Abstract
Cyclic AMP synthesized by Mycobacterium tuberculosis has been shown to play a role in pathogenesis. However, the high levels of intracellular cAMP found in both pathogenic and non-pathogenic mycobacteria suggest that additional and important biological processes are regulated by cAMP in these organisms. We describe here the biochemical characterization of novel cAMP-binding proteins in M. smegmatis and M. tuberculosis (MSMEG_5458 and Rv0998, respectively) that contain a cyclic nucleotide binding domain fused to a domain that shows similarity to the GNAT family of acetyltransferases. We detect protein lysine acetylation in mycobacteria and identify a universal stress protein (USP) as a substrate of MSMEG_5458. Acetylation of a lysine residue in USP is regulated by cAMP, and using a strain deleted for MSMEG_5458, we show that USP is indeed an in vivo substrate for MSMEG_5458. The Rv0998 protein shows a strict cAMP-dependent acetylation of USP, despite a lower affinity for cAMP than MSMEG_5458. Thus, this report not only represents the first demonstration of protein lysine acetylation in mycobacteria but also describes a unique functional interplay between a cyclic nucleotide binding domain and a protein acetyltransferase.
Keywords: Acetyl-CoA, Cyclic AMP (cAMP), Enzymes, Prokaryotic Signal Transduction, Protein Acylation, Mycobacterium, Universal Stress Protein
Introduction
Cyclic AMP is used as a signal for eliciting diverse cellular responses in both bacteria and higher eukaryotes. Nucleotide cyclases utilize ATP and GTP to make cAMP or cGMP, respectively, and of the six classes of nucleotide cyclases that have been described (1), the Class III adenylyl and guanylyl cyclases are encoded in many bacterial and almost all eukaryotic organisms (2). The genomes of pathogenic Mycobacterium tuberculosis, the causative agent of tuberculosis that accounts for almost 2 million deaths annually worldwide, harbor a number of genes that encode putative Class III nucleotide cyclases (3) and a single cyclic nucleotide phosphodiesterase (4), and many of these enzymes have been characterized biochemically as well as structurally (3–6). Most recently, it has been reported that cAMP production by the Rv0386 adenylyl cyclase is critical for allowing M. tuberculosis to establish itself within the mammalian host and cause disease (7). Thus, an understanding of the mechanisms underlying the production, utilization, and degradation of cAMP by M. tuberculosis may provide avenues for the identification of novel targets for drug development (8).
The genomes of non-pathogenic mycobacteria, such as Mycobacterium smegmatis, also encode multiple nucleotide cyclase genes, which could account for the high intracellular cAMP levels found in these bacteria (9). cAMP levels are modulated by various stress conditions in M. smegmatis (9), suggesting that cAMP participates in signaling within the bacterial cell, mediating its action by downstream effectors. The effectors of cAMP in mammalian cells have been well characterized and include protein kinase A (PKA),4 exchange protein activated by cAMP (EPAC), and the hyperpolarization-activated cyclic nucleotide-gated channels (10–11). The cyclic nucleotide (cNMP) binding domain in these proteins is conserved evolutionarily, and the best characterized protein with a cAMP binding domain in bacteria is the catabolite repressor protein, or CRP, that serves as a transcription factor, either enhancing or repressing transcription of a variety of genes in a cAMP-dependent manner (12–14). Binding of cAMP allosterically regulates the activity of these effector proteins.
Apart from CRP, no other downstream effector of cAMP in mycobacteria has been described. In this study we provide evidence for an entirely new signaling event mediated by cAMP and so far exclusive to mycobacteria. We show that the products of the Rv0998 gene and its ortholog in M. smegmatis, MSMEG_5458, are cNMP-binding proteins that share a significant similarity to the regulatory subunit of mammalian PKA. Remarkably, the cNMP domain in these proteins is fused to, and regulates the activity of, a GNAT-like protein acetyltransferase, which acetylates the epsilon amino group of a lysine residue in a Universal Stress Protein (USP, MSMEG_4207). This represents not only the first report of lysine acetylation in mycobacteria but also a novel effector function of cAMP in mycobacteria, which is perhaps reflective of the more complex cAMP/PKA/CREB/CBP pathway seen in eukaryotes (15).
EXPERIMENTAL PROCEDURES
Culturing of M. smegmatis and Preparation of Whole Cell Lysates
M. smegmatis strains were grown in 7H9TG (Middlebrook 7H9 broth supplemented with 0.2% glycerol and 0.05% Tween 80) at 37 °C, with shaking at 250 rpm, or on Middlebrook 7H10 agar. Cells were lysed by bead beating in buffer (10 mm Tris-Cl (pH 7.5), 100 mm NaCl, 10% glycerol, 5 μg/ml leupeptin, 5 μg/ml aprotinin, 5 μg/ml soybean trypsin inhibitor, 3 mm nicotinamide, and 1 μm trichostatin A). Nonidet P-40 and deoxycholate to final concentrations of 1% and 0.5%, respectively, were added to the lysate and mixing continued at 4 °C for 2 h, following which samples were centrifuged for 30 min at 4 °C at 13,000 × g. Supernatants were collected, and protein was estimated using a Micro BCA protein assay kit (Pierce).
Phylogenetic Analysis of the cNMP Binding Domain in Rv0998 and MSMEG_5458
The boundaries of the cNMP binding domain in MSMEG_5458 were identified by analysis in the Pfam database (pfam.sanger.ac.uk/). The sequence of the defined domain was used to identify orthologs in the SwissProt database using BLASTp. Individual full-length sequences of the hits thus obtained (>90) were then analyzed using Pfam to identify their cNMP binding domains. More than 156 domains were identified, because some proteins contained more than one cNMP binding domain in the full-length protein sequence. To this list of 156 sequences, the cNMP binding domain sequences of Rv0998, and the CRPs from M. tuberculosis and Escherichia coli were added, and all sequences were then aligned using ClustalW (EMBL). Sequences that were >80% identical were removed using JalView, and phylogenetic and molecular evolutionary analyses were conducted using MEGA version 4 (16).
Cloning and Mutagenesis of MSMEG_5458 and Rv0998
The list of primers used for PCR and mutagenesis are provided in supplemental Table S1. PCR was carried out on the genomic DNA of M. smegmatis mc2155 using MS5458FWD and MS5458RVS. The product was digested with BamHI and EcoRI and cloned into similarly digested pBKSII(+) to generate pBKS-MSMEG_5458. The clone was confirmed by sequencing (Macrogen, South Korea). A BamHI-EcoRV fragment from pBKS-MSMEG_5458 was cloned into BamHI-StuI-digested pPROEx-HTa to generate plasmid pPRO-MSMEG_5458. pPRO-MSMEG_5458 was digested with BamHI, end-filled, and digested with NotI, and the 1-kb fragment was cloned into SmaI-NotI-digested vector pGEX-6P-2 to generate pGEX-MSMEG_5458.
The Rv0998 gene was cloned using primers Rv0998FWD and Rv0998RVS. PCR was carried out on the genomic DNA of M. tuberculosis H37Rv, and the product was digested with EcoRI and XhoI and cloned into similarly digested pProEx-Htc to generate pPro-Rv0998. The clone was confirmed by sequencing (Macrogen). Plasmid pGEX-Rv0998 was generated by subcloning the EcoRI-XhoI fragment from pPRO-Rv0998 into similarly cut vector pGEX-6P-3.
Point mutations in MSMEG_5458 were generated by site-directed mutagenesis (17). pBKS-MSMEG_5458 was used as the template for mutation of Arg-95 and Glu-234 using primers MS5458R95K and MS5458E234A, respectively, to generate pBKS-MSMEG_5458R95K and MSMEG_5458E234A, respectively. Mutants were verified by sequencing (Macrogen). The BamHI-SnaBI fragment from pBKS-MSMEG_5458R95K was cloned into similarly digested pPro-MSMEG_5458 to generate pPro-MSMEG_5458R95K. The BamHI-KpnI fragment from pBKS-MSMEG_5458E234A was cloned into similarly digested pProEx-Hta to generate pPro-MSMEG_5458 E234A.
Expression and Purification of His-tagged Proteins
MSMEG_5458 or mutant proteins were expressed in the E. coli SP850 cyc− strain on induction using 500 μm isopropyl-β-thiogalactopyranoside. Cells were lysed by sonication in buffer containing 50 mm Tris-Cl (pH 8.2), 5 mm 2-mercaptoethanol (2-ME), 100 mm NaCl, 10% glycerol, 1 mm benzamidine, and 2 mm phenylmethylsulfonyl fluoride, followed by centrifugation at 30,000 × g. The supernatant was interacted with nickel-nitrilotriacetic acid beads (Agarose Bead Technologies, Spain), and bound protein was eluted with buffer containing 200 mm Tris-Cl (pH 8.2) buffer, 100 mm NaCl, 5 mm 2-ME, 300 mm imidazole, and 10% glycerol. Purified proteins were desalted into buffer containing 50 mm Tris-Cl (pH 8.2), 5 mm 2-ME, 50 mm NaCl, and 10% glycerol and stored in aliquots at −70 °C (supplemental Fig. S1a).
Gel Filtration and Dynamic Light Scattering
Gel filtration was carried out in buffer containing 50 mm Tris-Cl (pH 8.2), 5 mm 2-ME, 50 mm NaCl, and 10% glycerol at 4 °C at a flow rate of 200 μl/min using a Superose 12 column and an AKTA fast protein liquid chromatography system (Amersham Biosciences). The column was calibrated using commercially available gel filtration standards (thyroglobulin (670 kDa), bovine γ-globulin (158 kDa), chicken ovalbumin (44 kDa), equine myoglobin (17 kDa), and vitamin B12 (1.35 kDa)).
Dynamic light scattering experiments were performed on a DynaPro Molecular Sizing Instrument (Protein Solutions). Samples of purified MSMEG_5458 in the same buffer used for gel filtration were analyzed at concentrations of ∼1 mg/ml at 25 °C. Several measurements were taken at 277 K and analyzed using DYNAMICS Version 3.3 software (Protein Solutions). Data collection times of 10 s were used in all cases for a minimum of 15 acquisitions.
Cyclic Nucleotide Binding Assays
Cyclic nucleotide binding assays were carried out in a final reaction volume of 100 μl containing 25 mm Tris-Cl (pH 7.5), 100 mm NaCl, 10% glycerol, purified protein, and [3H]cAMP (59 Ci/mmol, PerkinElmer Life Sciences) in presence and absence of unlabeled cAMP. Reactions were incubated at 37 °C for 1 h and then filtered through nitrocellulose filters (0.45 μm), which were then washed with 5 ml of ice-cold buffer (10 mm Tris-Cl and 150 mm NaCl). The filters were dried, and bound radioactivity was measured by scintillation counting in a liquid scintillation counter (PerkinElmer Life Sciences). Cyclic nucleotide displacement assays were carried out with 200 ng (∼50 nm) of purified protein using a fixed concentration of labeled [3H]cAMP and varying amounts of unlabeled cAMP or analogs. Data were analyzed using GraphPad Prism 5, and values shown represent the mean ± S.E. of triplicate assays.
Western Blotting
Samples were electrophoresed on SDS-polyacrylamide gels (12% acrylamide, 1.2% bis-acrylamide) and transferred to a polyvinylidene difluoride membrane (Immobilon-P, Millipore). MSMEG_5458 and USP polyclonal antibodies were generated in the laboratory and used at a dilution of 1:5,000. Acetyl-lysine antibody (Cell Signaling Technology, cat. no. 9441S) was used at a dilution of 1:2500. To show specificity of the immunoreactive bands, acetyl-lysine antibodies were incubated with acetylated bovine serum albumin for 1 h prior to addition to the Western blot. This would adsorb antibodies specific to acetylated lysine residues, thereby preventing their interaction with the proteins bound to the membrane. Bound antibody was detected by enhanced chemiluminescence (GE Healthcare).
GST Pulldown Assays
GST-MSMEG_5458 and GST-Rv0998 proteins were expressed in the E. coli cyc− SP850 strain upon induction using 500 μm isopropyl-β-thiogalactopyranoside. Cells were lysed by sonication in a buffer containing 50 mm Tris-Cl (pH 8.2), 5 mm 2-ME, 100 mm NaCl, 10% glycerol, 5 mm DTT, 5 mm EDTA, 1 mm benzamidine, and 2 mm phenylmethylsulfonyl fluoride, followed by centrifugation at 30,000 × g. The supernatant was interacted with glutathione-agarose beads (Sigma), and the matrix was washed thrice with lysis buffer, thrice with Wash I buffer containing 50 mm Tris-Cl pH 8.2, 5 mm DTT, 100 mm NaCl, 5 mm EDTA, and 0.1% Triton X-100, and thrice with Wash II buffer containing 50 mm Tris-Cl (pH 8.2), 5 mm DTT, 100 mm NaCl, 5 mm EDTA, and 10% glycerol. GST or GST fusion proteins bound to glutathione beads were interacted with 2 mg of cytosolic proteins from M. smegmatis/M. bovis Bacillus Calmette-Guérin at 4 °C for 1 h, and the beads were washed five times with 1 ml of buffer containing 150 mm NaCl, 100 mm Tris-Cl (pH 8.2), 10% glycerol, 2 mm phenylmethylsulfonyl fluoride, 5 mm 2-ME, and 1 mm benzamidine. Bound proteins were analyzed on a 12% polyacrylamide gel.
Mass Spectrometry
Protein bands were excised from the gel and washed with wash solution (50 mm ammonium bicarbonate/acetonitrile, 1:1, v/v) repeatedly to remove the stain. The gel pieces were dehydrated in acetonitrile and then dried in a centrifugal evaporator (Heto). The gel piece was then treated with 1.5 mg/ml DTT in 100 mm ammonium bicarbonate, and then alkylated with 10 mg/ml iodoacetamide in 100 mm ammonium bicarbonate. Pieces were washed with wash solution and dehydrated again. Pieces were then rehydrated in 50 mm ammonium bicarbonate with ∼200 ng of modified porcine trypsin (Promega, Madison, WI), and digestion was continued at 37 °C for 12 h. Extraction buffer (60% acetonitrile and 0.1% trifluoroacetic acid) was added to the gel pieces to extract the peptides. The extract was pooled and concentrated in centrifugal evaporator. Equal volumes of sample and 2,5-dihydroxybenzoic acid matrix (1 μl each) were mixed and spotted onto a MALDI plate (MTP 384 ground steel, target plate). The samples were analyzed by MALDI-TOF (Ultraflex TOF/TOF, Bruker Daltonics, Germany). Flex Control software was used to acquire data using 25 KvA Reflector mode, N2 Laser, 337 nm, and 50 Hz. Each spectrum was the sum of the ion intensities from 300 laser shots. Flex Analysis 2.0 was used to analyze the spectra. The peptide mass fingerprint data were used to identify proteins using MASCOT on an M. smegmatis Proteomics Database. Analysis allowed for a single missed cleavage with a variable modification of acetylation and fixed modification of carbamidomethylation of cysteine residues, with mass tolerance of 1 Da.
Cloning and Mutagenesis of MSMEG_4207
MSMEG_4207 was cloned using primers MSMEG_4207FWD and MSMEG_4207RVS. PCR was carried out on the genomic DNA of M. smegmatis mc2155, and the product was digested with BamHI and XhoI and cloned into similarly digested pBKSII(+) to generate pBKS-MSMEG_4207. The clone was confirmed by sequencing (Macrogen). The fragment BamHI-XhoI from pBKS-MSMEG_4207 was cloned into similarly digested pPROEx-HTb to generate plasmid pPRO-MSMEG_4207.
The USPK104R mutant was generated using mutagenic primers MSMEG_4207K104R1 and MSMEG_4207K104R2. Wild-type and mutant proteins were expressed in E. coli BL21(DE3) and purified essentially as described for MSMEG_5458 (supplemental Fig. S1a).
In Vitro Acetylation of USP
Assays were carried out in a 20-μl total reaction volume containing 25 mm Tris-Cl, 100 mm NaCl, 5 mm EDTA, 30 μm acetyl-CoA, MSMEG_5458/Rv0998, and 2 μg of MSMEG_4207(USP) as a substrate, in the presence and absence of cAMP. Reactions were incubated at 25 °C for 10 min, stopped by boiling in SDS sample buffer, and analyzed by Western blotting with acetyl-lysine antibody and enhanced chemiluminescence using ECL+ reagent (GE Healthcare).
For quantitation of immunoreactivity in a Western blot assay was used. Enzymatic assays were performed with varying concentrations of MSMEG_5458, acetyl-CoA, and USP as indicated, and incubated for the times indicated at 25 °C. Reactions were terminated by addition of SDS sample dye and boiled, and equal aliquots were loaded onto a gel for subsequent Western blot analysis using the acetyl-lysine antibody and enhanced chemiluminescence. Images were acquired with the FluorChem Q MultiImage III system (Alpha Innotech), and image intensities were quantified using the AlphaView Q version 3.03 software. Values obtained were analyzed by GraphPad Prism 5 using built-in equations for enzyme kinetics.
For measuring the rate of USP acetylation by MSMEG_5458, an enzyme-coupled assay was adopted. Acetyltransferase activity was continuously measured by using a UV-visible spectrophotometer (Varian Cary Bio 100). The assay reaction mixtures contained 0.2 mm NAD, 0.2 mm thiamine pyrophosphate, 5 mm MgCl2, 1 mm DTT, 2.4 mm α-ketoglutarate, 50 μm acetyl-CoA, 50 μm MSMEG_4207, 0.03 unit of α-ketoglutarate dehydrogenase, 190 nm MSMEG_5458, 100 mm sodium acetate, 50 mm Bis-Tris, and 50 mm Tris borate buffer, pH 7.5, in a total volume of 150 μl. All assay components except MSMEG_5458 and MSMEG_4207 were incubated at 25 °C for 5 min, and the reaction was initiated by the addition of MSMEG_4207 and MSMEG_5458. The rates were analyzed continuously for 2 min by measuring NADH production at 340 nm.
Generation of the ΔMSMEG_5458 Strain
A strain with a deletion of the MSMEG_5458 gene (ΔMSMEG_5458) was generated using the suicide vector approach (18). The region 5′ to MSMEG_5458 consisting of ∼850 bases upstream was amplified by PCR using primers UPKOMS5458FWD and UPKOMS5458RVS, and a fragment of ∼830 bases downstream of the acetyltransferase region of MSMEG_5458 was amplified using DOWNKOMS5458FWD and DOWNKOMS5458RVS primers. The 5′ amplicon was cloned into pBKSII vector using HindIII and NotI sites, and the 3′ amplicon was cloned into pBKSII(+) vector using NotI and BamHI sites to generate plasmids pBKS-MS5458–5′KO and pBKS-MS5458–3′KO. DNA sequencing confirmed the identity of the cloned sequences. The HindIII-NotI cut pBKS-MS5458–5′KO and NotI-BamHI cut pBKS-MS5458–3′KO inserts were ligated into BamHI-HindIII cut p2NIL plasmid with kanamycin selection marker (kindly provided by Prof. Neil Stoker) to generate p2NIL-MS5458–5′3′KO plasmid. The PacI fragment from pGOAL19 containing three markers (β-galactosidase, hygromycin resistance, and sucrose susceptibility) was cloned into p2NIL-MS5458-5′3′KO to generate plasmid p2NIL-MS5458–5′3′KO-PacI. Plasmid p2NIL-MS5458–5′3′KO-PacI (1 μg) was electroporated into M. smegmatis mc2155, and single crossovers and double crossovers were obtained essentially as described (18). Double crossovers were further tested by genomic PCR and Southern blotting to obtain the ΔMSMEG_5458 strain.
To construct the ΔMSMEG_5458 strain complemented with the MSMEG_5458 gene driven by its own promoter, a KpnI-MscI fragment from pBKS-MS5458–5′KO, and the MscI-XbaI fragment from pPRO-MSMEG_5458 were cloned into KpnI and XbaI digested vector pMH94h (a kind gift of W. R. Bishai, Johns Hopkins Centre for Tuberculosis Research) to generate pMH-MSMEG_5458C, which was electroporated in the ΔMSMEG_5458 strain. Positive integrants (ΔMSMEG_5458Comp) carrying the required insert were screened by colony PCR and validated by Southern hybridization. PCRs were carried out with primers MS5458FWD and MS5458RVS on genomic DNA prepared from the wild-type strain and the ΔMSMEG_5458 and ΔMSMEG_5458Comp strains to confirm the presence of the deletion in the ΔMSMEG_5458 strain and the complementation of MSMEG_5458.
Southern Blot Analysis
Southern blotting was carried out by electrophoretic separation of 2 μg of genomic DNA of wild-type, knock-out, and complement strains digested with EcoRI and AgeI and by transferring the DNA to Hybond nylon membranes (Amersham Biosciences). The probe was prepared from a 150-bp fragment amplified using primers MS5458FWD and UPKOMS5458RVS using pMH-MSMEG_5458C plasmid as a template. The probe was end-labeled with γ-32P-labeled ATP catalyzed by polynucleotide kinase (New England Biolabs). The blot was probed at 60 °C for 16 h and washed once in 2× SSC (0.3 mm NaCl and 30 mm sodium citrate) for 15 min at 60 °C prior to exposure to a PhosphorImager.
Immunoprecipitation of MSMEG_4207
Whole cell lysates were used for immunoprecipitation. For pre-clearing, the lysates were incubated with normal rabbit IgG for 1 h at 4 °C. Protein G beads were added to the lysates and incubated for an additional 30 min. The beads were removed by centrifugation, and the pre-cleared supernatant was interacted with MSMEG_4207 IgG overnight at 4 °C. Protein G beads were added to the lysates and incubated for an additional 2 h. The beads were pelleted at 4 °C and washed thrice with wash buffer (10 mm Tris-Cl (pH 7.5), 100 mm NaCl, 0.1% Triton X-100) and twice with TBS (10 mm Tris-Cl (pH 7.5), 100 mm NaCl). The beads were then boiled in SDS sample buffer and subsequently analyzed by SDS-PAGE and Western blotting.
RESULTS
Identification of a Putative cAMP-regulated Acetyltransferase in Mycobacteria
Bioinformatic analysis had earlier identified putative cNMP-binding proteins encoded in mycobacterial genomes (19). One protein (Rv0998) caught our interest, because it contained a cNMP binding domain fused to a GNAT-like acetyltransferase domain. This protein is encoded as a full-length protein in all sequenced mycobacterial genomes, including that of M. leprae, whose genome has undergone a large degree of pseudogenization (supplemental Fig. S2). Interestingly, this domain fusion of a putative cNMP binding domain to an acetyltransferase domain is unique to mycobacteria, and we did not come across similar domain fusions in any other protein in databases.
Alignment of the N-terminal cNMP binding domain to the regulatory subunit of PKA showed that residues critical for cAMP binding in PKA (20–21) are conserved in Rv0998 and its ortholog in M. smegmatis, MSMEG_5458 (Fig. 1A). For example, residues involved in hydrogen bonding with the sugar phosphate moiety of cAMP in PKA RIIβ(A) (Glu-221, Gly-220, Arg-230, and Ala-223; 1NE6) are conserved in Rv0998 (Glu-89, Gly-88, Arg-98, and Ala-91) (10). Identical residues are found at equivalent positions in MSMEG_5458. The hydrogen bonds that these residues form are protected from the solvent in the regulatory subunit of PKA by hydrophobic residues that would also protect the cyclic nucleotide from hydrolysis by phosphodiesters. The hydrophobic residues that form this shield (Leu-222, Ala-232, Gly-183, Ile-180, and Val-179 in 1NE6) are conserved in Rv0998 (Ile-90, Ala-100, Gly-51, Leu-48, and Leu-47) and MSMEG_5458 (Ile-87, Ala-97, Gly-48, Ile-45, and Leu-44). In addition, a residue that contributes to a “hydrophobic sandwich” holding the adenine ring of cAMP in many cAMP binding domains (e.g. Val-182 and Val-300 in 1NE6; Val-564 in 1Q43; Ile-321 in 1CX4; Ile-57 in 3I59) is seen in the mycobacterial proteins (Val-67 in Rv0998; Val-64 in MSMEG_5458) (10). A phylogenetic tree of representative cNMP binding domains along with those from Rv0998 and MSMEG_5458 is shown in Fig. 1B, indicating that the mycobacterial proteins are related more closely to the A and B domains in cNMP-dependent protein kinases, than the cNMP binding domains in cyclic nucleotide gated channels, CRPs, and the little studied neuropathy target esterases (22).
FIGURE 1.
Sequence alignment of cNMP binding and acetyltransferase domains. A, multiple sequence alignment of the cNMP binding domains of Rv0998, MSMEG_5458, regulatory subunits of PKA (RIα, RIIα, and RIIβ), exchange protein activated by cAMP, hyperpolarization-activated cyclic nucleotide, and CRP from M. tuberculosis. The Protein Data Bank codes of structurally characterized proteins are shown. Asterisks represent important residues that are present in cNMP binding domains, which are discussed in the text. The black arrow represents the arginine (Arg-95 in MSMEG_5458) that is important for cAMP binding. Inset, a diagrammatic representation of the domain organization of MSMEG_5458/Rv0998. B, phylogenetic clustering of the cNMP binding domains in Rv0998 and MSMEG_5458. The gi numbers (supplemental Table S2, and in some cases, common names along with gi numbers) are shown, and A or B following the gi numbers identifies the first (N-terminal) or the second cNMP binding domain seen in the full-length protein sequences. PKA:A, cAMP binding domains in the regulatory subunits of PKA (A domain); GEF, guanine nucleotide exchange factor; PKA:B, cAMP binding domains in the regulatory subunits of PKA (B domain); PKG:A, cGMP binding domains in cGMP-dependent protein kinases (A domain); PKG:B, cGMP binding domains in cGMP-dependent protein kinases (B domain); CNG, cyclic nucleotide gated channels; NTE, neuropathy target esterase (also known as lysophospholipase NTE). Black dots indicate MSMEG_5458 and Rv0998. C, multiple sequence alignment of the acetyltransferase domain of Rv0998, MSMEG_5458, human GCN5 acetyltransferase, yeast histone acetyltransferase and tetrahymena GCN5 acetyltransferase. The Protein Data Bank codes of structurally characterized proteins are shown in the alignment. Asterisks represent important residues that are present in cNMP binding domains, which are discussed in the text. The black arrow identifies the glutamate residue important for acetyltransferase activity.
The C-terminal acetyltransferase domain in Rv0998 and MSMEG_5458 also shows conservation of critical amino acids that are important for acetyl-CoA binding and catalysis. Acetyl-CoA is bound in a cleft on the surface of structurally characterized acetyltransferases, and the cleft is composed of hydrophobic amino acids in Motifs A and D (23). These residues (Gln-184 through Gly-189 in 1YGH) are similar to those found at equivalent positions in Rv0998 and MSMEG_5458, and therefore could be implicated in acetyl-CoA binding (Fig. 1C). Importantly, a highly conserved catalytic residue (Glu-173 in 1YGH) is seen in Rv0998 (Glu-235) and MSMEG_5458 (Glu-234), along with additional hydrophobic residues (Ile-174 and Phe-176 in 1YGH) that are important in Motif A for acetyl-CoA binding (24, 25). This conservation of amino acid sequences strongly suggest that Rv0998 and MSMEG_5458 could function as cAMP-regulated acetyltransferases.
MSMEG_5458 Can Bind cAMP and Has Structural Features Similar to the Regulatory Subunit of PKA
We cloned the genes for the predicted proteins from both M. tuberculosis (Rv0998) and M. smegmatis (MSMEG_5458) and expressed them in an E. coli strain deleted for the sole adenylyl cyclase gene, to ensure that purified protein was obtained in a cAMP-free form (supplemental Fig. S1). Because expression of MSMEG_5458 was more robust, we proceeded with more detailed biochemical analysis. Gel filtration and dynamic light scattering indicated that MSMEG_5458 was a dimer (supplemental Fig. S1). MSMEG_5458 could bind cAMP, and binding could be competed with analogs of cAMP and cGMP (Fig. 2A). An EPAC-specific analog (8-pCPT-2′-O-methyl-cAMP) was unable to inhibit cAMP binding to MSMEG_5458, in agreement with the phylogenetic analysis that indicated the closer similarity of the cNMP binding domain in MSMEG_5458 to those found in protein kinases. MSMEG_5458 showed selectivity toward cAMP (IC50 100.2 ± 4 nm) versus cGMP (IC50 4223 ± 1415 nm, Fig. 2B). As was previously seen in the regulatory subunit of PKA (26), the (Sp)-cAMPS analog demonstrated a binding affinity similar to that of cAMP (IC50 65 ± 12.3 nm), whereas the Rp isomer was far poorer in terms of its interaction with MSMEG_5458 (IC50 23.1 ± 1.6 μm, Fig. 2B). Analysis of the data obtained showed that 1 mol of cAMP was bound to 1 mol of MSMEG_5458. Mutation of Arg-95 to a Lys compromised cAMP binding in MSMEG_5458, as was seen with an equivalent mutation in PKA (Fig. 2C) (27).
FIGURE 2.
cAMP binding to MSMEG_5458. A, displacement of [3H]cAMP from MSMEG_5458 (∼50 nm) by unlabeled cyclic nucleotides (10 μm). Values represent the mean ± S.E.; n = 3. B, displacement of [3H]cAMP (∼50 nm) from MSMEG_5458 with increasing concentrations of the indicated ligands; n = 3. Values represent the mean ± S.E. C, binding of [3H]cAMP (∼50 nm) to MSMEG_5458R95K. Values shown represent the mean ± S.E. of duplicate determinations in experiments repeated thrice.
Acetyltransferase Activity of MSMEG_5458
We then proceeded to study the GNAT-like domain in MSMEG_5458. Arylamine N-acetyltransferases have been characterized in mycobacteria (28), but MSMEG_5458 did not acetylate the aminoglycosides kanamycin or amikacin in biochemical assays. We therefore hypothesized that MSMEG_5458 could be a protein acetyltransferase. There has been no report to date for the acetylation of the ϵ-amino group in lysine in mycobacterial proteins. We performed Western blot analysis with antibodies specific to acetylated lysine, using lysates prepared from M. tuberculosis H37Rv, M. bovis Bacillus Calmette-Guérin, and M. smegmatis mc2. A number of immunoreactive bands in lysates prepared from all three strains was detected (Fig. 3A), which was absent when blotting was performed in the presence of acetylated bovine serum albumin (Fig. 3A), thereby indicating the specificity of the antibody. Thus, the detection of acetylated proteins in mycobacteria strengthened our hypothesis that the GNAT-like domain in MSMEG_5458 could acetylate proteins, because well characterized protein acetyltransferases show significant sequence similarity to the GNAT domain present in Rv0998 and MSMEG_5458 (Fig. 1C).
FIGURE 3.
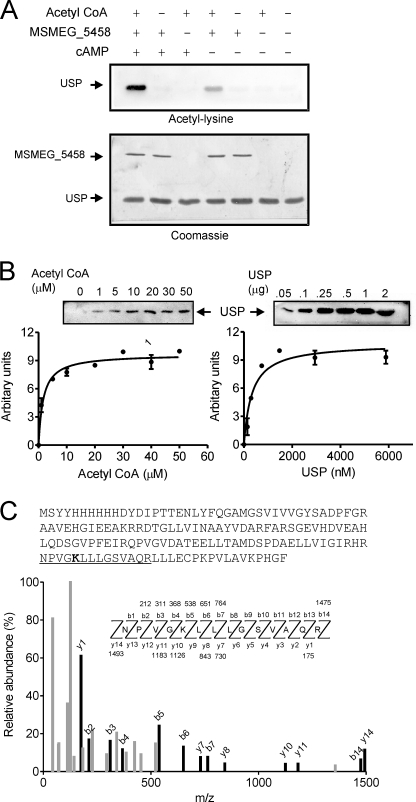
Protein acetylation in mycobacteria and identification of USP as an interacting partner of MSMEG_5458. A, whole cell lysates (50 μg) prepared from cultures of M. tuberculosis H37Rv, M. bovis Bacillus Calmette-Guérin, and M. smegmatis were separated by SDS-gel electrophoresis and Western blotting performed with acetyl-lysine antibodies in the presence or absence of acetylated bovine serum albumin. B, GST or GST-MSMEG_5458 bound to glutathione beads were incubated with cytosolic fractions prepared from M. smegmatis, in the presence or absence of 10 μm cAMP. Interacting proteins were resolved by SDS-PAGE and visualized by staining with Coomassie Brilliant Blue. One protein (*) was analyzed by mass spectrometry and identified to be MSMEG_4207 (USP). C, samples obtained after pulldown experiments under the conditions indicated were analyzed by Western blotting with acetyl-lysine antibodies. Experiments were repeated thrice.
To identify a putative protein substrate for MSMEG_5458, we utilized a pull-down approach and incubated GST-MSMEG_5458 fusion protein with lysates prepared from M. smegmatis, using GST as a control. Coomassie staining of the gel identified a band of Mr ∼ 14 kDa that appeared to be associated specifically with MSMEG_5458 (Fig. 3B). Western blotting performed with acetyl-lysine antibodies indicated that the protein was acetylated, although acetylation was not stimulated by cAMP (Fig. 3C). Peptide mass fingerprinting identified the protein as MSMEG_4207 (supplemental Fig. S3), annotated as a USP. USPs are found in diverse organisms from Archae and Eubacteria to yeast, fungi, and plants (29), and mycobacterial genomes encode a large number of USP-like proteins, although the functions of most of them are not clear. It is believed that USPs confer resistance to a wide variety of stress conditions, such as hypoxia and UV stress (30).
We cloned MSMEG_4207, expressed it in E. coli and utilized the purified protein to determine if it was a substrate of MSMEG_5458 using the in vitro acetyltransferase assays. Assays were performed in the presence of acetyl-CoA as the acetyl group donor, and acetylation of USP was monitored by Western blot analysis using acetyl-lysine antibodies. As seen in Fig. 4A, acetylation of USP was seen on incubation with MSMEG_5458 and acetyl-CoA, and acetylation increased in the presence of cAMP.
FIGURE 4.
Acetylation of USP and identification of the site of acetylation. A, USP (2 μg) was incubated alone, or in the presence of MSMEG_5458 (100 ng), 10 μm cAMP, and acetyl-CoA (30 μm) as indicated at 25 °C for 10 min, followed by Western blotting with acetyl-lysine antibodies (upper panel). Following blotting, the membrane was stained with Coomassie Brilliant Blue (lower panel). B, acetylation of USP was monitored in a Western blot based assay (“Experimental Procedures”). Left panel, assays performed in the presence of varying concentrations of acetyl-CoA with a fixed concentration of USP (25 μm). Right panel, assays performed in the presence of varying concentrations of USP and a fixed concentration of acetyl-CoA (30 μm). All assays were incubated at 25 °C for 10 min. Insets show representative blots obtained from the assays. Values represent the mean ± S.E. of assays performed thrice. C, tandem mass (MS/MS) spectrum of a tryptic peptide of mass/charge ratio (m/z) 1493.8 obtained from acetylated USP. Singly charged fragment ions marked in the spectrum in black represent peptide bond cleavage resulting in sequence information recorded from both the N and C termini (b- and y-type ions, respectively). This spectrum matched that of the peptide (underlined) in USP (sequence of purified protein showed as an inset), with mass shift in the b5 and y10 ions corresponding to acetylation at the lysine residue (bold in inset).
Using a direct and quantifiable gel-based assay, we verified that acetylation of USP by MSMEG_5458 was a bona fide catalysis by an enzyme. The formation of acetylated USP increased linearly with time and enzyme concentration (supplemental Fig. S4). As shown in Fig. 4B, the Km for acetyl-CoA was 10 ± 0.3 μm, and that for USP 335 ± 86 nm. Thus, we can conclude that MSMEG_5458 represents the first protein lysine acetyltransferase detected to date in mycobacteria.
To determine the site(s) for acetylation in USP, in vitro acetylated USP was separated from MSMEG_5458 by electrophoresis, and the band corresponding to USP was analyzed by mass spectrometry. Tandem mass spectrometry confirmed that a peptide of sequence NPVGKLLLGSVAQR (1493.8 Da) contained the acetylated lysine residue (Lys-104, Fig. 4C). As has been seen in diverse eukaryotic proteins, the Lys residue in USP is preceded by a flexible or small amino acid (31).
Allosteric Regulation of Acetylation by cAMP
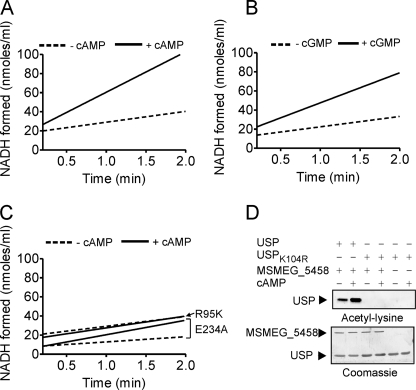
The presence of the cNMP binding domain fused to the acetyltransferase domain in these proteins, and an increase in formation of acetylated USP in the presence of cAMP (Fig. 4A) suggests that one domain may allosterically regulate the activity of the other. Acetyl-CoA did not affect the binding of cAMP to MSMEG_5458 (supplemental Fig. S1). We then determined the initial rate of acetylation of USP in the absence and presence of cAMP. We monitored acetylation rates in a coupled assay where the amount of CoA that is liberated following acetylation is measured by formation of reduced NADH from NAD+ (32). As shown in Fig. 5A, MSMEG_5458 could acetylate USP in the absence of cAMP (12.8 ± 1.2 nmol of NADH formed/min/ml), and the rate of acetylation was increased ∼2.4-fold in the presence of cAMP (31.2 ± 5.9 nmol of NADH formed/min/ml). cGMP was also able to increase the rate of acetylation by the wild-type protein (31.2 ± 0.9 nmol of NADH formed/min/ml in the presence of cGMP, Fig. 5B). Formation of NADH in the absence of enzyme varied from 1–5% of that seen with MSMEG_5458 in the absence of cAMP, reflecting the very low, non-enzymatic formation of NADH in the coupled assay.
FIGURE 5.
Initial rates of the acetyltransferase activity of MSMEG_5458. The acetyltransferase activities of wild type (A and B) and mutant MSMEG_5458 proteins (C) were measured using the coupled assay. MSMEG_5458 or mutant proteins (1 μg) were assayed in the presence of 30 μm acetyl-CoA and 50 μm USP. The initial rate of formation of NADH is shown, after subtracting the change in absorbance at 340 nm that is seen in assays performed in the absence of the enzymes, which was usually ∼1% of the change seen in the presence of enzyme. D, USPK104R (50 μm) was used as substrate for the acetyltransferase activity, in the presence or absence of cAMP (10 μm). Samples were subjected to Western blotting using acetyl-lysine antibodies, and either wild-type USP or USPK104R. Data shown are a representative of assays performed thrice.
This potentiation of acetyltransferase activity in the presence of cAMP was validated when assays were performed with MSMEG_5458R95K. The mutant protein showed no increase in the rate of acetylation in the presence of cAMP (Fig. 5C), presumably because of its inability to bind cAMP (13.1 ± 2.4 nmol of NADH formed/min/ml in the absence of cAMP; 12.2 ± 3.3 nmol of NADH formed/min/ml in the presence of cAMP). These results therefore demonstrate allosteric coupling between the cNMP binding and acetyltransferase domains in MSMEG_5458.
GNAT family members perform a sequential mechanism of acetyl transfer. An active site Glu acts as a base to assist in deprotonating the Lys residue allowing nucleophilic attack of the carbonyl carbon of acetyl-CoA, formation of a tetrahedral intermediate, and then collapse to the reaction products (33). Sequence alignment of the GNAT-like domain in MSMEG_5458 with structurally characterized protein acetyltransferases identified a Glu residue (Glu-234) that could act as the base at the active site (Fig. 1C). We generated a Glu-234 → Ala mutant and observed that the mutant protein showed a lower rate of acetylation of USP in the absence of cAMP (4.8 ± 0.6 nmol of NADH formed/min/ml, Fig. 5C) than the wild-type protein, but in the presence of cAMP, an increase in the rate of acetyltransferase activity could be detected (16.9 ± 1.3 nmol of NADH formed/min/ml). The Glu-234 → Ala mutant was able to bind cAMP as efficiently as the wild-type protein (supplemental Fig. S1).
We measured the rate of acetylation of a mutant USP, where the lysine residue that was acetylated in USP was mutated to an arginine (USPK104R). As shown in Fig. 5D, no acetylation of USPK104R was seen on Western blot analysis. These results demonstrate the absolute specificity of Lys-104 as a site for acetylation in USP by MSMEG_5458, in agreement with the results obtained from mass spectrometry.
USP Is a Substrate of MSMEG_5458 in Vivo
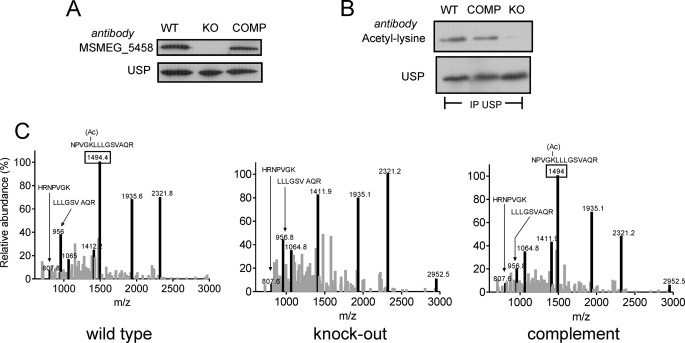
Although we have provided biochemical evidence to show that USP can be utilized as a substrate of MSMEG_5458 in vitro, it was important to determine if MSMEG_5458 could serve as a functional protein acetyltransferase and use USP as a substrate in vivo. We generated a strain of M. smegmatis deleted for MSMEG_5458 as well as complemented the strain by insertion (at the genomic att site) of the MSMEG_5458 gene driven by its own promoter (supplemental Fig. S5). We monitored expression of both MSMEG_5458 and USP in the three strains using antibodies specific to each of the proteins. No MSMEG_5458 was detected in the knock-out strain, and expression was restored in the complemented strain (Fig. 6A). USP levels were comparable in the wild-type and complemented strains (Fig. 6A).
FIGURE 6.
USP is a substrate of MSMEG_5458 in vivo. A, Western blot analysis of whole cell lysates prepared from wild type (WT), ΔMSMEG_5458 (KO), and ΔMSMEG_5458 complemented with wild-type MSMEG_5458 (COMP) strains using polyclonal MSMEG_5458 and USP antibodies. B, whole cell lysates from WT, KO, and COMP were immunoprecipitated with USP antibody and immunoprecipitates subjected to Western blot analysis with acetyl-lysine antibody followed by normalization with USP polyclonal antibodies. C, MALDI mass spectra of in-gel tryptic digests of immunoprecipitated USP obtained from WT, KO, and COMP cells. The peaks in black represent peptides from USP. The 1493.8-Da peptide (represented as a peak marked with a box) corresponds to the acetylated peptide and is present in USP from WT and COMP strains but absent in the KO strain.
The overall pattern of acetylation in the wild-type and knock-out strains was similar as detected by an acetyl-lysine immunoblot (data not shown). We immunoprecipitated USP from cultures of the three strains and subjected the immunoprecipitates to Western blot analysis using acetyl-lysine antibodies. No acetylated USP could be detected in the strain deficient for MSMEG_5458, but acetylation was restored in the complemented strain (Fig. 6B). To confirm these results, we subjected the immunoprecipitated USP to tryptic digestion, and MALDI analysis showed that whereas the acetylated peptide of mass 1493.8 Da was observed in the wild-type and complemented strains, no acetylated peptide was observed in the knock-out strain (Fig. 6C). The two peptides corresponding to cleavage after the unacetylated lysine residue were seen (807.8 and 957 Da). This clearly indicates that USP is indeed an in vivo substrate for MSMEG_5458 acetyltransferase.
Rv0998 Is a cAMP-dependent Acetyltransferase
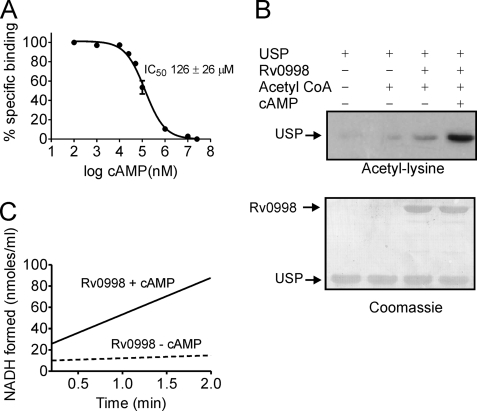
Expression of hexahistidine-tagged Rv0998 was poor in E. coli. However, sufficient amounts were obtained for biochemical characterization on expression as a fusion with GST, and subsequent cleavage of Rv0998 from the GST tag. Purified Rv0998 bound cAMP with an affinity ∼1000-fold lower than that of MSMEG_5458 (IC50 126 ± 26 μm, Fig. 7A). We attempted a pulldown assay approach to identify substrates from M. tuberculosis H37Rv using lysates prepared from M. bovis Bacillus Calmette-Guérin (which encodes a gene identical to Rv0998) but could not identify specifically interacting proteins.
FIGURE 7.
Biochemical properties of Rv0998. A, displacement of [3H]cAMP (∼50 nm) from Rv0998 (5 μg) with increasing concentrations of unlabeled cAMP. Values represent the mean ± S.E. with n = 3. B, acetylation of USP by Rv0998 was monitored by Western blot analysis with acetyl-lysine antibodies performed in the presence or absence of cAMP (1 mm, upper panel) followed by staining of the blot with Coomassie Brilliant Blue (lower panel). C, acetyltransferase activity of Rv0998 using the coupled assay, in the absence or presence of 1 mm cAMP. Data are representative of assays performed thrice.
An ortholog of MSMEG_4207 is not present in M. tuberculosis. Nevertheless, Western blot analysis using acetyl-lysine antibodies showed that Rv0998 acetylated USP significantly only in the presence of cAMP (Fig. 7B). These results were confirmed by the continuous assay where Rv0998 showed a low rate of acetylation of USP in the absence of cAMP (2.9 ± 0.3 nmol of NADH formed/min/ml) but the rate increased ∼10-fold in the presence of cAMP (29.9 ± 4 nmol of NADH formed/min/ml, Fig. 7C). Therefore, even though species-specific variability in the biochemical activities of these proteins is observed with respect to cAMP binding, the fact that USP serves as a substrate for Rv0998 in vitro suggests that there could be common in vivo substrates for these proteins in different mycobacteria.
DISCUSSION
The importance of cAMP in regulating biological processes has been conserved throughout evolution. What remains unique are downstream effectors coupled to cAMP. We now have identified a novel mechanism utilized by mycobacteria to potentially modify protein acetylation in a cAMP-responsive way.
Our study represents the first biochemical characterization of proteins containing a fusion of a GNAT-like domain with a cNMP domain, and provides a novel downstream effector function of cyclic nucleotides in mycobacteria. Such a fusion was not identified in an earlier analysis of the evolution of cNMP binding modules in both eukaryotes, and prokaryotic genome sequences obtained from Global Ocean Sampling (34), perhaps indicating that this domain fusion may be unique to mycobacteria.
In a phylogenetic analysis, MSMEG_5458 clusters with bacterial and fungal proteins, and based on a classification of the phosphate binding cassette, has residues that group it with fungal and alveolate regulatory subunits (35). The presence of a Ser/Thr residue at position 14 in the phosphate binding cassette (Ser-99 in Rv0998 and Thr-96 in MSMEG_5458) explains the observation that MSMEG_5458 binds to and is also regulated by cGMP (36). Whether cGMP has a physiological role in mycobacteria is uncertain, because the nucleotide cyclases encoded in mycobacterial genomes are predicted to be adenylyl cyclases, although some purified enzymes show low guanylyl cyclase activity (3).
The affinity of cAMP for MSMEG_5458 is in the range reported for the isolated A and B domains in the regulatory subunit of PKA (26). Given the high (∼100 μm to 1 mm) concentrations of intracellular cAMP in M. smegmatis (9), it is possible that MSMEG_5458 would almost always be completely saturated with cAMP. During evolution, this may have resulted in the production of a protein that shows only a 3-fold increase in substrate acetylation in the presence of cAMP. However, the very low acetyltransferase activity of Rv0998 in the absence of cAMP, and a dramatic increase in the presence, of cAMP, suggests that Rv0998 can be more tuned to the intracellular levels of cAMP and can modulate the levels of protein acetylation in the cell more precisely. It is conceivable that the actual rate of acetylation in vivo could be markedly different to that seen in vitro and could also be distinct for the two species.
A number of proteins containing acetyltransferase domains are found encoded in mycobacteria (37), and some have been characterized. For example, arylamine N-acetyltransferases, which catalyze the transfer of the acetyl group from acetyl-CoA to the free amino group of arylamines and hydrazines, can acetylate the antitubercular drug isoniazid, thus possibly preventing activation of the prodrug in the mycobacterial cell (28). Rv0802c exhibits the GNAT fold, and co-crystallization with succinyl-CoA suggested its preferential use as a substrate (38). We have observed that MSMEG_5458 could utilize butyryl-CoA as the substrate for acetylation of USP while the activity with succinyl-CoA was much lower. Some GNAT-like enzymes can acetylate proteins (usually histones are used as model substrates) as well as low molecular aminoglycosides (39). It is conceivable that MSMEG_5458 and Rv0998 could have non-proteinaceous substrates in mycobacteria, although both these proteins were inactive against aminoglycoside antibiotics in our assays. It may also be worth testing if newly described mycobacterial aminoglycoside acetyltransferases such as eis could acetylate proteins in addition to kanamycin (40), especially because a number of proteins appear to be acetylated in a MSMEG_5458-independent manner in M. smegmatis.
It is intriguing that the Glu-234 → Ala mutation in MSMEG_5458 reduced basal activity of the enzyme, but in the presence of cAMP, significant acetylation of USP was observed (Fig. 5C). In a recent study on the structure of the protein acetyltransferase from Sulfolobus, it was speculated that the process of inserting the lysine residue in a hydrophobic, substrate-binding pocket seen in protein acetyltransferase could significantly alter the pKa of the lysine residue, thereby allowing acetylation to proceed (41). Perhaps a similar mechanism is seen in MSMEG_5458 whereby cAMP binding could alter the hydrophobicity of the substrate binding site, thereby reducing the pKa of the lysine sufficiently to allow the acetylation reaction to occur, without the requirement of the Glu-234 residue to serve as a base (42).
The tight association of USP with MSMEG_5458 is surprising, because enzyme-substrate complexes are rarely identified by pulldown assay approaches in the absence of an inhibitor. Perhaps this is indicative of USP being a good substrate for MSMEG_5458, with a high affinity for the enzyme (supplemental Fig. S4). Indeed, USP may be in a stable complex with MSMEG_5458 in cells under steady-state conditions. As a corollary, tight binding of USP would preclude the binding of other substrates to MSMEG_5458, and thus USP may act as a regulator of protein acetylation brought about by MSMEG_5458.
In bacteria as well as eukaryotes, a number of enzymes involved in central metabolic pathways have recently been shown to be acetylated (43–44), suggesting a conserved means of regulation of intermediary metabolism. Protein acetylation in eukaryotes also regulates DNA-binding activity, protein-protein interaction, and protein stability via ubiquitination (45). In prokaryotes, there are reports of acetylation of the response regulator CheY (46) and acetyl-CoA synthetase (47). Indeed, Sulfolobus, an entire machinery for protein acetylation and deacetylation, has been described involving a Sir2-like deacetylase, the acetyltransferase protein acetyltransferase, and its substrate Alba, which is an acetylation-regulated DNA-binding protein (48–49). A global analysis of acetylated proteins in E. coli showed that a large number are metabolic enzymes, transcriptional/translational regulators, and stress response proteins (50). Such a proteome-wide study has not been performed in mycobacteria. Recent reports on pupylation, as a modification that occurs on lysine residues in mycobacterial proteins and regulates their stability (51, 52), suggest that a competition between acetylation and pupylation may occur on specific lysine residues in proteins as a means of regulating their stability within the cell. This of course would occur only if the sites of pupylation and acetylation overlap, and such studies would be of interest to pursue.
MSMEG_5458 is not an essential gene, but orthologs are conserved in all mycobacteria, suggesting a unique requirement for this protein product, perhaps under environmental conditions that are not achieved in laboratory cultures of M. smegmatis. Acetyl-CoA and NAD (or the NAD:NADH ratio) are key indicators of cellular energy status, and the requirement of acetyl-CoA for acetyltransferases and NAD for sirtuin-like enzymes indicates that protein lysine acetylation serves as a link that connects cellular energy levels with protein acetylation/deacetylation activity (53). In mycobacteria, a third player has a role to play, and that is cAMP. Intracellular cAMP levels if altered under certain conditions could modulate the effects brought about by the energy status of the cell and thereby regulate protein acetylation. The challenge ahead is to identify all acetylated proteins in mycobacteria and to determine how many of them are substrates for the Rv0998 family of proteins. Importantly, studies to elucidate the consequences of protein acetylation and deacetylation in the biology of these important bacteria are also required.
Supplementary Material
Acknowledgments
We acknowledge the technical assistance of Vaishnavi Ravikumar and Vani R. Iyer and thank members of the laboratory for useful discussions and careful reading of the manuscript. We also thank Sathisha K. in the Proteomics Facility, Molecular Biophysics Unit, Indian Institute of Science, for assistance with mass spectrometry.
This work was supported by the Department of Biotechnology, Government of India.

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. S1–S5 and Tables S1 and S2.
- PKA
- protein kinase A
- 2-ME
- 2-mercaptoethanol
- CREB
- cAMP response element-binding
- CBP
- CREB-binding protein
- cNMP
- cyclic nucleotide monophosphate
- CRP
- catabolite repressor protein
- DTT
- dithiothreitol
- GST
- glutathione S-transferase
- MALDI-TOF
- matrix-assisted laser desorption/ionization, time of flight
- USP
- universal stress protein
- Bis-Tris
- 2-[bis(2-hydroxyethyl)amino]-2-(hydroxymethyl)propane-1,3-diol
- (Sp)-cAMPS
- adenosine-3′,5′-cyclic monophosphorothioate, Sp isomer.
REFERENCES
- 1.Barzu O., Danchin A. (1994) Prog. Nucleic Acids Res. Mol. Biol. 49, 241–283 [DOI] [PubMed] [Google Scholar]
- 2.Linder J. U., Schultz J. E. (2003) Cell Signal. 15, 1081–1089 [DOI] [PubMed] [Google Scholar]
- 3.Shenoy A. R., Visweswariah S. S. (2006) FEBS Lett. 580, 3344–3352 [DOI] [PubMed] [Google Scholar]
- 4.Shenoy A. R., Sreenath N., Podobnik M., Kovacevic M., Visweswariah S. S. (2005) Biochemistry 44, 15695–15704 [DOI] [PubMed] [Google Scholar]
- 5.Podobnik M., Tyagi R., Matange N., Dermol U., Gupta A. K., Mattoo R., Seshadri K., Visweswariah S. S. (2009) J. Biol. Chem. 284, 32846–32857 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Shenoy A. R., Capuder M., Draskovic P., Lamba D., Visweswariah S. S., Podobnik M. (2007) J. Mol. Biol. 365, 211–225 [DOI] [PubMed] [Google Scholar]
- 7.Agarwal N., Lamichhane G., Gupta R., Nolan S., Bishai W. R. (2009) Nature 460, 98–102 [DOI] [PubMed] [Google Scholar]
- 8.Shenoy A. R., Visweswariah S. S. (2006) Trends Microbiol. 14, 543–550 [DOI] [PubMed] [Google Scholar]
- 9.Dass B. K., Sharma R., Shenoy A. R., Mattoo R., Visweswariah S. S. (2008) J. Bacteriol. 190, 3824–3834 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Berman H. M., Ten Eyck L. F., Goodsell D. S., Haste N. M., Kornev A., Taylor S. S. (2005) Proc. Natl. Acad. Sci. U.S.A. 102, 45–50 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Rehmann H., Wittinghofer A., Bos J. L. (2007) Nat. Rev. Mol. Cell Biol. 8, 63–73 [DOI] [PubMed] [Google Scholar]
- 12.Körner H., Sofia H. J., Zumft W. G. (2003) FEMS Microbiol. Rev. 27, 559–592 [DOI] [PubMed] [Google Scholar]
- 13.Gallagher D. T., Smith N., Kim S. K., Robinson H., Reddy P. T. (2009) J. Biol. Chem. 284, 8228–8232 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Stapleton M., Haq I., Hunt D. M., Arnvig K. B., Artymiuk P. J., Buxton R. S., Green J. (2010) J. Biol. Chem. 285, 7016–7027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wang L., Tang Y., Cole P. A., Marmorstein R. (2008) Curr. Opin. Struct. Biol. 18, 741–747 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Tamura K., Dudley J., Nei M., Kumar S. (2007) Mol. Biol. Evol. 24, 1596–1599 [DOI] [PubMed] [Google Scholar]
- 17.Shenoy A. R., Visweswariah S. S. (2003) Anal. Biochem. 319, 335–336 [DOI] [PubMed] [Google Scholar]
- 18.Parish T., Stoker N. G. (2000) Microbiology 146, 1969–1975 [DOI] [PubMed] [Google Scholar]
- 19.McCue L. A., McDonough K. A., Lawrence C. E. (2000) Genome Res. 10, 204–219 [DOI] [PubMed] [Google Scholar]
- 20.Kim C., Cheng C. Y., Saldanha S. A., Taylor S. S. (2007) Cell 130, 1032–1043 [DOI] [PubMed] [Google Scholar]
- 21.Taylor S. S., Kim C., Cheng C. Y., Brown S. H., Wu J., Kannan N. (2008) Biochim. Biophys. Acta 1784, 16–26 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Chang P. A., Wang Z. X., Long D. X., Qin W. Z., Wu Y. J. (2010) Mol. Cell Biochem. 339, 181–190 [DOI] [PubMed] [Google Scholar]
- 23.Trievel R. C., Rojas J. R., Sterner D. E., Venkataramani R. N., Wang L., Zhou J., Allis C. D., Berger S. L., Marmorstein R. (1999) Proc. Natl. Acad. Sci. U.S.A. 96, 8931–8936 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hodawadekar S. C., Marmorstein R. (2007) Oncogene 26, 5528–5540 [DOI] [PubMed] [Google Scholar]
- 25.Dyda F., Klein D. C., Hickman A. B. (2000) Annu. Rev. Biophys. Biomol. Struct. 29, 81–103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Dostmann W. R., Taylor S. S., Genieser H. G., Jastorff B., Døskeland S. O., Ogreid D. (1990) J. Biol. Chem. 265, 10484–10491 [PubMed] [Google Scholar]
- 27.Bubis J., Neitzel J. J., Saraswat L. D., Taylor S. S. (1988) J. Biol. Chem. 263, 9668–9673 [PubMed] [Google Scholar]
- 28.Sim E., Sandy J., Evangelopoulos D., Fullam E., Bhakta S., Westwood I., Krylova A., Lack N., Noble M. (2008) Curr. Drug Metab. 9, 510–519 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.O'Toole R., Williams H. D. (2003) Res. Microbiol. 154, 387–392 [DOI] [PubMed] [Google Scholar]
- 30.O'Toole R., Smeulders M. J., Blokpoel M. C., Kay E. J., Lougheed K., Williams H. D. (2003) J. Bacteriol. 185, 1543–1554 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Basu A., Rose K. L., Zhang J., Beavis R. C., Ueberheide B., Garcia B. A., Chait B., Zhao Y., Hunt D. F., Segal E., Allis C. D., Hake S. B. (2009) Proc. Natl. Acad. Sci. U.S.A. 106, 13785–13790 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Berndsen C. E., Denu J. M. (2005) Methods 36, 321–331 [DOI] [PubMed] [Google Scholar]
- 33.Tanner K. G., Trievel R. C., Kuo M. H., Howard R. M., Berger S. L., Allis C. D., Marmorstein R., Denu J. M. (1999) J. Biol. Chem. 274, 18157–18160 [DOI] [PubMed] [Google Scholar]
- 34.Kannan N., Wu J., Anand G. S., Yooseph S., Neuwald A. F., Venter J. C., Taylor S. S. (2007) Genome Biol. 8, R264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Canaves J. M., Taylor S. S. (2002) J. Mol. Evol. 54, 17–29 [DOI] [PubMed] [Google Scholar]
- 36.Weber I. T., Shabb J. B., Corbin J. D. (1989) Biochemistry 28, 6122–6127 [DOI] [PubMed] [Google Scholar]
- 37.Vagena E., Fakis G., Boukouvala S. (2008) Curr. Drug Metab. 9, 628–660 [DOI] [PubMed] [Google Scholar]
- 38.Vetting M. W., Errey J. C., Blanchard J. S. (2008) Acta Crystallogr. Sect. F Struct. Biol. Cryst. Commun. 64, 978–985 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Vetting M. W., Magnet S., Nieves E., Roderick S. L., Blanchard J. S. (2004) Chem. Biol. 11, 565–573 [DOI] [PubMed] [Google Scholar]
- 40.Zaunbrecher M. A., Sikes R. D., Jr., Metchock B., Shinnick T. M., Posey J. E. (2009) Proc. Natl. Acad. Sci. U.S.A. 106, 20004–20009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Brent M. M., Iwata A., Carten J., Zhao K., Marmorstein R. (2009) J. Biol. Chem. 284, 19412–19419 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Mehler E. L., Fuxreiter M., Simon I., Garcia-Moreno E. B. (2002) Proteins 48, 283–292 [DOI] [PubMed] [Google Scholar]
- 43.Wang Q., Zhang Y., Yang C., Xiong H., Lin Y., Yao J., Li H., Xie L., Zhao W., Yao Y., Ning Z. B., Zeng R., Xiong Y., Guan K. L., Zhao S., Zhao G. P. (2010) Science 327, 1004–1007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zhao S., Xu W., Jiang W., Yu W., Lin Y., Zhang T., Yao J., Zhou L., Zeng Y., Li H., Li Y., Shi J., An W., Hancock S. M., He F., Qin L., Chin J., Yang P., Chen X., Lei Q., Xiong Y., Guan K. L. (2010) Science 327, 1000–1004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Yang X. J., Seto E. (2008) Mol. Cell 31, 449–461 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Yan J., Barak R., Liarzi O., Shainskaya A., Eisenbach M. (2008) J. Mol. Biol. 376, 1260–1271 [DOI] [PubMed] [Google Scholar]
- 47.Starai V. J., Celic I., Cole R. N., Boeke J. D., Escalante-Semerena J. C. (2002) Science 298, 2390–2392 [DOI] [PubMed] [Google Scholar]
- 48.Marsh V. L., Peak-Chew S. Y., Bell S. D. (2005) J. Biol. Chem. 280, 21122–21128 [DOI] [PubMed] [Google Scholar]
- 49.Bell S. D., Botting C. H., Wardleworth B. N., Jackson S. P., White M. F. (2002) Science 296, 148–151 [DOI] [PubMed] [Google Scholar]
- 50.Zhang J., Sprung R., Pei J., Tan X., Kim S., Zhu H., Liu C. F., Grishin N. V., Zhao Y. (2009) Mol. Cell Proteomics 8, 215–225 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Pearce M. J., Mintseris J., Ferreyra J., Gygi S. P., Darwin K. H. (2008) Science 322, 1104–1107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Festa R. A., McAllister F., Pearce M. J., Mintseris J., Burns K. E., Gygi S. P., Darwin K. H.PLoS One 5, e8589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Yu J., Auwerx J. (2010) Pharmacol. Res. 62, 35–41 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.