]
- 17.Perez Mayorga M, Gromoll J, Behre HM, Gassner C, Nieschlag E, Simoni M. Ovarian response to follicle-stimulating hormone (FSH) stimulation depends on the FSHR genotype. J Clin Endocrinol Metab. 2000;85:3365–9. doi: 10.1210/jc.85.9.3365. [DOI] [PubMed] [Google Scholar]
- 18.Sudo S, Kudo M, Wada S, Sato O, Hsueh AJ, Fujimoto S. Genetic and functional analyses of polymorphism in the human FSHR gene. Mol Hum Reprod. 2002;8:893–9. doi: 10.1093/molehr/8.10.893. [DOI] [PubMed] [Google Scholar]
- 19.Behre HM, Greb RR, Mempel A, Sonntag B, Kiesel L, Kaltwasser P, et al. Significance of a common single nucleotide polymorphism in exon 10 of the follicle-stimulating hormone (FSH) receptor gene for the ovarian response to FSH: a pharmacogenetic approach to controlled ovarian hyperstimulation. Pharmacogenomics. 2005;15:451–6. doi: 10.1097/01.fpc.0000167330.92786.5e. [DOI] [PubMed] [Google Scholar]
- 20.Greb RR, Grieshaber K, Gromoll J, Sonntag B, Nieschlag E, Kiesel L, et al. A common single nucleotide polymorphism in exon 10 of the human follicle stimulating hormone receptor is a major determinant of length and hormonal dynamics of the menstrual cycle. J Clin Endocrinol Metab. 2005;90:4866–72. doi: 10.1210/jc.2004-2268. [DOI] [PubMed] [Google Scholar]
- 21.Loutradis D, Patsoula E, Minas V, Koussidis GA, Antsaklis A, Michalas S, et al. FSHR gene polymorphisms have a role for different ovarian response to stimulation in patients entering IVF/ICSI-ET programs. J Assist Reprod Genet. 2006;23:177–84. doi: 10.1007/s10815-005-9015-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jun JK, Yoon JS, Ku SY, Choi YM, Hwang KR, Park SY, et al. Follicle-stimulating hormone receptor gene polymorphism and ovarian responses to controlled ovarian hyperstimulation for IVF-ET. J Hum Genet. 2006;51:665–70. doi: 10.1007/s10038-006-0005-5. [DOI] [PubMed] [Google Scholar]
- 23.Achreker SK, Modi DN, Desai SK, Mangoli VS, Mangoli RV, Mahale SD. Follicle-stimulating hormone receptor polymorphism (Thr307Ala) is associated with variable ovarian response and ovarian hyper stimulation syndrome in Indian women. Fertil Steril. 2009;91:432–9. doi: 10.1016/j.fertnstert.2007.11.093. [DOI] [PubMed] [Google Scholar]
- 24.Achreker SK, Modi DN, Desai SK, Mangoli VS, Mangoli RV, Mahale SD. Poor ovarian response to gonadotropin stimulation is associated with FSHR polymorphism. Reprod Biomed Online. 2009;18:509–15. doi: 10.1016/S1472-6483(10)60127-7. [DOI] [PubMed] [Google Scholar]
- 25.Gromoll J, Bröcker M, Derwahl M, Höppner W. Detection of mutations in glycoprotein hormone receptors. Methods. 2000;21:83–97. doi: 10.1006/meth.2000.0977. [DOI] [PubMed] [Google Scholar]
- 26.Wunsch A, Ahda Y, Banaz-Yaşar F, Sonntag B, Nieschlag E, Simoni M, et al. Single-nucleotide polymorphisms in the promoter region influence the expression of the human follicle-stimulating hormone receptor. Fertil Steril. 2005;84:446–53. doi: 10.1016/j.fertnstert.2005.02.031. [DOI] [PubMed] [Google Scholar]
- 27.Innis A, Gelfand H, Sninsky J. PCR strategies. San Diego: Academic; 1995. p. 121. [Google Scholar]
- 28.Pooart J, Limpaiboon T, Lulitanond V. Improved non-isotopic PCR-SSCP for screening of p53 mutations. Clin Biochem. 1999;32:233–5. doi: 10.1016/S0009-9120(99)00002-8. [DOI] [PubMed] [Google Scholar]
- 29.Tong Y, Liao W, Roy A, Ng S. Absence of mutations in the coding regions of follicle-stimulating hormone receptor gene in Singapore Chinese women with premature ovarian failure and polycystic ovary syndrome. Horm Metab Res. 2001;33:221–6. doi: 10.1055/s-2001-14941. [DOI] [PubMed] [Google Scholar]
- 30.Layman LC, Amde S, Cohen DP, Jin M, Xie J. The Finnish follicle-stimulating hormone receptor gene mutation is rare in North American women with 46, XX ovarian failure. Fertil Steril. 1998;69:300–2. doi: 10.1016/S0015-0282(97)00480-9. [DOI] [PubMed] [Google Scholar]
- 31.Jiang M, Aittomäki K, Nilsson C, Pakarinen P, Iitiä A, Torresani T, et al. The frequency of an inactivating point mutation (566C–>T) of the human follicle-stimulating hormone receptor gene in four populations using allele-specific hybridization and time-resolved fluorometry. J Clin Endocrinol Metab. 1998;12:4338–43. doi: 10.1210/jc.83.12.4338. [DOI] [PubMed] [Google Scholar]
- 32.Nakayama T, Kuroi N, Sano M, Tabara Y, Katsuya T, Ogihara T, et al. Mutation of the follicle-stimulating hormone receptor gene 5′-untranslated region associated with female hypertension. Hypertension. 2006;48:512–8. doi: 10.1161/01.HYP.0000233877.84343.d7. [DOI] [PubMed] [Google Scholar]
- 33.Laven JS, Mulders AG, Suryandari DA, Gromoll J, Nieschlag E, Fauser BC, et al. Follicle-stimulating hormone receptor polymorphisms in women with normogonadotropic anovulatory infertility. Fertil Steril. 2003;8:986–92. doi: 10.1016/S0015-0282(03)01115-4. [DOI] [PubMed] [Google Scholar]
- 34.Forrest S, Cotton R, Landegren U, Southern E. How to find all those mutations. Nat Genet. 1995;10:375–6. doi: 10.1038/ng0895-375. [DOI] [PubMed] [Google Scholar]
- 35.Whitney EA, Lee A, Layman LC, Peak DB, Chan PJ. McDonough, PG. The follicle-stimulating hormone receptor gene is polymorphic in premature ovarian failure and normal controls. Fertil Steril. 1995;64:518–24. doi: 10.1016/s0015-0282(16)57786-3. [DOI] [PubMed] [Google Scholar]
- 36.Latronico AC, Arnhold IJ. Inactivating mutations of LH and FSHRs—from genotype to phenotype. Pediatr Endocrinol Rev. 2006;4:28–31. [PubMed] [Google Scholar]
- 37.Themmen APN, Huhtaniemi IT. Mutations of gonadotropins and gonadotropin receptors: elucidating the physiology and pathophysiology of pituitary-gonadal function. Endocr Rev. 2000;21:551–83. doi: 10.1210/er.21.5.551. [DOI] [PubMed] [Google Scholar]
- 38.Davis D, Liu X, Segaloff DL. Identification of the sites of N-linked glycosylation on the follicle-stimulating hormone (FSH) receptor and assessment of their role in FSHR function. Mol Endocrinol. 1995;2:159–70. doi: 10.1210/me.9.2.159. [DOI] [PubMed] [Google Scholar]