Abstract
Osteosarcoma is a primary bone malignancy that typically occurs during the pubertal growth spurt. Only a few small association studies have evaluated common germ line variation in individuals with osteosarcoma. The 8q24 chromosomal region contains several loci that are associated with risk of many different cancers. We conducted an association study of common single-nucleotide polymorphisms (SNPs) across 8q24 to explore the role this region may play in osteosarcoma risk. We genotyped 214 tag SNPs in 99 osteosarcoma cases and 1430 controls (65 controls from a hospital-based case–control study and 1365 controls from a population-based study). Additive, dominant and recessive genetic models were evaluated using unconditional logistic regression to estimate odds ratios (ORs) and 95% confidence intervals (CIs). Analyses of nine SNPs previously associated with cancer did not show strong statistically significant associations. Of the remaining 205 SNPs, 7 were statistically significant (P ≤ 0.05) in one or more genetic models; the most significant association was observed for the additive effect of the minor allele at rs896324 (OR 1.75, 95% CI 1.13–2.69, P = 0.01). This study suggests that several SNPs in 8q24 may be associated with osteosarcoma, but the susceptibility observed was modest. Future large studies of osteosarcoma genetic risk factors are warranted to improve our understanding of the genetic contribution to this cancer of adolescents and young adults.
Introduction
Osteosarcoma is a primary bone tumor that occurs primarily in adolescents and young adults (1). Patients with localized osteosarcoma at presentation have a 60–80% long-term survival rate but metastatic disease carries a poorer prognosis (2). Little is known about the etiology of osteosarcoma. Some studies suggest associations with height (3,4) and birth weight (5), but the data are inconsistent (6,7). Osteosarcoma occurs at increased frequencies among individuals with cancer predisposition syndromes, such as Li-Fraumeni Syndrome, retinoblastoma, Diamond–Blackfan anemia and Rothmund–Thomson Syndrome (8), but the genetic contribution to apparently sporadic osteosarcoma is not well understood.
A limited number of pilot studies have investigated common germ line genetic variation in osteosarcoma. One study noted higher frequencies of the FokI Ff genotype in the vitamin D receptor gene in osteosarcoma cases compared with controls but no association with single-nucleotide polymorphisms (SNPs) in the estrogen receptor or collagen Iα1 genes (7). Another study suggested a lower frequency of osteosarcoma in individuals with a tumor necrosis factor-alpha promoter variant (9). Strong associations with common SNPs in the TP53 gene were not noted in a pilot study of osteosarcoma (10). However, another study on the same case–control set identified statistically significant positive associations between a variant in the insulin-like growth factor 2 receptor gene (IGFR2), which alters methylation at that site and osteosarcoma risk (11). Two other studies suggested associations between polymorphisms in Fas (12) and MDM2 (13) and risk of osteosarcoma and high-grade osteosarcoma in females, respectively.
Genome-wide association studies (GWAS) have found associations between several SNPs at the chromosome 8q24 locus and the risk of different cancers, including prostate cancer (14–18), breast cancer (19,20), bladder cancer (21) and colorectal cancer (22). These SNPs appear to occur in different haplotype blocks in this region (23) and are located in an apparent ‘gene desert’. The MYC oncogene is ∼300 kb from the region associated with cancer in GWAS. The G allele of rs6983267 is preferentially amplified in colorectal cancer tumors and affects a binding site for the Wnt-regulated transcription factor 4 (24). It has also been shown that this region physically interacts with the MYC promoter (25,26) and that transcriptional enhancers in this region may be affected by the cancer-associated variants (27).
MYC is an oncogene with many functions that are still being elucidated, including transcriptional activation and repression (28). Mutations in MYC are not prevalent in cancer cells, instead MYC deregulation appears to occur through insertional mutagenesis, chromosomal translocations and gene amplification (28). MYC has been shown to be highly amplified in a subset of osteosarcomas (29,30) and is overexpressed in relapsed and metastatic osteosarcoma (31). A mouse transgenic osteosarcoma model demonstrated that brief inactivation of MYC results in sustained osteosarcoma regression and differentiation of malignant cells into mature osteocytes (32).
Genetic variation in the 8q24 chromosomal region may be associated with osteosarcoma because several SNPs in this region are associated with cancer and several studies have found amplification of the chromosome 8q24 region in osteosarcoma tumor tissues (33,34). In addition, the 8q24 SNPs may affect MYC function, which is thought to play a role in osteosarcoma pathogenesis. We genotyped 214 tag SNPs across 8q24 in 99 osteosarcoma cases and 1430 controls. We hypothesized that the nine SNPs previously associated with cancer had the highest prior probability of being associated with osteosarcoma and they were analyzed first. Subsequent analyses evaluated the remainder of the 8q24 SNPs.
Materials and methods
Study design
Osteosarcoma cases (n = 99) were derived from the Bone Disease and Injury Study of Osteosarcoma (5), which is a hospital-based prospective case–control study that collected blood samples and questionnaire data on individuals seen at orthopedic surgery departments in 10 USA medical centers between 1994 and 2000. Osteosarcoma patients were identified at the time of limb salvage surgery. There were no identified cases of Paget's disease of the bone in this study. Orthopedic controls from Bone Disease and Injury Study of Osteosarcoma (n = 65) consisted of individuals with benign tumors (26%) and other non-neoplastic conditions, such as inflammatory diseases, cysts and trauma, excluding those with hip fracture or osteoporosis. Institutional review boards at each of the medical centers approved the study protocol and informed consent was obtained from all study subjects. DNA was isolated from blood samples using standard procedures. This study was limited to individuals who were self-identified Caucasians in order to reduce potential effects of population stratification.
An additional 1365 cancer-free Caucasian control subjects were derived from the Prostate, Lung, Colorectal, Ovarian Cancer Screening Trial. Men and women, ages 55–74 years, were enrolled in the screening trial from 10 different centers in the USA between 1993 and 2001. All subjects included for this study were required to have completed a baseline questionnaire, provided a blood specimen and consented to participate in etiologic studies of cancer and related diseases. Controls were limited to Caucasians living in the continental USA without a diagnosis of colon adenoma or cancer at baseline. DNA was extracted from blood specimens using standard procedures. The institutional review boards at the National Cancer Institute and 10 screening centers approved the study.
Genotyping
Genotyping was conducted on a Custom Infinium® BeadChip (iSelect)™ from Illumina (San Diego, CA). The iSelect panel was created by investigators in the Division of Cancer Epidemiology and Genetics, National Cancer Institute, to target genetic variation in genes potentially important in carcinogenesis and cancer risk. Tag SNPs were identified from the HapMap CEU population assuming a minor allele frequency (MAF) ≥5% and a r2 threshold >0.80 using the Tagzilla module of the GLU software package (http://code.google.com/p/glu-genetics/) across the region of chromosome 8q24 previously identified in other GWAS as cancer risk loci. In this study, a total of 214 tag SNPs across the region of chromosome 8q24 from 128232156 to 128832477 were genotyped. The genotype completion rate was >95.1% for all SNPs. The concordance rate between duplicate samples on the iSelect panel was 99.5%. Only SNPs consistent with Hardy–Weinberg equilibrium among controls were analyzed in this study.
Statistical analyses
Unconditional logistic regression was used to estimate the odds ratio (OR) and 95% confidence intervals (CIs) for the strength of the association between osteosarcoma risk independently for each SNP, adjusting for sex. We evaluated the log-additive (which assumes an additive effect of each copy of the minor allele), codominant, dominant and recessive genetic inheritance models for each SNP in relationship to osteosarcoma case status. When there were no individuals homozygous for the minor allele, the codominant model was only used to evaluate the heterozygote compared with the homozygote of the common allele, and the other inheritance models also only compared these two categories. The homozygote of the common allele was used as the referent category for the additive and codominant models. The gene–dose effects for each SNP were estimated by a linear trend test by coding the genotypes based on the number of variant alleles (0, 1 and 2). Statistical analyses were performed using SAS software, version 9.1 (SAS Institute, Cary, NC) and PLINK software, version 1.06 (http://pngu.mgh.harvard.edu/purcell/plink/).
Statistical power was calculated using Quanto (35) using the log-additive, dominant and recessive models, 99 cases and 1430 controls, baseline population risk of 0.0000001 and type 1 error of 0.05. For the log-additive model, power was >80% to detect an OR of 1.82 assuming MAF of 10% and 1.53 assuming a MAF of 30%.
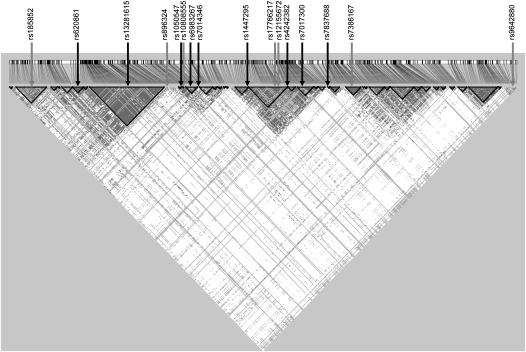
We estimated the correlation between SNPs (r2) with PLINK. We evaluated the linkage disequilibrium structure across the 8q24 chromosomal region using HapMap genotype data and Haploview version 4.1 (36,37). SNPs with MAFs of at least 1% present in the HapMap (release 27) Caucasian population (CEU) from Utah were used to create Figure 1.
Fig. 1.
Linkage disequilibrium on chromosome 8q24 in the HapMap Caucasian (CEU) population, release 27, determined using Haploview and SNPs with MAFs of at least 0.01. SNPs associated with other cancers are noted with the black arrows. Those associated with osteosarcoma in this study are noted with grey arrows. This region represents chromosome 8q24 nucleotides128342000–128807000. SNPs shown (left to right) are rs185852, rs620861, rs13281615, rs896324, rs10505477, rs10808555, rs6983267, rs7014346, rs1447295, rs17766217, rs12155672, rs4242382, rs7017300, rs7837688, rs7386167 and rs9642880.
Results
Subject characteristics
The characteristics of study participants, 99 osteosarcoma cases and 1430 controls, are shown in Table I. The median age of the 99 osteosarcoma cases was 20 years (range 8–80.4). There were 65 orthopedic controls derived from the Bone Disease and Injury Study of Osteosarcoma. Their median age was 18.5 years (range 7.2–68.5). Osteosarcoma cases and orthopedic controls had nearly equal numbers of males and females. The Prostate, Lung, Colorectal, Ovarian controls were older with a median age of 62 years (range 55–75). There were more males (63.9%) than females (36.1%) in the Prostate, Lung, Colorectal, Ovarian controls. All participants were self-identified Caucasians and from the continental USA.
Table I.
Characteristics of cases and controls
| N (%) male | N (%) female | Median age | Age range | Mean age (standard deviation) | Total N | |
| All controls | 907 (63.4) | 523 (36.6) | 62 | 7.2–75 | 60.9 (9.9) | 1430 |
| Orthopedic controls | 35 (53.8) | 30 (46.2) | 18.5 | 7.2–68.5 | 25 (15.2) | 65 |
| PLCO controls | 872 (63.9) | 493 (36.1) | 62 | 55–75 | 62.6 (5.2) | 1365 |
| OS cases | 56 (56.6) | 43 (43.4) | 20.4 | 8–80.4 | 26.6 (16.0) | 99 |
N, number of individuals; OS, osteosarcoma; PLCO, Prostate, Lung, Colon, Ovarian Cancer Cohort.
8q24 SNPs previously associated with other cancers
Our primary hypothesis was to test whether or not the nine SNPs associated with other cancers in the 8q24 chromosomal region (14,17,19,20,22) were also associated with osteosarcoma; therefore, we evaluated these genotypes first (Table II and Figure 1). Analyses of these nine SNPs did not show any strong statistically significant associations (Ptrend > 0.05). A positive association of borderline significance between osteosarcoma and individuals heterozygous at rs7017300 was noted (OR 1.56 and 95% CI 0.99–2.42); however, no statistically significant trend was observed (Ptrend = 0.087).
Table II.
Association of SNPs in the 8q24 chromosomal region and osteosarcoma by inheritance model
| SNP, location (references) | Genotype | No. control (%) | No. cases (%) | OR (95% CI) | P | Ptrend | Dominant P | Recessive P |
| 8q24 SNPs previously associated with other cancers | ||||||||
| rs620861, 128,404,855 (15,16) | G/G | 577 (40.4) | 49 (49.5) | 1 (ref) | ||||
| A/G | 652 (45.6) | 39 (39.4) | 0.70 (0.45–1.08) | 0.11 | ||||
| A/A | 201 (14.1) | 11 (11.1) | 0.64 (0.33–1.25) | 0.19 | 0.086 | 0.071 | 0.39 | |
| rs13281615, 128,424,800 (19) | A/A | 445 (33.2) | 37 (39) | 1 (ref) | ||||
| A/G | 656 (49) | 43 (45.3) | 0.79 (0.50–1.25) | 0.31 | ||||
| G/G | 238 (17.8) | 15 (15.8) | 0.77 (0.41–1.42) | 0.39 | 0.31 | 0.27 | 0.64 | |
| rs10505477, 128,476,625 (39,40) | G/G | 407 (28.5) | 24 (24.2) | 1 (ref) | ||||
| A/G | 696 (48.7) | 52 (52.5) | 1.27 (0.77–2.09) | 0.35 | ||||
| A/A | 326 (22.8) | 23 (23.2) | 1.19 (0.66–2.14) | 0.57 | 0.55 | 0.37 | 0.95 | |
| rs7014346, 128,493,974 (41,42) | G/G | 632 (44.3) | 37 (37.4) | 1 (ref) | ||||
| A/G | 614 (43) | 48 (48.5) | 1.35 (0.87–2.10) | 0.19 | ||||
| A/A | 182 (12.8) | 14 (14.1) | 1.30 (0.69–2.46) | 0.42 | 0.25 | 0.17 | 0.73 | |
| rs1447295, 128,554,220 (20,43,44) | C/C | 1165 (81.5) | 77 (77.8) | 1 (ref) | ||||
| A/C | 253 (17.7) | 22 (22.2) | 1.35 (0.82–2.21) | 0.24 | ||||
| A/A | 11 (0.8) | 0 (0) | 0 | 0.44 | 0.32 | 0.22 | ||
| rs6983267, 128,482,487 (20,40,45) | T/T | 389 (27.2) | 23 (23.2) | 1 (ref) | ||||
| G/T | 702 (49.1) | 51 (51.5) | 1.23 (0.74–2.04) | 0.43 | ||||
| G/G | 339 (23.7) | 25 (25.2) | 1.24 (0.69–2.22) | 0.47 | 0.47 | 0.39 | 0.75 | |
| rs4242382, 128,586,755 (46) | G/G | 1168 (81.7) | 77 (77.8) | 1 (ref) | ||||
| A/G | 249 (17.4) | 22 (22.2) | 1.37 (0.83–2.24) | 0.22 | ||||
| A/A | 12 (0.8) | 0 (0) | 0 | 0.43 | 0.31 | 0.2 | ||
| rs7017300, 128,594,450 (23) | A/A | 1077 (75.4) | 67 (67.7) | 1 (ref) | ||||
| A/C | 325 (22.7) | 31 (31.3) | 1.56 (0.99–2.42) | 0.051 | ||||
| C/C | 27 (1.9) | 1 (1) | 0.59 (0.08–4.38) | 0.60 | 0.17 | 0.087 | 0.48 | |
| rs7837688, 128,608,542 (23,47) | G/G | 1169 (81.8) | 78 (78.8) | 1 (ref) | ||||
| G/T | 248 (17.4) | 21 (21.2) | 1.29 (0.78–2.13) | 0.32 | ||||
| T/T | 12 (0.8) | 0 (0) | 0 | 0.57 | 0.43 | 0.21 | ||
| 8q24 SNPs associated with osteosarcoma | ||||||||
| rs185852, 128,362,638 | G/G | 934 (65.4) | 60 (60.6) | 1 (ref) | ||||
| A/G | 440 (30.8) | 31 (31.3) | 1.09 (0.70–1.71) | 0.69 | ||||
| A/A | 54 (3.8) | 8 (8.1) | 2.28 (1.04–5.02) | 0.039 | 0.13 | 0.35 | 0.043 | |
| rs896324, 128,465,694 | A/A | 1225 (85.7) | 76 (76.8) | 1 (ref) | ||||
| A/G | 191 (13.4) | 21 (21.2) | 1.79 (1.08–2.97) | 0.025 | ||||
| G/G | 13 (0.9) | 2 (2) | 2.75 (0.61-12.52) | 0.19 | 0.011 | 0.02 | 0.29 | |
| rs10808555, 128,478,693 | A/A | 679 (47.5) | 37 (37.4) | 1 (ref) | ||||
| A/G | 595 (41.6) | 52 (52.5) | 1.61 (1.04–2.48) | 0.033 | ||||
| G/G | 156 (10.9) | 10 (10.1) | 1.17 (0.57–2.40) | 0.67 | 0.19 | 0.05 | 0.78 | |
| rs17766217, 128,573,679 | T/T | 531 (37.2) | 47 (47.5) | 1 (ref) | ||||
| C/T | 692 (48.4) | 42 (42.4) | 0.69 (0.45–1.07) | 0.096 | ||||
| C/C | 206 (14.4) | 10 (10.1) | 0.56 (0.28–1.13) | 0.1 | 0.046 | 0.05 | 0.23 | |
| rs12155672, 128,576,206 | G/G | 366 (25.6) | 23 (23.2) | 1 (ref) | ||||
| A/G | 728 (50.9) | 42 (42.4) | 0.92 (0.55–1.56) | 0.76 | ||||
| A/A | 336 (23.5) | 34 (34.3) | 1.59 (0.92–2.76) | 0.098 | 0.079 | 0.6 | 0.022 | |
| rs7386167, 128,637,894 | G/G | 618 (43.2) | 37 (37.4) | 1 (ref) | ||||
| A/G | 626 (43.8) | 40 (40.4) | 1.07 (0.67–1.69) | 0.78 | ||||
| A/A | 186 (13) | 22 (22.2) | 1.95 (1.12–3.39) | 0.018 | 0.041 | 0.26 | 0.018 | |
| rs9642880, 128,787,250 | G/G | 416 (29.2) | 39 (39.4) | 1 (ref) | ||||
| G/T | 704 (49.4) | 42 (42.4) | 0.63 (0.40–0.99) | 0.048 | ||||
| T/T | 306 (21.5) | 18 (18.2) | 0.62 (0.35–1.11) | 0.11 | 0.065 | 0.034 | 0.43 | |
Statistically significant (P < 0.05) SNPs are shown in bold. The ORs and 95% CIs for the SNPs with P < 0.05 in the dominant or recessive models are noted in the text. For SNPs previously associated with other cancers, the representative references for the primary cancer association are cited. No., number; ref, referent genotype.
Additional SNPs associated with osteosarcoma
Since it is possible that other variants in the 8q24 region could be associated specifically with osteosarcoma but not other cancers, we conducted a comprehensive analysis of the additional 205 tag SNPs across 8q24 that were genotyped in this panel (Table II and Figure 1). The most significant association with osteosarcoma risk was observed for rs896324 (Ptrend = 0.01) with increased risks of 1.79 (95% CI 1.08–2.97) and 2.75 (95% CI 0.61–12.52) for the heterozygotes and variant homozygotes, respectively. The dominant model also resulted in a statistically significant association for rs896324 (OR 1.79, 95% CI 1.08–2.97, P = 0.025).
Three additional SNPs were associated with osteosarcoma in the dominant model: rs10808555 (OR 1.51, 95% CI 0.99–2.31), rs17766217 (OR 0.66, 95% CI 0.44–1.00) and rs9642880 (OR 0.63, 95% CI 0.41–0.96). The variant allele at rs10808555 was more common in the cases, whereas for the latter two SNPs, the variant allele were more common in the controls, providing a protective effect. None of these SNPs were in the same haplotype block (Figure 1) and were not correlated (r2 < 0.1).
The minor allele of rs738617 was associated with osteosarcoma most strongly in the recessive model (OR 1.89, 95% CI 1.15–3.11, P = 0.018); there were 22.2% of osteosarcoma cases homozygous for the variant allele compared with 13% of controls. A second association was noted in the recessive model at rs12155672 (OR 1.68, 95% CI 1.09–2.59, P = 0.022) with 34.3% of cases and 23.5% of controls being homozygous for the variant allele. A third positive association in the recessive genetic model was noted for rs185852 with OR 2.22 (95% CI 1.02–4.81, P = 0.043).
We evaluated the haplotype structure of the 8q24 chromosome region in order to better understand the location of these SNPs in relation to SNPs associated with other cancer. The SNPs genotyped in our study were tag SNPs and by definition have very little correlation or linkage disequilibrium. The majority of SNPs reported here reside in different haplotype blocks based on HapMap CEU data. However, rs17766217 (suggested a dominant association) and rs12155672 (suggested a recessive association) are located only 2527 bp apart and are in the same haplotype block in the HapMap CEU population. They were not correlated in our controls, r2 = 0.03.
Discussion
This study is one of a limited number of studies that have evaluated common genetic variation in osteosarcoma, a rare cancer. The common variant common disease hypothesis has been tested extensively and proven that there are numerous common SNPs associated with common diseases, for example, the association between variants in complement factor H and macular degeneration (48–51). The contribution of common genetic variants (i.e. MAF >1%) to rare diseases is not yet known. The role of rare variants in common or rare diseases is an area of active study (52). Rare cancers that occur at young ages, such as osteosarcoma, may have a stronger genetic component simply because there has been less time for environmental exposures to contribute to carcinogenesis.
We hypothesized that genetic variation in the chromosome 8q24 locus could play a role in osteosarcoma risk because this region is often amplified in osteosarcoma tumor tissues, contains MYC and GWAS of prostate, breast, colorectal and bladder cancer have identified specific SNPs in this region as cancer risk factors. The primary colorectal cancer risk variant, rs6983267, has recently been shown to indirectly affect MYC function through putative transcription factor-binding sites (24–26). Although MYC is still ∼330 kb away from r6983267, and even farther away from some of the other SNPs associated with cancer, it is the best candidate because of its important role in controlling cellular proliferation. It is amplified and overexpressed in many malignancies, including osteosarcoma. Notably, MYC inhibition resulted in differentiation of osteosarcoma cells to mature osteocytes (32).
We initially focused on SNPs with a high prior probability for association because they had been associated with other cancers. One SNP which was previously associated with prostate cancer (17), rs7017300, yielded a borderline association in the heterozygous state, but no significant trend was observed, suggesting that this may be a false positive association. Analyses were evaluated for multiple genetic models because prior studies of have not specifically implicated one inheritance model over another. Analyses of the dominant and recessive inheritance models did not identify any further associations between the nine SNPs previously associated with cancer and osteosarcoma.
We evaluated additional SNPs across 8q24 to explore the hypothesis that different regions in this area may be associated with risk of different cancers. These analyses identified three statistically significant associations in the additive model (rs896324, rs7386167 and rs17766217), three in the dominant model (rs896324, rs10808555 and rs9642880) and three in the recessive model (rs7386167, rs12155672 and rs185852). Although suggestive, we cannot rule out the possibility that these findings could be due to chance. The current study was limited by the number of cases. We augmented our statistical power by including a large number of controls and had sufficient power to detect effects as low as 1.5–1.8 assuming a type 1 error of 0.05. However, our power was limited due to multiple testing, and none of the SNPs remained statistically significant after adjustment for the number of tests performed. Therefore, although we observed some suggestive evidence of association between genetic variation at the 8q24 chromosomal locus and osteosarcoma, larger studies are required to confirm this finding. The role of MYC in osteosarcoma pathogenesis and presence in osteosarcoma tumors suggests that this locus may still play an important role in osteosarcoma.
In summary, this pilot study suggests that SNPs in the 8q24 region are not strongly associated with osteosarcoma but several SNPs with small effects may be present. Future large studies of osteosarcoma genetic risk factors are warranted to improve our understanding of the genetic contribution to this cancer for adolescents and young adults.
Funding
Intramural research program of the National Institutes of Health and the National Cancer Institute.
Acknowledgments
We are grateful to the Bone Disease and Injury Study of Osteosarcoma and Prostate, Lung, Colorectal, Ovarian participants for their valuable contributions.
Conflict of Interest Statement: None declared.
Glossary
Abbreviations
- CI
confidence interval
- GWAS
Genome-wide association studies
- MAF
minor allele frequency
- OR
odds ratio
- SNP
single-nucleotide polymorphism
Appendix
The National Osteosarcoma Etiology Study Group is represented by Michael A.Simon (University of Chicago), Marc C.Gebhardt (Massachusetts General Hospital), Mark T.Scarborough (Shands Medical Center, University of Florida), Steven Gitelis (Rush Presbyterian and St Lukes Medical Center), Jeffrey J.Eckardt (University of California at Los Angeles School of Medicine), James R.Neff (Nebraska Health System), Michael J.Joyce (Cleveland Clinic Foundation), Martin Malawer (Washington Cancer Institute), Michael McGuire, (Creighton University) and H.Clarke Anderson (University of Kansas Medical Center).
References
- 1.Mirabello L, et al. Osteosarcoma incidence and survival rates from 1973 to 2004: data from the Surveillance, Epidemiology, and End Results Program. Cancer. 2009;115:1531–1543. doi: 10.1002/cncr.24121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kager L, et al. Primary metastatic osteosarcoma: presentation and outcome of patients treated on neoadjuvant Cooperative Osteosarcoma Study Group protocols. J. Clin. Oncol. 2003;21:2011–2018. doi: 10.1200/JCO.2003.08.132. [DOI] [PubMed] [Google Scholar]
- 3.Longhi A, et al. Height as a risk factor for osteosarcoma. J. Pediatr. Hematol. Oncol. 2005;27:314–318. doi: 10.1097/01.mph.0000169251.57611.8e. [DOI] [PubMed] [Google Scholar]
- 4.Fraumeni JF., Jr. Stature and malignant tumors of bone in childhood and adolescence. Cancer. 1967;20:967–973. doi: 10.1002/1097-0142(196706)20:6<967::aid-cncr2820200606>3.0.co;2-p. [DOI] [PubMed] [Google Scholar]
- 5.Troisi R, et al. Perinatal factors, growth and development, and osteosarcoma risk. Br. J. Cancer. 2006;95:1603–1607. doi: 10.1038/sj.bjc.6603474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Operskalski EA, et al. A case-control study of osteosarcoma in young persons. Am. J. Epidemiol. 1987;126:118–126. doi: 10.1093/oxfordjournals.aje.a114643. [DOI] [PubMed] [Google Scholar]
- 7.Ruza E, et al. Analysis of polymorphisms of the vitamin D receptor, estrogen receptor, and collagen Ialpha1 genes and their relationship with height in children with bone cancer. J. Pediatr. Hematol. Oncol. 2003;25:780–786. doi: 10.1097/00043426-200310000-00007. [DOI] [PubMed] [Google Scholar]
- 8.Lindor NM, et al. Concise handbook of familial cancer susceptibility syndromes—second edition. J. Natl Cancer Inst. Monogr. 2008:1–93. doi: 10.1093/jncimonographs/lgn001. [DOI] [PubMed] [Google Scholar]
- 9.Patio-Garcia A, et al. Analysis of the human tumour necrosis factor-alpha (TNFalpha) gene promoter polymorphisms in children with bone cancer. J. Med. Genet. 2000;37:789–792. doi: 10.1136/jmg.37.10.789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Savage SA, et al. Germ-line genetic variation of TP53 in osteosarcoma. Pediatr. Blood Cancer. 2007;49:28–33. doi: 10.1002/pbc.21077. [DOI] [PubMed] [Google Scholar]
- 11.Savage SA, et al. Analysis of genes critical for growth regulation identifies Insulin-like Growth Factor 2 Receptor variations with possible functional significance as risk factors for osteosarcoma. Cancer Epidemiol. Biomarkers Prev. 2007;16:1667–1674. doi: 10.1158/1055-9965.EPI-07-0214. [DOI] [PubMed] [Google Scholar]
- 12.Koshkina NV, et al. Exploratory analysis of Fas gene polymorphisms in pediatric osteosarcoma patients. J. Pediatr. Hematol. Oncol. 2007;29:815–821. doi: 10.1097/MPH.0b013e3181581506. [DOI] [PubMed] [Google Scholar]
- 13.Toffoli G, et al. Effect of TP53 Arg72Pro and MDM2 SNP309 polymorphisms on the risk of high-grade osteosarcoma development and survival. Clin. Cancer Res. 2009;15:3550–3556. doi: 10.1158/1078-0432.CCR-08-2249. [DOI] [PubMed] [Google Scholar]
- 14.Gudmundsson J, et al. Genome-wide association study identifies a second prostate cancer susceptibility variant at 8q24. Nat. Genet. 2007;39:631–637. doi: 10.1038/ng1999. [DOI] [PubMed] [Google Scholar]
- 15.Yeager M, et al. Identification of a new prostate cancer susceptibility locus on chromosome 8q24. Nat. Genet. 2009;41:1055–1057. doi: 10.1038/ng.444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Al Olama AA, et al. Multiple loci on 8q24 associated with prostate cancer susceptibility. Nat. Genet. 2009;41:1058–1060. doi: 10.1038/ng.452. [DOI] [PubMed] [Google Scholar]
- 17.Yeager M, et al. Genome-wide association study of prostate cancer identifies a second risk locus at 8q24. Nat. Genet. 2007;39:645–649. doi: 10.1038/ng2022. [DOI] [PubMed] [Google Scholar]
- 18.Savage SA, et al. The evidence for prostate cancer risk loci at 8q24 grows stronger. J. Natl Cancer Inst. 2007;99:1499–1501. doi: 10.1093/jnci/djm186. [DOI] [PubMed] [Google Scholar]
- 19.Fletcher O, et al. Association of genetic variants at 8q24 with breast cancer risk. Cancer Epidemiol. Biomarkers Prev. 2008;17:702–705. doi: 10.1158/1055-9965.EPI-07-2564. [DOI] [PubMed] [Google Scholar]
- 20.Schumacher FR, et al. A common 8q24 variant in prostate and breast cancer from a large nested case-control study. Cancer Res. 2007;67:2951–2956. doi: 10.1158/0008-5472.CAN-06-3591. [DOI] [PubMed] [Google Scholar]
- 21.Kiemeney LA, et al. Sequence variant on 8q24 confers susceptibility to urinary bladder cancer. Nat. Genet. 2008;40:1307–1312. doi: 10.1038/ng.229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Tomlinson I, et al. A genome-wide association scan of tag SNPs identifies a susceptibility variant for colorectal cancer at 8q24.21. Nat. Genet. 2007;39:984–988. doi: 10.1038/ng2085. [DOI] [PubMed] [Google Scholar]
- 23.Yeager M, et al. Comprehensive resequence analysis of a 136 kb region of human chromosome 8q24 associated with prostate and colon cancers. Hum. Genet. 2008;124:161–170. doi: 10.1007/s00439-008-0535-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Tuupanen S, et al. The common colorectal cancer predisposition SNP rs6983267 at chromosome 8q24 confers potential to enhanced Wnt signaling. Nat. Genet. 2009;41:885–890. doi: 10.1038/ng.406. [DOI] [PubMed] [Google Scholar]
- 25.Pomerantz MM, et al. Evaluation of the 8q24 prostate cancer risk locus and MYC expression. Cancer Res. 2009;69:5568–5574. doi: 10.1158/0008-5472.CAN-09-0387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Pomerantz MM, et al. The 8q24 cancer risk variant rs6983267 shows long-range interaction with MYC in colorectal cancer. Nat. Genet. 2009;41:882–884. doi: 10.1038/ng.403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Jia L, et al. Functional enhancers at the gene-poor 8q24 cancer-linked locus. PLoS Genet. 2009;5:e1000597. doi: 10.1371/journal.pgen.1000597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Meyer N, et al. Reflecting on 25 years with MYC. Nat. Rev. Cancer. 2008;8:976–990. doi: 10.1038/nrc2231. [DOI] [PubMed] [Google Scholar]
- 29.Stock C, et al. Chromosomal regions involved in the pathogenesis of osteosarcomas. Genes Chromosomes Cancer. 2000;28:329–336. doi: 10.1002/1098-2264(200007)28:3<329::aid-gcc11>3.0.co;2-f. [DOI] [PubMed] [Google Scholar]
- 30.Ladanyi M, et al. Sporadic amplification of the MYC gene in human osteosarcomas. Diagn. Mol. Pathol. 1993;2:163–167. [PubMed] [Google Scholar]
- 31.Gamberi G, et al. C-myc and c-fos in human osteosarcoma: prognostic value of mRNA and protein expression. Oncology. 1998;55:556–563. doi: 10.1159/000011912. [DOI] [PubMed] [Google Scholar]
- 32.Jain M, et al. Sustained loss of a neoplastic phenotype by brief inactivation of MYC. Science. 2002;297:102–104. doi: 10.1126/science.1071489. [DOI] [PubMed] [Google Scholar]
- 33.Man TK, et al. Genome-wide array comparative genomic hybridization analysis reveals distinct amplifications in osteosarcoma. BMC Cancer. 2004;4:45. doi: 10.1186/1471-2407-4-45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Selvarajah S, et al. Genomic signatures of chromosomal instability and osteosarcoma progression detected by high resolution array CGH and interphase FISH. Cytogenet. Genome Res. 2008;122:5–15. doi: 10.1159/000151310. [DOI] [PubMed] [Google Scholar]
- 35.Gauderman WJ, et al. QUANTO 1.1: a computer program for power and sample size calculations for genetic-epidemiology studies. 2006 Ref Type: Computer Program. Available from http://hydra.usc.edu/gxe. [Google Scholar]
- 36.Barrett JC, et al. Haploview: analysis and visualization of LD and haplotype maps. Bioinformatics. 2005;21:263–265. doi: 10.1093/bioinformatics/bth457. [DOI] [PubMed] [Google Scholar]
- 37.Frazer KA, et al. A second generation human haplotype map of over 3.1 million SNPs. Nature. 2007;449:851–861. doi: 10.1038/nature06258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Klein RJ, et al. Complement factor H polymorphism in age-related macular degeneration. Science. 2005;308:385–389. doi: 10.1126/science.1109557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Meyer A, et al. Association of chromosomal locus 8q24 and risk of prostate cancer: a hospital-based study of German patients treated with brachytherapy. Urol. Oncol. 2009;27:373–376. doi: 10.1016/j.urolonc.2008.04.010. [DOI] [PubMed] [Google Scholar]
- 40.Gruber SB, et al. Genetic variation in 8q24 associated with risk of colorectal cancer. Cancer Biol. Ther. 2007;6 doi: 10.4161/cbt.6.7.4704. [DOI] [PubMed] [Google Scholar]
- 41.Poynter JN, et al. Variants on 9p24 and 8q24 are associated with risk of colorectal cancer: results from the Colon Cancer Family Registry. Cancer Res. 2007;67:11128–11132. doi: 10.1158/0008-5472.CAN-07-3239. [DOI] [PubMed] [Google Scholar]
- 42.Schafmayer C, et al. Investigation of the colorectal cancer susceptibility region on chromosome 8q24.21 in a large German case-control sample. Int. J. Cancer. 2009;124:75–80. doi: 10.1002/ijc.23872. [DOI] [PubMed] [Google Scholar]
- 43.Tenesa A, et al. Genome-wide association scan identifies a colorectal cancer susceptibility locus on 11q23 and replicates risk loci at 8q24 and 18q21. Nat. Genet. 2008;40:631–637. doi: 10.1038/ng.133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Severi G, et al. The common variant rs1447295 on chromosome 8q24 and prostate cancer risk: results from an Australian population-based case-control study. Cancer Epidemiol. Biomarkers Prev. 2007;16:610–612. doi: 10.1158/1055-9965.EPI-06-0872. [DOI] [PubMed] [Google Scholar]
- 45.Wang L, et al. Two common chromosome 8q24 variants are associated with increased risk for prostate cancer. Cancer Res. 2007;67:2944–2950. doi: 10.1158/0008-5472.CAN-06-3186. [DOI] [PubMed] [Google Scholar]
- 46.FitzGerald LM, et al. Analysis of recently identified prostate cancer susceptibility loci in a population-based study: associations with family history and clinical features. Clin. Cancer Res. 2009;15:3231–3237. doi: 10.1158/1078-0432.CCR-08-2190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Haiman CA, et al. Multiple regions within 8q24 independently affect risk for prostate cancer. Nat. Genet. 2007;39:638–644. doi: 10.1038/ng2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Berndt SI, et al. Pooled analysis of genetic variation at chromosome 8q24 and colorectal neoplasia risk. Hum. Mol. Genet. 2008 doi: 10.1093/hmg/ddn166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Edwards AO, et al. Complement factor H polymorphism and age-related macular degeneration. Science. 2005;308:421–424. doi: 10.1126/science.1110189. [DOI] [PubMed] [Google Scholar]
- 50.Haines JL, et al. Complement factor H variant increases the risk of age-related macular degeneration. Science. 2005;308:419–421. doi: 10.1126/science.1110359. [DOI] [PubMed] [Google Scholar]
- 51.Jakobsdottir J, et al. Susceptibility genes for age-related maculopathy on chromosome 10q26. Am. J. Hum. Genet. 2005;77:389–407. doi: 10.1086/444437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Schork NJ, et al. Common vs. rare allele hypotheses for complex diseases. Curr. Opin. Genet. Dev. 2009;19:212–219. doi: 10.1016/j.gde.2009.04.010. [DOI] [PMC free article] [PubMed] [Google Scholar]