Abstract
We have developed a stable analog for the ADP-insensitive phosphoenzyme intermediate with two occluded Ca2+ at the transport sites (E2PCa2) of sarcoplasmic reticulum Ca2+-ATPase. This is normally a transient intermediate state during phosphoenzyme isomerization from the ADP-sensitive to ADP-insensitive form and Ca2+ deocclusion/release to the lumen; E1PCa2 → E2PCa2 → E2P + 2Ca2+. Stabilization was achieved by elongation of the Glu40-Ser48 loop linking the Actuator domain and M1 (1st transmembrane helix) with four glycine insertions at Gly46/Lys47 and by binding of beryllium fluoride (BeFx) to the phosphorylation site of the Ca2+-bound ATPase (E1Ca2). The complex E2Ca2·BeF3− was also produced by lumenal Ca2+ binding to E2·BeF3− (E2P ground state analog) of the elongated linker mutant. The complex was stable for at least 1 week at 25 °C. Only BeFx, but not AlFx or MgFx, produced the E2PCa2 structural analog. Complex formation required binding of Mg2+, Mn2+, or Ca2+ at the catalytic Mg2+ site. Results reveal that the phosphorylation product E1PCa2 and the E2P ground state (but not the transition states) become competent to produce the E2PCa2 transient state during forward and reverse phosphoenzyme isomerization. Thus, isomerization and lumenal Ca2+ release processes are strictly coupled with the formation of the acylphosphate covalent bond at the catalytic site. Results also demonstrate the critical structural roles of the Glu40-Ser48 linker and of Mg2+ at the catalytic site in these processes.
Keywords: Bioenergetics, Calcium ATPase, Calcium Transport, Kinetics, Sarcoplasmic Reticulum, Site-directed Mutagenesis, Phosphate Analog, Phosphorylated Intermediate, Proteolysis, Structure and Function
Introduction
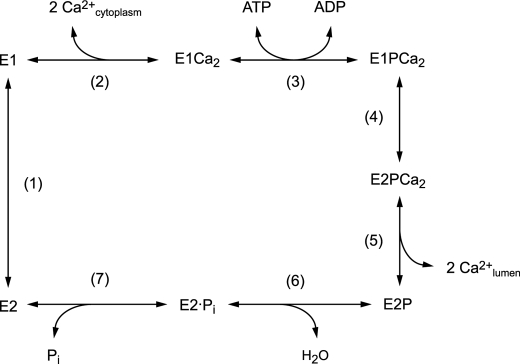
Sarcoplasmic reticulum Ca2+-ATPase (SERCA1a)2 catalyzes Ca2+ transport coupled with ATP hydrolysis (Fig. 1) (1–9). In the catalytic cycle the enzyme is activated by two cytoplasmic Ca2+ ions binding to the transport sites. It is then autophosphorylated at Asp351 by MgATP to produce the ADP-sensitive phosphoenzyme (E1P) that can react with ADP to regenerate ATP (steps 1–3). E1P formation results in Ca2+ occlusion at the transport sites (E1PCa2). Subsequent isomeric transition to an ADP-insensitive form (E2P), i.e. loss of ADP-sensitivity, results in Ca2+ deocclusion and release into the lumen (steps 4 and 5). This Ca2+-release process is very rapid so that an E2PCa2 intermediate state does not accumulate and in fact had never been found until we recently established its existence (10–13) and successfully trapped it for the first time (14). The Ca2+-free E2P is finally hydrolyzed to the inactive E2 state (steps 6 and 7). Mg2+ as the physiological catalytic cofactor is required for both phosphorylation and hydrolysis. The transport cycle is reversible. Thus, E2P can be formed from Pi in the presence of Mg2+ and absence of Ca2+. Subsequent Ca2+ binding to lumenal-oriented low affinity transport sites reverses the Ca2+-releasing step and the E1P to E2P isomerization.
FIGURE 1.
Ca2+-transport cycle of SERCA.
During EP isomerization/Ca2+-release (E1PCa2 → E2P + 2Ca2+), the A domain swings around parallel to the membrane plane (i.e. horizontal), whereas the A and P domains and M2 incline and tightly associate (Fig. 2) (15–25). We found that shortening of the A/M1′-linker by deletion of any single residue blocks E1PCa2 → E2PCa2 isomerization and E2P hydrolysis (26). On the other hand, its elongation by two or more glycine insertions markedly accelerates the isomerization and blocks Ca2+ deocclusion/release (E2PCa2 → E2P + 2Ca2+) (14). Thus, elongating the A/M1′-linker stabilized the normally transient intermediate state E2PCa2 (i.e. ADP-insensitive EP with occluded Ca2+) and showed that the length of this linker is critical for the structural changes that occur during E1PCa2 → E2PCa2 → E2P + 2Ca2+ and subsequent E2P hydrolysis (14, 26).
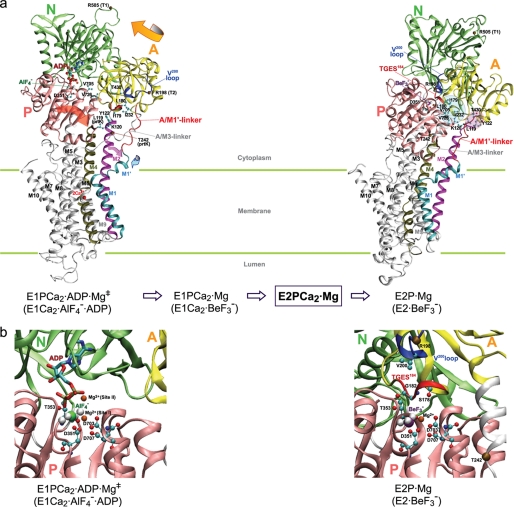
FIGURE 2.
Crystal structures of SERCA1a. The coordinates for structures E1Ca2·AlF4−·ADP (the analog of the transition state of phosphorylation, left) and E2·BeF3− (the analog of the E2P ground state, right) were obtained from Protein Data Bank (PDB accession code 1T5T (17) and 2ZBE (21), respectively). a, the cytoplasmic domains N (nucleotide binding), P (phosphorylation), A (actuator), 10 transmembrane helices (M1-M10), phosphorylation site Asp351, and TGES184 on the A domain are indicated. Cleavage sites by trypsin (T1 (Arg505) and T2 (Arg198 on the Val200 loop (DPRAVNQD203)) and by prtK (Leu119 on the top part of M2 and Thr242 on the A/M3-linker) are shown. Arrows indicate approximate motions of the A and P domains, M2, and M1′ from E1Ca2·AlF4−·ADP to E2·BeF3−. Note the large rotation of the A domain and the inclination of the P and A domains and M2. In the E2P state the A and P domains interact at three regions; at the 181TGES loop with the residues around Asp351, at the Val200 loop (Asp196-Asp203) with polar residues of the P domain, and at Leu119/Tyr122 on the top part of M2 with the A, P, and N domains. In E2·BeF3−(TG) (PDB accession code 2ZBF (21), supplemental Fig. S5), Leu119/Tyr122 produce the Tyr122-hydrophobic cluster with five other hydrophobic residues, Ile179/Leu180/Ile232 of the A domain and Val705/Val726 of the P domain. In E2·BeF3− without TG, the cluster structure is rather loose (as the side chains of Leu119/Tyr122 are pointing away from the hydrophobic cluster), but Leu119/Tyr122 produce a more extended interaction network involving Thr430 of the N domain and the hydrophobic cluster (see more details in supplemental Fig. S5). b, the catalytic site is enlarged, and the residues involved in the Mg2+ (site I) are depicted. The Val679-Lys686 region of the P domain is not depicted for simplicity (because it is positioned over the region of interest).
We have recently developed an E1Ca2·BeF3− complex as a stable analog of E1PCa2·Mg2+ (E1PCa2 with bound Mg2+ at the catalytic site) (27). Structural analysis of the analog and intermediate states suggests that formation of native E1PCa2·Mg2+ results in structural changes in the cytoplasmic and transmembrane domains due to configuration and ligation changes of the phosphate moiety (27). The Mg2+ bound at the catalytic site contributes to these structural changes (27). In fact, Ca2+ could not substitute for Mg2+ for formation of E1Ca2·BeF3−, and an attempt to substitute Ca2+ for Mg2+ destroyed the complex (27). It is well known that Ca2+ substitution of Mg2+ at the catalytic site markedly retards E1PCa2·Ca isomerization (28, 29), a step that includes rotation of the A domain.
Further understanding of the mechanism of EP processing via the transient E2PCa2 and of the critical roles of the A/M1′-linker and catalytic Mg2+ requires detailed characterization of the development of E2PCa2 and of factors contributing to its possible stabilization. A great advance would be the finding of an analog stable enough for crystallographic studies.
In this study we employed the mutant 4Gi-46/47 in which the A/M1′-linker is elongated by four glycine insertions at Gly46/Lys47 (14) and explored the formation of a stable structural analog of E2PCa2 using various configuration analogs of phosphate (BeFx/AlFx/MgFx) and catalytic cations (Mg2+/Mn2+/Ca2+). We found that BeFx is uniquely efficacious and that both mutant E1Ca2·BeF3− and mutant E2·BeF3− are capable of producing mutant E2Ca2·BeFx, most probably E2Ca2·BeF3−, and that Ca2+ can replace the catalytic Mg2+ when coming from the former species. The mutant complex E2Ca2·BeF3− is extremely stable even at 25 °C.
EXPERIMENTAL PROCEDURES
Mutagenesis and Expression
The pMT2 expression vector (30) carrying the mutant rabbit SERCA1a cDNA with four glycine residues inserted between Gly46 and Lys47 (4Gi-46/47) was constructed as described previously (14). Transfection of pMT2 DNA into COS-1 cells and preparation of microsomes from the cells were performed as described previously (31). The amount of expressed SERCA1a was quantified by a sandwich enzyme-linked immunosorbent assay (32). Expression levels of wild type SERCA1a and the mutants were 2–3% that of total microsomal proteins.
Metal Fluoride Treatment
Microsomes expressing the wild type or 4Gi-46/47 were treated at 25 °C for 30 min with BeFx, AlFx, and MgF42− as described previously (14, 23–25, 27, 33–36) and in the legends for Figs. 3–9 in detail.
FIGURE 3.
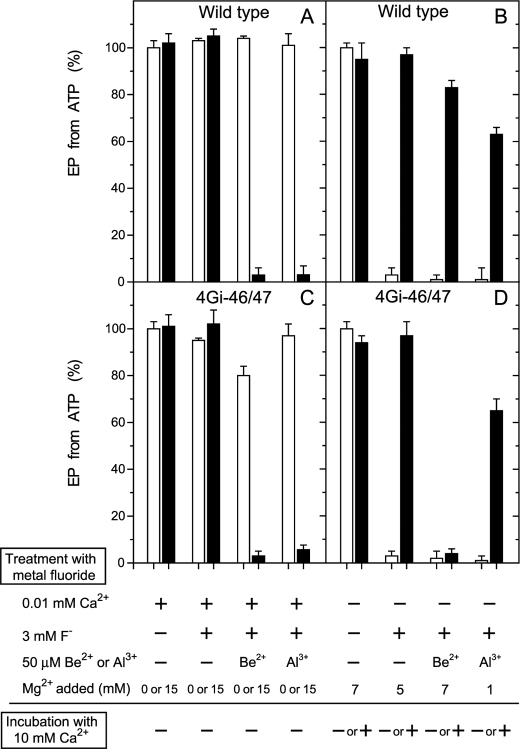
Inhibition of EP formation from ATP by metal fluoride. A and C, microsomes expressing the wild type or mutant 4Gi-46/47 (0.35 mg/ml) were treated at 25 °C for 30 min with metal fluoride in the presence of 0.01 mm Ca2+ (0.01 mm CaCl2 without EGTA) in 3 mm KF plus 50 μm BeSO4 or AlCl3, 0.1 m KCl, and 50 mm MOPS/Tris (pH 7) with (black bar) or without (white bar) 15 mm MgCl2. Subsequently, the samples were diluted 10-fold and phosphorylated at 0 °C for 15 s with 10 μm [γ-32P]ATP in 1 μm A23187, 0.1 mm Ca2+ (0.5 mm CaCl2 with 0.4 mm EGTA), 7 mm MgCl2, 0.1 m KCl, and 50 mm MOPS/Tris (pH 7), and the amount of EP formed was determined. The amount of EP formed with the wild type in the control sample, i.e. incubated without the fluoride compounds and Mg2+ (4.7 nmol/mg of the expressed SERCA1a), was normalized to 100%. The amount of EP formed with the mutant 4Gi-46/47 in the control sample was almost the same as that of wild type. B and D, microsomes were treated with metal fluoride in the absence of Ca2+ (1 mm EGTA without added CaCl2) and in the presence of the indicated concentrations of MgCl2. Subsequently, the samples were diluted 2.5-fold with a solution containing 1 μm A23187, 0.1 m KCl, 50 mm MOPS/Tris (pH 7), and EGTA (to give 1 mm, white bar) or CaCl2 (to give 10 mm Ca2+, black bar) and incubated at 25 °C for 1 h. The samples were then further diluted 10-fold and phosphorylated with 10 μm [γ-32P]ATP and 0.1 mm Ca2+ as in A and C, and the amount of EP formed was determined.
FIGURE 4.
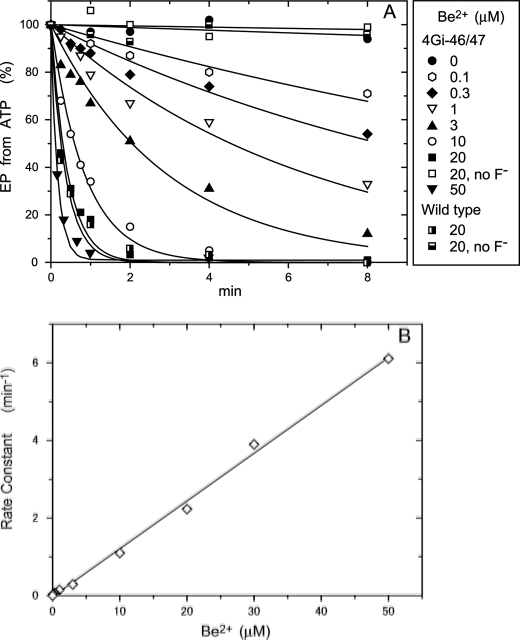
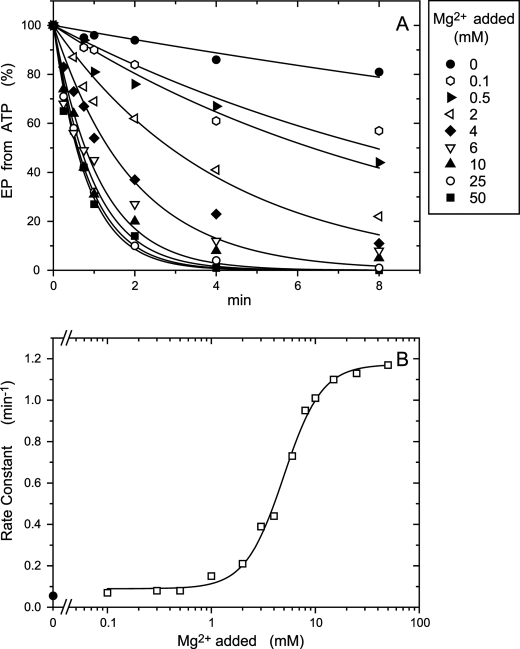
Be2+ dependence of the rate of EP inhibition by BeFx in 0.01 mm Ca2+. A, microsomes expressing the wild type or mutant 4Gi-46/47 were incubated for various periods in 0.01 mm Ca2+ and 1 mm KF with various concentrations of BeSO4 and otherwise as in Fig. 3, A and C, for BeFx-treatment. The samples were then diluted 10-fold and phosphorylated with 10 μm [γ-32P]ATP, and the amount of EP formed was determined, as in Fig. 3, A and C. Solid lines show the least squares fit to a single exponential. In B, the rate constants obtained in A were plotted versus the concentration of Be2+ added. The linear fit to the data gave a slope of 0.123 min−1μm−1.
FIGURE 5.
Mg2+ dependence of the rate of EP inhibition by BeFx in 0.01 mm Ca2+. A, microsomes expressing the mutant 4Gi-46/47 were incubated for various periods in 0.01 mm Ca2+, 1 mm KF, 10 μm BeSO4, and various concentrations of MgCl2 and otherwise as in Fig. 3, A and C, for BeFx treatment. The samples were then diluted 10-fold and phosphorylated with 10 μm [γ-32P]ATP, and the amount of EP formed was determined, as in Fig. 3, A and C. Solid lines show the least squares fit to a single exponential. In B the rate constants obtained in A were plotted versus the concentration of Mg2+ added. K0.5 for the Mg2+ activation and Hill coefficient obtained by fitting to the Hill equation (solid line) were 4.9 mm and 2.3, respectively.
FIGURE 6.
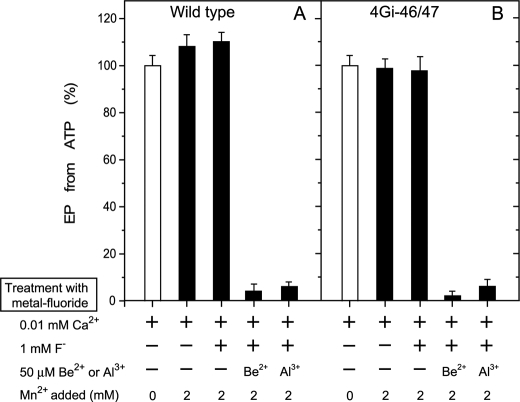
EP inhibition by Mn2+ and BeFx in 0.01 mm Ca2+ without Mg2+. Microsomes expressing the wild type or mutant 4Gi-46/47 were treated with 1 mm F− plus 50 μm Be2+ or Al3+ in 0.01 mm Ca2+ and in the absence (white bar) or presence (black bar) of 2 mm MnCl2 (in place of MgCl2) and otherwise as in Fig. 3, A and C. The samples were then diluted 10-fold and phosphorylated with 10 μm [γ-32P]ATP, and the amount of EP formed was determined as in Fig. 3, A and C.
FIGURE 7.
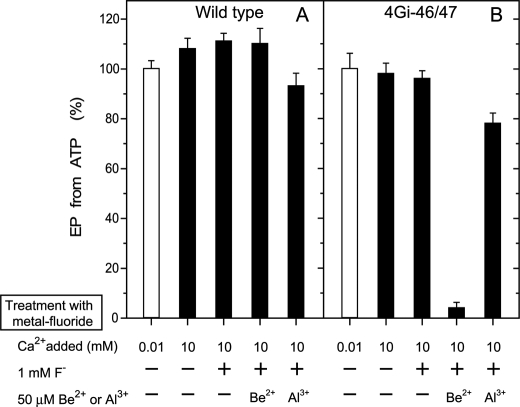
EP inhibition by 10 mm Ca2+ and BeFx without Mg2+ and Mn2+. Microsomes expressing wild type or mutant 4Gi-46/47 were treated with 1 mm F− plus 50 μm Be2+ or Al3+ in 0.01 or 10 mm CaCl2 without Mg2+ and Mn2+ and otherwise as in Fig. 3, A and C. The samples were then diluted 10-fold and phosphorylated with 10 μm [γ-32P]ATP, and the amount of EP was determined as in Fig. 3, A and C.
FIGURE 8.
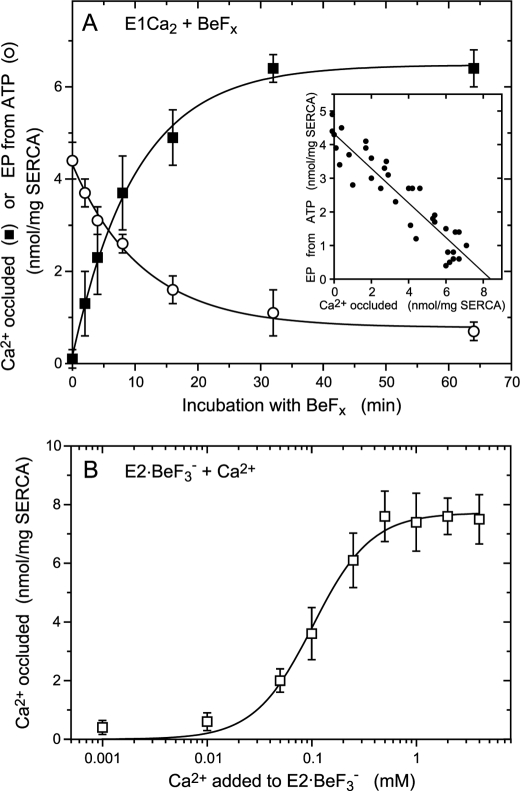
Ca2+ occlusion in E2Ca2·BeF3− of the mutant 4Gi-46/47 formed from E1Ca2 (A) and from E2·BeF3− (B). A, microsomes (0.2 mg/ml) expressing the mutant 4Gi-46/47 were incubated for various periods at 25 °C in 10 μl of a mixture containing 0.01 mm 45CaCl2, 1 mm KF, 1 μm BeSO4, 15 mm MgCl2, 0.1 m KCl, 50 mm MOPS/Tris (pH 7). The mixture was then diluted 200-fold at 0 °C with a washing solution containing 2 mm EGTA, 5 μm A23187, 0.1 m KCl, 7 mm MgCl2, and 50 mm MOPS/Tris (pH 7.0), subjected to membrane filtration, and washed rapidly with 6 ml of the washing solution for 4 s at 0 °C. For determination of EP, the above BeFx-incubation was made with 40Ca2+ instead of 45Ca2+ otherwise as above, and the sample was diluted 10-fold and phosphorylated with 10 μm [γ-32P]ATP at 0 °C for 15 s as in Fig. 3C. The sample was then further diluted 20-fold at 0 °C with the washing solution, immediately filtered as above, and washed rapidly with ice-cold trichloroacetic acid containing Pi. The EP level was not changed during the above sample handling because the decay of EP (E2PCa2) is almost completely blocked in the mutant (14). The amount of 45Ca2+ specifically bound and occluded (■) and that of E32P formed (○) in the expressed SERCA1a mutant were obtained by subtracting the background levels determined by including 1 μm TG in the BeFx incubation mixture. The values presented are the mean ± S.D. (n = 5). Inset, the amount of EP formed was replotted versus that of occluded Ca2+ with the BeFx treatment. The solid line represents the linear least squares fit. The y and x intercepts gave 4.3 and 8.4 nmol/mg of the expressed SERCA1a for the amounts of EP and of Ca2+ occluded, respectively. B, for formation of E2·BeF3−, microsomes (1 mg/ml) expressing the mutant 4Gi-46/47 were incubated at 25 °C for 30 min with 1 mm KF and 20 μm BeSO4 in 1 mm EGTA, 7 mm MgCl2, 50 mm LiCl, and 50 mm MOPS/Tris (pH 7). Then the mixture was diluted 2.5-fold with a solution containing 7 mm MgCl2, 50 mm LiCl, 50 mm MOPS/Tris (pH 7), 5 μm Ca2+ ionophore A23187, and various concentrations of 45CaCl2 to give the indicated final 45Ca2+ concentrations. After incubating at 25 °C for 1 min, the mixture was further diluted with 400-fold of the washing solution containing the excess EGTA, filtered, and washed with the washing solution as above. The amount of 45Ca2+ specifically bound and occluded in the SERCA1a was obtained by subtracting the nonspecific Ca2+ binding, which was determined without KF in the BeFx treatment mixture. In fitting to the Hill equation (solid line), the maximum amount of occluded Ca2+, K0.5 for the Ca2+ activation, and Hill coefficient were obtained as 7.7 nmol/mg of the expressed SERCA1a, 0.1 mm, and 1.6, respectively. The values presented are the mean ± S.D. (n = 7).
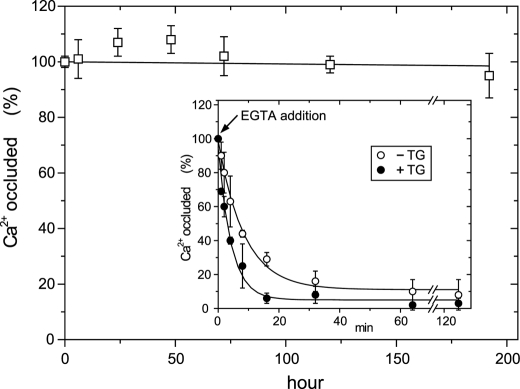
FIGURE 9.
Stability of E2Ca2·BeF3− of the mutant 4Gi-46/47. The complex E2Ca2·BeF3− was produced with the mutant 4Gi-46/47 for 30 min at 25 °C in 0.05 mm 45CaCl2, 1 mm KF, 50 μm BeSO4, and 15 mm MgCl2 and otherwise as in Fig. 8A. Then a small volume of A23187 was added to give 1 μm, and the incubation was further continued at 25 °C. At various times, the amount of 45Ca2+ specifically bound and occluded in the mutant was measured after an EGTA wash and by subtracting the background levels determined in the absence of F− in the incubation mixture and otherwise as in Fig. 8A. Inset, after the formation of E2Ca2·BeF3− as above, the sample was diluted 100-fold at 25 °C with a solution containing 1 μm A23187, 0.1 m KCl, 7 mm MgCl2, 2 mm EGTA, and 50 mm MOPS/Tris (pH 7.0) (without BeFx) in the absence (○) or presence (●) of 1 μm TG and incubated for various periods, and the amount of 45Ca2+ specifically bound and occluded in the mutant was obtained as above. The values presented are the mean ± S.D. (n = 7). Solid lines in the inset show the least squares fit to a single exponential, and the decay rate constants thus obtained are 7.0 (○) and 14.0 (●) h−1 without and with TG, respectively. In the main panel and inset, the amount of Ca2+ occluded in the complex E2Ca2·BeF3− at time 0 (immediately before starting the long incubation or the dilution) was normalized to 100%.
Formation of EP
Phosphorylation of SERCA1a in microsomes with [γ-32P]ATP was performed under conditions described in the legends for Figs. 3–8. The reactions were quenched with ice-cold trichloroacetic acid containing Pi. Precipitated proteins were separated by 5% SDS-PAGE at pH 6.0 according to Weber and Osborn (37). The radioactivity associated with the separated Ca2+-ATPase was quantified by digital autoradiography as described (38).
Ca2+ Occlusion in SERCA1a
Microsomes treated with metal fluoride were diluted with “washing solution” containing excess EGTA and then immediately filtered through a 0.45-μm nitrocellulose membrane filter (Millipore). The filter was washed extensively with the washing solution, and 45Ca2+ remaining on the filter was quantified. The amount of Ca2+ specifically bound to the transport sites of EP in the expressed SERCA1a was obtained by subtracting the amount of nonspecific Ca2+-binding, which was determined as described in the legends for Figs. 8 and 9. The Ca2+ occluded/mg of expressed SERCA1a protein was calculated from the amount of expressed SERCA1a and the amount of occluded Ca2+.
Limited Proteolysis and Western Blot Analysis
Major intermediates of the Ca2+-ATPase and their stable analogs were produced and subjected to structural analysis by limited proteolysis with trypsin and proteinase K (prtK) as described in the legends for supplemental Figs. S3 and S4. Proteolysis was terminated by 2.5% (v/v) trichloroacetic acid. The digests were subjected to SDS-PAGE (39) followed by Western blot analysis with IIH11 monoclonal antibody to the rabbit SERCA1a (Affinity Bioreagents), which recognizes an epitope between Ala199and Arg505 as described (14).
Miscellaneous
Protein concentrations were determined by the method of Lowry et al. (40) with bovine serum albumin as a standard. Data were analyzed by nonlinear regression using the program Origin (Microcal Software, Inc., Northampton, MA). Three-dimensional models of the enzyme were reproduced by the program VMD (41).
RESULTS
Inhibition of EP Formation by Metal Fluoride
The E1Ca2 state of wild type and mutant 4Gi-46/47 SERCA1a in 10 μm Ca2+ was treated with BeFx or AlFx and functionally analyzed. The ability to form EP from ATP (Fig. 3, A and C) and from Pi (data not shown) is almost completely lost in the presence of 15 mm Mg2+ but not in its absence. EP formation is not inhibited when F− treatment in 15 mm Mg2+ is made without Be2+ or Al3+. The results show that the E1Ca2 state of the mutant as well as of wild type forms stable complexes with BeFx and AlFx in the presence of Mg2+ but not with MgFx.
When the E2 state of wild type and mutant 4Gi-46/47 in the absence of Ca2+ was treated with BeFx, AlFx, and MgFx (in the absence of Be2+ and Al3+), the complexes E2·BeF3−, E2·AlF4−, and E2·MgF42−, respectively, are produced (14, 25), and EP formation from ATP (Fig. 3, B and D, open bars) and from Pi (data not shown) is almost completely inhibited. These complexes were then treated with 10 mm Ca2+ for 1 h in the presence of Ca2+ ionophore A23187 (black bars in Fig. 3, B and D). In the case of wild type, the ability to form EP is restored, consistent with the previous observation (25, 36) that a high concentration of Ca2+ in the presence of A23187 restores Ca2+-ATPase activity by destroying the complexes and converting the enzyme to E1Ca2. In mutant 4Gi-46/47, the Ca2+-induced restoration of EP formation is observed with E2·MgF42− and E2·AlF4− but not at all with E2·BeF3−. E2·BeF3− of the mutant is, thus, resistant to Ca2+. We previously found (14) that the transient intermediate E2PCa2 is produced and trapped in the mutant in the reverse direction of the pump cycle from E2P by Ca2+ binding from the lumenal side as well as in the forward direction from E1Ca2 through ATP-induced phosphorylation. Therefore, the complex produced in the mutant with BeFx is likely E2Ca2·BeF3−, an analog of E2PCa2 (as is in fact shown later in the Ca2+ binding and structural analyses in Fig. 8 and supplemental Figs. S3 and S4).
Kinetic Analysis of BeFx-induced Complex Formation
The E1Ca2 state of mutant 4Gi-46/47 was treated with various concentrations of Be2+ and 1 mm F− in 10 μm Ca2+ and 15 mm Mg2+, and the resulting species was analyzed (Fig. 4A). The presence of both Be2+ and F− (BeFx) but not F− without Be2+ or Be2+ (20 μm) without F− inhibits EP formation. The time courses of BeFx-induced inhibition follow first order kinetics. A plot of the inhibition rate constants versus Be2+ (BeFx) concentration is a straight line with no evidence of saturation within the experimental range, indicating that BeFx binding is the rate-determining step in the inhibition process (Fig. 4B). BeFx inhibits wild type at nearly the same rate as it does the mutant as seen at a representative 20 μm Be2+ with 1 mm F−.
In Fig. 5, the mutant E1Ca2 state in 10 μm Ca2+ was incubated with BeFx at various Mg2+ concentrations, and the level of inhibition of EP formation was determined. BeFx-induced inhibition is markedly accelerated with increasing Mg2+, giving a K0.5 value of 4.9 mm. The observed apparent Mg2+ affinity is consistent with those values obtained through phosphorylation of native Ca2+-ATPase (42–47) and for the formation of E1Ca2·BeF3− (E1PCa2·Mg2+ analog) (27), i.e. the Mg2+ binding affinity at the catalytic Mg2+ site (site I composed of Asp351/Thr353/Asp703 and the phosphate moiety (BeF3−)). Therefore, Mg2+ binding at site I is likely a prerequisite for BeFx binding and complex formation.
In Figs. 6 and 7, we further observed that the BeFx-induced complex formation from E1Ca2 in the mutant occurs with Mn2+ or Ca2+ in place of Mg2+. The K0.5 values are 1.4 mm for Mn2+ and 0.76 mm for Ca2+ (supplemental Figs. S1 and S2) and are consistent with such values for binding to the catalytic Mg2+ site (46, 48). In wild type the BeFx-induced E1Ca2·BeF3− formation, which inhibits EP formation occurs with Mn2+ but not with 10 mm Ca2+ in place of Mg2+ (Figs. 6 and 7). Thus, the complex formed from E1Ca2 with BeFx in the mutant 4Gi-46/47 (i.e. E2Ca2·BeF3−) is distinct from E1Ca2·BeF3− of wild type.
Interestingly, the Hill coefficient for the Mg2+ as well as Mn2+ and Ca2+ dependence for complex formation with BeFx (E2Ca2·BeF3−) in the mutant is nearly 2 (Fig. 5 and supplemental Figs. S1 and S2), suggesting the involvement of more than one metal ion. This is in contrast to the value 1 for E1Ca2·BeF3− formation with Mg2+ and Mn2+ in wild type (see supplemental Fig. 1 in Ref. 27).
AlFx produces the complex with the E1Ca2 state of the mutant 4Gi-46/47 as well as of wild type (E1Ca2·AlFx) with Mg2+ and Mn2+ but not with Ca2+ at the catalytic Mg2+ site (Figs. 3, 6, and 7). Therefore, in the mutant the complex with AlFx (E1Ca2·AlFx) is distinct from that with BeFx (E2Ca2·BeF3−) with respect to the strict preference of the divalent cation at the catalytic Mg2+ site.
Ca2+ Occlusion in the Mutant Complexed with BeFx
In Fig. 8A, the E1Ca2 state of the mutant 4Gi-46/47 in 10 μm 45Ca2+ and 15 mm Mg2+ was complexed with BeFx at a low concentration of Be2+ (1 μm) with 1 mm F− to slow complex formation. The amount of occluded 45Ca2+ was determined at various periods by membrane filtration with extensive washing with a solution containing excess EGTA and A23187. The loss of EP-forming ability with ATP decreases reciprocally and linearly with an increase in the amount of occluded Ca2+ (see the inset). The amount of occluded 45Ca2+ at the intercept of the abscissa, i.e. when all the ATPases are complexed with BeFx, is 8.4 nmol/mg of expressed SERCA1a mutant protein. The stoichiometry of the occluded Ca2+ is nearly 2 per phosphorylation site, which is 4.3 nmol/mg as determined from the intercept on the ordinate. Therefore, the complex formed with BeFx has two occluded Ca2+. When the mutant was incubated for 15 min with BeFx and 1.5 mm Mn2+ in place of Mg2+ under otherwise identical conditions, EP formation was completely inhibited, and the amount of occluded 45Ca2+ was 8.3 nmol/mg of expressed SERCA1a mutant protein, giving a stoichiometry of 2 per phosphorylation site (data not shown).
In Fig. 8B, we examined whether the complex E2Ca2·BeF3− can be produced from E2·BeF3− by lumenal Ca2+ binding, mimicking the reverse conversion E2P + 2Ca2+ → E2PCa2 (14). E2·BeF3− was first formed in the mutant in the absence of Ca2+ and then incubated for 1 min at 25 °C with various concentrations of 45Ca2+ in the presence of Ca2+ ionophore A23187. The amount of occluded 45Ca2+ was determined after a large dilution followed by filtration and extensive EGTA washing. The maximum amount of occluded 45Ca2+ is 7.7 nmol/mg of mutant SERCA1a protein and 1.8 times that of the phosphorylation site (4.3 nmol/mg), giving a stoichiometry of nearly 2. Thus, mutant E2Ca2·BeF3− is produced from mutant E2·BeF3− by the addition of Ca2+ in the presence of A23187.
K0.5 and the Hill coefficient observed in Fig. 8B are 0.1 mm and ∼2, respectively, i.e. very similar values to those observed during E2PCa2 formation from E2P and Ca2+ in the mutant (14). The observed low Ca2+ affinity is in agreement with the wild type property (25, 49) that E2·BeF3− as well as E2P have low affinity Ca2+ binding sites; that is, the lumenal-oriented transport sites. Importantly, E2Ca2·BeF3−/E2PCa2 formed in the mutant (either from E1Ca2 or from E2·BeF3−/E2P) is remarkably stable and virtually not in equilibrium with E1Ca2·BeF3−/E1PCa2 or E2·BeF3−/E2P, i.e. their formation is almost irreversible, as shown previously (14) and in this study. When Ca2+ comes from the cytoplasmic side for E2PCa2 formation from E1Ca2 with ATP (via E2 → E1Ca2 → E1PCa2 → E2PCa2) in the mutant, the apparent Ca2+ affinity is very high, with K0.5 = 0.14 μm (14), equal to the value for cytoplasmic Ca2+ binding at the transport sites in wild type. Also in the case of mutant E2Ca2·BeF3− formation from E1Ca2 with BeFx in Fig. 8A, 10 μm Ca2+ is obviously enough to saturate (even 1 μm Ca2+ saturates (data not shown)), suggesting a similar high Ca2+ affinity as in E2PCa2 formation from E1 + 2Ca2+.
Structures of Complexes Formed from E1Ca2 with Metal Fluoride
During the Ca2+ transport cycle, the A, P, and N domains move and reorganize substantially. These changes can be monitored by proteolytic patterns and resistance against trypsin and prtK (23, 24). Therefore, we applied proteolytic analyses to mutant E2Ca2·BeF3− to reveal the position of the domains and to establish whether it is a true structural E2PCa2 analog (supplemental Figs. S3 and S4 and Tables S1 and S2 and Refs. 54 and 55). All the various major intermediates and their analogs were formed from E1Ca2 in the mutant and wild type and then subjected to proteolyses. The results show that mutant E2Ca2·BeF3− has the same structure as that of mutant E2PCa2 and that this structural state is intermediate between wild type E1PCa2 (wild type E1Ca2·BeF3−) and Ca2+-free E2P (wild type as well as mutant E2·BeF3−) as described below. In mutant E2Ca2·BeF3− and in mutant E2PCa2, the T2 site Arg198 on the Val200 loop is completely resistant to trypsin, as in wild type E2P (E2·BeF3−), showing that the A domain has rotated from its position in E1PCa2 (E1Ca2·BeF3− of wild type) and is associated with the P domain at Arg198 of the Val200 loop.
In both wild type E1Ca2·BeF3− (E1PCa2) and wild type and mutant E2·BeF3− (E2P), Leu119 on the upper portion of M2 is completely resistant to prtK attack and is, thus, sterically protected as found previously (Refs. 25 and 27; see a detailed description and reasons for protection in supplemental Fig. S5 and Ref. 56). By contrast, in mutant E2Ca2·BeF3− and mutant E2PCa2, the prtK-site Leu119 is rapidly cleaved and, thus, exposed. Evidently Leu119/Tyr122 on M2 in mutant E2Ca2·BeF3− and mutant E2PCa2 have moved from their hidden position in E1PCa2 (E1Ca2·BeF3−) but are not yet buried again through interaction with the A and P domains as in E2P (E2·BeF3−), suggesting an intermediate structure. The results also reveal how critical the native length of the A/M1′-linker is for moving M2 and the A and P domains to realize the Ca2+-free state E2P (E2·BeF3−).
The proteolyses also reveal that wild type and mutant E1Ca2·AlFx are not structurally similar to wild type E1Ca2·BeF3− (E1PCa2) and mutant E2Ca2·BeF3− (E2PCa2). Interestingly, the rate of cleavage at the T2 site of mutant E1Ca2·AlFx is intermediate between that of wild type transition state (E1Ca2·AlFx/E1Ca2·AlF4−·ADP) and that of the E1PCa2 product state (E1Ca2·BeF3−), suggesting that the structure is also intermediate. Thus, elongation of the A/M1′-linker brought the E1Ca2·AlFx structure closer to that of wild type E1Ca2·BeF3−. Only BeFx, and not AlFx, produces a species analogous to the E2PCa2 structural state (E2Ca2·BeF3− via E1Ca2·BeF3−). This means that the phosphorylation reaction must have passed through the transition state to progress to the isomerization step.
In the mutant and wild type, the prtK-site Thr242 on the A/M3-linker is completely resistant in all the states E1Ca2·AlF4−·ADP/E1Ca2·AlFx, E1Ca2·BeF3− (E1PCa2), E2Ca2·BeF3− and E2PCa2, and E2·BeF3−/E2·AlF4−/E2·MgF42− (as shown previously with sarcoplasmic reticulum Ca2+-ATPase (23, 24)). The result indicates that in both mutant and wild type, the A/M3-linker is strained by the A-domain rotation perpendicular to the membrane plane upon E1PCa2 formation from E1Ca2 and remains taut during EP processing.
E2Ca2·BeF3− Formation from E2·BeF3− by Lumenal Ca2+ Binding
The Ca2+-free complexes E2·BeF3−, E2·AlF4−, and E2·MgF42− (the analogs of the E2P ground state, transition state, and product complex of E2P hydrolysis, respectively (25)) were first formed in mutant 4Gi-46/47 and wild type, with Mg2+ bound at the catalytic site, and subsequent proteolyses were performed with and without a 10 mm Ca2+ treatment in the presence of ionophore A23187 (supplemental Fig. S4 and Table S2). Under these conditions Ca2+-treated mutant E2·BeF3− exhibits complete resistance at the tryptic T2 site Arg198 and a fairly rapid prtK cleavage at Leu119 on the top of M2, exactly as in mutant E2PCa2 and E2Ca2·BeF3− produced from E1Ca2. These results agree with those in Fig. 3D where it is found that the ability to form EP is not restored by Ca2+ treatment of E2·BeF3−. Thus, E2Ca2·BeF3−, as the E2PCa2 analog, is produced from both E2·BeF3− and from E1Ca2 (mimicking lumenal Ca2+ binding to E2P in the reverse direction of the pump cycle and the forward ATP-induced EP formation and isomerization, respectively). On the other hand, mutant and wild type complexes E2·AlF4− and E2·MgF42− and wild type E2·BeF3− are destroyed by Ca2+ treatment as found previously with sarcoplasmic reticulum Ca2+-ATPase (25, 27).
Stability of Complex E2Ca2·BeF3−
In Fig. 9, E2Ca2·BeF3− was first produced from mutant E1Ca2 with BeFx in 50 μm 45Ca2+ and 15 mm Mg2+, then further incubated at 25 °C in the presence of these ligands, and the amount of occluded 45Ca2+ was determined. The results show that the complex E2Ca2·BeF3− of the mutant is perfectly stable even after 1 week. Proteolysis confirms that the structure remains unchanged during the incubation (data not shown). The stability of the complex was further tested by diluting into an EGTA-containing solution without BeFx, and the incubation was continued at 25 °C (see the inset). Ca2+ is slowly released with a rate constant of 7.0 h−1. The addition of thapsigargin (TG) to the diluent only doubles the rate of release, indicating that the transmembrane domain is fairly resistant to TG-induced structural perturbation. These decay rates are very similar to those of mutant E2PCa2 without and with TG addition, 9.7 and 27.3 h−1, respectively (14). Thus, in this respect also, mutant E2Ca2·BeF3− is analogous to mutant E2PCa2.
DISCUSSION
Mutant E2Ca2·BeF3− as an Analog of Native Transient State E2PCa2
Using our elongated A/M1′-linker mutant, we have developed the complex E2Ca2·BeFx, most probably E2Ca2·BeF3−, as a stable structural analog of the native transient state E2PCa2 (ADP-insensitive EP with two Ca2+ at the transport sites), an intermediate in EP isomerization and Ca2+ deocclusion/release. The complex E2Ca2·BeF3− has two occluded Ca2+ and is produced from both mutant E1Ca2 and mutant E2·BeF3−, mimicking native E2PCa2 formation from E1Ca2 after ATP-induced forward phosphorylation via E1PCa2 isomerization and in the reverse direction from E2P after lumenal Ca2+ binding. Mutant E2Ca2·BeF3− formation requires Mg2+ at the catalytic site as in native ATP- and Pi-induced EP formation. The disposition of the cytoplasmic domains in mutant E2Ca2·BeF3− is equivalent to that in E2PCa2 trapped with the mutant and intermediate between native E1PCa2·Mg2+ (E1Ca2·BeF3− of wild type) and native E2P·Mg2+ (E2·BeF3− of wild type and mutant). All these properties of mutant E2Ca2·BeF3− meet the requirements of a native E2PCa2 analog.
Importantly, AlFx and MgFx are not able to produce this E2PCa2 analog either from mutant E1Ca2 or from mutant E2·AlF4− and E2·MgF42−. Thus, BeFx is unique in this regard. The coordination chemistry of the beryllium in BeFx (BeF3−) allows it to directly ligate the aspartyl oxygen, thereby producing the same tetrahedral geometry as the covalent Asp351-acylphosphate, as seen in the atomic structure of the E2P ground state analog E2·BeF3− (21, 22). On the other hand, AlFx (AlF3 or AlF4−) mimics the transition state of phosphorylation and dephosphorylation as seen in structures E1Ca2·AlF4−·ADP and E2·AlF4− (17, 19, 22). MgF42− mimics Pi in the product complex E2·Pi after E2P hydrolysis as seen in structure E2·MgF42− (19). Our results taken together with the coordination chemistry show that the structural changes for EP isomerization and Ca2+ deocclusion/release in the forward and reverse reactions are strictly coupled with the particular configuration of the acylphosphate after formation of the covalent bond within the catalytic site. The product E1PCa2 state and the E2P ground state are ready for the changes, but the transition state structures are not.
Roles of A/M1′-linker and Structural Changes during EP Formation and Processing
The transient E2PCa2 state formed during EP processing and its analog E2Ca2·BeF3− were trapped and stabilized by elongation of the A/M1′-linker. As revealed by the proteolyses, in mutant E2Ca2·BeF3− and mutant E2PCa2, the A domain has already rotated parallel to membrane from its position in E1Ca2·BeF3− (E1PCa2·Mg) and has associated with the P domain at the Val200 loop. Because mutant E2PCa2 is ADP-insensitive (14), the outermost loop TGES184 of the A domain is most probably docked onto the Asp351 region, thereby blocking ADP access to the Asp351 acylphosphate (19). Thus, in mutant E2Ca2·BeF3− and mutant E2PCa2, the A domain is positioned above the P domain. On the other hand, the proteolyses also show that the spatial relationship of the top part of M2 (Leu119/Tyr122) with the P and A domains in mutant E2Ca2·BeF3− (equivalent to native E2PCa2·Mg) is intermediate between those of the wild type E1Ca2·BeF3− (native E1PCa2·Mg) and the wild type and mutant E2·BeF3− (native E2P·Mg). Thus, Leu119 (the prtK site) on the top part of M2 has broken its van der Waals contact with upper M4 seen in E1PCa2 but has not yet reached the P and A domains to form their interaction network at Leu119/Tyr122, i.e. the Tyr122 hydrophobic cluster has not formed (see supplemental Fig. S5 for its structure). This interaction network formed from Ile179/Leu180/Ile232 of the A domain, Val705/Val726 of the P domain, and Tyr122/Leu119 of M2 is actually critical for the E2P structure (11–13). Therefore, in E2Ca2·BeF3− and E2PCa2 stabilized by elongation of the A/M1′-linker, the inclining motions of domains and helix are not yet advanced enough to reach the E2P structure.
Deletion of any single residue in the A/M1′-linker, i.e. shortening it, completely blocks E1PCa2 isomerization to E2PCa2 (26). By contrast, its elongation markedly accelerates the isomerization and greatly stabilizes E2PCa2 blocking Ca2+ deocclusion/release from this transient state (14). These findings suggest that formation of the transient E2PCa2 state (mutant E2Ca2·BeF3−) from E1PCa2 (E1Ca2·BeF3−), strains the A/M1′-linker with the wild type/native length due to rotation and positioning of the A domain above the P domain, which in turn causes further movements of the A and P domains facilitating Ca2+ deocclusion/release (14) (see the schematic model in supplemental Fig. S6). The A and P domains incline more, as will M1/M2 and M4/M5 connected with these domains, favoring release of the Ca2+. This view agrees with the structural changes required for Ca2+ release described by Toyoshima et al. (19); the bending and movement of M4/M5 by inclination of the P domain is predicted to destroy the Ca2+ binding sites, and the inclination of M2 and M1 (as a V-shaped rigid body) will push the lower part of M4 via M1 and open the lumenal gate.
These domain and segmental motions associated with Ca2+ release will establish the interaction network at Leu119/Tyr122, the Tyr122 hydrophobic cluster, and stabilize the E2P structure with the lumenal gate open (11–13). The position of the two A-P domain interaction networks, with Leu119/Tyr122 at the lower part and Val200 loop on the upper part of the interface, seems particularly appropriate to stabilize the inclined A and P domains and helices and, therefore, the gate in an open state.
These cluster formations are also critical for producing the E2P catalytic site with hydrolytic ability (11–13). Therefore, in this mechanism E2P hydrolysis can only occur after Ca2+ release, ensuring energy coupling. The relative stability of native E2P may function as a brake to allow enough time for releasing Ca2+ and for refining the catalytic site for subsequent hydrolysis, e.g. appropriate positioning of TGES184 and Glu183-coordinated attacking water molecule.
Ca2+ Substitution of Mg2+ at the Catalytic Site
In the elongated A/M1′-linker mutant, Ca2+ as well as Mg2+ bound at the catalytic Mg2+ site is able to produce E2Ca2·BeF3− from E1Ca2 via E1Ca2·BeF3−. This binding of Ca2+ is also found when mutant E2PCa2 is formed from CaATP in the absence of Mg2+ (14). This is in sharp contrast to the situation in the wild type, where Ca2+ cannot substitute for Mg2+ at the catalytic site for E1Ca2·BeF3− formation. An attempt to substitute Ca2+ for Mg2+ actually destroys wild type E1Ca2·BeF3− (27). The extremely rapid isomerization of EP with bound Ca2+ at the Mg2+ site in the elongated A/M1′-linker mutant (E1PCa2·Ca → E2PCa2·Ca) is again very different to the markedly retarded E1PCa2·Ca isomerization in wild type (14). The atomic structures provide insights into why elongation of the linker allows Ca2+ to replace Mg2+ at the catalytic site.
In the atomic structures of E1Ca2·CaAMPPCP and E1Ca2·AlF4−·ADP described by Toyoshima et al. (18, 19), Mg2+ or Ca2+ ligation at the catalytic Mg2+ site I (Asp351/Thr353/Asp703 of the P domain and the phosphate moiety (or its analog); see Fig. 2) induces the P domain to bend and, thereby, the A domain to rotate upward, perpendicular to the membrane plane (see Figs. 4 and 5 in Ref. 18 and the schematic in supplemental Fig. S6). This A-domain rotation raises its junctions with the A/M1′-linker and the A/M3-linker. The strain imposed on the A/M3-linker in E1PCa2 probably drives the large horizontal rotation of the A domain during E1PCa2 to E2P isomerization (18, 19, 50, 51). In the stringent coordination chemistry, the ligation length is shorter in Mg2+ than in Ca2+ typically by 0.2 Å (e.g. 2.1 versus 2.3 Å (52, 53)). Therefore, Mg2+ ligation probably induces more P-domain bending and in consequence more upward swinging of the A domain, leading to a stronger pull from the A/M3-linker to effect the horizontal rotation of the A domain (27). This is substantiated by the finding that in wild type, E1PCa2·Mg2+ is rapidly isomerized, whereas in E1PCa2·Ca it is markedly retarded (28, 29).
The observed formation of E2Ca2·BeF3− and E2PCa2 (via very rapid E1PCa2 isomerization) from mutant E1Ca2 with Ca2+ or Mg2+ at the catalytic Mg2+ site shows that the poor Ca2+ effect on the A-domain upward rotation and subsequent horizontal rotation is relieved by elongation of the A/M1′-linker. Note again that the A-domain junction with the A/M1′-linker is raised by the upward movement of the A domain. It is, therefore, likely that in wild type, the A/M1′-linker is strained to some extent by this movement of the A domain on formation of E1PCa2. This possible strain is evidently not deleterious for wild type, but it becomes a serious energy barrier when the A/M1′-linker is shortened by deletion of any single residue as the deletions completely block E1PCa2 to E2PCa2 isomerization (26). Strain in the wild type A/M1′-linker in E1PCa2 is likely to be important as a build up to generating stronger strain during E1PCa2 to E2PCa2 isomerization. Thus, the strain of the A/M1′-linker seems to be imposed increasingly during E1PCa2 formation and the subsequent isomerization to E2PCa2, and this energy finally could be used for inducing structural changes for Ca2+ deocclusion and release.
E1Ca2·AlFx Formed from E1Ca2 in the Elongated A/M1′-linker Mutant
The proteolytic analyses reveal that in wild type organization of the cytoplasmic domains of the transition state analog E1Ca2·AlFx is identical to that of E1Ca2·AlF4−·ADP and has obviously not yet reached the product E1PCa2 state E1Ca2·BeF3−. Namely, during the reaction E1Ca2·AlF4−·ADP/E1Ca2·AlFx → E1Ca2·BeF3−, the A domain rotates partially in a horizontal direction and comes close to the P domain at tryptic T2 site Arg198 but is not completely engaged, so that it cannot produce the E2Ca2·BeF3− and E2·BeF3− states (Ref. 27 and see the schematic in supplemental Fig. S6). On the other hand, in the elongated A/M1′-linker mutant, the structure of E1Ca2·AlFx is intermediate between those of E1Ca2·AlF4−·ADP and E1Ca2·BeF3− of wild type as judged from the intermediate tryptic cleavage rate at Arg198. Thus, elongation of the A/M1′-linker partly relieves barriers to A-domain rotation, bringing the structure of E1Ca2·AlFx closer to that of E1Ca2·BeF3−. The finding agrees with our above postulate that the A/M1′-linker is strained by the A-domain upward movement during E1PCa2 (E1Ca2·BeF3−) formation from the transition state (E1Ca2·AlFx). In fact, because the length of the Asp351 O-phosphate bond in the transition state (as mimicked by AlFx) is obviously longer than that of the covalent acylphosphate bond (as mimicked by BeF3−), the transition state (AlFx) must exhibit less P-domain bending.
Lumenal Ca2+-induced E2Ca2·BeF3− Formation from E2·BeF3−
The observed reverse formation of E2Ca2·BeF3− (native E2PCa2) from mutant E2·BeF3− (E2P) through Ca2+ binding from the lumen shows that the lumenal gate (Ca2+ releasing pathway) is open in E2·BeF3− (E2P ground state immediately before Ca2+ binding). This is in contrast to the closed gate in E2·AlF4− and E2·MgF42− (25). Thus, lumenal gating is strictly coupled with the configuration change in the phosphate during E2P hydrolysis, thereby avoiding possible Ca2+ leakage (25). Note that in wild type, E2·BeF3− (open lumenal gate) formed with Mg2+ is converted to E1Ca2 + BeFx by Ca2+, because cycle reversal and subsequent Ca2+ substitution of Mg2+ at the catalytic site destabilizes E1Ca2·BeF3− as previously demonstrated (27). E2·AlF4− and E2·MgF42− (gates closed) in wild type and mutant were also decomplexed to E1Ca2 by Ca2+ but probably by the high Ca2+ concentration disrupting the lumenal and transmembrane regions, thereby destabilizing AlF4− and MgF42− ligation at the catalytic site.
Mg2+ Dependence of E2Ca2·BeF3− Formation from E1Ca2
The Mg2+ as well as Mn2+ or Ca2+ dependence of E2Ca2·BeF3− formation from mutant E1Ca2 (Fig. 5 and supplemental Figs. S1 and S2) exhibited a Hill coefficient of 2, which is in contrast to the value of 1 for wild type E1Ca2·BeF3− formation from E1Ca2 (27). The results suggest that one or more Mg2+ besides the one at catalytic Mg2+ site I is involved cooperatively in the E2Ca2·BeF3− formation from E1Ca2. In the atomic structures of E1Ca2·CaAMPPCP and E1Ca2·AlF4−·ADP, only one Mg2+ (or Ca2+) at site I is seen (in addition to the one coordinated with the nucleotide, which was predicted to aid phosphoryl transfer). Also, in the structures of E2·BeF3−, E2·AlF4−, and E2·MgF42−, only one Mg2+ is seen (at site I). Therefore, in E2Ca2·BeF3− (E2PCa2) formation a second (or more) Mg2+ may possibly be required only transiently and, together with the catalytic ion, aids the motions of N, P, and A domains and their gathering during the E1PCa2 isomerization to E2PCa2.
In summary, our previous (14, 26) and present studies show that the A/M1′-linker should be appropriately long for the E1PCa2 to E2PCa2 isomerization then short enough for the Ca2+ deocclusion/release from E2PCa2 and again appropriately long for E2P hydrolysis. Thus, the length of the A/M1′-linker in wild type is naturally designed to induce successive structural changes and motions of the cytoplasmic and transmembrane domains for these processes. These functions of the A/M1′-linker act in concert with the changing configuration of the phosphate and catalytic Mg2+ and the Asp351-phosphate bond length, with strength being critical in the formation of E2PCa2, a species poised to deliver Ca2+ to the lumen. The stable analogs, E1Ca2·BeF3− (27) and E2Ca2·BeF3− (this study) with bound Mg2+ could be critically important for obtaining atomic models of E1PCa2·Mg2+ and the hitherto elusive transient E2PCa2·Mg2+ intermediate for further understanding of the transport mechanism.
Supplementary Material
Acknowledgments
We thank Dr. David H. MacLennan, University of Toronto, for the generous gift of SERCA1a cDNA and Dr. Randal J. Kaufman, Genetics Institute, Cambridge, MA, for the generous gift of the expression vector pMT2. We are also grateful to Dr. Chikashi Toyoshima, University of Tokyo, for helpful discussions. We thank Dr. David B. McIntosh for reviewing and improving our manuscript.
This work was supported by a grant-in-aid for Scientific Research (C) (to T. D.) and (B) (to H. S.) from the Ministry of Education, Culture, Sports, Science, and Technology of Japan.

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. S1–S6 and Tables S1 and S2.
- SERCA1a
- adult fast-twitch skeletal muscle sarcoplasmic reticulum Ca2+-ATPase
- EP
- phosphoenzyme
- E1PCa2
- ADP-sensitive phosphoenzyme with occluded Ca2+
- E2PCa2
- ADP-insensitive phosphoenzyme with occluded Ca2+
- E2P
- ADP-insensitive phosphoenzyme
- TG
- thapsigargin
- MOPS
- 3-(N-morpholino)propanesulfonic acid
- prtK
- proteinase K
- AMPPCP
- adenosine 5′-(β,γ-methylene)triphosphate.
REFERENCES
- 1.Hasselbach W., Makinose M. (1961) Biochem. Z. 333, 518–528 [PubMed] [Google Scholar]
- 2.Ebashi S., Lipmann F. (1962) J. Cell Biol. 14, 389–400 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Inesi G., Sumbilla C., Kirtley M. E. (1990) Physiol. Rev. 70, 749–760 [DOI] [PubMed] [Google Scholar]
- 4.Møller J. V., Juul B., le Maire M. (1996) Biochim. Biophys. Acta 1286, 1–51 [DOI] [PubMed] [Google Scholar]
- 5.MacLennan D. H., Rice W. J., Green N. M. (1997) J. Biol. Chem. 272, 28815–28818 [DOI] [PubMed] [Google Scholar]
- 6.McIntosh D. B. (1998) Adv. Mol. Cell Biol. 23, 33–99 [Google Scholar]
- 7.Toyoshima C., Inesi G. (2004) Annu. Rev. Biochem. 73, 269–292 [DOI] [PubMed] [Google Scholar]
- 8.Toyoshima C. (2008) Arch. Biochem. Biophys. 476, 3–11 [DOI] [PubMed] [Google Scholar]
- 9.Toyoshima C. (2009) Biochim. Biophys. Acta 1793, 941–946 [DOI] [PubMed] [Google Scholar]
- 10.Kato S., Kamidochi M., Daiho T., Yamasaki K., Gouli W., Suzuki H. (2003) J. Biol. Chem. 278, 9624–9629 [DOI] [PubMed] [Google Scholar]
- 11.Yamasaki K., Daiho T., Danko S., Suzuki H. (2004) J. Biol. Chem. 279, 2202–2210 [DOI] [PubMed] [Google Scholar]
- 12.Wang G., Yamasaki K., Daiho T., Suzuki H. (2005) J. Biol. Chem. 280, 26508–26516 [DOI] [PubMed] [Google Scholar]
- 13.Yamasaki K., Wang G., Daiho T., Danko S., Suzuki H. (2008) J. Biol. Chem. 283, 29144–29155 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Daiho T., Yamasaki K., Danko S., Suzuki H. (2007) J. Biol. Chem. 282, 34429–34447 [DOI] [PubMed] [Google Scholar]
- 15.Toyoshima C., Nakasako M., Nomura H., Ogawa H. (2000) Nature 405, 647–655 [DOI] [PubMed] [Google Scholar]
- 16.Toyoshima C., Nomura H. (2002) Nature 418, 605–611 [DOI] [PubMed] [Google Scholar]
- 17.Sørensen T. L., Møller J. V., Nissen P. (2004) Science 304, 1672–1675 [DOI] [PubMed] [Google Scholar]
- 18.Toyoshima C., Mizutani T. (2004) Nature 430, 529–535 [DOI] [PubMed] [Google Scholar]
- 19.Toyoshima C., Nomura H., Tsuda T. (2004) Nature 432, 361–368 [DOI] [PubMed] [Google Scholar]
- 20.Olesen C., Sørensen T. L., Nielsen R. C., Møller J. V., Nissen P. (2004) Science 306, 2251–2255 [DOI] [PubMed] [Google Scholar]
- 21.Toyoshima C., Norimatsu Y., Iwasawa S., Tsuda T., Ogawa H. (2007) Proc. Natl. Acad. Sci. U.S.A. 104, 19831–19836 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Olesen C., Picard M., Winther A. M., Gyrup C., Morth J. P., Oxvig C., Møller J. V., Nissen P. (2007) Nature 450, 1036–1042 [DOI] [PubMed] [Google Scholar]
- 23.Danko S., Daiho T., Yamasaki K., Kamidochi M., Suzuki H., Toyoshima C. (2001) FEBS Lett. 489, 277–282 [DOI] [PubMed] [Google Scholar]
- 24.Danko S., Yamasaki K., Daiho T., Suzuki H., Toyoshima C. (2001) FEBS Lett. 505, 129–135 [DOI] [PubMed] [Google Scholar]
- 25.Danko S., Yamasaki K., Daiho T., Suzuki H. (2004) J. Biol. Chem. 279, 14991–14998 [DOI] [PubMed] [Google Scholar]
- 26.Daiho T., Yamasaki K., Wang G., Danko S., Iizuka H., Suzuki H. (2003) J. Biol. Chem. 278, 39197–39204 [DOI] [PubMed] [Google Scholar]
- 27.Danko S., Daiho T., Yamasaki K., Liu X., Suzuki H. (2009) J. Biol. Chem. 284, 22722–22735 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Shigekawa M., Wakabayashi S., Nakamura H. (1983) J. Biol. Chem. 258, 8698–8707 [PubMed] [Google Scholar]
- 29.Wakabayashi S., Shigekawa M. (1987) J. Biol. Chem. 262, 11524–11531 [PubMed] [Google Scholar]
- 30.Kaufman R. J., Davies M. V., Pathak V. K., Hershey J. W. B. (1989) Mol. Cell. Biol. 9, 946–958 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Maruyama K., MacLennan D. H. (1988) Proc. Natl. Acad. Sci. U.S.A. 85, 3314–3318 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Daiho T., Yamasaki K., Suzuki H., Saino T., Kanazawa T. (1999) J. Biol. Chem. 274, 23910–23915 [DOI] [PubMed] [Google Scholar]
- 33.Murphy A. J., Coll R. J. (1992) J. Biol. Chem. 267, 5229–5235 [PubMed] [Google Scholar]
- 34.Troullier A., Girardet J. L., Dupont Y. (1992) J. Biol. Chem. 267, 22821–22829 [PubMed] [Google Scholar]
- 35.Murphy A. J., Coll R. J. (1993) J. Biol. Chem. 268, 23307–23310 [PubMed] [Google Scholar]
- 36.Kubota T., Daiho T., Kanazawa T. (1993) Biochim. Biophys. Acta 1163, 131–143 [DOI] [PubMed] [Google Scholar]
- 37.Weber K., Osborn M. (1969) J. Biol. Chem. 244, 4406–4412 [PubMed] [Google Scholar]
- 38.Daiho T., Suzuki H., Yamasaki K., Saino T., Kanazawa T. (1999) FEBS Lett. 444, 54–58 [DOI] [PubMed] [Google Scholar]
- 39.Laemmli U. K. (1970) Nature 227, 680–685 [DOI] [PubMed] [Google Scholar]
- 40.Lowry O. H., Rosebrough N. J., Farr A. L., Randall R. J. (1951) J. Biol. Chem. 193, 265–275 [PubMed] [Google Scholar]
- 41.Humphrey W., Dalke A., Schulten K. (1996) J. Mol. Graph. 14, 33–38 [DOI] [PubMed] [Google Scholar]
- 42.de Meis L., Masuda H. (1974) Biochemistry 13, 2057–2062 [DOI] [PubMed] [Google Scholar]
- 43.Masuda H., de Meis L. (1973) Biochemistry 12, 4581–4585 [DOI] [PubMed] [Google Scholar]
- 44.Hasselbach W., Fassold E., Migala A., Rauch B. (1981) Fed. Proc. 40, 2657–2661 [PubMed] [Google Scholar]
- 45.González D. A., Ostuni M. A., Lacapère J. J., Alonso G. L. (2006) Biophys. Chem. 124, 27–34 [DOI] [PubMed] [Google Scholar]
- 46.Yamada S., Ikemoto N. (1980) J. Biol. Chem. 255, 3108–3119 [PubMed] [Google Scholar]
- 47.Kanazawa T. (1975) J. Biol. Chem. 250, 113–119 [PubMed] [Google Scholar]
- 48.Ogurusu T., Wakabayashi S., Shigekawa M. (1991) J. Biochem. 109, 472–476 [DOI] [PubMed] [Google Scholar]
- 49.de Meis L., Inesi G. (1982) J. Biol. Chem. 257, 1289–1294 [PubMed] [Google Scholar]
- 50.Möller J. V., Lenoir G., Marchand C., Montigny C., le Maire M., Toyoshima C., Juul B. S., Champeil P. (2002) J. Biol. Chem. 277, 38647–38659 [DOI] [PubMed] [Google Scholar]
- 51.Holdensen A. N., Andersen J. P. (2009) J. Biol. Chem. 284, 12258–12265 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Picard M., Jensen A. M., Sørensen T. L., Champeil P., Møller J. V., Nissen P. (2007) J. Mol. Biol. 368, 1–7 [DOI] [PubMed] [Google Scholar]
- 53.Peeraer Y., Rabijns A., Collet J. F., Van Schaftingen E., De Ranter C. (2004) Eur. J. Biochem. 271, 3421–3427 [DOI] [PubMed] [Google Scholar]
- 54.Juul B., Turc H., Durand M. L., Gomez de Gracia A., Denoroy L., Møller J. V., Champeil P., le Maire M. (1995) J. Biol. Chem. 270, 20123–20134 [DOI] [PubMed] [Google Scholar]
- 55.Lenoir G., Picard M., Gauron C., Montigny C., Le Maréchal P., Falson P., Le Maire M., Møller J. V., Champeil P. (2004) J. Biol. Chem. 279, 9156–9166 [DOI] [PubMed] [Google Scholar]
- 56.Seekoe T., Peall S., McIntosh D. B. (2001) J. Biol. Chem. 276, 46737–46744 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.